Abstract
Latent Epstein-Barr virus (EBV) is maintained by the virus replication origin oriP that initiates DNA replication with the viral oriP-binding factor EBNA1. However, it is not known whether oriP's replicator activity is regulated by virus proteins or extracellular signals. By using a transient replication assay, we found that a low level of expression of viral signal transduction activator latent membrane protein 1 (LMP1) suppressed oriP activity. The binding site of the tumor necrosis factor receptor-associated factor (TRAF) of LMP1 was essential for this suppressive effect. Activation of the TRAF signal cascade by overexpression of TRAF5 and/or TRAF6 also suppressed oriP activity. Conversely, blocking of TRAF signaling with dominant negative mutants of TRAF5 and TRAF6, as well as inhibition of a downstream signal mediator p38 MAPK, released the LMP1-induced oriP suppression. Furthermore, activation of TRAF6 signal cascade by lipopolysaccharides (LPS) resulted in loss of EBV from Burkitt's lymphoma cell line Akata, and inhibition of p38 MAPK abolished the suppressive effect of LPS. These results suggested that the level of oriP activity is regulated by LMP1 and extracellular signals through TRAF5- and TRAF6-mediated signal cascades.
Epstein-Barr virus (EBV) is related to Burkitt's lymphoma, T-cell lymphoma, gastric carcinoma, infectious mononucleosis, and opportunistic lymphoma in immunosuppressed patients (31), but resting memory B lymphocytes are normal cells infected latently with EBV in vivo (43, 44). During latent infection, the 170-kb EBV genome forms a circular plasmid DNA and is maintained by the 2.2-kb region oriP containing an origin of bidirectional DNA replication (62, 64). oriP is comprised of two EBNA1-binding elements, dyad symmetry (DS) and family of repeats (FR), separated by 960 bp. The DS element functions as a replication origin (16, 20, 43, 55, 65), and the FR element plays a major role in nuclear retention of the genome (26, 41). DNA replication from oriP (DS-dependent replication) requires only the viral oriP-binding protein EBNA1 and occurs once in a single S phase through a mechanism of replication licensing (21, 56, 62, 63). However, it is not known whether oriP's replicator activity is regulated by virus proteins or extracellular signals.
In contrast to these studies, in some EBV-positive lymphoma cell lines, replication of the EBV genome is initiated mostly in a broad initiation zone distant from oriP (DS-independent replication) (25). The occurrence of DS-independent replication was initially found in Raji and Daudi by 2D gel analysis (38) and then was demonstrated using the oriP-containing plasmid in several cell lines, including C33, HEK293, and P3HR1 (2, 35; unpublished data). Recently, Norio et al. (49) showed more direct evidence, using recombinant EBV virus, that the DS element is dispensable for EBV replication in BL30 and a P3HR1 clone. When DS-independent replication occurs, the DS-dependent replication from oriP is rare (38). The initiation region used for DS-independent replication may be preferentially used over oriP in lymphomas. Alternatively, the oriP activity may be negatively regulated by latent virus proteins expressing in these cell lines. To explore this possibility, we examined the effect of latent membrane protein 1 (LMP1) on oriP activity. LMP1 is an EBV integrated membrane protein that plays an essential role in immortalization of human B lymphocytes by EBV (29, 34, 45) and transforms rodent fibroblasts (3, 61). LMP1 induces activation of several signal mediators: NF-κB (22, 42), c-Jun amino-terminal kinase (JNK) (12, 32), extracellular signal-regulated kinases (ERKs) (52), p38 mitogen-activated protein kinase (MAPK) (13), and Janus kinase 3 (17). LMP1 has two C-terminal terminal activating regions, CTAR1/TES1 (amino acids [aa] 187 to 232) and CTAR2/TES2 (aa 351 to 386), which are responsible for activation of these signal mediators. CTAR1/TES1 contains the PxQxT motif that is a binding site for the tumor necrosis factor receptor-associated factors (TRAFs) (9, 22, 42, 46). TRAFs are the signal mediators of the cellular membrane receptors of TNFR and Toll/IR-1R superfamilies and initiate distinct but overlapping signal cascades (7, 23, 46, 47). Among the six TRAFs identified to date, TRAF1, TRAF2, TRAF3, and TRAF5 but not TRAF6 associate with the PxQxT motif of CTAR1/TES1 (5, 9, 10, 46, 52). TRAF2 also associates indirectly with CTAR2/TES2 via TRADD and RIP and mediates signal cascades leading to activation of NF-κB and JNK (12, 14, 24, 25, 32, 33, 58).
In this study, we showed that oriP activity is negatively regulated by the TRAF5-mediated signal initiated from LMP1 and the TRAF5- and TRAF6-mediated signals from cellular receptors. We also identified the p38 MAPK, a common downstream kinase in these signal cascades, as playing an important role in this negative regulation of EBV replication.
MATERIALS AND METHODS
Plasmids.
The oriP plasmid (KORI) containing the oriP region, the DS plasmid (KD11), the SV40 plasmid, and the internal control plasmid were described previously (55, 56). The expression plasmids of the LMP1 deletion derivatives were constructed from the LMP1 expression plasmid pNH-LMP1 (59). The plasmids expressing the LMP1 point mutants were described elsewhere (25). The expression plasmids of TRAF and their dominant negative mutants were also described elsewhere (27). A cDNA clone of mouse p38 MAPK was obtained by PCR from 15-day embryos using primers according to the p38 sequence (19) and was inserted into the expression vector pactEF (50). The dominant negative p38 mutant, p38AGF, was prepared by replacing the wild-type Thr180 (ACA) and Tyr182 (TAC) with an Ala (GCC) and a Phe (TTC), respectively, by oligonucleotide-directed mutagenesis.
Transient replication assay.
The oriP plasmid (2 μg) was transfected into HeLa/EB1 cells (2 × 106) (55) with the unmethylated control plasmid (1 μg) and the effector plasmid(s) by the calcium phosphate method. Transfection efficiency was estimated as 50% on average. After transfection, cells were cultured for 2 days in the experiments described in Fig. 1, 2, and 3, and for 3 days in the experiments described in Fig. 4, 5, and 6. DpnI digestion and Southern hybridization analysis were performed as described previously (56) using the oriP region (EcoRI-SacII) as a hybridization probe. The salt concentration in the DpnI reaction buffer was lowered to 50 mM in this study. Aliquots of about 1/10 of the extracts were used for a single DpnI assay. Radioisotope signals on Southern blots were analyzed quantitatively with a BAS2000 image analyzer (Fuji). The same membranes were rehybridized to detect the control plasmid. The hybridization signal of the DpnI-resistant oriP plasmid was normalized with the signal of the internal control plasmid in the same sample and was represented relatively to that of the vector-transfected sample. Expression of LMP1 and of EBNA1 was analyzed using a monoclonal antibody against LMP1 (S12) and a rabbit polyclonal antibody against EBNA1.
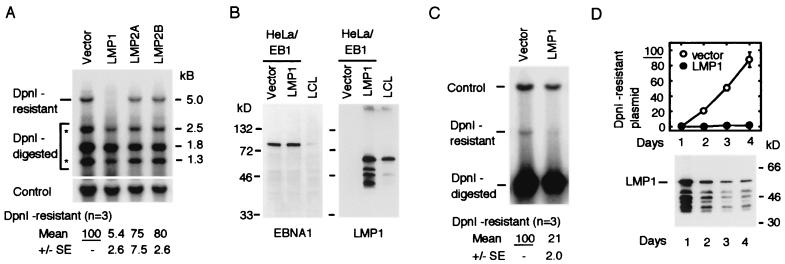
FIG. 1.
