Abstract
Plexiform neurofibromas (PNFs) are benign nerve tumors driven by loss of the NF1 tumor suppressor in Schwann cells. PNFs are rich in immune cells, but whether immune cells are necessary for tumorigenesis is unknown. We show that inhibition of stimulator of interferon gene (STING) reduces plasma CXCL10, tumor T cell and dendritic cell (DC) recruitment, and tumor formation. Further, mice lacking XCR-1+ DCs showed reduced tumor-infiltrating T cells and PNF tumors. Antigen-presenting cells from tumor-bearing mice promoted CD8+ T cell proliferation in vitro, and PNF T cells expressed high levels of CCL5, implicating T cell activation. Notably, tumors and nerve-associated macrophages were absent in Rag1−/−; Nf1f/f; DhhCre mice and adoptive transfer of CD8+ T cells from tumor-bearing mice restored PNF initiation. In this setting, PNF shrunk upon subsequent T cell removal. Thus, STING pathway activation contributes to CD8+ T cell–dependent inflammatory responses required for PNF initiation and maintenance.
Infiltrating T cells drive NF1 nerve tumor development.
INTRODUCTION
Inflammation is a hallmark of tumorigenesis (1) and a notable feature of the plexiform neurofibromas (PNFs). PNFs are peripheral nerve tumors that develop in 30 to 50% of individuals with neurofibromatosis type 1 (NF1) and can cause substantial morbidity (2). In mouse models, PNFs form months after loss of Nf1 in nerve Schwann cells (SCs). Over time, macrophages, mast cells, and, subsequently, small numbers of T cells and dendritic cells (DCs) are recruited to the paraspinal ganglia and nerves, after which PNFs develop (3–5). Each of these immune cell types also accumulates in human PNF, providing a microenvironment that fosters benign tumor growth (5–7). SCs are the only cells within neurofibromas that contain biallelic pathogenic variants in the NF1 tumor suppressor gene, which encodes a RAS–guanosine 5′-triphosphate (GAP), so that SCs have elevated levels of RAS-GTP (8). PNFs serve as a model of inflammation-driven tumors because immune cell infiltration and tumor formation are separable in time.
In the 1960s, mast cells were identified in neurofibromas and suggested to be important for tumor development [reviewed in (9, 10)]. Unexpectedly, however, mast cells were found to be dispensable for neurofibroma formation in murine models (11). It is now known that myeloid cells, largely, macrophages, are the dominant immune cells in neurofibromas (12); DCs and T cells are also present (5–7). A requirement for immune cells in PNF formation was underscored by the finding that, when Nf1 was lost in populations of SCs (Krox20-Cre;Nf1f/f), tumors formed after engrafting Nf1+/− hematopoietic cells (13). In addition, when Nf1 was lost in populations of SCs (P0-Cre;Nf1f/f), tumors formed after wound-related influx of wild-type (WT) or Nf1+/− immune cells (14). In the Nf1f/f; DhhCre model, all immune cells are WT at the Nf1 locus, and PNFs form in all mice. In this model, DC and T cells express the chemokine receptor CXCR3, and deletion of CXCR3 prevented PNF formation (15). Together, these gain- and loss-of-function studies strongly support the idea that mutant SCs require immune cells, likely DCs and/or T cells, for PNF initiation and growth. Whether T cells and/or DCs contribute to neurofibroma development has not been studied.
Given the importance of CXCR3 in PNF initiation, it is of interest that CXCR3-expressing cells migrate toward the CXCR3 ligands CXCL9 and/or CXCL10 and that CXCL10 is expressed by Nf1−/− SCs before tumor formation (3). The stimulator of interferon gene (STING) activation generally occurs when cells sense damage-related signals including double-stranded RNAs/DNAs, and STING activation promotes expression of cytokines including CXCL10 via induction of type I interferons (16, 17). Neurofibroma cells show evidence of damage, with an increase in SCs containing DNA damage foci compared to that in SCs in normal nerves (18). Damage likely occurs, at least, in part, because loss of Nf1 in SCs leads to elevated RAS-GTP, and acute elevation of RAS-GTP in cells can lead to cell damage, cell senescence, or cell death (16). We posited that, after SC-specific loss of NF1, STING activation increases cytokines and chemokines that modulate the tumor microenvironment via DC and/or T cell recruitment, resulting in tumorigenesis.
T cells have been implicated in promoting tumor formation. For example, CD8+ T cells induced proliferation of epithelial cells in benign prostatic hyperplasia (19). Conversely, T cell depletion delayed tumor formation in a mouse model of inflammation induced hepatocellular carcinoma (20). In addition, the absence of a subset of T cells (γδT cells) significantly decreased the incidence of chemically induced papilloma (21). Notably, reconstitution of tumor-prone mice with bone marrow lacking T cells (Rag2−/− or δTCR−/−) profoundly reduced tumor formation (22). How tumor cells recruit T cells and how T cells become activated in these models are poorly understood.
Through their T cell receptors, CD8+ T cells are primed by DCs presenting antigenic peptides in the context of major histocompatibility complex (MHC) molecules. Although peptides presented by class I MHC molecules are generally derived from inside the cell, specialized conventional type I DC (cDC1) cells have the capability of presenting extracellular antigens on class I MHC, via a process known as cross-presentation (23). In the context of tumors, cDC1 cells, marked by expression of Xcr-1, are highly efficient in the uptake and presentation of tumor associated antigens on class I MHC to tumor-specific CD8+ T cells (24). This “cross-priming” of T cells promotes their initial activation and enables their effector programs that normally facilitate tumor killing.
We show that Nf1 loss in SCs results in STING activation. STING activity contributes to production of CXCL10 and, therefore, to recruitment by SCs of CXCR3-expressing DC1, DC2, and T cells. Antigen-presenting cells (APCs) from tumor-bearing mice are sufficient to promote T cell proliferation and within tumors, T cells show evidence of activation. Last, we found that CD8 T cells are required for tumor formation and for tumor maintenance, demonstrating a previously unknown role for T cells in promoting tumorigenesis.
RESULTS
Activation of the STING pathway in PNF
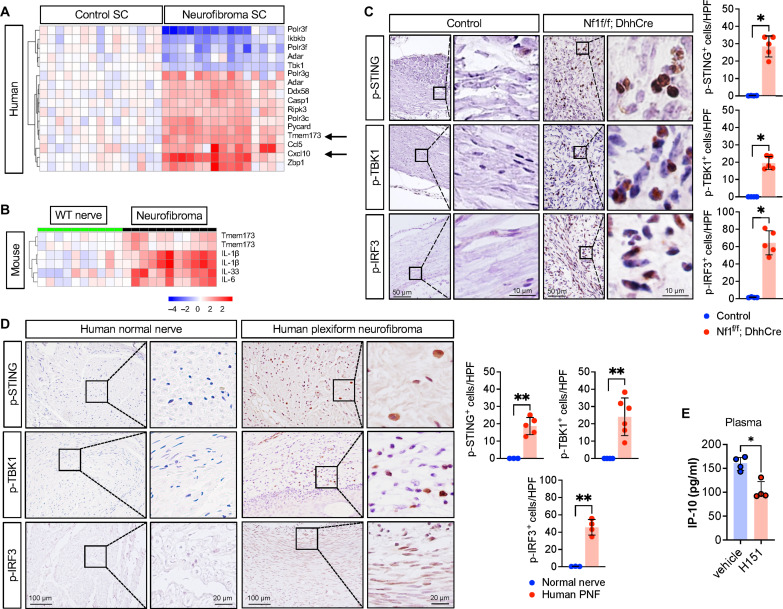
To determine whether STING is expressed in neurofibroma, we measured STING RNA and protein. Tmem173 (STING itself) mRNA was elevated in primary NF1−/− SCs cultured from human PNF, as compared to that in normal SCs (Fig. 1A). In mouse PNF tumors compared to that in normal nerve, Tmem173 was also increased (Fig. 1B). Activation of STING involves a cascade of signaling events, including STING phosphorylation on serine-366 and STING palmitoylation, which subsequently result in the phosphorylation of STING downstream effectors interferon regulatory factor 3 (IRF3) and TANK-binding kinase 1 (TBK1) (25). Validating activation of the STING pathway in PNF cells, we detected significant increase of phosphorylated (active) STING (p-STING), p-TBK1, and p-IRF3, by immunohistochemistry (IHC) in human and mouse PNF tissue sections, as compared to the low levels in normal nerve in each species (Fig. 1, C and D).
Fig. 1. STING activation in mice and human PNF.
(A) Human SC gene microarray show up-regulation of STING gene (Tmem173) and CXCL10. (B) Gene microarray of murine WT nerve or Nf1f/f; DhhCre PNF. (C) Immunohistochemistry (IHC) of mouse control nerve and PNF tissue section for phosphorylated STING (p-STING), TBK1 (p-TBK1), and IRF3 (p-IRF3), indicating the activation of STING signaling pathway. (D) Representative IHC images for p-STING, p-TBK1, and p-IRF3 staining and quantified in human normal and PNF. HPF, high-power field. (E) Serum IP-10 (CXCL10) in mice treated with vehicle or STING inhibitor H-151. *P < 0.05 and **P < 0.001.
Tumor-bearing mice also show elevated levels of CXCL10 in plasma. To test whether STING activity contributes to CXCL10 levels in neurofibroma-bearing mice, we used H-151, a potent and selective small-molecule inhibitor that specifically blocks the palmitoylation and clustering of STING (26). Treatment of tumor-bearing Nf1f/f; DhhCre mice with H-151 reduced plasma levels of CXCL10/IP10 by twofold (Fig. 1E). None of the six other factors tested was affected by STING pathway inhibition (fig. S1). These results show that STING is activated in PNF and that STING activity contributes to production of CXCL10, a DC and T cell chemoattractant.
