Abstract
In the canonical genetic code, many amino acids are assigned more than one codon. Work by us and others has shown that the choice of these synonymous codon is not random, and carries regulatory and functional consequences. Existing protein foundation models ignore this context-dependent role of coding sequence in shaping the protein landscape of the cell. To address this gap, we introduce cdsFM, a suite of codon-resolution large language models, including both EnCodon and DeCodon models, with up to 1B parameters. Pre-trained on 60 million protein-coding sequences from more than 5,000 species, our models effectively learn the relationship between codons and amino acids, recapitualing the overall structure of the genetic code. In addition to outperforming state-of-the-art genomic foundation models in a variety of zero-shot and few-shot learning tasks, the larger pre-trained models were superior in predicting the choice of synonymous codons. To systematically assess the impact of synonymous codon choices on protein expression and our models’ ability to capture these effects, we generated a large dataset measuring overall and surface expression levels of three proteins as a function of changes in their synonymous codons. We showed that our EnCodon models could be readily fine-tuned to predict the contextual consequences of synonymous codon choices. Armed with this knowledge, we applied EnCodon to existing clinical datasets of synonymous variants, and we identified a large number of synonymous codons that are likely pathogenic, several of which we experimentally confirmed in a cell-based model. Together, our findings establish the cdsFM suite as a powerful tool for decoding the complex functional grammar underlying the choice of synonymous codons.
1. Introduction
The canonical genetic code, the blueprint that links nucleic acid instructions to proteins, is highly degenerate. 18 of the 20 amino acids are encoded by more than one codon. These synonymous codons were long thought to be interchangeable as they encode the same amino acid. However, codon usage bias (CUB), which refers to the non-uniform use of synonymous codons, has long been known and studied [9, 35, 43, 49, 50, 64, 73]. Increasing evidence consistently shows organism-specific [35, 43, 50] or tissue-specific [6, 20, 65] patterns of codon usage in numerous species. In addition, a number of studies revealed an important role for codon usage in the regulation of gene expression and protein folding through various mechanisms[3, 7, 8, 13, 71, 78]. To name a few, synonymous codons have been shown to be influenced by the levels of cognate tRNA and tRNA gene copy numbers and therefore differentially impact translation elongation speed [43, 50, 64]. Moreover, more than 60 synonymous variants (mutations that do not alter the amino acid sequence) associated with diseases [19, 45], and over 450 linked to tumors [72], have been reported in ClinVar, highlighting the rich information encoded within nucleotide sequences beyond what is reflected in the amino acid sequence.
Protein language models have revolutionized our understanding of protein structures and functions by learning from a large number of protein sequences [1, 30, 33, 41, 46, 56, 68]. These models have become invaluable tools in fields such as protein engineering, synthetic biology and cancer therapeutics. However, the choice of synonymous codons that encode the protein sequence falls in the blind spot of these models. By ignoring these, protein language models miss critical regulatory and structural information present in coding sequences. There has also been a growing interest in developing language models that operate directly on DNA and RNA sequences [4, 17, 29, 39, 44, 60, 62, 90]. Both genomic and protein language models leverage self-supervised learning objectives to capture the underlying biological grammar of nucleotide and protein sequences , respectively, encompassing both coding information and regulatory elements. By doing so, they can be effectively deployed in downstream tasks where labeled data are limited, such as variant effect prediction [14, 22, 38, 88], gene expression prediction [2, 61], open reading frame (ORF) localization [57], protein function prediction [47, 83], and many other applications [36, 37, 48, 79–81].
To address the gap in our understanding of codon usage and better measure the impact of synonymous codons on protein expression and function, we generated a dataset of synonymous variants across three human surface proteins, and observed that while the amino acid sequence is conserved, the choice of synonymous codons can impact overall and surface expression of proteins. This is consistent with our earlier work demonstrating the link between codon usage, tRNA abundance and translation [26]. Therefore, synonymous codons are not always interchangeable; however, the underlying contextual grammar that governs the choice synonymous codons is not well-explored. In this study, we asked whether foundation models trained on coding sequences can capture the contextual interplay of synonymous codons that gives rise to these biological variations. For this, we developed a large suite of transformer models, including encoder and decoder architectures called EnCodon and DeCodon, respectively, at various scales. We pre-trained all models on a large corpus of 60 million coding sequences (CDS) from more than 5000 species aggregated from the NCBI Genomes database (referred to as NCBI CDS throughout the paper). We studied these pre-trained models to explore the biological concepts they capture at different scales. We also evaluated their performance on a variety of zero-shot and few-shot learning tasks related to gene function and post-transcriptional regulation. We then turned our focus to tasks related to the choice of synonymous codons. Our results both reflect the importance of synonymous codons on protein expression and the ability of our pre-trained large language models to capture their influence. By applying EnCodon and DeCodon models, zero-shot, to somatic variations observed in human cancers, we have nominated many synonymous variants that are likely cancer drivers; a form of pathogenicity that current protein-based therapeutic strategies cannot address. Taken together, our findings showcase the power of EnCodon and DeCodon models to capture the context-dependent function of synonymous codons and better represent the coding sequence beyond what is captured by existing DNA and protein models.
2. Results
2.1. Leveraging self-supervised learning to pre-train EnCodon and DeCodon
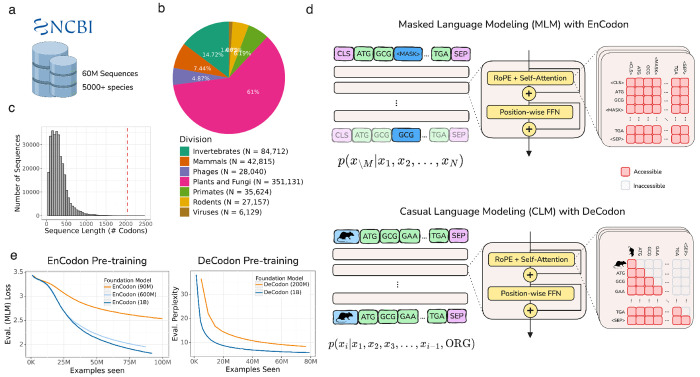
Both encoder and decoder transformer architectures can be effectively applied to coding sequences. Although bidirectional encoder transformers (BERT) are frequently the preferred choice for biological sequences [18, 29, 39, 44, 62, 90], causal language models have also demonstrated remarkable zero-shot and few-shot capabilities in this field [59, 60, 70]. Given this, along with the fact that translation is inherently directional, we chose to evaluate both architectures. We developed EnCodon and DeCodon models, codon-level language models using encoder and decoder architectures, respectively. These models were pre-trained on an aggregated dataset of 60 million coding sequences from 5,000 species, primarily composed of bacterial sequences (98.7%, or 59.4 million sequences), but also including mammals and primates (Figure 1a,b,c). Based on the length distribution of the coding sequences (Figure 1c), we set the maximum context size to 2048 codons for both EnCodon and DeCodon, ensuring that over 99.5% of all sequences would fit within this context window.
Figure 1: Overview of EnCodon and DeCodon:
a) Over 60 million coding sequences from 5000 species has been extracted from NCBI Genomes database and used to pre-train EnCodon and DeCodon foundation models. b) An overwhelming majority of the data (98.7%) is comprised of bacterial coding sequences. Pie chart depicting division makeup of non-bacterial coding sequences in NCBI is shown. c) Histogram of coding sequence lengths (number of codons) in NCBI Genomes database. We used 2048 as maximum sequence length supported by EnCodon and DeCodon based taking the shown distrbution into account to cover more than 99.8% of sequences. d) We pretrained EnCodon using masked language modeling (MLM) objective where parts of sequences were corrupted/masked and the model has to predict the true token at the positions given the rest of tokens (i.e. context). DeCodon is a conditional generative transformer model which provides controllable coding sequence generation by querying sequence organism as the very first input token. We pre-trained DeCodon with causal (autoregressive) language modeling objective on aggregated corpus of coding sequences where each sequence is prepended with a special organism token. Rotary Positional Self-Attention was used in both EnCodon and DeCodon blocks. e) 3 EnCodons and 2 DeCodons, differing in scale (i.e. number of trainable parameters) have been pre-trained for more than 1,000,000 optimization steps on the aggregated corpus from NCBI Genomes database.
During pre-training, two special tokens were added to each sequence: a <CLS> token (for EnCodon) or an organism-specific token (for DeCodon) was prepended and a <SEP> token was appended (Figure 1d). EnCodon was trained using the self-supervised objective of Masked Language Modeling (MLM), where a subset of codons was randomly masked, and the model was tasked with predicting the original codons using the contextual information provided by the unmasked tokens. In contrast, DeCodon used Causal Language Modeling (CLM), where the model generated the next codon based on the preceding context (Figure 1d). To evaluate the impact of model size on performance, we pre-trained three versions of EnCodon with 80 million, 620 million, and 1 billion parameters, and two versions of DeCodon with 200 million and 1 billion parameters. As shown in Figure 1e (and Supplementary Figure 3), the larger models achieved better performance, as indicated by lower MLM loss for EnCodon and lower perplexity for DeCodon. Furthermore, we observed that the sequencing embedding space learned by all models separate by domains of life, as visualized in Supplementary Figure 1. To address the significant imbalance in the training data—where 98.7% of the data consisted of bacterial sequences, while eukaryotic sequences were underrepresented, we conducted a second stage of pre-training focused on eukaryotic sequences to better adapt the models to these coding sequences. This adaptation significantly improved performance and generalizability for eukaryotic organisms, as demonstrated in subsequent experiments (Supplementary Figure 2,4). The eukaryotic-adapted versions of each pre-trained model are denoted with a superscript “Ada”, e.g., EnCodon (1B)Ada (see Methods 5.2).
2.2. Codon embeddings learned by EnCodon and DeCodon reflect the structure of the canonical genetic code
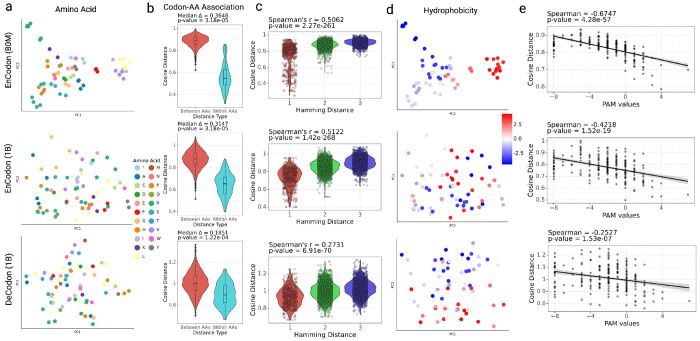
To better understand the biological principles learned by EnCodon and DeCodon models, we focused on their learned codon embeddings. We assessed the relationship between codons and the amino acids they encode (Figure 2a and Supplementary Figure 5a). As shown in Figure 2b and Supplementary Figure 5b, synonymous codons are embedded substantially closer to each other relative to non-synonymous pairs. In other words, the EnCodon and DeCodon models have learned the structure of the genetic code. Beyond synonymous codon embeddings, we observed that all pre-trained EnCodon and DeCodon models exhibited a consistently strong correlation (See Figure 2c and Supplementary Figure 5c) between the pairwise cosine distance of codon embeddings and the pairwise Hamming distance of their nucleotide sequences. This notable correlation indicates that all of our models have inherently captured “load minimization” [23, 25, 58], a key property of the canonical genetic code, without explicit supervision. Load minimization refers to the ability of the canonical genetic code to reduce the detrimental effects of mutations by ensuring that common mutations are less likely to cause significant changes in protein structure or function. This is measured and demonstrated using the physicochemical properties of amino acids, such as hydrophobicity, or directly from “Point Accepted Mutation” (PAM) matrices. As shown in Figure 2d–e and Supplementary Figure 5d–e, the learned codon embeddings and the cosine distances between them very well capture this load minimization principle.
