Abstract
Cellular communication network factor 2 (CCN2) is a secreted extracellular matrix-associated protein, and its aberrantly increased expression has been implicated in a diversity of diseases involving pathological processes of fibrosis, chronic inflammation, or tissue injury, which has promoted the evaluation of CCN2 as therapeutic targets for multiple disorders. However, human phenotypes associated with CCN2 deficiency have remained enigmatic; variants in CCN2 have not yet been associated with a human phenotype. Here, we collected families diagnosed with spondyloepimetaphyseal dysplasia (SEMD), and screened candidate pathogenic genes for families without known genetic causes using next-generation sequencing. We identified a monoallelic variant in signal peptide of CCN2 (NM_001901.2: c.65 G > C [p.Arg22Pro]) as the cause of SEMD in 14 subjects presenting with different degree of short stature, premature osteoarthritis, and osteoporosis. Affected subjects showed decreased serum CCN2 levels. Cell lines harboring the variant displayed decreased amount of CCN2 proteins in culture medium and an increased intracellular retention, indicating impaired protein secretion. And the variant weakened the stimulation effect of CCN2 on osteogenesis of bone marrow mesenchymal stem cells. Zebrafish ccn2a knockout model and osteoblast lineage-specific Ccn2-deficient mice (Ccn2fl/fl;Prx1Cre) partially recapitulated the phenotypes including low bone mass observed in affected subjects. Pathological mechanism implicated in the skeletal abnormality in Ccn2fl/fl;Prx1Cre mice involved decreased bone formation, increased bone resorption, and abnormal growth plate formation. Collectively, our study indicate that monoallelic variants in CCN2 lead to a human inherited skeletal dysplasia, and highlight the critical role of CCN2 in osteogenesis in human.
Subject terms: Bone, Metabolic bone disease
Introduction
Skeletal dysplasia is a group of human genetic disorders primarily involved musculoskeletal system.1 Spondyloepimetaphyseal dysplasia (SEMD) is among the most complicated subgroups of skeletal dysplasia, and is a collective term for clinically and genetically heterogeneous disorders marked by vertebral, epiphyseal, and metaphyseal abnormality.1,2 Bone deformity and premature osteoarthritis are usually the prominent clinical features of SEMD. In cases that SEMD is observed as a part of a syndrome, extraskeletal symptoms such as mental retardation, hypotonia, hypotrichosis, myopia, and atrophic scarring can also be predominant features. More than 20 subgroups of SEMD including Strudwick type, Aggrecan type, Matrilin type, Missouri type, and short limb-abnormal calcification type with different modes of inheritance have been described so far.1 No common molecular pathway has been identified yet in SEMD. Nevertheless, encoding proteins of a considerable part of established culprit genes are found to be components of extracellular matrix (ECM) or involved in processes of ECM metabolism.3–9 In these cases, variants in the pathogenic genes result in, for example, abnormal effects of protein on ECM, reduced protein secretion, or endoplasmic reticulum stress due to misfolded protein, leading to defects in the ECM structural framework or mediated biological signals necessary for skeletal growth and homeostasis.10,11 Substantial progresses have been made with respect to the genetic research in SEMD, however, the genetic heterogeneity as well as the paucity of families with multiple affected subjects have hindered the identification of new genetic causes for SEMD, hampering the molecular diagnosis and genetic counseling for a considerable number of affected families in clinical practice.
Cellular communication network factor 2 (CCN2), also known as connective tissue growth factor, is among established matricellular proteins, a group of non-structural proteins presented in ECM, and serves primarily regulatory roles.12–14 CCN2 was initially found to be mitogenic and chemotactic for fibroblast-like cells, and early studies were largely focused on its pathological significance in fibrotic disorders.15–17 In recent years, mounting evidence drawn from animal models and human patients have suggested the implication of aberrant CCN2 in a remarkable diversity of diseases besides organ fibrosis, such as diabetic nephropathy and retinopathy, carcinogenesis and tumor development, atherosclerosis, and arthritis, most of which are verified related to pathological processes of chronic inflammation or tissue injury.18,19 Emerging studies describe CCN2 as promising diagnostic or prognostic markers for these diseases.18 Moreover, potential therapeutic effect and safety of several drugs targeting CCN2 (i.e., FG-3019, OLX-10010, EXC-001) have been evaluated in a series of disorders, such as diabetic nephropathy, idiopathic pulmonary fibrosis, pancreatic cancer, and results drawn from these clinical trials are highly encouraging.20–23 In addition to the involvement of CCN2 in pathological conditions, it displays multiple physiological functions, and is involved in such as central nervous system development, spinal cord regeneration, and wound healing.17,19,24 Despite the many pivotal functions of CCN2 in both pathological and physiological situations, human phenotypes associated with CCN2 deficiency remain to be elucidated, and no human monogenic disorder has been associated with variants in CCN2 gene to date.
In present study, we collected families diagnosed with SEMD, and screened candidate pathogenic genes for families without known genetic causes using next-generation sequencing. Mechanism studies were further performed to explore underlying pathogenesis of the candidate genes.
Results
CCN2 variant shows complete cosegregation with clinical traits in a large family with SEMD
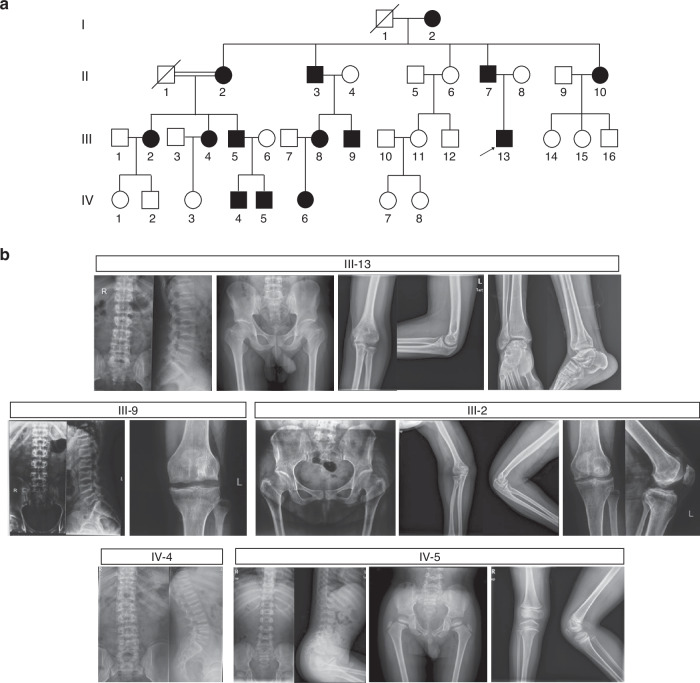
We detected a monoallelic variant (c.65 G > C, p.Arg22Pro) in CCN2 (NM_001901.2) in a four-generation Han Chinese family with an autosomal dominant form of SEMD, and a total of 14 subjects were affected (Fig. 1a). Affected individuals appeared asymptomatic at birth. The earliest changes were noticed at early childhood, and manifested as an abnormal walking or running gait. The majority of affected subjects suffered from limited flexion of knee, ankle, and elbow joint since their childhood. Joint pain and swelling in lower limbs developed around adolescence, gradually getting worse as they aged. The affected subjects also showed mild-to-severe disproportionate short stature with relative short lower limbs.25,26 Overt extraskeletal abnormalities were not observed. Radiographs prior to epiphyseal fusion displayed dysplastic epiphyses and irregular metaphyses of long bones, as well as bullet-shaped appearance of vertebral bodies with irregular epiphyses; while after epiphyseal fusion, exhibited platyspondyly with biconcave deformities of the central-posterior portion of vertebral bodies, irregular sclerosis of endplates, dysplasia acetabulum and proximal femur, and dysplasia of elbow and ankle joint with osteophyte formation, and in some subjects, showed obvious premature degenerative changes in weight-bearing joints that resembled radiographic features of osteoarthritis at elder ages, including decreased joint spaces, irregular articular surface, hyperostosis, subchondral cysts and bone sclerosis (Fig. 1b). Bone mineral density (BMD) measurement, which was qualified using dual-energy X-ray absorptiometry, showed osteoporosis or osteopenia in 6 of 8 adults.27 Detailed clinical features and radiographic findings were shown in Table 1, Table 2, and Supplementary Note. No significant abnormalities were detected in routine biochemical tests.
Fig. 1.
Pedigree and radiographical features of a large Chinese family with spondyloepimetaphyseal dysplasia (SEMD). a Pedigree of the family with SEMD. Squares and circles indicate males and females, respectively. Black symbols present affected individuals, and slashes deceased individuals. Arrow indicates the proband. b Radiographic abnormalities of subjects with SEMD. III-13: individual at age 25.7 years. Spine lateral shows platyspondyly and biconcave deformities of the central-posterior portion of vertebral bodies, with irregular sclerotic end plates. Pelvis shows mildly narrow iliac wings with irregular acetabulum and femoral head. Short and broad femoral necks are noted. Elbow and ankle joint show irregular proximal humerus and distal tibiae and fibulae, as well as hyperostosis. The joint spaces are preserved. III-9: individual at age 28.9 years. Platyspondyly with irregular sclerotic end plates, and biconcave vertebral bodies are noted. Knee joint shows mildly decreased joint spaces, irregular articular surface, osteophyte formation, together with sclerotic and cystic changes. III-2: individual at age 44.6 years. Femoral necks are short and broad, with irregular acetabuli and femoral head. Degenerative changes, including narrow joint cavity, hyperostosis, subchondral cysts and bone sclerosis are noted in hip, elbow and knee joints. IV-4: individual at age 15.1 years. Spine lateral shows flat vertebral bodies, with irregular epiphyses and mild scoliosis. IV-5: individual at age 4.6 years. Distinctive bullet-shaped appearance vertebral bodies and irregular epiphyses are noted. Pelvis and knee joint show irregular epiphyses and metaphyses of the long bones, together with acetabular abnormalities
Table 1.
