ABSTRACT
Visceral leishmaniasis (VL) is a neglected disease that is typical of tropical and subtropical parts of the world and is caused by the trypanosomatid Leishmania donovani complex. This disease is a multifactorial condition that involves parasitic, environmental, and immunogenetic characteristics. Genetic changes in genes encoding cytokines may be associated with changes in their expression and, consequently, with the development of clinical resistance or susceptibility to the disease. This systematic review and meta-analysis aimed to assess whether single nucleotide polymorphisms (SNPs) in interleukin genes influence the clinical consequences of visceral leishmaniasis infection. To this end, we carried out a systematic review and meta-analysis with structured searches in the EMBASE, PubMed, Scopus, SciELO, and Web of Science databases without time restrictions. Two independent reviewers examined the studies, performed data extraction, and assessed quality by assigning scores. If there were any discrepancies, a third reviewer with more experience was consulted. After the screening process, 28 articles were included in the systematic review and 9 in the final analysis of the meta-analysis. Statistical analyses were carried out using various genetic models. The odds ratio (OR) and corresponding 95% confidence intervals (CIs) were calculated to estimate the associations. Overall, the main clinical outcomes were classified as not associated or associated when they presented susceptibility, resistance, risk, or protective factors for the development of the disease. Associations between IFN-γ +874T/A polymorphisms in the dominant model (OR 1.64, 95% CI 1.13-2.38, I2 = 0%, p < 0.01) and heterozygous model (OR 1.72, 95% CI 1.15-2.57, I2 = 0%, p < 0.01) and IL-18 -137G/C in the recessive model (OR 1.33, 95% CI 1.02-1.71, I2 = 9%, p = 0.03) and VL were observed. For the IL-10 gene SNPs, there was no significant association. Our findings suggest that SNPs in the IFN-γ and IL-18 genes may be associated with the risk of developing VL.
Keywords: Kala-azar, Visceral leishmaniasis, Leishmania sp., Single base polymorphism, Systematic review
Background
Visceral leishmaniasis (VL) or kala-azar is a neglected disease typical of tropical and subtropical regions of the world. This disease is caused by trypanosomatid protozoa of the Leishmania donovani complex and can be fatal when early treatment is not administered [ 1- 2]. It is a vector-borne disease transmitted by sandflies of the genera Lutzomyia and Phlebotomus from the New and Old Worlds, respectively. According to the World Health Organization (WHO), there are 50 to 90,000 new cases annually; however, most of them are underreported, and only 25 to 45% of them are informed to the world health authority. More than 90% of these are concentrated in 10 countries: Brazil, China, Ethiopia, Eritrea, India, Kenya, Somalia, South Sudan, Sudan, and Yemen [ 2].
The development of the disease is induced by several factors, such as the environment, the pathogen, and host factors. Regarding the hosts, genetic and immune characteristics may trigger resistance or susceptibility to VL. Host genetic variations may be classified as changes in structure, copy number, transposon, insertion/deletion, or single nucleotide polymorphism (SNP) [ 3]. SNPs are single nucleotide changes that are present in more than 1% of the world's population. These factors may change the functions of promoter regions, inducing an increase or decrease in the production of gene transcripts or modifying the proteins themselves [ 3, 4].
Furthermore, the presence of polymorphisms in genes that encode interleukins may directly affect the type of immune response, interfere with their production levels, and, consequently, susceptibility or resistance to the disease [ 3- 5]. The intracellular location of the parasites triggers an immune response mediated by CD4+ lymphocytes; however, there is a duality, in which conventionally the Th1 profile is associated with resistance to the disease, with the presence of cytokines such as IFN-γ, IL-12, IL-2. The Th2 response is associated with susceptibility and with the synthesis of interleukins such as IL-4, IL-5, IL-10, and IL-13. The high production of Th1 cytokines may contribute to an exacerbated inflammatory response and to the pathogenesis of the disease. Therefore, this response is normally accompanied by regulatory T cells (Tregs), which synthesize IL-10 and TGF-β, for example, and act to modulate the immune response and prevent tissue damage. In addition, Th17 cells differentiate in the presence of TGF-β and IL-6 and synthesize interleukins such as IL-17 and IL-22, but their roles in the pathogenesis of this disease are still contradictory [ 6].
