Abstract
Pancreatic cancer is an aggressive cancer with silent symptoms and high mortality with less than 11% of the 5-year survival rate. Until now, the significance of genes as clinical biomarkers in the early stages of pancreatic cancer has not been fully understood. Hence, this study aims to reveal the significant genes in the early stages of pancreatic cancer using bioinformatic analysis and in vitro experiments, and to serve as clinical biomarkers for early detection. We used Cancer RNA-Seq Nexus database and identified one tumor suppressor gene (NAGK), and five oncogenes (FXYD3, ACTR1A, B3GNT3, SIGIRR, and EXOC1) that are significant in the early stages of pancreatic cancer. The expression of NAGK, FXYD3, ACTR1A, B3GNT3, SIGIRR, and EXOC1 were determined from the GEPIA, UALCAN, and HPA database. It has been shown that pancreatic cancer tumor dissemination is an event that can occur in early lesions, rather than being solely restricted in the developed primary tumor. Thus, the six hub genes that were differentially expressed between stage I and stage II of primary pancreatic cancer tumors were compared to metastasis-related genes (1938 genes) in the human cancer metastasis database (HCMDB), yielding two overlapped genes (B3GNT3 and FXYD3). To establish the expression correlation between these two specific genes with metastatic characteristics of the early stage of pancreatic cancer and migratory ability in pancreatic cancer cell lines, the expression patterns of B3GNT3 and FXYD3 were examined in four different migratory abilities of pancreatic cancer cell lines, including HPAC, BxPC-3, AsPC-1, and PANC-1, as well as the normal pancreatic duct epithelial cell line HPDE6-C7. The results displayed that the expression of the FXYD3 gene was dramatically increased with the migratory ability enhanced of four pancreatic cancer cell lines. Thus, in the follow-up study, we will demonstrate the functional role of FXYD3 in pancreatic cancer tumorigenesis. This study revealed that the FXYD3 may act as a significant oncogene in the early stage of pancreatic cancer.
Keywords: Pancreatic cancer, early stages, biomarker, metastasis, FXYD3
Introduction
Pancreatic cancer (PC) is still a high malignancy cancer with less than 11% of 5-year survival rate [1]. In Taiwan, the mortality rate of PC has elevated to 67.1% since 2012 and the ranking has increased from ninth to seventh in the top ten cancer list [1]. According to the 2022 cancer data from NIH, 12% of PC patients were diagnosed in the localized stage, 30% in the regional stage, and 51% in the distant stage. PC is commonly found in patients at the age above 55 years old associated with family history, smoking, or other pancreatic diseases [2,3]. PC is commonly diagnosed in the advanced stage due to the limited early diagnostic tools, and also the patients will experience silent symptoms including abdominal pain, digestive difficulties, loss of appetite, and sudden weight loss. This causes the patients to have less suspicion of developing PC, hence, late diagnosis.
Epithelial-to-mesenchymal transition (EMT) is a transition program of the cells from epithelial phenotype to mesenchymal phenotype [4]. It plays an important role in the progression and metastasis of pancreatic cancer [5]. This phenomenon was discovered in the In vivo study that the mouse model that developed pancreatic cancer from pancreatic inflammation within two months [6]. The authors suggested the primary tumor has entered the blood circulation and contributed to metastasis in other body parts, especially the liver [6,7]. However, a study discovered that the inhibition of the EMT-inducing transcription factor (Snail and Twist) did not affect tumor progression and migration [8]. Therefore, this highlights the need to identify the genes contributing to metastasis in the early stages of pancreatic cancer.
The increased discovery of diagnostic biomarkers improves the detection in the advanced and metastatic stages. In Taiwan, the approved tumor markers to measure PC are Carbohydrate Antigen 19-9 (CA19-9) and Carcinoembryonic antigen (CEA) [9,10]. These biomarkers are commonly used for PDAC patients at an advanced stage but are not sensitive to early detection. However, the markers’ level can also be affected by other factors, including other disease. Furthermore, not all patients will demonstrate an elevated level of these biomarkers.
It is urgent to determine the diagnostic biomarkers for the early stage of PC. This study used bioinformatic analysis and performed validation experiments to identify the significant gene in the early stage of pancreatic cancer.
Material and methods
Cancer RNA-Seq Nexus (CRN) database
Cancer RNA-Seq Nexus, a database of phenotype-specific transcriptome profiling in cancer cells [11]. This database was used to determine the significant tumor suppressor genes and oncogenes in the early stages (stage I and II) of pancreatic cancer.
Bioinformatic analysis
Gene Expression Profiling Interactive Analysis 2 (GEPIA2) was used to predict the gene expression across TCGA cancers with normal and tumor samples. The University of ALabama at Birmingham CANcer (UALCAN) data analysis Portal was used to demonstrate the correlation between gene expression and clinicopathological characteristics, including age, gender, cancer stage, tumor grade, nodal metastasis status, alcohol status, diabetes status, chronic status among the pancreatic cancer patients.
Immunohistochemistry (IHC) assay
Pancreas tissue microarray (PA2082a) was purchased from US Biomax (Rockville, MD, USA) which contained 96 cases with duplicate cores and was used for IHC assay. In brief, the slides were baked in pepsin for 15 minutes to retrieve antigens. Next, the slides were incubated with primary antibody against FXYD3 (Abcam, ab205534, 1:500) or B3GNT3 (Proteintech, 18098-1-AP, 1:100) for 16 hours at room temperature. This was followed by the incubation of secondary antibodies for 30 minutes. The slides were stained with DAB solution for 10 minutes followed by hematoxylin. The intensity of the slides was visualized using the Motic DSAssistant (4K) and quantified using the ImageJ quantification software. The staining intensity was calculated with the formula: (255 - mean grey value) × %Area.
