Abstract
Staphylococcus aureus poses a significant threat in both community and hospital settings due to its infective and pathogenic nature combined with its ability to resist the action of chemotherapeutic agents. Methicillin-resistant S. aureus (MRSA) represents a critical challenge. Metal-chelating thiosemicarbazones (TSCs) have shown promise in combating MRSA and while previous studies hinted at the antimicrobial potential of TSCs, their mechanisms of action against MRSA are still under investigation. We screened a chemical library for anti-staphylococcal compounds and identified a potent molecule named R91 that contained the NNSN structural motif found within TSCs. We identified that R91 and several structural analogs exhibited antimicrobial activity against numerous S. aureus isolates as well as other Gram-positive bacteria. RNAseq analysis revealed that R91 induces copper and oxidative stress responses. Checkerboard assays demonstrated synergy of R91 with copper, nickel, and zinc. Mutation of the SrrAB two-component regulatory system sensitizes S. aureus to R91 killing, further linking the oxidative stress response to R91 resistance. Moreover, R91 was found to induce hydrogen peroxide production, which contributed to its antimicrobial activity. Remarkably, no mutants with elevated R91 resistance were identified, despite extensive attempts. We further demonstrate that R91 can be used to effectively treat an intracellular reservoir of S. aureus in cell culture and can reduce bacterial burdens in a murine skin infection model. Combined, these data position R91 as a potent TSC effective against MRSA and other Gram-positive bacteria, with implications for future therapeutic development.
Keywords: MRSA, S. aureus, antibiotic, MOA, infection model, thiosemicarbazones
Staphylococcus aureus is a significant cause of community and hospital acquired infections, owing to the emergence of strains that display enhanced transmission, virulence, and/or resistance to multiple antibiotics (1). This pathogen is capable of causing a wide spectrum of infections, ranging from skin and soft tissue infections to life-threatening conditions such as bacteremia and endocarditis (1). Furthermore, the remarkable ability of S. aureus to acquire and upregulate antimicrobial resistance genes, coupled with the overuse and misuse of antibiotics, has led to a critical threat (2). Methicillin-resistant S. aureus (MRSA) is of particular concern. Beyond β-lactams, MRSA displays resistance to various antibiotic classes through mechanisms involving target modification, drug modification, and decreased cell membrane permeability (3). This abundance of resistance mechanisms has resulted in approximately 95% of MRSA clinical isolates being resistant to first-line antibiotics (4). Strains resistant to vancomycin and daptomycin, or “last resort” antibiotics, further complicate treatment, underscoring the urgency of discovering new antibiotics to address the growing threat of multidrug-resistant MRSA (MDR-MRSA) (2).
Metal ion homeostasis plays a critical role in bacterial infection and pathogenesis (5, 6). Metals such as iron, zinc, and copper are essential for various bacterial processes, including metabolism, DNA replication, and defense against oxidative stress induced by reactive oxygen species (ROS) (5, 6). Consequently, pathogens like S. aureus rely on tightly regulated metal ion acquisition and utilization systems to thrive within the host environment and evade host nutritional immunity mechanisms (7, 8).
Metal chelators, particularly thiosemicarbazones (TSCs), have emerged as a promising solution to combat MDR-MRSA (9, 10, 11, 12, 13). Characterized by their NNSN metal-binding structural motif (11), TSCs have been shown to interact with various metals, most commonly copper, and their antibacterial activity has often been demonstrated to be copper-dependent (11, 12). These chelators can mimic the host's nutritional immunity strategy by sequestering or chelating essential metals from bacteria, hindering vital bacterial processes that rely on these metals (11). Additionally, complexation with copper can deliver toxic levels of the metal to bacterial cells and increase membrane permeability to disrupt cell integrity and function (12).
Importantly, TSCs offer stability and selectivity in metal binding, reducing off-target toxic effects, and enhancing the specificity for bacterial targets (14, 15). Prior studies have reported the antimicrobial potential of TSCs (9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19), but their full spectrum of mechanisms of activity against MRSA remains an area of active research.
Here we report the identification of a TSC (named R91) that is highly effective in the low micromolar concentration range against MRSA, using high-throughput screening of bioactives from the Maybridge compound collection. Assessment of the activity of R91 and structurally related analogs against S. aureus isolates and other bacterial species demonstrated these compounds possess broad-spectrum antimicrobial activity against Gram-positive bacteria. RNAseq identified R91-induced copper and oxidative stress responses in S. aureus, highlighting its metal-associated antibacterial activities. Checkerboard assays demonstrated enhanced antibacterial activity of R91 when combined with copper, nickel, and zinc. Furthermore, we found that mutation of the SrrAB two-component regulatory system, which is involved in the response to oxidative stress, sensitized S. aureus to R91, providing an important link between sensing of ROS by SrrAB and resistance to R91. Importantly, despite numerous attempts using different strategies, we failed to identify S. aureus mutants with elevated resistance to R91. Finally, after demonstrating that R91 was nontoxic to eukaryotic cells (macrophages) at concentrations well above the minimum inhibitory concentration (MIC), we showed that R91 was effective in clearing intracellular macrophage infections. In subcutaneous (SQ) infections of mice, R91 reduced both lesion size and bacterial burden. Together, these findings advance our understanding of R91 as a representative TSC, offering important insight into its mechanisms of action and potential therapeutic applications.
Results
High-throughput screening and identification of an anti-staphylococcal TSC
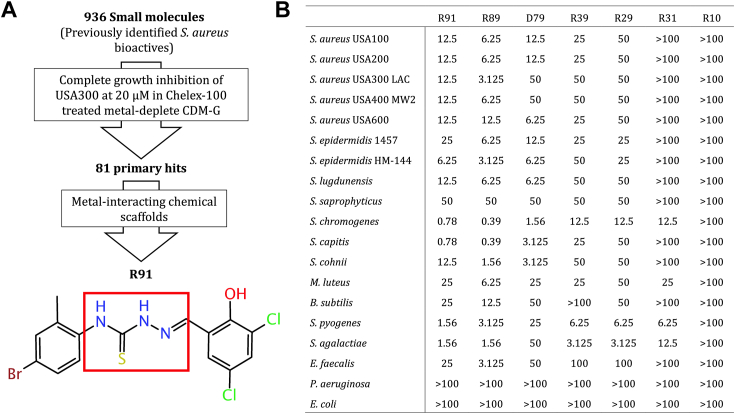
A high-throughput screen of 50,000 diverse small molecules sourced from the Maybridge a library against a methicillin-sensitive S. aureus (MSSA) strain in tryptic soy broth (TSB) at 10 μM previously identified S. aureus bioactives showing partial to complete growth inhibition of the tested MSSA under the screen conditions (20). Herein, we screened 936 of these bioactives against the S. aureus USA300 LAC strain, a widely circulating community-acquired MRSA strain in North America (21), at 20 μM in duplicate (Fig. 1A). We sought to enrich for compounds with the ability to enhance metal-related toxicity or interfere with determinants involved in metal ion homeostasis, which are more likely expressed under metal ion limitation rather than under metal-replete conditions (6). As such, we conducted the screen in a Chelex-100 resin-treated metal-deplete chemically defined medium containing glucose (CDM-G) (22), identifying 81 compounds that completely inhibited growth of S. aureus USA300 LAC. We filtered the hits based on chemical scaffolds known to interact with metals. We then prioritized R91 (see Fig. 1A), a TSC with an MIC of 20 μM against USA300 LAC in metal-deplete CDM-G, for further follow up. Importantly, the structure of R91 is unique compared to other reported small-molecule modulators of metal homeostasis (9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 23, 24, 25, 26). The ability of various TSC compounds to chelate metals and their potential as antimicrobials has been previously established, yet their mechanism of action remains undefined (9, 11, 12, 13, 16). In addition to R91, we selected other TSC structural analogs D79, R29, R39, R89, R31, and R10 (see Fig. S1), for subsequent testing to infer preliminary structure activity relationships.
Figure 1.
High-throughput screening identifies R91, a potent TSC with activity against MRSA USA300 LAC.A, the workflow undertaken in this study that identified R91 and the chemical structure of R91. A red box has been placed around the core NNSN structure, characteristic of all TSCs. B, MIC (μM) of TSCs against various Staphylococcus aureus isolates and other bacterial species was determined using the concentration of drug that completely inhibited growth as determined by A600 readings after 24 h of incubation in THY for Streptococcus, TSB for all other Gram-positive species, and LB for Gram-negatives. Strain designations and descriptions for the various bacterial species listed can be found in Table S1.
Antibacterial activities of TSCs on assorted bacteria
To assess the effect of R91 on bacterial viability outside of the above screening conditions, we evaluated the bactericidal activity of this TSC against USA300 LAC. Bacteria were co-inoculated with R91 at various concentrations in PBS before being incubated for 24 h at 37 °C. Subsequently, the number of viable bacteria was determined by drop plating onto tryptic soy agar (TSA) that was then incubated for an additional 24 h. These data revealed that, under the conditions used, only at concentrations ≥100 μM was R91 capable of significantly reducing S. aureus viability (Fig. S2A). To assess the efficacy of R91 against actively dividing cells, we substituted Mueller Hinton Broth (MHB) for PBS in our method. Similar trends were observed in MHB, with R91 inhibiting growth below 100 μM and reducing viability at concentrations ≥100 μM (Fig. S2B). Notably, although 100 μM of R91 decreased bacterial viability in MHB, the TSC was less effective than in PBS, suggesting a particular ability of R91 in targeting nondividing cells.
To further assess the utility of R91 and the other selected TSCs as antimicrobials, we assessed the antimicrobial potential of these compounds against a variety of clinically relevant bacteria (Fig. 1B). Of the TSCs examined, R89, R91, and D79 exhibited substantial antimicrobial activity against most of the Gram-positive bacteria examined, with MIC values ranging from less than 1 μM to 25 μM, except for Staphylococcus saprophyticus, which was more resistant. Interestingly, the MIC for D79 against representative USA300 and USA400 isolates was significantly higher than that of R89 and R91. In contrast, R10 and R31 exhibited no antimicrobial activity against most Gram-positive and Gram-negative bacteria, although notably, R31 was found to be effective against S. chromogenes, S. pyogenes, S. agalactiae, and M. luteus. R89 was identified as the analog with activity most comparable to R91 and thus was selected, along with R91, for stability and structure analysis. NMR spectroscopy and high-resolution mass spectrometry were used to confirm the chemical structures of R91 and R89 (Figs. S3–S11). Minimal time-dependent hydrolysis of the two TSCs was observed over 48 h, strongly suggesting that the active molecules were the TSCs (Figs. S12 and S13). Moreover, structural analysis revealed that TSCs with potent activity shared specific features, such as hydroxyl and halogen groups in the first aromatic ring. Additionally, an additional halogen in the 4′ position of the second aromatic ring further enhanced, but was not required for, antimicrobial activity. Despite the relatively comparable activity of R91 and R89 against USA300 LAC, we decided to use R91 as our representative TSC in follow-up experiments given that this TSC, but not the analogs, was identified in our initial high-throughput screening.
