Abstract
Professional phagocytes like neutrophils and macrophages tightly control what they consume, how much they consume, and when they move after cargo uptake. We show that plasma membrane abundance is a key arbiter of these cellular behaviors. Neutrophils and macrophages lacking the G-protein subunit Gβ4 exhibited profound plasma membrane expansion, accompanied by marked reduction in plasma membrane tension. These biophysical changes promoted the phagocytosis of bacteria, fungus, apoptotic corpses, and cancer cells. We also found that Gβ4-deficient neutrophils are defective in the normal inhibition of migration following cargo uptake. Sphingolipid synthesis played a central role in these phenotypes by driving plasma membrane accumulation in cells lacking Gβ4. In Gβ4 knockout mice, neutrophils exhibited enhanced phagocytosis of inhaled fungal conidia in the lung but also increased trafficking of engulfed pathogens to other organs. Taken together, these results reveal an unexpected, biophysical control mechanism central to myeloid functional decision-making.
One-sentence summary:
Plasma membrane abundance controls myeloid cell phagocytosis, migration, and in vivo responses against fungal pathogens.
INTRODUCTION
Professional phagocytes of the myeloid lineage, including neutrophils and macrophages, maintain homeostasis by clearing apoptotic corpses, cellular debris, and invading pathogens (1–4). Phagocytes take up cargo via phagocytosis, an evolutionarily conserved engulfment process by which cells surround objects and then internalize them. The importance of phagocytosis for proper multicellular physiology is highlighted by the consequences of its dysregulation: insufficient levels of phagocytic activity increase susceptibility to infection and aging, while both excessive and inadequate levels have been linked to autoimmunity, neurodegeneration, and atherosclerosis (5–9). Understanding the molecular pathways and cellular components that control phagocytosis will be critical for mitigating these conditions and also for harnessing professional phagocytes as immunotherapeutic agents to combat infections, cardiovascular disease, and cancer (10–13).
Phagocytosis is triggered by the recognition of cognate cargo ligands, such as phosphatidylserine, complement, and antibodies, by specific receptors on the phagocyte (6). Target recognition elicits dramatic remodeling of the cytoskeleton, which shapes the overlying plasma membrane into a phagocytic cup that embraces and then internalizes the cargo (14, 15). In carrying out their clearance function, myeloid phagocytes exhibit not only robust cargo uptake but also the ability to coordinate this activity with other cellular behaviors (16–20). Crosstalk between phagocytosis and cell motility is particularly well-established, with studies documenting an antagonistic relationship between cell migration and engulfment responses in multiple cell types (16–18). In neutrophils specifically, transient arrest following phagocytosis is thought to curtail the dissemination of intracellular microbes (21–23). Accordingly, efforts to modulate the activity of professional phagocytes will require a mechanistic understanding of not only phagocytosis itself but also its regulatory effects on other activities.
Most prior research on phagocytosis has focused on the biochemical mechanisms that control it, and as a result, much is now known about the molecules that mediate cargo recognition and the signal transduction pathways that drive cargo engulfment (24). Phagocytosis is also an intensely physical process (14, 25), implying that it might be subject to biophysical as well as biochemical modes of regulation. In that regard, it is interesting that the deformability of both cargo and underlying substrate have been shown to modulate the engulfment behavior of macrophages (26–31). Whether the architecture and mechanics of the phagocyte itself might also influence cargo uptake is not known, however, and how cell-intrinsic properties of this kind might affect functional crosstalk between phagocytosis and other cellular behaviors is completely unexplored.
The plasma membrane is a particularly interesting candidate mechanoregulator because it must be stretched to build a phagocytic cup and to accommodate increased cellular volume after engulfment (14). Prior work indicates that phagocytosis is both constrained by membrane tension and enhanced by the exocytosis of endomembranes (32–34), implying that the amount of plasma membrane available to a cell might establish its phagocytic capacity. In the present study, we demonstrate that loss of a specific heterotrimeric G-protein subunit, Gβ4, strongly increases the plasma membrane abundance of myeloid cells, leading to enhanced phagocytosis and impaired cross-regulation between phagocytosis and migration. This cellular phenotype markedly alters neutrophil function in vivo, driving enhanced consumption of microbes but also increased trafficking away from the site of infection after pathogen uptake. Hence, the functional potential of immune cells is dictated by their biophysical properties and architectural constraints.
RESULTS
Gβ4 deficiency enhances phagocytic responses against a wide range of targets
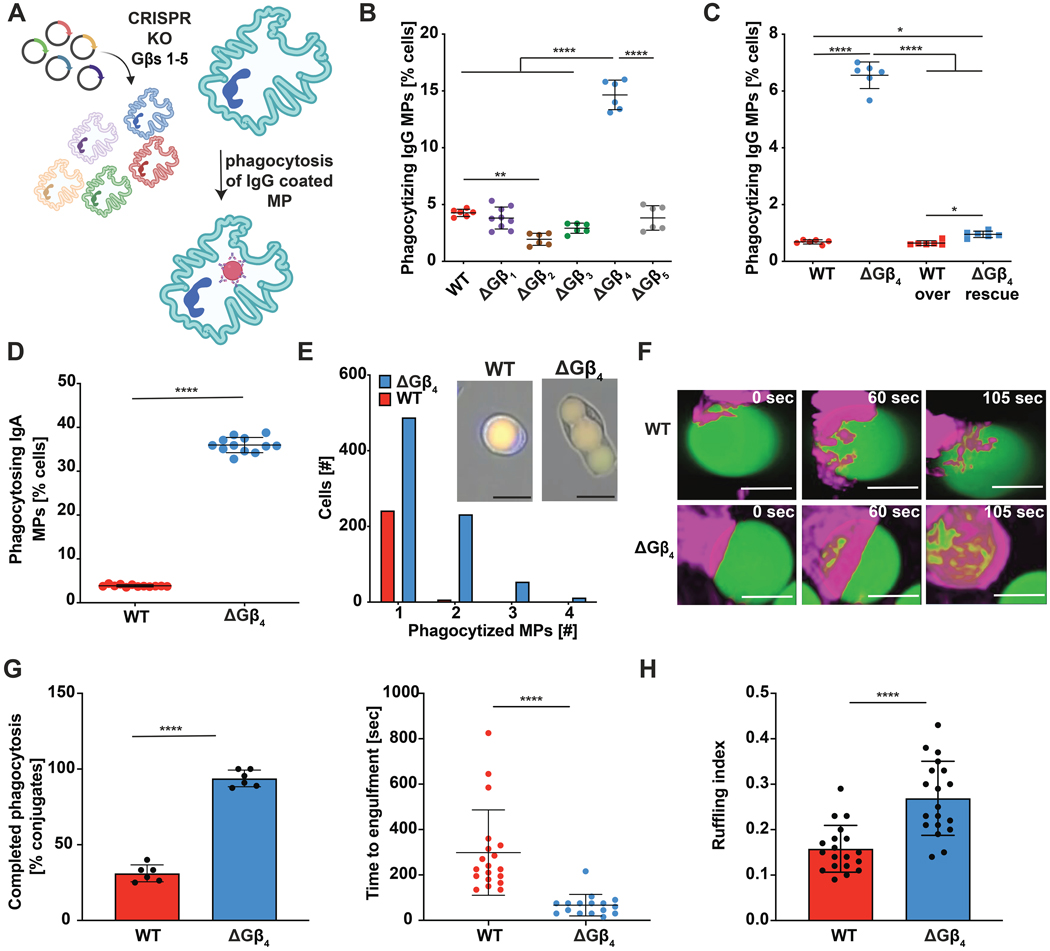
Heterotrimeric G-proteins regulate phagocytosis and migration in multiple myeloid cell types by transducing signals from seven-transmembrane G-protein coupled receptors (GPCRs) (35–42). GPCR activation induces dissociation of the G-protein α subunit (Gα) from the βγ subcomplex (Gβγ), freeing both components to bind and activate specific downstream effector molecules (43). Most prior research in this area has focused on Gα isoforms, while comparatively less is known about specific Gβ and Gγ subunits (44). Accordingly, we applied CRISPR-Cas9 to knock out each Gβ subunit in human neutrophil-like HL-60 cells (Fig. 1A and fig. S1A and B). Gene targeting was carried out in self-renewing HL-60 precursors, which were subsequently differentiated into neutrophil-like cells (called HL-60 neutrophils hereafter) by the addition of DMSO. All terminally differentiated cells were CD15+ CD16+ (fig. S1C), indicating that the genetic modifications we introduced did not prevent HL-60 progenitors from becoming neutrophils. Knockout and control HL-60 neutrophils were then subjected to phagocytosis assays to investigate potential effects on phagocytosis. As cargo for these experiments, we prepared 10 μm diameter IgG-coated polyacrylamide microparticles (MPs) (45, 46) derivatized with two fluorescent dyes: fluorescein isothiocyanate (FITC) and lissamine rhodamine B (LRB) (Fig. 1A and fig. S2). FITC, but not LRB, fluorescence is quenched in acidic phagolysosomes, leading to an emission ratio change that can be monitored by flow cytometry and fluorescence microscopy. In this manner, we identified Gβ subunits that either positively or negatively regulated phagocytosis. Consistent with prior work (38, 39), HL-60 neutrophils lacking Gβ2 exhibited a ~50% reduction in particle uptake (Fig. 1B). By contrast, depletion of Gβ4 induced a striking 3- to 4-fold increase in phagocytosis relative to wild type (WT) controls (Fig. 1B). This hyperphagic behavior resulted directly from loss of Gβ4, as re-expression of Gβ4 on the knock-out background restored phagocytosis to WT levels (Fig. 1C, fig. S3A). The Gβ4 knockout (ΔGβ4) phenotype was also cell-intrinsic, as it was readily apparent in both monocultures as well as cocultures containing a 1:1 mixture of WT and ΔGβ4 HL-60 neutrophils (Fig. 1D). Notably, phagosome acidification, which we measured using MPs conjugated to pH-sensitive pHrodo dyes, was unaffected by Gβ4 deficiency (fig. S3B), strongly suggesting that the hyperphagia we observed reflected increased uptake rather than accelerated phagosome maturation.
Fig. 1: 𝚫Gβ4 HL-60 neutrophils have increased phagocytic capacity.
A, Schematic of Gβ knockout workflow in HL-60 neutrophils and the MP-based phagocytosis assay. B, WT and ΔGβ HL-60 neutrophils were challenged with human IgG-coated MPs and phagocytosis quantified after 3 h. Data are mean ± s.d. C, WT and ΔGβ4 HL-60 neutrophils transfected with exogenous Gβ4 (WT over and ΔGβ4 rescue) or control lentivirus (WT and ΔGβ4) were challenged with human IgG-coated MPs and phagocytosis quantified after 3 h. Data are mean ± s.d. For B and C, n ≥ 6 for each sample, pooled from 3 biological replicates. One-way ANOVA, * p< 0.05, ** p< 0.01, and ****p< 0.0001. D, ΔGβ4 and WT HL-60 neutrophils were stained with different fluorophores, mixed 1:1, and then challenged with IgA-coated MPs. Graph shows phagocytic uptake after 3 h. Data are mean ± s.d. n = 12 for each sample, pooled from 3 biological replicates. Unpaired t test **** p< 0.0001. E, ΔGβ4 and WT HL-60 neutrophils were challenged with IgA coated MPs and imaged over 3 hours to assess per cell phagocytic capacity. Histogram shows the number of ΔGβ4 and WT cells that consumed 1, 2, 3, or 4 MPs during the experiment. Data pooled from 5 biological replicates. Inset shows sample images of WT and ΔGβ4 cells after MP uptake. Scale bars = 20 μm. F-G, WT and ΔGβ4 HL-60 neutrophils expressing F-tractin-mCherry (magenta) were fed IgG-coated MPs (green), and phagocytic uptake was monitored for 10–30 minutes. F, representative interactions are shown in time-lapse montage format. Scale bars = 5 μm. G, quantification of the frequency (left) and speed (right) of phagocytosis. Data points in the left graph correspond to mean values in each movie, while data points in the right graph denote individual cells. Bars denote mean ± s.d., pooled from 6 biological replicates. Unpaired t-test, ****p< 0.0001. H, Live videos of WT and ΔGβ4 HL-60 neutrophils expressing F-tractin (red) were analyzed for environmental probing. Graph shows quantification of ruffling index for both WT and ΔGβ4 HL-60 neutrophils. Data are mean ± s.d. n = 20 cells for each condition, pooled from 3 biological replicates. Unpaired t-test **** p< 0.0001.
Gβ4 deficiency markedly enhanced particle consumption on a per-cell basis. In wide-field imaging experiments, almost half of the phagocytic ΔGβ4 cells engulfed two or more MPs, whereas the vast majority of phagocytic WT cells took up just one (Fig. 1E). Super-resolution microscopy of individual phagocytic events revealed intriguing differences in engulfment dynamics. ΔGβ4 HL-60 neutrophils formed stereotypical phagocytic cups featuring a pronounced band of filamentous actin (F-actin) at the leading edge (Fig. 1F). This morphology was much less apparent in WT cells, which still engaged MP targets but only achieved full engulfment in less than 50% of conjugates. In contrast, ΔGβ4 HL-60 neutrophils completed over 90% of their phagocytic attempts and executed engulfment more than twice as fast as their wild-type counterparts (Fig. 1G, movie S1). To investigate whether this enhanced rate of phagocytosis reflected higher levels of environmental probing, we calculated the “ruffling index” of both ΔGβ4 and WT HL-60 neutrophils from live videos of fluorescently labeled cells (47, 48). ΔGβ4 cells exhibited a 2-fold increase in this ruffling index (Fig. 1H), indicative of increased physical activity and consistent with their enhanced phagocytic potential.
