Abstract
Retrovirus intasomes purified from virus-infected cells contain the linear viral DNA genome and integrase (IN). Intasomes are capable of integrating the DNA termini in a concerted fashion into exogenous target DNA (full site), mimicking integration in vivo. Molecular insights into the organization of avian myeloblastosis virus IN at the viral DNA ends were gained by reconstituting nucleoprotein complexes possessing intasome characteristics. Assembly of IN–4.5-kbp donor complexes capable of efficient full-site integration appears cooperative and is dependent on time, temperature, and protein concentration. DNase I footprint analysis of assembled IN-donor complexes capable of full-site integration shows that wild-type U3 and other donors containing gain-of-function attachment site sequences are specifically protected by IN at low concentrations (<20 nM) with a defined outer boundary mapping ∼20 nucleotides from the ends. A donor containing mutations in the attachment site simultaneously eliminated full-site integration and DNase I protection by IN. Coupling of wild-type U5 ends with wild-type U3 ends for full-site integration shows binding by IN at low concentrations probably occurs only at the very terminal nucleotides (<10 bp) on U5. The results suggest that assembly requires a defined number of avian IN subunits at each viral DNA end. Among several possibilities, IN may bind asymmetrically to the U3 and U5 ends for full-site integration in vitro.
Integration of the linear retrovirus DNA genome (∼10 kbp) into the host chromosome requires the viral integrase (IN) (for a review, see reference 4). Viral DNA and IN are found within cytoplasmic preintegration complexes (PIC) in newly virus-infected cells. IN removes a dinucleotide from the blunt-ended viral ends, producing 3′-OH recessed long terminal repeat (LTR) termini (15). Purified PIC from cells infected with murine leukemia virus (MLV) (3), Rous sarcoma virus (25), and human immunodeficiency virus type 1 (HIV-1) (13) can mediate the concerted insertion of viral DNA termini into exogenous target DNA in vitro.
Concerted insertion of the viral DNA termini by IN into the host chromosome (full-site integration) necessitates a close molecular interaction between the two termini. Molecular probing studies of the HIV-1 PIC suggest the two termini are held together with a protein bridge (30). The 3′-OH recessed LTR termini interact to form a functional complex for integration in vitro, termed an intasome (41, 42). The role cellular proteins have in PIC or intasomes is undefined (12, 24, 26, 41). When characterized by bacteriophage Mu-mediated PCR (MM-PCR) footprints, intasomes exhibit protection and enhancements near the termini (∼20 bp from the end) and an extended region of protection spanning several hundred base pairs from the ∼20th nucleotide, both requiring a functional IN (2, 7, 42). The need of the relatively large MM-PCR footprints in relationship to only the ∼15-bp terminal attachment (att) site sequence requirement for efficient integration in vivo is unknown (7).
Early attempts to reproduce the full-site integration reaction in vitro with linear DNA donors and IN required genetic selection or amplification of the full-site integration products for measurement (10, 14, 22). Progress in producing nucleoprotein complexes mediating more efficient full-site integration in vitro with purified virion and recombinant IN is evident (1, 6, 16, 17, 21, 37, 38). The donor-target products produced by the full-site integration reactions are defined by restriction enzyme analysis and agarose gel electrophoresis. Genetic isolation and DNA sequencing of individual recombinants have verified the host duplications observed upon concerted insertion of the viral DNA termini in vivo.
Assembly of nucleoprotein complexes generally requires a series of discrete steps (31, 33, 35, 36). For full-site integration, the assembly time is rapid (∼1 min) at 0°C using linear 480-bp LTR donors with avian myeloblastosis virus (AMV) IN (50 nM) (37). It was necessary to identify factors slowing the assembly process to investigate events at the molecular level. The 480-bp donors (15 ng) were replaced with 4.5-kbp donors (10 ng) containing the avian 330-bp LTRs at their termini (Fig. 1). With this modification, the number of LTR termini in the assembly mixture was decreased by a factor of ∼12, and subsequently, the concentration of IN was lowered proportionally. Different 4.5-kbp donors containing wild-type (wt) and att site mutations were used to investigate the molecular processes necessary for efficient and high-fidelity full-site integration. The assembly of IN-LTR donor complexes appears cooperative and is dependent on time, temperature, and protein concentration. DNase I footprint analysis of assembled nucleoprotein complexes revealed that wt U3 or gain-of-function (“G”) LTR ends are specifically protected by IN with the outer boundary mapping ∼20 nucleotides on each terminus. IN appears to bind only to the very terminal nucleotides (<10 bp) on wt U5. The results suggest that assembly of nucleoprotein complexes requires a defined number of IN subunits binding asymmetrically to the U3 and U5 ends for full-site integration in vitro.
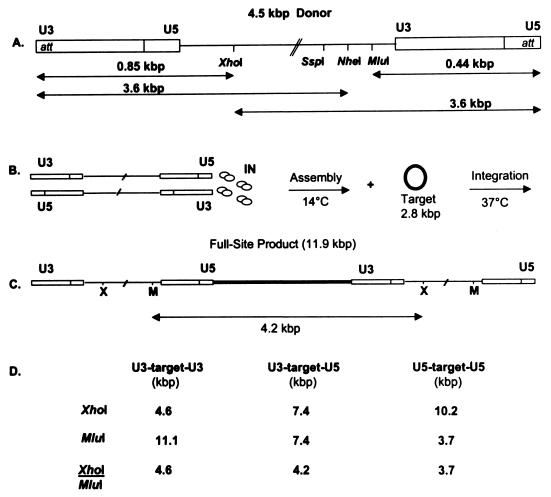
FIG. 1.
Schematic of the wt 4.5-kbp donor, the assembly and integration assay protocols, the full-site integration product, and the restriction enzyme fragments of full-site integration products. (A) The linear 4.5-kbp donor containing the wt U3 and wt U5 LTR termini is displayed. The att sites are identified at each end. XhoI, SspI, NheI, and MluI sites and the length of several fragments resulting from enzyme digestions are shown. (B) Two donors are assembled by IN displayed as four dimers. After assembly at 14°C, target (boldface type is added), followed by integration. (C) The linear full-site product (11.9 kbp) is the result of the concerted integration of two separate donors (bimolecular reaction) into a 2.8-kbp circular target DNA (bold line). MluI/XhoI-digestion produces a 4.2-kbp donor-target fragment containing U3 and U5 ends. (D) Only the labeled donor-target restriction fragments resulting from full-site integration are shown. The frequency of assembling two paired ends (U3-U3, U3-U5, U5-U5) to mediate bimolecular full-site integration is defined by Hardy-Weinberg distribution frequency. For example, when both LTR ends have equivalent integration activity, the distribution of the XhoI/MluI-digested full-site fragments will be 1:2:1 (Fig. 2, lane 4).
MATERIALS AND METHODS
Linear DNA donors.
A linear 4.5-kbp donor was constructed containing both the avian retrovirus wt U3 and wt U5 LTRs (each 330 bp long) (Fig. 1) (14). An NdeI site was produced at the circle junction of blunt-ended U3 and U5 ends by site-directed mutagenesis (Stratagene's QuikChange). NdeI digestion produces a linear 4.5-kbp donor containing 3′-OH recessed ends. Numbering of nucleotides was from the blunt end. The 5th and 6th nucleotides were modified at each terminus in several clones as indicated in the text. Each donor sequence was verified by DNA sequencing. The linear 3.4-kbp donor containing ∼30 bp of wt U5 and wt U3 sequences at its ends was described (14).
Labeling of donors.
The linear 4.5-kbp donors were 5′ end labeled using [γ-32P]ATP and polynucleotide kinase at the NdeI sites. The specific activities were ∼2,500 cpm (Cerenkov) per ng. For DNase I footprint analysis, the 5′-end-labeled 4.5-kbp donors were digested at either the SspI, NheI, XhoI, or MluI sites to produce different-size single-end-labeled LTR donors (Fig. 1). The fragments were isolated by agarose gel electrophoresis, electroeluted, and concentrated by a Centricon YM-30 filtering device. For nonspecific DNA, pGEM-3 (2.8 kbp) was digested with EcoRI and labeled. The labeled DNA was digested with NheI, and the single-end-labeled 2.5-kbp DNA was isolated for DNase I footprint analysis.