Figure 2: Codon Embedding Space Analysis for pre-trained EnCodons and DeCodons:
a) PCA visualization of codon embeddings learned by EnCodon (80M), EnCodon (1B), and DeCodon (1B) colored by Amino Acid. b) Violin plots of two cosine distance between pairwise synonynous against non-synonymous codons for the 3 models. c) Violin plot of two codon distance metrics i.e. cosine distance in learned embedding space and hamming distance between codon sequences for all possible pairs of codons annotated with spearman correlation between the two metrics. d) PCA visualization codon embeddings colored by their corresponding amino acid’s Hydrophobicity Index. e) Scatter-plot of pair-wise cosine distance between amino acids and their corresponding PAM250 entry score for pre-trained models.
As mentioned earlier, the larger models are more adept at distinguishing synonymous codons. This is reflected in their codon embeddings as well (See Figure 2a, Supplementary Figure 5f,g). Smaller EnCodon and DeCodon models showed better amino acid KNN purity scores (particularly for K values between 3 and 5) (Supplementary Figure 5f,g), indicating that synonymous codons are embedded closely together, and are therefore hard to distinguish. Conversely, larger models achieved significantly lower (better) Masked Language Model (MLM) losses during pre-training (See Figure 1e, Supplementary Figure 2a), by better distinguishing synonymous codons, in general moving beyond the simple structure of the genetic code. In other words, the smaller models have largely learned the mapping between the codons and the amino acids they encode, and therefore act as small protein language models with difficulty distinguishing synonymous codons. For instance, the Spearman correlation between the top 10 PC components of codon embeddings and the codon’s amino acid hydrophobicity index reveals that this information is captured in the first two PCs in smaller models. In contrast, the embedding space learned by larger models are less dominated by simple amino acid properties, with amino acid hydrophobicity captured in PC3 and beyond (See Supplementary Figure 2c). This observation highlights that larger models have learned a more sophisticated understanding of context-dependent usage of synonymous codons, and are expected to perform better in downstream tasks related to synonymous variants.
2.3. DeCodon generates functional organism-specific coding sequences
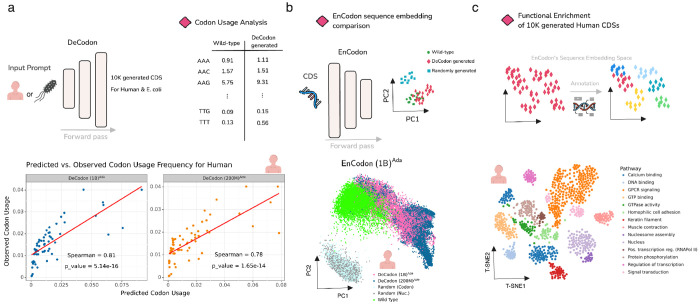
Since DeCodon models are generative, we sought to assess their learned knowledge of coding sequences by comparing model-generated sequences to natural ones. For this, we generated 10,000 coding sequences each for human and E. coli, respectively. Comparing average codon frequencies of wild-type and generated coding sequences, both DeCodon models showed strong Spearman correlations between the reference and generated sequences for both human (Figure 3a) and E. coli (Supplementary Figure 6c,d); two species with significantly different codon usage patterns. We then used our pre-trained EnCodon models to compare reference and generated sequence embeddings and we observed that sequences generated by DeCodon (1B) are notably better mixed with the reference cluster relative to randomly generated sequences (Figure 3b and Supplementary Figure 6a). Finally, we used a protein functional annotation tool called InterProScan to predict functional regions of the generated coding sequences. Using the computed sequence embeddings by EnCodon, we clustered the generated sequences, extracted enriched biological pathways in each cluster and annotated each cluster based on the most specific enriched pathway (Figure 3c, Supplementary Figure 6e,f,g). This observation highlights the ability of DeCodon models to generate coding sequences that capture a variety of learned protein sequence patterns.
Figure 3: DeCodon generates functional and organism-specific coding sequences:
a) DeCodon takes organism as input and generates a coding sequence specific to the queried species. We generated 10,000 coding sequences (CDS) for Human and E. coli species. Scatter plots of codon usage frequencies of wild-type (y-axis) and generated (x-axis) is shown for human annotated with spearman correlation and associated p-value. b) To further compare the generated CDS population with the wild-type, we generated two groups of randomly sampled CDSs and computed sequence embeddings of wild-type, DeCodon generated, and randomly generated groups. PCA visualization of sequence embeddings is shown for human-related coding sequences. c) Finally, we used protein functional annotation tools to test the functional enrichment of the sequence clusters in EnCodon embedding space. We used InterProScan to predict functional domains of human-generated CDSs by DeCodon (1B)Ada. T-SNE visualization of functionally annotated generated sequences by DeCodon (1B)Ada is shown where generated sequences were colored by their enriched biological pathway.
2.4. Codon Foundation Models perform zero-shot function prediction across DNA and RNA modalities
2.4.1. Clinical variant effect classification in human genome
One of the key benefits of foundation models is their ability to apply their learned principles to new tasks without the need for retraining. We therefore evaluated the zero-shot capabilities of our codon language models in a variety of biologically relevant downstream tasks. As the first task, we examined the ability of our models to identify pathogenic variants annotated for the human proteome. The assessment of variant pathogenicity is considered a suitable challenge due to the large combinatorial space of all potential variants and their interactions. Furthermore, the majority of clinically analyzed variants are classified as benign, thereby limiting the number of positive instances to learn from. Recently, language models trained on corpora of nucleotide or protein sequences (exclusively utilizing self-supervised learning objectives) have demonstrated exceptional predictive performance in downstream tasks such as variant pathogenicity classification.
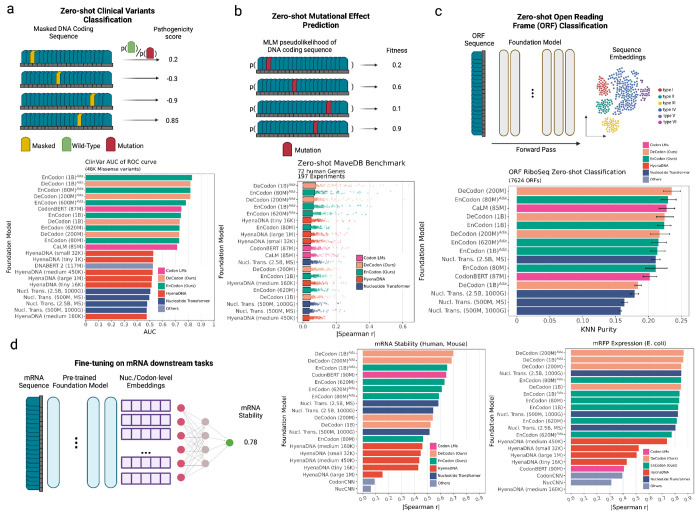
To perform zero-shot variant pathogenicity prediction, we calculated the pseudolikelihood (or sequence likelihood for decoder models) (See Methods 5.15.5) of 48,283 single nucleotide variants (SNVs) documented in ClinVar (see Figure 4a and Supplementary Figure 8, 9) using our language models and state-of-the-art nucleotide language models, including CaLM [62], CodonBERT [44], HyenaDNA [60], Nucleotide Transformer [18], and DNABERT 2 [90]. Upon evaluating the zero-shot pathogenicity prediction capabilities of the proposed language models on these clinically validated variants, we observed that, as expected, the Eukaryotic-adapted EnCodon and DeCodon exhibited superior performance relative to all other evaluated models (Figure 4a).
Figure 4: Downstream Benchmark of state-of-the-art nucleotide language models:
a) We tested zero-shot predictive capability of the language models on 48,042 filtered variants from ClinVar. For Encoder models like EnCodon, we masked the position of variant in coding sequences and computed the log-likelihood ratio (LLR) between mutated and wild-type codon/nucleotide/token. Concerning generative models like DeCodon, we defined the pathogenicity score of variant as difference between wild-type and mutated sequence likelihoods. The Area under the ROC curve (AUC) is shown as bars for each model colored by the architecture family used in the language model. b) We further compared the zero-shot capability of the language models on deep mutational scan studies (DMS). Spearman correlation is reported for each model as the correlation between the reported “fitness” score and predicted sequence likelihood (pseudolikelihood for encoder models). 197 Human DMS studies across 72 genes were used to assess the language models shown in the barplot where each dot is a single human DMS study. The bars are colored according to the architecture family used in the language model. c) We further tested the foundation models’ discriminative capability of sequence embedding space in localization of open-reading-frames (ORFs). We use a recently published collection of 7624 open-reading frames (ORFs) where sequences were annotated based on their genomic location. KNN Purity scores (using different Ks) of the extracted sequence embeddings were shown in bar plot where bars represent means showing as bars and lines as standard deviations. d) We used LoRA [34] technique for parameter-efficient fine-tuning of the language models on mRNA-related downstream tasks namely mRNA stability and mRFP expression prediction. For each downstream task, a barplot of Spearman correlation of the hold-out test set for all fine-tuned models on mRFP Expression (E. coli) and mRNA stability (Human and Mouse) tasks.
2.4.2. Predicting mutational effects on protein function measured by deep mutational scan (DMS) studies
We applied our CDS foundation models to predict the effect of mutations on protein function, utilizing data from deep mutational scanning (DMS) studies. These studies introduce comprehensive sets of mutations to protein sequences and experimentally measure their impact on fitness – a study-specific metric that quantifies protein functionality ([21]). To predict experimental fitness scores, similar to the previous task, we used pseudolikelihoods (or likelihoods for autoregressive models, See Methods 5.15.5) of codons or nucleotides generated by the models (Figure 4b). Our analysis was restricted to DMS studies that provided nucleotide information for wild-type sequences and their corresponding mutations. Therefore, we evaluated the foundation models on DMS studies across 7 different organisms (including human, E. coli, house mouse, chicken, etc), covering approximately 58,262 and 16,403 mutations, respectively (Figure 4b, Supplementary Figure 10 and 11).
The Eukaryotic-adapted codon language models consistently placed as the top five performers, showcasing the highest correlations for both human and E. coli datasets (Figure 4b, Supplementary Figure 10). This outcome indicates the efficacy of our adaptation technique and the versatility of our pre-trained models to specific organismal contexts. All coding sequence models, including CodonBERT (which ranked 6th), surpassed HyenaDNA and nucleotide transformer models. As expected, both the adapted and pretrained versions of our codon foundation models excelled in the E. coli benchmark (Supplementary Figure 10a). When comparing EnCodons with DeCodons, we consistently noted an increase in performance with larger DeCodon models, although the scale of improvement for both models were the same when the model size was fixed. These findings underscore the potential of our codon-level large language models in forecasting mutational impacts on protein functionality.
