Abstract
It is important to recognize the great diversity of monosaccharides commonly encountered in animals, plants, and microbes, as well as to organize them in a visually interesting style that also emphasizes their similarities and relatedness. This article discusses the nature of building blocks, monosaccharides, and monosaccharide derivatives—terms commonly used in discussing “glycomolecules” found in nature. To aid in awareness of monosaccharide diversity, here is presented a Periodic Table of Monosaccharides. The rationale is given for construction of the Table and the selection of 103 monosaccharides, which is largely based on those presented in the KEGG and SNFG websites of monosaccharides, and includes room to enlarge as new discoveries are made. The Table should have educational value and is intended to capture the attention and foster imagination of those not very familiar with glycosciences, and encourage researchers to delve deeper into this fascinating area.
Keywords: building block, derivative, monosaccharide, periodic table
Introduction
The building blocks of nature is a term often used to describe the precursors to life’s macromolecules. For nucleic acids, proteins, and lipids there are limited numbers, with 8 nucleotides for nucleic acids, 20–22 amino acids for proteins, along with several categories of lipid precursors (Marth 2008). For glycomolecules, which is any molecule in which carbohydrate building blocks are linked, the situation is more complicated (Herget et al. 2008; Marth 2008; Imperiali 2019; Seeberger 2022). The building blocks are termed monosaccharides but there are also monosaccharide derivatives that occur within glycomolecules as discrete, basic units with specific linkages. The term monosaccharide is derived from the Greek monos (single) and sacchar (sugar). Unlike the single type of peptide bond that links amino acids to each other, monosaccharides may have various linkages, including O-glycosidic, N-glycosidic, S-glycosidic, and C-glycosidic bonds (Varki et al. 2022). Remarkably, monosaccharide building blocks and their derivatives found within glycomolecules number in the hundreds, if not perhaps thousands, and unbelievably, not all of them are known (Imperiali 2019). But the terms building block, monosaccharide and monosaccharide derivative are not as clearly distinguished as one might imagine.
Features of Monosaccharides
Carbohydrates are often said to have a basic formula Cx(H2O)y, e.g. glucose (Glc) has the formula C6(H2O)6, also written as C6H12O6. But this chemical formula applies only to a small set of simple monosaccharides and does not apply to all monosaccharides, as many have a much more complicated formula. The textbook Essentials of Glycobiology states that “…all monosaccharides consist of a chain of chiral hydroxymethylene units, which terminates at one end with a hydroxymethyl group and at the other with either an aldehyde group (aldoses) or an α-hydroxy ketone group (ketoses)” (Seeberger 2022), which is a definition used here. The IUPAC nomenclature states that a monosaccharide is a poly(hydroxy) aldehyde or ketone with three or more carbon atoms (triose, tetrose, etc.), and can include a variety of derivatives such as amino, deoxy or carboxy forms (McNaught 1997) (Fig. 1). These two definitions would also appear to include monosaccharides and their derivatives. The Merriam Webster dictionary, however, defines a monosaccharide as a molecule that is not decomposable into a simpler monosaccharide by hydrolysis. Thus, there appear to be several definitions of monosaccharides. A separate nomenclature issue is the word sugar, which generally refers to simple monosaccharides or small-sized combinations of them, often with the implication that they have a sweet taste, which may be true for many monosaccharides, but not all have been tested. Sugar sweetness is usually defined by people, but sweetness depends on the organism tasting them. For example, cats lack expression of functional sweet taste receptors (Li et al. 2006).
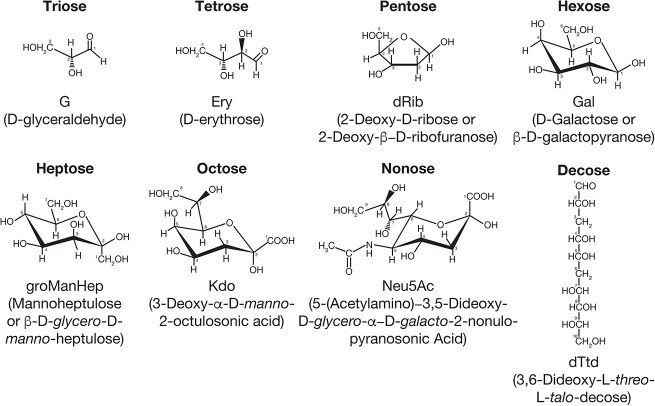
Fig. 1.
Examples of monosaccharides from triose to decose in terms of the number of carbons in their backbone structures, indicated. Many are depicted as in their ring structures as chair conformers and the decose is depicted as a straight chain.
While the definitions of monosaccharides or sugars may vary, their chemical nature is well known. Glucose has 6 carbons in its “backbone” and is a hexasaccharide, termed a hexose. As it can occur as an aldehyde, it is also termed an aldose (in biochemistry -ose refers to sugars). Fructose is an isomer of glucose and has the same chemical formula, but since it is a ketone, it is termed a ketose. N-acetylneuraminic (Neu5Ac) has the formula C11H19NO9, and although it has 11 carbons, its backbone structure has 9 carbons (2 carbons are in the acetyl derivative part), and hence, is termed a nonose. Neu5Ac is also defined as a nonulosonic acid (NulO) and is a member of a large family of 9-carbon α-keto acid sugars (Lewis et al. 2022; Fontana and Widmalm 2023). Thus, there is no single general chemical formula for any of the known building blocks of nature, including those for nucleic acids, proteins, lipids, or monosaccharides.
Interestingly, the monosaccharide D-glyceraldehyde, which is a triose and an aldose, is not a building block, i.e. it is not found directly in glycosidic linkage to other components within a larger macromolecule. However, D-glyceraldehyde holds a special place in the history of glycoscience, as in current nomenclature, all monosaccharide configurations are compared to that of D-glyceraldehyde in relation to the stereogenic carbon atom farthest from the carbonyl group (McNaught 1997), a convention arising from Fischer’s early discoveries about sugar enantiomers (D and L) (Jackson 2017). [The designation of D and L is historical but still useful to identify the enantiomers, e.g. D-glucose and L-glucose are non-superimposable mirror images, and allows us to avoid writing out IUPAC names including the stereochemical descriptors (R) and (S).]