Clinical features of subjects with the CCN2 variant
| Subject, Age (year)/Sexa | Waddling gait | Limited joint flexion | Limited joint extension | Joint swelling and pain | Height (SD)b/cm | Arm span/cm | U/L ratio | Head circumference/cm | Other |
|---|---|---|---|---|---|---|---|---|---|
| I-2, 80.2/F | + | N/A | N/A | + | 127.0 (−4.16) | N/A | N/A | N/A | kyphosis |
| II-2, 70.0/F | + | + | + | + | 144.0 (−1.38) | 152.0c | 1.0 | 55.0 | - |
| II-3, 67.2/M | + | N/A | N/A | + | 148.0 (−2.74) | N/A | N/A | N/A | N/A |
| II-7, 56.9/M | + | + | + | + | 145.0 (−3.43) | 158.5c | 1.1 | 56.0 | kyphosis, IJE, genu varum |
| II-10, 53.7/F | + | + | + | + | 140.1 (−2.71) | 148.0c | 1.1 | 55.5 | - |
| III-2, 44.6/F | + | + | - | + | 138.2 (−3.46) | 152.0 | 1.1 | 58.2 | genu valgum |
| III-4, 41.1/F | + | + | - | + | 140.2 (−3.11) | 149.0 | 1.1 | 56.5 | genu varum |
| III-5, 39.5/M | + | + | - | + | 154.7 (−2.26) | 165.5 | 1.1 | 59.0 | IJE |
| III-8, 33.3/F | + | + | - | + | 153.4 (−0.75) | 161.5 | 1.0 | 54.0 | joint stiffness, genu varum |
| III-9, 28.9/M | + | + | + | + | 161.6 (−1.15) | 177.0c | 1.0 | 58.0 | twice KAS |
| III-13, 25.7/M | + | + | - | + | 154.9 (−2.23) | 173.0 | 1.1 | 55.0 | - |
| IV-4, 15.1/M | + | + | - | + | 162.5 (−1.12) | 175.0 | 1.2 | 61.0 | DCF |
| IV-5, 4.6/M | + | - | - | - | 107.8 (0.02) | 106.5 | 1.4 | 53.0 | DCF |
| IV-6, 6.3/F | + | - | - | - | 118.6 (−0.17) | 120.5 | 1.1 | 49.0 | - |
F female, M male, SD standard deviation, U/L ratio upper segment/lower segment, IJE interphalangeal joint enlargement, KAS knee arthroscopic surgeries, DCF delayded closure of the fontanel, N/A not available, + denotes yes, – denotes unobserved
aRoman numerals indicate generation, and arabic numbers indicate the order of offspring within a generation
bSD, deviation from the mean height of the Chinese population25,26
cII-2, II-7, II-10, and III-9 have limited extension of elbow joints
Table 2.
Radiographic features and bone mineral density of subjects with the CCN2 variant
| Subject, Age (year)/ Sexa | Radiographic features | Bone mineral densityb | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Spine deformity | Abnormal metaphyses | Irregular epiphyses | OA like changes | Other | L1-4/ (g/cm2) | Z-scores/ T-scores | Neck/ (g/cm2) | Z-scores/ T-scores | Total/ (g/cm2) | Z-scores/ T-scores | |
| II-2, 70.0/F | platyspondyly | + | N/A | + | SFN | N/A | N/A | N/A | N/A | N/A | N/A |
| II-7, 56.9/M | platyspondyly | + | N/A | + | SFN | 0.734 | −2.4/−2.9 | 0.585 | −2.2/−3.0 | 0.615 | −2.6/−2.9 |
| II-10, 53.7/F | platyspondyly | + | N/A | + | SFN | 0.767 | −1.2/−2.9 | 0.689 | −0.6/−2.0 | 0.691 | −1.1/−2.2 |
| III-2, 44.6/F | platyspondyly, mild scoliosis | + | N/A | + | SFN | 0.875 | −2.4/−2.0 | 0.638 | −2.4/−2.4 | 0.756 | −1.8/−1.7 |
| III-4, 41.4/F | N/A | N/A | N/A | N/A | N/A | 0.842 | −1.7/−2.3 | 0.679 | −1.6/−2.1 | 0.800 | −0.9/−1.3 |
| III-5, 39.5/M | N/A | + | N/A | + | N/A | 0.701 | −2.7/−3.2 | 0.633 | −2.2/−2.7 | 0.678 | −2.3/−2.4 |
| III-8, 33.3/F | platyspondyly | + | N/A | - | - | 0.945 | −0.7/−1.4 | 0.640 | −1.9/−2.4 | 0.632 | −2.2/−2.6 |
| III-9, 28.9/M | platyspondyly, ISEP | + | N/A | + | SFN | 1.094 | 0.1/0.1 | 0.849 | −1.0/−1.0 | 0.893 | −0.8/−0.8 |
| III-13, 25.7/M | platyspondyly, ISEP | + | N/A | + | SFN | 1.053 | −0.3/−0.2 | 0.803 | −1.5/−1.3 | 0.829 | −1.3/−1.2 |
| IV-4, 15.1/M | platyspondyly, mild scoliosis | + | + | - | SFN | 0.861 | N/A | 0.797 | N/A | 0.798 | N/A |
| IV-5, 4.6/M | BSVB | + | + | - | IAPF | 0.416 | N/A | 0.576 | N/A | 0.553 | N/A |
| IV-6, 6.3/F | BSVB | + | + | - | IAPF | 0.574 | N/A | 0.537 | N/A | 0.467 | N/A |
F female, M male, OA osteoarthritis, L1-4 lumber spine 1–4, Neck femoral neck, Total total hip, ISEP irregular sclerotic end plates, BSVB bullet-shaped vertebral bodies, SFN short femoral necks, IAPF irregular acetabulum and proximal femur, N/A not available, + denotes yes, – denotes unobserved
aRoman numerals indicate generation, and arabic numbers indicate the order of offspring within a generation
bBone mineral density was qualified using dual-energy X-ray absorptiometry, and Z-scores/T-scores were calculated based on equipment-specific references27
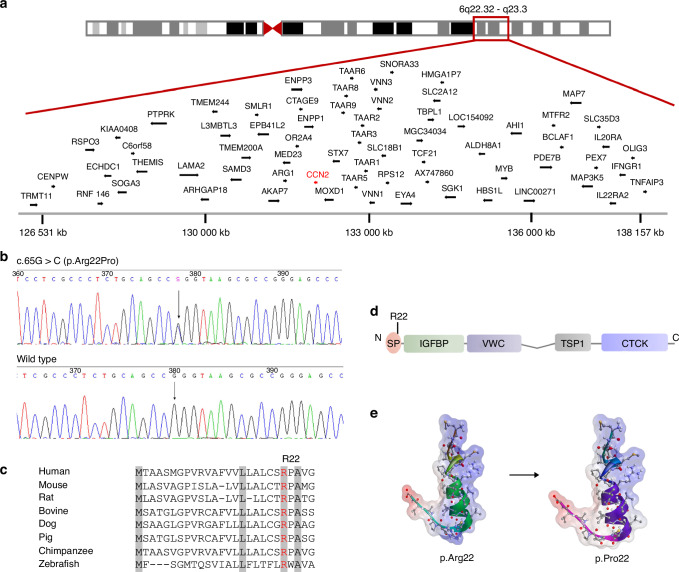
To identify genetic cause of the large Chinese family with SEMD, we used a combination of genome-wide linkage scan, whole-exome sequencing (WES), and Sanger sequencing. Genome-wide linkage analysis was first applied in 18 family members, including I-2, II-2, II-6, II-7, II-9, II-10, III-1, III-2, III-3, III-4, III-5, III-6, III-13, III-14, III-15, IV-2, IV-3, and IV-4. Linkage data was analyzed with a dominant model of inheritance, and revealed a single significant candidate region on chromosome 6q22.32-q23.3, flanked by markers kgp22768966 and rs9389536, with a maximum LOD score of 3.5 (Fig. 2a, Fig. S1). And we did not identify any genes known to cause SEMD in this region. WES was subsequently conducted in four affected (II-2, III-9, III-13, and IV-4) and two unaffected individuals (II-6 and III-15), and revealed 33 variants in 27 genes that were co-segregating with the clinical trait among these six subjects (Table S1). Meanwhile, eight variants were located in the linked region, and two variants of them, respective in CCN2 and FBXO30 gene, would potentially lead to coding-protein change. Sanger sequencing confirmed the existence of both variants, and primers used for PCR and direct sequencing are 5’-AGGTGGGGAGGAATGCGAGGA-3’ (forward), 5’-GCCGTCCAGCACGAGGCTCAC-3’ (reverse) for the variant in CCN2, and 5’-ATGGCTCTGAATTTGTGCAGT-3’ (forward), 5’-ACTGAGCGTAGTGAACGTCCT-3’ (reverse) for the variant in FBXO30. However, only the variant (c.65 G > C, p.Arg22Pro) in CCN2 (NM_001901.2) showed complete cosegregation with clinical traits in all available family members (n = 28), and was considered to be the excellent candidate causative variant for the disorder (Fig. 2b). To our knowledge, the variant in CCN2 is absent in current public databases of human sequence variants, including the 1000 Genomes Project, the Single Nucleotide Polymorphism database, the ESP, and the ExAC database, and it is also absent in the 750 control samples (1 500 alleles) enrolled in our study. The amino acid Arg22 residue is located in the signal peptide of CCN2, and is highly conserved across various species (Fig. 2c, d). Structural analysis showed elevated folding free energy leading to conformational changes in the mutant CCN2. Meanwhile, decreased isoelectric point of the signal peptide due to the mutant residue was indicated, which would probably impair the process of protein secretion (Fig. 2e).
Fig. 2.
