Abstract
The 3′-untranslated region (3′-UTR) of mouse hepatitis virus (MHV) RNA regulates the replication of and transcription from the viral RNA. Several host cell proteins have previously been shown to interact with this regulatory region. By immunoprecipitation of UV-cross-linked cellular proteins and in vitro binding of the recombinant protein, we have identified the major RNA-binding protein species as heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1). A strong hnRNP A1-binding site was located 90 to 170 nucleotides from the 3′ end of MHV RNA, and a weak binding site was mapped at nucleotides 260 to 350 from the 3′ end. These binding sites are complementary to the sites on the negative-strand RNA that bind another cellular protein, polypyrimidine tract-binding protein (PTB). Mutations that affect PTB binding to the negative strand of the 3′-UTR also inhibited hnRNP A1 binding on the positive strand, indicating a possible relationship between these two proteins. Defective-interfering RNAs containing a mutated hnRNP A1-binding site have reduced RNA transcription and replication activities. Furthermore, hnRNP A1 and PTB, both of which also bind to the complementary strands at the 5′ end of MHV RNA, together mediate the formation of an RNP complex involving the 5′- and 3′-end fragments of MHV RNA in vitro. These studies suggest that hnRNP A1-PTB interactions provide a molecular mechanism for potential 5′-3′ cross talks in MHV RNA, which may be important for RNA replication and transcription.
The genome of mouse hepatitis virus (MHV) is a single-stranded, linear, positive-sense, polyadenylated RNA of approximately 31 kb in length (22, 24, 39). All of the viral proteins are translated from subgenomic mRNAs, except those encoded by the first open reading frame, which are translated from the genome-sized RNA. In MHV-infected cells, six or seven subgenomic mRNAs are transcribed, all of which contain a common leader sequence derived from the 5′ end of the genome, as well as a common 3′ end including a 302- to 305-nucleotide (nt)-long untranslated region (3′-UTR) (21). When MHV is passaged at a high multiplicity of infection in cell culture, defective-interfering (DI) RNAs which represent deletion mutants of the MHV genome are frequently generated (33, 36). Since DI RNAs can replicate in the presence of a helper virus, they must retain all signals necessary for MHV genome replication.
Comparison of various MHV DI RNAs showed that all retain various lengths of both the 5′ and 3′ ends of the viral genome (34, 35, 37, 43). Mapping studies of MHV DI RNAs have further revealed that 400 to 859 nt from the 5′ end and 436 nt from the 3′ end of the genome are required for DI RNA replication (17, 29). However, only 55 nt from the 3′ end plus the poly(A) tail are required for synthesis of the negative strand of MHV RNA (30). Thus, it stands to reason that the replication signal from the 5′ end and the remaining replication signal (nt 55 to 436) from the 3′ end of the MHV genome are involved in synthesis of the positive-strand RNA. Moreover, since positive-strand RNA synthesis begins from the 5′ end of the genome, and the 3′ end will be the last region of the genome reached by the viral polymerase, the replication signal at the 3′ end likely interacts with signals at the 5′ end to exert its effect on RNA synthesis. Based on this reasoning, the 5′ and 3′ ends of the genome have been proposed to interact during viral RNA replication so that the 3′-end sequence can affect the initiation of RNA synthesis (20). Similar observations have also been made for the regulation of MHV subgenomic mRNA transcription (28). Typically, the regulatory sequence for the synthesis of a particular RNA strand resides on the complementary (template) strand; i.e., the signal for synthesizing the positive strand resides on the negative strand, and vice versa. However, the regulatory signals may also reside on the same strand. For example, in brome mosaic virus (BMV) and poliovirus, cis elements that affect synthesis of the positive-strand RNA are mapped to the 5′ end of the positive-strand RNA (2, 9, 40). Presumably this is due to the possibility that double-stranded replicative-form RNA is used as the template for positive-strand RNA synthesis. Thus, RNA synthesis may be regulated by 5′-3′-end interaction of both RNA strands.
Although 5′-3′-end interactions of MHV RNA have been suggested from functional studies (28), no apparent sequence complementarity exists between these two regions. Therefore, interaction between the 5′ and 3′ ends likely involves protein factors from either the host or the virus. Indeed, several host cell proteins have previously been found to bind to the transcription- and replication-regulatory regions of MHV RNA; these include heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1), binding to the minus-strand leader and minus-strand intergenic sequences (26, 53), and polypyrimidine tract-binding protein (PTB), binding to the positive-strand leader (25) and the complementary strand of the 3′-UTR (c3′-UTR) (11). Interestingly, the two regions that bind PTB are both 5′ ends of the positive- and negative-strand RNA, respectively. Another noticeable feature is that at the leader region, PTB and hnRNP A1 bind to the same stretch of nucleotides on opposite strands, which contains several UCUAA repeats (25, 26). Deletion of this UCUAA repeat sequence affects the binding of both hnRNP A1 and PTB to the leader region on opposite strands of the viral RNA (25, 26). Furthermore, deletion of the UCUAA repeats severely impaired RNA replication (25). The role of hnRNP A1 in MHV replication has recently been demonstrated by experiments showing that overexpression of hnRNP A1 facilitates MHV replication, whereas dominant-negative mutants of hnRNP A1 inhibit it (42).
In this study, we investigated the interaction between another regulatory region of MHV RNA, the 3′-UTR, and host cell proteins. We identified one of the binding proteins as hnRNP A1. Thus, just as PTB binds to the 5′ ends of both positive- and negative-strand RNA, hnRNP A1 binds to the 3′ ends of both strands. Furthermore, the hnRNP A1-binding region is complementary to one of the PTB-binding sites on the opposite strand. The mutation of this region not only impaired transcription of subgenomic RNA (11) but also affected the replication of a DI RNA as well. We further showed that hnRNP A1 and PTB can mediate interaction between the 5′ and 3′ ends of MHV RNA in vitro. Our results suggest that the interaction between the 5′ and 3′ ends of both positive- and negative-strand RNA through RNA-protein and protein-protein interactions may provide a molecular mechanism to bring the 5′ and 3′ ends of the MHV genome together to form RNA transcription and replication complexes (20).
