Abstract
An open reading frame (ORF) overlapping the amino-terminal portion of the Sendai virus (SeV) P ORF in the +1 frame produces a nested set of carboxy-coterminal proteins, C′, C, Y1, and Y2, which are referred to collectively as the C proteins. The C proteins are extremely versatile triple-role players; they counteract the antiviral action of interferons (IFNs), inhibit viral RNA synthesis, and are involved in virus assembly. In this study, we established HeLa cell lines stably expressing the C, Y1, and Y2 proteins individually and examined the capacities of these cells to circumvent the antiviral action of alpha/beta IFN (IFN-α/β) and IFN-γ and to inhibit viral transcription. The assay protocols included monitoring of IFN-α/β-mediated signaling by interferon-stimulated response element-driven reporter gene expression and of the antiviral state induced by IFN-α/β and IFN-γ and measurement of reporter gene expression from an SeV minigenome, as well as quantification of SeV primary transcripts. When necessary, the activities measured were carefully normalized to the expression levels of the respective C proteins in cells. The data obtained clearly indicate that the smallest protein, Y2, was as active as the C and Y1 proteins in both counteracting the antiviral action of IFNs and inhibiting viral transcription. The data further show that intracellular transexpression of either C, Y1, or Y2 rendered HeLa cells moderately or only poorly permissive for not only wild-type SeV but also 4C(−) SeV, which expressed none of the four C proteins. On the basis of these findings, the roles of SeV C proteins in the natural life cycle are discussed.
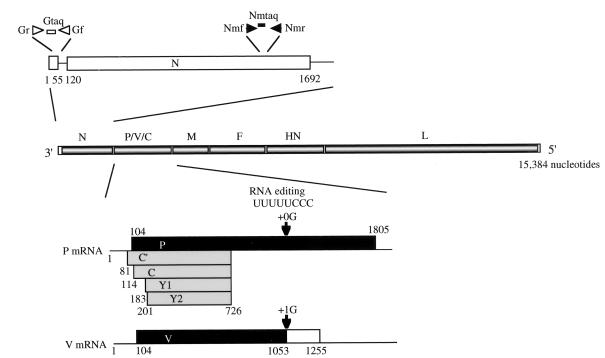
Sendai virus (SeV) is an enveloped virus with a linear, nonsegmented, negative-sense RNA genome of 15,384 nucleotides and belongs to the genus Respirovirus of the family Paramyxoviridae. The genome encodes, in 3′-to-5′ order, the nucleocapsid (N) protein, phospho (P) protein, matrix (M) protein, fusion (F) protein, hemagglutinin-neuraminidase (HA), and large (L) protein. Gene expression is usually monocistronic, generating a single mRNA which primarily directs a single translation product. However, P gene expression is exceptional because it gives rise to multiple protein species by a process known as RNA editing and by the use of an overlapping open reading frame (ORF), as shown in Fig. 1 (for a review, see reference 27).
FIG. 1.
Expression of P, V, and C proteins from the SeV P gene and positions of the primer-probe sets used in this study. The locations on the genome of the primer and probe pairs used to detect genome RNA and N mRNA are shown. The nucleotide numbering is from the 3′ end of the viral genome. For coding strategies of the various proteins, see the text. The numbers represent the nucleotide positions from the start site of the P mRNA.
In RNA editing, one nontemplated G residue is cotranscriptionally inserted at a specific position to generate the edited mRNA that encodes the protein termed V (43). The unedited mRNA that is the exact copy of the P gene encodes the P protein, the smaller subunit of RNA polymerase. The P and V proteins are therefore amino coterminal, while the −1 frame is used to generate the carboxyl-terminal half of the V protein (27). The V unique carboxyl-terminal region contains seven cysteine residues. These cysteine residues are highly conserved in paramyxovirus V proteins, form zinc finger motifs, and indeed bind Zn2+ (17, 33). To study the role of SeV V protein, we generated a series of V knockout viruses by reverse genetics and demonstrated that the V protein is completely dispensable for viral replication in tissue culture cells and devotes itself to maintaining high viral loads and causing severe pneumonia in mice (20). This luxury function required for in vivo pathogenesis was mapped to the V unique carboxyl-terminal portion (21) and associated with its zinc binding capacity (17).
An ORF overlapping the amino-terminal portion of the SeV P ORF in the +1 frame produces a nested set of carboxy-coterminal proteins called C′, C, Y1, and Y2 and referred to collectively as the C proteins. The translation of C′ is initiated on a non-AUG codon, ACG, at position 81, whereas the other three start on AUGs at positions 114, 183, and 201, respectively (13). All four C proteins are terminated at the same position, 726, as shown in Fig. 1. Among them, C is the major species expressed in infected cells at a molar ratio severalfold higher than the other three (27). It was possible to knock out all four C proteins, indicating that the C proteins are also categorically nonessential gene products (26). However, the replication of this knockout virus, named 4C(−), was greatly impaired even in tissue culture cells, partly because the C proteins were critically required for the assembly of major structural proteins into infectious progeny (14). SeV was previously found to be able to counteract the antiviral action of exogenous or endogenously produced interferons (IFNs) to allow superinfecting vesicular stomatitis virus (VSV), a highly IFN-sensitive virus, to grow vigorously even in the presence of exogenous added alpha IFN (IFN-α) (45). This anti-IFN action was found to be totally lost when all four C proteins were knocked out, indicating that it is the C proteins that encode the anti-IFN action (12). The great contribution of SeV C proteins to tissue culture replication and their anti-IFN action were also demonstrated by Garcin et al. (10). Earlier reports further showed that the SeV C protein bound the L protein, the larger subunit of polymerase, and would thereby inhibit viral mRNA synthesis (16). The replication of an SeV genome analogue, a minigenome, was also inhibited in cells expressing the C proteins (4). This inhibition appeared to be promoter specific, as antigenome replication was more profoundly inhibited than was genome replication (2). Thus, a presumable additional role of SeV C protein would be to optimize the intracellular levels of genome RNA, antigenome RNA, and mRNA (28).
These results indicate that the SeV P gene, consisting of only 1,893 nucleotides, encodes several categorically different functions and represents an extraordinary example of the fine and remarkable strategies a virus can contrive in order to extract as much genetic information as possible from a single gene (for a review, see reference 31). Particularly remarkable is the versatility of the C proteins, which are as short as only about 200 amino acids but responsible for anti-IFN action, viral RNA synthesis regulation, and virion assembly. Several important questions are now raised. These include whether or not the various C functions are interrelated with each other. At least the anti-IFN action and the contribution to virus assembly appeared to be roles independent of each other (14). The question of whether all four C proteins are equally active in each function also has to be addressed. Previously, it was suggested that C′ and C are active in RNA synthesis inhibition while Y1 and Y2 are inactive (4). Not only the 4C(−) virus but also the C′/C(−) virus exhibited greatly altered phenotypes, whereas silencing of the C′ ORF alone [C′(−) virus] did not appear to cause appreciable phenotype changes (26, 28). Therefore, we established cell lines stably expressing C, Y1, and Y2 individually to examine whether or not these three C proteins are equally active in counteracting IFN actions and inhibiting viral RNA synthesis. We demonstrated that even the smallest of them, Y2, is fully capable of both counteracting the antiviral action of IFNs and inhibiting viral RNA synthesis.