Therefore, the presence of SNPs in genes of interleukins may alter production levels and, consequently, the conditions for the development of the disease [ 3, 4, 5]. Thus, this study aimed to assess whether SNPs in interleukin genes influence the clinical consequences of visceral leishmaniasis by a systematic review and meta-analysis of all eligible studies, which may yield more accurate and robust estimates of the association between the polymorphisms and clinical outcome.
Methods
Registry protocol
In this systematic review, we adhered to the PRISMA 2020 guidelines and checklist -Preferred Reporting Items for Systematic Reviews and Meta-Analyses [ 7] to reduce the possibility of insertion of biases. Our methodology protocol was registered in the Prospective International Registry Platform for Systematic Reviews (PROSPERO/National Institute for Health Research - CRD42022350889) (Additional file 1).
Data sources and search strategy
A bibliographic survey of scientific articles was carried out between March and April 2024 without restriction on publication date. The databases used in the searches were EMBASE (Elsevier), PubMed (National Center for Biotechnology Information), Scopus (Elsevier), SciELO, and Web of Science (Clarivate). The search key and its MeSH terms were as follows: (“visceral leishmaniasis” OR “black fever” OR “fever-black” OR “kala-azar” OR “kala azar” OR “leishmaniasis, visceral” [Mesh]) AND (“gene polymorphism” OR “gene polymorphisms” OR “polymorphism, gene” OR “polymorphisms, gene” OR “genetic polymorphism” OR “genetic polymorphisms” OR “polymorphism (genetic)” OR “polymorphisms (genetics)” OR “polymorphisms, genetic” OR “polymorphism, genetic” [MESH]) AND (human OR “humans” [Mesh]) (Additional file 2). Hand-searching for studies was also carried out to identify studies not captured by the search key. The complete methodology with more details is available on the PROSPERO website (https://www.crd.york.ac.uk/prospero/display_record.php? ID=CRD42022350889) [ 8].
Eligibility criteria
Studies were considered eligible if they involved SNPs in genes encoding cytokines in individuals with VL. Unqualified papers were excluded if they met the following criteria: (i) research carried out with non-human VL; (ii) studies carried out with other types of leishmaniasis other than its visceral form; (iii) articles that focused only on bioinformatics analyses; and (iv) narrative reviews, systematic (with or without meta-analysis), editorials, conference abstracts, case reports, or books.
Study selection and data extraction
The references were imported into Rayyan online software [ 9] for data management. Two independent reviewers (AVBV and MRM) were responsible for the screening process. Initially, duplicates were removed. The first phase of selection was based on the analysis of titles and abstracts according to eligibility criteria. Subsequently, the second phase of the selection was based on the reading of the full texts of potentially eligible studies and reanalysis of the inclusion criteria. In the presence of any disagreements, a more experienced third reviewer (ZMM) assessed the data. Considering the eligibility criteria, the following data were extracted: (i) name(s) of the author(s), (ii) year of publication, (iii) polymorphism, (iv) SNP location, (v) genotyping method, and (vi) outcome. This extraction was performed independently by two reviewers (AVBV and MRM). In the presence of any discrepancies, a third reviewer (ZMM) checked the data.
Risk of bias and quality assessment
The selected studies were independently analyzed by two reviewers (AVBV and MRM) using the Standard Quality Assessment Criteria for Evaluation of Primary Research Papers from a Variety of Fields [ 10], a tool for systematically evaluating reviews. The protocol is composed of quantitative analysis, where 14 assessment items are used. The scores range from 0 to 2 points, in which “0” represents no, “1” represents partial, and “2” represents yes. The questions answered by N/A (not applicable) were not part of the calculation of the total points score (number of “yes” * 2 + number of “partials” * 1) or maximum (28 - number of “N/A” * 2). The quality of the studies was analyzed from the value of the final percentage; the higher this number was, the lower the risk of bias. In the presence of any disagreements, a third reviewer (ZMM) evaluated the studies.
Statistical analysis
The associations between polymorphisms and the development of VL were evaluated using comparative genetic models: dominant, recessive, allelic, homozygous, and heterozygous. The odds ratio (OR) values were used to test the association between each SNP and disease association in the studies. The 95% confidence intervals and p values < 0.05 were considered to indicate statistical significance. Cochran’s Q statistic and the I2 test were used to evaluate heterogeneity [ 11]. In the presence of high heterogeneity values (I2 > 50%), the random effects model was used; otherwise, the fixed effects model was applied [ 12]. The chi-square test was applied to examine the Hardy‒Weinberg equilibrium (HWE) in the healthy control groups in the different studies with PLINK v1.9 software. HWE was considered when p > 0.05. All tests in this meta-analysis were performed with RStudio software version 4.4.0 [ 13].
