Abstract
Intratumor heterogeneity reduces treatment efficacy and complicates our understanding of tumor progression and there is a pressing need to understand the functions of heterogeneous tumor cell subpopulations within a tumor, yet systems to study these processes in vitro are limited. Single-cell RNA sequencing (scRNA-seq) has revealed that some cancer cell lines include distinct subpopulations. Here, we present clusterCleaver, a computational package that uses metrics of statistical distance to identify candidate surface markers maximally unique to transcriptomic subpopulations in scRNA-seq which may be used for FACS isolation. With clusterCleaver, ESAM and BST2/tetherin were experimentally validated as surface markers which identify and separate major transcriptomic subpopulations within MDA-MB-231 and MDA-MB-436 cells, respectively. clusterCleaver is a computationally efficient and experimentally validated workflow for identification of surface markers for tracking and isolating transcriptomically distinct subpopulations within cell lines. This tool paves the way for studies on coexisting cancer cell subpopulations in well-defined in vitro systems.
Subject terms: Cell biology, Software, Cancer
Introduction
Intratumoral heterogeneity is a general term which describes the diversity of cell types and cell states within a tumor. Different cell types, including immune and stromal cells, are often found in tumor ecosystems1–6, and within a population of cancer cells there is genetic and non-genetic variation which further increases intratumoral heterogeneity7.
Cellular heterogeneity within tumors can alter tumor aggressiveness, promote metastasis, and is a known contributor to resistance against chemotherapy and targeted therapy8–11, making it one of the most significant factors in disease relapse and mortality. Single-cell sequencing technologies continue to reveal heterogeneity within primary patient tumor cells and within in vitro model systems12–15. This intratumoral heterogeneity can be interpreted from an eco-evolutionary perspective in that subpopulations of cells adapt, interact, mutate, proliferate, and perish in response to their environment16,17. While single-cell sequencing technologies have been crucial to revealing the existence of novel cancer cell subpopulations, these assays are often endpoint. To understand and probe into mechanisms which drive tumor heterogeneity and tumor progression more fully, there is a pressing need for tools which allow identification, isolation, and perturbation of coexisting tumor cell subpopulations.
Multiple methods have been developed which aim to find minimal sets of marker genes which can be used to define groups, primarily aimed at tools which have limited gene panels18–22. Several other methods have aimed to find 1 or 2 surface marker genes which will maximally separate subpopulations23–25; only one of these methods, COMET, demonstrated experimental validation. Most of these approaches have relied on determining an optimal expression threshold built on the assumption that RNA expression will directly correlate with protein expression, but this is not always guaranteed to occur. A highly expressed marker gene is not guaranteed to be a highly expressed protein26,27, nor guaranteed to be localized to the cell surface in a particular experimental model28. Furthermore, optimal thresholds based on RNA expression do not have a direct conversion to flow cytometry. Finally, many of these methods are not easily scalable across large datasets, as they either require computationally expensive statistical methods or are not readily compatible with standard single-cell programs such as scanpy29 and Seurat30 which are optimized for the storage and analysis of complicated datasets.
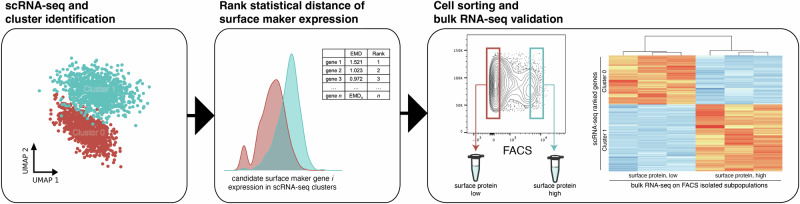
To address these problems, we developed “clusterCleaver”, a computationally efficient and scanpy compatible workflow in which the Earth Mover’s Distance (EMD)31,32, a measure of statistical distance, is applied to rank individual surface marker genes based on how well they separate transcriptomic clusters of cells in scRNA-seq data. We preprocessed single-cell data to be better suited for predicting surface markers which could enable separation with FACS and developed it as a package compatible with the popular single-cell gene expression package scanpy. To test and validate the clusterCleaver workflow (Fig. 1), multiplexed single-cell RNA-sequencing (scRNA-seq) was first performed on 5 breast cancer cell lines to identify cell lines with highly distinct transcriptomic subpopulations. Then, clusterCleaver was used on cell lines with distinct subpopulations to identify and rank candidate surface markers which could be used to physically separate transcriptomic clusters within each cell line. Top candidate markers were then experimentally screened with flow cytometry and subpopulations were immunostained and FACS-separated from the top hit for each. TagSeq, a bulk 3’ RNA-seq method33,34, and differential expression analysis was performed on isolated subpopulations to assess similarity with transcriptomic identities of the targeted scRNA-seq clusters.
Fig. 1. The clusterCleaver workflow.
Taking clustered single-cell RNA-sequencing data as input, clusterCleaver computes the EMD on predicted surface maker genes. Candidate surface marker genes are experimentally screened using flow cytometry, then surface staining antibodies which show subpopulation separation can be used to FACS isolate cell subpopulations. Transcriptomic identity of sorted cell subpopulations can be validated by performing bulk RNA-seq.