Identification of a pathogenic variant in CCN2 in the family with spondyloepimetaphyseal dysplasia (SEMD). a Schematic representation of chromosome 6 and a single overlapping region on chromosome 6q22.32–q23.3 identified by linkage analysis. b Sequencing chromatogram of a missense variant (c.65 G > C, arrow) in exon 1 in CCN2. The variant shows segregation with SEMD in all available family members. The base change predicts the replacement of the amino acid arginine in codon 22 with proline (p.Arg22Pro). c Alignments of the region of CCN2 from multiple species, showing the altered residue in red. Highly conserved amino acids are indicated in gray. d Schematic diagrams of protein domain organization of CCN2, along with the position of the identified variant. The molecular structure of CCN2 consists of a signal peptide (SP) and four conserved domains with sequence homologies to Insulin-like growth factor binding protein (IGFBP), von Willebrand factor type C domain (VWC), Thrombospondin type 1 repeat (TSP-1), and a carboxy-terminal cystine knot domain (CTCK). The amino acid residue Arg22 is located in the SP. e Structural model of SP of CCN2. Positive and negative charged regions are indicated in blue and red, respectively, and uncharged area is shown in white. The mutant residue Arg22 results in a decrease in positive charge region
CCN2 variant impairs CCN2 protein secretion and serum CCN2 levels in affected subjects are decreased
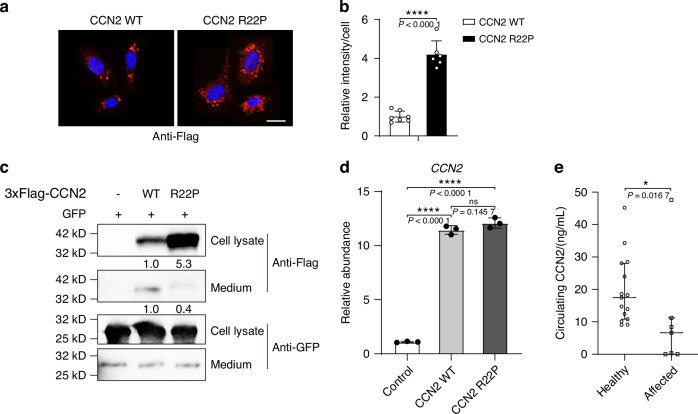
To assess whether the variant affect CCN2 secretion, we overexpressed Flag-tagged human wild-type or mutant (p.Arg22Pro) CCN2 in cell lines using plasmid vector, and respectively collected the culture medium and cells after transfection of 48 h. We detected a lower amount of CCN2 protein in the culture medium while an increased intracellular retention of the protein in cells transfected with the mutant CCN2, using immunofluorescence and western blot analysis, compared with cells transfected with the wild-type (Fig. 3a–c). We then performed RT-PCR to detect the levels of CCN2 mRNA, and found that the mutant transfected cells had similar increased levels of CCN2 mRNA with those of wild-type transfected cells (Fig. 3d). These findings indicated that the reduced secretion of CCN2 protein was most likely attributed to the detrimental effect of the variant on protein secretion. Consistent with the discoveries in the cell lines, serum CCN2 levels of affected subjects were significantly lower than those of healthy controls (Fig. 3e).
Fig. 3.
CCN2 R22P variant impairs CCN2 protein secretion. a, b Immunofluorescence analyses of intracellular distribution of Flag-tagged hCCN2 protein in C3H10T1/2 cells infected with plasmids encoding Flag-tagged wild-type or mutant hCCN2 (p.Arg22Pro) after 48 h. Scale bar = 10 μm. Data are represented as mean ± SD (n = 7 for wild-type, n = 6 for mutant). Student’s t test was used. ****P < 0.000 1. c Western blot analysis of Flag-tagged hCCN2 protein abundance in the cell lysates and culture medium of HEK293T cells transfected with indicated plasmids after 48 h. The relative expression of hCCN2 was monitored with the use of monoclonal antibody to Flag, showing bands of the expected size (~40 kD). d RT-PCR of total mRNA of CCN2 from HEK293T cells transfected with indicated plasmids after 48 h. Data are represented as mean ± SD, normalized by the housekeeping Gapdh. Student’s t test was used. ****P < 0.000 1; ns, not significant. e Serum CCN2 levels in affected subjects with SEMD (n = 7) and healthy controls (n = 15), data are represented as median with interquartile range. Mann–Whitney test was used. *P < 0.05
CCN2 variant weakens its stimulation effect on osteogenesis of bone marrow mesenchymal stem cells
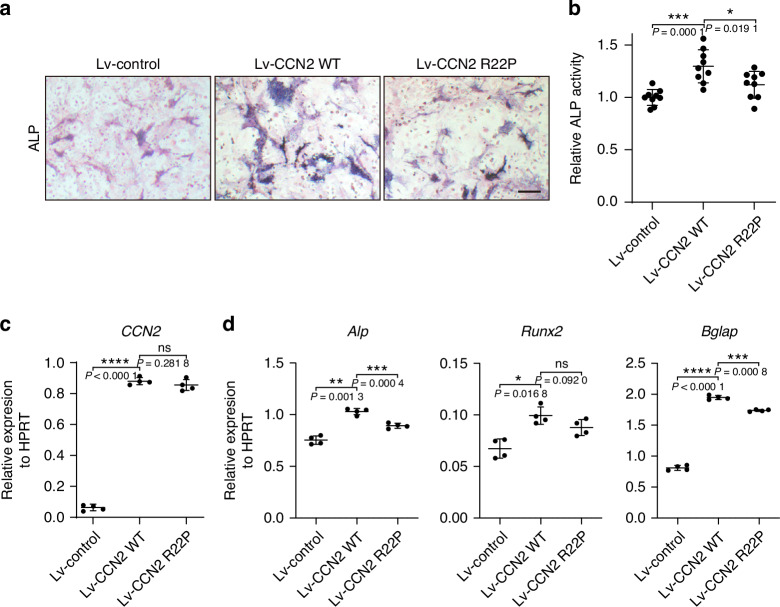
To further clarify the functional consequence of CCN2 variant, we overexpressed human wild-type or mutant CCN2 in mice bone marrow mesenchymal stem cells (BMSCs) using lentivirus. As shown in Fig. 4a, b, the osteoblast differentiation of the mutant CCN2 infected BMSCs were lower than those of wild-type infected cells, as determined by the ALP staining and activity quantification. Although the expression level of CCN2 of mutant infected cells increased to a similar extent to those of wild-type infected cells (Fig. 4c), the osteoblast differentiation marker genes Alp, Runx2, and Bglap increased to a less extent (Fig. 4d). These results indicated that CCN2 played a role in promoting osteoblast differentiation, and the identified variant in CCN2 reduce its effectiveness in this process.
Fig. 4.
CCN2 variant weakens its stimulation effect on osteogenesis of bone marrow mesenchymal stromal cells (BMSCs). a ALP staining results of osteoblast differentiation of BMSCs infected with indicated lentivirus overexpressing wild-type or mutant hCCN2 (p.Arg22Pro), and ZsGreen lentivirus was used as a control (n = 9). Scale bar = 10 μm. b ALP quantification results of BMSCs infected with indicated lentivirus at day 7. Data are represented as mean ± SD. Student’s t test was used (n = 9). *P < 0.05; ***P < 0.001. RT-PCR analysis of CCN2 (c) and Alp, Runx2, Bglap (d) expression at day 7 during BMSCs differentiation. Data are represented as mean ± SD. Student’s t test was used (n = 4). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.000 1; ns, not significant
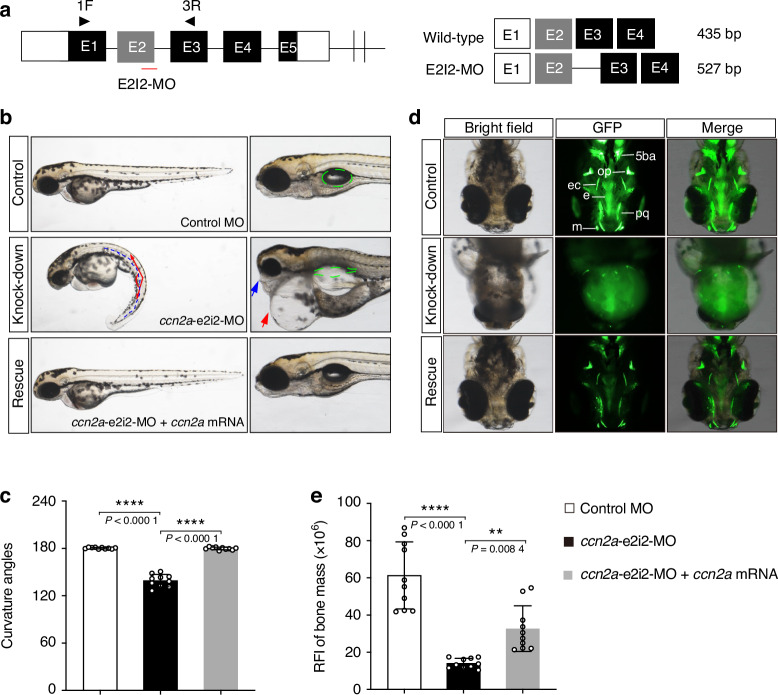
Ccn2a knockdown zebrafish show severe craniofacial skeleton dysplasia and decreased bone mass, and are rescued by ccn2a mRNA overexpression
Zebrafish exhibit a simple cartilage and bone development pattern which is most highly conserved among all vertebrates during evolution. Genetic studies have revealed similarities in developmental mechanisms underlying skeletal morphogenesis between zebrafish and human. These findings make zebrafish an excellent model to study molecular basis of cartilage and bone development.28,29 The protein sequence of zebrafish homolog, ccn2a, is 78.7% identical to human CCN2 protein. Therefore, we designed and validated specific morpholino oligonucleotides (MO) to knockdown the ccn2a in zebrafish to characterize the functional consequences of CCN2 deficiency in skeletal development (Fig. 5a, Fig. S2). The exon 2-intron 2 splice ccn2a MO (ccn2a-e2i2-MO) or the standard control MO was microinjected into fertilized one-cell stage embryos. And the zebrafish injected with ccn2a-e2i2-MO showed overt gross morphologic abnormalities at 2 days post-fertilization (dpf), including remarkable curved body axis, jaw defects, small eyes, pericardial oedema, and non-inflated swim bladder (Fig. 5b, c, Fig. S3). To further visualize the skeletal structures in zebrafish embryos, we performed calcein staining.30 At 5-dpf, fluorescent signals of well-developed craniofacial cartilage structures became apparent, and the ccn2a-e2i2-MO injected larvae showed severe dysplasia of craniofacial skeleton (Fig. 5d, Fig. S3). The craniofacial defects included the absence of Meckel’s cartilage, palatoquadrate, and fifth branchial arch, and the dysmorphic opercular bone, ectopterygoid, and ethmoid plate. The amount of stained mineralized tissue in mutant morphants was markedly reduced comparing with that of control, and quantification of the relative fluorescence intensity (RFI) of head skeleton bone mass showed significant decrease in ccn2a morphants (Fig. 5e). To confirm the skeleton dysplasia in ccn2a-e2i2-MO injected zebrafish were specially caused by ccn2a knockdown, we performed rescue experiments by co-injecting ccn2a-e2i2-MO with ccn2a mRNA, and observed that overexpression of ccn2a mRNA significantly rescued the skeleton malformation in ccn2a morphants (Fig. 5d, e). These findings suggest that ccn2a is essential for skeletal development in zebrafish.
