Abstract
The chemokine receptors CCR5 and CXCR4 act synergistically with CD4 in an ordered multistep mechanism to allow the binding and entry of human immunodeficiency virus type 1 (HIV-1). The efficiency of such a coordinated mechanism depends on the spatial distribution of the participating molecules on the cell surface. Immunoelectron microscopy was performed to address the subcellular localization of the chemokine receptors and CD4 at high resolution. Cells were fixed, cryoprocessed, and frozen; 80-nm cryosections were double labeled with combinations of CCR5, CXCR4, and CD4 antibodies and then stained with immunogold. Surprisingly, CCR5, CXCR4, and CD4 were found predominantly on microvilli and appeared to form homogeneous microclusters in all cell types examined, including macrophages and T cells. Further, while mixed microclusters were not observed, homogeneous microclusters of CD4 and the chemokine receptors were frequently separated by distances less than the diameter of an HIV-1 virion. Such distributions are likely to facilitate cooperative interactions with HIV-1 during virus adsorption to and penetration of human leukocytes and have significant implications for development of therapeutically useful inhibitors of the entry process. Although the mechanism underlying clustering is not understood, clusters were observed in small trans-Golgi vesicles, implying that they were organized shortly after synthesis and well before insertion into the cellular membrane. Chemokine receptors normally act as sensors, detecting concentration gradients of their ligands and thus providing directional information for cellular migration during both normal homeostasis and inflammatory responses. Localization of these sensors on the microvilli should enable more precise monitoring of their environment, improving efficiency of the chemotactic process. Moreover, since selectins, some integrins, and actin are also located on or in the microvillus, this organelle has many of the major elements required for chemotaxis.
Human immunodeficiency virus (HIV) therapies have been highly successful in slowing disease progression, increasing health and well-being, and prolonging life. However, viral resistance is now becoming common, and since most existing drugs target only two viral proteins, reverse transcriptase and protease, cross-resistance is a significant problem. One solution to the issue of resistance is development of new complementary therapies based on novel mechanisms of action. The discovery that the chemokine receptors CCR5 and CXCR4, in addition to CD4, are required for viral entry not only furthered understanding of the fusion and infection process but provided two new targets for therapeutic intervention (3, 12, 14, 17, 18, 22, 44).
The entry mechanism as currently understood is an ordered process in which the viral envelope protein, gp120, following interaction with CD4, undergoes a conformational change allowing binding to the appropriate chemokine receptor, CCR5 for macrophagetropic or R5 strains, and CXCR4 for T-cell-tropic or X4 strains. This second interaction produces a further conformational change in gp120, activating gp41 and thereby initiating fusion with the cell membrane and viral entry (33, 45, 51, 55, 58, 61, 62). Substantial human genetic evidence supports the pivotal role of CCR5 and the hypothesis that antagonists of the receptor will have antiviral activity (13, 27, 34, 49, 64). A 32-bp deletion in the coding region of the gene results in a frameshift, early termination of translation, and the lack of surface expression. Individuals homozygous for the Δ32 CCR5 allele are highly resistant to infection by HIV, while those who are heterozygous become infected but exhibit delayed disease progression. Importantly, the health of Δ32 homozygotes appears unimpaired, implying that CCR5 antagonists will be without mechanism of action based side effects.
CCR5 and CXCR4 are both G-protein-coupled receptors (GPCRs) (9, 22, 41, 48) and therefore attractive targets for conventional small molecule therapeutics, as there is a long history of success against this class of proteins. However, developing antagonists with antiviral activity may be challenging, as HIV, unlike classical ligands for GPCRs, is polyvalent. Each gp120-gp41 complex is a trimer, and there are many trimers on every viral particle (35). Thus, each virion has the potential to bind cooperatively to multiple receptors, a process which is more difficult to inhibit effectively than interaction of a monomeric ligand with a single binding site. In this regard, several mutations in CCR5 have been reported which dramatically lower affinity for monomeric gp120 but have much smaller effects on the receptor's ability to support infectivity (8, 52), a result consistent with polyvalent cooperative interaction.
Although the virus is polyvalent, efficient cooperative binding also depends on the spatial distribution of the cognate host cell components, CD4, CCR5, and CXCR4. A number of groups, using fluorescence microscopy, have found these molecules to be randomly distributed on the surface in an unstimulated state but to colocalize following addition of gp120 (2, 28, 59). Unfortunately, the resolution provided by light microscopy restricts the conclusions which can be drawn at the molecular level. To circumvent these limitations, we have carried out high-resolution immunogold electron microscopy. Here we report that CCR5, CXCR4, and CD4, rather than being randomly distributed, are preferentially located on cell surface microvilli in human macrophages and T cells, as well as in genetically engineered cells. Further, there is a tendency for each of the molecules to form homogenous microclusters, and while we have not observed mixed microclusters, separation between them is often less than a viral diameter.
MATERIALS AND METHODS
Antibodies.
Rabbit polyclonal antibodies directed against the N termini of the human chemokine receptors were generated using peptides with the sequences of residues 2-31-Nle for CCR5, 2-38-Nle for CXCR4, and 21-40-Nle for CCR2. Antibodies to the C terminus of CCR5 were produced using the sequence Cys-Nle-344-352. All peptides were coupled to thyroglobulin through their cysteine residues using the sulfo-m-maleimidobenzoyl-N-hydroxysulfosuccinimide ester method (1). Polyclonal receptor antibodies were produced by Pocono Rabbit Farm (Canadensis, Pa.) and were purified by protein A-Sepharose chromatography. Monoclonal antibodies (MAbs) to CCR5 (2D7) and CXCR4 (12G5) were obtained from PharMingen (San Diego, Calif.). MAbs against human CD4 (hCD4) (13B8.2 and MEM-115) and hCD8 (UCHT-4) were from Biodesign International (Kennebunk, Main). Additional MAbs against hCD4 were also obtained: OKT4 (Ortho Diagnostic Systems, Raritan, N.J.), Leu-3a (Becton Dickinson, Bedford, Mass.), MT-310 (DAKO International, Carpenteria, Calif.), and RPA-T4 (PharMingen). The MAb to hCD3 (UCHT1) was acquired from DAKO International. The human MAb 1b12, which recognizes the CD4-binding domain of gp 120 and neutralizes primary HIV isolates (11, 29), was generously provided by A. J. Conley, Merck Research Laboratories (West Point, Pa.).
Receptor cloning and expression.
Chemokine receptors were cloned from a human bone marrow DNA library, and sequences were verified. They were then subcloned into pBJNeo and stably expressed in CHO (CCR2, CCR5, and CXCR4), RBL2H3 (CCR1), or AML14D10.3 (CCR3) cells. CCR5 was also stably transfected into a HeLa line already expressing CD4 and CXCR4 (30) to produce the HeLa-C29 line.
Generation of CHO cells stably expressing YU2 gp143.
A 2.2-kb DNA fragment containing the tissue plasminogen activator leader sequence fused to the HIV strain YU2 gp143 sequence, codon optimized for mammalian expression, was subcloned into pBJ-Neo (a mammalian expression vector containing the cytomegalovirus immediate-early promoter). The plasmid was transfected into CHO-K1 cells with Lipofectamine (Life Technologies), and stable clones were selected with G418 (1 mg/ml).
Cell preparation.
Venous whole blood buffy coat or plasmaphoresed leukocytes from normal human donors was obtained from the New York Blood Center or the University of Pennsylvania Medical Center. Peripheral blood mononuclear cells (PBMCs) were separated by density centrifugation over lymphocyte separation medium (Organon Teknika Corporation, Durham, N.C.). Highly purified T cells (93 to 96% CD3+) were isolated from the PBMCs by E-rosetting with neuraminidase-treated sheep red blood cells, followed by density separation of the rosettes and lysis of the sheep red blood cells. The T cells were incubated overnight at 37°C in RPMI 1640 medium (CellGro; Mediatech, Inc., Herndon, Va.) supplemented with 10% heat-inactivated fetal calf serum (Sigma, St. Louis, Mo.), 2 mM glutamine, 1 mM sodium pyruvate, 100 μM nonessential amino acids, 20 mM HEPES, and 20 μg of gentamicin per ml in tissue culture flasks to remove contaminating adherent cells. Purified T cells were washed and incubated in flasks at 2 × 106/ml in medium containing 400 U/ml of recombinant human interleukin-2 (IL-2; Biosource International, Camarillo, Calif.) for 1 week. Such treatment increased the fraction of CCR5-bearing cells from approximately 15 to 25% to 30 to 50% as determined by immunostaining and fluorescence-activated cell sorting (FACS) analysis. Macrophages were prepared from freshly isolated PBMCs by allowing the cells (108 in 20 ml of RPMI 1640) to adhere to 15-cm-diameter tissue culture dishes for 1 to 2 h followed by removal of nonadherent cells. The adherent cells were kept in culture for 7 days prior to use, changing the medium every 3 days. HeLa cells were grown in Dulbecco modified Eagle medium and harvested for use in log phase.
