Abstract
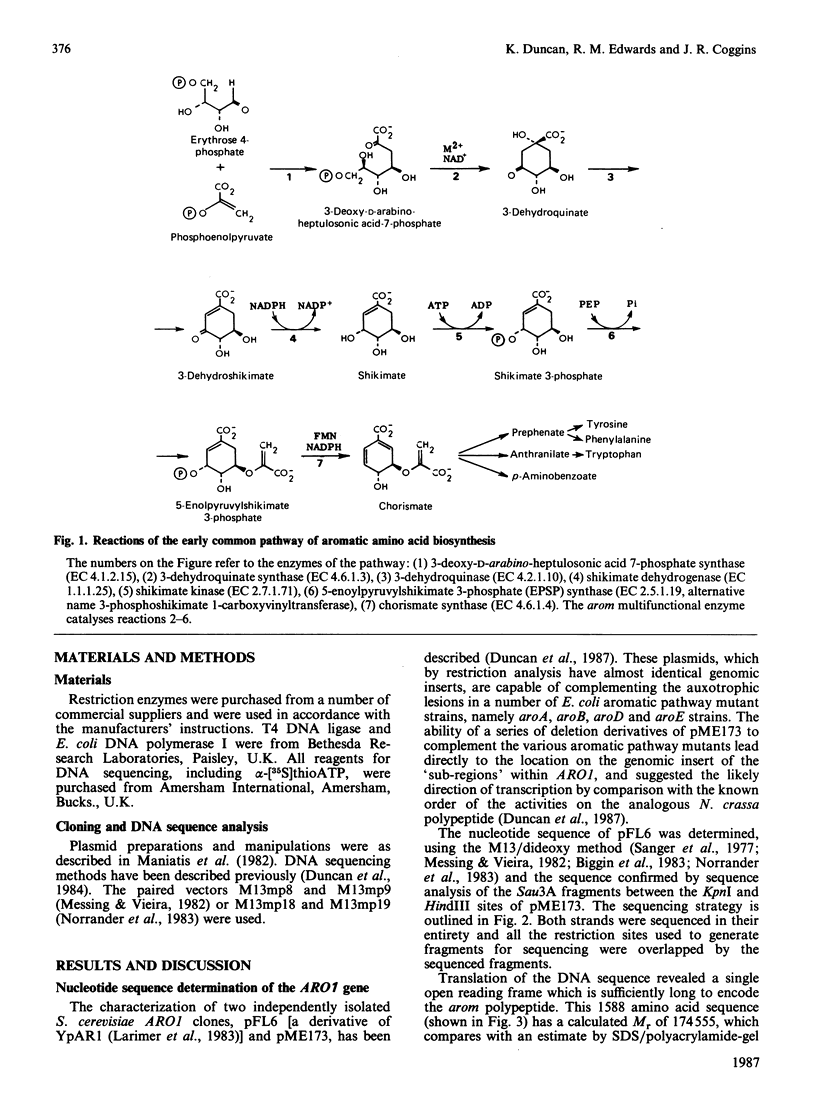
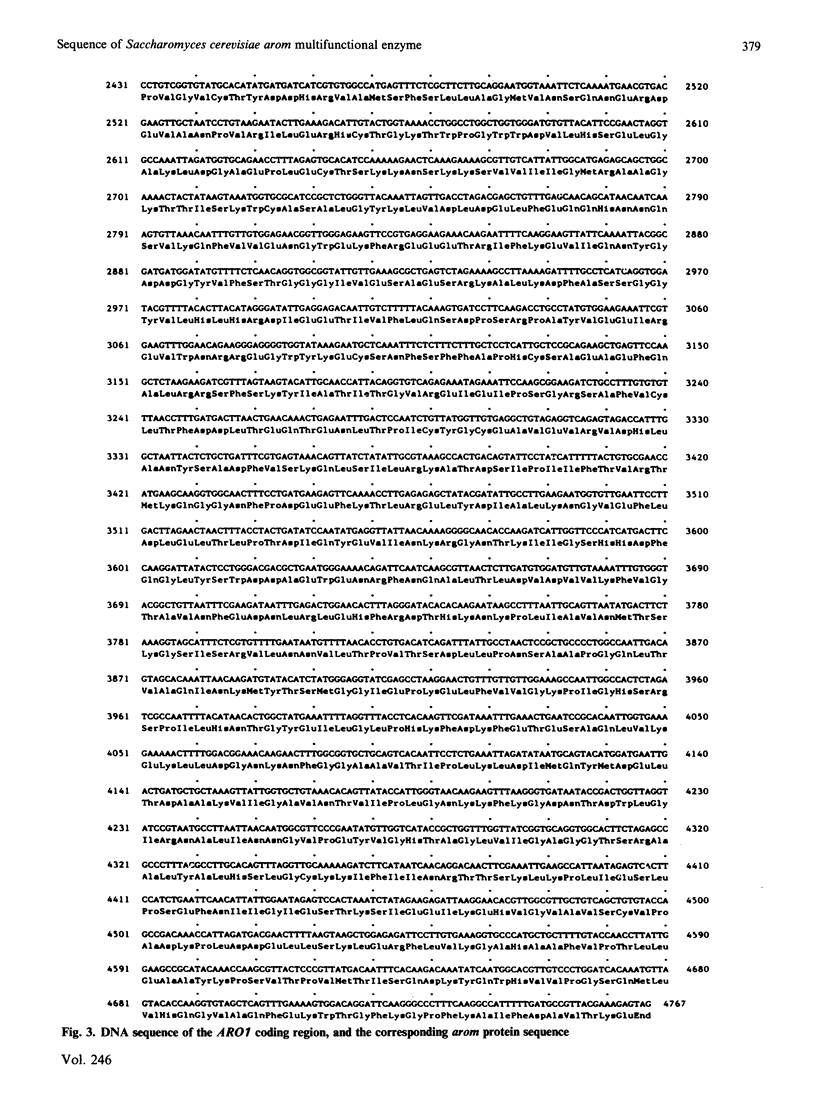
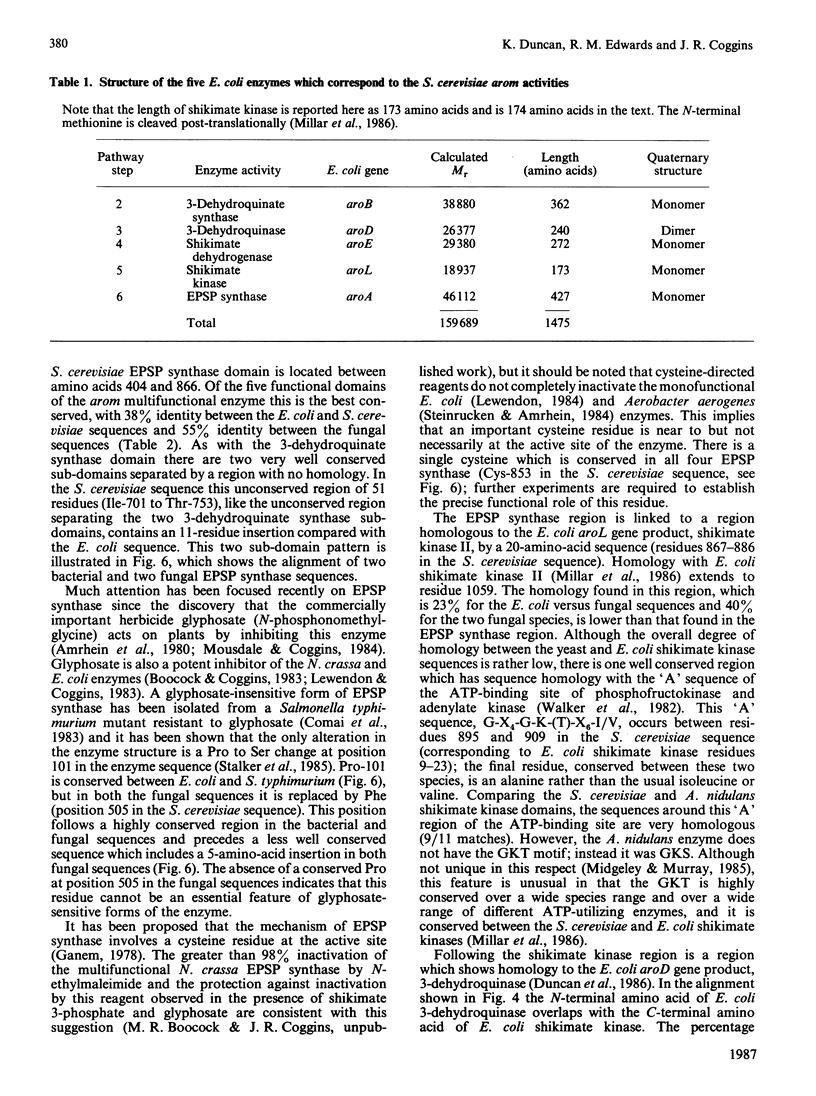
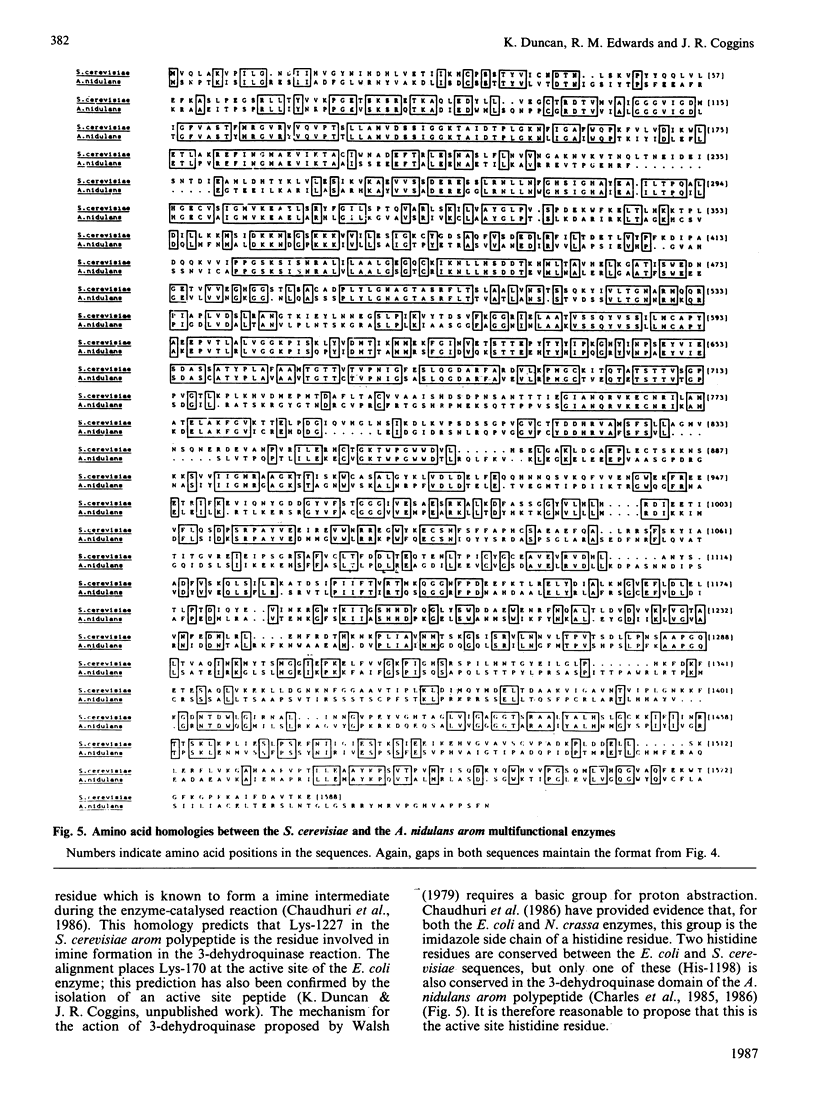
The nucleotide sequence of the Saccharomyces cerevisiae ARO1 gene which encodes the arom multifunctional enzyme has been determined. The protein sequence deduced for the pentafunctional arom polypeptide is 1588 amino acids in length and has a calculated Mr of 174555. Functional regions within the polypeptide chain have been identified by comparison with the sequences of the five monofunctional Escherichia coli enzymes whose activities correspond with those of the arom multifunctional enzyme. The observed homologies demonstrate that the arom polypeptide is a mosaic of functional domains and are consistent with the hypothesis that the ARO1 gene evolved by the linking of ancestral E. coli-like genes.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmed S. I., Giles N. H. Organization of enzymes in the common aromatic synthetic pathway: evidence for aggregation in fungi. J Bacteriol. 1969 Jul;99(1):231–237. doi: 10.1128/jb.99.1.231-237.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alton N. K., Buxton F., Patel V., Giles N. H., Vapnek D. 5'-Untranslated sequences of two structural genes in the qa gene cluster of Neurospora crassa. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1955–1959. doi: 10.1073/pnas.79.6.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfaiza J., Parsot C., Martel A., de la Tour C. B., Margarita D., Cohen G. N., Saint-Girons I. Evolution in biosynthetic pathways: two enzymes catalyzing consecutive steps in methionine biosynthesis originate from a common ancestor and possess a similar regulatory region. Proc Natl Acad Sci U S A. 1986 Feb;83(4):867–871. doi: 10.1073/pnas.83.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Berlyn M. B., Ahmed S. I., Giles N. H. Organization of polyaromatic biosynthetic enzymes in a variety of photosynthetic organisms. J Bacteriol. 1970 Nov;104(2):768–774. doi: 10.1128/jb.104.2.768-774.