Results
Identification of subpopulations within cell lines through scRNA-seq
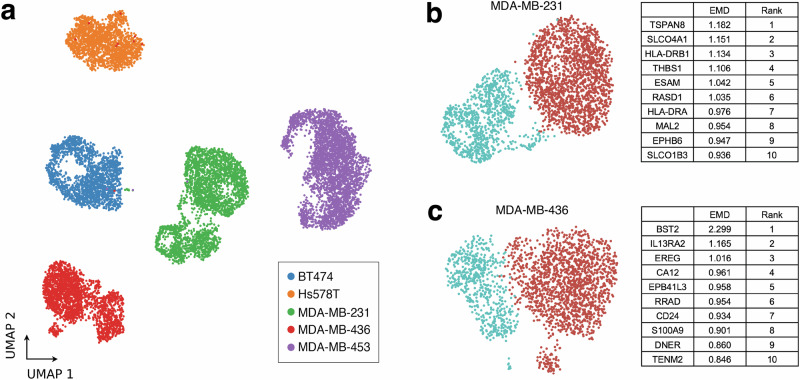
As previous studies have suggested the presence of multiple subpopulations within breast cancer cell lines13,35–39, multiplexed scRNA-seq was performed on 5 different unperturbed and early passage breast cancer cell lines (BT-474, MDA-MB-231, MDA-MB-436, MDA-MB-453, and Hs578T) to identify cell lines which contain distinct transcriptomic subpopulations (Fig. 2a). Leiden clustering was performed on each cell line (Supplementary Fig. 1a, Fig. 2b, c), then Pearson correlation coefficient (PCC) was calculated between Leiden clusters for each (Supplementary Fig. 1b). As clusters within MDA-MB-231 (PCC = 0.81) and MDA-MB-436 (PCC = 0.87) were the most dissimilar of the cell lines tested (BT-474 (PCC = 0.95), MDA-MB-453 (PCC = 0.95), Hs578T (PCC = 0.94)), MDA-MB-231 and MDA-MB-436 cells were chosen for testing and validation of the clusterCleaver workflow.
Fig. 2. Identification of multiple transcriptomic subpopulations in commonly used breast cancer cell lines.
a UMAP showing results of multiplexed scRNA-seq performed on BT-474, MDA-MB-231, MDA-MB-453, Hs578T, and MDA-MB-436 cells within 5 passages from ATCC. Leiden clustering and the top 10 EMD ranked candidate surface marker genes for separation of Leiden clusters for (b) MDA-MB-231 and (c) MDA-MB-436 cells.
Application of the Earth Mover’s Distance
Next, we applied the EMD to all genes within the scRNA-seq data ranked within the Cancer Surfaceome Atlas (TCSA)40 to identify candidate surface markers which could be used to physically separate unique transcriptomic clusters identified in scRNA-seq. EMD is a computationally efficient metric that compares two distributions. Intuitively, it can be thought of as the amount of work required to make two distributions equal41. Therefore, two distributions with low levels of overlap will have a relatively high EMD and highly overlapping distributions will have an EMD score close to 0. This property of EMD makes it highly suitable for quantitatively ranking marker genes in scRNA-seq with minimally overlapping expression distributions between transcriptomic clusters.
The EMD was applied to every candidate surface marker gene found both in the single-cell data and ranked within the TCSA database. TCSA provides a predicted surface score for each gene based on amalgamated data from nine different sources including experimental surface marker studies, computational protein conformation prediction, and prior database annotations40. TCSA surface marker scores are useful for filtering out genes unlikely to be surface expressed, but potential variation in mRNA translation and protein localization in different cell types highlights the need for experimental screening after running algorithms which depend on data built from other sources and cell types. The top candidate surface marker genes returned from clusterCleaver for MDA-MB-231 and MDA-MB-436 are summarized in Fig. 2b, c and a full list of EMD scores and rankings for each cell line can be found for each gene in Supplementary Data 1 and Supplementary Data 2.
Screening of candidate surface markers
The top ranked genes for MDA-MB-231 and MDA-MB-436 from the clusterCleaver workflow with commercially available fluorochrome-conjugated monoclonal antibodies were screened (Supplementary Table 1). On MDA-MB-231 cells, ESAM and TSPAN8 each identified distinct protein expression clusters by flow cytometry (Supplementary Fig. 2) but when immunostained in tandem, it was noted that TSPAN8 recognizes a subset of ESAM-high cells (Supplementary Fig. 2a). HLA-ABC and ITGA2/CD49b provided positive surface staining, but without clear delineation between subpopulations (Supplementary Fig. 2b, c). When stained in tandem with ESAM, HLA-ABC, and ITGA2/CD49b showed expected cluster-specific protein expression patterns, with the ESAM-low subpopulation displaying higher average expression of HLA-ABC and lower average expression of ITGA2/CD49b as expected from scRNA-seq measurements (Supplementary Fig. 2b, c). Tetherin (CD317), the protein product encoded by BST2, was found to be a surface marker for MDA-MB-436 cells which identifies two distinct protein expression clusters by flow cytometry (Supplementary Fig. 3). IL13RA2/CD213a2 and CA12 showed some positive surface staining (Supplementary Fig. 3a, b), while antibodies against GYPC and EREG failed to stain MDA-MB-436 cells (Supplementary Fig. 3c, d). From results of this surface marker screen, we selected ESAM as the top flow cytometry candidate surface marker for separating clusters in MDA-MB-231 cells, and BST2/tetherin for MDA-MB-436.
