Abstract
Lespedeza bicolor is a shrub plant that has been widely distributed in East Asia. The methanol extract from its LBR has been shown to exhibit anticancer and anti-bacterial effects. However, its anticancer efficacy in TNBC remains uncertain. This work aimed to study the anti-TNBC effect of LBR ethanol extract and its underlying mechanism. LBR triggered the cell death in TNBC through inhibiting cell proliferation, S-phase cell arrest, and induction of apoptosis. RNA-seq analysis revealed that the genes altered by LBR treatment were predominantly enriched in the cell adhesion. Notably, LBR inhibited phosphorylation and distribution of FAK. Furthermore, LBR demonstrated significant anticancer activity in xenograft tumors in mice through inhibiting cancer cell growth and inducing apoptosis. This work demonstrated the anticancer efficiency of LBR in TNBC without causing significant adverse effect, which providing a foundation for developing LBR based chemotherapeutic agents for breast cancer therapy.
Keywords: TNBC, Lespedeza bicolor, RNA-seq, FAK, cell proliferation, cell adhesion
Introduction
BC is accounted a second leading cause of cancer-associated mortality in women globally [1,2]. Four intrinsic subtypes are present: Luminal A, Luminal B, HER2, and TNBC [3,4]. Among BC subtypes, the treatment of TNBC is challenging due to absence of major receptors including ER, PGR, and HER2. Furthermore, TNBC is unresponsive to traditional endocrine therapies due to absence of these three receptors [5-7]. TNBC makes up 10-20% of all breast cancer instances and is characterized by its aggressive growth and significant metastatic potential, leading to 25% of BC deaths [8-10]. Currently, chemotherapy with cytotoxic agents remains the primary treatment for late-stage TNBC, but it inevitably leads to resistance and adverse effects [11-13]. Given the challenges of relapse and distant metastasis, new therapeutic agents are needed to markedly inhibit TNBC cell proliferation and invasiveness.
Medicinal plants are an essential for drug discovery, offering unique structural frameworks and profound bioactivity [14-16]. Numerous natural products offer complementary or alternative therapies for breast cancer (BC) [17-20]. An example is the ability of Astragalus membranaceus triggers apoptosis in BC cells by inhibiting the PI3K/Akt/mTOR signaling pathway [21]. Additionally, the Eclipta alba chloroform fraction induced apoptosis in BC cells via upregulation of Hsp60 [22]. Prunella vulgaris exhibited anti-proliferation and anti-metastasis properties in BC models, increasing cleaved caspase-3 and promoting nuclear DNA damage [23].
Lespedeza bicolor, a leguminous medicinal plant, has traditionally been used to address various health issues, including diabetes [24], inflammation [25], and cancer [26]. L. bicolor roots (LBR) are rich in prenylated polyphenolic compounds with activity against human prostate cancers and leukemia through multiple mechanisms involving tumor cell necrosis, apoptosis, cell cycle arrest, and prevention of tumor cell invasion and metastasis [27,28]. While LBR extracts impede TNBC cell line proliferation/migration, specific bioactive and migration mechanisms remain elusive. This work aimed to elucidate anti-proliferative and anti-migration effects of LBR in TNBC cells, via pharmacological models to illuminate potential anti-tumor mechanisms.
Materials and methods
Preparation of LBR extracts
The roots of L. bicolor was collected during June-2018 and were identified by H. S. Park (Medicinal Plant Garden in Boseong, Jeollanam-do, Republic of Korea). Furthermore, the identification of L. bicolor (voucher specimen: CNU-0602) was confirmed using a website http://www.theplantlist.org and stored at College of Pharmacy, Chonnam National University, Gwangju, Republic of Korea. The roots samples were dried and extracted with ethanol (100%) via ultrasonication overnight. Afterwards, the crude extract was collected and dried using a rotary evaporator and labelled as LBR.
LC-MS analysis
The LC MS analysis of LBR using an injection volume of 10 μL in Waters Arc HPLC system coupled with Waters Quattro Premier XE triple quadrupole MS. Chromatographic separation was conducted at 35°C using a column (XBridge® C18 (3.5 μm, 4.6 × 50 mm)). The formic acid (0.1%) in water (A) and formic acid (0.1%) in acetonitrile (B) were used as mobile phase, with an elution gradient pattern of 0-8 min, 10-90% (B) and 8-13 min, 100% (B). MS analysis was performed in full-scan mode (m/z 100-2000 Da) using ESI.
Cell lines and culture
Human TNBC cell line (BT549, MDA-MB-231, and MDA-MB-468) and human normal breast cell line (MCF-10A) sourced from Cell Bank of the Chinese Academy of Sciences in Shanghai, China. MDA-MB-231, MDA-MB-468 and MCF-10A were cultured in controlled environment (5% CO2 at 37°C) using DMEM with 10% FBS, while BT549 cells was grown in RPMI-1640.
Reagents, and antibodies
The ALT (Catalog No. C009-2-1) and AST (Catalog No. C010-2-1) test kit was received from Nanjing Jiancheng Bioengineering Institute, China. The Cr assay kit (Catalog No. ab204537) was obtained from Abcam (Boston, MA, USA). GAPDH antibodies (Catalog No. 60004-1-Ig) were sourced from Proteintech (Rosemont, IL, USA). Cell Signalling Technology (CST, Danvers, MA, USA) provided antibodies for γ-H2AX (Catalog No. 9718), FAK (Catalog No. 71433), and phosphor-FAK (Catalog No. 8556). Ki67 antibodies (Catalog No. ab15580) were obtained from Abcam. The anti-mouse lgG, HRP-linked antibody (Catalog No. 7076S) was obtained from Cell Signalling Technology.
Cell viability assay
Cytotoxicity effect of LBR was evaluated in BT549, MDA-MB-468, MDA-MB-231 and MCF-10A cell lines. Initially, the cells were seeded in a 96-well plate and exposed to the extract for 48 hours at 37°C. Post incubation, MTT (0.5 mg/mL; 20 μL/well) was added and incubated for 4 hours. Subsequently, 100 μL of DMSO was added to dissolve the accumulated formazan product. Absorbance was read at 490 nm by DTX880 microplate reader (Beckman Coulter, San Jose, CA, USA). The value for IC50 were calculated by Prism 8.0 GraphPad (La Jolla, CA, USA).
Colony formation assay
The BT549, MDA-MB-468, MDA-MB-231, and MCF-10A cells (500 cells/well) were seeded in 12-well plate and allowed to adhere overnight. After adhesion, LBR at 5, 10, and 20 µg/mL were treated to cells. Treatment continued until reach the countable colonies. Then, these colonies were rinsed with PBS for three times and then fixed in formaldehyde (4%) and stained with crystal violet (0.04%). Following a rinse with distilled H2O, the colonies observed by light microscopy. Colony counts were taken from three independent experiments.
Cell cycle analysis
To analyse the cell cycle arrest, the different concentration of LBR (0, 5, 10, or 20 µg/mL) were treated to BT549, MDA-MB-231, MDA-MB-468, and MCF-10A cells for 48 hours. Then, the cells were collected and washed with cold PBS, and then stained with PI analysed by a FACS (BD FACSCalibur; BD Biosciences, Cockeysville, MD, USA).
Cell apoptosis analysis
Apoptosis in BT549, MDA-MB-468, MDA-MB-231, and MCF-10A cells was evaluated following exposure to 10, 20, and 40 µg/mL of LBR for 48 hours. Treated cells were stained with the annexin V (FITC)/PI for 30 minutes at RT. The apoptosis level analysed by FlowJo software (version 10.8.1).
Alkaline comet assay
BT549, MDA-MB-231, and MDA-MB-468 cells treated with 0, 5, 10, or 20 µg/mL of LBR for 48 hours. Post-treatment, cells harvested, adjusted the cell density of 100 cells/μL, mixed with low melting point agarose, and spread on a CometSlide. Upon agarose solidification, the slides were submitted to cell lysis in lysis buffer at 4°C overnight. The slides then washed with an alkaline buffer for 20 minutes before undergoing electrophoresis (25 V and 300 mA) for 25 minutes. Following electrophoresis, the slides stained with PI (Cat. No.: KGA1813-50, KeyGEN, Guangdong, China). Then, fluorescence images were captured using fluorescence microscope. DNA damage was measured by olive tail moment and tail length.
IF assay
BT549, MDA-MB-231, and MDA-MB-468 cells were exposed to 5, 10, and 20 μg/mL LBR for 48 hours. After incubation, the cells were fixed in 4% PFA for 10 minutes, followed by permeabilization using Triton X-100 (0.5%) for 1 hour. Following three times washes with ice-cold PBS, the cells were kept at 4°C overnight with primary antibodies against FAK and 53bp1 in a dark room. Afterwards, the cells were exposed to a DyLight 488-conjugated secondary antibody (anti-rabbit). Cell nuclei counterstained with DAPI for 10 minutes. After washing the slides in PBS three times, the fluorescence was observed using a microscope (Nikon Ti).
