Abstract
Puumala virus (PUUV) is a negative-stranded RNA virus in the genus Hantavirus, family Bunyaviridae. In this study, detailed phylogenetic analysis was performed on 42 complete S segment sequences of PUUV originated from several European countries, Russia, and Japan, the largest set available thus far for hantaviruses. The results show that PUUV sequences form seven distinct and well-supported genetic lineages; within these lineages, geographical clustering of genetic variants is observed. The overall phylogeny of PUUV is star-like, suggesting an early split of genetic lineages. The individual PUUV lineages appear to be independent, with the only exception to this being the Finnish and the Russian lineages that are closely connected to each other. Two strains of PUUV-like virus from Japan form the most ancestral lineage diverging from PUUV. Recombination points within the S segment were searched for and evidence for intralineage recombination events was seen in the Finnish, Russian, Danish, and Belgian lineages of PUUV. Molecular clock analysis showed that PUUV is a stable virus, evolving slowly at a rate of 0.7 × 10−7 to 2.2 × 10−6 nt substitutions per site per year.
Puumala virus (PUUV) belongs to the genus Hantavirus of the family Bunyaviridae (12). Like other members of this family, PUUV is an enveloped virus with a segmented, single-stranded RNA genome of negative polarity. The large (L) segment of 6.5 kb encodes the viral RNA polymerase, the 3.7-kb medium (M) segment encodes the two surface glycoproteins, and the 1.8-kb small (S) segment encodes the nucleocapsid protein (N).
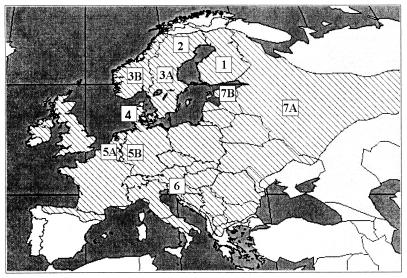
The natural host of PUUV is the bank vole, Clethrionomys glareolus, which belongs to the Arvicolinae subfamily of the Muridae family. The bank vole is found in most of Europe, excluding the Mediterranean coast and the northernmost areas (Fig. 1) (40). The virus causes a life-long persistent and asymptomatic infection in rodents (47). In contrast, in humans PUUV is pathogenic, causing nephropathia epidemica, a mild form of hemorrhagic fever with renal syndrome (HFRS) (8).
FIG. 1.
Map of Europe showing the distribution of the bank vole (40). PUUV sequences analyzed in this paper belong to the following seven distinct lineages: FIN (1), NSCA (2), SSCA (3) (sequences are from Sweden [A] and Norway [B]), DAN (4), BEL (5) (sequences are from Belgium [A] and Germany [B]), BAL (6), and RUS (7) (sequences are from Russia [A] and the Baltics [B]).
PUUV from the following countries has been genetically characterized (Fig. 1): Finland (45, 46, 48, 62), Sweden (24, 25, 38), Norway (38), Denmark (3), Russia (3, 16, 48, 49, 66), Belgium (6, 13), Austria (1, 5), and Germany (19, 43). So far, over 100 partial or complete PUUV sequences have been deposited in GenBank. These include 38 complete S segment sequences, 9 complete M segment sequences, and 2 complete L segment sequences. In general, the phylogenetic relationships between different hantaviruses and their rodent hosts mirror each other, supporting the idea of coevolution of the virus and its host (44). Furthermore, different strains of a given hantavirus type, including PUUV, show geographical clustering (48, 49) reflecting the sometime complicated history of host range movements. The most recent bank vole range changes are due to the recolonization of Europe and especially Fennoscandia after the last ice age 10,000 years ago (29). Earlier it has been shown that these migrations have had an impact on the evolution of PUUV (24, 38).
The main source of genetic variation of hantaviruses seems to be genetic drift, i.e., accumulation of base substitutions and deletions or insertions (3, 38, 48). Additionally, reassortment has been shown for some hantaviruses (20) and it has been proposed also for PUUV (45). It has been recently shown that recombination was involved in the evolution of Tula hantavirus (TULV) (55). There are indications that this mechanism is operating in PUUV as well (3, 13).
PUUV has been shown to form quasispecies populations in individual bank voles (48). In general, this feature is connected to a high potential for rapid evolution (11). Nevertheless, if the master genotype and phenotype have high fitness to the environment, the quasispecies populations might be stable for a long period of time. This is most probably the case for PUUV and other hantaviruses, which have been well adapted to their natural hosts since long ago.
Previous publications on PUUV evolution (1, 3, 13, 19, 24, 43, 45, 46, 48, 49) have focused mainly on specific geographical areas, including only a limited number of sequences, and all the phylogenetic analyses have been performed with distant matrix methods. Recently, a substantial set of new complete PUUV S segment sequences became available (references 3 and 13 and our unpublished results). In this study, for the first time we have attempted to analyze all the known complete S segment sequences of PUUV using different phylogenetic methods in order to gain more insight into the evolution of this virus. Special attention was paid to studying whether the concept of a molecular clock fits PUUV evolution or not and to the role of recombination in the evolution of this virus.
MATERIALS AND METHODS
The following PUUV S segment and N protein hantavirus sequences were analyzed. Sequences from strains of Finnish lineage (FIN) were strain Sotkamo, GenBank accession number X61035, Evo/12Cg/93, Z30702; Evo/13Cg/93, Z30703; Evo/14Cg/93, Z30704; Evo/15Cg/93, Z30705; Virrat/25Cg/95, Z69985; Karhumäki/Cg117/95, AJ238788; Kolodozero/Cg53/95, AJ238789; Gomselga/Cg4/95, AJ238790; Puumala/1324Cg/79, Z46942; and Pallasjarvi/63Cg/98, AJ314597. Sequences from strains from southern Scandinavia (SSCA) were Eidsvoll/1124v, AJ223368; Eidsvoll/Cg1138/87, AJ223369; Solleftea/Cg3/95, AJ223376; and Solleftea/Cg6/95, AJ223377. Sequences from strains from northern Scandinavia (NSCA) were Hundberget/Cg36/95, AJ223371; Mellansel/Cg47/94, AJ223374; Mellansel/Cg49/95, AJ223375; Tavelsjo/Cg81/94, AJ223380; Vindeln/L20Cg/83, Z48586; Vindeln/Cg4/94, AJ223381; and “Vranica”/Hällnäs, U14137. Sequences from Danish (DAN) strains were Fyn19, AJ238791; Fyn47, AJ238792; and Fyn131, AJ238793. Sequences from Russian (RUS) strains were Udmurtia/338Cg/92, Z30708; Udmurtia/444Cg/88, Z30706; Udmurtia/458Cg/88, Z30707; Udmurtia/894Cg/91, Z21497; Kazan, Z84204; Cg1820, M32750; P360, L11347; Baltic/49Cg/00, AJ314598; and Baltic/205Cg/00, AJ314599. Sequences from Belgian (BEL) strains were Cg13891, U22423; Cg-Erft, AJ238779; Thuin/33Cg/96, AJ277030; Montbliart/23Cg/96, AJ277031; Momignies/47Cg/96, AJ277032; Momignies/55Cg/96, AJ277033; and Couvin/59Cg/97, AJ277034. Sequences from Balkan (BAL) strains were Balkan/65Cg/00, AJ314600; and Balkan/78Cg/00, AJ314601. Sequences from Japanese (JPN) strains were Tobetsu-60Cr-93, AB010731; and Kamiiso-8Cr-95, AB010730. Sequences from other hantaviruses were TULV, strain Moravia02v (Z69991); Hantaan virus (HTNV), 76-118 (M14626); Dobrava, Dobrava (L41916); Saaremaa, Saaremaa/160V (AJ009773); Seoul, SR-11 (M34881); El Moro Canyon, RM-97 (U11427); New York, RI-1 (U11427); Sin Nombre, NM H10 (L25784); Bayou, Louisiana (L36929); Black Creek Canal (L39949); Laguna Negra, 510B (AF005727); Topografov, Ls136V (AJ011646); Khabarovsk, MF-43 (U35255); Prospect Hill, PH-1 (Z49098); and Andes, AH-1 (AF324902).