A monosaccharide found within macromolecules is often different from the monosaccharide that was initially incorporated into it, i.e. it may occur in the mature product as a monosaccharide derivative. In the biosynthesis of glycomolecules, a common donor is a nucleotide sugar derivative, e.g. UDP-Glc, UDP-GlcNAc, GDP-Man, CMP-Neu5Ac, etc. (Mikkola 2020) (monosaccharide abbreviations are presented in Figs 1 and 2). Interestingly, one key building block, iduronic acid (IdoA), does not occur as a nucleotide sugar. Rather, when D-glucuronic acid (GlcA) is incorporated from the donor UDP-GlcA into glycosaminoglycans, some D-GlcA residues are enzymatically converted to L-IdoA within the molecule (Lindahl et al. 1976). This conversion occurs in vertebrate D-GlcA acids in glycosaminoglycans, and also microbial polysaccharides (Raedts et al. 2011). L-IdoA may be further modified, e.g. to 2-sulfo-L-IdoA (Larnkjaer et al. 1995). Thus, while IdoA is a monosaccharide and building block, 2-sulfo-L-IdoA is a monosaccharide derivative. In fact, there are many sulfated monosaccharide building blocks in nature, including derivatives of GlcA, IdoA, glucosamine (GlcN), N-acetylglucosamine (GlcNAc) and galactose (Gal), e.g. 6-sulfo-N-sulfo-D-GlcN, 6-sulfo-D-GlcNAc, 3-sulfo-D-Gal, and 6-sulfo-D-Gal.
Fig. 2.
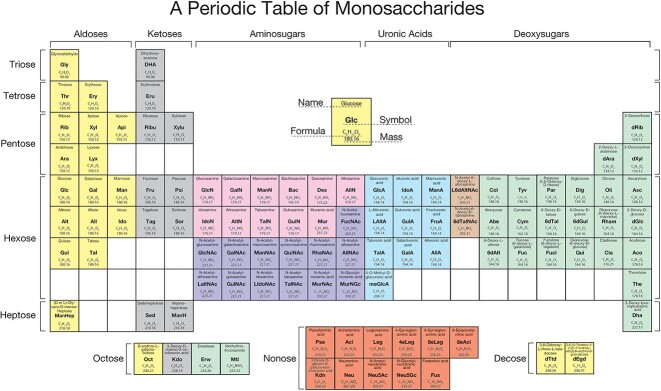
The monosaccharides and monosaccharide derivatives are depicted in a tabular form are the periodic table of monosaccharides. The groups of sugars are indicated across the top, and the periods of sugars based on the number of carbons in their backbone structure are indicated on the left side. For information about the groups and periods see the text. Each sugar is depicted with its name (in some cases the trivial names and in other cases more complete chemical names), along with an abbreviation for the sugar, its formula, and its average molar mass, as computed by either PubChem 2.1 or the molecular weight calculator at https://www.lenntech.com/calculators/molecular/molecular-weight-calculator.htm. As noted in the text, there is also a symbol nomenclature associated with many of these sugars, that may be accessed at the indicated sites. For the decoses, there do not yet appear to be abbreviations for the two here, so liberty was taken to designate them as dTtd and dEgd, respectively, for the uniformity of the table. Most if not all of the monosaccharides here are accessible at PubChem. There are also additional tools, bioinformatic resources, and repositories available to draw glycan structures and incorporate monosaccharides and their derivatives. A sampling of these resources can be found here (Campbell et al. 2014; Perez et al. 2015; Thieker et al. 2016; Tsuchiya et al. 2019; Kahsay et al. 2020; Lal et al. 2020; Mehta and Cummings 2020; York et al. 2020; Fujita et al. 2021; Cheng et al. 2023).
Such modifications of monosaccharides, especially when considered in regard to the conventional dictionary definitions of a monosaccharide, further blur the definitions of monosaccharides versus building blocks. For example, neither glucose nor glucosamine can be hydrolyzed to generate another monosaccharide; GlcNAc, however, can be deacetylated to generate glucosamine (Hackman 1954; Jiang et al. 2018), yet both are typically considered monosaccharides. In terms of monosaccharide derivatives, consider 9-O-acetyl-Neu5Ac, 4-O-acetyl-Neu5Ac, mannose-6-phosphate, heptose-4-phosphate, and xylose-2-phosphate [for the discussion below see references (Varki and Kornfeld 1980; Reitman and Kornfeld 1981; Yethon and Whitfield 2001; Iwersen et al. 2003; Yoshida-Moriguchi et al. 2013; Wen et al. 2014; Mandal et al. 2015; Zhu et al. 2016; Schauer and Kamerling 2018; Visser et al. 2021; Lewis et al. 2022; Lewis et al. 2023)]. Acetylation can occur on Neu5Ac within the donor nucleotide sugar, e.g. CMP-9-O-acetyl-Neu5Ac, by the acetyltransferase CASD1 that uses acetyl-CoA, which is imported into the Golgi apparatus; the acetyl group can migrate to generate 7-O-acetyl-Neu5Ac. However, 4-O-Acetyl-Neu5Ac is probably generated by a 4-O-acetyltransferase acting on Neu5Ac within glycans. Interestingly, 9-O-acetyl-Neu5Ac can be hydrolyzed to generate Neu5Ac. It is also interesting that there are many enzymes that can remove modifications of monosaccharides within mature glycans, e.g. the sulfatases (Sulfs) cause desulfation of 6-O-sulfated residues within heparan sulfate (HS) (Vives et al. 2014), and N-deacetylases/N-sulfotransferase can both deacetylate and sulfate amino groups on GlcNAc residues within HS (Sarrazin et al. 2011).
Oddly, by the definitions above, 9-O-acetyl-Neu5Ac, while a monosaccharide building block, might not be considered a monosaccharide. Similarly, mannose-6-phosphate is generated by phosphorylation of mannose within a glycan, and mannose-6-phosphate can be hydrolyzed to generate mannose. The generation of mannose-6-phosphate on different glycoproteins can occur either by donation of GlcNAc-1-P from UDP-GlcNAc to the 6-position of mannose on glycans of lysosomal hydrolases, or through an ATP-dependent kinase to phosphorylate mannose, as occurs within α-dystroglycan. Similarly, the lipopolysaccharide (LPS) core heptose(I) kinase RfaP adds phosphate to the O-4 position of the first heptose residue of the LPS inner core region. Lastly, the initiating xylose residue in the proteoglycan tetrasaccharide linkage region is phosphorylated by Fam20B using ATP. Altogether, we may consider some modifications of sialic acid as “biosynthetic” and that of mannose, heptose, and xylose as “post-biosynthetic”, i.e. after the monosaccharide is incorporated. These are some confusing terms, and alternatives could simply be “glycosylation” and “post-glycosylation”, respectively (Park 2019).