Isolation of subpopulations and validation of subpopulation transcriptomic identity
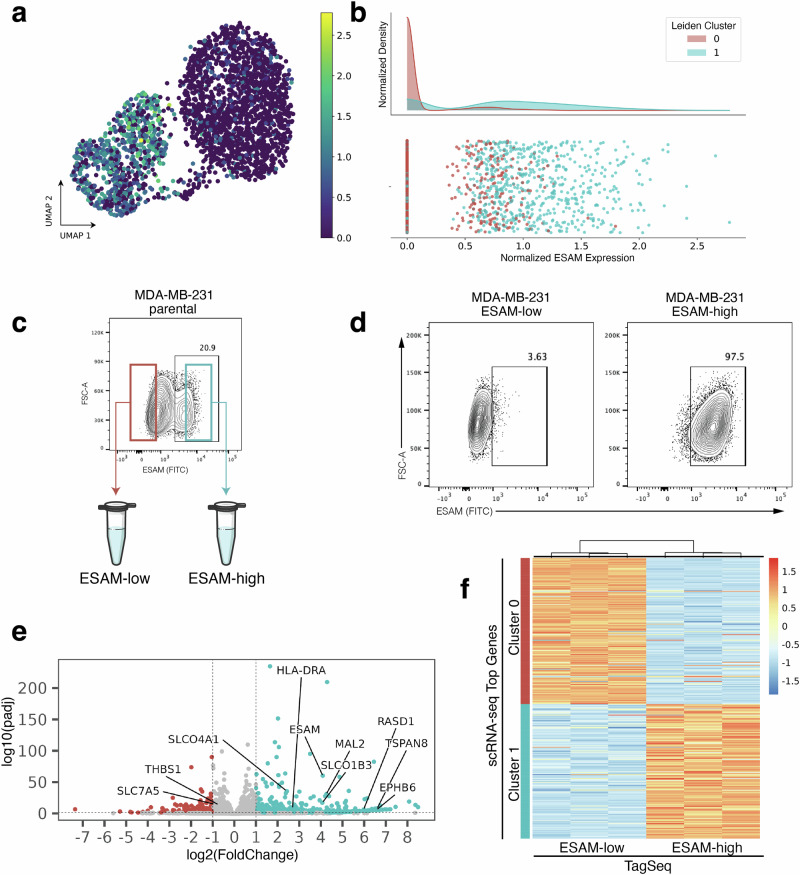
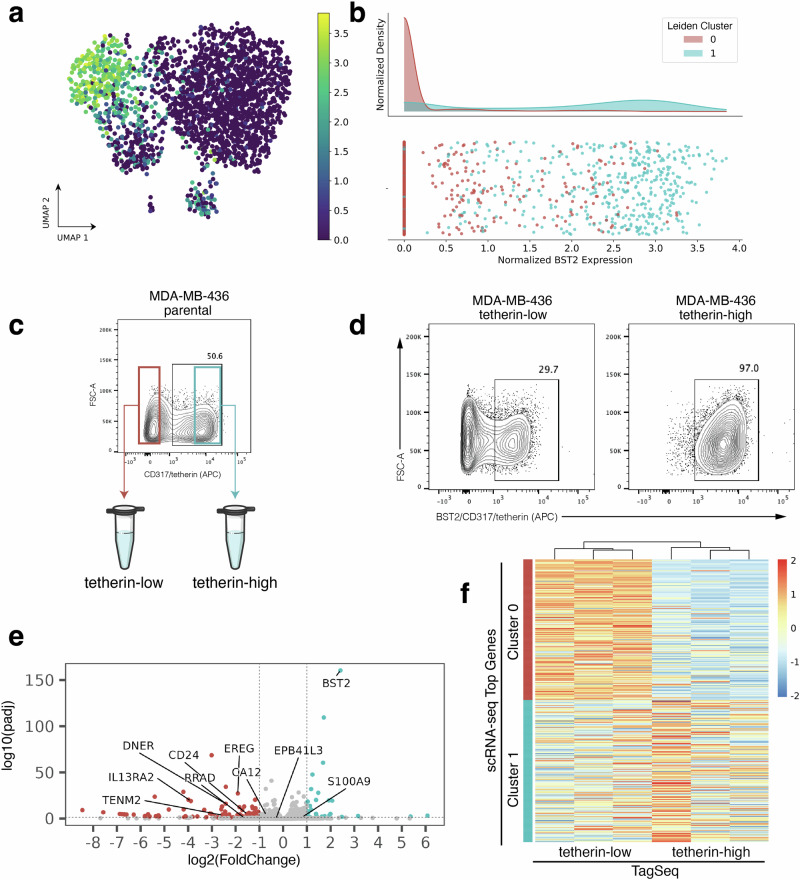
Returning to the goal of physically isolating subpopulations identified in scRNA-seq, we now asked whether the top protein surface markers identified using the EMD and screened with flow cytometry select for subpopulations with transcriptomic identities that match the respective scRNA-seq clusters. In the flow cytometry screen, immunostaining with ESAM (MDA-MB-231, Fig. 3a, b) and BST2/tetherin (MDA-MB-436, Fig. 4a, b) each revealed distinct subpopulations correlating to scRNA-seq cluster gene expression in their respective cell lines. FACS isolation was performed to enrich for ESAM-low/ESAM-high subpopulations of MDA-MB-231 cells (Fig. 3c) and tetherin-low/tetherin-high of the MDA-MB-436 cells (Fig. 4c).
Fig. 3. Experimental validation of ESAM as a surface marker for MDA-MB-231 subpopulations.
a scRNA-seq UMAP projection of MDA-MB-231 cells colored by ESAM expression. b Histogram of ESAM expression showing enrichment of cluster 1 for ESAM. c ESAM immunostaining on parental MDA-MB-231 cells. Cells with the lowest immunostaining were FACS enriched as “ESAM-low” and cells with the highest immunostaining as “ESAM-high”. d At the time of RNA collection (10 days post-FACS and expansion), sorted ESAM subpopulations each maintained around 97% purity as measured by ESAM immunostaining and flow cytometry analysis (ESAM-low, left; ESAM-high, right). e Volcano plot of differentially expressed genes from TagSeq performed on sorted subpopulations. Labels point to genes that were highly ranked in the scRNA-seq dataset using EMD. f Heatmap showing TaqSeq data for each FACS enriched subpopulation (ESAM-low, left; ESAM-high, right) plotted against ranked cluster genes from scRNA-seq (Cluster 0, top; Cluster 1, bottom).
Fig. 4. Experimental validation of BST2/tetherin as a surface marker for MDA-MB-436 subpopulations.
a scRNA-seq UMAP projection of MDA-MB-436 cells colored by BST2 expression. b Histogram of BST2 expression showing enrichment of cluster 1 for BST2. c BST2 encodes the protein product tetherin/CD317 and tetherin immunostaining reveals subpopulations within parental MDA-MB-436 cells. Cells with the lowest immunostaining were FACS enriched as “tetherin-low” and cells with the highest immunostaining as “tetherin-high” (d) At the time of RNA collection (13 days post-FACS and expansion), sorted the sorted tetherin-high subpopulation maintained 97% purity by tetherin immunostaining and flow cytometry analysis (d, right), however, the tetherin-low subpopulation dropped to 70% purity (d, left). e Volcano plot of differentially expressed genes from TagSeq performed on sorted subpopulations. Labels point to genes that were highly ranked in the scRNA-seq EMD. f Heatmap of TaqSeq data for each FACS enriched subpopulation (tetherin-low, left; tetherin-high, right) plotted against ranked cluster genes from scRNA-seq (Cluster 0, top; Cluster 1, bottom).
