Abstract
All positive-strand RNA viruses assemble their RNA replication complexes on intracellular membranes. Brome mosaic virus (BMV) replicates its RNA in endoplasmic reticulum (ER)-associated complexes in plant cells and the yeast Saccharomyces cerevisiae. BMV encodes RNA replication factors 1a, with domains implicated in RNA capping and helicase functions, and 2a, with a central polymerase-like domain. Factor 1a interacts independently with the ER membrane, viral RNA templates, and factor 2a to form RNA replication complexes on the perinuclear ER. We show that BMV RNA replication is severely inhibited by a mutation in OLE1, an essential yeast chromosomal gene encoding Δ9 fatty acid desaturase, an integral ER membrane protein and the first enzyme in unsaturated fatty acid synthesis. OLE1 deletion and medium supplementation show that BMV RNA replication requires unsaturated fatty acids, not the Ole1 protein, and that viral RNA replication is much more sensitive than yeast growth to reduced unsaturated fatty acid levels. In ole1 mutant yeast, 1a still becomes membrane associated, recruits 2a to the membrane, and recognizes and stabilizes viral RNA templates normally. However, RNA replication is blocked prior to initiation of negative-strand RNA synthesis. The results show that viral RNA synthesis is highly sensitive to lipid composition and suggest that proper membrane fluidity or plasticity is essential for an early step in RNA replication. The strong unsaturated fatty acid dependence also demonstrates that modulating fatty acid balance can be an effective antiviral strategy.
All positive-strand RNA viruses of eukaryotes studied to date have RNA replication complexes localized to intracellular membranes, often in association with infection-specific membrane proliferation and or vesiculation (28, 29, 38, 39, 41). Multiple results indicate that membrane association is important for viral RNA synthesis. In vitro synthesis of positive-strand RNAs of picornaviruses and nodaviruses depends on membranes (31, 48). Activation of the alphavirus Semliki Forest virus (SFV) RNA-capping enzyme requires lipids with anionic head groups (3). Cerulenin, an inhibitor of lipid synthesis, inhibits RNA replication by poliovirus and SFV (16, 34). Brefeldin A, an inhibitor of secretory vesicle formation, severely inhibits RNA replication by poliovirus and rhinovirus, though not by some other picornaviruses (11, 20, 30). Despite these results, the nature and function of membrane association in positive-strand RNA virus replication remain poorly understood.
Brome mosaic virus (BMV) is a representative member of the alphavirus-like superfamily of human, animal, and plant positive-strand RNA viruses. The BMV genome is composed of three RNAs. RNA1 and RNA2 respectively encode 1a and 2a, the only BMV proteins required for RNA replication. 1a and 2a interact (25) and contain three domains conserved with those of other superfamily members. 1a contains an N-terminal domain with m7G methyltransferase and m7GMP covalent binding activities required for capping viral RNA in vivo (1, 26) and a C-terminal DEAD box RNA helicase domain. 2a contains a central polymerase-like domain. 1a directs itself and 2a to the endoplasmic reticulum (ER) membrane to form replication complexes that colocalize with viral RNA synthesis (7, 37, 38). RNA3 encodes a cell-to-cell movement protein (3a) and the coat protein, which are dispensable for RNA replication. The 3′-end-proximal coat protein gene is not translatable from RNA3 but only from a subgenomic mRNA, RNA4, synthesized from negative-strand RNA3 (Fig. 1).
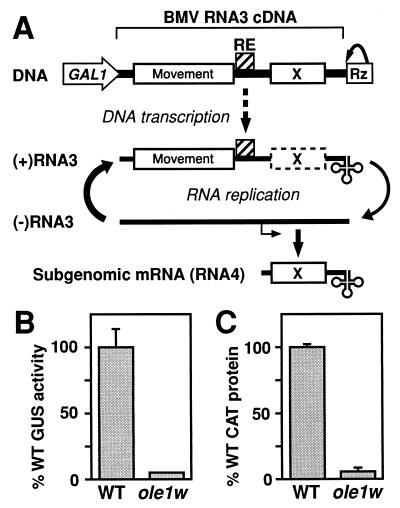
FIG. 1.
(A) Pathway for initiating BMV-directed, RNA-dependent RNA replication and subgenomic mRNA synthesis from DNA. (Top) cDNA-based RNA3 launching cassette including BMV noncoding regions (single lines), movement protein gene, intergenic replication enhancer (RE), 5′-end-flanking GAL1 promoter, and 3′-end-flanking hepatitis delta virus ribozyme (Rz). ORF X represents the BMV coat gene or its replacements, URA3, GUS, or CAT. On galactose induction, cellular RNA polymerase II synthesizes positive-strand RNA3 transcripts that serve as the templates for 1a- and 2a-directed RNA3 replication and subgenomic mRNA (RNA4) synthesis required to express the coat protein gene or its replacements. The bent arrow below negative-strand RNA3 represents the RNA4 start site. (B) BMV-directed GUS expression in wt YMI04 and mutant ole1w yeast. Yeast cells were grown in Gal-containing liquid medium for 48 h, and GUS activity per milligram of total protein was measured. Averages and standard deviations from three independent cultures of each yeast are shown. (C) BMV-directed CAT expression in wt YMI04 and mutant ole1w yeast transfected with B3CAT in vitro transcripts. Transfected spheroplasts were incubated 12 h in Gal medium, and CAT expression per milligram of total protein was measured. Data are presented as in panel B.
As well as in BMV's natural plant hosts, 1a and 2a direct RNA3 replication and subgenomic mRNA synthesis in the yeast Saccharomyces cerevisiae (24). This yeast system reproduces all known features of BMV RNA replication in plant cells, including localization of replication complexes to the perinuclear ER (7, 37). When RNA3 or its derivatives are introduced by transfection or in vivo DNA-dependent transcription into yeast expressing 1a and 2a, negative-strand RNA3 is synthesized and used as a template for RNA-directed synthesis of more positive-strand RNA3 and subgenomic mRNA to express the coat protein gene or its replacements (Fig. 1A). Replacing the coat gene with suitable reporter genes thus provides colony-selectable or -screenable phenotypes linked to BMV RNA replication (22, 24).
To identify cellular factors and functions required for BMV RNA replication, we screened for yeast mutants with defects in their support of BMV-directed gene expression. Here we describe the isolation and characterization of one such yeast mutant, herein designated ole1w yeast, which inhibited BMV RNA replication by more than 95%. The affected OLE1 gene encodes an integral ER membrane protein, Δ9 fatty acid desaturase, essential for conversion of saturated fatty acids (SFAs) to unsaturated fatty acids (UFAs) (44, 45). These UFAs are incorporated into membrane lipids and are major determinants of membrane fluidity and plasticity (10, 36, 40, 42, 47). We found that the OLE1 protein was not required for BMV RNA replication but that one or more steps between template recognition and initiation of viral RNA synthesis required UFAs at levels far above those required for yeast growth. Thus, the ole1 mutation reveals linkage between lipid composition and specific early steps in viral RNA replication. Through its ability to block RNA replication at a particular step, the ole1 mutation should be valuable for further study of RNA synthesis initiation and the membrane association of RNA replication. These results also suggest new directions for antiviral strategies.
MATERIALS AND METHODS
Plasmids.