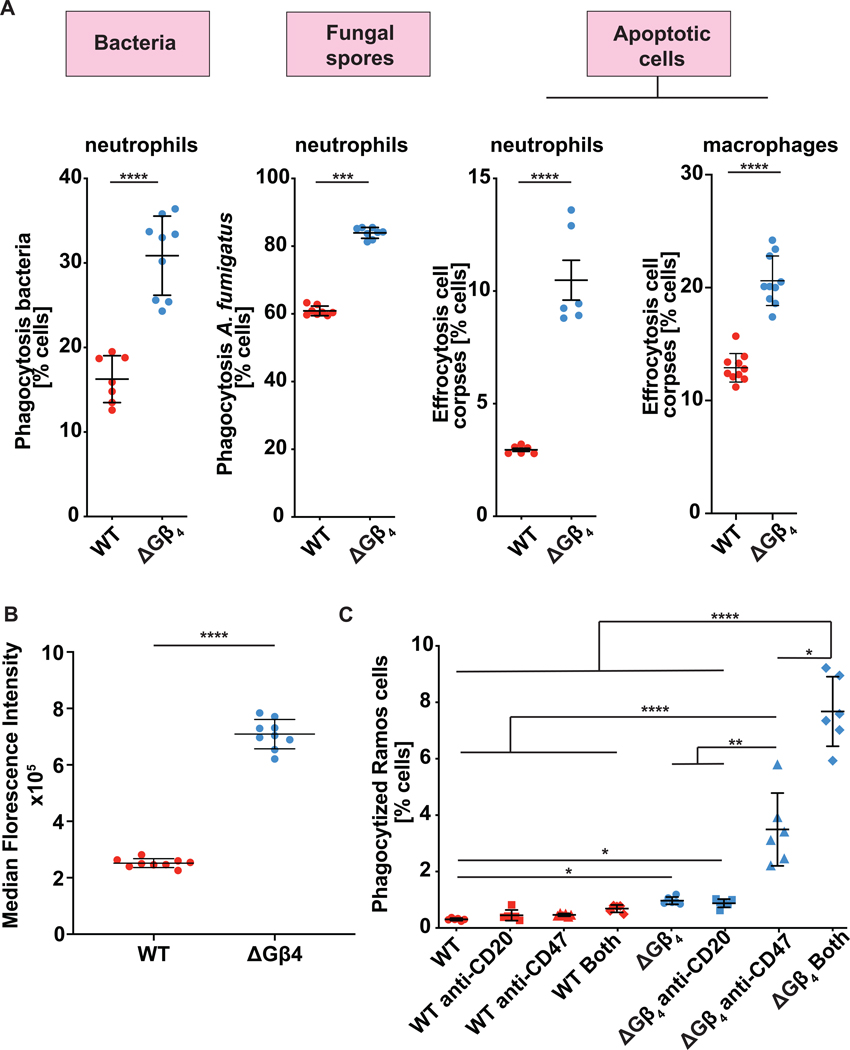
To further explore the scope of the ΔGβ4 phenotype, we challenged WT and ΔGβ4 HL-60 cells with MPs bearing alternative coatings, including phosphatidylserine (PS), complement, IgG, and IgA. ΔGβ4 cells phagocytosed 2-to 10-fold more cargo in each case, indicating that their hyperphagia was not limited to a specific uptake receptor (fig. S3C). Next, we measured the phagocytosis of four distinct biological targets: the gram-negative bacterium Pseudomonas aeruginosa, Staphylococcus aureus bioparticles, conidia from the fungus Aspergillus fumigatus, and apoptotic Jurkat T cell corpses (Fig. 2A, fig. S3D–E). These experiments utilized both HL-60 neutrophils and HL-60 derived macrophages (HL-60 macrophages), which we differentiated from progenitors using 12-O-tetradecanoylphorbol-13-acetate (TPA) (49, 50). ΔGβ4 cells outperformed WT controls in every case, further supporting the hypothesis that Gβ4 deficiency potentiates phagocytosis against diverse biological cargos. We also examined macropinocytosis, an F-actin-dependent process used by professional phagocytes to ingest antigen and other soluble factors (51). Although a similar fraction of WT and ΔGβ4 HL-60 neutrophils performed macropinocytosis, which we quantified by uptake of fluorescent 70 kDa dextran, ΔGβ4 cells took up substantially more cargo per cell (Fig. 2B). Hence, Gβ4 deficiency affects multiple cargo uptake mechanisms.
Fig. 2: 𝚫Gβ4 hyperphagia applies to a wide range of biological targets.
A, ΔGβ4 and WT HL-60 neutrophils or macrophages were challenged with fluorescently labeled Pseudomonas aeruginosa (left), Aspergillus fumigatus (center), or apoptotic Jurkat cell corpses (right), and phagocytosis quantified after 3 h. Data are mean ± s.d. n ≥ 7 for each sample, pooled from 3 biological replicates. Unpaired t-test *** p< 0.001, **** p< 0.0001. B, ΔGβ4 and WT HL-60 neutrophils were incubated with fluorescent 70 kDa dextran to measure macropinocytosis. Graph shows quantification of engulfed dextran. Data are mean ± s.d. n ≥ 9 for each sample, pooled from 3 biological replicates. Unpaired t-test **** p< 0.0001. C, ΔGβ4 and WT HL-60 neutrophils were challenged with RAMOS B lymphoma cells in the presence or absence of anti-CD20 and anti-CD47. Data are mean ± s.d. n ≥ 5 for each sample, pooled from 3 biological replicates. Two-way ANOVA, * p< 0.05, ** p< 0.01, **** p< 0.0001.
Next, we investigated whether loss of Gβ4 could augment the therapeutic phagocytosis of tumors. Cancer cells may evade immune detection and clearance by expressing CD47, a cell surface protein that has been proposed to function as a “don’t eat me” signal (52, 53). Antibodies or peptides against CD47 or its receptor, SIRPα, can stimulate an engulfment response either by blocking the “don’t eat me signal” or by opsonizing the cancer cell for phagocytosis (54–58). To adapt this approach to our experimental system, we challenged HL-60 neutrophils with Ramos B lymphoma target cells in the absence or presence of anti-CD47 blockade. Antibodies against the B cell marker CD20 were added to further promote phagocytosis via Fc receptor engagement. WT HL-60 neutrophils did not take up Ramos cells, and treatment with anti-CD47 and/or anti-CD20 failed to enhance their activity. In contrast, ΔGβ4 HL-60 neutrophils exhibited a low but measurable level of baseline phagocytosis, which increased 3-fold in the presence of anti-CD47, and even more so (8-fold) when anti-CD47 was combined with anti-CD20 (Fig. 2C). These results suggest that Gβ4 deficiency augments weak phagocytic responses against tumor cells.
Phosphoinositide 3-kinases (PI3K) promote phagocytosis, particularly the uptake of large cargos, by generating phosphatidylinositol-3,4,5-trisphosphate (PIP3) at the phagocytic cup (59, 60). The hyperphagic behavior of ΔGβ4 cells raised the possibility that they might have enhanced or otherwise modulated PI3K activity. During phagocytosis of IgG-coated MPs, however, WT and ΔGβ4 HL-60 neutrophils exhibited similar levels of PIP3 enrichment at the phagocytic cup, which we measured using the PIP3 biosensor PH-AKT-BFP (fig. S4A–B). Furthermore, inhibition of PI3K activity using the small molecule PIK90 abrogated MP uptake in both cell types (fig. S4C). Thus, Gβ4 deficiency neither enhances nor circumvents the need for PI3K signaling during phagocytosis.
Gβ4 deficiency alters cell migration and crosstalk between phagocytosis and motility
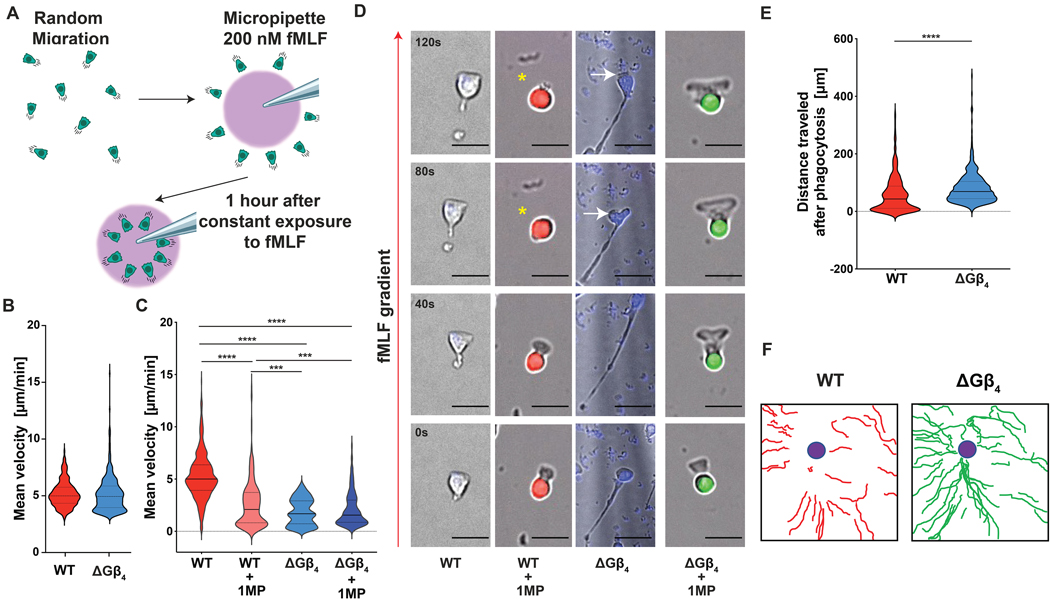
As proper coordination between cargo uptake and motility is essential for myeloid cell function (16–18), we next investigated the effects of Gβ4 on cell migration. To this end, we employed a chemotaxis assay in which HL-60 neutrophils were attached to fibronectin-coated glass and then presented with a point source of the peptide chemoattractant fMLF, applied via micropipette (Fig. 3A). Under these conditions, neutrophils adopt a fan-like morphology with a broad leading edge directed toward the point source and a thin uropod trailing behind. Both ΔGβ4 and WT cells were able to migrate up the fMLF gradient and moved at similar velocities during the first 15 minutes of each experiment (Fig. 3B). ΔGβ4 cells slowed substantially over the remainder of the experiment, however, leading to a reduction in mean velocity over the entire 1 hour time course (Fig. 3C, Supplementary Video 2). This reduced motility was associated with the formation of extended “tails” at the rear of migrating ΔGβ4 cells, suggestive of a defect in uropod retraction (Fig. 3D). Indeed, we observed multiple instances in which elongated ΔGβ4 cells appeared to be “struggling” against their own uropod to make forward progress (Fig. 3D). We obtained a similar set of results using HL-60 macrophages. In a uniform chemokinetic field of fMLF, ΔGβ4 HL-60 macrophages moved slower than their wild type counterparts and also formed elongated uropods during migration (fig. S5A–C). In some cases, ΔGβ4 macrophages exhibited multiple extended tails (fig. S5D), implying a defect in cell polarity. Collectively, these results indicate that Gβ4 deficiency impairs motility and alters migratory cell morphology in myeloid cells.
Fig. 3: Gβ4 deficiency alters cell migration and crosstalk between phagocytosis and motility.
A, Schematic of the micropipette-based chemotaxis assay. B-F, ΔGβ4 and WT HL-60 neutrophils were seeded onto fibronectin coated slides and exposed to 200 nM fMLF delivered through a micropipette over the course of an hour. Alternatively, ΔGβ4 and WT cells that had consumed 1 MP (WT + 1MP and ΔGβ4 + 1MP) were sorted, seeded at a 1:1 ratio on fibronectin coated glass, and exposed to 200 nM fMLF through a micropipette for one hour. B, Mean velocity of ΔGβ4 and WT HL-60 neutrophils during the first 15 minutes of the time course. C, Mean velocities of WT, WT + 1MP, ΔGβ4, and ΔGβ4 + 1MP cells were calculated from 3 separate 1 hour videos. D, Time-lapse montages of representative cells in each experimental group, with the red arrow indicating the direction of the fMLF gradient. Yellow asterisks denote a transient loss of migratory cell polarity in a WT + 1MP cell. White arrows indicate cell body regression in a ΔGβ4 cell with an extended uropod. Scale bars = 20 μm. E, Total distance travelled by WT + 1MP and ΔGβ4 + 1MP cells after 1 hour of exposure to 200 nM fMLF. F, Representative migration tracks for WT + 1MP and ΔGβ4 + 1MP over the course of 1 hour exposure to 200 nM fMLF. Violins in B, C and E encompass the entire distribution, with solid horizontal lines indicating the median and dotted lines indicating the upper and lower quartiles. *** p< 0.001 and **** p< 0.0001, calculated by unpaired t-test (E) or two-way ANOVA (C). n ≥ 174 for each sample, pooled from 3 biological replicates.
Next, we interrogated crosstalk between phagocytosis and migration by quantifying the motility of HL-60 neutrophils after uptake of one IgG-coated MP. WT and ΔGβ4 cells were exposed to MPs with distinct fluorescent labels (pHrodoRed (WT) versus pHrodoGreen (ΔGβ4)) to facilitate imaging and tracking in mixed samples. MP consumption slowed the movement of WT HL-60 neutrophils considerably relative to MP-less WT controls (Fig. 3C). By contrast, ΔGβ4 cells containing MPs moved at the same rate as their MP-lessΔGβ4 counterparts (Fig. 3C). Although the instantaneous speed of MP-bearing ΔGβ4 neutrophils remained slightly less than that of MP-bearing WT cells, their migration was more persistent, yielding significantly longer tracks that more closely approached the fMLF point source (Fig. 3E–F, Supplementary Video 2). These motility phenotypes were mirrored by changes in cell shape. In WT HL-60 neutrophils, phagocytosis appeared to hamper migratory polarization; cells periodically collapsed into a rounded configuration, and these morphological changes tended to coincide with stalls in motility (Fig. 3D, Supplementary Video 2). By contrast, ΔGβ4 morphology was normalized by MP uptake; MP-bearing cells formed persistent leading edges and lacked the extended uropods characteristic of the cargo-less state (Fig. 3D). Hence, Gβ4 depletion affects neutrophil migration in two ways: it reduces the speed of unencumbered cells, and it also impairs inhibitory cross-regulation between phagocytosis and motility.