Next, to check the transcriptomic identity of these isolated subpopulations, we expanded triplicate samples of each subpopulation after two rounds of FACS purification and then performed TagSeq, a bulk 3’ RNA-seq method33,34. At the time of RNA collection, parallel cells were immunostained and fixed to assess purity. We note that while the ESAM subpopulations of MDA-MB-231 cells and the tetherin-high subpopulation of the MDA-MB-436 cells maintained >97% purity (Figs. 3d and 4d), the tetherin-low subpopulation of the MDA-MB-436 cells were only 70% pure (Fig. 4d). The result has been repeated and is not due to sorting error. The inability of the MDA-MB-436-tetherin-low subpopulation to maintain purity may indicate a unique biological property of these cells which suggests future studies.
To first assess if isolated subpopulations were significantly different from each other, differential expression analysis was performed on TagSeq data. Analysis with DESeq2 revealed 2250 differentially expressed genes (FDR < 0.05) in the ESAM-separated MDA-MB-231 populations and 447 differentially expressed genes (FDR < 0.05) in the BST2/tetherin-separated MDA-MB-436 populations (Figs. 3e and 4e).
Finally, transcriptomic identity of FACS isolated subpopulations was compared to transcriptomic identity of targeted scRNA-seq clusters. Top scRNA-seq cluster-specific genes were calculated by applying a t-test between scRNA-seq clusters. Comparing the TagSeq data of the FACS isolated subpopulations against the top cluster-specific genes from scRNA-seq revealed that the transcriptome of the isolated subpopulations are well-matched to their expected scRNA-seq cluster identity (Figs. 3f and 4f). To quantify similarity between isolated subpopulations and scRNA-seq transcriptomic clusters, the top 50 differentially expressed genes for each subpopulation in TagSeq were then compared to the top 50 differentially expressed genes in scRNA-seq clusters. For MDA-MB-231, 47/50 genes (94%) overlapped between the isolated ESAM-high subpopulation and its target scRNA-seq cluster (2/50 with the non-target cluster), and 46/50 genes (92%) overlapped between the ESAM-low subpopulation and its target cluster (3/50 with the non-target cluster). For MDA-MB-436, 34/50 genes (68%) overlapped between the isolated BST2/tetherin-high subpopulation and its target scRNA-seq cluster (4/50 with the non-target cluster), and 38/50 genes (76%) overlapped between the BST2/tetherin-low subpopulation and its target cluster (5/50 with the non-target cluster). Discrepancies in overlapping differentially expressed genes in this analysis may arise from many sources not limited to instability of transcriptomic state, heterogeneity of gene expression within each scRNA-seq assigned transcriptomic cluster, differences in gene expression within the subpopulations when cultured together (as in scRNA-seq) compared to in isolation (for TagSeq), and from differences in the biotechnologies and computational pipelines used to obtain expression data (scRNA-seq vs. TagSeq). Despite these sources of error, we find a strong overlap of differentially expressed genes between isolated subpopulations and their targeted scRNA-seq cluster and weak overlap with the non-targeted cluster for each isolated subpopulation in both cell lines. These results suggest that clusterCleaver is a workflow which identifies surface marker genes which can be used to successfully enrich subpopulations from targeted scRNA-seq transcriptomic clusters.
Discussion
We have shown that clusterCleaver is a computationally efficient workflow that takes in scRNA-seq and applies the EMD to rank surface markers which can enrich for targeted transcriptomic subpopulations from heterogeneous populations of cancer cells.
The use of EMD has been previously used as a method for separating flow cytometry data directly42, but also it possesses distinct advantages over other proposed methods for identifying markers for scRNA-seq data. EMD is computationally efficient41, making it ideal for searching across large scRNA-seq datasets. However, its primary advantage is that it does not rely on finding an optimal threshold or otherwise implementing a loss function which accounts for the sensitivity and specificity of a given threshold. EMD instead can be sensitive to outliers, meaning that a cluster with a subset of cells which have distinctly high expression of a given gene can potentially be highly ranked. This is advantageous for identifying markers in cases where a large bimodal distribution cannot be found. This does introduce scenarios in which a gene with a long-tailed distribution may be ranked higher than a gene with minimal overlap. To account for this, clusterCleaver includes several visualization modules to facilitate domain-specific interpretation when choosing surface markers to screen in flow cytometry.
While clusterCleaver has many computational benefits, it does come with several caveats. Primarily, clusterCleaver functions optimally with data processed such that gene expression is bounded to be greater than or equal to 0. This means that regressed data, such as in cell cycle regression, may hamper results. Additionally, the current implementation of clusterCleaver only computes the EMD between two 1-dimensional distributions. However, the EMD between multivariate distributions is able to be calculated using estimations from the sliced-wasserstein distance43,44 as well as direct calculations45, but these methods come at a higher computational cost.
All surface marker prediction methods are ultimately limited in that they are forced to rely on external databases to predict surface expression of genes and do not check for commercial antibody availability. Several computational methods have been developed which attempt to predict protein expression from scRNA-seq data46–49 which could be implemented to better search for markers which are more reliably expressed. clusterCleaver uses genes ranked in TCSA40, but is an adaptable workflow that can rank any set of genes using the EMD. This allows clusterCleaver to use other curated surface marker datasets50 or user-provided gene lists. While this work focused on the application of clusterCleaver for isolating transcriptomic subpopulations found in cancer cell lines, it can be applied to any scRNA-seq data set in which transcriptomic clusters have been defined, such as the tumor microenvironment51,52, immune subsets, or cells found in other pathologies. As clusterCleaver is applied to annotated scRNA-seq data and annotations are highly dependent on the biology each data set represents, it is up to the user to ensure that scRNA-seq annotations match the expected biology and that the data is appropriately clustered before running clusterCleaver to prevent spurious outputs.
clusterCleaver is not limited to use in cancer cell lines. However, we highlight this application for its implications for past and future in vitro studies. Numerous studies have been performed on cell lines without knowledge of their underlying population structure, yet subpopulations of cells may behave disparately under different culture conditions and perturbations, skewing interpretation of bulk results. Subpopulations have been characterized in cell lines through different modalities35–38,53,54, but these studies may have isolated rare subsets of cells. In contrast, clusterCleaver delivers a workflow for isolating subpopulations starting from full knowledge of the transcriptomic diversity within cell lines by starting at scRNA-seq.