MATERIALS AND METHODS
Viruses and cells.
The plaque-cloned A59 strain (38) of MHV (MHV-A59) was used throughout this study. Viruses were propagated in DBT cells (a mouse astrocytoma cell line) (10).
Plasmids and PCR primers.
Sequences of the primers used in this study are listed in Table 1. The templates used are cDNA clones of the MHV DI RNAs: 25CAT (27), 25CATΔC (11), and 25CATsubsC (11).
TABLE 1.
Primers used for PCR amplification
| Primera | Sequenceb |
|---|---|
| Sense | |
| SP6-350 | 5′-ATTTAGGTGACACTATAGATAGATGATGGCGTAGTGCCAGATGGGTTAG-3′ |
| SP6-90 | 5′-ATTTAGGTGACACTATAGTTGCCCCCTGGGAAGAGC-3′ |
| SP6-172 | 5′-ATTTAGGTGACACTATAGTTGAAAGAGATTGCAAAATAG-3′ |
| SP6-260 | 5′-ATTTAGGTGACACTATAGAAAGTCGGGATAGGACAC-3′ |
| Antisense | |
| T7-0 | 5′-TAATACGACTCACTATAGGTGATTCTTCCAATTGGCCATGATCAACTTC-3′ |
| MHV(−)-90 | 5′-CGCCTATCGCCGTTC-3′ |
| MHV(−)-172 | 5′-ACTAATTGATACAGG-3′ |
| MHV(−)-260 | 5′-CTTTCTCGCGAGGGG-3′ |
Numbers in primer names denote nucleotide positions (counting from the 3′ end of MHV RNA) of the first MHV nucleotide.
T7 or SP6 promoter sequences are in boldface type.
In vitro transcription of PCR products.
For synthesizing various DI RNA fragments, PCR products made by using synthetic primers containing the SP6 promoter were used as templates for in vitro transcription. Briefly, PCR products (0.5 μg) amplified from DI cDNA plasmids were used in standard in vitro transcription reactions using SP6 RNA polymerase (Promega) according to the manufacturer's manual. [32P]UTP was added in some experiments. All RNA products were treated with RQ1 RNase-free DNase (1 U) (Promega) and passed through G-25 minicolumns (Eppendorf) to remove the unincorporated nucleotides.
UV cross-linking assay.
UV cross-linking assays were performed as previously described (7). Briefly, partially purified recombinant glutathione S-transferase (GST) fusion proteins (1 μg) or whole-cell extracts (30 μg) were incubated with in vitro-transcribed 32P-labeled RNA probes for 10 min at 30°C. The reaction mixtures were irradiated in a UV Stratalinker 2400 (Stratagene) for 10 min and then treated with RNase A (20 μg) for 15 min at 37°C. The reaction products were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 10% polyacrylamide gels.
Immunoprecipitation.
DBT whole-cell extract that had been UV cross-linked to 32P-labeled RNA was diluted with NETS buffer (50 mM Tris-HCl [pH 7.4], 5 mM EDTA, 1 mM dithiothreitol [DTT], 100 mM NaCl, 0.05% NP-40) to a final volume of 500 μl and mixed with various antibodies. After incubation at room temperature for 1 h, the immunocomplexes were immobilized on protein A-Sepharose 4B beads (Pharmacia) for 30 min. Extensive washing of the beads (five to six times) was performed before protein sample loading buffer was added to the beads. The mixtures were boiled for 3 min and resolved by SDS-PAGE. The monoclonal anti-hnRNP A1 antibody was kindly provided by Gideon Dreyfuss, University of Pennsylvania. The monoclonal anti-Sam68 antibody was purchased from Calbiochem. The anti-U170K antibody was purchased from American Research Products, Inc. The anti-Hsp90 antibody was purchased from Stressgen.
RNP complex formation assay.
The assay was modified according to the methods of Lamond and Sproat (23), Will et al. (49), and Zhang et al. (52). Briefly, biotin-labeled RNA, 32P-labeled probe, and protein components were incubated in a reaction buffer containing 5 U of RNasin (Promega), 25 mM KCl, 5 mM HEPES (pH 7.6), 2 mM MgCl2, 0.1 mM EDTA, 0.2% glycerol, and 2 mM DTT at 30°C for 1 h. Then preblocked streptavidin-agarose beads (Sigma) were added to the reaction mixture and incubated at 4°C for another hour. The beads were then washed five to six times with a wash buffer containing 20 mM HEPES (pH 7.9), 200 mM KCl, and 0.5% NP-40. RNAs bound to the streptavidin-agarose beads were eluted by a buffer consisting of 7 M urea, 350 mM NaCl, 10 mM Tris-HCl (pH 8.0), 10 mM EDTA, and 1% SDS. RNAs were further extracted with phenol-chloroform and precipitated with ethanol in the presence of yeast tRNA. The RNAs were resolved on a 6% polyacrylamide gel containing 7 M urea.
DI RNA transcription and transfection.
Plasmids 25CAT (27), 25CATΔC (11), and 25CATsubsC (11) were linearized by restriction endonuclease XbaI (New England Biolabs) and transcribed to RNA with T7 RNA polymerase (Promega) according to the manufacturer's manual. The RNAs were transfected into MHV-A59-infected DBT cells by the DOTAP method (Roche). Briefly, DBT cells were infected by MHV-A59 at a multiplicity of infection of 10. After 1 h, the virus-infected cells were transfected with the in vitro-transcribed DI RNAs, using the DOTAP method according to the manufacturer's manual.
Northern blot assay.