R91 induces copper and oxidative stress responses in S. aureus
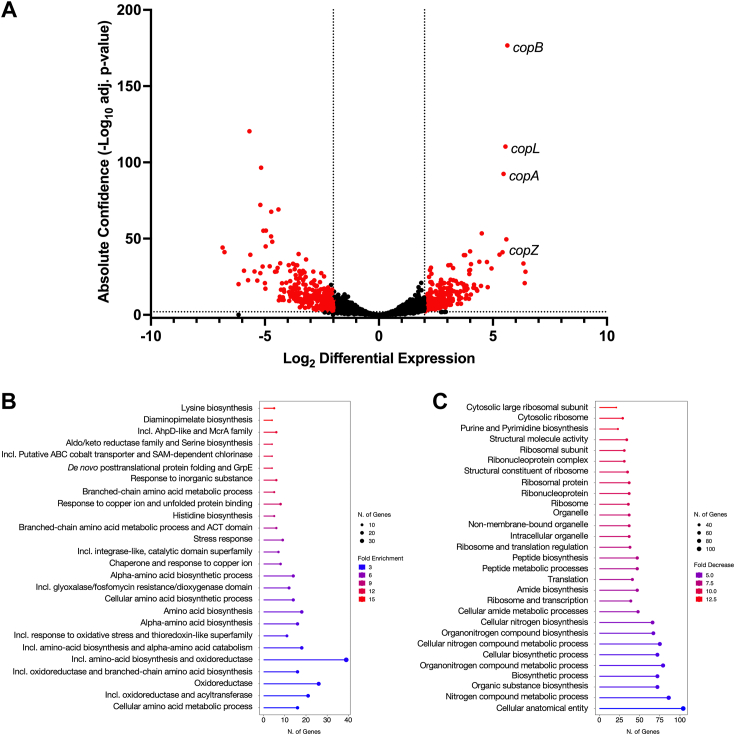
RNAseq was used to gain insight into the immediate transcriptional response of S. aureus to TSC exposure, using R91 as the representative molecule. Bacteria in early exponential growth phase were exposed to 0.5× MIC R91 for 30 min, at which time RNA was extracted and used for RNAseq analysis. The analysis of differentially expressed genes revealed that R91 exposure elicited a dramatic transcriptional response, as 267 genes were significantly upregulated while 269 genes were significantly downregulated (Fig. 2A). One prominent set of genes that were significantly upregulated in response to R91 exposure were the copA (29-fold), copZ (26-fold), copB (32-fold), and copL (30-fold) genes, which are associated with copper resistance (27). This upregulation suggests that exposure of S. aureus to R91 is activating a response similar to a high-copper stress response (27, 28, 29), despite the fact that copper was not added to the TSB growth medium for these experiments.
Figure 2.
R91 exposure results in the upregulation of copper homeostasis genes.A, volcano plot of RNAseq analysis of Staphylococcus aureus USA300 LAC in early exponential phase after 30 min exposure to sub-MIC R91 (3.125 μM). Results are from three independent experiments. Differentially expressed genes (absolute confidence >2, |log2 differential expression| >2) are colored in red. Detailed RNAseq data can be found in Table S3. Gene Ontology (GO) analysis of significantly upregulated (B) and downregulated (C) genes. Shown in (B and C) are the GO-enriched lollipop plots of differentially expressed pathways identified within the top 30 pathways. The GO terms corresponding to each pathway were initially filtered by a false discovery rate (FDR) of <0.05, before being selected by FDR and sorted for fold enrichment. The size of the dot represents the number of genes, and the color corresponds to the fold enrichment of the pathway.
Gene ontology (GO) analysis revealed several differentially regulated pathways and biological processes in response to R91 exposure (Fig. 2, B and C). Increased expression (>2-fold) of the CtsR (clpB, clpC, ctsR, mcsA, mcsB, clpP) and HrcA (dnaK, hrcA, grpE, groELS) regulons, which contain several protease and chaperone genes that are essential for resistance to heat shock and oxidative stress (30, 31), suggests activation of the stress response after R91 exposure. In particular, the upregulation of these regulons in the context of antimicrobial treatment has been associated with oxidative damage and accumulation of misfolded proteins (32). Consistent with the notion that R91 elicits an oxidative stress response, we also observed that R91 exposure induced expression of the katA gene (3.2-fold) encoding catalase and the sodA and sodM genes encoding superoxide dismutase (3- and 3.8-fold, respectively). Together, these results suggest a general oxidative stress response is activated upon R91 exposure.
Furthermore, R91 elicited decreased expression of pathways associated with purine and pyrimidine biosynthesis, ribosomal subunits, and protein translation genes (Fig. 2C), indicating induction of the stringent response (33). This shift in gene expression suggests a reallocation of bacterial resources toward stress response mechanisms over rapid growth in response to R91 exposure.
Metal-associated antibacterial activity of R91
Our RNAseq data revealed that exposure to R91 induced the expression of copper-detoxification genes. To explore whether bacteria deficient for these genes would display altered sensitivity to the antimicrobial effects of R91, we utilized mutants lacking either copAZ, copBL, or copAZ, and copBL and evaluated the MIC of R91 on these mutant strains. Surprisingly, none of the KO strains displayed an altered MIC for R91 (Fig. S14A), suggesting that these genes are not required for R91 activity or resistance. Nonetheless, R91 exposure did elicit a copper stress response, potentially implying a direct interaction between R91 and copper, supported by previous studies on the metal-binding abilities of TSCs (13, 14).
To rule out the possibility that the observed effects were due to copper contamination of R91, we performed inductively coupled plasma mass spectrometry (ICP-MS) analysis. Our results confirmed that R91 was not contaminated with copper during synthesis, with the measured copper concentration being approximately 1 ppb in a 115.4 μM solution in 1% dimethylsulfoxide (DMSO), equating to an approximate molar ratio of 1:9000 (Table S4). Additionally, we verified that R89, an analog with similar activity to R91, also had no detectable copper contamination, with a similar concentration of 1 ppb in a 139.6 μM solution in 1% DMSO, also yielding a molar ratio of approximately 1:9000 (Table S4). These findings suggest that activation of copper efflux-related genes in response to R91 exposure may be a result of the drug interacting with trace copper in the media and transporting it into the bacteria and not a result of copper contamination of the molecules themselves.
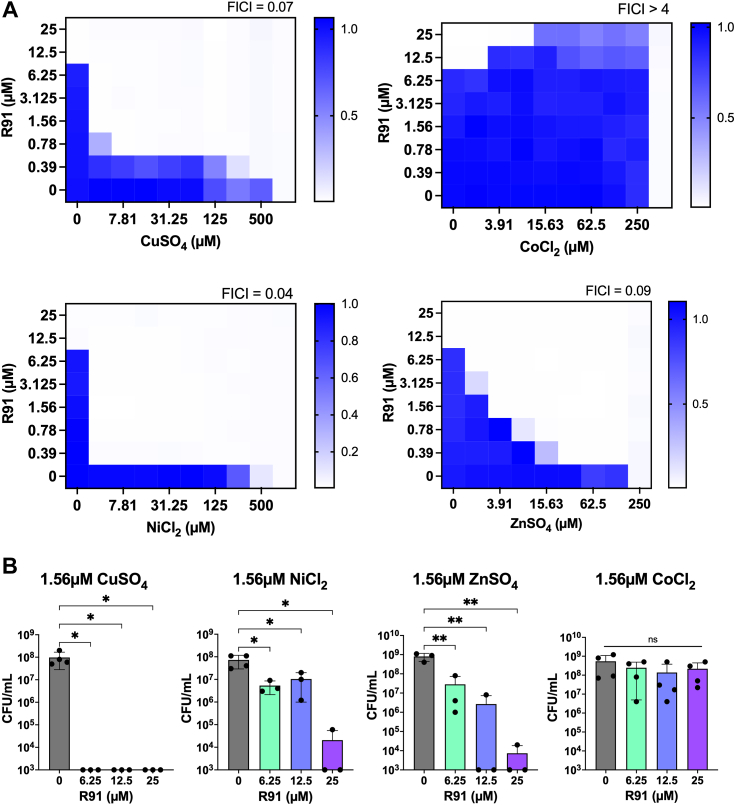
To further investigate the impact of R91 and metals on S. aureus growth, we assessed the synergy of R91 with copper and other metals using checkerboard assays and the fractional inhibitory concentration index (FICI) to define interaction types observed. Interestingly, iron, calcium, manganese, and magnesium had little effect on the activity of R91 (Fig. S15). However, the addition of copper did result in enhanced antibacterial activity of R91, indicated by synergy (FICI ≤ 0.5) between the two (Fig. 3A). Similar synergistic effects were observed with nickel (FICI = 0.04) and zinc (FICI = 0.09) (Fig. 3A). Notably, the synergy between R91 and copper, nickel, or zinc not only reduced the MIC of R91 but also induced a bactericidal effect at much lower compound concentrations (Fig. 3B). Despite this, the MIC of R91 remained unchanged between various Chelex-100 treated and untreated media (Fig. S14, B and C). These findings suggest that the combined action of R91 and specific metals significantly enhances the compound's antibacterial efficacy, but that the activity of R91 is not dependent on the presence of these metals under the conditions we have tested.
Figure 3.
Anti-MRSA activity of R91 is enhanced by Cu2+, Ni2+, and Zn2+. A, representative checkerboard assays of R91 with CuSO4, ZnSO4, NiCl2, or CoCl2 against Staphylococcus aureus USA300 LAC. A diagram corresponding to the setup of the checkerboards is displayed in the report by Bellio et al., 2021 (78). Growth was quantified by measuring absorbance at A600, and results are represented using heat maps, where a darker blue color corresponds to a greater bacterial density. Synergy is defined as FICI < 0.5, indifference as 0.5 < FICI ≥ 4, and antagonism as FICI > 4. The checkerboards were repeated three times independently and all replicates yielded similar results. B, bactericidal activity of R91 when in combination with synergistic metals. CFU/ml was quantified after bacterial exposure to R91 for 24 h. The limit of detection (LOD) was 103 CFU/ml. Data points below the limit of detection (LOD) were plotted at the LOD. Data are shown as the mean ± SD from at least three biological replicates. ∗p ≤ 0.05, ∗∗p ≤ 0.01 using a one-way ANOVA with Dunnett’s multiple comparison.
Interestingly, cobalt exhibited antagonism (FICI > 4) when in combination with R91 (Fig. 3A). This resulted in no decrease in CFU/ml of bacteria treated with both R91 and cobalt (Fig. 3B). While complexation of cobalt with TSCs has been reported (34), a change in conformation or coordination of R91 molecules may result in a loss of activity or target-binding ability.
Oxidative stress response is essential for resistance to R91
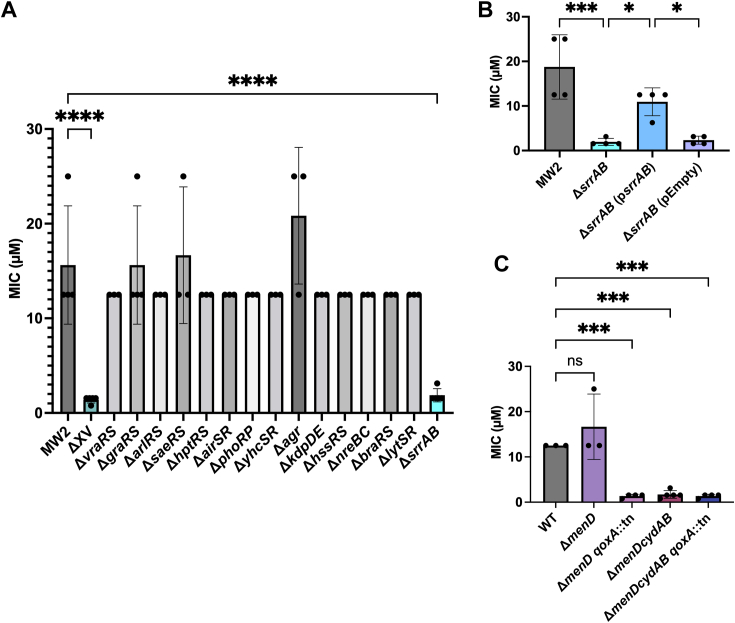
Two-component regulatory systems (TCRSs) enable bacteria to adapt and respond to environmental changes, including antibiotic exposure, by altering gene expression (35). Indeed, several staphylococcal TCRSs have been associated with antimicrobial resistance (36, 37). To begin to explore the role of TCRSs in resisting the antimicrobial activity of R91, we performed MIC analysis using a strain lacking all non-essential TCRSs (ΔXV) (38). This analysis revealed that ΔXV displayed heightened sensitivity to R91 as indicated by a 4-fold decrease in the MIC (Fig. 4A), suggesting involvement of these systems in resistance modulation. However, ΔXV did not exhibit hypersensitivity to R91 analogs, except for R31 (Fig. S16), indicating that the resistance mechanism may be specific to certain molecular structures or functional groups within the analogs.