Gβ4 deficiency alters lipid composition and plasma membrane abundance
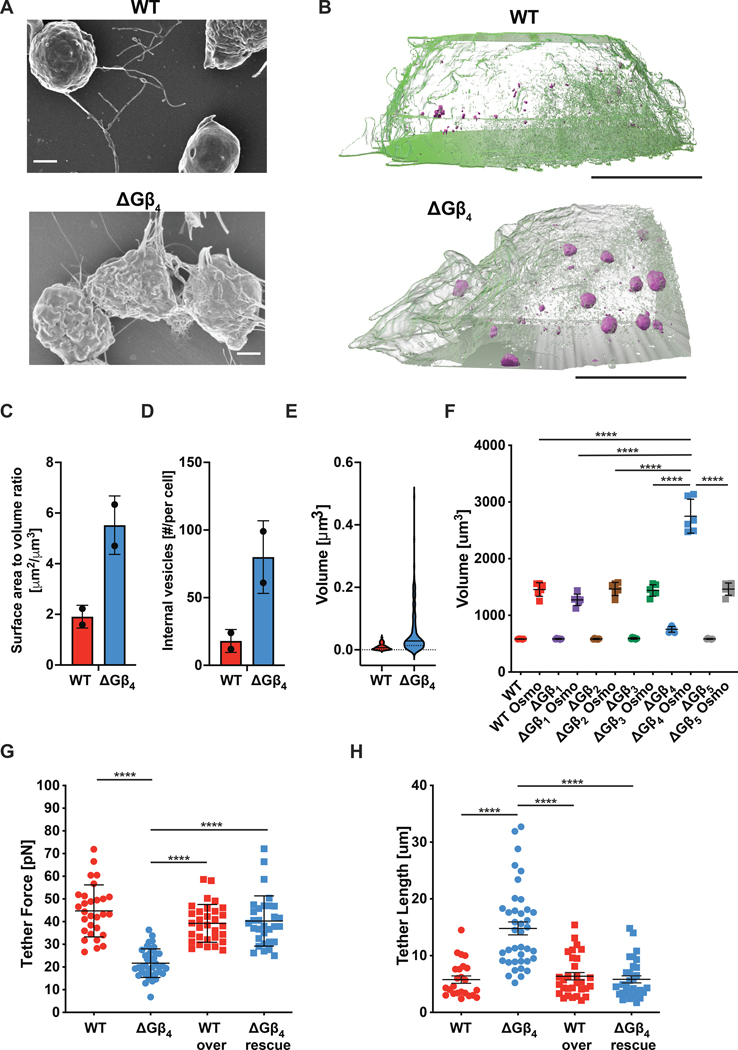
The striking uropod extension displayed by migrating ΔGβ4 HL-60 cells was suggestive of a substantial change in cellular architecture. Consistent with this notion, scanning electron microscopy (SEM) indicated that ΔGβ4 HL-60 cells had a more ruffled surface appearance than WT controls (Fig. 4A). To investigate this structural difference more closely, we performed Focused Ion Beam (FIB)-SEM, a method in which successive sections from the same sample are imaged by SEM and then used to generate a nanometer-resolution 3-D reconstruction (61). WT and ΔGβ4 HL-60 cells were osmium-stained to highlight lipid-rich cellular components, embedded in resin, and subjected to FIB-SEM at 40 nm sectioning. Supervised machine learning was used to define the plasma membrane in each image, followed by 3-D rendering (62). The resulting reconstructions revealed striking differences in plasma membrane configuration. Whereas the surface of WT cells was mostly smooth with small extensions, ΔGβ4 cells exhibited large flaps of plasma membrane projecting up to 10 μm from the cell body (Fig. 4B, Supplementary Video 3). The presence of these structures increased the surface to volume ratio of ΔGβ4 HL-60 neutrophils by a factor of two relative to WT controls (Fig. 4C). ΔGβ4 cells also contained larger and more numerous cytoplasmic lipid droplets, which appeared as high contrast compartments in osmium-stained FIB-SEM images (Fig. 4B, 4D–E). Using FM64 and BODIPY staining, we confirmed that loss of Gβ4 leads to excess accumulation of plasma membrane and lipid droplets, respectively (fig. S6A–C). Taken together, these data indicate that Gβ4 controls membrane architecture and lipid content in neutrophils.
Fig 4: ΔGβ4 HL-60 neutrophils have increased plasma membrane and decreased membrane tension.
A, Representative SEM images of WT and ΔGβ4 HL-60 neutrophils. Scale bars = 2 μm. Representative of 5 biological replicates. B-E, ΔGβ4 and WT HL-60 neutrophils stained with potassium ferrocyanide and osmium tetroxide were imaged by FIB-SEM (n = 2 of each cell type, from 1 biological replicate). B, 3-D reconstructions of representative cells, with plasma membrane shown in semi-transparent green and lipid droplets in solid magenta. Scale bars = 5 μm. C, Quantification of surface area to volume ratio. D, Quantification of lipid droplet number per cell. In C and D, data are mean ± s.d. E, Quantification of lipid droplet volume (n = 20 droplets in WT cells and 24 droplets in ΔGβ4 cells). Violins encompass the entire distribution, with solid horizontal lines indicating the median and dotted lines indicating the upper and lower quartiles. F, HL-60 neutrophils lacking the indicated Gβ subunits, along with WT controls, were subjected to osmotic shock to induce swelling to maximal volume. Graph shows volume measurements before and after osmotic shock (Osmo). Data are mean ± s.e.m. n = 6 for each sample, pooled from 3 biological replicates. One-way ANOVA ****p< 0.0001. G-H, Membrane tethers were generated from WT and ΔGβ4 HL-60 neutrophils transduced with exogenous Gβ4 (WT over and ΔGβ4 rescue) or control lentivirus (WT and ΔGβ4). G, Membrane tether forces. H, Membrane tether lengths. Data in G and H are mean ± s.d. n ≥ 29 for each sample, pooled from 5 biological replicates. Two-way ANOVA, ****p< 0.0001.
We considered the possibility that excess plasma membrane might concomitantly increase surface expression of Fc receptors and thereby enhance phagocytosis. ΔGβ4 and WT HL-60 neutrophils, however, expressed similar levels of FcγRI and FcγRII, and little to no FcγRIII (fig. S7A–C), arguing against a role for Fc receptor overexpression in the ΔGβ4 hyperphagic phenotype. SIRPα levels were similarly unaffected by Gβ4 deficiency (fig. S7D). ΔGβ4 and WT HL-60 neutrophils also contained nearly equivalent levels of F-actin, which we visualized by phalloidin staining, and most cells of either genotype exhibited canonical migratory cell morphology, defined by strong F-actin accumulation at the leading edge (fig. S7E–F). Hence, excess plasma membrane did not appear to cause cytoskeletal dysregulation in ΔGβ4 cells.
Having ruled out these potential mechanisms, we turned our attention to the biophysical and functional effects of Gβ4 deficiency on the plasma membrane itself. Acute osmotic shock swells cells to the limits of their plasma membrane capacity, thereby enabling comparisons of total surface area. ΔGβ4 HL-60 neutrophils were only slightly larger than their WT counterparts under isotonic conditions. Upon transfer to hypotonic medium, however, ΔGβ4 cells expanded to twice the size of controls (Fig. 4F). Assuming that swelled HL-60 neutrophils are spherical, this volume differential implies a ~60% increase in plasma membrane surface area. Excess plasma membrane would be expected to facilitate the formation of phagocytic cups and thereby promote engulfment. To quantify membrane mobilization during cup formation, we utilized a “frustrated phagocytosis” assay (63, 64) in which HL-60 neutrophils were applied to IgG-coated glass slides. Neutrophils and macrophages form flat, unresolved phagocytic cups on surfaces of this kind, which are easily visualized by total internal reflection fluorescence (TIRF) microscopy. Frustrated phagosomes formed by ΔGβ4 HL-60 neutrophils were substantially larger than those of WT controls (fig. S8), consistent with a role for increased membrane abundance in phagocytic cup formation.
Membrane tension is known to inhibit both phagocytosis and migration, presumably by mechanically constraining the formation of actin-based structures and/or activating inhibitory mechanosensory pathways (63). We speculated that, by increasing plasma membrane abundance, Gβ4 deficiency might reduce membrane tension and thereby attenuate these regulatory effects. To explore this possibility, we utilized an established approach in which a concanavalin-A-coated bead is adsorbed to the cell surface and then pulled away using an optical trap (65). The displaced bead remains attached to the cell via a thin membrane tether, which exerts a restoring force that is proportional to the square of membrane tension (66, 67). In this manner, we found that Gβ4 deficiency reduced tether forces by a factor of two in HL-60 neutrophils (Fig. 4G). ΔGβ4 cells also generated longer tethers then WT controls, indicating that they possessed larger plasma membrane reservoirs (Fig. 4H) (68). Rescue experiments confirmed that both phenotypes were specific to Gβ4 (Fig. 4G–H). In conjunction with the imaging and osmotic shock studies described above, these results suggest a central role for plasma membrane abundance in the regulation of neutrophil effector responses.
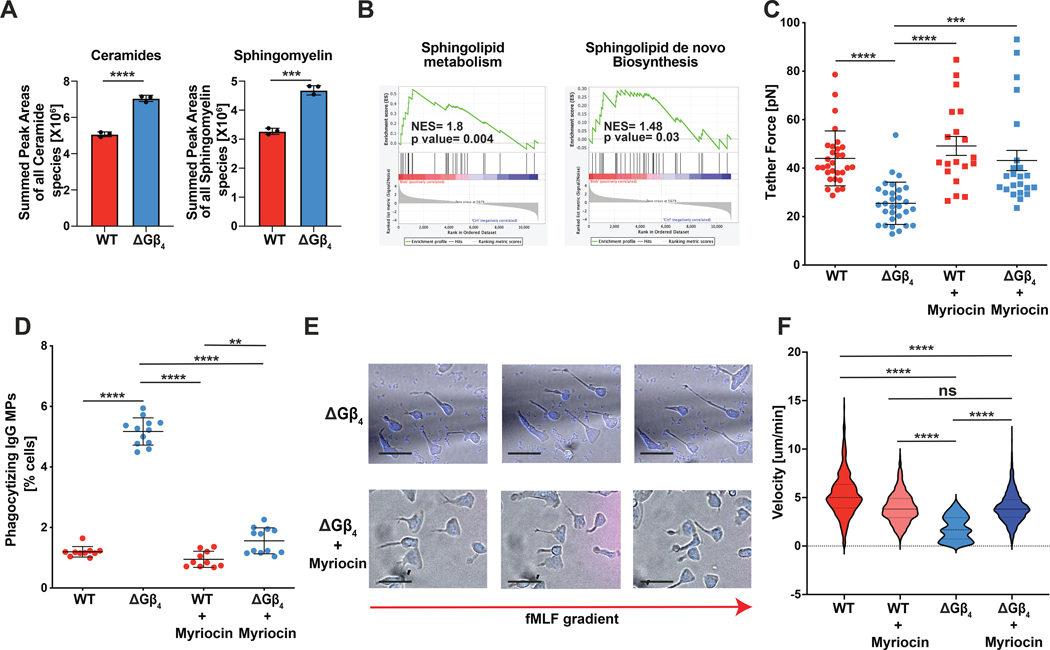
To explore potential mechanisms underlying the ΔGβ4 plasma membrane phenotype, we subjected ΔGβ4 and WT HL-60 neutrophils to comparative lipidomics (fig. S9A–B and table S1). As expected, ΔGβ4 cells contained more total lipids than WT controls on a per cell basis (fig. S9C). This increase did not apply equally to all lipid subtypes, however, as many species were unchanged in proportion between samples and some, like hexosylceramides, were more abundant in WT cells (fig. S9D). ΔGβ4 cells were distinguished by disproportionate enrichment of sphingolipids, particularly ceramide and sphingomyelin (Fig. 5A and fig. S9C), which are predominantly found in the plasma membrane. Using RNA-sequencing, we observed selective upregulation of sphingolipid synthesis genes in ΔGβ4 neutrophils after MP exposure (Fig. 5B), establishing an additional link between sphingolipids and the ΔGβ4 phenotype. To interrogate the role of sphingolipids in plasma membrane expansion more directly, we asked whether inhibition of sphingolipid synthesis could reverse the effects of Gβ4 deficiency. To this end, we applied the small molecule myriocin, a potent inhibitor of serine palmitoyltransferase, the first enzyme in the sphingolipid synthesis pathway (fig. S9E) (69, 70). Myriocin treatment during differentiation restored the membrane tension of ΔGβ4 HL-60 cells to WT levels (Fig. 5C), and it also largely reversed their hyperphagic behavior (Fig. 5D), their extended migratory morphology (Fig. 5E), and their motility defect (Fig. 5F). Notably, acute treatment of fully differentiated ΔGβ4 HL-60 cells with myriocin had no effect on MP uptake (fig. S9F), suggesting that the functionally relevant effects of Gβ4 signaling on sphingolipid synthesis occur during the differentiation phase. Collectively, these results indicate that the excess plasma membrane characteristic of ΔGβ4 cells is due, at least in part, to increased sphingolipid synthesis.
Fig 5: Gβ4 regulates plasma membrane expansion via sphingolipid synthesis.
A, Lipidomic quantification of ceramides (left) and sphingomyelin (right) in WT and ΔGβ4 HL-60 neutrophils. Data are mean ± s.d. n = 3, pooled from 3 biological replicates. Unpaired t-test, ***p< 0.001, ****p< 0.0001. B, ΔGβ4 and WT HL-60 neutrophils were compared by RNA-seq before and after consumption of IgG-coated MPs. GSEA plots are shown that associate the gene expression changes induced by MP uptake in ΔGβ4 cells with gene sets for sphingolipid metabolism (center) and de novo sphingolipid biosynthesis. NES = normalized enrichment score. C-F, WT and ΔGβ4 HL-60 neutrophils were differentiated in the presence or absence of myriocin and then subjected to biophysical and functional analysis. C, Quantification of membrane tension, determined by membrane tether pulling. Data represent mean ± s.d. n ≥ 19 for each sample, pooled from 5 biological replicates. Two-way ANOVA, *** p< 0.001 and **** p< 0.0001. D, Cells were challenged with IgG-coated MPs, and phagocytosis was quantified after 3 h. Data represent mean ± s.d. n ≥ 10 for each sample, pooled from 5 biological replicates. Two-way ANOVA, *** p< 0.001 and **** p< 0.0001. E, Representative images of myriocin treated and untreated ΔGβ4 HL-60 neutrophils. Scale bars = 10 μm. F, Migration velocities of myriocin treated ΔGβ4 and WT HL-60 neutrophils. Untreated ΔGβ4 and WT velocities from Fig. 3B are included for comparison. Data represent mean ± s.d. n ≥ 174 for each sample, pooled from 3 biological replicates. One-way ANOVA, ****p< 0.0001.