Currently, many studies which investigate eco-evolutionary dynamics in cancer in vitro rely on cocultures of cell lines from patients with different genetic backgrounds55,56 or mixes of drug-naïve cells with lab evolved drug resistant strains57–59. While studies like these have been paramount in unraveling complex cancer dynamics, these cocultured subpopulations may not accurately reflect the biology that is attempting to be modeled. To generate more physiologically relevant systems to study coexisting cancer cells, clusterCleaver was developed as a workflow to rank candidate flow cytometry surface markers which can be used to monitor populations changes in naturally coexisting subpopulations within a mixed population or separate out subpopulations of cells which once coexisted together for further investigation. In this study, we applied clusterCleaver to cancer cell lines and identified ESAM and BST2/tetherin as surface markers which can be used to isolate transcriptomically distinct subpopulations from MDA-MB-231 and MDA-MB-436 cells, respectively.
The ESAM-separated subpopulations of MDA-MB-231 cells were stable and showed strong transcriptomic agreement to their targeted scRNA-seq clusters. The BST2/tetherin-separated subpopulations of MDA-MB-436 cells had high agreement to their targeted transcriptomic clusters but did not match as strongly as the ESAM-separated subpopulations of MDA-MB-231 cells (~70% vs. >90% overlap of differentially expressed genes). While there are many potential sources of error, we reason that differences in differential gene expression may be primarily driven by heterogeneous BST2 expression within the BST2-high/tetherin-high targeted cluster (MDA-MB-436, Cluster 1) and potential instability of the BST2/tetherin-low cell state (Fig. 4d), compared to more even coverage of ESAM expression in the ESAM-high cluster (MDA-MB-231, Cluster 1) and apparent stability of the ESAM-high and ESAM-low cell states (Fig. 3d).
Genetic and non-genetic heterogeneity emerges within tumors and can also emerge within cancer cell lines7,16,60. The subpopulations identified and isolated from MDA-MB-231 and MDA-MB-436 cells in this study seem to show surprising dynamics, with the ESAM-separated subpopulations of MDA-MB-231 showing no signs of interconversion within the span of the experiment (Fig. 3d) but noting potential plasticity in the tetherin-low subpopulations of the MDA-MB-436 cells (Fig. 4d). Whether these subpopulations have emerged as stochastic cell states or as genotypically distinct clones is yet to be reconciled. Future studies into the biological characteristics of each of these subpopulations may help reveal common mechanisms of coexistence and help guide improved therapeutic strategies for heterogeneous tumors.
In conclusion, clusterCleaver is a computationally efficient method which can be applied to any clustered scRNA-seq data set to determine candidate surface markers for subpopulation tracking or isolation. We showed the development of clusterCleaver as a computational tool for ranking candidate surface markers in scRNA-seq and performed experimental validation of this tool with two commonly used breast cancer cell lines.
Methods
Cell culture
All cell lines were used within 5 passages from thawed ATCC stocks at the start of this experiment. The passage numbers provided by ATCC via Certificate of Analysis are as follows: MDA-MB-231 (ATCC p31), MDA-MB-436 (ATCC p20), MDA-MB-453 (ATCC p349), Hs578T (ATCC p52), and BT-474 (ATCC p89). MDA-MB-231 and MDA-MB-453 were maintained in high glucose DMEM (Sigma, D5796) supplemented with 1X Penn-Strep (ThermoFisher, 15140122) and 10% FBS (Sigma, F0926). MDA-MB-436 and Hs578T were maintained in high glucose DMEM (Sigma, D5796) supplemented with 1X Penn-Strep (ThermoFisher, 15140122), 10% FBS (Sigma, F0926), and 10 µg/mL insulin (ThermoFisher, 12585014). BT-474 cells were maintained in Richter’s modified MEM without phenol red (ThermoFisher, A1048801) supplemented with 1X Penn-Strep (ThermoFisher, 15140122), 10% FBS (Sigma, F0926), and 20 µg/mL insulin (ThermoFisher, 12585014). FACS isolated subpopulations were maintained in their parental media. MDA-MB-453 were passaged with 0.25% Trypsin-EDTA (ThermoFisher, 25200056), all other cell lines were passaged with 0.05% Trypsin-EDTA (ThermoFisher, 25300062) using standard protocols. A cell scraper was used to fully release MDA-MB-436 cells after 1 min of trypsinization.
scRNA-seq sample and library preparation
Cells were prepared for multiplexed scRNA-seq using the 10X Genomics 3ʹ CellPlex Kit (10X Genomics, 1000261). Briefly, MDA-MB-231 (p4), MDA-MB-436 (p4), MDA-MB-453 (p5), Hs578T (p4), and BT-474 (p3) were gently detached, neutralized, strained through a 40 µm cell strainer, then counted. 1e6 cells from each population were added to a 2 mL tube and washed with room temperature PBS supplemented with 0.04% BSA (ThermoFisher, 15260037). Each cell line was resuspended in 100 µL of a unique cell multiplexing oligo and incubated for 5 min before 3 rounds of washing with cold PBS plus 1% BSA. After washing, cells were counted, and two pools were made containing equal ratios of 3 cell lines each: (Pool A) MDA-MB-231, MDA-MB-453, BT-474, and (Pool B) MDA-MB-436 and Hs578T. Cells were dropped off to the UT Austin Genomic Sequencing and Analysis and 15,000 cells per pool were loaded into a Chromium Next GEM Single Cell 3’ Chip following standard 10X Genomics protocols.
