Abstract
Cortex glia in Drosophila central nervous system form a niche around neural cells for necessary signals to establish cross talk with their surroundings. These cells grow and expand their thin processes around neural cell bodies. Although essential for the development and function of the nervous system, how these cells make extensive and intricate connected networks remains largely unknown. In this study, we show that Cut, a homeodomain transcription factor, directly regulates the fate of the cortex glia, impacting neural stem cell (NSC) homeostasis. Focusing on the thoracic ventral nerve cord, we found that Cut is required for the normal growth and development of cortex glia and timely increase in DNA content through endocycle to later divide via acytokinetic mitosis. Knockdown of Cut in cortex glia significantly reduces the growth of cellular processes, the network around NSCs, and their progeny's cell bodies. Conversely, overexpression of Cut induces overall growth of the main processes at the expense of side ones. Whereas the Cut knockdown slows down the timely increase of DNA, the Cut overexpression results in a significant increase in nuclear size and volume and a 3-fold increase in DNA content of cortex glia. Further, we note that constitutively high Cut also interfered with nuclei separation during acytokinetic mitosis. Since the cortex glia form syncytial networks around neural cells, the finding identifies Cut as a novel regulator of glial growth and variant cell cycles to support a functional nervous system.
Keywords: cortex glia, variant cell cycles, Cut, Drosophila, polyploidy, neural stem cells
Introduction
Glial cells play a crucial role in maintaining and protecting the nervous system by promoting neural growth, facilitating communication, and providing nourishment and support for the overall homeostasis of neural stem cells (NSCs) and their progeny (Xiong and Montell 1995; Booth et al. 2000; Barres 2008; Trapp and Nave 2008; Stork et al. 2012). In addition, these cells form the blood–brain barrier and compartmentalize the central nervous system (CNS) into specialized domains in several invertebrates and vertebrates such as elasmobranch fishes (Oland and Tolbert 2003; Abbott 2005; Awasaki et al. 2008). It is particularly interesting to understand how glial cells form a dynamic environment around NSCs and their offspring during normal development and disease prospects. The mammalian and Drosophila glial cells are comparable and classified into functional groups based on morphology and molecular functions (Stork et al. 2012). In Drosophila, the 3 major classes of glia types based on morphology are surface, cortex, and neuropile-associated glia (Ito et al. 1995; Stork et al. 2012).
Cortex glia cells in Drosophila nervous system are born during the mid-embryonic stage. Their cell bodies are irregular and grow in multiple directions making massive networks around individual neural cell bodies for trophic and metabolic support (Ito et al. 1995; Dumstrei et al. 2003; Pereanu et al. 2005). In the ventral nerve cord (VNC) of the Drosophila larval nervous system, cortex glia are primarily located on the ventral and lateral sides. It is interesting to note that during the growth and remodeling period, the cortex glia nuclei adopt variant cell cycle processes to increase their DNA content and later divide through acytokinetic mitosis to form syncytium (Rujano et al 2022). These cells closely interact with neural cells and regulate their fate (Ito et al. 1995; Read et al. 2009; Dong et al. 2021). Similar to cortex glia, mammalian astrocytes also extend their processes toward the outer surface of synaptic neuropils (reviewed in Zhou et al. 2019). In order for glial processes to grow, insulin and PI3K signaling must be activated through the intake of nutrients (Spéder and Brand 2018; Yuan et al. 2020). In addition, Hippo, EGFR, and FGF signaling pathways also regulate cortex glia growth and remodeling (Read et al. 2009; Witte et al. 2009; Reddy and Irvine 2011; Avet-Rochex et al. 2012; Spéder and Brand 2018; Dong et al. 2021). Investigations continue how fine processes grow from the cortex glia cell body and find their correct path. We discovered that Cut, a homeodomain transcription factor, is critical in this process and plays multiple roles in the developmental growth of cortex glia to form a niche around neural cell bodies.
The Cut protein family is abundantly expressed in the developing CNS of both Drosophila and mammals, playing a crucial role in regulating neural identity and proper patterning. (Arya et al. 2019; Weiss et al. 2019). From an evolutionary and functional perspective, Cut is one of the most conserved proteins among all metazoans. It is known as CUX1 in humans and CUX1/2 in mice (Neufeld et al. 1992; Valarche et al. 1993; Yoon and Chikaraishi 1994; Quaggin et al. 1996; Tavares et al. 2000). Cut protein is well known for its diverse functions in determining cell specificity and identity in Drosophila and mammals and is expressed in multiple tissues (Liu et al. 1991; Andres et al. 1992; Liu and Jack 1992; Ludlow et al. 1996; Nepveu 2001; Zhai et al. 2012; Pitsouli and Perrimon 2013). It influences cell proliferation, migration, and response to DNA damage in various tissues (Coqueret et al. 1998; Michl et al. 2005; Cadieux et al. 2009; Vadnais et al. 2012; Pitsouli and Perrimon 2013) The protein Cut has multiple DNA-binding domains, resulting in various transcription factor isoforms (Bodmer et al. 1987; Blochlinger et al. 1988; Liu et al. 1991). Its isoforms have a single homeodomain and one or multiple Cut DNA-binding repeat domains. Depending on its interacting partners, Cut acts as a transcriptional activator or repressor of several genes during development (Yoon and Chikaraishi 1994; van Wijnen et al. 1996).
Consistent with its role as a gene regulator, Cut is also identified as a selector gene in several mammalian disorders, including cancer (Zhai et al. 2012). Loss of function mutations in Cut/CUX1 has been identified in several cancer types. At the same time, increases in CUX1 gene copy number and overexpression are also common in many cancers (Sansregret and Nepveu 2008; Ramdzan and Nepveu 2014). In the mouse, the CUX1 overexpression leads to multiorgan hyperplasia and tumor development after a long latency period (Ledford et al. 2002; vanden Heuvel et al. 2005). Since Cut performs a diverse function in several cellular processes and gene regulation, the detailed molecular understanding of its functional diversity is an area of active research.
This research reports that the Cut homeodomain transcription factor is crucial in regulating the growth and formation of the cortex glia network around NSCs and their progeny in the Drosophila CNS. Cut guides the cortex glia development in several ways. Cut levels affect the complex network of glial processes. Loss of Cut does not allow the cortex glia nuclei to timely increase their ploidy, which is needed for syncytium formation. On the other hand, continued high Cut expression in cortex glia increases their DNA ploidy to multifold, yet it also interferes with the normal acytokinetic mitosis by not allowing the nuclei to divide indicating a gain of function for Cut. It is the first report highlighting a novel role of Cut in regulating the growth and branching of cortex glia via controlling variant cell cycles.
Materials and methods
Fly stocks and genotypes
Drosophila melanogaster was reared at 22°C, and RNAi crosses were done at 29°C on a standard food medium containing sugar, agar, maize powder, and yeast. Appropriate fly crosses were set up following standard methods to obtain the progeny of desired genotypes. The following fly stocks are used in the experiments: Oregon R+ as wild-type, Repo-GAL4 (BL 7415), Cut-RNAi (BL 33967, strong), Cut-RNAi (BL29625, weak), UAS-Cut (II, referred in text as Cut-OE) (Norbert Perrimon, Harvard Medical School, MA, USA), Cyp4g15-GAL4 (39103), UAS-eGFP(II) (BL-5431), R54H02-GAL4 (BL-45784), R46H12-GAL4 (BL-50285), NP577-GAL4 (Awasaki et al. 2008), mCD8RFP (BL-27398), and Fly-FUCCI (BL-55121). The following genotype combinations are generated in the lab for the study: UAS-eGFP; Cyp4g15-GAL4, UAS-FUCCI; Cyp4g15-GAL4, mCD8RFP; Cyp4g15-GAL4, and UAS-eGFP; Cut-RNAi.
Survival assay
We used 3 strains (Oregon R+, Cyp4g15-GAL4, and Cut-RNAi) reared on standard agar–cornmeal–sugar–yeast food. We crossed 60 male and 90 virgin fruit flies. The Cyp4g15-GAL4 strain was used as males, and the Oregon R+ and Cut-RNAi strains were used as virgin females. The flies were isolated, fed, and reared for 2 days before being transferred to a food bottle for mating. We placed 20 males and 30 females in each replicate bottle and allowed them to mate for a day. After mating, the flies were transferred to a cage with an agar plate where they laid eggs for 12 h. The agar plates were changed daily. To analyze survival at the pupariation stage and eclosion, we collected synchronized first instar larvae and transferred them to food vials. We examined 11 replicates of 100 larvae each for the control and Cut-RNAi groups. The percentage of pupation and eclosion was calculated as described earlier (Kharat et al. 2020). At least 3 replicates of 100 larvae each were examined in each group for the survival assay, with 1–2 day-old imagoes placed in food vials. The recorded data were analyzed using GraphPad Prism 9 software for statistical analysis.
Immunostaining, confocal microscopy, and documentation
Larvae of the required age [early first instar (ALH 0–3 h), L2 (ALH 24–28 h), early third instar (ALH 48–54 h), mid-third instar (ALH 72 h), third instar (ALH 93–96 h)] and wandering larvae as late third instar (referred as LL3 throughout the text) from F1 progeny of respective crosses were selected, and CNSs were dissected in phosphate buffer (PBS 1× containing NaCl, KCl, Na2HPO4, KH2PO4, and pH 7.4), fixed in 4% paraformaldehyde for 30 min, rinsed thrice in 0.1% PBST (1×PBS and 0.1% TritonX-100), and incubated in blocking solution (0.1% TritonX-100, 0.1% BSA, 10% FCS, 0.1% deoxycholate, and 0.02% thiomersal) for 30 min, at room temperature. Samples were incubated in the required primary antibody at 4°C overnight. The antibodies used are mouse anti-Cut (1:10, Developmental Studies Hybridoma Bank, 2B10), mouse Anti-Repo (1:50, Developmental Studies Hybridoma Bank, 8D12), and rabbit anti-GFP antibody (1:200, Invitrogen, A-11122). For rat anti-Dpn antibody (1:150, ab195173, Abcam), the samples were kept for 2 consecutive overnights for better signal. The following day, the samples were rinsed thrice in 0.1% PBST and incubated with appropriate secondary antibody at 1:200 dilution overnight or for 2 h at room temperature. The secondary antibodies used are donkey anti-rat Alexa 488 (Invitrogen, A-21208), goat anti-mouse Alexa Fluor 568 (cat. No. A11004), and chicken anti-rabbit Alexa Fluor 488 (Invitrogen, A-21441). Following incubation with secondary antibodies, the samples were washed thrice in 0.1% PBST and counterstained with DAPI (1 µg/ml, Invitrogen, D1306) at 4°C overnight whenever needed. After the final washes in 0.1% PBST, the samples were mounted in DABCO (Sigma, D27802) for further analysis.