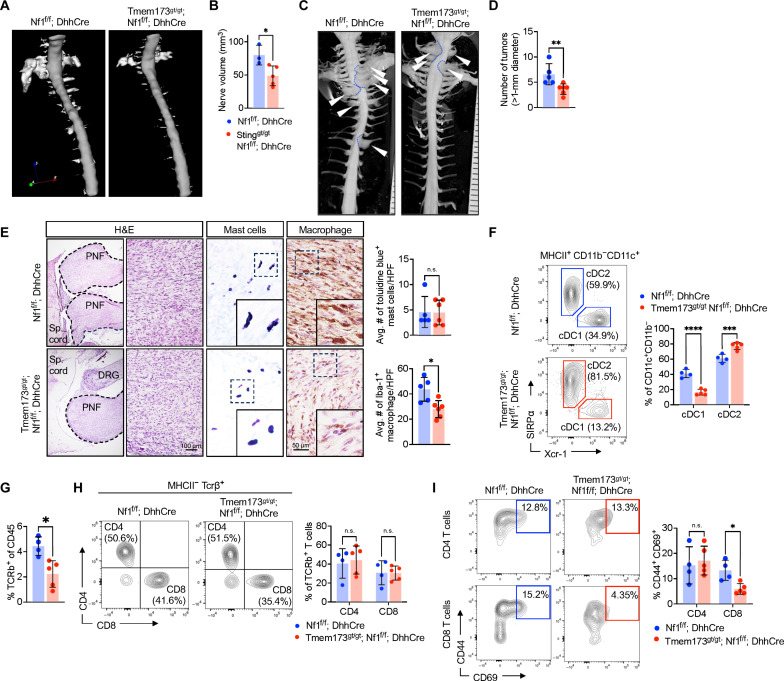
To test whether STING activity has a role in tumor initiation, we intercrossed Nf1f/f; DhhCre mice with Tmem173gt/gt mice. These mutant mice have a missense mutation, altering Iso-Arg at amino acid 199 in the C terminus of the STING protein, and loss of STING activity (27). To facilitate genotyping Tmem173gt/gt mutants, we developed a modified assay (fig. S2, A and B). At 4 months old, PNF can be detected by magnetic resonance imaging (MRI) in Nf1f/f; DhhCre mice. Four-month-old Tmem173gt/gt; Nf1f/f; DhhCre showed smaller and fewer PNF on MRI assessment; representative digital three-dimensional (3D) images of spinal cord with tumors within dorsal root ganglia (DRG) and nerves adjacent to the spinal cord are shown in Fig. 2A for mutants versus Nf1f/f; DhhCre controls. Figure 2B shows quantification, confirming significantly reduced tumor volume in PNF-bearing Nf1f/f; DhhCre mice lacking STING pathway activation versus controls. When these mice reached 7 months of age, they were euthanized. We confirmed reduced tumor number and tumor size by gross dissections and tumor measurements (Fig. 2, C and D). We performed histological analysis and found that the remaining tumors showed the spindled cells with wavy nuclei, in the presence of toluidine blue metachromatic mast cells and Iba-1–immunopositive macrophages that are consistent with PNF histopathology (Fig. 2E).
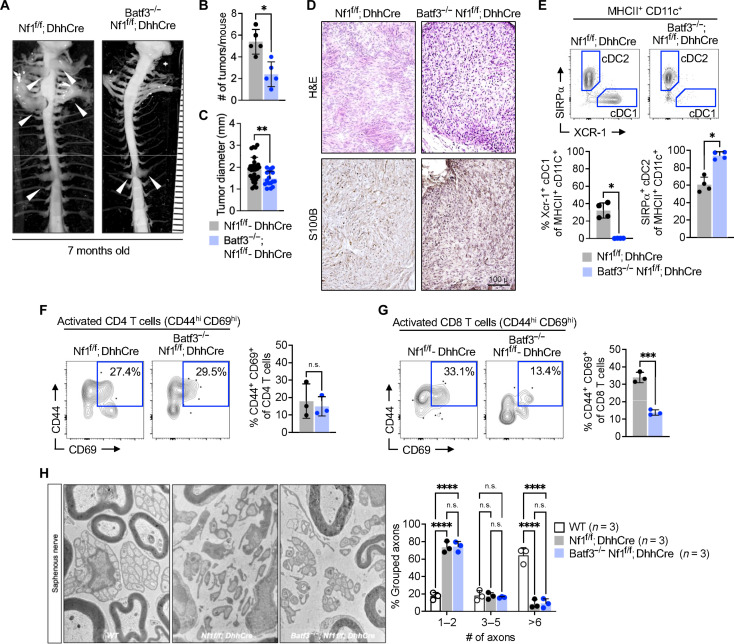
Fig. 2. Loss of STING attenuates PNF by reducing cDC1 and T cell infiltration.
(A) Representative digital three-dimensional (3D) rendering from MRI images of 4-month-old STING-sufficient Nf1f/f; DhhCre and STING-deficient STINGgt/gt; Nf1f/f; DhhCre. (B) Quantification of PNF volume. (C) Representative gross dissections of mice with WT STING and mutant STING at 7 months old. (D) Quantification of the number of tumors. (E) Representative hematoxylin and eosin (H&E) stain, toluidine blue stain for mast cells, and Iba-1 for macrophages. (F) PNF flow cytometer of Nf1f/f; DhhCre or STINGgt/gt Nf1f/f; DhhCre mice for MHCII+ CD11c+ CD172a− XCR-1+ cDC1 and MHCII+ CD11c+ CD172a+ XCR-1− cDC2 DCs, (G) TCRβ+ T cells and (H) CD4 and CD8 T cells with (I) represented analysis of activation markers (CD44 and CD69). Statistical significance (P < 0.05). n.s., not significant. Student’s t-test: *adj. *P < 0.05, **P < 0.001, ***P < 0.003, and ****P < 0.0001.
We next used multiparametric spectral flow cytometry of mouse tumors for improved definition of PNF cells. We dissected PNF from both Nf1f/f; DhhCre and Tmem173gt/gt; Nf1f/f; DhhCre mice and rapidly processed them by enzymatic and mechanical dissociation for analysis of inflammatory cells (fig. S3, A and B). Gating and panel design of flow cytometric data to analyze subpopulations of T cells and DC populations is shown in fig. S3 (C to E). The lack of functional STING significantly reduced tumor-residing cDC1 (MHCII+CD11c+CD11b−CD127a−XCR-1+), while cDC2 (MHCII+CD11c+CD11b+CD127a+XCR-1−) slightly increased as a percentage of CD11c+ DC (Fig. 2F). Lack of STING also significantly reduced TCRβ+ T cells (of total CD45+ cells) compared to STING-sufficient controls (Fig. 2G) but did not affect the proportions of CD4 and CD8 T cells (Fig. 2H). The activation profile (CD44+ CD69+) of CD8 T cells is significantly reduced in STING-deficient mice, whereas no effect on the activation of CD4 T cells (Fig. 2I). Together, these data indicate that STING activation contributes to the recruitment/accrual of cDC1 and T cells and to the initiation of PNF.
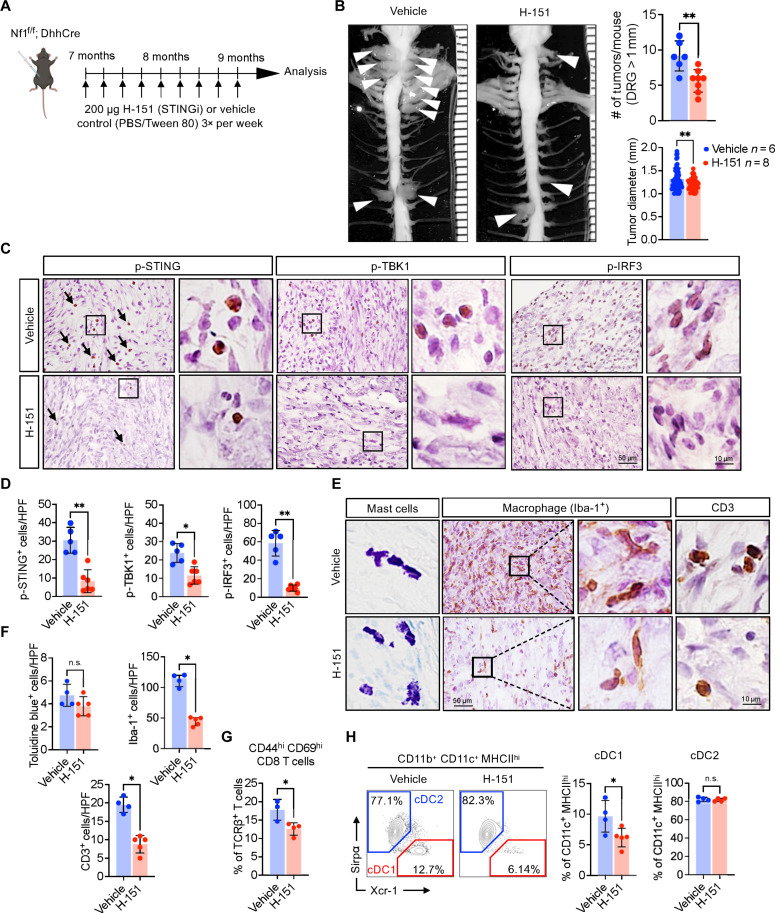
We next tested whether established tumors are affected by inhibition of STING activation. We randomly assigned tumor-bearing Nf1f/f; DhhCre mice to vehicle or H-151 treatment for 8 weeks (Fig. 3A). No signs of toxicity were identified, as determined by monitoring of body weight and mouse mobility (fig. S4). Significant reductions in tumor size and tumor number were quantified after gross dissection of mice treated with H-151 versus vehicle (Fig. 3B). IHC confirmed that H-151 significantly reduced numbers of p-STING positive cells in PNF; P-TBK1 and P-IRF3 were also significantly reduced (Fig. 3C). Numbers of mast cells did not differ between treatment groups in these PNF tissue sections. Iba-1+ macrophages and CD3+ T cells were each notably reduced in by STING inhibitor treatment (Fig. 3D). We performed flow cytometric analysis, which verified significantly reduced numbers of T cells in STING inhibitor–treated PNF. Residual T cells showed reduced activation, i.e., lower levels of CD44 and CD69 (Fig. 3E). The cDC1 population, but not the cDC2 population, was significantly reduced in H-151–treated PNF (Fig. 3F). Thus, targeting STING reduces plasma CXCL10 and reduces numbers of PNF inflammatory macrophages, cDC1, and activated T cells. Overall, STING pathway activation in PNF contributes to tumor initiation and tumor maintenance, likely via its downstream effects on immune cells.
Fig. 3. STING inhibition affects T cell activation and cDC1 infiltration.
(A) Schematic diagram of STING inhibitor (STINGi; H-151) treatment regimen. (B) Representative images of gross dissected paraspinal tumors from vehicle or H-151–treated mice and the corresponding graphical quantification of tumor numbers and size. (C) Representative images of IHC against p-STING, p-TBK1, and p-IRF3 and (D) quantified in vehicle versus H-151–treated mice. (E) Toluidine blue staining for mast cells, immunostaining of Iba-1 for detection of macrophages and CD3 for T cells in vehicle and H-151–treated groups and (F) quantified wherein each panel and each dot represents data from a single mouse, in which 5 HPF (high-power field) were analyzed per animal. (G and H) Flow cytometric quantification of activated (CD44hi CD69hi) CD8 T cells and cDC1 in vehicle or H-151–treated mice. Student’s t-test: *P < 0.05; **P < 0.001.