scRNA-seq analysis
Single-cell data was aligned to the GRCh38 (version refdata-gex-GRCh38-2020-A, 10X Genomics) and processed using cellranger’s (version 6.1.2) multi command. The data was then aggregated using cellranger’s aggr function. Cells were then loaded into scanpy (version 1.9.8). Quality control was done according to the scanpy’s single-cell best practices61. Automatic thresholding was done on the percentage of mitochondrial genes, the number of genes within a cell, or the total gene count within a cell using the median absolute deviation (MAD) as defined by Equation 1:
| 1 |
Where X is the expression of the given cell. Cells with a MAD greater than 5 were removed. Doublets were then removed using the package scDblFinder62. To normalize the data, we used the shifted logarithm technique as recommended by the single-cell best practices. Finally, we performed dimensionality reduction by finding highly variable genes, calculating the dataset’s principal components, computing a nearest neighbors graph, then calculating the UMAP representation. To remove cell cycle effects potentially biasing clustering, we regressed cell out cell cycle using gene lists from ref. 63. All clustering and UMAP visualization was done using the Leiden algorithm as implemented by scanpy on regressed gene expression data. All candidate marker searches were performed on non-regressed log-normalized gene expression data. Similarity between clusters was computed between each cluster within each cell line on the full concatenated dataset by taking the top 50 principal component values of the gene expression and calculating the Pearson correlation coefficient between all identified clusters.
Earth Mover’s Distance pre-processing
From experience, ideal surface markers are bimodally distributed between clusters, with one cluster having gene expression around 0. However, one case that often occurs is that both clusters will have a peak of 0 gene expression while one of the clusters will have a second peak of higher gene expression. Therefore, this gene is potentially a good candidate as many cells from the high-expressing gene cluster can still be theoretically isolated. However, this gene will not rank highly with the EMD because the clusters both have significant overlap around 0. To circumvent this, we removed gene expression levels of 0 in clusters which had higher average gene expression. This allowed us to find more genes which were candidate markers.
Flow cytometry and FACS
Cells gently detached and neutralized according to standard culture techniques. Cells were counted using a trypan blue exclusion automated cell counter. 0.2e6 cells were used for screening antibody labeling experiments. 1e6 cells were used for antibody validation and 2-5e6 cells were labeled for FACS cell sorting experiments. Cells were resuspended in cell staining buffer (PBS + 5 mM EDTA + 1% BSA + 1.6 mM NaOH + 0.01% sodium azide) and incubated in 1:100 diluted Zombie UV viability dye (Biolegend, 423107) or Zombie Violet viability dye (Biolegend, 423113) for 5 min on ice, then with the manufacturer recommended volume of antibody (antibody information in supplementary Table 1) for 20 min. Immunostained cells were washed 3 times with cell sorting buffer supplemented with 1:1000 diluted Zombie viability dye, then passed through a 40 µm cell strainer before flow analysis. For fixed cell preparation, cells were resuspended in 200 µL of 4% PFA in PBS for 15 min after the second wash, then washed 2 more times. Collection media for live cell sorting was prepared by supplementing complete media with 25 mM HEPES. Cells were sorted into 15 mL tubes containing 7 mL of collection media. Collected cells were spun down at 300 × g for 10 min, then plated in 50% conditioned media for 24 h before transitioning to fresh, complete media. Conditioned media (CM) was prepared from complete media incubated on a 70% confluent plate of parental cells for 24 h. CM was spun down at 500 × g for 10 min and supernatant was passed through a 0.22 µm filter. CM was diluted to 50% with fresh, complete media.
RNA collection for TagSeq
MDA-MB-231: MDA-MB-231-ESAM-low and MDA-MB-231-ESAM-high were FACS sorted 7 days before plating for RNA collection. 0.15e6 cells of MDA-MB-231 p8, MDA-MB-231 p13, MDA-MB-231-ESAM-low, MDA-MB-231-ESAM-high were plated across triplicate 6-well plates. Media was exchanged on plates after 24 h. 72 h after plating (10 days post-FACS), cells were 60–70% confluent, and RNA was collected using an in-plate lysis strategy following the Qiagen RNAeasy Mini protocol. MDA-MB-436: MDA-MB-436-tetherin-low and MDA-MB-436-tetherin-high were FACS sorted 11 days before plating for RNA collection. 0.84e6 cells of MDA-MB-436 p7, MDA-MB-436 p13, MDA-MB-436-tetherin-low, MDA-MB-436-tetherin-high were plated across triplicate 6-well plates. Media was exchanged on plates after 24 h. 48 h after plating (13 days post-FACS), cells were 60–70% confluent, and RNA was collected using an in-plate lysis strategy following the Qiagen RNAeasy Mini protocol. For all samples, RNA was eluted in 35 µL of nuclease-free water. RNA samples were quantified via Qubit, diluted, then submitted to the UT Austin Genomic Sequencing and Analysis Facility for 3’-Tag RNAseq (TagSeq) preparation.
TagSeq analysis
Raw FASTQ files were processed using nf-core/rnaseq (v3.14.0)64 using default settings. Data was aligned to GrCh38. Bias uncorrected counts were rounded and used as recommended with DESeq265 (v1.40.2) to generate differentially expressed genes and generate normalized expression values.
Supplementary information
Acknowledgements
We thank NIH F31CA268833 (to A.L.G.), R01CA226258, R01CA255536, and U01CA253540 (to A.B.) which provided funding that supported this project and the training of personnel involved. A.L.G. thanks undergraduates Lan Zheng, Drew Saunders, Chisom Iloegbunam, and Andrea Ramirez who helped this project succeed by providing concurrent support to other projects while this project was taking place. 10X Genomics Single Cell 3’ Gene Expression and TagSeq preparation were performed by the Genomic Sequencing and Analysis Facility at UT Austin, Center for Biomedical Research Support (RRID: SCR_021713). Flow cytometry and FACS was performed at the Center for Biomedical Research Support Microscopy and Imaging Facility at UT Austin (RRID: SCR_021756).