Full-site integration assay.
Conditions for assembling nucleoprotein complexes capable of producing full-site integration products significantly better than half-site integration products at low IN concentrations (<20 nM) were established. Assembly of IN with the 4.5-kbp donors (10 ng) occurred in the presence of 330 mM NaCl, 10 mM MgCl2, 3 mM dithiothreitol, 8% polyethylene glycol 6000, and 20 mM HEPES (pH 7.5) at 14°C. Organic solvents used previously in the integration assays were found to be inhibitory at low concentrations of AMV IN (37). The standard volume was 20 μl or multiples thereof. The concentration of IN, the time, and the temperature for assembly with donor were varied as indicated. The integration reactions were initiated by the addition of 50 ng of supercoiled DNA as target and immediately incubated at 37°C for only 5 or 10 min as indicated. Either pGEM-3 (2.8 kbp) or pUC19 (2.6 kbp) was used as a target. The target-to-donor molar ratio was three. The ratio of target (2.8 kbp) to donor (4.5 kbp) molecules was 8. The integration reactions were stopped by addition of sodium dodecyl sulfate and proteinase K, and the products were subjected to agarose gel electrophoresis. The amount of donor incorporated into the target was determined with a Molecular Dynamics PhosphorImager.
DNase I footprinting.
Double-end-labeled 4.5-kbp donors were digested with either XhoI, NheI, or SspI (Fig. 1A) to isolate single-end-labeled donors on agarose gels for DNase I footprinting. Titration experiments using DNase I with single-5′-end-labeled DNA were performed to determine the concentration of DNase I necessary to produce approximately one nick per DNA strand in assembly buffer. DNase I at 750 ng per ml was optimal. For assembly and for the DNase I footprint analyses, IN and the single-end-labeled 3.6-kbp donors were assembled at 14°C. An aliquot was removed from the mixture for measuring integration activity just prior to the addition of DNase I. The nucleoprotein complexes in the assembly mixture were further incubated with DNase I for 90 s at 14°C. The DNase I reactions were stopped by the addition of phenol. The DNase I-treated samples were subjected to denaturing 8, 10, 13, or 20% polyacrylamide gel electrophoresis depending on the required analysis. The dried gels (8 or 10%) were analyzed by a PhosphorImager and exposure to X-ray film.
Restriction analysis and genetic selection of donor-target recombinants.
The XhoI and MluI sites in the 4.5-kbp donors were used to characterize the donor-target products. pGEM-3 (2.8 kbp) or pUC19 (2.6 kbp) served as target DNA. The EcoRI site was destroyed in pUC19. The donor-target products produced with pUC19 were digested with EcoRI to isolate a unique 2.9-kbp fragment containing pUC19 with two flanking LTR termini on agarose gels. The purified fragment was ligated and transformed into Escherichia coli, and the plasmids from individual colonies were isolated and sequenced. The viral DNA-host junctions were sequenced as described (37).
Purification of AMV IN.
RESULTS
Full-site integration using linear 4.5 kbp donors with AMV IN.
Linear 4.5-kbp donors containing the U3 and U5 LTR termini (Fig. 1A) were used to investigate assembly conditions for full-site integration. Three different linear 4.5-kbp donors were used. The wt donor contained wt U3 (5′-ACTACAOH) and wt U5 (5′-GCTTCAOH) att site sequences at its 3′-OH recessed termini. Another donor was modified at both the U3 and U5 ends by changing the underlined 5th and 6th nucleotide positions to 5′-ACAACAOH and 5′-GCAACAOH, respectively. These att site sequences are defined as having a (“G”) mutation for integration (28, 39, 44). Lastly, a donor with att site mutations lacking significant integration activity contained 5′-ACCCCAOH and 5′-GCCCCAOH at its U3 and U5 ends, respectively.
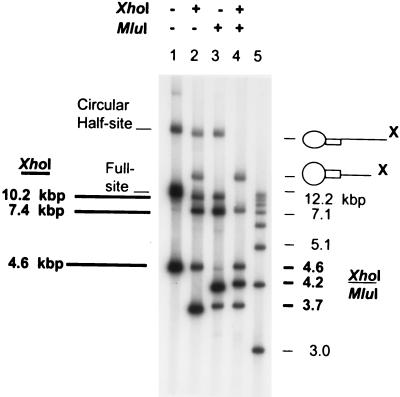
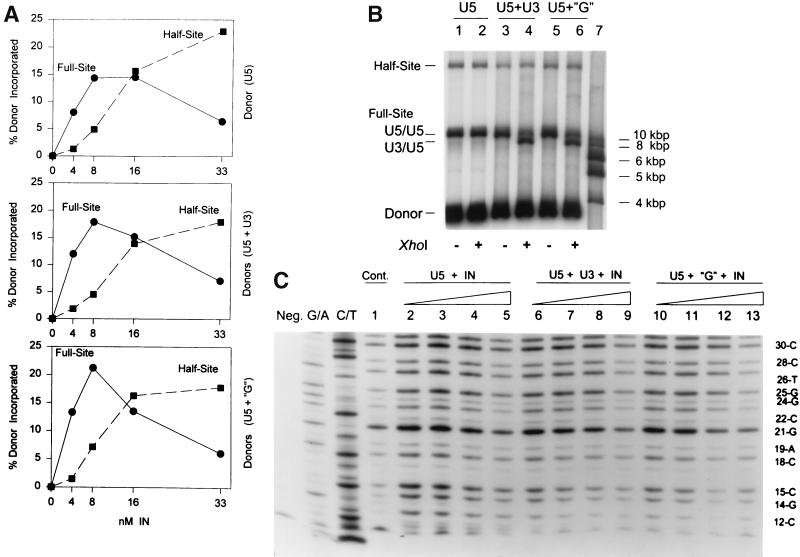
Two parameters established that IN produces the correct full-site integration products with the new 4.5-kbp donors. The full-site products contain two separate donor ends inserted in a concerted fashion into a circular DNA target (Fig. 1C). Restriction enzyme digestions of the 11.9-kbp full-site products by XhoI or MluI, or both, produced the anticipated DNA fragments (Fig. 1A, C, and D; Fig. 2). The donor contains the “G” LTR mutations at both the U3 and U5 termini. XhoI digestion of the 11.9-kbp products (Fig. 2, lane 1) produced the expected 10.2-, 7.4-, and 4.6-kbp fragments (Fig. 2, lane 2) (Fig. 1D). The fastest migrating band (3.6 kbp) in lane 2 is the digested donor (Fig. 1A). The two slowest migrating bands in lane 2 are half-site products (circular target with one inserted donor and different-size donor tails) (38). MluI digestion of the full-site products also produced the anticipated fragments (Fig. 2, lane 3; Fig. 1D). The combined XhoI/MluI digestions produced the expected 4.6-, 4.2-, and 3.7-kbp fragments (Fig. 2, lane 4). With the “G” donor containing nearly equivalent catalytic ends (39), the three XhoI/MluI full-site digestion fragments have near-molar ratios of 1:2:1 (Fig. 2, lane 4). The appropriate restriction patterns were also obtained with the 4.5-kbp donor containing wt U3 and wt U5 ends (data not shown). In summary, restriction analysis of the full-site products produced the anticipated fragments.
FIG. 2.
Restriction enzyme analysis of “G” 4.5-kbp donor-target full-site integration products. The circular half-site and full-site products (lane 1) are indicated on the left (lettering not in boldface type). The unincorporated donor on the 1.5% agarose gel is the fastest migrating band in lane 1. The XhoI restriction fragments in lane 2 resulting from the full-site products are indicated in boldface lettering (10.2, 7.4, and 4.6 kbp) on the left under XhoI (Fig. 1D). XhoI digestion of the circular half-site products (lane 2) results in circles with two different sizes of tails (X on right). MluI digestion (lane 3) produces the anticipated half-site and full-site fragments (not marked) (Fig. 1D). In lane 4, the combined XhoI/MluI digestion of the full-site product produces the anticipated 1:2:1 ratio of fragments migrating at 4.6, 4.2, and 3.7 kbp (Fig. 1D), respectively (at right, in boldface type). The two slowest migrating fragments in lane 4 are digested circular half-site products. Lane 5 contains molecular weight markers varying in size by ∼1 kbp (at right, not in boldface type).