DBT cells were infected with MHV-A59, and virus-infected cells were collected at 7 h postinfection. Cells were washed in ice-cold phosphate-buffered saline and collected into centrifuge tubes. Cells were resuspended in NTE buffer (150 mM NaCl, 50 mM Tris [pH 7.5], 1 mM EDTA) (200 μl for each 60-mm-diameter culture dish) containing 0.5% NP-40, 0.5 mM DTT, and 400 RNasin (U/ml; Promega) and left on ice for 15 min. After centrifugation, the supernatant was extracted with phenol-chloroform and precipitated with ethanol. The pellet was washed with 70% ethanol. After the RNA was dissolved in diethyl pyrocarbonate-treated water, approximately 5 μg of RNA was treated with glyoxal for 1 h at 50°C before loading onto a 1% agarose gel. After electrophoresis, RNA was transferred onto a nitrocellulose membrane (Hybond-C; Amersham), and the DI RNA was detected with an in vitro-transcribed, 32P-labeled RNA probe of 473 nt that represents sequences complementary to a region just downstream of the 5′ leader.
RESULTS
hnRNP A1 binds to the MHV 3′-UTR.
As previously shown (11), a protein of approximately 35 to 38 kDa (p35/38) was cross-linked to a 32P-labeled 350-nt RNA that contains the complete 3′-UTR of MHV RNA without a poly(A) tail, plus 48 nt from the 3′ end of the open reading frame encoding the viral nucleocapsid protein. Binding of p35/38 to this RNA was confirmed in the present study, although the overall pattern of the bound cellular proteins was slightly different (Fig. 1A, lane 1). Because the molecular weight of this protein is very similar to that of hnRNP A1, which has previously been shown to bind to the sequence complementary to the 5′ end of MHV RNA (26), we first investigated whether this 3′-UTR-binding protein was hnRNP A1. To this end, we performed an immunoprecipitation assay. After the DBT whole-cell extracts were UV cross-linked to the 32P-labeled 3′-UTR, various antibodies were added to the reaction mixture to precipitate the RNA-protein complex. Results in Fig. 1A showed that p35/38 was precipitated by a monoclonal antibody against hnRNP A1 (lane 2) but not by antibodies against U170K, Sam68, and Hsp90 (lanes 3 to 5). This result indicates that p35/38 is very likely hnRNP A1. Interestingly, the immunoprecipitation detected two protein species of similar molecular weights, which may represent hnRNP A1 isoforms or hnRNP A1-related species. Alternatively, they may represent degradation products of the same protein. To confirm the results of immunoprecipitation, GST fused to hnRNP A1 (GST-A1) was used in the UV cross-linking assay. Results in Fig. 1B show that recombinant GST-A1 (lane 2), but not GST (lane 1) can be UV cross-linked to the 3′-UTR. These results combined indicate that p35/38 is hnRNP A1.
FIG. 1.
Identification of p35/38 UV cross-linked to the MHV 3′-UTR. (A) Immunoprecipitation of UV-cross-linked cell extract. DBT whole-cell extract cross-linked to 32P-labeled 3′-UTR RNA was immunoprecipitated by various antibodies in lanes 2 to 5; lane 1 is the input of UV-cross-linked DBT whole-cell extract without immunoprecipitation (IP). The arrow indicates the position of p35/38. The immunoprecipitates were resolved on a SDS–10% polyacrylamide gel. (B) UV cross-linking of recombinant GST (lane 1) and GST-A1 (lane 2) to 32P-labeled MHV 3′-UTR. The arrow indicates the position of GST-A1. Positions of molecular weight standards (in kilodaltons) are shown on the left of each gel.
The interaction between hnRNP A1 and the MHV 3′-UTR is specific.
A competition assay was performed to determine the specificity of the interaction between hnRNP A1 and the MHV 3′-UTR. 32P-labeled 3′-UTR was UV cross-linked to GST-A1 in the presence of increasing molar excess amounts of cold homologous and heterologous competitors. Results in Fig. 2 showed that increasing amounts of the homologous competitor RNA (3′-UTR) reduced proportionally the binding of GST-A1 to the probe (lanes 1 to 4). At 20-fold molar excess, the homologous RNA reduced the binding to the 32P-labeled probe to almost 90% (lane 4). Significantly, the same molar excess of the cold negative-strand leader RNA reduced GST-A1 binding to the 3′-UTR to almost the same or even a slightly higher extent (lanes 5 to 8). This result is in agreement with the previous observation that the negative-strand leader RNA interacts with hnRNP A1 (26). In contrast, neither the positive-strand leader (lanes 9 to 12) nor the unrelated RNA transcribed from pBluescript (lanes 13 to 16) reduced hnRNP A1 binding to the 3′-UTR. The results of the competition assays showed that the interaction between hnRNP A1 and MHV 3′-UTR is specific.
FIG. 2.
Competition of RNA-A1 binding in a UV cross-linking assay. GST-hnRNP A1 was incubated with different amounts (fold excess over the labeled RNA) of various unlabeled RNAs and then UV cross-linked to 32P-labeled MHV 3′-UTR. Lanes 1 to 4, 3′-UTR; lanes 5 to 8, negative-strand leader; lanes 9 to 12, positive-strand leader; lanes 13 to 16, unrelated RNA transcribed from pBluescript. The arrow indicates the position of GST-A1.
The hnRNP A1-binding sites on the 3′-UTR are mapped to nt 90 to 170 and 260 to 350 from the 3′ end and opposite the PTB-binding sites on the c3′-UTR.
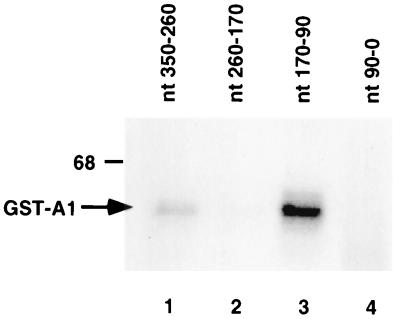
To define the binding site of hnRNP A1 within the MHV 3′-UTR, several RNA fragments representing different regions of the 3′-UTR were used in a UV cross-linking assay. As the results in Fig. 3 show, two hnRNP A1-binding sites were revealed. A strong hnRNP A1-binding site was mapped to nt 90 to 170 from the 3′ end of the MHV RNA (lane 3), and a weaker binding site was mapped to nt 260 to 350 from the 3′ end (lane 1).
FIG. 3.