Western blotting.
Recombinant cell lines expressing either CCR1, CCR2, CCR3, CXCR4, or CCR5 were dissolved in Laemmli sample buffer and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 12% acrylamide gels (Novex). Proteins from ∼5 × 104 to 1 × 105 cells/lane were transferred to polyvinylidene difluoride membranes, using a 25 mM Tris–192mM glycine buffer (pH 8.5) containing 0.02% sodium dodecyl sulfate. The membrane was washed, blocked with 5% (wt/vol) dry milk overnight at 4°C, incubated with primary immunoglobulin G (IgG; 1 to 5 μg/ml) for 1 h at room temperature, washed, and then incubated with a 1:2,000 dilution of horseradish peroxidase-conjugated goat anti-rabbit IgG (Zymed) for 30 min at room temperature. The membrane was washed and developed with ECL (enhanced chemiluminescence) reagents (Amersham).
Binding assays.
125I-labeled macrophage inflammatory protein 1α (MIP-1α; 2,200 μCi/mmol) was purchased from NEN Life Science Products. YU2 gp120 was generated as described previously (52), and JRFL gp120 was obtained from Primedica (Tarrytown, N.Y.). Conditions for assays in which the affinities of gp120 were measured by competition against MIP-1α have been described previously (52).
Immunofluorescence microscopy.
For immunofluorescence microscopy HeLa cells or purified monocytes were seeded onto glass coverslips pretreated with a 5-μg/ml solution of human fibronectin (Sigma) in 0.1 M NaHCO3 (pH 8.0). T cells were seeded onto coverslips coated with 10 μg of the anti-CD18 MAb IB4 per ml and permitted to adhere for 4 h prior to fixation. Fixation was performed with a freshly prepared solution of 3.5% paraformaldehyde (generated from paraformaldehyde powder; Fisher Scientific), 0.05% glutaraldehyde, and 0.1 M sucrose in Phosphate-buffered saline (PBS; pH 7.4) for 1 min at 4°C, followed by postfixation in Nakane's solution (a mixture of paraformaldehyde, lysine, and periodate) (38) for 1 h at 4°C. The coverslips were washed with PBS containing 0.5% bovine serum albumin (BSA), treated with clarified 5% nonfat dry milk followed by Fc blocking solution (Accurate Chemicals, Westbury, N.Y.), and stained with 10μg/ml solutions of various chemokine receptor antipeptide rabbit IgGs and 13B8.2 murine monoclonal anti-CD4 IgG. Blocking experiments were conducted to validate the specificity of the immunofluorescence labeling. Solutions of chemokine receptor peptide antibodies (10 μg/ml) were incubated with either the peptides used for immunization or irrelevant control peptides (each at 20 μg/ml) and clarified by centrifugation prior to immunolabeling. To control for the specificity of CD4 labeling, the CD4 MAb (10 μg/ml) was incubated with soluble CD4 (20 μg/ml) or gp 120 (20 μg/ml). Bound primary antibodies were detected by staining with a mixture containing 5 μg each of affinity-purified fluorescein isothiocyanate (FITC)-conjugated F(ab′)2 donkey anti-rabbit IgG and affinity purified tetramethyl rhodamine isothiocyanate F(ab′)2 donkey anti-mouse IgG (Jackson ImmunoResearch Laboratories, Avondale, Pa.) per ml. After washing in PBS and mounting in glycerin, the coverslips were studied and photographed at a magnification of ×100 with an Olympus Provus AX70 fluorescence microscope equipped with narrow-band filters for FITC and rhodamine isothiscyanate (RITC) and a dual-wavelength filter cube for observing FITC and RITC fluorescence simultaneously.
Scanning electron microscopy.
Purified human peripheral blood monocytes were cultured for 7 days on poly-l-lysine-treated coverslips, washed in PBS, and fixed overnight in a solution of 4% paraformaldehyde and 1% glutaraldehyde in PBS at 4°C, followed by additional washing and postfixation in 1% OsO4 for 15 min. The coverslips were then washed in distilled water, dehydrated in graded ethanols, critical point dried from CO2, mounted on stubs, carbon coated, and then sputter coated with gold-platinum. Samples were examined at a magnification of ×5,000 in an AMRAY 1000A scanning electron microscope.
Immunogold electron microscopy.
Following culture, HeLa cells, CHO cells, purified monocytes, or T cells were fixed as described for immunofluorescence microscopy. The macrophages and HeLa cells were fixed in situ and scraped up after fixation was completed. The other cell types and HeLa cells were also fixed in suspension. Fixed cell pellets were infiltrated in 2% low-gelling-temperature agarose, cryoprotected by overnight infiltration with 2.3 M sucrose and 50% polyvinylpyrrolidone in phosphate buffer (pH 7.2) (57), frozen by injection into liquid propane at −185°C in a KF-80 apparatus (Reichert Scientific Instruments, Buffalo, N.Y.), and stored under liquid nitrogen. To permit access of antibodies to all sectioned subcellular compartments, ultrathin (∼80-nm) frozen sections were cut with glass knives at −105°C (56) on a Reichert UCT ultramicrotome equipped with an FC-S cryoattachment and transferred to Formvar-coated 200-mesh hexagonal nickel grids. Sections were treated with a clarified solution of 5% nonfat dry milk and BSA buffer (1% BSA in PBS [pH 7.8] containing 0.1% sodium azide) for 30 min to block nonspecific binding, followed by overnight incubation in 5 to 10-μg/ml solutions of various rabbit anti-peptide IgGs or MAbs in BSA buffer at 4°C as previously described (53). For specificity controls, rabbit anti-chemokine receptor peptide IgGs (10 μg/ml) were absorbed with either the peptides used for immunization or irrelevant control peptides (each at 20 μg/ml) and clarified by centrifugation prior to immunolabeling. To validate the specificity of CD4 labeling, 10 μg of the CD4 MAb per ml was incubated with 20-μg/ml solutions of either CD4 or gp120. Other controls consisted of 10-μg/ml solutions of the corresponding preimmune rabbit IgGs or isotype-matched nonimmune MAbs in BSA buffer. After extensive washing in PBS, the bound primary rabbit IgGs were detected with a goat anti-rabbit 5-nm colloidal gold conjugate (GARG 5; Amersham), and the murine IgGs were labeled with goat anti-mouse 10-nm colloidal gold conjugate (GAMG10; Amersham), either alone or as mixtures in BSA buffer for double labeling. Human anti-gp120 1b12 IgG was detected with goat anti-human IgG 10-nm gold (GAHG10; Amersham). To control for possible differences in labeling efficiency, some experiments were conducted with goat anti-mouse 5-nm colloidal gold and goat anti-rabbit 10-nm colloidal gold conjugates (Amersham). The grids were then washed in PBS and fixed in 2% glutaraldehyde in PBS for 15 min, postfixed in 2% OsO4 in H2O for 20 min, stained with 2% uranyl acetate in H2O for 30 min, and embedded in 2% polyvinyl alcohol as described elsewhere (56). Grids were examined, and electron micrographs were taken in a JEOL 200CX electron microscope at 80 kV at initial magnifications of ×20,000 to ×35,000.
RESULTS
Binding affinity and infectivity do not correlate.