Multiple sequence alignments were prepared with ClustalX (60) using the following parameters: gap opening, 10; gap extension, 6.66; delay divergent sequences by 40%; DNA transition weight, 0.50; and no negative matrix.
Phylogenetic analysis.
The Wisconsin Package, version 10.2 (Genetics Computer Group, Madison, Wis.), was used for sequence entry and analysis. Nucleotide sequences were translated into amino acid sequences with SeqApp. PHYLIP was used to create phylogenetic trees using distance matrix (DM) methods (Fitch-Margoliash or neighbor joining) and maximum parsimony (MP) methods (15). These methods were applied for both the nucleotide and deduced amino acid sequences with 500 bootstrap replicates.
The TreePuzzle program was used to reconstruct phylogenetic trees using the maximum likelihood (ML) approach (57). Ten thousand puzzling steps were applied using the Hasegawa-Kishino-Yano (HKY) model of substitution (18). The transition/transversion ratio and nucleotide frequencies were estimated from the data set. Rate heterogeneity was applied using discrete gamma distribution with eight rate categories, and the shape parameter alpha was estimated from the data set.
Split decomposition analysis (4, 28) was performed with the program SplitsTree using the LogDet method for computing distances. This method presents conflicting evolutionary signals in the data set as a network instead of a dichotomously branching phylogenetic tree.
Similarity plots were created using Stuart Ray's SimPlot 2.5 (34). The window size was 200 to 300 nucleotides (nt) and the step size was 20 nt. Jukes-Cantor corrections were applied. Consensus sequences of PUUV lineages were used as query or reference sequences.
Rate of evolution.
To calculate the evolution rate of PUUV, the number of synonymous and nonsynonymous substitutions per 100 sites (dS and dN) was estimated using the program Diverge from the Genetics Computer Group Wisconsin package. Values were calculated comparing PUUV to two different hantavirus species, HTNV and TULV. Alternatively, the ML branch lengths derived from trees with contemporary tips (calculated using TreePuzzle) were used. Both the mean number of dS and the ML branch lengths were then divided by the time of divergence of rodents carrying these viruses to gain an estimate of the substitution rate.
RESULTS
Phylogeny of PUUV.
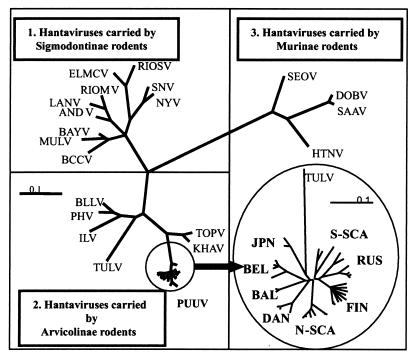
On a phylogenetic tree hantaviruses form three clades carried by Murinae, Arvicolinae, and Sigmodontinae rodents (Fig. 2). PUUV is placed within the second clade, which also includes TULV, Bloodland Lake, Prospect Hill, Isla Vista, and Khabarovsk viruses, all carried by voles, and Topografov virus, whose natural hosts are lemmings. TULV is used as an outgroup sequence in the phylogenetic analyses of PUUV in this paper.
FIG. 2.
A phylogenetic tree of hantavirus N protein sequences calculated using TreePuzzle (55). An enlarged phylogenetic tree of PUUV S-segment coding sequences created using FITCH of the PHYLIP package (15) is shown. The bootstrap support values for the PUUV lineages are given in Table 2.
The best collection of sequences is available for the S segment, which also seems to be a good representative of the whole PUUV genome (3). These sequences vary in length from 1,784 nt in strain CG1820 to 1,882 nt in strain Sollefteå-6 and contain an open reading frame coding for the N protein of 433 amino acids (aa). The 5′ noncoding region (NCR) in the positive strand is 42 nt in length and the 3′ NCR varies from 442 to 540 nt. Except for the last ≈100 nt, the S segment 3′ NCR of different strains could be aligned only within given genetic lineages of PUUV (3, 13, 38) and was therefore excluded from our analysis.
Phylogeny of PUUV S segment nucleotide sequences shows eight distinct genetic lineages, FIN, RUS, NSCA, SSCA, DAN, BEL, BAL, and JPN, which share a common ancient ancestor (Fig. 2). PUUV strains in each lineage are given in Table 1. The first seven lineages share a common more recent ancestor, while the JPN lineage occupies the most ancestral node. This lineage includes two wild-type strains recovered from tissue samples of Clethrionomys rufocanus trapped in Hokkaido (32). Being associated with a distinct host species, these strains cannot be strictly referred to as PUUV but rather are considered PUUV-like.
TABLE 1.