Thus, in broadly considering the definitions of the terms building blocks, monosaccharides, and monosaccharide derivatives, we may conclude that a building block is a monosaccharide or a monosaccharide derivative; in some cases, a monosaccharide or monosaccharide derivative may be a “precursor” and directly added to generate the final molecule, but in other cases the monosaccharide may be modified afterward. Thus, the number of building blocks and monosaccharide derivatives exceeds the number of monosaccharides.
The discussion above raises the important question, how many building blocks are there for the glycomolecules in nature? Two excellent reviews, by Marth (Marth 2008), and Herget et al., (Herget et al. 2008), tackled these issues. Marth recognized 32 monosaccharide building blocks, whereas Herget, et al estimated that the number of monosaccharides in nature is probably much larger—predicting 551 monosaccharides in bacteria alone. Many monosaccharides have been identified in surface polysaccharides of just the O and K antigens in E. coli (Dong et al. 2023).
A useful listing of monosaccharides provided by KEGG (Kyoto Encyclopedia of Gene and Genomes) systems biology lists ~100 monosaccharides that have been identified in pathways in many different organisms—see monosaccharide codes at https://www.genome.jp/kegg/catalog/codes2.html). The SNFG (Symbol Nomenclature for Glycans) at https://www.ncbi.nlm.nih.gov/glycans/snfg.html recognizes ~80 monosaccharides (Neelamegham et al. 2019), overlapping with KEGG. These sites also have the preferred/accepted symbol nomenclature for each monosaccharide or derivative, which is helpful in depicting their structures.
Our knowledge of monosaccharides, however, is constantly changing in terms of diversity and numbers, even in regard to monosaccharides found in mammalian glycans. For example, glucosamine, derived from the UDP-glucosamine donor, is a component of brain glycogen and less abundant in liver glycogen (Kirkman and Whelan 1986; Sun et al. 2021), whereas historically it was thought that glycogen contained only glucose, as often indicated in textbooks. Unmodified glucosamine also occurs in heparin (van den Born et al. 1995), and in GPI anchors (Ferguson and Williams 1988), but there it arises by deacetylation of incorporated GlcNAc. CDP-ribitol is a precursor to ribitol phosphate within the O-glycans of α-dystroglycan (Gerin et al. 2016; Praissman et al. 2016). In regard to ribose/ribitol, recall that ribose and deoxyribose are monosaccharides within RNA and DNA, respectively, and although we would not classify them as glycans, they are glycomolecules. Thus, while textbooks often indicate that there are only 10 building blocks for mammalian glycans (Glc, Gal, Man, Fuc, Xyl, GlcNAc, GalNAc, GlcA, IdoA, and sialic acids (Neu5Ac and non-human Neu5Gc)) (Varki et al. 2022), there are obviously additional ones, including glucosamine, ribose/ribitol phosphate, and deoxyribose, when one considers all the glycomolecules in mammals. Notably, while fructose is a common monosaccharide and metabolic intermediate in most organisms, it does not appear to be a building block in mammalian glycomolecules. By contrast, fructooligosaccharides generated by fructosyltransferases have been identified in other organisms (Choukade and Kango 2021), and inulin is a β-(2,1)fructan polysaccharide of fructose (Roberfroid 2005).
These considerations also raise the issue of monosaccharide linkages in glycomolecules and forms of monosaccharides themselves within them. The diversity of monosaccharides and linkages suggest a startling possibility of nearly infinite numbers of structures. Several years ago Laine presented calculations about the possible hexasaccharide isomers (Laine 1994), i.e. those with 6 monosaccharides linked to each other, and concluded that there are 1.05 × 1012 possible structures for a reducing hexasaccharide composed only of hexoses. Others have made similarly high predictions about the possible numbers of hexasaccharides when one includes aminosugars, deoxysugars, and hexoses (Hellerqvist 1990). Considering the great diversity of monosaccharides and unknown factors about their modifications, it is not possible to make such calculations for the large glycans found in nature. For example, when one considers a single Glc with its 4 chiral carbons, and not counting α versus β anomeric forms, there are 16 possible optical isomers (2n, where n is the number of chiral centers). These are the D-group of stereoisomers of glucose, which are D-glucose, D-allose, D-altrose, D-mannose, D-gulose, D-idose, D-galactose, and D-talose, and the L-group of stereoisomers. All such monosaccharides have been synthesized and are found in some type of natural glycomolecule. For example, L-gulose is a key component of the antibiotic bleomycin and key to its activity (Zhou et al. 2021). While compounds with D-talose are rare, 6-deoxy-D-talose is an LPS component of several microbes (Forsberg et al. 2000).
Mention should also be made of the unusual nature of sugar chemistry (Seeberger 2022). Most monosaccharides adopt a ring form with oxygen in the ring—a 5-member ring (termed a furanose) or a 6-member ring (termed a pyranose) to ease strain and lessen bond angles. For example, ribose and 2-deoxyribose are both pentoses as well as aldoses and occur as 5-member rings. Fructose and glucose are hexoses, but fructose is a ketose and occurs mainly in a 5-member furanose ring, whereas glucose is an aldose and occurs mainly in a 6-member ring. Glucose, for example, can cycle between furanose and pyranose forms, but the pyranose form is most energetically favorable, whereas the opposite is true for fructose. Galactose in mammalian glycans occurs in a pyranose form (galactopyranose, Galp), but galactose within polysaccharides of other organisms, such as arabinogalactan from Mycobacterium tuberculosis, can occur as galactofuranose (Galf) (Richards and Lowary 2009). The nonoses are ketoses, e.g. Neu5Ac, and typically occur as a 6-member ring. In solution a free monosaccharide undergoes mutarotation, and is in equilibrium between its straight chain aldehyde or ketone form; upon formation of its cyclic form it occurs as either a hemiacetal or hemiketal (ring-chain tautomerism) and adopts either an α- or β-configuration on the anomeric carbon (C1 for aldoses, C2 for ketoses). The shape-shifting nature of a monosaccharide in solution is prevented when it is incorporated with a glycosidic linkage within a glycomolecule. Interestingly, 7-member rings termed septanoses (6 carbons plus an oxygen in the ring) have been synthesized (Saha and Peczuh 2011) and have interesting properties, but whether they are regular or minor forms of sugars is unclear.