Figure 4.
Mutation of srrAB sensitizes Staphylococcus aureus USA400 MW2 to R91.A, MIC of R91 against two-component system mutants in an S. aureus USA400 MW2 background. Data are shown as the mean ± SD from at least three biological replicates. ∗∗∗∗p ≤ 0.0001 using a one-way ANOVA with Dunnett’s multiple comparisons. B, MIC of the S. aureus USA400MW2 background strain, ΔsrrAB, complement (psrrAB), and empty vector (pEmpty). Data are shown as the mean ± SD from at least three biological replicates. ∗p ≤ 0.05, ∗∗∗p ≤ 0.001 using a one-way ANOVA with a Šidák multiple comparison. C, MIC of ΔmenD and terminal electron acceptor mutants (qoxA::tn and ΔcydAB) in a ΔmenD background. MIC was determined in TSB after 24 h of growth using concentrations that completely inhibited growth, as determined by measurement of A600. Data are shown as the mean ± SD from at least three biological replicates. ∗∗∗p ≤ 0.001 using a one-way ANOVA with a Šidák multiple comparison.
Further MIC analyses on single TCRS mutants (38) identified heightened sensitivity in S. aureus lacking only the SrrAB TCRS, as indicated by the 4-fold decrease in the MIC (Fig. 4A). Complementation of the ΔsrrAB mutant by providing srrAB in trans resulted in the loss of heightened sensitivity (Fig. 4B), confirming that the SrrAB two-component system is responsible for the level of R91 resistance that is observed in S. aureus USA400 MW2. SrrAB is an oxygen-responsive regulator involved in biofilm formation and the oxidative stress response (39, 40, 41). Thus, the hypersensitivity of the ΔsrrAB mutant suggests that R91 may generate oxidative stress as part of its antibacterial mechanism.
In response to oxidative stress, SrrAB induces expression of the qox operon. In particular, qoxA is part of the quinol oxidase complex, contributing to respiratory electron transport (42). Therefore, we speculated that R91 may influence the oxidative stress response by interfering with the electron transport chain (ETC). CydAB, a cytochrome bd oxidase complex, and MenD, which is involved in the biosynthesis of menaquinone, are key components of the ETC and play essential roles in oxidative stress defense (43, 44). Thus, menD, cydAB, and qoxA mutants were used to investigate the effect of R91 on the ETC in S. aureus. Interestingly, while S. aureus deficient for menD did not exhibit increased sensitivity to R91, mutants deficient for cydAB, qoxA or cydAB, and qoxA, in a menD background were hypersusceptible to R91 (Fig. 4C). These data show that S. aureus lacking functional Qox and Cyd terminal oxidases in menD-deficient bacteria are hypersensitive to the antimicrobial effects of R91.
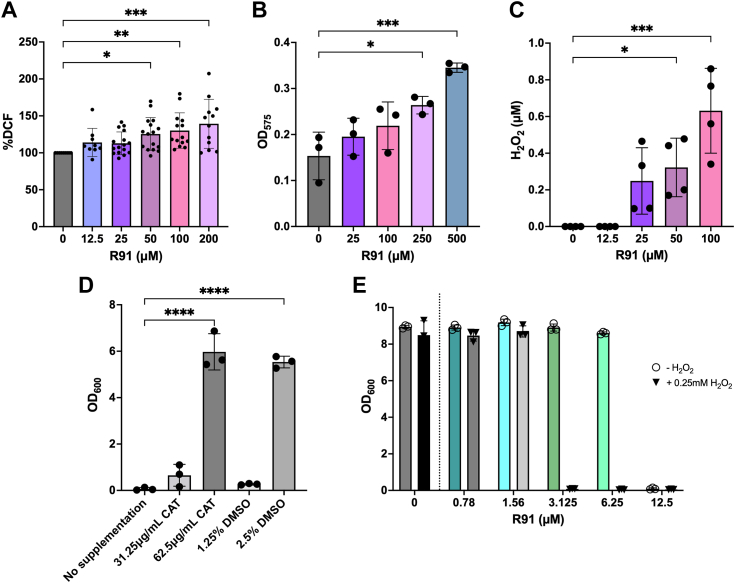
R91 elicits hydrogen peroxide formation
The upregulation of genes involved in oxidative stress (i.e. katA, sodA, and sodM) and the hypersensitivity of the aforementioned mutants to R91 prompted investigation of whether R91 induces formation of ROS. To this end, we employed dichlorodihydrofluorescein diacetate (DCFH-DA), a compound that is converted into the fluorescent product dichlorofluorescein upon exposure to ROS (Fig. 5A). Using this assay, we observed a significant increase in fluorescence after R91 exposure and the DCFH-DA fluorescence was directly proportional to the concentration of R91 used. Taken together, these data indicate that R91 induces ROS formation in S. aureus.
Figure 5.
R91 causes the production of hydrogen peroxide in Staphylococcus aureus.A, intracellular ROS levels in S. aureus USA300 LAC after treatment with R91. Bacteria were treated with DCFH-DA for 30 min after R91 exposure and fluorescence was measured at an excitation of 485 nm and an emission of 535 nm. Fluorescence was normalized to the A600 absorbances of the untreated controls. Data are shown as the mean ± SD of at least three biological replicates. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001 using a one-way ANOVA with Dunnett’s multiple comparison. B, intracellular superoxide (O2•−) levels in S. aureus USA300 LAC after exposure to R91 for 30 min. Nitro-blue tetrazolium (NBT) reduction was used to determine O2•− levels. Data are shown as the mean ± SD of at three independent experiments. ∗p ≤ 0.05, ∗∗∗p ≤ 0.001 using a one-way ANOVA with Dunnett’s multiple comparison. C, H2O2 levels in S. aureus USA300 LAC after R91 exposure. Bacteria were incubated with R91 for 1 h, before being lysed with lysostaphin and added to Amplex red and incubated for 30 min in the dark. Fluorescence was measured at an excitation of 560 nm and an emission of 590 nm. H2O2 (μM) was quantified using an equation generated from a linear regression of a set of H2O2 standards. Data are shown as the mean ± SD of three biological replicates. ∗p ≤ 0.05, ∗∗∗p ≤ 0.001 using a one-way ANOVA with Dunnett’s multiple comparison. D, A600 of S. aureus treated with R91 at the MIC was determined in the presence and absence of ROS scavengers, catalase (CAT) and dimethylsulfoxide (DMSO). A600 was determined after exposure to R91 and scavenger for 24 h. Data are shown as the mean ± SD of three biological replicates. ∗∗∗∗p ≤ 0.0001 using a one-way ANOVA with Dunnett’s multiple comparison. E, killing of S. aureus USA300 LAC by R91 and R91 with sublethal H2O2. A600 was read after exposure to 12.5 μM R91 ± 0.25 mM H2O2 for 24 h in TSB. Data are shown as the mean ± SD of three biological replicates.
We next sought to identify the nature of the ROS produced in response to exposure to R91. To achieve this, we employed a nitroblue tetrazolium (NBT) assay which can detect the presence of superoxide anions (O2-) through formazan formation. This assay revealed R91 concentration-dependent formazan production but only at concentrations significantly above the MIC (Fig. 5B). This suggests that O2- is likely not a major contributor to the anti-staphylococcal properties of R91.
Next, we considered whether R91 could induce the formation of hydrogen peroxide (H2O2), which can kill S. aureus (45). To address this, we utilized Amplex Red assays to measure H2O2 production in response to R91 treatment of S. aureus. In the absence of R91, the measurable level of H2O2 in S. aureus lysates was negligible. In contrast, in the presence of R91, a notable increase in Amplex Red fluorescence was detected (Fig. 5C), suggesting that R91 is causing the production of H2O2. Scavenger assays using catalase (for H2O2) and DMSO (for hydroxyl radicals) reinforced this notion as addition of either to R91-exposed cultures led to growth recovery at the MIC (Fig. 5D). Furthermore, indirect analysis of catalase activity using a foam formation assay (46), revealed that cultures exposed to R91 and subsequently to 30% hydrogen peroxide displayed enhanced foam formation (Fig. S17). Moreover, exposure to sub-inhibitory H2O2 concentrations (0.25 mM) reduced the MIC of R91 4-fold as compared to R91 alone (Fig. 5E). Finally, mutants lacking catalase (i.e. a katA::tn strain) were found to be hypersensitive to R91 and were killed at concentrations well below the bactericidal concentration of R91 against USA300 LAC (Fig. S18, A and B), while those with enhanced catalase expression (i.e. a perR::tn strain) (47) were more resistant to R91 killing, with no changes in CFU/ml at concentrations of R91 greater than 50 μM (Fig. S18C). Like the perR::tn mutant, the USA300 pKatA strain, which also overexpresses catalase, was found to be less sensitive to killing by R91, further confirming the importance of H2O2 detoxification mechanisms in R91 resistance (Fig. S18D). Taken together, these data establish that R91 induces ROS in the form of hydrogen peroxide and in response to R91, S. aureus induces the expression of active catalase enzyme that contributes to the basal level of resistance observed.
R91 treatment can prevent and eradicate biofilm formation
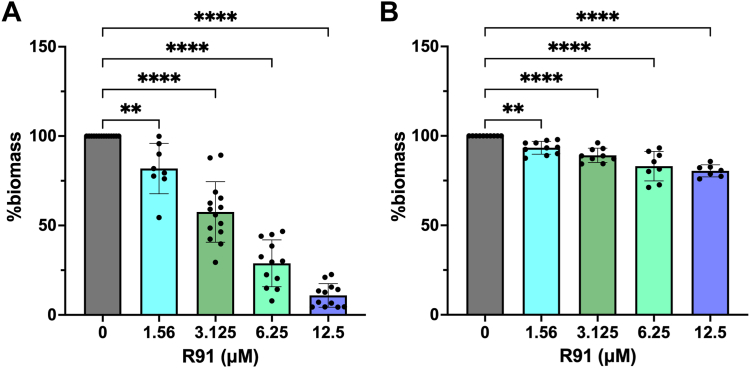
S. aureus biofilm formation is critically important in clinical settings due to its association with persistent infections and increased antibiotic resistance (48). Moreover, biofilm formation enables bacteria to evade host immunity and to resist conventional antibiotic treatments (48). We thus evaluated the prophylactic efficacy of R91 against S. aureus biofilm formation which involved statically culturing S. aureus with R91 for 24 h, followed by biofilm quantification. At a concentration of R91 4-fold below the established MIC, R91 significantly decreased biofilm formation as compared to untreated controls (Fig. 6A). This effect was further exacerbated at the MIC, suggesting R91 could be used to mitigate biofilm-formation in static culture.
Figure 6.
R91 prevents and eradicates biofilms. Biofilm formation in the presence and absence of R91 after (A) co-incubation of R91 and Staphylococcus aureus USA300 LAC for 24 h, (B) 24 h of R91 incubation after 24 h of initial biofilm formation. Biofilms were quantified using an A590 after crystal violet staining. %biomass was quantified by comparing the OD of the treated samples to the OD of the untreated samples. Data are shown as the mean ± SD of at least three independent experiments. ∗∗p ≤ 0.01, ∗∗∗∗p ≤ 0.0001 using a one-way ANOVA with Dunnett’s multiple comparison.