MATERIALS AND METHODS
Plasmids.
The DNA fragments encoding the C, Y1, and Y2 ORFs were obtained by PCR from a full-length cDNA clone of SeV, pSeV(+) (19). For the C ORF, two primers, CF (5′-GAATTCHindIIIaagcttGCC114ATGCCTTCATTCTTAAAG-3′) and CR (5′-GAATTCBamHIggatccCTA726TTACTCTTGCACTATGTG-3′), were used for PCR amplification. For the Y1 ORF, the Y1F (5′-GAATTCHindIII aagcttGCC183ATGTTATCGGATTCCTCG-3′) and CR primers were used, and for the Y2 ORF, the Y2F (5′-GAATTCHindIIIaagcttGCC201ATGCTGTCCTGTCGAGTG-3′) and CR primers were used. The initiation and termination codons are underlined and numbered in accordance with Fig. 1. Each region upstream of the initiation codon (italic letters) was modified to optimize for translation according to Kozak's rule (25). These fragments were cut with HindIII and BamHI (superscripts in the primers) and cloned into the same sites of plasmid pKS336 (provided by K. Sakai, National Institute of Infectious Diseases, Tokyo, Japan), a derivative of pEF-BOS (30). After verification of the sequences, plasmids encoding the C, Y1, and Y2 proteins, named pKS-C, pKS-Y1, and pKS-Y2, respectively, were used to establish stable transformants.
IFN-α/β-responsive plasmid pISRE-luci, containing three tandem repeat sequences of the ISRE (IFN-stimulated response element) fused to the firefly luciferase gene, was kindly provided by K. Ozato (National Institutes of Health). Control plasmid pRL-TK, containing the herpes simplex virus thymidine kinase promoter region fused to the Renilla luciferase gene, was purchased from Promega (Madison, Wis.). These two plasmids were used to monitor IFN-α/β-mediated signal transduction (44).
Transfection.
To establish stable transformants expressing the C, Y1, or Y2 protein, HeLa cells, maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum, were plated at a density of 106/9-cm-diameter dish. Ten micrograms of plasmid pKS-C, pKS-Y1, or pKS-Y2 was transfected into the cells using a mammalian transfection kit (Stratagene, La Jolla, Calif.). Two days later, the medium was replaced with DMEM containing blasticidin at 100 μg/ml. Colonies grown in the selection medium were picked up, propagated, and used throughout the study.
To monitor IFN-α/β-mediated signal transduction, 1 μg each of plasmids pISRE-luci and pRL-TK was cotransfected into parental and C-expressing cells grown in six-well plates (5 × 105/well) with the lipofection reagent DOTAP (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer's instructions. At 14 h posttransfection, the cells were incubated with or without IFN-α or -β at 1,000 IU/ml for 0, 2, 4, or 6 h. Portions of cell lysates were assayed for dual luciferase activities according to the manufacturer's (Promega) instructions using a luminometer (Luminos CT-9000D; Diaiatron, Tokyo, Japan). Relative expression levels were calculated by dividing firefly luciferase values by Renilla luciferase values.
Immunoblotting.
After being harvested with a cell scraper, cells were pelleted by centrifugation (1,500 × g, 5 min) and disrupted in lysis buffer (Promega). Portions of cell lysates were subjected to sodium dodecyl sulfate–15% (for C proteins) or 7.5% (for others) polyacrylamide gel electrophoresis. The proteins in the gels were electrotransferred onto polyvinylidene difluoride membranes (Millipore, Bedford, Mass.) and probed with anti-SeV C serum (26) or anti-STAT1α/β (Esc-346; Santa Cruz Biotechnology, Santa Cruz, Calif.) or anti-STAT2 (Esc-476; Santa Cruz Biotechnology) antibody.
Immunofluorescence assay.
Cells grown on chamber slide glasses (Biocoat; Becton Dickinson, Bedford, Mass.) were fixed with 10% formalin and permiabilized with 0.2% Triton X-100 in phosphate-buffered saline (PBS) as previously described (14). The cells were processed for indirect immunofluorescence assay using anti-C serum and goat anti-rabbit immunoglobulin antibody conjugated with fluorescein isothiocyanate (ICN Biomedicals, Aurora, Ohio) and observed under an inverted fluorescence microscope (Axiovert 135; Carl Zeiss, Jena, Germany).
Antiviral activity of IFNs.
Parental and C-expressing HeLa cells plated at a density of 5 × 104/well in 24-well plates were treated with human IFN-α, IFN-β, or IFN-γ at 0, 10, 100, or 1,000 IU/ml for 24 h. The cells were washed once with PBS and infected with encephalomyocarditis virus, Sindbis virus, or VSV at a multiplicity of infection (MOI) of 2 in 0.1 ml of serum-free medium for 60 min at 37°C. After removal of the virus-containing medium, the cells were further incubated in the same serum-free medium without IFN. For viral cytopathic effect assays, cells were fixed and stained with Coomassie brilliant blue when significant cytopathic changes developed. For the viral multiplication assay, VSV-infected cells and the culture supernatant were harvested after 24 h of incubation and lysed with sodium dodecyl sulfate and the lysates were immunoblotted with anti-VSV serum (provided by B. Gotoh, Fukui, Japan) (12).
Minigenome assay.
The RNA genome analog (minigenome), in which the luciferase ORF was substituted for the region beginning at the initiation codon for the N-ORF and ending at the stop codon of the L ORF in the context of the SeV genome, was transcribed from linearized pHVlucRT4(−) with a T7 transcription kit (AmpliScribe; Epicentre Technologies, Madison, Wis.) (19). In addition to the minigenome, plasmids pGEM-N, -P, and -L, carrying the SeV N, P, and L genes driven by the T7 promoter (provided by D. Kolakofsky, Geneva, Switzerland), and plasmid pRL-TK (used as an internal control) were cotransfected into the parental and respective C-expressing HeLa cells as previously described (19). Before transfection, cells were infected with recombinant vaccinia virus strain vTF7-3 expressing T7 polymerase (supplied by B. Moss, National Institutes of Health). Relative luciferase activities were measured at 20 h posttransfection as described above.