PUUV strains included in specific genetic lineages and amino acid signatures
| Lineage and PUUV strains | Amino acid signature |
|---|---|
| FIN | M262, D304, F388 |
| Puumala | |
| Sotkamo | |
| Virrat | |
| Karhumaki | |
| Kolodozero | |
| Gomselga | |
| Evo12 | |
| Evo13 | |
| Evo14 | |
| Evo15 | |
| Pallasjarvi63 | |
| RUS | T272, D302, S416a |
| Kazan | |
| U338 | |
| U444 | |
| U458 | |
| U894 | |
| CG 1820 | |
| P360 | |
| Baltic49 | |
| Baltic205 | |
| NSCA | D272, D429 |
| Tavelsjö | |
| Hundberget | |
| Mellansel47 | |
| Mellansel49 | |
| Vranica | |
| VindelnL20 | |
| SSCA | D9, A20 |
| Sollefteå3 | |
| Sollefteå6 | |
| Eidsvoll1124v | |
| Eidsvoll1138 | |
| DAN | L251, N256, N272 |
| Fyn131 | |
| Fyn47 | |
| Fyn19 | |
| BEL | Q64 |
| Montbliart23 | |
| Thuin33 | |
| Momignies55 | |
| Momignies47 | |
| Couvin59 | |
| Cg-Erft | |
| CG 13891 | |
| BAL | V236, P257 |
| Balkan65 | |
| Balkan78 | |
| JPN | K5, V68, A79, I126, I193, K258, P283 |
| Kamiiso | |
| Tobetsu |
The Baltic strains (49 and 205) do not contain this signature.
All genetic lineages of PUUV possess specific amino acid “signatures” (Table 1). In addition to signatures characteristic of the FIN and the RUS lineages, there are 2 aa residues (Val34 and Tyr61) shared by these lineages, indicating a closer relationship between them. The JPN lineage has the longest amino acid signature.
Comparison of the S segment sequence identities (reference 3 and our unpublished data) shows that the variation between the lineages ranges at the nucleotide level from 15 to 27%, with the smallest difference being observed between the RUS and the FIN lineages. The intralineage nucleotide diversity is 0.3 to 9.0% for all the lineages except SSCA, which shows diversity up to 13.4%, and RUS (15.6%). The SSCA lineage is actually formed by two sublineages constituted by strains from central Sweden and Norway, respectively, with the intrasublineage diversity ranging from 0.3 to 5.7% (38). The RUS lineage seems to be formed also by two sublineages formed by strains from the European part of Russia and the Baltics.
At the amino acid level, the interlineage variation translates to substantially lower values, of 0 to 7.8%, indicating that a strong purifying selection occurred at the N protein level. Notably, the PUUV N protein sequence diversity is higher than in other hantaviruses (23, 35) and in several cases even exceeds the cutoff level of 7% arbitrarily selected to define distinct hantavirus species (12).
The overall topology of the PUUV phylogenetic tree is star-like, suggesting an early split of the genetic lineages. In general, all these lineages are well supported (Table 2). Although the topology of all trees calculated with DM, MP, and ML is the same, the bootstrap support values vary. Both the MP and DM methods show different bootstrap support values depending on whether the calculations are based on nucleotide or amino acid sequences. MP gives more consistent results than the distance methods, but there are still values lower than 70% (Table 2), the widely accepted confidence limit (22). The support values calculated using the ML method are the most consistent.
TABLE 2.
Bootstrap support values for PUUV lineages calculated with different methods
| Lineage | Bootstrap value (%) for:
|
|||||
|---|---|---|---|---|---|---|
| DM methoda
|
MP method
|
ML method
|
||||
| aa | nt | aa | nt | aa | nt | |
| FIN | 89 | 53 | 90 | 86 | 93 | 91 |
| RUS | 63b | 97 | 83 | 87 | 83 | 93 |
| NSCA | 49b | 100 | 99 | 100 | 91 | 86 |
| SSCA | 60b | 95 | 71 | 92 | 98 | 94 |
| DAN | 98 | 100 | 98 | 100 | 79 | 89 |
| BEL | 69b | 100 | 68 | 100 | 60b | 58b |
| BLK | 95 | 100 | 98 | 100 | 95 | 96 |
| JPN | 100 | 100 | 100 | 100 | 83 | 87 |
| FIN plus RUS | 30b | 100 | 64b | 100 | 72 | 62b |
The DM method used was the neighbor-joining method.
Values below the confidentiality limit (70%).
The early split of PUUV genetic lineages manifested as the star-like topology of the trees infers that the relationships between distinct lineages remain obscure. The FIN and RUS lineages represent the only exception to this, being more closely connected to each other.
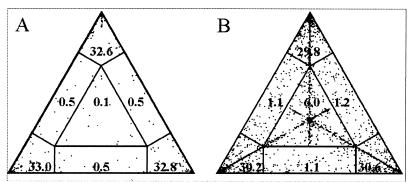
As the bootstrap support values sometimes varied depending on whether the calculations were based on nucleotide or amino acid sequences, this was further evaluated using the “likelihood mapping” option of the TreePuzzle program. This method can be used to visualize the phylogenetic content of a sequence alignment as follows. Different topologies of quartet trees are plotted in a triangle so that the corners represent the completely resolved, tree-like quartets; the sides represent quartets that are partially resolved and the center represents the unresolved quartets (56). It appeared that only a very low number (1.6%) of the quartets based on PUUV nucleotide sequences was partially or completely unresolved (Fig. 3); the corresponding number for the amino acid sequences is higher (9.4%) but is still low enough to consider the tree reconstruction to be accurate (56).
FIG. 3.
Likelihood mapping of PUUV nucleotide (A) and amino acid (B) sequences. Dots in the corners of triangles represent fully resolved tree topologies and dots in the center represent unresolved ones.
Molecular clock and the rate of PUUV evolution.
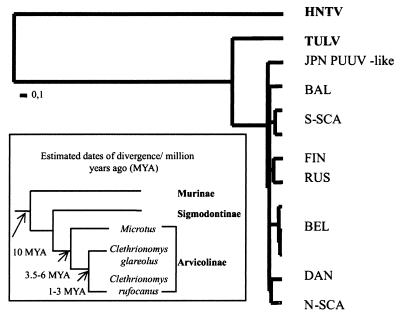
TreePuzzle was also used to perform a likelihood ratio test in order to check a molecular clock hypothesis. In our case, a clock-like tree did not pass the test when either all or only a few PUUV strains were considered assuming a uniform rate of nucleotide substitutions. Only when applying gamma distribution of rate heterogeneity was the clock no longer rejected (Fig. 4). The shape parameter alpha estimated from the data set is 0.23 ± 0.01, indicating that most of the sites evolve slowly but that a few sites have a moderate-to-high rate of evolution.
FIG. 4.
A phylogenetic tree of PUUV with contemporary tips and estimates of dates of divergence of the host rodents (9, 10, 30).
Passing the likelihood ratio test of the molecular clock showed that the data set can be used to estimate the evolution rate of PUUV. The mean values of dS and dN (the number of synonymous and nonsynonymous substitutions per 100 sites) calculated for all PUUV sequences were 88 ± 29 and 2.37 ± 0.83, respectively. Within a genetic lineage dS varied from 6 to 45, and between lineages it varied from 85 to 122. dS was highest within the SSCA and RUS lineages, probably reflecting the fact that these lineages actually contain well-separated sublineages; the corresponding values for dN were 0.1 to 1.2 and 0.1 to 3.8. In all cases, the dN/dS ratio was extremely low, indicating that positive selection is not the primary mechanism driving the evolution of this virus.