Western blot analysis
BT549 and MDA-MB-231 cells were treated with 5, 10, and 20 µg/mL LBR for 48 hours to evaluate its effect on FAK expression, after treatment, the protein was isolated using a lysis buffer at 4°C. The protein was separated by SDS-PAGE, then transferred onto a PVDF membrane (MilliporeSigma, Burlington, MA, USA). The membrane was incubated at 4°C overnight with primary antibodies against p-FAK (1:1000), FAK (1:1000), and GAPDH (1:50000). Following this, the membrane was washed and incubated with HRP-conjugated secondary antibodies at RT for 2 hours. The expression of protein was detected using a ECL substrate, and band densities were quantified with a protein blot detection system. Protein fold changes were calculated after normalization with GAPDH levels.
Transwell migration assay
After a 12-hour incubation BT549, MDA-MB-231, and MDA-MB-468 cells in serum-free medium, the cells were treated with 5, 10, and 20 µg/mL of LBR for another 12 hours. Subsequently, 3 × 104 cells transferred to the upper chamber of a Transwell plate (8.0 µm, 24 Cluster Plate; Costar, Glendale, AZ, USA), with the lower chamber containing 500 μL of medium supplemented with 10% FBS. 24 hours later, non-migratory cells, culture medium and Matrigel in the upper chamber were removed with a cotton swab while migrated cells in lower chamber were fixed in 1 mL of 4% PFA, then dyed with crystal violet (0.1%) and visualized using microscope.
Scratch assay
The cell line (BT549, MDA-MB-231, and MDA-MB-468) was cultured in 6-well plates until reach the 100 confluences. Then, a scratch was made on monolayer using a 200 μL of pipette tip. The cell debris was removed by washing with PBS, then cells were treated different concentration of LBR (0, 5, 10, and 20 μg/mL). Afterward, the migratory response was monitored and photographed at 0, 24, 48, and 72-hour intervals using a microscope (PrimoVert; Carl Zeiss, Oberkochen, Germany). Wound closure was quantified using ImageJ software (version 1.52a), with migration measured as the percentage of the cleared area initially created by the scratch that was subsequently filled by migrating cells.
RNA library construction and sequencing
The RNA was extracted from the LBR (5 µg/mL; for 48 hours) treated cells using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA). Next, paired-end transcriptome sequencing was conducted using an Illumina HiSeq 4000 at LC-BIO Technologies (Hangzhou, China) (GEO database number: GSE262226). DEGs with P < 0.05 were analyzed using DESeq2 software. Pathway and GO analysis were performed through the DAVID (P < 0.05 significant).
Survival analysis
The correlation between expression of key anti-TNBC targets in the LBR extract and the survival rate of breast cancer (BC) patients (n = 1086) was analyzed using the online tool Kaplan-Meier Plotter tool (http://kmplot.com/).
Xenograft assay
The animal research was approved by Institutional Animal Care and Use Committee at Wenzhou Medical University (wydw 2023-0294). The 4-week-old female BALB/c-nu nude mice were obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China) and acclimatized in a SPF environment for 7 days. The mice were randomly allocated into three groups (six mice/group), and injection sites were disinfected with 75% ethanol. Then, MDA-MB-231 cells (2 × 106 cells/mouse) were subcutaneously implanted into the right scapula region of the mice. Following tumor establishment over 14 days, treatments of 20 or 40 mg/kg LBR or saline were administered every other day for 20 days. Tumor growth was tacked by recording tumor volumes and weights every 2 days, using the formula: volume = tumor length × width2 × π/6. Upon experiment completion, mice were euthanized, and the tumors were collected, photographed, weighed, and preserved in 4% PFA.
H&E staining assay
The tumor tissues, kidneys, and livers from the mice were preserved in 4% PFA, then tissue samples were paraffin embedded and sectioned to slices (5-μm). Then stained with H&E (Boster Bioengineering Co., Ltd., Wuhan, China) and imaged under a light Nikon Ti microscope.
IHC staining assay
Immunohistochemical (IHC) staining was performed on tumor tissue sections (5-μm thick). After antigen retrieval in sodium citrate solution and PBS wash, the sections were treated with 3% H2O2 to inhibit endogenous peroxidase. Blocking was performed with 3% bovine serum albumin (BSA) for 30 minutes, flowed by overnight incubvation at 4°C with primary antibodies specific to Ki67, γ-H2AX, and FAK, all sourced from CST. Subsequently, the sections were incubated with an HRP-conjugated secondary antibody for 1 hour at RT. Images were then captured under the VS120 Virtual Slide Scanner (Nikon, Tokyo, Japan).
AST, ALT, and Cr tests
Whole blood was collected from the mice after sacrifice. Following centrifugation at 860× g for 15 minutes, the supernatant was utilized for quantifying plasma enzyme levels, including AST, ALT, and Cr.
Acute toxicity test
The acute toxicity test was sanctioned by the Institutional Animal Care and Use Committee of Wenzhou Medical University. ICR mice (25 g each) were divided into three groups, with six mice per group. Next, the mice received oral dose of LBR at concentrations of 100 and 300 mg/kg. Meanwhile, the control group was administered the equivalent volume of PBS. Observations were conducted daily for 7-day period, during which body weights were recorded every day. Finally, all mice were euthanized, and weights of key organs (heart, liver, lung, kidney, and spleen) were measured. Serum samples were collected, and levels of ALT, AST, and creatinine were determined using the Biobase® BK-400 automatic biochemistry analyzer (Biobase, Jinan, China).
Statistical analysis
The experiments were performed at least three times, and the results are expressed as mean ± standard deviation. The significance was evaluated by paired t-test. P < 0.05 was significant.
Results
Identification of the major secondary metabolites in LBR
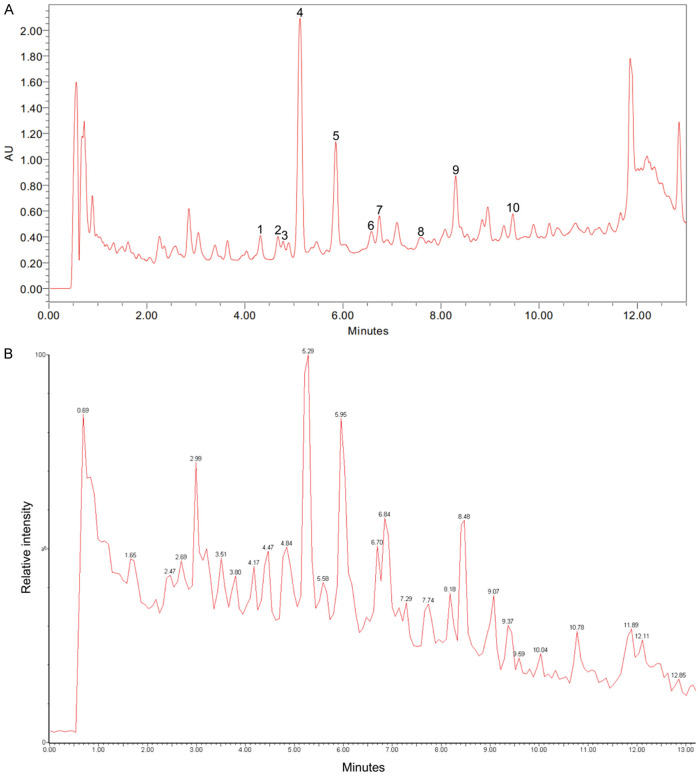
The obtained HPLC chromatogram and TIC are provided in Figure 1A and 1B, respectively. The major peaks in the LBR were further isolated to obtain pure compounds. Each compound’s structure was determined using NMR and mass spectrometry techniques. Ten main compounds were identified in LBR, including 1-methoxylespeflorin G11 (retention time [Rt] 4.32 minutes), 8-methoxybicolosin C (Rt 4.68 minutes), lesbicoumestan (Rt 4.69 minutes), 1-methoxyerythrabyssin II (Rt 5.13 minutes), bicolosin A (Rt 5.89 minutes), gangetin (Rt 6.58 minutes), 1-methoxyfolitenol (Rt 6.75 minutes), 2-geranyl-1-methoxylespeflorin G11 (Rt 7.57 minutes), 2-geranyl-1-methoxyerythrabyssin II (Rt 8.23 minutes), and 2-geranylbicolosin A (Rt 9.47 minutes) (see Table 1). The quantitative results of 10 isolated compounds are shown in Table S1, which indicated that pterocarpans are major compounds in the root extract. Additionally, the results indicated that 1-methoxyerythrabyssin II is the dominant pterocarpan in LBR (56.24 ± 0.71 mg/g). The calibration curves of 10 compounds showed good linearity (r2 > 0.9987) in the concentration ranges of 1.0-0.1 mg/mL (Table S2).
Figure 1.
Lespedeza bicolor root components (LBR) identified through LC-MS. A. High-performance liquid chromatography (HPLC) chromatogram of LBR. B. Total ion chromatogram (TIC) displaying LBR in the mode of positive ions.
Table 1.