Fig. 5.
Overexpression of ccn2a mRNA rescues morphological abnormalities and craniofacial phenotypes in ccn2a knockdown zebrafish. a Zebrafish ccn2a gene is targeted by specific morpholino antisense to prevent proper splicing of exon 2 (E2I2-MO). Primers 1 F and 3 R interrogate the presence of wild type transcripts or those in which intron 2 has been inserted. The diagram is a schematic depiction of the intron 2 inserted transcript in the E2I2-MO injected embryos (527 bp) as compared with control MO injected embryos (435 bp). b Representative lateral views of zebrafish at 2 days post-fertilization (dpf) injected with either control MO or ccn2a MO with or without ccn2a mRNA. The ccn2a morphants display curved body axis (blue dotted line), small eyes, pericardial oedema (red arrow), jaw defects (blue arrow), and non-inflated swim bladder (green circled area). Co-injection of nonmutant zebrafish ccn2a mRNA rescue the gross morphology in ccn2a morphants. c Quantification of the average curvature angle of zebrafish in each group at 2-dpf. Data are represented as mean ± SD (n = 10). One way ANOVA with Bonferroni’s post hoc multiple comparisons was used. ****P < 0.000 1. d Representative ventral view of the craniofacial skeleton of zebrafish in each group at 5-dpf labeled with calcein. 5ba, fifth branchial arch; op, opercular bone; ec, ectopterygoid; e, ethmoid plate; pq, palatoquadrate; m, Meckel’s cartilage. Co-injection of nonmutant zebrafish ccn2a mRNA significantly rescued head skeletal malformation in ccn2a morphants. e Quantification of the relative fluorescence intensity (RFI) of craniofacial skeleton bone mass of zebrafish in each group at 5-dpf (n = 10). Data are represented as mean ± SD. One way ANOVA with Bonferroni’s post hoc multiple comparisons was used. **P < 0.01; ****P < 0.000 1
Osteoblast lineage-specific Ccn2-deficient mice present with decreased bone mass
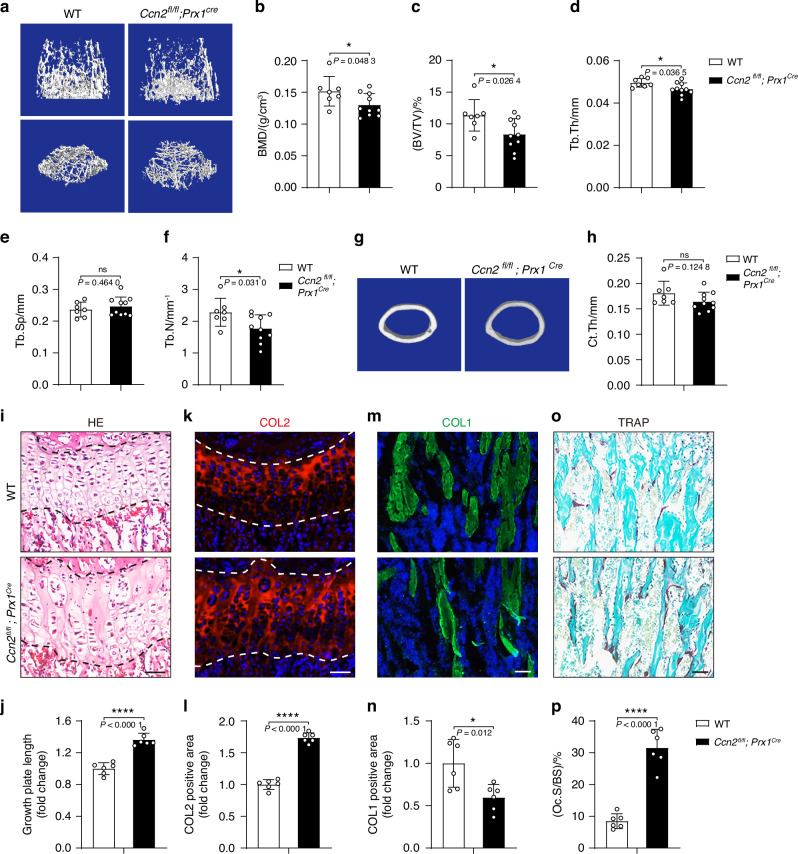
To understand the underlying molecular mechanism for the skeletal phenotypes of affected subjects with CCN2 deficiency, we determined whether the defects in skeletal development were caused by requirement of CCN2 in early mesenchymal progenitor cells. We knockout CCN2 in mesenchymal progenitors in mice (Ccn2fl/fl;Prx1Cre) by crossing Ccn2fl/fl mice with Prx1Cre mice, and the latter is a transgenic line that expressed Cre recombinase in a subset of early limb bud mesenchyme.31 To clarify the effect of CCN2 in skeletal system, we performed micro-CT to analyze the limbs isolated from the 6-week-old Ccn2fl/fl;Prx1Cre mice and corresponding control littermates. The Ccn2fl/fl;Prx1Cre mice displayed decreased femoral trabecular bone mass relative to their control littermates (Fig. 6a–h), with a reduction in BMD, bone volume fraction (BV/TV), thickness (Tb.Th) and number (Tb.N). And the measurements including trabecular separation (Tb.Sp) and cortical thickness (Ct.Th) of Ccn2fl/fl;Prx1Cre mice were similar to age-matched control littermates. To further understand how CCN2 deletion in mesenchymal progenitors caused the bone abnormality, we performed morphological analysis of femurs from 6-week-old mice. Hematoxylin and eosin (H/E) staining displayed distorted and enlarged of growth plate as shown in Fig. 6i, j. And immunofluorescence analysis for cartilage anabolism markers showed an expansion of Col2-positve area (Fig. 6k, l), and the number of the Col1α1-positive area were reduced in the Ccn2fl/fl;Prx1Cre mice (Fig. 6m, n). And the bone resorption as detected through TRAP staining showed an increased TRAP-positive area (Fig. 6o, p). These results suggested the contributions of cartilage and bone formation defects and increased bone resorption to bone abnormality in Ccn2fl/fl;Prx1Cre mice. With transcriptome sequencing of BMSCs from wild-type and Ccn2-deficient mice, we performed pathway analysis with the differentially expressed genes (DEGs) and found that the down-regulated genes were enriched in biological pathways like multicellular organism development, collagen fibril organization, skeletal system development, and limb joint morphogenesis (Fig. S4). The RNA-seq data provide additional support suggesting that CCN2 deficiency contributed to the skeletal deformities.
Fig. 6.
Deletion of CCN2 in mesenchymal progenitor cells leads to decreased bone mass. a–f Micro-CT image of trabecular bones of distal femurs isolated from 6-week-old wild-type and Ccn2fl/fl;Prx1Cre mice, and analysis for bone mineral density (BMD), bone volume per tissue volume (BV/TV), trabecular thickness (Tb.Th), trabecular spacing (Tb.Sp), and trabecular number (Tb.N). Data are represented as mean ± SD (n = 7 for wild-type, n = 10 for Ccn2fl/fl;Prx1Cre). Student’s t test was used. *P < 0.05; ns, not significant. g, h Micro-CT image of cortical bones of mid diaphysis of femurs isolated from 6-week-old mice, and analysis for cortical thickness (Ct.Th). Data are represented as mean ± SD (n = 7 for wild-type, n = 10 for Ccn2fl/fl;Prx1Cre). Student’s t test was used. ns, not significant. i, j Histology of growth plate in femurs of 6-week-old mice. The sections are stained with H/E. Scale bar = 50 μm. Data are represented as mean ± SD (n = 6). Student’s t test was used. **** P < 0.000 1. k, l Immunofluorescence analysis of Col2 of growth plate in femurs of 6-week-old mice. Scale bar = 50 μm. Data are represented as mean ± SD (n = 6). Student’s t test was used. ****P < 0.000 1. m, n Immunofluorescence analysis of Col1 in femurs of 6-week-old mice. Scale bar = 50 μm. Data are represented as mean ± SD (n = 6). Student’s t test was used.*P < 0.05. o, p TRAP staining in femurs of 6-week-old mice. Scale bar = 50 μm. Data are represented as mean ± SD (n = 6). Student’s t test was used. ****P < 0.000 1
Discussion
Herein we performed genetic analysis in SEMD families without known genetic causes, and reported a monoallelic missense variant in CCN2 (c.65 G > C, p.Arg22Pro) as the genetic cause for an autosomal dominant form of SEMD in 14 subjects. In silico analyses showed that the variant was located in the signal sequence of CCN2 and resulted in elevated folding free energy and decreased isoelectric point of the signal peptide. And the variant showed a deleterious effect on CCN2 processing and secretion, and was potentially implicated in impaired osteogenesis. We further generated zebrafish ccn2a knockout model and osteoblast lineage-specific Ccn2-deficient mice (Ccn2fl/fl;Prx1Cre) that recapitulated the defective skeleton phenotypes observed in SEMD subjects to a certain extent, especially the phenotype of low bone mass.
