Abstract
The intratumoral microbiome (TM) refers to the microorganisms in the tumor tissues, including bacteria, fungi, viruses, and so on, and is distinct from the gut microbiome and circulating microbiota. TM is strongly associated with tumorigenesis, progression, metastasis, and response to therapy. This paper highlights the current status of TM. Tract sources, adjacent normal tissue, circulatory system, and concomitant tumor co‐metastasis are the main origin of TM. The advanced techniques in TM analysis are comprehensively summarized. Besides, TM is involved in tumor progression through several mechanisms, including DNA damage, activation of oncogenic signaling pathways (phosphoinositide 3‐kinase [PI3K], signal transducer and activator of transcription [STAT], WNT/β‐catenin, and extracellular regulated protein kinases [ERK]), influence of cytokines and induce inflammatory responses, and interaction with the tumor microenvironment (anti‐tumor immunity, pro‐tumor immunity, and microbial‐derived metabolites). Moreover, promising directions of TM in tumor therapy include immunotherapy, chemotherapy, radiotherapy, the application of probiotics/prebiotics/synbiotics, fecal microbiome transplantation, engineered microbiota, phage therapy, and oncolytic virus therapy. The inherent challenges of clinical application are also summarized. This review provides a comprehensive landscape for analyzing TM, especially the TM‐related mechanisms and TM‐based treatment in cancer.
Keywords: analysis methods, immunotherapy, intratumoral microbiome, treatment application, tumor‐promotive and tumor‐suppressive mechanisms
Abbreviations
- AAVP
adeno‐associated virus/phage
- AhR
aryl hydrocarbon receptor
- AKK
Akkermansia muciniphila
- ALD
acetaldehyde
- ATM‐Chk2
ataxia telangiectasia mutated protein and checkpoint kinase 2
- Akt
protein kinase B
- B. thetaiotaomicron
Bacteroides thetaiotaomicron
- BFT
bacillus fragilis toxin
- CagA
cytotoxin‐associated gene A
- CAFs
cancer‐associated fibroblasts
- CDDL
bacterial cytidine deaminase
- ChIP
chromatin immunoprecipitation
- CLEM
correlative light and electron microscopy
- COX2
cyclooxygenase 2
- CRC
colorectal cancer
- CREB
cAMP response element‐binding protein
- CTLA‐4
cytotoxic T‐lymphocyte associated protein 4
- CTL
cytotoxic T lymphocyte
- CTX
cyclophosphamide
- DAMP
danger‐associated molecular patterns
- ddPCR
droplet digital PCR
- DDR
DNA damage response
- DSP
digital spatial profiling
- DSS
dextran sulfate sodium
- 3D
three‐dimensional
- ETS1
E26Transformation‐Specific Sequence 1
- E. coli
Escherichia coli
- EBV
Epstein‐Barr virus
- ECM
extracellular matrix
- E. hirae
Enterococcus hirae
- ELISA
enzyme‐linked immunosorbent assay
- ERK
extracellular regulated protein kinases
- ETBF
enterotoxigenic Bifidobacterium fragilis
- EVs
extracellular vesicles
- FAO
fatty acid oxidation
- FcγRs
Fcγ receptors
- FISH
fluorescence in situ hybridization
- Fap2
fibroblast activation protein‐2
- FMT
fecal microbiome transplantation
- GECs
gingival epithelial cells
- GM‐CSF
granulocyte‐macrophage colony‐stimulating factor
- HPV
human papilloma virus
- HSV‐1
herpes simplex virus type 1
- HSP27
Heat Shock Protein 27
- HTLV‐1
human T‐lymphotropic virus 1
- 3‐IAA
indole‐3‐acetic acid
- ICI
immune checkpoint inhibitors
- IL
interleukin
- I‐CZE
immuno‐capillary zone electrophoresis
- IHC
immunohistochemistry
- IOM
immuno‐oncology‐microbiome
- I3A
indole‐3‐aldehyde
- ICD
immunogenic cell death
- KSHV
Kaposi sarcoma‐associated herpesvirus
- LPS
lipopolysaccharide
- LTA
lipoteichoic acid
- MAPKs
mitogen‐activated protein kinases
- mAb
monoclonal antibody
- MCC
merkel cell carcinoma
- MEK
Mitogen‐Activated Protein Kinase
- MDSCs
myeloid‐derived suppressor cells
- MCPyV
merkel cell polyomavirus
- NF‐κB
nuclear factor κ‐B
- NATs
normal adjacent tissues
- NGS
next‐generation sequencing
- NO
Nitric Oxide
- NRPS
nonribosomal peptide megasynthases
- OAs
oncolytic adenoviruses
- OMT
oral microbiota transplantation
- OSCC
oral squamous cell carcinoma
- OMVs
outer membrane vesicles
- OVs
oncolytic viruses
- OVT
Oncolytic virus therapy
- PD‐1
Programmed Cell Death Protein 1
- PAR
Partitioning‐Defective
- PRRs
pattern recognition receptors
- P. aeruginosa
Pseudomonas aeruginosa
- pTh17
"pathogenic” helper T cells17
- PD‐L2
Programmed Death‐Ligand 2
- PDAC
pancreatic ductal adenocarcinoma
- PDOs
patient‐derived organoids
- PI3K
phosphoinositide 3 ‐ kinas
- PKS
polyketide megasynthases
- RIPA
rapid immuno‐filter paper assay
- RNI
reactive nitrogen intermediates
- RNS
reactive nit rogen species
- ROS
reactive oxygen species
- RAS
Rat Sarcoma
- RAF
Rapidly Accelerated Fibrosarcoma
- RGMb
Repulsive Guidance Molecule B
- rRNA
ribosomal RNA
- RT
radiotherapy
- RT‐qPCR
real‐time fluorescence quantitative polymerase chain reaction
- SCA
single‐cell analysis
- SCFAs
short‐chain fatty acids
- scRNA‐seq
single‐cell RNA sequencing
- SPRIA
solid‐phase radioimmunoassay
- ST
spatial transcriptome
- STAT
signal transducer and activator of transcription
- STING
stimulator of interferon genes
- TAMs
tumor‐associated macrophages
- TCA
tricarboxylic acid
- TCMA
the cancer microbiome atlas
- TLR4
toll‐like receptor 4
- TIGIT
T cell immunoglobulin and ITIM domain
- TEM
transmission electron microscopy
- TM
tumor microbiome (intratumoral microbiome)
- TME
tumor microenvironment
- U‐VEC
talimogene laherparepvec
1. BACKGROUND
Microorganisms have a rich history on Earth, dating back to some of the earliest forms of life [1]. They are among the oldest living organisms on Earth, playing an inestimable role in making the Earth's environment habitable for human habitation. Most microorganisms have specific common properties in their preference for the environment in which they live, such as the requirements for oxygen, nutrients, and temperature, which are either stringent or lenient. For a considerable duration, human‐focused microbiological research remained limited until the discovery of microorganisms within the human body during the 18th and 19th centuries. It was astonishing to find that the microbial population in humans, comprising fungi, bacteria, protozoa, viruses, phages, and other microorganisms, far outnumbered the count of eukaryotic cells. Microbes were also found in tumors, an environment abundant in nutrients, anaerobic, and suitable for microbial survival [2, 3]. Consequently, we realized that microbes might connect with human health and disease. However, due to the limitations of the research methods, we need to gain more knowledge of the intratumoral microbiome (TM) and its metabolites. Subsequently, next‐generation gene sequencing has allowed us to study TM more visually, and we have reconfirmed their widespread presence among tumor tissues. From studies that relied on the relationship between relatively large levels of gut microbes and gastrointestinal tumors [4], there has been a switch to studies of tumors with relatively minor microbial content.
This review focuses on hypotheses such as the origin of microorganisms within the tumor, the corresponding analytical techniques, tumor‐microbe interactions, pathogenic/oncogenic mechanisms, cancer therapy‐associated TM, and some controversies (Figure 1).
FIGURE 1.
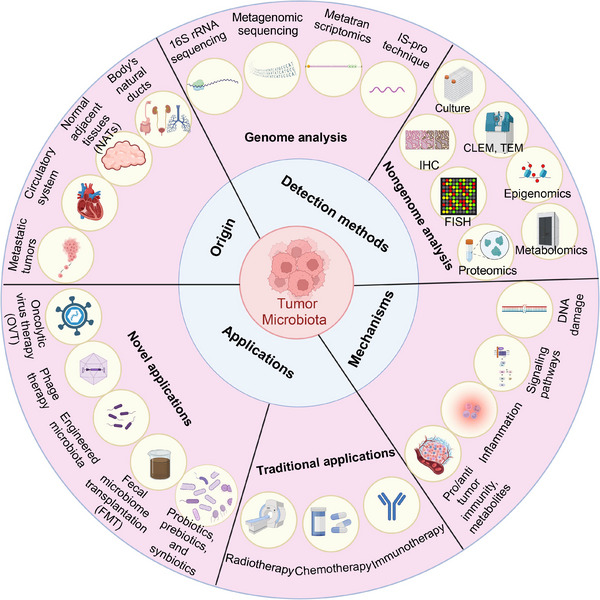
An overview of the advances in TM. Illustrating the origin of TM (Tract sources, adjacent normal tissue, circulatory system, and concomitant tumor co‐metastasis), the TM analysis methods (16SrRNA sequencing, shotgun metagenomic sequencing, metatranscriptomics, IS‐pro technique, immunohistochemistry, fluorescence in situ hybridization, proteomics, metabolomics, correlative light and electron microscopy and transmission electron microscopy, TM Culture, single cell analysis and spatial transcriptome, organoids and 3D technology, gene chip technology, nanotechnology, computational tool, molecular detection method based on viral nucleic acid, immunological method, and nucleic acid hybridization), the potential mechanism of how TM is involved in tumors (DNA damage, activation of oncogenic signaling pathways, influence cytokines and induce inflammatory responses, and the interaction with tumor microenvironment, and the promising directions of TM‐based treatment (immunotherapy, chemotherapy, radiotherapy, the application of probiotics/prebiotics/synbiotics, fecal microbiome transplantation, engineered microbiota, phage therapy, and oncolytic virus therapy). Abbreviations: CLEM, correlative light and electron microscopy; FISH, fluorescence in situ hybridization; FMT, fecal microbiome transplantation; IHC, immunohistochemistry; OVT, Oncolytic virus therapy; TEM, transmission electron microscopy; TM, tumor microbiome (intratumoral microbiome). Biorender supported the materials in Figure 1.
2. TM
There are about 4 × 1013 microbial cells in the human body, representing about 3 × 103 species. Among these, 97% are intracolonic bacteria, 2‐3% are extracolonic bacteria (proximal intestine, skin, lungs, tumor tissues, etc.), and the quantity and diversity of human infectious viruses and phages may be even greater [5, 6]. Many microorganisms, which far exceed the number of human somatic cells, are widely distributed throughout the human body, including within tumor tissues.
The concept of TM was introduced as early as the 19th century. TM refers to the microbiota in various tumor tissues, including bacteria, fungi, viruses, parasites, etc. [7]. Although few microbes have been confirmed to play a pathogenic role in cancer, a growing body of microbiota is demonstrated to be associated with specific cancer types and stages [8, 9]. Then, in 2020, over 1,500 samples of breast, lung, ovary, pancreas, melanoma, bone, and brain tumors were studied, and each kind was found to have a unique microbiome composition and diversity [1]. Additionally, evidence indicated that at least 33 major cancer types harbor specific intratumoral microbiomes, often organized within microniches [10]. These findings suggest that the microbiome could serve as a promising biomarker and therapeutic target for tumors and other diseases.
3. ORIGIN OF TM
Using various methods to detect bacteria, Nejman et al. [1] demonstrated the presence of bacteria in tumor cells and tumor‐infiltrating immune cells in various cancer settings. Given the body's triple barrier immune defense against foreign microorganisms, questions arise regarding the source of these microorganisms, their mechanisms of entry into tumors, evasion of immune clearance, and establishment of a stable microbiota within tumor subtype. Thus, addressing these questions could facilitate further research on the longitudinal chronology of tumorigenesis and microbial residence around the microbial‐host‐immune system and further hint at the causal relationship to clarify the mechanism of tumor promotion or suppression.
3.1. Gastrointestinal, respiratory, and urinary tract sources
In the body's natural ducts, such as the digestive tract, respiratory tract, and genitourinary tract, there is a mucosal barrier, which serves as the first barrier against foreign invasion and is the site where microbial aggregation most often occurs. Under normal circumstances, even though symbiotic microbial communities exist in the mucosa, they do not cause carcinogenic damage to the host organism. When some factors emerge, such as for the digestive tract and western diet, excessive nitrites disrupt the ecological balance of the mucosal symbiosis, destroy the mucosal barrier, colonize the epithelial tissues, and exert carcinogenic effects. Studies have identified the presence of microorganisms in tumors at the mucosal barrier, such as gastric cancer, colorectal cancer (CRC), lung cancer, urothelial tumors, and melanoma [1].
The bacterial biofilm discovered on oral squamous cell carcinoma (OSCC) had a higher overall abundance of total anaerobic and aerobic bacteria based on colony‐forming units, similar to data from the colon, compared to the intact digestive tract. The strains detected in CRC samples were identical to those isolated from the saliva of CRC patients, according to a study carried out simultaneously to identify isolates at the strain level by arbitrary primer Polymerase Chain Reaction (PCR). This finding supports the oral origin of Fusobacterium nucleatum (F. nucleatum) [11, 12]. The observation highlights the intricate relationship between tumor‐associated microbiota within the gastrointestinal tract and the origins of both oropharyngeal and colon cancers.
Additionally, the direct origin of the anatomical site is the more widely discussed part. After microbiome assessment of tumor samples from patients with pancreatic ductal adenocarcinoma (PDAC), one study found a predominance of the phyla Aspergillus, Bacteroides, and Synechococcus in the samples, while the Enterobacteriaceae (family), Pseudomonas (genus), and Elizabethan Aspergillus (genus) were particularly significant in the Aspergillus phylum, which may in part reveal the ability of Aspergillus to transfer from the intestine to the pancreas [12]. In pancreatic tumor samples, the γ‐anamorphic phylum, found in pancreatic tumor samples, is also considered to metastasize from the intestine to pancreatic tumors. The most likely route for this translocation is the pancreatic duct connected to the duodenum [13]. Pushalkar et al. [14] used oral gavage to implant fluorescently labeled enterococcus faecalis into wild‐type mice and directly observed that intestinal flora can migrate into the pancreas. Meanwhile, PDAC‐associated bacteria can originate from the gastrointestinal tract retrogradely [15], directly impacting the pancreatic microbial environment.
Moreover, as for urological tumors, one of the established risk factors for bladder cancer is a history of three or more urinary tract infections, most of which are Escherichia coli (E. coli) ‐associated urinary tract infections, suggesting that retrograde urinary tract infections may be closely related to bladder cancer [12].
3.2. Entry of the original microbiota in the adjacent normal tissue
In addition to the natural tract, normal adjacent tissues (NATs) are also considered the source of TM. Some researchers reported that adjacent “normal” tissue contains microbiota that may resemble TM [16]. In 2020, Nejman et al. [1] demonstrated that tumor tissues and their NATs had a similar microbial composition, while bacteria prevalence and metabolic‐associated enzymes significantly differed. For example, breast cancer has a higher diversity of bacteria and richness of enzymes related to anaerobic respiration [1]. That may illustrate that some specific microorganisms are indispensable in tumor formation. Nevertheless, some investigators believe that the similarity of the microbiota composition in tumor sites and NATs is due to the origin of microorganisms in NATs from tumor microenvironments [17]. Thus, it is uncertain whether NATs are one of the sources of intratumor microbes, and more evidence is required to clarify.
3.3. Through the circulatory system
A tumor microenvironment gradually forms neovascularization during progression and starts having an abundant blood supply. The most common metastasis, in general, is metastasis to the liver [19], followed by metastasis to the lung. When the mucosal barrier of the respiratory and digestive tract is damaged, some resident microorganisms may enter the circulatory system through the mucosa's rich and inflammatory blood vessels and flow to the site of the rich blood supply of the tumor [20]. In the meantime, the ecology of the microbiota at the mucosal barrier is dysregulated, and this ecological dysregulation can lead to impaired local, regional, and systemic immune responses, disruption of the mucosal barrier, translocation of intestinal bacteria to the mesentery lymph nodes (mLNs) and into the peripheral circulation, altered cytokine environment within the intestinal mucosa, the flow of mLNs to the inflammatory phenotype, activation of Th17 cells and effector T cells, leading to an influx of neutrophils and triggering severe inflammation in the local and systemic state [21]. Thus, the presence of microbes in the circulatory system and local microbial ecological dysregulation may be mutually causal, creating a vicious circle. The underlying mechanism is that mucosal barrier dysfunction promotes microbes to escape into the circulatory system [22, 23]. However, the detailed mechanism is still worth discussing.
3.4. Concomitant tumor co‐metastasis
For more metastatic tumors, one study by bacterial 16S ribosomal RNA (rRNA) gene sequencing confirmed (i) the presence of Clostridium species in paired primary metastases. (ii) a correlation between the relative abundance of Clostridium species in primary tumors and metastases. (iii) a dominant microbial genus in liver metastases corresponding to the dominant microbial genus in primary tumors, demonstrating paired microbiota stability between Clostridium‐positive primary metastases [24].
Nearly identical active Clostridium strains were found in matched primary and metastatic colorectal cancers, confirming the persistence of active Clostridium during metastasis and indicating that Clostridium may migrate to metastatic sites along with CRC cells [24]. Moreover, F. nucleatum bacteria and its associated microbiota persist in distant liver metastases from colorectal cancers [25], demonstrating that F. nucleatum may also co‐metastasize with the tumor [26].
To sum up, the source of TM mainly includes four aspects: the natural ducts, normal adjacent tissues, the circulatory system, and concomitant tumor co‐metastasis. At present, it is unclear how they enter the tumor, escape clearance by the immune system, and settle down to form a microbiota with a stable structure of tumor subtypes. More valuable research is worth exploring in the future.
4. METHODS OF TM ANALYSIS
Currently, a study by Fletcher et al. [27] has led to discussions about whether the intrinsic pancreatic mycobiome affects the initiation and development of PDAC. However, conclusive findings remain elusive due to the absence of standardized methods for generating and analyzing microbiome and sequencing data. Here, we will have a detailed introduction to the present detection methods of microbes [27]. Advancements in next‐generation sequencing (NGS) technology have revealed that tissues once believed to be sterile harbor a diverse array of microorganisms. 16S rRNA sequencing, metagenomic sequencing, metatranscriptomics, and Intergenic Spacer‐profiling (IS‐pro) technique have emerged as critical means of analysis for prokaryotes, viruses, fungi, and other microorganisms. In addition to the above genetic detection techniques, non‐genomic analysis, including proteomics, metabolomics, epigenomics, immunohistochemistry (IHC), fluorescence in situ hybridization (FISH), correlative light and electron microscopy (CLEM), transmission electron microscope (TEM), and culture, are also critical. Moreover, virus detection methods, cutting‐edge technologies, and some challenges will be described in detail in this chapter.
4.1. Genome analysis
4.1.1. 16S rRNA sequencing
16S rRNA sequencing is a widely used analytical method for bacteria and archaea [28, 29]. 16S rRNA is a component of the 30S subunit of the bacterial ribosome and possesses highly variable regions (V regions, which differ between species) and conserved regions (highly similar between species) [30]. The conserved regions reflect the affinities between biological species, while the variable areas represent the variation between species. Because of these characteristics, 16S rRNA sequencing can be applied to the research of community species composition, evolving relationships among species, and diversification of microbial populations in many tumor tissues, including intrahepatic cholangiocarcinoma [31], breast cancer [32], PDAC [1], CRC [33], etc. Additionally, 16S rRNA sequencing requires only specific sequences (rather than all sequences) to be detected, features the advantages of rapid detection and low cost, and enables to detect bacterial communities at species and strain level [30, 34]. However, 16S rRNA sequencing still has some deficiencies that need to be improved. For example, due to its relatively low resolution, 16S rRNA sequencing may be unable to distinguish between closely related species. It also potentially suffers from PCR amplification deviations and overstatement of diversity estimates [30].
4.1.2. Shotgun metagenomic sequencing
Shotgun sequencing is the off‐target sequencing of all genomes and can be used to analyze microbiota's taxonomic composition and potential function [35]. For example, Huang et al. [36] used shotgun metagenomic sequencing to assess compositional and functional microbiota profiling in CRC and the interaction of microorganisms such as Coprococcus with neoadjuvant chemoradiotherapy. Compared with 16S rRNA, it allows the detection of non‐bacterial microorganisms such as fungi, viruses, mycoplasma, etc. [37]. In particular, evidence shows that shotgun sequencing analysis of tumor genomes can identify considerably more virus‐positive cases [38]. The results of this analytical method are relatively more accurate, making it an indispensable technique for tumor microbiomes. However, this analysis method requires the detection of all gene sequences (including host normal and tumor cells) and is relatively time‐consuming, complicated, and high‐cost [39].
4.1.3. Metatranscriptomics
Metatranscriptomics, a subset of metagenomics, plays a crucial role in elucidating the gene expression profiles of complex microbial communities [40]. By providing insights into the expression of various genes, it enables researchers to understand the functional activities and mechanisms of microorganisms. Its high‐throughput capabilities make it invaluable for comprehensive analyses [41]. For example, Chai et al. [31] confirmed Paraburkholderia fungorum could inhibit the growth of intrahepatic cholangiocarcinoma through alanine, aspartate, and glutamate metabolism using transcriptomics and other analytical methods. Moreover, metatranscriptomics can also provide higher coverage and decrease the risk of artifacts. However, it still has some limitations, including instability of RNA molecules, high cost, and sensitivity to host RNA, especially rRNA contamination [40, 42, 43]. In the future, this technology integrated with single‐cell analysis technology may present a more valuable technique to study the interaction between tumor cells and intratumoral microorganisms.
4.1.4. IS‐pro technique
IS‐pro technique, an analytical technique similar to 16S rRNA sequencing, detects the microbial DNA, especially a universal ribosomal DNA region, the 16S‐23S rDNA intergenic spacer (IS) region, which is unique for each bacterial species [44]. According to studies, IS‐pro, in combination with rapid taxonomic categorization using phylum‐specific fluorescence labeling of PCR primers, can resolve bacterial taxa down to the species level. Additionally, a study revealed that IS‐pro analysis, compared to 16S rRNA sequencing technology, could speed up analyses, lower expenses, and keep the same level of profiling, opening the door to quick investigation of microbiota [45, 46].
4.2. Non‐genomic analysis
4.2.1. IHC
IHC is a traditional method that detects Gram‐positive and negative bacteria using antibodies against bacterial lipopolysaccharide (LPS) and lipoteichoic acid (LTA). This method can localize, characterize, and quantify bacteria by using specific antibodies labeled with chromogenic agents to react with the corresponding bacterial structure to visualize them chemically. This method is usually used to confirm the presence of TM, combined with other detection methods such as 16S rRNA sequencing and FISH [31, 47]. Generally, it is inexpensive and easy to perform but also has a high rate of false positives. For instance, it may yield LPS/LTA positivity following a prolonged period of bacterial phagocytosis, even when no viable bacteria are present [1].