Results of LC-MS analysis of LBR
| Peak | Compound name | RT (min) | m/z ([M+H]+) | Chemical formula |
|---|---|---|---|---|
| 1 | 1-Methoxylespeflorin G11 | 4.32 | 437.1730 | C26H28O6 |
| 2 | 8-Methoxybicolosin C | 4.68 | 453.3151 | C27H32O6 |
| 3 | Lesbicoumestan | 4.69 | 449.1729 | C26H24O7 |
| 4 | 1-Methoxyerythrabyssin II | 5.13 | 423.3520 | C26H30O5 |
| 5 | Bicolosin A | 5.89 | 437.1054 | C27H32O5 |
| 6 | Gangetin | 6.58 | 421.2116 | C26H28O5 |
| 7 | 1-Methoxyfolitenol | 6.75 | 421.1835 | C26H28O5 |
| 8 | 2-Geranyl-1-methoxylespeflorin G11 | 7.57 | 505.0522 | C31H36O6 |
| 9 | 2-Geranyl-1-methoxyerythrabyssin II | 8.23 | 491.0906 | C31H38O5 |
| 10 | 2-Geranylbicolosin A | 9.47 | 505.2710 | C32H40O5 |
LBR exhibits selective cytotoxicity against TNBC cells
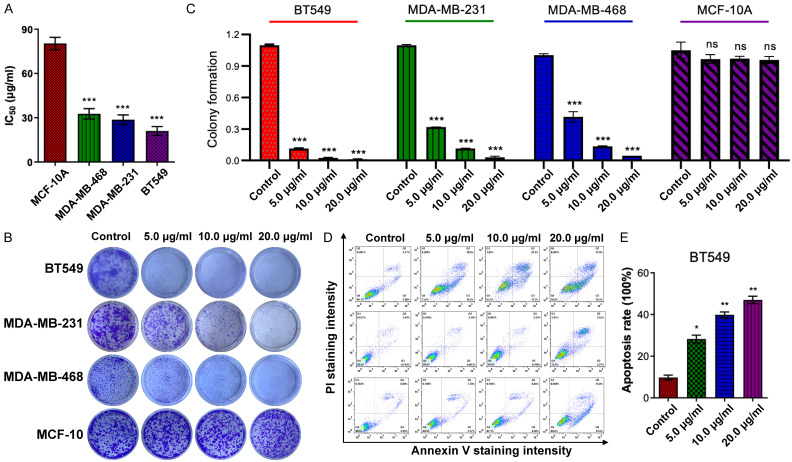
The anticancer efficacy of LBR was evaluated against three TNBC cell lines, MDA-MB-231, MDA-MB-468, and BT549 using the MTT assay to measure cell viability at various concentrations. After a 48-hour treatment, the LBR demonstrated dose-dependent inhibition of cell viability across all three TNBC cell lines, with IC50 values below 35 μg/mL. The determined IC50 values of LBR for the TNBC cell lines (MDA-MB-468, MDA-MB-231, and BT549) were 32.83 μg/mL, 29.4 μg/mL, and 20.28 μg/mL, respectively (Figure 2A). Notably, LBR’s cytotoxic effect on normal human breast epithelial MCF-10A cells (IC50 value of 80.33 μg/mL) was significantly lower compared to its effect on TNBC cells (Figure 2A), indicating a more specific inhibitory effect of LBR on TNBC cell proliferation rather than on normal human cells.
Figure 2.
L. bicolor root components (LBR) had a significant inhibitory effect on TNBC cell proliferation. A. Half-maximal inhibitory concentration (IC50) (mg/mL) values of LBR-treated cells were detected in MDA-MB-468, MDA-MB-231, and BT549 cells after treatment with LBR (0-100 μg/mL) for 48 h, respectively. B. The colony formation assay was performed after treatment with the indicated concentration of LBR for 24 h. C. Colony formation counting. D. MDA-MB-468, MDA-MB-231, and BT549 cells were stained with Annexin V/PI after LBR treatments for 24 h, and then analyzed by flow cytometry. E. Apoptosis rate in BT549 after treatment with LBR. Results are presented as mean ± standard deviation (SD), *P < 0.05 and **P < 0.01, vs. untreated group, n = 3.
To further examine LBR’s antiproliferative potential on TNBC cells, colony formation assays were conducted. The LBR treatment significantly inhibited MDA-MB-468, MDA-MB-231, and BT549 cells in a dose-dependent manner (Figure 2B, 2C). In contrast, colony formation in normal human breast epithelial MCF-10A cells remained unaffected under the same LBR treatment conditions. These results suggest that LBR displays potent cytotoxicity towards TNBC.
LBR triggers cell cycle arrest and promotes apoptosis in TNBC cells
The distribution of the cell cycle was assessed via flow cytometry to evaluate the effect of LBR on the cell cycle of BT549, MDA-MB-231, MDA-MB-468, and MCF-10A cells (Figure S1A). The results showed that LBR treatment enhanced the S-phase proportion in BT549 cells (from 36.3% to 43.5%) and reduced the G1-phase proportion (from 46.8% to 35.3%) (Figure S1B). Similar increments were observed in MDA-MB-231 and MDA-MB-468 cells (Figure S1C, S1D). Intriguingly, the cell distribution of MCF-10A remained unchanged with or without LBR treatment at the same concentration (Figure S1E).
We observed flattened and rounded cell morphologies following LBR treatment (Figure S2), prompting apoptosis assays in LBR-treated TNBC cell lines at 10, 20, and 40 µg/mL for 24 hours. These changes are often associated with cytoskeletal reorganization, cell shrinkage, chromatin condensation, and membrane blebbing, all of which are hallmarks of apoptosis [29]. After-staining with FITC-annexin V/PI binding buffer, flow cytometric analysis revealed LBR treatment significantly induced apoptosis in TNBC cells in a concentration-dependent manner (Figure 2D). At 40 µg/mL, LBR considerably increased apoptotic cell percentages (early and late stages) in BT549, MDA-MB-231, and MDA-MB-468 cell lines, reaching 48.6%, 28.8%, and 31.5% (Figures 2E, S3A and S3B), respectively. These results demonstrate LBR’s effective anti-proliferative action against breast cancer cells via apoptosis induction. Remarkably, compared to MDA-MB-231 and MDA-MB-468 cells, the BT549 cells exhibited higher sensitivity to LBR-induced apoptosis (Figure 2E).
LBR induces DNA damage in TNBC cells
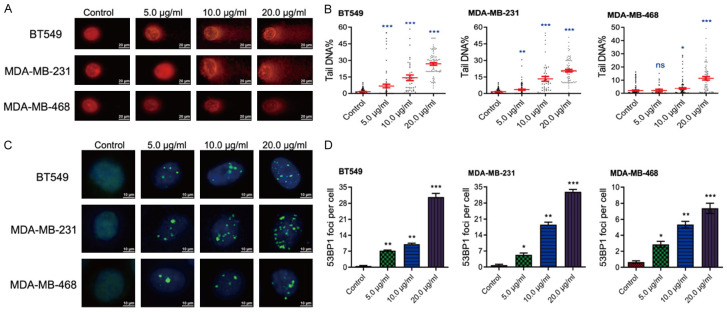
DNA is often considered a primary target of the antineoplastic actions of phytochemicals [30], and intense unrepaired DNA lesions can trigger cell apoptosis [31]. To determine whether LBR’s anticancer efficacy in TNBC is attributable to DNA damage modulations, alkaline comet assays were performed. These assays evaluated DNA integrity in both LBR-treated and untreated BT549, MDA-MB-231, and MDA-MB-468 cells (Figure S4). The results indicated that LBR, even at a lower concentration of 5 µg/mL, caused significant DNA double-strand or single-strand breaks, as evidenced by DNA fragments migrating from the nucleus, forming a ‘tail’ (Figure 3A). The prevalence of DNA fragmentation, induced by DNA damage, was quantitatively assessed by the tail DNA percentage, serving as an indicator of DNA damage post-LB treatment. As shown in Figure 2B, increasing LBR concentrations resulted in increased tail DNA content in all three TNBC cell lines, exceeding negative control levels. Furthermore, the data indicates a concentration-dependent induction of DNA damage by LBR (Figure 3B).
Figure 3.
LBR treatment induces DNA damage and DNA damage response in TNBC cells. A, B. Comet assay exhibiting nuclear staining image of the broken DNA tail distance for quantitative detection of DNA damage upon LBR treatment (n = 15). C, D. Immunofluorescence assay was used to show the 53BP1 foci in TNBC cells treated with indicated concentration of LBR. Quantification of the 53BP1 foci per cell was calculated.
To further investigate LBR’s role in the DNA damage response, immunofluorescence (IF) assays conducted using 53BP1, a known marker of DNA damage response [32]. The IF assays demonstrated a significant increase in 53BP1 foci within TNBC cell nuclei after a 48-hour LBR treatment (Figure 3C). Additionally, quantitative data showed LBR provoked the DNA damage response in a concentration-dependent manner (Figure 3D).
LBR inhibits TNBC cell-matrix adhesion and migration
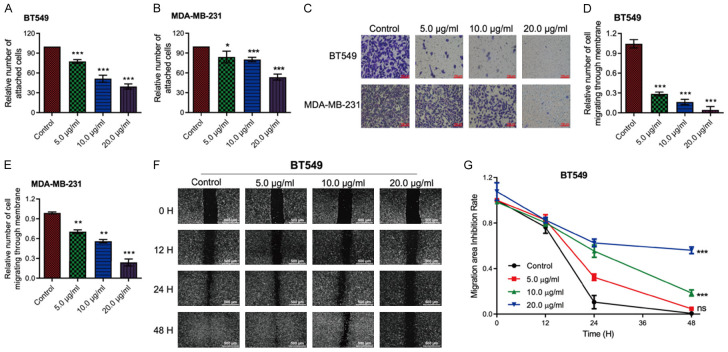
Adhesion assays were performed on LB-treated and untreated BT549, MDA-MB-231, and MDA-MB-468 cells. The results showed a significant, dose-dependent decrease in cell adhesion to the substrate in all three cell lines after LBR treatment. Even at a low concentration of 5 µg/mL, LBR mildly inhibited cell adhesions (Figures 4A, 4B and S5A). Cell-matrix adhesion has been shown to be closely associated with cell migration [33]. Subsequently, Transwell migration assays indicated LBR inhibited the cell migration of TNBC cells in a dose-dependent manner, especially in BT549 and MDA-MB-231 cells (Figures 4C-E, S5B, S5C).