CCN2 is a secretory protein consisting of an N-terminal signal peptide and four conserved modules, and is mainly secreted as an extracellular protein and interacts with a number of cell surface receptors, ECM components, and cytokines, fulling the common features as other matricellular proteins.17–19 Signal peptide of a secretory protein is generally short sequences in the amino terminus of newly synthesized proteins and direct the protein into the cell secretory pathway, and typically consists of three distinct and featured domains: a positively charged N-terminal region (n-region, 1–5 residues), a hydrophobic core (h-region, 7–15 residues), and a polar C-terminal region containing cleavage site (c-region, 3-7 residues).32,33 During the process of protein secretion, signal peptide is recognized by a targeting factor named signal recognition particle (SRP), and directs to the endoplasmic reticulum membrane for translocation, and the signal sequence is cleaved off at the luminal side of the endoplasmic reticulum membrane, and matured protein is finally transported outside the cell through Golgi.32,33 Several human mendelian metabolic bone disorders have been reported connected with variants in the signal sequences of pathogenic genes, such as CTSK for Pycnodysostosis,34,35 PTH for Hypoparathyroidism,36 and COL10A1 for Schmid metaphyseal chondrodysplasia.37 And the pathogenic basis of the variants in signal sequences are mainly connected with defects in the secretion of mutant protein through secretory pathway or pathological degradation of the mutant mRNA through a pathway named Regulation of Aberrant Protein Production (RAPP), and these two different consequences are demonstrated to be related to the position of the variant in signal sequence.38 It is indicated that the n-region plays roles in the protein docking and translocation, the h-region is the target for the SRP, and c-region influences the efficiency and accuracy of signal peptide cleavage.32,33 Therefore, variants in the n-region can interfere with protein translocation; variants in the h-region tend to prevent the interaction with SRP, triggering pathologically activation RAPP pathway and following mRNA degradation; variants in the c-region often prevent cleavage of the signal sequence, affecting protein secretion.38 In this study, the identified variant (c.65 G > C, p.Arg22Pro) is located in the signal peptide of CCN2. In silico analyses show that the signal sequence of CCN2 consists of 26 amino acids, and the variant is located in its c-region, and elevated folding free energy and decreased isoelectric point of the signal peptide due to the variant are implicated. We find a decreased secretion and an intracellular retention of CCN2 proteins in mutant transfected cells, with the CCN2 mRNA levels similar to those of wild-type transfected cells. These indicate the potential deleterious effect of the variant on protein processing and secretion. As CCN2 mainly acts as a secretory matricellular protein in ECM, the impaired process of protein secretion is most likely responsible for the disease. Limited evidence had ever indicated that CCN2 might display some intracellular function via endocytic pathways and thus affect gene transcription in the nucleus,39 therefore, further studies are required to exclude the possible existence of some unidentified intracellular function resulting from accumulated mutant CCN2, which may be also involved in the pathogenic mechanisms underlying the disorder. Due to the discovery that CCN2 can be cleaved into different fragments due to the presenting of multiple cleavage sites between different modules, and different fragments of CCN2 have been indicated with different biological roles,39,40 thus the human phenotypes associated with variants in different modules remain to be revealed in the future.
We observe defective skeletal development among the subjects with the CCN2 variant, including mild-to-severe disproportionate short stature with relative short lower limbs, limited joint flexion, premature osteoarthritis-like changes in weight-bearing joints, and low bone mass. CCN2 is among the group of regulatory proteins in ECM in various organs, with skeleton among the major expression sites during development.41 Previous in vivo studies demonstrated that CCN2 was strongly expressed in osteoblast lining metaphyseal trabeculae, an active osteogenesis site during the period of bone modeling.42 Through overexpressing CCN2 in BMSCs, we find that CCN2 caused an increase in ALP activity and increased mRNA levels of osteoblast-specific markers, suggesting that CCN2 lead to an enhanced osteoblast differentiation. This finding was consistent with retroviral vectors mediated CCN2 overexpression in ST-2 stromal cells, which also resulted in an increase in osteoblastogenesis.43 And the essential roles of CCN2 in MSCs osteogenesis was supported by findings of in vitro studies of MSCs with CCN2 knockout, which resulted in disruption in MSCs colony formation, inhibited MSCs proliferation, and enhanced adipocytes differentiation.44 We show that mutant CCN2 infected BMSCs have decreased osteogenesis compared with those of wild-type, indicating that impaired osteogenesis owing to the CCN2 variant is probably involved in the pathological mechanism underlying the SEMD. Our in vivo studies in zebrafish and mice models further provide evidence that CCN2 is required for skeletal development. Ccn2a-null zebrafish exhibit remarkable craniofacial skeleton dysplasia and decreased bone mass, and the abnormalities are reversed by overexpression of ccn2a mRNA. The zebrafish has recently emerged as a popular model for skeletal research due to the conservation of cell types in cartilage and bone, as well as the genetic pathways regulating their development between zebrafish and humans.28,29 Nevertheless, in zebrafish models of human disease, there is not always a one-to-one relationship in terms of the affected anatomical compartment or the degree of phenotypic severity, with potential mechanisms remaining to be elucidated.29,45 In present study, we did not observe notable dysmorphic facial phenotypes such as a prominent forehead, depressed nasal bridge, prominent cheekbone, cleft palate, or small jaw, as found in other certain types of SEMD, among affected subjects with CCN2 variant. However, delayed anterior fontanelle closing was observed in IV-5, and IV-4, a 15.1 year old boy, had a head circumference of 61.0 cm (normal range in adult: 54.0–58.0 cm). Currently, it remains challenging to determine whether the above findings are coincidental and to identify the potential clinical significance of our findings without more similar cases.
CCN2 has been demonstrated to display important roles in many cell types during the process of endochondral ossification and intramembranous ossification,13,14 two distinct processes of bone formation during embryogenesis in mammalian.46 Here, we also provide evidence that the requirement of CCN2 in early mesenchymal progenitor cells is potentially contributing to the molecular mechanism whereby CCN2 deficiency cause phenotypes observed in subject with SEMD. Ccn2 deletion in mesenchymal progenitors in mice (Ccn2fl/fl;Prx1Cre), partially recapitulate the clinical findings in SEMD, cause decreased bone mass. Moreover, it was reported that the Ccn2+/− mice also presented with osteopenia with a 30%–45% reduction in BV/TV,47 showing consistent phenotypes observed in Ccn2fl/fl;Prx1Cre mice in present study. Through the immunofluorescence analysis with Col1α1, we observe decreased bone formation in metaphyseal trabeculae in Ccn2fl/fl;Prx1Cre mice, and it is in line with the findings identified in vitro studies with MSCs. As the decreased bone mass can be the result of decreased bone formation, increased bone resorption, or a combination of both.48 We further examine the alternation in bone resorption using TRAP staining, and Ccn2fl/fl;Prx1Cre mice display an increase TRAP-positive area, indicating that the bone loss is also associated with an increase in bone resorption. It was previously demonstrated that Ccn2 knock out mice have fewer osteoclasts in the metaphyseal region.49 As we knock out Ccn2 in early mesenchymal progenitor cells in present study, the increase in TRAP-positive area was suspected as a secondary alternation. The underlying mechanism of the discrepant findings and of the uncoupling of bone formation and bone resorption in Ccn2fl/fl;Prx1Cre mice require further studies. Cartilage anabolism in growth plate determined by Col2 level through immunofluorescence analysis show that Col2-marked region is expanded in Ccn2fl/fl;Prx1Cre mice. These results suggest that the pathological mechanism underlying the skeletal phenotypes of Ccn2fl/fl;Prx1Cre mice involve decreased bone formation, increased bone resorption and abnormal cartilage formation. Bioinformatics analysis reveals that these down-regulated genes are enriched in some biological pathways, including multicellular organism development, collagen fibril organization, skeletal system development, and limb joint morphogenesis. The pathways enriched by KEGG pathway enrichment include cGMP-PKG signaling pathway, cAMP signaling pathway, both of which play important roles in skeletal remodeling;50 PI3K-Akt signaling pathway has been reported to regulate BMP2-induced MSCs osteogenic differentiation,51 promote fracture healing,52 and is essential for joint tissue metabolism and involved in the development of osteoarthritis;53 ECM-receptor interaction is also common in skeletal formation and regeneration through ECM dynamic interaction with osteoblasts and osteoclasts to regulate the new bone formation.54 And the protein-protein interaction network analysis of DEGs enrich in collagen fibril organization and skeletal system development. These data provide further evidence suggesting that CCN2 deficiency contributed to the skeletal deformities. However, it is noted that both the two mice models (Ccn2fl/fl;Prx1Cre mice and Ccn2+/−mice) display normal femoral length, and no obvious skeletal abnormalities except for the decreased bone mass are detected. Meanwhile, we do not observe obvious abnormalities in skeletal development or homeostasis in mice harboring the identified CCN2 variant (data not shown). CCN family have six members, and the remaining five CCN proteins have been found to be expressed during the process of bone formation and to have exhibiting similar or opposing roles to CCN2.13,55,56 The differences in skeletal phenotypic spectrum and severity due to CCN2 deficiency between genetic mouse models and human indicated the existence of differences in functional redundancy of CCN family members or other compensatory mechanisms for CCN2 deficiency in mice versus human. Alternatively, potential existence of differences in threshold effects of CCN2 deficiency for skeletal dysplasia between between mouse models and human remain to be clarified. The hypothesis of redundant or opposing roles of CCN proteins in skeletal development in mice is further supported by the finding that mice with CCN6 (formerly WISP3 [MIM: 603400]) knockout, the gene known to cause human progressive pseudorheumatoid dysplasia that is characterized by joint abnormalities, do not exhibit overt skeletal phenotypes.57,58 Mouse models are connected to the study of the functions of causative gene involved in human monogenic disease. However, these models may only replicate some aspects of the human disease phenotype, exhibit more severe phenotype, or have no clinical phenotype at all.59,60 Translating phenotypes across different species can be challenging due to potential differences in biology, genetic background, alternative pathways, or compensation mechanism.61–65 While the exact reasons for differences in skeletal phenotype due to CCN2 deficiency between species require further investigation, the detected discrepancies prompt us to look into the possible effects of modifier genes, alternative pathways, possible interacting proteins, and synergistic effects on the pathogenesis of SEMD, which will contribute to our understanding of biology with respect to the similarities and differences between species. In addition, despite that CCN2 has been demonstrated to display many critical functions in various physiological processes,14,17,19 affected subjects with CCN2 deficiency specifically exhibit skeletal abnormalities, and have no history of evident abnormalities in other organs, these indicating probable different sensitivities between skeleton and extra-skeletal organs to CCN2 deficiency in human. Although the comprehensive roles of CCN2 in human skeletal development remains to be further clarified, defective skeletal development owing to CCN2 deficiency is consistent with our understanding of pathogenesis underlying the SEMD.
In conclusion, present study therefore establish CCN2 as a pathogenic gene to SEMD spectrum, and uncover the human phenotypes causally associated with CCN2 deficiency. Given the process of skeletal development is potentially sensitive to CCN2 dosage reduction, the evaluation of skeleton alternations in subjects with CCN2-target therapies is potentially necessary in clinical settings. Further studies are required and expected to determine whether modulating CCN2 could provide a new therapeutic approach for osteoporosis.