4.2.2. FISH
FISH, which has long been a crucial tool for studying cultured microorganisms, uses a standard probe for bacterial 16S rRNA or conserved fungal 28S rRNA sequences that can be used to identify individual microbial cells directly. Due to the varying degrees of conservation of various rRNA regions, probes can be species‐specific or chosen based on various taxonomic levels [43]. So, this technique enables us to confirm the presence of microbes, including fungi and bacteria. For example, Cai et al. [31] used specific oligonucleotide probes to target bacterial DNA and found bacteria such as Klebsiella pneumoniae in intrahepatic cholangiocarcinoma. Recently, new techniques have made it possible to visualize and sort tiny amounts of bacteria, do single‐cell quantification, and better analyze particular microbial populations. These techniques include catalyzed reporter deposition‐FISH (CARD‐FISH) and highly phylogenetic resolution FISH (HiPR‐FISH) [48] and can be applied in this area.
4.2.3. Proteomics
Proteomics is the study of proteins interact with each other or other molecules and the roles they play within the organism [49]. Proteomic research provides an overall view of the processes underlying cellular processes at the protein level. Mass spectrometry coupled with liquid chromatography (LC‐MS) is one of the essential methods for these analyses and has become a powerful tool for TM research [50, 51, 52]. Broadly, proteomics can identify and quantify proteins that are differentially expressed between healthy and cancerous tissues, which prospectively reveal microbial pathogenic mechanisms and biomarkers of cancers [53]. With the data‐independent acquisition, targeted proteomics analysis, and immunoprecipitation, proteomics is becoming a promising functional, analytical technique for TM [43].
4.2.4. Metabolomics
The systematic identification and measurement of a biological system's small molecule metabolic byproducts at a given time are called “metabolomics” [54]. The goal of metabolomics in TM research is to characterize the metabolic variations and function of tumor microorganisms [55, 56]. As a result, metabolic analysis techniques may improve knowledge of the molecular pathways behind cancer development and the therapeutic response involving tumor microorganisms [57]. In addition, metabolites such as short‐chain fatty acids (SCFAs), bile acids, inosine, indole, etc., have been confirmed to be involved in the manipulation of the tumor microenvironment (TME), including immunity, inflammation, and signaling pathways, thus affecting tumorigenesis and treatment response [43, 58]. Researchers conducted spatially resolved metabolomics analysis to discover Akkermansia muciniphila‐associated metabolic features and anti‐tumor effect [59]. Metabolomics has the potential to better elucidate metabolite interactions between cancer cells and intratumoral microorganisms, facilitating the search for therapeutic targets.
4.2.5. Epigenomics
The study of phenotypic changes that do not include changes in the DNA sequence is called epigenetics. Epigenetic regulation is closely relevant to human diseases, notably cancer [60]. Specifically, histone glycation and aberrant methylation of DNA are demonstrated to be strongly associated with tumorigenesis and progression [61, 62]. Chromatin immunoprecipitation (ChIP), also known as binding site analysis, enables us to better understand epigenetic changes to the genome [63]. ChIP‐seq, ChIP in combination with NGS, is crucial to the (epi)genomic studies of both host and microbe cells [43] and has a great potential for studying the interactions of host and microbe cells.
4.2.6. CLEM and TEM
Using CLEM, it is possible to locate cells and molecules with excellent resolution and accuracy. CLEM combines the benefits of light microscopy and electron microscopy. The structural information, size distribution, and shape of nanoparticles consisting of lipids and proteins can be revealed using the high‐resolution technique known as TEM [64]. These techniques can verify the presence of microorganisms inside cancer cells and demonstrate the intracellular localization of microbes [65]. In addition, the researchers observed the morphology of the bacteria in conjunction with TEM and found that they could be encapsulated in lysosomes [31]. Integrating with fluorescent probes or specific nanoparticles, they can mark target molecules or cells or even lend to applications in single particle tracking (SPT) inside living cells [43, 65]. Therefore, CLEM and TEM may play a more excellent role in studies of intratumor microbiota.
4.2.7. TM culture
Although various technologies have been used to study TM, culturing is still an essential and significant method for microorganism research [64, 66]. Microbial culture contributes to describing new microbial species and enables us to obtain pure microbial culture for further research [67]. Nejman et al. [1] applied fluorescently labeled D‐alanine to culture slices from freshly resected human tumors and verified the presence, survival, and metabolically active bacteria in human breast tumors. Additionally, other studies have found that intratumor bacteria in fresh tumor tissue are alive by bacterial culture [31]. Recently, a method based on reverse genomics that can capture certain microbes by targeting specific protein epitopes has shown the potential to separate and culture intratumor microbes [68]. Moreover, promising organoid technology also provides the possibility for future tumor microbiota culturing [16, 43, 69].
4.3. Detection methods of viruses
In addition to the above methods, viruses, as much smaller microorganisms, require additional techniques to detect them, including molecular detection methods based on viral nucleic acid, immunological methods, nucleic acid hybridization, and gene chip technology.
4.3.1. Molecular detection method based on viral nucleic acid
Real‐time fluorescence quantitative polymerase chain reaction (RT‐qPCR) is a PCR technology that can quantitatively detect the amount of targeted gene amplification of the virus [70]. It has been developed into a widely used technology for various virus detection, with the advantages of reasonable specificity and high sensitivity [71, 72].
Droplet digital PCR (ddPCR) represents an innovative technology derived from reverse transcription quantitative PCR (RT‐qPCR) [73]. Both techniques operate on the principle of incorporating fluorescent dyes or fluorescently labeled oligonucleotide chains into the PCR system [74]. These labels bind to amplification products during PCR amplification, generating fluorescence upon excitation. By monitoring changes in fluorescence signals via a fluorescence signal detector, the number of copies of the target gene can be determined, providing insights into virus content [75, 76].
4.3.2. Immunological method
The immunological method is a virus detection based on the specific reaction of antigens and antibodies. The virus diagnosis using this method is accurate, sensitive, rapid, simple, and low‐cost. The immunological method mainly includes a hemagglutination inhibition test [77], complement fixation assay [78], neutralization test [79], enzyme‐linked immunosorbent assay (ELISA) [80], immunoprecipitation and immunoblotting [81], immunogold‐label assay [82], rapid immuno‐filter paper assay (RIPA) [83], immuno‐capillary zone electrophoresis (I‐CZE) [84], immuno‐PCR [85], and solid‐phase radioimmunoassay (SPRIA) [86].
4.3.3. Nucleic acid hybridization
Nucleic acid hybridization involves the annealing of two nucleic acid molecules, originating from different sources but sharing certain homology, to form heteroduplex molecules under denaturation conditions [87]. This process is commonly employed to fix the nucleic acid of interest onto a membrane, followed by hybridization with a nucleic acid probe. Subsequently, the hybridization complex is labeled and visualized. The specificity of detection relies on the extent of complementarity between the probe and the viral nucleic acid sequence. Nucleic acid hybridization demonstrates versatility in detecting various viral entities, including DNA viruses, RNA viruses, and viroids, with high sensitivity and specificity. It can be utilized in conjunction with reverse transcription polymerase chain reaction (RT‐PCR) for spot hybridization, intracellular in‐situ hybridization, DNA blot hybridization, RNA blot hybridization, and other methods, facilitating comprehensive virus detection [88, 89, 90, 91].
The advantages and disadvantages of different sequencing technologies are briefly summarized in Table 1.
TABLE 1.
Advantages and disadvantages of different detection technologies.
| Detection technologies | Detection materials | Advantage | Disadvantage | Refs |
|---|---|---|---|---|
| 16S rRNA sequencing | Sequencing of hypervariable 16S rRNA region (such as V3‐V4) allows classification of bacterial composition | Fast, inexpensive, enables the detection of bacterial communities at species and strain level | Limited accuracy, poor repeatability, unable to detect other species except bacteria, host genome contamination | [1, 16, 30, 31] |
| Shotgun sequencing | Whole genomic content of sample | Accurate, wide application, enables the functional analysis | Expensive, complicated, time‐consuming, host genome contamination | [35, 36, 37, 39] |
| Metatranscriptomics | Sequencing of transcribed bacterial RNA content of sample (RNAseq) | Provides whole gene expression | Short half‐life of mRNA, high cost and sensitivity to host RNA | [40, 42, 43] |
| Proteomics | Analysis of proteins in samples | Reveals potential interaction between microbes and host cells and carcinogenic mechanisms | Insufficient sensitivity, resolution, and accuracy in detecting differential protein expression, stringent conditions required for some protease analyses | [37, 43, 326] |
| Metabolomics | Global analysis of metabolites derived from microbes | High resolution, enables the in situ analysis, enables the construction of metabolic networks | Unable to observe dynamically | [43, 58, 59] |
| IS‐pro | Sequencing of 16S‐23S rRNA gene interspace regions | Fast, easy operation, enables the detection of bacterial communities at species level | Proprietary technology | [45, 46] |
| IHC | Antibody of microbial structure | Enables the reflection of microorganism presence | High false positive rate, sensitivity and specificity vary by species | [1, 327] |
| FISH | Probe against microbial ribosomal RNA (rRNA) | Species‐specific, enables the reflection of microorganism position | High requirement of microbial concentration | [43, 327] |
| CLEM, TEM and ECM | Slices of tissues | Shows presence and location of microorganism, presets the microbial morphology | Expensive, strict material requirement | [32, 43, 52, 65] |
| Culture | Refresh tissues | Shows presence of live bacteria | Insufficient culture methods | [16, 43, 69] |
| RT‐qPCR | DNA/RNA sequences | Quantifies the number of viruses, high specificity and sensitivity | Unable to test mutation genes | [71, 72] |
| DdPCR | Droplet nucleic acid samples | Visual response to the number of viruses | Limited throughput and complex operation | [75, 76] |
| Nucleic acid hybridization | Nucleic acid | High sensitivity and specificity | Complex operation | [88, 89, 90, 91] |
4.4. Cutting‐edge technology
4.4.1. Single‐cell analysis and spatial transcriptome
Single‐cell analysis (SCA) technology, studying the individual cell, is an emerging area to reveal the heterogeneity of single cells [92, 93]. They are integrating with other techniques at the single‐cell level to form single‐cell multi‐omics technologies, including single‐cell DNA sequencing, single‐cell RNA sequencing, single‐cell epigenetics, single‐cell proteomics, single‐cell metabolomics, etc. Single‐cell RNA sequencing (scRNA‐seq) is widely used in various fields and can reveal multiple cellular subpopulations and intratumoral transcriptional heterogeneity among cancer cells [94, 95, 96, 97]. Through single‐cell sequencing, investigators recognized immune cell heterogeneity. They identified secretory leukocyte protease inhibitors as an oncogene associated with cell viability and apoptosis, which offers a potential therapy target for pancreatic cancer [98]. However, scRNA‐seq presupposes tissues must be mechanically separated or enzymatically dissociated into single‐cell suspensions. This process inevitably loses primitive positional information and leads to a disruption of the intercellular communication network. Spatial transcriptome (ST) technology enables gene sequencing in situ in tissues to obtain spatial information on gene expression and compensates for the shortcomings of single‐cell technology. Simultaneous use of both techniques allows for transcriptional characterization of single cells in a local tissue context, discovers cell‐cell and molecular interaction in the TME, and clarifies the signaling pathway network [96]. For example, Galeano et al. [10] utilized targeted RNAscope‐fluorescence in situ hybridization (RNAscope‐FISH) imaging to confirm the heterogeneous spatial distribution of microorganisms and the unbiased 10x Visium spatial transcriptomics to further distinguish the spatial distribution and identity of the TM in the TME. Subsequently, the GeoMx digital spatial profiling (DSP) platform was used to present the expression profile of proteins that were related to anti‐tumor immunity and cancer progression. In addition, they introduced a single‐cell RNA‐sequencing method called invasion‐adhesion‐directed expression sequencing (INVADEseq), which targets a conserved region of intracellular bacterial 16S rRNA but does not affect the gene‐expression profile of host cells, showing the interactions and cellular functions of these host‐bacterial associations within the TME. They found that these intracellular bacteria enable the activation of transcriptional factors from the JUN and FOS families, which relate to cancer cell invasion, metastasis, DNA damage repair, and cell dormancy. Meanwhile, these invasive microorganisms secret specific interleukins and chemokines to recruit myeloid cells and induce inflammatory response through JAK‐STAT signaling, promoting T‐cell exclusion and tumor growth within the TME [10]. It was reported that intratumoral metabolic heterogeneity was specifically related to tumor immunosuppression microenvironment, confirmed by spatial transcriptomics [99]. Currently, scRNA‐seq and ST have been used in various cancers, including glioblastoma [100], squamous cell carcinoma [101], CRC [102], PDAC [97], breast cancer [103], etc. Besides, Wang et al. [104] integrated multiple ST slices to reconstruct 3 dimensional (3D) tissue architectures enabling us to better understand signal networks and biological processes. However, it is rarely reported for TM. Interestingly, a scRNA‐seq platform called massively‐parallel, multiplexed microbial sequencing (M3‐seq) has been created for bacteria that combines combinatorial cell indexing with post hoc rRNA depletion, which can profile bacterial cells of various species. [105] In the future, these technologies will be more used in this field and have great potential to unlock problems that cannot be solved.
4.4.2. Organoids and 3D technology
Organoid technology is spatially structured tissue analogs formed by in vitro 3D culture of adult stem cells or human pluripotent stem cells [106]. This technology can recapitulate the cellular heterogeneity, structure, and functions of human organs to the greatest extent possible and can be stably cultured for an extended period [107]. Nowadays, patient‐derived organoids (PDOs), which are organoids obtained by culturing patient biopsies, punctures, or surgically excised tissues in hydrogels for a specific time, have widely been studied for better oncology research [108]. A variety of PDOs have been constructed as organoid biobanks, including breast cancer [109], rectal cancer [110], PDAC [111], lung cancer [112], pancreatic cancer [113], ovarian cancer [114], glioblastoma [115], and head and neck squamous cell carcinoma [116], which enable to reflect histopathologic and molecular characteristics of cancers. Due to the features above, we consider organoids, especially PDOs, as potential models for further studies of the TM. In addition, organoid technology has been demonstrated to be capable of modeling the tumor immune microenvironment, applying it to cancer treatment studies, and predicting the therapeutic response, including immunotherapy [112, 115, 117, 118], chemotherapy [110, 114], and radiotherapy [110]. As such, this technique can see how microorganisms affect cancer treatments. Moreover, Puschhof et al. [119] introduced intestinal organoids and organ‐on‐a‐chip platforms. They described how they are used to study host‐microbiota interactions [32], which provide a theoretical basis for tumor microbiota research. 3D dual topographical tumor model, a viable experimental platform for investigating tumor invasion and identifying therapeutic targets against metastasis [120], and 3D‐printed microrobots [121] also have great potential to be utilized for TM research and merit more investigations.
4.4.3. Gene chip technology
Gene chip technology is also known as DNA chip, biochip (biochip), and microarray [122]. The principle is that the known biomolecular probe or gene probe is large‐scale or orderly arranged on the carrier, such as a small piece of silicon chip, and the biomolecular or gene sequence in the sample to be tested interacts and reacts in parallel. Under the excitation of a laser, the receiver collects the fluorescence spectrum signal, and the computer automatically analyzes and processes the data and reports the results. The advantage of gene chip technology is that it can simultaneously complete the detection and analysis of many sample DNA sequences, which solves many shortcomings of traditional nucleic acid hybridization technology [123, 124, 125].
4.4.4. Nanotechnology
Nanotechnology is increasingly crucial in TM research, including diagnosis, treatment, etc. For example, Yang et al. [126] proposed a strategy to sensitize bacteria for in vivo imaging by aggregating glucose polymer‐modified gold nanoparticles in bacterial cells to produce enhanced photoacoustic signals and even remarkable antibacterial activity [126].
4.4.5. Computational tool
Computational tools present an efficient analysis method for high‐throughput microbiome data. Zhu et al. [127] proposed CAMMiQ, a new computational tool that can identify microbes in high throughput sequencing samples and assess the abundance of each species or strain [127]. Besides, Wang et al. [128] designed a user‐friendly online platform aimed at advancing cancer‐related microbiome research. This platform enables users to browse, search, visualize, and download microbial abundance data from various tissues along with corresponding analysis results. Moreover, The Cancer Microbiome Atlas (TCMA), leveraging The Cancer Genome Atlas (TCGA), offers a curated collection of decontaminated microbial compositions from diverse tissues to facilitate intratumoral microbiome (TM) studies [129].
Overall, the field of TM research is advancing rapidly with the application of a wide range of technologies. However, many challenges, including sample acquisition, interference of contamination, and low repeatability, still need to be resolved. Recently, a large‐scale population study has even revealed that there is no common blood microbiome among healthy individuals [130]. Yet, cancer patients seem to contain a specific microbial population, which may contribute to early screening for cancer. In the future, multi‐omics and multi‐modal analysis is the way forward for TM detection technology and is strongly believed to facilitate the development of this field.
5. THE INVOLVEMENT OF MICROBIOTA IN CANCER
Microbiota has a dual role in oncology. The role of microbiota in tumorigenesis development may be of vital importance. First, it has been revealed that pathogenic microbes may mediate genetic material damage through genotoxins, cause chromosomal instability, or activate tumorigenesis‐related signaling pathways that directly trigger cancer [9]. Second, microbial ecological dysbiosis causes inflammation, especially chronic inflammation, promoting carcinogenesis, which was first proposed by the German pathologist Virchow more than 150 years ago [11]. Furthermore, given the fully recognized inextricable microbe‐immune system‐tumor relationship, pathogenic microorganisms can promote tumorigenesis and metastasis indirectly by suppressing the immune response.
Based on existing literature on the mechanisms by which TM participates in the pathogenesis and progression of tumors, we have summarized the effects on DNA damage, activation of cancer‐related signaling pathways, modulation of cytokines, and induction of inflammatory responses. Please refer to Table 2 for details.
TABLE 2.
Mechanisms of microbial carcinogenesis.
| Type | Mechanisms | Example | References |
|---|---|---|---|
| DNA damage | Directly damage DNA |
Colibactin is an unstable DNA alkylating agent produced by E.coli colonizing in colorectal polyps. It specifically targets adenine‐rich DNA regions, leading to interstrand cross‐linking and double‐strand breaks in human cells. This process is implicated in the progression of colorectal cancer. In OSCC, acetaldehyde produced by the oral mucosal microbiome, induces DNA damage by forming DNA adducts within oral epithelial cells. |
[12, 133, 135, 138, 328] |
| Inhibit DNA damage response (DDR) system |
Merkel cell polyomavirus (MCPyV) integrated into malignant Merkel cell carcinoma (MCC) cells impedes DDR activation and inactivates tumor suppressors RB via large T antigen. Both human T‐lymphotropic virus 1 (HTLV‐1) toxin and high‐risk HPV oncoprotein expression inhibit the DDR system, leading to associated carcinogenesis and eventual progression to malignancy. |
[139, 140, 141, 142] | |
| Activation of oncogenic signaling pathways | PI3K signaling pathway |
|
[131, 144‐146] |
| STAT signaling pathway | Prevotella, abundantly present in malignant oral epithelial cells, may be associated with gingival squamous cell carcinoma. It can suppress chemically induced intrinsic mitochondrial apoptosis by activating the JAK1/STAT3 and PI3K/Akt signaling pathways. | [138, 152, 153] | |
| WNT/β‐catenin signaling pathway |
|
[13, 156, 158, 160] | |
| Cytokines and inflammatory responses | Cytokines and inflammatory responses |
|
[12, 170] |
| Interaction with tumor microenvironment | Regulation of intratumoral immune cells |
|
[176, 179, 180, 182, 183, 184, 329] |
| Functions of microbial‐derived metabolites |
|
[23, 185] |
5.1. Damage to DNA
Many pathogenic microorganisms have evolved to produce compounds capable of causing DNA damage, cell cycle arrest, and genetic instability [131]. They are called genotoxins ‐ whether they directly cause DNA damage and chromosomal instability or reduce the DNA repair capacity of cells. The presence of microorganisms that produce this substance in the tumor microenvironment may directly increase the DNA mutations in the colonized tissues and accumulate to a certain level, eventually leading to cell growth dysregulation and tumor initiation [132]. The most typical one is E. coli. The manufacture of genotoxin colibactin is associated with the presence of a large genomic island named pks. pks E. coli strains exist in approximately 60% of CRC samples [132]. E. coli can invade the colonic mucus layer, colonize polyps (precancerous lesions), and encode for colibactins, an unstable DNA alkylating agent [133]. The active genotoxin contains α‐amino ketone, a positively charged functional group that enhances the affinity of E. coli for DNA [134]. In addition, colibactin can act in adenine‐rich DNA regions, causing DNA interstrand cross‐linking and double‐strand breaks in human cells. This DNA damage induces the phosphorylation of related proteins, which activates the ataxia telangiectasia mutated protein and checkpoint kinase 2 (ATM‐Chk2) signaling pathway, which leads to transient cell cycle arrest and cell swelling, finally inducing cytotoxicity, mutation, and promoting tumor formation [134, 135]. Meanwhile, with co‐colonizing microorganisms interacting, enterotoxigenic Bifidobacterium fragilis (ETBF) can degrade colonic mucus and promote colonization of pks+ E. coli, an ecological structure that helps the genotoxin colibactin reach colonic epithelial cells [133]. According to the above studies, pks+ E. coli may modify altered host genetic material while contributing to CRC initiation and progression [12]. In addition, Staphylococcus‐related epidermitis is also possibly caused by double‐stranded DNA breaks within the host cells [136]. It has also been revealed that the inflammatory environment triggered by microbial aberration within the tumor, factors that create genetic damage ‐ such as reactive oxygen species (ROS) and reactive nitrogen intermediates (RNI) ‐ can also damage DNA through direct disconnection or oxidation of guanine. ROS and RNI produced by immune activity during this intestinal inflammation may be more closely related to the pathogenesis of colitis‐associated colon cancer (CAC) than bacterial toxins that directly damage DNA [137]. Meanwhile, in OSCC, the genetic metabolite acetaldehyde produced by the oral mucosal microflora (including Streptococcus salivarius, S intermedius, S mittis, and non‐pathogenic Neisseria subspecies) is carcinogenic and causes DNA damage by forming DNA adducts in oral epithelial cells, and Streptococcus pharyngeus can trigger the increased synthesis of Nitric Oxide (NO) and cyclooxygenase 2 (COX2) leading to oral mucosal DNA damage [138].
In addition, the impairment of the DNA damage response (DDR) system is also recognized to be responsible for increased DNA damage and carcinogenesis (Figure 2A). Viruses easily cause DNA damage, manipulate the system, and lead to cancer development. Merkel cell polyomavirus (MCPyV) integrated into malignant Merkel cell carcinoma (MCC) cells impedes DDR activation and inactivates tumor suppressors gene RB via large T antigen [139, 140]. Both human T‐lymphotropic virus 1 (HTLV‐1) toxin and high‐risk human papilloma virus (HPV) oncoprotein expression also inhibit the DDR system, leading to associated carcinogenesis and eventual progression to malignancy [141, 142].