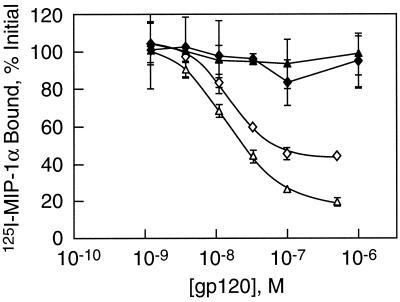
The interaction of CCR5 with the viral envelope protein gp120 is complex and involves at least two domains of the receptor, one in the core of the molecule and the second in its N terminus (21, 47, 52). During studies of the structural requirements for binding of gp120, we previously observed that substitution of arginine for glycine at position 163 in the core of CCR5 (top of transmembrane helix 4) drastically diminished the affinity of the interaction between the receptor and gp120 but had only a modest effect on the ability of the receptor to support infection (52). The role of the N terminus was assessed by preparing a series of N-terminal truncations. Figure 1 illustrates the impact of removing the first 10 residues on the affinity of CCR5 for gp120 from two R5 strains, YU2 and JRFL. As measured by their ability to inhibit the binding of the chemokine MIP-1α, both envelope proteins have 50% inhibitory concentrations of about 10 nM for the wild-type receptor. The N-terminal truncation reduces these affinities at least 100-fold. In contrast, it has been previously reported that Δ10 CCR5 is nearly as efficient at supporting infection as the wild-type receptor, the differences being only about two-fold (25). This result is consistent with data reported by Blanpain et al. for a Δ2–9 truncation of CCR5 (8). Although the binding assays may well be an imperfect mimic for the interaction between the virus and CCR5, taken together, these data demonstrate that infectivity is not tightly coupled to the energetics of the gp120-coreceptor interaction.
FIG. 1.
Δ10 CCR5 has greatly diminished affinity for gp120. The avidity of R5-tropic gp120s, JRFL (◊) or YU2 (▵), for the target receptor was measured by their ability to compete against 125I-MIP-1α in the presence of soluble CD4. Experiments were carried out with CHO cells expressing either wild-type CCR5 (open symbols) or Δ10 CCR5 (filled symbols).
The HIV virion contains multiple gp120 molecules, raising the possibility that interaction between the virus and host cell may involve multiple coreceptor molecules. Such cooperative binding provides a potential explanation for the lack of correlation between affinity and infectivity. The efficiency of this putative cooperativity depends, at least in part, on the spatial organization of both the coreceptors and CD4. To investigate these distributions, we carried out a series of high-resolution immunoelectron microscopic (immuno-EM) studies.
Antibody characterization.
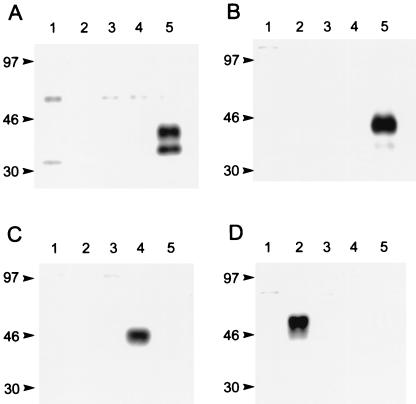
Rabbit polyclonal antibodies against the chemokine receptors were raised by immunization with peptides from the N and C termini of CCR5 and the N termini of CXCR4 and CCR2. As shown by Western blotting (Fig. 2), each antibody recognizes only the receptor from which its immunizing peptide was derived. The CD4 MAb 13B8.2 has been described elsewhere (2, 39, 50). In our hands, as shown by FACS analysis, 13B8.2 reacts with CD4+ but not CD8+ T cells. Moreover, since it intensely labeled CD4-expressing HeLa cells following fixation with our protocols, while other MAbs such as OKT4, Leu-3a, MT-310, RPA-4, and MEM-115 yielded only slight to moderate fluorescence (data not shown), 13B8.2 was used for all immunofluorescence and immuno-EM analyses of CD4.
FIG. 2.
Western blots demonstrate that each chemokine receptor IgG used for immuno-EM recognizes only the receptor from which its immunizing peptide was derived. Specificities were tested against extracts of whole cells expressing either hCCR1 (lane 1), hCCR2 (lane 2), hCCR3 (lane 3), hCXCR4 (lane 4), or hCCR5 (lane 5). Results are for the N-terminal CCR5 (R4603; A), C-terminal CCR5 (R4627; B), N-terminal CXCR4 (R5039; C), and N-terminal CCR2 (R4731; D) antibodies. Sizes are indicated in kilodaltons.
Immunofluorescence microscopy of chemokine receptors and CD4.
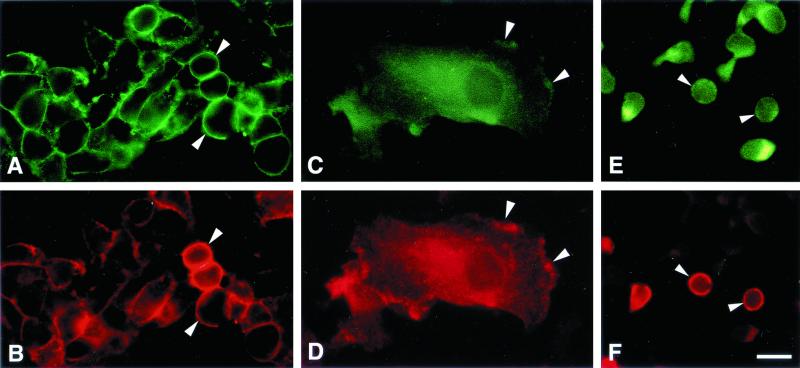
Initial experiments were carried out with the HeLa-C29 cell line since the overexpression of CCR5 and CD4 in these cells facilitated development and optimization of fixation and immunostaining methods. CCR5 and CD4 were broadly colocalized on the HeLa-C29 cell membrane, as shown by double-label immunofluorescence microscopy (Fig. 3A and B). Control preparations incubated with nonimmune rabbit IgG or mouse IgG1κ were not stained. In cultured human blood macrophages (Fig. 3C and D), CCR5 was often coincident with CD4 at the leading edge of the cell and on the dorsal cell surface. The cell surface distributions of CCR5, CXCR4, and CCR2 epitopes were all very similar and broadly coincident with CD4 patterns in these macrophages (not shown). CCR5 and CD4 were also broadly coincident on the surfaces of adherent CD4+ T cells (Fig. 3E and F).
FIG. 3.
Double-label immunofluorescence microscopy shows that CCR5 (A, C, and E) and CD4 (B, D, and F) are colocalized on the surfaces of HeLa-C29 cells (A and B), human macrophages (C and D), and human T cells (E and F) (bar = 20 μm). CCR5 was frequently found to be coincident with CD4 at the leading edge of human macrophages (arrowheads in panels C and D).
Immuno-EM of CCR5 and CD4 in HeLa-C29 cells.
Meaningful high-resolution studies require faithful preservation of the distribution and antigenicity of the chemokine receptors and CD4 and of the ultrastructural detail of all subcellular components. Simultaneously, epitopes of interest must be rendered accessible to their antibodies. To meet these criteria, the immuno-EM studies were performed using postsection immunolabeling of ultrathin cryosections of prefixed cells. Fixation prior to application of antibodies ensured that epitope distributions were not altered by the multivalent IgGs. Also, the cellular fixation, processing, and ultrathin cryosectioning were performed to minimize the extraction of membrane lipids and expose all sectioned subcellular epitopes, including molecules embedded in the cell surface glycocalyx, to access by immunoprobes and immunogold conjugates. Double-label experiments (5- and 10-nm immunogold probes) were used to assess the distributions of two components simultaneously.