The second parameter to establish full-site integration required sequencing of the donor-target junctions. The donor-target products from both wt and “G” donor reactions were digested with EcoRI (unique sites located in both LTRs). A 2.9-kbp donor-target fragment was isolated by agarose gel electrophoresis, ligated, and transformed into E. coli, and the plasmids from 20 colonies were sequenced. The avian host site duplications (5 to 7 bp, with the 6-bp duplication predominating) were present in all of the plasmids at the donor-target junctions with both donors. The data show IN mediates efficient full-site integration with a high fidelity with either the 4.5-kbp donors or the 480-bp donors (39).
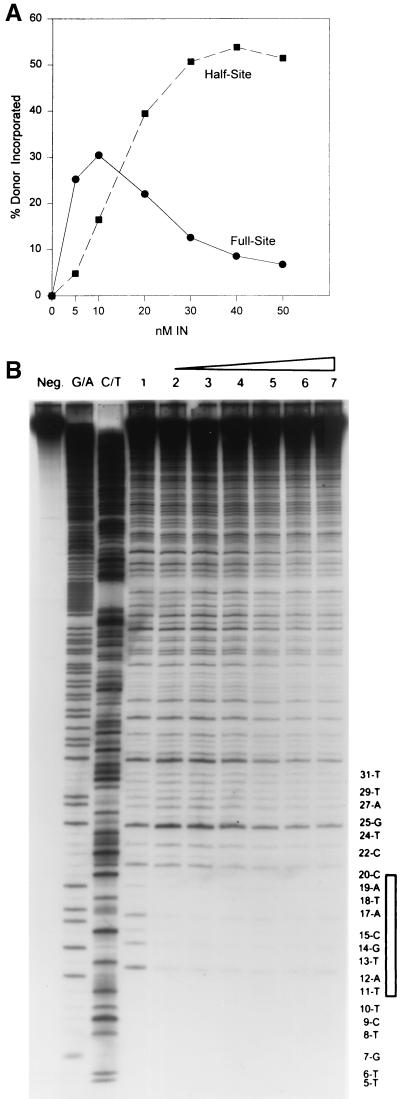
Assembly of nucleoprotein complexes mediating full-site integration is cooperative for protein concentration, is time-dependent, and is partially temperature dependent.
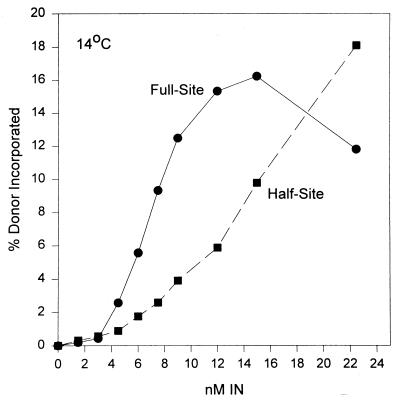
The assembly of IN-“G” donor complexes capable of full-site integration appears to be cooperative with respect to protein concentration at 14°C (Fig. 3). No full-site integration activity is observed at 14°C (data not shown). The assembly time was held constant at 10 min. After assembly, target was added and integration was allowed to proceed for only 5 min at 37°C, thus more closely reflecting the effects of the initial assembly events. A sigmoidal curve for incorporation of donor was apparent with respect to full-site integration but not half-site integration (Fig. 3). Similar kinetic data for both reactions were apparent upon assembly on ice for 10 min, although the total quantities for both products were decreased ∼25% at the same concentrations of IN. Similar cooperative interactions for assembly of complexes mediating full-site integration at 14°C were observed using the 4.5-kbp donor with wt U3 and U5 termini and the 3.4-kbp donor having only ∼30 bp of wt U3 and U5 LTR sequences at their termini (data not shown).
FIG. 3.
Assembly of IN-4.5 kbp donor complexes appears cooperative for full-site integration. Various concentrations of AMV IN (bottom) were assembled with the “G” 4.5-kbp donor at 14°C. The integration products were subjected to electrophoresis on a 1% agarose gel and quantified.
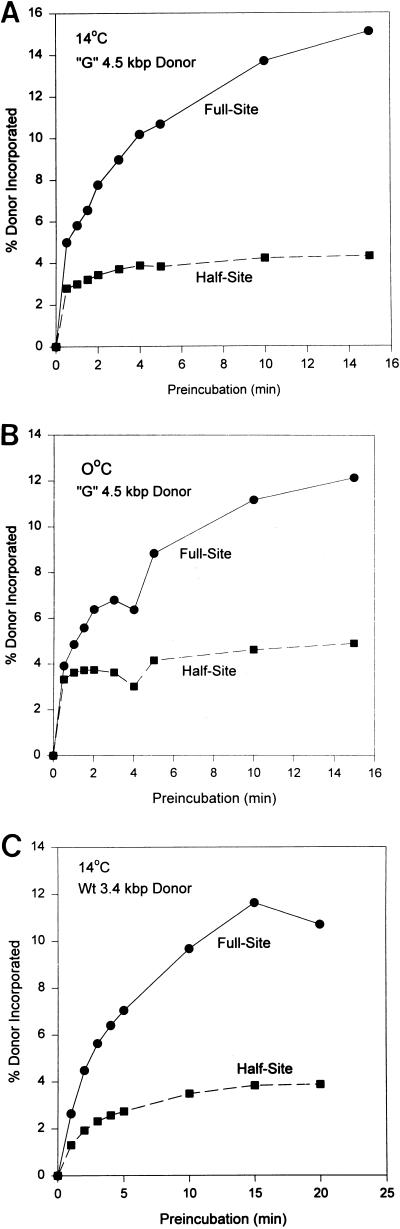
To differentiate further the assembly events for full-site and half-site integration, we investigated the effects of time and temperature. IN was added to the assembly mixture containing the “G” 4.5-kbp donor equilibrated at 14°C. The time of assembly was varied while the concentration of IN remained constant (6 nM) (Fig. 4A). The integration assays were again limited to 5 min at 37°C. The rate for forming nucleoprotein complexes capable of mediating full-site integration was time dependent, while half-site integration was essentially unchanged after 1 min (Fig. 4A). Varying the concentration of IN (4 to 15 nM) in a series of parallel assembly experiments with the 4.5-kbp “G” or wt donors also produced similar time-dependent assembly requirements for full-site integration but not half-site integration (data not shown). Similar assembly rates for nucleoprotein complexes were observed for both reactions with the “G” donor at 6 nM IN if the assembly temperature was at 0°C (Fig. 4B), although half-site integration was always proportionally higher at 0°C than at 14°C. Other studies revealed similar formation rates for both types of nucleoprotein complexes at 14°C in the absence of Mg2+ during assembly but more closely resembled the 0°C results observed in Fig. 4B (data not shown). Similar assembly results were also obtained if the 3.4-kbp donor containing ∼30 bp of wt U3 and wt U5 LTR sequences at its termini were assembled with 10 nM IN at 14°C (Fig. 4C).
FIG. 4.
Different assembly rates for nucleoprotein complexes mediating full-site and half-site integration. (A) IN was assembled with the “G” 4.5-kbp donor at 14°C. At various time intervals (bottom), aliquots were taken and added to tubes containing target DNA at room temperature. Following strand transfer at 37°C, the products were subjected to agarose gel electrophoresis and quantified. (B) The same assembly experiment was repeated as described in panel A, except assembly was on ice. (C) The same assembly experiment was repeated as described in panel A at 14°C, except IN was at 10 nM and the donor was the wt 3.4-kbp DNA containing ∼30 bp of wt U5 and wt U3 sequences at its ends.
In summary, the assembly of IN-donor complexes capable of mediating full-site integration appears to be cooperative for protein-DNA interactions and is sensitive to time and temperature. The assembly rates of nucleoprotein complexes for full-site events are significantly different from those observed for half-site events. For full-site integration, the assembly appears to be independent of LTR sequences mapping >30 bp from the termini. The results also suggest some or all of the nucleoprotein complexes responsible for half-site integration under these assembly conditions may not be precursors to nucleoprotein complexes involved in full-site integration.