In order to estimate the rate of nucleotide substitutions in PUUV evolution, the dS values between PUUV, Japanese PUUV-like viruses, TULV, and HTNV were calculated (Table 3). Assuming that the viruses coevolved with their hosts, the evolutionary rate of PUUV will be 1.9 × 10−7 to 2.2 × 10−6 synonymous nucleotide substitutions per site per year. The evolutionary rate of PUUV was also estimated using the ML branch lengths of the clock-like trees (Fig. 4). The values ranged from 0.7 × 10−7 to 1.1 × 10−6 nt per site per year depending on whether HTNV was included in the calculations in addition to PUUV and TULV (Table 3). The rate estimations obtained by these two methods gave encouragingly similar results, with all values being between 0.7 × 10−7 and 2.2 × 10−6 nt per site per year.
TABLE 3.
Substitution rate estimates based on the number of synonymous substitutions and ML branch lengths
| Viruses compared | dS (mean ± SD) | Divergence of host rodents (millions of yrs ago) | No. of synonymous substitutions/site/yr | Branch length | Divergence of host rodents (millions of yrs ago) | No. of nucleotide substitutions/site/yr |
|---|---|---|---|---|---|---|
| JPN PUUV-like vs PUUV | 110 ± 10 | 1–3 | 3.7 × 10−7–1.1 × 10−6 | 0.215–0.349 | 1–3 | 0.7 × 10−7–3.5 × 10−7 |
| TULV vs PUUV | 185 ± 36 | 3.5–6 | 2.5 × 10−7–2.2 × 10−6 | 0.823–1.689 | 3.5–6 | 1.4 × 10−7–4.8 × 10−7 |
| HTNV vs PUUV | 275 ± 48 | 10 | 2.3 × 10−7–3.3 × 10−7 | 10.37 | 10 | 1.1 × 10−6 |
| HTNV vs TULV | 222 ± 36 | 10 | 1.9 × 10−7–2.6 × 10−7 |
Recombination in PUUV evolution.
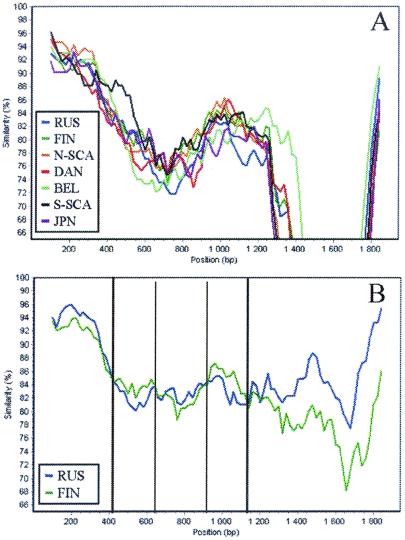
Recombination of hantaviruses has been shown for TULV (55) and suggested for PUUV as well (3, 13). To study this issue further, similarity plots were created to visualize the pattern of sequence similarity between the distinct PUUV lineages. These plots (Fig. 5A) revealed within the S-segment sequence some regions with higher-than-average similarity between the FIN and RUS lineages. This was particularly pronounced on the PUUV sequences from the Baltics within the RUS lineage (data not shown), suggesting a recombinant origin for these strains. Such a suggestion was further strengthened by our observation that inclusion of either one of these Baltic strains in a clock-like tree led to rejection of the molecular clock, just as would be expected from a recombinant sequence (54).
FIG. 5.
Similarity analysis based on consensus sequences of distinct PUUV lineages. The query sequence is the Balkan consensus sequence (A) or the Baltic consensus sequence (B).
A closer analysis has shown that two regions in the Baltic sequences, nt 440 to 630 and 940 to 1130, are in fact more similar to the FIN sequences (Fig. 5B). However, when trees were calculated for these regions, the Baltic sequences were not placed within the FIN lineage but formed a cluster of their own. A similar pattern has been seen earlier for the DAN lineage, which in one part was most closely related to the NSCA lineage while in another part it was most closely related to the SSCA lineage and thus was suggested to contain recombinant sequences (3).
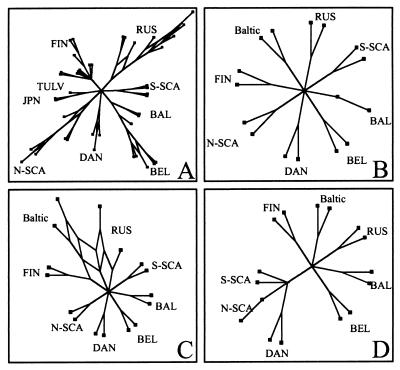
Since some evidence for recombination was revealed, we studied if a phylogenetic network showing the conflicting signals would illustrate the evolution of PUUV better than a phylogenetic tree. This was done using the program SplitsTree based on the split decomposition theory (4). On the SplitsTree (Fig. 6A), the RUS, BEL, NSCA, and DAN lineages are represented as networks, suggesting that they contain conflicting phylogenetic signals. This indicates that these lineages include sequences that might have undergone recombination during their evolution. Since the Baltic sequences were suspected to be recombinants between the FIN and RUS lineages, they were studied in more detail (Fig. 6B to D). At the 5′ end (nt 1 to 400) and the 3′ end (nt 1100 to 1344) of the S segment coding sequence the Baltic strains form a cluster of their own. In contrast, the middle section of the S segment (nt 400 to 1100) placed the Baltic sequences into a network which also includes FIN and RUS sequences. This finding provides further evidence that the Baltic sequences are recombinants of the ancestors of the FIN and RUS lineages.
FIG. 6.
(A) Splitstree based on the complete S-segment coding sequence of all PUUV strains. (B to D) Splitstrees based on partial sequences. (B) nt 1 to 400. (C) nt 401 to 1100. (D) nt 1101 to 1344. Two strains of each PUUV lineage are included together with two Baltic PUUV strains.
As for the networks seen in the BEL, NSCA, and DAN lineages, they are not contradictory to earlier suggestions on recombination within those lineages as well (3, 13).
DISCUSSION
Different phylogenetic methods show the same overall picture of PUUV evolution.
The earlier inferred PUUV phylogenies (1, 3, 13, 19, 24, 43, 45, 46, 48, 49) were obtained using DM methods. Since they are robust they are widely used, but they should be applied with careful validation. One of our goals was to compare different phylogenetic approaches in order to (i) confirm the earlier results and (ii) study the differences between these approaches and their effect on the results. Our analyses have shown that DM, MP, ML, and split decomposition methods all give the same overall picture of a star-like phylogeny of PUUV, assigning strains to their correct and well-supported phylogeographic genetic lineages. On the other hand, the connections between the lineages, with only one exception, are not pronounced, indicating their early split. The most ancestral position of the Japanese PUUV-like virus seems expected, since it originated from a distinct host species and cannot be strictly referred to as PUUV. Notably, unlike the bona fide PUUV, the Japanese variant appears to be nonpathogenic for humans (31).