Yeast 2μm plasmids, pB1CT19 (HIS3 marker gene) and pB2CT15 (LEU2 marker) (24), and centromeric plasmids, pB1YT3H (HIS3 marker) and pB2YT5 (LEU2 marker) (7), were used to express 1a and 2a from the ADH1 and GAL1 promoters, respectively. pB1YT3H was made by substituting the HIS3 gene for the URA3 gene in pB1YT3 (Y. Tomita and M. Ishikawa, unpublished results), a yeast centromeric plasmid with the 1a open reading frame (ORF) linked to the GAL1 promoter. All plasmids expressing RNA3 or its derivatives were derived from pB3RQ39, a centromeric plasmid with the TRP1 marker gene, as described previously (22). A yeast genomic DNA library, ATCC 77164, containing yeast strain YPH1 genomic DNA fragments (average size, 8.8 kb) in the centromeric vector pRS200 (TRP1 marker) (18) was used to identify the complementing gene. The ole1w mutation was isolated by using the gap repair method (12) to clone an HpaI-PacI DNA fragment containing the first 90% of the OLE1 ORF and 220 bp of 5′ noncoding sequence (Fig. 2B) from the mutant yeast into the vector pRS200. DNA sequencing was performed at the Automated DNA Sequencing Facility, University of Wisconsin Biotechnology Center.
FIG. 2.
(A) Schematic of a 5-kb region of yeast chromosome VII containing the OLE1 ORF (thick arrow), showing 2.9-kb fragment I, which complements BMV-directed GUS expression in ole1w yeast, and noncomplementing fragments II and III. Arrows show flanking ORFs. (B) Complementation of BMV-directed GUS expression in ole1w yeast by fragment I of panel A. wt and ole1w yeast cells were transformed with the yeast centromeric plasmid pRS200 carrying fragment I or with the empty plasmid vector. Transformants were grown, and GUS activity was measured as described in the legend to Fig. 1B. (C) The isogenic strain ole1wi, constructed by replacing the OLE1 gene in wt YMI04 with the ole1w gene from mutant yeast, reproduced the phenotype of the original ole1w mutant.
Yeast strains, cell growth, and transformation.
Yeast strain YPH500 (MATα ura3-52 lys2-801 ade2-101 tyr1-63 his3-200 leu2-1) and its derivatives (21) were used throughout, except that YMI06, a derivative of YPH499 (MATa ura3-52 lys2-801 ade2-101 trp1-63 his3-200 leu2-1), (21) was used for mating. YMI04, the parental strain for mutant isolation, was a YPH500 derivative containing chromosomally integrated B3URA3 and B3GUS expression cassettes and plasmids pB1CT19 and pB2CT15. ole1Δ::URA3 yeast was constructed by integrative transformation (12) of YM104 with the NheI-BsrG1 fragment of Fig. 2B with the EcoNI-PacI fragment, containing 400 bp of the 5′ noncoding sequence and 90% of the OLE1 ORF, replaced by the transcriptionally active URA3 gene. The isogenic strains ole1wiand ole1wi′ were constructed by integrative transformation of the NheI-BsrG1 fragment (Fig. 2B) containing the ole1w mutation into, respectively, ole1Δ::URA3 and an equivalent ole1Δ::URA3 derivative of YPH500. Correct integration was verified by Southern blot analysis.
Yeast cultures were grown at 30°C until mid-logarithmic phase (optical density at 600 nm = 0.5 to 0.7) in defined synthetic medium with relevant amino acids omitted to maintain selection for plasmids as described previously (22). Yeast cell pellets, harvested by low-speed centrifugation, were stored at −70°C until RNA or protein extraction. Tergitol Nonidet P-40 (1%) was added to the medium to solubilize fatty acids (44). Plasmid transformation was performed with a FROZEN-EZ yeast transformation kit (Zymo Research).
RNA transfection.
Capped in vitro RNA transcripts of B3CAT were synthesized from pB3CA101, spheroplasts were prepared from yeast grown for 24 h in Gal medium, and RNA transfections were performed as described previously (24).
GUS and CAT assays.
β-Glucuronidase (GUS) filter lifts and quantitative assays were performed as described previously (22). For chloramphenicol acetyltransferase (CAT) assays, yeast lysate was prepared as for the quantitative GUS assay but a different extraction buffer (50 mM Tris [pH 7.5], 5 mM EDTA, 0.1% N-lauroylsarcosine, 0.1% Triton X-100, and 1× protease inhibitors [0.5 mM phenylmethylsulfonyl fluoride, 2.5 mM benzamidine, 1 μg of pepstatin A per ml, and 2.5 μg each of aprotinin and leupeptin per ml]) was used. CAT protein levels were measured with a CAT enzyme-linked immunosorbent assay kit (Boehringer Mannheim), and total protein was determined with a Bradford protein assay kit (Bio-Rad) using bovine serum albumin as a concentration standard.
Western blotting.
Protein was prepared as for the CAT assays except that the extraction buffer was augmented with 20 mM 2-mercaptoethanol and a 2X solution of the protease inhibitors and the clarified cell lysate was supplemented with 1% sodium dodecyl sulfate and boiled for 5 min to inactivate the proteases. Total protein of each cell lysate was determined by a sodium dodecyl sulfate-tolerant Bio-Rad DC Protein Assay (Lowry assay). Equal amounts of total protein were electrophoresed and transferred to nylon membrane. 1a and 2a proteins were probed with corresponding antibodies and detected by chemiluminescence (38).
Northern blotting.
Total yeast RNA isolation, RNA concentration determination by absorbance at 260 nm, agarose-formaldehyde gel electrophoresis, and transfer to nylon membrane were performed as described previously (4, 24). Positive-strand RNA3 and RNA4 were detected with a 32P-labeled RNA probe complementary to their 3′ 200 bases. Negative-strand RNA3 was detected with a 32P-labeled RNA probe corresponding to the CAT gene (for B3CAT) or coat gene (for B3 and B3CPfs) coding sequence (24). Radioactive signals were measured with a Molecular Dynamics PhosphorImager.
RESULTS
Isolation of a yeast mutant strongly inhibiting BMV-directed gene expression.
To isolate mutants with a reduced ability to support BMV-directed gene expression, we used yeast strain YMI04 (21). YMI04 contains plasmids expressing BMV 1a and 2a from the constitutive ADH1 promoter and chromosomally integrated cassettes expressing B3URA3 and B3GUS from the galactose (Gal)-inducible GAL1 promoter. B3URA3 and B3GUS are BMV RNA3 derivatives with the coat gene replaced by the URA3 and GUS genes, respectively. URA3 or GUS expression requires both Gal to induce B3URA3 and B3GUS transcription and BMV 1a- and 2a-directed RNA replication and subgenomic mRNA synthesis (Fig. 1A).
To isolate mutants, UV-mutagenized YMI04 yeast cells were plated on Gal medium containing 0.1% 5-fluoroorotic acid to select against cells with BMV-directed URA3 expression. After 5 to 7 days, about 0.1% of the plated cells developed into colonies. Six thousand such colonies were examined for BMV-expressed GUS activity by filter lift assays. Three hundred isolates with blue color development lacking or delayed relative to that of wild-type (wt) YMI04 were selected and mated with YMI06, which contained no BMV sequences and had the mating type (MATa) opposite to that of YMI04 (MATα). Of the resulting 300 diploids, 100 showed restored GUS activity, implying that inhibition of BMV-directed GUS expression in the corresponding YMI04-derived parental haploids was due to recessive yeast chromosomal mutations complemented by the YMI06 genome. One such Gus− haploid isolate, in which BMV-directed GUS expression was reduced 20-fold, was chosen for further analysis. Complementation experiments showed that this mutation was independent of the previously described BMV-inhibiting yeast mutations mab1, -2, and -3 (21).