Materials and methods
Study approval
The human study was conducted in accordance with the principles of the Declaration of Helsinki, and was approved by the Ethics Committee of the Shanghai Jiao Tong University of Medicine Affiliated Sixth People’s Hospital. Written informed consent was received from participants or parents of minors prior to inclusion in the study. The zebrafish experiments were approved by the Institutional Animal Care and Use Committee of Shanghai Research Center for Model Organisms. And the zebrafish facility is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) International. The mice were bred and maintained under specific pathogen free conditions in the institutional animal facility of the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. All mice experiments were conducted in accordance with a protocol approved by the Animal Care and Use Committee of Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences.
Clinical and genetic analysis
Human subjects and clinical analysis
We investigated probands with SEMD and their available family members. All individuals underwent a thorough clinical inquiry, physical examination, biochemical and bone-densitometry analyses, and skeletal radiography. The diagnosis of SEMD was based on clinical manifestations and radiological findings. In addition, 750 healthy unrelated Chinese subjects were enrolled as controls.
Genome-wide linkage analysis
Genomic DNA was extracted and purified from peripheral blood leukocytes with traditional methods. Genotyping of the samples was performed using Illumina’s GenomeStudio Genotyping Module, with data collected according to Infinium HD Assay Super protocol. Multipoint parametric linkage analysis and haplotyping for a dominant inheritance model with complete penetrance were conducted by the MERLIN program, version 1.1.2. Putative protein-coding genes within candidate region were examined using UCSC Genome Browser on human Feb. 2009 (GRCh37/hg19) assembly (http://genome.ucsc.edu/).
Whole-exome sequencing
Qualified DNA sample was fragmented, ligated to adapters, amplified by ligation-mediated PCR, and hybridized to exome array for enrichment. Agilent 2100 Bioanalyzer and quantitative PCR were used to estimate the magnitude of enrichment of captured ligation-mediated PCR products. High-throughput sequencing were conducted for each qualified capture library loaded on Illumina Hiseq platforms. The derived data were then processed by Illumina base-calling Software for base-calling with default parameters, and were generated as paired-end reads. High quality reads were mapped to the human reference genome GRCh37 with Burrows-Wheeler Aligner (BWA V0.7.15), and duplicate reads were removed by Picard-tools (v2.5.0). Genome Analysis Toolkit (GATK) was used for local realignment around InDels and base quality score recalibration following best practices. HaplotypeCaller of GATK (v3.3.0) was applied for detection of genomic variation including SNPs and InDels, and high-confident variant calls were obtained using hard-filtering methods. SnpEff tool was conducted for a series of annotations for variants. Variants with an allele frequency >0.1% in the 1 000 Genomes Project database (http://www.1000genomes.org/), the ESP6500 (http://evs.gs.washington.edu/EVS/), and the ExAC database (http://exac.broadinstitute.org/) were excluded.
Sanger sequencing
The variants in candidate pathogenic genes, identified by combination of linkage analysis and WES, was confirmed by Sanger sequencing following PCR amplification in all available family members. Amplificated PCR products were purified with shrimp alkaline phosphatase (Promega) and exonuclease I (Epicentre). Sequencing reaction was performed using BigDye Terminator cycle sequencing ready reaction kit, version 3.1 (Applied Biosystems), and the products were detected and analyzed using an ABI Prism 3130 automated sequencer (Applied Biosystems) and Polyphred.
Cell culture
Mice BMSCs were isolated from tibia and femur bone marrow cavity of 4-week-old C57BL/6 mice: bone marrow was flushed out with pre-cooled PBS, followed by centrifugation and lysis of erythrocytes. The cell precipitates were gently resuspended and cultured in α-MEM supplemented with 10% FBS and 1% penicillin/streptomycin. HEK293T cells and C3H10T1/2 cells were purchased from the American Cell Type Culture Collection (ATCC), and HEK293T cells were cultured in DMEM containing 10% FBS and 1% penicillin/streptomycin, C3H10T1/2 cells were cultured in α-MEM containing 10% FBS and 1% penicillin/streptomycin. All the cells were cultured in a 5% CO2 humidified incubator at 37 °C.
Transfection and infection
Phage vector overexpressing Flag-tagged full-length human mutant CCN2 (p.Arg22Pro) or wild-type CCN2 were constructed. HEK293T cells were transfected with the wild-type or mutant vector together with pLenti-EGFP according to the Effectene Transfection protocol (QIAGEN, 301425), with pLenti-EGFP as a control. For lentivirus vector construction, we overexpressed ZsGreen, Flag-tagged full-length human mutant CCN2 or wild-type CCN2 using a pLenti vector (pLenti-hCCN2-3xFlag-ZsGreen-Puro). Lentivirus packaging was conducted according to the VSVG-delta 8.9 system. BMSCs or C3H10T1/2 were infected with lentivirus for 24 h, and treated with puromycin for 48 h. The ZsGreen lentivirus was used as a control.
In vitro osteoblast differentiation
For in vitro osteoblast differentiation, BMSCs treated with lentivirus were induced by α-MEM with 10% FBS, 1% penicillin/streptomycin, 50 μg/mL L-ascorbic acid (Sigma, A5960), and 1.08 mg/mL β-Glycerophosphate disodium salt hydrate (Sigma, G9422). On day 7, BMSCs were fixed with 4% paraformaldehyde (PFA) for 10 min and subjected to ALP staining according to the manual of BCIP/NBT Alkaline Phosphatase Color Development Kit (Beyotime, C3206). For ALP activity quantitative analysis, osteoblasts were incubated with Alamar Blue (Invitrogen) for 1 h and read with a luminometer (Envision) for cell number quantification. Then the osteoblasts were incubated with phosphatase substrate (Sigma, S0942) dissolved in the solution (6.5 mmol/L Na2CO3, 18.5 mmol/L NaHCO3, 2 mmol/L MgCl2) for 20 min. The ALP activity was read with a luminometer (Envision) at A405, and normalized as A405/Alamar Blue.
Protein secretion analysis
For CCN2 secretion analysis, HEK293T cells were transfected with indicated plasmids. Cells were harvested after 48 h and cells were lysed with 500 μL EBC buffer (50 mmol/L Tris, pH 7.5, 120 mmol/L NaCl, and 0.5% NP-40) supplemented with a protease inhibitor cocktail (MCE, HY-K0010) and centrifuged at 12 000 r/min. Culture medium was collected into 15 mL centrifuge tube and centrifuged at 2 000 r/min. Cell lysates and medium were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted with primary antibody against Flag (Sigma, F3165-5MG, 1:5 000) or GFP (Proteintech, 66002-1-Ig, 1:5 000) overnight at 4°C and secondary antibody of anti-mouse-immunoglobulins/HRP (Dako, P0260). After incubation with specific antibodies, we used enhanced chemiluminescence kit (Millipore) to detect the protein signals. Our method of quantification is to do a ratio between the destination bands and their matching internal reference bands after quantification. The ratio of each destination band is then normalized and presented below each destination band.
Immunofluorescence
For cell immunofluorescence analysis, C3H10T1/2 cells infected with indicated plasmids after 48 h were fixed in 4% PFA for 15 min and blocked in PBS with 3% horse serum and 0.3% Triton X-100 (Sangon Biotech, A110694-0500) for 30 min, then incubated with mouse-anti-Flag (Sigma, F3165-5MG, 1:200) overnight at 4 °C. Donkey anti-mouse-Cy3 (Jackson ImmunoResearch, 715-165-150, 1:1 000) was used as secondary antibody. DAPI (Sigma, D8417, 1:1 000) was used for counterstaining. Cells were mounted with anti-fluorescence mounting medium (Dako, S3023) and images were captured with a Leica SP8 confocal microscope.
RT-PCR analysis
Total RNA of cells was isolated in Trizol (Invitrogen) and reverse transcribed into cDNA using Transcriptor First strand cDNA synthesis kit (Roche). RT-PCR analysis was conducted with FastStart Universal SYBR Green Master (Roche) in a LightCycler480 PCR system (Roche). Relative expression of target genes was normalized to Gapdh or Hprt, and was calculated with delta-delta CT method. The primer used are listed as follows: hCCN2-qF, 5’-AAGGGCAAAAAGTGCATCCG-3’; hCCN2-qR, 5’-TCTTCTTCATGACCTCGCCG-3’; mGAPDH-qF: 5’-TTCCTACCCCCAATGTGTCC-3’; mGAPDH-qR, 5’-GGTCCTCAGTGTAGCCCAAG-3’; mAlp-qF, 5’-CGGGACTG GTACTCGGATAA-3’;mAlp-qR, 5’-ATTCCACGTCGGTTCTGTTC-3’; mCcn2-qF, 5’-CTGCAGGCTAGAGAAGCAGAG-3’; mCcn2-qR, 5’-GATGCACTTTTTGCCCTTCT-3’; mRunx2-qF, 5’-CCAACCGAGTCATTTAAGGCT-3’; mRunx2-qR, 5’-GCTCACGTCGCTCATCTTG-3’; mBglap-qF, 5’-GCAGCACAGGTCCTAAATAG-3’; mBglap-qR, 5’-GGGCAATAAGGTAGTGAACAG-3’; mHprt-qF, 5’-GTTAAGCAGTACAGCCCCAAA-3’; mHprt-qR, 5’-AGGGCATATCCAACAACAAACTT-3’.
Human serum CCN2 levels measurement
CCN2 protein levels in human serum were determined using Elisa kits (DRG Instruments GmbH, EIA-5195) following the manufacturer’s instructions. The intra-assay coefficients of variation (CVs) for CCN2 were 1.3% at a level of 110.1 ng/mL, 2.6% at a level of 49.9 ng/mL, 3.9% at a level of 129.7 ng/mL. The inter-assay CVs for CCN2 were 9.7% at a level of 27.2 ng/mL, 9.8% at a level of 15.0 ng/mL, 10.6% at a level of 31.0 ng/mL, 6.2% at a level of 32.2 ng/mL.
