Abstract
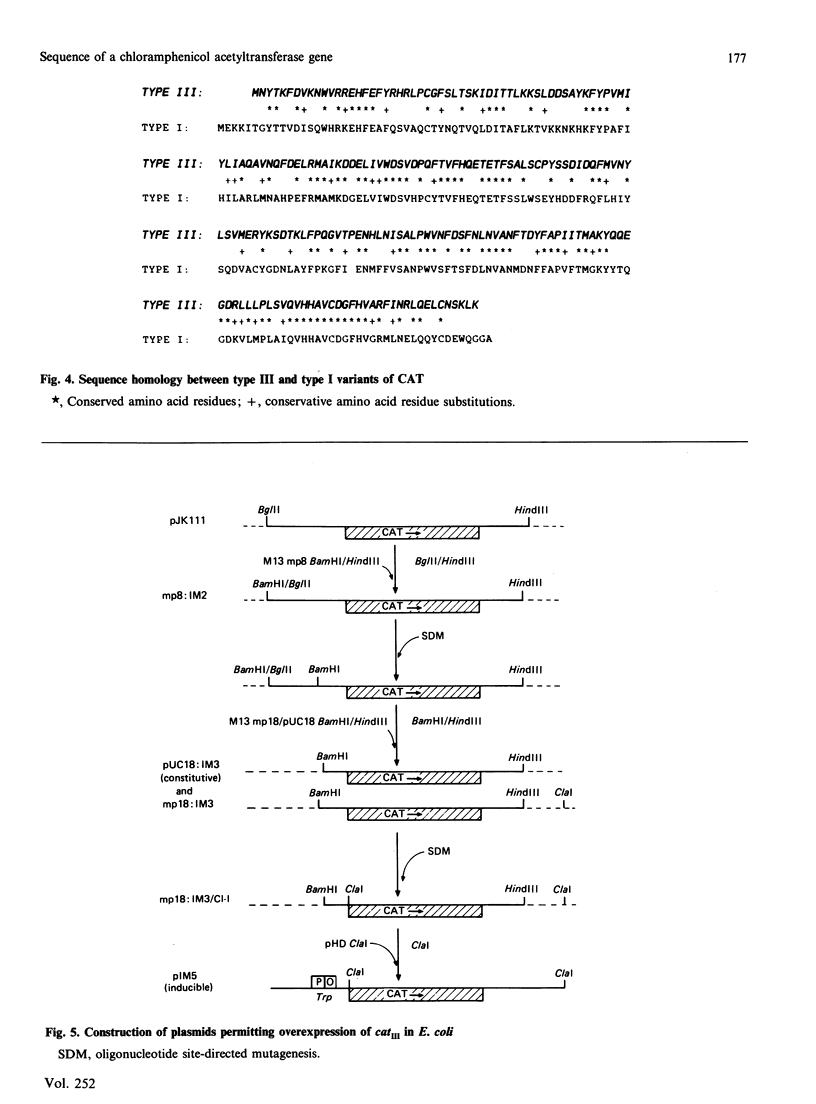
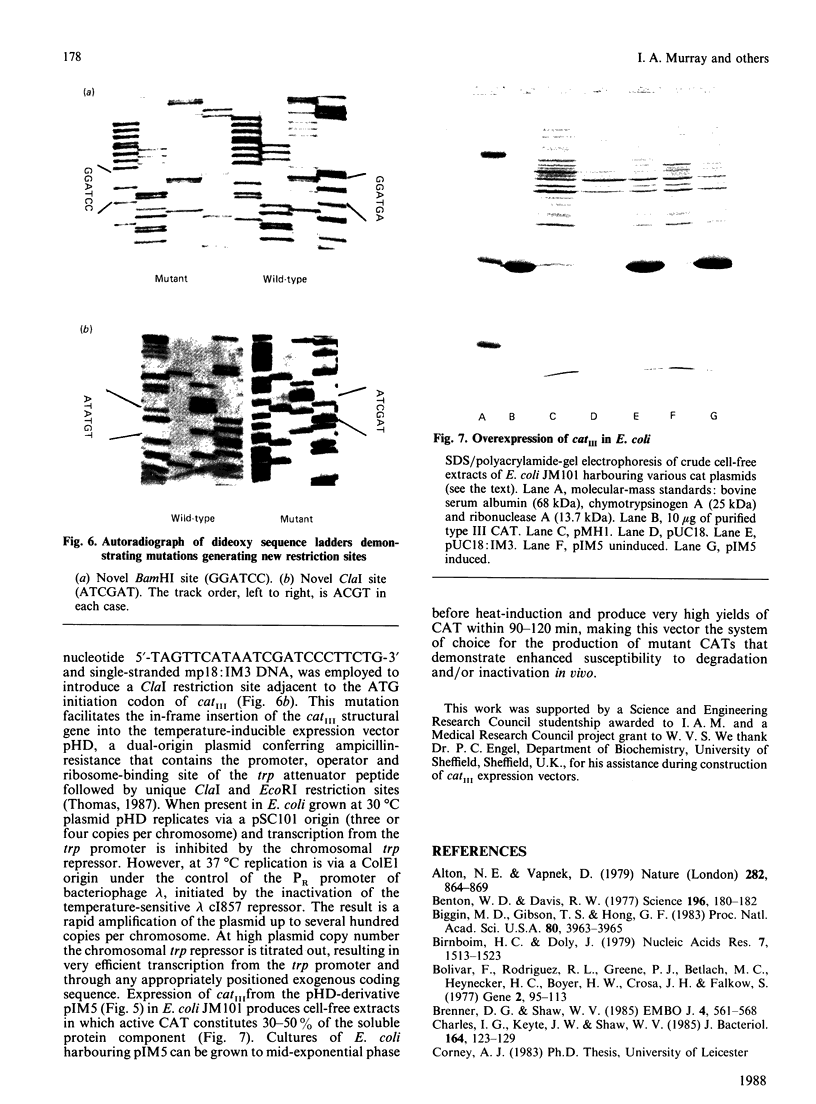
The gene catIII, encoding a type III enterobacterial chloramphenicol acetyltransferase, was cloned from the transmissible plasmid R387 into pBR322 and bacteriophage M13 mp8. Nucleotide sequence analysis of 1160 bp of DNA identified an open reading frame encoding a protein of 213 amino acid residues and a calculated molecular mass of 24965 Da. The predicted N-terminal sequence is identical with that determined by Edman degradation of chloramphenicol acetyltransferase purified from Escherichia coli harbouring R387. Sequences equivalent to the consensus motifs for initiation and rho-factor-independent termination of transcription in E. coli occur 5' and 3' to the catIII open reading frame. In contrast with the catI gene, present on transposon Tn9 and many enterobacterial plasmids, expression of catIII is not subject to cyclic AMP-mediated catabolite repression in vivo and there is no sequence in the 5' non-coding DNA that resembles that deduced as the consensus for the binding of cyclic AMP receptor protein. Unique restriction-endonuclease cleavage sites were introduced adjacent to the catIII reading frame by using oligonucleotide-directed mutagenesis to facilitate insertion into E. coli expression vectors. Fully active chloramphenicol acetyltransferase represents 30-50% of the soluble protein component of cell-free extracts of E. coli containing the appropriate plasmids.
Full text
PDF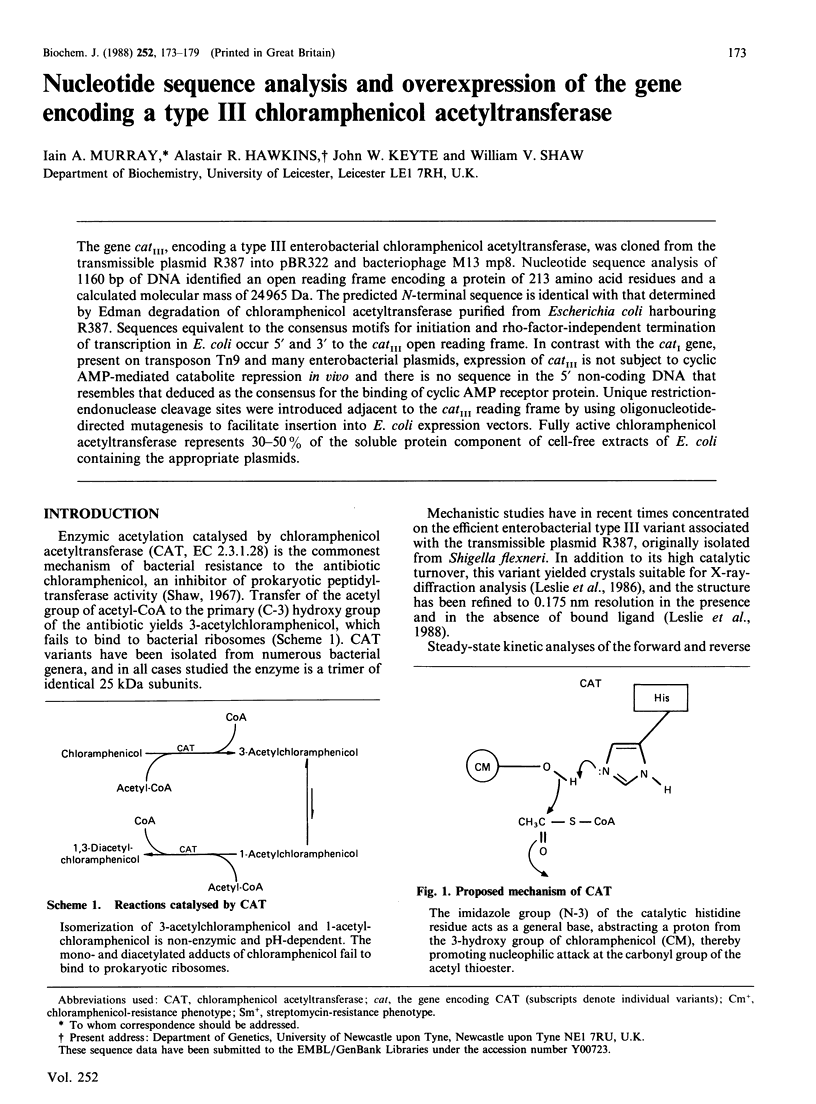




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alton N. K., Vapnek D. Nucleotide sequence analysis of the chloramphenicol resistance transposon Tn9. Nature. 1979 Dec 20;282(5741):864–869. doi: 10.1038/282864a0. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Brenner D. G., Shaw W. V. The use of synthetic oligonucleotides with universal templates for rapid DNA sequencing: results with staphylococcal replicon pC221. EMBO J. 1985 Feb;4(2):561–568. doi: 10.1002/j.1460-2075.1985.tb03665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles I. G., Keyte J. W., Shaw W. V. Nucleotide sequence analysis of the cat gene of Proteus mirabilis: comparison with the type I (Tn9) cat gene. J Bacteriol. 1985 Oct;164(1):123–129. doi: 10.1128/jb.164.1.123-129.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood C. R., Williams D. M., Lovett P. S. Nucleotide sequence of a Bacillus pumilus gene specifying chloramphenicol acetyltransferase. Gene. 1983 Oct;24(2-3):163–169. doi: 10.1016/0378-1119(83)90076-8. [DOI] [PubMed] [Google Scholar]
- Harwood J., Smith D. H. Catabolite repression of chloramphenicol acetyl transferase synthesis in E. coli K12. Biochem Biophys Res Commun. 1971 Jan 8;42(1):57–62. doi: 10.1016/0006-291x(71)90361-5. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleanthous C., Cullis P. M., Shaw W. V. 3-(Bromoacetyl)chloramphenicol, an active site directed inhibitor for chloramphenicol acetyltransferase. Biochemistry. 1985 Sep 24;24(20):5307–5313. doi: 10.1021/bi00341a006. [DOI] [PubMed] [Google Scholar]
- Kleanthous C., Shaw W. V. Analysis of the mechanism of chloramphenicol acetyltransferase by steady-state kinetics. Evidence for a ternary-complex mechanism. Biochem J. 1984 Oct 1;223(1):211–220. doi: 10.1042/bj2230211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Le Grice S. F., Matzura H., Marcoli R., Iida S., Bickle T. A. The catabolite-sensitive promoter for the chloramphenicol acetyl transferase gene is preceded by two binding sites for the catabolite gene activator protein. J Bacteriol. 1982 Apr;150(1):312–318. doi: 10.1128/jb.150.1.312-318.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie A. G., Liddell J. M., Shaw W. V. Crystallization of a type III chloramphenicol acetyl transferase. J Mol Biol. 1986 Mar 20;188(2):283–285. doi: 10.1016/0022-2836(86)90310-4. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y., Bender M. L. Methylation of histidine-57 in alpha-chymotrypsin by methyl p-nitrobenzenesulfonate. A new approach to enzyme modification. Biochemistry. 1970 Jan 20;9(2):259–267. doi: 10.1021/bi00804a011. [DOI] [PubMed] [Google Scholar]
- Packman L. C., Shaw W. V. Identification of "buried" lysine residues in two variants of chloramphenicol acetyltransferase specified by R-factors. Biochem J. 1981 Feb 1;193(2):525–539. doi: 10.1042/bj1930525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribnow D. Nucleotide sequence of an RNA polymerase binding site at an early T7 promoter. Proc Natl Acad Sci U S A. 1975 Mar;72(3):784–788. doi: 10.1073/pnas.72.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. V., Brenner D. G., LeGrice S. F., Skinner S. E., Hawkins A. R. Chloramphenicol acetyltransferase gene of staphylococcal plasmid pC221. Nucleotide sequence analysis and expression studies. FEBS Lett. 1985 Jan 1;179(1):101–106. doi: 10.1016/0014-5793(85)80200-3. [DOI] [PubMed] [Google Scholar]
- Shaw W. V., Sands L. C., Datta N. Hybridization of variants of chloramphenicol acetyltransferase specified by fi + and fi - R factors. Proc Natl Acad Sci U S A. 1972 Oct;69(10):3049–3053. doi: 10.1073/pnas.69.10.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. V. The enzymatic acetylation of chloramphenicol by extracts of R factor-resistant Escherichia coli. J Biol Chem. 1967 Feb 25;242(4):687–693. [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Siebenlist U. RNA polymerase unwinds an 11-base pair segment of a phage T7 promoter. Nature. 1979 Jun 14;279(5714):651–652. doi: 10.1038/279651a0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis: a simple method using two oligonucleotide primers and a single-stranded DNA template. DNA. 1984 Dec;3(6):479–488. doi: 10.1089/dna.1.1984.3.479. [DOI] [PubMed] [Google Scholar]