SeV infections.
The wild-type (Wt) and 4C(−) SeV (Z strain) recovered from the respective cDNAs (19, 26) were inoculated into the parental and C-expressing HeLa cells at an MOI of 5 per cell in the six-well plate. The infected cells were maintained in serum-free DMEM. The virus titers in the culture supernatants were measured at 0, 6, 12, 18, 24, and 30 h postinfection (p.i.).
Quantitative PCR assay.
Parental and C-expressing HeLa cells plated at a density of 5 × 105 per plate in 6-cm-diameter dishes were infected with Wt SeV at an MOI of 20 per cell. After viral adsorption for 1 h, the cells were washed twice with PBS and incubated for 5 or 10 h in MEM containing cycloheximide at 100 μg/ml to block de novo protein synthesis and to allow only the viral primary transcription catalyzed by polymerase in infecting viruses (22). To detect the genomic RNA, forward primer Gf (5′-21TGTATGGGATATGTAATGAAGTT43-3′), reverse primer Gr (5′-86ACCTGCTCCTCAGGGT71-3′), and TaqMan probe G-Taq (5′-Fam 65CTTTGACCCTAAAATCCT48 TAMRA-3′) were set between the leader and the N gene (Fig. 1). To detect the N mRNA, forward primer Nmf (5′-1383GACAATGCCGACATCGACC1401-3′), reverse primer Nmr (5′-1445ACCCCAACCCCTAGCGTC1428-3′), and TaqMan probe NTaq (5′-Fam 1403CGAAACAAAAGCCCCATGCGGACCA1426 TAMRA-3′) were used. The numbers at the both ends of the primers and probes represent nucleotide positions on the genome of the SeV Z strain (GenBank accession number X00087). Extracted RNAs were reverse transcribed either with primer Gf for the genomic RNA or with oligo(dT) primer for the mRNA using the TaqMan Gold reverse transcription-PCR reagents (Applied Biosystems Japan, Tokyo, Japan). Portions of cDNA were then amplified and quantitated using TaqMan PCR core reagents and an ABI PRISM 7700 (Applied Biosystems Japan) according to the manufacturer's recommendation. Plasmid pSeV(+) was used for the quantitative standard (19).
RESULTS
Establishment of stable cell lines expressing individual C proteins.
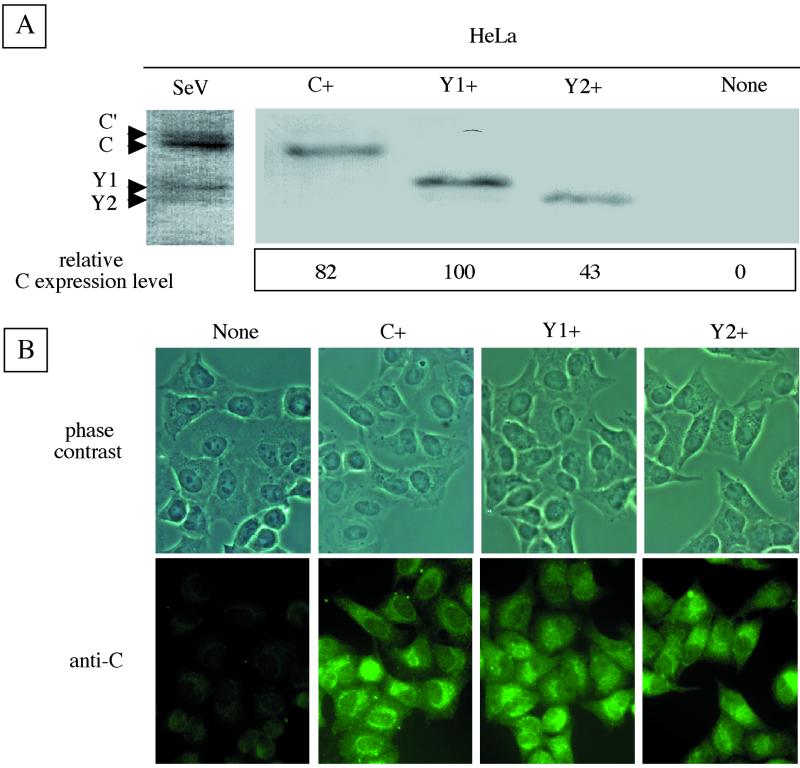
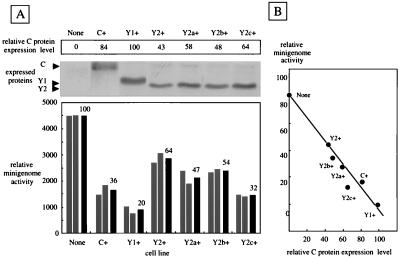
The DNA fragments encoding the C, Y1, and Y2 ORFs cloned into plasmid pKS336 were transfected into HeLa cells. Colonies grown in the selection medium were picked up and propagated. Several clones expressing C, Y1, or Y2 were obtained. As representatively shown in Fig. 2A, three clones named C+, Y1+, and Y2+ were found by immunoblotting with anti-C serum to express the C, Y1, and Y2 proteins, which comigrated with their counterparts expressed from SeV in the parental cells. Both Y1+ and Y2+ cells expressed higher levels of proteins Y1 and Y2 than did SeV-infected cells. The amounts of expressed proteins, quantitated by a densitometer, were somewhat variable. The Y1 expression level was the highest at 100, and compared with this, the relative levels were 82 for C and 43 for Y2 (Fig. 2A). These variations were pretty well maintained throughout this work. The intracellular expression of proteins C, Y1, and Y2 was visualized by immunofluorescent staining. Virtually all of the cells in culture appeared to express the respective C proteins (Fig. 2B). The distribution patterns within the cells were similar among the C, Y1, and Y2 proteins. They were distributed in the entire cytoplasm with enrichment at the perinuclear regions in some cell populations. Cells stably expressing the respective C, Y1, and Y2 proteins were thus readily established. They were propagated as efficiently as parental nontransfected HeLa cells (data not shown).
FIG. 2.
Expression of C, Y1, or Y2 protein in stable cell lines. Immunoblotting (A) and immunofluorescent staining (B) of C+, Y1+, and Y2+ cells are shown. All four C proteins expressed in SeV-infected HeLa cells were used as markers (SeV). Relative expression levels of C, Y1, and Y2 are shown (A, bottom). None indicates the parental HeLa cell line in this and subsequent figures.
Effects of various SeV C proteins on IFN-α/β-induced signaling.