Such vast numbers of monosaccharides and their derivatives can be bewildering to those interested in learning more about the glycosciences, and perhaps more simplified ways of exhibiting them could be helpful. One way to depict fundamental units of nature is in tabular form. For example, the genetic code and its codons may be depicted as a table of codons (Koonin and Novozhilov 2009), the standard DNA codon table TCAG, or the standard RNA codon table UCAG, and can also be depicted as a wheel, https://www.genome.gov/genetics-glossary/Genetic-Code.
A Periodic Table of Monosaccharides
The most famous table of building blocks is the Periodic Table of Elements. It was created in 1869 by Dmitri Mendeleev, but another was developed by Meyer (Gärditz 2023). One great feature of Mendeleev’s concept, in addition to ordering elements by atomic mass and their properties, was the prediction of then-unknown elements based on properties of known elements; such gaps were left to be filled in later. The Table was based on arranging the elements by their atomic weights and chemical similarities, and were listed as groups (I, II, III, etc.,) and periods (1, 2, 3, etc.,). The original form of the table was somewhat different from the modern form, which was created by Moseley in 1913, who reorganized it based on the atomic number (Egdell and Bruton 2020) [https://pubchem.ncbi.nlm.nih.gov/periodic-table/]. All of the first 118 elements have been identified and the gaps have been filled. There are important features to the modern Table, and the one commonly depicted identifies the elements in terms of their groups, periods, and similarities (often color coded), e.g. alkali metals, actinides, noble gasses, etc., along with their name, symbol, atomic number, and atomic mass. Such a depiction is highly informative, as it readily distinguishes unique features of elements to provide a context for comparing them.
Considering the diversity, yet similarities between monosaccharides and their derivatives, it is interesting to consider whether they may be placed within a type of periodic table. While the above discussion notes the possible existence of many different monosaccharides, for potentially depicting the monosaccharides in a periodic table, one has to choose the ones to include. There are many subjective things to consider, such as including those most commonly occurring, limiting the monosaccharides to a particular kingdom of life, choosing only those that cannot be hydrolyzed to another monosaccharide, omitting monosaccharide derivatives, etc. One approach is to choose a general definition of monosaccharides as described earlier and recognize those noted in the KEGG pathways and by the SNFG. This approach has some justification, as several unusual monosaccharides and derivatives found in microbes and invertebrates are included in these lists, which highlights the immense diversity of monosaccharides. Such an example is the unusual octose methylthiolincosamide (Mtl) (6′-amino-6′,8′-dideoxy-1′-thio-D-erythro-α-D-galactose) (Trefzer et al. 1999; Lin et al. 2014), found in the antimicrobial agent lincomycin A, the first lincosamide antibiotic isolated in 1962 from cultures of the actinomycete Streptomyces lincolnensis (Mason et al. 1962).
With this in mind, 103 monosaccharides are depicted in A Periodic Table of Monosaccharides (Fig. 2). It is possible to classify monosaccharides into 5 major groups - Aldoses, Ketoses, Aminosugars, Uronic acids, and Deoxysugars. The monosaccharides and derivatives are placed into periods based on the number of carbons in the backbone of the sugar, beginning with the 3-carbon Triose, followed by Tetrose, Pentose, Hexose, Heptose, Octose, Nonose, and Decose. These delineations naturally lead to the configuration of a Periodic Table of Monosaccharides (Fig. 2). The simple aldoses and ketoses do not have features found in the other 3 groups. Aminosugars, which may be either aldoses or ketoses, are defined as monosaccharides with at least one nitrogen, e.g. 2-doxy-N-acetyl; Uronic acids, which are typically aldoses, are defined as C-6 oxidized hexoses containing a carboxylate; and Deoxysugars, which may be aldoses or ketoses, are those with at least one OH group on a carbon replaced with H (as in, sugars lacking a hydroxyl group at a particular position, but not containing another substituent other than the H atom). Some monosaccharides may have overlapping group features, such as those that are both an aminosugar and a deoxysugar, and obviously, as mentioned some are aldoses or ketoses, but are not within the other 3 groups. Some monosaccharides have very unique features, e.g. apiose, which is listed as a pentose (Picmanova and Moller 2016), but is a unique branched chain sugar and one of its formal names is 3-C-(hydroxymethyl)-D-glycero-tetrose.
Colors are used to indicate groups and “subgroups”, for example the aminosugars include those that are amines and those that are N-acetyl derivatives. For grouping monosaccharides some liberties are taken in table placement, e.g. L6dAltNAc and 6dTalNAc are indicated with different colors, and are positioned between aminosugars and deoxysugars. Overall, as much as possible, a “dominant” feature was used in making that decision, with the amino modification being dominant over deoxy, e.g. FucNAc is included in the aminosugars and not in the deoxysugar group. In the Periodic Table of Elements, the term period, for which there are 7, refers to elements horizontally that increase in size by an additional proton; in the Periodic Table of Monosaccharides a period refers to the number of carbons in monosaccharides, tri- to decoses. Interestingly, there are 8 shown, but they are not numbered.
Considerations in Constructing the Table
Some liberties are obviously taken in assembling this Table, and they include the following.
(i) The different Groups are indicated at the top, and the Periods, based on the number of carbons in the backbone of the monosaccharides, on the left side, with the order of monosaccharides being generally based on putting the lower mass sugars at the top of the group. The number of columns of monosaccharides reflect an attempt at symmetry, as inspired by the Periodic Table of Elements. The organization flows from the relatively smaller number of tri- to pentoses compared to the hexoses and above. The rows of monosaccharides are chosen for symmetry. Attempts were made to develop “rules” for ordering each monosaccharides within groups and periods, but the incredible diversity of monosaccharides and their substituents did not permit assigning universal rules; furthermore, in draft preparations where rules were tested, the number of rules became both enormous and rather arbitrary. For some groups, like aldoses and ketoses, it might be possible to rationally organize them by rules of chirality of carbons moving along the carbohydrate backbone, but again, this was not readily applicable to other groups.