Images were acquired at the following confocal microscopes: Zeiss LSM-510 Meta at the Department of Zoology, Zeiss LSM-510 at ISLS BHU, and Leica SP8 STED facility at CDC, BHU. All the images were quantified using the Fiji/ImageJ software (NIH, USA). Images were assembled using Adobe Photoshop and MS PowerPoint softwares.
Image analysis using Fiji/ImageJ application
Confocal sections of cortex glia of the thoracic VNC (tVNC) were analyzed using the Fiji/ImageJ software (NIH) to measure number, area, and volume. For integrated intensity measurements in a region of interest, a freehand selection tool was used to mark the area manually.
The number of glia nuclei and NSCs were counted manually based on GAL4-driven GFP and Repo staining using the multipoint selection tool. To measure the nuclear size of cortex glia, GFP+, Repo+ was used to outline the area. The area outline was done using a freehand selection tool around Repo staining, followed by selecting an analyze-measure option. The chamber size was measured by outlining the cortex glia processes that are coming out from the cell body around 1 nucleus and forming a chamber around progeny using a freehand selection tool, and the size of the outlined area (µm2) was measured using the analyze-measure option.
For volume analysis of cortex glia, 1 specific threshold was chosen that fits best for the actual staining and was kept constant throughout the analysis. The thresholding procedure is used in image processing to select pixels of interest based on the GFP intensity of the pixel values. Thereafter, measure stack plugin was used to find the fluorescent area (GFP) of each cortex glia section of tVNC through the analyze-measure option where area, mean gray value, stack position, and limit to threshold checkboxes were selected. The area obtained was thus multiplied by the number of stack intervals (= 2) to find out the volume of cortex glia. Finally, the area covered was estimated by manually outlining the area occupied by tVNC.
The density of cortex glia processes was calculated through the plot profile and the surface profile options. The tools measure the signal intensities of cortex glia processes along the selected lines. The gray values indicate the intensities of cortex glia along a line, and the number of peaks denotes the number of cortex glia extensions.
The integrated density for DAPI in the cortex glia nuclei was measured by manually outlining each GFP+, Repo+ DAPI+ nucleus; the area outline was done using the freehand selection tool around Repo staining. The integrated density of DAPI was measured in each section of tVNC using the analyze-measure option. The confocal scanning parameters for each experimental setup were kept constant for all intensity quantifications. Controls were analyzed in parallel for each experimental set.
Statistical analysis was done using GraphPad Prism 9 software where a 2-tailed unpaired t-test was used to evaluate the statistical significance between the mean values of the 2 groups, using asterisk as follows: n with P < 0.05 (****P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05) considered statistically significant and if P > 0.05 considered nonsignificant.
Results
Cut expresses in glial subtypes and maintains niche around NSCs
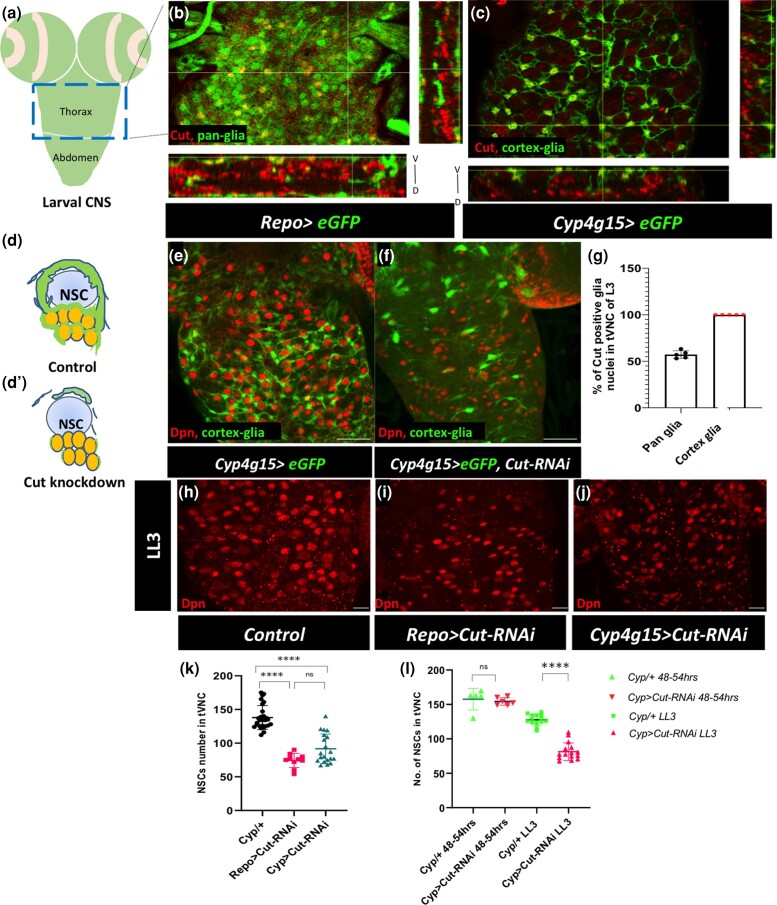
The transcription factor Cut is known to express in the Drosophila CNS and to regulate the developmental death of NSCs in the abdominal region of VNC (Arya et al. 2019). In addition to NSCs, Cut is expressed in several other cells in the larval CNS. Therefore, the larval CNS was stained with a Cut antibody to profile its expression in the glial cells. In this study, we focused on the tVNC to understand the role of Cut in glial morphogenesis (Fig. 1a). Since Cut and pan-glia marker Repo antibodies are both raised in mice and there is no commercial antibody for cortex glia, we used the UAS-GAL4 system to mark the glia with GFP using Repo-GAL4 (pan glia) and Cyp4g15-GAL4 (cortex glia) drivers (Fig. 1, b and c; Supplementary Fig. 1, d–i; Spéder and Brand 2018; Gonzalez-Gutierrez et al. 2019; Rujano et al. 2022). We counted the GFP+ and Cut+ nuclei in the tVNC to estimate the glia that express Cut. In the Repo > GFP tVNC, where GFP marks all the glial cells, around 55% of total GFP-positive nuclei were Cut positive (Fig. 1, b and g; Supplementary Fig. 1, d–f). On the other hand, when the numbers of GFP+ nuclei marking the cortex glia (Cyp4g15 > GFP) that also express Cut were counted, we found that all the tVNC cortex glia express Cut in late larval instar (Fig. 1, c and g; Supplementary Fig. 1, g–i).
Fig. 1.
Cut expresses in glial subtypes and maintains a niche around neural stem cells. a) Model of the larval CNS indicating the thoracic region and abdominal region of VNC. The blue box marks the late thoracic region of VNC (tVNC), shown in b–j). b and c) Single section view and orthogonal sections in xyz plane, and yxz plane shows the Cut expression (red) in pan glia (Repo > eGFP, b) and cortex glia (Cyp4g15 >eGFP, c). d) Model showing cortex glia (green) enwrapping NSC (sky blue) and its lineages (yellow) in control and d') disruption of the cortex glia network in the Cut knockdown. e) Well-developed glial extensions around NSCs (marked with Dpn in red) in control (Cyp4g15 > eGFP/+). f) Loss of Cut (Cyp4g15 > eGFP, Cut-RNAi) severely disrupts cortex glia and affects the distribution of NSCs. g) Quantification of the percentage of Cut positive glia nuclei in the control tVNC. h–j) NSC number and distribution (Dpn, red) in tVNC. h) Control (Repo-GAL4/+), i) pan glial Cut knockdown (Repo > Cut-RNAi), and j) cortex glia Cut knockdown (Cyp4g15 > Cut-RNAi). k) Quantification of NSCs loss upon the Cut knockdown with Repo-GAL4 and Cyp4g15-GAL4 compared with control. l) Quantification in ALH 48–54 h and LL3 to estimate the nonautonomous loss of NSCs upon the Cut knockdown in cortex glia. The statistical evaluation of significance based on unpaired t-test is marked with asterisks, ****P < 0.0001. Scale bar: 50 µm e and f), 20 µm h–j).
The cortex glia networks around neural cells establish the communication between the neural cells and their environment and are needed for the survival of newborn neurons (Freeman and Doe 2001; Dumstrei et al. 2003; Spéder and Brand 2018; Rujano et al. 2022). Selective knockdown of Cut expression in cortex glia with Cyp4g15-GAL4 driver resulted in substantial disruption of their cellular processes (compare Fig. 1d' with Fig. 1, d and f with the control Fig. 1e). We validated this finding with 2 separate Cut-RNAi lines (Supplementary Fig. 1, a and b). Since the cortex glia form a supporting system for the neural cells, the loss of glia and their network around neural bodies also affects NSCs. We used Deadpan (Dpn) as an NSC marker and noted that they were unusually clumped and were lost during development upon Cut ablation in cortex glia (Fig. 1, h–k). To ensure that the fewer NSCs observed at the late third instar larvae are not due to an early defect in their birth, we counted NSCs in the early instar stages. The number of NSCs in tVNC of control and Cut ablated cortex glia did not show any significant difference in the early third instar larvae (Fig. 1l). This indicates that NSC number was normal but declined later, possibly due to loss of the Cut defective cortex glia (Fig. 1l). Thus, the Cut defective cortex glia nonautonomously affects the NSC fate. We used 4 other cortex glia-specific drivers for the knockdown of Cut—R54H02, R46H12, and Np577 and found similar, significant nonautonomous loss of NSCs in tVNC (Supplementary Fig. 1c). We selected Cyp4g15-GAL4 for further studies. We conclude that Cut is expressed in all cortex glia and is essential for maintaining the glial niche around the NSCs and their progeny.
