Abstract
EBNA-LP-associated proteins were identified by sequencing proteins that immunoprecipitated with Flag epitope-tagged EBNA-LP (FLP) from lymphoblasts in which FLP was stably expressed. The association of EBNA-LP with Hsp70 (72/73) was confirmed, and sequences of DNA-PK catalytic subunit (DNA-PKcs), HA95, Hsp27, prolyl 4-hydroxylase α-1 subunit, α-tubulin, and β-tubulin were identified. The fraction of total cellular HA95 that associated with FLP was very high, while progressively lower fractions of the total DNA-PKcs, Hsp70, Hsp 27, α-tubulin, and β-tubulin specifically associated with EBNA-LP as determined by immunoblotting with antibodies to these proteins. EBNA-LP bound to two domains in the DNA-PKcs C terminus and DNA-PKcs associated with the EBNA-LP repeat domain. DNA-PKcs that was bound to EBNA-LP phosphorylated p53 or EBNA-LP in vitro, and the phosphorylation of EBNA-LP was inhibited by Wortmannin, a specific in vitro inhibitor of DNA-PKcs.
Epstein-Barr virus (EBV) is a human herpesvirus that initiates primary infection and replication in the oropharyngeal epithelium (62). EBV infection then spreads to B lymphocytes, which are largely nonpermissive for virus replication (47, 68). Based on in vitro studies of B-lymphocyte infection, the first EBV transcripts initiate within the viral long internal repeat (for reviews, see references 26 and 53). These transcripts are differentially spliced to encode two nuclear proteins, EBNA-LP and EBNA-2. These two proteins act in concert to activate transcription of cell and viral genes including the cellular c-myc, CD23, and cyclin D2 and the viral EBNA-3A, -3B, -3C, and -1 and latent infection membrane protein, LMP1 and -2, genes (1, 16, 23, 49, 61). These virus-encoded proteins cause the cell to enter S phase and proliferate indefinitely. In stably transformed B lymphocytes EBV expresses six EBNAs, two LMPs, two small RNA or EBERs, and transcripts from the BamHI A fragment (13, 18). Recombinant EBV reverse genetic studies indicate that EBNA-LP, EBNA-2, EBNA-3A, EBNA-3C, EBNA-1, and LMP1 are critical or essential for B-lymphocyte proliferation, while EBNA-3B, LMP2, EBERs, and most of the rest of the viral genome are not critical (8, 15, 24, 25, 27, 33, 34, 38–41, 43, 54, 55, 69).
The experiments described here investigate the associations of EBNA-LP with cellular proteins. EBNA-LP is unusual in that it is encoded mostly by repeating 66 and 132 b exons, which are derived from the EBV long internal repeat (10, 58, 73). The EBNA-LP open reading frame ends within 33 and 102 b exons that are transcribed from the unique DNA downstream of the long internal repeat. EBNA-LP lacks the ability to recognize specific DNA sequences and is dependent on interaction with EBNA-2 for promoter-specific transcriptional effects (16, 49). EBNA-2 has two essential domains (6, 7). One interacts with cellular, sequence-specific, DNA binding proteins, including RBP-Jκ (14, 17, 22, 32, 64, 78). The second is an acidic transcriptional activation domain that interacts with basal and activated transcription factors, with CREB binding protein and p300, and with a p100 nuclear protein that is a scaffolding protein for Pim-1 and c-myb (36, 70–72, 74). EBNA-LP markedly augments the transcriptional effects of EBNA-2 (16, 49). In fact, EBNA-LP markedly augments the activity of the EBNA-2 acidic domain when the latter domain is fused to the Gal4 DNA binding domain and expressed in cells that have a promoter with multiple upstream Gal-4 binding sites (16). Surprisingly, the 22- and 44-amino-acid repeating segments of EBNA-LP appear to be all that is required for coactivation with EBNA-2. Carboxyl-terminal truncation of EBNA-LP before the last 10 unique amino acids results in a transcriptionally inactive protein, while truncation of the entire 45-amino-acid unique sequence domain restores full activity in transient-transfection assays. These data are compatible with a model in which the EBNA-LP repeating domains mediate transcriptional activation and the last 45 residues regulate this activity. EBNA-LP is highly phosphorylated in G2/M, localizes to nuclear dot 10 or PML bodies, and associates with Hsp72/73 (28, 29, 42, 52, 59, 66). In vitro, casein kinase II can phosphorylate a serine in the unique EBNA-LP C terminus, whereas p34cdc2 can phosphorylate serines in the W2 repeat as well as the serine in the C terminus (28). Very little is currently know about the cellular proteins through which EBNA-LP coactivates transcription.
EBNA-LP-associated cellular proteins.
To facilitate the retrieval of EBNA-LP from cells and to minimize the potential effect of antibody in dissociating a cell protein from EBNA-LP, an exogenous Flag epitope was fused to the N terminus of EBNA-LP. Hygromycin-resistant, EBV-negative human B-lymphoma cells were selected that express Flag-epitope tagged EBNA-LP (FLP) after cotransfection with a simian virus 40 promoter and enhancer-FLP expression vector and an expression vector for hygromycin inactivation. Most of the hygromycin-resistant BJAB cell lines that were derived expressed FLP at levels that are 0.5 to 5 times the EBNA-LP level in the IB4, EBV-transformed, B-lymphoblast cell line. Despite an abnormally high level of EBNA-LP expression in some cell lines, cell growth was similar to that of parental BJAB cells. These data indicate that expression of EBNA-LP is not toxic to BJAB cells
Several liters of threefold FLP-overexpressing or parental BJAB cells were grown, and lysates were prepared from 2 × 109 to 3 × 109 cells of each type. Lysates were made by mixing the cells for 30 min at 4°C in 0.5% NP-40, isotonic NaCl, 50 mM Tris (pH 8.0), aprotinin (10 mg/ml), and 1 mM phenylmethylsulfonyl fluoride. The lysates were then clarified by spinning out the nuclei for 10 min at 1,000 × g. FLP was immune precipitated using M2 anti-Flag antibody that was conjugated to protein G-Sepharose beads (Sigma Chemical Company). The proteins that adsorbed to the beads were eluted with sodium dodecyl sulfate (SDS) buffer, boiled in SDS sample buffer, and resolved under denaturing conditions in 6.5, 7.5, or 12% polyacrylamide gels. The gels were stained with Coomassie brilliant blue to identify the proteins that had precipitated with EBNA-LP. As expected, FLP was the most abundant protein that precipitated with M2 beads from FLP-expressing BJAB cells and not from control BJAB cells (Fig. 1). The position of FLP was confirmed by immunoblotting with EBNA-LP-specific monoclonal antibody. The most abundant proteins associated with EBNA-LP were previously identified to be Hsp72 and hsc73 by microsequencing and immunoblotting (29, 42) and nearly stoichiometric amounts of HSP72 and HSC73 coimmunoprecipitated with FLP. The position of HSP72 was confirmed by immunoblotting with HSP72-specific antibody (Fig. 2). Six unknown proteins ranging in size from more than 250 kDa to less than 30 kDa were also identified as being substantially overrepresented in the FLP immunoprecipitate versus an M2 immunoprecipitate from BJAB cells that do not express FLP (Fig. 1). Proteins of similar size had been previously noted in immunoprecipitates from EBV-transformed lymphoblastoid cell lines (42), using the JF186 monoclonal antibody to EBNA-LP (12). Other coimmunoprecipitating proteins were close to the size of Rb or p53, proteins that had been tentatively identified as binding to the EBNA-LP repeat domain (67). Rb and p53 were absent from previous JF186 immunoprecipitates (42) and do not appear to physiologically interact with EBNA-LP (19).