Expression of LMP1 suppressed replication of the oriP plasmid in HeLa/EB1 cells. (A) Transient replication assay of the oriP plasmid. The oriP plasmid (2 μg), the control plasmid (1 μg), and the expression plasmid of LMP1, LMP2A, or LMP2B (0.5 μg) were transfected. Total amounts of plasmids were adjusted to 3.5 μg with the vector plasmid. Hirt's extracts were prepared 2 days after transfection, and plasmids were analyzed by DpnI digestion and Southern blot hybridization. The linearized DpnI-resistant plasmid (5.0 kb) and three DpnI-digested fragments (2.5, 1.8, and 1.3 kb) are shown. Two fragments indicated by asterisks (2.5 and 1.3 kb) are products of replication intermediates that were accumulated by the replication fork barrier at the FR element of oriP. Amounts of the DpnI-resistant plasmid (replicated plasmid) are normalized with that of the control plasmid and shown below. Data represent averages of three experiments with the standard errors (SE). (B) Expression of LMP1 and EBNA1 in transfected cells. Short polypeptides reacted with LMP1 antibody were digested products of LMP1. As a control, a similar number of LCL cells were analyzed in a parallel lane. (C) Replication assay of the DS plasmid. Experimental conditions are described above for panel A. Data represent averages of three experiments with the standard errors (SE). (D) Time course of the oriP plasmid replication. Transfected plasmids were the same as in panel A, and cells were collected for DpnI assay at the days indicated. A summary of two experiments is shown. Expression of LMP1 in one experiment is shown at the bottom. The same amount of total proteins was loaded on each lane.
FIG. 4.
Effects of TRAF expression on oriP activity. Shown are the results of a transient replication assay of the oriP plasmid. (A) Effects of single expression of TRAF. The oriP plasmid (2 μg), the control plasmid (0.5 μg), and the TRAF expression plasmid (4 μg) were transfected. Total amounts of plasmids were 6.5 μg. (B) Effects of coexpression of TRAFs. The oriP plasmid (2 μg), the control plasmid (0.5 μg), and the TRAF expression plasmids (4 μg each) were transfected. Total amounts of plasmids were 10.5 μg. (C) Effects of coexpression of TRAF and the dominant negative TRAF mutants, TRAF5DN and TRAF6DN. Normalized amounts of DpnI-resistant plasmid are shown below. Data represent averages of three experiments with standard errors (SE). Two fragments indicated by asterisks are products of replication intermediates.
[3H]thymidine incorporation of the LMP1/GFP expressing cells.
The pNH-LMP1 (0 to 4 μg) was transfected with the green fluorescent protein (GFP) expression plasmid pEGFP-C1 (4 μg) into HeLa/EB1 (2 × 106) in 100-mm dishes. Transfection efficiency was determined by counting GFP-expressing cells at 24 h after transfection. Then, cells were replated into 96-well plates (2,000 cells in 200 μl) and were cultured for 4 h in the presence of [3H]thymidine (1 μCi). Incorporation by the GFP/LMP1-expressing cells (2,000 cells) was calculated using the following equation: cpmG = [cpmS − (1 − f) × cpmO]/f, where cpmG is uptake by GFP-expressing cells; cpmS is total uptake by cells transfected with the GFP plasmid and the LMP1 plasmid; cpmO is total uptake by cells transfected with the GFP plasmid alone; and f is the ratio of GFP-expressing cells.
NF-κB activity.
The NF-κB reporter plasmid pκB-tkLuc (27) (2 μg) was transfected with pSV-β-gal (1 μg) and pNH-LMP1 (1 μg) into HeLa/EB1 (106) in 60-mm dishes. Luciferase activity was determined 2 days after transfection, and β-galactosidase activity was used as internal control.
Stimulation of Burkitt's lymphoma B cell lines.
Bacterial lipopolysaccharide (LPS) (Difco) (5 mg/ml) was added at 10 μg/ml to growing B cells (15 ml; 105 cells/ml). Two days after stimulation, cells (10 ml) were collected. Fresh culture medium containing LPS (10 ml) was added to the rest of the cells (5 ml), which were then cultured again for 2 days. Total DNA was prepared from these LPS-stimulated cells and unstimulated cells. The total DNA (4 μg) was digested with BamHI and analyzed by Southern hybridization methods using an EBV (B95-8) BamHI-C fragment for a probe. Hybridized signals were analyzed quantitatively with a BAS2000 image analyzer.
RESULTS
Expression of LMP1 induced suppression of oriP plasmid replication in HeLa/EB1 cells.
We have previously demonstrated that when the dam-methylated oriP plasmid is transfected into HeLa/EB1 cells, the DpnI-resistant replicated oriP plasmid is accumulated during 2 days after transfection (55, 56; Fig. 1A). When we analyzed the recovered oriP plasmid by DpnI digestion and Southern hybridization using the oriP region for a probe, we detected one linearized DpnI-resistant plasmid (5.0 kb) and five DpnI-digested fragments (2.5, 1.8, 1.3, 0.8, and 0.6 kb). The 0.8-kb and 0.6-kb fragments are not shown in the figures. Among these DpnI-digested fragments, the 1.8-kb, 0.8-kb, and 0.6-kb fragments were predicted from the restriction sites. The 2.5-kb and 1.3-kb fragments were the products of replication intermediates accumulated by a replication fork barrier at an FR element (16). Therefore, the amount of these replication intermediates was less than that of the 1.8-kb DpnI-digested fragment and related to the amount of replicated DpnI-resistant plasmid. To examine the effect of LMP1 expression on oriP activity, we cotransfected the LMP1 expression plasmid in this transient replication assay and found that expression of LMP1 significantly suppressed replication of the oriP plasmid (Fig. 1A). The amount of replicated plasmid was normalized to the amount of the internal control plasmid in the same sample and then was compared. The replicated plasmid in the LMP1-transfected cells was about 5% of that of the vector-transfected cells. We also examined the other latent membrane proteins LMP2A and LMP2B but observed only a weak suppression of oriP activity. Western analysis confirmed that expression of LMP1 did not affect expression of EBNA1, suggesting that insufficient expression of EBNA1 was not a cause of oriP suppression (Fig. 1B). Like the oriP plasmid, the DS plasmid containing only the replication origin was also sensitive to LMP1 expression, indicating that the origin element was responsible for LMP1-induced suppression (Fig. 1C). Suppressive effects of LMP1 continued for at least 4 days, while expression of LMP1 was highest 1 day after transfection and then decreased significantly at later time points (Fig. 1D). We then examined the dose dependency of the LMP1 plasmid for oriP suppression and found that the lowest dose of LMP1 plasmid (0.0625 μg) was significantly effective for oriP suppression (Fig. 2A). The amount of LMP1 plasmid required for 50% inhibition was 0.0625 μg for 2 × 106 cells (Fig. 2C). Fig. 2E shows the amount of LMP1 expressed in these transfected cells. When 0.5 μg of LMP1 plasmid was transfected into 2 × 106 cells, the amount of LMP1 was about equal to that expressed in an EBV-positive lymphoma cell line, Raji (results not shown).