Cut is required for the growth of the cortex glia processes and their branching
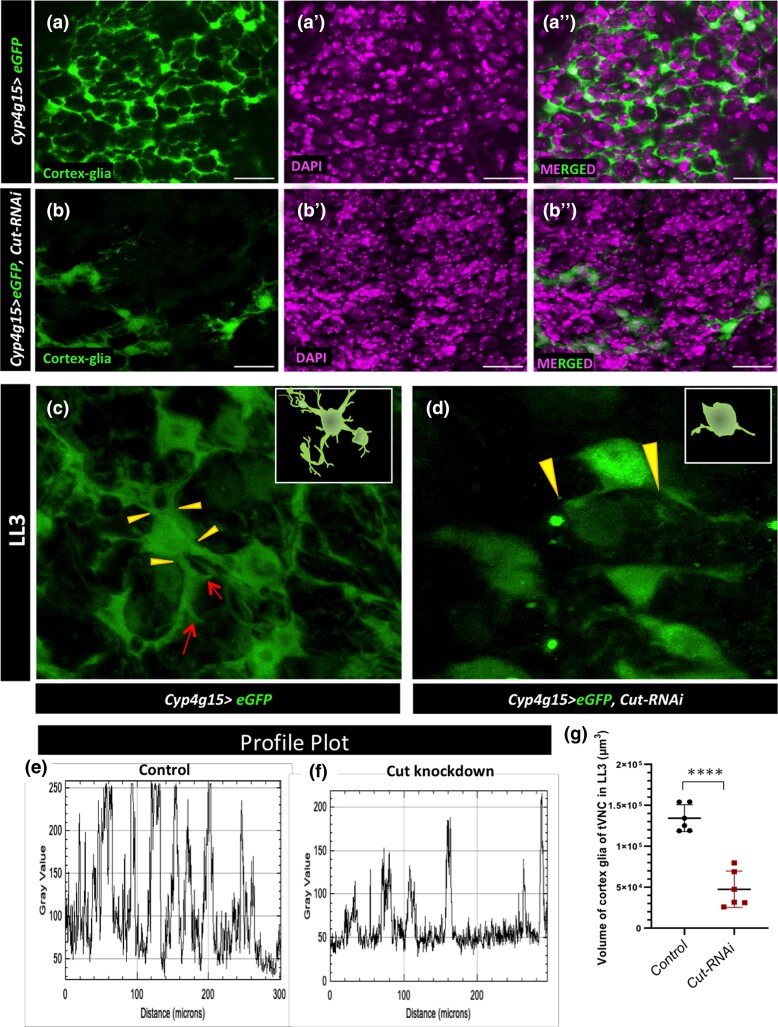
Following the above result that the Cut defective cortex glia cannot form a niche around the neural cells, we further explored the role of Cut in these cortex glia for their growth and chamber formation around neural cell bodies. Cortex glia continue to expand their cytoplasmic extensions from the late embryonic stage (Pereanu et al. 2005; Coutinho-Budd et al. 2017; Rujano et al. 2022). Therefore, as a first step, we checked the expression profile of the cortex glia GAL4 driver, Cyp4g15-GAL4, and found that it expresses from the late embryonic stage (Supplementary Fig. 2, i and i') and continues its expression through different larval stages. The GFP driven with the GAL4 line faithfully marks the glial network from the early to late developmental stages of larvae (Supplementary Fig. 2, i and i' with Repo, Figs. 2a and 3, a–h). As shown in Fig. 2, a and c (yellow arrowhead in Fig. 2c), several thick and thin cytoplasmic extensions emerge from cortex glia and spread their branches in multiple directions making a fine meshwork around individual neural cell bodies in the tVNC, known as trophospongium (Fig. 2c, red arrow). Recently, it has been shown that cortex glia are syncytial units, and different such units undergo homotypic fusion to allow an exchange of cytoplasmic proteins (Coutinho-Budd et al. 2017; Rujano et al. 2022). The Cut knockdown in the cortex glia leads to a massive loss of these lamelliform glial processes in the whole CNS (Fig. 2, b and b″; Supplementary Fig. 2, e and f″ for optic lobes and Supplementary Fig. 2, g, h″, and j for the abdominal region). Unlike the normal cortex glia, which extends the processes in several directions (Fig. 2c, yellow arrowheads), the Cut knockdown in these cortex glia leads to only 1–2 abnormal looking main cytoplasmic extensions (compare Fig. 2, c and d). To further compare the density of cortex glia extensions in the 2 genotypes, we used the ImageJ plot analysis tool to evaluate the number of peaks, indicating the number of cortex glia extensions. The Cut defective cortex glia show far less peaks than the control. Additionally, the intensities of peaks were also variable, indicating a reduction in cytoplasmic extensions (Supplementary Fig. 2, c and d). The plot profile along a line (Supplementary Fig. 2, a and b') in the tVNC also showed a loss of the glial network upon the Cut knockdown (Fig. 2, e and f). We also evaluated the volume of cortex glia in these genotypes through thresholding and network density, where the volume of Cut defective cortex glia was significantly reduced (Fig. 2g).
Fig. 2.
The Cut is required for cortex glia growth and branching. a and a″) Control (Cyp4g15 > eGFP/+) shows cortex glia niche enwrap the neural cells. b and b″) Cut defective cortex glia cannot form a niche around the neural cell bodies (green cortex glia, magenta DAPI). c) In Control (Cyp4g15 > eGFP/+), individual cortex glia cell body extends thick (yellow arrowheads) and lean processes (red arrows) in multiple directions; the inset is the model. d) Cut defective cortex glia (Cyp4g15 > eGFP, Cut-RNAi) with minimum extension growth. e and f) Plot profile along a straight line in tVNC showing that peaks and their intensities are more uniform and closer to each other in the control e) compared with Cut knockdown f), emphasizing the loss of glial network. g) Evaluation of the volume (µm3) of cortex glia in the LL3 tVNC of control (Cyp4g15 > eGFP/+), and Cut knockdown (Cyp4g15 > eGFP, Cut-RNAi) showing significantly reduced cortex glia volume upon Cut knockdown. Statistical evaluation of significance, ****P < 0.0001, based on unpaired t-test. Scale bar: 20 µm.
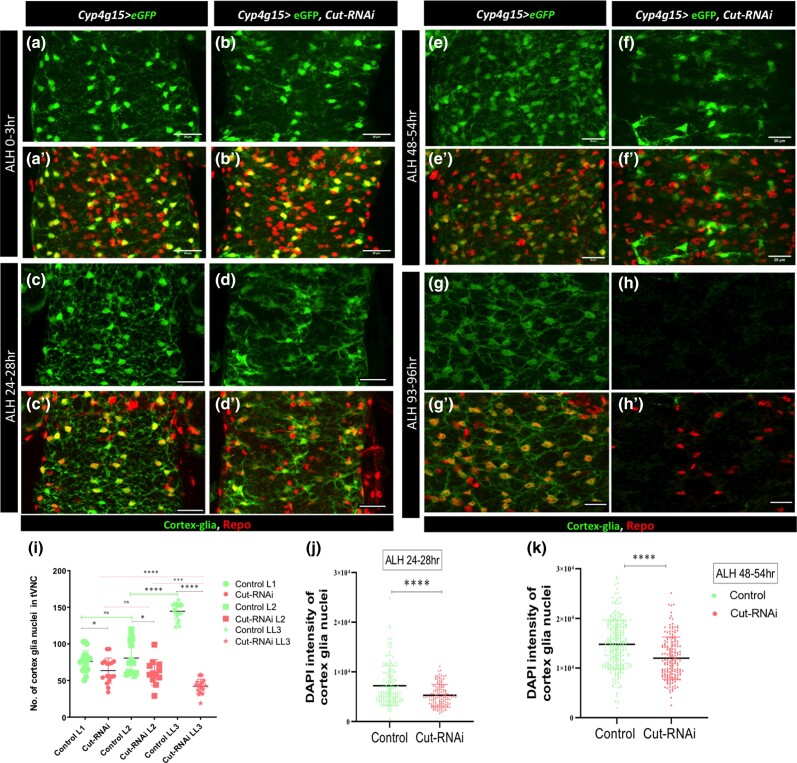
Fig. 3.
Cut defective cortex glia are unable to increase their nuclei number during development. a–k) Throughout the figure, the genotypes of control (Cyp4g15 > eGFP/+) and Cut knockdown (Cyp4g15 > eGFP, Cut-RNAi). a and b'’) Projection of ventral sections of only tVNC in the early first larval instar (L1, ALH 0–3 h), in a and a') control and b and b') Cut knockdown 3–4 cortex glia are visible in each thoracic hemisegment, 1 near the midline and 2 ventral, lateral ones are not clearly visible in all hemisegments. c and d') L2 (ALH 24–28 h) in c and c') control, the cortex glial nuclei remain largely unchanged compared with L1; however, the processes are more distinctly visible and infiltrate throughout tVNC. Upon Cut knockdown d and d'), the cortex glia nuclei appear to be a little less, and the cell processes are disrupted compared with control. e and f') L3 (ALH 48–54 h) in e and e') control, the number of cortex glial nuclei increases significantly, and the cell processes make a complex network. f and f') Cut defective cortex glia shows defect in the increase in nuclei number; they still resemble the L1/L2 pattern of nuclei distribution (compare f with a and c); the glia process remains stunted. g and h') Late L3 (ALH 93–96 h); in g and g') control, the nuclei number further increases, and the glia network are more like a honeycomb structure. Upon Cut knockdown h and h'), significant loss of cortex glia was noticed. i) Quantification of cortex glia nuclei in different larval timepoints (L1–L3); in L1, the number of nuclei in Cut defective cortex glia are slightly less compared with the control (P value 0.0207). The nuclei number increases significantly in control from L2 to L3 (****P value <0.0001). Upon Cut knockdown, the increase in nuclei number is largely hampered (*P value <0.0257), and in L3, several of them are lost (****P value <0.0001). j and k) Quantification of the integrated density of DAPI of individual cortex glia nucleus at j) ALH 24–28 h and k) ALH 48–54 h was evaluated (>5 CNS/genotype). The CNS was stained with Repo (glia), GFP (cortex glia), and DAPI (nucleus). j) DAPI content of cortex glia in control is more, compared with Cut knockdown (****P value <0.0001) at ALH 24–28 h. k) At ALH 48–54 h, the DNA content is more in control cortex glia compared Cut defective cortex glia (****P value <0.0001). The statistical evaluation of significance based on unpaired t-test. Scale bar: 20 µm.
The finding that the Cut knockdown prevents cortex glia from growing lamelliform extensions and filling gaps is significant. Although ablation of a few cortical glia causes the unaffected neighboring cortex glia to compensate by growing lamelliform extensions to efficiently fill the gaps (Coutinho-Budd et al. 2017), a general knockdown of Cut in all the cortex glia affects their growing capability, leading to the presence of spaces all over the tVNC. This clearly indicates that Cut is required for the normal growth and development of lamelliform extensions from the cell body. Upon the Cut knockdown, several of these defective glia could not undergo proper morphogenesis, and large spaces can be observed all over the tVNC.