2.4.3. Evaluation of Nucleotide Language Models for Human Open Reading Frame (ORF) Classification
We next sought to focus on tasks centered on the translational capacity of the transcriptome. Ribosome profiling (Ribo-seq) has significantly broadened our understanding of the translational output of the cell by uncovering numerous open reading frames (ORFs) in regions previously believed to be untranslated, such as long non-coding RNAs (lncRNAs) and untranslated regions (UTRs) of protein-coding genes [5, 12, 40, 52, 66, 84]. Mudge et al. [57] described a standardized, spatially-classified, and filtered collection of translated ORFs, integrating previously reported ORF databases. Given the multiple interpretative approaches possible for ORFs, the sequences were systematically classified based on their spatial relationships with existing gene annotations. Our objective was to evaluate our models’ predictive accuracy in classifying ORFs without further supervision (i.e., training). To this end, we employed the standardized catalog of 7,264 human Ribo-seq ORFs to assess the proficiency of embeddings from various models in ORF classification (Figure 4c). Accordingly, we reported the statistics of purity scores of the K-Nearest Neighbors (KNN) algorithm for each model across different K values, ranging from 5 to 50 (Figure 4d). The results indicated that our codon foundation models consistently outperformed all other tested language models, with DeCodon (200M) achieving the highest average KNN purity score of 24.57%, whereas the Nucleotide Transformer (2.5B, multi-species) achieved an average score of 21.45%, representing the best performance among the existing state-of-the-art language models.
2.4.4. Codon language models show superior performance on various mRNA downstream tasks
We further compared the foundation models’ performance across a few supervised mRNA prediction tasks namely mRNA stability (Human and Mouse) [2] and mRFP expression (E. coli) [61] prediction. Similar to zero-shot benchmarks, we benchmarked Nucleotide Transformer[11], HyenaDNA [60], CaLM [62], and CodonBERT [44] as prior methods alongside with two convolutional baselines i.e. NucCNN and CodonCNN (See 5.12). We used low-rank adaptation technique (LoRA) [34] to fine-tune the language models in the downstream tasks. Our benchmark shown in Figure 4d shows our EnCodon and DeCodon models outperform their counterparts in both mRNA stability and mRFP expression prediction tasks. More specifically, DeCodon (1B)Ada showed 5% improvement in mRNA stability prediction over Nucleotide Transformer (2nd best model), underscoring its superior understanding of mRNA dynamics.
2.5. EnCodon captures the effect of synonymous codon mutations on protein abundance levels
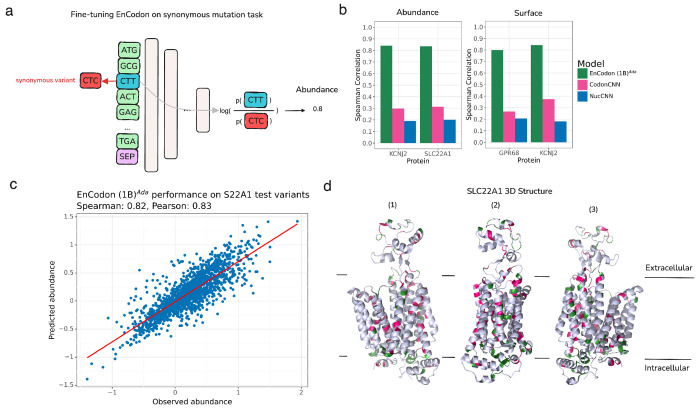
While most synonymous variations are considered neutral, there is mounting evidence that synonymous variations can also carry functional consequences [19, 71, 78]. Being able to discriminate between synonymous codons and better capture their possible context-dependent role in the translational output was a major motivation behind training EnCodon and DeCodon models. However, there are few instances of functional synonymous variants, and we currently lack large datasets evaluating synonymous codons. To address this gap, we generated a dataset of1054 synonymous coding variants across three membrane proteins with directly comparable phenotypes, namely SLC22A1, GPR68, and KCNJ2 [32, 51, 89]. While all datasets are on membrane proteins, they represent distinct protein architectures and functions, including a tetrameric 2 transmembrane cardiac potassium channel associated with defects associated with developmental disorders [82]; KCNJ2, a monomeric 12 transmembrane polyspecific cation transporter that has genetics variants associated with differential effects on drug metabolism [27]; and a proton sensing receptor, GPR68, a seven transmembrane with variants associated with chemotherapy-induced neuropathy [42]. We then systematically measured the overall abundance and surface expression of these variants. To test our models’ predictive performance, we held out 513 variants from SLC22A1 from the training dataset of the foundation models. Figure 5a depicts the fine-tuning procedure where the model takes wild-type sequence as input and log-likelihood ratio between wild-type and mutated codon at the mutated position is used to predict the abundance level of the protein (See 5.14). In other words, without the need to introduce any additional parameters to the pre-trained model, we repurposed the MLM head to model the variant’s effect. Comparing different pre-trained EnCodons (with or without adaptation) on holdout test variants in SLC22A1, consistent improvement in test performance is observed as the model gets larger (Supplementary Figure 12a and Supplementary Figure 13). As expected, the eukaryotic adaptation of the EnCodons also outperformed their non-adapted same-size model resulting in adapted EnCodon (1B) being the best performer which performed well across experimented proteins and hold-out test set (Figure 5b,c, Supplementary Figure 12a,b). The fine-tuned EnCodon model performs slightly better on surface expression than protein abundance in the one gene, KCNJ2, in which we have both measures for abundance and surface expression. This is likely because surface expression is dependent upon protein abundance but also includes effects that change whether the protein makes it to the surface.
Figure 5: EnCodon (1B)Ada generalizes well across unseen synonymous variants in membrane proteins.
a) We re-purposed the pre-trained language modeling classifier head for synonymous mutation effect modeling. Specifically, given a synonymous codon variant, we first compute the codon likelihoods (i.e. logits) for the variant’s position. Next, the log-ratio of wild-type codon against mutated codon is considered as the final variant’s effect prediction – protein abundance level or surface expression in this experiment. Notably, using no additional weights, the mutation’s effect on protein’s abundance measurement is modeled as the log-likelihood ratio between mutated and wild-type codon given the wild-type coding sequence in input. b) Spearman correlation between predicted and observed abundance (left bar plot) or surface expression (right bar plot) were shown for test synonymous variants in KCNJ2, SLC22A1, and GPR68 proteins. c) An test set of synonymous variants applied on SLC22A1 were held-out from the training data of the EnCodons. The scatter plot of predicted vs. observed abundance is shown for eukaryotic adapted EnCodon (1B) which showed as the top-performed compared to other fine-tuned EnCodons. d) After fine-tuning, we performed in-silico synonymous mutagenesis with the best-performing EnCodon model. We selected “critical” synonymous variants for which the predicted abundance was above 95-th (green) or below the 5-th quantile (pink). Next, extracted SLC22A1 extreme variants were overlayed in the protein’s 3D structure which is shown from 3 different angles.
We employed our top-performing fine-tuned EnCodon model to conduct in-silico synonymous codon mutagenesis, predicting the abundance levels for all possible synonymous coding variants within the protein sequences. Variants with exceptionally high or low abundance scores (i.e., critical variants) were selected for further analysis. To evaluate the spatial distribution of these critical variants within the 3D structures of the proteins studied, we applied Ripley’s K function [67], a well-known function that determines a spatial pattern (i.e. random, dispersed, or clustered) of certain points (in 3D structure) at a certain distance cut-off (See Methods 5.16). More specifically, the results revealed significant clustering of the identified “critical” variants for KCNJ2 and SLC22A1 at distances below 10 Å and 8 Å, respectively (Supplementary Figure 12d). The calculated p-values from Ripley’s test, compared against a null distribution (see Methods 5.16), indicate regions where the observed clustering significantly deviates from being randomly spread (i.e., p < 0.05, Supplementary Figure 12d).
Using a 9Å cutoff radius in the Ripley’s K function test, we demonstrated significant clustering of extreme variants in the 3D structures of SLC22A1 and KCNJ2 (see Methods 5.16). Despite having no information on protein folding and structure, the “critical” variants identified by EnCodon are predominantly located in functional elements of the 3D structure, such as α-helices or β-sheets, emphasizing the sequence-structure relationships that EnCodon captures in its learned representations (Figure 5d, Supplementary Figure 12c). The significant clustering observed at these short distances suggests that these variants are not randomly distributed.
2.6. Leveraging CDS foundation models to nominate and validate novel pathogenic synonymous variants
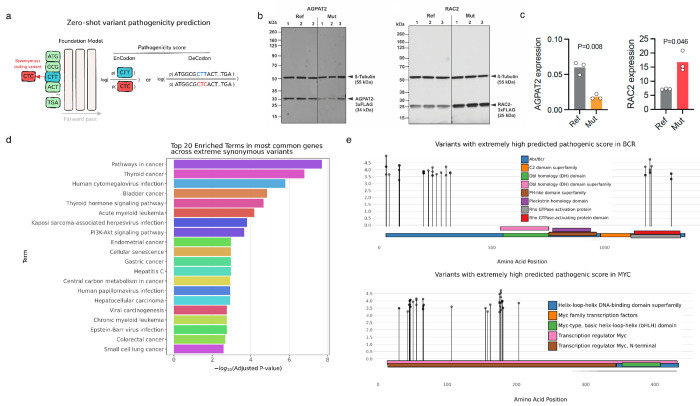
As previously shown and demonstrated here, synonymous codon usage can impact protein structure and gene expression through effects on translation efficiency, kinetics, elongation, mRNA stability, and co-translational protein folding [3, 7, 8, 13, 43]. Given the zero-shot performance of our CDS foundation models in predicting variant pathogenicity, and the ability of our models in distinguishing synonymous codons in mutational scans in the previous section, we set out to nominate previously unknown pathogenic synonymous variants. To this end, we computed pathogenicity scores (see Methods 5.15.5) for all synonymous coding variants in ClinVar that were previously labeled as variants of uncertain significance (VUS), using our best-performing codon foundation models (in zero-shot benchmarks), including EnCodon (1B)Ada and DeCodon (1B)AdaFigure 6a). We then selected four synonymous coding variants with extremely low predicted pathogenicity scores (less than the 5th percentile, i.e., within the highly pathogenic region) and two control variants with extremely high predicted scores (greater than the 95th percentile, i.e., within the highly benign region) for experimental testing (see Methods 5.8). Our experimental results demonstrated significant changes in protein expression levels for two of the four nominated synonymous variants with extreme predicted pathogenic scores by our DeCodon (1B)Ada model: NM_006412.4(AGPAT2):c.702C>T and NM_002872.5(RAC2):c.501C>T (Figure 6b,c and Supplementary Figure 12a). These findings provide compelling evidence not only revealing the post-translational roles of synonymous codon variants but also underscoring the broader significance of these variants in regulating protein expression. Furthermore, this demonstrates the caability of our DeCodon models to capture such nuanced patterns in synonymous codon usage, emphasizing the potential utility of our model in guiding future research and clinical interpretations of synonymous variants, offering new insights into their functional consequences.
Figure 6: Pathogenic synonymous variants predicted by codon foundation models cause notable changes in gene expression levels:
a) We use different scoring fashions for pathogenicity of synonymous variants. Specifically, we report log likelihood ratio between wild-type and mutated codons for EnCodon and log ratio of wild-type and mutated sequence likelihoods for DeCodon models. To nominate variants for experimental validation, We fetched all synonymous variants from ClinVar and COSMIC Census Mutations databases and computed pathogenicity scores for all of our pre-trained EnCodons and DeCodons. Next, we selected synonymous variants with extremely pathogenic scores (above 99th quantile of the distribution). b) Experimental measurement of gene expression levels for AGPAT2 and RAC2 are shown for wild-type vs. mutated coding sequence. Each dot denotes an individual replicates and bars show the average expression across replicates. c) Immunoblots of FLAG-tagged AGPAT2 and RAC2 protein variants expressed in HEK293T cells, showing three biological replicates for both wild-type (Ref) and mutated (Mut) sequences. β-tubulin signal is used as a loading control. d) Geneset Enrichment Analysis of top 20 abundant genes with highest number of “pathogenic” variants with extreme scores. d) Lollipop plot of extreme variants predicted for BCR and MYC are shown across protein sequence annotated by functional domains extracted from InterPro.