FIGURE 2.
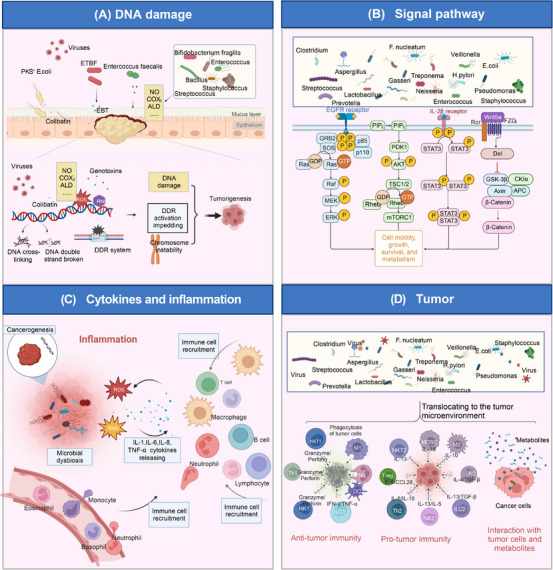
Involvement of microbiota in cancer. Microbiota is involved in cancer and mainly affects carcinogenesis or cancer prevention through the four aspects. (A) Tumorigenesis of microbial damage to DNA. Microbiota dysbiosis is often related to tumor initiation. Pathogenic microorganisms produce more compounds (for example, NO, COX2, acetaldehyde) capable of causing DNA damage, chromosomal instability, impairment of the DNA damage response (DDR) system, etc., thus causing tumorigenesis. For instance, pks E. coli can invade the mucus layer and encode an unstable DNA alkylating agent, colibatin, causing DNA interstrand cross‐linking and double‐strand breaks. Other microbiota, including ETBF, Enterococcus faecallis, etc., secret genotoxins and lead to reactive oxygen species (ROS) and reactive nitrogen intermediates (RNI) production, eventually promoting DNA damage. (B) Activation of oncogenic signaling pathways. Microbes activate the PI3K signaling pathway, STAT signaling pathway, Wnt/β‐catenin signaling pathway, etc., influencing essential activities such as cell motility, growth, survival, and metabolism, resulting in carcinogenesis. (C) Inflammatory carcinogenesis caused by microbial dysbiosis. Microbial dysbiosis results in ROS and reactive nit ogen species (RNS) production, immune cell recruitment, and inflammatory microenvironment formation. Chronic inflammation promotes tumorigenesis and progression. (D) Interaction with the tumor microenvironment. Microorganisms translocate to the tumor microenvironment (TME), exerting pro‐tumor and anti‐tumor effects through enhancement of anti‐tumor immunity, suppression of anti‐tumor immunity, and interaction with tumor cells and metabolite production. Abbreviations: ALD, acetaldehyde; APC, antigen presenting cell; COX2, cyclooxygenase 2; DDR, DNA damage response; 3D, three‐dimensional; ETBF, enterotoxigenic Bifidobacterium fragilis; EBT, enterotoxigenic bacterial toxin; MDSC, marrow‐derived suppressor cell; PI3K, phosphoinositide 3 ‐ kinas; ROS, reactive oxygen species; RNI, reactive nitrogen intermediates; RNS, reactive nit rogen species; STAT, signal transducer and activator of transcription; TME, tumor microenvironment. Biorender supported the materials in Figure 2.
It should be noted that whether the effect of DNA damage caused by pathogenic microorganisms forms mutations sufficient to cause cancer is still a matter of doubt that needs to be solved by extensive research.
5.2. Activate oncogenic signaling pathways
Signaling pathways influence essential activities such as cell motility, growth, survival, and metabolism in normal or tumor tissues. Extensive studies have been conducted to find that activation of human cancer‐related signaling pathways is an essential mechanism in tumorigenesis and development, and they are mostly signaling pathways involved in cell survival, growth, and proliferation (Figure 2B).
5.2.1. PI3K signaling pathway
Phosphoinositide 3 ‐ kinas, briefly named PI3K, is a class of lipid kinases involved in cellular functions, including cell proliferation, growth, differentiation, migration, and survival. A joint genetic event, PIK3CA amplification, was identified in various tumor types, including lung cancer, cervical cancer and other tumors [143]. There were significant differences in the composition of the lower respiratory transcriptome of lung cancer patients compared to controls, including upregulation of the PI3K signaling pathway [144]. The lower airways of lung cancer patients are enriched with oral taxa (veillonella and streptococcus), and elevated pathogenic microorganisms of the above genera were associated with PI3K upregulation; in vitro assays, airway epithelial cells exposed to veillonella, prevotella, and streptococcus also showed PI3K signaling pathway upregulation [144]. Genomic signature activation of the phosphoinositide 3‐kinas (PI3K) pathway was significantly increased in lung cancer smokers and cytologically normal bronchial airways with dysplastic lesions. Deregulating this pathway is considered an early, reversible event in lung carcinogenesis [145]. Thus, the above accumulation of oral commensal bacteria in the lower respiratory tract may lead to upregulation of the PI3K pathway, thereby promoting lung carcinogenesis [131]. As reported, viral oncoproteins E5, E6, and E7 of HPV target the PI3K pathway and promote cell division, causing tumor initiation and progression [146]. Other oncogenic viruses, such as HTLV‐1 [147], Kaposi sarcoma‐associated herpesvirus (KSHV) [148], MCPyV [149], etc., have been demonstrated to engage this pathway to develop tumor formation.
5.2.2. JAK‐STAT signaling pathway
The Janus kinase/signal transducers and activators of the transcription signaling pathway, also called the JAK‐STAT signaling pathway, is thought to play a vital role in almost all cytokine‐driven signaling and is critical in controlling cell cycle progression and apoptosis [150]. As growth factor dysregulation is often central to cellular transformation in many cancer cells, STAT sustained activation holds importance in human tumorigenesis [151]. It has been investigated that Porphyromonas gingivalis is often found in subgingival plaque and is present in large numbers in malignant oral epithelial cells, which may be associated with gingival squamous cell carcinoma [152, 153]. It inhibits chemically induced endogenous mitochondrial apoptosis in gingival epithelial cells (GECs) by activating the JAK1/STAT3 and PI3K/Akt (protein kinase B) signaling pathways [138]. Similarly, it is now clear that enterotoxin‐producing mycobacterium fragilis can induce colitis and colon tumors in adenomatous polyposis coli (Apc)multiple intestinal neoplasia (Min)/+ mice (a mouse model) by triggering the Th17 inflammatory response. Besides, ETBF also presents a unique role of acquired immunity in colon carcinogenesis through the selective activation of STAT3 [9, 154].
5.2.3. WNT/β‐catenin signaling pathway
The WNT/β‐catenin signaling pathway, which regulates cell stemness, polarity, and growth, is key in embryonic development, postnatal progression, and dynamic homeostasis of adult tissues. Abnormal β catenin signaling pathway can promote the transcription of oncogenes and tumor progression [155]. This signaling pathway occurs altered in many malignancies, including gastric cancer, CRC, and other cancers, which may be interfered with by some cancer‐associated bacteria [156, 157]. H. pylori infection can promote gastric carcinogenesis through various mechanisms. Some strains can express cytotoxin‐associated gene A (CagA) protein, which is injected directly into the cytoplasm of host cells and induces tumor progression by regulating β‐catenin to induce cancer [13, 156]. By activating multiple kinases, F. nucleatum invokes the proliferation of oral epithelial cells, among which F.nucleatum can activate β‐catenin by producing adherent FadA that binds to E‐cadherin [138]. Significant upregulation of FadA gene expression associated with F. nucleatum was found in colon cancer tissues compared to controls, and enterotoxic fragile Bacillus enrichment, which stimulates E‐calmodulin cleavage via Btf, also leads to β‐catenin activation [12, 156]. Additionally, viruses, including KSHV [158, 159], Epstein‐Barr virus (EBV) [158], HTLV‐1 [160], etc., are reported to regulate the WNT/β‐catenin signaling pathway, thereby increasing cell proliferation and promoting tumorigenesis.
5.2.4. ERK signaling pathway
Microorganisms enable the direct or indirect activation of extracellular regulated protein kinases (ERK) signaling in tumor cells. The ERK signaling pathway is often called a conserved Rat Sarcoma (RAS)‐ Rapidly Accelerated Fibrosarcoma (RAF)‐ Mitogen‐Activated Protein Kinase (MEK)‐ERK signaling cascade. MEK‐ERK is activated by phosphorylated MAPKK(RAF)‐MEK and then enters the nucleus, regulating transcription factors and gene expression related to cell growth and proliferation [161]. Overall, the ERK signaling pathway [162] influences tumor initiation by regulating cell growth and proliferation [17].
5.2.5. Other signaling pathway
The STING signaling pathway, activated by microbiota‐derived agonists such as c‐di‐AMP, can induce IFN‐γ secretion and enhance DC/NK cell crosstalk, thereby establishing the STING‐type I IFN‐NK/DC axis and regulating melanoma therapy [163]. The RhoA/ROCK signaling pathway and the PERK signaling pathway are implicated in reorganizing the actin cytoskeleton or modulating endoplasmic reticulum stress, thus promoting tumor cell proliferation and metastasis [32, 164, 165]. Additionally, the α5‐nicotinic acetylcholine receptors (α5‐nAChRs)‐Notch signaling pathway has been shown to facilitate the proliferation, migration, and invasiveness of melanoma [166]. In summary, TM can have a significant impact on cancer development. Understanding the complex interactions between TM and cancer is an active area of research with implications for cancer prevention, diagnosis, and treatment.
5.3. Influence cytokines and induce inflammatory responses
More than 150 years ago, German pathologist Virchow proposed that inflammation promotes carcinogenesis [11]. Much clinical and epidemiological evidence suggests that chronic inflammation is a risk factor for various tumors, especially in gastrointestinal malignancies, such as esophageal, gastric, hepatic, pancreatic, and colorectal cancers [167]. Many studies have shown that ecological dysregulation of the local bacterial community leads to a chronic pro‐inflammatory immune response. It has been revealed that the key to linking inflammation and cancer is the abnormal transcription of genes encoding inflammatory mediators, growth factors, transfer proteins, angiogenic factors, genomic instability and damage, and malfunctioning epigenetic control [168].
Nuclear factor κ‐B (NF‐κB) is one of the downstream regulators of intracellular receptors important in inflammation. Its ability to regulate the expression levels of pro/anti‐inflammatory cytokines that have a critical role in tumor cell survival helps explain the relationship between inflammation and cancer at the molecular level. Its activation occurs as an essential feature of bacterial‐associated tumor development [138, 169] (Figure 2C). Since H. pylori lacks any direct genotoxic components and has a substantial correlation with gastric cancer, it is believed that it causes cancer indirectly by triggering a persistent inflammatory response rather than directly through any known virulence mechanisms [12].
Among Bacteroides fragilis strains, a toxin‐producing ETBF can induce colonic inflammation associated with diarrhea, inflammatory bowel disease, and cancer [12]. Bacillus fragilis toxin (BFT), a zinc‐dependent metalloproteinase toxin, cleaves e‐calmodulin, triggering the activation of MAPKs and NF‐κB pathway, which increases chemokine IL‐8 secretion and attracts polymorphonuclear cell aggregation [12, 170]. The simultaneous F. nucleatum aggregation and significant upregulation of FadA gene expression in human colon cancer support, to some extent, the mechanism that the F. nucleatum uses the virulence factor FadA to bind to the extracellular structural domain of E‐cadherin to activate toll‐like receptor 4 (TLR4)‐activated NF‐κB signaling and induce the proliferation of colon cancer cells [12].
Upon infection of the host by Gram‐negative bacilli, their transendothelial release of endotoxins, such as LPS, which bind to pattern recognition receptors (PRRs), one of which includes TLRs, mainly TLR4, thereby activating the production of inflammation‐associated cytokines via the NF‐κB signaling pathway to activate the production of inflammation‐associated cytokines [138]. In addition to lipopolysaccharide, the flagellum of Pseudomonas aeruginosa (P. aeruginosa), cytotoxins such as ExoU have potent inflammatory activity, recruiting neutrophils while activating the NF‐κB signaling pathway [138]. Porphyromonas gingivalis induces matrix metalloproteinase‐9 (pro‐MMP‐9) overexpression by upregulating ERK1/2‐E26 Transformation‐Specific Sequence 1 (ETS1), p38/Heat Shock Protein 27 (HSP27), and Partitioning‐Defective (PAR)/NF‐κB pathways. Porphyromonas gingivalis induces overexpression of related receptors in oral epithelial cells by increasing the production of interleukin (IL)‐1, IL‐6, IL‐8, and TNF‐α, which leads to chronic inflammation [138]. Experiments in dextran sulfate sodium (DSS)‐induced colitis mice suggest that barrier disruption, microbes, or microbial products (such as endotoxin and nucleic acids) activate TLR signaling in mucosal macrophages, which produce several tumor‐promoting cytokines, including TNF, which exert their oncogenic effects via NF‐κB. In addition to endotoxins and nucleic acids, the commensal‐ Immunoglobulin G (IgG) immune complex activates NF‐κB and NOD‐like Receptor Family Pyrin Domain Containing 3 (NLRP3) inflammatory vesicles via Fcγ receptors (FcγRs) [171].
5.4. Interact with TME
The TME comprises immune cells, cancer‐associated fibroblasts, tumor microbes, microbial products, cytokines, chemokines, and extracellular matrix (ECM) around tumor cells [172]. TME were once considered bystanders of tumorigenesis but are now recognized to play critical roles in cancer pathogenesis. Nowadays, it is preferred to believe that microorganisms reprogram the TME by translocating into the intratumoral niche, thereby influencing tumorigenesis and progression [22, 173, 174] (Figure 2D).
5.4.1. Microbial regulation of intratumoral immune cells
The immuno‐oncology‐microbiome (IOM) axis has been proposed because some researchers contend that cancers rarely produce directly by microorganisms but are more frequently mediated by the host's immune system [9]. Local microorganisms have been demonstrated to influence local immune surveillance in addition to causing inflammation by reducing antitumor immune responses. This kind of immunosuppression has been seen in lung cancer mouse models and humans with colon cancer [131].
In the intestine, the largest immune organ, microbial mechanisms can manipulate the non‐hematopoietic and hematopoietic components of the intestinal epithelial barrier, regulate primary and secondary lymphoid organ activity, and modulate the immune tone of the TME [9]. In the lungs of vancomycin/neomycin‐aerosolized mice, where intratumoral bacteria load was remarkably decreased, a reduction in regulatory T cells and enhanced activation of T and NK cells were associated with a significant reduction of melanoma B16 lung metastasis, suggesting that intra‐tumor microbes may suppress anti‐tumor immunity and indirectly promoting lung tumor metastasis [152, 175].
F. nucleatum and H. pylori can suppress T‐cell activity [176]. The binding of F. nucleatum fibroblast activation protein‐2 (Fap2) protein to the human inhibitory receptor T cell immunoglobulin and ITIM domain (TIGIT) protects tumors from immune cell attacks [177]. Using the mutant library from the nucleus accumbens, it was discovered that the direct interaction of the Nucleus inhibited the NK cells' harmful effect of accumbens Fap2 protein with TIGIT. TIGIT is also shown to be expressed by tumor‐infiltrating lymphocytes, and F. nucleatum suppresses T‐cell activation by way of FAP2. By using the Fap2 protein of the Nucleus pulposus to block immune cell activity via TIGIT, the tumor can evade the immune system, according to the previous findings [178].
In a murine model of 4‐nitroquinoline‐1 oxide (4NQO)‐induced carcinogenesis, Porphyromonas gingivalis invasion of oral lesions increased oral lesion diversity, while in vitro observations revealed increased infiltration of CD11b+ myeloid cells and myeloid suppressor cells in oral lesions, and Porphyromonas gingivalis may facilitate tumor progression by expressing chemokines and chemokine ligands, IL‐6 and IL‐8 and other cytokines to recruit bone marrow‐derived suppressor cells (MDSCs) to drive tumor progression [179]. Currently, Liu et al. [37] also found that intratumoral mycobiome Aspergillus sydowii could promote lung adenocarcinoma development by inducing myeloid‐derived suppressor cells (MDSCs) expansion.
In addition to suppressing anti‐tumor immunity, TM also enhances anti‐tumor immunity. Oncolytic viruses or bacteria can specifically target the TME and lys tumor cells, inducing antitumor response [180, 181]. Additionally, engulfed microorganisms such as Fusobacterium and Treponema in the macrophage cell cluster are presumed to activate TNF, INF, and Janus Kinase (JAK)‐STAT signaling pathways to produce interleukins and inflammatory responses. Conversely, the absence of microbiota will skew the TME towards pro‐tumorigenic macrophage [163]. Microbiota such as Bifidobacterium breve and Enterococcus hirae were reported to activate anti‐tumor T cells through bacterial peptides and antigen mimicry to enhance antigen presence and cross‐reactivity [182, 183, 184]. Neutrophils with F. nucleatum are observed to reduce their migration capabilities in response to bacterial infection, which influences the infiltration of neutrophils into the TME. Moreover, increased translocation of pathogenic gram‐negative taxa, including Proteobacteria, Fusobacteria, etc., affirms that lipopolysaccharides and flagellins can bind to specific TLRs and activate tolerogenic macrophages in the TME [14].
5.4.2. Microbial regulation of tumor cells and production of metabolites
The TM is reported to interact with the cancer cells directly. Some intratumor bacteria can reorganize the actin cytoskeleton of cancer cells [59]. Similarly, invasive bacteria F. nucleatum enables change in how infected cancer cells move and increases single‐cell migration capabilities [10].
Microbial‐derived metabolites such as inosine, bile acids, SCFAs, enzymes, etc., are demonstrated to influence the TME, thereby manipulating cancer progression. Microbial‐derived bile acids have been shown to modulate natural killer cells and play an essential role in the initiation and progression of hepatic cancer [185]. An isoform of cytidine deaminase expressed by γ‐transforming bacilli in the TME can convert the chemotherapeutic drug activity [15]. Azurin secreted by Pseudomonas aeruginosa enables to induce apoptosis in tumor cells, whereas aldolase A released by cancer cells promotes P. aeruginosa adhesion and colonization in cancer cells [186, 187]. Lam et al. [163] revealed in the TME that microbiota‐derived products are needed to program the innate immune. For example, STING agonists c‐di‐AMP derived microbiota, such as Akkermansia muciniphila, can trigger the STING‐IFN‐DC/NK axis to promote anti‐tumor immunity in TME [188]. Inosine released by Bifidobacterium pseudolongum can promote the activation of T cells in tumor tissue [189]. Additionally, butyrate could increase the expression of IFN‐γ and granzyme B in CD8+ T cells and regulate glycolysis, tricarboxylic acid (TCA) cycle, and fatty acid oxidation (FAO) in antitumor effector cells [190, 191, 192]. Recently, tryptophan catabolite indole‐3‐aldehyde (I3A) derived from Lactobacillus reuteri acts through CD8 T cell‐specific aryl hydrocarbon receptor (AhR) signaling to promote IFNγ‐production to promote the anti‐tumor immunity of TME [23]. Collectively, microbial metabolites play a crucial role in TME reprogramming.
We have summarized the main mechanisms of microbial carcinogenesis in Table 2. As can be seen, the microbiome in the TME can be either suppressive or tumor‐supporting. Some studies have proposed the IOM axis, setting a broad context for studying the mechanisms [9]. However, there is still a long way to go, and more studies must be conducted. In addition to the described mechanisms above, some studies have revealed that microbiota can impact cancer via outer membrane vesicles (OMVs) [193, 194]. The mechanisms of microbial carcinogenesis are intricate, and perhaps only a tiny fraction of them have been thoroughly understood.
6. TM‐INVOLVED TREATMENT AND APPLICATION
As countless studies show, the microbiome strongly relates to oncology treatment. Cancer therapy, including chemotherapy, radiotherapy, and immunotherapy, has been demonstrated to be influenced by a variety of microbiomes. In addition, novel microbial applications containing probiotics, fecal microbiome transplantation (FMT), engineered microbiota, and bacteriophage are increasingly employed in tumor prevention, treatment, and drug delivery.
6.1. Immunotherapy
Immunotherapy is a treatment approach that employs the body's immune system as a breakthrough to regulate and activate the body's immune system. There are four main categories: immune checkpoint inhibitors (ICIs), tumor vaccines, cellular immune cell therapy, and non‐specific immunomodulators. It is reported that microorganisms can influence the therapeutic effect of these immunotherapies. And this section mainly focuses on the history and mechanisms of which the microbiome plays a role in ICIs.
In 2015, Sivan et al. [195] found that melanoma mice with different commensal bacteria differed in tumor growth, and this discrepancy could be eliminated by cohousing and FMT. He further demonstrated that the intestinal symbiotic bacterium Bifidobacterium enhances the antitumor immune effect of TME and the efficacy of PD‐L1 antibody treatment. In the same year, Vétizou et al. [196] also revealed a key role for Bacteroides thetaiotaomicron (B. thetaiotaomicron) and B. fragilis in the immunostimulatory effects of cytotoxic T‐lymphocyte associated protein 4 (CTLA‐4) blockade. Subsequently, researchers proposed intestinal flora as a marker for immunotherapy [197, 198]. Furthermore, research indicates that in a subset of melanoma, FMT and anti‐Programmed Cell Death Protein 1 (PD‐1) reprogrammed the tumor microenvironment and altered the gut microbiome to overcome anti‐PD‐1 resistance [199], which may be attributed to the STING‐IFN‐I‐NK/DC axis [163]. In 2022, Akkermansia muciniphila (AKK) bacteria were recognized to predict NSCCL prognosis independently, and numerous studies were conducted with the bacterium [200, 201]. Generally, the microbiota has a high potential to become an adjuvant immunotherapy therapy, attracting many researchers to conduct extensive studies.
Although the mechanisms by which microbes influence immunotherapy are still unclear, some hypotheses have been proposed (Figure 3).
Signal pathway: Lam et al. [163] demonstrated that microbiota‐derived STING (stimulator of interferon genes) agonists such as c‐di‐AMP induce type I IFN (IFN‐I) production by intratumoral monocytes, modulating macrophage polarization and NK‐DC crosstalk, triggering a positive feedback loop and promoting antitumor immunity. In addition, Shi et al. [174] observed that Bifidobacterium bifidum promotes anti‐CD47 immunotherapy in an STING and interferon‐dependent manner in the TME.