Activation of leukocytes in the neurofibroma microenvironment
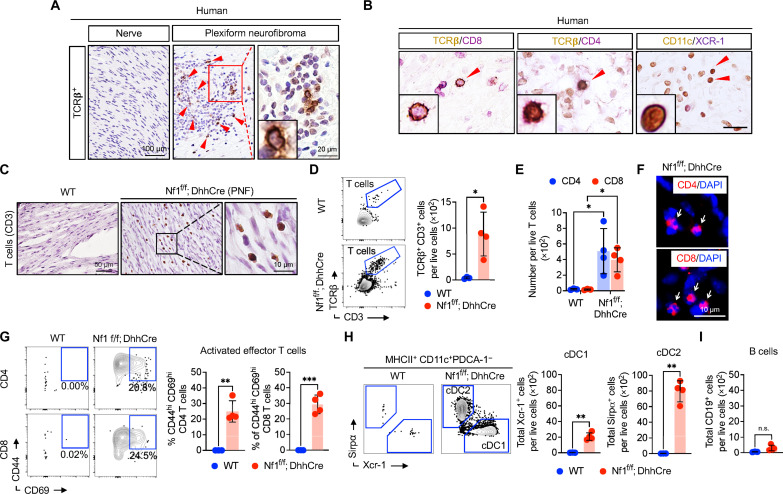
T cells and DCs are rare in normal human nerve specimens. Double-labeling IHC identified TCRβ+CD4+ and TCRβ+CD8+ T cells and CD11c+XCR-1+ DC1s in human PNF tissue sections (Fig. 4, A and B). In mouse Nf1f/f; DhhCre model PNF sections, CD3+ T cells also increased as compared to those in tissue sections from WT nerve, in agreement with prior studies (Fig. 4C). Multiparametric spectral flow cytometry of mouse tumors enabled T cell and DC quantification. Control DRG/nerves contained few TCRβ+ T cells, whereas numerous TCRβ+ T cells were isolated from neurofibromas (Fig. 4D); T cells accounted for 2 to 5% of live cells in PNF. Roughly half of PNF T cells were CD4+ and the other half were CD8+ as monitored by flow cytometry (Fig. 4E). Immunostaining of tumor sections confirmed this result (Fig. 4F). PNF T cells in Nf1f/f; DhhCre neurofibromas were activated/memory T cells, based on high expression of CD44. A significant fraction of both CD4+ and CD8+ T cells also expressed CD69, an early, transiently expressed, activation marker (Fig. 4G).
Fig. 4. Lymphocyte infiltration and activation in Nf1f/f; DhhCre neurofibroma mice.
(A) Representative IHC images of human nerve or neurofibroma biopsies for the detection of TCRβ T cells and (B) double labeling for TCRβ (brown) with CD4 or CD8 (red) T cells and CD11c (brown) and XCR-1 (red) for DCs. (C) Representative IHC images WT DRG or Nf1f/f; DhhCre neurofibroma. (D) Representative flow cytometric analysis of TCRβ+ CD3+ T cells and quantification. (E) CD4 and CD8 T cells subtypes and in PNF tissue corresponding flow cytometric quantification. (F) Immunofluorescence showing CD4 and CD8 T cells subtypes in neurofibroma tissues. (G) Representative flow cytometric analysis of antigen primed and activation marker CD44high CD69high of CD4 or CD8 T cell subpopulation from a WT DRG or Nf1f/f; DhhCre neurofibromas and graphical quantification showing frequency of effector T cells indicated by CD44+ CD69+ expression of CD4 or CD8 T cells. (H) Flow cytometric analysis of conventional DCs gated from MHCII+ and CD11c+ cells in WT DRG or Nf1f/f; DhhCre neurofibromas with corresponding quantification of Xcr-1+ (cDC1) and Sirpα+ (cDC2). (I) Flow cytometric quantification of B cells marked by CD19+ cells in Nf1f/f; DhhCre neurofibroma compared to those in WT DRG. Data represent means ± SD; t test, P < 0.05. DAPI, 4′,6-diamidino-2-phenylindole. Student’s t-test: *adj. *P < 0.05, **P < 0.001, and ***P < 0.003.
DCs are key initiators of T cell responses. DCs act by presenting antigen to promote the priming and proliferation of naïve T cells (28). In tumors, cDC1s are a subpopulation of DCs that are critical for “cross-presentation” of tumor associated antigens that promote tumor specific CD8+ T cell responses including cell proliferation and cell activation. PDCA1−;MHCII+;CD11c+ DCs were elevated in PNF, and some plasmacytoid DCs (PDCA1+;CD11c−), which we did not study. Of tumor CD11c+ DCs, 30% were Xcr-1+ cDC1, and 70% were Sirp+-expressing cDC2 (Fig. 4H). CD19+ B cells were rare in Nf1f/f; DhhCre, albeit marginally detectable versus controls (Fig. 4I). Thus, CD4+ and CD8+ T cells expressing TCRβ and CD11c+;XCR-1+ cDC1 are present in human and mouse PNF.
Deficiency of cDC1 APCs attenuates PNF formation
cDC1 maturation and maintenance is dependent on the transcription factor Batf3 (29). Given our identification of cDC1 cells in NF tumors by flow cytometry and the reduction in cDC1 by STING antagonism, we tested the role of cDC1 in neurofibroma formation by intercrossing Nf1f/+; DhhCre and Nf1f/f mice with Batf3−/− mice (cDC1-deficient) mice. Gross dissection of 7-month-old Batf3−/−; Nf1f/f; DhhCre mice revealed attenuated PNF (Fig. 5A); tumor number and tumor size were each significantly reduced versus Nf1f/f; DhhCre controls (Fig. 5, B and C); tumors that did form were PNFs (Fig. 5D). We confirmed the expected absence of cDC1 cells (CD11c+CD103+Xcr-1+ MHCIIhigh) in Batf3−/−; Nf1f/f; DhhCre mice using flow cytometry (Fig. 5E). Activated CD44+CD69+CD4+ T cells were not obviously perturbed in Batf3−/−; Nf1f/f; DhhCre PNF versus Nf1f/f; DhhCre controls (Fig. 5F). Notably, however, activated CD44+CD69+CD8+ T cells were reduced by nearly threefold in Batf3−/−; Nf1f/f; DhhCre PNF (Fig. 5G). Thus, like STING loss of function, loss of cDC1 partially blocks PNF initiation. In contrast, the disruption of nerve SC–nerve axon interaction that is characteristic of PNF was unaffected in Batf3−/−; Nf1f/f; DhhCre mice (Fig. 5H).
Fig. 5. Deficiency of T cell cross-presenting cDC1 cell attenuated neurofibroma formation.
(A) Representative image of gross dissection of the spinal cord with attached DRG or tumors of Nf1f/f; DhhCre mice with heterozygous or homozygous loss of Batf3 transcription factor necessary for cDC1 development. (B and C) Quantification of the number of tumors and size in Nf1f/f; DhhCre mice with or without cDC1 subset deficiency. (D) Representative H&E and S100B staining of DRG/tumors from Nf1f/f; DhhCre or Batf3−/−; Nf1f/f; DhhCre mice. (E) Flow cytometric analysis of cDC1 (Xcr-1+) and cDC2 (SIRPα+) population in Nf1f/f; DhhCre or Batf3−/−; Nf1f/f; DhhCre tumors. (F and G) Representative flow cytometry and quantification of activated (CD44+ CD69+) CD4 or CD8 T cells. (H) Electron micrographs of the saphenous nerve of 7-month-old WT control, Nf1f/f; DhhCre neurofibroma, and Batf3−/−; Nf1f/f; DhhCre mice and quantification of intact Remak bundle. Scale bar, 100 μm. Student’s t-test: *adj. *P < 0.05, ***P < 0.003, and ****P < 0.0001.
CD8 T cells from neurofibroma-bearing mice proliferate in response to APC
As loss of cDC1 cells decreased T cell number and T cell activation in PNF, we reasoned that DC might stimulate CD8+ T cell responses in Nf1f/f; DhhCre mice. To test this, we cultured carboxyfluorescein diacetate succinimidyl ester (CFSE)–labeled CD8+ T cells from either WT or Nf1f/f; DhhCre mice with APCs from WT or Nf1f/f; DhhCre mouse spleen (Fig. 6A) and assessed the loss of CFSE staining as an indicator of T cell proliferation. WT T cells showed background levels of proliferation in response to APCs from either WT or Nf1f/f; DhhCre mice (Fig. 6B). In contrast, CD8+ T cells from tumor-bearing Nf1f/f; DhhCre mice had >2× higher proliferation in response to APCs from Nf1f/f; DhhCre or WT APCs (Fig. 6B). Combined, these data show that CD8 T cells from tumor-bearing mice are primed to proliferate in response to APC and suggest that T cells activated during PNF formation might contribute to tumor development.
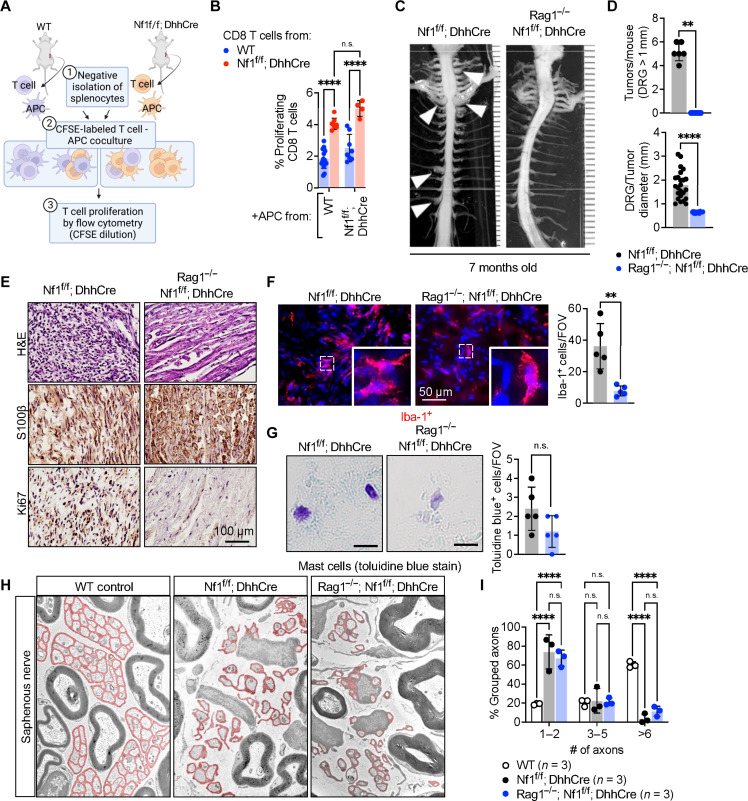
Fig. 6. Absence of adaptive immune cells neurofibroma development.