Mapping of hnRNP A1-binding sites on the MHV 3′-UTR. The 3′-UTR (350 nt) was separated into four fragments as indicated above the gel. 32P-labeled in vitro transcript of each fragment was subjected to UV cross-linking with GST-A1. Numbering of the nucleotides is from the 3′ end of MHV RNA for convenience. The position of the 68-kDa size marker is shown on the left.
Previous studies in our laboratory identified two PTB-binding sites on the c3′-UTR: a strong binding site at nt 53 to 149, and a weak binding site at nt 270 to 307 (11). Comparison of the hnRNP A1-binding site on the 3′-UTR and the PTB-binding site on the c3′-UTR revealed that both the strong and weak hnRNP A1-binding sites and the corresponding PTB-binding sites are at the complementary sites of the opposite RNA strands. Further mapping within the strong PTB-binding site showed that two stretches of pyrimidine nucleotides at positions 77 to 82 (stretch A) and 132 to 136 (stretch C; AGAAG) from the 3′ end of the c3′-UTR are important for PTB binding (11). The latter (stretch C) is within the site complementary to the strong hnRNP A1-binding site on the 3′-UTR (nt 90 to 170). We thus examined whether nt 132 to 136 are also responsible for hnRNP A1 binding to the 3′-UTR. Two MHV 3′-UTR RNAs, with stretch C substituted by AGATT (subsC mutant) and deleted (ΔC), were used for UV cross-linking assays with GST-A1. Results in Fig. 4 show that the subsC and ΔC mutations of 3′-UTR RNA reduced hnRNP A1 binding to 15.3 and 8.0%, respectively, of the wild-type RNA level. Previous studies had shown that the corresponding substitution and deletion mutations in the complementary strand (c3′-UTR) reduced PTB binding to 13.1 and 19.7%, respectively (11). Therefore, there is a rough correlation between the extents of PTB binding to the c3′-UTR and those of hnRNP A1 binding to the 3′ UTR. We conclude that within the strong hnRNP A1- and PTB-binding sites, hnRNP A1 and PTB bind to the complementary sequences on the positive and negative strands, respectively.
FIG. 4.
Identification of nucleotides from positions 170 to 90 that are important for hnRNP A1 binding. (A) Computer-predicted secondary structure of the 3′-most 299 nt of MHV viral RNA, generated by the Mulfold2 program (55). (B) Expanded structure of nt 52 to 150. Stretch C nucleotides are marked. (C) UV cross-linking assay using wild-type (lane 1), subsC (lane 2), and ΔC (lane 3) mutants with GST-A1.
hnRNP A1 and PTB mediate the 5′-3′-end interaction of both positive- and negative-strand RNA in vitro.
The results shown above indicate that hnRNP A1 binds to the 3′-UTR, whereas a previous study showed that PTB binds to the MHV leader at the 5′-UTR (25). Since hnRNP A1 and PTB have been shown to form parts of a spliceosome complex (3), we were interested in knowing whether the interaction between hnRNP A1 and PTB allows the MHV leader and 3′-UTR to form a complex. We used an in vitro RNP complex formation assay for this purpose. MHV leader RNA labeled with biotin and MHV 3′-UTR labeled with [32P]UTP were incubated with GST-A1 or GST-PTB, separately or together, at 30°C; then streptavidin-agarose beads were added to pull down the biotin-labeled MHV leader. 32P-labeled RNAs that complexed with the biotin-labeled RNA were eluted and examined by denaturing PAGE. Results in Fig. 5A showed that in the presence of both GST-A1 and GST-PTB (lane 5), the 32P-labeled 3′-UTR was pulled down along with the biotin-labeled 5′-UTR, indicating a possible interaction between the MHV leader and MHV 3′-UTR through protein-RNA and protein-protein interactions. GST alone (lane 2), hnRNP A1 alone (lane 3), or PTB alone (lane 4) did not induce the formation of similar RNP complexes. In addition, biotin-labeled leader RNA by itself did not interact with 32P-labeled 3′-UTR directly (lane 1). Finally, 32P-labeled 3′-UTR did not bind to the streptavidin-agarose beads directly (lane 6).
FIG. 5.
RNP complex formation between 5′ and 3′ ends of both strands. (A) RNP complex formation between the leader and 3′-UTR RNA (positive strand). Various proteins or protein combinations were incubated with 32P-labeled MHV 3′-UTR and biotin-labeled leader sequence. RNP complexes were precipitated by streptavidin-agarose beads. RNA was extracted from the beads and resolved on a 6% denaturing polyacrylamide gel. (B) RNP complex formation between negative-strand leader [Leader(−)] and c3′-UTR.
We performed a similar experiment using negative-strand RNA, since PTB and hnRNP A1 also bind to the 5′ and 3′ ends, respectively, of this RNA. Using biotin-labeled negative-strand leader and 32P-labeled c3′-UTR, we showed that these two RNAs could form an RNP complex through hnRNP A1 and PTB (Fig. 5B, lane 5). hnRNP A1 without PTB also induced a small amount of RNP complex (lane 3). This result is consistent with previous findings that a small amount of hnRNP A1 also binds to the c3′-UTR (8) and that hnRNP A1 can interact with itself (5). PTB alone (lane 4) or hnRNP A1 and PTB in the absence of biotin-labeled 5′-UTR RNA (lane 6) did not induce 5′-3′-end interaction. These results combined indicate that PTB and hnRNP A1 mediate specific interactions between the 5′ and 3′ ends of MHV RNA of both positive and negative strands. It should be noted that the poly(A) sequence was not included in the study of the positive-strand 5′-3′-end interaction and the poly(U) sequence was not included in the study of the negative-strand 5′-3′-end interaction. Therefore, these interactions did not involve the poly(A) sequence.