IFN-α and IFN-β are involved in innate responses as a major host defense mechanism and establish the antiviral state through the induction of IFN-stimulated gene (ISG) products (36, 40). Binding of IFN-α/β to the specific receptor triggers the activation of receptor-associated tyrosine kinases Jak1 and Tyk2, followed by the phosphorylation of transcription factors STAT1 and STAT2 (7). Phosphorylated STAT1 and STAT2 form a heterodimer, associate with p48, forming ISG factor 3 (ISGF3), and migrate to the nucleus. ISGF3 activates the transcription of genes regulated by the ISRE within their promoters (38, 41).
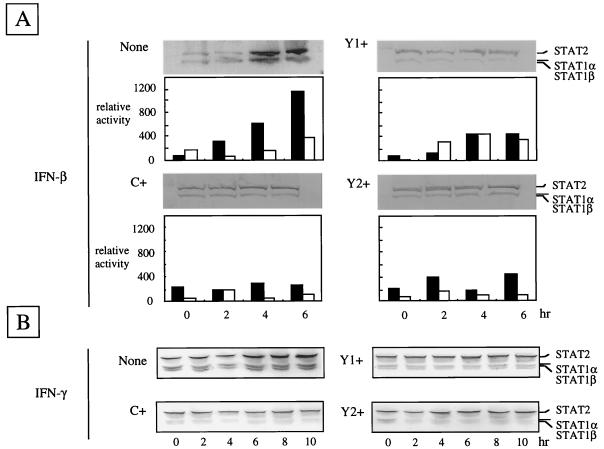
To see the effects of the C, Y1, and Y2 proteins on the IFN-mediated signaling, IFN-α/β-responsive plasmid pISRE-luc and control plasmid pRL-TK were cotransfected into C+, Y1+, and Y2+ cells, as well as parental cells. As shown in Fig. 3A, the relative luciferase activities calculated by dividing firefly luciferase values by Renilla luciferase values increased linearly with incubation time (0 to 6 h) in IFN-β-treated parental cells. In parallel with this, the levels of the ISG products STAT1α/β and STAT2 were found to increase when the aliquots of the cell lysates were analyzed by immunoblotting using the mixture of specific sera. In contrast, little or no appreciable signal transduction or stimulation of STAT1α/β and STAT2 synthesis was found in C+, Y1+, and Y2+ cells. Essentially the same results were obtained when cell lysates were immunoblotted with anti-IRF-1 or anti-PKR serum or when cells were treated with IFN-α (data not shown). These findings indicated clearly that the C, Y1, and Y2 proteins block IFN-α/β-mediated signaling equally well without the aid of any other SeV proteins.
FIG. 3.
Relative luciferase activities and expression of STAT1 and STAT2 in parental and C-expressing HeLa cells. Reporter luciferase plasmid pISRE-luci driven by the ISRE and control luciferase plasmid pRL-TK driven by the tk promoter were transfected (for details, see Materials and Methods). Relative luciferase activities were measured at 0, 2, 4, and 6 h of incubation with (solid bars) or without (open bars) INF-β. The amounts of STAT1α/β and STAT2 proteins in the presence of IFN-β (A) or IFN-γ (B) at the times indicated are shown in each immunoblot.
Abrogation of the IFN-α/β-induced antiviral state by any of the three C proteins.
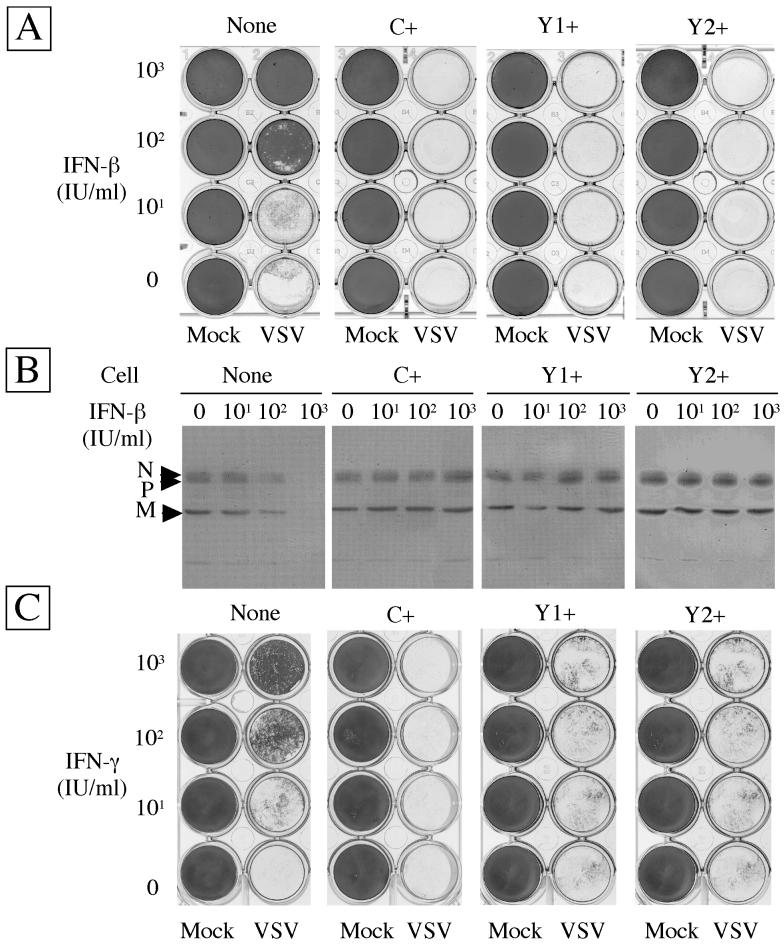
To see the effect of the C, Y1, and Y2 proteins on establishment of the antiviral state, cells preincubated with IFN-β at 0, 10, 100, or 1,000 IU/ml for 24 h were challenged with VSV. As shown in Fig. 4A, IFN-β alone did not cause any apparent cytopathogenicity in any of the cells tested under these conditions. All of the cells that had received no IFN-β were detached from the plates by subsequent VSV infection. The parental cells were protected from a VSV-induced cytopathic effect by IFN-β pretreatments at 100 and 1,000 IU/ml. In contrast, C+, Y1+, and Y2+ cells were totally detached by VSV infection even after pretreatment with IFN-β at 1,000 U/ml. Essentially the same results were obtained when IFN-α was used instead of IFN-β and when encephalomyocarditis virus or Sindbis virus was used instead of VSV (data not shown).
FIG. 4.
IFN-induced antiviral states of parental and C-expressing HeLa cells. Cells were incubated for 24 h with various concentrations of IFN-β (A and B) or IFN-γ (C) as indicated. Subsequently, they were challenged with VSV or mock infected (Mock). Cells which survived the challenge infection and were attaching to the plates were fixed and stained (A and C). Viral multiplication was detected by immunoblotting with anti-VSV serum (B). Viral proteins are indicated on the left.