We next investigated whether R91 could dissipate preformed biofilms. Here, biofilms were allowed to form over 24 h at which time R91 was added to the culture medium for an additional 24 h (Fig. 6B). These experiments revealed that at the MIC, R91 significantly reduced pre-formed biofilm. This anti-biofilm effect was also observed using R91 concentrations up to 10-fold below the MIC (Fig. 6B).
Prolonged exposure to R91 does not produce TSC-resistant bacteria
To identify the R91 target(s) and to explore its mechanism of action, we next sought to identify S. aureus mutants that had increased resistance to R91. To this end, we screened the Nebraska Transposon Mutant Library (NTML), which is comprised of 1920 nonessential transposon mutants in an S. aureus USA300 background. Using this approach, we failed to identify any mutant that displayed R91 resistance. In a parallel effort, using disk diffusion assays, we attempted to isolate spontaneous resistant colonies from within zones of inhibition; however, even after 5 days of incubation, resistant colonies failed to grow within the zones of inhibition. We continued this pursuit and next performed a 14-days serial passage experiment where S. aureus bacteria were exposed to 0.5× MIC of R91; however, resistant colonies were never identified (Fig. S19). In contrast, USA300 LAC exposed to 0.5× MIC ciprofloxacin quickly developed resistance, with stable resistance emerging after 5 days of subculturing indicating the approach, in principle, can produce antibiotic resistant bacteria (Fig. S19). Next, these experiments were extended out to 50 days, and despite this prolonged exposure to sub-MIC R91, resistance was never observed. Attempts to induce resistance in the presence of copper and with the copper efflux-deficient mutant ΔcopAZB also failed to produce resistant colonies. Taken together, these data indicate that resistance to R91 in S. aureus does not readily occur, at least under the conditions employed here.
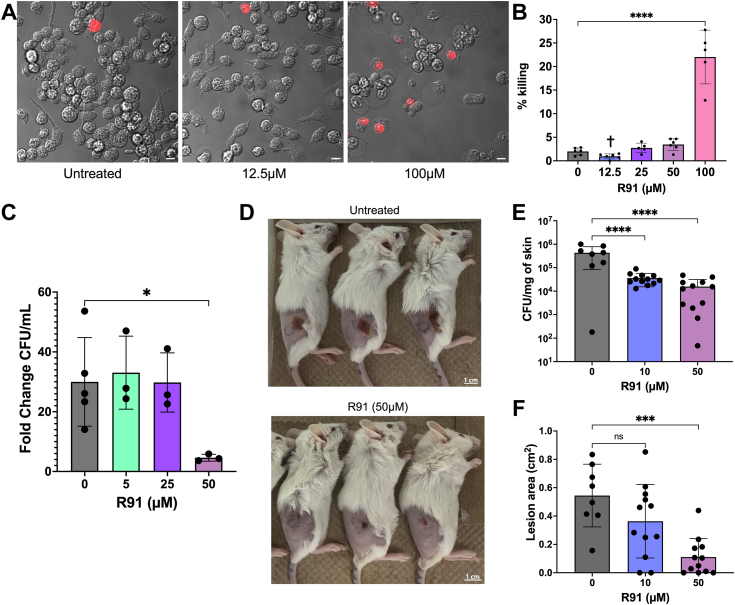
R91 can kill S. aureus residing within macrophages
Several TSCs are currently approved for anti-cancer and anti-Mtb treatments (49, 50). Despite this, their use is often impeded by poor water solubility, leading to challenges in bioavailability and potential toxicity (51, 52). To investigate whether R91 was noncytotoxic, we conducted experiments using RAW 264.7 macrophages as a model eukaryotic immune cell. Macrophages were either exposed to R91 or treated with an equivalent volume of DMSO for 24 h and then stained with propidium iodide (PI) to evaluate cell death (Fig. 7A). Quantification of dead cells through PI staining (Fig. 7B) revealed that R91 demonstrated noncytotoxic behavior at concentrations up to 4× greater than the MIC of the compound for S. aureus. However, at 100 μM (8× the MIC), cytotoxic effects were observed. Importantly, most R91 analogs were found to be cytotoxic at the concentration required to inhibit USA300 LAC (Fig. S20). While R89 was an exception to this, cytotoxicity occurred at levels notably below that of R91, and this analog was found to be cytotoxic at concentrations required to inhibit several non-USA300 strains of S. aureus, as well as other Gram positive bacteria.
Figure 7.
R91 is noncytotoxic and abrogates infections in macrophages and in a skin infection model. R91 was incubated with RAW 264.7 macrophages in RPMI supplemented with 5% fetal bovine serum (FBS). The MIC of R91 in the RPMI for Staphylococcus aureus USA300 LAC is 6.25 μM. A, after 24 h, the cells were stained with propidium iodide (PI) and imaged using a widefield microscope at 63× magnification with all scale bars representing 10 μm (B) and the %killing of macrophages was calculated using the number of PI-stained cells compared to the total number of cells. The MIC (μM) of R91 against USA300 LAC is indicated by †. Data are shown as the mean ± SD of at least five independent experiments ∗∗∗∗p ≤ 0.0001 using a one-way ANOVA with Dunnett’s multiple comparison. C, R91 was added to S. aureus–infected macrophages 1.5 hpi. The cells were then lysed at 12 hpi and CFUs were quantified to determine the ability of R91 to inhibit bacterial replication intracellularly. Data are shown as the mean ± SD of three independent experiments. ∗p ≤ 0.05 using a one-way ANOVA with Dunnett’s multiple comparison. D, representative images showing skin lesions 3 dpi. Mice were subcutaneously co-injected with S. aureus USA300 LAC and R91. An additional dose of R91 was given at 1 dpi. The mice were sacrificed at 3 dpi and (E) CFU/mg of homogenized lesion and (F) lesion sizes were quantified. Data are shown as the mean ± SD of three independent experiments. ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001 using a one-way ANOVA with Dunnett’s multiple comparison.
S. aureus exhibits the ability to survive and reproduce within both murine and human macrophages (53, 54, 55, 56, 57, 58). The ability to treat these intracellular reservoirs is an attractive feature of novel anti-staphylococcal agents. Thus, RAW 264.7 macrophages already infected with USA300 LAC were treated with up to 50 μM R91, which was already established to be noncytotoxic towards macrophages. These experiments revealed that akin to untreated macrophages, infected macrophages treated with R91 below 50 μM failed to restrict S. aureus growth. In contrast, at 50 μM R91, the ability of the bacteria to grow inside macrophages was significantly impaired as indicated by the reduced fold change in CFU/ml at 12hpi as compared to the other conditions (Fig. 7C). These data indicate that R91 can antagonize S. aureus growth from within the model macrophage.
R91 reduces lesion size and bacterial burden in a skin infection model
As a first step towards determining whether the anti-MRSA efficacy of R91 could be observed in vivo, we conducted murine SQ skin infection experiments using S. aureus USA300 LAC. This strain is a leading cause of skin and soft tissue infections in humans and is widely used within the research community for its ability to cause SQ infections in mice. Here, a mixture of USA300 LAC and R91 (mixed immediately prior to SQ injection) or untreated bacteria alone were injected SQ into the flanks of mice that had been depilated 24 h prior to infection (Fig. 7D). At 72 h post-infection, lesion sizes and the bacterial burden for individual lesions were measured. These experiments revealed that R91 caused significant reduction in both bacterial burden (Fig. 7E) and in lesion size (Fig. 7F) as compared to untreated animals in this model.
Discussion
MRSA is a high-priority pathogen of concern due to its ability to both acquire and upregulate antimicrobial resistance genes. The escalating incidence of MDR-MRSA emphasizes the urgent need for the discovery of novel antibiotics. TSCs, with their unique and pleiotropic mechanisms of action, hold promise in addressing the challenges posed by the increasing prevalence of MDR-MRSA. Here, using a high-throughput screen, we aimed to identify novel antimicrobials that were effective against MRSA and disrupted metal homeostasis. A number of antimicrobials that act through the modulation of S. aureus metal homeostasis have been previously reported (23, 24, 25, 26). We therefore chose to focus specifically on TSCs because they not only interact with metals to enhance metal-related toxicity (59) but also exhibit high potency against S. aureus and target a variety of cellular processes, making them promising candidates for novel antimicrobial strategies (9, 10, 11, 12, 13, 16, 19, 60). Nevertheless, further investigation is warranted to elucidate the full scope of their antimicrobial activity and potential application in combating MRSA infection.
In our high-throughput screen of the Maybridge bioactives library, we identified a TSC with an unexplored scaffold and high efficacy, “R91” (Fig. 1A), and using this compound and related analogs, we have begun to characterize their activity. Similar to R91, analogs R89, R29, R39, and D79 were effective against all Gram-positive bacteria tested, with the exception of S. saprophyticus (Fig. 1B). Conceivably, given that R91 induces H2O2 production (Fig. 5), the observed S. saprophyticus resistance is likely due to the fact it carries a second catalase gene in its genome that could endow this species with an enhanced ability to detoxify H2O2 (61). Notably, while analogs R31 and R10 were found to be ineffective against most Gram-positive bacteria and all Gram-negative bacteria tested, R31 was capable of inhibiting S. chromogenes, S. pyogenes, S. agalactiae, and M. luteus. Collectively, this suggests that TSCs as a class of antimicrobials can have an extensive range of activity and indicates that particular TSCs could be used to address distinct bacterial species and strains. The efficacy of these analogs against various staphylococcal species allowed for the identification of key structural moieties that appear to be associated with antimicrobial activity. Indeed, the presence of a halogen and a hydroxyl group on the first aromatic group is required for TSC antimicrobial function, indicating these are likely required for target binding. Phenol moieties within TSCs have been predicted to facilitate interactions with copper (62). Furthermore, we observed increased activity with the presence of an additional halogen group, as has been shown previously (10, 63). As halogenated antimicrobials are known for their enhanced lipophilicity and ability to cross the cellular membrane (64), it is possible that TSCs possessing additional halogens have enhanced antimicrobial activity due to increased intracellular concentrations.
While the specific cellular target for R91 is not known, we provide compelling evidence showing that trace metals are intertwined with its activity. Indeed, the RNAseq data (Fig. 2A) demonstrate that genes, copAZ and copBL, involved in copper detoxification are induced upon S. aureus exposure to R91, despite the fact that we observed activity of R91 without the addition of exogenous copper and in metal-deplete media (Figs. 1A and S14B). Why R91 induces these copper-specific genes is presently unclear, but conceivably, if R91 directly binds copper, entry of the drug into the cell with trace copper from the media may cause a rapid increase in intracellular copper. Regardless, there exists an important relationship between R91 and copper as evidenced by the fact that copper and the TSC synergize to produce enhanced anti-staphylococcal activity (Fig. 3).