2.6.1. Pathogenic synonymous variants identified by cdsFMs are predominantly enriched in cancer-related genes
To expand our dataset of predicted synonymous variants, we scored COSMIC Census Mutation database [75], focusing on those with extreme pathogenic variants predicted by all our CDS foundation models (See Figure 6a). Specifically, we isolated 3365 synonymous coding variants with extremely high pathogenicity scores and performed gene-set enrichment analysis on their associated genes. Given that COSMOS reports on somatic variations in cancer, we expect identifying synonymous mutations that are enriched in known drivers. As expected, the analysis revealed a significant enrichment in cancer-related pathways (Figure 6c), underscoring the potential impact of these synonymous variants on cancer biology.
For instance, BCR and MYC, which are well-established oncogenic drivers across many cancers [10, 24, 28, 31, 74], harbor 64 of these extreme variants. Lollipop plots, annotated with InterPro domains, illustrate the distribution and functional implications of these variants. The observed spatial clustering of these synonymous mutations further highlights their biological function on the expression of these proteins (Figure 6d, Supplementary Figure 14b,c,d,e,f). Using this approach, we have significantly expanded the list of likely pathogenic synonymous mutations in cancer.
Discussion
Synonymous codons are often underappreciated and underexplored. Although the vast majority of synonymous variants are silent, many are likely pathogenic and functional. Similarly, synonymous codons are not interchangeable in synthetic biology and can be used to tune the expression of the same underlying protein. The absence of reliable machine learning models to help elucidate the context-dependent role of codons was the main motivation for building EnCodon and DeCodon models. As we embarked on this, other groups have also released codon-resolution or codon-aware models. CodonBERT, with its ability to handle codon-level inputs, has shown strong potential in certain tasks, particularly those involving sequence-level transformations. cdsBERT, on the other hand, has demonstrated proficiency in capturing protein-level semantic information from coding sequences. However, these models are an order of magnitude smaller than our largest models, are trained on fewer tokens, and are limited to the BERT architecture. As we demonstrated here, the larger models are crucial to capture the context dependence of synonymous codons. More importantly, the DeCodon models perform superior to EnCodon models in a number of key downstream tasks.
In this study, we have demonstrated the versatility and effectiveness of our suite of codon-based foundation models, EnCodon and DeCodon, in comparison to a wide range of genomic language models of various scales (i.e., differing in the number of trainable parameters) across a variety of downstream tasks relevant to coding sequences. Synonymous codons fall in the blind spot of protein language models and as we demonstrated here, genomic foundation models have not capture the codon structure in the coding regions. This is likely because coding sequences are a small minority of sequences that the genomic models are trained on. Here, we showed that codon-resolution large-scale language models extend beyond the basic codon-amino acid associations to capture more intricate codon-codon relationships, which proved highly beneficial for tasks like synonymous variant effect prediction.
In this study, we successfully confirmed two out of four synonymous variants predicted as pathogenic by our EnCodon and DeCodon models. This 50% confirmation rate is likely an underestimate given the variety of ways beyond expression by which synonymous variants may impact protein function, e.g. by impacting the folding dynamics. Our broader data generation effort for measuring the impact of synonymous codons on protein expression, further highlights the importance of capturing the functional consequences synonymous variants. Our results not only highlight the potential of EnCodon and DeCodon in practical genomic applications but also provide additional evidence for the heterogeneous effects of synonymous codons on gene regulation and expression.
In this study, we included both masked and causal language modeling, and found that the latter showed superior performance particularly in downstream few-shot tasks. This suggests that the causal modeling approach of DeCodon offers more flexible and informative context-aware representations. At first glance, this may be counterintuitive as having access to the entirety of the sequence context should provide more information. However, other models, such as Evo [59] or LoRNASH [70], have similarly shown success in learning biological sequences via causal language modeling. The added benefit of these models is that they are generative in nature, and the coding sequences they generate can be studied to further understand the biological concepts the models have internalized.
In conclusion, our suite of codon foundation models, namely EnCodon and DeCodon, provides a powerful toolkit for advancing our understanding of synonymous codons. Given the challenges of generating deep mutational scanning at scale for synonymous codons, these models are essential for better nomination of likely functional variants. This knowledge, and the extent to which it impacts human diseases, will also reshape how we consider therapeutic modalities, as some mutations will not have an impact on protein sequence, yet it can still impact expression level or activity.
5. Methods
5.1. Pretraining data
For pretraining, we aggregated coding sequences (CDS) from all available species in the NCBI Genomes database. We utilized the most recent genomic annotation files (GTF files) available for each organism to ensure accuracy and completeness. The resulting dataset, which we refer to as NCBI CDS data, comprises a total of 60 million sequences from over 5,000 species. This comprehensive collection serves as a robust foundation for training models on a diverse set of genetic sequences.
5.2. Eukaryotic adaptation data
Due to overwhelming representation of bacterial coding sequences in the NCBI CDS dataset, we curated a separate dataset of eukaryotic coding sequences for adaptation of our pre-trained models. The dataset contains 567,281 coding sequences from 227 eukaryotic species, including human, mouse, fruit fly, and zebrafish. This dataset was used to fine-tune the pre-trained models on eukaryotic sequences, enabling them to better capture the nuances of eukaryotic codon usage.
5.3. ClinVar dataset
We obtained the latest variant summary file in TSV format (variant_summary.txt.gz) from the ClinVar database, which initially contained 2,366,650 GRCh38-aligned variants. From this dataset, we extracted coding single-nucleotide variants (SNVs) with a review status of 2+ stars, indicating higher confidence in clinical interpretation. These SNVs were then mapped to the GRCh38 RefSeq reference genome to extract corresponding coding sequences. Two versions of the preprocessed ClinVar dataset were created: v0.1 and v0.2. Version v0.2 includes all variants from v0.1 and additional variants with uncertain clinical significance (VUS). v0.1 was employed for zero-shot benchmarking of language models, while v0.2 was used to identify candidate synonymous variants for experimental validation. Detailed statistics of the dataset versions are provided in Supplementary Table 2.
5.4. MaveDB Collection
We retrieved 3,153 experimental datasets from the MaveDB database using its API, which included their corresponding score set CSV files. To focus on coding sequences, we filtered out experiments lacking nucleotide-level information for the target sequence or its variants. Additionally, we restricted the dataset to single-nucleotide variants (SNVs), resulting in a final collection of 1,911,200 variants across 13 species. The species included Human (n = 519,771), Chicken (n = 122,460), Ocean pout (n = 73,578), House mouse (n = 27,608), E. coli (n = 14,831), Thale cress (n = 14,817), Fruit fly (n = 9,192), Antarctic eel pout (n = 8,229), African clawed frog (n = 4,347), Viviparous blenny (n = 4,089), Pig (n = 3,969), Norway rat (n = 3,768), and Ocean sunfish (n = 3,639).
5.5. Membrane protein abundance and surface expression deep mutational scanning
Deep mutational scanning datasets for membrane proteins were generated as previously reported in more detail within manuscripts [32, 51, 89]. However, in all cases, the oligo-based library generation pipeline, DIMPLE, is used to make variant libraries, and these libraries are BxBI-mediated integrated in stable landing pad cell HEK293T cell lines [54, 55]. These library containing cell lines were into four populations by fluorescent cell sorting based on a massively parallel measure for protein abundance and/or surface expression. For the protein abundance assay, cells containing mutational libraries were sorted based on a split fluorescent protein complementation [53], whereas for surface expression screens cells were sorted based on fluorescent antibodies that recognize extracellular exposed epitopes [16]. Once these cells are separated, DNA is extracted, PCR-amplified, fragmented, and prepared for sequencing by Illumina Nextera kits and sequenced on an Illumina Novaseq 6000 short sequencer. Variant effect scores were generated using Enrich2 [69].
5.6. Open Reading Frame (ORF) Data
We obtained a collection of 7,264 translated open reading frames (ORFs) from the supplementary material provided by [57], which were filtered and derived from Ribo-seq studies. Specifically, we concatenated ORFs from two Excel sheets titled S2. PHASE I Ribo-seq ORFs and S3. Single-study Ribo-seq ORFs. The ORF locations were then mapped to the GRCh38 RefSeq genome to ensure consistency with our other datasets.
5.7. mRNA Stability Dataset
For the mRNA stability downstream benchmark, we utilized a preprocessed collection of mRNA decay rate datasets provided by [2]. This dataset encompasses 39 human and 27 mouse transcriptome-wide studies, collectively containing 26,725 mRNA sequences (12,981 for human and 13,744 for mouse) with reported half-life measurements. This data is crucial for understanding mRNA stability and its implications in gene expression regulation.
5.8. Synonymous protein variants Dataset
5.8.1. Cloning of plasmids expressing synonymous protein variants
For restriction enzyme cloning, custom double-stranded DNA sequences (Twist Biosciences) encoding synonymous protein variants were designed as follows: 5’-AAGCTG-GCTAGC (NheI)-GCCACC (Kozak sequence)-Variable Coding Sequence-GGT GGA GGC GGT AGC (GGGS linker)- GACTACAAGGAC-CACGACGGCGATTATAAGGATCACGACATCGACTACAAAGACGACGATGACAAG (3xFLAG)- XXX (Gene-specific Stop Codon)-GAATTC (EcoRI)-TGCAGA-3’. The exact DNA sequences for every synonymous protein variant used for cloning can be found in Supplemntary Table (other details can be found in Supplementary Table 3). DNA was double-digested with NheI-HF (New England Biolabs, #R3131L) and EcoRI-HF (New England Biolabs, #R3101L) for 30 min at 37°C, and heat-inactivated for 20 min at 80°C. pcDNA3.1 vector (Thermo Fisher Scientific, #V79020) was double-digested with NheI-HF and EcoRI-HF for 30 min at 37°C, dephosphorylated by addition of Quick-CIP (New England Biolabs, #M0525L) (10 min at 37°C), and heat-inactivated for 20 min at 80°C. For ligation in 1x rCutSmart™ Buffer (New England Biolabs, #B6004S) supplemented with 1 mM ATP, 50 ng crude digested pcDNA3.1 vector was combined with a 7-times molar excess of crude digested insert and incubated for 30 min at room temperature after addition of 1 μ L Quick Ligase (New England Biolabs, #M2200). NEB stable competent E. coli (New England Biolabs, #C3040H) were transformed according to manufacturer’s instructions. Bacteria were grown on LB-agar plates supplemented with 0.1 mg/mL carbenicillin overnight at 37°C. LB supplemented with 0.1 mg/mL ampicillin was inoculated with single colonies and incubated in a shaking incubator at 37°C overnight. Plasmid DNA was extracted using ZymoPURE™ Plasmid Miniprep Kit (Zymo Research, #D4210) and eluted in nuclease-free water. The plasmid DNA concentration was measured using a NanoDrop™ One spectrophotometer (Invitrogen) by measuring the absorbance at 260 nm, and the plasmids were fully sequenced by Nanopore sequencing (Quintara Biosciences).