FIG. 1.
Coomassie brilliant blue-stained polyacrylamide gels of Flag antibody immunoprecipitates (Flag-IP) from BJAB B lymphoblasts that were stably converted to FLP expression (lane marked BFLP) or from negative-control BJAB B lymphoblasts (lane marked B). Cells were lysed in nonionic detergent, and extracts were immunoprecipitated using M2 Flag antibody-coupled beads. Proteins bound to the beads were eluted in SDS sample buffer; resolved in 6.5, 7.5, or 12% polyacrylamide gels; and stained with Coomassie brilliant blue. FLP and Hsp72/Hsc73 were confirmed by immunoblotting. Other proteins of >250, 95, 65, 55, 53, and 30 kDa that were consistently present in immunoprecipitates from FLP-expressing BJAB cells and absent in control immunoprecipitates from nonexpressing BJAB cells—designated I, II, III, IV, V, and VI—were excised from the gels and identified by amino acid sequence. MW, molecular weight, markers (in thousands).
FIG. 2.
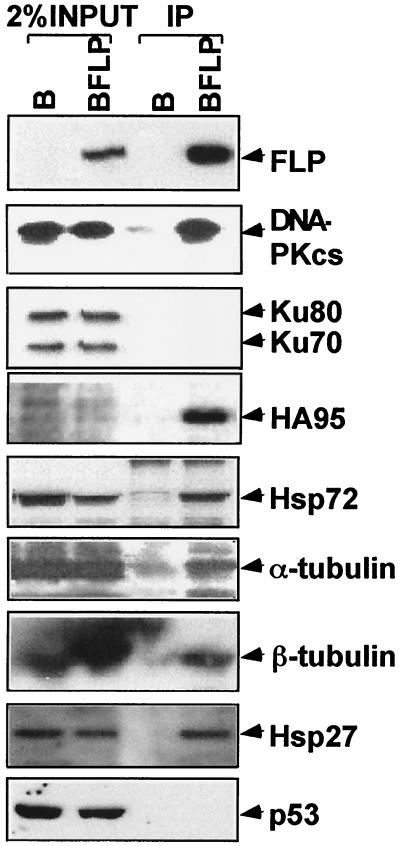
DNA-PKcs, HA95, Hsp72, α-and β-tubulin, and Hsp27 specifically associate with EBNA-LP. The specificity and relative efficiency of coimmunoprecipitation of DNA-PKcs, HA95, Ku70 and 80, HSP72, α-and β-tubulin, Hsp27, or p53 with FLP were evaluated by comparing the abundance of these proteins in M2 immunoprecipitates from FLP-expressing and negative-control cell lysates with the abundance of the proteins in 2% of the cell lysates. The proteins were detected by immunoblotting with specific antibodies.
Slices containing each of the six EBNA-LP coimmunoprecipitating proteins were excised from the gel, and the proteins were cleaved in gel by trypsin. The digested peptides were separated, and multiple peptides were sequenced by microcapillary high-performance liquid chromatography–ion trap–tandem mass spectrometry. The protein larger than 250 kDa yielded sequences (TVGALQVLGTEAQSSLLK and LLLQGEADQSLLTFDIK) of DNA-PK catalytic subunit (DNA-PKcs). The 95-kDa protein yielded sequences (GENPFTDSPEE and ADFLQEYVTNK) of a putative homologue of AKAP95 that has been designated HA95 (50). The 65-kDa sequence (LQDTYNLDTDTISK) matched the prolyl 4-hydroxylase α subunit. The 55- and 53-kDa sequences (TIGGGDDSFNTFFSETGAGK, TIQFVDWPTGFK and ISVYYNEATGGK, NSSYFVEWIPNN) identified α- and β-tubulin. The 27-kDa sequence (LATQSNEITIPVTFESR) matched Hsp27.
DNA-PKcs, HA95, HSP72, α- and β-tubulin, and Hsp27 specifically associate with EBNA-LP.
Antibodies that can detect DNA-PKcs (Santa Cruz), HA95 (50), Hsp70 (Santa Cruz), α- and β-tubulin (Amersham), Hsp27 (Santa Cruz), p53 (Upstate Biotechnology), Ku 70 and 80 (Santa Cruz), and EBNA-LP (JF186) in immunoblots were used to evaluate the specificity and fraction of total cellular protein that are associated with the FLP immunoprecipitate. Approximately 10% of FLP was in a typical immunoprecipitate as is apparent from comparison of the Flag immunoprecipitate lane in Fig. 2 with 2% of the BJAB-FLP (BFLP) cell lysate. DNA-PKcs, HA95, Hsp72, α- and β-tubulin, and Hsp27 specifically immunoprecipitated with FLP from BJAB-FLP cells and were not in M2 antibody immune precipitates from BJAB cells that did not express FLP. Compare these proteins in immune precipitates from BFLP and B cells in Fig. 2.