Clearly, the effects of R91 on the cell are complex as evidenced by the differential expression of genes linked to general stress responses and altered biosynthetic pathways, in addition to copper intoxication. Indeed, transcriptomic analysis (Fig. 2, B and C) reveals a stringent response in S. aureus triggered by R91, with downregulation of nucleotide biosynthesis and protein translation pathways possibly indicating a trade-off between growth and stress responses (33). The stringent response is also characterized by an upregulation in branched-chain amino acid biosynthesis and genes related to oxidative stress resistance (33), and upregulation of genes involved in both processes is observed after acute R91 exposure. Furthermore, the transcriptomic response of S. aureus to R91 closely mirrors the response of the pathogen to lapachol, a naphthoquinone antibiotic that induces a quinone and oxidative stress response in S. aureus (65). As with lapachol, R91 strongly induced expression of the CtsR, HrcA, PerR, NsrR, MhqR, and QsrK operons. Specifically, the PerR regulon responds to hydrogen peroxide, leading to the de-repression of genes involved in the oxidative stress response (66). This suggests R91 may induce oxidative stress and protein damage, as indicated by the heightened expression of genes controlling protein quality control machinery (ctsR and hrcA).
| Consistent with the notion that R91 induces oxidative stress, S. aureus bacteria deficient for srrAB, qoxA, and cydAB all display hypersensitivity to R91 (Fig. 4). Previous studies have demonstrated that strains lacking the SrrAB two-component system are hypersensitive to H2O2-induced killing (41). Furthermore, SrrAB co-regulates expression of genes involved in the response to oxidative stress, including katA, ahpC, and dps (40, 67). This activation of oxidative stress response genes is linked to the ability of SrrAB to sense impaired electron flow and correspondingly modulate expression of the qoxABCD and cydAB genes required for cytochrome biosynthesis and assembly (39). |
Excessive ROS production can result in oxidative stress, causing damage to cellular components such as proteins, lipids, and DNA (59). ROS production has been associated with the bactericidal effects of certain antibiotics (68, 69). As such, we believe our data support the notion that ROS production is a supplementary mechanism of R91 that is enhanced in the presence of copper or at elevated concentrations of R91. Previously, copper complexes and other antimicrobials have been shown to specifically induce H2O2 production in bacterial cells (68, 70). Given the RNAseq results, as well as the hypersensitivity of mutants deficient in their response to oxidative stress, we postulated that R91 might cause the generation of H2O2. In support of this, using both direct and indirect methods, we provide compelling evidence showing that R91 induces ROS in the form of H2O2 and conceivably H2O2 contributes to the antimicrobial activity of this TSC (Fig. 5).
Biofilms, a key virulence mechanism employed by bacteria like S. aureus, pose significant challenges clinically due to their ability to shield bacteria from antimicrobials and evade the immune system, leading to chronic infections and treatment failures (39, 41, 48). Notably, R91 was found to be capable of preventing biofilm formation, as well as disrupting established biofilms, even at concentrations 4-fold below the established MIC (Fig. 6).
The two-component system SrrAB plays a role in biofilm formation by regulating the ica operon, which is responsible for polysaccharide intracellular adhesin production (41). Additionally, copper and ROS antagonize biofilm formation by repressing genes essential for biofilm development, such as sae, agr, and eap (28, 71). The anti-biofilm effect of R91, coupled with the H2O2 production caused by this TSC, aligns with the ROS-linked effect of other antibiotics on biofilm formation (72, 73). The mechanism behind the action of R91 on biofilms may involve H2O2 production or direct targeting of SrrAB, warranting further investigation into this TSC’s role in resisting R91.
The multifaceted mechanism of action exhibited by R91 is likely a key factor preventing the development of resistance by MRSA. S. aureus is well-known for its propensity to develop resistance to antibiotics (74). Therefore, it is crucial that new antimicrobials are designed with mechanisms that minimize the likelihood of selecting for mutants displaying resistance. Remarkably, even after up to 50 days of serial passage in the presence of R91, both with and without copper, S. aureus bacteria displaying heightened resistance to R91 could not be acquired (Fig. S19). These findings underscore the potential for R91 as an effective treatment option.
Encouraged by the lack of resistance development, we proceeded to assess the clinical efficacy of R91 by evaluating its ability to clear intracellular infections and to treat murine skin infection. S. aureus is known to reside, replicate, and persist within macrophages, leading to chronic infections (53). Thus, it is essential for new antibiotics targeting S. aureus to be capable of entering the macrophage and targeting these intracellular reservoirs. R91 demonstrated efficacy against intracellular macrophage infections, albeit only at levels above the MIC, but still below cytotoxic concentrations (Fig. 7C). The utility of R91 to treat SQ skin infections in vivo was also assessed and revealed that prophylactic R91 treatment effectively reduced the bacterial burden within skin lesions and reduced lesion size (Fig. 7, E and F). Future experiments will focus on the ability of this TSC in ameliorating other types of infection and investigate the post-infection treatment efficacy of this compound.
Together, our data demonstrate the potent efficacy of the TSC R91 against MDR MRSA, revealing a multifaceted mechanism of action that includes prevention of biofilm formation and induction of reactive oxygen species production. The lack of resistance development to R91 and its effectiveness in treating intracellular infections and skin lesions underscore its potential as a versatile and promising antimicrobial agent for combating antibiotic-resistant S. aureus infections.
Experimental procedures
Bacterial strains and plasmids
Bacterial strains and plasmids are listed in Table S1. All strains were cultured at 37 °C with shaking at 200 rpm, except for Streptococcal species which were grown without shaking. All Staphylococcal species and strains, as well as most Gram-positive species were grown in TSB or on TSA plates, unless otherwise indicated. Streptococcal species were grown in Todd Hewitt Broth supplemented with 1% yeast extract (THY) and on THY agar plates. Escherichia coli and Pseudomonas aeruginosa were grown in LB or on LB agar.
High-throughput screen
S. aureus strain USA300 LAC was inoculated into either Chelex 100–treated CDM-G (chemically-defined medium with 0.4% glucose) (75) or CDM-G with 30 μM FeSO4 in 96-well plates to a final A600 of 0.001. These plates contained 20 μM of each of the 936 compounds sourced from MayBridge that were previously identified as bioactives against MSSA (20). The plates were then incubated with shaking for 24 h at 37 °C followed by A600 determination. The screens followed general screening protocols that have been described previously (76). The primary hits were then confirmed for activity using broth dilution in Chelex 100–treated CDM-G then TSB using S. aureus USA300 LAC.
Determination of MIC
Bacteria were washed and normalized to an A600 of 1 before being inoculated into TSB, LB, or THY containing various concentrations of TSC (dissolved in DMSO) to a final A600 of 0.01. Cultures were grown for 24 h at 37 °C with shaking, before the A600 was measured. The exception to this was S. pyogenes and S. agalactiae, which were grown without shaking. The concentration of TSC that completely inhibited growth was determined to be the MIC.
Concentration-dependent bactericidal effect of R91
An overnight culture of S. aureus was inoculated 1:100 into PBS or MHB containing R91 and/or metal at concentrations ranging from 6.25 μM to 200 μM. After incubation for 24 h at 37 °C, 10 μl drops of a serial dilution were plated onto TSA. The plates were then incubated for an additional 24 h and CFU/ml were determined.
Chemical characterization of R91 and R89
NMR spectra were obtained in D2O or D2O/DMSO-d6, using a 600 MHz Bruker Ascend 600 instrument. NMR chemical shifts are reported in ppm and are calibrated against the residual solvent signals of DMSO-d6 (2.5 ppm). Coupling constants (J) are given in Hz. Electrospray ionization mass spectrometry was performed using a Bruker microOTOF 11 spectrometer operating in negative ion mode.
R91
1H NMR (600 MHz, DMSO-d6) δ 11.97 (s, 1H), 10.14 (br s, 1H), 10.08 (s, 1H), 8.44 (s, 1H), 8.20 (s, 1H), 7.55 (d, J = 2.6 Hz, 1H), 7.53 (d, J = 2.4 Hz, 1H), 7.42 (dd, J = 8.4, 2.4 Hz, 1H), 7.20 (d, J = 8.4 Hz, 1H), 2.22 (s, 3H). 13C NMR (600 MHz, DMSO-d6) δ 178.4, 151.2, 139.2, 138.4, 138.1, 133.2, 132.9, 131.6, 130.3, 129.3, 125.5, 124.7, 123.2, 120.0, 18.0. HRMS: calcd [M]- (C15H12BrCl2N3OS):430.9261 Found (ESI): 430.9136
R89
1H NMR (600 MHz, DMSO-d6) δ 11.94 (s, 1H), 10.24 (s, 1H), 10.16 (br s, 1H), 8.45 (s, 1H), 8.19 (s, 1H), 7.55 (d, J = 15.7 Hz, 1H), 7.52–7.47 (m, 2H), 7.25–7.18 (m, 2H). 13C NMR (600 MHz, DMSO-d6) δ 177.2, 160.3 (d, J = 241 Hz), 151.22, 138.8, 135.90, 135.89, 130.4, 129.1, 125.7, 124.6, 123.11, 115.3 (d, J = 23 Hz). 19F NMR (600 MHz, DMSO-d6) −116.84 (tt, J = 9.0, 5.1 Hz). HRMS: calcd [M]- (C14H10Cl2FN3OS): 356.9905 Found (ESI): 356.9911.
Stability test of R91 and R89
R91 (5 mg) and R89 (5 mg) were each dissolved in 0.6 ml of 5:1 phosphate buffered D2O (0.01 M, pH 7.4):DMSO-d6 (5:1). The solutions were incubated at 37 °C and 1H NMR spectra were recorded at various time points over 48 h.
RNAseq and GO analysis
An overnight culture of USA300 LAC was inoculated 1:100 into TSB and grown to an A600 of 1.5 at 37 °C with shaking. 3.125 μM of R91 or an equal volume of DMSO was added to the treated cultures and all cultures were then grown for an additional 30 min. The bacteria were then normalized to an A600 of 3, pelleted, and treated with RNA-protect before freezing at −80 °C overnight. RNA was extracted using the QIAGEN RNAEasy kit. Briefly, cell pellets were thawed in 750 μl TE buffer (pH 8), before being resuspended in 500 μg/ml of lysostaphin. The pellets were then incubated at 37 °C for 1 h. RLT buffer and ethanol were added, and the solution was added to the RNeasy mini spin column to extract RNA according to the manufacturer’s protocol. DNAse treatment was performed using the TURBO DNA Free kit (Invitrogen) for 2 × 30 min, followed by inactivation with the supplied DNase Inactivation Reagent. Cleaned RNA was stored in sodium acetate and ethanol at −20 °C overnight. RNA sequencing was performed by the Microbial Genome Sequencing Center in Pittsburgh. Data analysis, including read mapping and differential expression, were performed by the Microbial Genome Sequencing Center in Pittsburgh. Data analysis was conducted using the Geneious Prime software package.
GO analysis was conducted using ShinyGO (77). Differentially expressed genes (absolute confidence >2, log2 differential expression >2) were sorted by a false discovery rate of <0.05 before being selected by FDR and sorted for fold enrichment.
Metal-associated activity and dependence of R91
The synergy of R91 with metals was determined by checkerboard assays as previously reported (78). A clear diagram of checkerboard organization and set up can be found in the above citation. Briefly, R91 and various metals were serially diluted down each axis of a 96-well plate. The wells were then inoculated with USA300 LAC to obtain a final A600 of 0.01. The plates were subsequently incubated for 24 h at 37 °C with shaking. The FICIs were calculated using the following formula:
| FICI= (MICDrugA in combination/MICDrugA alone) + (MICDrug B in combination/MICDrugB alone) |
where synergy is defined as a FICI ≤ 0.5, indifference as 0.5 <FICI ≤ 4, and antagonism is defined as FICI >4.
ICP-MS of compounds R91 and R89
ICP-MS analysis was carried out in the Louisiana Animal Disease Diagnostic Laboratory. Briefly, R89 and R91 were dissolved to 5 mg/ml in DMSO. Each compound was diluted 100× into Ultrapure water (VWR International) with 2% trace metal grade HNO3 (VWR) in triplicate. To control for DMSO metal content, a vehicle control consisting of 1% DMSO and 2% trace metal grade HNO3 in Ultrapure water was analyzed in triplicate. Hundred microliters of each sample was analyzed for 24 Mg, 43Ca, 55Mn, 59Co, and 63Cu on a PerkinElmer NexION 2000 interfaced with a PerkinElmer Micro DX autosampler (PerkinElmer) running Syngistix Software version 3.3 (PerkinElmer). Instrument voltages were optimized empirically through a tune just prior to analyzing the samples. Quantification was performed by comparing the counts per second of each analyte to an external calibration standard (VWR) containing 1000, 100, 10, or 1 ppb of each metal.