Results
Flow of included studies
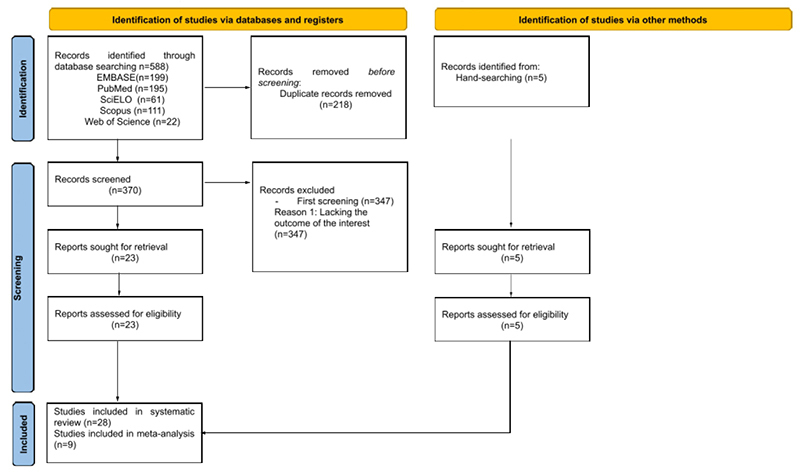
The study selection process is summarized in the flow diagram of the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) shown in Figure 1. The initial search yielded 588 articles (199 in EMBASE, 195 in PubMed, 61 in SciELO, 111 in Scopus, and 22 in Web of Science). Of these, 218 were excluded because they were duplicated. After this exclusion, 370 articles remained and passed to the first screening (reading of titles and abstracts), in which the inclusion criteria were verified and 347 were excluded for not following the objective of the study. Subsequently, full texts of the 23 previously selected studies were read, all of which were included because they fully met the inclusion criteria. Manual searches were also carried out in the reference lists of previously selected articles, with 5 studies included using this methodology. At the end of the entire selection process, 28 articles were included in this systematic review, 9 of which were included in the meta-analysis stage.
Figure 1. Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flow diagram displaying the systematic search and review process.
Description of the studies
The characteristics of all included articles are shown in Table 1. The selected articles were published from 2002 to 2021. Case-control studies, with comparison groups (VL carriers, asymptomatic and healthy) and using family members were carried out in 14, 11, and 3 articles, respectively. Sample sizes ranged from small groups of 15 cases to studies with more than 1000 individuals divided between families. The study areas of the inserted articles were Brazil = 3, India = 4, Iran = 15, Iraq = 3, Morocco = 1, Mexico = 1 and Sudan = 1.
Table 1. Details of studies that evaluated polymorphisms in VL-associated cytokine genes.
| References | Country | Species of protozoan | Genotyping method | SNP location | Outcome | Polymorphism |
|---|---|---|---|---|---|---|
| Moravejet al. [14] | Iran | Leishmania infantum | PCR-RFLP | Exon | Associated [Susceptibility (-511CC genotype and CC haplotype (-511/+3953) and resistance (-511 TT genotype) and tde TT haplotype (-511/+3953)] | +3953 (IL-1β) |
| Promoter | − 511 (IL-1β) | |||||
| Ahmadiet al. [15] | Iran | Leishmania infantum | PCR-RFLP | 3'UTR | Associated (risk) | rs1126647 (IL-8) |
| Hajilooiet al. [16] | Iran | Leishmania infantum | AS-PCR | Promoter | Associated | rs4073 (IL-8) |
| Fradeet al. [17] | Brazil | Leishmania infantum | PCR-RFLP | Promoter | Not associated | rs4073 (IL-8) |
| Fradeet al. [17] | Brazil | Leishmania infantum | PCR-RFLP | Promoter | Associated [susceptibility (-509 T allele)] | rs1800469 (TGFB1) |
| Exon | rs1800470 (TGFB1) | |||||
| Kalaniet al. [18] | Iran | Leishmania infantum | PCR-RFLP | 3' UTR | Not associated | +1188 (IL-12) |
| Kalaniet al. [18] | Iran | Leishmania infantum | AS-PCR | Intron | Associated [susceptibility (AT genotype) and resistance (TT resistance)] | rs2430561 (IFN-γ) |
| Kalaniet al. [18] | Iran | Leishmania infantum | PCR-RFLP | Promoter | Not associated | rs1800872 (IL-10) |
| rs1800871 IL-10) | ||||||
| rs1800896 (IL-10) | ||||||
| Zahra’a et al. [19] | Iraq | Leishmania infantum | Cytokine CTS-PCR-SSP Tray Kit | Promoter | Not associated | rs3212227 (IL-12B) |
| Rasouli et al. [20] | Iran | Leishmania infantum | PCR-RFLP | Exon | Associated (Risk) | 13.687 (IL-15) |
| Intron | Not associated | 367 (IL-15) | ||||