Gβ4 controls membrane abundance and phagocytosis in primary myeloid cells
Having demonstrated that Gβ4 deficiency induces hyperphagia in HL-60 neutrophils and macrophages, we next investigated whether this phenotype would manifest in primary cells. By applying CRISPR-Cas9 targeting to the ER-HoxB8 immortalized murine progenitor system (22, 71, 72) (fig. S10A–B), we were able to generate primary-like neutrophils lacking Gβ4, along with controls expressing non-targeting guide (g)RNA. Both ΔGβ4 and WT ER-HoxB8 neutrophils expressed equivalent levels of CD11b and Ly6G after differentiation (fig. S10c), indicating that Gβ4 is dispensable for the acquisition of neutrophil fate. As with ΔGβ4 HL-60 cells, ΔGβ4 ER-HoxB8 neutrophils displayed clear signs of plasma membrane dysregulation, including reduced membrane tension and increased membrane tether length in optical trap experiments (fig. S10D–E). ΔGβ4 ER-HoxB8 neutrophils also expanded to nearly twice the volume of WT controls upon transfer to hypotonic medium (fig. S10F). In phagocytosis assays, both WT and ΔGβ4 ER-HoxB8 neutrophils failed to consume IgG-coated MPs, likely because primary murine neutrophils are substantially smaller than HL-60 neutrophils and therefore unable to accommodate larger (~10 μm diameter) cargos. When challenged with smaller (2 μm diameter) S. aureus bioparticles, however, ΔGβ4 ER-HoxB8 neutrophils more than doubled the uptake of WT controls (fig. S10G).
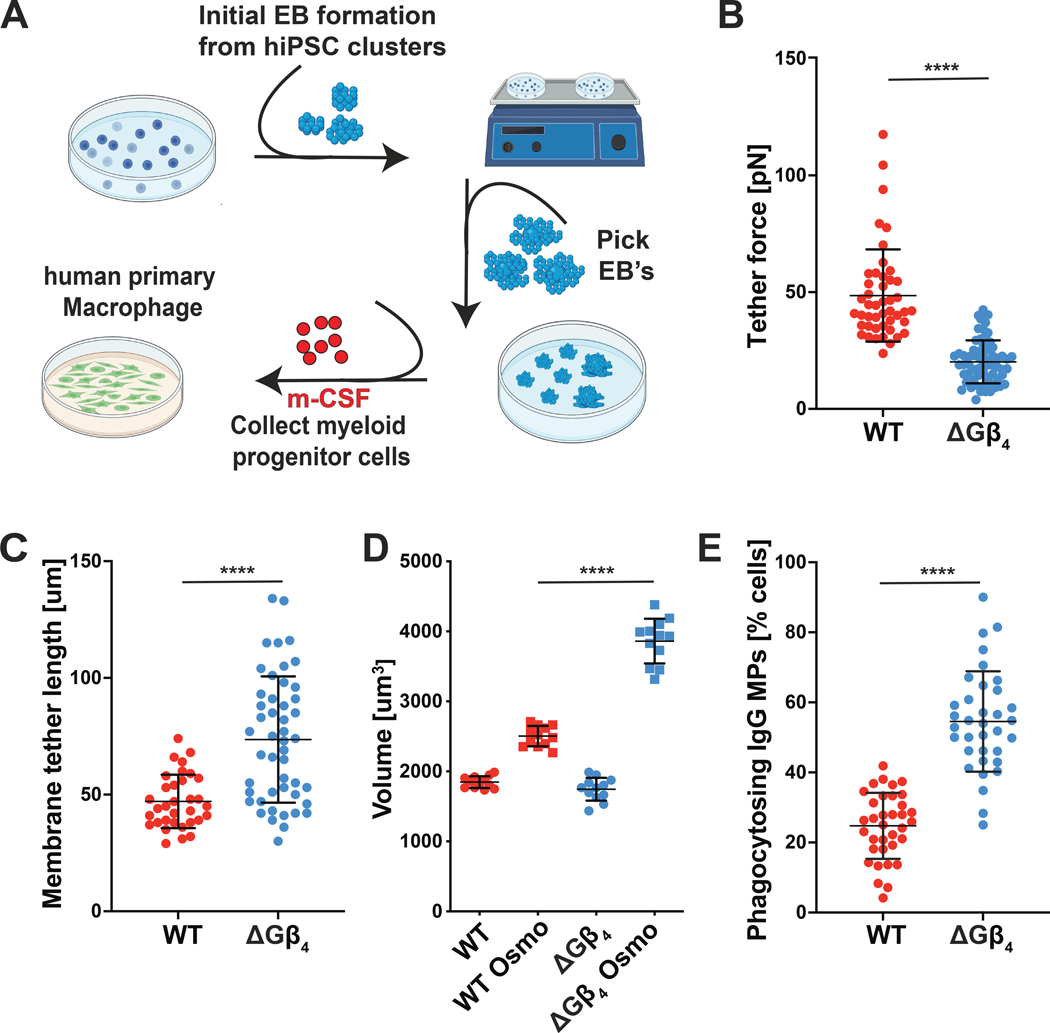
To assess the role of Gβ4 signaling in primary macrophages, we prepared WT and ΔGβ4 macrophages from human induced pluripotent stem cells (hiPSCs) (Fig. 6A and fig. S11A)(73–75). CRISPR-Cas9 was used to target the GNB4 locus (encoding Gβ4) at the hiPSC stage, and WT control hiPSCs were generated in parallel using a nontargeting gRNA (fig. S11B). Granulocyte monocyte progenitor (GMP) cells derived from these hiPSC lines were then differentiated into mature CD11b+ CD14+ CD68+ macrophages. Importantly, Gβ4 deficiency did not affect the efficacy of this differentiation protocol (fig. S11C). In optical trap experiments, ΔGβ4 macrophages generated weaker tether forces and longer membrane tethers than WT controls (Fig. 6B–C). ΔGβ4 macrophages also became substantially larger than their WT counterparts in hypotonic medium, despite being similarly sized under isotonic conditions (Fig. 6D). These phenotypes, which mirrored our results in the HL-60 and ER-HoxB8 systems, were strongly suggestive of excess plasma membrane accumulation. Finally, we evaluated phagocytic capacity and found that Gβ4 deficiency markedly increased the uptake of IgG-coated MPs (Fig. 6E). We conclude that the Gβ4 pathway regulates plasma membrane abundance and phagocytosis in primary neutrophils and macrophages.
Fig 6: Gβ4 deficiency boosts primary macrophage phagocytosis.
A-D, Isogenic ΔGβ4 and WT macrophages were derived from hiPSCs and subjected to biophysical and functional assays. A, Schematic of embryoid body generation from hiPSCs and differentiation of primary macrophages from GMPs. B-C, Membrane tethers were generated from WT and ΔGβ4 macrophages using an optical trap. B, Quantification of membrane tether force. C, Quantification of membrane tether length. Data in B-C represent mean ± s.d. n ≥ 46 for each sample, pooled from 5 biological replicates. D, ΔGβ4 and WT macrophages were osmotically shocked and allowed to expand to their full volume to assess total plasma membrane. Data represent mean ± s.d. n = 12 for each sample, pooled from 3 biological replicates. E, WT and ΔGβ4 macrophages were challenged with IgG-coated MPs for 2 h and phagocytic uptake quantified by florescence microscopy. Data represent mean ± s.d., pooled from 5 biological replicates. Unpaired t-test (B, C, E) or two-way ANOVA (D), **** p< 0.0001.
Gβ4 regulates anti-fungal immunity in vivo
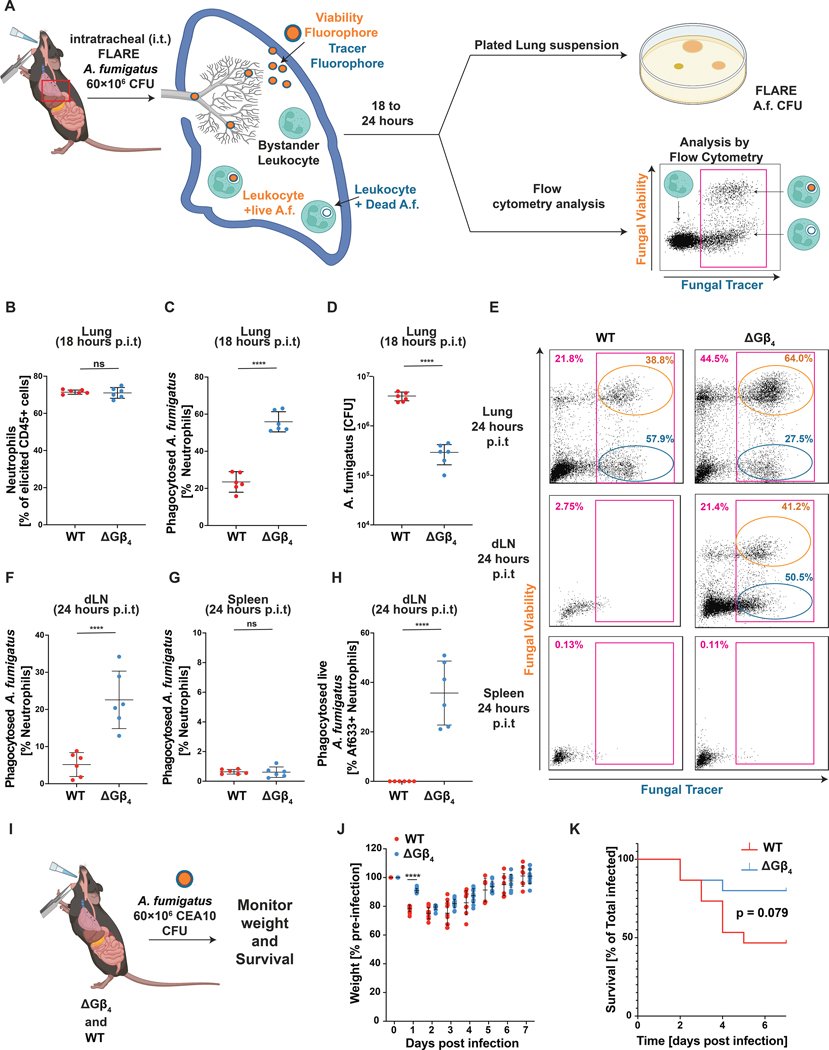
Phagocytes are the first line of defense against microbes in multiple epithelial tissues (3, 13, 76). To evaluate the role of Gβ4-dependent membrane allocation during this early phase of immunity, we generated mice with a targeted deletion of exon 4 of the Gnb4 locus (fig. S12A–B). This modification prematurely terminated the Gnb4 open reading frame, leading to constitutive Gβ4 deficiency. Mice homozygous for this deletion (Gnb4−/−) and WT (Gnb4+/+) littermate controls were infected intratracheally with Aspergillus fumigatus, a fungal pathogen that elicits the robust recruitment of phagocytically active neutrophils to the lung (77–79). To enable flow cytometric detection of fungal uptake by these cells, we employed Fluorescent Aspergillus Reporter (FLARE) conidia that expressed dsRed, a degradable fluorescent protein, and were also labeled with Alexa Fluor (AF)633, a non-degradable small molecule dye. Cells that take up FLARE conidia become AF633+dsRed+, while cells that have killed the phagocytized fungi are AF633+dsRed− (Fig. 7A) (21). FLARE A. fumigatus infection induced similar levels of neutrophil recruitment to the lungs of WT and Gnb4−/− animals, with CD11b+Ly6G+SiglecF− cells accounting for ~70% of CD45+ infiltrates in both experimental groups (Fig. 7B). However, fungal phagocytosis was almost 3 times higher in Gnb4−/− neutrophils (Fig. 7C), mirroring the hyperphagic phenotype seen in HL-60 cells, ER-HoxB8 neutrophils, and hiPSC-derived macrophages lacking Gβ4. Among these phagocytic cells, the frequency of fungal killing was unaffected by Gβ4 deficiency (fig. S12C), consistent with the interpretation that Gβ4 signaling modulates cargo uptake but not subsequent acidification of the phagolysosome. Gnb4−/− animals also exhibited a ~10-fold reduction in fungal CFU in the lung relative to WT controls at the 18 hour time point (Fig. 7D), indicating that the increased conidial phagocytosis conferred by loss of Gβ4 improved fungal clearance during early stage infection.
Fig. 7: Gβ4 deficiency boosts phagocytic responses during in vivo fungal infection.
A-H, WT and Gnb4−/− mice infected with FLARE A. fumigatus were assessed for in vivo phagocytosis and fungal infection after 18 h or 24 h. A, FLARE conidia express dsRed and are labeled with AF633. Phagocytes become dsRed+AF633+ upon conidial uptake, and then transition to dsRed−AF633+ as internalized conidia are destroyed. B, Neutrophil infiltration into the lung, 18 h post-infection. C, Quantification of A. fumigatus phagocytosis by lung neutrophils at the 18 h time point. D, Fungal CFU in lung extracts at the 18 h time point. Data in B-D represent mean ± s.d. n = 6 biological replicates. Unpaired t test **** p< 0.0001. E, Representative flow cytometry plots showing neutrophils (CD45+, CD11b+, and Ly6G+) containing FLARE A. fumigatus in the lungs, dLN, and spleen 24 h post-infection (magenta gates). Gates for neutrophils with live (orange) and dead (blue) conidia are shown. F, Quantification of neutrophils bearing engulfed FLARE A. fumigatus in the dLN, 24 h post-infection. G, Quantification of neutrophils bearing engulfed FLARE A. fumigatus in the spleen, 24 h post-infection. H, Quantification of the neutrophils in the dLN containing live A. fumigatus, 24 h post-infection. Data in F-H represent mean ± s.d. n = 6 biological replicates. Unpaired t test **** p< 0.0001. I-K, Gnb4−/− and WT mice were challenged with CEA10 A. fumigatus. I, Schematic diagram of the experimental approach. J-K, Weight (J) and survival (K) of infected mice was monitored over the course of 7 days. Data represent mean ± s.d. n = 15 biological replicates. Statistical analysis in J performed by unpaired t test, with **** p< 0.0001. Statistical analysis in K performed by Logrank test.