IN-DNA complexes capable of full-site integration produce DNase I footprints with an outer boundary mapping ∼20 bp from the LTR end.
The molecular probing of IN-donor complexes provided information regarding the physical association of IN with the viral LTR termini. This association was examined under the same conditions as required for assembly to investigate structure-functional relationships necessary for full-site integration. DNase I was chosen as the mildest molecular probe. The physical interactions of IN with functional and nonfunctional LTR donors for integration were analyzed.
The single 5′-end labeled 3.6 kbp DNA fragment (Fig. 1A) containing “G” sequences on the U3 end was assembled with varying concentrations of IN for 15 min at 14°C. After assembly of the IN-DNA complexes, aliquots were taken to measure integration activity (10 min at 37°C) (Fig. 5A), and the rest of the samples were subjected to DNase I digestion for 90 s at 14°C prior to being stopped with phenol (Fig. 5B). Full-site integration dominates at low concentrations, while half-site integration is preferred at higher IN concentrations (Fig. 5A). At 5 nM IN, the full-site integration reaction is approximately fivefold more than the half-site reaction.
FIG. 5.
Full-site integration is correlated to DNase I protection by IN at the LTR termini. (A) The single-end-labeled 3.6-kbp fragment containing “G” sequences on the U3 LTR end was assembled with IN at different concentrations. (B) The rest of the assembly mixtures in A were subjected to DNase I digestion. The DNase I-treated samples were subjected to electrophoresis on a denaturing 8% polyacrylamide gel. Equivalent counts were loaded per lane. The far left lane contains the untreated 3.6-kbp DNA (Neg.). The two adjacent lanes are G/A and C/T chemical reactions. The concentrations of IN in lanes 1 to 7 were 0, 5, 10, 20, 30, 40, and 50 nM, respectively. The nucleotide positions numbered from the blunt-end are marked (right) and the area protected by IN (vertical rectangle) mapping ∼20 bp from the end is shown. Chemical markers have one less nucleotide than the adjacent fragments produced by DNase I. The dried gel was exposed to X-ray film for 10 days without an intensifying screen.
The assembled complexes tested for integration activity (Fig. 5A) were also subjected to parallel DNase I footprint analysis to examine the physical association of IN at the LTR ends. At 5 and 10 nM IN (Fig. 5B, lanes 2 and 3), a specific region of protection at the “G” donor termini was observed whose outer boundary mapped to ∼20 nucleotides from the end in comparison to the naked “G” donor subjected to DNase I without IN (Fig. 5B, lane 1). The protection is from approximately position 11 (T) to position 20 (C). DNase I is sensitive to position relative to the ends of the DNA and will not nick closer than ∼10 bp with high frequency. Starting at 20 nM IN (Fig. 5B, lane 4) and particularly at higher IN concentrations (lanes 5 to 7), an extended pattern of partial DNase I protection is observed which almost encompasses the entire length of visualized DNA fragments (∼80 nucleotides past position 20). The results suggest that a specific size of footprint, which correlates to full-site integration activity, is produced by IN at lower concentrations (Fig. 5A).
Several other observations are apparent with the DNase I footprint analysis. At the lower IN concentrations (Fig. 5B, lanes 2 to 4), there were no observable adjacent or other internal regions of protection produced by IN after the 20th nucleotide relative to the same DNA treated with DNase I without IN (lane 1). Instead, there appears to be enhanced DNase I nicking at several nucleotides within the next ∼10 internal positions. PhosphorImager analysis of the DNase I protection patterns revealed IN protects DNA from position 11 to 20 by a factor of approximately five or more (Fig. 5B [compare lane 1 with lanes 2 and 3], 6, and 8). IN at concentrations 20 nM or more (Fig. 5B, lanes 4 to 7) did not alter this protection pattern below position 20, suggesting that this protected region was also specific (see Fig. 7 for nonfunctional donor). Lastly, assembled IN-donor complexes treated with DNase I were selected by nitrocellulose filters to determine if the purified complexes possessed different footprints. After DNase I treatment, the nucleoprotein complexes were immediately filtered onto nitrocellulose filters, washed with assembly buffer, eluted with sodium dodecyl sulfate buffer, and examined on denaturing gels. The same pattern of DNase I protection (∼20 bp from the end) by IN shown in Fig. 5B (lanes 2 to 4) was observed (data not shown).
FIG. 7.
Lack of DNase I protection by IN with a nonfunctional LTR donor. IN was assembled with a 3.6-kbp fragment containing a mutation (5′-CCCCAOH) at the U5 LTR end. Lane 1 contains naked DNA treated with DNase I. Lanes 2 to 5 contain 3, 5, 10, and 20 nM IN, respectively. Labeling is the same as in Fig. 5.
In summary, comparing the full-site integration activity (Fig. 5A) to the parallel DNase I footprint (Fig. 5B) suggests a specific set of IN subunits binding cooperatively to LTR ends is necessary for full-site integration. The protected region maps to ∼20 nucleotides from the “G” LTR end. The outer boundary is specifically observed at lower IN concentrations (<20 nM) and is associated with full-site and not half-site integration activity.
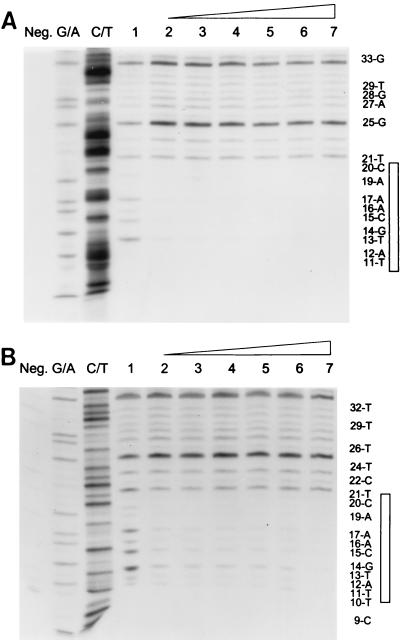
Stability and specificity of assembled IN-donor complexes capable of full-site integration are correlated to functional att site sequences.
Additional time studies were undertaken to show the stable physical association of IN with the 3.6-kbp “G” and wt U3 LTR donor ends (Fig. 6A and B, respectively). AMV IN was assembled at 14°C with each donor, and aliquots were analyzed for full-site integration and by DNase I footprint analysis. Both donors allowed IN to form a stable complex (15 min to 6 h) whose outer boundary maps ∼20 bp from the end of the DNA. For the “G” donor reaction, the percent donor incorporated for full-site integration was 24, 27, 31, 32, 28, and 27, and for half-site integration it was 6, 6, 8, 7, 6, and 7, respectively (Fig. 6A, lanes 2 to 7). For the wt U3 reaction, the percent donor incorporated for full-site integration was 4, 6, 8, 13, 16, and 16, and for half-site integration it was 1, 2, 2, 4, 3, and 4, respectively (Fig. 6B, lanes 2 to 7).
FIG. 6.
Highly stable DNase I footprints associated with IN-donor complexes for full-site integration. (A) IN (5 nM) was assembled with the 3.6-kbp U3 donor containing “G” sequences. Assembly times were 15, 30, 60, 120, 240, and 360 min in lanes 2 to 7, respectively. Lane 1 contains no IN. Labeling is the same as in Fig. 5. (B) IN (10 nM) was assembled with the single-end-labeled 3.6-kbp fragment containing wt U3 sequences for the same times. Labeling is the same as in Fig. 5.