(ii) The table does not include every known monosaccharide and their derivatives, e.g. 9-O-acetyl-Neu5Ac, while obviously a building block, is not listed here as a separate monosaccharide derivative. In fact, for sialic acid there are known to be at least 50 naturally occurring derivatives (Schauer and Kamerling 2018). These can differ dramatically, which include acetylation, methylation, lactylation, lactonization, and sulfation (Sato and Kitajima 2019). Perhaps in the future, others may construct another type of periodic table for sialic acids, which might be equally complex to the one depicted here. In addition, sulfated derivatives of monosaccharides, such as 2-sulfo-L-iduronic acid, are also not included. There is obvious, but unavoidable, subjectivity in such decisions, and it is anticipated that the Table may enlarge in the future. In addition, programs can be developed online to link the monosaccharides within the Table to all known derivatives.
(iii) Some of the monosaccharides in the Table are considered “rare sugars”, defined as those of low abundance in nature. They can be produced by different approaches, e.g. hydrolysis of natural products, enzymatic transformations of other monosaccharides, and chemical synthesis (Granstrom et al. 2004; Mijailovic et al. 2021). Among the rare sugars are D-psicose, D-tagatose, and D-lyxose. There is much interest in such sugars in terms of their potential health effects upon human consumption (Ahmed et al. 2022), and there is increasing interest in them as potential sugar substitutes (Mooradian et al. 2017).
(iv) The structures of the monosaccharides and their derivatives are not depicted here, but if one goes to the KEGG and SNFG sites, a simple click on the names will take readers to either the KEGG site or the National Library of Medicine PubChem site at https://pubchem.ncbi.nlm.nih.gov/, respectively, where detailed information is available. In addition, the symbol nomenclature of the SNFG is not used in the Table, but is available in the links herein, as the colored symbols clash with the colors of each group. At the National Center for Functional Glycomics (NCFG) website, these symbols and links will be incorporated online into this Periodic Table of Monosaccharides (https://research.bidmc.org/ncfg/).
(v) Many monosaccharides have well-known common or trivial names and these are used wherever possible, e.g. glucose, psicose, etc. But each monosaccharide also has multiple names. For example, the names for psicose in the D-configuration include D-ribo-Hex-2-ulose, D-ribo-2-Hexulose, and others. Thus, the entire list of names cannot be provided in the Table. One can track the complete chemical name for such monosaccharides, based on the above references and sources. In that vein, the abbreviations for the monosaccharides used here represent a commonly used one for each, but there are some uncertainties. For example, glyceraldehyde, the simplest triose, is abbreviated simply as Gly, in keeping with a minimal three letter code format, mannoheptulose is abbreviated ManH, but in other studies it is sometimes abbreviated MH, and N-acetylmuramic acid is abbreviated MurNAc, but is also often abbreviated NAM. In addition, the abbreviations for many rare monosaccharides are not adopted uniformly in the field, and some are just 3 letters but others are longer. A discussion of abbreviations may be found in the IUPAC literature (McNaught 1997).
(vi) The D- versus L-configurations of sugars are not indicated per se, but in some cases the abbreviation and name includes the stereoisomer. Such is the case for N-acetyl-6-deoxy-L-altrosamine (L6dAltNAc) as an L-isomer, but the D-isomer of N-acetyl-6-deoxy-D-talosamine is abbreviated 6dTalNAc. For some sugars, such as L-fucose and L-iduronic acid, they commonly occur as the L-isomer, and this may be understood, but not indicated here. For other monosaccharides, they may exist as D- or L-isomers in glycomolecules, and would be noted in the name of the molecule. However, for some monosaccharides, especially the deoxysugars, the names may indicate that feature, e.g. 6dGul, whereas for others, such as Rham, they do not. This is in line with the historical nature of naming in the field. In addition, the formula mass of each monosaccharide is indicated, and illustrate the isomeric nature of monosaccharides.Additionally, while beyond the scope of this article, many of these monosaccharides in the Table have been identified as nucleotide sugar precursors for the generation of glycomolecules, but research in this area is relatively incomplete (Van Overtveldt et al. 2020). All of the monosaccharides that are found in mammalian glycomolecules, except L-iduronic acid, have been identified as nucleotide sugar derivatives. Many others are found in microorganisms and plants. For example, the unusual nucleotide GDP-colitose is a donor for the α1,2-colitosyltransferase in E. coli 055:h7 (Wu et al. 2016), the gene synthesizing CDP-paratose was identified in Salmonella typhi, rfbS (Hallis et al. 1998), and GDP-D-erythro-α-D-gluco-octose is involved in lincomycin A biosynthesis (Lin et al. 2014). Finally, while many of the simple monosaccharides are commercially available, this is unclear for many of the more complex monosaccharides.
(vii) There are “gaps” in the Table, and these are indicated by unfilled boxes; it might be predicted that the gaps in the Table would include new monosaccharides. An issue might arise as to where to place a new or additional monosaccharide, given the ordering based on the masses. Thus, if lower mass monosaccharides were added, that may reorder the listing. Certainly, with greater inspections and proof-of-structures, new monosaccharides will be found and added to the Table over time. For example, the Table includes the 9-carbon fusaminic acid (Fus) (5-acetamidino-3,5,9-trideoxy-L-glycero-L-gluco-non-2-ulosonic acid), which was discovered in the LPS of Fusobacterium nucleatum (Vinogradov et al. 2017; Wei et al. 2019), and the unusual 8-carbon monosaccharide erwiniose (Erw) (3,6,8-trideoxy-4-C-(R-1-hydroxyethyl)-D-gulo-octose), which occurs in Pectobacterium versatile and Pectobacterium atrosepticum (Gorshkov et al. 2017; Kowalczyk et al. 2023). However, there are some rare monosaccharides not presently included in the table, e.g. the undecosamine named hikosamine (4-amino-4-deoxy-D-glycero-D-galacto-D-gluco-undecose) (Uchida and Das 1973; Fujino et al. 2021), and caryophyllose (3,6,10-trideoxy-4-C-(D-glycero-1-hydroxyethyl)-D-erythro-D-gulo-decose), a rare branched sugar present in the LPS of Pseudomonas caryophylli (Rombouts et al. 2009).
(viii) Finally, in terms of differentiation, the Table represents a visual depiction of groups and sorting of the major monosaccharide subtypes, and is not meant to predict or infer chemical reactivity, as is the case for Mendeleev’s Table.