A significant fraction of the total cellular HA95 was associated with FLP, as is evident from the enrichment for HA95 in the BFLP immunoprecipitate versus 2% of the BFLP or B cell lysate, using rabbit antiserum to HA95 (50) to identify HA95 in immunoblots (Fig. 2). Indeed, HA95 is as enriched in the FLP immunoprecipitate versus the cell lysate as is FLP. Versus 2% of the lysate, FLP and HA95 were about fivefold enriched in the Flag immunoprecipitate, whereas DNA-PKcs was about twofold enriched, Hsp72 and Hsp27 were about equal to the lysate, and α-and β-tubulin were less abundant in the Flag immunoprecipitate than in the lysate (Fig. 2). Thus, these data indicate that most of the cellular HA95 and a significant fraction of DNA-PKcs can associate with EBNA-LP in B lymphoma cells. Although Hsp72, Hsp27, and α- and β-tubulin are specifically and significantly represented in the FLP immunoprecipitate, these are abundant cell proteins, and a smaller fraction of these proteins is associated with FLP.
Antibodies to prolyl 4-hydroxylase were not available to study the extent of its association with FLP. Prolyl 4-hydroxylase is an endoplasmic reticulum resident protein required for hydroxylation of proline residues in collagen. Although prolyl 4-hydroxylase can affect tissue invasion (44, 56) its relevance to EBNA-LP's intracellular effects is not obvious and the association with EBNA-LP was not further pursued.
DNA-PKcs is highly associated with Ku 70 and 80 (5, 35, 65). EBNA-LP might therefore associate with Ku 70 and 80 through DNA-PKcs. However, immunoblots with Ku 70 and 80 antibody did not detect Ku 70 and 80 in the EBNA-LP immunoprecipitates (Fig. 2). The Ku 70 and 80 and DNA-PKcs complex is usually stable through immunoprecipitation (65). Therefore, FLP probably specifically associates with free DNA-PKcs.
The potential associations of EBNA-LP with p53, pRb, p107, and p130 (67) were also reevaluated (Fig. 2 and data not shown). Despite the specific detection of p53, pRb, p107, and p130 in cell lysates, p53, pRb, p107, and p130 were not detected in the FLP immunoprecipitates.
The EBNA-LP repeat domain interacts with at least two sequences in the DNA-PKcs C terminus.
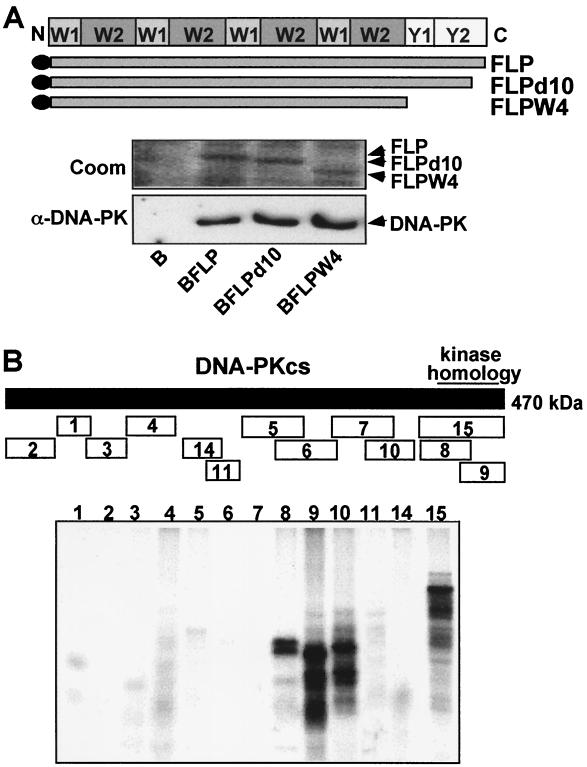
The EBNA-LP open reading frame is composed of repeating W1 and W2 exons that are spliced from the EBV internal BamHI W repeats and unique 3′ Y1 and Y2 exons (Fig. 3A). To identify the domain(s) within EBNA-LP that mediates interaction with DNA-PKcs, BJAB stable cell lines converted to expression of FLP and with deletions of the C-terminal 10 (FLPd10) or C-terminal 45 (FLPW4) amino acids were established and used to compare the efficiency of DNA-PK association with wild-type or mutant EBNA-LP. As shown in Fig. 3A, DNA-PKcs associated with FLPd10 and FLPW4 as efficiently as with wild-type FLP. These data indicate that DNA-PKcs associates with the W1-W2 repeat domain of EBNA-LP as was previously demonstrated for Hsp70 (42).
FIG. 3.
Mapping of the EBNA-LP and DNA-PKcs interacting domains. (A) The association of DNA-PKcs with wild-type or C-terminally truncated EBNA-LP was evaluated by determining the extent to which DNA-PKcs coimmunoprecipitated with wild-type FLP or FLP with a deletion of the last 10 (FLPd10) or all 45 (FLPd45 or FLPW4) unique residues from lysates of BJAB cells that stably express the respective protein or negative control cells. A schematic diagram of the repeat and unique domains of EBNA-LP is shown above the data. Equal amounts of wild-type or mutant FLPs were immunoprecipitated as shown in the photograph of a Coomassie-stained (Coom) gel. The amount of coimmunoprecipitating DNA-PKcs was evaluated by immunoblotting (α-DNA-PK). (B) Polypeptide fragments of DNA-PKcs as indicated in the schematic diagram were in vitro translated in the presence of [35S]methionine and incubated at 4°C with 2 μg of FLP adsorbed onto M2-Sepharose beads. After elution from beads, bound polypeptides were analyzed on SDS–10% gel and subjected to fluorography at a low temperature as shown in the bottom panel.