| Associated (Risk) | 267 (IL-15) | |||||
| 3' UTR | Not associated | 14,035 (IL-15) | ||||
| Kumar et al. [21] | India | Leishmania donovani | PCR-RFLP | Promoter | Associated [protective (G allele)] | rs1946519 (IL-18) |
| Not associated | rs187238(IL-18) | |||||
| Codon region | Not associated | rs549908 (IL-18) | ||||
| Moravej et al [22] | Iran | Leishmania infantum | PCR-RFLP | Promoter | Not associated | rs187238(IL-18) |
| Codon region | rs549908 (IL-18) | |||||
| Ahmadpour et al. [23] | Iran | Leishmania infantum | ARMS-PCR | Promoter | Associated [resistance (CC genotype)] | rs1946518 (IL-18) |
| Not associated | rs187238(IL-18) | |||||
| Babaloo et al. [24] | Iran | Leishmania infantum | ARMS-PCR | Promoter | Not associated | rs2227473 (IL-22) |
| Kalani et al. [25] | Iran | Leishmania infantum | PCR-RFLP | Intron | Not associated | rs2227501 (IL-22) |
| Intron | Associated (susceptibility AG genotype) | rs2227513 (IL-22) | ||||
| 3' UTR | Associated [allele A and the AA can be considered as a protective factor against the development] | rs1026786 (IL-22) | ||||
| Karplus et al. [26] | Brazil | Leishmania infantum | PCR-RFLP | Promoter | Associated | -307 (TNF) |
| Ejghal et al. [27] | Morocco | Leishmania infantum | PCR-RFLP | Promoter | Not associated | rs1800629 (TNF-α) |
| Intron | Not associated | rs361525 (TNF-β) | ||||
| Ortiz-Flores et al. [28] | Mexico | Leishmania infantum | PCR-RFLP | Promoter | Not associated | -308(TNF) |
| Not associated | -238(TNF) | |||||
| Maghsood et al. [29] | Iran | Leishmania infantum | ARMS-PCR | Intron | Not associated | rs2430561 (IFN-γ) |
| Al-Bashier [30] | Iraq | Leishmania infantum | ARMS-PCR | Intron | Associated [risk factor (allele A)] | rs2430561 (IFN-γ) |
| Al-Bashier [30] | Iraq | Leishmania infantum | ARMS-PCR | Promoter | Not associated | rs1800896 (IL-10) |
| Mishra et al. [31] | India | Leishmania donovani | PCR-Sequencing | Promoter | Not associated | rs2243250(IL-4) |
| rs2070874(IL-4) | ||||||
| Intron | rs79071878 (IL-4) | |||||
| Mohamed et al. [32] | Sudan | Leishmania donovani | PCR-Sequencing | Intron | Associated | IL4RP2 |
| IL4RP1 | ||||||
| Jerônimo et al. [33] | Brazil | Leishmania infantum | TaqMan SNP genotyping | Promoter | Associated | rs2070874 (IL-4) |
| Hajilooi et al. [34] | Iran | Leishmania infantum | PCR-RFLP | Promoter | Associated [Risk factor (C/T)] | rs1800871 (IL-10) |
| Ahmed et al. [35] | Iraq | Leishmania infantum | Cytokine CTS-PCR-SSP Tray Kit | Promoter | Not associated | rs1800896 (IL-10) |
| rs1800871 (IL-10 | ||||||
| rs1800872 (IL-10) | ||||||
| Hajilooi et al. [36] | Iran | Leishmania infantum | PCR-RFLP | Promoter | Associated 472. [Risk factor (A/G)] | rs1800896 (IL-10) |
| Mishra et al. [37] | India | Leishmania donovani | PCR-Sequencing | Intron | Associated (risk-reducing G allele and the AG and GG genotypes related to protection) | rs1518111(IL-10) |
| Intron | rs1554286 (IL-10) | |||||
| 3'UTR | rs3024496 (IL-10) | |||||
| 3'UTR | rs3024498 (IL-10) | |||||
| Hamidi et al. [38] | Iran | Leishmania infantum | ARMS-PCR | Promoter | Associated [susceptibility (-509 T allele)] | rs1800469 (TGFB1) |
| Rasouli et al. [39] | Iran | Leishmania infantum | PCR-RFLP | Promoter | Associated (resistance) | rs3819024 (IL-17-A) |
| Intron | Associated (resistance) | rs3819025 (IL-17A) | ||||
| Intron | Associated (resistance) | rs8193038 (IL-17A) | ||||
| 3’UTR | Associated [susceptibility (AA genotype and A allele)] | rs3748067 (IL-17A) | ||||