Cut defective cortex glia are unable to increase their nuclei number during development
To better understand the defect of cortex glia in tVNC during development and its time of onset, we examined CNS at different time points during larval stages. In the early first larval instar (L1, ALH 0–3 h), there is typically 4–5 cortex glia in each thoracic hemisegment arranged in a ventrolateral pattern (Ito et al. 1995). One cortex gila can be seen near the midline, 2 are placed ventrally, and 1 or 2 are placed laterally (Fig. 3, a and a'). The extensions coming out of the cortex glia are very fine at this stage. In L2 (ALH 24–28 h), the extensions are more distinctly visible and infiltrate throughout tVNC. However, the number of cortex glial nuclei remains largely unchanged compared with L1 (compare Fig. 3, c and c' with Fig. 3, a and a'; Ito et al. 1995). The Cut knockdown in cortex glia impacts the networking of cellular processes and nuclei number as early as ALH 24–28 h (Fig. 3, d and d'). Even though the number of cortex glia nuclei do not show severe change and are comparable with L1 of the Cut knockdown, the connection of cellular processes is severely affected, and fine networks are mostly missing at this point (Fig. 3, d and i arrow).
The number of cortex glia nuclei typically increases from L2 onward by employing different forms of cell cycle, which includes endocycle, endomitosis, and acytokinetic mitosis (Fig. 3i; Rujano et al. 2022). Furthermore, a fusion of cortex glia results in the formation of a complex pattern of nuclei and cellular network (Coutinho-Budd et al. 2017; Rujano et al. 2022; (Fig. 3, e, e', and i). Strikingly, the Cut knockdown affects the increase in nuclei number in L3 (48–96 h; compare Fig. 3, e and e' with Fig. 3, f and f' and Fig. 3, g and g′ with Fig. 3, h and h'). It is interesting to note that at this stage, the nuclei pattern in the Cut knockdown of tVNC resembles more of the L1/L2 arrangement rather than the L3 controls of the same age (compare Fig. 3, f and f' with Fig. 3, c and c' and Fig. 3, e and e'); the cell bodies appear flatten and unconnected.
Normally, cortex glia show a significant increase in nuclear volume from L2 onward and undergo endoreplication (Rujano et al. 2022). In literature, there are different overlapping definitions of polyploidization, endoreplication, and endocycle. We consider endocycle as a bigger umbrella with variant cell cycles, including endoreplication and endomitosis (Edgar and Orr-Weaver 2001; Zielke et al. 2013), and will refer to the process as endocycle from here onward. We further investigate whether Cut plays a role in increasing the DNA in cortex glia. We measured the DNA content through the integrated density of DAPI of each cortex glia nuclei labeled with GFP and Repo (DAPI+, GFP+, Repo+) using ImageJ. The nuclear content of cortex glia shows a sharp increase from ALH 24 to ALH 48 h as shown before (Fig. 3, j and k). We observed that upon the Cut knockdown, the integrated density of DAPI does not show the kind of increase as seen in the control group (Fig. 3, j and k). This indicates that low Cut levels hinder the timely increase of DNA in cortex glia.
We further checked how Cut knockdown in the cortex glia affects an organism's survival. There were indications of delay in progression through different developmental stages although this delay did not reach statistical significance (Supplementary Fig. 3, a and b). In adult organisms, a decrease in Cut resulted in a significant reduction in average life span compared with control animals (Supplementary Fig. 3b). Since the Cut knockdown does not show severe consequences on the overall life span of the organism, it may be due to a late and progressive effect on cortex glia morphogenesis. A very severe effect on glial growth upon Cut depletion occurs later in larval life (Fig. 3, d–h), which may allow the organism to survive into adulthood after passing the critical stage of NSC activation during the larval life. We hypothesized that Cut is required for timely cortex glia growth and increase in its DNA content that is required for syncytium formation during later stages.
Overexpression of Cut enhances the growth of cortex glia main branches at the cost of side extensions
During Drosophila development, Cut influences the complexity of the neuronal dendritic arbor in the peripheral nervous system, determines the identity of wrapping glia, and influences the formation of cellular processes (Grueber et al. 2003; Bauke et al. 2015). Furthermore, the Cut protein level also regulates the branching pattern of dendrites in various neuronal classes in the peripheral nervous system differentially (Grueber et al. 2003). Therefore, we asked whether constitutively increased Cut levels in cortex glia would affect their growth and branching patterns.
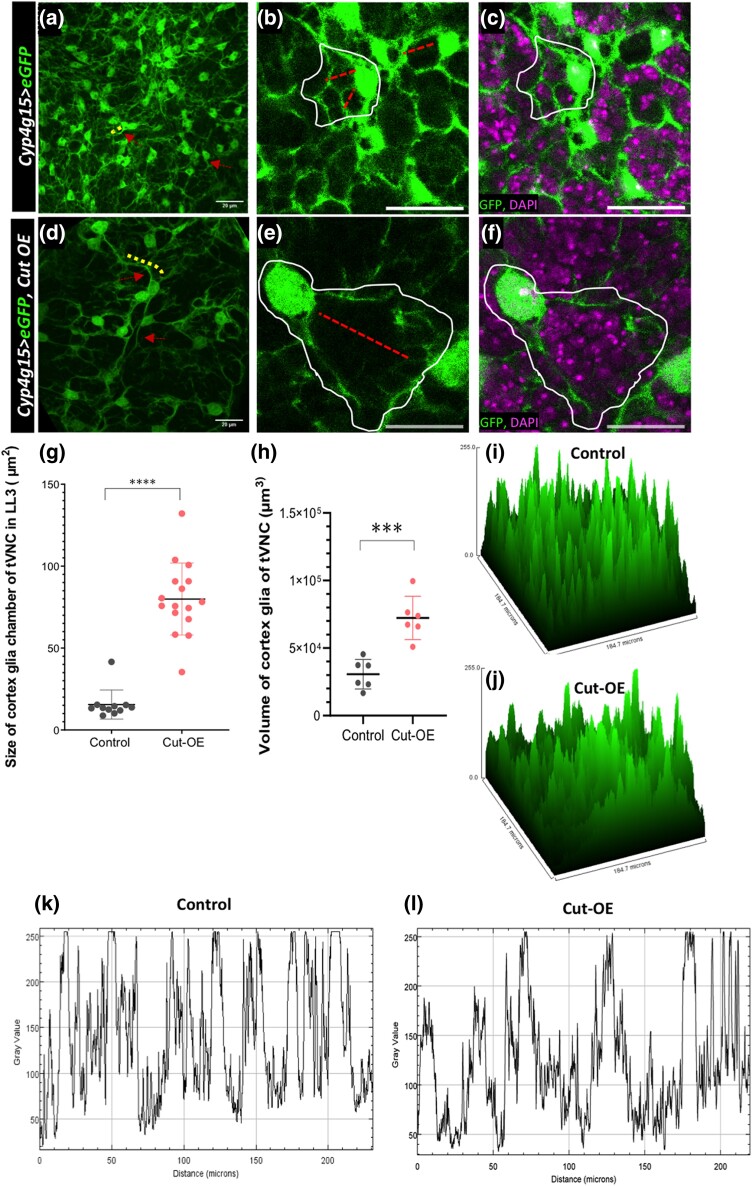
As discussed in earlier sections, the cortex glial cells make a complex network of processes around the neural cells. These processes come in various lengths and thicknesses. Some originate directly from the cell body as the main branch, while others stem from the main branch as side processes with varying thicknesses. We found that the Cut in the cortex glia remarkably affects the cellular processes in several ways. While the Cut knockdown resulted in stunted growth of cortex glia processes as noted above (Figs. 2 and 3), its overexpression led to longer main extensions but with little side branches (see Fig. 4, a and d, red arrows and yellow dotted lines marking main extensions). Note in the control where one of the marked main extensions coming out of the cell body is shorter (Fig. 4a, red arrow, yellow line); in the Cut overexpressing glia, it grows thicker and longer (Fig. 4d, red arrow, yellow line). To validate the visual interpretation, we measured the total volume of cortex glia and found an apparent increase in overall glial volume upon the Cut overexpression (Fig. 4h). Since the Cut overexpressing glia have more extended processes, the cortex glia chambers are also larger and globular (compare Fig. 4, b and c with Fig. 4, e, f, and g). We consider a chamber to be an area that is surrounded by thick extensions coming out from one cytoplasmic body with fine extensions covering neural cell bodies. The chamber size was measured by outlining the cortex glia processes coming out from the cytoplasm around one nucleus and forming an enclosure around progeny. Further, to check the extent of loss of side fine processes upon Cut overexpression, we compare the density of main vs side extensions through the surface and plot profile along a line. The loss of side extensions upon Cut overexpression is very evident as the profile of Cut overexpressing glia have broader peaks, away from each other, compared with the control, where the peaks are more uniform, sharp, and close to each other (Fig. 4, i–l). Thus, these observations all together propose that high Cut levels are sufficient to influence the overall growth of cortex glial processes. In addition, the Cut level affects the glial network's complexity by increasing the growth of main processes and retard the growth of side extensions.
Fig. 4.
The overexpression of Cut in cortex glia enhances the growth of cell body and main branches at the cost of terminal extensions. a–c) Control (Cyp4g15 > eGFP/+): a) cortex glia cell bodies (red arrows) and dense main extension (yellow lines) can be seen, b) the cortex glia chambers (white encircle) are small and compact (red line to show the area covered), and c) same image as in b) with DAPI showing single DAPI neural nuclei in the encapsulation. d–f) Cut overexpressing cortex glia (Cyp4g15 > eGFP, Cut-OE). Compare d–f) with control a–c) where cell bodies are less dense and big, and their cytoplasmic extensions are longer and thicker; the main cortex glia branches are significantly longer d), making bigger glial chambers e), and encapsulating several neural bodies marked with DAPI f). g and h) Quantifications in the control (Cyp4g15 > eGFP/+) and Cut overexpression (Cyp4g15 > eGFP, Cut-OE). g) Size of the cortex glia chamber, total chambers counted 5/tVNC (n = 3). h) Total cortex glia volume in tVNC (n > 5). i and j) The surface plot of cortex glia in tVNC to compare the intensity and extensions of Cut overexpressing cortex glia cells with control. In Control i), there are more peaks representing cell and their extensions. In contrast, upon Cut overexpression, j) the number of peaks is less and of variable intensities showing the loss of cortex glia side extensions. k and l) Plot profile along a line in the tVNC showing the peaks and their intensities in control k) and Cut overexpression l), again emphasizing the loss of fine glial network. The statistical evaluation of significance ****P < 0.0001, ***P < 0.001, and **P < 0.01 based on unpaired t-test. Scale bar: 20 µm.