Figure 4.
L. bicolor root components (LBR) suppress invasion and migration of TNBC cells. A, B. Cell adhesion assays were performed on BT549 and MDA-MB-231 cells. C. Transwell assays were used to analyze the invasion of BT549 and MDA-MB-231 cells. D, E. Quantification of cell invasion. F. The inhibition effect of BT549 cell migration was detected by scratch assay at 0 h, 12 h, 24 h, and 48 h. G. Quantification of BT549 cell migration. Data are displayed as mean ± standard deviation (SD) (n = 3), *P < 0.05 or **P < 0.01.
Furthermore, scratch assays were used to evaluate the migration rates of BT549 and MDA-MB-468 cells at 0, 24, 48, and 72-hour post-treatment with the indicated LBR concentrations. The results showed LBR significantly impeded cancer cell migration in a dose-dependent manner, particularly for BT549 cells (Figure 4F). Consistently, after 72 hours, the control group scratch wounds were almost completely covered by migrated BT549 cells, while 20 µg/mL LBR treatment yielded only 53.6% coverage (Figure 4G). Similar migration inhibition occurred in LBR-treated MDA-MB-468 cells (Figure S5D and S5E).
LBR regulated FAK-mediated signaling pathway in TNBC cells
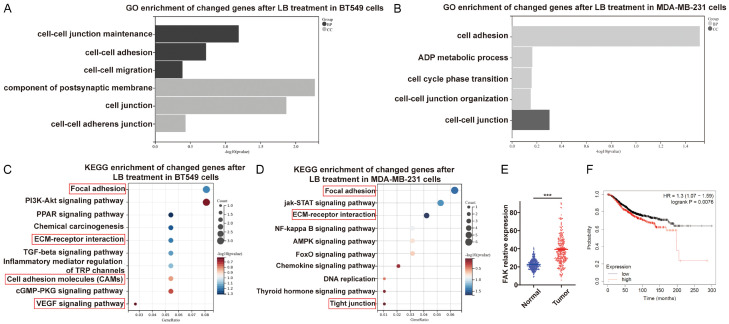
To investigate the underlying anti-TNBC molecular mechanisms of LBR, RNA-seq transcriptome analysis was conducted on BT549 and MDA-MB-231 cells treated with LBR, compared to untreated controls. The analysis revealed DEGs compared to the controls. Subsequently, a comprehensive bioinformatics approach was applied to further understand the biological functions and molecular mechanisms of DEGs. GO enrichment of all DEGs revealed various BPs, mainly related to cell regulation, adhesion, and migration. Regarding CC, enriched DEGs were mainly associated with cell junction and cell-cell adhesion junction (Figure 5A and 5B). KEGG pathway enrichment analysis demonstrated DEGs significantly participate in cell adhesion-related pathways including focal adhesion, ECM-receptor interaction, cell adhesion molecules, VEGF signalling pathway, and tight junction (Figure 5C and 5D). These findings strongly suggest LBR’s inhibitory effect on TNBC cell growth and migration may be primarily mediated through modulation of the focal adhesion pathway. FAK as a non-receptor tyrosine kinase with key roles in regulating cell proliferation, adhesion, and migration [34,35]. Consequently, we conducted expression and prognosis analysis of the FAK protein. We noted FAK expression was higher in breast cancer cells than normal cells (Figure 5E). Additionally, utilizing the Kaplan-Meier Plotter database, we analyzed the correlation between FAK expression and survival in TNBC. This revealed high FAK expression was associated with poorer prognosis in TNBC patients (Figure 5F). Subsequently, we explored whether LBR achieved potent anti-TNBC activity affecting FAK expression, phosphorylation, and distribution.
Figure 5.
Exploration of potential mechanisms of L. bicolor root components (LBR) in TNBC cells via RNA-Seq network analysis. A, C. Top biological process (BP) and cellular component (CC) in Gene Ontology (GO) analysis for BT549 and MDA-MB-231 cells. Item ranking based on p-value. B, D. Bubble diagram of Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis for potential signaling pathways in LBR-treated BT549 and MDA-MB-231 cells. The bubble size indicates the pathway target number. The bubble color represents the p-value magnitude, with a redder hue indicating a smaller p-value and higher enrichment degree. E. Expression levels of focal adhesion kinase (FAK) in TNBC versus non-cancerous tissues. F. Overall survival analysis of FAK in TNBC.
LBR inhibits the phosphorylation of FAK and reduced the pseudopodia area in TNBC
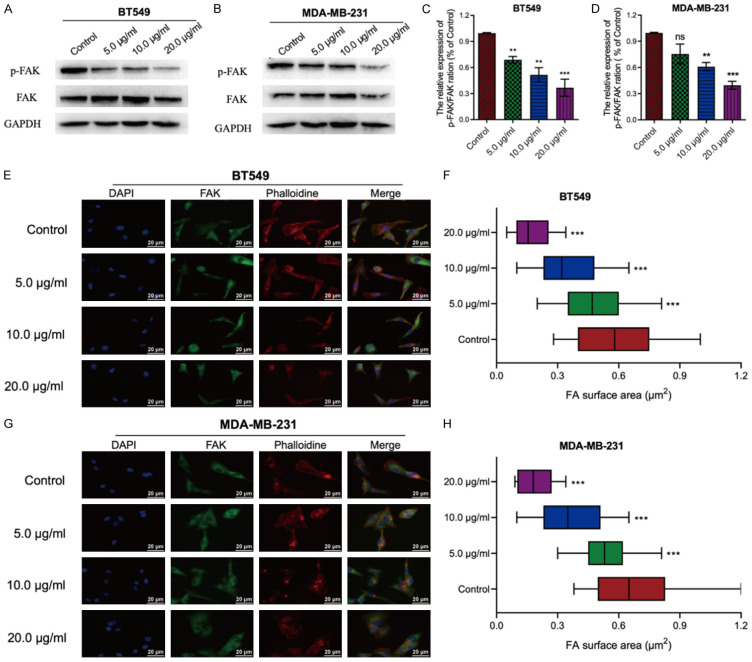
Western blot analysis revealed a significant decrease in the expression of both FAK and its phosphorylated form (p-FAK) in BT549 and MDA-MB-231 cells following LBR treatment compared to untreated cells (Figure 6A and 6B). Notably, the inhibition of FAK phosphorylation by LBR was dose-dependent (Figure 6C and 6D). Similarly, IF assay results revealed decreased FAK expression at the LBR-treated BT549 and MDA-MB-231 cells, with FAK localized in the cell nucleus (Figure 6E and 6G). Moreover, LBR treatment led to dose-dependent reduction in the pseudopodia area of TNBC cells (Figure 6F and 6H). These results indicate LBR inhibits TNBC cell migration by suppressing FAK phosphorylation and altering the FAK distribution.
Figure 6.
L. bicolor root components (LBR) suppress BT549 and MDA-MB-231 migration by regulating F-actin expression and focal adhesion kinase (FAK) inactivation. A-D. Cells were treated with 0.01% dimethyl sulfoxide (control) or LBR at 5, 10, and 20 μg/mL for 48 h and the phosphorylation levels of FAK were determined by western blotting. E, G. Immunofluorescence staining of 4’,6-diamidino-2-phenylindole (DAPI) (blue), anti-FAK antibody (green), and phalloidin (red) in BT549 and MDA-MB-231 cells. F, H. Quantification of the FA surface area. Values indicate the average ± standard deviation from three independent experiments. *P < 0.05 and **P < 0.01 compared with the control group.