5.8.2. Western blotting of synonymous protein variants
HEK293T cells (ATCC, #CRL-3216) were maintained in DMEM (Gibco, #11965-092) supplemented with 10% FBS (Avantor, #97068-085), under standard tissue culture conditions (37°C, 5% CO2) and were passaged every 2–3 days. To express synonymous protein variants, HEK293T cells were seeded in a 24-well format (75,000 cells per well), and after 24 hours transfected with 0.5 μ g variant-encoding plasmid using TransIT transfection reagent (Mirus Bio, #MIR6606). 24 hours after transfection, the cell culture media was changed, and cells were harvested after incubating for another 24 hours. Cells were lysed in RIPA buffer (Thermo Fisher Scientific, #89901) supplemented with 1x Halt protease inhibitor cocktail (Thermo Fisher Scientific, #78425) by shaking for 30 min at 4°C. After centrifugation for 30 min (4°C) at 14,000 x g, the supernatant was collected and the protein concentration was determined by BCA assay (Thermo Fisher Scientific, #23225). Protein samples were stored at −20°C. After denaturing samples for 5 min at 95°C in 1x NuPAGE LDS loading buffer (Invitrogen, #NP0007) supplemented with 50 mM DTT, 5 μ g of total protein per sample and a pre-stained protein ladder (LI-COR Biosciences, #928-60000) were separated on a NuPage Bis-Tris Mini Protein Gel, 4–12% (Invitrogen, #NP0322BOX) for 1 hour at 120 V in 1x NuPAGE™ MES SDS running buffer (Thermo Fisher Scientific, #NP0002). Protein bands were transferred onto a PVDF membrane (Invitrogen, #IB34001) using the iBlot3 Western Blot Transfer System (Invitrogen). After blocking the membrane in SuperBlock blocking buffer (Thermo Fisher Scientific, #37537) for 10 min at room temperature, it was incubated overnight at 4°C with rabbit anti-FLAG monoclonal antibody (Cell Signaling, #14793) and mouse anti-β-tubulin monoclonal antibody (Sino Biological, #100109-MM05T), both diluted 1:2,000 in blocking buffer. The membrane was washed three times for 10 min each in 1x TBS (Thermo Fisher Scientific, #J62938.K3-T) supplemented with 0.1% (v/v) Tween-20 (TBS-T), followed by incubation at room temperature for 30 min with anti-rabbit IgG DyLight 800 4X PEG conjugate (Cell Signaling, #5151) and anti-mouse IgG DyLight 680 conjugate (Cell Signaling, #5470), both diluted 1:5,000 in blocking buffer. Membranes were washed two times for 5 min in TBS-T, followed by a 5 min wash in TBS without Tween-20, and the fluorescence signal was recorded on an Odyssey DLx Imager (LI-COR) and analyzed in Image Studio (LI-COR Biosciences, version 5.2.5). Protein expression levels were calculated by normalizing the anti-FLAG signal (800 nm channel) to the corresponding anti-β-tubulin signal (700 nm channel).
5.9. COSMIC Census Mutations Database
We acquired the Mutant Census v100 file from the COSMIC database [75], which details coding mutations in genes listed in the Cancer Gene Census. The raw dataset comprised 1,949,478 coding mutations. We filtered the data to retain only single-nucleotide variants (SNVs) and mapped their locations to the GRCh38 RefSeq genome. The final processed dataset included 244,400 coding variants, forming a key resource for studying mutational impacts in cancer.
5.10. Foundation Models
5.10.1. EnCodon
Architecture
The architecture of EnCodon is inspired by the RoFormer as described in [76] (see 5.11). It is fundamentally based on the transformer encoder model, which incorporates multiple layers of self-attention mechanisms followed by position-wise feed-forward networks but similar to RoFormer, we use rotary positional encoding (RoPE) instead of absolute or relative PE approaches. EnCodon is a large language model designed to provide contextual codon-level hidden representations (i.e., where ) for any given input coding sequence (i.e., ) of length L.
The EnCodon model comprises three primary components: a learnable vocabulary embedding layer, a stack of transformer encoder blocks, and a language modeling head. The input coding sequence x is represented as a vector of L token IDs—integers that represent either a codon or a special token. The EnCodon vocabulary includes 69 tokens: 64 codons and 5 special tokens, namely [CLS], [SEP], [PAD], [MASK], and [UNK]. [CLS] and [SEP] are control tokens prepended and appended to each input coding sequence, marking the start and end of the sequence, respectively. The [PAD] token is used for padding during batched training or inference, [UNK] is used for unknown codons, and [MASK] is used for masking during training. It is important to note that the model is not provided with any information regarding the nucleotide composition of the codons; the only input information consists of token IDs. Consequently, the model does not have prior knowledge of the nucleotide differences between codons during training. Mathematically, EnCodon model takes the tokenized input sequence xi, which contains L token IDs:
| (1) |
The embedding layer processes x to produce the embedding tensor E, defined as:
| (2) |
Subsequently, the embedding tensor E is passed through multiple transformer encoder blocks, which follow the architecture described in [76]. Each block consists of a rotary attention layer, which applies rotary positional encoding (see 5.11) followed by a self-attention operation (see 5.10.3), and a position-wise feed-forward network. We employed the “Sub-LayerNorm” approach proposed by [86] to enhance expressivity and utilized a scalable, theoretically-derived initialization strategy. Detailed hyperparameters for each pre-trained EnCodon model are provided in Supplementary Table 1.
Pretraining Procedure
For pretraining, EnCodon uses the Masked Language Modeling (MLM) objective, defined as:
where M represents the set of masked positions, xi is the true token at position i, and denotes the input sequence with tokens at positions in M masked. This loss function encourages the model to accurately predict the original tokens based on the surrounding context. The EnCodon foundation models were pretrained on 2 NVIDIA H100 GPUs for 2 weeks. The models were implemented using PyTorch [63] and the HuggingFace [87] framework was used for pretraining.
5.10.2. DeCodon
Architecture
DeCodon is a controllable generative language model specifically designed for the codon-level generation of coding sequences. Similar to EnCodon, the architecture of DeCodon is similar to the RoFormer [76] but also utilizing Extrapolatable Position Embedding (XPOS) [77] in rotary attention layers (See 3a). XPOS has recently shown significant improvement in the generalization of mega-scale language models like PaLM[15] on very long sequences without sacrificing models’ performance on shorter sequences. We used sequences’ organism as the conditional information for DeCodon making the generation controllable. Similar to EnCodon, DeCodon operates on input sequences of codons, but it is optimized for sequence generation tasks rather than contextual encoding.
The DeCodon model consists of three main components: a learnable vocabulary embedding layer, a stack of transformer decoder blocks, and a sequence generation head. The input to DeCodon is a sequence of L token IDs, where each ID corresponds to a codon, specie, or a special token from a vocabulary that includes the 5124 tokens: 64 codons, 5 special tokens, and 5055 organism tokens. The model is autoregressive, meaning that it generates each token in the sequence one at a time, conditioned on the previously generated tokens in addition to the organism of interest.
Given an input sequence of length L:
| (3) |
The embedding layer converts this input sequence into an embedding tensor E, defined as:
| (4) |
This embedding tensor is then passed through a series of transformer decoder blocks. Each block consists of a masked self-attention layer, where attention is only computed over previous tokens in the sequence, followed by a position-wise feed-forward network. The use of rotary positional encoding (see 5.11) allows the model to effectively capture the sequential dependencies between codons. As in EnCodon, we employed the “Sub-LayerNorm” approach [86] for improved expressivity and model stability. The final output is produced by the sequence generation head, which predicts the next token in the sequence based on the transformer outputs.
Training Procedure
DeCodon model is trained using a standard causal (i.e. autoregressive) language modeling (CLM) objective, which aims to maximize the likelihood of the target sequence given the input sequence:
where denotes the sequence of tokens preceding the i-th token. s also denotes the sequence’s specie. This loss encourages the model to generate codon sequences that are contextually and sequentially accurate. The DeCodon model was pretrained on the same dataset and hardware as EnCodon, utilizing PyTorch [63] and the HuggingFace [87] framework.
5.10.3. Self-Attention Operation
The self-attention mechanism, proposed in [85], is a data-driven operation that quantifies the information flow between all possible pairs of tokens in an input sequence. This mechanism forms as the core component of the Transformer architecture.
Mathematically, let be the input sequence of N token representations, where each . The self-attention operator first transforms the input token representations into three different representations called query, key, and value:
| (5) |
| (6) |
| (7) |
where are learned parameter matrices.
The attention scores are then computed as:
| (8) |
where represent the query, key, and value matrices derived from the input representations. The scaling factor is introduced to mitigate the effect of large dot products in high-dimensional spaces.
This mechanism allows the model to weigh the importance of different tokens within the input sequence, regardless of their positions. The output of the self-attention operation is a weighted sum of the value vectors, where the weights are determined by the compatibility between the query and key vectors.
In practice, multi-head attention is often employed, which involves applying multiple sets of query, key, and value projections in parallel:
| (9) |
where each head is computed as:
| (10) |
Here, and are learned parameters, and h is the number of attention heads.
The self-attention mechanism enables the model to capture complex dependencies and relationships within the input sequence, contributing significantly to the success of Transformer-based models in various natural language processing tasks.
5.11. Rotary Positional Encoding (RoPE)
Self-attention is a position-invariant operation, suggesting the need to encode positional information explicitly. Several approaches have been proposed to incorporate positional information in self-attention which can be categorized into absolute, relative, and hybrid PE methods. Rotary Positional Encodings (RoPE), recently proposed by [76], is a positional encoding (PE) approach that is a combination of relative and absolute PE approaches. RoPE has showed consistent improvement in various natural language understanding and computer vision tasks making it a common practical choice for positional information encoding which motivated us to use them in EnCodon and DeCodon architectures.
5.12. Baselines
5.12.1. NucCNN
NucCNN is a convolutional neural network (CNN) model with a nucleotide vocabulary embeddings. We used 5 convolutional layers with kernel sizes of 3, 5, 7, 9, and 11, each followed by a max-pooling layer. The model was trained using the same dataset as EnCodon and DeCodon in the benchmarked tasks, with the same hyperparameters. The model was implemented using PyTorch and the HuggingFace framework.
5.12.2. CodonCNN
CodonCNN is almost identical to NucCNN, but it uses a codon vocabulary instead of nucleotides. The model was trained using the same dataset as EnCodon and DeCodon in the benchmarked tasks, with the same hyperparameters. The model was implemented using PyTorch and the HuggingFace framework.
5.13. Sequence Embeddings
The proposed EnCodon and DeCodon models generate codon-level representations, where each codon in the input sequence is represented by a d-dimensional vector. For ‘sequence embedding,’ we calculate the average of all codon-level embeddings in the EnCodon models, while for the DeCodon models, we use the representation of the last codon in the sequence.
5.14. Fine-tuning scheme for synonymous variant prediction
To fine-tune the pre-trained EnCodon language models for the synonymous variant prediction task, we reused the pre-trained language model head without adding any new layers or parameters. Specifically, for each synonymous variant, EnCodon takes the wild-type coding sequence as input and computes the log-likelihood ratio between the wild-type and mutated codon at the variant position (See Figure 5a). This log-likelihood ratio is used as the predicted score for the variant. The model was fine-tuned using the Huber loss function, with a learning rate of 1e-5, over 5 epochs.
5.15. Metrics
5.15.1. KNN Purity
KNN purity evaluates clustering quality by measuring the homogeneity of KNN clusters with respect to their ground-truth labels. It uses the K-nearest neighbors algorithm to assess how well cluster points match their true labels. The KNN purity score is mathematically defined as P:
| (11) |
where is the set of unique labels, Cl is the set of points with label l, and K is the number of nearest neighbors. The score ranges from 0 to 1; a higher score indicates more homogeneity. A score of 1 means perfect clustering (all points in a cluster share the same label), while a lower score indicates greater heterogeneity.
5.15.2. Spearman correlation
Spearman’s rank correlation coefficient is a non-parametric measure that assesses the strength and direction of a monotonic relationship between two variables. Mathematically, for two variables X and Y, the Spearman correlation coefficient, ρ, is given by the formula:
| (12) |
where is the difference between the ranks of each pairs of values and n is the number of data points. It is important to mention that this correlation is robust to outliers and skewed data, making it suitable for datasets where traditional parametric assumptions do not hold.