(A) Schematic diagram of the T cell proliferation assay: CD8+ T cells were purified by negative selection using a Miltenyi kit. T cells from tumor-bearing mice were labeled with CFSE and mixed with APCs from WT or tumor-bearing Nf1f/f; DhhCre mice. Four days later, cells were harvested and assessed for CFSE dilution. Graph shows the percent of CD8+ T cells that had proliferated (CFSE low). (B) Quantification on proliferating CD8 T cells from WT mice cocultured with APC from WT or Nf1f/f; DhhCre mice. (C) CD8 T cell proliferation from Nf1f/f; DhhCre mice cocultured with APC from WT or Nf1f/f; DhhCre mice. (C) Representative images of gross dissections of mouse spinal cord with the associated neurofibromas of the dorsal root in Nf1f/f; DhhCre mice not observed in Rag1−/−; Nf1f/f; DhhCre mice that lack adaptive immune cells at 7 months of age. (D) Quantification of the number of tumors observed per mouse and tumor diameters between Nf1f/f; DhhCre and Rag1−/−; Nf1f/f; DhhCre mice. (E) Representative tissue histochemistry of DRGs/PNF in Nf1f/f; DhhCre or Rag1−/−; Nf1f/f; DhhCre stained with H&E, S100β, and Ki67. (F) Immunofluorescence of Iba-1+ macrophages in DRG or neurofibromas, and the number of Iba-1+ per field of view (FOV). (G) Toluidine blue stain for detection of mast cells, and the number of toluidine blue–positive mast cells per field of view. (H) Electron micrographs of the saphenous nerve of 7-month-old WT control, Nf1f/f; DhhCre neurofibroma, and Rag1−/−; Nf1f/f; DhhCre mice. (I) Quantification of intact Remak bundle. *P < 0.05; Student’s t-test: **adj. **P < 0.001; ***P < 0.003, and ****P < 0.0001.
The absence of adaptive immune cells prevents neurofibroma formation
To formally test the requirement of lymphocytes in neurofibroma formation, we intercrossed mice from our Nf1f/f; DhhCre colony to Rag1−/− mice, which lack B and T cells due to the role of Rag1 in B and T cell development (30). Gross dissection of 7-month-old Rag1+/−; Nf1f/f; DhhCre littermate controls showed an average of five paraspinal tumors per mouse, typical for the PNF model at this time point. Notably, in contrast, there were no visible neurofibromas in Rag1−/−; Nf1f/f; DhhCre mice (Fig. 6, C and D). There was also no tumor pathology present in Rag1−/−; Nf1f/f; DhhCre DRG/nerve tissue sections; numbers of Ki67+ proliferating cells were reduced to low normal levels, and S100b+ SCs showed normal morphology (Fig. 6E). This study demonstrates that T and/or B cells are required for neurofibroma initiation. Of note, Iba-1+ macrophages were reduced to normal levels, suggesting a lack of sustained inflammation in the absence of B and T cells in the Rag mutant mice (Fig. 6F). No significant reduction in mast cells was observed (Fig. 6G). As in the Batf3 knockout, the disrupted axon-SC bundle phenotype (Remak bundles) characteristic of Nf1f/f; DhhCre nerve was not rescued in Rag1−/−; Nf1f/f; DhhCre nerves (Fig. 6, H and I).
CD8 T cells promote neurofibroma formation in Rag1−/−; Nf1f/f; DhhCre mice
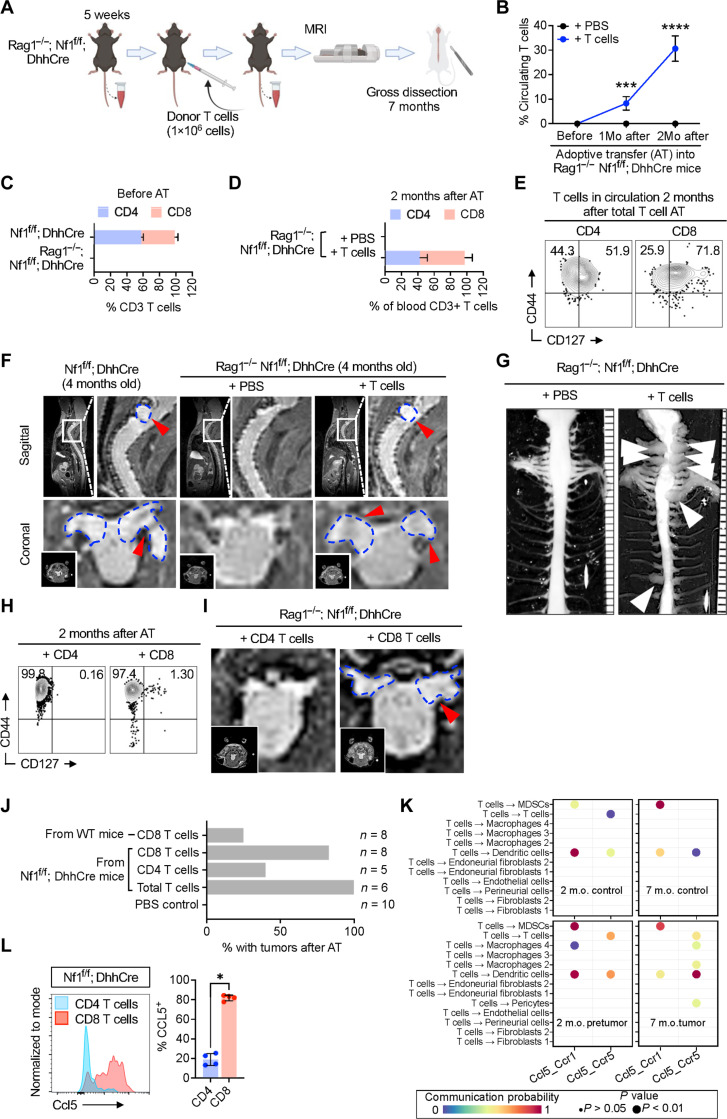
To test whether T cells are sufficient to promote neurofibroma formation, after Nf1 loss in SCs, we adoptively transferred T cells isolated from the spleens of tumor-bearing Nf1f/f; DhhCre mice into Rag1−/−; Nf1f/f; DhhCre mice that lack circulating CD3 T cells (Fig. 7A and fig. S5, A and B). We confirmed increasing T cells in circulation after grafting (Fig. 7, B and C). Isolated T cells for grafting were > 90% CD3+ (fig. S5C); and almost half were CD4+ and the other half were CD8+ (Fig. 7C). Transferred CD4 and CD8 T cells persisted in the blood of recipient animals for at least 2 months (fig. S5D and Fig. 7D) and maintained their CD44+ high memory phenotype (Fig. 7E). We acquired MRI images 3 months after vehicle or CD3+ T cell transfer. No tumors were visible in Rag1−/−; Nf1f/f; DhhCre mice injected with phosphate-buffered saline (PBS; n = 10) or T cells from WT mice (n = 8). MRI revealed paraspinal tumors in each Rag1−/−; Nf1f/f; DhhCre mouse adoptively transferred with T cells (Fig. 7F). The development of neurofibromas was confirmed by gross dissection of spinal cord and nerve from 7-month-old mice (6 months after T cell transfer); tumors were present only in those mice containing transferred T cells (Fig. 7G).
Fig. 7. T cell adoptive transfer promoted neurofibroma formation.
(A) Schematic of CD8 T cell depletion. (B) Flow cytometric quantification in the proportion of CD4 or CD8 of the total blood CD3 T cells in Nf1f/f; DhhCre compared to that in Rag1−/−; Nf1f/f; DhhCre. (C) Percentage of CD3 T cells at 1 and 2 months (1Mo and 2Mo, respectively) after adoptive transfer. (D) Percentage of CD4 and CD8 T cells present in circulation after adoptive transfer. (E) Representative flow cytometric analysis of blood CD3 T cells for the markers CD44 and CD127 at 2 months old after total T cell AT. (F) Representative MRI image of Nf1f/f; DhhCre neurofibroma bearing mice and Rag1−/−; Nf1f/f; DhhCre that were adoptively transferred with T cells or PBS as control. (G) Representative images of gross dissection of spinal cord with the associated neurofibromas of the dorsal root in 7-month-old Rag1−/−; Nf1f/f; DhhCre after adoptive transfer of pan-T cells. (H) Representative flow cytometric analysis for CD44 and CD127 of blood from Rag1−/−; Nf1f/f; DhhCre mice at 2 months old after adoptive transfer (AT) of CD8 or CD4 T cells. (I) Representative MRI image of Rag1−/−; Nf1f/f; DhhCre that were adoptively transferred with CD4 T cells or CD8 T cells alone. (J) Quantification of T cell recipient Rag1−/−; Nf1f/f; DhhCre mice that developed neurofibromas after adoptively transferred with or without total T cells or CD4 or CD8 T cells. (K) Predicted gene expression of CCL5 and its receptor CCR5 in immune cells of PNF tissue. m.o., months old. (L) CCL5 protein expression determined by flow cytometry of PNF tissue. Student’s t-test: *adj. *P < 0.05, ***P < 0.003, and ****P < 0.0001..
To test whether CD4 and/or CD8 T cells enable neurofibroma formation, we enriched CD8+ or CD4+ T cells from spleens of tumor-bearing mice (fig. S5, E and F). We then adoptively transferred 2 × 106 cells of each type into separate 1-month-old Rag1−/−; Nf1f/f; DhhCre recipient mice and monitored over time by flow cytometry (fig. S5G). By 2 months after adoptive transfer of CD4 T cells, >94% circulating effector CD4 T cells were present and nearly 0.2% were activated (CD44+CD127+); after adoptive transfer of CD8 T cells, 97% of circulating T cells were effector CD8+ T cells and over 1% are activated (CD44+CD127+) (Fig. 7H). We acquired MRI images 3 months after adoptive transfer (Fig. 7I); two of the eight recipients of CD8 T cells taken from non–tumor-bearing mice develop tumors, whereas six of the eight recipients of CD8+ T cells obtained from tumor-bearing donor mice formed tumors (Fig. 7J). While three of the five recipients of CD4+ T cells did not form tumors, the blood in CD4 cell recipients contained not only CD4+ T cells but also 14.2% CD8+ cells at this time point, suggesting in vivo amplification of residual CD8 T cells. Thus, CD8+ T cells are sufficient to initiate neurofibroma formation.