LBR inhibited the growth and metastasis of TNBC xenograft tumors in mice
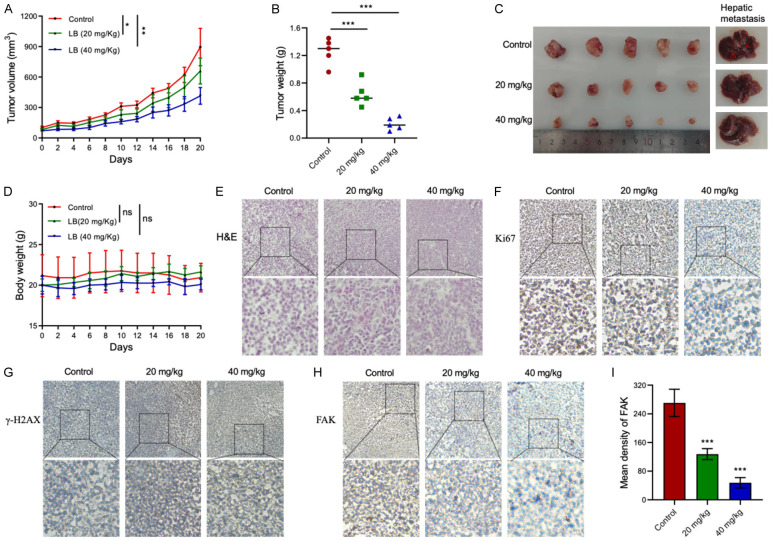
Motivated by promising in vitro findings, we developed an MDA-MB-231 xenograft tumor model in female BALB/c mice to evaluate the in vivo anti-tumor effects of LBR. Two weeks after inoculating the mice with cancer cells, the mice were administered intraperitoneal injections of LBR at 20 mg/kg and 40 mg/kg every other day. Although tumors were visible in the LBR-treated groups, after 20 days of treatment, tumor size and weight were significantly inhibited in treated group compared to untreated group (control group, mean ± standard deviation = 1.258 ± 0.142 g; 20 mg/kg treatment group, mean ± SD = 0.642 ± 0.126 g; 40 mg/kg treatment group, mean ± SD = 0.208 ± 0.074 g; P < 0.05) (Figure 7A-C). Importantly, there was no significant difference in body weight among the different groups of mice (Figure 7D), suggesting LBR treatment did not negatively impact the overall health and body weight of mice.
Figure 7.
Lespedeza bicolor root components (LBR) suppress xenograft tumor growth and hepatic metastasis in vivo. A. Tumor volume examined by caliper measurements from treatment beginning, n = 6 per group. B. Tumor weight in different treatment groups. C. Tumors collected at treatment end in different treatment groups. D. Body weight of mice in different treatment groups. E. Pathological section via hematoxylin and eosin (H&E) stain. F-H. Immunohistochemical (IHC) analysis of Ki67, γ-H2AX, and focal adhesion kinase (FAK) expression in tumor tissues (control versus LBR treatment group) in the TNBC mouse model, respectively. Magnification is 20× at the top and 40× at the bottom. I. Quantitative result of FAK density in different treatment groups. Data are presented as mean ± standard deviation (SD), n ≥ 3 (*P < 0.05, **P < 0.01, ***P < 0.001).
H&E staining showed LBR treatment disrupted nuclear integrity and the cell membranes of tumor cells in a dose-dependent manner (Figure 7E), indicating LBR induces cell death in tumor tissues. Furthermore, the number of Ki-67 positive cells (a marker of proliferation) [36] in tumor sections from LBR-treated mice was significantly decreased (Figure 7F), while γ-H2AX, a marker of DNA damage [37], was increased in LBR-treated tumor tissues compared to that of the untreated mice (Figure 7G). Consistent with in vitro results, the expression of FAK, which is crucial in cancer invasion and metastasis [38], was dose-dependently decreased in the LBR-treated groups compared to that of the untreated groups (Figure 7H and 7I). This consistency between in vitro and in vivo results further confirms LBR’s potential as a therapeutic agent against TNBC via targeting FAK.
LBR exhibited a high safety margin
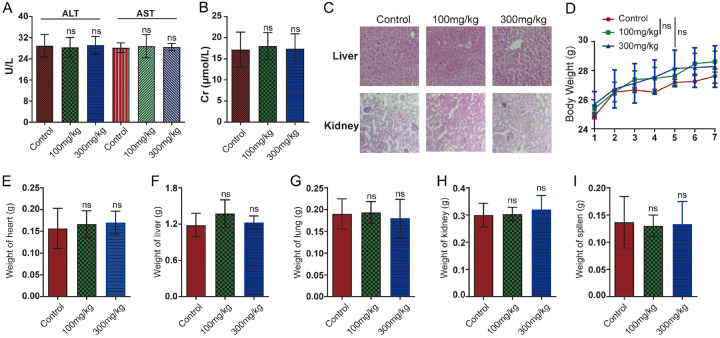
To evaluate the in vivo toxicity of LBR, serum levels of ALT and AST were analyzed in ICR mice with or without LBR treatment. The ALT and AST levels showed no significant difference between the LBR-treated and control groups (Figure 8A). Additionally, Cr is a key marker for evaluating renal function [39]. As shown in Figure 8B, LBR does not impair renal function in rats. Further, histopathological examination of liver and kidney tissues from LBR-treated mice revealed no toxicity signs compared to those of the untreated control mice (Figure 8C). The safety profile of LBR was further assessed through acute toxicity tests, where mice were orally administered LBR. There was no notable difference in body weight gain between the control group and mice treated with LBR, even at a dose of 300 mg/kg (Figure 8D). Moreover, throughout the experiment, visual observations showed no significant behavioral changes or abnormal clinical signs in any of the mice. Additionally, the weight of essential organs such as the heart, liver, lungs, kidneys, and spleen in LBR-treated mice was similar to that of the control group, indicating no adverse effects on these organs (Figure 8E-I). These results collectively suggest that LBR does not exhibit significant toxicity in xenografted mice at the administered doses.
Figure 8.
Evaluation of acute toxicity of Lespedeza bicolor root components (LBR) in mice. Six to eight-week-old male BALB/c-nu nude mice were treated with LBR (100 mg/kg and 300 mg/kg) or the saline control. A, B. Serum levels of liver injury markers alanine transferase (ALT), aspartate aminotransferase (AST), and creatinine (Cr) were measured. C. Liver tissues were stained with hematoxylin and eosin (H&E) for histological analysis. D. Body weight was monitored throughout the treatment period. E-I. Changes in heart, liver, lung, kidney, and spleen weight were assessed during LBR treatment. Data are represented as mean ± standard deviation (SD) (n = 6); *P < 0.05 and **P < 0.01 compared with the control group.
Discussion
The lack of ER/PR/HER2 receptor in TNBC is less responsive to conventional treatments, such as chemotherapy and radiation therapy. As a result, certain patients with TNBC have shown positive response to immunotherapy, specifically checkpoint inhibitors like PD-1/PD-L1 inhibitors [40]. However, not all TNBC patients respond to immunotherapy, and it may lead to immune-related adverse effects [41].
Natural products are widely used in cancer treatment both alone and in conjunction with chemotherapy drugs [42-44]. Natural products offer reduced toxicity and a broad spectrum of anticancer and chemoprotective properties [45]. The multiple active ingredients in natural compounds can produce additive or synergistic effects by simultaneously targeting various pathways [46,47]. Medicinal plants active against BC can inhibit epithelial-mesenchymal transition (EMT), enhance apoptosis, and prevent invasion, offering potential as targeted therapies suppressing cancer metastasis pathways [48,49]. For instance, dandelion leaf extract effectively targets BC cells via an ERK-dependent pathway [50]. Withaferin A, a key constituent of Withania somnifera, inhibits TNBC cell proliferation and β-tubulin expression [51]. Artemisia annua extract, rich in compounds like flavonolignans, prolongs ERK1/2 activation and augments lysosome quantity and size, suggesting autophagy induction [52]. L. bicolor, a medicinal plant, exhibits antioxidant, anti-inflammatory, anticancer, and antimicrobial activities [53-56]. However, the effects and mechanisms of LBR on TNBC are inadequately explored, with limited research on cancer markers, genomic targets, and organismal toxicity. In this study, we demonstrate LBR inhibits TNBC cell proliferation and migration in vitro and in vivo. Moreover, the IC50 of the LBR extract was compared with other effective plant extracts reported against TNBC cancer cells (Table 2). The results indicated that the anticancer effects of different plant extracts against TNBC varied depending on the extraction methods and the plants used. Among the plants tested, Holothuria scabra, Ruellia tuberosa, and Lespedeza bicolor (LBR) exhibited IC50 values below 50 μg/mL. These findings highlight the significant anticancer potential of LBR.
Table 2.
Comparison of IC50 of various plant extracts against triple negative breast cancer cell lines
| Plant name | Extraction methods | Model | Pathway | IC50 | Reference |
|---|---|---|---|---|---|
| Citrus hystrix | Hexane | MDA-MB-231 | Induces apoptosis | 317.6 μg/mL | [72] |
| Annona cherimmola | Ethanol | MDA-MB-231 | Induces apoptosis | 555.3 μg/mL | [73] |
| Holothuria scabra | 95% ethanol | MDA-MB-231 | Regulate Akt/mTOR/HIF-1 axis | 11.8 μg/mL | [74] |
| Garcinia quaesita | Hexane | bCSCs | Induces apoptosis | 57.5 μg/mL | [75] |
| Origanum syriacum | 80% ethanol | MDA-MB-231 | Induces apoptosis | 875 μg/mL | [76] |
| Antenoron Filiforme | Ethanol | MDA-MB-231, MDA-MB-453 | Skp2/p21 | 149.7 μg/mL in MDA-MB-231, 34.26 μg/mL in MDA-MMB-453 | [77] |
| BALB/c | |||||
| Ziziphus nummularia | Ethanol | MDA-MB-231 | Induce autophagy and apoptosis | 662.4 μg/mL | [78] |
| Ruellia tuberosa | Methanol | MDA-MB-231, 4T1 | Induces apoptosis | 23.8 μg/mL | [79] |
| BALB/c mouse | |||||
| Lespedeza bicolor | Ethanol | BT549, MDA-MB-231, and MDA-MB-468 | Regulate FAK pathways | MDA-MB-468, MDA-MB-231, and BT549 were 32.83 μg/mL, 29.4 μg/mL, and 20.28 μg/mL, respectively | Present study |
| Balb/c mouse |
In terms of mechanisms, RNA-seq and network pharmacology analysis identified several key pathways and targets, notably cell adhesion. Given TNBC’s malignant nature and associated low survival rates due to high invasiveness and metastatic tendency [57,58], targeting cell adhesion is vital. Further experimental research revealed that LBR regulates the proliferation and migration of TNBC cells through affecting FAK phosphorylation and distribution (Figure 5). As a non-receptor tyrosine protein kinase, FAK has emerged as a crucial regulator in the focal adhesion pathway [59,60], integrating cell growth and cell-matrix adhesion signals, and driving cancer invasion and metastasis, thus becoming a significant target in anti-tumor drug development [61,62]. Evidence demonstrated that FAK influences cell growth and migration through multiple pathways, including PI3K, MAPK, GTPase, and p53 [63-66]. Additionally, FAK linked to TNBC’s pathogenesis and progression [67,68]. Likewise, our studies also demonstrated that FAK overexpression in TNBC is associated with poor prognosis (Figure 4F). Collectively, these results suggest that the anti-TNBC mechanism of LBR involved in FAK-related signaling pathway inhibition.