FIG. 2.
The LMP1 plasmid dose dependency of oriP suppression. (A) Transient replication assay of the oriP plasmid. The oriP plasmid (2 μg), the control plasmid (1 μg), and the LMP1 plasmid (0, 0.0625, 0.25, 1, and 4 μg) were transfected. Total amounts of plasmids were 7 μg. Two fragments indicated by asterisks are products of replication intermediates. (B) Transient replication assay of the SV40 plasmid. The SV40 plasmid (2 μg) was transfected with other plasmids as described above for panel A. (C) Summary of experiments described in panels A and B. Data represent averages of two experiments with standard errors (SE). (D) Expression of LMP1 and EBNA1 in the transfected cells. (E) [3H]thymidine incorporation of the LMP1/GFP expressing cells. The LMP1 plasmid was transfected with pEGFP-C1 (4 μg). Data represent averages of three experiments with standard errors (SE).
A low level of LMP1 expression that induced oriP suppression did not inhibit cell growth.
Several studies have demonstrated that high levels of LMP1 expression inhibit cell growth (11, 18, 30). To monitor the growth inhibitory effect of LMP1 in our transient replication assay, we used the SV40 plasmid containing SV40 origin and the T-antigen gene (56). The SV40 plasmid was transfected with the LMP1 plasmid and its replication was analyzed at 2 days after transfection (Fig. 2B). In contrast to the oriP plasmid, a lower dose of the LMP1 plasmid (0.0625 and 0.25 μg) did not suppress the SV40 plasmid replication. With a higher dose of LMP1 plasmid (1 and 4 μg), replication of the SV40 plasmid was suppressed by 60 and 40%, respectively, suggesting that the growth inhibitory effect of LMP1 appeared at these doses. To confirm this result by another assay, [3H]thymidine incorporation by LMP1-expressing cells was examined. We transfected the same amount of the GFP plasmid (4 μg) with several different amounts of the LMP1 plasmid (0 to 4 μg) into HeLa/EB1. 3H incorporation by the GFP-expressing cells was estimated from the ratio of GFP-positive cells and 3H incorporation by total cells. 3H incorporation by GFP-positive cells was not affected with lower doses of LMP1 plasmid (0.0625 and 0.25 μg) but was reduced by 71 and 46% with higher doses (1 and 4 μg) (Fig. 2E). Thus, a lower level of LMP1 expression that induced oriP suppression did not inhibit cell growth.
The TRAF-binding motif of LMP1 was mostly responsible for induction of oriP suppression.
To determine the signal cascade leading to oriP suppression, we examined the LMP1 domain responsible for induction of oriP suppression. An LMP1 mutant, LMP1Δ(351-386), had a deletion of CTAR2 (aa 351 to 386) but retained CTAR1 (aa 187 to 232). Expression of LMP1Δ(351-386) suppressed oriP replication to the same extent as that of the wild-type molecule (Fig. 3A). Similarly, LMP1Δ(212-386), in which CTAR2 and most of CTAR1 were deleted but the PxQxT motif (aa 204 to 208) was retained, also showed the wild-type function. However, complete deletion of CTAR1 and CTAR2, including the PxQxT motif in LMP1Δ(187-386), eliminated most of the suppressive effect, and the internal deletion of CTAR1 in LMP1Δ(187-351) showed only a weak suppressive effect. This indicated the importance of the PxQxT motif for oriP suppression, which was further confirmed using LMP1 point mutants. The LMP1 mutant LMP1(PQT→AAA) had amino acid substitutions Pro204 to Ala, Gln206 to Ala, and Thr208 to Ala in the PxQxT motif and did not bind TRAFs (32, 33). As shown in Fig. 3A, LMP1(PQT→AAA) lost most of the suppressive effect.
FIG. 3.
Mutational analysis of LMP1 domains for oriP suppression. (A) Transient replication assay of the oriP plasmid. The oriP plasmid (2 μg), the control plasmid (1 μg), and the LMP1 plasmid (0.5 μg) were transfected. Total amounts of plasmids were 3.5 μg. The amino acid numbers in parentheses indicate the region deleted from LMP1. Amino acid substitutions in the LMP1 point mutants are also shown in parentheses. Normalized amounts of DpnI-resistant plasmid are shown below. Data represent averages of three experiments with standard errors (SE). Activation of NF-κB activity by these LMP1 mutants was examined using the pκB-Luc luciferase reporter plasmid. Data represent averages of two experiments with standard errors (SE). (B) Expression of LMP1 and its point mutants. The same amount of total protein was loaded in each lane. (C) Structure of LMP1.
We also examined Tyr384 in CTAR2, the amino acid residue important for binding of the TRADD-TRAF2 complex (14, 24, 33). Unexpectedly, the point mutant LMP1(Y384G) showed complete loss of the suppressive effect, although CTAR2 was not required for most of the LMP1's suppressive effect (Fig. 3A). Western analysis confirmed expression of a similar level of LMP1 protein (Fig. 3B). As the Y384G mutation abolishes the binding of the TRADD-TRAF2 complex to the CTAR2 domain, this result suggested that the absence of the TRADD-TRAF2 complex on the CTAR2 domain may have induced a large conformational change in LMP1 and interfered with the function of CTAR1.
Overexpression of TRAF5 and TRAF6 suppressed oriP activity.
TRAF1, TRAF2, TRAF3, and TRAF5 associate with the PxQxT motif of CTAR1 (5, 9, 10, 46, 53). To identify the TRAF-mediated signal cascade leading to oriP suppression, we overexpressed each TRAF in the absence of LMP1 and examined its effect on oriP activity. TRAF expression vectors used in this experiment were constructed with the same mammalian expression plasmid, and cells were collected 3 days after transfection because transfection of a large amount of plasmids reduced the efficiency of oriP replication. By this transient replication assay, we found that overexpression of TRAF5, but not of TRAF2 or TRAF3, reduced oriP activity by 60% (Fig. 4A). We also examined TRAF6, although it did not bind to the PxQxT motif, and found that overexpression of TRAF6 suppressed oriP activity to a similar extent as TRAF5.
Because TRAF2 or TRAF3 might work synergistically with TRAF5 and TRAF6, we examined combinations of TRAFs in a similar assay. However, coexpression of TRAF2 with TRAF5 or TRAF6 showed suppressive effects similar to those of TRAF5 and TRAF6 alone, indicating that TRAF2 did not interfere with the functions of TRAF5 or TRAF6 (Fig. 4B). Coexpression of TRAF3 reduced TRAF5-induced oriP suppression but it did not affect TRAF6-induced suppression. In contrast, when TRAF5 and TRAF6 were coexpressed, their suppressive effects were added and the oriP activity was reduced by 10%. This additive effect was not observed when one of TRAF5 and TRAF6 had a deletion in the effector domain (Fig. 4C). We also examined expression of TRAF1. Like TRAF2, expression of TRAF1 neither suppressed oriP activity nor interfered with the suppressive effects of TRAF5 and TRAF6 (results not shown).
oriP was activated by inhibition of the TRAF-mediated signaling in HeLa/EB1.