Given that ΔGβ4 HL-60 neutrophils failed to arrest their motility after cargo uptake in vitro (Fig. 3C–F), we speculated that Gnb4−/− phagocytes might exhibit altered trafficking behavior in vivo. Accordingly, we examined the organ distribution of phagocytic WT and Gnb4−/− neutrophils 24 hours after FLARE infection (Fig. 7A). In WT mice, neutrophils bearing AF633-labeled material were found predominantly in the lungs, with small numbers in the draining lymph nodes (dLNs) and essentially none in the spleen and blood (Fig. 7E–G, fig. S12D). These observations were consistent with the expectation that phagocytosis inhibits motility after conidial uptake in the lungs, thereby preventing the dissemination of neutrophils with internalized pathogens to other organs. By contrast, we observed a substantial number of AF633+ neutrophils in the dLNs of Gnb4−/− animals (Fig. 7E–F), suggesting that Gβ4 deficiency at least partially relieves the brake on motility applied by phagocytosis. Notably, whereas essentially all AF633+ neutrophils in WT dLNs were DsRed−, indicating that they had degraded their internalized conidia, a substantial fraction of DsRed+AF633+ neutrophils were observed in Gnb4−/− dLNs (Fig. 7E, H). Given that Gβ4 deficiency does not affect phagolysosome maturation (fig. S3B), these results strongly suggest that Gnb4−/− neutrophils continue to migrate after conidial uptake, enabling them to reach proximal organs like the dLN before their cargo is broken down. We conclude that Gβ4 deficiency not only increases the phagocytic capacity of myeloid cells in vivo but also disrupts the cross-regulatory relationship between phagocytosis and migration.
To investigate whether the altered behavior of Gnb4−/− neutrophils affected the overall efficacy of anti-fungal immunity, we infected Gnb4−/− and WT mice with A. fumigatus CEA10, a clinically derived strain with increased pathogenicity in mice (Fig. 7I) (80). WT mice experienced acute weight loss in the first 72 hours after CEA10 administration and half succumbed to the infection within the first week (Fig. 7J–K). Both weight loss and mortality were attenuated in Gnb4−/− animals, indicating that loss of Gβ4 protects mice from the adverse effects of A. fumigatus infection.
Discussion
Our results identify Gβ4 as a critical regulator of plasma membrane abundance in myeloid cells and demonstrate that the plasma membrane exerts biophysical control over phagocytic capacity and functional crosstalk. Because the plasma membrane must be mobilized to build protrusive cellular structures, limiting its abundance provides a simple mechanism for not only constraining the magnitude of a given architectural response (e.g. phagocytosis) but also enabling cross-regulation between responses (e.g. phagocytosis inhibiting migration). This scarcity model is consistent with prior work documenting antagonism between uptake behaviors, such as phagocytosis and macropinocytosis, and migration (16–18). Thus, the mechanisms governing plasma membrane allocation in myeloid cells effectively dictate their functional potential.
Notably, the plasma membrane-dependent control mechanism studied here appears to most strongly affect the phagocytosis of large (~10 um diameter) cargos, such as apoptotic cells and cancer cells, while having less impact on the uptake of small entities like bacteria. This distinction likely reflects the fact that engulfing large, unbroken targets with high surface area places a disproportionate burden on plasma membrane mobilization. The importance of Gβ4 for regulating this type of phagocytosis suggests that it may be particularly relevant for processes that involve the clearance of large eukaryotic cells, such as anti-tumor immunosurveillance and the maintenance of tissue homeostasis.
Although lipid droplets are clearly augmented in ΔGβ4 cells, their role in myeloid cell hyperphagia remains unclear. Prior work has associated increased lipid droplet formation with reduced, rather than enhanced, phagocytosis (81–83). In these studies, however, lipid droplet growth either occurred downstream of a genetic defect in phagocytosis itself or was induced by cell-extrinsic nutritional changes, complicating direct comparisons with our findings. Lipid droplets almost exclusively contain triglycerides (84), which must be processed into amphipathic lipids in intracellular organelles before being incorporated into the plasma membrane. As such, it seems highly unlikely that lipid droplets would augment the plasma membrane directly during phagocytosis. That being said, it has been shown that cells utilize lipid droplets to buffer membrane depletion and fatigue under conditions of metabolic stress and protein misfolding (84–87). Hence, it is tempting to speculate that the enlarged lipid droplets present in ΔGβ4 phagocytes might enable these cells to replenish the plasma membrane after phagocytosis, thereby facilitating serial consumption.
The striking functional gains exhibited by ΔGβ4 neutrophils and macrophages raise the question of why Gβ4-dependent control of membrane abundance evolved in the first place. One obvious answer is that tight cross-inhibition between phagocytosis and migration is essential for immune function, at least in some contexts. Indeed, we found that Gnb4−/− neutrophils not only take up more A. fumigatus conidia in vivo but also traffic them out of the lung to the dLN. While increased fungal dispersion did not appear to compromise immunity in our experiments, one could imagine that a pathogen more capable of resisting or escaping the phagolysosome could exploit the dysregulated migration of Gβ4-deficient phagocytes in order to spread.
Although we cannot at present rule out a role for Gβ4 in the acute regulation of phagocytosis and motility, we favor a model in which the Gβ4 pathway acts during myeloid cell differentiation to dictate the size and composition of the plasma membrane, which then serves as a master mechanoregulator of morphology and effector responses in the terminally differentiated state (fig. S13). This model is consistent with our observations that ΔGβ4 neutrophils and macrophages exhibit architectural abnormalities (Fig. 4 and fig. S6) and also that acute myriocin treatment fails to reverse the ΔGβ4 phagocytosis phenotype (fig. S9F). Conceptually, the regulation of plasma membrane abundance by Gβ4 signaling is quite analogous to how the unfolded protein response sets the size of the endoplasmic reticulum, and therefore the capacity to process misfolded proteins, under conditions of cell stress (88). We postulate that differential Gβ4 expression among myeloid progenitors might promote functional diversification by enabling the formation of differentiated subsets with a spectrum of plasma membrane-defined phagocytic and migratory setpoints. This type of diversity could enable the innate immune system to engage effectively with a wide variety of homeostatic and microbial challenges.
Gβ4 mutations have been associated with Charcot-Marie-Tooth disease (CMTD) (89–92), a hereditary neurological disorder characterized by the progressive demyelination of peripheral nerves. Although the pathogenesis of this disease is generally thought to arise from cell-intrinsic glial dysfunction, macrophages routinely patrol peripheral nerves and are well-positioned to induce autoimmune neuropathy (93, 94). Interestingly, macrophage depletion has been shown to attenuate neurodegeneration in mouse CMTD models (95–97). In light of these reports, our data raise the intriguing possibility that dysregulated macrophage phagocytosis might contribute to at least some forms of CMTD.
The identification of Gβ4 signaling and sphingolipid synthesis as key regulators of phagocytic capacity in myeloid cells reveals heretofore unexplored avenues for enhancing innate immunity in therapeutic contexts. By targeting the architectural basis of cargo uptake, one could potentially modulate phagocyte activity in a manner that is agnostic to specific targets. We anticipate that an approach like this would be particularly useful for treating systemic microbial infections and for enhancing the anti-tumor potential of chimeric antigen receptor (CAR)-macrophages (10, 98). Exploring these possibilities in translationally relevant experimental systems will be an interesting topic for future research.
Materials and Methods
Study design
The goal of this study was to determine how Gβ4 signaling affects myeloid cell phagocytosis and migration. In vitro assays were used to measure phagocytic uptake and chemotaxis. FIB-SEM was employed for ultrastructural analysis of the cell membrane, and optical trap methodology used to quantify membrane tension. We also applied lipidomics/mass spectrometry and RNA-sequencing to investigate the molecular basis of the ΔGβ4 phenotype. Experimental sample sizes were not predetermined, and there were no predefined study end points. Experiments were not randomized, and investigators were not blinded during data acquisition and analysis. In general, experiments were performed at least 3 times (3 biological replicates). Specific information about replication is included in the figure legends. Data analysis protocols are detailed below and in the Supplementary Materials.
HL-60 genetic manipulation
Gβ1-Gβ5 were deleted from HL-60 progenitors using the IDT Alt-R CRISPR-Cas9 system (IDT). Briefly, crRNAs specific for Gβ1-Gβ5 (see Table S2 for sequence details) were resuspended in sterile Duplex Buffer (IDT) to a final concentration of 200 μM. Each crRNA was then mixed with tracrRNA-ATTO 550 (IDT) at a 1:1 ratio and annealed using a thermocycler (BioRad). sgRNA:Cas9 RNP’s were formed by mixing 0.3 μL of 36 μM Cas9 protein (IDT) with annealed sgRNAs at a 1:1 ratio followed by room temperature incubation for 10–20 minutes. HL-60 progenitor cells at a density of 2.2 × 106 (>95% viability) were washed with PBS and then transferred into 200 μL resuspension buffer (Buffer R, Invitrogen). The cells were then mixed with RNPs, loaded into an electroporation cuvette (ThermoFisher Scientific) with E2 buffer, and electroporated at 1350 V for 35 s. Electroporated cells were recovered in 2 mL of RPMI with 20% FBS, 1% Pen/Strep and placed into the incubator at 37 °C, 5% CO2 for 24–48 hours. Subsequently, ATTO 550 positive cells were single cell sorted into 96-well plates using a FACS Aria 3. Cell clones were screened for Gβ gene expression after 2–3 weeks. For screening, cells were fixed and permeabilized using BD fixation and permeabilization buffers, followed by staining using primary antibodies against Gβ isoforms (Table S3) at a dilution 1:100 for 30 min at 4 °C. Cells were then stained with goat anti-rabbit AF647 antibody (ThermoFisher Scientific) for 20 min at room temperature before flow cytometric analysis on a CytoFlex LX machine (Beckman).
To overexpress Gβ4, a GNB4 gene block (IDT) containing homology arms to the lentiviral vector pLVX-Puro (CloneTech) was inserted into XhoI-digested pLVX-Puro using the In-Fusion HD Kit (Takara Bio). The homology arms were 5’-ctaccggactcagatctcga-3’ at the N-terminus of GNB4 and 5’-tcgagctcatcgggatcccgctcgacta-3’ at the C-terminus. The resulting plasmid was combined with plasmids encoding (1) gag-pol from HIV53 and (2) appropriate viral glycoproteins (VSV-G), and the mixture then transfected into HEK293T lentiX cells (Takara Bio) using the Xtremegene transfection reagent (Roche Applied Science). Lentivirus was collected 24 and 48 hours post transfection and stored at −80 °C until use. For transduction, 0.5 to 1 × 106 HL-60 progenitor cells were mixed with lentivirus preparations (1 mL of virus supplemented with polybrene and HEPES) and centrifuged at 1400 × g at 37 °C for 2 hours in either 24-well or 6-well polystyrene plates (Corning). Subsequently, 1 mL of RPMI complete media (10% FBS, 1% Pen/Strep) was added to the cells, followed by overnight incubation at 37 °C, 5% CO2. Cells were then placed in 0.5 μg/mL puromycin for 14 days to select for successfully transduced cells. HL-60 progenitors transduced with lentivirus derived from empty pLVX-Puro were prepared in parallel as controls. To express F-tractin-mCherry in HL-60 cells, DNA encoding the F-tractin-mCherry fusion was amplified from a C1-F-tractin-mCherry plasmid (Addgene, #155218) and then subcloned into the lentiviral vector pLVX-M-Puro (CloneTech) by Gibson reaction. Lentivirus production and transduction was performed as described above. To express PH-AKT-BFP in HL60 cells, DNA encoding the PH domain of AKT was amplified from a PH-AKT-Venus plasmid (Plasmid #85223) and DNA encoding BFP from a pLentiCas9-BFP (Plasmid #78545). The fragments were then subcloned into the lentiviral vector pHR using the Gibson approach.
Flow cytometric MP phagocytosis assay
Neutrophils stained with either Hoechst or CellVue Maroon (both from ThermoFisher) were seeded into fibronectin-coated 24 well plates at a density of 150,000 cells per well. FITC/LRB labeled MPs coated with a phagocytic target ligand (e.g. IgG) were then added at a 1:2 (MP:cell) ratio, followed by incubation at 37 °C, 5% CO2 for 3 hours. In certain experiments, neutrophils were pretreated with 1μM PIK90 (Tocris Biosciences) for 30 min prior to MP addition and maintained in 1μM PIK90 for the duration of the 3 hour coincubation. Cells were then removed from the surface by trypsinization and transferred into FACS tubes for analysis using a Cytoflex LX machine (Beckman). Analysis was performed by first gating out free floating MPs and then identifying LRB+FITClow cells, indicative of successful uptake and acidification of MPs. In each sample, the total number of LRB+FITClow cells was normalized to the total number of live cells to generate a % phagocytosis metric. In experiments using pHrodoGreen or pHrodoRed dyed MPs, the % phagocytosis metric was calculated using pHrodoBright cells (in FITC or PE channels) as the numerator and total cells as the denominator. In experiments comparing ΔGβ4 and wild type HL-60 cells in the same sample, each cell type was labeled with a different dye (either Hoechst or CellVue Maroon, both from ThermoFisher) to distinguish them during endpoint flow cytometric analysis. Dyes were switched between experiments to control for potential effects of the dyes on phagocytosis.
Widefield imaging of MP phagocytosis
Neutrophils or macrophages were stained using Hoechst and/or CellVue Maroon (both from ThermoFisher) and seeded into fibronectin coated 8-well chamber slides (ibidi) at a density of 100,000 or 50,000 cells per well, respectively. FITC/LRB labeled IgG-coated MPs were then added at a 1:2 (MP:cell) ratio, and the samples imaged on a Zeiss Axiovert microscope using a 20× objective lens for at least 2 hours at 1 min intervals. DAPI, FITC, and TRITC images were collected at each time point. Phagocytosed MPs were identified in Fiji by a reduction in FITC signal due to phagolysosome acidification. % phagocytosis was determined by dividing the number of phagocytes containing acidified MPs by the total number of phagocytes in each frame.