To identify the components of DNA-PKcs that bind to EBNA-LP, 15 different 35S-labeled polypeptides, representing the entire DNA-PKcs open reading frame were synthesized by in vitro transcription-translation (21) (Fig. 3B). Each in vitro translation reaction produced a polypeptide of the expected size by SDS-polyacrylamide gel electrophoresis (PAGE) (data not shown), and equal amounts of 35S-labeled polypeptides were incubated at 4°C with equal amounts of FLP adsorbed onto M2 beads. Polypeptides 8, 9, and 15 that overlap in the C-terminal kinase homology domain of DNA-Pkcs bound most efficiently to EBNA-LP, while the adjacent, nonoverlapping, polypeptide 10 bound almost as well. The binding of polypeptide 10 is most likely due to its most-C-terminal third, since polypeptide 7, which overlapped with the N-terminal two-thirds of 10, did not bind to FLP. These data indicate that at least two separate domains in the DNA-PKcs C terminus are able to bind to FLP. The DNA-PKcs kinase homology domain had been shown to interact with the c-Abl nuclear tyrosine kinase and Ku (21). Although the DNA-PKcs kinase homology domain is a large domain, the binding of both EBNA-LP and Ku to this domain could be related to the finding mentioned above that EBNA-LP-associated DNA-PKcs is not associated with Ku. This domain of DNA-PKcs may associate with EBNA-LP or Ku but not with both.
EBNA-LP immunoprecipitates contain active DNA-PKcs, and EBNA-LP autophosphorylation is suppressed by the specific DNA-PKcs inhibitor Wortmannin.
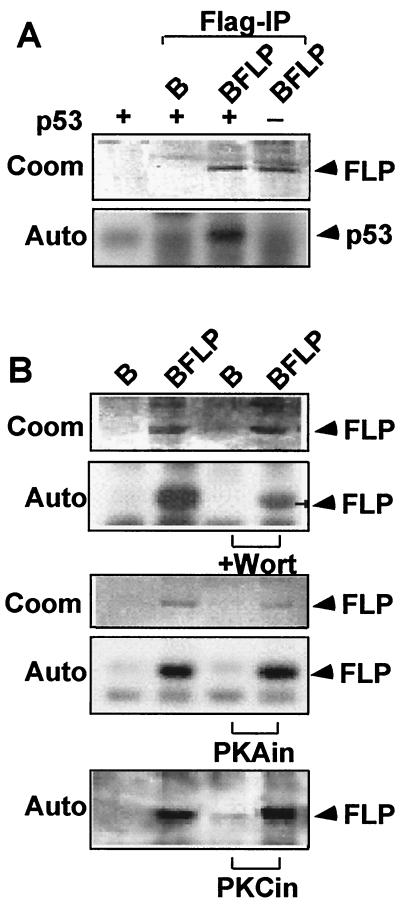
DNA-PK phosphorylates a number of transcription factors, in vitro, including the p53 tumor suppressor protein. This phosphorylation activates p53 in response to DNA damage (77). To evaluate whether EBNA-LP-associated DNA-PKcs is active, glutathione S-transferase (GST)-p53 was used as a substrate in an in vitro kinase assay with a buffer consisting of 25 mM HEPES (pH 7.5), 75 mM KCl, 10 mM MgCl2, 10 mM MnCl2, 0.4 mM EGTA, 0.2 mM EDTA, 1 mM dithiothreitol, 50 μM ATP, and [γ-32P]ATP. GST-p53 was phosphorylated, in vitro, by FLP immunoprecipitates on M2 beads, but not by immunoprecipitates from cells that do not express FLP (Fig. 4A). These data suggest that DNA-PKcs that is associated with FLP can be active in p53 phosphorylation.
FIG. 4.
EBNA-LP is associated with and phosphorylated by DNA-PK. (A) DNA-PKcs phosphorylation of GST-p53 is used to assay EBNA-LP-associated DNA-PKcs activity. FLP adsorbed onto M2 beads from lysates of FLP-expressing BJAB cells was incubated in kinase buffer with purified recombinant GST-p53 (100 ng) or control protein and [γ-32P]ATP. 32P-labeled GST-p53 was analyzed for phosphorylation level by PAGE followed by exposure to X-ray film (Auto). Coomassie brilliant blue (Coom) was used to estimate FLP abundance. (B) FLP adsorbed onto on M2 beads from BFLP lysates was incubated with [γ-32P]ATP under in vitro kinase assay conditions in the presence (+Wort) or absence of 50 μM Wortmannin, a specific in vitro inhibitor of DNA-PK, or with 5 μM PKA or PKC inhibitory peptides (PKAin or PKCin, respectively). (Sigma-Aldrich). Coomassie brilliant blue is used to estimate protein levels, and phosphorylation was estimated by autoradiography.
Since FLP-associated DNA-PKcs appeared to be active in p53 phosphorylation, we considered the possibility that EBNA-LP might also be phosphorylated by DNA-PKcs. A similar in vitro DNA-PKcs [γ-32P]ATP kinase assay was done with DNA-PKcs and FLP adsorbed onto M2 beads from BFLP cell lysates. FLP was strongly phosphorylated in the in vitro kinase reaction (Fig. 4B and data not shown). Wortmannin is a specific inhibitor of DNA-PKcs in vitro (20). Reactions were therefore done in the presence and absence of Wortmannin to further test whether the phosphorylation of FLP was due to DNA-PKcs. FLP phosphorylation, in vitro, was about 70% inhibited by 50 μM Wortmannin (Fig. 4B). Thus, DNA-PKcs probably has a significant role in EBNA-LP phosphorylation. As a control for the possible presence of protein kinase A (PKA) or PKC, reactions were done in the presence or absence of specific PKA or PKC inhibitory peptides (Sigma-Aldrich). These inhibitors did not affect FLP phosphorylation (Fig. 4B).
These experiments identify DNA-PKcs, HA95, prolyl 4-hydroxylase α, α-and β-tubulin, and hsp27 as proteins that may associate with EBNA-LP in cells or during the process of cell lysis and confirm an extensive association of EBNA-LP with HA95, Hsp72/Hsc73, Hsp27, and DNA-PKcs. HA95 and DNA-PKcs are of obvious interest since they and EBNA-LP are largely nuclear. Thus, these associations are more likely to exist in vivo rather than to occur after lysis (5, 50).