Concluding Remarks
In conclusion, there are a great many monosaccharides, and thus there is a need to organize our information about these important building blocks of glycomolecules. While this is a modest effort in that direction, it may serve as a guide to highlight the relationships among the monosaccharides and their derivatives, their groupings, diversity, and similarities, as well as the monosaccharides yet to be discovered. Finally, this effort may inspire the imagination of individuals not yet familiar with the glycosciences, and prompt them to inquire a bit deeper.
Supplementary Material
Acknowledgments
The author acknowledges the helpful discussions with members of his laboratory, and in particular discussions with Sandra Cummings, Jamie Heimburg-Molinaro, Maxence Noel, and Akul Mehta.
Funding
This work was supported by the National Center for Functional Glycomics (https://research.bidmc.org/ncfg/) (NIH grant R24GM137763).
Conflict of interest statement: The author declares no conflicts of interest in preparation of this article.
References
- Ahmed A, Khan TA, Dan Ramdath D, Kendall CWC, Sievenpiper JL. Rare sugars and their health effects in humans: a systematic review and narrative synthesis of the evidence from human trials. Nutr Rev. 2022:80(2):255–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den BornJ, Gunnarsson K, Bakker MA, Kjellén L, Kusche-Gullberg M, Maccarana M, Berden JH, Lindahl U. Presence of N-unsubstituted glucosamine units in native heparan sulfate revealed by a monoclonal antibody. J Biol Chem. 1995:270(52):31303–31309. [DOI] [PubMed] [Google Scholar]
- Campbell MP, Peterson R, Mariethoz J, Gasteiger E, Akune Y, Aoki-Kinoshita KF, Lisacek F, Packer NH. UniCarbKB: building a knowledge platform for glycoproteomics. Nucleic Acids Res. 2014:42(D1):D215–D221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T, Ono T, Shiota M, Yamada I, Aoki-Kinoshita KF, Bolton EE. Bridging glycoinformatics and cheminformatics: integration efforts between GlyCosmos and PubChem. Glycobiology. 2023:33(6):454–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choukade R, Kango N. Production, properties, and applications of fructosyltransferase: a current appraisal. Crit Rev Biotechnol. 2021:41(8):1178–1193. [DOI] [PubMed] [Google Scholar]
- Dong A, Liu C, Hua X, Yu Y, Guo Y, Wang D, Liu X, Chen H, Wang H, Zhu L. Bioinformatic analysis of structures and encoding genes of Escherichia coli surface polysaccharides sheds light on the heterologous biosynthesis of glycans. BMC Genomics. 2023:24(1):168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egdell RG, Bruton E. Henry Moseley, X-ray spectroscopy and the periodic table. Philos Trans A Math Phys Eng Sci. 2020:378(2180):20190302. [DOI] [PubMed] [Google Scholar]
- Ferguson MA, Williams AF. Cell-surface anchoring of proteins via glycosyl-phosphatidylinositol structures. Annu Rev Biochem. 1988:57(1):285–320. [DOI] [PubMed] [Google Scholar]
- Fontana C, Widmalm G. Primary structure of glycans by NMR spectroscopy. Chem Rev. 2023:123(3):1040–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg LS, Bhat UR, Carlson RW. Structural characterization of the O-antigenic polysaccharide of the lipopolysaccharide from rhizobium etli strain CE3. J Biol Chem. 2000:275(25):18851–18863. [DOI] [PubMed] [Google Scholar]
- Fujino H, Nagatomo M, Inoue M. Total syntheses of Hikosamine and Hikizimycin. J Org Chem. 2021:86(23):16220–16230. [DOI] [PubMed] [Google Scholar]
- Fujita A, Aoki NP, Shinmachi D, Matsubara M, Tsuchiya S, Shiota M, Ono T, Yamada I, Aoki-Kinoshita KF. The international glycan repository GlyTouCan version 3.0. Nucleic Acids Res. 2021:49(D1):D1529–D1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gärditz KF. The poetry of the universe, the periodic table, and the scientific progress: a review of new studies on the periodic table of the elements. Found Chem. 2023:25(2):269–283. [Google Scholar]
- Gerin I, Ury B, Breloy I, Bouchet-Seraphin C, Bolsee J, Halbout M, Graff J, Vertommen D, Muccioli GG, Seta N, et al. ISPD produces CDP-ribitol used by FKTN and FKRP to transfer ribitol phosphate onto α-dystroglycan. Nat Commun. 2016:7(1):11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorshkov V, Islamov B, Mikshina P, Petrova O, Burygin G, Sigida E, Shashkov A, Daminova A, Ageeva M, Idiyatullin B, et al. Pectobacterium atrosepticum exopolysaccharides: identification, molecular structure, formation under stress and in planta conditions. Glycobiology. 2017:27(11):1016–1026. [DOI] [PubMed] [Google Scholar]
- Granstrom TB, Takata G, Tokuda M, Izumori K. Izumoring: a novel and complete strategy for bioproduction of rare sugars. J Biosci Bioeng. 2004:97(2):89–94. [DOI] [PubMed] [Google Scholar]
- Hackman RH. Studies on chitin. I. Enzymic degradation of chitin and chitin esters. Aust J Biol Sci. 1954:7(2):168–178. [DOI] [PubMed] [Google Scholar]
- Hallis TM, Lei Y, Que NL, Liu H. Mechanistic studies of the biosynthesis of paratose: purification and characterization of CDP-paratose synthase. Biochemistry. 1998:37(14):4935–4945. [DOI] [PubMed] [Google Scholar]
- Hellerqvist CG. Linkage analysis using Lindberg method. Methods Enzymol. 1990:193:554–573. [DOI] [PubMed] [Google Scholar]
- Herget S, Toukach PV, Ranzinger R, Hull WE, Knirel YA, von der LiethCW. Statistical analysis of the bacterial carbohydrate structure data base (BCSDB): characteristics and diversity of bacterial carbohydrates in comparison with mammalian glycans. BMC Struct Biol. 2008:8(1):35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperiali B. Bacterial carbohydrate diversity—a brave new world. Curr Opin Chem Biol. 2019:53:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwersen M, Dora H, Kohla G, Gasa S, Schauer R. Solubilisation and properties of the sialate-4-O-acetyltransferase from guinea pig liver. Biol Chem. 2003:384(7):1035–1047. [DOI] [PubMed] [Google Scholar]