5.15.3. Pearson correlation
Pearson correlation coefficient, r, is a measure of linear correlation between two random variants X and Y:
| (13) |
where Xi, and Yi are individual data points and and are sample averages of samples drawn from X and Y, respectively. The correlation ranges from −1 (negative linear relationship) to +1 (positive linear relationship) and 0 indicates no linear relationship.
5.15.4. Synonymous Codon Confusion
Synonymous Codon Confusion is a amino acid level metric, measuring the how often a langauge model incorrectly choose synonymous codon over each other. The metric can be computed directly from the confusion matrix of a codon-level language model. Mathematically speaking:
| (14) |
where is the row-normalized confusion matrix of a codon-level lagnuage model.
5.15.5. variant effect predicted score (VEP)
Focusing on the language models zero-shot applicability on variant effect prediction tasks, we used different formulations for encoder and decoder langauge models. Specifically, concerning encoder models, we computed the masked lanuage modeling (MLM) pseudolikelihood ratio between wild-type and mutated codon/nucleotide/token at the position of mutation. Mathematically speaking, we formulate VEP as follows:
| (15) |
where and are wild-type and mutated coding sequences, respectively. k indicates the variant’s token-level position in the coding sequence depending on the tokenization used for the encoder language model. For example, we used variant’s codon position to compute the log-likelihood ratio (i.e. VEP) score with our EnCodons.
Concerning decoder language models, we reported the difference between sequence likelihood scores (SL) computed for the wild-type and mutated coding sequences. In constrast to encoder language models, we don’t take the variant’s location information into account for computing the variant effect predicted (VEP) score:
| (16) |
where xi denotes the i-th position in the input sequence of length L and p(.) is the output probability at the i-th position computed by the decoder lanaguge model. Finally, we reported the difference between the two computed sequence likelihoods (SL) as the predicted score:
| (17) |
5.16. Spatial Clustering Analysis Using Ripley’s K Functions (L)
To investigate the spatial distribution of extreme variants within the 3D structure of various proteins, we applied Ripley’s K function, a statistical method designed to detect clustering or dispersion of points in a spatial domain [67]. Ripley’s K function in three dimensions is defined as:
where λ represents the density of points, r is the distance threshold, and E[⋅] denotes the expectation.
For each protein in the experiment, we performed a radius range r from 0 to 50 Å with 100 discrete intervals. The statistical significance of clustering was assessed using a permutation test with 10,000 iterations. Specifically, we generated 10,000 sets of randomly chosen variants (Vr) as the “null distribution” with the same number of points as the observed “critical” synonymous variants (Vc) for the protein. Then, we first extracted the Cα atom coordinates of the amino acids of each set of points from the protein PDB structure using the Bio.PDB module in Python. Next, we computed pairwise Euclidean distances between the Cα atoms and calculated normalized Ripley’s K function at various radius r using:
where n is the total number of points, and I(⋅) is an indicator function that equals 1 if the condition inside is true. We calculated Ripley’s K function at 10000 different distances ranging from 0 to 100Å. Finally, The observed K values were compared against the null distribution to compute p-values, representing the probability that the observed clustering occurred by chance:
| (18) |
Supplementary Material
4. Acknowledgments
We thank Brian Plosky and Chiara Ricci-Tam for their valuable comments and insights during the preparation of this manuscript. HG is an Arc Core Investigator and this study was in part supported by the Arc Institute. WCM acknowledges support from a Howard Hughes Medical Institute Hanna Gray Fellowship, and UCSF Quantitative Biosciences Institute Fellowship, and is a Chan Zuckerberg Biohub San Francisco Investigator. MKH and CBM are supported by NIGMS 5T32GM139786 and 1F32GM152977.
Footnotes
3 Code Availability
Code and models are accessible at https://github.com/goodarzilab/cdsFM. Additionally, pre-trained models have been made available on HuggingFace [87].
References
- [1].Abramson Josh, Adler Jonas, Dunger Jack, Evans Richard, Green Tim, Pritzel Alexander, Ronneberger Olaf, Willmore Lindsay, Ballard Andrew J, Bambrick Joshua, Bodenstein Sebastian W, Evans David A, Hung Chia-Chun, O’Neill Michael, Reiman David, Tunyasuvunakool Kathryn, Wu Zachary, Žemgulytė Akvilė, Arvaniti Eirini, Beattie Charles, Bertolli Ottavia, Bridgland Alex, Cherepanov Alexey, Congreve Miles, Cowen-Rivers Alexander I, Cowie Andrew, Figurnov Michael, Fuchs Fabian B, Gladman Hannah, Jain Rishub, Khan Yousuf A, Low Caroline M R, Perlin Kuba, Potapenko Anna, Savy Pascal, Singh Sukhdeep, Stecula Adrian, Thillaisundaram Ashok, Tong Catherine, Yakneen Sergei, Zhong Ellen D, Zielinski Michal, Žídek Augustin, Bapst Victor, Kohli Pushmeet, Jaderberg Max, Hassabis Demis, and Jumper John M. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature, 630(8016):493–500, June 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Agarwal Vikram and Kelley David R.. The genetic and biochemical determinants of mrna degradation rates in mammals. Genome Biology, 23(1), November 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Agashe Deepa, Martinez-Gomez N. Cecilia, Drummond D. Allan, and Marx Christopher J.. Good codons, bad transcript: Large reductions in gene expression and fitness arising from synonymous mutations in a key enzyme. Molecular Biology and Evolution, 30(3):549–560, December 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Avsec Žiga, Agarwal Vikram, Visentin Daniel, Ledsam Joseph R, Grabska-Barwinska Agnieszka, Taylor Kyle R, Assael Yannis, Jumper John, Kohli Pushmeet, and Kelley David R. Effective gene expression prediction from sequence by integrating long-range interactions. Nat. Methods, 18(10):1196–1203, October 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Calviello Lorenzo, Mukherjee Neelanjan, Wyler Emanuel, Zauber Henrik, Hirsekorn Antje, Selbach Matthias, Landthaler Markus, Obermayer Benedikt, and Ohler Uwe. Detecting actively translated open reading frames in ribosome profiling data. Nature Methods, 13(2):165–170, December 2015. [DOI] [PubMed] [Google Scholar]
- [6].Camiolo Salvatore, Farina Lorenzo, and Porceddu Andrea. The relation of codon bias to tissue-specific gene expression inarabidopsis thaliana. Genetics, 192(2):641–649, October 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Carlini David B.. Experimental reduction of codon bias in the drosophila alcohol dehydrogenase gene results in decreased ethanol tolerance of adult flies. Journal of Evolutionary Biology, 17(4):779–785, July 2004. [DOI] [PubMed] [Google Scholar]
- [8].Carlini David B and Stephan Wolfgang. In vivointroduction of unpreferred synonymous codons into the drosophilaadhgene results in reduced levels of adh protein. Genetics, 163(1):239–243, January 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chaney Julie L and Clark Patricia L. Roles for synonymous codon usage in protein biogenesis. Annu. Rev. Biophys., 44(1):143–166, February 2015. [DOI] [PubMed] [Google Scholar]
- [10].Chen Hui, Liu Hudan, and Qing Guoliang. Targeting oncogenic myc as a strategy for cancer treatment. Signal Transduction and Targeted Therapy, 3(1), February 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chen Jiayang, Hu Zhihang, Sun Siqi, Tan Qingxiong, Wang Yixuan, Yu Qinze, Zong Licheng, Hong Liang, Xiao Jin, King Irwin, et al. Interpretable rna foundation model from unannotated data for highly accurate rna structure and function predictions. arXiv preprint arXiv:2204.00300, 2022. [Google Scholar]
- [12].Chen Jin, Brunner Andreas-David, Cogan J. Zachery, Nuñez James K., Fields Alexander P., Adamson Britt, Itzhak Daniel N., Li Jason Y., Mann Matthias, Leonetti Manuel D., and Weissman Jonathan S.. Pervasive functional translation of noncanonical human open reading frames. Science, 367(6482):1140–1146, March 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chen Siyu, Li Ke, Cao Wenqing, Wang Jia, Zhao Tong, Huan Qing, Yang Yu-Fei, Wu Shaohuan, and Qian Wenfeng. Codon-resolution analysis reveals a direct and context-dependent impact of individual synonymous mutations on mrna level. Molecular Biology and Evolution, 34(11):2944–2958, August 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cheng Jun, Novati Guido, Pan Joshua, Bycroft Clare, Žemgulytė Akvilė, Applebaum Taylor, Pritzel Alexander, Wong Lai Hong, Zielinski Michal, Sargeant Tobias, Schneider Rosalia G., Senior Andrew W., Jumper John, Hassabis Demis, Kohli Pushmeet, and Avsec Žiga. Accurate proteome-wide missense variant effect prediction with alphamissense. Science, 381(6664), September 2023. [DOI] [PubMed] [Google Scholar]
- [15].Chowdhery Aakanksha, Narang Sharan, Devlin Jacob, Bosma Maarten, Mishra Gaurav, Roberts Adam, Barham Paul, Chung Hyung Won, Sutton Charles, Gehrmann Sebastian, Schuh Parker, Shi Kensen, Tsvyashchenko Sasha, Maynez Joshua, Rao Abhishek, Barnes Parker, Tay Yi, Shazeer Noam, Prabhakaran Vinodkumar, Reif Emily, Du Nan, Hutchinson Ben, Pope Reiner, Bradbury James, Austin Jacob, Isard Michael, Gur-Ari Guy, Yin Pengcheng, Duke Toju, Levskaya Anselm, Ghemawat Sanjay, Dev Sunipa, Michalewski Henryk, Garcia Xavier, Misra Vedant, Robinson Kevin, Fedus Liam, Zhou Denny, Ippolito Daphne, Luan David, Lim Hyeontaek, Zoph Barret, Spiridonov Alexander, Sepassi Ryan, Dohan David, Agrawal Shivani, Omernick Mark, Dai Andrew M., Pillai Thanumalayan Sankaranarayana, Pellat Marie, Lewkowycz Aitor, Moreira Erica, Child Rewon, Polozov Oleksandr, Lee Katherine, Zhou Zongwei, Wang Xuezhi, Saeta Brennan, Diaz Mark, Firat Orhan, Catasta Michele, Wei Jason, Meier-Hellstern Kathy, Eck Douglas, Dean Jeff, Petrov Slav, and Fiedel Noah. Palm: Scaling language modeling with pathways, 2022. [Google Scholar]
- [16].Coyote-Maestas Willow, He Yungui, Myers Chad L., and Schmidt Daniel. Domain insertion permissibility-guided engineering of allostery in ion channels. Nature Communications, 10(1), January 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dalla-Torre Hugo, Gonzalez Liam, Mendoza-Revilla Javier, Carranza Nicolas Lopez, Grzywaczewski Adam Henryk, Oteri Francesco, Dallago Christian, Trop Evan, de Almeida Bernardo P., Sirelkhatim Hassan, Richard Guillaume, Skwark Marcin, Beguir Karim, Lopez Marie, and Pierrot Thomas. The nucleotide transformer: Building and evaluating robust foundation models for human genomics. bioRxiv, 2023. [DOI] [PubMed] [Google Scholar]