CCN2 knock-down studies in Zebrafish
Zebrafish care, maintenance, and microinjections
Adult wild-type zebrafish (AB strain) were maintained at 28.5 °C on a 14 h light/10 h dark cycle,66 and 5 to 6 pairs of them were set up for nature mating every time. The generated embryos were maintained at 28.5 °C in fish water (0.2% Instant Ocean Salt in deionized water), and were washed and staged according to previous studies.67 For ccn2a gene knock-down experiment, the ccn2a-e2i2-MO (5’-AACAGCCAAGATCCTTACCTGTGCA-3’) and the standard control MO (5’-CCTCTTACCTCAGTTACAATTTATA-3’), was designed (Gene Tools, http://www.gene-tools.com/) and microinjected into fertilized one-cell stage embryos.68 RT-PCR analysis and Sanger sequencing were performed to confirm the efficacy of the ccn2a-e2i2-MO. Total RNA of 30 to 50 embryos per group was extracted and reverse transcribed into cDNA. The primers of ccn2a spanning exon 1 and exon 3 were 5’-CTCTGCTGTTCCTGACTTTCTT-3’ (forward) and 5’-ACTTCCCAGGCACTTTCAC-3’ (reverse). The primers of ef1α, used as the internal control, were 5’-GGAAATTCGAGACCAGCAAATAC-3’ (forward) and 5’-GATACCAGCCTCAAACTCACC-3’ (reverse). For rescue experiments, 4 ng ccn2a-e2i2-MO was co-injected with 100 pg pcDNA3.1 containing nonmutant zebrafish ccn2a cDNA per embryo. The coding region of the wild-type zebrafish ccn2a was synthed by Sangon Biotech and subcloned into pcDNA3.1 vector (Invitrogen).
Calcein staining and image acquisition
Zebrafish at 5-dpf were firstly washed with fish water and immersed in 0.2% calcein solution for 10 min. And the zebrafish were then rinsed thoroughly in fish water again and anaesthetized with 0.016% MS-222 (tricaine methanesulfonate, Sigma-Aldrich). Finally, the zebrafish were oriented on ventral side and mounted with 3% methylcellulose (Sigma-Aldrich) in a depression slide for observation by fluorescence microscopy (Nikon SMZ 1500) and subsequently photographed with digital cameras. Qualification of the RFI of head skeleton bone mass was performed using morphometric analysis (NIS-Elements D3.1). To optimally visualize the expression patterns, a subset of images was adjusted for levels, brightness, contrast, hue and saturation (Adobe Photoshop 7.0 software). A total of 10 zebrafish for each group were quantified and averaged.
Ccn2fl/fl;Prx1Cre mice model analysis
Mice model generation
Ccn2fl/fl mice bearing loxP sites flanking exons 1–5 of the Ccn2 gene were provided by GemPharmatech (Nanjing, China). The Prx1Cre mice strain was a gift from Andrew McMahon. Ccn2fl/fl mice were crossed with Prx1Cre mice to generate Ccn2fl/fl;Prx1Cre mice. All mice analyzed were maintained on the C57BL/6 background.
Micro-CT analysis
Left femurs isolated from age- and sex-matched 6-week-old Ccn2fl/fl;Prx1Cre mice and control littermates were fixed in 70% ethanol and scanned using SkyScan-1176 micro-computed tomography (μCT) (Bruker micro CT, Belgium) system. Scans were performed using 8.96 μm voxel size, 50 KV, 500 μA and 0.4 degrees rotation step (180 degrees angular range). The 1.6 version of NRecon software (Bruker) was used for three-dimensional (3D) reconstruction and viewing of images, and 3D photos of bones were performed using 3.3.0 version of CTvox software (Bruker). After 3D reconstruction, the 1.13 version of CTan software (Bruker) was used for bone analysis, and the segmentation were range from 85/120 to 255/255. For trabecular bone, micro-CT evaluation was performed on a 2 mm region of metaphyseal spongiosa in the distal femur, located 0.5 mm above the growth plate. For cortical bone, measurements were performed on a 0.5 mm region of the mid diaphysis of the femur. Trabecular bone measurements included BMD, BV/TV, Tb.Th, Tb.N and Tb.Sp. Cortical bone measurements included Ct.Th.
Histological staining, immunofluorescence, and immunohistochemistry
Femur and tibiae bones from mice were fixed with 4% PFA for 48 h at 4 °C, decalcified in 10% EDTA for 2 weeks, dehydrated in alcohol, cleared with xylene, and embedded in paraffin. Each sample was sectioned sagittally using microtome (Lexica Microsystems Nussle GmbH) at a thickness of 8 μm for staining. H/E staining were performed using a standard protocol. For TRAP staining, sections were stained with the Leukocyte Acid Phosphatase Kit (Sigma, 387A-1KT) for 1 h at 37 °C following the manufacturers’ instructions. For immunofluorescence, sections were blocked in PBS with 10% horse serum and 0.3% Triton X-100 (Sangon Biotech, A110694-0500) for 1 h and then stained overnight at 4 °C with rabbit-anti-Collagen II (Abcam, ab34712) or rabbit-anti-Collagen I (Abcam, ab21286). Secondary antibodies were used according to the species of the primary antibody, and DAPI (Sigma, D8417) was used for counter staining. Slides were mounted with anti-fade fluorescence mounting medium (Dako, S3023). Images were acquired with microscope (Olympus BX51, Tokyo, Japan).
RNA sequencing and analysis
We collected femurs and tibias of wild-type and Ccn2-deficient mice, removed the surrounding soft tissues, and isolated BMSCs: bone marrow was flushed out with pre-cooled PBS, followed by centrifugation and lysis of erythrocytes. The cell precipitates were gently resuspended in culture medium (α-MEM supplemented with 10% FBS and 1% penicillin/streptomycin), seeded (2 × 106 cells per dish) in 100-mm culture dishes, and incubated at 37 °C under 5% CO2 conditions. After 48 h, nonadherent cells were washed away with PBS, adherent cells were cultured for another 5 days. We collected the cells and obtained RNA for transcriptome sequencing (fold change greater than 2.0 and P value less than 0.05). The data were analyzed on the free online platform of Majorbio Cloud Platform (www.majorbio.com).
Statistics
Statistical analysis was performed using the GraphPad Prism 9 software (GraphPad Software). Cell-based experiments were performed at least twice. Animals were randomized into different groups and at least 3 mice were used for each group, unless otherwise stated. All quantitative data are presented as mean ± SD. Student’s t test, one-way ANOVA, and Mann–Whitney test was used, as appropriate, for statistical evaluations of group comparisons. Statistical significance is indicated by *, where P < 0.05, **, where P < 0.01, ***, where P < 0.001, ****, where P < 0.000 1, and all tests were two-sided.
Supplementary information
BONERES-03199R3. Revised Supplementary Appendix_Shanshan Li
Acknowledgements
We are very grateful to all the subjects for their excellent collaboration. We acknowledge technical assistance from Shanghai Genesky Biotechnology, and Shenzhen Huada Gene Research Institute. This work was funded by the National Key Research and Development Program of China (2018YFA0800801), the National Basic Research Program of China (2014CB942903), the National Natural Science Foundation of China (81900807), Shanghai Research Center for Endocrine and Metabolic Diseases (2022ZZ01002).
Author contributions
Z.Z. and W.Z. designed the study. S.L. (Shanshan Li) and S.L. (Shufa Li) obtained and managed clinical data from study subjects. R.S., S.L. (Shanshan Li), J.Z., Q.D., P.L., Z.W., and S.X. performed and analyzed functional assays. Z.Z., B.L., and S.L. (Shanshan Li) conducted sequencing data analyses. Z.Z., W.Z., B.L., and L.C. provided critical guidance for functional researches and genetic analyses. Z.Z. supervised the studies. S.L. (Shanshan Li) and R.S. wrote the manuscript. All authors contributed to the final version of the manuscript.
Data availability
Data that support the findings of this study are present in the paper and/or the Supplementary Materials, and additional data related to this paper are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Shanshan Li, Rui Shao, Shufa Li
Contributor Information
Weiguo Zou, Email: zouwg94@sibcb.ac.cn.
Zhenlin Zhang, Email: zhangzl@sjtu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41413-024-00364-2.