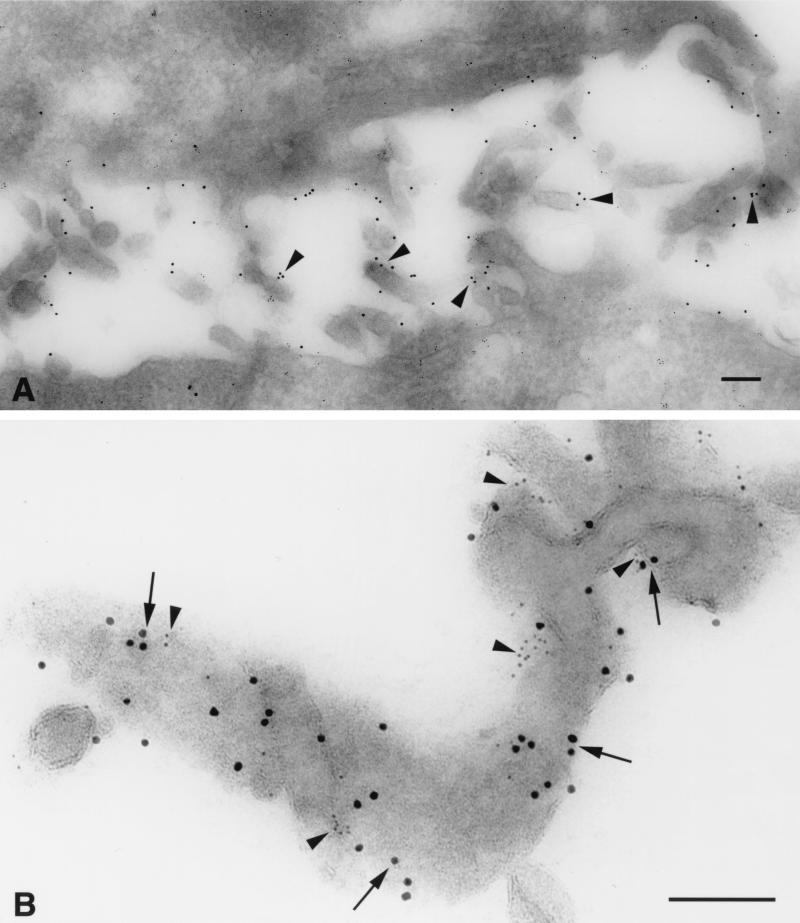
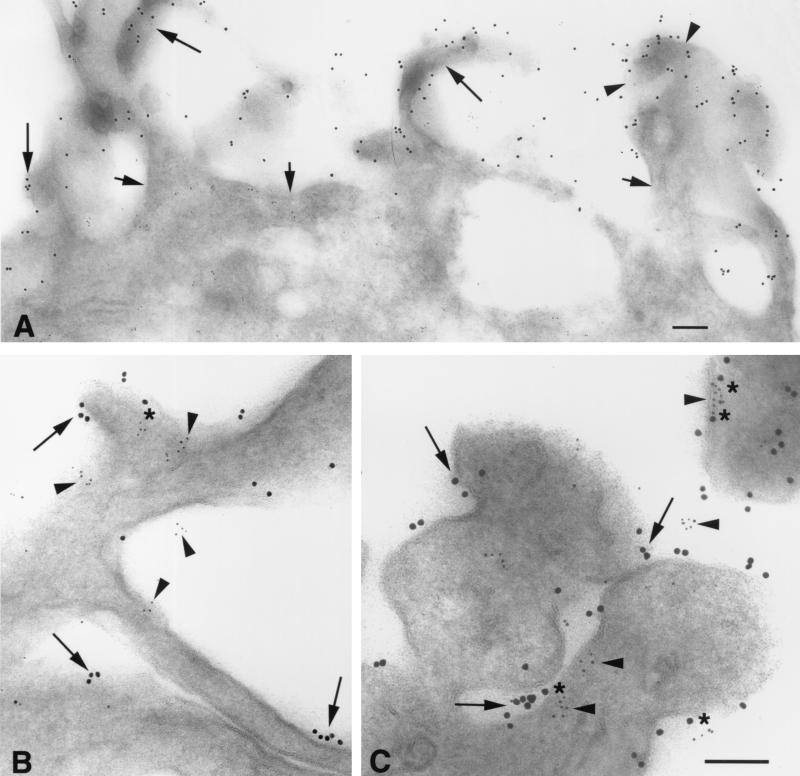
In contrast to observations made with immunofluoresence microscopy, immuno-EM studies revealed that the chemokine receptors and CD4 were nonrandomly distributed on the cell surface. As illustrated for CCR5 (Fig. 4A), these molecules were preferentially localized on microvilli. Moreover, higher-magnification images using double labeling revealed that both CCR5 (10-nm gold particles) and CD4 (5-nm gold) were frequently localized in microclusters on the microvilli (Fig. 4B). The microclusters were largely homogeneous, as there was little mixing of CCR5 and CD4 within a cluster. Some of the CCR5 and CD4 molecules and clusters were located within ∼5 to 10 nm of each other, a distance substantially less than the 100-nm diameter of the HIV capsid. Comparable labeling patterns were obtained with both the N-terminal and C-terminal CCR5 peptide antibodies and with MAb 2D7.
FIG. 4.
Immuno-EM detects homogeneous microclusters of CCR5 and CD4 concentrated on the microvilli of HeLa-C29 cells. (A) CCR5 is preferentially localized on cell surface microvilli (arrowheads; 10-nm anti-rabbit immunogold). (B) Prominent homogeneous microclusters of CCR5 (arrows; 10-nm anti-rabbit immunogold) and CD4 (arrowheads; 5-nm anti-mouse immunogold) are closely apposed on the surface membranes of microvilli. The central arrowhead depicts a linear array of CD4 epitopes situated within 10 nm of a CCR5 immunogold particle in the outer membrane glycocalyx. Bars = 100 nm.
Multiple approaches were used to validate the specificity of the immunogold labeling. First, substitution of nonimmune rabbit IgG or mouse IgG1κ for the first antibody ablated staining. Second, preincubation of the antibody with the immunizing peptide, but not an irrelevant peptide, completely blocked labeling with anti-CCR5 antibodies. Similarly, pretreatment of the CD4 MAb with recombinant human CD4 inhibited the immunogold labeling, whereas gp120 (YU2) did not (data not shown). Further validation was obtained using CHO cell lines stably transfected with either CCR5, CXCR4, or CCR2. Antibodies to CCR5 labeled only the line expressing CCR5 (again in microclusters on microvilli), while antibodies raised against the N terminus of CXCR4 or CCR2 stained only the appropriate cell lines (data not shown).
Scanning electron microscopy.
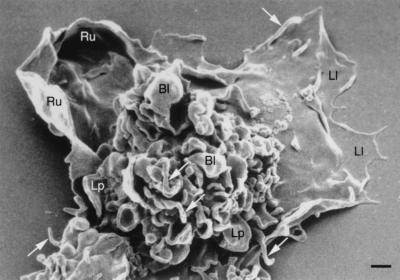
While the HeLa-C29 line served as a model system, the focus of our studies was the distribution of CCR5, CXCR4, and CD4 on primary cells. Since macrophages have a very complex surface, scanning electron microscopy was conducted to help interpret the immuno-EM ultrathin frozen sections to follow. The outer membrane of these cells was highly pleomorphic and exhibited numerous structures reflecting intense cell surface activity: microvilli, blebs, lamellipodia, ruffling membranes, and leading lamellae (Fig. 5).
FIG. 5.
Scanning electron micrograph of a typical cultured human blood macrophage. The cells exhibit structures reflecting intense surface membrane activity: prominent microvilli (white arrows), blebs (Bl), lamellipodia (Lp), ruffling membranes (Ru), and leading lamellae (Ll). Bar = 1 μm.
Immuno-EM of CCR5, CXCR4, CCR2, and CD4 in macrophages.