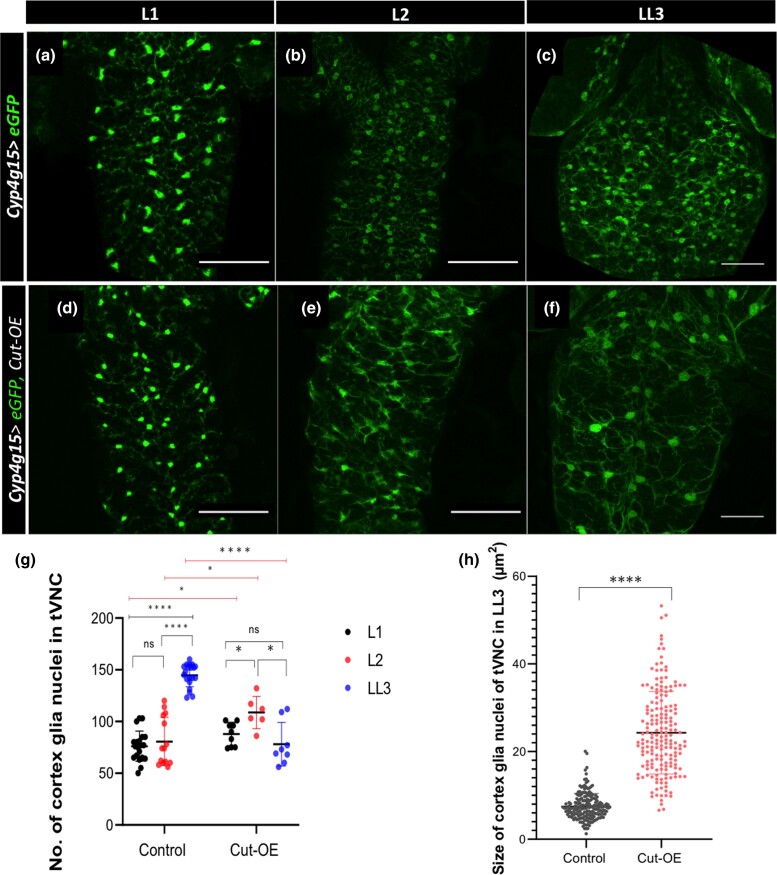
Constitutive activation of Cut increases DNA content in cortex glia
The number of cortex glia nuclei in the tVNC progressively increase from L1 to LL3 stages (Fig. 5, a–c; Coutinho-Budd et al. 2017; Rujano et al. 2022). The Cut overexpressing cortex glia in tVNC increases their nuclei number from the L1 to late L2 stage (compare Fig. 5, d and e with Fig. 5, a, b, and h). On the contrary, while this number increases further in the control tVNC in the late L3 stage (Fig. 5, b, c, and g), the rise is not seen in the Cut overexpressing cortex glia nuclei number (Fig. 5, e, f, and g). Instead of an increase in the nuclei number, the cortex glia constitutively expressing Cut increase their nuclei size (compare Fig. 5h). We observe a 3-fold increase in nuclei size upon Cut overexpression (Fig. 5i). Furthermore, we observed that the distribution pattern of cortex glia nuclei in the LL3 tVNC of Cut overexpressing CNS is more similar to the control L1/L2 tVNC instead of LL3 (compare Fig. 5f with Fig. 5, a and b), although the pattern is deformed due to their unexpected loss (Fig. 5f).
Fig. 5.
Cut regulates endomitosis of cortex glia. a–f) Confocal projections of the ventral few sections are shown for clarity. Compare the pattern of cortex glia distribution from L1 to LL3 in the tVNC of control (Cyp4g15 > eGFP/+) a–c) with Cut overexpressing (Cyp4g15 > eGFP, Cut-OE) d–f). L1 and L2 control tVNC a and b), the nuclei are arranged more or less in a vertical line pattern (nuclei number drastically increased in LL3 with brighter cell bodies). The distribution of cortex glia in the L1 of Cut overexpression is more or less similar to control (compare a with d), whereas in L2, it gets a bit distorted (compare b with e). A striking difference is seen in Cut overexpressing LL3 f) from LL3 control c). The pattern of GFP-positive cortex glia upon Cut overexpression appears more like L1/L2 of controls (compare d with a, b). No increase in the GFP+ nuclei is observed in the Cut overexpressing tVNC (compare f with c). g) Further, quantification of cortex glia nuclei from L1 to LL3 in control, and Cut-OE clearly shows that in Cut overexpression, the nuclei number does not increase from L2 to LL3 (n > 5). h) The size of the cortex glia nuclei (Repo+, GFP+) was measured by counting 20 nuclei/tVNC (n = 5). The statistical evaluation of significance **** P < 0.0001, ***P < 0.001, **P < 0.01, and *P < 0.05 based on unpaired t-test using GraphPad Prism 9 software. Scale bar: 50 µm in a–f).
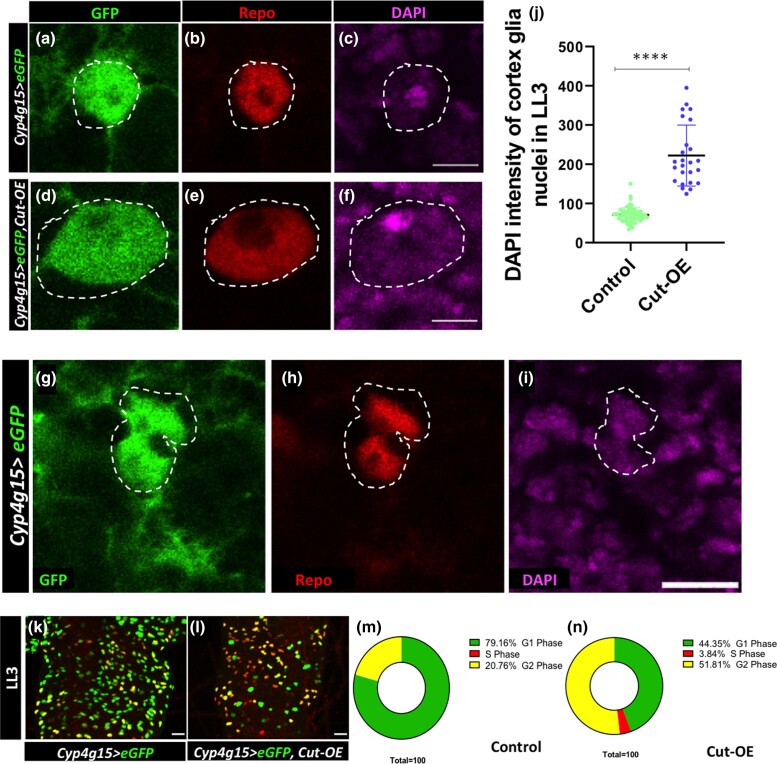
Since we see a similar defect in the Cut knockdown where the nuclei number does not increase, we set out to check the DNA content upon Cut overexpression. We measured the DNA content through the integrated density of DAPI of each cortex glia nuclei labeled with GFP and Repo (DAPI+, GFP+, Repo+) through ImageJ. Strikingly, these nuclei showed a 2-to-3-fold increase in DNA content upon Cut overexpression when compared with control (Fig. 6, compare Fig. 6c with Fig. 6, f and j). Thus, the Cut overexpressing cortex glia continue to synthesize the DNA, which confirms that Cut is necessary and sufficient to increase the DNA content.
Fig. 6.
Constitutive activation of Cut increases DNA content in cortex glia. a–i) Cortex glia nuclei of LL3 are marked with GFP (green), Repo (red), and DAPI (magenta). a–c) Control cortex glia (Cyp4g15 > eGFP/+) nuclei of LL3 tVNC. d–f) Overexpression of Cut (Cyp4g15 > eGFP, Cut-OE) results in a significant increase in the size of cortex glia nucleus (compare control a–c with d–f Cut-OE). g–i) Control cortex glia appears to be separating during endomitosis. j) DNA quantification by the integrated density of DAPI in each cortex glia nucleus [>20 nuclei counted per CNS (n = 3) showing a significant increase in DNA content upon Cut overexpression]. k–n) Cell cycle stages of cortex glia nuclei in LL3 detected with Fly-FUCCI in k) control (Cyp4g15 > eGFP/+) and (Cyp4g15 > eGFP, Cut-OE) l). k–n) Fluorescence representing G1 (green), S (red), and G2/M (yellow). m and n) Control count of nuclei in different cell cycle stages shows that the majority (79.16%) of nuclei are in G1, and rest (20.76%) are in G2/M, whereas the large number of Cut overexpression nuclei are still in S phase (3.83%) and also in G2/M phase (51.81%). Scale bar 5 µm in a–i) and 20 µm in k—m). The statistical evaluation of significance ****P < 0.0001, and nsP < 0.183 based on unpaired t-test.
To get more details about the cell cycle, we used the Fly-FUCCI tool to discriminate among G1, S, and G2/M phases of the cell cycle in the Cut overexpressing cortex glia. It utilizes fluorescently labeled degrons derived from Cyclin B and E2F1 proteins, which are degraded by APC/C and CRL4cdt2 proteins from mid-mitosis to the onset of the S phase, respectively (Zielke et al. 2014). At ALH 96 h, most (79.16%) cortex glia in control appear in the G1 phase (Fig. 6, k and m). While upon the Cut overexpression, a significantly high number of nuclei were still in S (3.83%) and G2/M phases (51.81%) (Fig. 6, l and n). Together, it indicates that a large fraction of nuclei in the Cut overexpressing glia continue the cell cycle instead of resting at G1, unlike the control (compare Fig. 6n with Fig. 6m). Therefore, we propose that the homeodomain protein Cut is required for the cortex glia nuclei to increase their DNA content. Probably, the nucleus divides when Cut levels go down. The continued high Cut levels interfere with acytokinetic mitosis resulting in a single large nucleus in a cell.