Micropipette migration assay
Differentiated ΔGβ4 and WT HL-60 neutrophils were stained with Hoechst (MilliporeSigma) for 20 min at 25 °C and allowed to attach to fibronectin-coated 35 mm glass-bottom cover slips for 1 hour (250,000 cells per dish). Samples were washed to remove nonadherent cells and left in 3 mL of complete RPMI media prior to imaging. A femptotip (Eppendorf) micropipette loaded with sterile filtered RPMI containing 2% fatty acid free BSA, 1% Pen/Strep, 200 nM fMLF, and trace AF647 dye was attached to a FemptoJet 4i (Eppendorf), and the micropipette tip positioned and the center of the imaging frame. Pressure (400 psi) was then applied to dispense the chemoattractant mixture and serial images (40 s intervals between frames) collected over the course of an hour at 20× magnification using a Ti Eclipse microscope with a CSU-0W1 Yokogawa camera (Nikon) at 37 °C, 5% CO2. To measure migration after MP uptake, ΔGβ4 and WT HL-60 cells were preincubated with pHrodoGreen or pHrodoRed labeled MPs, respectively, for 3 hours. Cells that had taken up MPs during this time were FACS sorted based on green (ΔGβ4) or red (WT) fluorescence. A 1:1 mixture of these cells was then applied to fibronectin coated cover slips (250,000 cells per dish) and chemotaxis toward a point source of fMLF measured as described above.
FIB-SEM imaging and analysis
Cells were plated on Aclar coverslips coated with fibronectin and then fixed with 2% glutaraldehyde, 2 mM CaCl2 in 0.08 M sodium cacodylate buffer, pH 7.2. This primary fixation was followed by a ROTO protocol (Reduced Osmium-Thiocarbohydrazide-Osmium) as follows: cells were incubated for one hour in 1% OsO4, 1.25% potassium ferrocyanide in 0.1 M sodium cacodylate buffer on ice, washed with buffer, and then incubated with 1% thiocarbohydrazide in water for 12 minutes. After washing, cells were treated with 1% Osmium tetroxide in 0.1 M cacodylate buffer for 30 min on ice. The samples were then dehydrated using a graded series of ethanol solutions and embedded in Eponate 12. Sample blocks were trimmed and then mounted on an SEM sample holder using double-sided carbon tape (EMS). Colloidal silver paint (EMS) was used to electrically ground the sides of the resin block. The entire surface of the specimen was then sputter coated with a thin layer (5 nm) of gold/palladium. The sample was imaged using immersion, TLD, back-scattered electron (BSE) mode on an FEI Helios Nanolab 650 microscope. Images were recorded after each round of ion beam milling using the SEM beam at 2.0 keV and 0.10 nA current with a working distance of <5 mm. The ion beam was held at 30 keV, with a milling current of 80 pA. Data acquisition occurred through automation using Auto Slice and View G3 software. Raw images were 4,096 × 2,048 pixels, with 20–50 nm slices viewed at a −38° cross-sectional angle. Each raw image had a horizontal field width (HFW) of 10–15 μm with an XY pixel size of 2–4 nm and a 40 nm Z-step size. Images were aligned using the image processing programs in IMOD. All segmentation was performed using semi-manual thresholding and manual annotation of a test set (10% of data) using the LabKit machine learning plug-in for Imaris. 3-D renderings, reconstructions, surface/volume calculations, and internal vesicle volume calculations were performed using Imaris (BitPlane).
Myriocin sphingolipid synthesis inhibition assays
For acute myriocin treatment, HL-60 cells on the 5th day of DMSO differentiation were treated with a final concentration of 50 nM myriocin (Millipore Sigma Aldrich) for 12 hours prior to phagocytosis or C-trap experiments. For long term inhibition of sphingolipid synthesis, HL-60 progenitor cells were incubated over the 5 day DMSO differentiation in the presence of 50 nM myriocin. Treated cells were then used in phagocytosis, migration, or C-trap experiments. No observable effect on viability was observed in either treatment regime.
Osmotic shock experiments
HL-60 cells or terminally differentiated primary human macrophages (derived from hiPSC cells) were suspended in 200 μL complete RPMI (10% FBS, 1.5% DMSO, 1% Pen/Strep) or RPMI 1650 (h-mCSF, 10% FBS, 1% Pen/Strep), respectively, and then applied to a Vi Cell Blue (Beckman Coulter, MA) cell counter to measure cell diameter. The samples were then osmotically shocked for 2 min or 4 min respectively by adding 180 μL of sterile ddH2O to 20 μL of suspended cells (final concentration of 2 × 105 cells per mL) before loading onto the same device. Each measurement was performed using 9 biological replicates. All recorded samples had >90% viability during the assay, as monitored by trypan blue (ThermoFisher) exclusion.
Membrane tension measurements
Plasma membrane tension was quantified using a C-Trap optical trapping device (Lumicks BV, Netherlands). An IR laser beam (50 mW, 1064 nm) was tightly focused through a series of mirrors, beam expanders and a high numerical aperture objective lens (63×/1.2 NA, Nikon Instruments) to form a steerable optical trap. Cells were immobilized inside an Ibidi μ-slide (Ibidi GmbH, Germany) treated with 200 μg/mL fibronectin (Thermo/Sigma). To measure plasma membrane tension, polystyrene beads (2.2 μm, Spherotech Inc, IL) were coated with concanavalin A (50 μg/mL, Thermo/Sigma) and added to the cell culture medium inside the slide. Beads were momentarily placed in contact with the cell membrane, and tethers were then extruded by moving the bead away from the cell perpendicularly at a speed of 2 μm/s. Force measurements were made using the Lumicks Bluelake software suite by capturing the exiting trapping light with a high numerical aperture condenser lens (63×/1.45, oil immersion, Zeiss AB, Germany) and measuring bead displacement in the trap with position-sensitive detectors through back focal plane interferometry. Membrane tether breaking was documented as a sharp discontinuity in tether force during tether extrusion, with breaking distance measured from simultaneously collected brightfield images using FIJI. Data analysis was performed using Python 3.8.0.
Lipidomics
Frozen cell pellets were thawed and extracted using a modified Folch protocol (99). In brief, samples were resuspended in 300 μL of methanol containing SPLASH LIPIDOMIX (Avanti Polar Lipids) as internal standards, vortexed, and then mixed with 600 μL of chloroform. 180 μL of water was added to each tube to induce phase separation, and after mixing, samples were centrifuged at 16,000 × g for 5 min at 4 °C. Subsequently, the lower, chloroform layer was collected and the aqueous layer re-extracted using 450 μL of chloroform:methanol:water (3:48:47 v/v/v). The lower, chloroform layer was collected and pooled with the previous extract. Samples were then dried under nitrogen at 40 °C and resuspended in 100 μL of 90:10 methanol:chloroform. Lipid profiling was performed using an Agilent 6546 Q-TOF mass spectrometer in positive and negative ionization modes, coupled to a ZORBAX Eclipse Plus C-18 Column (100 mm × 2.1, 1.8 μm particle size, Agilent). Mobile Phase A consisted of 10 mM ammonium formate in 50:30:20 water:acetonitrile:isopropanol. Mobile Phase B consisted of 10 mM ammonium formate in 1:9:90 water:acetonitrile:isopropanol. LC gradient conditions were: 0 min at 0% B; 2.7 min at 45% B; 2.8 min at 53% B; 9 min at 65% B; 9.10 min at 89% B; 11 min at 92% B; 11.10 min 100% B; 12 min at 10% B; 15 min at 10% B. Other LC parameters were: flow rate at 0.4 mL/min, column temperature at 60 °C and injection volume was 5 μL. MS source parameters included: gas temp: 200 °C; gas flow: 10 L/min; nebulizer pressure: 50 psig; sheath gas temp: 300 °C; sheath gas flow: 12 L/min; Vcap: 3000 V; nozzle voltage: 0 V; fragmentor: 150V. For lipid annotation, 5 injections of iterative MS/MS acquisition were performed on a pooled lipid extract in both positive and negative polarity. To identify lipid species, iterative MS/MS acquisition data in positive and negative polarity was processed using Agilent Lipid Annotator software 1.0 (100). Subsequent targeted feature extraction and peak integration was performed using Skyline (101). For statistical analysis, the Welch T-test was used for pairwise comparisons between wild type and ΔGβ4 groups with Benjamini-Hochberg FDR correction.
Generation of Gnb4−/− mice
The animal protocols used for this study were approved by the Institutional Animal Care and Use Committee of MSKCC. Two sgRNA sequences (5’-cgtcaaaatatcgcaagtgc-3’) and (5’-aggtgtcagatcaaacc-3’) targeting exon 4 of the Gnb4 locus (IDT) were injected together with purified Cas9 protein (IDT) into C57BL/6 zygotes, which were then transferred into C57BL/6 pseudopregnant females. Founder animals were screened for deletion of the entire exon 4 and the presence of an early stop codon in exon 5, and then bred to homozygosity. Routine genotyping PCRs were performed using the following forward (5’-ggagaacagctagtactcttaac-3’) and reverse (5’-aaaagtatttattagcagtatc-3’) primers. The resulting amplicons for WT and Gnb4 KO mutant alleles are 1360 bp and 305 bp respectively.
A. fumigatus FLARE intratracheal mouse fungal infections
Gnb4−/− and WT mice were infected by intratracheal (i.t.) administration of 60 × 106 FLARE Af293 A. fumigatus (see Supplementary Materials and Methods for staining protocol) in 50 μL. WT mice were also infected with unlabeled A. fumigatus to serve as analysis controls (see below). 18 hours post infection, mice were euthanized and their lungs harvested into 5 mL of digestion buffer (PBS with 5% FBS, 0.1 mg/mL DNAse I, and Type IV collagenase (Worthington) at 2.2 mg/mL). Tissue homogenization was performed using a MACSTm Octo Dissociator for 55 s at 1302 rpm, followed by slow rotation at 37 °C for 40 min. A final mechanical homogenization was performed for 37 s at 2079 rpm, after which all samples were diluted with 5 mL of PBS + 5% FBS, filtered through a 100 μm pore size cell strainer, and centrifuged at 300 × g for 5 min at 4 °C. The pellet was resuspended in 2 mL of ACK lysis buffer (BD) and incubated for 15 min to achieve red blood cell lysis. After quenching in 4 mL of RPMI-1640 +10% FBS, cells were centrifuged at 300 × g for 5 min at 4 °C, followed by resuspension in cold FACS buffer (PBS + 5% FBS). To assess dissemination at 24 hours post-infection, lungs, dLN, spleen, and 200 μL of blood were harvested from each mouse. Lungs were processed as described above. dLN and spleen were homogenized and filtered through a 100 μm pore size cell strainer. Subsequently, the spleen single cell suspension was centrifuged and resuspended in 2 mL of ACK lysis buffer (BD) and incubated for 15 min to achieve red blood cell lysis. After quenching in 4 mL of RPMI-1640 +10% FBS, cells were centrifuged at 300 × g for 5 min at 4 °C, followed by resuspension in cold FACS buffer (PBS + 5% FBS). Blood samples were diluted into 2 mL of ACK lysis buffer (BD) containing 100 mM EDTA and incubated for 15 min to achieve red blood cell lysis. After quenching in 4 mL of RPMI-1640 +10% FBS, cells were centrifuged at 300 × g for 5 min at 4 °C, followed by resuspension in cold FACS buffer (PBS + 5% FBS). Aliquots of 2 × 106 cells from each organ were dispensed into round bottom 96 well plates (Corning) and then stained for 20 min at 4 °C with a viability dye (Tonbo Ghostdye Violet 510 1:250) along with antibodies against Ly6G (BUV395, 1:100), CD11b (BUV805, 1:100), Siglec F (BV650, 1:100), CD45 (BV785), and CD11c (PE-Cy7, 1:100) (Table S4). Samples were then applied to a CytoFlex LX flow cytometer and analyzed using FlowJo software. Neutrophils were identified as CD11b+Ly6G+SiglecF−CD11c− cells. % phagocytosis was determined by dividing the total number of AF633+ neutrophils in each sample by the total number of neutrophils. % FLARE killing was expressed as the number of AF633+dsRed- neutrophils over the total number of AF633+ neutrophils in each sample. Gating for AF633+ neutrophils was facilitated using unlabeled neutrophils extracted from mice infected with unlabeled A. fumigatus. To quantify fungal infection, lung suspensions were diluted 50-fold and 50 μL of each sample was plated on Sabouraud dextrose agar (ThermoFisher). CFUs were quantified after 2 d of incubation at 37 °C. For survival experiments, both Gnb4−/− and WT mice were infected with 65 × 106 CFU CEA10 A. fumigatus. Weight and survival were monitored over the course of 7 d.
Statistical analysis
Figure panels show either representative images or data pooled from all biological replicates. T-, ANOVA, and Logrank testing was performed as indicated using Prism. Error bars denote SD or SEM as described in the figure legends.
Supplementary Material
Acknowledgements:
The authors thank the Molecular Cytology Core Facility, the Cell Metabolism Core Facility, the Flow Cytometry Core Facility, and the Integrated Genomics Operation at MSKCC for technical support; the New York Structural Biology Center for assistance with FIB-SEM; and members of the M.H., O.D.W., and J.G.C. labs for advice.
Funding:
This work was supported in part by the NIH (R01-AI087644 to M.H., R35-GM118167 to O.D.W., R01-AI45073 to J.G.C., R25-AI40472 to J.R.C, R01-NS115715 to F.G., R01-AI130345 to F.G., and P30-CA008748 to MSKCC), the National Science Foundation (2019598 and 1548297 to O.D.W.), the Donald B. and Catherine C. Marron Cancer Metabolism Center (J.R.C.), the Schmidt Science Fellows Program (B.Y.W.), the Cancer Research Institute (B.Y.W.), and the MSKCC Basic Research Postdoc Innovation Award (B.Y.W.).
Footnotes
Competing interests: B.Y.W., O.D.W., J.G.C., and M.H. have applied for a patent on the discovery that knockout or null mutations in Gβ4 leads to increased phagocytosis in professional phagocytic cells (PCT/US2024/019787). J.G.C. is a member of the Scientific advisory Board of BeBio Pharma and consults for Lycia therapeutics and DrenBio inc.
Data and materials availability:
RNAseq data has been deposited with the BioSample (SAMN41132906), Bioproject (PRJNA1106430), and SRA (PRJNA1106430) repositories. Custom MATLAB code may be found at Github (https://github.com/benjaminywiner/) or Zenodo (10.5281/zenodo). Tabulated data underlying the figures is provided in Data file S1. All other data needed to support the conclusions of the paper are present in the paper itself or the Supplementary Materials. Gnb4−/− mice, hiPSC cell lines, and all other reagents are available via material transfer agreement. Requests for reagents should be addressed to B.Y.W. (winerb@mskcc.org) or M.H. (husem@mskcc.org).