Given the finding of about 3% of DNA-PKcs in the FLP immunoprecipitate and a 10% efficiency of FLP immunoprecipitation, we estimate that about 30% of the cellular DNA-PKcs is associated with overexpressed EBNA-LP. DNA-PKcs could therefore have a role in EBNA-LP transcriptional or survival effects. DNA-PKcs can phosphorylate EBNA-LP, as is evident by Wortmannin inhibition of EBNA-LP phosphorylation, in vitro. DNA-PKcs associates with the repeat domain of EBNA-LP, and the repeat domain is implicated in transcriptional activation (16). In preliminary experiments, autophosphorylation of FLPW4 in vitro was similar to that of FLP and was similarly inhibited by Wortmannin. DNA-PKcs is also a large protein that could mediate the interaction of EBNA-LP with other nuclear proteins, including proteins involved in transcription, repair, or recombination. Thus, the activity of EBNA-LP could be affected by association with DNA-PKcs or by DNA-PKcs-mediated phosphorylation of EBNA-LP or EBNA-LP-associated proteins. EBNA-LP may also affect DNA-PKcs interactions with other proteins. DNA-PKcs is essential for VDJ type recombinations, for double-strand DNA repair (4), and for normal telomere maintenance (3). DNA-PKcs also has effects on transcription mediated by thyroid hormone receptor binding protein (30) and on apoptosis mediated by p53 (75). Since Wortmannin is not specific for DNA-PKcs in vivo, further understanding of the in vivo role of DNA-PKcs in EBNA-LP's effects will require comparison of EBV and EBNA-LP's effects in cells from healthy humans with the effects in cells from humans that lack DNA-PKcs.
Most of the HA95 in cells is associated with overexpressed EBNA-LP. Thus, EBNA-LP is likely to be affected by HA95 and to substantially alter or redirect the activity of HA95. HA95 is a recently discovered nuclear protein that is homologous to and a tandem gene duplication with AKAP95 (50). AKP95 is a nuclear protein that associates during mitosis with the RII subunit of PKA and targets PKA to the condensed chromatin spindle region (11). AKAP95 is important for chromosome condensation during mitosis (63). HA95 tracks with AKAP95 to the mitotic spindle but does not bind to the PKA RII subunit or to AKAP95; its role in mitosis is uncertain (50). HA95 was independently discovered through a yeast two-hybrid screen with RNA helicase A. HA95 can increase expression by improving function of a constitutive transport element (37, 76). Hence, EBNA-LP effects in transcriptional coactivation could be in part due to EBNA-LP alteration of HA95's effect on the transport of RNAs with a functional CTE.
The most abundant cellular proteins that specifically coimmunoprecipitate with EBNA-LP are Hsp72/Hsc73 and α- and β-tubulin. Tubulin has been noted to interact with viral and cellular oncoproteins and with regulatory components of the cell cycle apparatus (45, 57). More recently, the association of c-myc with tubulin has become more interesting with the mapping of the tubulin interacting domain to the c-myc N terminus and the finding that mutations in c-myc T-58 correlate with hyperstabilization, increased phosphorylation, disrupted interaction with α-tubulin, and increased transforming capacity (48, 57). c-myc association with α-tubulin is physiologically disrupted by mitosis-specific c-myc hyperphosphorylation. EBNA-LP also undergoes mitosis specific hyperphosphorylation and the effect on EBNA-LP activity has not been assessed (28). EBNA-LP coactivation with EBNA-2 of viral and cellular latency-associated promoters (16, 49) may be affected by cell cycle-specific factors since LMP1 levels fall in Raji cells under conditions of growth arrest (2).
Hsp27 is also quite specifically associated with EBNA-LP. HSP27 is primarily cytoplasmic in location (60) and is involved in heat shock-induced translational inhibition (9), in Cox-2 transcript stabilization (31), and in inhibition of caspase-3 activation (51). However, Hsp27 can translocate to the nucleus upon insult-induced stress (46) and nuclear Hsp27 could modulate EBNA-LP effects on cell growth in response to cell stress.
Acknowledgments
This research was supported by grant number CA47006 from the National Cancer Institute, National Institutes of Health, of the United States Public Health Service.
REFERENCES
- 1.Alfieri C, Birkenbach M, Kieff E. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology. 1991;181:595–608. doi: 10.1016/0042-6822(91)90893-g. [DOI] [PubMed] [Google Scholar]
- 2.Allday M J, Farrell P J. Epstein-Barr virus nuclear antigen EBNA3C/6 expression maintains the level of latent membrane protein 1 in G1-arrested cells. J Virol. 1994;68:3491–3498. doi: 10.1128/jvi.68.6.3491-3498.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey S M, Meyne J, Chen D J, Kurimasa A, Li G C, Lehnert B E, Goodwin E H. DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc Natl Acad Sci USA. 1999;96:14899–14904. doi: 10.1073/pnas.96.26.14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blunt T, Finnie N J, Taccioli G E, Smith G C, Demengeot J, Gottlieb T M, Mizuta R, Varghese A J, Alt F W, Jeggo P A, et al. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 5.Carter T, Vancurova I, Sun I, Lou W, DeLeon S. A DNA-activated protein kinase from HeLa cell nuclei. Mol Cell Biol. 1990;10:6460–6471. doi: 10.1128/mcb.10.12.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen J I, Kieff E. An Epstein-Barr virus nuclear protein 2 domain essential for transformation is a direct transcriptional activator. J Virol. 1991;65:5880–5885. doi: 10.1128/jvi.65.11.5880-5885.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen J I, Wang F, Kieff E. Epstein-Barr virus nuclear protein 2 mutations define essential domains for transformation and transactivation. J Virol. 1991;65:2545–2554. doi: 10.1128/jvi.65.5.2545-2554.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen J I, Wang F, Mannick J, Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci USA. 1989;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuesta R, Laroia G, Schneider R J. Chaperone hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes Dev. 2000;14:1460–1470. [PMC free article] [PubMed] [Google Scholar]