Because overexpression of TRAF5 and TRAF6 induced oriP suppression, we next examined the effects of inhibition of the TRAF-mediated signaling. Under normal culture conditions, the TRAF-mediated signal cascade was activated at a low level. We inhibited this basal activity of TRAF signaling by expressing the dominant negative mutant of TRAF, TRAFDN, which had deletions in the amino-terminal effector domain. When TRAF5DN or TRAF6DN was expressed, oriP replication was moderately activated, by 126 or 150% (Fig. 5A). Coexpression of TRAF5DN and TRAF6DN showed further activation, by 242%, indicating that the effects of TRAF5DN and TRAF6DN were added like those of TRAF5 and TRAF6. This result confirmed that TRAF5- and TRAF6-mediated signal cascades negatively regulated oriP activity and also indicated that oriP was sensitive to even a basal level of signaling in HeLa/EB1 cells.
FIG. 5.
Effects of dominant negative TRAF expression on oriP activity. Shown are the results of a transient replication assay of the oriP plasmid. (A) Effects of dominant negative TRAF (TRAFDN) in the absence of LMP1 expression. The oriP plasmid (2 μg), the control plasmid (0.5 μg), and the expression plasmids of TRAFDN (4 μg each) were transfected. Total amounts of plasmids were 10.5 μg. (B) Effects of dominant negative TRAFDN on LMP1-induced suppression. The oriP plasmid (2 μg), the control plasmid (0.5 μg), the TRAF expression plasmids (4 μg each), and LMP1Δ(212-386) (0.25 μg) were transfected. Total amounts of plasmids were 10.75 μg. Normalized amounts of DpnI-resistant plasmid are shown at the bottom. Data represent averages of three experiments with standard errors (SE). Two fragments indicated by asterisks are products of replication intermediates.
Interestingly, expression of TRAF3DN also activated oriP replication by 150%, although overexpression of TRAF3 did not suppress oriP. Coexpression of TRAF3DN with TRAF5DN or TRAF6DN showed further activation, by 215 and 325%, respectively. An in vitro study indicated that TRAF5 does not bind to the PxQxT motif directly, but that TRAF3 forms TRAF3-TRAF5 hetero-oligomers through the amino-terminal effector domain and mediates binding of TRAF5 to the motif (51). Because TRAF3DN lacked the effector domain, TRAF3DN competitively inhibited the binding of TRAF3-TRAF5 hetero-oligomers to the PxQxT motif and inhibited TRAF5-mediated signaling. Therefore, TRAF3DN was functionally similar to TRAF5DN.
We also examined the effects of TRAFDN on LMP1-induced suppression. Single expression of TRAF3DN or TRAF5DN did not affect the LMP1Δ(212-386)-induced suppression of oriP activity, but coexpression of both mutants partially released the suppression, confirming that TRAF5 mediated the LMP1-induced signal for oriP suppression (Fig. 5B). Although LMP1 does not bind TRAF6, TRAF6DN also partially released oriP suppression when it was coexpressed with TRAF3DN or TRAF5DN. This suggested that the LMP1-induced (TRAF5-mediated) signal and the TRAF6-mediated signal had a common downstream mediator for oriP suppression.
The p38 MAPK regulated oriP activity.
Because expression of LMP1, TRAF5, or TRAF6 induced activation of p38 MAPK in HeLa cells (4, 13; unpublished data), the kinase was a candidate common signal mediator for oriP regulation. We examined whether p38 MAPK was involved in the signal cascade leading to oriP suppression. Under the condition that oriP replication was suppressed by about 35% with LMP1, expression of the dominant negative mutant of p38 MAPK, p38AGF, released the LMP1-induced oriP suppression by about 90% (Fig. 6A). Similarly, treatment of cells with the specific inhibitor of p38 MAPK, SB203580 (20 μM), also released oriP suppression. In contrast, the wild-type p38 MAPK did not affect oriP replication. Because p38AGF and SB203580 did not stimulate [3H]thymidine incorporation, these results suggested that p38 MAPK was a downstream mediator of LMP1 for oriP suppression. Furthermore, in the absence of LMP1, expression of p38AGF also activated oriP activity by 195%, and treatment with SB208530 showed further activation, by 315%. This indicated that oriP activity was negatively regulated by p38 MAPK and that its basal level activity in HeLa/EB1 cells reduced oriP replication by threefold.
FIG. 6.
Examination of the signal mediators that affect oriP activity. Shown are the results of a transient replication assay of the oriP plasmid. (A) The p38 MAPK. The oriP plasmid (2 μg), the control plasmid (0.5 μg), LMP1Δ(212-386) (0.25 μg), and the expression plasmid of p38 MAPK or the dominant negative mutant of p38 MAPK, p38AGF (8 μg), were transfected. Total amounts of plasmids were 10.75 μg. The specific inhibitor for SB208350 was added into the culture medium at a concentration of 20 μM. (B) Ras and MEK. The oriP plasmid (2 μg), the control plasmid (0.5 μg), one of the expression plasmids of LMP1 (0.5 μg), c-H-ras (4 μg), the dominant active ras 12Vras (4 μg), or the constitutively active mutant of MEK1, MEK1EE (4 μg), were transfected. Total amounts of plasmids were 6.5 μg. Normalized amounts of DpnI-resistant plasmid are shown at the bottom. Data represent averages of three experiments with standard errors (SE). Two fragments indicated by asterisks are products of replication intermediates.
We next examined the involvement of the ras-raf1-MEK-ERK signal pathway in oriP regulation (52). Expression of the dominant positive mutant of H-ras, 12Vras, and the constitutively active mutant of MEK1, MEK1EE, were reported to induce activation of ERKs. However, expression of 12Vras and MEK1EE did not suppress oriP activity (Fig. 6B). Thus, activation of the ERK signal pathway did not induce oriP suppression. We also analyzed NF-κB activation by LMP1 mutants and compared it with their ability to induce oriP suppression. LMP1Δ(351-386) and LMP1Δ(212-386) suppressed oriP replication as effectively as the wild type but these mutants did not activate NF-κB (Fig. 3). In contrast, LMP1Δ(187-351) and LMP1(PQT→AAA) suppressed oriP only weakly but they activated NF-κB similarly to the wild-type LMP1. These results indicated that distinct domains of LMP1 induced activation of NF-κB and oriP suppression. Similarly, CTAR2 and the region between CTAR1 and CTAR2 were essential for activation of JNK and Janus kinase 3, respectively (12, 17, 32), but both regions were dispensable for oriP suppression (Fig. 3).
Activation of the TRAF6-mediating signal cascade by LPS resulted in loss of the EBV genome from Akata.