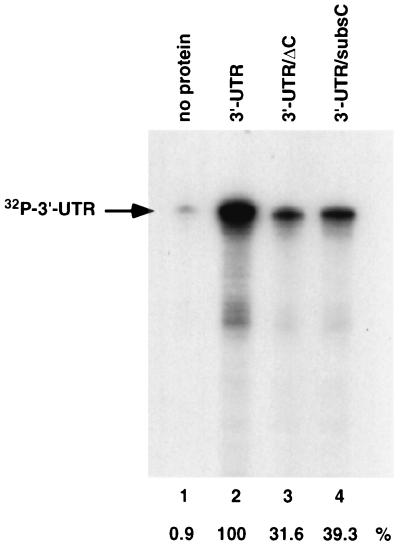
To confirm that the interaction between the MHV 5′ leader and 3′-UTR is mediated through hnRNP A1, mutant 3′-UTR RNAs with deletion (ΔC); or substitution (subsC) of the hnRNP A1-binding site were used in the in vitro RNP complex formation assay. Results in Fig. 6 showed that the ΔC (lane 3) and subsC (lane 4) mutants reduced the formation of the RNP complex between the 5′ leader and the 3′-UTR to 31.6 and 39.3%, respectively, of the wild-type RNA level. These values were slightly higher than the corresponding extents of hnRNP A1 binding to these respective RNAs, probably because the RNA used in this assay also contains a weak hnRNP A1-binding site at nt 260 to 350 from the 3′ end. (In contrast, the RNA used in the protein binding assay in Fig. 4 included only the strong hnRNP A1-binding site). These results suggest that the mutations that affected hnRNP A1 binding also affected the 5′-3′-end interactions.
FIG. 6.
RNP complex formation using mutant MHV 3′-UTR. ΔC (lane 3) and subsC (lane 4) mutants and wild-type (lane 2) 3′-UTR RNA were used in an RNP complex formation assay as described for Fig. 5. The arrow indicates the position of 32P-labeled 3′-UTR. Relative efficiencies of RNP complex formation assay are shown at the bottom.
We also used the DBT whole-cell extract in lieu of the recombinant GST-hnRNP A1 and GST-PTB in the RNP complex formation assay. The results showed that the cell extract also caused the formation of the MHV 5′-3′-end complex, although the nonspecific background of RNP complex formation was higher than when the recombinant proteins were used (data not shown). These data suggest that the endogenous cellular proteins could promote the 5′-3′-end interaction of MHV RNA, although we could not establish that this interaction was mediated by hnRNP A1 and PTB in the cells.
Mutations that affect hnRNP A1 binding to the 3′-UTR affects replication of a DI RNA.
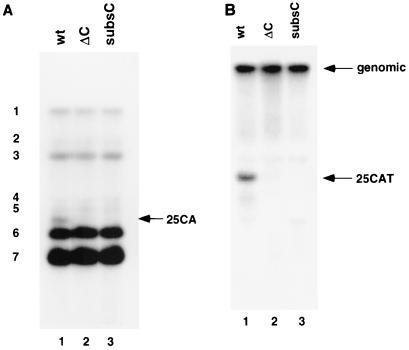
Previous results have shown that ΔC and subsC mutations on the 3′-UTR can affect the subgenomic mRNA transcription from an inserted intergenic (IG) sequence in the DI construct 25CAT RNA (11). In this study, we further investigated whether the same mutations also affect the replication efficiency of the same DI construct. 25CAT wild-type RNA and 25CAT-based DI RNAs with deletion (ΔC) or substitution (subsC) of the hnRNP A1-binding site were transfected into A59-infected DBT cells. Supernatant was collected at 16 h postinfection and used to infect DBT cells. At 7 h postinfection, virus-infected cells were collected and cytoplasmic RNAs were extracted for Northern blot analysis using a probe that detects all viral RNA species. Results in Fig. 7A showed that the DI RNAs with the ΔC (lane 2) and subsC (lane 3) mutation were almost undetectable. To confirm this result, we performed another Northern blot analysis using a probe complementary to the sequence downstream of the leader RNA that detects only the genomic and DI RNAs. Results in Fig. 7B show that these two mutations severely affected the ability of DI RNA to replicate. This result is consistent with the previous report that deletion of nt 129 to 139 from the 3′-UTR of the DI RNA of MHV-JHM strain inhibited replication of the RNA (31). These studies combined suggest that the hnRNP A1- and PTB-binding sites in the 3′-UTR are important for MHV DI RNA replication.
FIG. 7.
Northern blot analysis of replication of DI RNAs harboring mutations at the hnRNP A1-binding sites. (A) Northern blot analysis of the replication of wild-type (wt; lane 1), ΔC (lane 2), and subsC (lane 3) 25CAT DI RNAs detected by a probe complementary to the 3′-UTR of MHV RNA. The arrow indicates the position of 25CAT DI RNA; the seven mRNA species from the helper virus are indicated at the left. (B) Northern blot analysis using a probe complementary to the 5′-end of MHV RNA just downstream of the leader sequence. Arrows indicate the positions of genomic RNA and 25CAT DI RNA.
DISCUSSION
The 5′ and 3′ ends of viral RNA are important for viral RNA replication. One obvious reason is that these end sequences provide the signals for initiation of synthesis of the respective complementary strands. For example, the 3′ end of a positive-strand RNA regulates the synthesis of the negative strand, and the 5′-end sequence regulates the synthesis of the positive strand. However, an increasing body of evidence has shown that the 3′-end sequence of a positive-strand RNA may also be involved in the regulation of positive-strand RNA synthesis. For example, the 3′-UTR sequences are important in regulating positive-strand RNA synthesis of BMV (19) and synthesis of satellite RNA C associated with turnip crinkle virus (8) and MHV (28, 29). In order for the 3′-end sequence to influence synthesis of its own RNA, which starts from the 5′ end, it needs to interact with the 5′ end of RNA. Indeed, the 5′-3′-end interactions of viral RNA have been shown to be necessary for viral RNA synthesis of influenza virus (6, 32, 54) and vesicular stomatitis virus (14, 47, 48). Long-distance interactions between well-separated RNA regions have also been reported for potato virus X (15, 16). In these cases, there is sequence complementarity between the both ends of the RNA to allow direct RNA-RNA interactions. When sequence complementarity between the two ends of RNA is not sufficient to direct this 5′-3′ interaction, protein factors may facilitate such interactions. Furthermore, the 5′-end sequence of a positive-strand RNA, as well as the complementary sequence on the negative strand, can affect positive-strand RNA synthesis, as shown for poliovirus and BMV (2, 19). Thus, direct or indirect RNA-RNA interactions involving both ends of the template strand and product strand may be important for the regulation of viral RNA synthesis.