Discussion
In this study, we report a novel role of the homeodomain transcription factor, Cut, in the growth and development of cortex glia. Cortex glia undergo variant cell cycles to form syncytium and a complex reticular network around the neural cells. We observed that loss of Cut interferes with the growth of cell processes and timely increase of DNA content through endocycle, thus limiting these nuclei to enter acytokinetic mitosis and formation of syncytium. On the other hand, continued high Cut levels allow cortex glia to increase their DNA content multifold. We also discovered that despite increased DNA content, the constitutively high Cut levels interfere with acytokinetic mitosis by not allowing these nuclei to separate. We have identified that Cut acts as a novel player in autonomously regulating the dynamic growth of cortex glia around neural cells by controlling cellular morphogenesis and variant cell cycles.
Cut, a novel regulator of cortex glia morphogenesis
The transcription factor Cut is known to play several roles in the context of cell type and tissue-dependent manner. Acting as a selector gene, it regulates the identity of cells and their growth during development (Krupp et al. 2005). The differential level of Cut and its mammalian homolog, CUX1/2, controls the complexity of dendritic branching, the number of dendritic spines, and tracheal development (Grueber et al. 2003; Cubelos et al. 2010; Pitsouli and Perrimon 2013). Similarly, Cut instructs the growth of cellular processes of the wrapping glia in the peripheral nervous system and acts downstream of FGF signaling has been observed in pancreatic cancer (Bauke et al. 2015). In the case of cortex glia, we see that Cut activation enhances the overall growth and thickness of main cortex glia extensions but at the cost of side branching. Our data add that Cut controls the growth of cortex glia and endocycle.
It is significant that when cortex glia are ablated in a restricted area, the neighboring cortex glia extend their processes and fill the gaps (Coutinho-Budd et al. 2017; Hirase et al. 2022). However, the Cut defective cortex glia do not show such growth, thus resulting in visible wide gaps in the glial trophospongium in the larval CNS. Therefore, we conclude that Cut is required and sufficient for the growth of cortex glia processes. However, the signaling pathways and the domain through which Cut functions await further investigation.
Cortex glia processes start growing around the NSCs and neurons of the larval nervous system after receiving the nutritional signals via an activation of PI3K/Akt signaling (Spéder and Brand 2018; Yuan et al. 2020). Since the Cut defective cortex glia show severely hampered growth of cytoplasmic extensions even as early as L2, we think that Cut may function within the PI3K/Akt signaling pathway in glial cells in a manner analogous to what has been observed in pancreatic cancer cells (Ripka et al. 2010). Other known regulators of cortex glia growth are FGF signaling, components of the membrane fusion machinery, and secreted neurotrophin Spätzle 3 (Read et al. 2009; Witte et al. 2009; Reddy and Irvine 2011; Avet-Rochex et al. 2012; Coutinho-Budd et al. 2017; Read 2018; Spder and Brand 2018; Dong et al. 2021). However, the broad phenotype of Cut activation differs from FGF activation in the cortex glia. The ectopic activation of FGF signaling induces cortex glia overgrowth with a robust increase in nuclei number and overall cell surface area (Avet-Rochex et al. 2012). We found that the overexpression of Cut does not enhance the nuclei number of cortex glia. Instead, it resulted in giant nuclei with higher DNA content. Furthermore, the overexpression of FGF receptor htlACT in cortex glia makes a heavy network of finer cortex glia extensions (Avet-Rochex et al. 2012). In contrast, Cut overexpression causes a widespread reduction in the finer extensions. Thus, the cytoplasmic growth of fine extension in the overexpression of Cut vs FGF signaling in cortex glia also has different outcomes and may act through distinct mechanisms.
Regulation of ploidy is essential in the glial cells to control their growth
The size of a cell is important and plays a crucial role during development. Several differentiated cells undergo ploidy changes to increase their size or metabolic output. For example, germline nurse cells, salivary glands, fat bodies, gut, and trachea show varying levels of endopolyploidy (Lilly and Spradling 1996). Interestingly, neurons and glia in invertebrate and vertebrate adult brains compensate for aging-related cell loss or other damages by undergoing polyploidy (Losick et al. 2013, 2016; Tamori and Deng 2013; Cohen et al. 2018; Nandakumar et al. 2020). On the other hand, endomitosis is also a way to increase the cell size through which cells undergo incomplete mitosis and increase the number of nuclei per cell. A few examples of the same are known in Drosophila and mammalian systems (Unhavaithaya and Orr-Weaver 2012; Britton and Edgar 1998; Edgar and Orr-Weaver 2001; Taniguchi et al. 2012; Rios et al. 2016; Von Stetina et al. 2018; Windmueller et al. 2020). Several reports from independent labs strongly indicate that cortex glia increase their nuclei number by undergoing variant cell cycles (Coutinho-Budd et al. 2017; Yuan et al. 2020; Rujano et al. 2022). A recent study shows that cortex glia make syncytium and complex networks by undergoing endocycle and eventually acytokinetic mitosis (Rujano et al. 2022). The syncytial cortex glial units also show fusion and form complex compartments to facilitate exchange (Rujano et al. 2022). Cut regulates the growth of cortex glia by regulating the complexity of cytoplasmic processes and interferes with endocycling. The cortex glia having Cut knockdown shows a clear defect in DNA increase and remains stuck in the earlier development stage. Their nuclei do not divide or enter the acytokinetic mitosis. Interestingly, the overexpression of Cut clearly shows an increase in DNA content, yet these nuclei do not divide. Thus, in cortex glia, the overexpression of Cut does not show the routine increase of nuclei number from the L2 to LL3 stage as seen in the control tVNC; instead, it increases the ploidy in nuclei. In Drosophila oogenesis, follicular cells undergo endocycle to increase their DNA content. To ensure proper development, the downregulation of Cut is necessary to switch from mitosis to endocycle (Sun and Deng 2005). Interestingly, the role of Cut in cortex glia cells is different, as high levels of Cut promote DNA increase per nucleus. This suggests that Cut may have diverse and tissue-specific effects on the cell cycle.
Aneuploidy is one of the most common phenomena seen in several tumors (Rajagopalan and Lengauer 2004), and Cut overexpression is also (Sansregret and Nepveu 2008) reported in diverse classes of cancer (Sansregret and Nepveu 2008; Ramdzan and Nepveu 2014). There are several tumors, including glioma, where CUX1 overexpression has been reported and considered among driver mutation (Sansregret and Nepveu 2008; Kaur et al. 2018; Feng et al. 2021; Griesmann et al. 2021; Xu et al. 2021). An in vitro study shows that mammalian CUX1 has the potential to cause chromosomal instability (Sansregret et al. 2011). Our findings highlight that Cut regulates DNA content of cortex glia. It would be interesting to see whether the genetic combination of Cut with any other tumor driver identified in glioma could make the nuclei unstable and lead to cancer in the brain. In conclusion, our finding that Cut regulates cortex glia growth and variant cell cycles is of significant importance for development and disease prospects.
Supplementary Material
Acknowledgments
We are grateful to Profs. S. C. Lakhotia, R. Raman, P. Sinha, and A. Mukherjee and Drs A. Verma, B. C. Mondal, and G. Pandey for critically reviewing the manuscript, insightful comments, and valuable suggestions throughout. We also acknowledge Prof. A. Nepveu, Prof. M. Freeman, and Dr Jaeda C. Coutinho-Budd for their communication, which helped us develop and refine our hypothesis. We also thank N. Perrimon, T. Awasaki, and Bloomington Drosophila Stock Center (BDSC), NIH P40OD018537, for providing fly stocks. FlyBase release (e.g. FB2022_03) used for any data obtained/analysed (Gramates et al. 2022). The national facility of Zeiss 510 Meta confocal microscopy at the Department of Zoology, and ISLS BHU, Leica SP8 STED confocal microscopy facility at CDC, BHU are also acknowledged. Finally, we thank the Cytogenetics Laboratory, Department of Zoology, for all the instrumentation facilities.
Contributor Information
Vaishali Yadav, Cytogenetics Laboratory, Department of Zoology, Institute of Science, Banaras Hindu University, Varanasi 221005, India.
Ramkrishna Mishra, Cytogenetics Laboratory, Department of Zoology, Institute of Science, Banaras Hindu University, Varanasi 221005, India.
Papri Das, Cytogenetics Laboratory, Department of Zoology, Institute of Science, Banaras Hindu University, Varanasi 221005, India.
Richa Arya, Cytogenetics Laboratory, Department of Zoology, Institute of Science, Banaras Hindu University, Varanasi 221005, India.
Data availability
Fly lines are obtained from Bloomington Stock Center, USA, and genetic combinations created are available upon request. The authors affirm that all other data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material available at GENETICS online.
Funding
This work was supported by grants from the Department of Biotechnology (DBT) Ministry of Science and Technology, India (BT/RLF/Re-Entry/30/2015), DST-SERB (ECR/2018/002837), DST-SERB (CRG/2022/006350), and BHU-IoE to R.A., providing SERB-JRF/SRF and BHU Non-NET-RET fellowship to V.Y., and University Grants Commission (UGC), India for providing NET-JRF to P.D.
Author contributions
All authors contributed to the article. V.Y. performed the animal experiments, image acquisition, design of the quantification workflow, statistical analysis, and made the figure panels under the supervision of R.A. Part of the quantification of Figs. 1k and 3j and Supplementary Fig. 3, a and b is done by R.M. under V.Y. and R.A. supervision. P.D. has reanalyzed the quantification of Figs. 4g and 6j under the supervision of V.Y. and R.A.