To gain insight into how T cells drive tumor formation, we analyzed expression of cytokines and growth factors in T cells, using an existing single-cell RNA sequencing (scRNA-seq) dataset (GSE181985), analyzed using CellChat. The STING-inducible cytokine CCL5 was the only cytokine predicted to be up-regulated selectively in T cells, not in other cell types. CCL5 was increased in Nf1f/f; DhhCre DRG/nerve versus WT controls in 2-month-old mice, before tumor formation [when CXCL10 is first detectable; (15)] and at 7 months after tumors have formed (Fig. 7K). CCL5 (RANTES) is a STING-inducible cytokine that binds to the CCR5 receptor. CellChat predicted CCR5-responsive DCs, macrophages, and myeloid-derived suppressor cells (MDSC) in Nf1f/f; DhhCre DRG/nerve before tumor formation and DCs, macrophages, and MDSC plus pericytes and T cells themselves in tumors. Neither CD8 nor CD4 T cells from PNF showed significant expression of interleukin-17, interferon-γ, or tumor necrosis factor–α (not shown). In contrast, CCL5 protein expression was robust in CD8 T cells from PNF monitored using flow cytometry (Fig. 7L). CD4 T cells did not contain CCL5 protein. The expression of CCL5 protein, as opposed to mRNA, is a marker of T cell response to antigen (31), supporting the idea that PNF T cells are primed by antigen in vivo and further suggest that this T cell–produced chemokine might contribute to the failure to resolve inflammation and tumor formation that occur in the setting of NF1 loss.
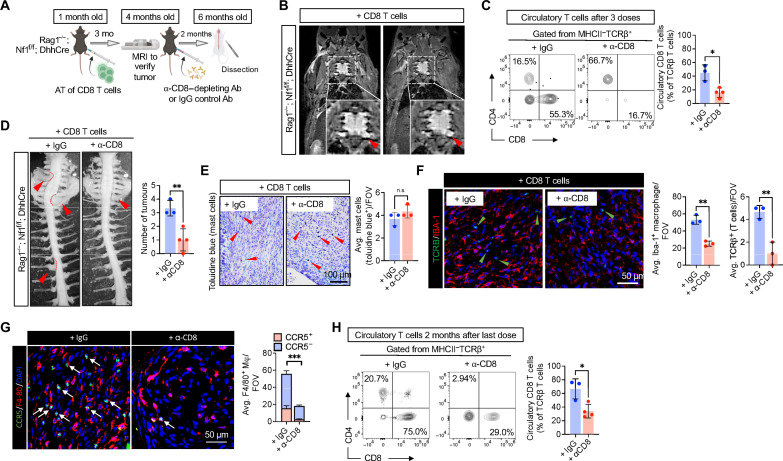
Last, we tested whether CD8 T cells are relevant for PNF maintenance, using the adoptive transfer model. After their adoptive transfer had driven development of NF1 tumors in Rag1−/−; Nf1f/f; DhhCre mice, we then depleted CD8 T cells with anti-CD8 antibody (Fig. 8A). We confirmed tumor development via MRI scan in T/B cell–deficient Rag1−/−; Nf1f/f; DhhCre mice 3 months after adoptive transfer of CD8 T cells (Fig. 8B). Then, each mouse was treated with either monoclonal anti-CD8 antibody or immunoglobulin G (IgG) control weekly for 3 weeks, by which time circulating CD8 T cells had been significantly depleted as assessed by flow cytometry (Fig. 8C). During antibody-mediated T cell depletion, antibiotic therapy was necessary for mouse survival. We euthanized mice and examined PNF by gross dissection when mice were 6 months old. All Rag1−/−; Nf1f/f; DhhCre reconstituted with CD8 T cells and treated with IgG control antibody had multiple paraspinal tumors. In contrast, those mice in which CD8 T cells had been depleted showed significantly attenuated numbers of PNF (Fig. 8D). Residual tumors showed histological features of neurofibroma (fig. S6A). Numbers of toluidine blue+ mast cells did not change (Fig. 8E). CD8 T cell depletion significantly reduced Iba-1+ macrophages as well as TCRβ+ T cells in PNF in tumor sections (Fig. 8F). The number of CCR5+ F4/80 macrophages is significantly reduced by CD8 T cell depletion (Fig. 8G), while CCR5 expression did not colocalized with Iba-1 macrophages (fig. S6B). Thus, although residual CD8 T cells were present in circulation at the time of sacrifice (threefold reduced relative to IgG controls; Fig. 8H), depletion of primed, activated CD8 T cells is necessary for PNF initiation and maintenance.
Fig. 8. CD8 T cell depletion attenuates PNF formation.
(A) Experimental design of CD8 T cell depletion, in which, after adoptive transfer of CD8 T cells in Rag1−/−; Nf1f/f DhhCre and confirming PNF development, CD8 antibody (Ab) or IgG control were administered once a week at 250 μg per mouse. (B) MRI images of Rag1−/−; Nf1f/f; DhhCre after CD8 T cell adoptive transfer confirming PNF development. (C) Flow cytometry of circulatory T cells after three treatment doses. (D) Gross dissection of mice after 2 months from treatment of CD8 antibody revealing reduced PNF formation. (E) Toluidine blue staining for mast cells of nerve or PNF sections. (F) Immunofluorescence of TCRβ for T cells and Iba-1 for macrophages in nerve or PNF. (G) Immunofluorescence of CCR5 (green) expressing F4/80 macrophages (red). (H) Flow cytometry of circulatory T cells after at the 6-month-old endpoint indicates reduced CD8 T cells mice treated with CD8 depletion antibody compared to that in IgG-treated control. Student’s t-test: *adj. *P < 0.05, **P < 0.001, and ***P < 0.003.
DISCUSSION
This study identifies the innate immune regulator STING as increased in NF1 SCs. STING contributes to CXCL10 elevation and, thus, to T cell infiltration and T cell activation in PNF. In the Nf1f/f; DhhCre model of neurofibroma formation studied here, all immune cells are WT at the Nf1 locus. Therefore, Nf1-mutant SCs recruit WT immune cells that support PNF initiation and growth (32). Our series of genetic crosses revealed a contribution of WT cDC1 to tumor number and size and demonstrate that mice lacking T cells have no tumors. Thus, infiltrating T cells, a component of the adaptive immune system, are necessary for the development of benign NF1-associated PNF.
Our data are consistent with prior work showing that inflammation contributes to neurofibroma development (3, 11) and extends it by demonstrating that T cells are the critical immune cell type driving tumor formation. The data reveal a pathway in which loss of NF1 in SCs leads to SC activation of STING, which itself promotes expression of STING inducible cytokines. One such cytokine, CXCL10, drives recruitment of DCs and T cells that ultimately drive neurofibroma development, likely through T cell recognition of antigen. In contrast to effects on tumors, the absence of adaptive immune cells failed to restore abnormal Remak bundle structure that is a characteristic of nerves and tumors containing SCs lacking NF1 (33, 34). Therefore, some effects of cell Nf1 loss of function in vivo are independent of tumor-driving components of the adaptive immune system. Intriguingly, in contrast, the deletion of Cxcr3 in the Nf1f/f; DhhCre mice prevented tumor formation and rescued nerve ultrastructure (15). In addition, as STING inhibition or deletion only partially blocked neurofibroma initiation and growth, additional inputs must cooperate with STING activation after an initiating Nf1 insult.
We suggest that stressed Nf1−/− SCs, via STING, increase expression of CXCL10, which recruits T cells and DCs, enabling PNF initiation, given that STING activation contributes to circulating CXCL10 protein levels, in which pharmacologic suppression of STING in established tumors reduces PNF number and size. Genetic deletion of STING also reduced tumor number and size. Consistent with our findings, in a model of inflammation-induced skin carcinogenesis, after DMBA application, STING became activated, preceding papilloma formation, and STING activation resulted in production of cytokines and inflammation (35). These results suggest the generalizability of a STING-tumor initiation pathway across tumor types and emphasize the potential utility of STING pathway antagonism at early stages in cell transformation. STING antagonism is also being tested to block cancer-related metastasis (25, 36). In contrast, however, STING agonists are being tested clinically in efforts to activate immune cells, including effector T lymphocytes, for antitumor effects (37). In a murine model of malignant peripheral nerve sheath tumor, a recent study showed that STING agonist potentiated the antitumor effects of anti-PD1 (38). How activating or inactivating the STING pathway affects specific stages of transformation to malignancy needs further study.
Flow cytometry analysis identified a MHCIIhi; CD11b−; CD11c+;Xcr-1+ cDC1 population among PNF immune cells. cDC1s are key regulators of tumor antigen trafficking and CD8 T cell recruitment/activation (23, 39–41). The deficiency of Batf3+ cDC1 cells attenuated PNF formation and reduced tumor number. Batf3 deficiency also resulted in decreased TCRβ+ CD8+ T cells and T cell activation. This finding is consistent with a role for cDC1 cells in promoting CD8+ T cell responses (42). Batf3 may also contribute to memory CD8+ T cell survival, so future experiments will need to distinguish between roles of Batf3 in cDC1 versus T cells during PNF formation.
In PNF, Batf3-dependent cDC1 might present antigens to cause activation of T cells in neurofibroma, as cDC1s are APCs that efficiently cross-present antigen to T cells, stimulating their proliferation. However, given that the deficiency of cDC1 cells did not completely prevent neurofibroma formation, additional APCs may also contribute to cross-presentation of antigen to T lymphocytes. cDC2 can also cross-prime T cells (43), so deletion of both cDC1 and cDC2 cells may be required to mimic the marked effects of T cell loss on PNF initiation and growth. Other DC or macrophage populations may also present antigen, albeit at lower levels than do cDC1. In addition, in peripheral nerve inflammation, neuropathic disorder, and after injury, SCs themselves are capable APCs and might augment and/or compensate for the loss of cDC1 cells (44–46).