- 10.Dillner J, Kallin B, Alexander H, Ernberg I, Uno M, Ono Y, Klein G, Lerner R A. An Epstein-Barr virus (EBV)-determined nuclear antigen (EBNA5) partly encoded by the transformation-associated Bam WYH region of EBV DNA: preferential expression in lymphoblastoid cell lines. Proc Natl Acad Sci USA. 1986;83:6641–6645. doi: 10.1073/pnas.83.17.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eide T, Coghlan V, Orstavik S, Holsve C, Solberg R, Skalhegg B S, Lamb N J, Langeberg L, Fernandez A, Scott J D, Jahnsen T, Tasken K. Molecular cloning, chromosomal localization, and cell cycle-dependent subcellular distribution of the A-kinase anchoring protein, AKAP95. Exp Cell Res. 1998;238:305–316. doi: 10.1006/excr.1997.3855. [DOI] [PubMed] [Google Scholar]
- 12.Finke J, Rowe M, Kallin B, Ernberg I, Rosen A, Dillner J, Klein G. Monoclonal and polyclonal antibodies against Epstein-Barr virus nuclear antigen 5 (EBNA-5) detect multiple protein species in Burkitt's lymphoma and lymphoblastoid cell lines. J Virol. 1987;61:3870–3878. doi: 10.1128/jvi.61.12.3870-3878.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fries K L, Sculley T B, Webster-Cyriaque J, Rajadurai P, Sadler R H, Raab-Traub N. Identification of a novel protein encoded by the BamHI A region of the Epstein-Barr virus. J Virol. 1997;71:2765–2771. doi: 10.1128/jvi.71.4.2765-2771.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grossman S R, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc Natl Acad Sci USA. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammerschmidt W, Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature. 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 16.Harada S, Kieff E. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J Virol. 1997;71:6611–6618. doi: 10.1128/jvi.71.9.6611-6618.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 18.Hitt M M, Allday M J, Hara T, Karran L, Jones M D, Busson P, Tursz T, Ernberg I, Griffin B E. EBV gene expression in an NPC-related tumour. EMBO J. 1989;8:2639–2651. doi: 10.1002/j.1460-2075.1989.tb08404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inman G J, Farrell P J. Epstein-Barr virus EBNA-LP and transcription regulation properties of pRB, p107 and p53 in transfection assays. J Gen Virol. 1995;76:2141–2149. doi: 10.1099/0022-1317-76-9-2141. [DOI] [PubMed] [Google Scholar]
- 20.Izzard R A, Jackson S P, Smith G C. Competitive and noncompetitive inhibition of the DNA-dependent protein kinase. Cancer Res. 1999;59:2581–2586. [PubMed] [Google Scholar]
- 21.Jin S, Kharbanda S, Mayer B, Kufe D, Weaver D T. Binding of Ku and c-Abl at the kinase homology region of DNA-dependent protein kinase catalytic subunit. J Biol Chem. 1997;272:24763–24766. doi: 10.1074/jbc.272.40.24763. [DOI] [PubMed] [Google Scholar]
- 22.Johannsen E, Koh E, Mosialos G, Tong X, Kieff E, Grossman S R. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by Jκ and PU.1. J Virol. 1995;69:253–262. doi: 10.1128/jvi.69.1.253-262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaiser C, Laux G, Eick D, Jochner N, Bornkamm G W, Kempkes B. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J Virol. 1999;73:4481–4484. doi: 10.1128/jvi.73.5.4481-4484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaye K M, Izumi K M, Kieff E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kempkes B, Pich D, Zeidler R, Sugden B, Hammerschmidt W. Immortalization of human B lymphocytes by a plasmid containing 71 kilobase pairs of Epstein-Barr virus DNA. J Virol. 1995;69:231–238. doi: 10.1128/jvi.69.1.231-238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2343–2396. [Google Scholar]
- 27.Kim O J, Yates J L. Mutants of Epstein-Barr virus with a selective marker disrupting the TP gene transform B cells and replicate normally in culture. J Virol. 1993;67:7634–7640. doi: 10.1128/jvi.67.12.7634-7640.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitay M K, Rowe D T. Cell cycle stage-specific phosphorylation of the Epstein-Barr virus immortalization protein EBNA-LP. J Virol. 1996;70:7885–7893. doi: 10.1128/jvi.70.11.7885-7893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitay M K, Rowe D T. Protein-protein interactions between Epstein-Barr virus nuclear antigen-LP and cellular gene products: binding of 70-kilodalton heat shock proteins. Virology. 1996;220:91–99. doi: 10.1006/viro.1996.0289. [DOI] [PubMed] [Google Scholar]
- 30.Ko L, Cardona G R, Chin W W. Thyroid hormone receptor-binding protein, an LXXLL motif-containing protein, functions as a general coactivator. Proc Natl Acad Sci USA. 2000;97:6212–6217. doi: 10.1073/pnas.97.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lasa M, Mahtani K R, Finch A, Brewer G, Saklatvala J, Clark A R. Regulation of cyclooxygenase 2 mRNA stability by the mitogen-activated protein kinase p38 signaling cascade. Mol Cell Biol. 2000;20:4265–4274. doi: 10.1128/mcb.20.12.4265-4274.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laux G, Adam B, Strobl L J, Moreau-Gachelin F. The Spi-1/PU.1 and Spi-B ets family transcription factors and the recombination signal binding protein RBP-J kappa interact with an Epstein-Barr virus nuclear antigen 2 responsive cis-element. EMBO J. 1994;13:5624–5632. doi: 10.1002/j.1460-2075.1994.tb06900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee M A, Diamond M E, Yates J L. Genetic evidence that EBNA-1 is needed for efficient, stable latent infection by Epstein-Barr virus. J Virol. 1999;73:2974–2982. doi: 10.1128/jvi.73.4.2974-2982.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee M A, Yates J L. BHRF1 of Epstein-Barr virus, which is homologous to human proto-oncogene bcl2, is not essential for transformation of B cells or for virus replication in vitro. J Virol. 1992;66:1899–1906. doi: 10.1128/jvi.66.4.1899-1906.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lees-Miller S P, Chen Y R, Anderson C W. Human cells contain a DNA-activated protein kinase that phosphorylates simian virus 40 T antigen, mouse p53, and the human Ku autoantigen. Mol Cell Biol. 1990;10:6472–6481. doi: 10.1128/mcb.10.12.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leverson J D, Koskinen P J, Orrico F C, Rainio E M, Jalkanen K J, Dash A B, Eisenman R N, Ness S A. Pim-1 kinase and p100 cooperate to enhance c-Myb activity. Mol Cell. 1998;2:417–425. doi: 10.1016/s1097-2765(00)80141-0. [DOI] [PubMed] [Google Scholar]