This original mutant strain is herein designated ole1w yeast because, as shown below, the causal mutation that inhibits BMV RNA replication maps to the yeast OLE1 gene. w is an allele designation to distinguish this mutation from other ole1 mutations. ole1w yeast grew normally. Its doubling time in defined Gal medium, about 5 h, paralleled that of wt YMI04 yeast. Nevertheless, BMV-directed gene expression was strongly inhibited: GUS activity per milligram of total protein in extracts of ole1w yeast averaged 5% of that of wt YMI04 yeast (Fig. 1B). To determine if this inhibition was due to defective DNA-directed transcription or nucleocytoplasmic transport of B3GUS RNA3, these nuclear steps were bypassed by transfecting ole1w yeast with in vitro transcripts of B3CAT, an RNA3 derivative with the coat gene replaced by the CAT gene. Since the ratio of CAT expression in ole1w yeast to that in wt yeast was equal to that for GUS (Fig. 1B to C), cytoplasmic steps of BMV RNA synthesis must be inhibited in ole1w yeast.
The yeast OLE1 gene complements the mutant defect in BMV-directed gene expression.
To identify genes able to complement this recessive defect in supporting BMV-directed gene expression, ole1w yeast cells were transformed with a yeast genomic DNA library carried by the shuttle vector pRS200, which bears the yeast TRP1 gene (18). Of 20,000 transformants screened by filter lift assays for BMV-directed GUS activity, 5 reproducibly showed wt blue color development. From each of these transformants, a pRS200-based plasmid was isolated by its ability to permit E. coli auxotrophic strain KC8 to grow on medium lacking tryptophan (4). Each of these plasmids complemented the ole1w mutation when it was retransformed into ole1w yeast. Sequencing both ends of the yeast genomic DNA in these plasmids revealed two overlapping fragments of yeast chromosome VII: bases 397187 to 406757 and bases 398499 to 407045. The 8.25-kb region common to both fragments contained five ORFs of 100 or more codons and two tRNA genes.
By deletion mapping and filter lift assays for BMV-directed GUS activity, complementing activity was assigned to a 2.9-kb NheI-BsrG1 fragment containing only the OLE1 ORF (Fig. 2A). When transformed into ole1w yeast, this fragment restored BMV-directed GUS expression to wt levels (Fig. 2B). Moreover, the complete OLE1 gene was required for full complementation (Fig. 2A).
To determine whether OLE1 was the originally mutated gene or an extragenic suppressor, the ole1w gene was cloned from the mutant yeast by gap repair and used to replace the OLE1 gene in wt YMI04 yeast by integrative transformation. The resulting ole1wi isogenic strain reproduced the original ole1w mutant phenotype, inhibiting BMV-directed GUS expression to 5% of that of the wt, and this phenotype was suppressed by a plasmid bearing the wt OLE1 gene (Fig. 2C).
To identify the causal mutation in the ole1w allele, restriction fragments were exchanged between the mutant and wt OLE1 genes and the recombinant plasmids were tested for the ability to complement ole1w yeast. The mutant phenotype was mapped to a 280-bp DNA fragment encoding Arg167-Leu262 of the OLE1-encoded protein, Ole1p. DNA sequencing of this region in the wt and mutant genes revealed a single A-to-G substitution, causing a Tyr212 (TAT)-to-Cys (TGT) substitution in Ole1p.
UFAs restore BMV-directed gene expression in ole1w and ole1Δ yeast.
Ole1p is the Δ9 fatty acid desaturase, a 510-amino-acid (57-kDa) integral ER membrane protein that converts saturated palmitic (16:0) and stearic (18:0) acids into unsaturated palmitoleic (16:1) and oleic (18:1) acids (Fig. 3A). These UFAs exist in yeast cells primarily (>95% of the time) as acy1 chains of membrane phospholipids and are important determinants of membrane fluidity and other physical properties. Transcriptional and posttranscriptional regulation of OLE1 by UFAs, SFAs, and other conditions are largely responsible for regulating the UFA/SFA ratio and thus membrane fluidity (9, 17). As the only enzyme converting SFAs to UFAs, OLE1 is essential for yeast growth in media lacking UFAs (44). Δ9 fatty acid desaturase is also the rate-limiting, initial enzyme for UFA synthesis in animals (32).
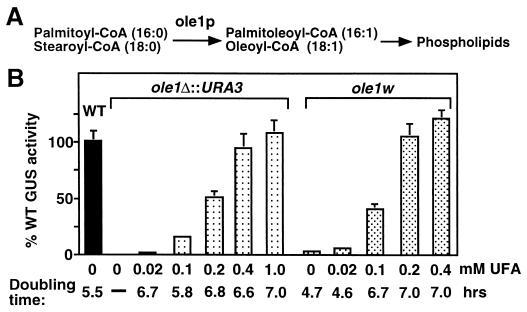
FIG. 3.
(A) Pathway of unsaturated fatty acid synthesis and incorporation into membrane phospholipids. Ole1p, Δ9 fatty acid desaturase, synthesizes palmitoleoyl coenzyme A (CoA) and oleoyl-CoA by introducing a double bond between C-9 and C-10 of palmitoyl-CoA and stearoyl-CoA, respectively. (B) UFAs restore BMV-directed GUS expression in yeast lacking OLE1 (ole1Δ::URA3) and ole1w yeast. wt YMI04, ole1Δ::URA3, and ole1w yeast cells were grown in defined Gal media containing the indicated amounts of UFA (an equimolar mixture of palmitoleic and oleic acids; see Results) until mid-log phase. GUS activity was measured as described in the legend to Fig. 1B. Cell doubling time was calculated from the increase in A600 during log-phase growth.
Because BMV RNA synthesis is also associated with yeast ER membranes (37), the function and localization of Ole1p suggested two possible explanations for the inhibition of BMV-directed gene expression in mutant yeast. First, the ole1w mutation might alter the level of UFAs in yeast membranes, which might inhibit BMV RNA replication, subgenomic mRNA synthesis, or both through effects on membrane fluidity or other physical properties. In keeping with this hypothesis, the ole1w mutation (Tyr212 to Cys) is located in the predicted catalytic domain of Ole1p (45). Alternatively, Ole1p itself, as an integral membrane protein, may be required as an anchor for the BMV RNA replication complex on the ER.
To determine whether BMV-directed gene expression required Ole1p itself or only UFAs, we used integrative transformation to delete the OLE1 ORF of wt YMI04 yeast and replace it with the URA3 gene, creating ole1Δ::URA3 yeast. As expected, ole1Δ::URA3 yeast was unable to grow in medium lacking UFAs (Fig. 3B). The growth of ole1Δ::URA3 yeast and its ability to support BMV-directed gene expression were then tested in medium supplemented with increasing amounts of UFA. UFA was provided as an equimolar mixture of the Ole1p products palmitoleic and oleic acids, which results in a cellular fatty acid composition similar to that in unsupplemented wt yeast (6). UFA (0.02 to 0.1 mM) was sufficient to restore ole1Δ::URA3 yeast to growth with a wt doubling time, but BMV-directed GUS expression remained inhibited to 5 to 15% of wt levels (Fig. 3B). Higher UFA levels progressively improved BMV-directed GUS expression, with nearly wt levels being restored by 0.4 mM UFA. Thus, UFAs but not Ole1p were important for BMV-directed GUS expression. The ability of ole1 mutant yeast to grow with substantially reduced UFA levels is consistent with the finding that UFA levels in wt yeast membranes are five- to ninefold higher than required for growth under optimal conditions (19, 44). The excess UFA is thought to provide extra membrane fluidity required to adapt to environmental changes such as a fall in temperature. Consistent with this, ole1w and ole1wi yeast lost viability within a few days in storage at 4°C while wt yeast was stable for several weeks.