Cell culture
Human pancreatic duct epithelial cell line (HPDE6-C7) and pancreatic cancer cell lines (AsPC-1, BXPC-3, PANC-1, HPAC) were used. HPDE6-C7 was cultured in Keratinocyte-SFM supplemented with 1% antimycotic. PANC-1 and HPAC were cultured in DMEM with an additional 10% fetal bovine serum (FBS), 1% Penicillin-Streptomycin, 1% L-glutamine, 1% sodium pyruvate, 1% MEM NEAA. AsPC-1 and BXPC-3 were cultured in RPMI 1640 with an additional 10% fetal bovine serum (FBS), 1% Penicillin-Streptomycin, 1% L-glutamine, 1% sodium pyruvate, 1% MEM NEAA, and 2.5 g/L glucose. All cell lines were kept in a constant temperature incubator at 37°C and 5% CO2.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA isolation from cells was completed by GenezolTM TriRNA Pure Kit. NANODrop 2000 system was used to determine the RNA concentration. According to the kit instructions of miScript II RT kit and PrimeScript RT Master Mix, mRNA was transcribed into cDNA. cDNAs were used as the templates for the qPCR. EnTURBO SYBRGreen PCR SuperMix (ELK Biotechnology) was used for qPCR based on the provided protocol. The three-step amplification procedure was performed as follows: 1 cycle of pre-denaturation at 95°C for 30 sec; 40 cycles of denaturation at 95°C for 5 sec, annealing at 55°C for 30 sec, extension at 72°C for 30 sec; 1 cycle of melting at 95°C for 10 sec, 65°C for 60 sec, and 95°C for 1 sec. The primers are listed in Table 1.
Table 1.
List of primer sequences
| Gene | Primer sequences (5’→3’) | |
|---|---|---|
|
| ||
| Forward | Reverse | |
| NAGK | GTTTTCTTCTGGCGCTGACC | GCGCTATAGTCCATGGGGAG |
| ACTR1A | CTTTCCAAACTATGTGGGCCG | AGAAAACTTCGGCAGCTCGT |
| FXYD3 | TTGTGTTCCTGGCAGGCTTT | CTGGATGGTGACCGGACTT |
| B3GNT3 | TGCTCAACGGGGATGATGAC | GATCAGTTGCCCCACGAAGA |
| EXOC1 | ACCAAATGATGAACGCCTGC | ACAGCAAGATCTCGAAGGGC |
| SIGIRR | CACTGAAGTCTATGGGGCCTT | ACGTTGAGACGGCACTTGAC |
| Vimentin | GGACCAGCTAACCAACGACA | AAGGTCAAGACGTGCCAGAG |
| E-Cadherin | GCTGGACCGAGAGAGTTTCC | CAAAATCCAAGCCCGTGGTG |
| GAPDH | GTCTCCTCTGACTTCAACAGCG | ACCACCCTGTTGCTGTAGCCAA |
Human Cancer Metastasis database and EMTome analysis
Human Cancer Metastasis database (HCMDB) [12], metastasis-related gene database, and EMTome [13], EMT-related gene database. HCMDB (1938 genes) and EMTome (936 genes) were used to predict the metastasis gene in pancreatic cancer.
Pathway enrichment analysis from the GEO database
Two Gene Expression Omnibus (GEO) datasets of pancreatic cancer, GSE15471 (36 pairs of normal and tumor tissue samples) and GSE16515 (36 tumor samples and 16 normal samples) were selected to perform the pathway enrichment pathway. Log2FC>1 and P<0.05 were set as the cutoff point. Enrichr was used to screen the biological pathway via the Molecular Signatures Database (MSigDb) [14]. The enrichment pathway was plotted using ImageGP.
Cell transfection
siRNAs of FXYD3 were obtained from Purigo (Taiwan) (Table 2). AsPC-1 or PANC-1 cells were separately transfected with siRNAs at concentrations of 50 nM and 100 nM for 48 hours using the INTERFERin transfection reagent (Polypus Transfection, New York, NY, USA) by following the manufacturer’s protocol.
Table 2.
Sequences of small interfering RNAs (siRNAs) of FXYD3
| siRNAs | Sense (5’-3’) | Anti-sense (5’-3’) |
|---|---|---|
| siFXYD3_1 | UUCUAGGUCAUUGGCGUCC | GGACGCCAAUGACCUAGAA |
| siFXYD3_3 | UAUCUUCUAGGUCAUUGGC | GCCAAUGACCUAGAAGAUA |
Western blot
After transfection, protein extraction from cells was completed by using RIPA lysis at 30 minutes and absorbance was determined at 595 nM. The extracted protein samples were loaded into 10% and 15% SDS-PAGE and transferred onto PVDF membranes for 1 hour. After 1 hour of blocking with 5% BSA solution, the membranes were incubated with primary antibodies overnight at 4°C. The primary antibodies against FXYD3 (Abcam, ab205534, 1:1000), Vimentin (Cell Signaling, 3932, 1:1000), E-cadherin (BD Transduction, 610182, 1:5000), and β-actin (Proteintech, 66009-1-Ig, 1:10000) were used. On the next day, the membranes were washed with TBST at 10 minutes three times and incubated with secondary antibodies for 1 hour. The protein bands were visualized using the ECL detection reagent. ImageJ quantification software was used to analyze and quantify the band intensity.
Cell viability assay
After 48 hours of cell transfection, the PC cell lines were collected and resuspended in the complete culture medium. The cells were seeded at the density of 2 × 104 cells per well in 96-well plates and incubated for 48 hours and 72 hours at 37°C. After the incubation, the culture medium was removed. The cells were stained with crystal violet for 1 hour at room temperature. The purple formazan products were solubilized by using the citrate buffer. The absorbance was examined at 595 nm.
Colony formation assay
After 48 hours of cell transfection, the PC cell lines were collected and resuspended in the complete culture medium. The cells were seeded at the density of 4 × 103 cells per well in six-well plates and incubated for 8 days at 37°C. Every two days, the culture medium was replaced. After the incubation, the culture medium was removed. The cells were stained with crystal violet for 1 hour at room temperature. The cell colonies were recorded using ImageJ quantification software.
Wound healing assay
After 48 hours of cell transfection, the PC cell lines were collected and resuspended in the complete culture medium. The cells were seeded in 12-well plates with Ibidi culture insert 2 wells (Ibidi) and incubated for 24 hours at 37°C. Once the cell was attached, the culture inserts were removed. The 12-well plates were filled with 1 mL complete culture medium. The cells were observed using a light microscope at different times. The area of cell gaps was recorded using ImageJ quantification software. The percentage of wound closure was calculated with the formula: (length at initial - length at end)/(length at initial) × 100%.