References
- 1.Unger, S. et al. Nosology of genetic skeletal disorders: 2023 revision. Am. J. Med. Genet. A.191, 1164–1209 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cormier-Daire, V. Spondylo-epi-metaphyseal dysplasia. Best Pract. Res. Clin. Rheumatol.22, 33–44 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Tiller, G. E. et al. Dominant mutations in the type II collagen gene, COL2A1, produce spondyloepimetaphyseal dysplasia, Strudwick type. Nat. Genet.11, 87–89 (1995). [DOI] [PubMed] [Google Scholar]
- 4.Tompson, S. W. et al. A recessive skeletal dysplasia, SEMD aggrecan type, results from a missense mutation affecting the C-type lectin domain of aggrecan. Am. J. Hum. Genet.84, 72–79 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman, K. L. et al. Mutations in the region encoding the von Willebrand factor A domain of matrilin-3 are associated with multiple epiphyseal dysplasia. Nat. Genet.28, 393–396 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Kennedy, A. M. et al. MMP13 mutation causes spondyloepimetaphyseal dysplasia, Missouri type (SEMD(MO). J. Clin. Invest.115, 2832–2842 (2005). [DOI] [PMC free article] [PubMed]
- 7.Bargal, R. et al. Mutations in DDR2 gene cause SMED with short limbs and abnormal calcifications. Am. J. Hum. Genet.84, 80–84 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egunsola, A. T. et al. Loss of DDRGK1 modulates SOX9 ubiquitination in spondyloepimetaphyseal dysplasia. J. Clin. Invest.127, 1475–1484 (2017). [DOI] [PMC free article] [PubMed]
- 9.Cho, S. Y. et al. BGN mutations in X-linked spondyloepimetaphyseal dysplasia. Am. J. Hum. Genet.98, 1243–1248 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bateman, J. F., Boot-Handford, R. P. & Lamande, S. R. Genetic diseases of connective tissues: cellular and extracellular effects of ECM mutations. Nat. Rev. Genet.10, 173–183 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Krishnan, Y. & Grodzinsky, A. J. Cartilage diseases. Matrix Biol.71–72, 51–69 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murphy-Ullrich, J. E. & Sage, E. H. Revisiting the matricellular concept. Matrix Biol.37, 1–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giusti, V. & Scotlandi, K. CCN proteins in the musculoskeletal system: current understanding and challenges in physiology and pathology. J. Cell. Commun. Signal.15, 545–566 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu, M., Peng, D., Lan, T., Wei, Y. & Wei, X. Multifunctional regulatory protein connective tissue growth factor (CTGF): a potential therapeutic target for diverse diseases. Acta Pharm. Sin. B.12, 1740–1760 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonnylal, S. et al. Selective expression of connective tissue growth factor in fibroblasts in vivo promotes systemic tissue fibrosis. Arthritis Rheum.62, 1523–1532 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brigstock, D. R. Connective tissue growth factor (CCN2, CTGF) and organ fibrosis: lessons from transgenic animals. J. Cell. Commun. Signal.4, 1–4 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramazani, Y. et al. Connective tissue growth factor (CTGF) from basics to clinics. Matrix Biol.68–69, 44–66 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Jun, J. I. & Lau, L. F. Taking aim at the extracellular matrix: CCN proteins as emerging therapeutic targets. Nat. Rev. Drug. Discov.10, 945–963 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubota, S. & Takigawa, M. Cellular and molecular actions of CCN2/CTGF and its role under physiological and pathological conditions. Clin. Sci.128, 181–196 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Adler, S. G. et al. Phase 1 study of anti-CTGF monoclonal antibody in patients with diabetes and microalbuminuria. Clin. J. Am. Soc. Nephrol.5, 1420–1428 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raghu, G. et al. FG-3019 anti-connective tissue growth factor monoclonal antibody: results of an open-label clinical trial in idiopathic pulmonary fibrosis. Eur. Respir. J.47, 1481–1491 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Richeldi, L. et al. Pamrevlumab, an anti-connective tissue growth factor therapy, for idiopathic pulmonary fibrosis (PRAISE): a phase 2, randomised, double-blind, placebo-controlled trial. Lancet Respir. Med.8, 25–33 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Shen, Y. W., Zhou, Y. D., Luan, X. & Zhang, W. D. Blocking CTGF-mediated tumor-stroma interplay in pancreatic cancer. Trends Mol. Med.26, 1064–1067 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Mokalled, M. H. et al. Injury-induced ctgfa directs glial bridging and spinal cord regeneration in zebrafish. Science354, 630–634 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang, X. G. et al. Study on weight and height of the Chinese people and the differences between 1992 and 2002. Zhonghua. Liu. Xing. Bing. Xue. Za. Zhi.26, 489–493 (2005). [PubMed] [Google Scholar]
- 26.Li, H., Ji, C., Zong, X. & Zhang, Y. Height and weight standardized growth charts for Chinese children and adolescents aged 0 to 18 years. Zhonghua. Er. Ke. Za. Zhi.47, 487–492 (2009). [PubMed] [Google Scholar]
- 27.Zeng, Q. et al. The prevalence of osteoporosis in China, a Nationwide, Multicenter DXA Survey. J. Bone Miner. Res.34, 1789–1797 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Dietrich, K. et al. Skeletal biology and disease modeling in Zebrafish. J. Bone Miner. Res.36, 436–458 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Kwon, R. Y., Watson, C. J. & Karasik, D. Using zebrafish to study skeletal genomics. Bone126, 37–50 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du, S. J., Frenkel, V., Kindschi, G. & Zohar, Y. Visualizing normal and defective bone development in zebrafish embryos using the fluorescent chromophore calcein. Dev. Biol.238, 239–246 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Logan, M. et al. Expression of Cre recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis.33, 77–80 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Owji, H. et al. A comprehensive review of signal peptides: structure, roles, and applications. Eur. J. Cell. Biol.97, 422–441 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Gutierrez Guarnizo, S. A. et al. Pathogenic signal peptide variants in the human genome. NAR Genom. Bioinform.5, lqad093 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donnarumma, M. et al. Molecular analysis and characterization of nine novel CTSK mutations in twelve patients affected by pycnodysostosis. Hum. Mutat.28, 524 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Fujita, Y. et al. Novel mutations of the cathepsin K gene in patients with pycnodysostosis and their characterization. J. Clin. Endocrinol. Metab.85, 425–431 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Sunthornthepvarakul, T., Churesigaew, S. & Ngowngarmratana, S. A novel mutation of the signal peptide of the preproparathyroid hormone gene associated with autosomal recessive familial isolated hypoparathyroidism. J. Clin. Endocrinol. Metab.84, 3792–3796 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Chan, D., Ho, M. S. & Cheah, K. S. Aberrant signal peptide cleavage of collagen X in Schmid metaphyseal chondrodysplasia. Implications for the molecular basis of the disease. J. Biol. Chem.276, 7992–7997 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Tikhonova, E. B., Karamysheva, Z. N., von Heijne, G. & Karamyshev, A. L. Silencing of aberrant secretory protein expression by disease-associated mutations. J. Mol. Biol.431, 2567–2580 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leguit, R. J., Raymakers, R. A. P., Hebeda, K. M. & Goldschmeding, R. CCN2 (Cellular Communication Network factor 2) in the bone marrow microenvironment, normal and malignant hematopoiesis. J. Cell. Commun. Signal.15, 25–56 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abd El Kader, T. et al. The regenerative effects of CCN2 independent modules on chondrocytes in vitro and osteoarthritis models in vivo. Bone59, 180–188 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Friedrichsen, S. et al. CTGF expression during mouse embryonic development. Cell. Tissue Res.312, 175–188 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Safadi, F. F. et al. Expression of connective tissue growth factor in bone: its role in osteoblast proliferation and differentiation in vitro and bone formation in vivo. J. Cell. Physiol.196, 51–62 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Smerdel-Ramoya, A., Zanotti, S., Deregowski, V. & Canalis, E. Connective tissue growth factor enhances osteoblastogenesis in vitro. J. Biol. Chem.283, 22690–22699 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Battula, V. L. et al. Connective tissue growth factor regulates adipocyte differentiation of mesenchymal stromal cells and facilitates leukemia bone marrow engraftment. Blood122, 357–366 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willems, B. et al. The Wnt co-receptor Lrp5 is required for cranial neural crest cell migration in Zebrafish. PLoS. One10, e0131768 (2015). [DOI] [PMC free article] [PubMed]
- 46.Salhotra, A., Shah, H. N., Levi, B. & Longaker, M. T. Mechanisms of bone development and repair. Nat. Rev. Mol. Cell. Biol.21, 696–711 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Canalis, E., Zanotti, S., Beamer, W. G., Economides, A. N. & Smerdel-Ramoya, A. Connective tissue growth factor is required for skeletal development and postnatal skeletal homeostasis in male mice. Endocrinology151, 3490–3501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dirckx, N., Moorer, M. C., Clemens, T. L. & Riddle, R. C. The role of osteoblasts in energy homeostasis. Nat. Rev. Endocrinol.15, 651–665 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ivkovic, S. et al. Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development. Development130, 2779–2791 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim, S. M., Yuen, T., Iqbal, J., Rubin, M. R. & Zaidi, M. The NO-cGMP-PKG pathway in skeletal remodeling. Ann. N. Y. Acad. Sci.1487, 21–30 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao, S. et al. PI3K-Akt signaling regulates BMP2-induced osteogenic differentiation of mesenchymal stem cells (MSCs): a transcriptomic landscape analysis. Stem. Cell. Res.66, 103010 (2023). [DOI] [PubMed] [Google Scholar]
- 52.Dong, J., Xu, X., Zhang, Q., Yuan, Z. & Tan, B. The PI3K/AKT pathway promotes fracture healing through its crosstalk with Wnt/β-catenin. Exp. Cell. Res.394, 112137 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Sun, K. et al. The PI3K/AKT/mTOR signaling pathway in osteoarthritis: a narrative review. Osteoarthr. Cartil.28, 400–409 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Alcorta-Sevillano, N., Macías, I., Infante, A. & Rodríguez, C. I. Deciphering the relevance of bone ECM signaling. Cells9, 2630 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kawaki, H. et al. Differential roles of CCN family proteins during osteoblast differentiation: involvement of Smad and MAPK signaling pathways. Bone49, 975–989 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Kawaki, H. et al. Cooperative regulation of chondrocyte differentiation by CCN2 and CCN3 shown by a comprehensive analysis of the CCN family proteins in cartilage. J. Bone Miner. Res.23, 1751–1764 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hurvitz, J. R. et al. Mutations in the CCN gene family member WISP3 cause progressive pseudorheumatoid dysplasia. Nat. Genet.23, 94–98 (1999). [DOI] [PubMed] [Google Scholar]
- 58.Kutz, W. E., Gong, Y. & Warman, M. L. WISP3, the gene responsible for the human skeletal disease progressive pseudorheumatoid dysplasia, is not essential for skeletal function in mice. Mol. Cell. Biol.25, 414–421 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Elsea, S. H. & Lucas, R. E. The mousetrap: what we can learn when the mouse model does not mimic the human disease. Ilar. J.43, 66–79 (2002). [DOI] [PubMed] [Google Scholar]
- 60.Pound, P. & Ritskes-Hoitinga, M. Is it possible to overcome issues of external validity in preclinical animal research? Why most animal models are bound to fail. J. Transl. Med.16, 304 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thysen, S., Luyten, F. P. & Lories, R. J. Targets, models and challenges in osteoarthritis research. Dis. Model. Mech.8, 17–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jänne, P. A. et al. Functional overlap between murine Inpp5b and Ocrll may explain why deficiency of the murine ortholog for OCRL1 does not cause Lowe syndrome in mice. J. Clin. Invest.101, 2042–2053 (1998). [DOI] [PMC free article] [PubMed]
- 63.Raben, N., Nagaraju, K., Lee, E. & Plotz, P. Modulation of disease severity in mice with targeted disruption of the acid alpha-glucosidase gene. Neuromuscul. Disord.10, 283–291 (2000). [DOI] [PubMed] [Google Scholar]
- 64.Ohshima, T. et al. alpha-Galactosidase a deficient mice: a model of Fabry disease. Proc. Natl. Acad. Sci. USA94, 2540–2544 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leslie, N. D., Yager, K. L., McNamara, P. D. & Segal, S. A mouse model of galactose-1-phosphate uridyl transferase deficiency. Biochem. Mol. Med.59, 7–12 (1996). [DOI] [PubMed] [Google Scholar]
- 66.Westerfield, M. (eds) The Zebrafish Book: A Guide for the Laboratory use of Zebrafish (Oregon Univ. Press, 2000).
- 67.Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B. & Schilling, T. F. Stages of embryonic development of the zebrafish. Dev. Dyn.203, 253–310 (1995). [DOI] [PubMed] [Google Scholar]
- 68.Nasevicius, A. & Ekker, S. C. Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet.26, 216–220 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BONERES-03199R3. Revised Supplementary Appendix_Shanshan Li
Data Availability Statement
Data that support the findings of this study are present in the paper and/or the Supplementary Materials, and additional data related to this paper are available from the corresponding author upon reasonable request.