Our in vivo studies showed LBR treatment markedly slowed tumor progression in MDA-MB-231 xenograft mice without affecting weight. Notably, this correlated with reduced Ki-67 and FAK, and increased γ-H2AX expression. Ki-67 is a cancer prognostic marker [69], γ-H2AX serves as an early DNA damage response marker [70], and FAK is involved in cancer-related angiogenesis and the EMT process [71]. Collectively, this suggests that LBR inhibits FAK function, leading to decrease interactions between cells and the extracellular matrix, ultimately resulting in reduced levels of the Ki67 cell proliferation marker. At the same time, LBR may directly or indirectly cause DNA damage, resulting in an increase in γ-H2AX and triggering repair mechanisms. If damage is irreparable, programmed cell death (apoptosis) triggered. Thus, LBR may induce tumor cell death by the combined effects of disrupting cytoskeleton-matrix interactions, inhibiting proliferation, and causing DNA damage, consistent with in vitro results.
Conclusion
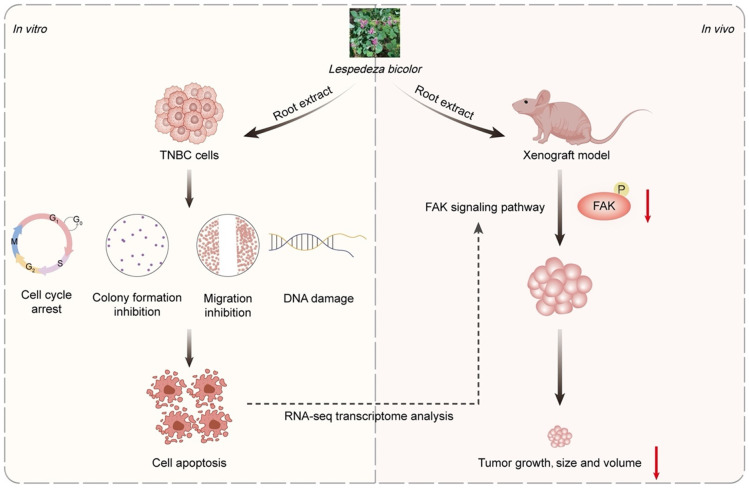
In conclusion, our results indicate that LBR inhibits the proliferation of TNBC in both in vivo and in vitro models by regulating DNA damage, apoptosis and the FAK signalling pathway (Figure 9). Additionally, acute toxicity studies have shown that oral administration of LBR does not result in any significant differences in body weight gain, behavioural changes, or abnormal clinical signs in any of the mice. Moreover, the weights and histopathological analysis of organs (heart, liver, lungs, kidneys, and spleen) were comparable to the control group, indicating no adverse effects on these organs. Collectively, these results suggest that LBR exhibits negligible toxicity in xenografted mice at the administered doses while significantly inhibiting the TNBC growth. Therefore, these findings emphasized that LBR could considered a potential option for developing novel agent for treatment of TNBC.
Figure 9.
Molecular mechanisms of L. bicolor root components (LBR) in TNBC.
Acknowledgements
This research was supported by the Korean government (MSIT) (grant number: NRF-2022R1C1C1009626).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Arnold M, Morgan E, Rumgay H, Mafra A, Singh D, Laversanne M, Vignat J, Gralow JR, Cardoso F, Siesling S, Soerjomataram I. Current and future burden of breast cancer: global statistics for 2020 and 2040. Breast. 2022;66:15–23. doi: 10.1016/j.breast.2022.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilkinson L, Gathani T. Understanding breast cancer as a global health concern. Br J Radiol. 2022;95:20211033. doi: 10.1259/bjr.20211033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin L, Duan JJ, Bian XW, Yu SC. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020;22:61. doi: 10.1186/s13058-020-01296-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russnes HG, Lingjærde OC, Børresen-Dale AL, Caldas C. Breast cancer molecular stratification: from intrinsic subtypes to integrative clusters. Am J Pathol. 2017;187:2152–2162. doi: 10.1016/j.ajpath.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 5.Pal SK, Childs BH, Pegram M. Triple negative breast cancer: unmet medical needs. Breast Cancer Res Treat. 2011;125:627–636. doi: 10.1007/s10549-010-1293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao H, He G, Yan S, Chen C, Song L, Rosol TJ, Deng X. Triple-negative breast cancer: is there a treatment on the horizon? Oncotarget. 2017;8:1913–1924. doi: 10.18632/oncotarget.12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marra A, Trapani D, Viale G, Criscitiello C, Curigliano G. Practical classification of triple-negative breast cancer: intratumoral heterogeneity, mechanisms of drug resistance, and novel therapies. NPJ Breast Cancer. 2020;6:54. doi: 10.1038/s41523-020-00197-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aysola K, Desai A, Welch C, Xu J, Qin Y, Reddy V, Matthews R, Owens C, Okoli J, Beech DJ, Piyathilake CJ, Reddy SP, Rao VN. Triple negative breast cancer - an overview. Hereditary Genet. 2013;2013(Suppl 2):001. doi: 10.4172/2161-1041.S2-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baranova A, Krasnoselskyi M, Starikov V, Kartashov S, Zhulkevych I, Vlasenko V, Oleshko K, Bilodid O, Sadchikova M, Vinnyk Y. Triple-negative breast cancer: current treatment strategies and factors of negative prognosis. J Med Life. 2022;15:153–161. doi: 10.25122/jml-2021-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gluz O, Liedtke C, Gottschalk N, Pusztai L, Nitz U, Harbeck N. Triple-negative breast cancer--current status and future directions. Ann Oncol. 2009;20:1913–1927. doi: 10.1093/annonc/mdp492. [DOI] [PubMed] [Google Scholar]
- 11.Nedeljković M, Damjanović A. Mechanisms of chemotherapy resistance in triple-negative breast cancer-how we can rise to the challenge. Cells. 2019;8:957. doi: 10.3390/cells8090957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13:674–690. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isakoff SJ, Mayer EL, He L, Traina TA, Carey LA, Krag KJ, Rugo HS, Liu MC, Stearns V, Come SE, Timms KM, Hartman AR, Borger DR, Finkelstein DM, Garber JE, Ryan PD, Winer EP, Goss PE, Ellisen LW. TBCRC009: a multicenter phase II clinical trial of platinum monotherapy with biomarker assessment in metastatic triple-negative breast cancer. J. Clin. Oncol. 2015;33:1902–1909. doi: 10.1200/JCO.2014.57.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nasim N, Sandeep IS, Mohanty S. Plant-derived natural products for drug discovery: current approaches and prospects. Nucleus (Calcutta) 2022;65:399–411. doi: 10.1007/s13237-022-00405-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atanasov AG, Waltenberger B, Pferschy-Wenzig EM, Linder T, Wawrosch C, Uhrin P, Temml V, Wang L, Schwaiger S, Heiss EH, Rollinger JM, Schuster D, Breuss JM, Bochkov V, Mihovilovic MD, Kopp B, Bauer R, Dirsch VM, Stuppner H. Discovery and resupply of pharmacologically active plant-derived natural products: a review. Biotechnol Adv. 2015;33:1582–1614. doi: 10.1016/j.biotechadv.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chopra B, Dhingra AK. Natural products: a lead for drug discovery and development. Phytother Res. 2021;35:4660–4702. doi: 10.1002/ptr.7099. [DOI] [PubMed] [Google Scholar]
- 17.Sarin N, Engel F, Kalayda GV, Mannewitz M, Cinatl J Jr, Rothweiler F, Michaelis M, Saafan H, Ritter CA, Jaehde U, Frötschl R. Cisplatin resistance in non-small cell lung cancer cells is associated with an abrogation of cisplatin-induced G2/M cell cycle arrest. PLoS One. 2017;12:e0181081. doi: 10.1371/journal.pone.0181081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ko EY, Moon A. Natural products for chemoprevention of breast cancer. J Cancer Prev. 2015;20:223–231. doi: 10.15430/JCP.2015.20.4.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta N, Kumar H, Gupta S, S M B, Saini K. A concise review on natural products and their derivatives for breast cancer treatment. Chem Biodivers. 2023;20:e202300688. doi: 10.1002/cbdv.202300688. [DOI] [PubMed] [Google Scholar]