PCR and construct generation
Genomic DNA was isolated from S. aureus USA400 MW2 (for srrAB) and S. aureus USA300 LAC (for katA) by phenol-chloroform extraction. Primers containing a KpnI or SacI restriction site were used to amplify srrAB and katA (see Table S2 for primers). The amplified product was ligated into pALC2073 that was also digested using KpnI and SacI. The ligated product was transformed into E. coli DH5α. Plasmids isolated from E. coli were then passaged through S. aureus RN4220, and subsequent plasmids isolated from this strain were introduced into electrocompetent USA400 ΔsrrAB or USA300 LAC.
Markerless deletions of menD and cydAB were constructed using the pKOR1 system, as previously described (79). Briefly, upstream and downstream regions flanking the menD and cydAB regions were PCR amplified (see Table S2 for primers) and recombined into pKOR1. The resulting vectors were then passaged through S. aureus RN4220 before being introduced into electrocompetent USA300 LAC. Allelic replacement of menD and cydAB was confirmed using PCR and DNA sequencing. The qoxA::tn mutant was taken from the NTML library and mobilized using standard phage transduction. Phage lysate was prepared from the donor strain using phage 80α and recipient strains (ΔmenD and ΔmenD ΔcydAB) were infected. Transductants were selected for using erythromycin and confirmed using PCR.
Detection of ROS formed by R91 treatment of S. aureus
Dichlorodihydrofluorescein diacetate
DCFH-DA was used to look at the production of general reactive oxygen species using previously described methods, with modifications (73). USA300 LAC was subcultured to an A600 of 1 and grown for 1 h at 37 °C with shaking. The culture was split into 1 ml aliquots and R91 was added at various concentrations. The cultures were then grown for an additional 2 h. The bacteria were then washed twice with PBS and normalized to an A600 of 1 in 500 μl of PBS. The solutions were then diluted 1:10 and 135 μl of each diluted culture was plated in a 96-well plate. Twenty microliters of 100 μM DCFH-DA was added to each well and the plate was incubated at 37 °C for 30 min, protected from light. Fluorescence was read at an excitation of 485 nm and emission of 535 nm and corrected for background fluorescence of an uninoculated control. General ROS production was quantified by normalizing the fluorescence of treated cells to untreated cells.
Amplex Red
The Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit from Thermo Fisher Scientific was used to detect H2O2 production after R91 exposure according to the manufacturer’s protocol and previously established methods with modifications (68). USA300 LAC was grown to early exponential phase (A600 ∼ 1.5) before R91 at various concentrations was added for 1 h. Bacteria were washed and normalized to an A600 of 1 in PBS and the pellet was resuspended in 50 μl of lysostaphin (0.01 μg/μl) for 30 min at 37 °C to lyse the cells. Fifty microliters of Amplex Red working solution, prepared according to the manufacturer’s instructions, was added to the lysed cells and incubated at room-temperature for 30 min, protected from light. The H2O2 standard curve was prepared according to the manufacturer’s instructions. Fluorescence was read at an excitation of 545 nm and an emission of 590 nm, and background fluorescence was corrected for using an untreated control. H2O2 produced after R91 exposure was determined by comparing background-corrected fluorescence of treated samples to the H2O2 standard curve generated.
Nitroblue tetrazolium
Nitroblue tetrazolium (NBT) assays were performed as previously described (80). Briefly, various concentrations of R91 were added to 0.1 ml of USA300 LAC at an A600 of 0.1 in Hank’s balanced salt solution. 0.5 ml of NBT (1 mg/ml) was added and the solutions were incubated at 37 °C for 30 min. 0.1 ml of 0.1 M HCl was added, and the tubes were centrifuged at 1500g for 10 min. Pellets were treated with 0.6 ml DMSO to extract the reduced NBT (formazan) product. 0.8 ml of PBS was added and the absorbance was read at 575 nm to quantify intracellular superoxide production.
Hydrogen peroxide and scavenger supplementation
MICs were performed as described above, with the addition of 0.25 mM H2O2, and various concentrations of catalase and DMSO to either synergize or antagonize the activity of R91.
Foam-based determination of catalase activity
Catalase activity was assessed visually using foam height, as previously described (81), with modifications. USA300 LAC was grown to early exponential phase (A600 = 1.5) before R91 or hydrogen peroxide at various concentrations were added, and the bacteria were grown for an additional hour. Cells were normalized to an A600 of 2 in 1 ml of PBS after incubation. 100 μL of 1× Triton-X (in PBS) and 100 μL of 30% hydrogen peroxide were added and mixed thoroughly. The tubes were then incubated at room temperature for 15 min to allow the reaction to complete. Foam height was measured using a ruler and normalized to the level of foam produced by an untreated control.
Screening assays for resistance to R91
Evolutionary adaptation
Overnight cultures of S. aureus were inoculated to an A600 of 0.01 in fresh TSB in the presence and absence of either 0.5× MIC R91 (6.25 μM) or ciprofloxacin (25 μM). After 24 h of incubation, the cultures were again subcultured in the presence and absence of R91 or CIP, and this process was repeated for up to 50 days. MICs were taken from days 0 to 14 to determine resistance development in both the R91-exposed and CIP-exposed cultures. The concentration of CIP was increased as resistance development was observed. 50 μM of copper was added where indicated.
Disk diffusion
Overnight cultures of S. aureus were normalized to an A600 of 1. Sterile cotton swabs were used to spread plate this culture onto TSA or Mueller-Hinton agar. Sterile paper disks were infused with 10 μL of R91 with various concentrations and allowed to dry before being placed on top of the bacteria. Plates were then incubated for up to 72 h at 37 °C.
NTML screen
The NTML library is composed of 1920 nonessential transposon mutants in an S. aureus USA300 background (82). Transposon mutants were inoculated into TSB in 96 well plates from frozen stock cultures. The mutants were then grown at 37 °C for 24 h with shaking. Mutants were then subcultured 1:100 into 96 well plates with fresh TSB containing R91 at either 1.56 μM or 50 μM to screen for hypersusceptible (decreased MIC) or resistant (increased MIC). The plates were then incubated for 24 h at 37 °C with shaking. Growth was determined spectrophotometrically at A600.
Biofilm assays
Biofilm assays were performed as previously described (83) with modifications. Two hundred microliters of TSB, supplemented with 0.4% w/v glucose, was inoculated in triplicate with a 1:100 dilution of an overnight culture of USA300 LAC into a 96-well plate. For pre-treatment, various concentrations of R91 were co-incubated with the bacteria. The plate was then statically incubated at 37 °C for 24 h. For pre-formed biofilms, the above is followed, without the addition of R91. After 24 h, the supernatant was replaced with fresh TSB + 0.4% glucose containing various concentrations of R91. The plates were then incubated for an additional 24 h. Following incubation, the supernatant was removed, and the biofilms were washed three times with PBS. Biofilms were then fixed by drying the plate inverted at 42 °C for 1 h. Subsequently, crystal violet (0.4% w/v) was used to stain the cells for 15 min. Excess stain was removed by washing with distilled H2O twice. The remaining crystal violet was dissolved in glacial acetic acid (10% v/v), and the absorbance was measured at 595 nm. Absorbance values were normalized by subtracting the background absorbance of untreated/inoculated wells and further normalized using the absorbance of the untreated USA300 LAC strain, setting it to 1 or representing 100% growth.
R91 cytotoxicity assays
RAW 264.7 macrophages were re-seeded into flasks in RPMI media supplemented with 5% fetal bovine serum. The media contained either DMSO (vehicle control) or various concentration of R91. The cells were incubated for 24 h and then stained with PI to evaluate cell death. Percentage killing was quantified by determining the number of PI-stained cells, relative to the total number of cells in the image obtained at 63× magnification using a widefield microscope. Image analyses and cropping were performed using FIJI.
S. aureus infection of macrophages
Efficacy of R91 on intracellular reservoirs of S. aureus was determined in a manner as previously described (56). Briefly, R91, at concentrations determined to be noncytotoxic, was added to in vitro infections. USA300 LAC was used to infect RAW 264.7 macrophages at an MOI of 10 and incubated for 1.5 h before being treated with 100 μg/ml gentamicin for 1 h to eliminate extracellular bacteria. R91 was subsequently added to the media outside of the infected macrophages, to determine if the compounds were able to restrict the growth of intracellular S. aureus. The cells with and without the compounds were then lysed using 0.1% (v/v) Triton X-100 in sterile PBS at 12 h post infection and compared to a sample lysed at the 1.5-h time point immediately after gentamicin treatment to determine the level of bacterial replication by plating on TSA for CFU counts. The fold change in growth was determined by dividing the number of bacteria measured at 12 h post-infection by the number of bacteria that were measured at 1.5 post-infection.
Mouse SQ abscess model
The mouse study (Animal Use Protocol #2021-090) was approved by the University of Western Ontario Animal Care Committee. Female Balb/c mice were used for this study and were housed in a dedicated mouse facility. The procedure used closely followed that which we have previously published (84). Briefly, eight-to-ten-week-old female Balb/c mice were shaved and naired 24 h prior to infection. Overnight cultures (grown in TSB) of USA300 LAC were subcultured to A600 of 0.05 in TSB and grown to A600 of 2.0 to 2.2. Bacterial cells were pelleted and washed in PBS twice. Bacterial cells were then normalized to an A600 of ∼1.9 (∼4.5–5 × 108 CFUs). Epilated flanks of mice were injected subcutaneously with 50 μl of the bacterial suspension or ∼ 2.25 to 2.5 × 107 CFU in each flank. For control mice, the bacteria were left untreated, whereas for mice receiving R91, the bacteria suspension was treated with R91 at a final concentration of 10 or 50 μM immediately before injection. Mice that received R91 treatment were injected again with 50 μl of PBS containing R91 at 10 or 50 μM 1-day post-infection. The control group received only PBS 1-day post-infection. Infected mice were monitored daily for 3 days and sacrificed at 72 h post infection. Lesions were excised in PBS with 0.1% (v/v) Triton X-100, homogenized, and serially diluted before being plated onto TSA to enumerate CFUs. For each lesion, the bacterial burden was normalized to the mass of the excised lesion. Lesion sizes were analyzed using the ROI measurement tool within the FIJI software package.
Statistical analyses
All statistical analyses were performed using the Prism9 software package (GraphPad). The number of biological replicates and meaning of error bars can be found in the figure legends. Computational analyses with respect to the RNAseq are described above.
Data availability
RNAseq data can be found in the GEO repository under accession number GSE266471. The authors declare that the data supporting the findings of this study are available within the paper and its supporting Supplementary information files.
Supporting information
This article contains supporting information (38, 82).
A document containing Figures S1–S20 and Tables S1 and S2.
Table S3. Excel file containing gene expression levels from RNAseq analysis.
Table S4: Excel file containing ICP-MS output report of metal analysis.
Conflicts of interest
The authors declare that they have no conflicts of interests with the contents of this article.
Acknowledgments
We thank E. Brown and M. Farha from McMaster University for providing the list of hits identified in a previous screen against MSSA, I. Lasa for providing the two-component system knockout mutants and D. Mach for providing S. epidermidis 1457.
Author contributions
A. G., R. S. F., C. S., W. N. B., E. R. G., O. M. E., and D. E. H. writing–review and editing; A. G. writing–original draft; A. G. and R. S. F. visualization; A. G., R. S. F., and E. R. G. validation; A. G., D. W. W., R. S. F., P. R., C. S., A. A., W. N. B., and O. M. E. methodology; A. G., D. W. W., R. S. F., P. R., C. S., A. A., W. N. B., and O. M. E. investigation; A. G. and D. E. H. formal analysis; A. G. and D. E. H. data curation; A. G. and D. E. H. conceptualization; E. R. G. and D. E. H. supervision; E. R. G., O. M. E., and D. E. H. funding acquisition; D. E. H. resources; D. E. H. project administration.