Given the results suggesting that activation of TRAF5- and TRAF6-mediating signal cascades suppresses replication of the oriP plasmid in HeLa/EB1 cells, we next examined whether the same signalings negatively affected EBV replication in the infected cells. To see the suppression of oriP activity, it was essential that the EBV genome was maintained predominantly by the DS-dependent replication from oriP in the infected cells. A Burkitt's lymphoma cell line, Akata, showed the latency type I phenotype and did not express LMP1. In addition, the spontaneous loss of Akata EBV was also reported (54). Therefore it was very likely that Akata EBV was maintained by the DS-dependent replication from oriP. To activate TRAF-mediated signal cascades, we used LPS. It was shown that LPS activated cells through Toll-like receptors, and TRAF6 was the signaling mediator from Toll-like receptors to NF-κB and MAPKs (1, 28, 39, 40). We cultured growing Akata cells (105 cells/ml) in the presence of 10 μg of LPS/ml for 2 or 4 days. To examine the copy number of the Akata EBV genome, total DNA was prepared from these cells and was analyzed by Southern blot hybridization using the same amount of DNA (4 μg) and a BamHI-C fragment for a probe. As shown in Fig. 7A, Akata EBV decreased significantly after LPS stimulation. Quantitative analysis indicated that EBV DNA was decreased by 28% during 4 days of LPS stimulation. In a control experiment, we examined another Burkitt's lymphoma cell line, Raji. Raji EBV DNA was maintained by the replication initiated in a region out of oriP (DS-independent replication) (38), and oriP was not used for replication origin, presumably because the cell expressed LMP1. As we expected, activation of the TRAF6 signal cascade by LPS did not reduce the copy number of Raji EBV, suggesting that LPS stimulation suppressed the oriP activity in Akata. We also examined Daudi EBV replication for another control. Like Akata, Daudi did not express LMP1 but Daudi EBV was replicated by both DS-dependent and DS-independent mechanisms (38). LPS stimulation resulted in only a little loss of Daudi EBV. These results suggested that activation of the TRAF6 signal cascade suppressed EBV replication in the infected cells when EBV was maintained by the DS-dependent replication from oriP. To confirm that p38 MAPK mediated the signal cascade leading to the suppression of Akata EBV replication, Akata cells were stimulated with LPS in the presence of SB208350. As shown in Fig. 7B, when p38 MAPK was inhibited, Akata EBV was not lost by LPS stimulation. We also examined the effects of SB208350 on replication of the EBV genome in infected cells. In contrast to HeLa/EB1, in which initial accumulation of the replicated oriP plasmid was increased by inhibiting the basal activity of p38 MAPK, similar treatment of the EBV-infected cells for 4 days did not increase the copy number of Akata and Raji EBV. Similar results were also obtained with LCLs and AG876 cell lines (data not shown).
FIG. 7.
LPS simulation resulted in loss of Akata EBV. (A) LPS stimulation of EBV-infected B cell lines. Cells (105 cells/ml) were stimulated with 10 μg of LPS/ml for 2 or 4 days. Total DNA of these stimulated and unstimulated cells (4 μg) was digested with BamHI and was analyzed by Southern blot hybridization using an EBV(B95-8) BamHI-C fragment for a probe, which cross-hybridized with BamHI-W (3.1 kb). For the loading controls, EtBr-staining images of agarose gels are shown in the middle panel. Hybridization signals were measured and shown as a relative copy number in the lower panel. Raji and Daudi EBVs were replicated by the DS-independent mechanism (16). (B) The effects of SB208350 on EBV replication. EBV-infected B cell lines (105/ml) were cultured in the presence of the specific inhibitor for p38 MAPK SB208350 (20 μM) with or without LPS stimulation (5 μg/ml) for 4 days. Total DNA (4 μg) was prepared and analyzed as described above for panel A. Hybridization signals of the BamHI-C fragment are shown in the left panels. EtBr-staining images of agarose gels are shown in the right panels.
DISCUSSION
We demonstrated that the replicator activity of oriP was negatively regulated by the TRAF5-mediated signal cascade from LMP1 and the TRAF5 and TRAF6 signal cascades from cellular receptors. This negative regulation was shown in the transient replication assay of the oriP plasmid and was also demonstrated in the analysis of Akata EBV replication.
While the DS element of oriP initiates DNA replication, the FR element of oriP functions as a replication terminator where two replication forks proceeding to opposite directions meet and a round of DNA replication is completed (16). After bidirectional replication is initiated from the DS element, one replication fork proceeds through most of the EBV plasmid, and the other fork proceeds only a short distance, directly to the FR element in the opposite direction. Two-dimensional gel analysis showed that this replication fork, after a short distance, was stopped by a replication fork barrier at the FR element, and the replication intermediates were accumulated (16). In the transient replication assay of the oriP plasmid replication, we found that two DpnI-digested oriP fragments (2.5 and 1.3 kb) were not predicted from the DpnI sites in oriP. Amounts of these fragments are roughly related to that of the DpnI-resistant oriP plasmid. Longer enzyme digestion and use of excess enzymes did not reduce these products, indicating that the fragments were not products of incomplete digestion of DpnI. Furthermore, these fragments were not detected in the samples when the oriP plasmid was transfected into replication-incompetent HeLa cells (55) and the DS plasmid lacking the FR element was transfected into HeLa/EB1 cells (Fig. 1B). From these results, we considered that these DpnI-sensitive fragments are products of replication intermediates that were accumulated by the replication fork barrier at the FR element.
The LMP1 expression required to suppress 90% of oriP activity was almost equal to that in Raji cells. This indicated that oriP activity was sensitive enough to be suppressed by LMP1 expressed in EBV-infected cells. TRAF1, TRAF2, TRAF3, and TRAF5 bind to the PxQxT motif of LMP1 (5, 9, 10, 46, 53). TRAF1 participates in the antiapoptotic activity of LMP1 (6, 60) and does not mediate regulation of oriP. TRAF2 and TRAF5 initiate signal cascades which are overlapped in the activation of NF-κB and JNK. However, only TRAF5 regulates oriP activity through p38 MAPK. Thus, activation of the signal cascade leading to oriP suppression is a TRAF5-specific function. TRAF3 facilitates the function of TRAF5 by binding the TRAF3-TRAF5 hetero-oligomer to LMP1 (51). In addition, TRAF3 may mediate its own signal cascade, because overexpression of TRAF3 reverses the TRAF5-induced oriP suppression. This suggests the importance of balance between TRAF3 and TRAF5 in this signal transduction. TRAF6 binds to CD40, RANK, and p75 NGFR and also associates with IL-1R indirectly. As with TRAF5, overexpression of TRAF6 activates NF-κB, JNK, and p38 MAPK. Our results suggest that p38 MAPK is a common downstream mediator for oriP suppression in the signal cascades activated by LMP1, TRAF5, and TRAF6.
Eliopoulos et al. (13) showed that both CTAR1 and CTAR2 of LMP1 contribute to activation of p38 MAPK. In contrast, our results showed that CTAR1 contributed mostly for oriP suppression and the contribution of CTAR2 was only a part. There are four isozymes of p38 MAPK, p38α, p38β, p38γ, and p38δ. Among these isozymes, p38α and p38β are sensitive to SB203580 and appear to mediate distinct functions (15). A difference in the contribution of CTAR1 and CTAR2 to p38 activation and oriP suppression may be explained by identifying the p38 isozyme that is activated by LMP1 and suppresses oriP activity. The p38 MAPK is activated by phosphorylation by MAPK kinases (MAPKK) MAKK3 and MAKK6. These MAPKKs are activated by a group of MAPKK kinases (MAPKKK). Since MAPKKKs also activate the signal pathways leading to JNK, there may be cross-talk between the signal pathways leading to p38 MAPK and JNK (8). This suggests that the signal pathway initiated from CTAR2 and leading to JNK activation may also contribute to oriP suppression. Our results showed that LMP1 mutants lacking CTAR1 but retaining CTAR2, LMP1Δ(187-351), induced oriP suppression weakly. A mechanism by which p38 MAPK suppresses oriP activity is not yet known. The p38 MAPK may modify EBNA1 directly or indirectly in nuclei.