Pathway enrichment analysis from the RNA-sequencing of FXYD3 knockdown
RNA-sequencing (RNA-seq) libraries were constructed using samples of FXYD3 knockdown against control. The reads were mapped to reference genome (hg38) by Hisat2 (version 2.2.1) and htseq-count by HTSeq (version 0.6.0). The candidate genes were assessed by DESeq2 (version 1.42.1). Genes with Log2FC>2 or Log2FC<-2 were known as the differential expressed genes (DEGs). Enrichr was used to screen the biological pathway via Molecular Signatures Database (MSigDb) [14]. The enrichment pathway was plotted using ImageGP.
Statistical analysis
All data were reported as mean ± standard deviation. GraphPad Prism 8.0 software was used to perform statistical analysis with a two-tailed Student’s t-test. P<0.05 was determined as a significant value.
Results
NAGK, ACTR1A, B3GNT3, EXOC1, FXYD3, SIGIRR are significant genes in early stages of pancreatic cancer
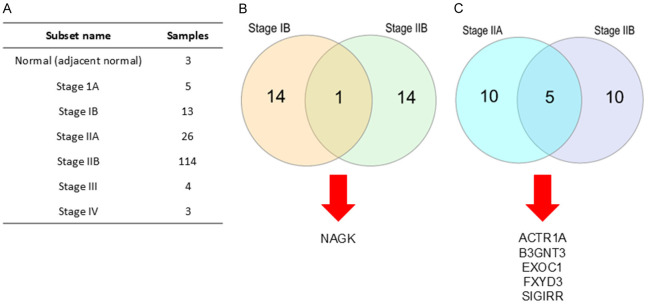
The significant genes in the early stages of pancreatic cancer were analyzed using TCGA data from the CRN database (Figure 1A). In the group of tumor suppressor genes, the top 15 genes in stages IB and IIB were determined. We found out that the N-acetylglucosamine kinase (NAGK) was intersected as a significant tumor suppressor gene (Figure 1B). In the group of oncogenes, the top 15 genes in stages IIA and IIB were also determined. We found the Beta-1,3-N-acetylglucosaminyltransferase 3 (B3GNT3), Single Ig IL-1-related receptor (SIGIRR), Exocyst Complex Component 1 (EXOC1), FXYD domain-containing ion transport regulator 3 (FXYD3), Actin Related Protein 1A (ACTR1A) were intersected as the significant oncogenes (Figure 1C). These findings predicted that NAGK, ACTR1A, B3GNT3, EXOC1, FXYD3, and SIGIRR are significant genes in the early stages of pancreatic cancer.
Figure 1.
The RNA-seq data of 165 PC samples and 3 normal pancreas samples from TCGA were obtained from the CRN database. (A) The dataset consisted 7 subsets and 168 samples. The top 15 intersected genes from early stages samples and normal samples were analyzed and classified into (B) tumor suppressor genes and (C) oncogenes.
Upregulated NAGK, ACTR1A, B3GNT3, EXOC1, FXYD3, SIGIRR with poor survival outcome in pancreatic cancer
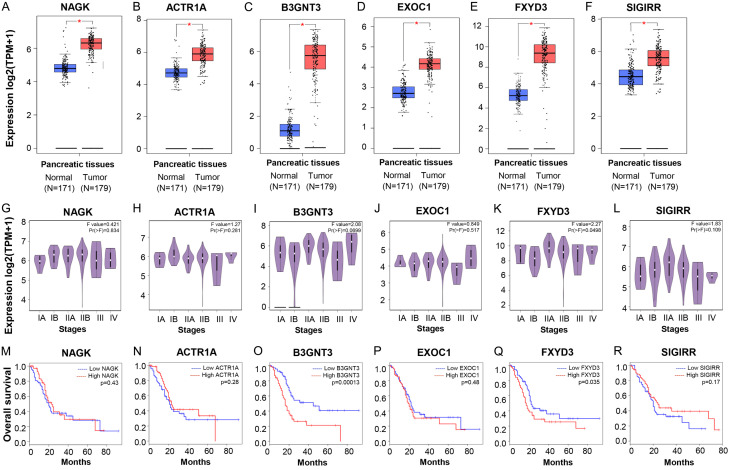
The six significant genes were observed in the GEPIA2 database. As the oncogenes, ACTR1A, B3GNT3, EXOC1, FXYD3, and SIGIRR were significantly upregulated in tumor tissue (P<0.05) compared to normal tissue (Figure 2B-F). In addition, the database suggested that the ACTR1A (stage IIB), B3GNT3 (stage IIB), EXOC1 (stage IIB), FXYD3 (stage IIB), and SIGIRR (stage IB, IIA, IIB) were significantly expressed in early stages pancreatic cancer (Figure 2H-L). Based on the overall survival, the patients with high B3GNT3 (P=0.00013) and high FXYD3 (P=0.0035) had significantly low survival rates while patients with high SIGIRR (P=0.17), high EXOC1 (P=0.48), and high ACTR1A (P=0.28) had no significant survival rates (Figure 2N-R). Surprisingly, as a predicted tumor suppressor gene in the CRN database, NAGK was upregulated in tumor tissue (P<0.05) and significantly expressed in stage IIB (Figure 2A, 2G). However, NAGK expression has no significant survival rate (Figure 2M). Taken together, these results suggested the high expression of ACTR1A, B3GNT3, EXOC1, FXYD3, and SIGIRR in the early stages of pancreatic cancer.
Figure 2.
The expression of NAGK, ACTR1A, B3GNT3, EXOC1, FXYD3, SIGIRR compared with normal and tumor pancreas tissue based on the TCGA and GTEx data by GEPIA. A-F. Boxplot expression. G-L. Substage expression. M-R. Overall survival.