T cell activity is regulated by the complex interactions of T cells with DCs and other APCs. These interactions are mediated by both soluble cytokines and cell-cell complexes (47). Spleen cells from tumor-bearing mice promoted expansion of T cells in vitro, suggesting that antigens play roles in neurofibroma formation. In a murine model of Nf1-driven low-grade glioma, depletion of CD8+ T cells attenuated tumor cell proliferation; the authors suggested that T cells produce cytokines that indirectly support tumor growth (48). It is plausible that antigens and DCs also play roles in these benign Nf1-driven nervous system tumors. The idea that antigens or antibodies are involved in tumor formation was suggested by seminal studies of (49). In that study, however, transfer of B cells or serum from tumor-bearing mice, not transfer of T cells, enabled chronic inflammation and tumor cell proliferation in a model of HPV16-driven skin carcinogenesis. Thus, diverse tumor types may use divergent mechanisms to sustain immune cell activation for tumor initiation.
We presented four lines of evidence supporting antigen priming of PNF T cells. First, culture of T cells from Nf1f/f; DhhCre mice showed evidence of proliferation in response to APC, a marker of primed T cells responding to antigen. Second, within PNF, CD8+ T cells expressed high levels of CD44, an activation/memory marker, as well as CD69, another acute activation marker. Third, PNF tumor-resident CD8+ T cells expressed CCL5 protein. This is suggestive of recent TCR stimulation as quiescent memory T cells express high levels of CCL5 mRNA, but not protein, unless given a TCR stimulus (31). Fourth, after adoptive transfer of CD4 and CD8+ T cells into Rag1−/− mice, CD8+ T cells expanded and became preferentially enriched over their CD4+ T cell counterparts. Combined, these data strongly support the scenario whereby CD8+ T cells primed during development of NF1 tumors undergo activation, clonal expansion, and acquisition of effector function in a manner that promotes tumor development.
Expression of CCL5 by PNF T cells is also consistent with literature, suggesting that STING modulates inflammation via altering cytokine/chemokine production, contributing to tumor development (50, 51). When CCL5-expressing T cells are absent in Rag1−/−; Nf1f/f; DhhCre mice, we observed a profound decrease in nerve macrophages and the resolution of nerve inflammation. PNF CCL5 (RANTES) may contribute to the T cell–driven maintenance of macrophages in PNF through known RANTES effects on cell migration and matrix remodeling and/or altering immune cell phenotypes such as polarizing macrophages toward immunosuppressive phenotypes (52). CCL5 is also present in an Nf1 low-grade glioma model, albeit produced by microglia, and depletion of CD3+ or CD8+ T cells using blocking antibodies attenuated tumor cell proliferation in that system, suggesting relevance of CCL5 and T cells to other NF1-mutant tumors (48). The CCL5/CCR5 axis may be targetable as numerous clinical trials of antagonists are ongoing (53). This is important as suppressing all T cell functions is not advisable, and selective modulation of T cell function aimed at slowing or ablating tumor formation might be tolerable in the setting of predisposition to neurofibroma.
The absence of T cells abrogates PNF formation, and the adoptive transfer of CD8+ T cells from tumor-bearing mice elicits PNF formation. These findings are consistent with pro-tumorigenic functions of CD8 T cells described in some models of low-grade or premalignant tumor types (19–21, 48, 54). Notably, however, pro-tumorigenic roles of CD4+ T cells have been described in other tumorigenesis models (55–58). In neurofibromas, it remains necessary to formally exclude roles of CD4 T cells in tumorigenesis, given that transplantation of >98% pure CD4 T cells into Rag1−/−; Nf1f/f; DhhCre mice resulted in expansion of a residual CD8+ cell population. However, given that CD8+ T cells alone promoted neurofibroma formation, we conclude that CD8+ T cells are sufficient to promote the transition from nerve inflammation to PNF formation. In other models of tumor initiation, it will be of interest to study the early roles of STING and antigen and the effects on tumor myeloid cells.
In conclusion, T cells play a critical role in promoting the formation of NF1-associated PNFs and for driving the expansion of an inflammatory environment. Identification of molecules involved in T cell activation and trafficking and whether T cell activation is dependent on specific antigens or cytokines warrant further study. Intercepting the pro-inflammatory cooperation of immune cells, including APC–T cell signals that contribute to the formation of tumors is anticipated to provide new therapeutic approaches toward management and/or prevention of NF1 tumors.
MATERIALS AND METHODS
Mice
All animal procedures were approved by the Intuitional Animal Care and Use Committee at Cincinnati Children’s Hospital Medical Center. Mice were housed in temperature- and humidity-controlled facilities with free access to food and water, on a 12-hour light-dark cycle. The mouse lines Rag1−/− (B6.129S7-Rag1tm1Mom/J) and Batf3−/− (B6.129S(C)-Batf3tm1Kmm/J) on a C57BL/6 background were acquired from the Jackson Laboratory. These lines were crossed to an in-house Nf1f/f and Nf1f/+; DhhCre mice (32), which backcrossed >8 generations onto a C57BL/6 background to generate experimental mice for this study. Mice of both sexes were used in all experiments.
Genotyping
A small amount of tissue was harvested from mouse toes, and, then, genomic DNA was prepared with the Direct Mouse Genotyping Kit (APExBio, K1025) according to the manufacturer’s instruction. Next, ~200 ng of genomic DNA was used for genotyping polymerase chain reaction (PCR) with the Q5 high-fidelity polymerase (New England Biolabs, M0492S). For the STING knockout cross, PCR–restriction fragment length polymorphism assay was established with forward primer 5′-CATGCGGCAGCTCCTTGAC-3′ and reverse primer 5′-GGATCACGCCTGTCTCACAGA-3′. Then, PCR product was digested by Bst CI (New England Biolabs, R064S) at 50°C for 30 min, followed by the electrophoresis in ~2% agarose. For Rag1−/− and Batf3−/−, standard PCR was followed as recommended by the Jackson Laboratory. The Nf1 flox and DhhCre genotyping was performed as previously described (32).
Adoptive transfer of purified T cells
Adoptive transfer was performed at 5 weeks after birth of Rag1−/−; Nf1f/f; DhhCre recipient mice. T cells from donor spleens of Nf1f/f; DhhCre mice were purified using cell-specific negative selection isolation kit (Pan T cells, catalog no. 130-095-130; CD4 T cells, catalog no. 130-104-454; and CD8 T cells, catalog no. 130-104-075; Miltenyi Biotec) and resuspended in PBS under sterile conditions. The purity of each T cell population (>95%) was confirmed by flow cytometry before intraperitoneal injection.
Blood sampling
To assess the efficiency of T cell engraftment, blood samples were collected by tail vein bleed into pre-coated EDTA tubes containing 500 μl of PBS, 1 week before the adoptive transfer and at 1 and 2 months after adoptive transfer. Blood lymphocytes (1 × 106 cells) were examined by flow cytometry and analyzed using FlowJo (v10).
T cell depletion using antibodies
Mice with verified tumors were injected intraperitoneally with 250 μg of anti IgG2b (BioXCell, catalog no. BE090) or anti-CD8 clone 2.43 (BioXCell, cat. no. BE0061) in PBS three times per week for 8 weeks.
Histological assays
IHC was performed on tissue sections of human biopsy of PNFs obtained from incidental to therapeutic surgery as fixed specimen from our pathology core. To clear paraffin, sections were incubated in three changes of xylenes; rehydrated sequentially in 100, 95, and 70% ethanol (EtOH); and rinsed in water. Sections were then boiled for 10 min in 0.1 M citrate buffer (pH 6.0) and were cooled at room temperature for 30 min; then, sections were rinsed in water then in PBS, incubated in 30% hydrogen peroxide for 10 min, rinsed in PBS, permeabilized in PBS with 0.3% Triton X-100, and blocked in 10% normal goat serum (NGS) with 0.3% Triton X-100 for half an hour. Sections were then incubated in primary antibody overnight at 4°C (see below the source and catalog numbers and dilutions of antibodies used). Next day, sections were rinsed in buffer, incubated in secondary antibodies [either biotinylated goat and rabbit (Vector Labs, catalog no. BA-1000) or goat anti mouse (Vector Labs, catalog no. BA-9200)] at a dilution of 1:200 (in NGS with Triton X-100) for 1 hour, rinsed in buffer, and incubated in ABC solution (Vector Labs, catalog no. PK-6100) for 1 hour. Sections then were rinsed in PBS, incubated in 3,3′-diaminobenzidine (DAB) (Vector Labs, catalog no. SK-4100) for 5 min, rinsed in double-distilled water, counterstained with hematoxylin, dehydrated with graded EtOH, cleared in xylenes, and cover slipped in histomount.
Double labeling
For double labeling, sections were processed with DAB for first primary antibody, rinsed in buffer, blocked in NGS with Triton X-100 for 1 hour, and then incubated with the second primary antibody overnight. The next day, sections were rinsed and incubated in second secondary antibody and then in ABC solution. After ABC, sectioned were rinsed and incubated with vector peroxidase substrate (VIP) (Vector labs), rinsed and counterstained with hematoxylin, dehydrated with graded EtOH, cleared in xylenes, and cover glassed using histomount.
Antibodies
The following antibodies were used: phospho-STING (Ser366) (E9A9K) rabbit monoclonal antibody (mAb) (Cell Signaling Technology), phospho-STING (Ser365) (D1C4T) rabbit mAb (no. 62912, Cell Signaling Technology), phospho-STING (Ser366) (E9A9K) rabbit mAb (no. 50907, Cell Signaling Technology), phospho-IRF3 (Ser396) (4D4G) rabbit mAb (Cell Signaling Technology), phospho-TBK1/NAK (Ser172) (D52C2) XP rabbit mAb (Cell Signaling Technology), phospho-IRF3 (Ser396) (D6O1M) rabbit mAb (no. 29047S), F4/80 (BM8.1) rat mAb (Cell Signaling Technology), mouse CCR5 antibody (MAB6138-SP, Thermo Fisher Scientific), CoraLite488 anti-mouse TCRβ (H57-597) (CL488-65106, Proteintech), anti-CD4 rabbit polyclonal antibody (BS-0647R, Bioss Antibodies), anti-CD8A rabbit polyclonal antibody (BS-0648R, Bioss Antibodies), XCR1 (D2F8T) rabbit mAb (no. 44665S, Cell Signaling Technology), CD103/INGAE (integrin alpha e) (EP206) rabbit mAb (no. 95835S, Cell Signaling Technology), anti-CD8 antibody, rabbit monoclonal (clone SP16) (SAB5500074, Sigma-Aldrich), CD3ε (E4T1B) XP Rabbit mAb (no. 78588, Cell Signaling Technology), and Iba-1 rabbit polyclonal (no. 019-19741, Wako).