Recently, it has been shown that poly(A)-binding protein can interact with the bovine coronavirus 3′ poly(A) sequence and that the poly(A) sequence is required for bovine coronavirus DI replication (44). We have examined the possible contribution of the poly(A) tail to the 5′-3′-end interaction of MHV RNA in our RNP complex formation assay. Using an uncapped MHV 3′-UTR plus 23 poly(A) residues in the presence of DBT whole-cell extract, we found that the presence of a poly(A) sequence did not have a significant effect on RNP complex formation involving the uncapped 5′and 3′ ends of MHV RNA (data not shown). Thus, the binding of PTB and hnRNP A1 to the 3′-UTR is sufficient for the 5′-3′-end interaction. Poly(A)-binding protein likely also contributes to the potential interaction of the 5′ and 3′ ends of MHV RNA in vivo since it has been shown to interact with the translation initiation factor eIF4G, which in turn interacts with the cap-binding protein eIF4E (45) to induce the looping of mRNA (46).
In this report, we demonstrated that hnRNP A1 can specifically bind to the 3′-UTR of the MHV RNA. From this study combined with other three reports of host cell proteins that bind to the MHV RNA regulatory regions (11, 25, 26), an interesting picture has emerged: for the MHV positive-strand RNA, PTB binds to the 5′ end and hnRNP A1 binds to the 3′ end. For the negative strand, the same pattern holds: PTB binds to the 5′ end (i.e., the c3′-UTR) and hnRNP A1 binds to the 3′ end (i.e., the minus-strand leader). Thus, the same two proteins (hnRNP A1 and PTB) bind to both ends of both positive- and negative-strand RNA. Interestingly, both ends of hepatitis A virus RNA also bind the same set of cellular proteins (18). The 5′ and 3′ ends of MHV RNA have been suggested from functional studies to interact (20, 28). Since there is no obvious sequence complementarity between the 5′ and 3′ ends, these interactions are probably mediated by the proteins that bind to the two regions. In this study, hnRNP A1 and PTB indeed were shown to mediate the formation of an RNP complex involving the both ends of MHV RNA. The 3′-UTR of MHV RNA has already been shown to be important in both MHV RNA replication and subgenomic mRNA transcription (17, 28, 29). Our present study further showed that the binding sites of PTB and hnRNP A1 are the crucial elements for viral RNA synthesis. Previous reports have detected several host cell proteins that can interact with the 3′-UTR of MHV RNA (31, 50, 51). Liu et al. found that among them, a 40-kDa protein bound to an 11-nt region at nt 154 to 129 from the 3′ end of MHV RNA and that this site was important for MHV DI RNA replication (31). This binding site coincides with the strong hnRNP A1-binding site reported in this study. Although this 40-kDa protein was not identified, results of the present study suggest that it is hnRNP A1.
It is interesting that hnRNP A1 and PTB bind to the precisely complementary sequences on opposite strands of MHV RNA. The hnRNP A1-binding site on the negative-strand leader RNA has previously been mapped to the complementary sequence of the UCUAA pentanucleotide repeats (26), while the PTB-binding site on the positive-strand leader RNA has been mapped to the UCUAA repeats (25). In this report, the PTB- and hnRNP A1-binding sites on the 3′ UTR were also mapped to complementary sequences, including a region of 5 nt (132 to 136 nt from the 3′ end of the MHV genome) on the opposite strands. On the other hand, the PTB- and hnRNP A1-binding sequences at the 5′ and 3′ ends of the same strand are not complementary. Since the hnRNP A1-binding motif is not very stringent (1, 4), this result is not surprising. Nevertheless, the finding that these two proteins bind to the complementary sites suggests that they may have cooperative activities. Indeed, hnRNP A1 and PTB are part of the spliceosome complex, which is involved in alternative RNA splicing (3), indicating that they can bring different splicing donor and acceptor RNA sites together. It is possible that when hnRNP A1 and PTB interact with each other, they can promote the annealing of long complementary strands. They may also promote reverse reactions to unwind the complementary strands. Indeed, we have previously shown that the binding of PTB altered the conformation of the c3′-UTR of MHV RNA (11). hnRNP A1 has also been shown to promote both strand annealing and helix destabilizing (41).
In the RNP complex formation assay for negative-strand RNA, there was a small degree of complex formation when hnRNP A1 alone was used (Fig. 5B, lane 3). This is consistent with the finding that hnRNP A1 binds weakly to MHV c3′-UTR (11); the dimerization of hnRNP A1, which binds to both the 5′ and 3′ ends of the negative-strand RNA (26), allows these two RNA elements to interact. However, the efficiency of RNP complex formation mediated by hnRNP A1 and PTB was clearly higher than that mediated by hnRNP A1 alone (comparing lanes 3 and 5 in Fig. 5B).
Although the functional roles of hnRNP A1 binding to the MHV 3′-UTR in vivo were not directly demonstrated in this study, recent results in our laboratory did suggest the importance of hnRNP A1 in MHV replication (42): expression of dominant-negative mutants of hnRNP A1 inhibited the replication of MHV, whereas overexpression of the wild-type hnRNP A1 enhanced MHV replication. Many cellular proteins have now been shown to bind to RNAs of various viruses (reviewed in reference 20), and some of the host proteins have been shown to affect virus replication. For example, in a BMV replication system in yeast (13), several host cell proteins were shown to be involved in the replication of BMV RNA (12). Further understanding of the roles of hnRNP A1 and PTB will likely contribute to our knowledge of the mechanisms of viral RNA synthesis.
ACKNOWLEDGMENTS
We thank Daphne Shimoda for editorial assistance.