Literature cited
- Abbott NJ. 2005. Dynamics of CNS barriers: evolution, differentiation, and modulation. Cell Mol Neurobiol. 25(1):5–23. doi: 10.1007/S10571-004-1374-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres V, Bernardo N-G, Vijak M. 1992. Clox, a mammalian homeobox gene related to Drosophila cut, encodes DNA-binding regulatory proteins differentially expressed during development. Development. 116:321–334. doi: 10.1242/dev.116.2.321. [DOI] [PubMed] [Google Scholar]
- Arya R, Gyonjyan S, Harding K, Sarkissian T, Li Y, Zhou L, White K. 2019. A Cut/cohesin axis alters the chromatin landscape to facilitate neuroblast death. Development (Cambridge). 146:9. doi: 10.1242/dev.166603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avet-Rochex A, Kaul AK, Gatt AP, McNeill H, Bateman JM. 2012. Concerted control of gliogenesis by InR/TOR and FGF signalling in the Drosophila post-embryonic brain. Development. 139(15):2763–2772. doi: 10.1242/dev.074179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasaki T, Lai SL, Ito K, Lee T. 2008. Organization and postembryonic development of glial cells in the adult central brain of Drosophila. J Neurosci. 28(51):13742–13753. doi: 10.1523/JNEUROSCI.4844-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barres BA. 2008. The mystery and magic of glia: a perspective on their roles in health and disease. Neuron. 60(3):430–440. doi:10.1016/J.NEURON.2008.10.013/ATTACHMENT/A48AE9A5-41E7-4DAA-B6BB-75687BD12A10/MMC1.PDF. [DOI] [PubMed] [Google Scholar]
- Bauke AC, Sasse S, Matzat T, Klämbt C. 2015. A transcriptional network controlling glial development in the Drosophila visual system. Development (Cambridge). 142(12):2184–2193. doi: 10.1242/dev.119750. [DOI] [PubMed] [Google Scholar]
- Blochlinger K, Bodmer R, Jack J, Jan LY, Jan YN. 1988. Primary structure and expression of a product from cut, a locus involved in specifying sensory organ identity in Drosophila. Nature. 333(6174):629–635. doi: 10.1038/333629A0. [DOI] [PubMed] [Google Scholar]
- Bodmer R, Barbel S, Sheperd S, Jack JW, Jan LY, Jan YN. 1987. Transformation of sensory organs by mutations of the cut locus of D. melanogaster. Cell. 51(2):293–307. doi: 10.1016/0092-8674(87)90156-5. [DOI] [PubMed] [Google Scholar]
- Booth GE, Kinrade EFV, Hidalgo A. 2000. Glia maintain follower neuron survival during Drosophila CNS development. Development. 127(2):237–244. doi: 10.1242/DEV.127.2.237. [DOI] [PubMed] [Google Scholar]
- Britton JS, Edgar BA. 1998. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development. 125(11):2149–2158. doi: 10.1242/dev.125.11.2149. [DOI] [PubMed] [Google Scholar]
- Cadieux C, Kedinger V, Yao L, Vadnais C, Drossos M, Paquet M, Nepveu A. 2009. Mouse mammary tumor virus p75 and p110 CUX1 transgenic mice develop mammary tumors of various histologic types. Cancer Res. 69(18):7188–7197. doi: 10.1158/0008-5472.CAN-08-4899. [DOI] [PubMed] [Google Scholar]
- Cohen E, Allen SR, Sawyer JK, Fox DT. 2018. Fizzy-related dictates A cell cycle switch during organ repair and tissue growth responses in the Drosophila hindgut. Elife. 7:e38327. doi: 10.7554/eLife.38327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coqueret O, Bérubé G, Nepveu A. 1998. The mammalian Cut homeodomain protein functions as a cell-cycle-dependent transcriptional repressor which downmodulates p21(WAF1/CIP1/SDI1) in S phase. EMBO J. 17(16):4680–4694. doi: 10.1093/emboj/17.16.4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho-Budd Jaeda C, Sheehan AE, Freeman MR. 2017. The secreted neurotrophin spätzle 3 promotes glial morphogenesis and supports neuronal survival and function. Genes Dev. 31(20):2023–2038. doi: 10.1101/gad.305888.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubelos B, Sebastián-Serrano A, Beccari L, Calcagnotto ME, Cisneros E, Kim S, Dopazo A, Alvarez-Dolado M, Redondo JM, Bovolenta P, et al. 2010. Cux1 and Cux2 regulate dendritic branching, spine morphology, and synapses of the upper layer neurons of the cortex. Neuron. 66(4):523–535. doi: 10.1016/j.neuron.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q, Zavortink M, Froldi F, Golenkina S, Lam T, Cheng LY. 2021. Glial Hedgehog signalling and lipid metabolism regulate neural stem cell proliferation in Drosophila. EMBO Rep. 22(5):e52130. doi: 10.15252/embr.202052130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumstrei K, Wang F, Hartenstein V. 2003. Role of DE-cadherin in neuroblast proliferation, neural morphogenesis, and axon tract formation in Drosophila larval brain development. J Neurosci. 23(8):3325–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Orr-Weaver TL. 2001. Endoreplication cell cycles: review more for less. Cell. 105:297–306. [DOI] [PubMed] [Google Scholar]
- Feng F, Zhao Z, Zhou Y, Cheng Y, Wu X, Heng X. 2021. CUX1 facilitates the development of oncogenic properties via activating Wnt/β-catenin signaling pathway in glioma. Front Mol Biosci. 8:764. doi: 10.3389/fmolb.2021.705008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman MR, Doe CQ. 2001. Asymmetric prospero localization is required to generate mixed neuronal/glial lineages in the Drosophila CNS. Development. 128(20):4103–4112. doi: 10.1242/dev.128.20.4103. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Gutierrez A, Ibacache A, Esparza A, Barros LF, Sierralta J. 2019. Monocarboxylate transport in Drosophila larval brain during low and high neuronal activity. bioRxiv 610196. 10.1101/610196, preprint: not peer reviewed. [DOI] [PubMed]
- Griesmann H, Mühl S, Riedel J, Theuerkorn K, Sipos B, Esposito I, Vanden Heuvel GB, Michl P. 2021. CUX1 enhances pancreatic cancer formation by synergizing with KRAS and inducing MEK/ERK-dependent proliferation. Cancers (Basel). 13(10):2462. doi: 10.3390/cancers13102462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramates LS, Agapite J, Attrill H, Calvi BR, Crosby M, dos Santos G, Goodman JL, Goutte-Gattat D, Jenkins V, Kaufman T, et al. 2022. FlyBase: a guided tour of highlighted features. Genetics. 220(4):iyac035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Nung Jan Y. 2003. Different levels of the homeodomain protein Cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell. 112(6):805–818. [DOI] [PubMed] [Google Scholar]
- Hirase H, López-Hidalgo M, Klämbt C, Coutinho-Budd J, Salazar G, Ross G, Maserejian AE. 2022. Quantifying glial-glial tiling using automated image analysis in Drosophila. Front Cell Neurosci. 16:826483. doi: 10.3389/fncel.2022.826483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Urban J, Technau GM. 1995. Distribution, classification, and development of Drosophila glial cells in the late embryonic and early larval ventral nerve cord. Rouxs Arch Dev Biol. 204(5):284–307. doi: 10.1007/BF02179499. [DOI] [PubMed] [Google Scholar]
- Kaur S, Ramdzan ZM, Guiot MC, Li L, Leduy L, Ramotar D, Sabri S, Abdulkarim B, Nepveu A. 2018. CUX1 stimulates APE1 enzymatic activity and increases the resistance of glioblastoma cells to the mono-alkylating agent temozolomide. Neuro Oncol. 20(4):484–493. doi: 10.1093/neuonc/nox178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharat P, Sarkar P, Mouliganesh S, Tiwary V, Priya VBR, Sree NY, Annapoorna HV, Saikia DK, Mahanta K, Thirumurugan K. 2020. Ellagic acid prolongs the lifespan of Drosophila melanogaster. Geroscience. 42(1):271–285. doi: 10.1007/s11357-019-00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp JJ, Yaich LE, Wessells RJ, Bodmer R. 2005. Identification of genetic loci that interact with cut during Drosophila wing-margin development. Genetics. 170:1775–1795. doi: 10.1534/genetics.105.043125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledford AW, Brantley JG, Kemeny G, Foreman TL, Quaggin SE, Igarashi P, Oberhaus SM, Rodova M, Calvet JP, vanden Heuvel GB. 2002. Deregulated expression of the homeobox gene Cux-1 in transgenic mice results in downregulation of p27(kip1) expression during nephrogenesis, glomerular abnormalities, and multiorgan hyperplasia. Dev Biol. 245(1):157–171. doi: 10.1006/DBIO.2002.0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly MA, Spradling AC. 1996. The Drosophila endocycle is controlled by Cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev. 10(19):2514–2526. [DOI] [PubMed] [Google Scholar]
- Liu S, Jack J. 1992. Regulatory interactions and role in cell type specification of the Malpighian tubules by the cut, Krüppel, and caudal genes of Drosophila. Dev Biol. 150(1):133–143. doi: 10.1016/0012-1606(92)90013-7. [DOI] [PubMed] [Google Scholar]
- Liu S, McLeod E, Jack J. 1991. Four distinct regulatory regions of the cut locus and their effect on cell type specification in Drosophila. Genetics. 127(1):151–159. doi: 10.1093/GENETICS/127.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick VP, Fox DT, Spradling AC. 2013. Polyploidization and cell fusion contribute to wound healing in the adult Drosophila epithelium. Curr Biol. 23(22):2224–2232. doi: 10.1016/j.cub.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick VP, Jun AS, Spradling AC, Dr M. 2016. Wound-induced polyploidization: regulation by hippo and JNK signaling and conservation in mammals. PLoS One. 11:e0151251. doi: 10.1371/journal.pone.0151251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow C, Choy R, Blochlinger K. 1996. Functional analysis of Drosophila and mammalian cut proteins in flies. Dev Biol. 178(1):149–159. doi: 10.1006/DBIO.1996.0205. [DOI] [PubMed] [Google Scholar]
- Michl P, Ramjaun AR, Pardo OE, Warne PH, Wagner M, Poulsom R, D’Arrigo C, Ryder K, Menke A, Gress T, et al. 2005. CUTL1 is a target of TGFβ signaling that enhances cancer cell motility and invasiveness. Cancer Cell. 7(6):521–532. doi: 10.1016/j.ccr.2005.05.018. [DOI] [PubMed] [Google Scholar]
- Nandakumar S, Grushko O, Buttitta LA. 2020. Polyploidy in the adult Drosophila brain. Elife. 9:1–25. doi: 10.7554/ELIFE.54385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepveu A. 2001. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene. 270(1–2):1–15. doi: 10.1016/S0378-1119(01)00485-1. [DOI] [PubMed] [Google Scholar]
- Neufeld EJ, Skalnik DG, Lievens PMJ, Orkin SH. 1992. Human CCAAT displacement protein is homologous to the Drosophila homeoprotein, cut. Nat Genet. 1(1):50–55. doi: 10.1038/NG0492-50. [DOI] [PubMed] [Google Scholar]