In addition, the inhibitory effect of IFN on viral multiplication was assessed by immunoblotting. As shown in Fig. 4B, VSV multiplied almost equally in parental and C-expressing cells not treated with IFN. In the parental cells, partial and complete inhibitions of VSV multiplication by IFN, at 100 and 1,000 IU/ml, respectively, were observed. In contrast, VSV multiplication in C+, Y1+, or Y2+ cells was perfectly maintained even at the highest concentration of IFN used. These results were consistent with those obtained in the cytopathological experiment (Fig. 4A), indicating that the cell detachment was indeed caused by VSV multiplication in infected cells and that the C, Y1, and Y2 proteins were equally active in abrogating the IFN-β-induced antiviral state.
Effects of SeV C proteins on the IFN-γ-induced antiviral state.
Binding of IFN-γ to the receptor triggers the activation of receptor-associated tyrosine kinases Jak1 and Jak2, followed by phosphorylation of STAT1. Phosphorylated STAT1 forms a homodimer and migrates to the nucleus to function as the gamma-activated factor which binds to gamma-activated sequence elements in the promoters (7, 29). To see if the C, Y1, and Y2 proteins would also be able to counteract the IFN-γ-induced antiviral state, STAT1α/β and STAT2 expression was again examined following exposure of the cells to IFN-γ. As shown in Fig. 3B, induction of these genes was clearly visible at 6 to 10 h in the parental control cells while no such induction took place in either C+, Y1+, or Y2+ cells. Accordingly, the antiviral state was seen only for the parental cells with IFN-γ at 100 to 1,000 IU/ml while C+, Y1+, or Y2+ cells appeared to be fully susceptible to VSV (Fig. 4C). These results clearly indicated that the smallest of the SeV C proteins, Y2, was fully capable of counteracting the IFN-γ-mediated signaling and abrogating the antiviral state.
Effects of various C proteins on viral growth.
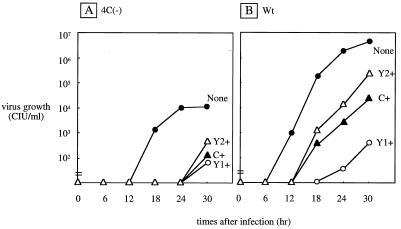
The 4C(−) SeV strain, which expresses none of the four C proteins, was previously found to grow very poorly in CV1 cells, with a peak titer several hundredfold lower than that of Wt SeV. This growth impairment was due, at least in part, to disturbance of the virion assembly step because of the lack of C proteins (14). CV1 cells are known to produce IFN-α/β, but Vero cells do not. We found that the growth of the 4C(−) virus was improved to some extent in Vero cells. We were now interested in whether the various C-expressing cell lines would complement the replication defect of the 4C(−) virus. As shown in Fig. 5A, no such helper function for 4C(−) virus replication was found in any C-expressing cell line. Rather, virus growth was strongly suppressed. The growth of Wt SeV was also compared in the parental and C-expressing cell lines, and essentially the same growth impairment was found in the C-expressing cells (Fig. 5B). These results clearly indicated that the transexpressed C, Y1, or Y2 protein renders HeLa cells moderately or only poorly permissive, especially for SeV. This property of C proteins was obviously attributable to their inhibitory effect on SeV RNA synthesis (see below).
FIG. 5.
SeV multiplication in parental and C-expressing HeLa cells. Cells were infected with 4C(−) (A) or Wt (B) SeV. The virus titers in the culture supernatants were determined at the times indicated.
Effects of various C proteins on viral RNA synthesis.
The first property described for SeV C proteins was inhibition of viral RNA synthesis in a cotransfection system of a genome analogue (minigenome) with N-, P-, and L-expressing plasmids as well as a C-expressing plasmid (4). The inhibitory activity was found for C′ and C but not for Y1 and Y2. These findings may be partly consistent with the phenotype of C′/C knockout SeV showing accumulation of more viral RNAs, compared to parental SeV (28). However, this phenotype of RNA accumulation was not so significant for our C′/C knockout virus (26). Here, a minigenome with the luciferase reporter gene was cotransfected with supporting plasmids pGEM-N, -P, and -L, as well as control plasmid pRL-TK, into C+, Y1+, and Y2+ cells. The minigenome was replicated and transcribed by the N, P, and L proteins expressed from the pGEM plasmids by the T7 polymerase supplied by coinfecting recombinant vaccinia virus strain vTF7.3 (19). In the pGEM-P plasmid, the expression of C proteins had been silenced by replacing its translational initiation site with the influenza virus hemagglutinin tag (4). The amount of transcription derived from the minigenome was monitored by measuring luciferase activities. Potential fluctuations in transfection efficiency and other experimentation were normalized to the level of Renilla luciferase expression from pRL-TK.
The relative luciferase activities in C+ cells were about one-third (mean, 36%) of those in parental cells, confirming the RNA synthesis-inhibiting property of SeV C protein (Fig. 6A). Inhibition was greater in Y1+ cells, although Y1 is shorter than C by 22 amino acids. On the other hand, Y2 is shorter than Y1 by six amino acids, and the inhibition by Y2 was less significant than that by Y1 and C. These variable inhibition levels might either reflect the possible differences in the intrinsic capacity of the individual C proteins or be due to their different expression levels (43% for Y2 and 84% for C relative to Y1; Fig. 6A, top). To test these alternatives, three more stable clones (Y2a+, Y2b+, and Y2c+) expressing the Y2 protein were used for a minigenome assay in which the Y2 expression levels were varied (58, 48, and 64%, respectively). The relative levels of luciferase expression from the minigenome in these three lines were 47, 54, and 32%, respectively (Fig. 6A, bottom). The C expression level was inversely proportional to the minigenome-derived luciferase level (Fig. 6B). These results strongly suggest that C, Y1, and Y2 are equally active in inhibiting viral RNA synthesis.
FIG. 6.
Reporter gene expression from the SeV minigenome in parental and C-expressing HeLa cells. In addition to C+, Y1+, and Y2+ cells, three other Y2-expressing lines, Y2a+, Y2b+, and Y2c+, were used. (A) Expression of the respective C proteins in these cells, determined by immunoblotting, along with their relative expression levels (top). Relative luciferase activities expressed from the minigenome in the various cells are shown along with the mean value (black bars) for two measurements (bottom). (B) Relationship between the luciferase activity from the minigenome and the intracellular expression level of each C protein.
Effects of various C proteins on SeV transcription.