- [18].Dalla-Torre Hugo, Gonzalez Liam, Revilla Javier Mendoza, Carranza Nicolas Lopez, Grywaczewski Adam Henryk, Oteri Francesco, Dallago Christian, Trop Evan, Sirelkhatim Hassan, Richard Guillaume, et al. The nucleotide transformer: Building and evaluating robust foundation models for human genomics. bioRxiv, pages 2023–01, 2023. [DOI] [PubMed] [Google Scholar]
- [19].Dhindsa Ryan S., Wang Quanli, Vitsios Dimitrios, Burren Oliver S., Hu Fengyuan, DiCarlo James E., Kruglyak Leonid, MacArthur Daniel G., Hurles Matthew E., and Petrovski Slavé. A minimal role for synonymous variation in human disease. The American Journal of Human Genetics, 109(12):2105–2109, December 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dittmar Kimberly A., Goodenbour Jeffrey M., and Pan Tao. Tissue-specific differences in human transfer rna expression. PLoS Genetics, 2(12):e221, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Esposito Daniel, Weile Jochen, Shendure Jay, Starita Lea M., Papenfuss Anthony T., Roth Frederick P., Fowler Douglas M., and Rubin Alan F.. Mavedb: an open-source platform to distribute and interpret data from multiplexed assays of variant effect. Genome Biology, 20(1), November 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Frazer Jonathan, Notin Pascal, Dias Mafalda, Gomez Aidan, Min Joseph K., Brock Kelly, Gal Yarin, and Marks Debora S.. Disease variant prediction with deep generative models of evolutionary data. Nature, 599(7883):91–95, October 2021. [DOI] [PubMed] [Google Scholar]
- [23].Freeland S. J. and Hurst L. D.. Load minimization of the genetic code: history does not explain the pattern. Proceedings of the Royal Society of London. Series B: Biological Sciences, 265(1410):2111–2119, November 1998. [Google Scholar]
- [24].Gabay M., Li Y., and Felsher D. W.. Myc activation is a hallmark of cancer initiation and maintenance. Cold Spring Harbor Perspectives in Medicine, 4(6):a014241–a014241, June 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].GOODARZI H, NAJAFABADI H, NEJAD H, and TORABI N. The impact of including trna content on the optimality of the genetic code. Bulletin of Mathematical Biology, 67(6):1355–1368, November 2005. [DOI] [PubMed] [Google Scholar]
- [26].Goodarzi Hani, Nguyen Hoang C.B., Zhang Steven, Dill Brian D., Molina Henrik, and Tavazoie Sohail F.. Modulated expression of specific trnas drives gene expression and cancer progression. Cell, 165(6):1416–1427, June 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Goswami Srijib, Gong Li, Giacomini Kathleen, Altman Russ B., and Klein Teri E.. Pharmgkb summary: very important pharmacogene information for slc22a1. Pharmacogenetics and Genomics, 24(6):324–328, June 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].GROFFEN J, STEPHENSON J, HEISTERKAMP N, DEKLEIN A, BARTRAM C, and GROSVELD G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell, 36(1):93–99, January 1984. [DOI] [PubMed] [Google Scholar]
- [29].Hallee Logan, Rafailidis Nikolaos, and Gleghorn Jason P.. cdsbert - extending protein language models with codon awareness. bioRxiv, 2023. [Google Scholar]
- [30].Hayes Thomas, Rao Roshan, Akin Halil, Sofroniew Nicholas J., Oktay Deniz, Lin Zeming, Verkuil Robert, Tran Vincent Q., Deaton Jonathan, Wiggert Marius, Badkundri Rohil, Shafkat Irhum, Gong Jun, Derry Alexander, Molina Raul S., Thomas Neil, Khan Yousuf, Mishra Chetan, Kim Carolyn, Bartie Liam J., Nemeth Matthew, Hsu Patrick D., Sercu Tom, Candido Salvatore, and Rives Alexander. Simulating 500 million years of evolution with a language model. bioRxiv, 2024. [Google Scholar]
- [31].Heisterkamp Nora, Stam Kees, Groffen John, de Klein Annelies, and Grosveld Gerard. Structural organization of the bcr gene and its role in the ph translocation. Nature, 315(6022):758–761, June 1985. [DOI] [PubMed] [Google Scholar]
- [32].Howard Matthew K., Hoppe Nicholas, Huang Xi-Ping, Macdonald Christian B., Mehrota Eshan, Grimes Patrick Rockefeller, Zahm Adam, Trinidad Donovan D., English Justin, Coyote-Maestas Willow, and Manglik Aashish. Molecular basis of proton-sensing by g protein-coupled receptors. April 2024. [DOI] [PubMed] [Google Scholar]
- [33].Hsu Chloe, Verkuil Robert, Liu Jason, Lin Zeming, Hie Brian, Sercu Tom, Lerer Adam, and Rives Alexander. Learning inverse folding from millions of predicted structures. ICML, 2022. [Google Scholar]
- [34].Hu Edward J., Shen Yelong, Wallis Phillip, Allen-Zhu Zeyuan, Li Yuanzhi, Wang Shean, Wang Lu, and Chen Weizhu. Lora: Low-rank adaptation of large language models, 2021. [Google Scholar]
- [35].Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Molecular Biology and Evolution, 2(1):13–34, 01 1985. [DOI] [PubMed] [Google Scholar]
- [36].Ilzhöfer Dagmar, Heinzinger Michael, and Rost Burkhard. Seth predicts nuances of residue disorder from protein embeddings. Frontiers in Bioinformatics, 2, October 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Indriani Fatma, Mahmudah Kunti Robiatul, Purnama Bedy, and Satou Kenji. Prottrans-glutar: Incorporating features from pre-trained transformer-based models for predicting glutarylation sites. Frontiers in Genetics, 13, May 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jagota Milind, Ye Chengzhong, Albors Carlos, Rastogi Ruchir, Koehl Antoine, Ioannidis Nilah, and Song Yun S.. Cross-protein transfer learning substantially improves disease variant prediction. Genome Biology, 24(1), August 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ji Yanrong, Zhou Zhihan, Liu Han, and Davuluri Ramana V. DNABERT: pre-trained Bidirectional Encoder Representations from Transformers model for DNA-language in genome. Bioinformatics, 37(15):2112–2120, 02 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ji Zhe, Song Ruisheng, Regev Aviv, and Struhl Kevin. Many lncrnas, 5’utrs, and pseudogenes are translated and some are likely to express functional proteins. eLife, 4, December 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Jumper John, Evans Richard, Pritzel Alexander, Green Tim, Figurnov Michael, Ronneberger Olaf, Tunyasuvunakool Kathryn, Bates Russ, Žídek Augustin, Potapenko Anna, Bridgland Alex, Meyer Clemens, Kohl Simon A A, Ballard Andrew J, Cowie Andrew, Romera-Paredes Bernardino, Nikolov Stanislav, Jain Rishub, Adler Jonas, Back Trevor, Petersen Stig, Reiman David, Clancy Ellen, Zielinski Michal, Steinegger Martin, Pacholska Michalina, Berghammer Tamas, Bodenstein Sebastian, Silver David, Vinyals Oriol, Senior Andrew W, Kavukcuoglu Koray, Kohli Pushmeet, and Hassabis Demis. Highly accurate protein structure prediction with AlphaFold. Nature, 596(7873):583–589, August 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Khan Zia, Jung Min, Crow Megan, Mohindra Rajat, Maiya Vidya, Kaminker Joshua S., Hackos David H., Chandler G. Scott, McCarthy Mark I., and Bhangale Tushar. Whole genome sequencing across clinical trials identifies rare coding variants in gpr68 associated with chemotherapy-induced peripheral neuropathy. Genome Medicine, 15(1), June 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Komar A A. Synonymous codon usage—a guide for co-translational protein folding in the cell. Mol. Biol., 53(6):777–790, November 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Li Sizhen, Moayedpour Saeed, Li Ruijiang, Bailey Michael, Riahi Saleh, Miladi Milad, Miner Jacob, Zheng Dinghai, Wang Jun, Balsubramani Akshay, Tran Khang, Zacharia Minnie, Wu Monica, Gu Xiaobo, Clinton Ryan, Asquith Carla, Skalesk Joseph, Boeglin Lianne, Chivukula Sudha, Dias Anusha, Montoya Fernando Ulloa, Agarwal Vikram, Bar-Joseph Ziv, and Jager Sven. Codonbert: Large language models for mrna design and optimization. bioRxiv, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lin Brian C., Kaissarian Nayiri M., and Kimchi-Sarfaty Chava. Implementing computational methods in tandem with synonymous gene recoding for therapeutic development. Trends in Pharmacological Sciences, 44(2):73–84, February 2023. [DOI] [PubMed] [Google Scholar]
- [46].Lin Zeming, Akin Halil, Rao Roshan, Hie Brian, Zhu Zhongkai, Lu Wenting, Smetanin Nikita, Costa Allan dos Santos, Fazel-Zarandi Maryam, Sercu Tom, Candido Sal, et al. Language models of protein sequences at the scale of evolution enable accurate structure prediction. bioRxiv, 2022. [Google Scholar]
- [47].Littmann Maria, Heinzinger Michael, Dallago Christian, Olenyi Tobias, and Rost Burkhard. Embeddings from deep learning transfer go annotations beyond homology. Scientific Reports, 11(1), January 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Littmann Maria, Heinzinger Michael, Dallago Christian, Weissenow Konstantin, and Rost Burkhard. Protein embeddings and deep learning predict binding residues for various ligand classes. Scientific Reports, 11(1), December 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Liu Yi. A code within the genetic code: codon usage regulates co-translational protein folding. Cell Commun. Signal., 18(1):145, September 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Liu Yi. A code within the genetic code: codon usage regulates co-translational protein folding. Cell Commun. Signal., 18(1):145, September 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Macdonald Christian B., Nedrud David, Grimes Patrick Rockefeller, Trinidad Donovan, Fraser James S., and Coyote-Maestas Willow. Dimple: deep insertion, deletion, and missense mutation libraries for exploring protein variation in evolution, disease, and biology. Genome Biology, 24(1), February 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Martinez Thomas F., Chu Qian, Donaldson Cynthia, Tan Dan, Shokhirev Maxim N., and Saghatelian Alan. Accurate annotation of human protein-coding small open reading frames. Nature Chemical Biology, 16(4):458–468, December 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Matreyek Kenneth A., Starita Lea M., Stephany Jason J., Martin Beth, Chiasson Melissa A., Gray Vanessa E., Kircher Martin, Khechaduri Arineh, Dines Jennifer N., Hause Ronald J., Bhatia Smita, Evans William E., Relling Mary V., Yang Wenjian, Shendure Jay, and Fowler Douglas M.. Multiplex assessment of protein variant abundance by massively parallel sequencing. Nature Genetics, 50(6):874–882, May 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Matreyek Kenneth A, Stephany Jason J, Chiasson Melissa A, Hasle Nicholas, and Fowler Douglas M. An improved platform for functional assessment of large protein libraries in mammalian cells. Nucleic Acids Research, October 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Matreyek Kenneth A., Stephany Jason J., and Fowler Douglas M.. A platform for functional assessment of large variant libraries in mammalian cells. Nucleic Acids Research, 45(11):e102–e102, March 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Meier Joshua, Rao Roshan, Verkuil Robert, Liu Jason, Sercu Tom, and Rives Alexander. Language models enable zero-shot prediction of the effects of mutations on protein function. bioRxiv, 2021. [Google Scholar]