Clinicopathological characteristics of NAGK, ACTR1A, B3GNT3, EXOC1, FXYD3, SIGIRR in pancreatic cancer
The expression of NAGK, ACTR1A, B3GNT3, EXOC1, FXYD3, SIGIRR, and the correlation with clinicopathological characteristics in pancreatic cancer patients was determined using the UALCAN data analysis Portal (Table 3). NAGK expression was significantly correlated with age (41-80 years old), gender (Female P=2.14E-05, Male P=2.31E-06), cancer stage (Stage 1 P=1.18E-04, Stage 2 P=3.98E-06), nodal metastasis (N0 P=1.48E-06, N1 P=1.88E-05), alcohol history (Yes P=1.42E-04, No P=2.01E-04), diabetes history (Yes P=8.40E-04, No P=3.09E-07), pancreatitis status (Yes P=1.40E-04, No P=1.59E-05). B3GNT3 expression was significantly correlated with age (above 41 years old), gender (Female P=5.01E-11, Male P=1.09E-13), cancer stage (Stage 2 P=7.64E-11), nodal metastasis (N0 P=6.30E-12, N1 P=1.27E-11), alcohol history (Yes P=4.32E-09, No P=1.11E-06), diabetes history (Yes P=2.65E-09, No P=1.20E-11), pancreatitis status (Yes P=3.99E-05, No P=4.42E-11). FXYD3 expression was significantly correlated with age (above 41 years old), gender (Female P=1E-12, Male P=1.62E-12), cancer stage (Stage 2 P=1E-12), nodal metastasis (N0 P=2.78E-11, N1 P=1.62E-12), alcohol history (Yes P=2.32E-06, No P=5.85E-07), diabetes history (Yes P=1.09E-07, No P=1E-12), pancreatitis status (Yes P=1.01E-04, No P=1.62E-12). Referring to the nodal metastasis status, two oncogenes, B3GNT3 and FXYD3, were highly significant (P<0.05). These results suggested the overexpression of B3GNT3 and FXYD3 may be associated with tumor metastasis in the early stage of pancreatic cancer.
Table 3.
Clinicopathological characteristics of NAGK, ACTR1A, B3GNT3, EXOC1, FXYD3, SIGIRR in pancreatic cancer patients by UALCAN
| Parameters | p value | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| NAGK | ACTR1A | B3GNT3 | EXOC1 | FXYD3 | SIGIRR | ||
| Age | 21-40 | 0.007 | 0.950 | 0.321 | 0.384 | 0.356 | 0.803 |
| 41-60 | 1.54E-05 | 0.864 | 1.87E-12 | 0.737 | 2.38E-13 | 0.655 | |
| 61-80 | 2.71E-06 | 0.602 | 8.78E-12 | 0.844 | <1E-12 | 0.722 | |
| 81-100 | 0.003 | 0.387 | 3.94E-05 | 0.697 | 7.51E-05 | 0.426 | |
| Gender | Female | 2.14E-05 | 0.824 | 5.01E-11 | 0.958 | <1E-12 | 0.552 |
| Male | 2.31E-06 | 0.665 | 1.09E-13 | 0.941 | 1.62E-12 | 0.783 | |
| Cancer stage | 1 | 1.18E-04 | 0.520 | 0.073 | 0.650 | 0.033 | 0.675 |
| 2 | 3.98E-06 | 0.634 | 7.64E-11 | 0.995 | <1E-12 | 0.723 | |
| 3 | 0.079 | 0.776 | 0.113 | 0.316 | 0.061 | 0.428 | |
| 4 | 0.006 | 0.942 | 0.063 | 0.499 | 0.038 | 0.320 | |
| Tumor grade | 1 | 0.862 | 0.323 | 0.101 | 0.979 | 0.204 | 0.209 |
| 2 | 0.937 | 0.640 | 0.027 | 0.815 | 0.889 | 0.107 | |
| 3 | 0.939 | 0.674 | 0.019 | 0.317 | 0.034 | 0.108 | |
| 4 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Nodal Metastasis status | N0 | 1.48E-06 | 0.914 | 6.30E-12 | 0.805 | 2.78E-11 | 0.998 |
| N1 | 1.88E-05 | 0.656 | 1.27E-11 | 0.970 | 1.62E-12 | 0.564 | |
| Alcohol history | Yes | 1.42E-04 | 0.634 | 4.32E-09 | 0.459 | 2.32E-06 | 0.915 |
| No | 2.01E-04 | 0.361 | 1.11E-06 | 0.962 | 5.85E-07 | 0.924 | |
| Diabetes history | Yes | 8.40E-04 | 0.717 | 2.65E-09 | 0.607 | 1.09E-07 | 0.827 |
| No | 3.09E-07 | 0.749 | 1.20E-11 | 0.920 | <1E-12 | 0.789 | |
| Chronic pancreatitis status | Yes | 1.40E-04 | 0.496 | 3.99E-05 | 0.677 | 1.01E-04 | 0.425 |
| No | 1.59E-05 | 0.682 | 4.42E-11 | 0.781 | 1.62E-12 | 0.673 | |
FXYD3 and B3GNT3 are highly expressed in the early stages of pancreatic cancer
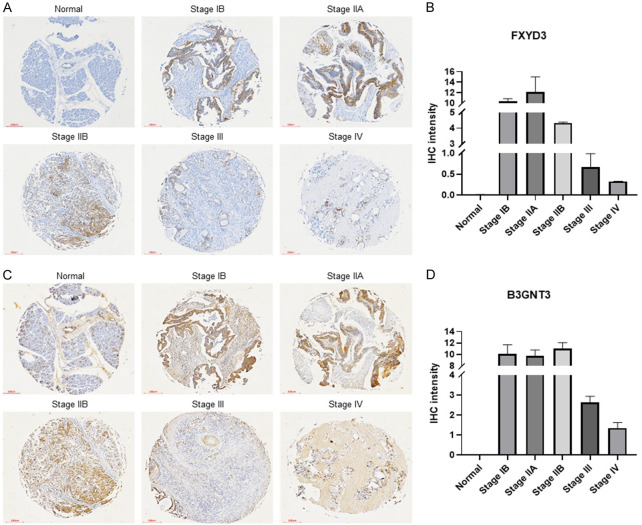
To determine the immunohistochemistry-based protein expression of the genes in pancreatic cancer, an IHC assay was performed. The assay demonstrated the high protein expression of FXYD3 (Figure 3A, 3B) and B3GNT3 (Figure 3C, 3D) in early stages (stage IB, stage IIA, stage IIB) compared to the normal pancreas tissue. To further confirm the protein expression in patients, the human protein atlas (HPA) database was used (Figure S1). The protein expression level was manually calculated with the intensity of the antibody stain (negative, weak, moderate, strong). The results demonstrated the protein expression level of FXYD3 (25% weak intensity, 58% moderate intensity, 17% strong intensity) was highly detected in pancreatic tissue tissues while the protein expression level of B3GNT3 (42% negative intensity, 42% moderate intensity, 17% strong intensity) was not significantly expressed. The predicted tumor suppressor gene, NAGK, showed 75% negative intensity of protein expression level in pancreatic cancer tissue (Figure S1). These results suggested that the FXYD3 and B3GNT3 may play an important role in the early detection of pancreatic cancer.