Metabolites: Inosine, SCFAs, bile acids, and indoles are believed to be involved in microbial regulation of immunotherapeutic processes. 1) Inosine: Inosine enhances the immunogenicity of tumor cells and provides an alternative carbon source for CD8+ T cells [202, 203] and both AKK and Bifidobacterium pseudolongum, which are capable of producing inosine, have been shown to have antitumor effects [204]. 2) SCFAs: SCFAs and ICIs have been hot research topics recently. SCFAs can inhibit tumor cell differentiation, induce apoptosis, promote anti‐tumor response, and provide a carbon source for immune cells. According to one study, butyric acid can cause CRC cells to undergo apoptosis by suppressing the expression of genes that control histone deacetylase. Another study showed that butyric acid could increase the expression of IFN‐γ and granzyme B in CD8+ T cells and regulate glycolysis, TCA cycle, and FAO in antitumor effector cells to improve the efficiency of ICI [190, 202]. 3) indole and tryptophan: indole, a product of tryptophan metabolism, has been shown to play a role in ICI therapies in several studies. Hezaveh et al. [205] showed that indole activated the AhR activity, which directed the polarization of macrophages, inhibited inflammatory T‐cell infiltration, and promoted its growth. Deletion of AhR from bone marrow cells or pharmacological inhibition of AhR reduced pancreatic tumor growth and improved the efficacy of immune checkpoint blockade. Of note, recently, Bender et al. [23] have provided the opposite result. The study elucidates how the tryptophan metabolite indole‐3‐aldehyde (I3A), produced by intratumoral Lactobacillus reuteri, promotes IFN production in a cAMP Response Element‐Binding Protein (CREB)‐dependent way and enhances ICI therapy in advanced melanoma patients. Moreover, providing a tryptophan‐enriched diet or intratumoral injection of I3A also generated the mimetic effect [22]. Similarly, Lactobacillus gallinarum‐derived indole‐3‐carboxylic acid (3‐ICA) has recently been reported to boost anti‐PD1 efficacy in colorectal cancer [206]. These controversial results may be due to the cancer types and different indole derivatives; more evidence is required to elucidate. 4) Others: Metabolites, such as bile, cardiac acid, and succinic acid, have also been reported to enable modulating immunotherapy. For instance, Jiang et al. [207] recently showed that succinic acid from F. nucleatum reduced CRC patients' sensitivity to anti‐PD‐1 therapy by affecting CD8+ T cell‐mediated antitumor immunity.
Immunogenicity: commensal microbiota can also enhance the antitumor effects in TME through autoantigenic epitopes with immune cell recognition and antigen presentation [202, 208]. However, there is currently no consensus on the specific mechanism of microbial modulation of immunotherapy, and more experiments need to be conducted to investigate it.
FIGURE 3.
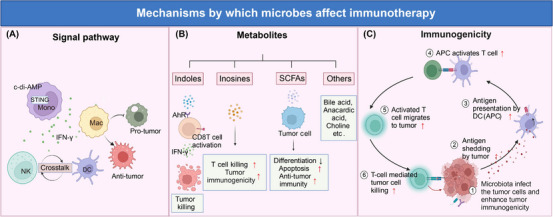
Mechanisms by which microbes affect immunotherapy. Microorganisms affect immunotherapy mainly in three ways: modulating signal pathways, producing metabolites, and enhancing immunogenicity. (A) modulating signal pathways: Microbiota‐derived agonists c‐di‐AMP modulate STING of monocyte, induce IFN‐γ secretion, bolster DC/NK cell crosstalk, and promote anti‐tumor macrophage activation. (B) metabolites producing: Many microbial metabolites, including indole, inosine, SCFAs, etc., have anti‐tumor effects. Indole activates CD8+ T cells via binding AhR and increases IFN‐γ releasing, thus enhancing tumor killing. Inosine promotes T‐cell killing and enhances tumor immunogenicity. Moreover, SCFAs inhibit tumor cell differentiation, induce apoptosis, and promote anti‐tumor response. (C) enhancing immunogenicity: Commensal microbiota increases tumor antigen shedding and antigen presentation, activating more T cells and migrating to the tumor, causing more tumor cell killing. Abbreviations: APC, antigen presenting cell; DC, dendritic cells; Mac, macrophage; NK, natural killer cell; SCFAs, short‐chain fatty acids; STING, stimulator of interferon genes. Biorender supported the materials in Figure 3.
Immunotherapy such as PD‐1/PD‐L1 inhibitors has been a boon for many patients with progressive tumors (non‐small cell carcinoma, renal cell carcinoma, melanoma), with only 10%‐40% of patients achieving remission or partial remission [199, 202]. Different microbial species may play different roles in different tumors, and favorable and unfavorable strains and metabolites are briefly listed in Table 3. Currently, research on microbial and immunotherapy is in full swing. But many questions need to be addressed. Whether metabolites reach specific tissues through various circulations? Do bacteria specifically enter the TME first and then exert their effects? Are butyrate promoters or inhibitors of immunotherapy? Do other fungi or viruses have a role in immunotherapy? Is treatment with antibiotic before immunotherapy detrimental or beneficial [14, 198, 200]? More studies are needed to demonstrate these controversies. Moreover, studies have recently shown that microbes can overcome microbiome‐dependent resistance to PD‐1 pathway inhibitors via blocking Programmed Death‐Ligand 2 (PD‐L2)–Repulsive Guidance Molecule B (RGMb) interactions, which informed a possible therapeutic strategy to overcome resistance to immunotherapy [209]. Microbiology and immunotherapy are promising areas of research. In the future, studies will focus more on the impact of microbes on immunotherapy efficacy and resistance, bringing hope for more patients to be treated.
TABLE 3.
Classification of microbe strains and metabolites.
| Antitumor effect | Microbe strains | Metabolites |
|---|---|---|
| Favorable | Bifidobacterium, Lactobacillus, Streptococcus thermophiles, Enterococcus hirae, Bacteroides fragilis, Faecalibacterium, Akkermansia muciniphila, Gemmiger, Roseburia, Faecalibaculum rodentium, Olsenella. | Indole, inosine, c‐di‐AMP, butyrate, niacin, vitamin B, indole‐3‐carboxylic acid. |
| Unfavorable | γ‐transforming bacilli, Faecalibacterium, Helicobacter pylori, Fusobacterium nucleatum, Bacteroides fragilis, Epstein‐Barr virus, Lactobacillus iners. | SCFA, succinic acid, secondary bile acid (DCA), kynurenine, L‐lactate. |
6.2. Chemotherapy
Chemotherapy, the current predominant therapeutic approach for most tumors in the intermediate and advanced phases [210], is considered the final guardian of survivorship for a large population of cancer patients. Chemoresistance constitutes an essential cause of mortality for many cancer patients [211]. Numerous investigations have found that tumor microbes are responsible for chemotherapy resistance and are predicted to be an attractive target for improving chemotherapy efficacy [15].
The major mechanisms of microbial action on chemotherapeutic resistance are as follows. 1) Metabolism: Microorganisms influence the active form of chemotherapeutic drugs by modulating the various specific enzymes. In a murine model of colorectal tumors, γ‐transforming bacilli within the tumor metabolized the active gemcitabine (2 ‘,2 ’ ‐difluorodeoxycytidine) into its inactive form 2 ‘, 2 ’‐difluorodeoxyuridine by bacterial enzyme cytidine deaminase (CDDL), and this chemoresistance can be reversed by the antibiotic ciprofloxacin [15]. Additionally, microbial β‐glucuronidases from the gut microbiome enable to reactivate the intestinal inactive metabolite of irinotecan into active form, which has been associated with adverse drug reactions such as severe diarrhea. Targetting this microbial enzyme potentially affects its therapeutic efficacy and ameliorates side effects [212]. Indole‐3‐acetic acid (3‐IAA), a molecule generated from the microbiota, has recently been recognized as a crucial amplifier of the response to chemotherapy in pancreatic cancer in humanized gnotobiotic mouse models [213]. 2) immunity and inflammation: microbial entrance into secondary lymphoid organs activates immune cells and regulates the efficacy of chemotherapeutic drugs through inflammatory responses and antitumor immunity. Using a murine model, Viaud et al. [214] discovered that cyclophosphamide induces translocation of specific species of Gram‐positive bacteria into sub‐lymphatic organs and stimulates memory Th1 cell immune response by “pathogenic” helper T cells17 (pTh17). The pTh17 response was reduced in germ‐free mice or mice in which Gram‐positive bacteria were eliminated with antibiotics, and their tumors showed resistance to cyclophosphamide [214]. Two gut commensal species, enterococcus hirae and Barnesiella intestinihominis were reported to influence the antitumoral activity of cyclophosphamide (CTX) by lowering regulatory T cells and inducing appropriate anticancer cytotoxic T lymphocyte (CTL) responses and oral gavage with Enterococcus hirae (E. hirae) clone selectively restored the CTX‐mediated antitumor effects [215]. 3) Autophagy: In a study by Iida et al. [216] comparing colorectal cancer (CRC) patients who relapsed after chemotherapy with those who did not, it was found that the quantity of F. nucleatum was increased in relapsed patients. Moreover, it was demonstrated that F. nucleatum impacts CRC through the Toll‐like receptor 4 (TLR4) and myeloid differentiation primary response 88 (MYD88) pathway, resulting in the selective suppression of miR‐18a* and miR‐4802 expression. This, in turn, activates autophagy, thereby promoting chemoresistance in CRC patients. Furthermore, the integrity of the symbiotic microbiota is essential for optimal responses to cancer therapy. Dysbiosis of the microbiota has been shown to impair subcutaneous tumor responses to CpG‐oligonucleotide immunotherapy and platinum‐based chemotherapy. Additionally, apart from bacteria, the mycobiome also plays a detrimental role in chemotherapy. Aykut et al. [217] identified that fungal ablation enhances the efficacy of gemcitabine‐based chemotherapy.
A diversity of microbes has both positive and negative effects on chemotherapy. The application of precise and personalized microbial adjuvant therapy may be able to reverse chemotherapy resistance and optimize the therapeutic effect of chemotherapy.
6.3. Radiotherapy (RT)
RT, which uses ionizing radiation to produce cytotoxic effects on tumor cells, remains one of the main therapeutic approaches for many progressive solid malignancies [218]. Recently, it has been shown that commensal microorganisms such as intestinal flora can influence the efficacy and prognosis of radiation therapy. It has been suggested that the interaction between radiation therapy and commensal microorganisms is reciprocal [219, 220, 221]; that is to say, radiation therapy affects the composition and abundance of commensal microorganisms, especially intestinal flora. In contrast, commensal microorganisms influence the efficacy and prognosis of radiation therapy and have the potential to become a prognostic indicator for malignant tumors. However, the exact mechanisms involved remain unclear.
Vancomycin, an antibiotic limited to the gut and mostly targets gram‐positive bacteria, was discovered by Uribe‐Herranz et al. [222] in 2019 to enhance the RT‐induced antitumor immune response and tumor growth suppression. In 2021, Shiao et al. [223] used a cocktail of antibiotics to eliminate commensal bacteria, leading to the growth and expansion of commensal fungi and this manipulation can render radiotherapy less effective. In the same year, Dong et al. [220] used metronidazole to eliminate Fusobacterium nucleatum, significantly improving the effectiveness of radiotherapy. In addition, Guo et al. [224] used high‐dose radiotherapy to screen elite survivors. They found that two tryptophan pathway metabolites, 1H‐indole‐3‐carboxaldehyde (I3A) and kynurenic acid, could provide long‐term radioprotection in vivo [224]. Teng et al. [225] revealed that Bacteroides vulgatus‐mediated nucleotide biosynthesis dampened rectal cancer patients' responsiveness to chemoradiotherapy. Additionally, researchers have found that tumoral Lactobacillus iners, a L‐lactate‐producing lactic acid bacteria, enable to induce chemoradiation resistance through efficient L‐lactic acid production and metabolic remodeling in tumors [226]. Some studies have proposed that dietary therapy [227], antibiotic application [228], FMT [229], and oral microbiota transplantation (OMT) [230] are all promising adjuvant methods to improve radiotherapy.
However, for this area of microorganisms and radiotherapy, the composition of various beneficial bacteria and related mechanisms still needs further research. Based on improving radiotherapy and reducing damage, the optimal combination and application of disease‐specific and individualized probiotics will be an essential research direction in the future.
6.4. Probiotics, prebiotics, and synbiotics
In the early twentieth century, Bulgarians with a long life were found to maintain a diet of fermented dairy products, suggesting that fermented dairy products may alter the microorganisms in the gut to affect human health [231, 232]. Subsequently, ‘probiotics’ was proposed by several sources to describe those active substances that are beneficial to human health [233]. In 1989, Roy Fuller et al. [234] redefined probiotics as “a live microbial dietary supplement that produces beneficial effects in the host”. After the 1990s, prebiotics (indigestible food components that selectively stimulate the growth of one or several beneficial bacteria in the colon and thus improve host health) [235, 236] and synbiotics (a combination of prebiotics and probiotics) [236] have been gradually coming into view. Investigators have demonstrated their benefits in treating infant diarrhea, lactose intolerance, elevated blood lipids, ulcerative colitis, and eczema. Some probiotics, such as Lactobacilli, have been revealed to play a significant role in cancer prevention [237], stimulating research on probiotics and CRC. Moreover, the emergence of high‐throughput technology and next‐generation gene sequencing enabled people to realize that microorganisms exist not only in the intestine but also in tumors, exerting an important role in tumor development, treatment, and therapy response. In addition, molecular biology and gene engineering applications have been propelling new chapters for this area, appearing with targeted and engineered probiotics, tailored microorganisms, and so on.
The CRC preventive ability of lactic acid bacteria was discovered 20 years ago, and since then, enormous studies, from animal experiments to human clinical studies, have been conducted and reported [238]. Other microorganisms, including Bifidobacteria, yeast, and AKK, were also beneficial to cancer prevention [239]. The mechanisms of probiotics mainly include the following aspects. 1) reshaping in intestinal microflora (decrease in harmful bacteria such as F. nucleatum and increase in beneficial bacteria such as bifidobacteria); 2) regulation in intestinal conditions (reducing the activity of oncogenic enzymes, altering intestinal physicochemical level, decreasing carcinogens and elevation in anticancer substances); 3) enhancement of intestinal barrier function (such as mucus secretion and improvement of tight junction strength); 4) enhancement of host immunity (increasing cytokines secretion and immune cell infiltration); 5) regulation of signal pathway(such as tyrosine inhibition pathway); 6) regulation of metabolic conditions in TME; 7) modulation of tumor cell apoptosis [236, 239, 240, 241, 242, 243, 244, 245, 246]. Of note, recently, a probiotic bacterial strain called Lactobacillus reuteri, has been reported to move from the small intestine into the TME of solid tumors, according to a method described by Bender et al. [23], enhances anti‐tumor immunity, and facilitates ICI therapy via its metabolite indole‐3‐aldehyde (I3A) through I3A‐AhR‐CD8+ CTL axis. Meanwhile, Zhang et al. [247] demonstrate that a probiotic strain, Lactobacillus plantarum L168, and its metabolite indole‐3‐lactic acid can accelerate IL12a production in dendritic cells and change chromatin accessibility, thereby enhancing the function of tumor‐infiltrating CD8+ T cells. In addition, the mechanism of action of probiotics and synbiotics has been widely studied. In addition to promoting probiotics' growth, they can modulate xenobiotic metabolizing enzyme activity, alter gene expression in the intestine, and regulate autoimmunity. Synbiotics can modify the colonic bacterial ecosystem and improve metabolic activity, downregulating inducible NO‐synthase and cycloxygenase‐2 enzymes [248]. However, there is no conclusive mechanism for the tumorigenic effects of probiotics, prebiotics, and synbiotics, and different strains and tumors may have specific regulatory mechanisms.
Probiotics, prebiotics, and synbiotics have significant commercial potential in people's lives and may play a more critical role in the future, especially in the fight against cancer. However, since the beginning of the popularity of probiotics, there has been an ongoing debate, with some studies claiming that probiotics, prebiotics, and symbiotics are not effective for cancer prevention [236] and that probiotic metabolite short‐chain fatty acids such as butyrate did not differ significantly between trials and controls in terms of anti‐inflammatory and anti‐cancer effects [249]. These controversial results may be due to the strain‐specific nature of the commensal flora, and future tailored probiotic protocols may break through in this regard. In addition, the significance of probiotics in preoperative and postoperative cancer management has been studied and refuses to be ignored. Studies have shown that preoperative use of probiotics can reduce the chance of postoperative infection [238], and the application of probiotics can reduce chemoradiotherapy‐related diarrhea [250, 251], and also enhance the efficacy of immunotherapy [246]. In addition, the gene editing of probiotics as targeted transport vectors, combined with nanotechnology and fluorescence technology, can also advance the development of probiotics (Figure 4A).
FIGURE 4.
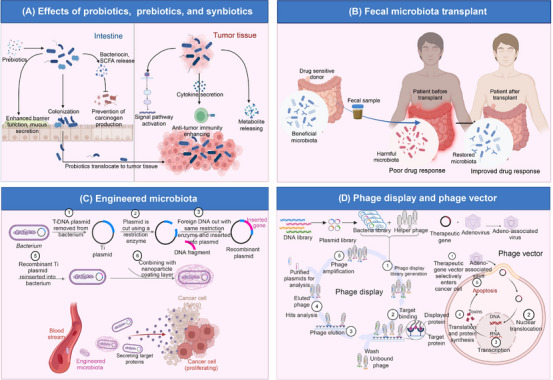
Mechanisms of probiotics, fecal microbiome transplantation, engineered microbiota, and phage therapy. (A) Effect of probiotics. Prebiotics could promote the growth of probiotics. Probiotics colonize in the intestine and reshape the microflora. These beneficial microbiotas could regulate intestinal conditions (increasing anticancer substances and decreasing carcinogens). In addition, they also enhance the intestinal barrier function. Some probiotics enable translocation to tumor tissue and regulate signal pathways, inflammation, and metabolic conditions. (B) Fecal microbiome transplantation. Fecal microbiota from drug‐sensitive patients can overcome drug resistance and improve the anti‐tumor effects in patients with poor drug response through microbiota remodeling. (C) Engineered microbiota. The microbiota is engineered and equipped with the target gene and the nanoparticle coating layer. The engineered microbiota easily spreads into the bloodstream and the tumor tissue and then secretes target molecules that help kill tumors. (D) Phage display and phage vector. Phage display requires five steps: polypeptide‐displayed phage libraries generation and amplification, target molecule binding, removing unbound and nonspecific phages, elution of target‐bound phages and infection of host bacteria, and replication. These infective bacteria help in the early diagnosis, treatment, and prevention of cancer vaccines. Phages such as adeno‐associated viruses combined with therapeutic genes could enter the tumor cell and translocate nuclei into DNA. These genes enable the expression of toxins that cause apoptosis in tumor cells. Abbreviations: SCFAs, short‐chain fatty acids; FMT, fecal microbiome transplantation. Biorender supported the materials in Figure 4.
6.5. FMT
In recent years, FMT as an emerging therapy has shown surprising results in treating intestinal diseases, including recurrent Clostridium difficile infection, active ulcerative colitis, and extraintestinal diseases such as diabetes, Parkinson's disease, and ankylosing spondylitis [252, 253, 254, 255, 256]. In addition, microbiota transplantation has been demonstrated to have preventive and therapeutic effects on cancer. Rosshart et al. [254] transplanted the gut flora of wild mice into laboratory mice, which enhanced the ability of laboratory mice to resist inflammation‐induced CRC. Riquelme et al. [257] revealed that the survival time of patients with PDAC is related to intratumoral microbial composition and diversity. FMT can remodel gut microflora, inhibit mouse tumor growth, and improve the effectiveness of immunotherapy for cancer [257]. Meanwhile, investigators transplanted feces from patients who responded well to PD‐1 treatment into animals or humans and found this could improve the anti‐tumor effects of PD‐1 treatment and even overcome drug resistance in patients [199, 258]. The main way in which FMT influences immunotherapy is by regulating the tumor immune microenvironment, including both innate and adaptive immunity. FMT has been found to increase the infiltration of antigen‐presenting cells, such as dendritic cells, into the tumor microenvironment, which leads to the release of chemokines and activation of T cells [258]. Additionally, studies have shown that FMT could overcome the drug resistance to anti‐PD‐1 therapy in patients with melanoma, possibly by boosting CD8+ T cell activation and reducing the frequency of IL‐8‐expressing myeloid cells in the TME [199]. Furthermore, aside from reprogramming the tumor immune microenvironment, FMT's therapeutic effects have also been linked to microbial metabolites. Studies have shown that SCFAs, specifically acetate, are associated with improved anti‐tumor efficacy in FMT studies. Acetate may provide energy for intestinal epithelial cells, resulting in more mucin secretion, increasing mucus layer thickness and enhancing the epithelial barrier, thereby suppressing tumor growth [259]. In addition, acetate accumulation has been shown to potentially stimulate IFN‐secretion of CD8+ T cells and enhance its tumor‐killing property [259]. The other metabolites, such as amino acid histidine [260] and bile acids [261], have also been shown to be related to the efficacy of FMT. However, further research is needed to understand these mechanisms fully. In conclusion, FMT is known to be capable of reprogramming the intestinal microbiome and generating anti‐tumor effects through various metabolites and regulation of the tumor immune microenvironment.
It has been reported that there is no statistical difference between the clinical effects of fresh stool transplantation and freeze‐dried stool capsules and frozen solution capsules in Clostridium difficile infection [262], and future clinical practice may prefer to use a frozen stool for fecal transplantation. In the future, establishing a fecal transplantation donor bank, optimizing frozen stool technology, and forming a standard stool transplantation treatment process will be more favorable for the development of FMT. However, FMT's mechanism of action, ethical issues, and treatment protocols are still ambiguous, and more high‐quality randomized controlled trials and clinical studies are required to clarify these controversies (Figure 4B).
6.6. Engineered microbiota
Two decades ago, scientists showed that genetically modified non‐pathogenic strains of the bacterial species Bifidobacterium, Salmonella, and clostridia might be used to target cancer therapy towards the hypoxic and necrotic zones typical of solid human tumors [2, 3, 263]. The engineered microorganisms display many functions, serving as monotherapy or complementing other anticancer therapies, prevention vaccines or diagnostic signals, and even medicine delivery vectors and imageable therapeutic probes. Owing to self‐replication characteristics, bacteria combined with synthetic biotechnology could be developed as H2O2 biosynthesizers for tumor therapy [264]. Additionally, these nontoxic and highly selective engineered microorganisms can enhance the efficacy of radiotherapy and chemotherapy, such as enhancing tumor radiosensitivity by sustaining oxygen supply and converting inactive chemotherapeutics into active ones [265, 266]. Meanwhile, it is well known that immunotherapy has brought tremendous well‐being to a wealth of advanced cancer patients [267]. However, these therapeutics face significant challenges due to harmful deep tissue penetration and primary or secondary resistance. Tumor‐targeted engineered bacteria may offer an option. A range of related studies shows immunotherapy, including PD‐1/PD‐L1 monoclonal antibody (mAb) therapy equipped with these microscopic ‘robotic factories’, is amenable to improving the therapeutic effects [268, 269, 270, 271]. In addition to what has been mentioned, engineered bacteria serve as an imageable therapeutic probe and deliver medicine as selective vectors [272]. Furthermore, researchers also designed an oral delivery of engineered probiotics to indicate the presence of liver metastasis of tumors by producing easily detectable signals in urine [273, 274]. Besides treatment and diagnosis, microorganisms are engineered to develop as vaccines for prevention. Bacteria with nanoparticle coating layers demonstrated that they can promote the dissemination of the bacteria into the blood and improve antigen expression and anti‐tumor immunity effectively [275]. In addition, integrating with other technologies such as nanotechnology, bioorthogonal chemistry, and fluorescent techniques represents a new perspective for diagnosis and treatment detection [276, 277]. Yet now, the engineered microbiota mechanisms have not been entirely made out. Apoptosis and autophagy of tumor cells and immunogenicity enhancement in the tumor microenvironment may explain some of them, which have been illustrated in detail in Chen's article [278]. While human trials have been conducted [279], the majority of studies remain at the animal stage. In the future, it is anticipated that more studies involving humans will be carried out. This will enable the confirmation of safe dosages and precise manipulation of bacteria. The development of therapy using these bacteria is expected to contribute significantly to our arsenal in the fight against cancer, especially as more tumor‐targeting microbes are intelligently developed and enter clinical testing (Figure 4C).