Tumor analysis
MRI data were collected on a 7-T Bruker BioSpec system equipped with gradients at 400 G/cm, and tumor burden was calculated (59). To quantify tumor numbers, gross dissection of perfusion-fixed mice was performed using a stereoscope (Leica). A tumor was defined as a mass of the DRG or nerve roots, with a diameter greater than 1 mm, measured perpendicular to DRG/nerve roots. Alternatively, tumor and nerve roots were measured in 3D in MRI images using the image computing platform software 3D Slicer (59).
Sample preparation for flow cytometry
Paraspinal tumors or DRG were dissected from intracardially ice-cold PBS-perfused mice and chopped finely and then subjected to enzyme and mechanical dissociation. Briefly, finely cut DRG/tumor tissue was digested with the digestion enzyme mixture: collagenase A (10 mg/ml), collagenase type IV (10 mg/ml), soybean trypsin inhibitor (100 μg/ml), deoxyribonuclease I (250 U), and calcium chloride (5 mM) for 30 min at 37C with shaking. Digestion was terminated in 10% fetal calf serum, and mechanical dissociation via syringe needles was carried out on ice. Cells in suspension cells were filtered through 100-μm and then 70-μm strainers.
Flow cytometry
Spectral flow cytometry analysis was performed on single-cell suspensions. Autologous spleen or beads were used as controls. The following viability and antibodies against mouse were used: TCRβ BV711 (catalog no. 109243, BioLegend), CD8a PerCP, CD4 BUV395 (catalog no. 563790, BD Horizon), CD11b AF700 (catalog no. 101222, BioLegend), CD11c ef450 (catalog no. 48-0114-82, Thermo Fisher Scientific), MHC II V500 (catalog no. 562366, BD Horizon), CD44 BUV737 (catalog no. 612799, BD Horizon), CD69 BV605 (catalog no. 104530, BioLegend), CD19 BV785 (catalog no. 115543, BioLegend), Gr-1 APC–cyanine 7 (Cy7) (catalog no. 108423, BioLegend), PDCA-1 phycoerythrin (PE) (catalog no. 127023, BioLegend), Xcr-1 BV650, Sirpα PE-Cy7, and Trem2 APC. Samples were measured by Aurora cell multiparametric flow analyzer (Cytek Biosciences, Fremont, CA) and analyzed with FlowJo (v10) or SpectroFlo software.
Electron microscopy
Mice were perfusion fixed with a 4% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M phosphate buffer at pH 7.4. Saphenous nerve was dissected and postfixed overnight, then transferred to 0.175 M cacodylate buffer, osmicated, dehydrated, and embedded in Embed 812 (Ladd Research Industries). Ultrathin sections were stained in uranyl acetate and lead citrate and viewed on a Hitachi H-7600 electron microscope.
T cell/APC in vitro coculture and proliferation assay
CD8+ T cells were enriched using a CD8+ T cell–negative selection column from Miltenyi Biotec. T cells from control or tumor-bearing (Nf1f/f; DhhCre) mice were labeled with CFSE and mixed with APCs from WT or tumor-bearing mice. APCs were purified from spleens of control WT or tumor-bearing Nf1f/f; DhhCre mice by removal of pan-T cells using a T cell–positive selection column from Miltenyi Biotec. Four days later, cells were harvested and assessed for CFSE dilution by flow cytometer.
RNA-seq data analysis
CellChat (v1.6.0; www.cellchat.org) objects were created from four Seurat (version 3.1.2; https://satijalab.org/seurat) objects [2- and 7-month-old WT mouse DRG controls, 2-month-old mouse neurofibroma pretumor (Nf1f/f; DhhCre), and 7-month-old mouse neurofibroma tumor (Nf1f/f; DhhCre)] extracted from the 10x Genomics scRNA-seq data (5). The “secreted signaling interactions” sub-database (for mouse) was chosen to infer the cell state–specific communications. CellChat identifies overexpressed ligands or receptors in one cell group and then identifies overexpressed ligand-receptor interactions if either ligand or receptor is overexpressed. CellChat infers the biologically significant cell-cell communication by assigning each interaction with a probability value and performing a permutation test. These steps create a complex cell-cell communication networks with assigned the communication probability scores. After inferring aggregated cell-cell networks, we removed autocrine interactions and focused on cell-cell interactions where T cells and immune/stromal cells participate as sources (i.e., ligand expressing) and targets (i.e., receptor expressing), respectively. The P value of 0.05 was chosen to extract significant ligand-receptor interactions for each sample set. Dot plots were generated to show the permutation P value and the communication probability of each interaction.
Statistical analysis
Statistical significance was evaluated using two-tailed student’s t test, one-way analysis of variance (ANOVA), or two-way ANOVA when appropriate. The differences between individual groups were tested by ANOVA analyses using Tukey’s or Dunnett’s multiple comparisons test as indicated. A P value of <0.05 was considered statistically significant. Data are presented as the means ± SD of at least three independent biological replicates or experiments. Graphical visualization and analyses were performed using GraphPad Prism 9.
Acknowledgments
Funding: This work was supported by NIH NS28840 and W81XWH-19-1-0816 (to N.R.) and by W81XWH-20-1-0116 (to J.P.).
Author contributions: N.R., D.A.H., and J.P. conceived the study. J.P. drafted the manuscript. N.R. and D.A.H. revised the manuscript. All authors approved the final version. J.P., S.A.D., and M.T. carried out mouse husbandry and flow cytometry. M.T. and S.A.H. developed the flow cytometry antibody panel for PNF. L.H. developed mouse genotyping. T.A.R. prepared and quantified IHC and electron microscopy images. K.C. analyzed gene datasets and CellChat data.
Competing interests: The authors declare that they have no competing interests. Some research, unrelated to this study, in the N.R. lab, is funded by Revolution Medicines, Healyx, and Boehringer-Ingelheim.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
REFERENCES AND NOTES
- 1.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Gutmann D. H., Ferner R. E., Listernick R. H., Korf B. R., Wolters P. L., Johnson K. J., Neurofibromatosis type 1. Nat. Rev. Dis. Primers. 3, 17004 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Fletcher J. S., Pundavela J., Ratner N., After Nf1 loss in Schwann cells, inflammation drives neurofibroma formation. Neurooncol. Adv. 2, i23–i32 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang C., McKay R. M., Le L. Q., Tumorigenesis in neurofibromatosis type 1: Role of the microenvironment. Oncogene 40, 5781–5787 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kershner L. J., Choi K., Wu J., Zhang X., Perrino M., Salomonis N., Shern J. F., Ratner N., Multiple Nf1 Schwann cell populations reprogram the plexiform neurofibroma tumor microenvironment. JCI Insight 7, e154513 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farschtschi S., Park S. J., Sawitzki B., Oh S. J., Kluwe L., Mautner V. F., Kurtz A., Effector T cell subclasses associate with tumor burden in neurofibromatosis type 1 patients. Cancer Immunol. Immunother. 65, 1113–1121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haworth K. B., Arnold M. A., Pierson C. R., Choi K., Yeager N. D., Ratner N., Roberts R. D., Finlay J. L., Cripe T. P., Immune profiling of NF1-associated tumors reveals histologic subtype distinctions and heterogeneity: Implications for immunotherapy. Oncotarget 8, 82037–82048 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ratner N., Miller S. J., A RASopathy gene commonly mutated in cancer: The neurofibromatosis type 1 tumour suppressor. Nat. Rev. Cancer 15, 290–301 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottfried O. N., Viskochil D. H., Couldwell W. T., Neurofibromatosis type 1 and tumorigenesis: Molecular mechanisms and therapeutic implications. Neurosurg. Focus 28, E8 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Staser K., Yang F. C., Clapp D. W., Mast cells and the neurofibroma microenvironment. Blood 116, 157–164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao C. P., Booker R. C., Brosseau J. P., Chen Z., Mo J., Tchegnon E., Wang Y., Clapp D. W., Le L. Q., Contributions of inflammation and tumor microenvironment to neurofibroma tumorigenesis. J. Clin. Invest. 128, 2848–2861 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prada C. E., Jousma E., Rizvi T. A., Wu J., Dunn R. S., Mayes D. A., Cancelas J. A., Dombi E., Kim M.-O., West B. L., Bollag G., Ratner N., Neurofibroma-associated macrophages play roles in tumor growth and response to pharmacological inhibition. Acta Neuropathol. 125, 159–168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang F. C., Ingram D. A., Chen S., Zhu Y., Yuan J., Li X., Yang X., Knowles S., Horn W., Li Y., Zhang S., Yang Y., Vakili S. T., Yu M., Burns D., Robertson K., Hutchins G., Parada L. F., Clapp D. W., Nf1-dependent tumors require a microenvironment containing Nf1+/−- and c-kit-dependent bone marrow. Cell 135, 437–448 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ribeiro S., Napoli I., White I. J., Parrinello S., Flanagan A. M., Suter U., Parada L. F., Lloyd A. C., Injury signals cooperate with Nf1 loss to relieve the tumor-suppressive environment of adult peripheral nerve. Cell Rep. 5, 126–136 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Fletcher J. S., Wu J., Jessen W. J., Pundavela J., Miller J. A., Dombi E., Kim M. O., Rizvi T. A., Chetal K., Salomonis N., Ratner N., Cxcr3-expressing leukocytes are necessary for neurofibroma formation in mice. JCI Insight 4, e98601 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Courtois-Cox S., Genther Williams S. M., Reczek E. E., Johnson B. W., McGillicuddy L. T., Johannessen C. M., Hollstein P. E., MacCollin M., Cichowski K., A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell 10, 459–472 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dou Z., Ghosh K., Vizioli M. G., Zhu J., Sen P., Wangensteen K. J., Simithy J., Lan Y., Lin Y., Zhou Z., Capell B. C., Xu C., Xu M., Kieckhaefer J. E., Jiang T., Shoshkes-Carmel M., Tanim K., Barber G. N., Seykora J. T., Millar S. E., Kaestner K. H., Garcia B. A., Adams P. D., Berger S. L., Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550, 402–406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu Y., Choi K., Wu J., Andreassen P. R., Dexheimer P. J., Keddache M., Brems H., Spinner R. J., Cancelas J. A., Martin L. J., Wallace M. R., Legius E., Vogel K. S., Ratner N., NF1 patient missense variants predict a role for ATM in modifying neurofibroma initiation. Acta Neuropathol. 