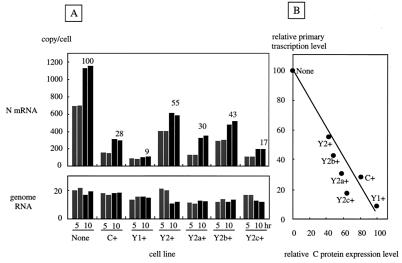
It should be noted that any function of viral proteins required in the natural life cycle can be bypassed in minigenome systems because their expression from transfected plasmids is no longer under the control of a virus-specific promoter and polymerase (31, 32). Thus, we further attempted to evaluate the inhibitory activity of the C, Y1, and Y2 proteins in the context of viral infection of each cell line. Since viral replication in infected cells does not take place without de novo protein synthesis, it was possible to compare the amounts of primary transcripts by incubating infected cells with cycloheximide. Wt SeV was inoculated into parental and C-expressing HeLa cells at an MOI of 20 per cell in the presence of cycloheximide. At 5 and 10 h p.i., both genome RNA and N mRNA were quantified by TaqMan PCR with specific primer-probe sets (Fig. 1).
As shown in Fig. 7A, the copy numbers of viral genome RNA were calibrated to be 10 to 20 per cell at 5 h p.i. and did not increase in the subsequent incubation up to 10 h p.i. but rather decreased slightly. These results not only confirmed complete inhibition of genome replication but also suggested initiation of infection of these different cell lines with nearly equal numbers of infecting viruses. On the other hand, N mRNA reached nearly 700 copies/cell at 5 h p.i. and increased to 1,100 copies/cell by 10 h p.i. in the parental cell line (Fig. 7A). This primary transcription was more or less inhibited in cells expressing the various C proteins. The levels of transcripts were also plotted against the relative expression levels of the various C proteins, and again a good inverse correlation was obtained (Fig. 7B). These data were consistent with those obtained with the minigenome (Fig. 6) and confirmed the notion that C, Y1, and Y2 are equally active in inhibiting viral RNA synthesis. These results, taken together, strongly suggest that, although essential to full-level virus production, the C proteins can be inhibitory if they are present before the natural life cycle starts. It was therefore not surprising that the growth of not only the Wt but also the 4C(−) strain was strongly suppressed in C+, Y1+, and Y2+ cells (Fig. 5).
FIG. 7.
Quantification of SeV primary transcripts in parental and C-expressing HeLa cells. (A) Cells were infected with Wt SeV at an MOI of 20 per cell and incubated for 5 (gray bars) or 10 (black bars) h in the presence of cycloheximide (100 μg/ml), and total RNAs were extracted. The copy numbers of N mRNA (top) and genome RNA (bottom) were then quantified by TaqMan PCR with specific primers and probes. Relative copy numbers measured at 0 h p.i. are shown above the black bars. (B) Relationship between the relative copy number of N mRNA and the relative levels of intracellular C proteins.
DISCUSSION
The SeV C′, C, Y1, and Y2 proteins are carboxyl-coterminal proteins of 215, 204, 181, and 175 amino acids, respectively. In this work, we established HeLa cell lines stably expressing the C, Y1, and Y2 proteins individually without difficulty, although apoptotic activity was suggested for the C protein (15, 18). We have shown that these cell lines are equally active in counteracting the signal transduction and the resulting antiviral state induced by both IFN-α/β and IFN-γ. Thus, it was concluded that the smallest protein, Y2, encodes the full amount of information required for these functions. During the preparation of this report, Garcin et al. reported the use of a similar approach to address whether all four C proteins would be equally active in blocking IFN-mediated signal transduction using various C protein mutants (11). It was shown that Y1 and Y2 expression can exert the blocking function. However, it was not determined whether either of them is sufficient or both are required. Furthermore, their studies used a transfection protocol of these genes which allowed only transient expression of the respective proteins in some cell populations and therefore could not address the question of whether the blocking of signaling by Y1 and Y2 or by any other mutants indeed circumvented the antiviral state using an indicator virus such as VSV. Thus, our present study provides the first straightforward evidence of circumvention of the IFN-α/β- and IFN-γ-mediated antiviral state by the smallest of the four SeV C proteins, Y2.
IFNs represent a major aspect of innate immunity against viral infections, and many viruses are known to have acquired an anti-IFN function to counteract the antiviral action of IFNs (1, 5, 34, 37, 39, 46). How SeV C proteins can play this role remains an important question to be answered (24). SV5 is a member of the genus Rubulavirus and does not encode C proteins (27). It is the V protein that plays this role in the case of SV5 (6, 47). The SV5 V protein appears to target STAT1 to proteasome-mediated degradation (3, 23). Garcin et al. reported that STAT1 was rendered unstable in some SeV C-expressing cells (10) but not in others (11). No such instability of STAT1 was observed in our SeV C-expressing cells (Fig. 3), indicating that the degradation of STAT1 by C proteins requires some additional cellular factors. C proteins are encoded by the members of the genera Respirovirus and Morbillivirus. The amino acid sequences are pretty well conserved within each genus but divergent between the two genera (31). However, C proteins are all highly basic, with isoelectric points of around 10. NS1 of influenza A viruses also encodes anti-IFN action and is also basic (8, 42). It may be interesting to learn whether these paramyxovirus and influenza virus basic proteins interact with molecules such as Jak1 and STAT1, which are commonly phosphorylated in the IFN-α/β- and IFN-γ-mediated signaling pathways (41).
Our present work further demonstrated that C, Y1, and Y2 are equally active in inhibiting viral transcription. This conclusion is not compatible with the notion that RNA synthesis inhibition was attributable to C′, C, or both but not to Y1 and/or Y2 (4) but is in good agreement with our previous observation that increased viral mRNA and protein synthesis is characteristic of the 4C(−) virus but not of the C′/C(−) virus (26). More remarkable than these was the inability of the intracellularly transexpressed C proteins to complement 4C(−) virus replication (Fig. 5). Rather, accumulation of C proteins within cells appeared to be a strong impediment to the successful initiation of replication of the infecting 4C(−) virus, as well as Wt SeV. This also explains why the C frames had to be silenced in pGEM-P to initiate the generation of infectious SeV entirely from cDNA (9, 19). On the other hand, extremely impaired replication of the 4C(−) virus in normal cells without transexpressed C proteins indicates a critical requirement of C proteins for the normal life cycle of SeV (14, 26). Whatever the basis of this requirement is, optimization of intracellular levels of transcripts, genome and antigenome, or something else, C proteins must play this role in a highly coordinated fashion within the natural life cycle. Their overexpression, relative to their target components, could restrict ongoing viral replication. When the target levels become higher, the C protein levels may have to be increased accordingly. Although the expression levels were significantly different (100 versus 43), C+ and Y2+ cells were able to counteract the antiviral action of IFNs equally well. Regarding RNA synthesis inhibition, however, Y2+ was less efficient than C+. The higher the Y2 protein level, the greater the inhibition. Therefore, anti-IFN action can be achieved by a dose of C proteins lower than that required for their RNA-inhibiting function. It appears to be reasonable that C proteins are incorporated into virions in a quite low copy number (about 40 copies per virion) (35) so that they do not inhibit the onset of viral RNA synthesis but are likely sufficient to exert an anti-IFN action. Late in viral infection, on the other hand, they are expressed abundantly to prevent viral RNA accumulation in excess.