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlyn M. B., Giles N. H. Organization of enzymes in the polyaromatic synthetic pathway: separability in bacteria. J Bacteriol. 1969 Jul;99(1):222–230. doi: 10.1128/jb.99.1.222-230.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode R., Birnbaum D. Aggregation und Trennbarkeit der Enzyme des Shikimat-Pathway bei Hefen. Z Allg Mikrobiol. 1981;21(6):417–422. doi: 10.1002/jobm.3630210602. [DOI] [PubMed] [Google Scholar]
- Boocock M. R., Coggins J. R. Kinetics of 5-enolpyruvylshikimate-3-phosphate synthase inhibition by glyphosate. FEBS Lett. 1983 Apr 5;154(1):127–133. doi: 10.1016/0014-5793(83)80888-6. [DOI] [PubMed] [Google Scholar]
- Case M. E., Giles N. H. Partial enzyme aggregates formed by pleiotropic mutants in the arom gene cluster of Neurospora crassa. Proc Natl Acad Sci U S A. 1971 Jan;68(1):58–62. doi: 10.1073/pnas.68.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catcheside D. E., Storer P. J., Klein B. Cloning of the ARO cluster gene of Neurospora crassa and its expression in Escherichia coli. Mol Gen Genet. 1985;199(3):446–451. doi: 10.1007/BF00330757. [DOI] [PubMed] [Google Scholar]
- Charles I. G., Keyte J. W., Brammar W. J., Hawkins A. R. Nucleotide sequence encoding the biosynthetic dehydroquinase function of the penta-functional arom locus of Aspergillus nidulans. Nucleic Acids Res. 1985 Nov 25;13(22):8119–8128. doi: 10.1093/nar/13.22.8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles I. G., Keyte J. W., Brammar W. J., Smith M., Hawkins A. R. The isolation and nucleotide sequence of the complex AROM locus of Aspergillus nidulans. Nucleic Acids Res. 1986 Mar 11;14(5):2201–2213. doi: 10.1093/nar/14.5.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S., Coggins J. R. The purification of shikimate dehydrogenase from Escherichia coli. Biochem J. 1985 Feb 15;226(1):217–223. doi: 10.1042/bj2260217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S., Lambert J. M., McColl L. A., Coggins J. R. Purification and characterization of 3-dehydroquinase from Escherichia coli. Biochem J. 1986 Nov 