Supplementing the original ole1w yeast with UFAs also restored BMV-directed GUS expression (Fig. 3B), implying that the original mutant phenotype was caused by reduced desaturase activity. ole1w yeast required less UFA supplementation than its ole1Δ::URA3 counterpart to restore a similar level of BMV-directed GUS expression. This finding is consistent with the fact that ole1w yeast cells were isolated and grow normally on defined medium lacking UFAs (see above) and so must retain sufficient desaturase activity for cell growth. When either ole1 mutant was grown in high levels of UFA, some increase in doubling time was noted. However, a similar result was seen with wt yeast and mild inhibitory effects of UFAs on yeast growth have been reported previously (49).
1a and 2a protein accumulation and membrane association in mutant yeast.
To facilitate the viral RNA accumulation experiments described below, we made an additional isogenic yeast strain, ole1wi′, bearing the ole1w allele but lacking the chromosomally integrated B3URA3 and B3GUS expression cassettes of YMI04 and ole1wi. This ole1wi′ strain allowed study of wt RNA3 and RNA3 derivatives introduced on plasmids, while avoiding interference from B3URA3 and B3GUS RNAs in Northern blot analysis of BMV RNA replication products. The initial BMV RNA template used was B3CAT, which combines an easily assayed reporter gene with higher accumulations of BMV RNA replication products than are found with B3GUS.
wt and ole1wi′ yeast were transformed with plasmids expressing B3CAT, 1a, and 2a. With ADH1-expressed 1a and 2a, ole1wi′ yeast showed wt 1a protein accumulation and slightly reduced 2a protein accumulation (Fig. 4A, lanes 1 to 3). Since 2a levels can be reduced substantially without inhibiting BMV RNA replication (13), it was unclear if this reduction contributed to the ole1w RNA replication phenotype. To resolve this, we tested plasmids expressing 1a and 2a from the GAL1 promoter which yield higher levels of and more stable 1a and 2a expression in yeast (12). As intended, GAL1-promoted expression increased 1a and 2a accumulation in wt yeast, and these higher 1a and 2a levels were reproduced in ole1wi′ yeast with or without UFA supplementation (Fig. 4A, lanes 4 to 6).
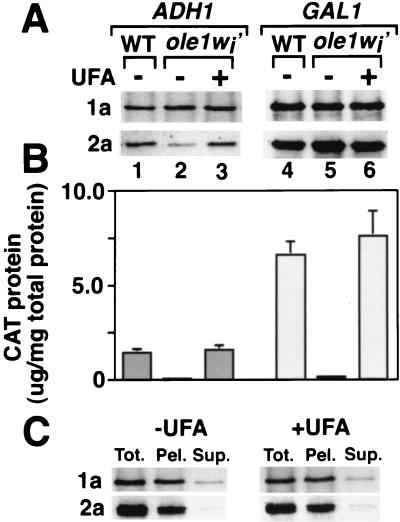
FIG. 4.
(A) Western blot analysis of 1a and 2a protein accumulation in wt and ole1wi′ yeast containing a plasmid expressing B3CAT and either ADH1-promoted 1a and 2a expression plasmids (lanes 1 to 3) or GAL1-promoted 1a and 2a expression plasmids (lanes 4 to 6). Yeast was grown to mid-log phase in Gal medium containing no UFA (−) or 0.2 mM UFA (+). Cell lysates were prepared, and equal amounts of total protein were electrophoresed and Western blotted as described in Materials and Methods. (B) BMV-directed CAT expression in the yeast cells described in the legend to panel A. (C) Distribution of 1a and 2a between membrane and soluble cytoplasmic fractions in ole1wi′ yeast with or without the UFA supplementation. ole1wi′ yeast cells expressing GAL1-promoted 1a, 2a, and RNA3 were harvested at mid-log phase, and total protein (Tot.) was extracted. A portion of the lysate was centrifuged at 10,000 × g to yield pellet (Pel.) and supernatant (Sup.) fractions. The same percentage of each fraction was analyzed by electrophoresis and Western blotting.
BMV-directed CAT expression in ole1wi′ yeast with ADH1-expressed 1a and 2a was 5% of that of the wt (Fig. 4B, left side), duplicating the original ole1w phenotype (Fig. 1C). Adding 0.2 mM UFA to the medium restored 2a accumulation and CAT expression to wt levels. The GAL1-promoted increase in 1a and 2a accumulation coincided with a fivefold increase in BMV-directed CAT expression in wt yeast and UFA-supplemented ole1wi′ yeast (Fig. 4B). However, despite wt 1a and 2a levels, CAT expression in unsupplemented ole1wi′ yeast with GAL1-promoted 1a and 2a was only 2% of that in the wt (Fig. 4B). Thus, the ole1w mutation inhibited BMV-directed gene expression at one or more steps after 1a and 2a protein production. To provide equal levels of 2a accumulation in wt and ole1 mutant yeast, all subsequent experiments were performed with GAL1-expressed 1a and 2a.
Confocal microscopy and cell fractionation show that 1a and 2a are associated with ER membrane in wt yeast that replicates BMV RNA (7, 37). To determine if the ole1w mutation inhibited membrane association of 1a and 2a, ole1wi′ spheroplasts with GAL1-expressed 1a, 2a, and RNA3 were osmotically lysed, membranes were pelleted at 10,000 × g, and Western blotting was used to examine the distribution of 1a and 2a between the membrane and soluble cytoplasmic fractions (7). As shown in Fig. 4C, patterns of distribution were identical in ole1wi′ yeast with or without the UFA supplementation and identical to that in wt yeast (7). Thus, the ole1w mutation did not impede membrane association of 1a or its ability to direct 2a to membrane.
Inhibited accumulation of BMV RNA replication products in ole1wi′ yeast.
To determine whether inhibition of BMV-directed gene expression by ole1w mutation was due to a defect in subgenomic mRNA (RNA4) synthesis or translation, we measured B3CAT RNA4 accumulation in wt and ole1wi′ yeast. Positive-strand RNA4 accumulation in ole1wi′ yeast was only 2% of that of the wt (Fig. 5, lanes 1 to 3), fully accounting for the reduction of BMV-directed CAT protein expression (Fig. 4B). Similar inhibition of positive- and negative-strand B3CAT genomic RNA (RNA3) accumulation was seen in ole1wi′ yeast (7 and 5% of wt levels). All of these viral RNA accumulation defects were suppressed by medium supplementation with 0.2 mM UFA (Fig. 5, lanes 2 to 3).
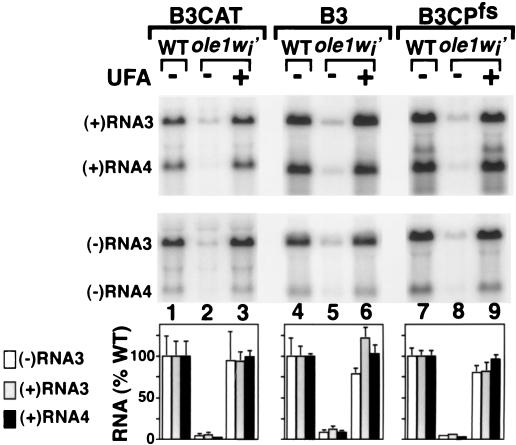
FIG. 5.