The 5th, 6th, and 7th nucleotides from the ends of the avian U3 and U5 LTRs have a major influence on the ability of IN to mediate half-site and full-site integration (9, 39). The U5 LTR end (5′-GCTTCAOH) of the 4.5-kbp donor (Fig. 1A) was modified to 5′-GCCCCAOH. The single-end-labeled 3.6-kbp fragment containing the CC mutation was assembled with various concentrations of IN. Aliquots were taken for integration activity and DNase I protection analysis. At 20 nM IN with the DNase I footprint (Fig. 7, lane 5), there was ∼1% incorporation of the U5 donor containing the CC mutation into each of the half-site and full-site integration products (10-min reaction) (data not shown). No specific protection by IN with an outer boundary mapping at ∼20 nucleotides from the end was evident with the CC mutant donor (Fig. 7, lanes 1 to 5). Increasing the concentration of IN to 45 nM also resulted in no distinct footprint on the U5 end with the CC mutation (data not shown). The U3 end containing the CC mutation at the fifth and sixth nucleotide also lacks significant integration activity. Finally, IN was also preincubated at 14°C with a 2.4-kbp nonspecific DNA fragment containing a 5′-end-labeled EcoRI restriction site. There was neither integration activity nor evidence of a specific DNase I protection pattern by IN (10 to 30 nM) (data not shown).
In summary, IN is capable of forming a very stable complex having a specific physical association within the first ∼20 bp on either the wt U3 or “G” donor ends for full-site integration. This specific association appears to be related to att site sequences capable of efficiently mediating integration activity in vitro.
Formation of the outer protected boundary at ∼20 bp by IN on the “G” donor end with target present.
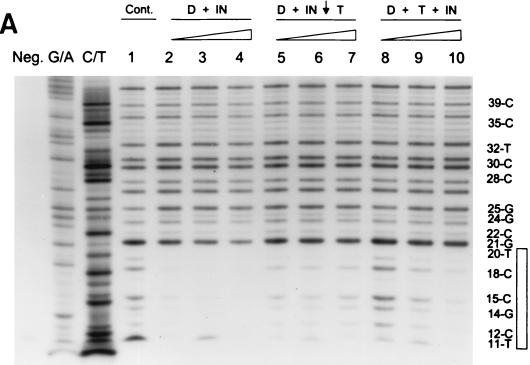
Earlier attempts to reconstitute full-site integration employed the preincubation of donor and target together with IN (10, 14, 22). We investigated whether AMV IN was capable of forming the same specific DNase I protection patterns at the LTR ends with target present during assembly and when assembled IN-donor complexes were challenged with target. The single-end-labeled 3.6-kbp fragment with the U5 end containing the “G” mutation (Fig. 1A) was used as the donor.
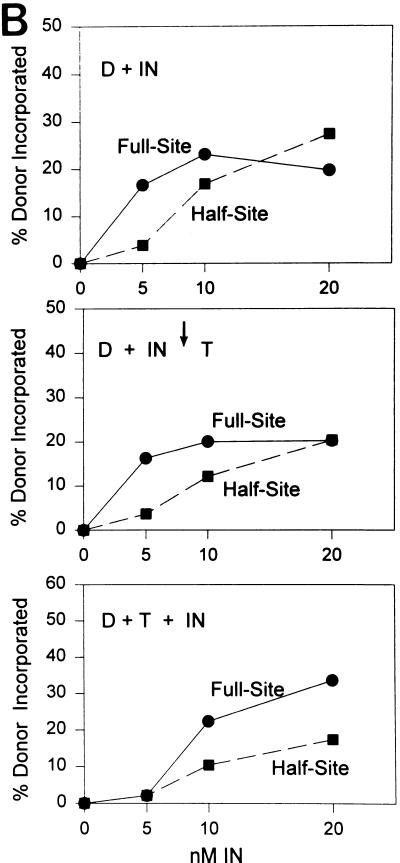
Assembly of AMV IN (5 to 20 nM) with the U5 end containing the “G” mutation for 45 min at 14°C produced the usual DNase I protection pattern mapping ∼20 bp from the end (Fig. 8A, lanes 1 to 4), and this protection was correlated with full-site integration activity (Fig. 8B, top). These assembled IN-donor complexes were also challenged with the normal concentration of target used to measure integration activity. The target was further incubated with the assembled complexes for 10 min at 14°C prior to DNase I digestion. The DNase I protection pattern (Fig. 8A, lanes 5 to 7) and the full-site integration activity (Fig. 8B, middle) were slightly reduced relative to those of the unchallenged controls (Fig. 8A, lanes 2 to 4). Finally, IN was assembled on the donor ends in the presence of target. The donor and target were preincubated together with IN for 45 min at 14°C prior to DNase I digestion. Little DNase I protection (Fig. 8A, lane 8) and integration activity (Fig. 8B, bottom) were apparent at 5 nM IN. However at 20 nM IN, the ∼20-bp DNase I protection pattern (Fig. 8A, lane 10) as well as a significant increase in full-site integration activity was observed (Fig. 8B, bottom).
FIG. 8.
Assembly of IN-donor complexes with target present and challenge of assembled IN-donor complexes with target. (A) IN was assembled with single-end-labeled 3.6-kbp fragment (10 ng) containing “G” sequences on the U5 end. Lanes 2 to 4, 5 to 7, and 8 to 10 contain 5, 10, and 20 nM IN, respectively. In lanes 2 to 4, IN was assembled on the donor DNA only prior to taking an aliquot for integration (Fig. 8B, top) and DNase I treatment. In lanes 5 to 7, IN was assembled with donor and then challenged with target as indicated by the arrow (50 ng per 20 μl). An aliquot was taken for integration activity (Fig. 8B, middle), and the remaining sample treated with DNase I. In lanes 8 to 10, donor, target, and IN were assembled together prior to assaying for integration activity (Fig. 8B, bottom) and DNase I treatment. Labeling is the same as in Fig. 5. (B) Strand transfer activity of samples shown in panel A.
The results suggest IN is capable of forming stable complexes with donor ends for full-site integration in the presence of target and the assembled IN-DNA complexes are relatively stable in the presence of competitor DNA. The DNase I protection pattern observed at the U5 end containing the “G” mutation appears nearly identical to the pattern observed at the U3 end containing the “G” mutation (Fig. 5 and 6).
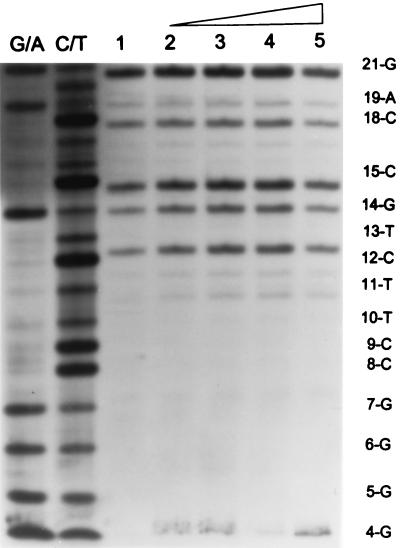
Asymmetric binding at the wt U5 end by IN for full-site integration in comparison to the “G” and wt U3 ends.
Asymmetric recognition of DNA binding sites by proteins for DNA recombination events is common. Two different approaches were used to address this possibility for IN-LTR interactions. We investigated whether IN was capable of producing the same DNase I protection pattern observed when two wt U3 or two “G” LTR ends were coupled together (Fig. 5, 6, and 8) when two wt U5 ends were coupled together for full-site integration. We also investigated the physical association of IN with wt U5 ends coupled to either wt U3 or “G” donor ends for full-site integration (28).
In the first approach, IN was assembled with the 3.6-kbp fragment containing the wt U5 end for 45 min at 14°C prior to strand transfer for 10 min at 37°C. IN mediated efficient full-site integration at low concentrations (4 and 8 nM) (Fig. 9A, top), with full-site products being in the majority in comparison to half-site products (Fig. 9B, lane 1). At these low concentrations, no DNase I protection pattern with a distinct boundary at approximately the 20th nucleotide position with wt U5 was observed (Fig. 9C, lanes 1 to 5). Repeated attempts to demonstrate a distinct DNase I footprint at the wt U5 end were negative. At higher IN concentrations (Fig. 9C, lanes 4 and 5), only a general partial decrease of the entire DNase I footprint was evident relative to what was observed with IN at lower concentrations (lanes 2 and 3).
FIG. 9.