6.7. Phage therapy
Viruses are among the most abundant biological entities on Earth, with an estimated number of about 1030 in nature, as reported by The Database of Useful Biological Numbers. They are ubiquitous and found in various environments such as soil, bacteria, and even within human internal organs [280]. Bacteriophage, one kind of virus, also called phage, refers to parasitic viruses on microorganisms such as bacteria, fungi, archaea, and spirochete. Due to specific bacterial infections, phages have drawn much attention in recent years as an alternative to anti‐bacterial therapy, especially antibiotic‐resistant infections. Besides this, phages are suitable for novel nanostructure and biomaterials exploiting and promoting the development of biomedicine, such as molecular targeting, cancer diagnosis and treatment, drug and gene delivery, and so on [281]. In this account, bacteriophages in malignancies, including remarkable advances, will be described. Decades ago, phages were discovered to be accumulated in tumor tissues and suppressed the tumor's growth [282]. Subsequently, these microorganisms were found to be capable of binding to tumor cells or interacting with fibroblasts within TME. Until now, significant advancements have been made on phages and two novel technologies have been introduced into this area: phage display and vectors.
Phage display technology is a biotechnology that inserts DNA sequences of exogenous proteins or polypeptides into the appropriate positions of structural genes of phage capsid proteins so that the exogenous genes are expressed with the exogenous (poly) peptides on the surface of phage particles [284]. Phage display is powerful for screening and isolating target‐specific peptides. Five steps are required to obtain a bacteriophage with specific peptides: polypeptide‐displayed phage libraries construction and amplification, target molecule capturing, removing unbound and nonspecific phages, elution of target‐bound phages and infection of host bacteria, and replication.
Peptides with high specificity and affinity usually target tumor microenvironment (TME), tumor‐associated macrophages (TAMs), and cancer‐associated fibroblasts (CAFs), which are conducive to early diagnosis, treatment and prevention vaccines for a variety of cancers [285]. M13 filamentous bacteriophage modified in place of antibodies as receptors enable early cancer detection directly on bodily fluids [286]. In addition, tumor‐targeting peptides, including radiolabeled Peptides, bioluminescent and fluorescently labeled peptides, and nanoparticles‐based peptides, are used to diagnose enormous malignancies described in detail in Li et al.’s article [287]. Moreover, angiogenin is a protein overexpressed and secreted by tumors that trigger angiogenesis to promote tumor growth. Filamentous fd phage could be engineered to express angiogenin‐binding peptide and tumor‐homing peptide on its surface by phage display, capable of first homing to tumor tissue and then capturing angiogenin to suppress tumor growth, resulting in targeted cancer therapy without side effects [288]. Gurung S et al. [289] demonstrated that phage display‐identified PD‐L1‐binding peptides, including PD‐L1 Peptide (Pep)‐1 and PD‐L1 Pep‐2, reinvigorate T‐cell activity and inhibit tumor progression. In addition to diagnosis and treatment, phages can produce preventive and therapeutic vaccination, taking HER2‐displaying M13 Bacteriophages aiming at breast cancer [290].
Phages as delivery vectors combining nanoparticles and medicine reveal new cancer image and drug delivery perspectives. First, because they are tiny, nanoparticles can pass past endothelial cells and infiltrate the tumor microenvironment [291]. Numerous nanoparticles have distinctive magnetic and optical characteristics that allow multivalent targeting of one or more biomarkers [287]. For example, carbon, polymer, and gold nanoparticles are demonstrated to show cancer images and early real‐time detection [292, 293]. In addition, conjugation of bacteriophage Qβ virus‐like particle and 9‐NHAc‐GD2 mimic (an antigen deviation) as a potential anticancer vaccine could produce robust and long‐lasting IgG responses in mice and canines [294]. Recently, a phage vector adeno‐associated virus/phage (AAVP) has gained tremendous popularity due to its characteristics of delivering therapeutic genes to tumors precisely and efficiently. AAVP combines a mammalian transgene cassette of adeno‐associated virus 2 (AAV2) and an fUSE‐5 (peptide display) vector derived from the fd bacteriophage genome [295]. AAVP is designed as an excellent delivery system for various cancers described before [282]. A new peptide called CSP3 was discovered by Xiao et al. [296] and demonstrated that the peptide could be coupled with nanomaterials and chemotherapeutics to create a targeted vehicle for the delivery of therapeutic drugs against cervical cancer.
As a promising tumor therapeutic, phage therapy deserves more clinical trials to confirm its safe dosage and concrete mechanism for better use (Figure 4D).
6.8. Oncolytic virus therapy (OVT)
OVs are classified as naturally occurring or genetically modified viruses that multiply only in cancer cells and kill them without affecting healthy cells [297]. OVs generate benefits in the TME by selectively replicating in tumor cells, delivering different eukaryotic transgenic payloads, inducing immunogenic cell death, and enhancing anticancer immunity [298]. Generally, OVs encompass most DNA and RNA viruses that are naturally cancer‐selected or can be engineered [299]. Coxsackievirus, measles, Maraba, Newcastle disease, polio, and reovirus are a few examples of oncolytic RNA viruses. Adenovirus, herpes simplex, and poxviruses are often employed oncolytic DNA viruses [300]. However, clinical studies mostly use DNA viruses because their biology is better understood. Mechanistically, OVs play a role in two main ways. One is an oncolytic effect whereby OVs infect and replicate in selective tumor cells, eventually leading to tumor lysis. The other is the induction of anti‐tumor immunity [301]. From the details, OVs aim at selective tumor cells and result in immunogenic cell death (ICD), releasing progeny viral particles to reinfect neighboring tumor cells, viral and tumor‐associated antigens, danger‐associated molecular patterns (DAMP), cytokines, and interferons, thereby recruiting and activating DCs, NK cells, and T cells, thus enhancing the anti‐tumor immunity [301] (Figure 5).
FIGURE 5.
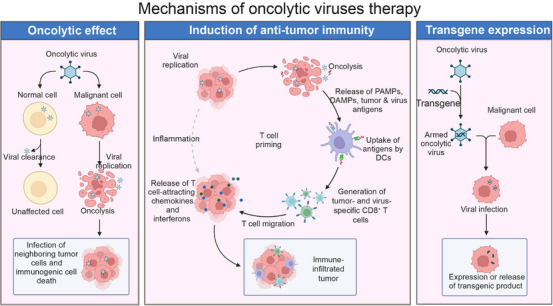
Mechanisms of oncolytic virus therapy. Oncolytic viruses (OVs) produce effects in two main ways. One is an oncolytic effect whereby OVs infect and replicate in selective tumor cells, eventually leading to tumor lysis. The other is that OVs aim at selective tumor cells and result in oncolysis, releasing viral and tumor‐associated antigens, danger‐associated molecular patterns (DAMPs), pathogen‐associated molecular patterns (PAMPs), T cell attracting cytokines and interferons, thereby recruiting and activating DCs, NK cells, and T cells, thus enhancing the anti‐tumor immunity. OVs also have a property that can express and release transgenic products as vectors in malignant cells. Abbreviations: DAMPs, danger‐associated molecular patterns; DC, dendritic cells; NK, natural killer cell; OVs, oncolytic viruses; PAMPs, pathogen‐associated molecular patterns. Biorender supported the materials in Figure 5.
Currently, four OVs and one non‐oncolytic virus have been authorized to treat cancer in different parts of the world [298]. They are talimogene laherparepvec (T‐VEC), H101, ECHO‐7 (Echovirus), Teserpaturev, and one non‐oncolytic virus (Nadofaragene firadenovec). Approved by the United States Food and Drug Administration (FDA) in 2015 as the first oncolytic virus drug in the United States, T‐VEC is a second‐generation oncolytic Herpes Simplex Virus type 1 (HSV‐1) engineered to express human granulocyte‐macrophage colony‐stimulating factor (GM‐CSF), primarily for advanced melanoma and has recently been clinically reported for other various cancers such as triple‐negative breast cancer [301, 302]. China SFDA approved H101, an E1B‐deleted adenovirus, in 2005 for the treatment of head and neck cancer and esophageal cancer [303]. H101 can replicate in p53‐deficient tumor cells [297, 303]. ECHO‐7 (rigvir), a native and oncolytic enterovirus, was once approved for the treatment of melanoma but has now been discontinued [298, 304]. Teserpaturev, also named G47Δ, is a triple‐mutated, third‐generation HSV1‐based OV for patients with malignant gliomas approved in Japan in 2021 [297, 298, 305]. In addition to OVs, a non‐oncolytic adenovirus (Nadofaragene firadenovec) was approved by the FDA in December 2022, which encodes IFNα‐2b for the treatment of bacillus Calmette‐Guerin (BCG)‐unresponsive, non‐muscle invasive bladder cancer (NMIBC) [298]. Other drugs, such as JX‐594 (an engineered vaccinia virus), CG0070 (an oncolytic adenovirus), Reolysin (wild‐type reovirus), etc., are currently in clinical development [297]. In addition, OVs are designed to arm various molecules. GM‐CSF, a widely used immunomodulatory cytokine in OVs, can promote DC migration and maturation, eventually leading to enhanced priming of T cell responses [306]. Meanwhile, IL‐2, IL‐12, IL‐18, chemokines, cytokine, immune‐activating ligands, bispecific T cell engager (BiTE) molecules, etc, are reported to be utilized by Ovs [307, 308, 309]. Furthermore, some novel technologies are applied to OVT and show potential for further virotherapy. Fares et al. [310] utilize neural stem cells (NSCs) targeting glioma cells to deliver oncolytic adenovirus in treating newly diagnosed malignant glioma. Extracellular vesicles (EVs), promoted as promising vehicles for delivering therapeutic cargo, are intended to be capsid‐free for releasing oncolytic adenoviral DNA [311]. Liposome‐cloaked oncolytic adenoviruses (OAs) conjugated onto tumor‐homing Escherichia coli (E. coli‐lipo‐OAs) are proven to be more enriched in non‐small cell lung cancer compared with intravenously injected bare Oas [312]. Applying nanotechnology enhances tumor targeting and oncolytic activity of HSV‐1 virus [313].
Moreover, OVT has been adopted as a monotherapy or adjuvant therapy for many cancers, including advanced melanoma [314], glioma [315], breast cancer [316], hepatocellular carcinoma [317], pancreatic cancer [309], and so on. Notably, OVs are used as monotherapy and preferentially combined with other therapeutics. Preclinical and clinical studies have shown that OVT is well‐tolerated and benefits cancer patients when combined with traditional therapies such as chemotherapy [318, 319]. PD‐1/PD‐L1 therapy has been widely used around the world. However, studies show that OVs can attract effector T cells and induce PD‐L1 expression on the tumor's cancer and immune cells, thereby bolstering the PD‐1/PD‐L1 therapeutic efficacy [314, 320, 321]. The combination of OVT and CTLA‐4 inhibitors appears to have greater efficacy in advanced melanoma patients [322, 323]. The pattern of OV‐ICB combinations transforms immunologically ‘cold’ tumors into ‘hot’ ones, provides better outcomes than monotherapy [324], and will promisingly be a popular therapeutic shortly. Wing and Watanabe's studies demonstrated that OVT significantly overcomes the limitation of monotherapy and enhances the efficacy of CAR‐T cell therapy [309, 325].
To sum up, OVT is generally recognized as a promising emerging cancer treatment and is believed to occupy a more important position in tumor therapeutics in the foreseeable future. Although OVT has the characteristics of tumor‐specific and non‐pathogenic, special drug pharmacokinetics, maneuverability, and low possibility for resistance [299], there are still some challenges required to overcome, such as the risks of viral leakage and unintentional transmission, stringent transport and storage conditions, special OVs injection method, etc. [298]. Besides, systematical neutralizing antibodies and local complement immunity also limit OVT. In the future, more studies are needed to address these problems.
7. PERSPECTIVES AND PROSPECT
In a relatively short time, the microbiome field has shed important light on the symbiotic relationships between human physiology and disease. One of the most pernicious, poorly understood, and complex human illnesses is cancer, which is influenced by global shifts in the microbiome makeup. Identifying human diversity in cancer development, progression, and treatment response may be possible by investigating the causal and molecular relationships between cancers and symbiotic bacteria. In this review, we discussed the advances in TM analysis, potential mechanisms of microbiome‐involved tumor development, and novel microbiota applications for tumor therapy.
TM has been widely associated with chemotherapy response, radiotherapy response, and especially immunotherapy response. Besides, TM is considered an emerging and thriving research direction that offers unparalleled possibilities for tumor therapy of inestimable value. However, the causal role of the microbiome in cancer treatment responses has not been fully established due to a lack of uniform methods, including variations in sample selection, sample collection, and technology. Regarding sample selection, to obtain objective research results and prevent skewed findings, various samples should be gathered and examined because the content and richness of TM from different sources are different. Regarding sample collection, any sample contamination (contaminant DNA and cross‐contamination) would significantly impede microbial research due to the low biomass of TM. Randomized sample types, wearing clean suits, using DNA extraction blank controls, using unique redundant barcodes, etc., could reduce sample contamination. Regarding technology, several methods have been applied to study TM. They are mainly classified into genome sequencing methods (16S rRNA, shotgun metagenomic sequencing, metatranscriptomics, and IS‐pro technique), non‐genome sequencing methods (IHC, FISH, Proteomics, Metabolomics, CLEM, and TEM, TM culture), and cutting‐edge technology (Single‐cell analysis and spatial transcriptome, organoids and 3D technology, nanotechnology, computational tool). As for virus detection, the molecular detection method based on viral nucleic acid, immunological method, and nucleic acid hybridization are generally used. Epigenetics and SCA techniques are generally used to analyze host‐microbiome interactions. Such technical diversity leads to heterogeneity of data resources and accessibility issues. Even when comparing studies that focus on the same type of cancer, it can be difficult, to say the least, to identify overarching, comprehensible themes. Therefore, a standard operating procedure should be proposed and recognized.
Besides, there needs to be more longitudinal studies and clinical trials. As genetics, dietary patterns, age, sex, race, and geographical variations exist as biological variances, a multi‐center, longitudinal, collaborative effort to study the microbiome in cancer, combined with tumor, blood, and fecal collection, multi‐omics data generation, and combined with experimental contamination control to conduct a synchronous meta‐analysis of existing cancer data sets is significant. Such data sets are uniform and may be able to identify global microbial drivers in cancer pathogenesis and treatment.
Interactions are prevalent in the tumor microenvironment. Notably, since the microbiome could widely affect tumor cells, tumor cells should impact the microbiome. Tumor cells can release various factors that can alter the composition and function of the microbiome. For example, tumor cells can release cytokines that promote certain bacteria's growth while inhibiting others' growth. Additionally, tumor cells can alter the pH and oxygen levels in the TME, which can also affect the microbiome. Correspondingly, the microbiome can adapt to the TME in several ways. For example, certain bacteria may be able to thrive in the low‐oxygen, acidic conditions found in tumors. Additionally, the microbiome might possess the ability to immune escape as immune cells could release chemicals that affect the growth and composition of the microbiome. The exact mechanisms by which tumor cells affect the microbiome and the microbiome adapts to the TME are still being studied. However, in tumor microecology, the shared goal of microbes and cancer cells is survival. They are either in a competitive relationship or a symbiotic or commensal one, and any relationship in which the cancer cells gain influences through the presence of bacteria may provide a potential new avenue for cancer treatment.
In addition, the mechanism of how bacteria enter tumor cells has yet to be fully understood. However, some studies suggest that certain bacteria can use specific receptors on the surface of tumor cells to enter them. Other studies suggest that bacteria can enter tumor cells through endocytosis, a process where the cell membrane engulfs the bacteria and brings it inside the cell.
Lastly, ideal cell/animal models are urgently needed for research about TM. Several approaches can be taken. One approach uses genetically engineered mice with specific mutations that mimic human tumors. Another approach is to use patient‐derived xenografts, which involve transplanting human tumor tissue into immunodeficient mice. Additionally, in vitro cell culture models can be used to study the interactions between bacteria and tumor cells.
To sum up, some controversies and challenges required to be addressed: i) the definition of the TM; ii) how to formulate unified and feasible detection methods; iii) whether the relation between TM and tumor could be unraveled to causality; iv); the mechanism of how bacteria enter tumor cells; v) how to establish ideal models applied to TM research.
The development of tailored screening techniques and microbiome‐based therapeutics to enhance patient care and treatment outcomes may be guided by further study into the modification of the microbiome in the pathophysiology and carcinogenesis of cancer. Additionally, saliva, blood, and fecal microbiological analyses may become screening methods for certain cancers. There are countless research opportunities to inform and direct new clinical practice standards in cancer due to the growing significance of the microbiome in a range of human cancers. We firmly believe that TM‐based treatment will soon become a safe and individual‐tailored therapeutic option for cancer patients.
8. CONCLUSIONS
Overall, the review underscores the significance of the TM in tumorigenesis and treatment response. By expanding our understanding beyond the gut microbiome, researchers and clinicians can harness the potential of the TM to advance cancer diagnosis, treatment, and patient care.
AUTHOR CONTRIBUTIONS
Yuan Cheng, Quan Cheng, and Guodong Liu conceived the study, supervised the manuscript, and revised the manuscript. Quan Cheng, Hao Zhang, and Guodong Liu provided funding support. Li Fu and Hao Zhang drafted and revised the manuscript. Xinwen Leiliang, Chunrun Qu, Wantao Wu, Rong Wen, Ning Huang, and Qiuguang He conducted literature retrieval and revised the manuscript. All authors have read and approved the final manuscript.
CONFLICT OF INTEREST STATEMENT
All authors declare that they have no competing interests.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
ACKNOWLEDGMENTS
Biorender supported the material in graphics in this article. This work was supported by the National Natural Science Foundation of China (No. 82372943, 82303610), Hunan Provincial Natural Science Foundation of China (No. 2022JJ20095), Hunan Youth Science and Technology Talent Project (No. 2023RC3074), China Postdoctoral Science Foundation (No. 2023MD734131), Chongqing Postdoctoral Science Foundation (No. CSTB2023NSCQBHX0002), Kuanren Talents Program of the second affiliated hospital of Chongqing Medical University, and Chongqing Postdoctoral Research Special Funding Project (2023CQBSHTB3095).
Zhang H, Fu L, Leiliang X, Qu C, Wu W, Wen R, et al. Beyond the Gut: The intratumoral microbiome's influence on tumorigenesis and treatment response. Cancer Commun. 2024;44:1130–1167. 10.1002/cac2.12597
Hao Zhang and Li Fu authors contributed equally to this work.
Contributor Information
Quan Cheng, Email: chengquan@csu.edu.cn.
Guodong Liu, Email: 304678@hospital.cqmu.edu.cn.
Yuan Cheng, Email: chengyuan@hospital.cqmu.edu.cn.
DATA AVAILABILITY STATEMENTS
Not applicable.