- Oland LA, Tolbert LP. 2003. Key interactions between neurons and glial cells during neural development in insects. Annu Rev Entomol. 48(1):89–110. doi: 10.1146/annurev.ento.48.091801.112654. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Shy D, Hartenstein V. 2005. Morphogenesis and proliferation of the larval brain glia in Drosophila. Dev Biol. 283(1):191–203. doi: 10.1016/j.ydbio.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Pitsouli C, Perrimon N. 2013. The homeobox transcription factor cut coordinates patterning and growth during Drosophila airway remodeling. Sci Signal. 6(263):ra12. doi: 10.1126/scisignal.2003424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaggin SE, vanden Heuvel GB, Golden K, Bodmer R, Igarashi P. 1996. Primary structure, neural-specific expression, and chromosomal localization of Cux-2, a second murine homeobox gene related to Drosophila cut. J Biol Chem. 271(37):22624–22634. doi: 10.1074/JBC.271.37.22624. [DOI] [PubMed] [Google Scholar]
- Rajagopalan H, Lengauer C. 2004. Aneuploidy and cancer. Nature. 432(7015):338–341. doi: 10.1038/NATURE03099. [DOI] [PubMed] [Google Scholar]
- Ramdzan ZM, Nepveu A. 2014. CUX1, a haploinsufficient tumour suppressor gene overexpressed in advanced cancers. Nat Rev Cancer. 14(10):673–682. doi: 10.1038/nrc3805. [DOI] [PubMed] [Google Scholar]
- Read RD. 2018. Pvr receptor tyrosine kinase signaling promotes post-embryonic morphogenesis, and survival of glia and neural progenitor cells in Drosophila. Development (Cambridge). 145(23):dev164285. doi: 10.1242/dev.164285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read RD, Cavenee WK, Furnari FB, Thomas JB. 2009. A Drosophila model for EGFR-Ras and PI3K-dependent human glioma. PLoS Genet. 5(2):e1000374. doi: 10.1371/journal.pgen.1000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy BVVG, Irvine KD. 2011. Regulation of Drosophila glial cell proliferation by Merlin-Hippo signaling. Development. 138(23):5201–5212. doi: 10.1242/dev.069385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios AC, Fu NY, Jamieson PR, Pal B, Whitehead L, Nicholas KR, Lindeman GJ, Visvader JE. 2016. Essential role for a novel population of binucleated mammary epithelial cells in lactation. Nat Commun. 7(1):11400. doi: 10.1038/NCOMMS11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripka S, Neesse A, Riedel J, Bug E, Aigner A, Poulsom R, Fulda S, Neoptolemos J, Greenhalf W, Barth P, et al. 2010. CUX1: target of Akt signalling and mediator of resistance to apoptosis in pancreatic cancer. Gut. 59(8):1101–1110. 10.1136/GUT.2009.189720. [DOI] [PubMed] [Google Scholar]
- Rujano MA, Briand D, Ðeli B, Marc J, Spéder P. 2022. An interplay between cellular growth and atypical fusion defines morphogenesis of a modular glial niche in Drosophila. Nat Commun. 13(1):1–25. doi: 10.1038/s41467-022-32685-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansregret L, Nepveu A. 2008. The multiple roles of CUX1: insights from mouse models and cell-based assays. Gene. 412(1–2):84–94. doi: 10.1016/J.GENE.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Sansregret L, Vadnais C, Livingstone J, Kwiatkowski N, Awan A, Cadieux C, Leduy L, Hallett MT, Nepveu A. 2011. Cut homeobox 1 causes chromosomal instability by promoting bipolar division after cytokinesis failure. Proc Natl Acad Sci U S A. 108(5):1949–1954. doi: 10.1073/pnas.1008403108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spéder P, Brand AH. 2018. Systemic and local cues drive neural stem cell niche remodelling during neurogenesis in Drosophila. Elife. 7:e30413. doi: 10.7554/eLife.30413.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork T, Bernardos R, Freeman MR. 2012. Analysis of glial cell development and function in Drosophila. Cold Spring Harb Protoc. 7(1):1–17. doi: 10.1101/pdb.top067587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Deng WM. 2005. Notch-dependent downregulation of the homeodomain gene cut is required for the mitotic cycle/endocycle switch and cell differentiation in Drosophila follicle cells. Development. 132(19):4299–4308. doi: 10.1242/dev.02015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamori Y, Deng WM. 2013. Tissue repair through cell competition and compensatory cellular hypertrophy in postmitotic epithelia. Dev Cell. 25(4):350–363. doi: 10.1016/j.devcel.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, Kokuryo A, Imano T, Minami R, Nakagoshi H, et al. 2012. Binucleation of Drosophila adult male accessory gland cells increases plasticity of organ size for effective reproduction. Biol Syst Open Access. 1:e101. doi: 10.4172/2329-6577.1000e101. [DOI] [Google Scholar]
- Tavares AT, Tsukui T, Belmonte JCI. 2000. Evidence that members of the Cut/Cux/CDP family may be involved in AER positioning and polarizing activity during chick limb development. Development. 127(23):5133–5144. doi: 10.1242/DEV.127.23.5133. [DOI] [PubMed] [Google Scholar]
- Trapp BD, Nave KA. 2008. Multiple sclerosis: an immune or neurodegenerative disorder? Annu Rev Neurosci. 31(1):247–269. doi: 10.1146/annurev.neuro.30.051606.094313. [DOI] [PubMed] [Google Scholar]
- Unhavaithaya Y, Orr-Weaver TL. 2012. Polyploidization of glia in neural development links tissue growth to blood–brain barrier integrity. Genes Dev. 26(1):31–36. doi: 10.1101/GAD.177436.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadnais C, Davoudi S, Afshin M, Harada R, Dudley R, Clermont PL, Drobetsky E, Nepveu A. 2012. CUX1 transcription factor is required for optimal ATM/ATR-mediated responses to DNA damage. Nucleic Acids Res. 40(10):4483–4495. doi: 10.1093/nar/gks041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valarche I, Tissier-Seta JP, Hirsch MR, Martinez S, Goridis C, Brunet JF. 1993. The mouse homeodomain protein Phox2 regulates N cam promoter activity in concert with Cux/CDP and is a putative determinant of neurotransmitter phenotype. Development. 119:881–896. doi: 10.1242/dev.119.3.881. [DOI] [PubMed] [Google Scholar]
- vanden Heuvel GB, Brantley JG, Alcalay NI, Sharma M, Kemeny G, Warolin J, Ledford AW, Pinson DM. 2005. Hepatomegaly in transgenic mice expressing the homeobox gene Cux-1. Mol Carcinog. 43(1):18–30. doi: 10.1002/MC.20091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijnen AJ, van Gurp MF, de Ridder MC, Tufarellit C, Last TJ, Birnbaum M, Vaughan PS, Giordanot A, Krek W, Neufeldt EJ, et al. 1996. CDP/Cut is the DNA-binding subunit of histone gene transcription factor HiNF-D: a mechanism for gene regulation at the G1/S phase cell cycle transition point independent of transcription factor E2F (proliferation/gene expression/cyclin-dependent kinase/tumor suppressor). Biochemistry. 93:11516–11521. doi: 10.1073/pnas.93.21.11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Stetina JR, Frawley LE, Unhavaithaya Y, Orr-Weaver TL. 2018. Variant cell cycles regulated by Notch signaling control cell size and ensure a functional blood-brain barrier. Development (Cambridge). 145(3):dev157115. doi:10.1242/DEV.157115/19236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S, Melom JE, Ormerod KG, Zhang YV, Littleton JT. 2019. Glial Ca2+ signaling links endocytosis to K+ buffering around neuronal somas to regulate excitability. Elife. 8:e44186. doi: 10.7554/eLife.44186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windmueller R, Leach JP, Babu A, Zhou S, Morley MP, Wakabayashi A, Petrenko NB, Viatour P, Morrisey EE. 2020. Direct comparison of mononucleated and binucleated cardiomyocytes reveals molecular mechanisms underlying distinct proliferative competencies. Cell Rep. 30(9):3105–3116.e4. doi: 10.1016/J.CELREP.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte HT, Jeibmann A, Klämbt C, Paulus W. 2009. Modeling glioma growth and invasion in Drosophila melanogaster. Neoplasia. 11(9):882–888. doi: 10.1593/neo.09576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W-C, Montell C. 1995. Defective glia induce neuronal apoptosis in the repo visual system of Drosophila. Neuron. 14(3):581–590. doi: 10.1016/0896-6273(95)90314-3. [DOI] [PubMed] [Google Scholar]
- Xu A, Wang X, Luo J, Zhou M, Yi R, Huang T, Lin J, Wu Z, Xie C, Ding S, et al. 2021. Overexpressed P75CUX1 promotes EMT in glioma infiltration by activating β-catenin. Cell Death Dis. 12(1):1–15. doi: 10.1038/s41419-021-03424-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SO, Chikaraishi DM. 1994. Isolation of two E-box binding factors that interact with the rat tyrosine hydroxylase enhancer. J Biol Chem. 269(28):18453–18462. doi: 10.1016/s0021-9258(17)32330-x. [DOI] [PubMed] [Google Scholar]
- Yuan X, Sipe Conor W, Suzawa M, Bland ML, Siegrist SE. 2020. Dilp-2-mediated PI3-kinase activation coordinates reactivation of quiescent neuroblasts with growth of their glial stem cell niche. PLoS Biol. 18(5):e3000721. doi: 10.1371/journal.pbio.3000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z, Ha N, Papagiannouli F, Hamacher-Brady A, Brady N, Sorge S, Bezdan D, Lohmann I. 2012. Antagonistic regulation of apoptosis and differentiation by the cut transcription factor represents a tumor-suppressing mechanism in Drosophila. PLoS Genet. 8(3):e1002582. doi: 10.1371/journal.pgen.1002582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Zuo YX, Jiang RT. 2019. Astrocyte morphology: diversity, plasticity, and role in neurological diseases. CNS Neurosci Ther. 25(6):665–673. doi: 10.1111/cns.13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke N, Edgar BA, DePamphilis ML. 2013. Endoreplication. Cold Spring Harb. Perspect. Biol. 5(1):a012948. doi: 10.1101/cshperspect.a012948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke N, Korzelius J, vanStraaten M, Bender K, Schuhknecht GFP, Dutta D, Xiang J, Edgar BA. 2014. Fly-FUCCI: a versatile tool for studying cell proliferation in complex tissues. Cell Rep. 7(2):588–598. doi: 10.1016/j.celrep.2014.03.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Fly lines are obtained from Bloomington Stock Center, USA, and genetic combinations created are available upon request. The authors affirm that all other data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplemental material available at GENETICS online.