References
- 1.Wang J, Hossain M, Thanabalasuriar A, Gunzer M, Meininger C, Kubes P, Visualizing the function and fate of neutrophils in sterile injury and repair. Science 358, 111–116 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Peiseler M, Kubes P, More friend than foe: the emerging role of neutrophils in tissue repair. J Clin Invest 129, 2629–2639 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Witter AR, Okunnu BM, Berg RE, The Essential Role of Neutrophils during Infection with the Intracellular Bacterial Pathogen Listeria monocytogenes. J Immunol 197, 1557–1565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pylaeva E, Korschunow G, Spyra I, Bordbari S, Siakaeva E, Ozel I, Domnich M, Squire A, Hasenberg A, Thangavelu K, Hussain T, Goetz M, Lang KS, Gunzer M, Hansen W, Buer J, Bankfalvi A, Lang S, Jablonska J, During early stages of cancer, neutrophils initiate anti-tumor immune responses in tumor-draining lymph nodes. Cell Rep 40, 111171 (2022). [DOI] [PubMed] [Google Scholar]
- 5.Li C, Ebrahimi A, Schluesener H, Drug pipeline in neurodegeneration based on transgenic mice models of Alzheimer’s disease. Ageing Res Rev 12, 116–140 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Gordon S, Phagocytosis: An Immunobiologic Process. Immunity 44, 463–475 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Yanuck SF, Microglial Phagocytosis of Neurons: Diminishing Neuronal Loss in Traumatic, Infectious, Inflammatory, and Autoimmune CNS Disorders. Front Psychiatry 10, 712 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schrijvers DM, De Meyer GR, Herman AG, Martinet W, Phagocytosis in atherosclerosis: Molecular mechanisms and implications for plaque progression and stability. Cardiovasc Res 73, 470–480 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Abdolmaleki F, Farahani N, Gheibi Hayat SM, Pirro M, Bianconi V, Barreto GE, Sahebkar A, The Role of Efferocytosis in Autoimmune Diseases. Front Immunol 9, 1645 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alvey C, Discher DE, Engineering macrophages to eat cancer: from “marker of self” CD47 and phagocytosis to differentiation. J Leukoc Biol 102, 31–40 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dooling LJ, Andrechak JC, Hayes BH, Kadu S, Zhang W, Pan R, Vashisth M, Irianto J, Alvey CM, Ma L, Discher DE, Cooperative phagocytosis of solid tumours by macrophages triggers durable anti-tumour responses. Nat Biomed Eng, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrechak JC, Dooling LJ, Discher DE, The macrophage checkpoint CD47 : SIRPalpha for recognition of ‘self’ cells: from clinical trials of blocking antibodies to mechanobiological fundamentals. Philos Trans R Soc Lond B Biol Sci 374, 20180217 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johansson C, Kirsebom FCM, Neutrophils in respiratory viral infections. Mucosal Immunol 14, 815–827 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaumouille V, Waterman CM, Physical Constraints and Forces Involved in Phagocytosis. Front Immunol 11, 1097 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mylvaganam S, Freeman SA, Grinstein S, The cytoskeleton in phagocytosis and macropinocytosis. Curr Biol 31, R619–R632 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Bretou M, Saez PJ, Sanseau D, Maurin M, Lankar D, Chabaud M, Spampanato C, Malbec O, Barbier L, Muallem S, Maiuri P, Ballabio A, Helft J, Piel M, Vargas P, Lennon-Dumenil AM, Lysosome signaling controls the migration of dendritic cells. Sci Immunol 2, (2017). [DOI] [PubMed] [Google Scholar]
- 17.Chabaud M, Heuze ML, Bretou M, Vargas P, Maiuri P, Solanes P, Maurin M, Terriac E, Le Berre M, Lankar D, Piolot T, Adelstein RS, Zhang Y, Sixt M, Jacobelli J, Benichou O, Voituriez R, Piel M, Lennon-Dumenil AM, Cell migration and antigen capture are antagonistic processes coupled by myosin II in dendritic cells. Nat Commun 6, 7526 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo Y, Isaac BM, Casadevall A, Cox D, Phagocytosis inhibits F-actin-enriched membrane protrusions stimulated by fractalkine (CX3CL1) and colony-stimulating factor 1. Infect Immun 77, 4487–4495 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Branzk N, Lubojemska A, Hardison SE, Wang Q, Gutierrez MG, Brown GD, Papayannopoulos V, Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat Immunol 15, 1017–1025 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yipp BG, Petri B, Salina D, Jenne CN, Scott BN, Zbytnuik LD, Pittman K, Asaduzzaman M, Wu K, Meijndert HC, Malawista SE, de Boisfleury Chevance A, Zhang K, Conly J, Kubes P, Infection-induced NETosis is a dynamic process involving neutrophil multitasking in vivo. Nat Med 18, 1386–1393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jhingran A, Mar KB, Kumasaka DK, Knoblaugh SE, Ngo LY, Segal BH, Iwakura Y, Lowell CA, Hamerman JA, Lin X, Hohl TM, Tracing conidial fate and measuring host cell antifungal activity using a reporter of microbial viability in the lung. Cell Rep 2, 1762–1773 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hopke A, Scherer A, Kreuzburg S, Abers MS, Zerbe CS, Dinauer MC, Mansour MK, Irimia D, Neutrophil swarming delays the growth of clusters of pathogenic fungi. Nat Commun 11, 2031 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamza B, Wong E, Patel S, Cho H, Martel J, Irimia D, Retrotaxis of human neutrophils during mechanical confinement inside microfluidic channels. Integr Biol (Camb) 6, 175–183 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uribe-Querol E, Rosales C, Phagocytosis: Our Current Understanding of a Universal Biological Process. Front Immunol 11, 1066 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vorselen D, Labitigan RLD, Theriot JA, A mechanical perspective on phagocytic cup formation. Curr Opin Cell Biol 66, 112–122 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Jain N, Moeller J, Vogel V, Mechanobiology of Macrophages: How Physical Factors Coregulate Macrophage Plasticity and Phagocytosis. Annu Rev Biomed Eng 21, 267–297 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Adlerz KM, Aranda-Espinoza H, Hayenga HN, Substrate elasticity regulates the behavior of human monocyte-derived macrophages. Eur Biophys J 45, 301–309 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Blakney AK, Swartzlander MD, Bryant SJ, The effects of substrate stiffness on the in vitro activation of macrophages and in vivo host response to poly(ethylene glycol)-based hydrogels. J Biomed Mater Res A 100, 1375–1386 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okamoto T, Takagi Y, Kawamoto E, Park EJ, Usuda H, Wada K, Shimaoka M, Reduced substrate stiffness promotes M2-like macrophage activation and enhances peroxisome proliferator-activated receptor gamma expression. Exp Cell Res 367, 264–273 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Underhill DM, Goodridge HS, Information processing during phagocytosis. Nat Rev Immunol 12, 492–502 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sosale NG, Rouhiparkouhi T, Bradshaw AM, Dimova R, Lipowsky R, Discher DE, Cell rigidity and shape override CD47’s “self”-signaling in phagocytosis by hyperactivating myosin-II. Blood 125, 542–552 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bajno L, Peng XR, Schreiber AD, Moore HP, Trimble WS, Grinstein S, Focal exocytosis of VAMP3-containing vesicles at sites of phagosome formation. J Cell Biol 149, 697–706 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braun V, Fraisier V, Raposo G, Hurbain I, Sibarita JB, Chavrier P, Galli T, Niedergang F, TI-VAMP/VAMP7 is required for optimal phagocytosis of opsonised particles in macrophages. EMBO J 23, 4166–4176 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hackam DJ, Rotstein OD, Sjolin C, Schreiber AD, Trimble WS, Grinstein S, v-SNARE-dependent secretion is required for phagocytosis. Proc Natl Acad Sci U S A 95, 11691–11696 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peracino B, Borleis J, Jin T, Westphal M, Schwartz JM, Wu L, Bracco E, Gerisch G, Devreotes P, Bozzaro S, G protein beta subunit-null mutants are impaired in phagocytosis and chemotaxis due to inappropriate regulation of the actin cytoskeleton. J Cell Biol 141, 1529–1537 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wen X, Xu X, Sun W, Chen K, Pan M, Wang JM, Bolland SM, Jin T, G-protein-coupled formyl peptide receptors play a dual role in neutrophil chemotaxis and bacterial phagocytosis. Mol Biol Cell 30, 346–356 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan M, Xu X, Chen Y, Jin T, Identification of a Chemoattractant G-Protein-Coupled Receptor for Folic Acid that Controls Both Chemotaxis and Phagocytosis. Dev Cell 36, 428–439 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen A, Jeng EE, Voorhies M, Symington J, Ali N, Rodriguez RA, Bassik MC, Sil A, Genome-scale CRISPR screening reveals that C3aR signaling is critical for rapid capture of fungi by macrophages. PLoS Pathog 18, e1010237 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamber RA, Nishiga Y, Morton B, Banuelos AM, Barkal AA, Vences-Catalán F, Gu M, Fernandez D, Seoane JA, Yao D, Liu K, Lin S, Spees K, Curtis C, Jerby-Arnon L, Weissman IL, Sage J, Bassik MC, Inter-cellular CRISPR screens reveal regulators of cancer cell phagocytosis. Nature 597, 549–554 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamada KM, Sixt M, Mechanisms of 3D cell migration. Nat Rev Mol Cell Biol 20, 738–752 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Schwarz J, Bierbaum V, Vaahtomeri K, Hauschild R, Brown M, de Vries I, Leithner A, Reversat A, Merrin J, Tarrant T, Bollenbach T, Sixt M, Dendritic Cells Interpret Haptotactic Chemokine Gradients in a Manner Governed by Signal-to-Noise Ratio and Dependent on GRK6. Curr Biol 27, 1314–1325 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Sarris M, Sixt M, Navigating in tissue mazes: chemoattractant interpretation in complex environments. Curr Opin Cell Biol 36, 93–102 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Clapham DE, Neer EJ, G protein beta gamma subunits. Annu Rev Pharmacol Toxicol 37, 167–203 (1997). [DOI] [PubMed] [Google Scholar]
- 44.Kankanamge D, Tennakoon M, Karunarathne A, Gautam N, G protein gamma subunit, a hidden master regulator of GPCR signaling. J Biol Chem 298, 102618 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vorselen D, Wang Y, de Jesus MM, Shah PK, Footer MJ, Huse M, Cai W, Theriot JA, Microparticle traction force microscopy reveals subcellular force exertion patterns in immune cell-target interactions. Nat Commun 11, 20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vorselen D, Barger SR, Wang Y, Cai W, Theriot JA, Gauthier NC, Krendel M, Phagocytic ‘teeth’ and myosin-II ‘jaw’ power target constriction during phagocytosis. Elife 10, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Araki N, Johnson MT, Swanson JA, A role for phosphoinositide 3-kinase in the completion of macropinocytosis and phagocytosis by macrophages. J Cell Biol 135, 1249–1260 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bohdanowicz M, Schlam D, Hermansson M, Rizzuti D, Fairn GD, Ueyama T, Somerharju P, Du G, Grinstein S, Phosphatidic acid is required for the constitutive ruffling and macropinocytosis of phagocytes. Mol Biol Cell 24, 1700–1712, S1701–1707 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mishra AK, Rodriguez M, Torres AY, Smith M, Rodriguez A, Bond A, Morrissey MA, Montell DJ, Hyperactive Rac stimulates cannibalism of living target cells and enhances CAR-M-mediated cancer cell killing. Proc Natl Acad Sci U S A 120, e2310221120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rovera G, Santoli D, Damsky C, Human promyelocytic leukemia cells in culture differentiate into macrophage-like cells when treated with a phorbol diester. Proc Natl Acad Sci U S A 76, 2779–2783 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Z, Roche PA, Macropinocytosis in phagocytes: regulation of MHC class-II-restricted antigen presentation in dendritic cells. Front Physiol 6, 1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chao MP, Weissman IL, Majeti R, The CD47-SIRPalpha pathway in cancer immune evasion and potential therapeutic implications. Curr Opin Immunol 24, 225–232 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Logtenberg MEW, Scheeren FA, Schumacher TN, The CD47-SIRPalpha Immune Checkpoint. Immunity 52, 742–752 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willingham SB, Volkmer JP, Gentles AJ, Sahoo D, Dalerba P, Mitra SS, Wang J, Contreras-Trujillo H, Martin R, Cohen JD, Lovelace P, Scheeren FA, Chao MP, Weiskopf K, Tang C, Volkmer AK, Naik TJ, Storm TA, Mosley AR, Edris B, Schmid SM, Sun CK, Chua MS, Murillo O, Rajendran P, Cha AC, Chin RK, Kim D, Adorno M, Raveh T, Tseng D, Jaiswal S, Enger PO, Steinberg GK, Li G, So SK, Majeti R, Harsh GR, van de Rijn M, Teng NN, Sunwoo JB, Alizadeh AA, Clarke MF, Weissman IL, The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc Natl Acad Sci U S A 109, 6662–6667 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jalil AR, Hayes BH, Andrechak JC, Xia Y, Chenoweth DM, Discher DE, Multivalent, Soluble Nano-Self Peptides Increase Phagocytosis of Antibody-Opsonized Targets while Suppressing “Self” Signaling. ACS Nano 14, 15083–15093 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodriguez PL, Harada T, Christian DA, Pantano DA, Tsai RK, Discher DE, Minimal “Self” peptides that inhibit phagocytic clearance and enhance delivery of nanoparticles. Science 339, 971–975 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kitai Y, Ishiura M, Saitoh K, Matsumoto N, Owashi K, Yamada S, Muromoto R, Kashiwakura JI, Oritani K, Matsuda T, CD47 promotes T-cell lymphoma metastasis by up-regulating AKAP13-mediated RhoA activation. Int Immunol 33, 273–280 (2021). [DOI] [PubMed] [Google Scholar]