1;239(3):699–704. doi: 10.1042/bj2390699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements J. M., Roberts C. F. Transcription and processing signals in the 3-phosphoglycerate kinase (PGK) gene from Aspergillus nidulans. Gene. 1986;44(1):97–105. doi: 10.1016/0378-1119(86)90047-8. [DOI] [PubMed] [Google Scholar]
- Coggins J. R., Boocock M. R., Campbell M. S., Chaudhuri S., Lambert J. M., Lewendon A., Mousdale D. M., Smith D. D. Functional domains involved in aromatic amino acid biosynthesis. Biochem Soc Trans. 1985 Apr;13(2):299–303. doi: 10.1042/bst0130299. [DOI] [PubMed] [Google Scholar]
- Comai L., Sen L. C., Stalker D. M. An Altered aroA Gene Product Confers Resistance to the Herbicide Glyphosate. Science. 1983 Jul 22;221(4608):370–371. doi: 10.1126/science.221.4608.370. [DOI] [PubMed] [Google Scholar]
- Da Silva A. J., Whittington H., Clements J., Roberts C., Hawkins A. R. Sequence analysis and transformation by the catabolic 3-dehydroquinase (QUTE) gene from Aspergillus nidulans. Biochem J. 1986 Dec 1;240(2):481–488. doi: 10.1042/bj2400481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Farabaugh P. J., Fink G. R. The nucleotide sequence of the HIS4 region of yeast. Gene. 1982 Apr;18(1):47–59. doi: 10.1016/0378-1119(82)90055-5. [DOI] [PubMed] [Google Scholar]
- Duncan K., Chaudhuri S., Campbell M. S., Coggins J. R. The overexpression and complete amino acid sequence of Escherichia coli 3-dehydroquinase. Biochem J. 1986 Sep 1;238(2):475–483. doi: 10.1042/bj2380475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler E., Schultz G. Localization, purification, and characterization of shikimate oxidoreductase-dehydroquinate hydrolyase from stroma of spinach chloroplasts. Plant Physiol. 1985 Sep;79(1):212–218. doi: 10.1104/pp.79.1.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertner F. H., Ericson M. C., DeMoss J. A. Catalytic facilitation in vitro by two multienyzme complexes from Neurospora crassa. J Biol Chem. 1970 Feb 10;245(3):595–600. [PubMed] [Google Scholar]