Figure 3.
Immunohistochemistry stain of FXYD3 and B3GNT3 expression. A. TMA slides of FXYD3 expression. B. IHC intensity of FXYD3 expression. C. TMA slides of B3GNT3 expression. D. IHC intensity of B3GNT3 expression. Bar =100 µm.
Gene expressions of NAGK, ACTR1A, B3GNT3, EXOC1, FXYD3, SIGIRR in pancreatic cancer cell lines
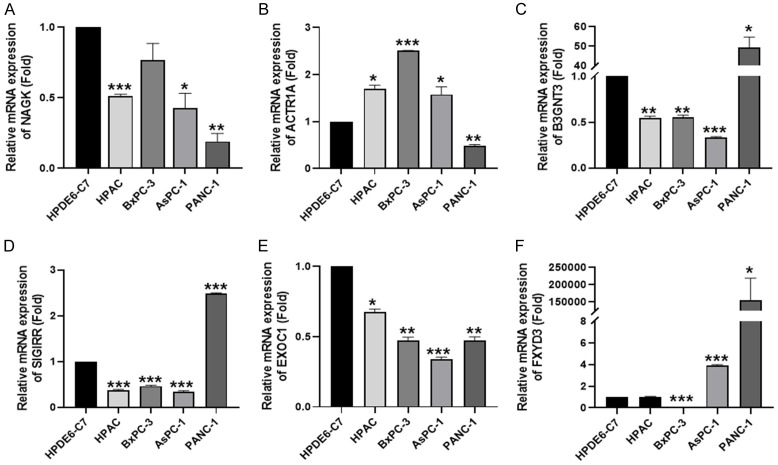
To understand the gene expression of NAGK, B3GNT3, SIGIRR, EXOC1, FXYD3, and ACTR1A in the pancreatic cancer cell lines, RT-qPCR analysis was used (Figure 4). The low NAGK expression in pancreatic cancer cell lines did not match with the bioinformatic analysis (Figure 2A, 2G, 2M). The high expression of B3GNT3, SIGIRR, and FXYD3 in pancreatic cancer cell lines indicated that they are oncogenes in pancreatic cancer. Considering that AsPC-1 and PANC-1 cell lines have a migratory ability [15], it is suggested that B3GNT3, SIGIRR, and FXYD3 may play a metastatic role in pancreatic cancer.
Figure 4.
The gene expression of NAGK, ACTR1A, B3GNT3, EXOC1, FXYD3, SIGIRR in pancreatic cancer cell line. Expression of (A) NAGK, (B) ACTR1A, (C) B3GNT3, (D) EXOC1, (E) FXYD3, (F) SIGIRR in HPDE6-C7 and pancreatic cancer cell lines (HPAC, BxPC-3, AsPC-1, PANC-1). Statistical significance is indicated by *P<0.05, **P<0.02, ***P<0.001.
B3GNT3 and FXYD3 contribute to metastasis role in pancreatic cancer
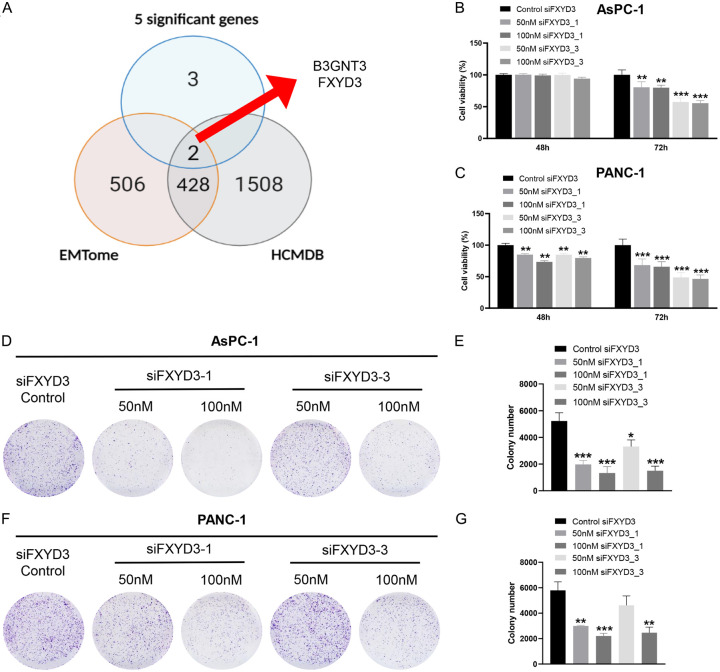
To determine the metastasis role of oncogenes in pancreatic cancer, we used the HCMDB (1938 genes) and EMTome (936 genes) to predict the metastasis gene in pancreatic cancer. B3GNT3 and FXYD3 were found in the two databases as the metastatic genes in different cancers including colorectal cancer and breast cancer (Figure 5A). Besides that, two GEO datasets (GSE15471 and GSE16515) containing B3GNT3 and FXYD3 revealed the significant enrichment of biological role in epithelial-mesenchymal transition (EMT) (Figure S2). This finding suggested that the B3GNT3 and FXYD3 may act as the metastatic genes in pancreatic cancer.
Figure 5.
FXYD3 knockdown inhibited the cell proliferation. (A) The prediction of EMT genes from EMTome and HCMDB. The cell viability of (B) AsPC-1 and (C) PANC-1 collected after 48 h and 72 h. The cell colonies of (D, E) AsPC-1 and (F, G) PANC-1. Statistical significance is indicated by *P<0.05, **P<0.02, ***P<0.001.