Author contributions
T.A.J.: Conceptualization, Methodology, Software, Formal Analysis, Data Curation, Writing—Original Draft, Writing—Review & Editing, Visualization. A.L.G.: Conceptualization, Methodology, Validation, Investigation, Data Curation, Writing—Original Draft, Writing—Review & Editing, Visualization. D.M.: Methodology, Writing—Review & Editing. A.B.: Resources, Writing—Review & Editing, Supervision, Project administration, Funding acquisition.
Data availability
TagSeq data has been deposited in the Gene Expression Omnibus (GEO) under accession code GSE268250. Single cell data was deposited under accession code GSE268249. Flow cytometry FCS files are available upon request.
Code availability
Notebooks used to process transcriptomic data and generate figures can be accessed at www.github.com/brocklab/clusterCleaver-analysis. The clusterCleaver package and instructions for installation and usage can be found at www.github.com/brocklab/clusterCleaver.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Andrea L. Gardner, Tyler A. Jost.
Supplementary information
The online version contains supplementary material available at 10.1038/s41540-024-00441-6.
References
- 1.Regev, A. et al. The human cell atlas. Elife6, e27041 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Domínguez Conde, C. et al. Cross-tissue immune cell analysis reveals tissue-specific features in humans. Science376, eabl5197 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rozenblatt-Rosen, O. et al. The human tumor atlas network: charting tumor transitions across space and time at single-cell resolution. Cell181, 236–249 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eraslan, G. et al. Single-nucleus cross-tissue molecular reference maps toward understanding disease gene function. Science376, eabl4290 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The Tabula Sapiens Consortium A multiple-organ, single-cell transcriptomic atlas of humans. Science376, eabl4896 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karlsson, M. et al. A single–cell type transcriptomics map of human tissues. Sci. Adv.7, eabh2169 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brock, A., Chang, H. & Huang, S. Non-genetic heterogeneity—a mutation-independent driving force for the somatic evolution of tumours. Nat. Rev. Genet.10, 336–342 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Burrell, R. A., McGranahan, N., Bartek, J. & Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature501, 338–345 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Koren, S. & Bentires-Alj, M. Breast tumor heterogeneity: source of fitness, hurdle for therapy. Mol. Cell60, 537–546 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Lindström, L. S. et al. Intratumor heterogeneity of the estrogen receptor and the long-term risk of fatal breast cancer. J. Natl Cancer Inst.110, 726–733 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawson, D. A., Kessenbrock, K., Davis, R. T., Pervolarakis, N. & Werb, Z. Tumour heterogeneity and metastasis at single-cell resolution. Nat. Cell Biol.20, 1349–1360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karaayvaz, M. et al. Unravelling subclonal heterogeneity and aggressive disease states in TNBC through single-cell RNA-seq. Nat. Commun.9, 3588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dave, A. et al. The breast cancer single-cell atlas: defining cellular heterogeneity within model cell lines and primary tumors to inform disease subtype, stemness, and treatment options. Cell Oncol.46, 603–628 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinker, G. S. et al. Pan-cancer single-cell RNA-seq identifies recurring programs of cellular heterogeneity. Nat. Genet.52, 1208–1218 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang, C. A. et al. Ontogeny and vulnerabilities of drug-tolerant persisters in HER2+ breast cancer. Cancer Discov.12, 1022–1045 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pisco, A. O. et al. Non-Darwinian dynamics in therapy-induced cancer drug resistance. Nat. Commun.4, 2467 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howard, G. R., Johnson, K. E., Rodriguez Ayala, A., Yankeelov, T. E. & Brock, A. A multi-state model of chemoresistance to characterize phenotypic dynamics in breast cancer. Sci. Rep.8, 12058 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ranjan, B. et al. DUBStepR is a scalable correlation-based feature selection method for accurately clustering single-cell data. Nat. Commun.12, 5849 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ibrahim, M. M. & Kramann, R. genesorteR: feature ranking in clustered single cell data. 676379 Preprint at 10.1101/676379 (2019).
- 20.Gregory, W., Sarwar, N., Kevrekidis, G., Villar, S. & Dumitrascu, B. MarkerMap: nonlinear marker selection for single-cell studies. npj Syst. Biol. Appl.10, 1–12 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen, X., Chen, S. & Thomson, M. Minimal gene set discovery in single-cell mRNA-seq datasets with ActiveSVM. Nat. Comput Sci.2, 387–398 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson, M. E., Riva, S. G. & Cvejic, A. SMaSH: a scalable, general marker gene identification framework for single-cell RNA-sequencing. BMC Bioinform.23, 328 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, R., Banjanin, B., Schneider, R. K. & Costa, I. G. Detection of cell markers from single cell RNA-seq with sc2marker. BMC Bioinform.23, 276 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delaney, C. et al. Combinatorial prediction of marker panels from single-cell transcriptomic data. Mol. Syst. Biol.15, e9005 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazzara, S. et al. CombiROC: an interactive web tool for selecting accurate marker combinations of omics data. Sci. Rep.7, 45477 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buccitelli, C. & Selbach, M. mRNAs, proteins and the emerging principles of gene expression control. Nat. Rev. Genet.21, 630–644 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Schukken, K. M. & Sheltzer, J. M. Extensive protein dosage compensation in aneuploid human cancers. Genome Res.32, 1254–1270 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu, Y.-Y., Zhou, H., Murphy, R. F. & Shen, H.-B. Consistency and variation of protein subcellular location annotations. Proteins89, 242 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol.19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramdas, A., Garcia, N. & Cuturi, M. On Wasserstein two sample testing and related families of nonparametric tests. Entropy19, 47 (2017).
- 32.Rubner, Y., Tomasi, C. & Guibas, L. J. The Earth Mover’s Distance as a metric for image retrieval. Int. J. Comput. Vis. 40, 99–121 (2000).