Further support for the distinct genetic lineages comes from the amino acid signatures which can be assigned to each of them as well as from levels of intra- and interlineage variation. Sequence analysis also shows that the ratio of synonymous substitutions to nonsynonymous substitutions is extremely high, suggesting a neutral mode of evolution (17). As no immunological pressure seems to operate in the rodent host (37), which is the main evolutionary scene of PUUV, mechanisms like sampling, population bottlenecks, and founder effects (63) have more likely contributed to the formation of the distinct PUUV lineages. These events could be linked to the well-known temporal population fluctuations of the bank vole (42).
The different methods used for inferring phylogenies are based on different evolutionary assumptions. Although they give the same overall picture of PUUV evolution, the bootstrap support values observed in MP and DM methods differ depending on whether the calculations were based on nucleotide or amino acid sequences. This suggests that the chosen models for correcting distances, Kimura's two-parameter model of nucleotide substitutions or the Dayhoff's model for amino acid substitutions, fail to adequately describe the evolutionary processes in PUUV. MP gives more consistent support values than distance methods, but still not all the values reach the confidence limit of 70% (22). In general, MP performs best when the number of actual sequence changes is small, and thus this method might be affected by the fact that PUUV sequences are highly divergent.
ML is often considered the best method for inferring phylogenies due to an explicit evolutionary model implemented in it (59). The practical problem is that ML is very time-consuming, and with a large data set, the use of traditional ML programs is usually not convenient. We therefore performed ML analysis using the TreePuzzle program, which speeds up the calculations by considering only four sequences at a time. Our phylogenetic reconstruction of PUUV evolution confirmed that the ML approach gives the most consistent results, with the support values for distinct genetic lineages on the trees based on nucleotide and amino acid sequences being essentially the same.
Molecular clock and a slow evolution of hantaviruses.
A molecular clock has not yet been reported for hantaviruses. Here, for the first time, the clock assumption was tested using the ML ratio test (14). It should be emphasized that the explicit evolutionary model implemented in the ML approach allows for more detailed analysis of PUUV S sequences that differ in many aspects from an average sequence set. There exists an extremely high dS/dN ratio, and the transition/transversion ratio (3.5) is unusually high. Furthermore, different sites seem to evolve at different rates and there is a bias in the overall nucleotide composition of the viral genome (e.g., the A content is 33% while the C content is only 19%). The S-segment coding and noncoding regions are functioning under different pressures that seem to be unequal for different parts of the coding region as well. For instance, in the N protein there is a hypervariable region (aa 233 to 275) carrying epitopes recognized by both monoclonal antibodies and human patient sera (36, 61). Within this region, the rate of nonsynonymous substitutions is unusually high, suggesting that positive selection may favor amino acid replacements (26). However, the ratio of dN/dS does not exceed 1, which would be supportive of a positive selection (41). Instead, the nonsynonymous changes seem to occur at random (26, 27), leading to the conclusion that this region is more likely under low functional or structural constraints.
Taking these factors into account, it is not surprising that the molecular clock test on PUUV S sequences is passed only when the gamma distribution of rate heterogeneity among sites is applied. The estimated substitution rate ranged from 0.7 × 10−7 to 2.2 × 10−6 nt per site per year. This slow rate of evolution is indirectly supported by the low value (0.23) of the shape parameter alpha of the gamma distribution. Here it should be stressed that our estimations of the substitution rates are based on present knowledge on evolution of the rodent hosts. Paleontological records and other molecular data range widely, and there are also controversial reports on the phylogenies of rodents (9, 10, 30, 51). This leads to different estimations of the PUUV evolutionary rate ranging approximately 20-fold. Notably, the rate of evolution of the M segment estimated for nine complete sequences known so far (3.7 × 10−7 to 8.7 × 10−7 nt per site per year) is in the range determined for the S segment. Thus, both genes of PUUV evolve at a similar rate, and the overall conclusion that PUUV is evolving rather slowly seems to be well justified.
The nucleotide substitution rate in hantavirus evolution presented in this paper correlates well with the estimation based on the separation of Old World and New World Microtus voles of 2.41 × 10−7 to 2.68 × 10−7 nt per site per year (26). It is also comparable with the rates suggested for other stable RNA viruses like human T-cell lymphotropic virus type 2 in tribes infected in areas of endemicity (1.71 × 10−7 to 7.31 × 10−7) (52) and hepatitis G virus (9 × 10−6) (58). These slowly evolving viruses infect their primary hosts (humans) persistently and are well adapted to them. Thus, they remain most of the time in equilibrium close to an adaptive peak, as the vast majority of new mutations would probably decrease the fitness of the virus (52). In contrast, the substitution rates estimated for more rapidly evolving RNA viruses such as human immunodeficiency virus (HIV) and hepatitis C virus are much higher, being on the order of 10−2 to 10−5 nt per site per year (2, 33).
Some key events in the PUUV history may now be dated, albeit with not very high precision, based on our estimation of its substitution rate. Thus, the Japanese PUUV-like strains seemed to diverge from the branch leading to PUUV not later than 100,000 years ago (YA). This may be related to the last Weichselian glaciation of the northern hemisphere starting about 115,000 YA. It looks like hypothetical founder populations of the distinct PUUV lineages were established not later than 85,000 YA, and the present lineages were geographically separated during the last deglaciation, which started 21,000 to 17,000 YA. The retreating glacial ice sheet left behind several immigration routes for flora and fauna to colonize the revealed land, greatly affecting the evolution of these species (21), including bank voles carrying PUUV. This is reflected in the closer connections of some PUUV lineages and the existence of clearly distinct sublineages.
Recombination (genetic shift) in hantavirus evolution.
For many RNA viruses, recombination has been shown to be an important feature of their evolution (for review, see reference 64). These include, e.g., the well-established case of HIV (39) as well as more recent reports on enteroviruses (53) and dengue virus (65). The first evidence of recombination in negative-stranded RNA viruses has been reported for TULV (55). In our study several indications of recombination in PUUV were seen. First, the molecular clock analysis indirectly suggested that out data set includes recombinant sequences, since the evolutionary clock was rejected when a large set of sequences was used in the calculations. Recently, it has been shown that even a few recombination events can lead to rejection of the clock (54). Indeed, when the Baltic PUUV sequences of a suspected recombination origin were excluded from the calculations, the molecular clock was no longer rejected. Second, as some lineages are represented by networks, they probably include recombinant sequences (64). Third, similarity analysis of the Baltic strains revealed that two regions of their S segment have higher-than-average similarity to the FIN PUUV strains, while other regions are most closely related to the RUS strains. This pattern may be interpreted as evidence of recombination between these two lineages of PUUV. A similar pattern has been seen earlier for the DAN PUUV strains (3).