We confirmed that TRAF-mediated signaling suppressed oriP activity in EBV-infected B cells (Fig. 7). Using EBV-positive Burkitt's lymphoma cell line Akata, we showed that activation of the TRAF6-mediated signal cascade with LPS decreased the copy number of EBV by 28% after stimulation for 4 days (Fig. 7). Loss of 72% of the genome copy during three cell cycles corresponded to 74% suppression of EBV replication in each cell cycle, indicating that suppression with LPS stimulation was significant. The specificity of this suppression of EBV replication was shown by the result that LPS stimulation did not suppress replication of Raji EBV that was maintained by the DS-independent replication. Furthermore, we showed that p38 MAPK was involved in suppression of both EBV and the oriP plasmid replication, suggesting that the same mechanism regulated negatively the oriP activity of Akata EBV and the oriP plasmid in HeLa/EB1 cells. Consistent with these results, spontaneous loss of the EBV genome was observed in the EBV-positive Burkitt's lymphoma cell lines Akata and Mutu (54, 57).
In HeLa/EB1 cells, the p38 MAPK was activated at a low level under normal culture conditions, which was enough to suppress the initial accumulation of the replicated oriP plasmid by 30% (Fig. 5 and 6). This negative pressure imposed on oriP activity may cause constant loss of the oriP plasmid from transfected cells for longer cultures, which was reported in several studies (41, 56, 62). In contrast to HeLa/EB1, inhibition of p38 MAPK by SB208350 under unstimulated conditions did not increase Akata EBV in a short time (4 days). We speculated that the basal activity of p38 MAPK was lower in Akata than in HeLa/EB1 and may suppress Akata EBV replication only slightly in normal culture conditions. This is also consistent with the observation that spontaneous loss of Akata EBV (54) was a relatively slow process compared to LPS-induced loss (Fig. 7A).
Latently infected EBV appears to replicate by two distinct mechanisms, DS-dependent replication and DS-independent replication. The DS-dependent replication is initiated from the DS element of oriP and requires EBNA1 for initiation of DNA replication (62, 64). In contrast, DS-independent replication is initiated in a broad region out of oriP and EBNA1 functions only for maintenance of the EBV chromosome (38). DS-independent replication appears to be performed by cellular replication factors without EBNA1 and is activated only in certain cell lines (unpublished data). DS-dependent and DS-independent mechanisms are not mutually exclusive and occur simultaneously, as is observed in Daudi. Another significant difference in these replication mechanisms is in their sensitivity to LMP1- and TRAF-induced signaling. As we showed in this study, activation of these signal cascades suppressed DS-dependent replication but not DS-independent replication. Based upon this knowledge of EBV replication, it is possible to make several speculations about latent infection of EBV. When latent EBV is replicated mainly by the DS-dependent mechanism, activation of TRAF5 and TRAF6 signal cascades or induction of LMP1 expression suppresses DS-dependent replication and the copy number of the EBV genome in infected cells may decrease. Normal cells infected latently with EBV in vivo are resting memory B cells (43, 44), which are eventually activated by CD4+ T cells. Upon activation, TRAF5- and TRAF6-mediated signaling are initiated from CD40, TNFRII, and IL-1R. Therefore, when the EBV-infected B cell is latency phenotype I (EBNA1-only cells), it is likely that oriP activity is suppressed in activated B cells and EBV may be reduced or eventually lost from activated B cells. In contrast, when latent EBV is maintained predominantly by DS-independent replication, expression of LMP1 does not suppress latent EBV replication. Therefore, this type of infected cell can express LMP1 continuously. In vitro experiments have shown that continuous expression of LMP1 induced immortalization and the transformation phenotype in cultured cells (3, 34, 45, 61). Therefore, activation of DS-independent replication may facilitate lymphoproliferative disorders. It is unknown why some cell lines activate DS-independent replication of EBV but others do not. Because most B cell lines that were infected with EBV in vitro are latency phenotype III and expressing LMP1, activation of DS-independent replication may be related to immortalization of cells.
The DS-independent mechanism of EBV replication is apparently important in establishing the latent infection status in vitro, because LMP1 is expressed in both EBV-infected peripheral blood mononuclear cells and immortalized LCL clones established later. However, EBV can promote cell growth without expression of LMP1 by expressing virus-encoded poly(A)− RNA EBER (36, 37). Interestingly, when EBV-infected cell lines are prepared using normal gastric epithelial cells, the EBV-infected epithelial cells do not express LMP1 (48). Therefore, the DS-dependent replication from oriP may also play an important role during infection and establishing of the latent state in nonlymphoid cells.
ACKNOWLEDGMENTS
We thank E. Kieff for the S12 antibody and cDNA clones of TRAF1, TRAF2, and TRAF3, W. Hammerschmidt and A. Kieser for the LMP1 mutant plasmids, G. Mosialos for the TRAF1 clone, H. Nakano for TRAF5 clones, J. Inoue for TRAF6 clone, and H. Kitayama for ras mutant clones.