Northern blot analysis of RNA3 and RNA4 accumulation in wt and ole1wi′ yeast containing plasmids directing GAL1-promoted expression of 1a, 2a, and the indicated RNA3 derivatives. Yeast cells were grown as described for Fig. 4. Total RNA was prepared, and equal amounts of total RNA were electrophoresed and Northern blotted as described in Materials and Methods. Because negative-strand RNA3 accumulates to 30- to 100-fold-lower levels than those of positive-strand RNA3 in wt yeast, negative-strand blots were printed at a higher intensity to facilitate visualization. Negative-strand RNA3 (open bars), positive-strand RNA3 (shaded bars), and positive-strand RNA4 (filled bars) accumulations were measured with a PhosphorImager (Molecular Dynamics) and normalized to that of wt yeast. The histogram shows averages and standard deviations from three experiments.
Since B3CAT is not a natural BMV RNA replication template, we also tested wt RNA3 replication in ole1wi′ yeast. As shown in Fig. 5, lanes 4 to 6, negative- and positive-strand RNA3 and positive-strand RNA4 accumulations in unsupplemented ole1wi′ yeast were 9, 13, and 9% of wt levels. The slightly larger accumulations of wt RNA3 and RNA4 in ole1wi′ yeast (13 and 9% of wt levels) relative to levels of B3CAT RNA3 and RNA4 (7 and 2% of wt levels) could be due to expression of small amounts of coat protein, which selectively encapsidates and stabilizes BMV RNAs (27). To explore this, we tested B3CPfs, in which coat protein expression was eliminated by a 4-base frameshifting insertion immediately after the initiating AUG codon and simultaneous mutation of the second in-frame AUG codon to AUC (46). As shown in Fig. 5, lanes 7 to 9, B3CPfs RNA3 and RNA4 accumulated in ole1wi′ yeast to 7 and 3% of wt levels, implying that coat protein was largely responsible for increased wt RNA3 and -4 accumulations relative to those in B3CAT. To eliminate any effects of coat protein on RNA3 stability and accumulation (see above), B3CPfs and its derivatives were used in all subsequent experiments.
Normal 1a-induced RNA3 stabilization in ole1wi′ yeast.
In wt yeast lacking 2a, 1a acts through the cis-acting intergenic replication enhancer (RE) of positive-strand RNA3 (Fig. 1A) to dramatically increase the stability and accumulation of RNA3 transcripts while inhibiting their translation (23). Multiple results, including parallel inhibitory and stimulatory effects of RE mutations on 1a-induced RNA3 stabilization and RNA3 replication, indicate that these 1a-induced effects reflect the initial recruitment of RNA3 templates from translation to RNA replication (12, 46). To better determine the stage at which RNA3 replication was inhibited in ole1wi′ yeast, we tested for 1a-induced stimulation of RNA3 transcript accumulation in ole1wi′ yeast.
In the absence of 1a and 2a, plasmid-derived, positive-strand RNA3 transcripts accumulated to equal levels in ole1wi′ yeast with or without the UFA supplementation that suppresses the ole1w phenotype (Fig. 6, lanes 1 to 2). Thus, the ole1w mutation did not affect DNA-dependent synthesis or accumulation of RNA3 transcripts. In the presence of 1a, RNA3 accumulation increased 16-fold in ole1wi′ yeast, again independently of UFA supplementation (lanes 3 to 4). Thus, 1a-induced stimulation of RNA3 accumulation was also not inhibited by the ole1w mutation. Nevertheless, RNA3 replication and subgenomic mRNA synthesis in ole1wi′ yeast remained strongly dependent on UFAs (lanes 5 to 6).
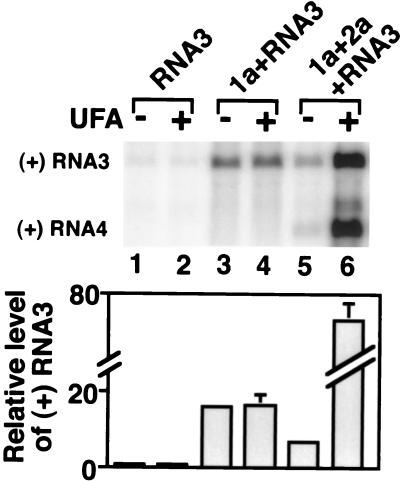
FIG. 6.
Northern blot analysis of BMV positive-strand RNA3 accumulation in ole1wi′ yeast expressing the indicated BMV components. The B3CPfs derivative of RNA3 was used to avoid the effects of coat protein on RNA accumulation (see Results). Yeast cells were grown, and positive-strand RNA3 accumulation was analyzed as described for Fig. 5. The histogram shows averages and standard deviations for positive-strand RNA3 accumulation, normalized to the level in UFA-supplemented ole1wi′ yeast (lane 2), from three experiments. Thus, the histogram scale represents the fold increase in RNA3 accumulation in the presence of 1a (lanes 3 to 4) or 1a plus 2a (lanes 5 to 6), relative to the level of DNA-directed transcription in the absence of 1a and 2a (lanes 1 and 2).
Unexpectedly, the ole1w-dependent inhibition of RNA3 replication in lane 5 revealed that less positive-strand RNA3 accumulated in the presence of 1a and 2a than with 1a alone (lanes 3 to 4). Further results below (Fig. 7) show that, when RNA3 replication is inhibited by a cis-acting mutation, this 2a-induced decrease in RNA3 accumulation occurs even when the ole1w mutant phenotype is suppressed by UFA supplementation. Thus, 2a interference with 1a stabilization of RNA3 appears to be a normal feature of 1a and 2a interaction. Since 2a protein interacts directly with 1a (25) and since 2a mRNA is derived from BMV RNA2, another RNA replication template, either 2a or its mRNA, might competitively inhibit RNA3 interaction with 1a.
FIG. 7.
Inhibition of negative-strand RNA3 synthesis in ole1wi′ yeast. (A) Schematic of B3(5′GAL, CPfs) and its parent B3CPfs, indicating cis-acting elements required for template recruitment (RE), negative-strand initiation, and positive-strand initiation. B3(5′GAL, CPfs) was constructed by replacing the complete viral 5′ NCR of B3CPfs with the 5′ NCR of yeast GAL1 mRNA. (B) Northern blot analysis of positive-strand RNA3 accumulation in wt and ole1wi′ yeast expressing the indicated BMV components. Yeast cells were grown, and the amount of positive-strand RNA3 in each sample was analyzed as described for Fig. 5. (C) Northern blot analysis of negative-strand RNA3 accumulation in wt and ole1wi′ yeast expressing 1a, 2a, and B3(5′GAL, CPfs). The histogram shows averages and standard deviations for negative-strand RNA3 accumulation, normalized to that in wt yeast, from three experiments.
Inhibition of negative-strand RNA3 synthesis in ole1wi′ yeast.
The negative-strand RNA3 synthesis pathway in yeast is not saturated by DNA-transcribed positive-strand RNA3 templates, so that negative-strand RNA3 accumulation is stimulated by RNA-dependent amplification of positive-strand RNA3 templates (22). Consequently, due to the cyclical nature of wt RNA3 replication (Fig. 1A), the reduced negative-strand accumulation in ole1wi′ yeast (Fig. 5) is consistent either with direct inhibition of negative-strand synthesis or with a primary defect in positive-strand synthesis, reducing the templates available for negative-strand synthesis.