- Giles N. H., Case M. E., Partridge C. W., Ahmed S. I. A gene cluster in Nuerospora crassa coding for an aggregate of five aromatic synthetic enzymes. Proc Natl Acad Sci U S A. 1967 Oct;58(4):1453–1460. doi: 10.1073/pnas.58.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles N. H., Partridge C. W., Ahmed S. I., Case M. E. The occurrence of two dehydroquinases in Neurospora crassa, one constitutive and one inducible. Proc Natl Acad Sci U S A. 1967 Nov;58(5):1930–1937. doi: 10.1073/pnas.58.5.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henner D. J., Hoch J. A. The Bacillus subtilis chromosome. Microbiol Rev. 1980 Mar;44(1):57–82. doi: 10.1128/mr.44.1.57-82.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Repeated DNA sequences upstream from HIS1 also occur at several other co-regulated genes in Saccharomyces cerevisiae. J Biol Chem. 1983 Apr 25;258(8):5238–5247. [PubMed] [Google Scholar]
- Horowitz N. H. On the Evolution of Biochemical Syntheses. Proc Natl Acad Sci U S A. 1945 Jun;31(6):153–157. doi: 10.1073/pnas.31.6.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinghorn J. R., Hawkins A. R. Cloning and expression in Escherichia coli K-12 of the biosynthetic dehydroquinase function of the arom cluster gene from the eucaryote, Aspergillus nidulans. Mol Gen Genet. 1982;186(1):145–152. doi: 10.1007/BF00422927. [DOI] [PubMed] [Google Scholar]
- Kirschner K., Bisswanger H. Multifunctional proteins. Annu Rev Biochem. 1976;45:143–166. doi: 10.1146/annurev.bi.45.070176.001043. [DOI] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J. M., Boocock M. R., Coggins J. R. The 3-dehydroquinate synthase activity of the pentafunctional arom enzyme complex of Neurospora crassa is Zn2+-dependent. Biochem J. 1985 Mar 15;226(3):817–829. doi: 10.1042/bj2260817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimer F. W., Morse C. C., Beck A. K., Cole K. W., Gaertner F. H. Isolation of the ARO1 cluster gene of Saccharomyces cerevisiae. Mol Cell Biol. 1983 Sep;3(9):1609–1614. doi: 10.1128/mcb.3.9.