This work is supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, Nakanishi K, Akira S. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 2.Aiyar A, Tyree C, Sugden B. The plasmid replicon of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element. EMBO J. 1998;17:6394–6403. doi: 10.1093/emboj/17.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baichwal V R, Sugden B. Transformation of Balb 3T3 cells by the BNLF-1 gene of Epstein-Barr virus. Oncogene. 1988;2:461–467. [PubMed] [Google Scholar]
- 4.Baud V, Liu Z-G, Bennett B, Suzuki N, Xia Y, Karin M. Signaling by proinflammatory cytokines: oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev. 1999;13:1297–1308. doi: 10.1101/gad.13.10.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodeur S R, Cheng G, Baltimore D, Thorly-Lawson D A. Localization of the major NF-κB activating site and the sole TRAF3 binding site of LMP-1 defines two distinct signaling motifs. J Biol Chem. 1997;272:19777–19784. doi: 10.1074/jbc.272.32.19777. [DOI] [PubMed] [Google Scholar]
- 6.Cahir-McFarland E D, Davidson D M, Schauer S L, Duong J, Kieff E. NF-kappa B inhibition causes spontaneous apoptosis in Epstein-Barr virus-transformed lymphoblastoid cells. Proc Natl Acad Sci USA. 2000;97:6055–6060. doi: 10.1073/pnas.100119497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng G, Cleary A M, Ye Z, Hong D I, Lederman S, Baltimore D. Involvement of CRAF1, a relative of TRAF, in CD40 signaling. Science. 1995;267:1494–1498. doi: 10.1126/science.7533327. [DOI] [PubMed] [Google Scholar]
- 8.Davis R J. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 9.Devergne O, Hatzivassiliou E, Izumi K M, Kaye K M, Kleijnen M F, Kieff E, Mosialos G. Role of the TRAF binding site and NF-κB activation in Epstein-Barr virus latent membrane protein 1-induced cell gene expression. Mol Cell Biol. 1996;16:7098–7108. [Google Scholar]
- 10.Devergne O, McFarland E C, Mosialos G, Izumi K M, Ware C F, Kieff E. Role of the TRAF binding site and NF-κB activation in Epstein-Barr virus latent membrane protein 1-induced cell gene expression. J Virol. 1998;72:7900–7908. doi: 10.1128/jvi.72.10.7900-7908.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eliopoulos A G, Dawson C W, Mosialos G, Floettmann J E, Rowe M, Armitage R J, Dawson J, Zapata J M, Kerr D J, Wakelam M O J, Reed J C, Kieff E, Young L S. CD40-induced growth inhibition in epithelial cells is mimicked by Epstein-Barr virus-encoded LMP1: involvement of TRAF3 as a common mediator. Oncogene. 1996;13:2243–2254. [PubMed] [Google Scholar]
- 12.Eliopoulos A G, Young L S. Activation of the c-Jun N-terminal kinase (JNK) pathway by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) Oncogene. 1998;16:1731–1742. doi: 10.1038/sj.onc.1201694. [DOI] [PubMed] [Google Scholar]
- 13.Eliopoulos A G, Gallagher N J, Blake S M S, Dawson C W, Young L S. Activation of the p38 mitogen-activated protein kinase pathway by Epstein-Barr virus-encoded latent membrane protein 1 coregulates interleukin-6 and interleukin-8 production. J Biol Chem. 1999;274:16085–16096. doi: 10.1074/jbc.274.23.16085. [DOI] [PubMed] [Google Scholar]
- 14.Eliopoulos A G, Blake S M S, Floettmann J E, Rowe M, Young L S. Epstein-Barr virus-encoded latent membrane protein 1 activates the JNK pathway through its extreme C terminus via a mechanism involving TRADD and TRAF2. J Virol. 1999;73:1023–1035. doi: 10.1128/jvi.73.2.1023-1035.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enslen H, Brancho D M, Davis R J. Molecular determinants that mediate selective activation of p38 MAP kinase isoforms. EMBO J. 2000;19:1301–1311. doi: 10.1093/emboj/19.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gahn T A, Schildkraut C L. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989;58:527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- 17.Gires O, Kohlhuber F, Kilger E, Baumann M, Kieser A, Kaiser C, Zeidler R, Scheffer B, Ueffing M, Hammerschmidt W. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. EMBO J. 1999;18:3064–3073. doi: 10.1093/emboj/18.11.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammerschmidt W, Sugden B, Baichwal V. The transforming domain of the latent membrane protein of Epstein-Barr virus is toxic to cells when expressed at high levels. J Virol. 1996;63:2469–2475. doi: 10.1128/jvi.63.6.2469-2475.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J, Lee J D, Bibbs L, Ulevitch R J. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science. 1994;265:808–811. doi: 10.1126/science.7914033. [DOI] [PubMed] [Google Scholar]
- 20.Harrison S, Fisenne K, Hearing J. Sequence requirements of the Epstein-Barr virus latent origin of DNA replication. J Virol. 1994;68:1913–1925. doi: 10.1128/jvi.68.3.1913-1925.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirai K, Shirakata M. Replication licensing of the EBV oriP minichromosome. Curr Top Microbiol Immunol. 2001;258:13–33. doi: 10.1007/978-3-642-56515-1_2. [DOI] [PubMed] [Google Scholar]
- 22.Huen D S, Henderson S A, Croom C D, Rowe M. The Epstein-Barr virus latent membrane protein-1 (LMP1) mediates activation of NF-κB and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene. 1995;10:549–560. [PubMed] [Google Scholar]
- 23.Ishida T, Mizushima S I, Azuma S, Kobayashi N, Tojo T, Suzuki K, Aizawa S, Watanabe T, Mosialos G, Kieff E, Yamamoto T, Inoue J. Identification of TRAF6, a novel tumor necrosis factor receptor-associated factor protein that mediates signaling from an amino-terminal domain of the CD40 cytoplasmic region. J Biol Chem. 1996;271:28745–28748. doi: 10.1074/jbc.271.46.28745. [DOI] [PubMed] [Google Scholar]
- 24.Izumi K M, Kieff E D. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activates NF-κB. Proc Natl Acad Sci USA. 1997;94:12529–12597. doi: 10.1073/pnas.94.23.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Izumi K M, McFarland E C, Ting A T, Riley E A, Seed B, Kieff E D. The Epstein-Barr virus oncoprotein latent membrane protein 1 engages the tumor necrosis factor receptor-associated proteins TRADD and receptor-interacting protein (RIP) but does not induce apoptosis or require RIP for NF-κB activation. Mol Cell Biol. 1999;19:5759–5767. doi: 10.1128/mcb.19.8.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jankelevich S, Kolman J L, Bodnar J W, Miller G. A nuclear matrix attachment region organizes the Epstein-Barr viral plasmid in Raji cells into a single DNA domain. EMBO J. 1992;11:1165–1176. doi: 10.1002/j.1460-2075.1992.tb05157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kashiwada M, Shirakata Y, Inoue J, Nakano H, Okazaki K, Okumura K, Yamamoto T, Nagaoka H, Takemori T. Tumor necrosis factor receptor-associated factor 6 (TRAF6) stimulates extracellular signal regulated kinase (ERK) activity in CD40 signaling along a ras-independent pathway. J Exp Med. 1998;187:237–244. doi: 10.1084/jem.187.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 1999;11:115–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- 29.Kaye K M, Izumi K M, Kieff E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaykas A, Sugden B. The amino-terminus and membrane-spanning domain of LMP-1 inhibit cell proliferation. Oncogene. 2000;19:1400–1410. doi: 10.1038/sj.onc.1203365. [DOI] [PubMed] [Google Scholar]
- 31.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, Chanock P M, Monath T P, Melnick J L, Roizman B, Straus S E, editors. Virology. Philadelphia, Pa: Raven; 1996. pp. 2343–2396. [Google Scholar]