Outer boundary of ∼20 bp DNase I protection by IN is not observed at wt U5 end for full-site integration. (A) Top panel: IN was assembled with the wt U5 end on the 3.6-kbp fragment, aliquots were assayed for integration activity, and the percent donor incorporated into target was determined. The concentrations of IN are indicated at the bottom of each panel, and the donors are indicated on the right. Middle panel: the U5 donor was mixed with unlabeled 2.9-kbp fragment containing the wt U3 end prior to assembly with IN as described for panel A. Bottom panel: same as the middle panel except the 2.9-kbp U3 fragment contained the “G” mutation. (B) Aliquots of each of the reaction mixtures described for panel A produced at 4 and 8 nM IN were pooled. The donor-target products were (+) or were not (−) subjected to XhoI digestion. Lanes 1 and 2 contain the U5-U5 products (A, top), lanes 3 and 4 contain the U5-wt U3 products (A, middle), and lanes 5 and 6 contain the U5-U3 with the “G” mutation products (A, bottom). The full-site U5-U5 marker (left) identifies two U5 end products while the U3-U5 marker identifies products of either the U5-wt U3 (lane 4) or the U5-U3 with the “G” mutation (lane 6). Size markers are in lane 7 (right). (C) DNase I footprinting of assembled IN-donor complexes as indicated in panel A. Labeling is the same as in Fig. 5 except lanes 2 to 5, 6 to 9, and 10 to 13 contain 4, 8, 16, and 32 nM IN, respectively. The combinations of donors are shown at the top.
Possibly, the assembly of IN on wt U5 ends required the presence of target to produce a DNase I protection pattern mapping to approximately the 20th nucleotide. No distinct DNase I footprints were obtained at wt U5 donor ends if assembly occurred in the presence of target (data not shown). As shown previously with the “G” donor (Fig. 8), IN (20 nM) was capable of assembling with the 3.6-kbp wt U5 donor in the presence of target (45 min at 14°C) in an efficient manner (22 and 21% full-site and half-site products in 10 min at 37°C), even though no distinct DNase I footprint was evident at the wt U5 end.
In the second approach, we investigated whether a distinct DNase I protection pattern could be produced by IN when a wt U5 end was coupled to either wt U3 or “G” donor ends for full-site integration. Equal amounts (5 ng each) of the single-end-labeled 3.6-kbp wt U5 fragment were assembled together with either unlabeled 2.9-kbp wt U3 fragment or the same U3 fragment containing the “G” mutation. The 3.6-kbp U5 fragment was produced by XhoI digestion while SspI digestion produced the 2.9-kbp U3 fragments containing the XhoI site (Fig. 1A). From PhosphorImager analysis, approximately 73% of the full-site integration product produced with the labeled wt U5 end was coupled to either unlabeled U3 or “G” donor ends (Fig. 9A, middle and bottom, respectively). This conclusion was reached because the full-site products produced with only the labeled wt U5 donor in comparison to the wt U5 donor coupled with either unlabeled wt U3 or “G” donor results in different size restriction fragments after XhoI digestion (Fig. 9B, compare lane 2 with lanes 4 and 6, respectively). Coupling of the labeled wt U5 end with either the wt U3 donor or the U3 donor with the “G” mutation did not appear to significantly modify the DNase I footprints on wt U5 (Fig. 9C, lanes 6 to 9 and lanes 10 to 13, respectively) in comparison to the footprint obtained with only two wt U5 ends (Fig. 9C, lanes 2 to 5). At 4 or 8 nM IN, no distinct protection of DNA from nucleotides ∼11 to 20 was evident even though full-site integration was the predominant reaction (Fig. 9C, lanes 2 and 3, 6 and 7, and 10 and 11, respectively). Partial protection of this DNA region and the adjacent DNA was evident at higher IN concentrations. Similar results were also obtained if the ratio of the labeled wt U5 (3.5 ng) to unlabeled wt U3 and “G” donors (6.5 ng) was used during assembly (data not shown).
The second approach was further modified to determine the physical association of IN upon coupling U3 and U5 termini for full-site integration. As a positive control for this approach, the labeled donor was now the 2.9-kbp U3 fragment containing the “G” mutation (1.5 ng) while the unlabeled donor was the 3.6-kbp fragment with the wt U5 end (8.5 ng). Assembly was with IN (5 to 15 nM) for 45 min at 14°C. As shown previously by restriction analysis (Fig. 9B), 90% of the observed full-site integration reaction at 5 nM IN was the result of coupling U3 and U5 ends together (data not shown). As anticipated, DNase I footprint analysis showed IN protected the labeled U3 end with a defined outer boundary ∼20 nucleotides for the end (data not shown) as previously observed upon coupling of two U3 ends together for full-site integration (Fig. 5, 6, and 8).
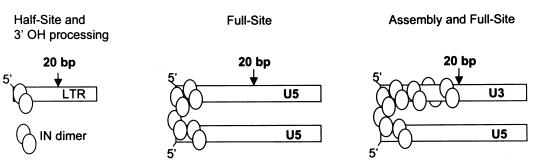
In summary, the results suggest IN binds within the first ∼10 nucleotides on the wt U5 ends (beyond the detection level of the DNase I probe) for full-site integration. The lack of DNase I protection by IN at lower concentrations with a clear boundary at approximately the 20th nucleotide on the wt U5 ends coupled to either the wt U3 ends or the “G” ends suggests IN binds asymmetrically at the ends of the viral DNA to mediate full-site integration (Fig. 10).
FIG. 10.
Models for multimerization of IN. (Left) IN is represented as a dimer, and a dimer is required for the 3′-OH processing step and half-site integration (4). The location of IN is only an approximation in relationship to the 20-bp marker. (Middle) A tetramer per LTR end is predicted for full-site integration (5, 20). (Right) The model predicts an unknown number of avian IN subunits that forms multimers on the U3 end with a predicted tetramer on the U5 end.
DISCUSSION
The reconstitution of nucleoprotein complexes with AMV IN and linear 4.5-kbp donors capable of mediating full-site integration in vitro has revealed novel insights into assembly requirements and the physical association of IN at the donor LTR ends. The apparent cooperative binding of IN dimers at the LTR ends results in the formation of multimers producing a region of DNase I protection whose outer boundary maps to approximately the 20th nucleotide position on wt U3 and LTR ends containing the “G” mutation. Interestingly, the binding of IN to the wt U5 end appears to be only near the terminal att sequences (<10 bp) for full-site integration. The asymmetric binding of AMV IN to the U3 and U5 LTR ends suggests a model in which IN has multiple roles for assembly of nucleoprotein complexes capable of full-site integration in vitro (Fig. 10).
The assembly of nucleoprotein complexes for full-site integration at 14°C is significantly different from those for half-site integration with respect to time and protein concentrations (Fig. 3 to 5). At 5 nM IN, the ratio of IN dimer to 4.5-kbp donor ends is 15. The requirements for full-site integration suggest an ordered set of events being performed by IN. These events may be related to formation of a specific number of multimers at the LTR ends as well as to holding together the two donors in a stable structure necessary for the concerted insertion of the termini into the target (Fig. 10). Both events may require different activation energies for the protein-protein and protein-DNA interactions envisioned necessary for assembly. The assembly process appears to be independent of internal LTR sequences (>30 bp from the end Fig. 4C) and can occur in the presence of target (Fig. 8). The time-dependent assembly process of avian intasome-like complexes in vitro mirrors the slow formation of MLV intasomes after virus infection (41) and some of the assembly processes associated with the phage transposase-Mu DNA complexes in vitro (31, 36). Further studies will be needed to determine the number of subunits, the role of the individual subunits, and the role of the three functional domains of IN (4) in assembly of intasome-like complexes. Similar in vitro studies are actively being investigated for assembling Tn5 and Mu transposase-DNA complexes (43).
For full-site integration, purified retrovirus intasomes and nucleoprotein complexes assembled at low AMV IN concentrations catalyze equivalent incorporation of viral DNA into target (percent incorporated) in vitro. DNase I footprints of the reconstituted AMV IN-donor complexes have revealed new insights into the physical association of IN at the viral ends with some limitations. DNase I is not capable of efficiently nicking DNA closer than ∼10 bp from the end. As with the MM-PCR footprinting technique used to investigate retrovirus intasomes, DNase I footprinting is also restricted to observations obtained with the 5′-labeled end of the donor (nonprocessed strand). The most striking similarity between the two integration systems is that the protection of sequences by AMV IN possesses a clear boundary ∼20 bp from the end (Fig. 5, 6, and 8) while the boundary for MM-PCR protection and enhancements for both MLV and HIV-1 intasomes is also ∼20 bp from the end (7, 41, 42). Both the reconstituted intasome-like complexes and the purified retrovirus intasomes require active att site sequences for assembly, protection, and full-site integration activity.