This work was supported by research grant AI19244 from the National Institutes of Health. M. M. C. Lai is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Abdul-Manan N, Williams K R. hnRNP A1 binds promiscuously to oligoribonucleotides: utilization of random and homo-oligonucleotides to discriminate sequence from base-specific binding. Nucleic Acids Res. 1996;24:4063–4070. doi: 10.1093/nar/24.20.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andino R, Rieckhof G E, Achacoso P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bothwell A L, Ballard D W, Philbrick W M, Lindwall G, Maher S E, Bridgett M M, Jamison S F, Garcia-Blanco M A. Murine polypyrimidine tract binding protein. Purification, cloning, and mapping of the RNA binding domain. J Biol Chem. 1991;266:24657–24663. [PubMed] [Google Scholar]
- 4.Burd C G, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cartegni L, Maconi M, Morandi E, Cobianchi F, Riva S, Biamonti G. hnRNP A1 selectively interacts through its Gly-rich domain with different RNA-binding proteins. J Mol Biol. 1996;259:337–348. doi: 10.1006/jmbi.1996.0324. [DOI] [PubMed] [Google Scholar]
- 6.Fodor E, Pritlove D C, Brownlee G G. The influenza virus panhandle is involved in the initiation of transcription. J Virol. 1994;68:4092–4096. doi: 10.1128/jvi.68.6.4092-4096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furuya T, Lai M M C. Three different cellular proteins bind to the complementary sites on the 5′-end positive- and 3′-end negative strands of mouse hepatitis virus RNA. J Virol. 1993;67:7215–7222. doi: 10.1128/jvi.67.12.7215-7222.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan H, Song C, Simon A E. RNA promoters located on (−)-strands of a subviral RNA associated with turnip crinkle virus. RNA. 1997;3:1401–1412. [PMC free article] [PubMed] [Google Scholar]
- 9.Herold J, Andino R. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J Virol. 2000;74:6394–6400. doi: 10.1128/jvi.74.14.6394-6400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirano N, Fujiwara K, Hino S, Matsumoto M. Replication and plaque formation of mouse hepatitis virus (MHV-2) in mouse cell line DBT culture. Arch Gesamte Virusforsch. 1974;44:298–302. doi: 10.1007/BF01240618. [DOI] [PubMed] [Google Scholar]
- 11.Huang P, Lai M M C. Polypyrimidine tract-binding protein binds to the complementary strand of the mouse hepatitis virus 3′ untranslated region, thereby altering RNA conformation. J Virol. 1999;73:9110–9116. doi: 10.1128/jvi.73.11.9110-9116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishikawa M, Diez J, Restrepo-Hartwig M, Ahlquist P. Yeast mutations in multiple complementation groups inhibit brome mosaic virus RNA replication and transcription and perturb regulated expression of the viral polymerase-like gene. Proc Natl Acad Sci USA. 1997;94:13810–13815. doi: 10.1073/pnas.94.25.13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janda M, Ahlquist P. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell. 1993;72:961–970. doi: 10.1016/0092-8674(93)90584-d. [DOI] [PubMed] [Google Scholar]
- 14.Keene J D, Schubert M, Lazzarini R A. Terminal sequences of vesicular stomatitis virus RNA are both complementary and conserved. J Virol. 1979;32:167–174. doi: 10.1128/jvi.32.1.167-174.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim K H, Hemenway C L. Long-distance RNA-RNA interactions and conserved sequence elements affect potato virus X plus-strand RNA accumulation. RNA. 1999;5:636–645. doi: 10.1017/s1355838299982006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim K H, Hemenway C L. Mutations that alter a conserved element upstream of the potato virus X triple block and coat protein genes affect subgenomic RNA accumulation. Virology. 1997;232:187–197. doi: 10.1006/viro.1997.8565. [DOI] [PubMed] [Google Scholar]
- 17.Kim Y-N, Jeong Y S, Makino S. Analysis of cis-acting sequences essential for coronavirus defective interfering RNA replication. Virology. 1993;197:53–63. doi: 10.1006/viro.1993.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kusov Y Y, Weitz M, Dollenmeier G, Gauss-Muller V, Siegl G. RNA-protein interactions at the 3′ end of the hepatitis A virus RNA. J Virol. 1996;70:1890–1897. doi: 10.1128/jvi.70.3.1890-1897.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lahser F C, Marsh L E, Hall T C. Contributions of the brome mosaic virus RNA-3 3′-nontranslated region to replication and translation. J Virol. 1993;67:3295–3303. doi: 10.1128/jvi.67.6.3295-3303.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai M M C. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology. 1998;244:1–12. doi: 10.1006/viro.1998.9098. [DOI] [PubMed] [Google Scholar]
- 21.Lai M M C, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai M M C, Stohlman S A. The RNA of mouse hepatitis virus. J Virol. 1978;26:236–242. doi: 10.1128/jvi.26.2.236-242.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamond A, Sproat B S. Isolation and characterization of ribonucleoprotein complexes. In: Higgins S J, Hames B D, editors. RNA processing: a practical approach. New York, N.Y: Oxford University Press; 1996. pp. 103–140. [Google Scholar]
- 24.Lee H-J, Shieh C-K, Gorbalenya A E, Koonin E V, La Monica N, Tuler J, Bagdzyahdzhyan A, Lai M M-C. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H-P, Huang P, Park S, Lai M M C. Polypyrimidine tract-binding protein binds to the leader RNA of mouse hepatitis virus and serves as a regulator of viral transcription. J Virol. 1999;73:772–777. doi: 10.1128/jvi.73.1.772-777.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H-P, Zhang X, Duncan R, Comai L, Lai M M C. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc Natl Acad Sci USA. 1997;94:9544–9549. doi: 10.1073/pnas.94.18.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao C-L, Lai M M C. Requirement of the 5′-end genomic sequence as an upstream cis-acting element for coronavirus subgenomic mRNA transcription. J Virol. 1994;68:4727–4737. doi: 10.1128/jvi.68.8.4727-4737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin Y-J, Zhang X, Wu R-C, Lai M M C. The 3′-untranslated region of the coronavirus RNA is required for subgenomic mRNA transcription from a defective interfering RNA. J Virol. 1996;70:7236–7240. doi: 10.1128/jvi.70.10.7236-7240.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Y-J, Lai M M C. Deletion mapping of a mouse hepatitis virus defective-interfering RNA reveals the requirement of an internal and discontiguous sequence for replication. J Virol. 1993;67:6110–6118. doi: 10.1128/jvi.67.10.6110-6118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin Y-J, Liao C L, Lai M M C. Identification of the cis-acting signal for minus-strand RNA synthesis of a murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J Virol. 1994;68:8131–8140. doi: 10.1128/jvi.68.12.8131-8140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Q, Yu W, Leibowitz J L. A specific host cellular protein binding element near the 3′-end of mouse hepatitis virus genomic RNA. Virology. 1997;232:74–85. doi: 10.1006/viro.1997.8553. [DOI] [PubMed] [Google Scholar]