To block RNA-dependent positive-strand RNA synthesis and test negative-strand RNA synthesis directly, the wt BMV 5′ noncoding region (NCR) of B3CPfs was replaced with the 5′ NCR of the yeast GAL1 mRNA in an expression plasmid designated B3(5′GAL, CPfs) (Fig. 7A). The resulting B3(5′GAL, CPfs) transcript retained the RE region and, like wt RNA3, showed a strong 1a-dependent increase in accumulation (Fig. 7B, lanes 1 to 4). Moreover, as expected, B3(5′GAL, CPfs) directed UFA-dependent subgenomic mRNA synthesis (Fig. 7B, lane 6). However, even in UFA-supplemented yeast, coexpression of 1a and 2a did not produce the dramatic further increase in positive-strand RNA3 accumulation seen for B3CPfs and wt RNA3 (Fig. 7B, lanes 5 to 6). Rather, with or without UFA supplementation, positive-strand RNA3 accumulation in the presence of 1a and 2a was less than with 1a alone (Fig. 7B, lanes 3 to 6). Thus, B3(5′GAL, CPfs) RNA3 supported little or no BMV-directed positive-strand RNA3 synthesis, confirming prior results that the wt RNA3 5′ NCR contains signals required for positive-strand synthesis (22).
Thus, for B3(5′GAL, CPfs), the only templates for negative-strand RNA3 synthesis were provided by GAL1-promoted DNA transcription, which was unaffected by the ole1wi′ mutation (Fig. 7B, lanes 1 to 2). Accordingly, in wt yeast (2) and in UFA-supplemented ole1wi′ yeast (Fig. 7C and data not shown), the 5′ GAL substitution reduced negative-strand RNA3 accumulation to 15% of that of replicating wt RNA3. More importantly, in unsupplemented ole1wi′ yeast, negative-strand RNA accumulation for B3(5′ GAL, CPfs) was reduced a further 10-fold relative to that from the same template in wt yeast or UFA-supplemented ole1wi′ yeast (Fig. 7C). Thus, in unsupplemented ole1wi′ yeast, BMV RNA replication was inhibited at or before negative-strand RNA3 synthesis.
DISCUSSION
The studies presented here show that BMV RNA replication in yeast is severely inhibited by mutation of OLE1, an essential yeast gene encoding the Δ9 fatty acid desaturase required for unsaturated fatty acids synthesis. UFA supplementation of an engineered ole1 deletion strain showed that BMV RNA replication did not require the Ole1 protein but rather required UFA levels well above those required for yeast cell growth. These results demonstrate in vivo the functional importance of lipids for BMV RNA replication and, as discussed below, imply an intimate and potentially dynamic relationship between RNA replication factors and the lipid bilayer.
The RNA replication defect in ole1w mutant yeast was traced to a narrow interval in early replication. In ole1w yeast, RNA replication factor 1a carried out several normal functions. 1a still became membrane associated and directed the membrane association of 2a (Fig. 4C). The 2a-independent ability of 1a to stabilize RNA3 transcripts, a function strongly linked to selection of RNA3 templates for replication (12, 46), was also unimpaired in ole1wi′ yeast (Fig. 6). Nevertheless, negative-strand RNA3 synthesis was reduced to 10% or less of that of the wt (Fig. 7C). Thus, BMV RNA synthesis was inhibited after initial recognition of the positive-strand RNA3 template but at or before the first phase of RNA synthesis, i.e., negative-strand RNA synthesis. While this defect in negative-strand synthesis is sufficient to explain the overall reduction in BMV RNA replication, the results do not rule out additional defects in later steps of positive-strand RNA3 and subgenomic mRNA synthesis. For flock house virus, e.g., complete in vitro replication of viral RNA and positive-strand synthesis in particular depends on glycerophospholipids (48). Also, the capping functions of the alphavirus SFV nsP1 are activated by lipids, with a requirement for anionic head groups (3). While BMV may be subject to similar influences from polar head groups of membrane lipids, the results presented here show that BMV RNA replication is also highly sensitive to the fatty acid composition of the lipid bilayer.
Recently, BMV RNA replication was also found to be inhibited by mutation of the yeast gene LSM1 (12). LSM1 and OLE1 show many disparate characteristics and appear to be involved in distinct aspects of BMV RNA replication. Unlike OLE1, LSM1 is dispensable for yeast growth in minimal medium at 30°C, though it is required at 37°C. The LSM1-encoded protein, Lsm1p, is not membrane associated but distributed throughout the cytoplasm. Lsm1p is not a biosynthetic enzyme but rather is related to RNA splicing factors and implicated in the metabolism of viral and cellular mRNAs, including the transition of mRNAs from translation to other fates such as degradation and replication (5, 12). Accordingly, LSM1 mutation inhibits 1a-induced stabilization of RNA3, which is unimpaired in ole1wi′ mutants (Fig. 6). These results, isolation of additional BMV-inhibiting yeast mutations, and other findings suggest that many if not most steps in viral RNA replication depend on distinct host factors (12, 21).
UFA dependence of RNA replication.
Cerulenin, an inhibitor of lipid synthesis, inhibits RNA replication by poliovirus and the alphavirus SFV (16, 34). While alternate interpretations cannot be ruled out due to cerulenin's ability to inhibit processes other than lipid synthesis (33), this inhibition of RNA replication suggests a possible requirement for continued lipid and/or membrane synthesis. The inhibition of BMV RNA replication in ole1w yeast, however, is not due to a general block of lipid or membrane synthesis. Ole1p is the desaturase that converts newly synthesized SFAs to UFAs. When UFA levels in yeast are limited by ole1 mutations, membrane synthesis proceeds at normal rates but the UFA/SFA ratio in membrane phospholipids drops (44). Moreover, our experiments showed that ole1w yeast cells had a normal growth rate and size, and this did not change when the cells expressed 1a, 2a, and RNA3.
The UFA/SFA ratio affects many membrane-associated functions because of its strong effect on membrane fluidity and other physical properties (14, 42). wt BMV RNA replication required approximately five times more UFA supplementation than did normal growth of mutant yeast (Fig. 3), suggesting that optimal assembly or function of the RNA replication complex requires a highly fluid membrane. After membrane association, rapid diffusion might be required for 1a, 2a, or another replication factor to locate a required interaction partner before being trapped in a competing nonproductive interaction. During replication, rotation or translation of membrane-associated RNA replication factors might be required for RNA unwinding, translocation along RNA templates, or necessary cyclical alterations in protein-protein interactions.
In addition to kinetic effects, reduced UFA levels may also impede BMV RNA synthesis by perturbing the form or stability of replication factor interactions. Under conditions of reduced UFA levels, increased lipid packing density and membrane microviscosity tend to displace membrane-associated proteins farther into the aqueous phase, altering their potential for interacting with other factors and the position of such interactions relative to the membrane (8, 42). Since introducing a cis double bond shifts lipids from a cylindrical to a more cone-shaped profile, UFAs also influence membrane curvature and flexibility (40). Modulating any of these parameters may impede the functional interaction of 1a, 2a, viral RNA, or host components with each other. Since the ole1w mutation did not inhibit 1a association with membrane or 1a-directed membrane association of 2a (Fig. 4C), the required interaction of the N-terminal 120 amino acids of 2a with the 1a C-terminal helicase-like domain (7) was not affected in ole1w yeast. However, other 1a-2a interactions required for later RNA replication steps may be perturbed. For example, BMV RNA replication also depends on an independent interaction between 1a and the central 2a polymerase domain (43).