- Jackson CM. Emil Fischer and the "art of chemical experimentation". Hist Sci. 2017:55(1):86–120. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Lv X, Liu Y, Shin HD, Li J, Du G, Liu L. Biocatalytic production of glucosamine from N-acetylglucosamine by diacetylchitobiose deacetylase. J Microbiol Biotechnol. 2018:28(11):1850–1858. [DOI] [PubMed] [Google Scholar]
- Kahsay R, Vora J, Navelkar R, Mousavi R, Fochtman BC, Holmes X, Pattabiraman N, Ranzinger R, Mahadik R, Williamson T, et al. GlyGen data model and processing workflow. Bioinformatics. 2020:36(12):3941–3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkman BR, Whelan WJ. Glucosamine is a normal component of liver glycogen. FEBS Lett. 1986:194(1):6–11. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Novozhilov AS. Origin and evolution of the genetic code: the universal enigma. IUBMB Life. 2009:61(2):99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk A, Babinska-Wensierska W, Lojkowska E, Kaczynski Z. The structure of O-polysaccharide isolated from the type strain of Pectobacterium versatile CFBP6051(T) containing an erwiniose—higher branched monosaccharide. Carbohydr Res. 2023:524:108743. [DOI] [PubMed] [Google Scholar]
- Laine RA. A calculation of all possible oligosaccharide isomers both branched and linear yields 1.05 × 10(12) structures for a reducing hexasaccharide: the isomer barrier to development of single-method saccharide sequencing or synthesis systems. Glycobiology. 1994:4(6):759–767. [DOI] [PubMed] [Google Scholar]
- Lal K, Bermeo R, Perez S. Computational tools for drawing, building and displaying carbohydrates: a visual guide. Beilstein J Org Chem. 2020:16:2448–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larnkjaer A, Hansen SH, Ostergaard PB. Isolation and charaterization of hexasaccharides derived from heparin. Analysis by HPLC and elucidation of structure by 1H NMR. Carbohydr Res. 1995:266(1):37–52. [DOI] [PubMed] [Google Scholar]
- Lewis AL, Chen X, Schnaar RL, Varki A. Sialic acids and other nonulosonic acids. In: Varki A, Cummings RD, Esko JD, et al., editors. Essentials of glycobiology. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2022. pp. 185–204 [Google Scholar]
- Lewis AL, Toukach P, Bolton E, Chen X, Frank M, Lutteke T, Knirel Y, Schoenhofen I, Varki A, Vinogradov E, et al. Cataloging natural sialic acids and other nonulosonic acids (NulOs), and their representation using the symbol nomenclature for glycans. Glycobiology. 2023:33(2):99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li W, Wang H, Bayley DL, Cao J, Reed DR, Bachmanov AA, Huang L, Legrand-Defretin V, Beauchamp GK, et al. Cats lack a sweet taste receptor. J Nutr. 2006:136(7):1932S–1934S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CI, Sasaki E, Zhong A, Liu HW. In vitro characterization of LmbK and LmbO: identification of GDP-d-erythro-α-d-gluco-octose as a key intermediate in lincomycin A biosynthesis. J Am Chem Soc. 2014:136(3):906–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl U, Jacobsson I, Hook M, Backstrom G, Feingold DS. Biosynthesis of heparin. Loss of C-5 hydrogen during conversion of D-glucuronic to L-iduronic acid residues. Biochem Biophys Res Commun. 1976:70(2):492–499. [DOI] [PubMed] [Google Scholar]
- Mandal C, Schwartz-Albiez R, Vlasak R. Functions and biosynthesis of O-acetylated sialic acids. Top Curr Chem. 2015:366:1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marth JD. A unified vision of the building blocks of life. Nat Cell Biol. 2008:10(9):1015–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason DJ, Dietz A, DeBoer C. Lincomycin, a new antibiotic. I. Discovery and biological properties. Antimicrob Agents Chemother. 1962:554–559. [Google Scholar]
- McNaught AD. International Union of Pure and Applied Chemistry and International Union of Biochemistry and Molecular Biology. Joint Commission on Biochemical Nomenclature. Nomenclature of carbohydrates. Carbohydr Res. 1997:297(1):1–92. [DOI] [PubMed] [Google Scholar]
- Mehta AY, Cummings RD. GlycoGlyph: a glycan visualizing, drawing and naming application. Bioinformatics. 2020:36(11):3613–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijailovic N, Nesler A, Perazzolli M, Ait Barka E, Aziz A. Rare sugars: recent advances and their potential role in sustainable crop protection. Molecules. 2021:26(6):1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola S. Nucleotide sugars in chemistry and biology. Molecules. 2020:25(23):5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooradian AD, Smith M, Tokuda M. The role of artificial and natural sweeteners in reducing the consumption of table sugar: a narrative review. Clin Nutr ESPEN. 2017:18:1–8. [DOI] [PubMed] [Google Scholar]
- Neelamegham S, Aoki-Kinoshita K, Bolton E, Frank M, Lisacek F, Lutteke T, O'Boyle N, Packer NH, Stanley P, Toukach P, et al. Updates to the symbol nomenclature for glycans guidelines. Glycobiology. 2019:29(9):620–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SS. Post-glycosylation modification of sialic acid and its role in virus pathogenesis. Vaccines (Basel). 2019:7(4):171. 10.3390/vaccines7040171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez S, Sarkar A, Rivet A, Breton C, Imberty A. Glyco3D: a portal for structural glycosciences. Methods Mol Biol. 2015:1273:241–258. [DOI] [PubMed] [Google Scholar]
- Picmanova M, Moller BL. Apiose: one of nature's witty games. Glycobiology. 2016:26(5):430–442. [DOI] [PubMed] [Google Scholar]
- Praissman JL, Willer T, Sheikh MO, Toi A, Chitayat D, Lin YY, Lee H, Stalnaker SH, Wang S, Prabhakar PK, et al. The functional O-mannose glycan on α-dystroglycan contains a phospho-ribitol primed for matriglycan addition. Elife. 2016:5:e14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raedts J, Kengen SW, van der OostJ. Occurrence of L-iduronic acid and putative D-glucuronyl C5-epimerases in prokaryotes. Glycoconj J. 2011:28(2):57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitman ML, Kornfeld S. Lysosomal enzyme targeting. N-acetylglucosaminylphosphotransferase selectively phosphorylates native lysosomal enzymes. J Biol Chem. 1981:256(23):11977–11980. [PubMed] [Google Scholar]