The retrovirus intasomes appear to have near-symmetrical MM-PCR footprints, while the reconstituted AMV IN-donor complexes have asymmetrical DNase I footprints at their DNA ends. Another dissimilarity between the systems is the lack of DNase I enhancements within the AMV IN protected sequences (∼11 to 20 nucleotides from the end on wt U3 and LTR ends with the “G” mutation) while the HIV-1 intasome MM-PCR major enhancements mapped to two regions (at nucleotides 8, 9, and 13 on wt U3 and at nucleotides 9, 11, and 16 on wt U5) (2). On only three random occasions with numerous analyses, there were additional fragments produced by DNase I digestion migrating near position 9 or 10, or both, when AMV IN was bound to either the “G” or U3 donor ends which were not observed with the naked DNA treated with DNase I (Fig. 5 to 9). The significance of the occasionally observed enhanced DNase I cleavage near these positions and its relationship to a possible transient assembly intermediate is under investigation. Further studies are needed to unravel the organization of IN at the viral DNA ends in both systems.
Mutagenesis studies of retrovirus att sequences have established that the first ∼7 to 12 nucleotides from the ends are necessary and sufficient for near maximum integration activity in vivo and for 3′-OH processing and half-site integration in vitro (2, 4, 7, 27). These terminal nucleotides interact with specific residues on IN (11, 20). Mutagenesis of avian retrovirus LTR sequences has shown the 5th, 6th, and 7th nucleotides play critical roles for controlling half-site and full-site integration activities and for coupled communications between two LTR termini by IN in trans for full-site integration (9, 39).
The DNase I protection experiments in this report define the physical association of AMV IN within the wt U3 and wt U5 LTR termini for assembly and full-site integration. The minimum physical association of IN at low concentrations (<10 nM) with wt U5 appears to be limited to fewer than 10 nucleotides from the U5 end (Fig. 10, middle) even when coupled to the wt U3 end for full-site integration (Fig. 10, right). We cannot rule out a different model wherein a different array of multimers on the U5 end occurs when coupled to wt U3, particularly at higher IN concentrations at which partial protection is observed (Fig. 9C, lanes 6 to 9 and 10 to 13). However, binding of IN at the very end of wt U5 (<10 nucleotides) appears sufficient for assembly and efficient full-site integration because no DNase I protection is observed between nucleotides ∼10 to 30 (Fig. 9C, lanes 2 to 5, and A, top) (data not shown). Other molecular probes may reveal IN interactions at the very end (<10 bp) of U5. AMV IN at low concentrations is capable of forming an extended nucleoprotein complex at the wt U3 ends or U3 and U5 ends containing the “G” mutation with a well-defined outer boundary of DNase I protection mapping ∼20 nucleotides from the LTR end (Fig. 5, 6, and 8). The reasons for the apparent asymmetry of IN binding at the wt U3 and wt U5 ends for assembly and full-site integration is unknown. Interestingly, HIV-1 IN possibly recognizes the U3 and U5 att sites independently in vivo (27).
The extended association of IN with several LTR termini appears to be directly related to at least the 5th and 6th nucleotides (underlined below), because wt U5 lacks the extended protection (Fig. 9) while the U5 end containing the “G” mutation has the extended protection (Fig. 8). It is interesting to speculate that these nucleotides (<8 nucleotides from the end) allow IN at lower concentrations to nucleate at the very terminus and that this initial event mediates further recruitment of IN at higher concentrations (Fig. 5B). The possible requirement effect requires wt U3 (5′-CTACAOH) or “G” (5′-CAACAOH) ends, while the effect is not apparent with wt U5 (5′-CTTCAOH) or CC mutant (5′-CCCCAOH) ends. The catalytic strand specificity by IN (39) suggests IN has the capacity to distinguish A · T from T · A possibly, in a minor DNA groove capacity (23, 40).
At higher IN concentrations (>20 nM), the gradually increasing partial DNase I footprints observed past the 20th nucleotide position may represent either specific or nonspecific multimer formation (Fig. 5B). IN may possess different DNA binding modes depending upon protein concentration and solution conditions, possibly similar to the single-stranded DNA binding protein from E. coli (34). Direct visualization of assembled IN-donor complexes and IN-donor-target complexes (Fig. 8) may reveal insights into the physical structure of these complexes, the role of the asymmetric binding of IN at the LTR ends, and the ability of IN to loop DNA (8, 18). IN is involved in forming and maintaining critical protein-protein and protein-DNA contacts within the retrovirus PIC or intasome structures (7, 30, 41). Efforts to identify other internal viral sequences (up to ∼150 bp from the end) protected by AMV IN (<20 nM) by DNase I footprinting were unsuccessful. At either low or moderate IN concentrations, intramolecular looping of DNA by IN (transient or stable) may facilitate physical interactions between two donors, thereby enhancing interactions of donor ends for full-site integration in vitro. In contrast, at high IN concentrations, a large percentage of IN-donor complexes are nonspecific in nature, thereby decreasing proper pairing of LTR ends necessary for full-site integration but not single LTR ends for half-site integration (Fig. 5).
Studies with the MLV and HIV-1 PIC and their respective intasomes suggest IN has multiple functions in these structures (7, 42). The minimum IN structure for 3′-OH processing and half-site integration is a dimer (4) (Fig. 10, left), with the predicted minimum structure of two tetramers required for full-site integration (5, 20) (Fig. 10, middle). IN bound to two LTR appears to communicate in trans for integration in vivo (32) and for full-site integration in vitro (1, 28, 38). AMV IN apparently forms a multimeric structure encompassing ∼20 nucleotides on the wt U3 LTR (Fig. 10, right) whose function in assembly and full-site integration has yet to be defined. The simultaneous use of reconstituted intasome-like complexes with site-directed mutagenesis of IN may provide insights into the multiple structural-functional relationships of IN for assembly and full-site integration.
ACKNOWLEDGMENTS
This work was supported by a National Cancer Institute grant (CA16312).
We thank Roger Chiu and Sapna Sinha for reviewing the manuscript.