REFERENCES
- 1. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type‐specific intracellular bacteria. Science. 2020;368(6494):973–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lemmon MJ, van Zijl P, Fox ME, Mauchline ML, Giaccia AJ, Minton NP, et al. Anaerobic bacteria as a gene delivery system that is controlled by the tumor microenvironment. Gene Ther. 1997;4(8):791–796. [DOI] [PubMed] [Google Scholar]
- 3. Yazawa K, Fujimori M, Amano J, Kano Y, Taniguchi S. Bifidobacterium longum as a delivery system for cancer gene therapy: selective localization and growth in hypoxic tumors. Cancer Gene Ther. 2000;7(2):269–274. [DOI] [PubMed] [Google Scholar]
- 4. Alexeev EE, Lanis JM, Kao DJ, Campbell EL, Kelly CJ, Battista KD, et al. Microbiota‐Derived Indole Metabolites Promote Human and Murine Intestinal Homeostasis through Regulation of Interleukin‐10 Receptor. Am J Pathol. 2018;188(5):1183–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, et al. IL‐6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis‐associated cancer. Cancer Cell. 2009;15(2):103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016;14(8):e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cullin N, Antunes CA, Straussman R, Stein‐Thoeringer CK, Elinav E. Microbiome and cancer. Cancer Cell. 2021;39(10):1317–1341. [DOI] [PubMed] [Google Scholar]
- 8. Yuan L, Yang P, Wei G, Hu X, Chen S, Lu J, et al. Tumor microbiome diversity influences papillary thyroid cancer invasion. Commun Biol. 2022;5(1):864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sepich‐Poore GD, Zitvogel L, Straussman R, Hasty J, Wargo JA, Knight R. The microbiome and human cancer. Science. 2021;371(6536):eabc4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Galeano Nino JL, Wu H, LaCourse KD, Kempchinsky AG, Baryiames A, Barber B, et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature. 2022;611(7937):810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Irfan M, Delgado RZR, Frias‐Lopez J. The Oral Microbiome and Cancer. Front Immunol. 2020;11:591088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Knippel RJ, Drewes JL, Sears CL. The Cancer Microbiome: Recent Highlights and Knowledge Gaps. Cancer Discov. 2021;11(10):2378–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matson V, Chervin CS, Gajewski TF. Cancer and the Microbiome‐Influence of the Commensal Microbiota on Cancer, Immune Responses, and Immunotherapy. Gastroenterology. 2021;160(2):600–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018;8(4):403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Geller LT, Barzily‐Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357(6356):1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Picardo SL, Coburn B, Hansen AR. The microbiome and cancer for clinicians. Crit Rev Oncol Hematol. 2019;141:1–12. [DOI] [PubMed] [Google Scholar]
- 17. Yang L, Li A, Wang Y, Zhang Y. Intratumoral microbiota: roles in cancer initiation, development and therapeutic efficacy. Signal Transduct Target Ther. 2023;8(1):35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen A, Neuwirth I, Herndler‐Brandstetter D. Modeling the Tumor Microenvironment and Cancer Immunotherapy in Next‐Generation Humanized Mice. Cancers (Basel). 2023;15(11):2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao W, Dai S, Yue L, Xu F, Gu J, Dai X, et al. Emerging mechanisms progress of colorectal cancer liver metastasis. Front Endocrinol (Lausanne). 2022;13:1081585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ma PJ, Wang MM, Wang Y. Gut microbiota: A new insight into lung diseases. Biomed Pharmacother. 2022;155:113810. [DOI] [PubMed] [Google Scholar]
- 21. Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. 2017;17(4):219–232. [DOI] [PubMed] [Google Scholar]
- 22. Pereira MS, Kriegel MA. Translocating Lactobacillus torments tumors via tryptophan catabolism. Cell. 2023;167(6):1481–1494 e18. [DOI] [PubMed] [Google Scholar]
- 23. Bender MJ, McPherson AC, Phelps CM, Pandey SP, Laughlin CR, Shapira JH, et al. Dietary tryptophan metabolite released by intratumoral Lactobacillus reuteri facilitates immune checkpoint inhibitor treatment. Cell. 2023;186(9):1846–1862 e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358(6369):1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chassaing B, Kumar M, Baker MT, Singh V, Vijay‐Kumar M. Mammalian gut immunity. Biomed J. 2014;37(5):246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wei X, Chen Y, Jiang X, Peng M, Liu Y, Mo Y, et al. Mechanisms of vasculogenic mimicry in hypoxic tumor microenvironments. Mol Cancer. 2021;20(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fletcher AA, Kelly MS, Eckhoff AM, Allen PJ. Revisiting the intrinsic mycobiome in pancreatic cancer. Nature. 2023;620(7972):E1–E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Patel JB. 16S rRNA gene sequencing for bacterial pathogen identification in the clinical laboratory. Mol Diagn. 2001;6(4):313–321. [DOI] [PubMed] [Google Scholar]
- 29. Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer KH, et al. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol. 2014;12(9):635–645. [DOI] [PubMed] [Google Scholar]
- 30. Johnson JS, Spakowicz DJ, Hong BY, Petersen LM, Demkowicz P, Chen L, et al. Evaluation of 16S rRNA gene sequencing for species and strain‐level microbiome analysis. Nat Commun. 2019;10(1):5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chai X, Wang J, Li H, Gao C, Li S, Wei C, et al. Intratumor microbiome features reveal antitumor potentials of intrahepatic cholangiocarcinoma. Gut Microbes. 2023;15(1):2156255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fu A, Yao B, Dong T, Chen Y, Yao J, Liu Y, et al. Tumor‐resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell. 2022;185(8):1356–1372 e26. [DOI] [PubMed] [Google Scholar]
- 33. Liu W, Zhang X, Xu H, Li S, Lau HC, Chen Q, et al. Microbial Community Heterogeneity Within Colorectal Neoplasia and its Correlation With Colorectal Carcinogenesis. Gastroenterology. 2021;160(7):2395–2408. [DOI] [PubMed] [Google Scholar]
- 34. Clarridge JE, 3rd . Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004;17(4):840–862, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Quince C, Walker AW, Simpson JT, Loman NJ, Segata N. Corrigendum: Shotgun metagenomics, from sampling to analysis. Nat Biotechnol. 2017;35(12):1211. [DOI] [PubMed] [Google Scholar]
- 36. Huang X, Chen C, Xie W, Zhou C, Tian X, Zhang Z, et al. Metagenomic Analysis of Intratumoral Microbiome Linking to Response to Neoadjuvant Chemoradiotherapy in Rectal Cancer. Int J Radiat Oncol Biol Phys. 2023;117(5):1255–1269. [DOI] [PubMed] [Google Scholar]
- 37. Liu NN, Yi CX, Wei LQ, Zhou JA, Jiang T, Hu CC, et al. The intratumor mycobiome promotes lung cancer progression via myeloid‐derived suppressor cells. Cancer Cell. 2023;41(11):1927–1944 e9. [DOI] [PubMed] [Google Scholar]
- 38. Zapatka M, Borozan I, Brewer DS, Iskar M, Grundhoff A, Alawi M, et al. The landscape of viral associations in human cancers. Nat Genet. 2020;52(3):320–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou X, Kandalai S, Hossain F, Zheng Q. Tumor microbiome metabolism: A game changer in cancer development and therapy. Front Oncol. 2022;12:933407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mukherjee A, Reddy MS. Metatranscriptomics: an approach for retrieving novel eukaryotic genes from polluted and related environments. 3 Biotech. 2020;10(2):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ojala T, Kankuri E, Kankainen M. Understanding human health through metatranscriptomics. Trends Mol Med. 2023;29(5):376–389. [DOI] [PubMed] [Google Scholar]
- 42. Xie Y, Xie F, Zhou X, Zhang L, Yang B, Huang J, et al. Microbiota in Tumors: From Understanding to Application. Adv Sci (Weinh). 2022;9(21):e2200470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang N, Kandalai S, Zhou X, Hossain F, Zheng Q. Applying multi‐omics toward tumor microbiome research. Imeta. 2023;2(1):e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Budding AE, Grasman ME, Lin F, Bogaards JA, Soeltan‐Kaersenhout DJ, Vandenbroucke‐Grauls CM, et al. IS‐pro: high‐throughput molecular fingerprinting of the intestinal microbiota. FASEB J. 2010;24(11):4556–4564. [DOI] [PubMed] [Google Scholar]
- 45. Budding AE, Hoogewerf M, Vandenbroucke‐Grauls CM, Savelkoul PH. Automated Broad‐Range Molecular Detection of Bacteria in Clinical Samples. J Clin Microbiol. 2016;54(4):934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Singer M, Koedooder R, Bos MP, Poort L, Schoenmakers S, Savelkoul PHM, et al. The profiling of microbiota in vaginal swab samples using 16S rRNA gene sequencing and IS‐pro analysis. BMC Microbiol. 2021;21(1):100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guo W, Zhang Y, Guo S, Mei Z, Liao H, Dong H, et al. Tumor microbiome contributes to an aggressive phenotype in the basal‐like subtype of pancreatic cancer. Commun Biol. 2021;4(1):1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Matturro B, Rossetti S, Leitão P. CAtalyzed Reporter Deposition Fluorescence In Situ Hybridization (CARD‐FISH) for Complex Environmental Samples. Methods Mol Biol. 2021;2246:129–140. [DOI] [PubMed] [Google Scholar]
- 49. Zhang L, Xiao D, Cheng K. Proteomic analysis of microbial infections. Molecular Medical Microbiology. Elsevier; 2024. p. 1951–1963. [Google Scholar]
- 50. Wu AH, French D. Implementation of liquid chromatography/mass spectrometry into the clinical laboratory. Clin Chim Acta. 2013;420:4–10. [DOI] [PubMed] [Google Scholar]
- 51. Qian X, Zhang HY, Li QL, Ma GJ, Chen Z, Ji XM, et al. Integrated microbiome, metabolome, and proteome analysis identifies a novel interplay among commensal bacteria, metabolites and candidate targets in non‐small cell lung cancer. Clin Transl Med. 2022;12(6):e947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Alharbi RA. Proteomics approach and techniques in identification of reliable biomarkers for diseases. Saudi J Biol Sci. 2020;27(3):968–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Al‐Amrani S, Al‐Jabri Z, Al‐Zaabi A, Alshekaili J, Al‐Khabori M. Proteomics: Concepts and applications in human medicine. World J Biol Chem. 2021;12(5):57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Idle JR, Gonzalez FJJCm. Metabolomics. Cell Metab. 2007;6(5):348–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bauermeister A, Mannochio‐Russo H, Costa‐Lotufo LV, Jarmusch AK, Dorrestein PC. Mass spectrometry‐based metabolomics in microbiome investigations. Nat Rev Microbiol. 2022;20(3):143–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bhosle A, Wang Y, Franzosa EA, Huttenhower C. Progress and opportunities in microbial community metabolomics. Curr Opin Microbiol. 2022;70:102195. [DOI] [PubMed] [Google Scholar]
- 57. Tang J. Microbial metabolomics. Curr Genomics. 2011;12(6):391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Daliri EB, Wei S, Oh DH, Lee BH. The human microbiome and metabolomics: Current concepts and applications. Crit Rev Food Sci Nutr. 2017;57(16):3565–3576. [DOI] [PubMed] [Google Scholar]
- 59. Zhu Z, Cai J, Hou W, Xu K, Wu X, Song Y, et al. Microbiome and spatially resolved metabolomics analysis reveal the anticancer role of gut Akkermansia muciniphila by crosstalk with intratumoral microbiota and reprogramming tumoral metabolism in mice. Gut Microbes. 2023;15(1):2166700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cheng Y, He C, Wang M, Ma X, Mo F, Yang S, et al. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct Target Ther. 2019;4:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang L, Wang R, Xie Z. The roles of DNA methylation on the promotor of the Epstein‐Barr virus (EBV) gene and the genome in patients with EBV‐associated diseases. Appl Microbiol Biotechnol. 2022;106(12):4413–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zheng Q, Omans ND, Leicher R, Osunsade A, Agustinus AS, Finkin‐Groner E, et al. Reversible histone glycation is associated with disease‐related changes in chromatin architecture. Nat Commun. 2019;10(1):1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lüdtke TH, Wojahn I, Kleppa MJ, Schierstaedt J, Christoffels VM, Künzler P, et al. Combined genomic and proteomic approaches reveal DNA binding sites and interaction partners of TBX2 in the developing lung. Respir Res. 2021;22(1):85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Barhoum A, Luisa García‐Betancourt M. Chapter 10 ‐ Physicochemical characterization of nanomaterials: size, morphology, optical, magnetic, and electrical properties. In: Barhoum A, Makhlouf ASH, editors. Emerging Applications of Nanoparticles and Architecture Nanostructures. Elsevier; 2018. p. 279–304. [Google Scholar]
- 65. Pope I, Tanner H, Masia F, Payne L, Arkill KP, Mantell J, et al. Correlative light‐electron microscopy using small gold nanoparticles as single probes. Light Sci Appl. 2023;12(1):80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Huang Z, Mo S, Yan L, Wei X, Huang Y, Zhang L, et al. A Simple Culture Method Enhances the Recovery of Culturable Actinobacteria From Coastal Sediments. Front Microbiol. 2021;12:675048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lewis WH, Tahon G, Geesink P, Sousa DZ, Ettema TJG. Innovations to culturing the uncultured microbial majority. Nat Rev Microbiol. 2021;19(4):225–240. [DOI] [PubMed] [Google Scholar]
- 68. Cross KL, Campbell JH, Balachandran M, Campbell AG, Cooper CJ, Griffen A, et al. Targeted isolation and cultivation of uncultivated bacteria by reverse genomics. Nat Biotechnol. 2019;37(11):1314–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. LeSavage BL, Suhar RA, Broguiere N, Lutolf MP, Heilshorn SC. Next‐generation cancer organoids. Nat Mater. 2022;21(2):143–159. [DOI] [PubMed] [Google Scholar]
- 70. Sule WF, Oluwayelu DO. Real‐time RT‐PCR for COVID‐19 diagnosis: challenges and prospects. Pan Afr Med J. 2020;35(Suppl 2):121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ahmed W, Bivins A, Metcalfe S, Smith WJM, Ziels R, Korajkic A, et al. RT‐qPCR and ATOPlex sequencing for the sensitive detection of SARS‐CoV‐2 RNA for wastewater surveillance. Water Res. 2022;220:118621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen L, Chen H, Ye J, Ge Y, Wang H, Dai E, et al. Intratumoral expression of interleukin 23 variants using oncolytic vaccinia virus elicit potent antitumor effects on multiple tumor models via tumor microenvironment modulation. Theranostics. 2021;11(14):6668–6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kojabad AA, Farzanehpour M, Galeh HEG, Dorostkar R, Jafarpour A, Bolandian M, et al. Droplet digital PCR of viral DNA/RNA, current progress, challenges, and future perspectives. J Med Virol. 2021;93(7):4182–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Galimberti S, Balducci S, Guerrini F, Del Re M, Cacciola R. Digital Droplet PCR in Hematologic Malignancies: A New Useful Molecular Tool. Diagnostics (Basel). 2022;12(6):1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Li CL, Ho MC, Lin YY, Tzeng ST, Chen YJ, Pai HY, et al. Cell‐Free Virus‐Host Chimera DNA From Hepatitis B Virus Integration Sites as a Circulating Biomarker of Hepatocellular Cancer. Hepatology. 2020;72(6):2063–2076. [DOI] [PubMed] [Google Scholar]
- 76. Zhao MH, Liu W, Zhang X, Zhang Y, Luo B. Epstein‐Barr virus miR‐BART2‐5p and miR‐BART11‐5p regulate cell proliferation, apoptosis, and migration by targeting RB and p21 in gastric carcinoma. J Med Virol. 2023;95(1):e28338. [DOI] [PubMed] [Google Scholar]
- 77. Pedersen JC. Hemagglutination‐inhibition test for avian influenza virus subtype identification and the detection and quantitation of serum antibodies to the avian influenza virus. Methods Mol Biol. 2008;436:53–66. [DOI] [PubMed] [Google Scholar]
- 78. Sethi J, Pei D, Hirshaut Y. Choice and specificity of complement in complement fixation assay. J Clin Microbiol. 1981;13(5):888–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Westhaus S, Rabenau HF. Neutralization Assay for SARS‐CoV‐2 Infection: Plaque Reduction Neutralization Test. Methods Mol Biol. 2022;2452:353–360. [DOI] [PubMed] [Google Scholar]
- 80. Tabatabaei MS, Ahmed M. Enzyme‐Linked Immunosorbent Assay (ELISA). Methods Mol Biol. 2022;2508:115–134. [DOI] [PubMed] [Google Scholar]
- 81. Burckhardt CJ, Minna JD, Danuser G. Co‐immunoprecipitation and semi‐quantitative immunoblotting for the analysis of protein‐protein interactions. STAR Protoc. 2021;2(3):100644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Granzow H, Klupp BG, Mettenleiter TC. Entry of pseudorabies virus: an immunogold‐labeling study. J Virol. 2005;79(5):3200–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tanaka S, Nishii H, Ito S, Kameya‐Iwaki M, Sommartya P. Detection of Cymbidium Mosaic Potexvirus and Odontoglossum Ringspot Tobamovirus from Thai Orchids by Rapid Immunofilter Paper Assay. Plant Dis. 1997;81(2):167–170. [DOI] [PubMed] [Google Scholar]
- 84. Eun AJ, Wong SM. Detection of cymbidium mosaic potexvirus and odontoglossum ringspot tobamovirus using immuno‐capillary zone electrophoresis. Phytopathology. 1999;89(6):522–528. [DOI] [PubMed] [Google Scholar]
- 85. Ryazantsev DY, Voronina DV, Zavriev SK. Immuno‐PCR: Achievements and Perspectives. Biochemistry (Mosc). 2016;81(13):1754–1770. [DOI] [PubMed] [Google Scholar]
- 86. Trent DW, Harvey CL, Qureshi A, LeStourgeon D. Solid‐phase radioimmunoassay for antibodies to flavivirus structural and nonstructural proteins. Infect Immun. 1976;13(5):1325–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhang DY, Chen SX, Yin P. Optimizing the specificity of nucleic acid hybridization. Nat Chem. 2012;4(3):208–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Shirasawa H, Tomita Y, Kubota K, Kasai T, Sekiya S, Takamizawa H, et al. Detection of human papillomavirus type 16 DNA and evidence for integration into the cell DNA in cervical dysplasia. J Gen Virol. 1986;67(Pt 9):2011–2015. [DOI] [PubMed] [Google Scholar]
- 89. Pan ST, Chang WS, Murphy M, Martinez A, Chuang SS. Cutaneous peripheral T‐cell lymphoma of cytotoxic phenotype mimicking extranodal NK/T‐cell lymphoma. Am J Dermatopathol. 2011;33(2):e17–e20. [DOI] [PubMed] [Google Scholar]
- 90. Huang X‐M, Wei SG, Wang LF. Reversal of malignant phenotype of human hepatoma cells by antisense: c‐ets‐2, c‐myc and N‐ras. Zhonghua Zhongliu Zazhi. 1994;16(4):243–246. [PubMed] [Google Scholar]
- 91. Manjunath N, Kaur H, Bala S, Kaur R, Bhargava V, Rath GK, et al. Detection of herpes simplex virus type 2 DNA in uterine cervix lesions using cloned Bgl II N fragment of HSV‐2 DNA as a probe. Indian J Med Res. 1988;87:127–133. [PubMed] [Google Scholar]
- 92. Chen X, Yang Z. Chapter 3 ‐ Biosensors for single‐cell metabolomic characterization. In: Chen J, Lu Y, editors. Biosensors for Single‐Cell Analysis. Academic Press; 2022. p. 37–70. [Google Scholar]
- 93. Yasuga H, Shoji K, Koiwai K, Kawano R. New Sensing Technologies: Microtas/NEMS/MEMS. In: Narayan R, editor. Encyclopedia of Sensors and Biosensors (First Edition). Oxford: Elsevier; 2023. p. 526–540. [Google Scholar]
- 94. Ghaddar B, Biswas A, Harris C, Omary MB, Carpizo DR, Blaser MJ, et al. Tumor microbiome links cellular programs and immunity in pancreatic cancer. Cancer Cell. 2022;40(10):1240–1253 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wu F, Fan J, He Y, Xiong A, Yu J, Li Y, et al. Single‐cell profiling of tumor heterogeneity and the microenvironment in advanced non‐small cell lung cancer. Nat Commun. 2021;12(1):2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Longo SK, Guo MG, Ji AL, Khavari PA. Integrating single‐cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat Rev Genet. 2021;22(10):627–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Moncada R, Barkley D, Wagner F, Chiodin M, Devlin JC, Baron M, et al. Integrating microarray‐based spatial transcriptomics and single‐cell RNA‐seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat Biotechnol. 2020;38(3):333–342. [DOI] [PubMed] [Google Scholar]
- 98. Ai B, Liang Y, Yan T, Lei Y. Exploration of immune cell heterogeneity by single‐cell RNA sequencing and identification of secretory leukocyte protease inhibitor as an oncogene in pancreatic cancer. Environ Toxicol. 2024. [DOI] [PubMed] [Google Scholar]
- 99. Liu Z, Zhang Z, Zhang Y, Zhou W, Zhang X, Peng C, et al. Spatial transcriptomics reveals that metabolic characteristics define the tumor immunosuppression microenvironment via iCAF transformation in oral squamous cell carcinoma. Int J Oral Sci. 2024;16(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Jain S, Rick JW, Joshi RS, Beniwal A, Spatz J, Gill S, et al. Single‐cell RNA sequencing and spatial transcriptomics reveal cancer‐associated fibroblasts in glioblastoma with protumoral effects. J Clin Invest. 2023;133(5):e147087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ji AL, Rubin AJ, Thrane K, Jiang S, Reynolds DL, Meyers RM, et al. Multimodal Analysis of Composition and Spatial Architecture in Human Squamous Cell Carcinoma. Cell. 2020;182(2):497–514 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Qi J, Sun H, Zhang Y, Wang Z, Xun Z, Li Z, et al. Single‐cell and spatial analysis reveal interaction of FAP(+) fibroblasts and SPP1(+) macrophages in colorectal cancer. Nat Commun. 2022;13(1):1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wu SZ, Al‐Eryani G, Roden DL, Junankar S, Harvey K, Andersson A, et al. A single‐cell and spatially resolved atlas of human breast cancers. Nat Genet. 2021;53(9):1334–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wang G, Zhao J, Yan Y, Wang Y, Wu AR, Yang C. Construction of a 3D whole organism spatial atlas by joint modelling of multiple slices with deep neural networks. Nat Mach Intell. 2023;5(11):1200–1213. [Google Scholar]
- 105. Wang B, Lin AE, Yuan J, Novak KE, Koch MD, Wingreen NS, et al. Single‐cell massively‐parallel multiplexed microbial sequencing (M3‐seq) identifies rare bacterial populations and profiles phage infection. Nat Microbiol. 2023;8(10):1846–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Tang XY, Wu S, Wang D, Chu C, Hong Y, Tao M, et al. Human organoids in basic research and clinical applications. Signal Transduct Target Ther. 2022;7(1):168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Yang S, Hu H, Kung H, Zou R, Dai Y, Hu Y, et al. Organoids: The current status and biomedical applications. MedComm (2020). 2023;4(3):e274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Yang R, Yu Y. Patient‐derived organoids in translational oncology and drug screening. Cancer Lett. 2023;562:216180. [DOI] [PubMed] [Google Scholar]
- 109. Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, et al. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell. 2018;172(1‐2):373–386 e10. [DOI] [PubMed] [Google Scholar]
- 110. Ganesh K, Wu C, O'Rourke KP, Szeglin BC, Zheng Y, Sauve CG, et al. A rectal cancer organoid platform to study individual responses to chemoradiation. Nat Med. 2019;25(10):1607–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Below CR, Kelly J, Brown A, Humphries JD, Hutton C, Xu J, et al. A microenvironment‐inspired synthetic three‐dimensional model for pancreatic ductal adenocarcinoma organoids. Nat Mater. 2022;21(1):110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Kim M, Mun H, Sung CO, Cho EJ, Jeon HJ, Chun SM, et al. Patient‐derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat Commun. 2019;10(1):3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Tiriac H, Belleau P, Engle DD, Plenker D, Deschenes A, Somerville TDD, et al. Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer Discov. 2018;8(9):1112–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kopper O, de Witte CJ, Lohmussaar K, Valle‐Inclan JE, Hami N, Kester L, et al. An organoid platform for ovarian cancer captures intra‐ and interpatient heterogeneity. Nat Med. 2019;25(5):838–849. [DOI] [PubMed] [Google Scholar]