- 20.Levitsky DO, Dembitsky VM. Anti-breast cancer agents derived from plants. Nat Prod Bioprospect. 2014;5:1–16. doi: 10.1007/s13659-014-0048-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou R, Chen H, Chen J, Chen X, Wen Y, Xu L. Extract from Astragalus membranaceus inhibit breast cancer cells proliferation via PI3K/AKT/mTOR signaling pathway. BMC Complement Altern Med. 2018;18:83. doi: 10.1186/s12906-018-2148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arya RK, Singh A, Yadav NK, Cheruvu SH, Hossain Z, Meena S, Maheshwari S, Singh AK, Shahab U, Sharma C, Singh K, Narender T, Mitra K, Arya KR, Singh RK, Gayen JR, Datta D. Anti-breast tumor activity of Eclipta extract in-vitro and in-vivo: novel evidence of endoplasmic reticulum specific localization of Hsp60 during apoptosis. Sci Rep. 2015;5:18457. doi: 10.1038/srep18457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo H, Zhao L, Li Y, Xia B, Lin Y, Xie J, Wu P, Liao D, Zhang Z, Lin L. An in vivo and in vitro assessment of the anti-breast cancer activity of crude extract and fractions from Prunella vulgaris L. Heliyon. 2022;8:e11183. doi: 10.1016/j.heliyon.2022.e11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee H, Kim SY, Lim Y. Lespedeza bicolor extract supplementation reduced hyperglycemia-induced skeletal muscle damage by regulation of AMPK/SIRT/PGC1α-related energy metabolism in type 2 diabetic mice. Nutr Res. 2023;110:1–13. doi: 10.1016/j.nutres.2022.12.007. [DOI] [PubMed] [Google Scholar]
- 25.Lee SJ, Hossaine MD, Park SC. A potential anti-inflammation activity and depigmentation effect of Lespedeza bicolor extract and its fractions. Saudi J Biol Sci. 2016;23:9–14. doi: 10.1016/j.sjbs.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sami U. Methanolic extract from Lespedeza bicolor: potential candidates for natural antioxidant and anticancer agent. J Tradit Chin Med. 2017;37:444–451. [PubMed] [Google Scholar]
- 27.Dyshlovoy SA, Tarbeeva D, Fedoreyev S, Busenbender T, Kaune M, Veselova M, Kalinovskiy A, Hauschild J, Grigorchuk V, Kim N, Bokemeyer C, Graefen M, Gorovoy P, von Amsberg G. Polyphenolic compounds from lespedeza bicolor root bark inhibit progression of human prostate cancer cells via induction of apoptosis and cell cycle arrest. Biomolecules. 2020;10:451. doi: 10.3390/biom10030451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang B, Kim S, Kim Y, Qiu Y, Lee CM, Lai Y, Liu Z, Wang K, Cho N. 1-methoxyerythrabyssin II induces autophagy in leukemia cells via PI3K/Akt/mTOR pathways. Planta Med. 2023;89:1204–1214. doi: 10.1055/a-2114-0980. [DOI] [PubMed] [Google Scholar]
- 29.Chaudhry GES, Akim AM, Sung YY, Muhammad TST. Cancer and apoptosis. Apoptosis and Cancer: Methods and Protocols. Springer; 2022. pp. 191–210. [DOI] [PubMed] [Google Scholar]
- 30.Choudhari AS, Mandave PC, Deshpande M, Ranjekar P, Prakash O. Phytochemicals in cancer treatment: from preclinical studies to clinical practice. Front Pharmacol. 2020;10:1614. doi: 10.3389/fphar.2019.01614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Zio D, Cianfanelli V, Cecconi F. New insights into the link between DNA damage and apoptosis. Antioxid Redox Signal. 2013;19:559–571. doi: 10.1089/ars.2012.4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta A, Hunt CR, Chakraborty S, Pandita RK, Yordy J, Ramnarain DB, Horikoshi N, Pandita TK. Role of 53BP1 in the regulation of DNA double-strand break repair pathway choice. Radiat Res. 2014;181:1–8. doi: 10.1667/RR13572.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conway JRW, Jacquemet G. Cell matrix adhesion in cell migration. Essays Biochem. 2019;63:535–551. doi: 10.1042/EBC20190012. [DOI] [PubMed] [Google Scholar]
- 34.Tapial Martínez P, López Navajas P, Lietha D. FAK structure and regulation by membrane interactions and force in focal adhesions. Biomolecules. 2020;10:179. doi: 10.3390/biom10020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall JE, Fu W, Schaller MD. Chapter five - focal adhesion kinase: exploring fak structure to gain insight into function. In: Jeon KW, editor. International Review of Cell and Molecular Biology. Academic Press; 2011. pp. 185–225. [DOI] [PubMed] [Google Scholar]
- 36.Pathmanathan N, Balleine RL. Ki67 and proliferation in breast cancer. J Clin Pathol. 2013;66:512–516. doi: 10.1136/jclinpath-2012-201085. [DOI] [PubMed] [Google Scholar]
- 37.Mah LJ, El-Osta A, Karagiannis TC. gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24:679–686. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- 38.Sulzmaier FJ, Jean C, Schlaepfer DD. FAK in cancer: mechanistic findings and clinical applications. Nat Rev Cancer. 2014;14:598–610. doi: 10.1038/nrc3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gowda S, Desai PB, Kulkarni SS, Hull VV, Math AA, Vernekar SN. Markers of renal function tests. N Am J Med Sci. 2010;2:170–173. [PMC free article] [PubMed] [Google Scholar]
- 40.Tavares DF, Chaves Ribeiro V, Andrade MAV, Moreira Cardoso-Júnior L, Rhangel Gomes Teixeira T, Ramos Varrone G, Lopes Britto R. Immunotherapy using PD-1/PDL-1 inhibitors in metastatic triple-negative breast cancer: a systematic review. Oncol Rev. 2021;15:497. doi: 10.4081/oncol.2021.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dixon-Douglas J, Loi S. Immunotherapy in early-stage triple-negative breast cancer: where are we now and where are we headed? Curr Treat Options Oncol. 2023;24:1004–1020. doi: 10.1007/s11864-023-01087-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nisar S, Masoodi T, Prabhu KS, Kuttikrishnan S, Zarif L, Khatoon S, Ali S, Uddin S, Akil AA, Singh M, Macha MA, Bhat AA. Natural products as chemo-radiation therapy sensitizers in cancers. Biomed Pharmacother. 2022;154:113610. doi: 10.1016/j.biopha.2022.113610. [DOI] [PubMed] [Google Scholar]
- 43.Talib WH, Alsayed AR, Barakat M, Abu-Taha MI, Mahmod AI. Targeting drug chemo-resistance in cancer using natural products. Biomedicines. 2021;9:1353. doi: 10.3390/biomedicines9101353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee YK, Bae K, Yoo HS, Cho SH. Benefit of adjuvant traditional herbal medicine with chemotherapy for resectable gastric cancer. Integr Cancer Ther. 2018;17:619–627. doi: 10.1177/1534735417753542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newman DJ, Cragg GM. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod. 2020;83:770–803. doi: 10.1021/acs.jnatprod.9b01285. [DOI] [PubMed] [Google Scholar]
- 46.Yang Z, Zhang Q, Yu L, Zhu J, Cao Y, Gao X. The signaling pathways and targets of traditional Chinese medicine and natural medicine in triple-negative breast cancer. J Ethnopharmacol. 2021;264:113249. doi: 10.1016/j.jep.2020.113249. [DOI] [PubMed] [Google Scholar]
- 47.Caesar LK, Cech NB. Synergy and antagonism in natural product extracts: when 1 + 1 does not equal 2. Nat Prod Rep. 2019;36:869–888. doi: 10.1039/c9np00011a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anwar S, Malik JA, Ahmed S, Kameshwar VA, Alanazi J, Alamri A, Ahemad N. Can natural products targeting emt serve as the future anticancer therapeutics? Molecules. 2022;27:7668. doi: 10.3390/molecules27227668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hashemi M, Arani HZ, Orouei S, Fallah S, Ghorbani A, Khaledabadi M, Kakavand A, Tavakolpournegari A, Saebfar H, Heidari H, Salimimoghadam S, Entezari M, Taheriazam A, Hushmandi K. EMT mechanism in breast cancer metastasis and drug resistance: revisiting molecular interactions and biological functions. Biomed Pharmacother. 2022;155:113774. doi: 10.1016/j.biopha.2022.113774. [DOI] [PubMed] [Google Scholar]
- 50.Sigstedt SC, Hooten CJ, Callewaert MC, Jenkins AR, Romero AE, Pullin MJ, Kornienko A, Lowrey TK, Slambrouck SV, Steelant WFA. Evaluation of aqueous extracts of Taraxacum officinale on growth and invasion of breast and prostate cancer cells. Int J Oncol. 2008;32:1085–1090. [PubMed] [Google Scholar]