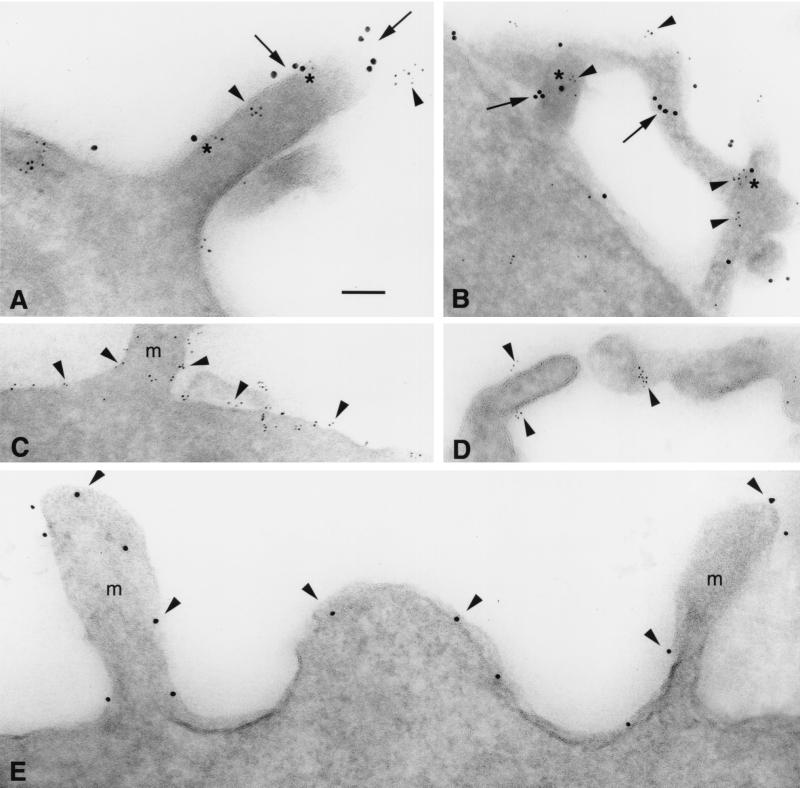
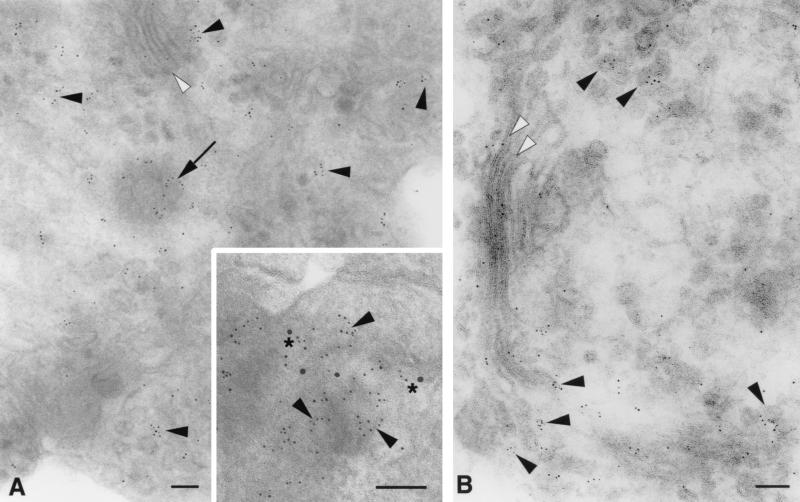
Adherent cultures of human blood macrophages were fixed in situ and processed for immuno-EM. Numerous cell surface microvilli, blebs, and lamellipodia exhibiting CCR5 and CD4 epitopes were found in ultrathin frozen sections of these cells (Fig. 6). As in the HeLa cells, CD4 was concentrated on the surface membranes of microvilli, frequently in microclusters (Fig. 6A). Double labeling illustrates that both CCR5 and CD4 were localized on the outer membranes of microvilli (Fig. 6B) and blebs (Fig. 6C), often in homogeneous microclusters. These microclusters were often closely apposed (within 5 to 10 nm). In additional double-labeling experiments, homogeneous microclusters of CXCR4 or CCR2 were observed to be closely associated with microclusters of CD4 on the surfaces of blebs, ruffling membranes, and lamellipodia, as well as on microvilli (not shown).
FIG. 6.
CCR5 and CD4 form homogeneous microclusters on microvilli of human blood macrophages detected by immuno-EM. (A) CD4 (10-nm immunogold) is concentrated on microvilli (long arrows) and blebs (arrowheads), while little staining is apparent on the cell surface membrane (short arrows). Ultrathin cryosections through microvilli (B) and blebs (C) exhibit homogeneous microclusters of CCR5 (arrowheads; 5-nm immunogold) and CD4 (arrows; 10-nm immunogold) localized on their surface membranes; asterisks indicate closely apposed CCR5 and CD4. A complex of CCR5 and CD4 (C, upper right corner) contains two loci of CD4 epitopes (asterisks) closely flanking an elongated CCR5 aggregate on the cell membrane (arrowhead). Bars = 100 nm.
Localization of chemokine receptors and CD4 in T cells.
As shown in Fig. 7, IL-2-stimulated T cells, fixed in suspension, exhibited numerous microvilli. As observed with other cell types, CD4 and the chemokine receptors CCR5 and CXCR4 were preferentially localized on the microvilli. Again, these molecules tend be found in homogeneous microclusters which are often closely associated (∼10 nm apart). This can be seen in Fig. 7A for the CCR5-CD4 combination and in Fig. 7B for CXCR4-CD4. Interestingly, the distribution of CD8 was very similar to that of CD4, with CD8 microaggregates localized predominantly on the surface membranes of microvilli (Fig. 7D). As counterexamples to this pattern of distribution, CD3 is distributed over the entire cell surface including the microvilli, although it too has a tendency to cluster (Fig. 7C), while gp143 (from R5 strain YU2) expressed in CHO cells is randomly distributed over the entire cell surface and is unclustered (Fig. 7E).
FIG. 7.
Immuno-EM exhibits homogeneous microclusters of CCR5, CXCR4, and CD4 on primary human T cells. (A) T-cell microvilli exhibit homogeneous microaggregates of CCR5 (arrowheads; 5-nm immunogold) and CD4 (arrows; 10-nm immunogold); asterisks indicate closely apposed CCR5 and CD4 epitopes. (B) CXCR4 (arrowheads; 5-nm immunogold) exhibits similar homogeneous microclusters, closely apposed (at asterisks) to CD4 (arrows; 10-nm immunogold) on T-cell microvilli. (C) CD3 (arrowheads) is localized on T-cell microvilli (m) and over the entire cell surface membrane. (D) CD8 preferentially labels T-cell microvilli as small aggregates (arrowheads) and is not detected on the surface membrane. (E) gp120 epitopes (arrowheads; 10-nm immunogold) appear unclustered and randomly distributed on microvilli (m) and the cell membranes of CHO cells expressing 105 YU2 gp143 copies per cell (labeled with 1b12, a human MAb to gp120). Bar (applies to all panels) = 100 nm.
Presence of CCR5 and CXCR4 in separate microclusters.
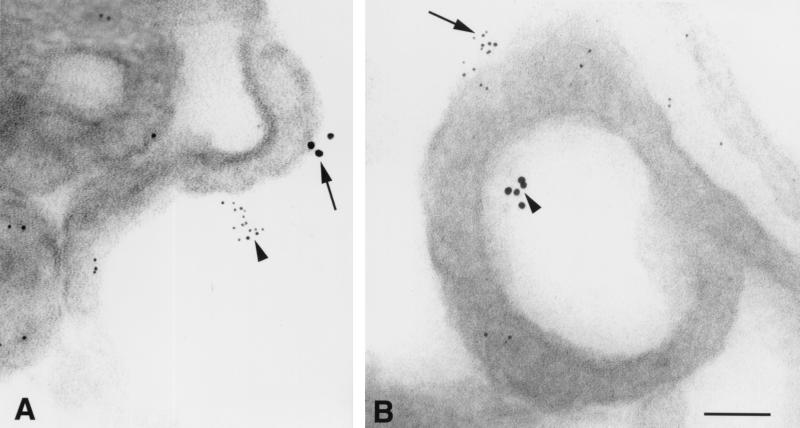
When cryosections of macrophages or T cells were double labeled with antibodies recognizing two different chemokine receptors (i.e., CCR5 and CXCR4 or CCR2 and CCR5), staining for each chemokine receptor was segregated as homogeneous microclusters of immunogold particles in both the cytoplasm and at the cell surface; mixed clusters were never observed. Homogeneous microclusters of CCR5 and CXCR4 were located within ∼200 nm of each other on microvilli and lamellipodia (Fig. 8); very similar patterns of CCR5 and CXCR4 labeling were observed using either rabbit anti-peptide IgGs or MAbs to detect these chemokine receptors.
FIG. 8.