- 115. Jacob F, Salinas RD, Zhang DY, Nguyen PTT, Schnoll JG, Wong SZH, et al. A Patient‐Derived Glioblastoma Organoid Model and Biobank Recapitulates Inter‐ and Intra‐tumoral Heterogeneity. Cell. 2020;180(1):188–204 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Driehuis E, Kretzschmar K, Clevers H. Establishment of patient‐derived cancer organoids for drug‐screening applications. Nat Protoc. 2020;15(10):3380–3409. [DOI] [PubMed] [Google Scholar]
- 117. Yuki K, Cheng N, Nakano M, Kuo CJ. Organoid Models of Tumor Immunology. Trends Immunol. 2020;41(8):652–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell. 2018;175(7):1972–1988 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Puschhof J, Pleguezuelos‐Manzano C, Clevers H. Organoids and organs‐on‐chips: Insights into human gut‐microbe interactions. Cell Host Microbe. 2021;29(6):867–878. [DOI] [PubMed] [Google Scholar]
- 120. Su CY, Burchett A, Dunworth M, Choi JS, Ewald AJ, Ahn EH, et al. Engineering a 3D collective cancer invasion model with control over collagen fiber alignment. Biomaterials. 2021;275:120922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Dabbagh SR, Sarabi MR, Birtek MT, Seyfi S, Sitti M, Tasoglu S. 3D‐printed microrobots from design to translation. Nat Commun. 2022;13(1):5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Zimdahl H, Hübner N. Gene Chip Technology and Its Application to Molecular Medicine. Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine. 2005:650–655. [Google Scholar]
- 123. Chan LC, Kalyanasundram J, Leong SW, Masarudin MJ, Veerakumarasivam A, Yusoff K, et al. Persistent Newcastle disease virus infection in bladder cancer cells is associated with putative pro‐survival and anti‐viral transcriptomic changes. BMC Cancer. 2021;21(1):625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Yen CJ, Ai YL, Tsai HW, Chan SH, Yen CS, Cheng KH, et al. Hepatitis B virus surface gene pre‐S(2) mutant as a high‐risk serum marker for hepatoma recurrence after curative hepatic resection. Hepatology. 2018;68(3):815–826. [DOI] [PubMed] [Google Scholar]
- 125. Chen EC, Miller SA, DeRisi JL, Chiu CY. Using a pan‐viral microarray assay (Virochip) to screen clinical samples for viral pathogens. J Vis Exp. 2011;(50):2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Yang Y, Chu B, Cheng J, Tang J, Song B, Wang H, et al. Bacteria eat nanoprobes for aggregation‐enhanced imaging and killing diverse microorganisms. Nat Commun. 2022;13(1):1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Zhu K, Schaffer AA, Robinson W, Xu J, Ruppin E, Ergun AF, et al. Strain level microbial detection and quantification with applications to single cell metagenomics. Nat Commun. 2022;13(1):6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Wang P, Zhang S, He G, Du M, Qi C, Liu R, et al. microbioTA: an atlas of the microbiome in multiple disease tissues of Homo sapiens and Mus musculus. Nucleic Acids Res. 2023;51(D1):D1345–D1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Dohlman AB, Arguijo Mendoza D, Ding S, Gao M, Dressman H, Iliev ID, et al. The cancer microbiome atlas: a pan‐cancer comparative analysis to distinguish tissue‐resident microbiota from contaminants. Cell Host Microbe. 2021;29(2):281–298 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Tan CCS, Ko KKK, Chen H, Liu J, Loh M, Consortium SGKH, et al. No evidence for a common blood microbiome based on a population study of 9,770 healthy humans. Nat Microbiol. 2023;8(5):973–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Wong‐Rolle A, Wei HK, Zhao C, Jin C. Unexpected guests in the tumor microenvironment: microbiome in cancer. Protein Cell. 2021;12(5):426–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Jiang M, Yang Z, Dai J, Wu T, Jiao Z, Yu Y, et al. Intratumor microbiome: selective colonization in the tumor microenvironment and a vital regulator of tumor biology. MedComm (2020). 2023;4(5):e376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359(6375):592–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Wilson MR, Jiang Y, Villalta PW, Stornetta A, Boudreau PD, Carra A, et al. The human gut bacterial genotoxin colibactin alkylates DNA. Science. 2019;363(6428):eaar7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Allen J, Sears CL. Impact of the gut microbiome on the genome and epigenome of colon epithelial cells: contributions to colorectal cancer development. Genome Med. 2019;11(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Urbaniak C, Gloor GB, Brackstone M, Scott L, Tangney M, Reid G. The Microbiota of Breast Tissue and Its Association with Breast Cancer. Appl Environ Microbiol. 2016;82(16):5039–5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Irrazabal T, Thakur BK, Kang M, Malaise Y, Streutker C, Wong EOY, et al. Limiting oxidative DNA damage reduces microbe‐induced colitis‐associated colorectal cancer. Nat Commun. 2020;11(1):1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Chattopadhyay I, Verma M, Panda M. Role of Oral Microbiome Signatures in Diagnosis and Prognosis of Oral Cancer. Technol Cancer Res Treat. 2019;18:1533033819867354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. DeCaprio JA. Molecular Pathogenesis of Merkel Cell Carcinoma. Annu Rev Pathol. 2021;16:69–91. [DOI] [PubMed] [Google Scholar]
- 140. Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, et al. T antigen mutations are a human tumor‐specific signature for Merkel cell polyomavirus. Proc Natl Acad Sci U S A. 2008;105(42):16272–16277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Wallace NA, Khanal S, Robinson KL, Wendel SO, Messer JJ, Galloway DA. High‐Risk Alphapapillomavirus Oncogenes Impair the Homologous Recombination Pathway. J Virol. 2017;91(20):e01084–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Nicot C. HTLV‐I Tax‐Mediated Inactivation of Cell Cycle Checkpoints and DNA Repair Pathways Contribute to Cellular Transformation: “A Random Mutagenesis Model”. J Cancer Sci. 2015;2(2). 10.13188/2377-9292.1000009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Peng Y, Wang Y, Zhou C, Mei W, Zeng C. PI3K/Akt/mTOR Pathway and Its Role in Cancer Therapeutics: Are We Making Headway? Front Oncol. 2022;12:819128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Tsay JJ, Wu BG, Badri MH, Clemente JC, Shen N, Meyn P, et al. Airway Microbiota Is Associated with Upregulation of the PI3K Pathway in Lung Cancer. Am J Respir Crit Care Med. 2018;198(9):1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Gustafson AM, Soldi R, Anderlind C, Scholand MB, Qian J, Zhang X, et al. Airway PI3K pathway activation is an early and reversible event in lung cancer development. Sci Transl Med. 2010;2(26):26ra5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Zhang L, Wu J, Ling MT, Zhao L, Zhao KN. The role of the PI3K/Akt/mTOR signalling pathway in human cancers induced by infection with human papillomaviruses. Mol Cancer. 2015;14:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Olagnier D, Sze A, Bel Hadj S, Chiang C, Steel C, Han X, et al. HTLV‐1 Tax‐mediated inhibition of FOXO3a activity is critical for the persistence of terminally differentiated CD4+ T cells. PLoS Pathog. 2014;10(12):e1004575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Chang HH, Ganem D. A unique herpesviral transcriptional program in KSHV‐infected lymphatic endothelial cells leads to mTORC1 activation and rapamycin sensitivity. Cell Host Microbe. 2013;13(4):429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Mullen PJ, Christofk HR. The Metabolic Relationship Between Viral Infection and Cancer. Annual Review of Cancer Biology. 2022;6(1):1–15. [Google Scholar]
- 150. Xue C, Yao Q, Gu X, Shi Q, Yuan X, Chu Q, et al. Evolving cognition of the JAK‐STAT signaling pathway: autoimmune disorders and cancer. Signal Transduct Target Ther. 2023;8(1):204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Bromberg J, Darnell JE, Jr . The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19(21):2468–2473. [DOI] [PubMed] [Google Scholar]
- 152. Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL, Jr . Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25(2):134–144. [DOI] [PubMed] [Google Scholar]
- 153. Katz J, Onate MD, Pauley KM, Bhattacharyya I, Cha S. Presence of Porphyromonas gingivalis in gingival squamous cell carcinoma. Int J Oral Sci. 2011;3(4):209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor‐immune microenvironment. Cell Host Microbe. 2013;14(2):207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Gao Y, Huang E, Zhang H, Wang J, Wu N, Chen X, et al. Crosstalk between Wnt/beta‐catenin and estrogen receptor signaling synergistically promotes osteogenic differentiation of mesenchymal progenitor cells. PLoS One. 2013;8(12):e82436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Garrett WS. Cancer and the microbiota. Science. 2015;348(6230):80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Xu J, Prosperi JR, Choudhury N, Olopade OI, Goss KH. beta‐Catenin is required for the tumorigenic behavior of triple‐negative breast cancer cells. PLoS One. 2015;10(2):e0117097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Jha HC, Banerjee S, Robertson ES. The Role of Gammaherpesviruses in Cancer Pathogenesis. Pathogens. 2016;5(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Angelova M, Ferris M, Swan KF, McFerrin HE, Pridjian G, Morris CA, et al. Kaposi's sarcoma‐associated herpesvirus G‐protein coupled receptor activates the canonical Wnt/β‐catenin signaling pathway. Virol J. 2014;11:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Ma G, Yasunaga J, Fan J, Yanagawa S, Matsuoka M. HTLV‐1 bZIP factor dysregulates the Wnt pathways to support proliferation and migration of adult T‐cell leukemia cells. Oncogene. 2013;32(36):4222–4230. [DOI] [PubMed] [Google Scholar]
- 161. Samatar AA, Poulikakos PI. Targeting RAS‐ERK signalling in cancer: promises and challenges. Nat Rev Drug Discov. 2014;13(12):928–942. [DOI] [PubMed] [Google Scholar]
- 162. Kim HJ, Bar‐Sagi D. Modulation of signalling by Sprouty: a developing story. Nat Rev Mol Cell Biol. 2004;5(6):441–450. [DOI] [PubMed] [Google Scholar]
- 163. Lam KC, Araya RE, Huang A, Chen Q, Di Modica M, Rodrigues RR, et al. Microbiota triggers STING‐type I IFN‐dependent monocyte reprogramming of the tumor microenvironment. Cell. 2021;184(21):5338–5356.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Bobrovnikova‐Marjon E, Grigoriadou C, Pytel D, Zhang F, Ye J, Koumenis C, et al. PERK promotes cancer cell proliferation and tumor growth by limiting oxidative DNA damage. Oncogene. 2010;29(27):3881–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Wang Y, Alam GN, Ning Y, Visioli F, Dong Z, Nor JE, et al. The unfolded protein response induces the angiogenic switch in human tumor cells through the PERK/ATF4 pathway. Cancer Res. 2012;72(20):5396–5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Dang N, Meng X, Qin G, An Y, Zhang Q, Cheng X, et al. alpha5‐nAChR modulates melanoma growth through the Notch1 signaling pathway. J Cell Physiol. 2020;235(11):7816–7826. [DOI] [PubMed] [Google Scholar]
- 167. Wang D, DuBois RN. Immunosuppression associated with chronic inflammation in the tumor microenvironment. Carcinogenesis. 2015;36(10):1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Cutolo M, Paolino S, Pizzorni C. Possible contribution of chronic inflammation in the induction of cancer in rheumatic diseases. Clin Exp Rheumatol. 2014;32(6):839–847. [PubMed] [Google Scholar]
- 169. Bhatelia K, Singh K, Singh R. TLRs: linking inflammation and breast cancer. Cell Signal. 2014;26(11):2350–2357. [DOI] [PubMed] [Google Scholar]
- 170. Valguarnera E, Wardenburg JB. Good Gone Bad: One Toxin Away From Disease for Bacteroides fragilis. J Mol Biol. 2020;432(4):765–785. [DOI] [PubMed] [Google Scholar]
- 171. Shalapour S, Karin M. Cruel to Be Kind: Epithelial, Microbial, and Immune Cell Interactions in Gastrointestinal Cancers. Annu Rev Immunol. 2020;38:649–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Chen Y, Liu B, Wei Y, Kuang DM. Influence of gut and intratumoral microbiota on the immune microenvironment and anti‐cancer therapy. Pharmacol Res. 2021;174:105966. [DOI] [PubMed] [Google Scholar]
- 173. Choi Y, Lichterman JN, Coughlin LA, Poulides N, Li W, Del Valle P, et al. Immune checkpoint blockade induces gut microbiota translocation that augments extraintestinal antitumor immunity. Sci Immunol. 2023;8(81):eabo2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Shi Y, Zheng W, Yang K, Harris KG, Ni K, Xue L, et al. Intratumoral accumulation of gut microbiota facilitates CD47‐based immunotherapy via STING signaling. J Exp Med. 2020;217(5):e20192282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Le Noci V, Guglielmetti S, Arioli S, Camisaschi C, Bianchi F, Sommariva M, et al. Modulation of Pulmonary Microbiota by Antibiotic or Probiotic Aerosol Therapy: A Strategy to Promote Immunosurveillance against Lung Metastases. Cell Rep. 2018;24(13):3528–3538. [DOI] [PubMed] [Google Scholar]
- 176. Ohadian Moghadam S, Momeni SA. Human microbiome and prostate cancer development: current insights into the prevention and treatment. Front Med. 2021;15(1):11–32. [DOI] [PubMed] [Google Scholar]
- 177. Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Abed J, Emgard JE, Zamir G, Faroja M, Almogy G, Grenov A, et al. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor‐Expressed Gal‐GalNAc. Cell Host Microbe. 2016;20(2):215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Wen L, Mu W, Lu H, Wang X, Fang J, Jia Y, et al. Porphyromonas gingivalis Promotes Oral Squamous Cell Carcinoma Progression in an Immune Microenvironment. J Dent Res. 2020;99(6):666–675. [DOI] [PubMed] [Google Scholar]
- 180. Zhang B, Cheng P. Improving antitumor efficacy via combinatorial regimens of oncolytic virotherapy. Mol Cancer. 2020;19(1):158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Wang Y, Guo W, Wu X, Zhang Y, Mannion C, Brouchkov A, et al. Oncolytic Bacteria and their potential role in bacterium‐mediated tumour therapy: a conceptual analysis. J Cancer. 2019;10(19):4442–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Kalaora S, Nagler A, Nejman D, Alon M, Barbolin C, Barnea E, et al. Identification of bacteria‐derived HLA‐bound peptides in melanoma. Nature. 2021;592(7852):138–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Fluckiger A, Daillère R, Sassi M, Sixt BS, Liu P, Loos F, et al. Cross‐reactivity between tumor MHC class I‐restricted antigens and an enterococcal bacteriophage. Science. 2020;369(6506):936–942. [DOI] [PubMed] [Google Scholar]
- 184. Bessell CA, Isser A, Havel JJ, Lee S, Bell DR, Hickey JW, et al. Commensal bacteria stimulate antitumor responses via T cell cross‐reactivity. JCI Insight. 2020;5(8):e135597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, et al. Gut microbiome‐mediated bile acid metabolism regulates liver cancer via NKT cells. Science. 2018;360(6391):eaan5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Parida S, Siddharth S, Xia Y, Sharma D. Concomitant analyses of intratumoral microbiota and genomic features reveal distinct racial differences in breast cancer. NPJ Breast Cancer. 2023;9(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Choi JK, Naffouje SA, Goto M, Wang J, Christov K, Rademacher DJ, et al. Cross‐talk between cancer and Pseudomonas aeruginosa mediates tumor suppression. Commun Biol. 2023;6(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Lam KC, Araya RE, Huang A, Chen Q, Di Modica M, Rodrigues RR, et al. Microbiota triggers STING‐type I IFN‐dependent monocyte reprogramming of the tumor microenvironment. Cell. 2021;184(21):5338–5356 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Mager LF, Burkhard R, Pett N, Cooke NCA, Brown K, Ramay H, et al. Microbiome‐derived inosine modulates response to checkpoint inhibitor immunotherapy. Science. 2020;369(6510):1481–1489. [DOI] [PubMed] [Google Scholar]
- 190. Drobner JC, Lichtbroun BJ, Singer EA, Ghodoussipour S. Examining the Role of Microbiota‐Centered Interventions in Cancer Therapeutics: Applications for Urothelial Carcinoma. Technol Cancer Res Treat. 2023;22:15330338231164196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Kang X, Liu C, Ding Y, Ni Y, Ji F, Lau HCH, et al. Roseburia intestinalis generated butyrate boosts anti‐PD‐1 efficacy in colorectal cancer by activating cytotoxic CD8(+) T cells. Gut. 2023;72(11):2112–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Lu C, Liu Y, Ali NM, Zhang B, Cui X. The role of innate immune cells in the tumor microenvironment and research progress in anti‐tumor therapy. Front Immunol. 2022;13:1039260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Chmiela M, Walczak N, Rudnicka K. Helicobacter pylori outer membrane vesicles involvement in the infection development and Helicobacter pylori‐related diseases. J Biomed Sci. 2018;25(1):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Gonzalez MF, Diaz P, Sandoval‐Borquez A, Herrera D, Quest AFG. Helicobacter pylori Outer Membrane Vesicles and Extracellular Vesicles from Helicobacter pylori‐Infected Cells in Gastric Disease Development. Int J Mol Sci. 2021;22(9):4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Sivan A, Corrales L, Hubert N, Williams JB, Aquino‐Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti‐PD‐L1 efficacy. Science. 2015;350(6264):1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA‐4 blockade relies on the gut microbiota. Science. 2015;350(6264):1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti‐PD‐1 efficacy in metastatic melanoma patients. Science. 2018;359(6371):104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Derosa L, Hellmann MD, Spaziano M, Halpenny D, Fidelle M, Rizvi H, et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non‐small‐cell lung cancer. Ann Oncol. 2018;29(6):1437–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin JM, Morrison RM, et al. Fecal microbiota transplant overcomes resistance to anti‐PD‐1 therapy in melanoma patients. Science. 2021;371(6529):595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Derosa L, Routy B, Thomas AM, Iebba V, Zalcman G, Friard S, et al. Intestinal Akkermansia muciniphila predicts clinical response to PD‐1 blockade in patients with advanced non‐small‐cell lung cancer. Nat Med. 2022;28(2):315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Bae M, Cassilly CD, Liu X, Park SM, Tusi BK, Chen X, et al. Akkermansia muciniphila phospholipid induces homeostatic immune responses. Nature. 2022;608(7921):168–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Lu Y, Yuan X, Wang M, He Z, Li H, Wang J, et al. Gut microbiota influence immunotherapy responses: mechanisms and therapeutic strategies. J Hematol Oncol. 2022;15(1):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Wang T, Gnanaprakasam JNR, Chen X, Kang S, Xu X, Sun H, et al. Inosine is an alternative carbon source for CD8(+)‐T‐cell function under glucose restriction. Nat Metab. 2020;2(7):635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Oh M, Zhang L. DeepGeni: deep generalized interpretable autoencoder elucidates gut microbiota for better cancer immunotherapy. Sci Rep. 2023;13(1):4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Hezaveh K, Shinde RS, Klötgen A, Halaby MJ, Lamorte S, Ciudad MT, et al. Tryptophan‐derived microbial metabolites activate the aryl hydrocarbon receptor in tumor‐associated macrophages to suppress anti‐tumor immunity. Immunity. 2022;55(2):324–340.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Fong W, Li Q, Ji F, Liang W, Lau HCH, Kang X, et al. Lactobacillus gallinarum‐derived metabolites boost anti‐PD1 efficacy in colorectal cancer by inhibiting regulatory T cells through modulating IDO1/Kyn/AHR axis. Gut. 2023;72(12):2272–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Jiang SS, Xie YL, Xiao XY, Kang ZR, Lin XL, Zhang L, et al. Fusobacterium nucleatum‐derived succinic acid induces tumor resistance to immunotherapy in colorectal cancer. Cell Host Microbe. 2023;31(5):781–797 e9. [DOI] [PubMed] [Google Scholar]
- 208. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti‐PD‐1 immunotherapy in melanoma patients. Science. 2018;359(6371):97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Park JS, Gazzaniga FS, Wu M, Luthens AK, Gillis J, Zheng W, et al. Targeting PD‐L2‐RGMb overcomes microbiome‐related immunotherapy resistance. Nature. 2023;617(7960):377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Anand U, Dey A, Chandel AKS, Sanyal R, Mishra A, Pandey DK, et al. Cancer chemotherapy and beyond: Current status, drug candidates, associated risks and progress in targeted therapeutics. Genes Dis. 2023;10(4):1367–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Ramos A, Sadeghi S, Tabatabaeian H. Battling Chemoresistance in Cancer: Root Causes and Strategies to Uproot Them. Int J Mol Sci. 2021;22(17):9451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Guthrie L, Gupta S, Daily J, Kelly L. Human microbiome signatures of differential colorectal cancer drug metabolism. NPJ Biofilms Microbiomes. 2017;3:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Tintelnot J, Xu Y, Lesker TR, Schönlein M, Konczalla L, Giannou AD, et al. Microbiota‐derived 3‐IAA influences chemotherapy efficacy in pancreatic cancer. Nature. 2023;615(7950):168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillère R, Hannani D, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342(6161):971–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Daillère R, Vétizou M, Waldschmitt N, Yamazaki T, Isnard C, Poirier‐Colame V, et al. Enterococcus hirae and Barnesiella intestinihominis Facilitate Cyclophosphamide‐Induced Therapeutic Immunomodulatory Effects. Immunity. 2016;45(4):931–943. [DOI] [PubMed] [Google Scholar]
- 216. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342(6161):967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. 2019;574(7777):264–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Pomella S, Cassandri M, Melaiu O, Marampon F, Gargari M, Campanella V, et al. DNA Damage Response Gene Signature as Potential Treatment Markers for Oral Squamous Cell Carcinoma. Int J Mol Sci. 2023;24(3):2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Liu J, Liu C, Yue J. Radiotherapy and the gut microbiome: facts and fiction. Radiat Oncol. 2021;16(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Dong J, Li Y, Xiao H, Cui M, Fan S. Commensal microbiota in the digestive tract: a review of its roles in carcinogenesis and radiotherapy. Cancer Biol Med. 2021;19(1):43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Reis Ferreira M, Pasto A, Ng T, Patel V, Guerrero Urbano T, Sears C, et al. The microbiota and radiotherapy for head and neck cancer: What should clinical oncologists know? Cancer Treat Rev. 2022;109:102442. [DOI] [PubMed] [Google Scholar]
- 222. Uribe‐Herranz M, Rafail S, Beghi S, Gil‐de‐Gómez L, Verginadis I, Bittinger K, et al. Gut microbiota modulate dendritic cell antigen presentation and radiotherapy‐induced antitumor immune response. J Clin Invest. 2020;130(1):466–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Shiao SL, Kershaw KM, Limon JJ, You S, Yoon J, Ko EY, et al. Commensal bacteria and fungi differentially regulate tumor responses to radiation therapy. Cancer Cell. 2021;39(9):1202–1213.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Guo H, Chou WC, Lai Y, Liang K, Tam JW, Brickey WJ, et al. Multi‐omics analyses of radiation survivors identify radioprotective microbes and metabolites. Science. 2020;370(6516):eaay9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Teng H, Wang Y, Sui X, Fan J, Li S, Lei X, et al. Gut microbiota‐mediated nucleotide synthesis attenuates the response to neoadjuvant chemoradiotherapy in rectal cancer. Cancer Cell. 2023;41(1):124–138.e6. [DOI] [PubMed] [Google Scholar]