139, 157–174 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y., Hu S., Liu J., Cui Y., Fan Y., Lv T., Liu L., Li J., He Q., Han W., Yu W., Sun Y., Jin J., CD8+ T cells promote proliferation of benign prostatic hyperplasia epithelial cells under low androgen level via modulation of CCL5/STAT5/CCND1 signaling pathway. Sci. Rep. 7, 42893 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Endig J., Buitrago-Molina L. E., Marhenke S., Reisinger F., Saborowski A., Schutt J., Dual role of the adaptive immune system in liver injury and hepatocellular carcinoma development. Cancer Cell 30, 308–323 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Roberts S. J., Ng B. Y., Filler R. B., Lewis J., Glusac E. J., Hayday A. C., Tigelaar R. E., Girardi M., Characterizing tumor-promoting T cells in chemically induced cutaneous carcinogenesis. Proc. Natl. Acad. Sci. U.S.A. 104, 6770–6775 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arwert E. N., Lal R., Quist S., Rosewell I., van Rooijen N., Watt F. M., Tumor formation initiated by nondividing epidermal cells via an inflammatory infiltrate. Proc. Natl. Acad. Sci. U.S.A. 107, 19903–19908 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bottcher J. P., C. Reis e Sousa., The role of type 1 conventional dendritic cells in cancer immunity. Trends Cancer 4, 784–792 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wculek S. K., Cueto F. J., Mujal A. M., Melero I., Krummel M. F., Sancho D., Dendritic cells in cancer immunology and immunotherapy. Nat. Rev. Immunol. 20, 7–24 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Chen C., Xu P., Cellular functions of cGAS-STING signaling. Trends Cell Biol. 33, 630–648 (2023). [DOI] [PubMed] [Google Scholar]
- 26.Haag S. M., Gulen M. F., Reymond L., Gibelin A., Abrami L., Decout A., Heymann M., van der Goot F. G., Turcatti G., Behrendt R., Ablasser A., Targeting STING with covalent small-molecule inhibitors. Nature 559, 269–273 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Sauer J. D., Sotelo-Troha K., von Moltke J., Monroe K. M., Rae C. S., Brubaker S. W., Hyodo M., Hayakawa Y., Woodward J. J., Portnoy D. A., Vance R. E., The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 79, 688–694 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mildner A., Jung S., Development and function of dendritic cell subsets. Immunity 40, 642–656 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Lukowski S. W., Rodahl I., Kelly S., Yu M., Gotley J., Zhou C., Millard S., Andersen S. B., Christ A. N., Belz G., Frazer I. H., Chandra J., Absence of Batf3 reveals a new dimension of cell state heterogeneity within conventional dendritic cells. iScience 24, 102402 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mombaerts P., Iacomini J., Johnson R. S., Herrup K., Tonegawa S., Papaioannou V. E., RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68, 869–877 (1992). [DOI] [PubMed] [Google Scholar]
- 31.Swanson B. J., Murakami M., Mitchell T. C., Kappler J., Marrack P., RANTES production by memory phenotype T cells is controlled by a posttranscriptional, TCR-dependent process. Immunity 17, 605–615 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Wu J., Williams J. P., Rizvi T. A., Kordich J. J., Witte D., Meijer D., Stemmer-Rachamimov A. O., Cancelas J. A., Ratner N., Plexiform and dermal neurofibromas and pigmentation are caused by Nf1 loss in desert hedgehog-expressing cells. Cancer Cell 13, 105–116 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng H., Chang L., Patel N., Yang J., Lowe L., Burns D. K., Zhu Y., Induction of abnormal proliferation by nonmyelinating schwann cells triggers neurofibroma formation. Cancer Cell 13, 117–128 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Erlandson R. A., Woodruff J. M., Peripheral nerve sheath tumors: An electron microscopic study of 43 cases. Cancer 49, 273–287 (1982). [DOI] [PubMed] [Google Scholar]
- 35.Ahn J., Xia T. L., Konno H., Konno K., Ruiz P., Barber G. N., Inflammation-driven carcinogenesis is mediated through STING. Nat. Commun. 5, 5166 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berger G., Marloye M., Lawler S. E., Pharmacological modulation of the STING pathway for cancer immunotherapy. Trends Mol. Med. 25, 412–427 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Decout A., Katz J. D., Venkatraman S., Ablasser A., The cGAS-STING pathway as a therapeutic target in inflammatory diseases. Nat. Rev. Immunol. 21, 548–569 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Somatilaka B. N., Madana L., Sadek A., Chen Z., Chandrasekaran S., McKay R. M., Le L. Q., STING activation reprograms the microenvironment to sensitize NF1-related malignant peripheral nerve sheath tumors for immunotherapy. J. Clin. Invest. 134, e176748 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engelhardt J. J., Boldajipour B., Beemiller P., Pandurangi P., Sorensen C., Werb Z., Egeblad M., Krummel M. F., Marginating dendritic cells of the tumor microenvironment cross-present tumor antigens and stably engage tumor-specific T cells. Cancer Cell 21, 402–417 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts E. W., Broz M. L., Binnewies M., Headley M. B., Nelson A. E., Wolf D. M., Kaisho T., Bogunovic D., Bhardwaj N., Krummel M. F., Critical Role for CD103+/CD141+ dendritic cells bearing CCR7 for tumor antigen trafficking and priming of T cell immunity in melanoma. Cancer Cell 30, 324–336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spranger S., Dai D., Horton B., Gajewski T. F., Tumor-residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell 31, 711–23.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ataide M. A., Komander K., Knopper K., Peters A. E., Wu H., Eickhoff S., Gogishvili T., Weber J., Grafen A., Kallies A., Garbi N., Einsele H., Hudecek M., Gasteiger G., Holzel M., Vaeth M., Kastenmuller W., BATF3 programs CD8+ T cell memory. Nat. Immunol. 21, 1397–1407 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Ruhland M. K., Roberts E. W., Cai E., Mujal A. M., Marchuk K., Beppler C., Nam D., Serwas N. K., Binnewies M., Krummel M. F., Visualizing synaptic transfer of tumor antigens among dendritic cells. Cancer Cell 37, 786–99.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hartlehnert M., Derksen A., Hagenacker T., Kindermann D., Schafers M., Pawlak M., Kieseier B. C., Horste G. M. Z., Schwann cells promote post-traumatic nerve inflammation and neuropathic pain through MHC class II. Sci. Rep. 7, 12518 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horste G. M. Z., Heidenreich H., Lehmann H. C., Ferrone S., Hartung H. P., Wiendl H., Kieseier B. C., Expression of antigen processing and presenting molecules by Schwann cells in inflammatory neuropathies. Glia 58, 80–92 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wekerle H., Schwab M., Linington C., Meyermann R., Antigen presentation in the peripheral nervous system: Schwann cells present endogenous myelin autoantigens to lymphocytes. Eur. J. Immunol. 16, 1551–1557 (1986). [DOI] [PubMed] [Google Scholar]
- 47.Jain A., Pasare C., Innate control of adaptive immunity: Beyond the three-signal paradigm. J. Immunol. 198, 3791–3800 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo X., Pan Y., Xiong M., Sanapala S., Anastasaki C., Cobb O., Dahiya S., Gutmann D. H., Midkine activation of CD8+ T cells establishes a neuron-immune-cancer axis responsible for low-grade glioma growth. Nat. Commun. 11, 2177 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Visser K. E., Korets L. V., Coussens L. M., De novo carcinogenesis promoted by chronic inflammation is B lymphocyte dependent. Cancer Cell 7, 411–423 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Vatner R. E., Janssen E. M., STING, DCs and the link between innate and adaptive tumor immunity. Mol. Immunol. 110, 13–23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barber G. N., STING: Infection, inflammation and cancer. Nat. Rev. Immunol. 15, 760–770 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aldinucci D., Borghese C., Casagrande N., The CCL5/CCR5 axis in cancer progression. Cancer 12, 1765 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Halama N., Zoernig I., Berthel A., Kahlert C., Klupp F., Suarez-Carmona M., Suetterlin T., Brand K., Krauss J., Lasitschka F., Lerchl T., Luckner-Minden C., Ulrich A., Koch M., Weitz J., Schneider M., Buechler M. W., Zitvogel L., Herrmann T., Benner A., Kunz C., Luecke S., Springfeld C., Grabe N., Falk C. S., Jaeger D., Tumoral immune cell exploitation in colorectal cancer metastases can be targeted effectively by anti-CCR5 therapy in cancer patients. Cancer Cell 29, 587–601 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Kwong B. Y., Roberts S. J., Silberzahn T., Filler R. B., Neustadter J. H., Galan A., Reddy S., Lin W. M., Ellis P. D., Langford C. F., Hayday A. C., Girardi M., Molecular analysis of tumor-promoting CD8+ T cells in two-stage cutaneous chemical carcinogenesis. J. Invest. Dermatol. 130, 1726–1736 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ortiz M. L., Kumar V., Martner A., Mony S., Donthireddy L., Condamine T., Seykora J., Knight S. C., Malietzis G., Lee G. H., Moorghen M., Lenox B., Luetteke N., Celis E., Gabrilovich D., Immature myeloid cells directly contribute to skin tumor development by recruiting IL-17–producing CD4+ T cells. J. Exp. Med. 212, 351–367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.den Boer A. T., van Mierlo G. J., Fransen M. F., Melief C. J., Offringa R., Toes R. E., CD4+ T cells are able to promote tumor growth through inhibition of tumor-specific CD8+ T-cell responses in tumor-bearing hosts. Cancer Res. 65, 6984–6989 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Nishikawa H., Kato T., Tawara I., Takemitsu T., Saito K., Wang L., Ikarashi Y., Wakasugi H., Nakayama T., Taniguchi M., Kuribayashi K., Old L. J., Shiku H., Accelerated chemically induced tumor development mediated by CD4+CD25+ regulatory T cells in wild-type hosts. Proc. Natl. Acad. Sci. U.S.A. 102, 9253–9257 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeNardo D. G., Barreto J. B., Andreu P., Vasquez L., Tawfik D., Kolhatkar N., Coussens L. M., CD4+ T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell 16, 91–102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fedorov A., Beichel R., Kalpathy-Cramer J., Finet J., Fillion-Robin J. C., Pujol S., Bauer C., Jennings D., Fennessy F., Sonka M., Buatti J., Aylward S., Miller J. V., Pieper S., Kikinis R., 3D Slicer as an image computing platform for the quantitative imaging network. Magn. Reson. Imaging 30, 1323–1341 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6