- [57].Mudge Jonathan M, Ruiz-Orera Jorge, Prensner John R, Brunet Marie A, Calvet Ferriol, Jungreis Irwin, Gonzalez Jose Manuel, Magrane Michele, Martinez Thomas F, Schulz Jana Felicitas, Yang Yucheng T, Albà M Mar, Aspden Julie L, Baranov Pavel V, Bazzini Ariel A, Bruford Elspeth, Martin Maria Jesus, Calviello Lorenzo, Carvunis Anne-Ruxandra, Chen Jin, Couso Juan Pablo, Deutsch Eric W, Flicek Paul, Frankish Adam, Gerstein Mark, Hubner Norbert, Ingolia Nicholas T, Kellis Manolis, Menschaert Gerben, Moritz Robert L, Ohler Uwe, Roucou Xavier, Saghatelian Alan, Weissman Jonathan S, and van Heesch Sebastiaan. Standardized annotation of translated open reading frames. Nat. Biotechnol., 40(7):994–999, July 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Najafabadi Hamed Shateri, Goodarzi Hani, and Torabi Noorossadat. Optimality of codon usage in escherichia coli due to load minimization. Journal of Theoretical Biology, 237(2):203–209, November 2005. [DOI] [PubMed] [Google Scholar]
- [59].Nguyen Eric, Poli Michael, Durrant Matthew G, Thomas Armin W, Kang Brian, Sullivan Jeremy, Ng Madelena Y, Lewis Ashley, Patel Aman, Lou Aaron, Ermon Stefano, Baccus Stephen A, Hernandez-Boussard Tina, Re Christopher, Hsu Patrick D, and Hie Brian L. Sequence modeling and design from molecular to genome scale with evo. February 2024. [DOI] [PubMed] [Google Scholar]
- [60].Nguyen Eric, Poli Michael, Faizi Marjan, Thomas Armin, Birch-Sykes Callum, Wornow Michael, Patel Aman, Rabideau Clayton, Massaroli Stefano, Bengio Yoshua, Ermon Stefano, Baccus Stephen A., and Ré Chris. Hyenadna: Long-range genomic sequence modeling at single nucleotide resolution, 2023. [Google Scholar]
- [61].Nieuwkoop Thijs, Terlouw Barbara R, Stevens Katherine G, Scheltema Richard A, de Ridder Dick, van der Oost John, and Claassens Nico J. Revealing determinants of translation efficiency via whole-gene codon randomization and machine learning. Nucleic Acids Research, 51(5):2363–2376, 01 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Outeiral Carlos and Deane Charlotte M. Codon language embeddings provide strong signals for use in protein engineering. Nat. Mach. Intell., 6(2):170–179, February 2024. [Google Scholar]
- [63].Paszke Adam, Gross Sam, Chintala Soumith, Chanan Gregory, Yang Edward, DeVito Zachary, Lin Zeming, Desmaison Alban, Antiga Luca, and Lerer Adam. Automatic differentiation in pytorch. 2017. [Google Scholar]
- [64].Plotkin Joshua B and Kudla Grzegorz. Synonymous but not the same: the causes and consequences of codon bias. Nat. Rev. Genet., 12(1):32–42, January 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Plotkin Joshua B., Robins Harlan, and Levine Arnold J.. Tissue-specific codon usage and the expression of human genes. Proceedings of the National Academy of Sciences, 101(34):12588–12591, August 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Raj Anil, Wang Sidney H, Shim Heejung, Harpak Arbel, Li Yang I, Engelmann Brett, Stephens Matthew, Gilad Yoav, and Pritchard Jonathan K. Thousands of novel translated open reading frames in humans inferred by ribosome footprint profiling. eLife, 5, May 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Ripley B. D.. Modelling spatial patterns. Journal of the Royal Statistical Society Series B: Statistical Methodology, 39(2):172–192, January 1977. [Google Scholar]
- [68].Rives Alexander, Meier Joshua, Sercu Tom, Goyal Siddharth, Lin Zeming, Liu Jason, Guo Demi, Ott Myle, Zitnick C. Lawrence, Ma Jerry, and Fergus Rob. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. PNAS, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Rubin Alan F., Gelman Hannah, Lucas Nathan, Bajjalieh Sandra M., Papenfuss Anthony T., Speed Terence P., and Fowler Douglas M.. A statistical framework for analyzing deep mutational scanning data. Genome Biology, 18(1), August 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Saberi Ali, Choi Benedict, Wang Sean, Naghipourfar Mohsen, Namini Arsham Mikaeili, Ramani Vijay, Emad Amin, Najafabadi Hamed S., and Goodarzi Hani. A long-context rna foundation model for predicting transcriptome architecture. August 2024. [Google Scholar]
- [71].Scott Alexandra, Petrykowska Hanna M., Hefferon Timothy, Gotea Valer, and Elnitski Laura. Functional analysis of synonymous substitutions predicted to affect splicing of the cftr gene. Journal of Cystic Fibrosis, 11(6):511–517, December 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Sharma Yogita, Miladi Milad, Dukare Sandeep, Boulay Karine, Caudron-Herger Maiwen, Groß Matthias, Backofen Rolf, and Diederichs Sven. A pan-cancer analysis of synonymous mutations. Nature Communications, 10(1), June 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Sharp Paul M., Tuohy Therese M.F., and Mosurski Krzysztof R.. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Research, 14(13):5125–5143, 07 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Shtivelman E, Gale RP, Dreazen O, Berrebi A, Zaizov R, Kubonishi I, Miyoshi I, and Canaani E. bcr-abl rna in patients with chronic myelogenous leukemia. Blood, 69(3):971–973, March 1987. [PubMed] [Google Scholar]
- [75].Sondka Zbyslaw, Bamford Sally, Cole Charlotte G, Ward Sari A, Dunham Ian, and Forbes Simon A. The COSMIC cancer gene census: describing genetic dysfunction across all human cancers. Nat. Rev. Cancer, 18(11):696–705, November 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Su Jianlin, Lu Yu, Pan Shengfeng, Murtadha Ahmed, Wen Bo, and Liu Yunfeng. Roformer: Enhanced transformer with rotary position embedding, 2021. [Google Scholar]
- [77].Sun Yutao, Dong Li, Patra Barun, Ma Shuming, Huang Shaohan, Benhaim Alon, Chaudhary Vishrav, Song Xia, and Wei Furu. A length-extrapolatable transformer, 2022. [Google Scholar]
- [78].Supek Fran, Miñana Belén, Valcárcel Juan, Gabaldón Toni, and Lehner Ben. Synonymous mutations frequently act as driver mutations in human cancers. Cell, 156(6):1324–1335, March 2014. [DOI] [PubMed] [Google Scholar]
- [79].Teufel Felix, Armenteros José Juan Almagro, Johansen Alexander Rosenberg, Gíslason Magnús Halldór, Pihl Silas Irby, Tsirigos Konstantinos D., Winther Ole, Brunak Søren, von Heijne Gunnar, and Nielsen Henrik. Signalp 6.0 predicts all five types of signal peptides using protein language models. Nature Biotechnology, 40(7):1023–1025, January 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Thumuluri Vineet, Armenteros José Juan Almagro, Johansen Alexander Rosenberg, Nielsen Henrik, and Winther Ole. Deeploc 2.0: multi-label subcellular localization prediction using protein language models. Nucleic Acids Research, 50(W1):W228–W234, April 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Thumuluri Vineet, Martiny Hannah-Marie, Armenteros Jose J Almagro, Salomon Jesper, Nielsen Henrik, and Johansen Alexander Rosenberg. Netsolp: predicting protein solubility in escherichia coli using language models. Bioinformatics, 38(4):941–946, November 2021. [DOI] [PubMed] [Google Scholar]
- [82].Tristani-Firouzi Martin, Jensen Judy L., Donaldson Matthew R., Sansone Valeria, Meola Giovanni, Hahn Angelika, Bendahhou Said, Kwiecinski Hubert, Fidzianska Anna, Plaster Nikki, Fu Ying-Hui, Ptacek Louis J., and Tawil Rabi. Functional and clinical characterization of kcnj2 mutations associated with lqt7 (andersen syndrome). Journal of Clinical Investigation, 110(3):381–388, August 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Unsal Serbulent, Atas Heval, Albayrak Muammer, Turhan Kemal, Acar Aybar C., and Doğan Tunca. Learning functional properties of proteins with language models. Nature Machine Intelligence, 4(3):227–245, March 2022. [Google Scholar]
- [84].van Heesch Sebastiaan, Witte Franziska, Schneider-Lunitz Valentin, Schulz Jana F., Adami Eleonora, Faber Allison B., Kirchner Marieluise, Maatz Henrike, Blachut Susanne, Sandmann Clara-Louisa, Kanda Masatoshi, Worth Catherine L., Schafer Sebastian, Calviello Lorenzo, Merriott Rhys, Patone Giannino, Hummel Oliver, Wyler Emanuel, Obermayer Benedikt, Mücke Michael B., Lindberg Eric L., Trnka Franziska, Memczak Sebastian, Schilling Marcel, Felkin Leanne E., Barton Paul J.R., Quaife Nicholas M., Vanezis Konstantinos, Diecke Sebastian, Mukai Masaya, Mah Nancy, Oh Su-Jun, Kurtz Andreas, Schramm Christoph, Schwinge Dorothee, Se-bode Marcial, Harakalova Magdalena, Asselbergs Folkert W., Vink Aryan, de Weger Roel A., Viswanathan Sivakumar, Widjaja Anissa A., Gärtner-Rommel Anna, Milting Hendrik, dos Remedios Cris, Knosalla Christoph, Mertins Philipp, Landthaler Markus, Vingron Martin, Linke Wolfgang A., Seidman Jonathan G., Seidman Christine E., Rajewsky Nikolaus, Ohler Uwe, Cook Stuart A., and Hubner Norbert. The translational landscape of the human heart. Cell, 178(1):242–260.e29, June 2019. [DOI] [PubMed] [Google Scholar]
- [85].Vaswani Ashish, Shazeer Noam, Parmar Niki, Uszkoreit Jakob, Jones Llion, Gomez Aidan N., Kaiser Lukasz, and Polosukhin Illia. Attention is all you need, 2023. [Google Scholar]
- [86].Wang Hongyu, Ma Shuming, Huang Shaohan, Dong Li, Wang Wenhui, Peng Zhiliang, Wu Yu, Bajaj Payal, Singhal Saksham, Benhaim Alon, Patra Barun, Liu Zhun, Chaudhary Vishrav, Song Xia, and Wei Furu. Foundation transformers, 2022. [Google Scholar]
- [87].Wolf Thomas, Debut Lysandre, Sanh Victor, Chaumond Julien, Delangue Clement, Moi Anthony, Cistac Pierric, Rault Tim, Louf Rémi, Funtowicz Morgan, and Brew Jamie. Huggingface’s transformers: State-of-the-art natural language processing. CoRR, abs/1910.03771, 2019. [Google Scholar]
- [88].Wu Yingzhou, Liu Hanqing, Li Roujia, Sun Song, Weile Jochen, and Roth Frederick P.. Improved pathogenicity prediction for rare human missense variants. The American Journal of Human Genetics, 108(10):1891–1906, October 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Yee Sook Wah, Macdonald Christian B., Mitrovic Darko, Zhou Xujia, Koleske Megan L., Yang Jia, Silva Dina Buitrago, Grimes Patrick Rockefeller, Trinidad Donovan D., More Swati S., Kachuri Linda, Witte John S., Delemotte Lucie, Giacomini Kathleen M., and Coyote-Maestas Willow. The full spectrum of slc22 oct1 mutations illuminates the bridge between drug transporter biophysics and pharmacogenomics. Molecular Cell, 84(10):1932–1947.e10, May 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Zhou Zhihan, Ji Yanrong, Li Weijian, Dutta Pratik, Davuluri Ramana, and Liu Han. Dnabert-2: Efficient foundation model and benchmark for multi-species genome, 2023. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.