The question of why not only field isolates but also laboratory strains of SeV encode or have to encode four different C proteins also remains to be answered. To address this question, on-off expression of each C protein, allowing fine tuning of its intracellular level, is necessary. Using this system, it will be possible to define how much each C protein complements strain 4C(−) replication and whether or not all four C proteins are equally active as chaperons in guiding viral structural proteins into the assembly pathway (14). Generation of additional C knockout viruses and their characterization both in cells and in host animals will also help to answer this question. Our previous results demonstrated that the C′/C(−) virus vigorously expressed Y1 and Y2 in cells but was still greatly impaired in replication capability, not only in vivo but also in vitro (26). Thus, the coding strategy used to express four C proteins appears to have been maintained by SeV.
ACKNOWLEDGMENTS
We thank K. Sakai, K. Ozato, and B. Gotoh for providing plasmids pKS336 and pISRE-luci and anti-VSV serum, respectively. We also thank K. Takeuchi, M. Hishiyama, and A. Masumi for technical help and suggestions.
This work was supported by research grants from the Ministry of Education, Sports, Culture, Science, and Technology, Japan, and from the Bio-oriented Technology Research Advancement Institution (BRAIN), Japan. Y.O. is a recipient of a BRAIN fellowship.
REFERENCES
- 1.Atreya P L, Kulkarni S. Respiratory syncytial virus A2 is resistant to the antiviral effects of type I interferons and human MxA. Virology. 1999;261:227–241. doi: 10.1006/viro.1999.9835. [DOI] [PubMed] [Google Scholar]
- 2.Cadd T, Garcin D, Tapparel C, Itoh M, Homma M, Roux L, Curran J, Kolakofsky D. The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J Virol. 1996;70:5067–5074. doi: 10.1128/jvi.70.8.5067-5074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chin Y E, Kitagawa M, Kuida K, Flavell R A, Fu X-Y. Activation of the STAT signaling pathway can cause expression of caspase 1 and apoptosis. Mol Cell Biol. 1997;17:5328–5337. doi: 10.1128/mcb.17.9.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curran J, Marq J-B, Kolakofsky D. The Sendai virus nonstructural C proteins specifically inhibit viral mRNA synthesis. Virology. 1992;189:647–656. doi: 10.1016/0042-6822(92)90588-g. [DOI] [PubMed] [Google Scholar]
- 5.Didcock L, Young D F, Goodbourn S, Randall R E. Sendai virus and simian virus 5 block activation of interferon-responsive genes: importance for virus pathogenesis. J Virol. 1999;73:3125–3133. doi: 10.1128/jvi.73.4.3125-3133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Didcock L, Young D F, Goodbourn S, Randall R E. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J Virol. 1999;73:9928–9933. doi: 10.1128/jvi.73.12.9928-9933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durbin J E, Hackenmiller R, Simon M C, Levy D E. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 8.García-Sastre A, Egorov A, Matassov D, Brandt S, Levy D E, Durbin J E, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 9.Garcin D, Pelet T, Calain P, Roux L, Curran J, Kolakofsky D. A highly recombinogenic system for the recovery of infectious Sendai paramyxovirus from cDNA: generation of a novel copy-back nondefective interfering virus. EMBO J. 1995;14:6087–6094. doi: 10.1002/j.1460-2075.1995.tb00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcin D, Latorre P, Kolakofsky D. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J Virol. 1999;73:6559–6565. doi: 10.1128/jvi.73.8.6559-6565.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcin D, Curran J, Kolakofsky D. Sendai virus C proteins must interact directly with cellular components to interfere with interferon action. J Virol. 2000;74:8823–8830. doi: 10.1128/jvi.74.19.8823-8830.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gotoh B, Takeuchi K, Komatsu T, Yokoo J, Kimura Y, Kurotani A, Kato A, Nagai Y. Knockout of the Sendai virus C gene eliminates the viral ability to prevent the interferon-α/β-mediated responses. FEBS Lett. 1999;459:205–210. doi: 10.1016/s0014-5793(99)01241-7. [DOI] [PubMed] [Google Scholar]
- 13.Gupta K C, Patwardhan S. ACG, the initiator codon for Sendai virus C protein. J Biol Chem. 1988;263:8553–8556. [PubMed] [Google Scholar]
- 14.Hasan M K, Kato A, Muranaka M, Yamaguchi R, Sakai Y, Hatano I, Tashiro M, Nagai Y. Versatility of the accessory C proteins of Sendai virus: contribution to virus assembly as an additional role. J Virol. 2000;74:5619–5628. doi: 10.1128/jvi.74.12.5619-5628.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heylbroeck C, Balachandran S, Servant M J, DeLuca C, Barber G N, Lin R, Hiscott J. The IRF-3 transcription factor mediates Sendai virus-induced apoptosis. J Virol. 2000;74:3781–3792. doi: 10.1128/jvi.74.8.3781-3792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horikami S M, Hector R E, Smallwood S, Moyer S A. The Sendai virus C protein binds the L polymerase protein to inhibit viral RNA synthesis. Virology. 1997;235:261–270. doi: 10.1006/viro.1997.8702. [DOI] [PubMed] [Google Scholar]
- 17.Huang C, Kiyotani K, Fujii Y, Fukuhara N, Kato A, Nagai Y, Yoshida T, Sakaguchi T. Involvement of the zinc-binding capacity of Sendai virus V protein in viral pathogenesis. J Virol. 2000;74:7834–7841. doi: 10.1128/jvi.74.17.7834-7841.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itoh M, Hotta H, Homma M. Increased induction of apoptosis by a Sendai virus mutant is associated with attenuation of mouse pathogenicity. J Virol. 1998;72:2927–2934. doi: 10.1128/jvi.72.4.2927-2934.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato A, Sakai Y, Shioda T, Kondo T, Nakanishi M, Nagai Y. Initiation of Sendai virus multiplication from transfected cDNA or RNA with negative or positive sense. Genes Cells. 1996;1:569–579. doi: 10.1046/j.1365-2443.1996.d01-261.x. [DOI] [PubMed] [Google Scholar]