Funding and additional information
This work was funded by an operating grant from the Canadian Institutes of Health Research to D. E. H. (PJT-183848). O. M. E. acknowledges a Canada Research Chair award in Chemogenomics and Antimicrobial Research and an establishment grant from Saskatchewan Health Research Foundation. Work by C. S. and E. R. G. was supported by NSERC grant RGPIN-2021-03950.
Reviewed by members of the JBC Editorial Board. Edited by Chris Whitfield
Supporting information
References
- 1.Tong S.Y.C., Davis J.S., Eichenberger E., Holland T.L., Fowler V.G. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mancuso G., Midiri A., Gerace E., Biondo C. Bacterial antibiotic resistance: the most critical pathogens. Pathogens. 2021;10:1310. doi: 10.3390/pathogens10101310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munita J.M., Arias C.A. Mechanisms of antibiotic resistance. Microbiol. Spectr. 2016;4 doi: 10.1128/microbiolspec.VMBF-0016–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaur D.C., Chate S.S. Study of antibiotic resistance pattern in methicillin resistant Staphylococcus aureus with special reference to newer antibiotic. J. Glob. Infect. Dis. 2015;7:78–84. doi: 10.4103/0974-777X.157245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murdoch C.C., Skaar E.P. Nutritional immunity: the battle for nutrient metals at the host–pathogen interface. Nat. Rev. Microbiol. 2022;20:657–670. doi: 10.1038/s41579-022-00745-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandrangsu P., Rensing C., Helmann J.D. Metal homeostasis and resistance in bacteria. Nat. Rev. Microbiol. 2017;15:338–350. doi: 10.1038/nrmicro.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassat J.E., Skaar E.P. Metal ion acquisition in Staphylococcus aureus: overcoming nutritional immunity. Semin. Immunopathol. 2012;34:215–235. doi: 10.1007/s00281-011-0294-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheldon J.R., Laakso H.A., Heinrichs D.E. Iron acquisition strategies of bacterial pathogens. Microbiol. Spectr. 2016 doi: 10.1128/microbiolspec.VMBF-0010-2015. [DOI] [PubMed] [Google Scholar]
- 9.Crawford C.L., Dalecki A.G., Perez M.D., Schaaf K., Wolschendorf F., Kutsch O. A copper-dependent compound restores ampicillin sensitivity in multidrug-resistant Staphylococcus aureus. Sci. Rep. 2020;10:8955. doi: 10.1038/s41598-020-65978-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X.-M., Guo H., Li Z.-S., Song F.-H., Wang W.-M., Dai H.-Q., et al. Synthesis and evaluation of isatin-β-thiosemicarbazones as novel agents against antibiotic-resistant Gram-positive bacterial species. Eur. J. Med. Chem. 2015;101:419–430. doi: 10.1016/j.ejmech.2015.06.047. [DOI] [PubMed] [Google Scholar]
- 11.Delpe-Acharige A., Zhang M., Eschliman K., Dalecki A., Covarrubias-Zambrano O., Minjarez-Almeida A., et al. Pyrazolyl thioureas and carbothioamides with an NNSN motif against MSSA and MRSA. ACS Omega. 2021;6:6088–6099. doi: 10.1021/acsomega.0c04513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haeili M., Moore C., Davis C.J.C., Cochran J.B., Shah S., Shrestha T.B., et al. Copper complexation screen reveals compounds with potent antibiotic properties against methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2014;58:3727–3736. doi: 10.1128/AAC.02316-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh V., Palakkeezhillam V.N.V., Manakkadan V., Rasin P., Valsan A.K., Kumar V.S., et al. Recent developments on the potential biological applications of transition metal complexes of thiosemicarbazone derivatives. Polyhedron. 2023;245 [Google Scholar]
- 14.Gupta S., Singh N., Khan T., Joshi S. Thiosemicarbazone derivatives of transition metals as multi-target drugs: a review. Results Chem. 2022;4 [Google Scholar]
- 15.Netalkar P.P., Netalkar S.P., Revankar V.K. Transition metal complexes of thiosemicarbazone: synthesis, structures and invitro antimicrobial studies. Polyhedron. 2015;100:215–222. [Google Scholar]
- 16.Beckford F.A., Webb K.R. Copper complexes containing thiosemicarbazones derived from 6-nitropiperonal: antimicrobial and biophysical properties. Spectrochimica Acta A: Mol. Biomol. Spectrosc. 2017;183:158–171. doi: 10.1016/j.saa.2017.04.057. [DOI] [PubMed] [Google Scholar]
- 17.Dalecki A.G., Malalasekera A.P., Schaaf K., Kutsch O., Bossmann S.H., Wolschendorf F. Combinatorial phenotypic screen uncovers unrecognized family of extended thiourea inhibitors with copper-dependent anti-staphylococcal activity. Metallomics. 2016;8:412–421. doi: 10.1039/c6mt00003g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bisceglie F., Bacci C., Vismarra A., Barilli E., Pioli M., Orsoni N., et al. Antibacterial activity of metal complexes based on cinnamaldehyde thiosemicarbazone analogues. J. Inorg. Biochem. 2020;203 doi: 10.1016/j.jinorgbio.2019.110888. [DOI] [PubMed] [Google Scholar]
- 19.D’Agostino I., Mathew G.E., Angelini P., Venanzoni R., Angeles Flores G., Angeli A., et al. Biological investigation of N-methyl thiosemicarbazones as antimicrobial agents and bacterial carbonic anhydrases inhibitors. J. Enzyme Inhib. Med. Chem. 2022;37:986–993. doi: 10.1080/14756366.2022.2055009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bæk K.T., Jensen C., Farha M.A., Nielsen T.K., Paknejadi E., Mebus V.H., et al. A Staphylococcus aureus clpX mutant used as a unique screening tool to identify cell wall synthesis inhibitors that reverse β-lactam resistance in MRSA. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.691569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linz M.S., Mattappallil A., Finkel D., Parker D. Clinical impact of Staphylococcus aureus skin and soft tissue infections. Antibiotics. 2023 doi: 10.3390/antibiotics12030557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheldon J.R., Marolda C.L., Heinrichs D.E. TCA cycle activity in Staphylococcus aureus is essential for iron-regulated synthesis of staphyloferrin A, but not staphyloferrin B: the benefit of a second citrate synthase. Mol. Microbiol. 2014;92:824–839. doi: 10.1111/mmi.12593. [DOI] [PubMed] [Google Scholar]
- 23.Juttukonda L.J., Beavers W.N., Unsihuay D., Kim K., Pishchany G., Horning K.J., et al. A small-molecule modulator of metal homeostasis in gram-positive pathogens. mBio. 2020;11 doi: 10.1128/mbio.02555-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crawford C.L., Dalecki A.G., Narmore W.T., Hoff J., Hargett A.A., Renfrow M.B., et al. Pyrazolopyrimidinones, a novel class of copper-dependent bactericidal antibiotics against multi-drug resistant S. aureus. Metallomics. 2019;11:784–798. doi: 10.1039/c8mt00316e. [DOI] [PubMed] [Google Scholar]
- 25.Kaul L., Abdo A.I., Coenye T., Krom B.P., Hoogenkamp M.A., Zannettino A.C.W., et al. The combination of diethyldithiocarbamate and copper ions is active against Staphylococcus aureus and Staphylococcus epidermidis biofilms in vitro and in vivo. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.999893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma B., Shukla S., Rattan R., Fatima M., Goel M., Bhat M., et al. Antimicrobial agents based on metal complexes: present situation and future prospects. Int. J. Biomater. 2022;2022 doi: 10.1155/2022/6819080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosario-Cruz Z., Eletsky A., Daigham N.S., Al-Tameemi H., Swapna G.V.T., Kahn P.C., et al. The copBL operon protects Staphylococcus aureus from copper toxicity: CopL is an extracellular membrane–associated copper-binding protein. J. Biol. Chem. 2019;294:4027–4044. doi: 10.1074/jbc.RA118.004723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baker J., Sitthisak S., Sengupta M., Johnson M., Jayaswal R.K., Morrissey J.A. Copper stress induces a global stress response in Staphylococcus aureus and represses sae and agr expression and biofilm formation. Appl. Environ. Microbiol. 2010;76:150–160. doi: 10.1128/AEM.02268-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tarrant E., P Riboldi G., McIlvin M.R., Stevenson J., Barwinska-Sendra A., Stewart L.J., et al. Copper stress in Staphylococcus aureus leads to adaptive changes in central carbon metabolism. Metallomics. 2019;11:183–200. doi: 10.1039/c8mt00239h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michel A., Agerer F., Hauck C.R., Herrmann M., Ullrich J., Hacker J., et al. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 2006;188:5783–5796. doi: 10.1128/JB.00074-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fleury B., Kelley W.L., Lew D., Götz F., Proctor R.A., Vaudaux P. Transcriptomic and metabolic responses of Staphylococcus aureus exposed to supra-physiological temperatures. BMC Microbiol. 2009;9:76. doi: 10.1186/1471-2180-9-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loi V.V., Busche T., Preuß T., Kalinowski J., Bernhardt J., Antelmann H. The AGXX® antimicrobial coating causes a thiol-specific oxidative stress response and protein S-bacillithiolation in Staphylococcus aureus. Front. Microbiol. 2018;9:3037. doi: 10.3389/fmicb.2018.03037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reiß S., Pané-Farré J., Fuchs S., François P., Liebeke M., Schrenzel J., et al. Global analysis of the Staphylococcus aureus response to mupirocin. Antimicrob. Agents Chemother. 2012;56:787–804. doi: 10.1128/AAC.05363-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aly A.A., Abdallah E.M., Ahmed S.A., Rabee M.M., Bräse S. Transition metal complexes of thiosemicarbazides, thiocarbohydrazides, and their corresponding carbazones with Cu(I), Cu(II), Co(II), Ni(II), Pd(II), and Ag(I)—a review. Molecules. 2023;28:1808. doi: 10.3390/molecules28041808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tierney A.R., Rather P.N. Roles of two-component regulatory systems in antibiotic resistance. Future Microbiol. 2019;14:533–552. doi: 10.2217/fmb-2019-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang S.-J., Bayer A.S., Mishra N.N., Meehl M., Ledala N., Yeaman M.R., et al. The Staphylococcus aureus two-component regulatory system, GraRS, senses and confers resistance to selected cationic antimicrobial peptides. Infect. Immun. 2012;80:74–81. doi: 10.1128/IAI.05669-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu S., Lin K., Liu Y., Zhang H., Lei L. Two-component signaling pathways modulate drug resistance of Staphylococcus aureus (Review) Biomed. Rep. 2020;13:5. doi: 10.3892/br.2020.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villanueva M., García B., Valle J., Rapún B., Ruiz de los Mozos I., Solano C., et al. Sensory deprivation in Staphylococcus aureus. Nat. Commun. 2018;9:523. doi: 10.1038/s41467-018-02949-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kinkel T.L., Roux C.M., Dunman P.M., Fang F.C. The Staphylococcus aureus SrrAB two-component system promotes resistance to nitrosative stress and hypoxia. mBio. 2013;4:e00696-13. doi: 10.1128/mBio.00696-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mashruwala A.A., Boyd J.M. The Staphylococcus aureus SrrAB regulatory system modulates hydrogen peroxide resistance factors, which imparts protection to aconitase during aerobic growth. PLoS One. 2017;12 doi: 10.1371/journal.pone.0170283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mashruwala A.A., van de Guchte A., Boyd J.M. Impaired respiration elicits SrrAB-dependent programmed cell lysis and biofilm formation in Staphylococcus aureus. eLife. 2017;6 doi: 10.7554/eLife.23845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hammer N.D., Schurig-Briccio L.A., Gerdes S.Y., Gennis R.B., Skaar E.P. CtaM is required for menaquinol oxidase aa3 function in Staphylococcus aureus. mBio. 2016;7:e00823-16. doi: 10.1128/mBio.00823-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borisov V.B., Siletsky S.A., Nastasi M.R., Forte E. ROS defense systems and terminal oxidases in bacteria. Antioxidants (Basel) 2021;10:839. doi: 10.3390/antiox10060839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Painter K.L., Strange E., Parkhill J., Bamford K.B., Armstrong-James D., Edwards A.M. Staphylococcus aureus adapts to oxidative stress by producing H2O2-resistant small-colony variants via the SOS response. Infect. Immun. 2015;83:1830–1844. doi: 10.1128/IAI.03016-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Repine J.E., Fox R.B., Berger E.M. Hydrogen peroxide kills Staphylococcus aureus by reacting with staphylococcal iron to form hydroxyl radical. J. Biol. Chem. 1981;256:7094–7096. [PubMed] [Google Scholar]