- 33.Meyer, E., Aglyamova, G. V. & Matz, M. V. Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-Seq procedure. Mol. Ecol.10.1111/j.1365-294X.2011.05205.x (2011). [DOI] [PubMed]
- 34.Lohman, B. K., Weber, J. N. & Bolnick, D. I. Evaluation of TagSeq, a reliable low‐cost alternative for RNA seq. Mol. Ecol. Resour.16, 1315–1321 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Hapach, L. A. et al. Phenotypic heterogeneity and metastasis of breast cancer cells. Cancer Res.81, 3649–3663 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hapach, L. A. et al. Phenotypically sorted highly and weakly migratory triple negative breast cancer cells exhibit migratory and metastatic commensalism. Breast Cancer Res.25, 102 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen, Y. et al. Detecting heterogeneity in and between breast cancer cell lines. Cancer Converg.4, 1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sirois, I. et al. A unique morphological phenotype in chemoresistant triple-negative breast cancer reveals metabolic reprogramming and PLIN4 expression as a molecular vulnerability. Mol. Cancer Res.17, 2492–2507 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Enciso-Benavides, J. et al. Biological characteristics of a sub-population of cancer stem cells from two triple-negative breast tumour cell lines. Heliyon7, e07273 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu, Z. et al. The cancer surfaceome atlas integrates genomic, functional and drug response data to identify actionable targets. Nat. Cancer2, 1406–1422 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pele, O. & Werman, M. Fast and robust Earth Mover’s Distances. in 2009IEEE 12th International Conference on Computer Vision 460–467. 10.1109/ICCV.2009.5459199 (2009).
- 42.Orlova, D. Y. et al. Earth Mover’s Distance (EMD): a true metric for comparing biomarker expression levels in cell populations. PLoS One11, e0151859 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flamary, R. et al. POT: python optimal transport. J. Mach. Learn. Res.22, 1–8 (2021). [Google Scholar]
- 44.Bonneel, N., Rabin, J., Peyré, G. & Pfister, H. Sliced and radon wasserstein barycenters of measures. J. Math. Imaging Vis.51, 22–45 (2015). [Google Scholar]
- 45.Peyré, G. & Cuturi, M. Computational Optimal Transport: With Applications to Data Science. FNT in Machine Learning 11, 355–607 (2019).
- 46.Lakkis, J. et al. A multi-use deep learning method for CITE-seq and single-cell RNA-seq data integration with cell surface protein prediction and imputation. Nat. Mach. Intell.4, 940–952 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu, F., Wang, S., Dai, X., Mundra, P. A. & Zheng, J. Ensemble learning models that predict surface protein abundance from single-cell multimodal omics data. Methods189, 65–73 (2021). [DOI] [PubMed] [Google Scholar]
- 48.Zhou, S., Li, Y., Wu, W. & Li, L. scMMT: a multi-use deep learning approach for cell annotation, protein prediction and embedding in single-cell RNA-seq data. Brief. Bioinforma.25, bbad523 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou, Z., Ye, C., Wang, J. & Zhang, N. R. Surface protein imputation from single cell transcriptomes by deep neural networks. Nat. Commun.11, 651 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu, C. et al. CellMarker 2.0: an updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic Acids Res.51, D870–D876 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Azizi, E. et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell174, 1293–1308.e36 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang, K., Dong, M., Li, C. & Sheng, J. Unraveling heterogeneity of tumor cells and microenvironment and its clinical implications for triple negative breast cancer. Front. Oncol. 11, 557477 (2021). [DOI] [PMC free article] [PubMed]
- 53.Yang, E. Y., Howard, G. R., Brock, A., Yankeelov, T. E. & Lorenzo, G. Mathematical characterization of population dynamics in breast cancer cells treated with doxorubicin. Front. Mol. Biosci.9, 972146 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Howard, G. R., Jost, T. A., Yankeelov, T. E. & Brock, A. Quantification of long-term doxorubicin response dynamics in breast cancer cell lines to direct treatment schedules. PLoS Comput. Biol.18, e1009104 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Freischel, A. R. et al. Frequency-dependent interactions determine outcome of competition between two breast cancer cell lines. Sci. Rep.11, 4908 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodriguez Messan, M. et al. Predicting the results of competition between two breast cancer lines grown in 3-D spheroid culture. Math. Biosci.336, 108575 (2021). [DOI] [PubMed] [Google Scholar]
- 57.Maltas, J. et al. Drug dependence in cancer is exploitable by optimally constructed treatment holidays. Nat. Ecol. Evol.8, 147–162 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nam, A. et al. Dynamic phenotypic switching and group behavior help non-small cell lung cancer cells evade chemotherapy. Biomolecules12, 8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaznatcheev, A., Peacock, J., Basanta, D., Marusyk, A. & Scott, J. G. Fibroblasts and alectinib switch the evolutionary games played by non-small cell lung cancer. Nat. Ecol. Evol.3, 450–456 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gutierrez, C. et al. Multifunctional barcoding with ClonMapper enables high-resolution study of clonal dynamics during tumor evolution and treatment. Nat. Cancer2, 758–772 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heumos, L. et al. Best practices for single-cell analysis across modalities. Nat. Rev. Genet.24, 550–572 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Germain, P.-L., Lun, A., Meixide, C. G., Macnair, W. & Robinson, M. D. Doublet identification in single-cell sequencing data using scDblFinder. F1000Res10, 979 (2021). [DOI] [PMC free article] [PubMed]
- 63.Tirosh, I. et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature539, 309–313 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patel, H. et al. nf-core/rnaseq: nf-core/rnaseq v3.14.0-Hassium Honey Badger. Zenodo, 10471647 https://github.com/nf-core/rnaseq?tab=readme-ov-file#citations (2024).
- 65.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TagSeq data has been deposited in the Gene Expression Omnibus (GEO) under accession code GSE268250. Single cell data was deposited under accession code GSE268249. Flow cytometry FCS files are available upon request.
Notebooks used to process transcriptomic data and generate figures can be accessed at www.github.com/brocklab/clusterCleaver-analysis. The clusterCleaver package and instructions for installation and usage can be found at www.github.com/brocklab/clusterCleaver.