- 20.Kato A, Kiyotani K, Sakai Y, Yoshida T, Nagai Y. The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis. EMBO J. 1997;16:578–587. doi: 10.1093/emboj/16.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato A, Kiyotani K, Sakai Y, Yoshida T, Shioda T, Nagai Y. Importance of the cysteine-rich carboxyl-terminal half of V protein for Sendai virus pathogenesis. J Virol. 1997;71:7266–7272. doi: 10.1128/jvi.71.10.7266-7272.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato A, Kiyotani K, Hasan M K, Shioda T, Sakai Y, Yoshida T, Nagai Y. Sendai virus gene start signals are not equivalent in reinitiation capacity: moderation at the fusion protein gene. J Virol. 1999;73:9237–9246. doi: 10.1128/jvi.73.11.9237-9246.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King P, Goodbourn S. STAT1 is inactivated by a caspase. J Biol Chem. 1998;273:8699–8704. doi: 10.1074/jbc.273.15.8699. [DOI] [PubMed] [Google Scholar]
- 24.Komatsu T, Takeuchi K, Yokoo J, Tanaka Y, Gotoh B. Sendai virus blocks alpha interferon signaling to signal transducers and activators of transcription. J Virol. 2000;74:2477–2480. doi: 10.1128/jvi.74.5.2477-2480.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozak M. Selection of initiation sites by eucaryotic ribosomes: effect of inserting AUG triplets upstream from the coding sequence for preproinsulin. Nucleic Acids Res. 1984;12:3873–3893. doi: 10.1093/nar/12.9.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurotani A, Kiyotani K, Kato A, Shioda T, Sakai Y, Mizumoto K, Yoshida T, Nagai Y. The paramyxovirus, Sendai virus, C proteins are categorically nonessential gene products but silencing their expression severely impairs viral replication and pathogenesis. Genes Cells. 1998;2:111–124. doi: 10.1046/j.1365-2443.1998.00170.x. [DOI] [PubMed] [Google Scholar]
- 27.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. New York, N.Y: Lippincott-Raven Press; 1996. pp. 1177–1204. [Google Scholar]
- 28.Latorre P, Cadd T, Itoh M, Curran J, Kolakofsky D. The various Sendai virus C proteins are not functionally equivalent and exert both positive and negative effects on viral RNA accumulation during the course of infection. J Virol. 1998;72:5984–5993. doi: 10.1128/jvi.72.7.5984-5993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meraz M A, White J M, Sheehan K C, Bach E A, Rodig S J, Dighe A S, Kaplan D H, Riley J K, Greenlund A C, Campbell D, Carver-Moore K, DuBois R N, Clarks R, Aguet M, Schreiber R D. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 30.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagai Y. Paramyxovirus replication and pathogenesis. Reverse genetics transforms understanding. Rev Med Virol. 1999;9:83–99. doi: 10.1002/(sici)1099-1654(199904/06)9:2<83::aid-rmv244>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 32.Nagai Y, Kato A. Paramyxovirus reverse genetics is coming of age. Microbiol Immunobiol. 1999;43:613–624. doi: 10.1111/j.1348-0421.1999.tb02448.x. [DOI] [PubMed] [Google Scholar]
- 33.Paterson R G, Leser G P, Shaughnessy M A, Lamb R A. The paramyxovirus SV5 V protein binds two atoms of zinc and is a structural component of virions. Virology. 1995;208:121–131. doi: 10.1006/viro.1995.1135. [DOI] [PubMed] [Google Scholar]
- 34.Ploegh H L. Viral strategies of immune evasion. Science. 1998;280:248–253. doi: 10.1126/science.280.5361.248. [DOI] [PubMed] [Google Scholar]
- 35.Portner A, Murti K G, Seyer J M, Beachey E H, Kingsbury D W. Localization and characterization of Sendai virus nonstructural C and C′ proteins by antibodies against synthetic peptides. Virus Res. 1986;6:109–121. doi: 10.1016/0168-1702(86)90043-2. [DOI] [PubMed] [Google Scholar]
- 36.Samuel C E. Antiviral actions of interferon: interferon regulated cellular proteins and their surprisingly selective antiviral activities. Virology. 1991;183:1–11. doi: 10.1016/0042-6822(91)90112-o. [DOI] [PubMed] [Google Scholar]
- 37.Schlender J, Bossert B, Buchholz U, Conzelmann K-K. Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J Virol. 2000;74:8234–8242. doi: 10.1128/jvi.74.18.8234-8242.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sen G C, Ransohoff R M. Interferon induced antiviral actions and their regulation. Adv Virus Res. 1993;43:57–102. doi: 10.1016/s0065-3527(08)60083-4. [DOI] [PubMed] [Google Scholar]
- 39.Smith G L. Virus strategies for evasion of the host response to infection. Trends Microbiol. 1994;2:81–88. doi: 10.1016/0966-842x(94)90539-8. [DOI] [PubMed] [Google Scholar]
- 40.Staeheli P. Interferon induced proteins and the antiviral state. Adv Virus Res. 1990;38:147–200. doi: 10.1016/s0065-3527(08)60862-3. [DOI] [PubMed] [Google Scholar]
- 41.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 42.Talon J, Horvath C M, Polley R, Basler C F, Muster T, Palese P, García-Sastre A. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J Virol. 2000;74:7989–7996. doi: 10.1128/jvi.74.17.7989-7996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vidal S, Curran J, Kolakofsky D. A stuttering model for paramyxovirus P mRNA editing. EMBO J. 1990;9:2017–2022. doi: 10.1002/j.1460-2075.1990.tb08330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang I M, Blanco J C, Tsai S Y, Tsai M J, Ozato K. Interferon regulatory factors and TFIIB cooperatively regulate interferon-responsive promoter activity in vivo and in vitro. Mol Cell Biol. 1996;16:6313–6324. doi: 10.1128/mcb.16.11.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yokoo J, Gotoh B, Komatsu T, Takeuchi K, Miyadai T. Replication-incompetent Sendai virus can suppress the antiviral action of type I interferon. Arch Virol. 1999;144:1043–1055. doi: 10.1007/s007050050568. [DOI] [PubMed] [Google Scholar]
- 46.Yokosawa N, Kubota T, Fujii N. Poor induction of interferon induced 2′,5′-oligoadenylate synthetase (2-5 AS) in cells persistently infected with mumps virus is caused by decrease of STAT-1 alpha. Arch Virol. 1998;143:1985–1992. doi: 10.1007/s007050050434. [DOI] [PubMed] [Google Scholar]
- 47.Young D F, Didcock L, Goodbourn S, Randall R E. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology. 2000;269:383–390. doi: 10.1006/viro.2000.0240. [DOI] [PubMed] [Google Scholar]