FXYD3 knockdown inhibits the proliferation and migration of pancreatic cancer cells
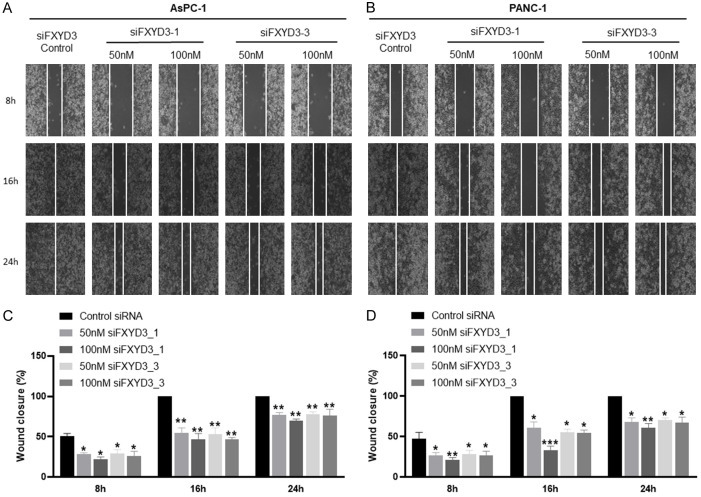
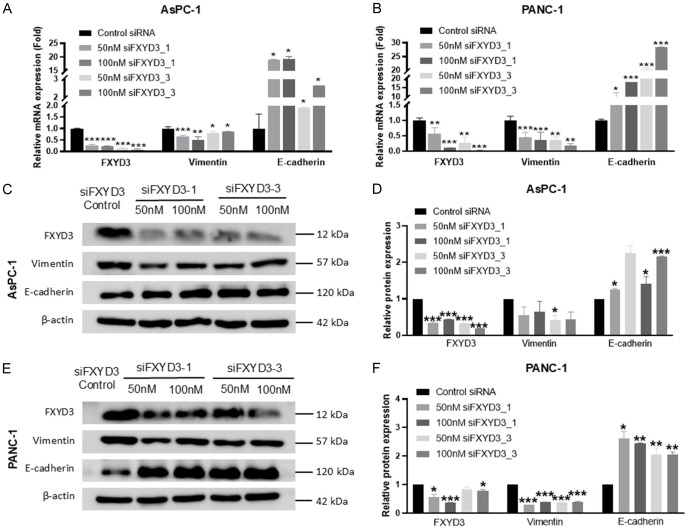
With the high expression in both migratory cell lines (AsPC-1 and PANC-1 cells), the role of FXYD3 was further determined using the experimental method. The AsPC-1 and PANC-1 cell lines were transfected with a control group and FXYD3 knockdown groups (siFXYD3_1 50 nM and siFXYD3_1 100 nM, siFXYD3_3 50 nM and siFXYD3_3 100 nM). The cell viability assay demonstrated a decrease in cell proliferation in the FXYD3 knockdown groups of AsPC-1 and PANC-1 cells after 72 hours (Figure 5B, 5C). However, no decrease or slight decrease was observed after 48 hours. Furthermore, the colony formation assay demonstrated that the number of colony cells was low in the FXYD3 knockdown groups compared to the control group in AsPC1 (Figure 5D, 5E) and PANC-1 cells (Figure 5F, 5G). The wound healing assay indicated that the migration of AsPC-1 (Figure 6A, 6B) and PANC-1 cells (Figure 6C, 6D) was inhibited by the FXYD3 knockdown as compared to the control groups. Next, the expression of FXYD3 in the PC cells was measured using RT-qPCR and western blot analysis. The results confirmed the FXYD3 knockdown in AsPC-1 and PANC-1 cells (Figure 7A, 7B). To understand the EMT role of FXYD3, the expression of epithelial marker (E-cadherin) and mesenchymal marker (Vimentin) were analyzed in the FXYD3 knockdown groups. The results confirmed the low expression of Vimentin and high expression of E-cadherin in FXYD3 knockdown groups compared to the control group in AsPC-1 (Figure 7C, 7D) and PANC-1 cells (Figure 7E, 7F). Taken together, FXYD3 plays an important role in pancreatic cancer by promoting cell proliferation and migration.
Figure 6.
FXYD3 knockdown inhibited the cell migration. The wound closure of (A, C) AsPC-1 and (B, D) PANC-1 observed at different time. Statistical significance is indicated by *P<0.05, **P<0.02, ***P<0.001. Bar =100 µm.
Figure 7.
FXYD3 knockdown inhibited the EMT. The relative mRNA expression of FXYD3, Vimentin, E-cadherin in (A) AsPC1 and (B) PANC-1. The relative protein expression of FXYD3, Vimentin, E-cadherin in (C, D) AsPC1 and (E, F) PANC-1. Statistical significance is indicated by *P<0.05, **P<0.02, ***P<0.001.
RNA-sequencing analysis of FXYD3
The mechanism of FXYD3 in cancers including pancreatic cancer remains unclear. To further look into the mechanisms, RNA-sequencing (RNA-seq) of FXYD3 knockdown samples was performed. There was a total of 292 differential expressed genes with 93 downregulated and 199 upregulated (Figure S3A). The pathways are mainly involved in the epithelial-mesenchymal transition, p53 pathway, spermatogenesis, and peroxisome (Figure S3B). Further EMT pathway analysis showed the related genes including LAMA1, DPYSL3, LOXL1, FBLN5, DCN, and LOXL2 (Figure S3C). The finding of the EMT pathway is consistent with the enrichment pathway obtained from the GEO datasets (Figure S2). This result suggested that FXYD3 may play a role in cell migration to promote EMT.
Discussion
Pancreatic cancer is still a high mortality death cancer with limited biomarker detection for early-stage diagnosis. In this study, six genes (NAGK, ACTR1A, B3GNT3, EXOC1, FXYD3, SIGIRR) were determined as significant genes in the early stages of pancreatic cancer. The bioinformatic analysis using the TCGA database revealed a strong correlation between poor overall survival rates in pancreatic cancer patients and the high expression of B3GNT3 and FXYD3. In addition, the mRNA analysis reported low expression of NAGK while high expression of ACTR1A, B3GNT3, FXYD3, and SIGIRR in pancreatic cancer cell lines. These data revealed the importance of B3GNT3 and FXYD3 as the EMT genes in the early stages of pancreatic cancer.
The bioinformatics and mRNA analysis of NAGK expression contradicts the results. The CRN database described NAGK as a tumor suppressor gene, however, the GEPIA database demonstrated that NAGK is highly expressed in tumor samples. To further understand the role of NAGK, the mRNA expression of NAGK was analyzed using the RT-qPCR. Surprisingly, NAGK expression is low in pancreatic cancer cells compared to normal pancreas cells. The low NAGK expression in the current study conflicts with prior studies [16,17]. The presence of NAGK in the hexosamine biosynthesis pathway (HBP) contributed to the development of pancreatic tumors. We suggested the low expression of NAGK in this study may be caused by the presence of GFAT1 for glucose supply [17].