- 37.Li J, Tang H, Mullen T M, Westberg C, Reddy T R, Rose D W, Wong-Staal F. A role for RNA helicase A in post-transcriptional regulation of HIV type 1. Proc Natl Acad Sci USA. 1999;96:709–714. doi: 10.1073/pnas.96.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Longnecker R, Miller C L, Miao X Q, Marchini A, Kieff E. The only domain which distinguishes Epstein-Barr virus latent membrane protein 2A (LMP2A) from LMP2B is dispensable for lymphocyte infection and growth transformation in vitro; LMP2A is therefore nonessential. J Virol. 1992;66:6461–6469. doi: 10.1128/jvi.66.11.6461-6469.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Longnecker R, Miller C L, Miao X Q, Tomkinson B, Kieff E. The last seven transmembrane and carboxy-terminal cytoplasmic domains of Epstein-Barr virus latent membrane protein 2 (LMP2) are dispensable for lymphocyte infection and growth transformation in vitro. J Virol. 1993;67:2006–2013. doi: 10.1128/jvi.67.4.2006-2013.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Longnecker R, Miller C L, Tomkinson B, Miao X Q, Kieff E. Deletion of DNA encoding the first five transmembrane domains of Epstein-Barr virus latent membrane proteins 2A and 2B. J Virol. 1993;67:5068–5074. doi: 10.1128/jvi.67.8.5068-5074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mannick J B, Cohen J I, Birkenbach M, Marchini A, Kieff E. The Epstein-Barr virus nuclear protein encoded by the leader of the EBNA RNAs is important in B-lymphocyte transformation. J Virol. 1991;65:6826–6837. doi: 10.1128/jvi.65.12.6826-6837.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mannick J B, Tong X, Hemnes A, Kieff E. The Epstein-Barr virus nuclear antigen leader protein associates with hsp72/hsc73. J Virol. 1995;69:8169–8172. doi: 10.1128/jvi.69.12.8169-8172.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marchini A, Tomkinson B, Cohen J I, Kieff E. BHRF1, the Epstein-Barr virus gene with homology to Bc12, is dispensable for B-lymphocyte transformation and virus replication. J Virol. 1991;65:5991–6000. doi: 10.1128/jvi.65.11.5991-6000.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsui H, Kubochi K, Okazaki I, Yoshino K, Ishibiki K, Kitajima M. Collagen biosynthesis in gastric cancer: immunohistochemical analysis of prolyl 4-hydroxylase. J Surg Oncol. 1999;70:239–246. doi: 10.1002/(sici)1096-9098(199904)70:4<239::aid-jso8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 45.Maxwell S A, Ames S K, Sawai E T, Decker G L, Cook R G, Butel J S. Simian virus 40 large T antigen and p53 are microtubule-associated proteins in transformed cells. Cell Growth Differ. 1991;2:115–127. [PubMed] [Google Scholar]
- 46.Mehlen P, Mehlen A, Guillet D, Preville X, Arrigo A P. Tumor necrosis factor-alpha induces changes in the phosphorylation, cellular localization, and oligomerization of human hsp27, a stress protein that confers cellular resistance to this cytokine. J Cell Biochem. 1995;58:248–259. doi: 10.1002/jcb.240580213. [DOI] [PubMed] [Google Scholar]
- 47.Niedobitek G, Hamilton-Dutoit S, Herbst H, Finn T, Vetner M, Pallesen G, Stein H. Identification of Epstein-Barr virus-infected cells in tonsils of acute infectious mononucleosis by in situ hybridization. Hum Pathol. 1989;20:796–799. doi: 10.1016/0046-8177(89)90075-0. [DOI] [PubMed] [Google Scholar]
- 48.Niklinski J, Claassen G, Meyers C, Gregory M A, Allegra C J, Kaye F J, Hann S R, Zajac-Kaye M. Disruption of Myc-tubulin interaction by hyperphosphorylation of c-Myc during mitosis or by constitutive hyperphosphorylation of mutant c-Myc in Burkitt's lymphoma. Mol Cell Biol. 2000;20:5276–5284. doi: 10.1128/mcb.20.14.5276-5284.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nitsche F, Bell A, Rickinson A. Epstein-Barr virus leader protein enhances EBNA-2-mediated transactivation of latent membrane protein 1 expression: a role for the W1W2 repeat domain. J Virol. 1997;71:6619–6628. doi: 10.1128/jvi.71.9.6619-6628.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orstavik S, Eide T, Collas P, Han I O, Tasken K, Kieff E, Jahnsen T, Skalhegg B S. Identification, cloning and characterization of a novel nuclear protein, HA95, homologous to A-kinase anchoring protein 95. Biol Cell. 2000;92:27–37. doi: 10.1016/s0248-4900(00)88761-4. [DOI] [PubMed] [Google Scholar]
- 51.Pandey P, Farber R, Nakazawa A, Kumar S, Bharti A, Nalin C, Weichselbaum R, Kufe D, Kharbanda S. Hsp27 functions as a negative regulator of cytochrome c-dependent activation of procaspase-3. Oncogene. 2000;19:1975–1981. doi: 10.1038/sj.onc.1203531. [DOI] [PubMed] [Google Scholar]
- 52.Petti L, Sample C, Kieff E. Subnuclear localization and phosphorylation of Epstein-Barr virus latent infection nuclear proteins. Virology. 1990;176:563–574. doi: 10.1016/0042-6822(90)90027-o. [DOI] [PubMed] [Google Scholar]
- 53.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2397–2446. [Google Scholar]
- 54.Robertson E, Kieff E. Reducing the complexity of the transforming Epstein-Barr virus genome to 64 kilobase pairs. J Virol. 1995;69:983–993. doi: 10.1128/jvi.69.2.983-993.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robertson E S, Tomkinson B, Kieff E. An Epstein-Barr virus with a 58-kilobase-pair deletion that includes BARF0 transforms B lymphocytes in vitro. J Virol. 1994;68:1449–1458. doi: 10.1128/jvi.68.3.1449-1458.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rocnik E F, Chan B M, Pickering J G. Evidence for a role of collagen synthesis in arterial smooth muscle cell migration. J Clin Investig. 1998;101:1889–1898. doi: 10.1172/JCI1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakamuro D, Prendergast G C. New Myc-interacting proteins: a second Myc network emerges. Oncogene. 1999;18:2942–2954. doi: 10.1038/sj.onc.1202725. [DOI] [PubMed] [Google Scholar]