Another variety of genetic shift, reassortment, has been demonstrated for some members of the genus Bunyavirus (50). In the genus Hantavirus it has been shown for Sin Nombre virus (20) and suggested for PUUV as well (45). Thus, both mechanisms seem to play a role in the evolution of PUUV in cooperation with the classical genetic drift.
Detailed knowledge of the rate and mode of genetic variation is essential for understanding how hantaviruses induce disease in humans as well as for molecular epidemiology. The pattern of PUUV evolution revealed in this study (the low rate of evolution obeying the molecular clock and the early split of genetic lineages and recombination) would most probably be applicable to other hantavirus types, such as Hantaan, Dobrava, Sin Nombre, and the like, thus advancing the research of these severe human pathogens.
ACKNOWLEDGMENTS
We thank Heikki Henttonen, Olli Vapalahti, and Vincent Moulton for their helpful comments.
This study was supported by grants from the Academy of Finland, by EC grant QLK2-CT-1999-01119, and by the Sigrid Jusélius Foundation, Helsinki, Finland.
REFERENCES
- 1.Aberle S W, Lehner P, Ecker M, Aberle J H, Arneitz K, Khanakah G, Radda A, Radda I, Popow-Kraupp T, Kunz C, Heinz F X. Nephropathia epidemica and Puumala virus in Austria. Eur J Clin Microbiol Infect Dis. 1999;18:467–472. doi: 10.1007/s100960050325. [DOI] [PubMed] [Google Scholar]
- 2.Allain J P, Dong Y, Vandamme A M, Moulton V, Salemi M. Evolutionary rate and genetic drift of hepatitis C virus are not correlated with the host immune response: studies of infected donor-recipient clusters. J Virol. 2000;74:2541–2549. doi: 10.1128/jvi.74.6.2541-2549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asikainen K, Hänninen T, Henttonen H, Niemimaa J, Laakkonen J, Andersen H K, Bille N, Leirs H, Vaheri A, Plyusnin A. Molecular evolution of Puumala hantavirus in Fennoscandia: phylogenetic analysis of strains from two recolonization routes, Karelia and Denmark. J Gen Virol. 2000;81:2833–2841. doi: 10.1099/0022-1317-81-12-2833. [DOI] [PubMed] [Google Scholar]
- 4.Bandelt H-J, Dress A W M. Split decomposition: a new and useful approach to phylogenetic analysis of distance data. Mol Phylogenet Evol. 1992;1:242–252. doi: 10.1016/1055-7903(92)90021-8. [DOI] [PubMed] [Google Scholar]
- 5.Bowen M D, Gelbmann W, Ksiazek T G, Nichol S T, Nowotny N. Puumala virus and two genetic variants of Tula virus are present in Austrian rodents. J Med Virol. 1997;53:174–181. doi: 10.1002/(sici)1096-9071(199710)53:2<174::aid-jmv11>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 6.Bowen M D, Kariwa H, Rollin P E, Peters C J, Nichol S T. Genetic characterization of a human isolate of Puumala hantavirus from France. Virus Res. 1995;38:279–289. doi: 10.1016/0168-1702(95)00058-x. [DOI] [PubMed] [Google Scholar]
- 7.Brummer-Korvenkontio M, Vaheri A, Hovi T, von Bonsdorff C H, Vuorimies J, Manni T, Penttinen K, Oker-Blom N, Lahdevirta J. Nephropathia epidemica: detection of antigen in bank voles and serologic diagnosis of human infection. J Infect Dis. 1980;141:131–134. doi: 10.1093/infdis/141.2.131. [DOI] [PubMed] [Google Scholar]
- 8.Brummer-Korvenkontio M, Vapalahti O, Henttonen H, Koskela P, Kuusisto P, Vaheri A. Epidemiological study of nephropathia epidemica in Finland, 1989–96. Scand J Infect Dis. 1999;31:427–435. doi: 10.1080/00365549950163941. [DOI] [PubMed] [Google Scholar]
- 9.Catzeflis F M, Aquilar J-P, Jaeger J-J. Muroid rodents: phylogeny and evolution. TREE. 1992;7:122–126. doi: 10.1016/0169-5347(92)90146-3. [DOI] [PubMed] [Google Scholar]
- 10.Conroy C J, Cook J A. MtDNA evidence for repeated pulses of speciation within arvicoline and murid rodents. J Mamm Evol. 1999;6:221–245. [Google Scholar]
- 11.Domingo E, Holland J J. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 12.Elliott R M, Bouloy M, Calisher C H, Goldbach R, Moyer J T, Nichol S T, Pettersson R, Plyusnin A, Schmaljohn C S. Family Bunyaviridae. In: Van Regenmortel M H V, Fauquet C M, Bishop D H L, Carsten E B, Estes M K, Lemon S M, Maniloff J, Mayo M A, McGeoch D J, Pringle C R, Wickner R B, editors. Virus taxonomy. Seventh report of the International Committee on Taxonomy of Viruses. San Diego, Calif: Academic Press; 2000. pp. 599–621. [Google Scholar]
- 13.Escutenaire S, Chalon P, Heyman P, Van der Auwera G, Van der Groen G, Verhagen R, Thomas I, Karelle-Bui L, Vaheri A, Pastoret P-P, Plyusnin A. Genetic characterization of Puumala hantavirus strains from Belgium: evidence for a distinct phylogenetic lineage. Virus Res. 2001;74:1–15. doi: 10.1016/s0168-1702(00)00224-0. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- 15.Felsenstein J. PHYLIP—Phylogeny Inference Package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 16.Giebel L B, Stohwasser R, Zoller L, Bautz E K, Darai G. Determination of the coding capacity of the M genome segment of nephropathia epidemica virus strain Hallnas B1 by molecular cloning and nucleotide sequence analysis. Virology. 1989;172:498–505. doi: 10.1016/0042-6822(89)90192-x. [DOI] [PubMed] [Google Scholar]
- 17.Gojobori T, Moriyama E N, Kimura M. Molecular clock of viral evolution, and the neutral theory. Proc Natl Acad Sci USA. 1990;87:10015–10018. doi: 10.1073/pnas.87.24.10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 19.Heiske A, Anheier B, Pilaski J, Volchkov V E, Feldmann H. A new Clethrionomys-derived hantavirus from Germany: evidence for distinct genetic sublineages of Puumala viruses in Western Europe. Virus Res. 1999;61:101–112. doi: 10.1016/s0168-1702(99)00024-6. [DOI] [PubMed] [Google Scholar]