B3GNT3 has been well studied as an oncogene in different types of cancer including non-small cell lung cancer, pancreatic cancer, cervical cancer, and gynecologic cancer [18-22]. According to several studies, the high expression of B3GNT3 in pancreatic cancer was linked to tumor growth and a poor prognosis [19,22]. B3GNT3 was also found to be involved in epithelial-to-mesenchymal transition (EMT) [22]. The findings were consistent with the present study.
Unlike the B3GNT3, the expression of ACTR1A, EXOC1, and SIGIRR showed inconsistent results. These genes showed high expression in none or one pancreatic cancer cell. Due to the experimental validations being inconsistent with the predicted findings from the bioinformatic analysis, these genes are not further discussed in this study.
FXYD3 has been investigated in various cancers including breast cancer, pancreatic cancer, cervical cancer, and colon cancer [23-28]. FXYD3 is highly expressed in these cancers and can promote proliferation, migration, and invasion. However, the mechanism is still unclear. To the best of our knowledge, this is the first study using bioinformatic analysis and experiments to reveal FXYD3 as an early-stage oncogene in pancreatic cancer.
A previous study performed by Peng et al. successfully demonstrated the proliferation, migration, and invasion of FXYD3 in BxPC-3 and CAPAN2 cells [25]. However, we determined the expression of FXYD3 was highly expressed in PANC-1 and AsPC-1 cells while low expression in BxPC-3 cells. By confirming the high expression of FXYD3 in metastatic cell lines, this clearly stated the role of FXYD3 as a metastatic gene [29,30]. The FXYD3 knockdown inhibits the EMT activity by causing the cancer cells to have tight adhesion in pancreatic cancer [31]. Besides that, the pathway enrichment analysis illustrated that FXYD3-related genes were significantly related to the epithelial-mesenchymal transition pathway. We suggested that FXYD3 is a highly metastatic gene in the early stage of pancreatic cancer.
Conclusion
The tumor suppressor gene, NAGK, and the oncogenes, ACTR1A, B3GNT3, EXOC1, FXYD3, SIGIRR were found to be significant in the early stages of pancreatic cancer. FXYD3 is associated with poor outcomes in pancreatic cancer patients at early stages. In addition, it is involved in cell proliferation and migration. This study demonstrated that the FXYD3 is predicted as a biomarker of early-stage pancreatic cancer.
Acknowledgements
This research was funded by Department of Medical Research, Tri-Service General Hospital, National Defense Medical Center (grant numbers: TSGH-E-112208 and TSGH-E-113300; awarded to Y.-F.W.), the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan (grant numbers: DP2-111-21121-01-C-01-03, DP2-TMU-112-C-05, and DP2-TMU-113-C-05), the Health and Welfare Surcharge of Tobacco Products of Taiwan (Hualien Tzu Chi Hospital Joint Cancer Center; grant numbers: MOHW111-TDU-B-221-014013 and MOHW112-TDU-B-221-124013; awarded to K.-H.L.), National Science and Technology Council (grant numbers: NSTC 112-2320-B-038-058 and NSTC 113-2320-B-038-013; awarded to K.-H.L.), and Taipei Medical University Research Center of Cancer Translational Medicine (Featured Areas Research Center Program, within the framework of the Higher Education Sprout Project by the Taiwanese Ministry of Education).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Lee HA, Chen KW, Hsu CY. Prediction model for pancreatic cancer-a population-based study from NHIRD. Cancers (Basel) 2022;14:882. doi: 10.3390/cancers14040882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 3.Hu JX, Zhao CF, Chen WB, Liu QC, Li QW, Lin YY, Gao F. Pancreatic cancer: a review of epidemiology, trend, and risk factors. World J Gastroenterol. 2021;27:4298–4321. doi: 10.3748/wjg.v27.i27.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fares J, Fares MY, Khachfe HH, Salhab HA, Fares Y. Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct Target Ther. 2020;5:28. doi: 10.1038/s41392-020-0134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filip S, Vymetalkova V, Petera J, Vodickova L, Kubecek O, John S, Cecka F, Krupova M, Manethova M, Cervena K, Vodicka P. Distant metastasis in colorectal cancer patients-do we have new predicting clinicopathological and molecular biomarkers? A comprehensive review. Int J Mol Sci. 2020;21:5255. doi: 10.3390/ijms21155255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, Reichert M, Beatty GL, Rustgi AK, Vonderheide RH, Leach SD, Stanger BZ. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Usón PLS Junior, Tolentino FDS, Santos VM, Rother ET, Maluf FC. The impact of metastatic sites in advanced pancreatic adenocarcinoma, systematic review and meta-analysis of prospective randomized studies. PLoS One. 2020;15:e0230060. doi: 10.1371/journal.pone.0230060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS, Kalluri R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu YF, Liu TW, Shan YS, Chen JS, Li CP, Ho CL, Hsieh RK, Hwang TL, Chen LT, Ch’ang HJ Taiwan Cooperative Oncology Group pancreatic cancer study group. Carbohydrate Antigen 19-9 response to initial adjuvant chemotherapy predicts survival and failure pattern of resected pancreatic adenocarcinoma but not which patients are suited for additional adjuvant chemoradiation therapy: from a prospective randomized study. Int J Radiat Oncol Biol Phys. 2023;117:74–86. doi: 10.1016/j.ijrobp.2023.02.061. [DOI] [PubMed] [Google Scholar]
- 10.van Manen L, Groen JV, Putter H, Vahrmeijer AL, Swijnenburg RJ, Bonsing BA, Mieog JSD. Elevated CEA and CA19-9 serum levels independently predict advanced pancreatic cancer at diagnosis. Biomarkers. 2020;25:186–193. doi: 10.1080/1354750X.2020.1725786. [DOI] [PubMed] [Google Scholar]