While negative-strand RNA synthesis was strongly dependent on UFAs in vivo, a preformed, template-dependent negative-strand RNA synthesis activity can be solubilized from membranes of BMV-infected plant cells or yeast expressing 1a, 2a, and RNA3 (35). Thus, the UFA requirement may lie in assembly of a functional RNA synthesis complex. Alternatively, in vivo UFA dependence and membrane association of negative-strand RNA synthesis may relate to functions missing from the solubilized, in vitro negative-strand synthesis activity. Anomalous characteristics of the in vitro system include low efficiency of template usage (<0.1% of added template) and a lack of response to the intercistronic replication enhancer, which in vivo directs 1a-dependent RNA3 stabilization and stimulates negative-strand RNA3 synthesis and RNA3 replication approximately 100-fold (35, 46).
While unsaturated oleic and/or palmitoleic acids were required for BMV RNA replication, oleic acid disrupts poliovirus RNA replication in HeLa cells (15) or HeLa cell extracts (31). These results may be related to more complex effects of oleic acid on HeLa cells. Supplementing ole1 mutant yeast with oleic acid, palmitoleic acid, or other UFAs yields a direct increase in membrane glycerophospholipids containing these UFAs (44). However, treating of HeLa cells with oleic acid resulted in major changes in the synthesis of many lipids, including dramatic increases in the synthesis of cholesterol and other neutral lipids, a reduced phosphatidylserine/phosphatidylcholine ratio, and other changes (15). Similarly, in HeLa cell extracts, oleic acid inhibited in vitro translation as well as poliovirus RNA replication (31).
In conclusion, we find that BMV RNA replication is strongly dependent on UFA levels in vivo. When UFAs were limited, ER-associated RNA replication was blocked after 1a and 2a membrane association and RNA3 template recognition and stabilization but before negative-strand RNA synthesis. The ability to use the ole1w mutation to block RNA replication at this stage should help to elucidate the early events in initiation of RNA synthesis. Dependence of BMV RNA replication on UFA levels in particular implies a requirement for host membrane fluidity, suggesting that the membrane is not just a static anchoring site for RNA replication complexes. Accordingly, further study of ole1w yeast should help to illuminate the nature and function of membrane association in RNA replication in positive-strand RNA viruses.
Since membrane-associated RNA replication appears to be a universal feature of positive-strand RNA viruses of eukaryotes, the replication of other viruses in this class may also be dependent on the fatty acid compositions of membrane lipids. Thus, while different host cells and viruses may function optimally over different lipid composition ranges, the finding that BMV RNA replication is much more sensitive than normal yeast cell growth to reduced levels of UFAs suggests that genetic or pharmacological approaches to modulate the lipid composition of host membranes may provide useful antiviral strategies.
ACKNOWLEDGMENTS
We thank Yukio Tomita for pB1YT3, Michael Sullivan for pB3(5′ GAL, CPfs), Amine Noueiry and Juana Diez for valuable experimental advice, and Cindy Dorner for excellent technical assistance.
This work was supported by the National Institutes of Health through grant GM35072. P.A. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Ahola T, Ahlquist P. Putative RNA capping activities encoded by brome mosaic virus: methylation and covalent binding of guanylate by replicase protein 1a. J Virol. 1999;73:10061–10069. doi: 10.1128/jvi.73.12.10061-10069.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahola T, den Boon J, Ahlquist P. Helicase and capping enzyme active site mutations in brome mosaic virus protein 1a cause defects in template recruitment, negative-strand RNA synthesis, and viral RNA capping. J Virol. 2000;74:8803–8811. doi: 10.1128/jvi.74.19.8803-8811.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahola T, Lampio A, Auvinen P, Kaariainen L. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J. 1999;18:3164–3172. doi: 10.1093/emboj/18.11.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 5.Boeck R, Lapeyr B, Brown C E, Sachs A B. Capped mRNA degradation intermediates accumulate in the yeast spb8–2 mutant. Mol Cell Biol. 1998;18:5062–5072. doi: 10.1128/mcb.18.9.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bossie M A, Martin C E. Nutritional regulation of yeast Δ-9 fatty acid desaturase activity. J Bacteriol. 1989;171:6409–6413. doi: 10.1128/jb.171.12.6409-6413.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J, Ahlquist P G. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J Virol. 2000;74:4310–4318. doi: 10.1128/jvi.74.9.4310-4318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherry R J, Muller U, Holenstein C, Heyn M P. Lateral segregation of proteins induced by cholesterol in bacteriorhodopsin-phospholipid vesicles. Biochim Biophys Acta. 1980;596:145–151. doi: 10.1016/0005-2736(80)90179-0. [DOI] [PubMed] [Google Scholar]
- 9.Choi J Y, Stukey J, Hwang S Y, Martin C E. Regulatory elements that control transcription activation and unsaturated fatty acid-mediated repression of the Saccharomyces cerevisiae OLE1 gene. J Biol Chem. 1996;271:3581–3589. doi: 10.1074/jbc.271.7.3581. [DOI] [PubMed] [Google Scholar]
- 10.Cronan J E J, Gelmann E P. Physical properties of membrane lipids: biological relevance and regulation. Bacteriol Rev. 1975;39:232–256. doi: 10.1128/br.39.3.232-256.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuconati A, Molla A, Wimmer E. Brefeldin A inhibits cell-free, de novo synthesis of poliovirus. J Virol. 1998;72:6456–6464. doi: 10.1128/jvi.72.8.6456-6464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diez J, Ishikawa M, Kaido M, Ahlquist P. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc Natl Acad Sci USA. 2000;97:3913–3918. doi: 10.1073/pnas.080072997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dinant S, Janda M, Kroner P A, Ahlquist P. Bromovirus RNA replication and transcription require compatibility between the polymerase- and helicase-like viral RNA synthesis proteins. J Virol. 1993;67:7181–7189. doi: 10.1128/jvi.67.12.7181-7189.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emmerson P J, Clark M J, Medzihradsky F, Remmers A E. Membrane microviscosity modulates mu-opioid receptor conformational transitions and agonist efficacy. J Neurochem. 1999;73:289–300. doi: 10.1046/j.1471-4159.1999.0730289.x. [DOI] [PubMed] [Google Scholar]
- 15.Guinea R, Carrasco L. Effects of fatty acids on lipid synthesis and viral RNA replication in poliovirus-infected cells. Virology. 1991;185:473–476. doi: 10.1016/0042-6822(91)90802-i. [DOI] [PubMed] [Google Scholar]
- 16.Guinea R, Carrasco L. Phospholipid biosynthesis and poliovirus genome replication, two coupled phenomena. EMBO J. 1990;9:2011–2016. doi: 10.1002/j.1460-2075.1990.tb08329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gyorfy Z, Horvath I, Balogh G, Domonkos A, Duda E, Maresca B, Vigh L. Modulation of lipid unsaturation and membrane fluid state in mammalian cells by stable transformation with the Δ9-desaturase gene of Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1997;237:362–366. doi: 10.1006/bbrc.1997.7141. [DOI] [PubMed] [Google Scholar]