- 226. Colbert LE, El Alam MB, Wang R, Karpinets T, Lo D, Lynn EJ, et al. Tumor‐resident Lactobacillus iners confer chemoradiation resistance through lactate‐induced metabolic rewiring. Cancer Cell. 2023;41(11):1945–1962 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Eaton SE, Kaczmarek J, Mahmood D, McDiarmid AM, Norarfan AN, Scott EG, et al. Exploiting dietary fibre and the gut microbiota in pelvic radiotherapy patients. Br J Cancer. 2022;127(12):2087–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Poonacha KNT, Villa TG, Notario V. The Interplay among Radiation Therapy, Antibiotics and the Microbiota: Impact on Cancer Treatment Outcomes. Antibiotics (Basel). 2022;11(3):331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Cui M, Xiao H, Li Y, Zhou L, Zhao S, Luo D, et al. Faecal microbiota transplantation protects against radiation‐induced toxicity. EMBO Mol Med. 2017;9(4):448–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Xiao H, Fan Y, Li Y, Dong J, Zhang S, Wang B, et al. Oral microbiota transplantation fights against head and neck radiotherapy‐induced oral mucositis in mice. Comput Struct Biotechnol J. 2021;19:5898–5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Mackowiak PA. Recycling metchnikoff: probiotics, the intestinal microbiome and the quest for long life. Front Public Health. 2013;1:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Cavaillon JM, Legout S. Centenary of the death of Elie Metchnikoff: a visionary and an outstanding team leader. Microbes Infect. 2016;18(10):577–594. [DOI] [PubMed] [Google Scholar]
- 233. Suez J, Zmora N, Segal E, Elinav E. The pros, cons, and many unknowns of probiotics. Nat Med. 2019;25(5):716–729. [DOI] [PubMed] [Google Scholar]
- 234. Fuller R. History and development of probiotics. In: Fuller R, editor. Probiotics: The scientific basis. Dordrecht: Springer Netherlands; 1992. p. 1–8. [Google Scholar]
- 235. Lim CC, Ferguson LR, Tannock GW. Dietary fibres as “prebiotics”: implications for colorectal cancer. Mol Nutr Food Res. 2005;49(6):609–619. [DOI] [PubMed] [Google Scholar]
- 236. Geier MS, Butler RN, Howarth GS. Probiotics, prebiotics and synbiotics: a role in chemoprevention for colorectal cancer? Cancer Biol Ther. 2006;5(10):1265–1269. [DOI] [PubMed] [Google Scholar]
- 237. Chong ES. A potential role of probiotics in colorectal cancer prevention: review of possible mechanisms of action. World J Microbiol Biotechnol. 2014;30(2):351–374. [DOI] [PubMed] [Google Scholar]
- 238. Gianotti L, Morelli L, Galbiati F, Rocchetti S, Coppola S, Beneduce A, et al. A randomized double‐blind trial on perioperative administration of probiotics in colorectal cancer patients. World J Gastroenterol. 2010;16(2):167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. de Moreno de LeBlanc A, Matar C, Perdigon G. The application of probiotics in cancer. Br J Nutr. 2007;98(Suppl 1):S105–S110. [DOI] [PubMed] [Google Scholar]
- 240. Commane DM, Shortt CT, Silvi S, Cresci A, Hughes RM, Rowland IR. Effects of fermentation products of pro‐ and prebiotics on trans‐epithelial electrical resistance in an in vitro model of the colon. Nutr Cancer. 2005;51(1):102–109. [DOI] [PubMed] [Google Scholar]
- 241. Kumar M, Kumar A, Nagpal R, Mohania D, Behare P, Verma V, et al. Cancer‐preventing attributes of probiotics: an update. Int J Food Sci Nutr. 2010;61(5):473–496. [DOI] [PubMed] [Google Scholar]
- 242. Chen CC, Lin WC, Kong MS, Shi HN, Walker WA, Lin CY, et al. Oral inoculation of probiotics Lactobacillus acidophilus NCFM suppresses tumour growth both in segmental orthotopic colon cancer and extra‐intestinal tissue. Br J Nutr. 2012;107(11):1623–1634. [DOI] [PubMed] [Google Scholar]
- 243. Uccello M, Malaguarnera G, Basile F, D'Agata V, Malaguarnera M, Bertino G, et al. Potential role of probiotics on colorectal cancer prevention. BMC Surg. 2012;12(Suppl 1):S35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Noor S, Ali S, Riaz S, Sardar I, Farooq MA, Sajjad A. Chemopreventive role of probiotics against cancer: a comprehensive mechanistic review. Mol Biol Rep. 2023;50(1):799–814. [DOI] [PubMed] [Google Scholar]
- 245. Rafter J. Probiotics and colon cancer. Best Pract Res Clin Gastroenterol. 2003;17(5):849–859. [DOI] [PubMed] [Google Scholar]
- 246. Shi L, Sheng J, Chen G, Zhu P, Shi C, Li B, et al. Combining IL‐2‐based immunotherapy with commensal probiotics produces enhanced antitumor immune response and tumor clearance. J Immunother Cancer. 2020;8(2):e000973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Zhang Q, Zhao Q, Li T, Lu L, Wang F, Zhang H, et al. Lactobacillus plantarum‐derived indole‐3‐lactic acid ameliorates colorectal tumorigenesis via epigenetic regulation of CD8(+) T cell immunity. Cell Metab. 2023;35(6):943–960 e9. [DOI] [PubMed] [Google Scholar]
- 248. Raman M, Ambalam P, Kondepudi KK, Pithva S, Kothari C, Patel AT, et al. Potential of probiotics, prebiotics and synbiotics for management of colorectal cancer. Gut Microbes. 2013;4(3):181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Liong MT. Roles of probiotics and prebiotics in colon cancer prevention: Postulated mechanisms and in‐vivo evidence. Int J Mol Sci. 2008;9(5):854–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Redman MG, Ward EJ, Phillips RS. The efficacy and safety of probiotics in people with cancer: a systematic review. Ann Oncol. 2014;25(10):1919–1929. [DOI] [PubMed] [Google Scholar]
- 251. Tian Y, Li M, Song W, Jiang R, Li YQ. Effects of probiotics on chemotherapy in patients with lung cancer. Oncol Lett. 2019;17(3):2836–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368(5):407–415. [DOI] [PubMed] [Google Scholar]
- 253. Xu MQ, Cao HL, Wang WQ, Wang S, Cao XC, Yan F, et al. Fecal microbiota transplantation broadening its application beyond intestinal disorders. World J Gastroenterol. 2015;21(1):102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, et al. Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell. 2017;171(5):1015–1028 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Yao B, Cai Y, Wang W, Deng J, Zhao L, Han Z, et al. The Effect of Gut Microbiota on the Progression of Intervertebral Disc Degeneration. Orthop Surg. 2023;15(3):858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Wang L, Wei Z, Pan F, Song C, Peng L, Yang Y, et al. Case report: Fecal microbiota transplantation in refractory ankylosing spondylitis. Front Immunol. 2023;14:1093233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell. 2019;178(4):795–806 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Baruch EN, Youngster I, Ben‐Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal microbiota transplant promotes response in immunotherapy‐refractory melanoma patients. Science. 2021;371(6529):602–609. [DOI] [PubMed] [Google Scholar]
- 259. Wang Z, Qin X, Hu D, Huang J, Guo E, Xiao R, et al. Akkermansia supplementation reverses the tumor‐promoting effect of the fecal microbiota transplantation in ovarian cancer. Cell Rep. 2022;41(13):111890. [DOI] [PubMed] [Google Scholar]
- 260. Quesada‐Vázquez S, Castells‐Nobau A, Latorre J, Oliveras‐Cañellas N, Puig‐Parnau I, Tejera N, et al. Potential therapeutic implications of histidine catabolism by the gut microbiota in NAFLD patients with morbid obesity. Cell Rep Med. 2023;4(12):101341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Bustamante JM, Dawson T, Loeffler C, Marfori Z, Marchesi JR, Mullish BH, et al. Impact of Fecal Microbiota Transplantation on Gut Bacterial Bile Acid Metabolism in Humans. Nutrients. 2022;14(24):5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Zheng L, Ji YY, Wen XL, Duan SL. Fecal microbiota transplantation in the metabolic diseases: Current status and perspectives. World J Gastroenterol. 2022;28(23):2546–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Pawelek JM, Low KB, Bermudes D. Tumor‐targeted Salmonella as a novel anticancer vector. Cancer Res. 1997;57(20):4537–4544. [PubMed] [Google Scholar]
- 264. Fan JX, Peng MY, Wang H, Zheng HR, Liu ZL, Li CX, et al. Engineered Bacterial Bioreactor for Tumor Therapy via Fenton‐Like Reaction with Localized H(2) O(2) Generation. Adv Mater. 2019;31(16):e1808278. [DOI] [PubMed] [Google Scholar]
- 265. Huang C, Wang FB, Liu L, Jiang W, Liu W, Ma W, et al. Hypoxic Tumor Radiosensitization Using Engineered Probiotics. Adv Healthc Mater. 2021;10(10):e2002207. [DOI] [PubMed] [Google Scholar]
- 266. Liu SC, Minton NP, Giaccia AJ, Brown JM. Anticancer efficacy of systemically delivered anaerobic bacteria as gene therapy vectors targeting tumor hypoxia/necrosis. Gene Therapy. 2002;9(4):291–296. [DOI] [PubMed] [Google Scholar]
- 267. Bai RL, Chen NF, Li LY, Cui JW. A brand new era of cancer immunotherapy: breakthroughs and challenges. Chin Med J (Engl). 2021;134(11):1267–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Zhou S, Gravekamp C, Bermudes D, Liu K. Tumour‐targeting bacteria engineered to fight cancer. Nat Rev Cancer. 2018;18(12):727–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. He L, Yang H, Tang J, Liu Z, Chen Y, Lu B, et al. Intestinal probiotics E. coli Nissle 1917 as a targeted vehicle for delivery of p53 and Tum‐5 to solid tumors for cancer therapy. J Biol Eng. 2019;13:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Shi LL, Sheng JY, Wang ML, Luo H, Zhu J, Zhang BX, et al. Combination Therapy of TGF‐beta Blockade and Commensal‐derived Probiotics Provides Enhanced Antitumor Immune Response and Tumor Suppression. Theranostics. 2019;9(14):4115–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Tang Q, Sun S, Wang P, Sun L, Wang Y, Zhang L, et al. Genetically Engineering Cell Membrane‐Coated BTO Nanoparticles for MMP2‐Activated Piezocatalysis‐Immunotherapy. Adv Mater. 2023:e2300964. [DOI] [PubMed] [Google Scholar]
- 272. Nguyen VH, Kim HS, Ha JM, Hong Y, Choy HE, Min JJ. Genetically engineered Salmonella typhimurium as an imageable therapeutic probe for cancer. Cancer Res. 2010;70(1):18–23. [DOI] [PubMed] [Google Scholar]
- 273. Danino T, Prindle A, Kwong GA, Skalak M, Li H, Allen K, et al. Programmable probiotics for detection of cancer in urine. Sci Transl Med. 2015;7(289):289ra84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Chowdhury S, Castro S, Coker C, Hinchliffe TE, Arpaia N, Danino T. Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat Med. 2019;25(7):1057–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Hu Q, Wu M, Fang C, Cheng C, Zhao M, Fang W, et al. Engineering nanoparticle‐coated bacteria as oral DNA vaccines for cancer immunotherapy. Nano Lett. 2015;15(4):2732–2739. [DOI] [PubMed] [Google Scholar]
- 276. Yi W, Xiao P, Liu X, Zhao Z, Sun X, Wang J, et al. Recent advances in developing active targeting and multi‐functional drug delivery systems via bioorthogonal chemistry. Signal Transduct Target Ther. 2022;7(1):386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Xiao Y, Wang D, Luo B, Chen X, Yao Y, Song C, et al. In‐situ synthesis of melanin in tumor with engineered probiotics for hyperbaric oxygen‐synergized photothermal immunotherapy. Nano Today. 2022;47:101632. [Google Scholar]
- 278. Chen J, Li T, Liang J, Huang Q, Huang JD, Ke Y, et al. Current status of intratumour microbiome in cancer and engineered exogenous microbiota as a promising therapeutic strategy. Biomed Pharmacother. 2022;145:112443. [DOI] [PubMed] [Google Scholar]
- 279. Roberts NJ, Zhang L, Janku F, Collins A, Bai RY, Staedtke V, et al. Intratumoral injection of Clostridium novyi‐NT spores induces antitumor responses. Sci Transl Med. 2014;6(249):249ra111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Camarillo‐Guerrero LF, Almeida A, Rangel‐Pineros G, Finn RD, Lawley TD. Massive expansion of human gut bacteriophage diversity. Cell. 2021;184(4):1098–1109.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Cao B, Yang M, Mao C. Phage as a Genetically Modifiable Supramacromolecule in Chemistry, Materials and Medicine. Acc Chem Res. 2016;49(6):1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Petrov G, Dymova M, Richter V. Bacteriophage‐Mediated Cancer Gene Therapy. Int J Mol Sci. 2022;23(22):14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Jones KM, Karanam B, Jones‐Triche J, Sandey M, Henderson HJ, Samant RS, et al. Phage Ligands for Identification of Mesenchymal‐Like Breast Cancer Cells and Cancer‐Associated Fibroblasts. Front Oncol. 2018;8:625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Pande J, Szewczyk MM, Grover AK. Phage display: concept, innovations, applications and future. Biotechnol Adv. 2010;28(6):849–858. [DOI] [PubMed] [Google Scholar]
- 285. Saw PE, Song EW. Phage display screening of therapeutic peptide for cancer targeting and therapy. Protein Cell. 2019;10(11):787–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Bhasin A, Drago NP, Majumdar S, Sanders EC, Weiss GA, Penner RM. Viruses Masquerading as Antibodies in Biosensors: The Development of the Virus BioResistor. Acc Chem Res. 2020;53(10):2384–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Li C, Li J, Xu Y, Zhan Y, Li Y, Song T, et al. Application of Phage‐Displayed Peptides in Tumor Imaging Diagnosis and Targeting Therapy. Int J Pept Res Ther. 2021;27(1):587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Li Y, Qu X, Cao B, Yang T, Bao Q, Yue H, et al. Selectively Suppressing Tumor Angiogenesis for Targeted Breast Cancer Therapy by Genetically Engineered Phage. Adv Mater. 2020;32(29):e2001260. [DOI] [PubMed] [Google Scholar]
- 289. Gurung S, Khan F, Gunassekaran GR, Yoo JD, Poongkavithai Vadevoo SM, Permpoon U, et al. Phage display‐identified PD‐L1‐binding peptides reinvigorate T‐cell activity and inhibit tumor progression. Biomaterials. 2020;247:119984. [DOI] [PubMed] [Google Scholar]
- 290. Wang J, Lamolinara A, Conti L, Giangrossi M, Cui L, Morelli MB, et al. HER2‐Displaying M13 Bacteriophages induce Therapeutic Immunity against Breast Cancer. Cancers (Basel). 2022;14(16):4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Sindhwani S, Syed AM, Ngai J, Kingston BR, Maiorino L, Rothschild J, et al. The entry of nanoparticles into solid tumours. Nat Mater. 2020;19(5):566–575. [DOI] [PubMed] [Google Scholar]
- 292. de la Zerda A, Bodapati S, Teed R, May SY, Tabakman SM, Liu Z, et al. Family of enhanced photoacoustic imaging agents for high‐sensitivity and multiplexing studies in living mice. ACS Nano. 2012;6(6):4694–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Li J, Rao J, Pu K. Recent progress on semiconducting polymer nanoparticles for molecular imaging and cancer phototherapy. Biomaterials. 2018;155:217–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Wu X, Ye J, DeLaitsch AT, Rashidijahanabad Z, Lang S, Kakeshpour T, et al. Chemoenzymatic Synthesis of 9NHAc‐GD2 Antigen to Overcome the Hydrolytic Instability of O‐Acetylated‐GD2 for Anticancer Conjugate Vaccine Development. Angew Chem Int Ed Engl. 2021;60(45):24179–24188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Yata T, Lee EL, Suwan K, Syed N, Asavarut P, Hajitou A. Modulation of extracellular matrix in cancer is associated with enhanced tumor cell targeting by bacteriophage vectors. Mol Cancer. 2015;14:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Xiao L, Ma N, He H, Li J, Cheng S, Yang Q, et al. Development of a novel drug targeting delivery system for cervical cancer therapy. Nanotechnology. 2019;30(7):075604. [DOI] [PubMed] [Google Scholar]
- 297. Fukuhara H, Ino Y, Todo T. Oncolytic virus therapy: A new era of cancer treatment at dawn. Cancer Science. 2016;107(10):1373–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Shalhout SZ, Miller DM, Emerick KS, Kaufman HL. Therapy with oncolytic viruses: progress and challenges. Nat Rev Clin Oncol. 2023;20(3):160–177. [DOI] [PubMed] [Google Scholar]
- 299. Chiocca EA, Rabkin SD. Oncolytic viruses and their application to cancer immunotherapy. Cancer Immunol Res. 2014;2(4):295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Hennessy ML, Bommareddy PK, Boland G, Kaufman HL. Oncolytic Immunotherapy. Surg Oncol Clin N Am. 2019;28(3):419–430. [DOI] [PubMed] [Google Scholar]
- 301. Conry RM, Westbrook B, McKee S, Norwood TG. Talimogene laherparepvec: First in class oncolytic virotherapy. Hum Vaccin Immunother. 2018;14(4):839–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Soliman H, Hogue D, Han H, Mooney B, Costa R, Lee MC, et al. Oncolytic T‐VEC virotherapy plus neoadjuvant chemotherapy in nonmetastatic triple‐negative breast cancer: a phase 2 trial. Nat Med. 2023;29(2):450–457. [DOI] [PubMed] [Google Scholar]
- 303. Garber K. China approves world's first oncolytic virus therapy for cancer treatment. J Natl Cancer Inst. 2006;98(5):298–300. [DOI] [PubMed] [Google Scholar]
- 304. Doniņa S, Strēle I, Proboka G, Auziņš J, Alberts P, Jonsson B, et al. Adapted ECHO‐7 virus Rigvir immunotherapy (oncolytic virotherapy) prolongs survival in melanoma patients after surgical excision of the tumour in a retrospective study. Melanoma Res. 2015;25(5):421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Todo T, Ito H, Ino Y, Ohtsu H, Ota Y, Shibahara J, et al. Intratumoral oncolytic herpes virus G47∆ for residual or recurrent glioblastoma: a phase 2 trial. Nat Med. 2022;28(8):1630–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Bommareddy PK, Patel A, Hossain S, Kaufman HL. Talimogene Laherparepvec (T‐VEC) and Other Oncolytic Viruses for the Treatment of Melanoma. Am J Clin Dermatol. 2017;18(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Ylosmaki E, Cerullo V. Design and application of oncolytic viruses for cancer immunotherapy. Curr Opin Biotechnol. 2020;65:25–36. [DOI] [PubMed] [Google Scholar]
- 308. Russell L, Swanner J, Jaime‐Ramirez AC, Wang Y, Sprague A, Banasavadi‐Siddegowda Y, et al. PTEN expression by an oncolytic herpesvirus directs T‐cell mediated tumor clearance. Nat Commun. 2018;9(1):5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Watanabe K, Luo Y, Da T, Guedan S, Ruella M, Scholler J, et al. Pancreatic cancer therapy with combined mesothelin‐redirected chimeric antigen receptor T cells and cytokine‐armed oncolytic adenoviruses. JCI Insight. 2018;3(7):e99573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Fares J, Ahmed AU, Ulasov IV, Sonabend AM, Miska J, Lee‐Chang C, et al. Neural stem cell delivery of an oncolytic adenovirus in newly diagnosed malignant glioma: a first‐in‐human, phase 1, dose‐escalation trial. Lancet Oncol. 2021;22(8):1103–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Saari H, Turunen T, Lõhmus A, Turunen M, Jalasvuori M, Butcher SJ, et al. Extracellular vesicles provide a capsid‐free vector for oncolytic adenoviral DNA delivery. J Extracell Vesicles. 2020;9(1):1747206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Sun M, Yang S, Huang H, Gao P, Pan S, Cheng Z, et al. Boarding Oncolytic Viruses onto Tumor‐Homing Bacterium‐Vessels for Augmented Cancer Immunotherapy. Nano Lett. 2022;22(12):5055–5064. [DOI] [PubMed] [Google Scholar]
- 313. Howard FHN, Al‐Janabi H, Patel P, Cox K, Smith E, Vadakekolathu J, et al. Nanobugs as Drugs: Bacterial Derived Nanomagnets Enhance Tumor Targeting and Oncolytic Activity of HSV‐1 Virus. Small. 2022;18(13):e2104763. [DOI] [PubMed] [Google Scholar]
- 314. Bommareddy PK, Aspromonte S, Zloza A, Rabkin SD, Kaufman HL. MEK inhibition enhances oncolytic virus immunotherapy through increased tumor cell killing and T cell activation. Sci Transl Med. 2018;10(471):eaau0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Saha D, Martuza RL, Rabkin SD. Macrophage Polarization Contributes to Glioblastoma Eradication by Combination Immunovirotherapy and Immune Checkpoint Blockade. Cancer Cell. 2017;32(2):253–267 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316. Bourgeois‐Daigneault MC, Roy DG, Aitken AS, El Sayes N, Martin NT, Varette O, et al. Neoadjuvant oncolytic virotherapy before surgery sensitizes triple‐negative breast cancer to immune checkpoint therapy. Sci Transl Med. 2018;10(422):eaao1641. [DOI] [PubMed] [Google Scholar]
- 317. Chen Z, Xie H, Hu M, Huang T, Hu Y, Sang N, et al. Recent progress in treatment of hepatocellular carcinoma. Am J Cancer Res. 2020;10(9):2993–3036. [PMC free article] [PubMed] [Google Scholar]
- 318. Muthana M, Rodrigues S, Chen YY, Welford A, Hughes R, Tazzyman S, et al. Macrophage delivery of an oncolytic virus abolishes tumor regrowth and metastasis after chemotherapy or irradiation. Cancer Res. 2013;73(2):490–495. [DOI] [PubMed] [Google Scholar]
- 319. Villalona‐Calero MA, Lam E, Otterson GA, Zhao W, Timmons M, Subramaniam D, et al. Oncolytic reovirus in combination with chemotherapy in metastatic or recurrent non‐small cell lung cancer patients with KRAS‐activated tumors. Cancer. 2016;122(6):875–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Liu Z, Ravindranathan R, Kalinski P, Guo ZS, Bartlett DL. Rational combination of oncolytic vaccinia virus and PD‐L1 blockade works synergistically to enhance therapeutic efficacy. Nat Commun. 2017;8:14754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321. Ribas A, Dummer R, Puzanov I, VanderWalde A, Andtbacka RHI, Michielin O, et al. Oncolytic Virotherapy Promotes Intratumoral T Cell Infiltration and Improves Anti‐PD‐1 Immunotherapy. Cell. 2017;170(6):1109–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Puzanov I, Milhem MM, Minor D, Hamid O, Li A, Chen L, et al. Talimogene Laherparepvec in Combination With Ipilimumab in Previously Untreated, Unresectable Stage IIIB‐IV Melanoma. J Clin Oncol. 2016;34(22):2619–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Franklin C, Livingstone E, Roesch A, Schilling B, Schadendorf D. Immunotherapy in melanoma: Recent advances and future directions. Ejso. 2017;43(3):604–611. [DOI] [PubMed] [Google Scholar]
- 324. Harrington K, Freeman DJ, Kelly B, Harper J, Soria JC. Optimizing oncolytic virotherapy in cancer treatment. Nat Rev Drug Discov. 2019;18(9):689–706. [DOI] [PubMed] [Google Scholar]
- 325. Wing A, Fajardo CA, Posey AD, Jr. , Shaw C, Da T, Young RM, et al. Improving CART‐Cell Therapy of Solid Tumors with Oncolytic Virus‐Driven Production of a Bispecific T‐cell Engager. Cancer Immunol Res. 2018;6(5):605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326. Shi W, Wang Y, Xu C, Li Y, Ge S, Bai B, et al. Multilevel proteomic analyses reveal molecular diversity between diffuse‐type and intestinal‐type gastric cancer. Nat Commun. 2023;14(1):835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327. Narunsky‐Haziza L, Sepich‐Poore GD, Livyatan I, Asraf O, Martino C, Nejman D, et al. Pan‐cancer analyses reveal cancer‐type‐specific fungal ecologies and bacteriome interactions. Cell. 2022;185(20):3789–3806 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328. Nougayrede JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, et al. Escherichia coli induces DNA double‐strand breaks in eukaryotic cells. Science. 2006;313(5788):848–851. [DOI] [PubMed] [Google Scholar]
- 329. Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 Protein of Fusobacterium nucleatum to Human Inhibitory Receptor TIGIT Protects Tumors from Immune Cell Attack. Immunity. 2015;42(2):344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