- 46.Wang Y., Hougaard A.B., Paulander W., Skibsted L.H., Ingmer H., Andersen M.L. Catalase expression is modulated by vancomycin and ciprofloxacin and influences the formation of free radicals in Staphylococcus aureus cultures. Appl. Environ. Microbiol. 2015;81:6393–6398. doi: 10.1128/AEM.01199-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horsburgh M.J., Ingham E., Foster S.J. In Staphylococcus aureus, Fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 2001;183:468–475. doi: 10.1128/JB.183.2.468-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng Q., Tang X., Dong W., Sun N., Yuan W. A review of biofilm formation of Staphylococcus aureus and its regulation mechanism. Antibiotics. 2023;12:12. doi: 10.3390/antibiotics12010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pavan F.R., da S Maia P.I., Leite S.R.A., Deflon V.M., Batista A.A., Sato D.N., et al. Thiosemicarbazones, semicarbazones, dithiocarbazates and hydrazide/hydrazones: anti – Mycobacterium tuberculosis activity and cytotoxicity. Eur. J. Med. Chem. 2010;45:1898–1905. doi: 10.1016/j.ejmech.2010.01.028. [DOI] [PubMed] [Google Scholar]
- 50.Kalinowski D.S., Quach P., Richardson D.R. Thiosemicarbazones: the new wave in cancer treatment. Future Med. Chem. 2009;1:1143–1151. doi: 10.4155/fmc.09.80. [DOI] [PubMed] [Google Scholar]
- 51.Hall M.D., Salam N.K., Hellawell J.L., Fales H.M., Kensler C.B., Ludwig J.A., et al. Synthesis, activity and pharmacophore development for isatin-β-thiosemicarbazones with selective activity towards multidrug resistant cells. J. Med. Chem. 2009;52:3191–3204. doi: 10.1021/jm800861c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Savjani K.T., Gajjar A.K., Savjani J.K. Drug solubility: importance and enhancement techniques. ISRN Pharm. 2012;2012 doi: 10.5402/2012/195727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flannagan R.S., Heit B., Heinrichs D.E. Intracellular replication of Staphylococcus aureus in mature phagolysosomes in macrophages precedes host cell death, and bacterial escape and dissemination. Cell Microbiol. 2016;18:514–535. doi: 10.1111/cmi.12527. [DOI] [PubMed] [Google Scholar]
- 54.Flannagan R.S., Heit B., Heinrichs D.E. Antimicrobial mechanisms of macrophages and the immune evasion strategies of Staphylococcus aureus. Pathogens. 2015;4:826–868. doi: 10.3390/pathogens4040826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flannagan R.S., Kuiack R.C., McGavin M.J., Heinrichs D.E. Staphylococcus aureus uses the GraXRS regulatory system to sense and adapt to the acidified phagolysosome in macrophages. mBio. 2018;9 doi: 10.1128/mBio.01143-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.El-Halfawy O.M., Czarny T.L., Flannagan R.S., Day J., Bozelli J.C., Kuiack R.C., et al. Discovery of an antivirulence compound that reverses β-lactam resistance in MRSA. Nat. Chem. Biol. 2020;16:143–149. doi: 10.1038/s41589-019-0401-8. [DOI] [PubMed] [Google Scholar]
- 57.Flannagan R.S., Heinrichs D.E. Macrophage-driven nutrient delivery to phagosomal Staphylococcus aureus supports bacterial growth. EMBO Rep. 2020;21 doi: 10.15252/embr.202050348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bertrand B.P., Shinde D., Thomas V.C., Whiteley M., Ibberson C.B., Kielian T. Metabolic diversity of human macrophages: potential influence on Staphylococcus aureus intracellular survival. Infect. Immun. 2024;92 doi: 10.1128/iai.00474-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Orta-Rivera A.M., Meléndez-Contés Y., Medina-Berríos N., Gómez-Cardona A.M., Ramos-Rodríguez A., Cruz-Santiago C., et al. Copper-based antibiotic strategies: exploring applications in the hospital setting and the targeting of Cu regulatory pathways and current drug design trends. Inorganics. 2023;11:252. doi: 10.3390/inorganics11060252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Toan V.N., Thanh N.D., Khuyen V.H., Tu L.T.C., Tri N.M., Huong N.T.T. N-(2,3,4,6-tetra-O-acetyl-β-d-glucopyranosyl)thiosemicarbazones of 6-alkoxy-2-oxo-2H-chromene-4-carbaldehydes: synthesis, evaluation of their antibacterial, anti-MRSA, antifungal activity, and docking study. Med. Chem. Res. 2021;30:743–759. [Google Scholar]
- 61.Gaupp R., Ledala N., Somerville G.A. Staphylococcal response to oxidative stress. Front. Cell Infect. Microbiol. 2012;2:33. doi: 10.3389/fcimb.2012.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petrasheuskaya V., A T., Kiss M., Dömötör O., Holczbauer T., V. May N., et al. Salicylaldehyde thiosemicarbazone copper complexes: impact of hybridization with estrone on cytotoxicity, solution stability and redox activity. New J. Chem. 2020;44:12154–12168. [Google Scholar]
- 63.El-Sharief M.A., Abbas S.Y., El-Bayouki K.A.M., El-Gammal E.W. Synthesis of thiosemicarbazones derived from N-(4-hippuric acid)thiosemicarbazide and different carbonyl compounds as antimicrobial agents. Eur. J. Med. Chem. 2013;67:263–268. doi: 10.1016/j.ejmech.2013.06.031. [DOI] [PubMed] [Google Scholar]
- 64.Faleye O.S., Boya B.R., Lee J.-H., Choi I., Lee J. Halogenated antimicrobial agents to combat drug-resistant pathogens. Pharmacol. Rev. 2024;76:90–141. doi: 10.1124/pharmrev.123.000863. [DOI] [PubMed] [Google Scholar]
- 65.Linzner N., Fritsch V.N., Busche T., Tung Q.N., Loi V.V., Bernhardt J., et al. The plant-derived naphthoquinone lapachol causes an oxidative stress response in Staphylococcus aureus. Free Radic. Biol. Med. 2020;158:126–136. doi: 10.1016/j.freeradbiomed.2020.07.025. [DOI] [PubMed] [Google Scholar]
- 66.Ji C.-J., Kim J.-H., Won Y.-B., Lee Y.-E., Choi T.-W., Ju S.-Y., et al. Staphylococcus aureus PerR is a hypersensitive hydrogen peroxide sensor using iron-mediated histidine oxidation. J. Biol. Chem. 2015;290:20374–20386. doi: 10.1074/jbc.M115.664961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oogai Y., Kawada-Matsuo M., Komatsuzawa H. Staphylococcus aureus SrrAB affects susceptibility to hydrogen peroxide and Co-existence with Streptococcus sanguinis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0159768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dwyer D.J., Belenky P.A., Yang J.H., MacDonald I.C., Martell J.D., Takahashi N., et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kohanski M.A., Dwyer D.J., Hayete B., Lawrence C.A., Collins J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 70.Hoshino N., Kimura T., Hayakawa F., Yamaji A., Ando T. Bactericidal activity of catechin-copper (II) complexes against Staphylococcus aureus compared with Escherichia coli. Lett. Appl. Microbiol. 2000;31:213–217. doi: 10.1046/j.1365-2672.2000.00800.x. [DOI] [PubMed] [Google Scholar]
- 71.Arce Miranda J.E., Sotomayor C.E., Albesa I., Paraje M.G. Oxidative and nitrosative stress in Staphylococcus aureus biofilm. FEMS Microbiol. Lett. 2011;315:23–29. doi: 10.1111/j.1574-6968.2010.02164.x. [DOI] [PubMed] [Google Scholar]
- 72.Paul P., Chakraborty P., Chatterjee A., Sarker R.K., Dastidar D.G., Kundu T., et al. 1,4-Naphthoquinone accumulates reactive oxygen species in Staphylococcus aureus: a promising approach towards effective management of biofilm threat. Arch. Microbiol. 2021;203:1183–1193. doi: 10.1007/s00203-020-02117-1. [DOI] [PubMed] [Google Scholar]
- 73.Khodade V.S., Sharath Chandra M., Banerjee A., Lahiri S., Pulipeta M., Rangarajan R., et al. Bioreductively activated reactive oxygen species (ROS) generators as MRSA inhibitors. ACS Med. Chem. Lett. 2014;5:777–781. doi: 10.1021/ml5001118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Foster T.J. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 2017;41:430–449. doi: 10.1093/femsre/fux007. [DOI] [PubMed] [Google Scholar]
- 75.Kaiser J.C., Omer S., Sheldon J.R., Welch I., Heinrichs D.E. Role of BrnQ1 and BrnQ2 in branched-chain amino acid transport and virulence in Staphylococcus aureus. Infect. Immun. 2015;83:1019. doi: 10.1128/IAI.02542-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zlitni S., Blanchard J.E., Brown E.D. In: Cell-Based Assays for High-Throughput Screening: Methods and Protocols. Clemons P.A., Tolliday N.J., Wagner B.K., editors. Humana Press; Totowa, NJ: 2009. High-throughput screening of model bacteria; pp. 13–27. [DOI] [Google Scholar]
- 77.Ge S.X., Jung D., Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36:2628–2629. doi: 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bellio P., Fagnani L., Nazzicone L., Celenza G. New and simplified method for drug combination studies by checkerboard assay. MethodsX. 2021;8 doi: 10.1016/j.mex.2021.101543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bae T., Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2006;55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 80.Páez P.L., Becerra M.C., Albesa I. Effect of the association of reduced glutathione and ciprofloxacin on the antimicrobial activity in Staphylococcus aureus. FEMS Microbiol. Lett. 2010;303:101–105. doi: 10.1111/j.1574-6968.2009.01867.x. [DOI] [PubMed] [Google Scholar]
- 81.Iwase T., Tajima A., Sugimoto S., Okuda K., Hironaka I., Kamata Y., et al. A simple assay for measuring catalase activity: a visual approach. Sci. Rep. 2013;3:3081. doi: 10.1038/srep03081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fey P.D., Endres J.L., Yajjala V.K., Widhelm T.J., Boissy R.J., Bose J.L., et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio. 2013;4:e00537-12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goncheva M.I., Flannagan R.S., Sterling B.E., Laakso H.A., Friedrich N.C., Kaiser J.C., et al. Stress-induced inactivation of the Staphylococcus aureus purine biosynthesis repressor leads to hypervirulence. Nat. Commun. 2019;10:775. doi: 10.1038/s41467-019-08724-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chin D., Goncheva M.I., Flannagan R.S., Deecker S.R., Guariglia-Oropeza V., Ensminger A.W., et al. Coagulase-negative staphylococci release a purine analog that inhibits Staphylococcus aureus virulence. Nat. Commun. 2021;12:1887. doi: 10.1038/s41467-021-22175-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNAseq data can be found in the GEO repository under accession number GSE266471. The authors declare that the data supporting the findings of this study are available within the paper and its supporting Supplementary information files.