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewendon A., Coggins J. R. Purification of 5-enolpyruvylshikimate 3-phosphate synthase from Escherichia coli. Biochem J. 1983 Jul 1;213(1):187–191. doi: 10.1042/bj2130187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden J., Coggins J. R. The subunit structure of the arom multienzyme complex of Neurospora crassa. A possible pentafunctional polypeptide chain. Biochem J. 1977 Mar 1;161(3):599–607. doi: 10.1042/bj1610599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Midgley C. A., Murray N. E. T4 polynucleotide kinase; cloning of the gene (pseT) and amplification of its product. EMBO J. 1985 Oct;4(10):2695–2703. doi: 10.1002/j.1460-2075.1985.tb03989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar G., Coggins J. R. The complete amino acid sequence of 3-dehydroquinate synthase of Escherichia coli K12. FEBS Lett. 1986 May 5;200(1):11–17. doi: 10.1016/0014-5793(86)80501-4. [DOI] [PubMed] [Google Scholar]
- Millar G., Lewendon A., Hunter M. G., Coggins J. R. The cloning and expression of the aroL gene from Escherichia coli K12. Purification and complete amino acid sequence of shikimate kinase II, the aroL-gene product. Biochem J. 1986 Jul 15;237(2):427–437. doi: 10.1042/bj2370427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miozzari G. F., Yanofsky C. Gene fusion during the evolution of the tryptophan operon in enterobacteriaceae. Nature. 1979 Feb 8;277(5696):486–489. doi: 10.1038/277486a0. [DOI] [PubMed] [Google Scholar]
- Nakanishi N., Yamamoto M. Analysis of the structure and transcription of the aro3 cluster gene in Schizosaccharomyces pombe. Mol Gen Genet. 1984;195(1-2):164–169. doi: 10.1007/BF00332740. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Polley L. D. Purification and characterization of 3-dehydroquinate hydrolase and shikmate oxidoreductase. Evidence for a bifunctional enzyme. Biochim Biophys Acta. 1978 Sep 11;526(1):259–266. doi: 10.1016/0005-2744(78)90310-8. [DOI] [PubMed] [Google Scholar]