- Richards MR, Lowary TL. Chemistry and biology of galactofuranose-containing polysaccharides. Chembiochem. 2009:10(12):1920–1938. [DOI] [PubMed] [Google Scholar]
- Roberfroid MB. Introducing inulin-type fructans. Br J Nutr. 2005:93(Suppl 1):S13–S25. [DOI] [PubMed] [Google Scholar]
- Rombouts Y, Burguiere A, Maes E, Coddeville B, Elass E, Guerardel Y, Kremer L. Mycobacterium marinum lipooligosaccharides are unique caryophyllose-containing cell wall glycolipids that inhibit tumor necrosis factor-alpha secretion in macrophages. J Biol Chem. 2009:284(31):20975–20988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha J, Peczuh MW. Synthesis and properties of septanose carbohydrates. Adv Carbohydr Chem Biochem. 2011:66:121–186. [DOI] [PubMed] [Google Scholar]
- Sarrazin S, Lamanna WC, Esko JD. Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol. 2011:3(7):a004952. 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato C, Kitajima K. Chapter one - sialic acids in neurology. In: Baker DC, editors. Sialic acids, part II: biological and biomedical aspects. New York: Elsevier; 2019. pp. 1–64 [Google Scholar]
- Schauer R, Kamerling JP. Exploration of the sialic acid world. Adv Carbohydr Chem Biochem. 2018:75:1–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeberger PH. Monosaccharide diversity. In: Varki A, Cummings RD, Esko JD, et al., editors. Essentials of glycobiology. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2022. pp. 21–32 [Google Scholar]
- Sun RC, Young LEA, Bruntz RC, Markussen KH, Zhou Z, Conroy LR, Hawkinson TR, Clarke HA, Stanback AE, Macedo JKA, et al. Brain glycogen serves as a critical glucosamine cache required for protein glycosylation. Cell Metab. 2021:33(7):1404–1417.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieker DF, Hadden JA, Schulten K, Woods RJ. 3D implementation of the symbol nomenclature for graphical representation of glycans. Glycobiology. 2016:26(8):786–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trefzer A, Salas JA, Bechthold A. Genes and enzymes involved in deoxysugar biosynthesis in bacteria. Nat Prod Rep. 1999:16(3):283–299. [DOI] [PubMed] [Google Scholar]
- Tsuchiya S, Yamada I, Aoki-Kinoshita KF. GlycanFormatConverter: a conversion tool for translating the complexities of glycans. Bioinformatics. 2019:35(14):2434–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida K, Das BC. Hikosamine, a novel C11 aminosugar component of the antibiotic hikizimycin. Biochimie. 1973:55(5):635–636. [DOI] [PubMed] [Google Scholar]
- Van Overtveldt S, Da Costa M, Gevaert O, Joosten HJ, Beerens K, Desmet T. Determinants of the nucleotide specificity in the carbohydrate epimerase family 1. Biotechnol J. 2020:15(11):e2000132. [DOI] [PubMed] [Google Scholar]
- Varki A, Kornfeld S. Identification of a rat liver alpha-N-acetylglucosaminyl phosphodiesterase capable of removing ``blocking'' alpha-N-acetylglucosamine residues from phosphorylated high mannose oligosaccharides of lysosomal enzymes. J Biol Chem. 1980:255(18):8398–8401. [PubMed] [Google Scholar]
- Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Mohnen D, Kinoshita T, Packer NH, Prestegard JH, et al. Essentials of glycobiology. 4th ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2022 [PubMed] [Google Scholar]
- Vinogradov E, St. Michael F, Cox AD. The structure of the LPS O-chain of Fusobacterium nucleatum strain 25586 containing two novel monosaccharides, 2-acetamido-2,6-dideoxy-l-altrose and a 5-acetimidoylamino-3,5,9-trideoxy-gluco-non-2-ulosonic acid. Carbohydr Res. 2017:440-441:10–15. [DOI] [PubMed] [Google Scholar]
- Visser EA, Moons SJ, Timmermans S, de JongH, Boltje TJ, Büll C. Sialic acid O-acetylation: from biosynthesis to roles in health and disease. J Biol Chem. 2021:297(2):100906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives RR, Seffouh A, Lortat-Jacob H. Post-synthetic regulation of HS structure: the yin and Yang of the Sulfs in cancer. Front Oncol. 2014:3:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R, Liu H, Li X. De novo synthesis of novel bacterial monosaccharide fusaminic acid. J Antibiot (Tokyo). 2019:72(6):420–431. [DOI] [PubMed] [Google Scholar]
- Wen J, Xiao J, Rahdar M, Choudhury BP, Cui J, Taylor GS, Esko JD, Dixon JE. Xylose phosphorylation functions as a molecular switch to regulate proteoglycan biosynthesis. Proc Natl Acad Sci U S A. 2014:111(44):15723–15728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Zhao G, Li T, Qu J, Guan W, Wang J, Ma C, Li X, Zhao W, Wang PG, et al. Biochemical characterization of an α1,2-colitosyltransferase from Escherichia coli O55:H7. Glycobiology. 2016:26(5):493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yethon JA, Whitfield C. Purification and characterization of WaaP from Escherichia coli, a lipopolysaccharide kinase essential for outer membrane stability. J Biol Chem. 2001:276(8):5498–5504. [DOI] [PubMed] [Google Scholar]
- York WS, Mazumder R, Ranzinger R, Edwards N, Kahsay R, Aoki-Kinoshita KF, Campbell MP, Cummings RD, Feizi T, Martin M, et al. GlyGen: computational and informatics resources for glycoscience. Glycobiology. 2020:30(2):72–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida-Moriguchi T, Willer T, Anderson ME, Venzke D, Whyte T, Muntoni F, Lee H, Nelson SF, Yu L, Campbell KP. SGK196 is a glycosylation-specific O-mannose kinase required for dystroglycan function. Science. 2013:341(6148):896–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Ye W, Cao Y, Wang M, Qi D, Liao G, Li H, Huang W, Chen W, Wang X, et al. A gulose moiety contributes to the belomycin (BLM) disaccharide selective targeting to lung cancer cells. Eur J Med Chem. 2021:226:113866. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Venzke D, Walimbe AS, Anderson ME, Fu Q, Kinch LN, Wang W, Chen X, Grishin NV, Huang N, et al. Structure of protein O-mannose kinase reveals a unique active site architecture. Elife. 2016:5:e22238. 10.7554/eLife.22238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.