- 20.Henderson W W, Monroe M C, St. Jeor S C, Thayer W P, Rowe J E, Peters C J, Nichol S T. Naturally occurring Sin Nombre virus genetic reassortants. Virology. 1995;214:602–610. doi: 10.1006/viro.1995.0071. [DOI] [PubMed] [Google Scholar]
- 21.Hewitt G M. Post-glacial recolonization of European biota. Biol J Linnean Soc. 1999;68:87–112. [Google Scholar]
- 22.Hillis D M, Bull J J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 1993;42:182–192. [Google Scholar]
- 23.Hjelle B, Jenison S A, Goade D E, Green W B, Feddersen R M, Scott A A. Hantaviruses: clinical, microbiologic, and epidemiologic aspects. Crit Rev Clin Lab Sci. 1995;32:469–508. doi: 10.3109/10408369509082592. [DOI] [PubMed] [Google Scholar]
- 24.Horling J, Lundkvist A, Jaarola M, Plyusnin A, Tegelström H, Persson K, Lehväslaiho H, Hornfeldt B, Vaheri A, Niklasson B. Distribution and genetic heterogeneity of Puumala virus in Sweden. J Gen Virol. 1996;77:2555–2562. doi: 10.1099/0022-1317-77-10-2555. [DOI] [PubMed] [Google Scholar]
- 25.Horling J, Lundkvist A, Persson K, Mullaart M, Dzagurova T, Dekonenko A, Tkachenko E, Niklasson B. Detection and subsequent sequencing of Puumala virus from human specimens by PCR. J Clin Microbiol. 1995;33:277–282. doi: 10.1128/jcm.33.2.277-282.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes A L, Friedman R. Evolutionary diversification of protein-coding genes of hantaviruses. Mol Biol Evol. 2000;17:1558–1568. doi: 10.1093/oxfordjournals.molbev.a026254. [DOI] [PubMed] [Google Scholar]
- 27.Hughes A L, Ota T, Nei M. Positive Darwinian selection promotes charge profile diversity in the antigen binding cleft of class I major histocompatibility complex molecules. Mol Biol Evol. 1990;7:515–524. doi: 10.1093/oxfordjournals.molbev.a040626. [DOI] [PubMed] [Google Scholar]
- 28.Huson D H. SplitsTree: a program for analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 29.Jaarola M, Tegelström H, Fredga K. Colonization history in Fennoscandian rodents. Biol J Linnean Soc. 1999;68:113–127. [Google Scholar]
- 30.Kaneko Y, Nakata K, Saitoh T, Stenseth N C, Bjornstad O N. The biology of the vole Clethrionomys rufocanus: a review. Res Pop Ecol. 1998;40:21–37. [Google Scholar]
- 31.Kariwa H, Yoshimatsu K, Araki K, Chayama K, Kumada H, Ogino M, Ebihara H, Murphy M E, Mizutani T, Takashima I, Arikawa J. Detection of hantaviral antibodies among patients with hepatitis of unknown etiology in Japan. Microbiol Immunol. 2000;44:357–362. doi: 10.1111/j.1348-0421.2000.tb02506.x. [DOI] [PubMed] [Google Scholar]
- 32.Kariwa H, Yoshizumi S, Arikawa J, Yoshimatsu K, Takahashi K, Takashima I, Hashimoto N. Evidence for the existence of Puumula-related virus among Clethrionomys rufocanus in Hokkaido, Japan. Am J Trop Med Hyg. 1995;53:222–227. doi: 10.4269/ajtmh.1995.53.222. [DOI] [PubMed] [Google Scholar]
- 33.Leitner T, Albert J. The molecular clock of HIV-1 unveiled through analysis of a known transmission history. Proc Natl Acad Sci USA. 1999;96:10752–10757. doi: 10.1073/pnas.96.19.10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lole K S, Bollinger R C, Paranjape R S, Gadkari D, Kulkarni S S, Novak N G, Ingersoll R, Sheppard H W, Ray S C. Full-length human immunodeficiency virus type I genomes from subtype c-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez N, Padula P, Rossi C, Miguel S, Edelstein A, Ramirez E, Franze-Fernandez M T. Genetic characterization and phylogeny of Andes virus and variants from Argentina and Chile. Virus Res. 1997;50:77–84. doi: 10.1016/s0168-1702(97)00053-1. [DOI] [PubMed] [Google Scholar]
- 36.Lundkvist A, Bjorsten S, Niklasson B, Ahlborg N. Mapping of B-cell determinants in the nucleocapsid protein of Puumala virus: definition of epitopes specific for acute immunoglobulin G recognition in humans. Clin Diagn Lab Immunol. 1995;2:82–86. doi: 10.1128/cdli.2.1.82-86.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lundkvist A, Niklasson B. Bank vole monoclonal antibodies against Puumala virus envelope glycoproteins: identification of epitopes involved in neutralization. Arch Virol. 1992;126:93–105. doi: 10.1007/BF01309687. [DOI] [PubMed] [Google Scholar]
- 38.Lundkvist A, Wiger D, Hörling J, Sjölander K B, Plyusnina A, Mehl R, Vaheri A, Plyusnin A. Isolation and characterization of Puumala hantavirus from Norway: evidence for a distinct phylogenetic sublineage. J Gen Virol. 1998;79:2603–2614. doi: 10.1099/0022-1317-79-11-2603. [DOI] [PubMed] [Google Scholar]
- 39.Malim M H, Emerman M. HIV-1 sequence variation: drift, shift, and attenuation. Cell. 2001;104:469–472. doi: 10.1016/s0092-8674(01)00234-3. [DOI] [PubMed] [Google Scholar]
- 40.Mitchell-Jones A J, Amori G, Bogdanowicz W, Krystufek B, Reijnders P J H, Spitzenberger F, Stubbe M, Thissen J B M, Vohralik V V, Zima J. Clethrionomys glareolus. In: Mitchell-Jones A J, editor. The atlas of European mammals. London, United Kingdom: T & AD Poyser Natural History; 1999. pp. 212–213. [Google Scholar]
- 41.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;12:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 42.Niklasson B, Hornfeldt B, Lundkvist A, Bjorsten S, Leduc J. Temporal dynamics of Puumala virus antibody prevalence in voles and of nephropathia epidemica incidence in humans. Am J Trop Med Hyg. 1995;53:134–140. doi: 10.4269/ajtmh.1995.53.134. [DOI] [PubMed] [Google Scholar]
- 43.Pilaski J, Feldmann H, Morzunov S, Rollin P E, Ruo S L, Lauer B, Peters C J, Nichol S T. Genetic identification of a new Puumala virus strain causing severe hemorrhagic fever with renal syndrome in Germany. J Infect Dis. 1994;170:1456–1462. doi: 10.1093/infdis/170.6.1456. [DOI] [PubMed] [Google Scholar]