- 18.Halbrook J, Hoekstra M F. Mutations in the Saccharomyces cerevisiae CDC1 gene affect double-strand-break-induced intrachromosomal recombination. Mol Cell Biol. 1994;14:8037–8050. doi: 10.1128/mcb.14.12.8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henry S A. Membrane lipids of yeast: biochemical and genetic studies. In: Strathern J, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces: metabolism and gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. pp. 101–149. [Google Scholar]
- 20.Irurzun A, Perez L, Carrasco L. Involvement of membrane traffic in the replication of poliovirus genomes: effects of brefeldin A. Virology. 1992;191:166–175. doi: 10.1016/0042-6822(92)90178-r. [DOI] [PubMed] [Google Scholar]
- 21.Ishikawa M, Diez J, Restrepo-Hartwig M, Ahlquist P. Yeast mutations in multiple complementation groups inhibit brome mosaic virus RNA replication and transcription and perturb regulated expression of the viral polymerase-like gene. Proc Natl Acad Sci USA. 1997;94:13810–13815. doi: 10.1073/pnas.94.25.13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishikawa M, Janda M, Krol M A, Ahlquist P. In vivo DNA expression of functional brome mosaic virus RNA replicons in Saccharomyces cerevisiae. J Virol. 1997;71:7781–7790. doi: 10.1128/jvi.71.10.7781-7790.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janda M, Ahlquist P. Brome mosaic virus RNA replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc Natl Acad Sci USA. 1998;95:2227–2232. doi: 10.1073/pnas.95.5.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janda M, Ahlquist P. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell. 1993;72:961–970. doi: 10.1016/0092-8674(93)90584-d. [DOI] [PubMed] [Google Scholar]
- 25.Kao C C, Ahlquist P. Identification of the domains required for direct interaction of the helicase-like and polymerase-like RNA replication proteins of brome mosaic virus. J Virol. 1992;66:7293–7302. doi: 10.1128/jvi.66.12.7293-7302.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong F, Sivakumaran K, Kao C C. The N-terminal half of the brome mosaic virus 1a protein has RNA capping-associated activities: specificity for GTP and S-adenosylmethionine. Virology. 1999;259:200–210. doi: 10.1006/viro.1999.9763. [DOI] [PubMed] [Google Scholar]
- 27.Krol M A, Olson N H, Tate J, Johnson J E, Baker T S, Ahlquist P. RNA-controlled polymorphism in the in vivo assembly of 180-subunit and 120-subunit virions from a single capsid protein. Proc Natl Acad Sci USA. 1999;96:13650–13655. doi: 10.1073/pnas.96.24.13650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackenzie J M, Jones M K, Westaway E G. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J Virol. 1999;73:9555–9567. doi: 10.1128/jvi.73.11.9555-9567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magliano D, Marshall J A, Bowden D S, Vardaxis N, Meanger J, Lee J Y. Rubella virus replication complexes are virus-modified lysosomes. Virology. 1998;240:57–63. doi: 10.1006/viro.1997.8906. [DOI] [PubMed] [Google Scholar]
- 30.Maynell L A, Kirkegaard K, Klymkowsky M W. Inhibition of poliovirus RNA synthesis by brefeldin A. J Virol. 1992;66:1985–1994. doi: 10.1128/jvi.66.4.1985-1994.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molla A, Paul A V, Wimmer E. Effects of temperature and lipophilic agents on poliovirus formation and RNA synthesis in a cell-free system. J Virol. 1993;67:5932–5938. doi: 10.1128/jvi.67.10.5932-5938.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ntambi J M. Regulation of stearoyl-CoA desaturase by polyunsaturated fatty acids and cholesterol. J Lipid Res. 1999;40:1549–1558. [PubMed] [Google Scholar]
- 33.Oda T, Wu H C. Cerulenin inhibits the cytotoxicity of ricin, modeccin, Pseudomonas toxin, and diphtheria toxin in brefeldin A-resistant cell lines. J Biol Chem. 1993;268:12596–12602. [PubMed] [Google Scholar]
- 34.Perez L, Guinea R, Carrasco L. Synthesis of Semliki Forest virus RNA requires continuous lipid synthesis. Virology. 1991;183:74–82. doi: 10.1016/0042-6822(91)90119-v. [DOI] [PubMed] [Google Scholar]
- 35.Quadt R, Ishikawa M, Janda M, Ahlquist P. Formation of brome mosaic virus RNA-dependent RNA polymerase in yeast requires coexpression of viral proteins and viral RNA. Proc Natl Acad Sci USA. 1995;92:4892–4896. doi: 10.1073/pnas.92.11.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rattray J B, Schibeci A, Kidby D K. Lipids of yeasts. Bacteriol Rev. 1975;39:197–231. doi: 10.1128/br.39.3.197-231.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Restrepo-Hartwig M, Ahlquist P. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J Virol. 1999;73:10303–10309. doi: 10.1128/jvi.73.12.10303-10309.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Restrepo-Hartwig M A, Ahlquist P. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J Virol. 1996;70:8908–8916. doi: 10.1128/jvi.70.12.8908-8916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaad M C, Jensen P E, Carrington J C. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 1997;16:4049–4059. doi: 10.1093/emboj/16.13.4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneiter R, Kohlwein S D. Organelle structure, function, and inheritance in yeast: a role for fatty acid synthesis? Cell. 1997;88:431–434. doi: 10.1016/s0092-8674(00)81882-6. [DOI] [PubMed] [Google Scholar]
- 41.Sethna P B, Brian D A. Coronavirus genomic and subgenomic minus-strand RNAs copartition in membrane-protected replication complexes. J Virol. 1997;71:7744–7749. doi: 10.1128/jvi.71.10.7744-7749.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shinitzky M. Membrane fluidity and cellular functions. In: Shinitzky M, editor. Physiology of membrane fluidity. Vol. 1. Boca Raton, Fla: CRC Press; 1984. pp. 1–51. [Google Scholar]
- 43.Smirnyagina E, Lin N S, Ahlquist P. The polymerase-like core of brome mosaic virus 2a protein, lacking a region interacting with viral 1a protein in vitro, maintains activity and 1a selectivity in RNA replication. J Virol. 1996;70:4729–4736. doi: 10.1128/jvi.70.7.4729-4736.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stukey J, McDonough V M, Martin C E. Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J Biol Chem. 1989;264:16537–16544. [PubMed] [Google Scholar]
- 45.Stukey J, McDonough V M, Martin C E. The OLE1 gene of Saccharomyces cerevisiae encodes the Δ9 fatty acid desaturase and can be functionally replaced by the rat stearoyl-CoA desaturase gene. J Biol Chem. 1990;265:20144–20149. [PubMed] [Google Scholar]
- 46.Sullivan M L, Ahlquist P. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J Virol. 1999;73:2622–2632. doi: 10.1128/jvi.73.4.2622-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Rest M E, Kamminga A H, Nakano A, Anraku Y, Poolman B, Konings W N. The plasma membrane of Saccharomyces cerevisiae: structure, function, and biogenesis. Microbiol Rev. 1995;59:304–322. doi: 10.1128/mr.59.2.304-322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu S X, Ahlquist P, Kaesberg P. Active complete in vitro replication of nodavirus RNA requires glycerophospholipid. Proc Natl Acad Sci USA. 1992;89:11136–11140. doi: 10.1073/pnas.89.23.11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang S, Skalsky Y, Garfinkel D J. MGA2 or SPT23 is required for transcription of the Δ9 fatty acid desaturase gene, OLE1, and nuclear membrane integrity in Saccharomyces cerevisiae. Genetics. 1999;151:473–483. doi: 10.1093/genetics/151.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]