CCR5 and CXCR4 are localized in separate microclusters on human macrophages. (A) Arrowhead shows a homogeneous microcluster of CXCR4 stained with an N-terminal rabbit anti-peptide IgG, and the arrow depicts a separate microaggregate of CCR5 labeled with MAb 2D7; both clusters are localized on a single microvillus. (B) A CCR5 microaggregate is labeled with a C-terminal rabbit antipeptide IgG (arrow), while a CXCR4 microcluster is stained with MAb 12G5 (arrowhead); both are located on the same lamellipodium. Bar = 100 nm.
CCR5 microclusters are localized in the Golgi apparatus.
CCR5 microaggregates were also detected in small rounded secretory vesicles of the Golgi apparatus, with minimal labeling in the Golgi cysternae; curvilinear arrays of CCR5 epitopes were sometimes observed at the periphery of these vesicles (Fig. 9). In other favorable sections which provided tangential views (inset), curvilinear assemblies of CCR5 epitopes were found in the dense cortical cytoplasm in close association with CD4. These CCR5-containing secretory vesicles are probably about to fuse, or are in the process of fusing, with the cell membrane. Similar distributions of CCR5 labeling were observed in the Golgi bodies of HeLa-C29 cells (Fig. 9B) and T cells (not shown). These patterns are indicative of the synthesis and transport of preformed CCR5 microclusters to the cell surface.
FIG. 9.
Microclusters of CCR5 are localized in Golgi secretory vesicles in human macrophages and HeLa-C29 cells. (A) Cryosection through the Golgi apparatus of a human macrophage labeled for CCR5 with 5-nm immunogold. Microaggregates of CCR5 epitopes are localized as clusters (arrowheads) or curvilinear arrays (arrow) in Golgi secretory vesicles. Little CCR5 label is present in the Golgi cysternae (white arrowhead). (Inset) Grazing section through membrane blebs at the macrophage surface double labeled for CCR5 and CD4. Curvilinear aggregates of CCR5 epitopes (arrowheads; 5-nm immunogold) are associated with a dense submembranous cortex and with CD4 labeling (asterisks; 10-nm immunogold). This image suggests that CCR5-positive secretory vesicles are fusing with the cell membrane. (B) Localization of CCR5 (5-nm immunogold) in the Golgi apparatus of a HeLa-C29 cell. Microclusters of CCR5 are concentrated in Golgi secretory vesicles (arrowheads), while reduced CCR5 labeling is detected in the Golgi cysternae (white arrowheads). Bars = 100 nm.
DISCUSSION
Studies from several laboratories have concluded that the chemokine receptors, most notably CCR5, CXCR4, and CCR2, as well as CD4 are, in the absence of stimulation, relatively uniformly and randomly distributed on the cell surface (2, 28, 59). However, these investigations employed optical imaging techniques which lack the resolution necessary to provide spatial information at near molecular levels. In this study, we used immunogold electron microscopy to show that all four molecules, rather than being uniformly distributed, are preferentially located on microvilli. We find the same distribution on all the cell types studied, including (i) HeLa cells engineered to express CCR5, CXCR4, and CD4 (ii) CHO cells expressing either CCR5, CXCR4, or CCR2, (iii) IL-2-stimulated human T cells, and (iv) monocyte-derived human macrophages. The distribution of receptors is also independent of the overall level of receptor expression over a range of at least 20- to 50-fold, as the HeLa line used in this study expressed 106 CCR5 molecules/cell, compared to approximately 2 × 104 to 5 × 104 receptors/cell for the macrophages. Distributions of these molecules on the microvilli are nonrandom as well. Rather, the receptors, as depicted by individual gold particles, often appear in small clusters. Moreover, as shown by double-label immuno-EM, these cell surface microclusters are homogeneous, as there is little intermingling of different receptor types within a single cluster. This can be seen for CCR5 and CXCR4 in Fig. 8 and is also true for combinations of CCR5 plus CCR2 and CXCR4 plus CCR2 (unpublished observations).
The efficiency of a mechanism, like viral entry, which requires interaction of three components (gp120/gp41, CD4, and coreceptor) is greatly enhanced if the process can be reduced to two sequential two-body interactions. The distributions and microclustering of CD4 and coreceptors facilitate such a reduction. Viral gp120-gp41 complexes exist as trimers (35), and since many of the CD4 molecules form cell surface microclusters, it is likely that the initial interaction between the virus and cell will be cooperative, with multiple CD4 molecules binding to several gp120-gp41 complexes. In effect, the virus becomes a tethered ligand. The energetic stabilization provided by the polyvalent interaction should prolong the lifetime of the virus-cell complex, thereby increasing the probability of productive interaction with a coreceptor. This probability is further enhanced by localization of both CD4 and the coreceptors on microvilli, and in particular by their close apposition as CCR5 and CXCR4 molecules are often found within a viral diameter of many of the CD4 microclusters. Such a cooperative mechanism is likely to be rather insensitive to coreceptor-gp120 energetics, a hypothesis consistent with our findings, as well as those of others (6, 16, 25, 52), that the affinity of CCR5 for gp120 can be substantially lowered with minimal effect on infectivity.
A number of basic questions about the entry mechanism remain to be addressed. How many gp120 molecules within a trimer must interact with CD4 and coreceptor to activate the trimer? How many trimers must be activated to allow viral entry? Recently, it has been reported that interaction with six CCR5 molecules is necessary for viral infection (31), a number which suggests that activation of more than one trimer is required. While there are still no data on the requirements for trimer activation, if multiple gp120 molecules must bind to host components, microclustering of CD4 and the coreceptors should greatly enhance the process.
While many of the microclusters are within close proximity (5 to 10 nm, less than the diameter of a viral capsid), the distributions of CD4 and the chemokine receptors, as revealed by immuno-EM, are clearly separate. The lack of constitutive colocalization is consistent with conclusions from previous lower-resolution studies of CD4 and CXCR4 using optical microscopy where little overlap between the two molecules was observed until stimulation with X4 envelope protein (28, 59). It is also consistent with biochemical studies which demonstrated coimmunoprecipitation of CD4 and CXCR4 only in the presence of an X4 gp120 (33). Our data suggest a similar lack of constitutive association between CD4 and CCR5, as we see few CCR5 and CD4 molecules sufficiently close to one another to allow direct physical interaction. These results are not entirely consistent with the findings of Xiao et al. (63), who suggest a constitutive physical association between CCR5 and CD4 because they can coimmunoprecipitate the two in the absence of an R5 gp120. However, it is unclear from the work of Xiao et al. what fractions of the two receptors actually coimmunoprecipitate, and although some care was taken to show specificity, the coimmunoprecipitation may be indirect. While it can be argued that our failure to see molecules close enough to allow physical association may be due to steric hindrance between anti-CCR5 and anti-CD4 molecules, we believe this unlikely since similar labeling patterns were observed using different antibodies recognizing either the extracellular N terminus or the intracellular C terminus of CCR5.
Although recent efforts on CCR5 and CXCR4 have focused largely on their roles as HIV coreceptors, the major biological function of chemokine receptors is regulation of leukocyte trafficking during normal homeostasis and in immune and inflammatory responses (5, 36, 43). The receptors act as sensors to detect concentration gradients of chemokines, providing directional information to the cells which express them and thus leading them toward the source of the mediator. Localization of the sensors on microvilli and ruffling membranes should enable the cells to more precisely monitor their environment, improving the efficiency of the chemotactic process. Efficiency might be further enhanced if some or all of the signaling induced by receptor activation was local rather than global, enabling a single microvillus to act as a unit. Indeed, just such a model has been proposed to govern Ca2+ signaling and actin polymerization in nonexcitable cells (32). It is noteworthy that in addition to major components of the Ca2+ signaling pathway, other elements of the adhesion system necessary for the chemotactic process are also located on microvilli. These include the selectins (10), which mediate the initial rolling of leukocytes along the vascular endothelium, the integrins α4β7 and α4β1, which generate firm attachments and the traction necessary for the cells to extravasate (7, 20), and the actin/myosin cytoskeleton, which generates forward protrusion (26, 54).