- 58.Sample J, Hummel M, Braun D, Birkenbach M, Kieff E. Nucleotide sequences of mRNAs encoding Epstein-Barr virus nuclear proteins: a probable transcriptional initiation site. Proc Natl Acad Sci USA. 1986;83:5096–5100. doi: 10.1073/pnas.83.14.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sauter M, Boos H, Hirsch F, Mueller-Lantzsch N. Characterization of a latent protein encoded by the large internal repeats and the BamHI Y fragment of the Epstein-Barr virus (EBV) genome. Virology. 1988;166:586–590. doi: 10.1016/0042-6822(88)90530-2. [DOI] [PubMed] [Google Scholar]
- 60.Schlesinger M, Ashburner M, Tissieres A. Heat shock from bacteria to man. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 61.Sinclair A J, Palmero I, Peters G, Farrell P J. EBNA-2 and EBNA-LP cooperate to cause G0 to G1 transition during immortalization of resting human B lymphocytes by Epstein-Barr virus. EMBO J. 1994;13:3321–3328. doi: 10.1002/j.1460-2075.1994.tb06634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sixbey J W, Nedrud J G, Raab-Traub N, Hanes R A, Pagano J S. Epstein-Barr virus replication in oropharyngeal epithelial cells. N Engl J Med. 1984;310:1225–1230. doi: 10.1056/NEJM198405103101905. [DOI] [PubMed] [Google Scholar]
- 63.Steen R L, Cubizolles F, Le Guellec K, Collas P. A kinase-anchoring protein (AKAP)95 recruits human chromosome-associated protein (hCAP)-D2/Eg7 for chromosome condensation in mitotic extract. J Cell Biol. 2000;149:531–536. doi: 10.1083/jcb.149.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Strobl L J, Hofelmayr H, Stein C, Marschall G, Brielmeier M, Laux G, Bornkamm G W, Zimber-Strobl U. Both Epstein-Barr viral nuclear antigen 2 (EBNA2) and activated Notch1 transactivate genes by interacting with the cellular protein RBP-J kappa. Immunobiology. 1997;198:299–306. doi: 10.1016/s0171-2985(97)80050-2. [DOI] [PubMed] [Google Scholar]
- 65.Suwa A, Hirakata M, Takeda Y, Jesch S A, Mimori T, Hardin J A. DNA-dependent protein kinase (Ku protein-p350 complex) assembles on double-stranded DNA. Proc Natl Acad Sci USA. 1994;91:6904–6908. doi: 10.1073/pnas.91.15.6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Szekely L, Pokrovskaja K, Jiang W Q, de The H, Ringertz N, Klein G. The Epstein-Barr virus-encoded nuclear antigen EBNA-5 accumulates in PML-containing bodies. J Virol. 1996;70:2562–2568. doi: 10.1128/jvi.70.4.2562-2568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Szekely L, Selivanova G, Magnusson K P, Klein G, Wiman K G. EBNA-5, an Epstein-Barr virus-encoded nuclear antigen, binds to the retinoblastoma and p53 proteins. Proc Natl Acad Sci USA. 1993;90:5455–5459. doi: 10.1073/pnas.90.12.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tierney R J, Steven N, Young L S, Rickinson A B. Epstein-Barr virus latency in blood mononuclear cells: analysis of viral gene transcription during primary infection and in the carrier state. J Virol. 1994;68:7374–7385. doi: 10.1128/jvi.68.11.7374-7385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tomkinson B, Robertson E, Kieff E. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol. 1993;67:2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tong X, Drapkin R, Reinberg D, Kieff E. The 62- and 80-kDa subunits of transcription factor IIH mediate the interaction with Epstein-Barr virus nuclear protein 2. Proc Natl Acad Sci USA. 1995;92:3259–3263. doi: 10.1073/pnas.92.8.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tong X, Wang F, Thut C J, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain can interact with TFIIB, TAF40, and RPA70 but not with TATA-binding protein. J Virol. 1995;69:585–588. doi: 10.1128/jvi.69.1.585-588.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tong X, Yalamanchili R, Harada S, Kieff E. The EBNA-2 arginine-glycine domain is critical but not essential for B-lymphocyte growth transformation; the rest of region 3 lacks essential interactive domains. J Virol. 1994;68:6188–6197. doi: 10.1128/jvi.68.10.6188-6197.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang F, Petti L, Braun D, Seung S, Kieff E. A bicistronic Epstein-Barr virus mRNA encodes two nuclear proteins in latently infected, growth-transformed lymphocytes. J Virol. 1987;61:945–954. doi: 10.1128/jvi.61.4.945-954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang L, Grossman S R, Kieff E. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc Natl Acad Sci USA. 2000;97:430–435. doi: 10.1073/pnas.97.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang S, Guo M, Ouyang H, Li X, Cordon-Cardo C, Kurimasa A, Chen D J, Fuks Z, Ling C C, Li G C. The catalytic subunit of DNA-dependent protein kinase selectively regulates p53-dependent apoptosis but not cell-cycle arrest. Proc Natl Acad Sci USA. 2000;97:1584–1588. doi: 10.1073/pnas.97.4.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Westberg C, Yang J P, Tang H, Reddy T R, Wong-Staal F. A novel shuttle protein binds to RNA helicase A and activates the retroviral constitutive transport element. J Biol Chem. 2000;275:21396–21401. doi: 10.1074/jbc.M909887199. [DOI] [PubMed] [Google Scholar]
- 77.Woo R A, McLure K G, Lees-Miller S P, Rancourt D E, Lee P W. DNA-dependent protein kinase acts upstream of p53 in response to DNA damage. Nature. 1998;394:700–704. doi: 10.1038/29343. [DOI] [PubMed] [Google Scholar]
- 78.Yalamanchili R, Tong X, Grossman S, Johannsen E, Mosialos G, Kieff E. Genetic and biochemical evidence that EBNA 2 interaction with a 63-kDa cellular GTG-binding protein is essential for B lymphocyte growth transformation by EBV. Virology. 1994;204:634–641. doi: 10.1006/viro.1994.1578. [DOI] [PubMed] [Google Scholar]