REFERENCES
- 1.Aiyar N, Hindmarsh P, Skalka A M, Leis J. Concerted integration of linear retroviral DNA by the avian sarcoma virus integrase in vitro: dependence on both long terminal repeat termini. J Virol. 1996;70:3571–3580. doi: 10.1128/jvi.70.6.3571-3580.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown H E, Chen H, Engelman A. Structure-based mutagenesis of the human immunodeficiency virus type 1 DNA attachment site: effects on integration and cDNA synthesis. J Virol. 1999;73:9011–9020. doi: 10.1128/jvi.73.11.9011-9020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown P, Bowerman B, Varmus H, Bishop J. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 4.Brown P O. Integration. In: Coffin J M, Hughes S J, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1998. pp. 161–203. [Google Scholar]
- 5.Cai M, Zheng R, Caffrey M, Craigie R, Clore G M, Gronenborn A M. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat New Biol. 1997;4:567–577. doi: 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- 6.Carteau S, Gorelick R J, Bushman F D. Coupled integration of human immunodeficiency virus type 1 cDNAs ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J Virol. 1999;73:6670–6679. doi: 10.1128/jvi.73.8.6670-6679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H, Wei S Q, Engelman A. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type 1 intasome. J Biol Chem. 1999;274:17358–17364. doi: 10.1074/jbc.274.24.17358. [DOI] [PubMed] [Google Scholar]
- 8.Cherepanov P, Surratt D, Toelen J, Pluymers W, Griffith J, De Clercq E, Debyser Z. Activity of recombinant HIV-1 integrase on mini-HIV DNA. Nucleic Acids Res. 1999;27:2202–2210. doi: 10.1093/nar/27.10.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu R, Grandgenett D P. Avian retrovirus DNA internal attachment site requirements for full-site integration in vitro. J Virol. 2000;74:8292–8298. doi: 10.1128/jvi.74.18.8292-8298.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craigie R, Fujiware T, Bushman F D. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 11.Esposito D, Craigie R. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein-DNA interactions. EMBO J. 1998;17:5832–5843. doi: 10.1093/emboj/17.19.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farnet C, Bushman F D. HIV-1 cDNA integration: requirement of HMG 1(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 13.Farnet C M, Haseltine W A. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J Virol. 1991;65:1910–1915. doi: 10.1128/jvi.65.4.1910-1915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitzgerald M, Vora A C, Zeh W, Grandgenett D P. Concerted integration of viral DNA termini by purified avian myeloblastosis virus integrase. J Virol. 1992;66:6257–6263. doi: 10.1128/jvi.66.11.6257-6263.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujiwara T, Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 16.Goodarzi G, Im G, Brackmann K, Grandgenett D P. Concerted integration of retrovirus-like DNA by human immunodeficiency virus type 1 integrase. J Virol. 1995;69:6090–6097. doi: 10.1128/jvi.69.10.6090-6097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodarzi G, Pursley M, Felock P, Witmer M, Hazuda D, Brackmann K, Grandgenett D P. Efficiency and fidelity of full-site integration reactions using recombinant simian immunodeficiency virus integrase. J Virol. 1999;73:8104–8111. doi: 10.1128/jvi.73.10.8104-8111.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grandgenett D P, Inman R, Vora A C, Fitzgerald M. Comparison of DNA binding and integration half-site selection by avian myeloblastosis virus integrase. J Virol. 1993;67:2628–2636. doi: 10.1128/jvi.67.5.2628-2636.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grandgenett D P, Vora A C, Schiff R. A 32,000-dalton nucleic acid-binding protein from avian retrovirus cores possesses DNA endonuclease activity. Virology. 1978;89:119–132. doi: 10.1016/0042-6822(78)90046-6. [DOI] [PubMed] [Google Scholar]
- 20.Heuer T, Brown P O. Photo-crosslinking-studies suggest a model for the architecture of an active human immunodeficiency virus type 1 integrase-DNA complex. Biochemistry. 1998;37:6667–6678. doi: 10.1021/bi972949c. [DOI] [PubMed] [Google Scholar]
- 21.Hindmarsh P, Ridky T, Reeves R, Andrake M, Skalka A M, Leis J. HMG protein family members stimulate human immunodeficiency virus type 1 and avian sarcoma virus concerted DNA integration in vitro. J Virol. 1999;73:2994–3003. doi: 10.1128/jvi.73.4.2994-3003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katz R A, Merkel J, Kulkosky J, Leis J, Skalka A M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 23.Kielkopf C L, White S, Szewczyk J W, Turner J M, Baird E E, Dervan P B, Rees D C. A structural basis for recognition of A · T and T · A base pairs in the minor groove of B-DNA. Science. 1998;282:111–115. doi: 10.1126/science.282.5386.111. [DOI] [PubMed] [Google Scholar]
- 24.Lee M S, Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. Proc Natl Acad Sci USA. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee Y, Coffin J. Relationship of avian retrovirus DNA synthesis to integration in vitro. Mol Cell Biol. 1991;11:1419–1430. doi: 10.1128/mcb.11.3.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Farnet C M, Anderson W F, Bushman F D. Modulation of activity of Moloney murine leukemia virus preintegration complexes by host factors in vitro. J Virol. 1998;72:2125–2131. doi: 10.1128/jvi.72.3.2125-2131.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masuda T, Kuroda M J, Harada S. Specific and independent recognition of U3 and U5 att sites by human immunodeficiency virus type 1 integrase in vivo. J Virol. 1998;72:8396–8402. doi: 10.1128/jvi.72.10.8396-8402.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCord M, Chiu R, Vora A C, Grandgenett D P. Retrovirus DNA termini bound by integrase communicate in trans for full-site integration in vitro. Virology. 1999;259:392–401. doi: 10.1006/viro.1999.9782. [DOI] [PubMed] [Google Scholar]
- 29.McCord M, Stahl J, Mueser T C, Hyde C C, Vora A C, Grandgenett D P. Purification of recombinant Rous sarcoma virus integrase possessing physical and catalytic properties similar to virion-derived integrase. Protein Purif Expr. 1998;14:167–177. doi: 10.1006/prep.1998.0954. [DOI] [PubMed] [Google Scholar]
- 30.Miller M D, Farnet C M, Bushman F D. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizuuchi M, Baker T, Muzuuchi K. Assembly of the active form of the transposase-Mu DNA complex: a critical control point in Mu transposition. Cell. 1992;70:303–311. doi: 10.1016/0092-8674(92)90104-k. [DOI] [PubMed] [Google Scholar]
- 32.Murphy J E, Goff S P. A mutation at one end of Moloney murine leukemia virus DNA blocks cleavage of both ends by the viral integrase in vivo. J Virol. 1992;66:5092–5095. doi: 10.1128/jvi.66.8.5092-5095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Namgoong S Y, Harshey R M. The same two monomers within a MuA tetramer provide the DDE domains for the strand cleavage and strand transfer steps of transposition. EMBO J. 1998;17:3775–3785. doi: 10.1093/emboj/17.13.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raghunathan S, Kozlov A G, Lohman T M, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat Struct Biol. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 35.Record M T, Ha J H, Fisher M. Analysis of equilibrium and kinetic measurements to determine thermodynamic origins of stability, specificity and mechanism of formation of site-specific complexes between proteins and helical DNA. Methods Enzymol. 1991;208:291–343. doi: 10.1016/0076-6879(91)08018-d. [DOI] [PubMed] [Google Scholar]
- 36.Surette M, Chaconas G. The Mu transpositional enhancer can function in trans: requirement of the enhancer for synapsis but not strand cleavage. Cell. 1992;68:1101–1108. doi: 10.1016/0092-8674(92)90081-m. [DOI] [PubMed] [Google Scholar]
- 37.Vora A C, Grandgenett D P. Assembly and catalytic properties of retrovirus integrase-DNA complexes capable of efficiently performing concerted integration. J Virol. 1995;69:7483–7488. doi: 10.1128/jvi.69.12.7483-7488.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vora A C, McCord M, Fitzgerald M, Inman R B, Grandgenett D P. Efficient concerted integration of retrovirus-like DNA in vitro by avian myeloblastosis virus integrase. Nucleic Acids Res. 1994;22:4454–4461. doi: 10.1093/nar/22.21.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vora A C, McCord M, Goodarzi G, Stahl S J, Mueser T C, Hyde C C, Grandgenett D P. Avian retrovirus U3 and U5 DNA inverted repeats: role of nonsymmetrical nucleotides in promoting full-site integration by purified virion and bacterial recombinant integrases. J Biol Chem. 1997;272:23938–23945. doi: 10.1074/jbc.272.38.23938. [DOI] [PubMed] [Google Scholar]
- 40.Wang T, Balakrishnan M, Jonsson C B. Major and minor groove contacts in retroviral integrase-LTR interactions. Biochemistry. 1999;38:3624–3632. doi: 10.1021/bi982124i. [DOI] [PubMed] [Google Scholar]
- 41.Wei S Q, Mizuuchi K, Craigie R. A large nucleoprotein assembly at the viral DNA mediates retroviral DNA integration. EMBO J. 1997;16:7511–7520. doi: 10.1093/emboj/16.24.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei S Q, Mizuuchi K, Craigie R. Footprints on the viral DNA ends in Moloney murine leukemia virus preintegration complexes reflect a specific association with integrase. Proc Natl Acad Sci USA. 1998;95:10535–10540. doi: 10.1073/pnas.95.18.10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams T L, Baker T A. Transposase team puts a headlock on DNA. Science. 2000;289:73–74. doi: 10.1126/science.289.5476.73. [DOI] [PubMed] [Google Scholar]
- 44.Zhou H, Rainey G J, Wong S K, Coffin J M. Substrate sequence selection by retroviral integrase. J Virol. 2001;75:1359–1370. doi: 10.1128/JVI.75.3.1359-1370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]