The microclusters may also be relevant to the function of chemokine receptors. While GPCRs have long been thought to be monomeric, evidence has slowly been accumulating that these receptors, like those of other classes (24, 60), may require oligomerization in order to signal (23, 37, 40). Recently presented data indicate that CCR2 signals in an oligomeric rather than monomeric state (46). The clusters we observe are consistent with this possibility.
The finding that both the chemokine receptors and CD4 are often found in homogeneous microclusters raises a number of intriguing questions. Where, when, and how does the clustering originate? What are the molecular mechanisms which maintain this organization, and why are the clusters homogeneous? Our data suggest that clustering may well occur shortly after synthesis and prior to insertion of the receptors in the cellular membrane, as the phenomenon is already visible in small spherical secretory vesicles of the trans-Golgi apparatus. These clusters appear to be precursors to the cell surface microclusters since both are homogeneous in composition and similar in size. The homogeneous microclustering of chemokine receptors in small spherical cytoplasmic vesicles is analogous to the observations of Orci et al. (42), who found that different proteins segregate into separate Golgi vesicles. It is likely that receptor containing Golgi-derived vesicles fuse with the cell membrane, delivering a preformed cluster to the cellular surface. Interactions with the actin cytoskeleton may occur during transport and might be driven by association with as yet unknown adaptor proteins as suggested for CD2 (19), providing a mechanism for directing the receptor clusters to the actin-containing microvilli.
Finally, the tendency of the chemokine receptors and CD4 to form homogeneous clusters which are frequently closely apposed has important implications for development of receptor antagonists with antiviral activity. As discussed above, as a result of cooperative interaction between gp120-gp41 complexes on the viral capsid with clusters of CD4, the virus is likely to become a tethered ligand, kinetically favoring interaction with the coreceptor. Although it is clear that coreceptor inhibitors can be developed (4, 15), the geometry and cooperativity of the system indicate that special properties will be required for potent antiviral activity.
REFERENCES
- 1.Aithal H N, Knigge K M, Kartha S, Czyzewski E A, Toback F G. An alternate method utilizing small quantities of ligand for affinity purification of monospecific antibodies. J Immunol Methods. 1988;112:63–69. doi: 10.1016/0022-1759(88)90034-8. [DOI] [PubMed] [Google Scholar]
- 2.Alfano M, Schmidtmayerova H, Amella C A, Pushkarsky T, Bukrinsky M. The B-oligomer of pertussis toxin deactivates CC chemokine receptor 5 and blocks entry of M-tropic HIV-1 strains. J Exp Med. 1999;190:597–605. doi: 10.1084/jem.190.5.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 4.Baba M, Nishimura O, Kanzaki N, Okamoto M, Sawada H, Iizawa Y, Shiraishi M, Aramaki Y, Okonogi K, Ogawa Y, Meguro K, Fujino M. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc Natl Acad Sci USA. 1999;96:5698–5703. doi: 10.1073/pnas.96.10.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baggiolini M, Dewald B, Moser B. Human chemokines: an update. Annu Rev Immunol. 1997;15:675–705. doi: 10.1146/annurev.immunol.15.1.675. [DOI] [PubMed] [Google Scholar]
- 6.Baik S S, Doms R W, Doranz B J. HIV and SIV gp120 binding does not predict coreceptor function. Virology. 1999;259:267–273. doi: 10.1006/viro.1999.9779. [DOI] [PubMed] [Google Scholar]
- 7.Berlin C, Bargatze R F, Campbell J J, von Andrian U H, Szabo M C, Hasslen S R, Nelson R D, Berg E L, Erlandsen S L, Butcher E C. α4 integrins mediate lymphocyte attachment and rolling under physiologic flow. Cell. 1995;80:413–422. doi: 10.1016/0092-8674(95)90491-3. [DOI] [PubMed] [Google Scholar]
- 8.Blanpain C, Doranz B J, Vakili J, Rucker J, Govaerts C, Baik S S, Lorthioir O, Migeotte I, Libert F, Baleux F, Vassart G, Doms R W, Parmentier M. Multiple charged and aromatic residues in CCR5 amino-terminal domain are involved in high affinity binding of both chemokines and HIV-1 Env protein. J Biol Chem. 1999;274:34719–34727. doi: 10.1074/jbc.274.49.34719. [DOI] [PubMed] [Google Scholar]
- 9.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 10.Bruehl R E, Springer T A, Bainton D F. Quantitation of L-selectin distribution on human leukocyte microvilli by immunogold labeling and electron microscopy. J Histochem Cytochem. 1996;44:835–844. doi: 10.1177/44.8.8756756. [DOI] [PubMed] [Google Scholar]
- 11.Burton D R, de Barbas C F, Persson M A, Koenig S, Chanock R M, Lerner R A. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci USA. 1991;88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 13.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O'Brien S J. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. . (Erratum, 274:1069.) [DOI] [PubMed] [Google Scholar]
- 14.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 15.Donzella G A, Schols D, Lin S W, Este J A, Nagashima K A, Maddon P J, Allaway G P, Sakmar T P, Henson G, De Clercq E, Moore J P. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat Med. 1998;4:72–77. doi: 10.1038/nm0198-072. [DOI] [PubMed] [Google Scholar]
- 16.Doranz B J, Baik S S, Doms R W. Use of a gp120 binding assay to dissect the requirements and kinetics of human immunodeficiency virus fusion events. J Virol. 1999;73:10346–10358. doi: 10.1128/jvi.73.12.10346-10358.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 18.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 19.Dustin M L, Olszowy M W, Holdorf A D, Li J, Bromley S, Desai N, Widder P, Rosenberger F, van der Merwe P A, Allen P M, Shaw A S. A novel adaptor protein orchestrates receptor patterning and cytoskeletal polarity in T-cell contacts. Cell. 1998;94:667–677. doi: 10.1016/s0092-8674(00)81608-6. [DOI] [PubMed] [Google Scholar]
- 20.Erlandsen S L, Hasslen S R, Nelson R D. Detection and spatial distribution of the beta 2 integrin (Mac-1) and L-selectin (LECAM-1) adherence receptors on human neutrophils by high-resolution field emission SEM. J Histochem Cytochem. 1993;41:327–333. doi: 10.1177/41.3.7679125. [DOI] [PubMed] [Google Scholar]
- 21.Farzan M, Choe H, Vaca L, Martin K, Sun Y, Desjardins E, Ruffing N, Wu L, Wyatt R, Gerard N, Gerard C, Sodroski J. A tyrosine-rich region in the N terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J Virol. 1998;72:1160–1164. doi: 10.1128/jvi.72.2.1160-1164.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 23.Hebert T E, Moffett S, Morello J P, Loisel T P, Bichet D G, Barret C, Bouvier M. A peptide derived from a β2-adrenergic receptor transmembrane domain inhibits both receptor dimerization and activation. J Biol Chem. 1996;271:16384–16392. doi: 10.1074/jbc.271.27.16384. [DOI] [PubMed] [Google Scholar]
- 24.Heldin C H. Dimerization of cell surface receptors in signal transduction. Cell. 1995;80:213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 25.Hill C M, Kwon D, Jones M, Davis C B, Marmon S, Daugherty B L, DeMartino J A, Springer M S, Unutmaz D, Littman D R. The amino terminus of human CCR5 is required for its function as a receptor for diverse human and simian immunodeficiency virus envelope glycoproteins. Virology. 1998;248:357–371. doi: 10.1006/viro.1998.9283. [DOI] [PubMed] [Google Scholar]
- 26.Horwitz A R, Parsons J T. Cell migration—movin' on. Science. 1999;286:1102–1103. doi: 10.1126/science.286.5442.1102. [DOI] [PubMed] [Google Scholar]