- 32.Luo G, Luytjes W, Enami M, Palese P. The polyadenylation signal of influenza virus RNA involves a stretch of uridines followed by the RNA duplex of the panhandle structure. J Virol. 1991;65:2861–2867. doi: 10.1128/jvi.65.6.2861-2867.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makino S, Fujioka N, Fujiwara K. Structure of the intracellular defective viral RNAs of defective interfering particles of mouse hepatitis virus. J Virol. 1985;54:329–336. doi: 10.1128/jvi.54.2.329-336.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makino S, Shieh C-K, Keck J G, Lai M M C. Defective interfering particles of murine coronavirus: mechanism of transcription of defective viral RNA. Virology. 1988;163:104–111. doi: 10.1016/0042-6822(88)90237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makino S, Shieh C-K, Soe L H, Baker S C, Lai M M C. Primary structure and translation of a defective-interfering RNA of murine coronavirus. Virology. 1988;166:550–560. doi: 10.1016/0042-6822(88)90526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Makino S, Taguchi F, Fujiwara K. Defective interfering particles of mouse hepatitis virus. Virology. 1984;133:9–17. doi: 10.1016/0042-6822(84)90420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makino S, Yokomori K, Lai M M C. Analysis of efficiently packaged defective-interfering RNAs of murine coronavirus: localization of a possible RNA-packaging signal. J Virol. 1990;64:6045–6053. doi: 10.1128/jvi.64.12.6045-6053.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manaker R A, Piczak C V, Miller A A, Stanton M F. A hepatitis virus complicating studies with mouse leukemia. J Natl Cancer Inst. 1961;27:29–51. [PubMed] [Google Scholar]
- 39.Pachuk C J, Bredenbeek P J, Zoltick P W, Spaan W J M, Weiss S R. Molecular cloning of the gene encoding the putative polymerase of mouse hepatitis coronavirus strain A59. Virology. 1989;171:141–148. doi: 10.1016/0042-6822(89)90520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pogue G P, Hall T C. The requirement for a 5′ stem-loop structure in brome mosaic virus replication supports a new model for viral positive-strand RNA initiation. J Virol. 1992;66:674–684. doi: 10.1128/jvi.66.2.674-684.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Portman D S, Dreyfuss G. RNA annealing activities in HeLa nuclei. EMBO J. 1994;13:213–221. doi: 10.1002/j.1460-2075.1994.tb06251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi S T, Huang P, Li H-P, Lai M M C. Heterogeneous nuclear ribonucleoprotein A1 regulates RNA synthesis of a cytoplasmic virus. EMBO J. 2000;19:4701–4711. doi: 10.1093/emboj/19.17.4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spaan W, Cavanagh D, Horzinek M C. Coronaviruses: structure and genome expression. J Gen Virol. 1988;69:2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- 44.Spagnolo J F, Hogue B G. Host protein interactions with the 3′ end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J Virol. 2000;74:5053–5065. doi: 10.1128/jvi.74.11.5053-5065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tarun S Z J, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 46.Wells S E, Hillner P E, Vale R D, Sachs A B. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 47.Wertz G W, Whelan S, LeGrone A, Ball L A. Extent of terminal complementarity modulates the balance between transcription and replication of vesicular stomatitis virus RNA. Proc Natl Acad Sci USA. 1994;91:8587–8591. doi: 10.1073/pnas.91.18.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whelan S P, Wertz G W. Regulation of RNA synthesis by the genomic termini of vesicular stomatitis virus: identification of distinct sequences essential for transcription but not replication. J Virol. 1997;73:297–306. doi: 10.1128/jvi.73.1.297-306.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Will C L, Kastner B, Luehrmann R. Analysis of ribonucleoprotein interactions. In: Higgins S J, Hames B D, editors. RNA processing: a practical approach. New York, N.Y: Oxford University Press; 1996. pp. 141–177. [Google Scholar]
- 50.Yu W, Leibowitz J L. A conserved motif at the 3′ end of mouse hepatitis virus genomic RNA required for host protein binding and viral RNA replication. Virology. 1995;214:128–138. doi: 10.1006/viro.1995.9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu W, Leibowitz J L. Specific binding of host cellular proteins to multiple sites within the 3′ end of mouse hepatitis virus genomic RNA. J Virol. 1995;69:2016–2023. doi: 10.1128/jvi.69.4.2016-2023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X, Li H-P, Xue W, Lai M M C. Formation of a ribonucleoprotein complex of mouse hepatitis virus involving heterogeneous nuclear ribonucleoprotein A1 and transcription-regulatory elements of viral RNA. Virology. 1999;264:115–124. doi: 10.1006/viro.1999.9970. [DOI] [PubMed] [Google Scholar]
- 53.Zhang X M, Lai M M C. Interactions between the cytoplasmic proteins and the intergenic (promoter) sequence of mouse hepatitis virus RNA: correlation with the amounts of subgenomic mRNA transcribed. J Virol. 1995;69:1637–1644. doi: 10.1128/jvi.69.3.1637-1644.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng H, Palese P, Garcia-Sastre A. Nonconserved nucleotides at the 3′ and 5′ ends of an influenza A virus RNA play an important role in viral RNA replication. Virology. 1996;217:242–251. doi: 10.1006/viro.1996.0111. [DOI] [PubMed] [Google Scholar]
- 55.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]