- 11.Li JR, Sun CH, Li W, Chao RF, Huang CC, Zhou XJ, Liu CC. Cancer RNA-Seq Nexus: a database of phenotype-specific transcriptome profiling in cancer cells. Nucleic Acids Res. 2016;44:D944–951. doi: 10.1093/nar/gkv1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng G, Ma Y, Zou Y, Yin A, Li W, Dong D. HCMDB: the human cancer metastasis database. Nucleic Acids Res. 2018;46:D950–D955. doi: 10.1093/nar/gkx1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vasaikar SV, Deshmukh AP, den Hollander P, Addanki S, Kuburich NA, Kudaravalli S, Joseph R, Chang JT, Soundararajan R, Mani SA. EMTome: a resource for pan-cancer analysis of epithelial-mesenchymal transition genes and signatures. Br J Cancer. 2021;124:259–269. doi: 10.1038/s41416-020-01178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM, Jeon M, Ma’ayan A. Gene set knowledge discovery with enrichr. Curr Protoc. 2021;1:e90. doi: 10.1002/cpz1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deer EL, González-Hernández J, Coursen JD, Shea JE, Ngatia J, Scaife CL, Firpo MA, Mulvihill SJ. Phenotype and genotype of pancreatic cancer cell lines. Pancreas. 2010;39:425–435. doi: 10.1097/MPA.0b013e3181c15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell S, Mesaros C, Izzo L, Affronti H, Noji M, Schaffer BE, Tsang T, Sun K, Trefely S, Kruijning S, Blenis J, Blair IA, Wellen KE. Glutamine deprivation triggers NAGK-dependent hexosamine salvage. Elife. 2021;10:e62644. doi: 10.7554/eLife.62644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim PK, Halbrook CJ, Kerk SA, Radyk M, Wisner S, Kremer DM, Sajjakulnukit P, Andren A, Hou SW, Trivedi A, Thurston G, Anand A, Yan L, Salamanca-Cardona L, Welling SD, Zhang L, Pratt MR, Keshari KR, Ying H, Lyssiotis CA. Hyaluronic acid fuels pancreatic cancer cell growth. Elife. 2021;10:e62645. doi: 10.7554/eLife.62645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W, Hou T, Niu C, Song L, Zhang Y. B3GNT3 expression is a novel marker correlated with pelvic lymph node metastasis and poor clinical outcome in early-stage cervical cancer. PLoS One. 2015;10:e0144360. doi: 10.1371/journal.pone.0144360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong K, Zhao Y, Xia L, Jiang H, Xu M, Zheng J. B3GNT3: a prognostic biomarker associated with immune cell infiltration in pancreatic adenocarcinoma. Oncol Lett. 2021;21:159. doi: 10.3892/ol.2020.12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J, Guo Z, Yuan S, Li H, Luo S. Upregulation of B3GNT3 is associated with immune infiltration and activation of NF-kappaB pathway in gynecologic cancers. J Reprod Immunol. 2022;152:103658. doi: 10.1016/j.jri.2022.103658. [DOI] [PubMed] [Google Scholar]
- 21.Gao L, Zhang H, Zhang B, Zhu J, Chen C, Liu W. B3GNT3 overexpression is associated with unfavourable survival in non-small cell lung cancer. J Clin Pathol. 2018;71:642–647. doi: 10.1136/jclinpath-2017-204860. [DOI] [PubMed] [Google Scholar]
- 22.Zhuang H, Zhou Z, Zhang Z, Chen X, Ma Z, Huang S, Gong Y, Zhang C, Hou B. B3GNT3 overexpression promotes tumor progression and inhibits infiltration of CD8+ T cells in pancreatic cancer. Aging (Albany NY) 2020;13:2310–2329. doi: 10.18632/aging.202255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin M, Zhang H, Yang J, Zheng Z, Liu K. Expression mode and prognostic value of FXYD family members in colon cancer. Aging (Albany NY) 2021;13:18404–18422. doi: 10.18632/aging.203290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng X, Gao J, Cai C, Zhang Y. LncRNA LINC01503 aggravates the progression of cervical cancer through sponging miR-342-3p to mediate FXYD3 expression. Biosci Rep. 2020;40:BSR20193371. doi: 10.1042/BSR20193371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng Y, Zeng X, Lian M, Wang Y. FXYD3 promotes the proliferation, migration, and invasion of pancreatic cancer cells by regulating the cGMP-PKG signaling pathway. Mol Cell Toxicol. 2022;18:371–381. [Google Scholar]
- 26.Kayed H, Kleeff J, Kolb A, Ketterer K, Keleg S, Felix K, Giese T, Penzel R, Zentgraf H, Buchler MW, Korc M, Friess H. FXYD3 is overexpressed in pancreatic ductal adenocarcinoma and influences pancreatic cancer cell growth. Int J Cancer. 2006;118:43–54. doi: 10.1002/ijc.21257. [DOI] [PubMed] [Google Scholar]
- 27.Friess H, Ding J, Kleeff J, Fenkell L, Rosinski JA, Guweidhi A, Reidhaar-Olson JF, Korc M, Hammer J, Buchler MW. Microarray-based identification of differentially expressed growth- and metastasis-associated genes in pancreatic cancer. Cell Mol Life Sci. 2003;60:1180–1199. doi: 10.1007/s00018-003-3036-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi JI, Rim JH, Jang SI, Park JS, Park H, Cho JH, Lim JB. The role of Jagged1 as a dynamic switch of cancer cell plasticity in PDAC assembloids. Theranostics. 2022;12:4431–4445. doi: 10.7150/thno.71364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen WH, Horoszewicz JS, Leong SS, Shimano T, Penetrante R, Sanders WH, Berjian R, Douglass HO, Martin EW, Chu TM. Human pancreatic adenocarcinoma: In vitro and In vivo morphology of a new tumor line established from ascites. In Vitro. 1982;18:24–34. doi: 10.1007/BF02796382. [DOI] [PubMed] [Google Scholar]
- 30.Liu Z, Zhu L, Qin H, Li D, Xie Z, Ke X, Zhao Q. Re-expression of cell adhesion molecule inhibits growth and induces apoptosis of human pancreatic cancer cell line PANC-1. J Huazhong Univ Sci Technolog Med Sci. 2011;31:762–767. doi: 10.1007/s11596-011-0673-z. [DOI] [PubMed] [Google Scholar]
- 31.Kyuno D, Takasawa A, Kikuchi S, Takemasa I, Osanai M, Kojima T. Role of tight junctions in the epithelial-to-mesenchymal transition of cancer cells. Biochim Biophys Acta Biomembr. 2021;1863:183503. doi: 10.1016/j.bbamem.2020.183503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.