- 27.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau N R, Phair J, Ho D D, Koup R A. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 28.Iyengar S, Hildreth J E K, Schwartz D H. Actin-dependent receptor colocalization required for human immunodeficiency virus entry into host cells. J Virol. 1998;72:5251–5255. doi: 10.1128/jvi.72.6.5251-5255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kessler J A, II, McKenna P M, Emini E A, Chan C P, Patel M D, Gupta S K, Mark III G E, Barbas III C F, Burton D R, Conley A J. Recombinant human monoclonal antibody IgG1b12 neutralizes diverse human immunodeficiency virus type 1 primary isolates. AIDS Res Hum Retroviruses. 1997;13:575–582. doi: 10.1089/aid.1997.13.575. [DOI] [PubMed] [Google Scholar]
- 30.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuhmann S E, Platt E J, Kozak S L, Kabat D. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J Virol. 2000;74:7005–7015. doi: 10.1128/jvi.74.15.7005-7015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lange K. Microvillar Ca++ signaling: a new view of an old problem. J Cell Physiol. 1999;180:19–34. doi: 10.1002/(SICI)1097-4652(199907)180:1<19::AID-JCP3>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 33.Lapham C K, Ouyang J, Chandrasekhar B, Nguyen N Y, Dimitrov D S, Golding H. Evidence for cell-surface association between fusin and the CD4-gp120 complex in human cell lines. Science. 1996;274:602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- 34.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 35.Lu M, Blacklow S C, Kim P S. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 36.Luster A D. Chemokines—chemotactic cytokines that mediate inflammation. N Engl J Med. 1998;338:436–445. doi: 10.1056/NEJM199802123380706. [DOI] [PubMed] [Google Scholar]
- 37.Maggio R, Vogel Z, Wess J. Coexpression studies with mutant muscarinic/adrenergic receptors provide evidence for intermolecular “cross-talk” between G-protein-linked receptors. Proc Natl Acad Sci USA. 1993;90:3103–3107. doi: 10.1073/pnas.90.7.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McLean I W, Nakane P K. Periodate-lysine-paraformaldehyde fixative. A new fixation for immunoelectron microscopy. J Histochem Cytochem. 1974;22:1077–1083. doi: 10.1177/22.12.1077. [DOI] [PubMed] [Google Scholar]
- 39.McMichael A J. Leukocyte typing III: white cell differentiation antigens. New York, N.Y: Oxford University Press; 1987. [Google Scholar]
- 40.Monnot C, Bihoreau C, Conchon S, Curnow K M, Corvol P, Clauser E. Polar residues in the transmembrane domains of the type 1 angiotensin II receptor are required for binding and coupling. Reconstitution of the binding site by co-expression of two deficient mutants. J Biol Chem. 1996;271:1507–1513. doi: 10.1074/jbc.271.3.1507. [DOI] [PubMed] [Google Scholar]
- 41.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J L, Arenzana-Seisdedos F, Schwartz O, Heard J M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. . (Erratum, 384:288.) [DOI] [PubMed] [Google Scholar]
- 42.Orci L, Stamnes M, Ravazzola M, Amherdt M, Perrelet A, Sollner T H, Rothman J E. Bidirectional transport by distinct populations of COPI-coated vesicles. Cell. 1997;90:335–349. doi: 10.1016/s0092-8674(00)80341-4. [DOI] [PubMed] [Google Scholar]
- 43.Premack B A, Schall T J. Chemokine receptors: gateways to inflammation and infection. Nat Med. 1996;2:1174–1178. doi: 10.1038/nm1196-1174. [DOI] [PubMed] [Google Scholar]
- 44.Raport C J, Gosling J, Schweickart V L, Gray P W, Charo I F. Molecular cloning and functional characterization of a novel human CC chemokine receptor (CCR5) for RANTES, MIP-1β, and MIP-1α, J. Biol Chem. 1996;271:17161–17166. doi: 10.1074/jbc.271.29.17161. [DOI] [PubMed] [Google Scholar]
- 45.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez-Frade J M, Vila-Coro A J, de Ana A M, Albar J P, Martinez A C, Mellado M. The chemokine monocyte chemoattractant protein-1 induces functional responses through dimerization of its receptor CCR2. Proc Natl Acad Sci USA. 1999;96:3628–3633. doi: 10.1073/pnas.96.7.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rucker J, Samson M, Doranz B J, Libert F, Berson J F, Yi Y, Smyth R J, Collman R G, Broder C C, Vassart G, Doms R W, Parmentier M. Regions in beta-chemokine receptors CCR5 and CCR2b that determine HIV-1 cofactor specificity. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 48.Samson M, Labbe O, Mollereau C, Vassart G, Parmentier M. Molecular cloning and functional expression of a new human CC-chemokine receptor gene. Biochemistry. 1996;35:3362–3367. doi: 10.1021/bi952950g. [DOI] [PubMed] [Google Scholar]
- 49.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 50.Sattentau Q J, Dalgleish A G, Weiss R A, Beverley P C. Epitopes of the CD4 antigen and HIV infection. Science. 1986;234:1120–1123. doi: 10.1126/science.2430333. [DOI] [PubMed] [Google Scholar]
- 51.Sattentau Q J, Moore J P, Vignaux F, Traincard F, Poignard P. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J Virol. 1993;67:7383–7393. doi: 10.1128/jvi.67.12.7383-7393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siciliano S J, Kuhmann S E, Weng Y, Madani N, Springer M S, Lineberger J E, Danzeisen R, Miller M D, Kavanaugh M P, DeMartino J A, Kabat D. A critical site in the core of the CCR5 chemokine receptor required for binding and infectivity of human immunodeficiency virus type 1. J Biol Chem. 1999;274:1905–1913. doi: 10.1074/jbc.274.4.1905. [DOI] [PubMed] [Google Scholar]
- 53.Singer I I, Scott S, Chin J, Bayne E K, Limjuco G, Weidner J, Miller D K, Chapman K, Kostura M J. The interleukin-1 beta-converting enzyme (ICE) is localized on the external cell surface membranes and in the cytoplasmic ground substance of human monocytes by immuno-electron microscopy. J Exp Med. 1995;182:1447–1459. doi: 10.1084/jem.182.5.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smilenov L B, Mikhailov A, Pelham R J, Marcantonio E E, Gundersen G G. Focal adhesion motility revealed in stationary fibroblasts. Science. 1999;286:1172–1174. doi: 10.1126/science.286.5442.1172. [DOI] [PubMed] [Google Scholar]
- 55.Thali M, Moore J P, Furman C, Charles M, Ho D D, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tokuyasu K T. Application of cryoultramicrotomy to immunocytochemistry. J Microsc. 1986;143:139–149. doi: 10.1111/j.1365-2818.1986.tb02772.x. [DOI] [PubMed] [Google Scholar]
- 57.Tokuyasu K T. Use of poly(vinylpyrrolidone) and poly(vinyl alcohol) for cryoultramicrotomy. Histochem J. 1989;21:163–171. doi: 10.1007/BF01007491. [DOI] [PubMed] [Google Scholar]
- 58.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 59.Ugolini S, Moulard M, Mondor I, Barois N, Demandolx D, Hoxie J, Brelot A, Alizon M, Davoust J, Sattentau Q J. HIV-1 gp120 induces an association between CD4 and the chemokine receptor CXCR4. J Immunol. 1997;159:3000–3008. [PubMed] [Google Scholar]
- 60.Weiss A, Schlessinger J. Switching signals on or off by receptor dimerization. Cell. 1998;94:277–280. doi: 10.1016/s0092-8674(00)81469-5. [DOI] [PubMed] [Google Scholar]
- 61.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 62.Wyatt R, Kwong P D, Desjardins E, Sweet R W, Robinson J, Hendrickson W A, Sodroski J G. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 63.Xiao X, Wu L, Stantchev T S, Feng Y R, Ugolini S, Chen H, Shen Z, Riley J L, Broder C C, Sattentau Q J, Dimitrov D S. Constitutive cell surface association between CD4 and CCR5. Proc Natl Acad Sci USA. 1999;96:7496–7501. doi: 10.1073/pnas.96.13.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zimmerman P A, Buckler-White A, Alkhatib G, Spalding T, Kubofcik J, Combadiere C, Weissman D, Cohen O, Rubbert A, Lam G, Vaccarezza M, Kennedy P E, Kumaraswami V, Giorgi J V, Detels R, Hunter J, Chopek M, Berger E A, Fauci A S, Nutman T B, Murphy P M. Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: studies in populations with contrasting clinical phenotypes, defined racial background, and quantified risk. Mol Med. 1997;3:23–36. [PMC free article] [PubMed] [Google Scholar]