- 58.Nagata S, Segawa K, Sensing and clearance of apoptotic cells. Curr Opin Immunol 68, 1–8 (2021). [DOI] [PubMed] [Google Scholar]
- 59.Cox D, Tseng CC, Bjekic G, Greenberg S, A requirement for phosphatidylinositol 3-kinase in pseudopod extension. J Biol Chem 274, 1240–1247 (1999). [DOI] [PubMed] [Google Scholar]
- 60.Schlam D, Bagshaw RD, Freeman SA, Collins RF, Pawson T, Fairn GD, Grinstein S, Phosphoinositide 3-kinase enables phagocytosis of large particles by terminating actin assembly through Rac/Cdc42 GTPase-activating proteins. Nat Commun 6, 8623 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ritter AT, Shtengel G, Xu CS, Weigel A, Hoffman DP, Freeman M, Iyer N, Alivodej N, Ackerman D, Voskoboinik I, Trapani J, Hess HF, Mellman I, ESCRT-mediated membrane repair protects tumor-derived cells against T cell attack. Science 376, 377–382 (2022). [DOI] [PubMed] [Google Scholar]
- 62.Wang G, Luo X, Gu R, Yang S, Qu Y, Zhai S, Zhao Q, Li K, Zhang S, PyMIC: A deep learning toolkit for annotation-efficient medical image segmentation. Comput Methods Programs Biomed 231, 107398 (2023). [DOI] [PubMed] [Google Scholar]
- 63.Masters TA, Pontes B, Viasnoff V, Li Y, Gauthier NC, Plasma membrane tension orchestrates membrane trafficking, cytoskeletal remodeling, and biochemical signaling during phagocytosis. Proc Natl Acad Sci U S A 110, 11875–11880 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kovari DT, Wei W, Chang P, Toro JS, Beach RF, Chambers D, Porter K, Koo D, Curtis JE, Frustrated Phagocytic Spreading of J774A-1 Macrophages Ends in Myosin II-Dependent Contraction. Biophys J 111, 2698–2710 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dai J, Sheetz MP, Mechanical properties of neuronal growth cone membranes studied by tether formation with laser optical tweezers. Biophys J 68, 988–996 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bo L, Waugh RE, Determination of bilayer membrane bending stiffness by tether formation from giant, thin-walled vesicles. Biophys J 55, 509–517 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heinrich V, Waugh RE, A piconewton force transducer and its application to measurement of the bending stiffness of phospholipid membranes. Ann Biomed Eng 24, 595–605 (1996). [DOI] [PubMed] [Google Scholar]
- 68.Raucher D, Sheetz MP, Characteristics of a membrane reservoir buffering membrane tension. Biophys J 77, 1992–2002 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Glaros EN, Kim WS, Wu BJ, Suarna C, Quinn CM, Rye KA, Stocker R, Jessup W, Garner B, Inhibition of atherosclerosis by the serine palmitoyl transferase inhibitor myriocin is associated with reduced plasma glycosphingolipid concentration. Biochem Pharmacol 73, 1340–1346 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Wadsworth JM, Clarke DJ, McMahon SA, Lowther JP, Beattie AE, Langridge-Smith PR, Broughton HB, Dunn TM, Naismith JH, Campopiano DJ, The chemical basis of serine palmitoyltransferase inhibition by myriocin. J Am Chem Soc 135, 14276–14285 (2013). [DOI] [PubMed] [Google Scholar]
- 71.Greene CJ, Nguyen JA, Cheung SM, Arnold CR, Balce DR, Wang YT, Soderholm A, McKenna N, Aggarwal D, Campden RI, Ewanchuk BW, Virgin HW, Yates RM, Macrophages disseminate pathogen associated molecular patterns through the direct extracellular release of the soluble content of their phagolysosomes. Nat Commun 13, 3072 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khoyratty TE, Ai Z, Ballesteros I, Eames HL, Mathie S, Martin-Salamanca S, Wang L, Hemmings A, Willemsen N, von Werz V, Zehrer A, Walzog B, van Grinsven E, Hidalgo A, Udalova IA, Distinct transcription factor networks control neutrophil-driven inflammation. Nat Immunol 22, 1093–1106 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang H, Reilly MP, Human Induced Pluripotent Stem Cell-Derived Macrophages for Unraveling Human Macrophage Biology. Arterioscler Thromb Vasc Biol 37, 2000–2006 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vaughan-Jackson A, Stodolak S, Ebrahimi KH, Browne C, Reardon PK, Pires E, Gilbert-Jaramillo J, Cowley SA, James WS, Differentiation of human induced pluripotent stem cells to authentic macrophages using a defined, serum-free, open-source medium. Stem Cell Reports 16, 3093 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alsinet C, Primo MN, Lorenzi V, Bello E, Kelava I, Jones CP, Vilarrasa-Blasi R, Sancho-Serra C, Knights AJ, Park JE, Wyspianska BS, Trynka G, Tough DF, Bassett A, Gaffney DJ, Alvarez-Errico D, Vento-Tormo R, Robust temporal map of human in vitro myelopoiesis using single-cell genomics. Nat Commun 13, 2885 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Giese MA, Hind LE, Huttenlocher A, Neutrophil plasticity in the tumor microenvironment. Blood 133, 2159–2167 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guo Y, Kasahara S, Jhingran A, Tosini NL, Zhai B, Aufiero MA, Mills KAM, Gjonbalaj M, Espinosa V, Rivera A, Luster AD, Hohl TM, During Aspergillus Infection, Monocyte-Derived DCs, Neutrophils, and Plasmacytoid DCs Enhance Innate Immune Defense through CXCR3-Dependent Crosstalk. Cell Host Microbe 28, 104–116 e104 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mircescu MM, Lipuma L, van Rooijen N, Pamer EG, Hohl TM, Essential role for neutrophils but not alveolar macrophages at early time points following Aspergillus fumigatus infection. J Infect Dis 200, 647–656 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Espinosa V, Dutta O, Heung LJ, Wang K, Chang YJ, Soteropoulos P, Hohl TM, Siracusa MC, Rivera A, Cutting Edge: Neutrophils License the Maturation of Monocytes into Effective Antifungal Effectors. J Immunol 209, 1827–1831 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Monod M, Paris S, Sarfati J, Jaton-Ogay K, Ave P, Latge JP, Virulence of alkaline protease-deficient mutants of Aspergillus fumigatus. FEMS Microbiol Lett 106, 39–46 (1993). [DOI] [PubMed] [Google Scholar]
- 81.Wei W, Zhang L, Xin W, Pan Y, Tatenhorst L, Hao Z, Gerner ST, Huber S, Juenemann M, Butz M, Huttner HB, Bahr M, Fitzner D, Jia F, Doeppner TR, TREM2 regulates microglial lipid droplet formation and represses post-ischemic brain injury. Biomed Pharmacother 170, 115962 (2024). [DOI] [PubMed] [Google Scholar]
- 82.Pang XW, Chu YH, Zhou LQ, Chen M, You YF, Tang Y, Yang S, Zhang H, Xiao J, Deng G, Wang W, Shang K, Qin C, Tian DS, Trem2 deficiency attenuates microglial phagocytosis and autophagic-lysosomal activation in white matter hypoperfusion. J Neurochem 167, 489–504 (2023). [DOI] [PubMed] [Google Scholar]
- 83.Castoldi A, Sanin DE, van Teijlingen Bakker N, Aguiar CF, de Brito Monteiro L, Rana N, Grzes KM, Kabat AM, Curtis J, Cameron AM, Caputa G, Antonio de Souza T, Souto FO, Buescher JM, Edwards-Hicks J, Pearce EL, Pearce EJ, Saraiva Camara NO, Metabolic and functional remodeling of colonic macrophages in response to high-fat diet-induced obesity. iScience 26, 107719 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Walther TC, Farese RV Jr., Lipid droplets and cellular lipid metabolism. Annu Rev Biochem 81, 687–714 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Farese RV Jr., Walther TC, Glycerolipid Synthesis and Lipid Droplet Formation in the Endoplasmic Reticulum. Cold Spring Harb Perspect Biol 15, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jarc E, Petan T, Lipid Droplets and the Management of Cellular Stress. Yale J Biol Med 92, 435–452 (2019). [PMC free article] [PubMed] [Google Scholar]
- 87.Olzmann JA, Carvalho P, Dynamics and functions of lipid droplets. Nat Rev Mol Cell Biol 20, 137–155 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hetz C, Zhang K, Kaufman RJ, Mechanisms, regulation and functions of the unfolded protein response. Nat Rev Mol Cell Biol 21, 421–438 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lassuthova P, Safka Brozkova D, Neupauerova J, Krutova M, Mazanec R, Seeman P, Confirmation of the GNB4 gene as causal for Charcot-Marie-Tooth disease by a novel de novo mutation in a Czech patient. Neuromuscul Disord 27, 57–60 (2017). [DOI] [PubMed] [Google Scholar]
- 90.Soong BW, Huang YH, Tsai PC, Huang CC, Pan HC, Lu YC, Chien HJ, Liu TT, Chang MH, Lin KP, Tu PH, Kao LS, Lee YC, Exome sequencing identifies GNB4 mutations as a cause of dominant intermediate Charcot-Marie-Tooth disease. Am J Hum Genet 92, 422–430 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hsu YH, Lin KP, Guo YC, Tsai YS, Liao YC, Lee YC, Mutation spectrum of Charcot-Marie-Tooth disease among the Han Chinese in Taiwan. Ann Clin Transl Neurol 6, 1090–1101 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kwon HM, Kim HS, Kim SB, Park JH, Nam DE, Lee AJ, Nam SH, Hwang S, Chung KW, Choi BO, Clinical and Neuroimaging Features in Charcot-Marie-Tooth Patients with GNB4 Mutations. Life (Basel) 11, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zigmond RE, Echevarria FD, Macrophage biology in the peripheral nervous system after injury. Prog Neurobiol 173, 102–121 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kolter J, Kierdorf K, Henneke P, Origin and Differentiation of Nerve-Associated Macrophages. J Immunol 204, 271–279 (2020). [DOI] [PubMed] [Google Scholar]
- 95.Klein D, Patzko A, Schreiber D, van Hauwermeiren A, Baier M, Groh J, West BL, Martini R, Targeting the colony stimulating factor 1 receptor alleviates two forms of Charcot-Marie-Tooth disease in mice. Brain 138, 3193–3205 (2015). [DOI] [PubMed] [Google Scholar]
- 96.Klein D, Groh J, Weishaupt A, Martini R, Endogenous antibodies contribute to macrophage-mediated demyelination in a mouse model for CMT1B. J Neuroinflammation 12, 49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Carenini S, Maurer M, Werner A, Blazyca H, Toyka KV, Schmid CD, Raivich G, Martini R, The role of macrophages in demyelinating peripheral nervous system of mice heterozygously deficient in p0. J Cell Biol 152, 301–308 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sloas C, Gill S, Klichinsky M, Engineered CAR-Macrophages as Adoptive Immunotherapies for Solid Tumors. Front Immunol 12, 783305 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Folch J, Lees M, Sloane Stanley GH, A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226, 497–509 (1957). [PubMed] [Google Scholar]
- 100.Koelmel JP, Li X, Stow SM, Sartain MJ, Murali A, Kemperman R, Tsugawa H, Takahashi M, Vasiliou V, Bowden JA, Yost RA, Garrett TJ, Kitagawa N, Lipid Annotator: Towards Accurate Annotation in Non-Targeted Liquid Chromatography High-Resolution Tandem Mass Spectrometry (LC-HRMS/MS) Lipidomics Using A Rapid and User-Friendly Software. Metabolites 10, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Adams KJ, Pratt B, Bose N, Dubois LG, St John-Williams L, Perrott KM, Ky K, Kapahi P, Sharma V, MacCoss MJ, Moseley MA, Colton CA, MacLean BX, Schilling B, Thompson JW, Alzheimer’s Disease Metabolomics C, Skyline for Small Molecules: A Unifying Software Package for Quantitative Metabolomics. J Proteome Res 19, 1447–1458 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Murao S, Gemmell MA, Callaham MF, Anderson NL, Huberman E, Control of macrophage cell differentiation in human promyelocytic HL-60 leukemia cells by 1,25-dihydroxyvitamin D3 and phorbol-12-myristate-13-acetate. Cancer Res 43, 4989–4996 (1983). [PubMed] [Google Scholar]
- 103.Punt PJ, Oliver RP, Dingemanse MA, Pouwels PH, van den Hondel CA, Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 56, 117–124 (1987). [DOI] [PubMed] [Google Scholar]
- 104.Mikkelsen L, Sarrocco S, Lubeck M, Jensen DF, Expression of the red fluorescent protein DsRed-Express in filamentous ascomycete fungi. FEMS Microbiol Lett 223, 135–139 (2003). [DOI] [PubMed] [Google Scholar]
- 105.Wang GG, Calvo KR, Pasillas MP, Sykes DB, Hacker H, Kamps MP, Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nat Methods 3, 287–293 (2006). [DOI] [PubMed] [Google Scholar]
- 106.Miller JD, Ganat YM, Kishinevsky S, Bowman RL, Liu B, Tu EY, Mandal PK, Vera E, Shim JW, Kriks S, Taldone T, Fusaki N, Tomishima MJ, Krainc D, Milner TA, Rossi DJ, Studer L, Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell 13, 691–705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lachmann N, Ackermann M, Frenzel E, Liebhaber S, Brennig S, Happle C, Hoffmann D, Klimenkova O, Luttge D, Buchegger T, Kuhnel MP, Schambach A, Janciauskiene S, Figueiredo C, Hansen G, Skokowa J, Moritz T, Large-scale hematopoietic differentiation of human induced pluripotent stem cells provides granulocytes or macrophages for cell replacement therapies. Stem Cell Reports 4, 282–296 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Quann EJ, Merino E, Furuta T, Huse M, Localized diacylglycerol drives the polarization of the microtubule-organizing center in T cells. Nat Immunol 10, 627–635 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNAseq data has been deposited with the BioSample (SAMN41132906), Bioproject (PRJNA1106430), and SRA (PRJNA1106430) repositories. Custom MATLAB code may be found at Github (https://github.com/benjaminywiner/) or Zenodo (10.5281/zenodo). Tabulated data underlying the figures is provided in Data file S1. All other data needed to support the conclusions of the paper are present in the paper itself or the Supplementary Materials. Gnb4−/− mice, hiPSC cell lines, and all other reagents are available via material transfer agreement. Requests for reagents should be addressed to B.Y.W. (winerb@mskcc.org) or M.H. (husem@mskcc.org).