- 44.Plyusnin A, Morzunov S P. Virus evolution and genetic diversity of hantaviruses and their rodent hosts. Curr Top Microbiol Immunol. 2001;256:47–75. doi: 10.1007/978-3-642-56753-7_4. [DOI] [PubMed] [Google Scholar]
- 45.Plyusnin A, Hörling J, Kanerva M, Mustonen J, Cheng Y, Partanen J, Vapalahti O, Kukkonen S K, Niemimaa J, Henttonen H, Niklasson B, Lundkvist A, Vaheri A. Puumala hantavirus genome in patients with nephropathia epidemica: correlation of PCR positivity with HLA haplotype and link to viral sequences in local rodents. J Clin Microbiol. 1997;35:1090–1096. doi: 10.1128/jcm.35.5.1090-1096.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plyusnin A, Mustonen J, Asikainen K, Plyusnina A, Niemimaa J, Henttonen H, Vaheri A. Analysis of Puumala hantavirus genome in patients with nephropathia epidemica and rodent carriers from the sites of infection. J Med Virol. 1999;59:397–405. doi: 10.1002/(sici)1096-9071(199911)59:3<397::aid-jmv21>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 47.Plyusnin A, Vapalahti O, Vaheri A. Hantaviruses: genome structure, expression and evolution. J Gen Virol. 1996;77:2677–2687. doi: 10.1099/0022-1317-77-11-2677. [DOI] [PubMed] [Google Scholar]
- 48.Plyusnin A, Vapalahti O, Lehväslaiho H, Apekina N, Mikhailova T, Gavrilovskaya I, Laakkonen J, Niemimaa J, Henttonen H M, Brummer-Korvenkontio M, et al. Genetic variation of wild Puumala viruses within the serotype, local rodent populations and individual animal. Virus Res. 1995;38:25–41. doi: 10.1016/0168-1702(95)00038-r. [DOI] [PubMed] [Google Scholar]
- 49.Plyusnin A, Vapalahti O, Ulfves K, Lehvaslaiho H, Apekina N, Gavrilovskaya I, Blinov V, Vaheri A. Sequences of wild Puumala virus genes show a correlation of genetic variation with geographic origin of the strains. J Gen Virol. 1994;75:405–409. doi: 10.1099/0022-1317-75-2-405. [DOI] [PubMed] [Google Scholar]
- 50.Pringle C R. Genetics and genome segment reassortment. In: Elliott R M, editor. The Bunyaviridae. New York, N.Y: Plenum Press; 1996. pp. 189–226. [Google Scholar]
- 51.Robinson M, Catzeflis F, Briolay J, Mouchiroud D. Molecular phylogeny of rodents, with special emphasis on murids: evidence from nuclear gene LCAT. Mol Phylogenet Evol. 1997;8:423–434. doi: 10.1006/mpev.1997.0424. [DOI] [PubMed] [Google Scholar]
- 52.Salemi M, Lewis M, Egan J F, Hall W W, Desmyter J, Vandamme A M. Different population dynamics of human T cell lymphotropic virus type II in intravenous drug users compared with endemically infected tribes. Proc Natl Acad Sci USA. 1999;96:13253–13258. doi: 10.1073/pnas.96.23.13253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Santti J, Hyypiä T, Kinnunen L, Salminen M. Evidence of recombination among enteroviruses. J Virol. 1999;73:8741–8749. doi: 10.1128/jvi.73.10.8741-8749.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schierup M H, Hein J. Recombination and the molecular clock. Mol Biol Evol. 2000;17:1578–1579. doi: 10.1093/oxfordjournals.molbev.a026256. [DOI] [PubMed] [Google Scholar]
- 55.Sibold C, Meisel H, Kruger D H, Labuda M, Lysy J, Kozuch O, Pejcoch M, Vaheri A, Plyusnin A. Recombination in Tula hantavirus evolution: analysis of genetic lineages from Slovakia. J Virol. 1999;73:667–675. doi: 10.1128/jvi.73.1.667-675.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strimmer K, von Haeseler A. Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc Natl Acad Sci USA. 1997;94:6815–6819. doi: 10.1073/pnas.94.13.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strimmer K, von Haeseler A. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 58.Suzuki Y, Katayama K, Fukushi S, Kageyama T, Oya A, Okamura H, Tanaka Y, Mizokami M, Gojobori T. Slow evolutionary rate of GB virus C/hepatitis G virus. J Mol Evol. 1999;48:383–389. doi: 10.1007/pl00006482. [DOI] [PubMed] [Google Scholar]
- 59.Swofford D L, Olsen G J, Waddel P J, Hillis D M. Phylogenetic inference. In: Hillis D M, Moritz G, Mable B K, editors. Molecular systematics. Sunderland, Mass: Sinauer Associates, Inc. Publishers; 1996. pp. 407–509. [Google Scholar]
- 60.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vapalahti O, Kallio-Kokko H, Närvänen A, Julkunen I, Lundkvist A, Plyusnin A, Lehväslaiho H, Brummer-Korvenkontio M, Vaheri A, Lankinen H. Human B-cell epitopes of Puumala virus nucleocapsid protein, the major antigen in early serological response. J Med Virol. 1995;46:293–303. doi: 10.1002/jmv.1890460402. [DOI] [PubMed] [Google Scholar]
- 62.Vapalahti O, Kallio-Kokko H, Salonen E M, Brummer-Korvenkontio M, Vaheri A. Cloning and sequencing of Puumala virus Sotkamo strain S and M RNA segments: evidence for strain variation in hantaviruses and expression of the nucleocapsid protein. J Gen Virol. 1992;73:829–838. doi: 10.1099/0022-1317-73-4-829. [DOI] [PubMed] [Google Scholar]
- 63.Weaver S C, Rico-Hesse R, Scott T W. Genetic diversity and slow rates of evolution in New World alphaviruses. Curr Top Microbiol Immunol. 1992;176:99–117. doi: 10.1007/978-3-642-77011-1_7. [DOI] [PubMed] [Google Scholar]
- 64.Worobey M, Holmes E C. Evolutionary aspects of recombination in RNA viruses. J Gen Virol. 1999;80:2535–2543. doi: 10.1099/0022-1317-80-10-2535. [DOI] [PubMed] [Google Scholar]
- 65.Worobey M, Rambaut A, Holmes E C. Widespread intra-serotype recombination in natural populations of dengue virus. Proc Natl Acad Sci USA. 1999;96:7352–7357. doi: 10.1073/pnas.96.13.7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xiao S Y, Spik K W, Li D, Schmaljohn C S. Nucleotide and deduced amino acid sequences of the M and S genome segments of two Puumala virus isolates from Russia. Virus Res. 1993;30:97–103. doi: 10.1016/0168-1702(93)90019-j. [DOI] [PubMed] [Google Scholar]