- Rines H. W., Case M. E., Giles N. H. Mutants in the arom gene cluster of Neurospora crassa specific for biosynthetic dehydroquinase. Genetics. 1969 Apr;61(4):789–800. doi: 10.1093/genetics/61.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K. E., Roth J. R. Linkage map of Salmonella typhimurium, Edition VI. Microbiol Rev. 1983 Sep;47(3):410–453. doi: 10.1128/mr.47.3.410-453.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. D., Coggins J. R. Isolation of a bifunctional domain from the pentafunctional arom enzyme complex of Neurospora crassa. Biochem J. 1983 Aug 1;213(2):405–415. doi: 10.1042/bj2130405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinrücken H. C., Amrhein N. 5-Enolpyruvylshikimate-3-phosphate synthase of Klebsiella pneumoniae. 1. Purification and properties. Eur J Biochem. 1984 Sep 3;143(2):341–349. doi: 10.1111/j.1432-1033.1984.tb08378.x. [DOI] [PubMed] [Google Scholar]
- Strauss A. The genetic fine structure of the complex locus aro3 involved in early aromatic amino acid biosynthesis in Schizosaccharomyces pombe. Mol Gen Genet. 1979;172(3):233–241. doi: 10.1007/BF00271722. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch G. R., Gaertner F. H. Coordinate activation of a multienzyme complex by the first substrate. Evidence for a novel regulatory mechanism in the polyaromatic pathway of Neurospora crassa. Arch Biochem Biophys. 1976 Feb;172(2):476–489. doi: 10.1016/0003-9861(76)90101-6. [DOI] [PubMed] [Google Scholar]
- Welch G. R., Gaertner F. H. Influence of an aggregated multienzyme system on transient time: kinetic evidence for compartmentation by an aromatic-amino-acid synthesizing complex of Neurospora crassa. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4218–4222. doi: 10.1073/pnas.72.11.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalkin H., Paluh J. L., van Cleemput M., Moye W. S., Yanofsky C. Nucleotide sequence of Saccharomyces cerevisiae genes TRP2 and TRP3 encoding bifunctional anthranilate synthase: indole-3-glycerol phosphate synthase. J Biol Chem. 1984 Mar 25;259(6):3985–3992. [PubMed] [Google Scholar]
- Zalkin H., Yanofsky C. Yeast gene TRP5: structure, function, regulation. J Biol Chem. 1982 Feb 10;257(3):1491–1500. [PubMed] [Google Scholar]