- 51.Antony ML, Lee J, Hahm ER, Kim SH, Marcus AI, Kumari V, Ji X, Yang Z, Vowell CL, Wipf P, Uechi GT, Yates NA, Romero G, Sarkar SN, Singh SV. Growth arrest by the antitumor steroidal lactone withaferin A in human breast cancer cells is associated with down-regulation and covalent binding at cysteine 303 of β-tubulin. J Biol Chem. 2014;289:1852–1865. doi: 10.1074/jbc.M113.496844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lang SJ, Schmiech M, Hafner S, Paetz C, Werner K, El Gaafary M, Schmidt CQ, Syrovets T, Simmet T. Chrysosplenol d, a flavonol from artemisia annua, induces ERK1/2-mediated apoptosis in triple negative human breast cancer cells. Int J Mol Sci. 2020;21:4090. doi: 10.3390/ijms21114090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tarbeeva DV, Pislyagin EA, Menchinskaya ES, Berdyshev DV, Kalinovskiy AI, Grigorchuk VP, Mishchenko NP, Aminin DL, Fedoreyev SA. Polyphenolic compounds from Lespedeza bicolor protect neuronal cells from oxidative stress. Antioxidants (Basel) 2022;11:709. doi: 10.3390/antiox11040709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tarbeeva DV, Fedoreyev SA, Veselova MV, Blagodatski AS, Klimenko AM, Kalinovskiy AI, Grigorchuk VP, Berdyshev DV, Gorovoy PG. Cytotoxic polyphenolic compounds from Lespedeza bicolor stem bark. Fitoterapia. 2019;135:64–72. doi: 10.1016/j.fitote.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 55.Paing YMM, Valencia M, Cho N, Lee SH. Preventive effect of 1-methoxylespeflorin G11 on nitrite release in lipopolysaccharide-stimulated glial cells. Nat Prod Commun. 2023;18:1934578X231176150. [Google Scholar]
- 56.Ren C, Li Q, Luo T, Betti M, Wang M, Qi S, Wu L, Zhao L. Antioxidant polyphenols from Lespedeza bicolor Turcz. Honey: anti-inflammatory effects on lipopolysaccharide-treated RAW 264.7 macrophages. Antioxidants (Basel) 2023;12:1809. doi: 10.3390/antiox12101809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Almansour NM. Triple-negative breast cancer: a brief review about epidemiology, risk factors, signaling pathways, treatment and role of artificial intelligence. Front Mol Biosci. 2022;9:836417. doi: 10.3389/fmolb.2022.836417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Magna M, Hwang GH, McIntosh A, Drews-Elger K, Takabatake M, Ikeda A, Mera BJ, Kwak T, Miller P, Lippman ME, Hudson BI. RAGE inhibitor TTP488 (Azeliragon) suppresses metastasis in triple-negative breast cancer. NPJ Breast Cancer. 2023;9:59. doi: 10.1038/s41523-023-00564-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan X, Yan Y, Song B, Zhu S, Mei Q, Wu K. Focal adhesion kinase: from biological functions to therapeutic strategies. Exp Hematol Oncol. 2023;12:83. doi: 10.1186/s40164-023-00446-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Provenzano PP, Keely PJ. The role of focal adhesion kinase in tumor initiation and progression. Cell Adh Migr. 2009;3:347–350. doi: 10.4161/cam.3.4.9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chuang HH, Zhen YY, Tsai YC, Chuang CH, Hsiao M, Huang MS, Yang CJ. FAK in cancer: from mechanisms to therapeutic strategies. Int J Mol Sci. 2022;23:1726. doi: 10.3390/ijms23031726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murphy JM, Rodriguez YAR, Jeong K, Ahn EE, Lim SS. Targeting focal adhesion kinase in cancer cells and the tumor microenvironment. Exp Mol Med. 2020;52:877–886. doi: 10.1038/s12276-020-0447-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao X, Guan JL. Focal adhesion kinase and its signaling pathways in cell migration and angiogenesis. Adv Drug Deliv Rev. 2011;63:610–615. doi: 10.1016/j.addr.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bahar ME, Kim HJ, Kim DR. Targeting the RAS/RAF/MAPK pathway for cancer therapy: from mechanism to clinical studies. Signal Transduct Target Ther. 2023;8:455. doi: 10.1038/s41392-023-01705-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fu W, Hall JE, Schaller MD. Focal adhesion kinase-regulated signaling events in human cancer. Biomol Concepts. 2012;3:225–240. doi: 10.1515/bmc-2011-0049. [DOI] [PubMed] [Google Scholar]
- 66.Ebata T, Hirata H, Kawauchi K. Functions of the tumor suppressors p53 and Rb in actin cytoskeleton remodeling. Biomed Res Int. 2016;2016:9231057. doi: 10.1155/2016/9231057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pan MR, Hou MF, Ou-Yang F, Wu CC, Chang SJ, Hung WC, Yip HK, Luo CW. FAK is required for tumor metastasis-related fluid microenvironment in triple-negative breast cancer. J Clin Med. 2019;8:38. doi: 10.3390/jcm8010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jeon M, Hong S, Cho H, Park H, Lee SM, Ahn S. Targeting FAK/PYK2 with SJP1602 for anti-tumor activity in triple-negative breast cancer. Curr Issues Mol Biol. 2023;45:7058–7074. doi: 10.3390/cimb45090446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jing Y, Zhou Q, Zhu H, Zhang Y, Song Y, Zhang X, Huang X, Yang Y, Ni Y, Hu Q. Ki-67 is an independent prognostic marker for the recurrence and relapse of oral squamous cell carcinoma. Oncol Lett. 2019;17:974–980. doi: 10.3892/ol.2018.9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ivashkevich A, Redon CE, Nakamura AJ, Martin RF, Martin OA. Use of the γ-H2AX assay to monitor DNA damage and repair in translational cancer research. Cancer Lett. 2012;327:123–133. doi: 10.1016/j.canlet.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yoon H, Dehart JP, Murphy JM, Lim ST. Understanding the roles of FAK in cancer: inhibitors, genetic models, and new insights. J Histochem Cytochem. 2015;63:114–128. doi: 10.1369/0022155414561498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ho Y, Suphrom N, Daowtak K, Potup P, Thongsri Y, Usuwanthim K. Anticancer effect of citrus hystrix DC. Leaf extract and its bioactive constituents citronellol and, citronellal on the triple negative breast cancer MDA-MB-231 cell line. Pharmaceuticals (Basel) 2020;13:476. doi: 10.3390/ph13120476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Younes M, Ammoury C, Haykal T, Nasr L, Sarkis R, Rizk S. The selective anti-proliferative and pro-apoptotic effect of A. cherimola on MDA-MB-231 breast cancer cell line. BMC Complement Med Ther. 2020;20:343. doi: 10.1186/s12906-020-03120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yurasakpong L, Apisawetakan S, Pranweerapaiboon K, Sobhon P, Chaithirayanon K. Holothuria scabra extract induces cell apoptosis and suppresses Warburg effect by down-regulating Akt/mTOR/HIF-1 axis in MDA-MB-231 breast cancer cells. Nutr Cancer. 2021;73:1964–1975. doi: 10.1080/01635581.2020.1814825. [DOI] [PubMed] [Google Scholar]
- 75.Colamba Pathiranage V, Lowe JN, Rajagopalan U, Ediriweera MK, Senathilake K, Piyathilaka P, Tennekoon KH, Samarakoon SR. Hexane extract of Garcinia quaesita fruits induces apoptosis in breast cancer stem cells isolated from triple negative breast cancer cell line MDA-MB-231. Nutr Cancer. 2021;73:845–855. doi: 10.1080/01635581.2020.1773511. [DOI] [PubMed] [Google Scholar]
- 76.Mesmar J, Abdallah R, Hamade K, Baydoun S, Al-Thani N, Shaito A, Maresca M, Badran A, Baydoun E. Ethanolic extract of Origanum syriacum L. leaves exhibits potent anti-breast cancer potential and robust antioxidant properties. Front Pharmacol. 2022;13:994025. doi: 10.3389/fphar.2022.994025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liao Y, Li S, An J, Yu X, Tan X, Gui Y, Wang Y, Huang L, Zhou S, Wang D. Ethyl acetate extract of Antenoron Filiforme inhibits the proliferation of triple negative breast cancer cells via suppressing Skp2/p21 signaling axis. Phytomedicine. 2023;116:154856. doi: 10.1016/j.phymed.2023.154856. [DOI] [PubMed] [Google Scholar]
- 78.Abdallah R, Shaito AA, Badran A, Baydoun S, Sobeh M, Ouchari W, Sahri N, Eid AH, Mesmar JE, Baydoun E. Fractionation and phytochemical composition of an ethanolic extract of Ziziphus nummularia leaves: antioxidant and anticancerous properties in human triple negative breast cancer cells. Front Pharmacol. 2024;15:1331843. doi: 10.3389/fphar.2024.1331843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guha S, Talukdar D, Mandal GK, Mukherjee R, Ghosh S, Naskar R, Saha P, Murmu N, Das G. Crude extract of Ruellia tuberosa L. flower induces intracellular ROS, promotes DNA damage and apoptosis in triple negative breast cancer cells. J Ethnopharmacol. 2024;332:118389. doi: 10.1016/j.jep.2024.118389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.