- 32.Kieser A, Kilger E, Gires O, Ueffing M, Kolch W, Hammerschmidt W. Epstein-Barr virus latent membrane protein-1 triggers AP-1 activity via the c-Jun N-terminal kinase cascade. EMBO J. 1997;16:6478–6485. doi: 10.1093/emboj/16.21.6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kieser A, Kaiser C, Hammerschmidt W. LMP1 signal transduction differs substantially from TNF receptor 1 signaling in the molecular functions of TRADD and TRAF2. EMBO J. 1999;18:2511–2521. doi: 10.1093/emboj/18.9.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kilger E, Kieser A, Baumann M, Hammerschmidt W. Epstein-Barr virus-mediated B-cell proliferation is dependent upon latent membrane protein 1, which simulates an activated CD40 receptor. EMBO J. 1998;17:1700–1709. doi: 10.1093/emboj/17.6.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirchmaier A L, Sugden B. Rep∗: a viral element that can partially replace the origin of plasmid DNA synthesis of Epstein-Barr virus. J Virol. 1998;72:4657–4666. doi: 10.1128/jvi.72.6.4657-4666.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitagawa, N., M. Goto, K. Kurozumi, S. Maruo, M. Fukayama, T. Naoe, M. Yasukawa, K. Hino, T. Suzuki, S. Todo, and K. Takada. Epstein-Barr virus-encoded poly(A)− RNA supports Burkitt's lymphoma growth through interleukin-10. EMBO J. 19:6742–6750. [DOI] [PMC free article] [PubMed]
- 37.Komano J, Moruo S, Koizumi K, Oda T, Takada K. Oncogenic role of Epstein-Barr virus-encoded RNAs in Burkitt's lymphoma cell line Akata. J Virol. 1999;73:9827–9831. doi: 10.1128/jvi.73.12.9827-9831.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Little R, Schildkraut C. Initiation of latent DNA replication in the Epstein-Barr virus genome can occur at sites other than the genetically defined origin. Mol Cell Biol. 1995;15:2893–2903. doi: 10.1128/mcb.15.5.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lomaga M A, Yeh W-C, Sarosi I, Duncan G S, Furlonger C, Ho A, Morony S, Capparelli C, Van G, Kafman S, van der Heiden A, Itie A, Wakeham A, Khoo W, Sasaki T, Cao Z, Penninger J M, Paige C J, Lacey D L, Dunstan C R, Boyle W J, Goeddel D V, Mak T W. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 1999;13:1015–1024. doi: 10.1101/gad.13.8.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Medzhitov R, Preston-Hurlburt P, Kopp E, Stadlen A, Chen C, Ghosh S, Janeway C A., Jr MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol Cell. 1998;2:253–258. doi: 10.1016/s1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- 41.Middleton T, Sugden B. Retention of plasmid DNA in mammalian cells is enhanced by binding of the Epstein-Barr virus replication protein. J Virol. 1994;68:4067–4071. doi: 10.1128/jvi.68.6.4067-4071.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell T, Sugden B. Stimulation of NF-κB-mediated transcription by mutant derivatives of the latent membrane protein of Epstein-Barr virus. J Virol. 1995;69:2968–2976. doi: 10.1128/jvi.69.5.2968-2976.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyashita E M, Yang B, Lam K M, Crawford D H, Thorley-Lawson D A. A novel form of Epstein-Barr virus latency in normal B cells in vivo. Cell. 1995;80:593–601. doi: 10.1016/0092-8674(95)90513-8. [DOI] [PubMed] [Google Scholar]
- 44.Miyashita E M, Yang B, Babcock B J, Thorley-Lawson D A. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. J Virol. 1997;71:4882–4891. doi: 10.1128/jvi.71.7.4882-4891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moorthy R K, Thorley-Lawson D A. All three domains of the Epstein-Barr virus-encoded latent membrane protein LMP-1 are required for transformation of rat-1 fibroblasts. J Virol. 1993;67:1638–1646. doi: 10.1128/jvi.67.3.1638-1646.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 47.Nakano H, Oshima H, Chung W, Williams A L, Ware C F, Yagita H, Okumura K. TRAF5, an activator of NF-kappaB and putative signal transducer for the lymphotoxin-beta receptor. J Biol Chem. 1996;271:14661–14664. doi: 10.1074/jbc.271.25.14661. [DOI] [PubMed] [Google Scholar]
- 48.Nishikawa J, Imai S, Oda T, Kojima T, Okita K, Takada K. Epstein-Barr virus promotes epithelial cell growth in the absence of EBNA2 and LMP1 expression. J Virol. 1999;73:1286–1292. doi: 10.1128/jvi.73.2.1286-1292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Norio P, Schildkraut C L, Yates J L. Initiation of DNA replication within oriP is dispensable for stable replication of the latent Epstein-Barr virus chromosome after infection of established cell lines. J Virol. 2000;74:8563–8574. doi: 10.1128/jvi.74.18.8563-8574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okazaki K, Sagata N. MAP kinase activation is essential for oncogenic transformation of NIH3T3 cells by Mos. Oncogene. 1995;10:1149–1157. [PubMed] [Google Scholar]
- 51.Pullen S S, Miller H G, Everdeen D S, Dang T T A, Crute J J, Kehry M R. CD40-tumor necrosis factor receptor-associated factor (TRAF) interactions: regulation of CD40 signaling through multiple TRAF binding sites and TRAF hetero-oligomerization. Biochemistry. 1998;37:11836–11845. doi: 10.1021/bi981067q. [DOI] [PubMed] [Google Scholar]
- 52.Roberts M L, Cooper N R. Activation of a ras-MAPK-dependent pathway by Epstein-Barr virus latent membrane protein 1 is essential for cellular transformation. Virology. 1998;240:93–99. doi: 10.1006/viro.1997.8901. [DOI] [PubMed] [Google Scholar]
- 53.Sandberg M, Hammerschmidt W, Sugden B. Characterization of LMP-1's association with TRAF1, TRAF2, and TRAF3. J Virol. 1997;71:4649–4656. doi: 10.1128/jvi.71.6.4649-4656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shimizu N, Tanabe-Tochikura A, Kuroiwa Y, Takada K. Isolation of Epstein-Barr virus (EBV)-negative cell clones from the EBV-positive Burkitt's lymphoma (BL) line Akata: malignant phenotypes of BL cells are dependent on EBV. J Virol. 1994;68:6069–6073. doi: 10.1128/jvi.68.9.6069-6073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shirakata M, Hirai K. Identification of minimal oriP of Epstein-Barr virus required for DNA replication. J Biochem. 1998;123:175–181. doi: 10.1093/oxfordjournals.jbchem.a021907. [DOI] [PubMed] [Google Scholar]
- 56.Shirakata M, Imadome K, Hirai K. Requirement of replication licensing for the dyad symmetry element-dependent replication of the Epstein-Barr virus oriP minichromosome. Virology. 1999;263:42–54. doi: 10.1006/viro.1999.9965. [DOI] [PubMed] [Google Scholar]
- 57.Srinivas S K, Sample J T, Sixbey J W. Spontaneous loss of viral episomes accompanying Epstein-Barr virus reactivation in a Burkitt's lymphoma cell line. J Infect Dis. 1998;177:1705–1709. doi: 10.1086/517427. [DOI] [PubMed] [Google Scholar]
- 58.Sylla B S, Hung S C, Davidson D M, Hatzivassiliou E, Malinin N, Wallach D, Gilmore T D, Kieff E, Mosialos G. Epstein-Barr virus-transforming protein latent infection membrane protein 1 activates transcription factor NF-κB through a pathway that includes the NF-κB-inducing kinase and the IκB kinases IKKα and IKKβ. Proc Natl Acad Sci USA. 1998;95:10106–10111. doi: 10.1073/pnas.95.17.10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takanashi M, Li J, Shirakata M, Mori S, Hirai K. Tumorigenicity of mouse BALB/c 3T3 fibroblast cells which express Epstein-Barr virus-encoded LMP1 and show normal growth phenotypes in vitro is correlated with loss of transforming growth factor-β1-mediated growth inhibition. Arch Virol. 1999;144:241–257. doi: 10.1007/s007050050501. [DOI] [PubMed] [Google Scholar]
- 60.Wang C, Mayo M W, Korneluk R G, Goedel D V, Baldwin A S., Jr NF-κB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 1998;281:1680–1683. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- 61.Wang D, Liebowitz D, Kieff E. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell. 1985;43:831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- 62.Yates J L, Wallen N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 63.Yates J L, Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol. 1991;65:483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yates J L. Epstein-Barr virus DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 751–773. [Google Scholar]
- 65.Yates J L, Camiolo S M, Bashaw J M. The minimal replicator of Epstein-Barr virus oriP. J Virol. 2000;74:4512–4522. doi: 10.1128/jvi.74.10.4512-4522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]