| Khatonier et al. [40] | India | Leishmania donovani | PCR-RFLP | Promoter | Associated [susceptibility (rs8193036TT genotype and the T allele)] | rs2275913 (IL-17A) |
| rs8193036 (IL-17A) | ||||||
| Sadighi et al. [41] | Iran | Leishmania infantum | ARMS-PCR | 3’UTR | Not associated | rs1974226 (IL-17A) |
AS-PCR: allele-specific polymerase chain reaction, ARMS-PCR: amplification refractory mutation system-polymerase chain reaction, PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism.
The methods used for genotyping the individuals in each study varied and included the following: AS-PCR: allele-specific polymerase chain reaction (2); ARMS-PCR: amplification refractory mutation system-polymerase chain reaction (7); cytokine CTS-PCR-SSP Tray Kit (2); PCR-Sequencing (3); PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism (17); and TaqMan SNP genotyping (1).
The SNPs studied are from different gene regions, such as 3 exons, 13 introns, 22 promoters, 8 3'UTRs, and 2 codon regions. For 22 SNPs, there was no association with the development of VL. In 24 of them, there was the presence of an outcome of disease (susceptibility, resistance, and risk or protective factor). Most studies (23) were carried out where the species L. infantum is endemic. In the other studies (5), the parasite responsible for infection was L. donovani.
SNPs associated with clinical outcomes of the disease
- 511T/C and +3953 T/C polymorphisms
Moravej et al. [ 14] concluded that the -511CC genotype and CC haplotype (-511/+3953) of the IL-1β gene were related to susceptibility to the development of the disease. In contrast, the -511 TT genotype and the TT haplotype (-511/+3953) were associated with resistance factors in the Iranian population. Furthermore, the serum levels of this interleukin were greater in individuals with the CC genotype than in those with the TT genotype.
rs1126647 and rs4073 polymorphisms
Ahmadi et al. [ 15], when studying Iranian individuals, concluded that genotype +2767 A/A of the IL-8 gene is associated with positive regulation of this cytokine in patients with VL. Furthermore, Hajilooi et al. [ 16], when evaluating the same population, concluded that the TT genotype in the -251 T/A region is considered a risk factor for the development of VL. In Brazilians, there was no association between this SNP and the disease [ 17].
+1188 and -1188 polymorphisms
In a study carried out by Kalani et al. [ 18] using the +1188 region of the IL-12 gene, there was no significant association between polymorphisms and disease development in an Iranian population. However, Zahra'a et al. [ 19], when evaluating the -1188 C/A of IL-12B, concluded that although this SNP was not associated with resistance or susceptibility, the serum levels of this interleukin were greater in patients with the CC genotype than in those with the other genotypes.
267 and 13687 polymorphisms
Rasouli et al. [ 20] concluded that SNPs at positions 267 and 13687 of the IL-15 gene were associated with the risk of visceral leishmaniasis in Iranians.
rs1946519, rs1946518, rs187238 polymorphisms
Kumar et al. [ 21], when evaluating rs1946519 (-656) of the IL-18 gene, concluded that the G allele may be associated with protection against VL in the Indian population; however, Moravej et al. [ 22], when evaluating the same region, concluded that the T allele may be a resistance factor in Iranian individuals. Furthermore, Ahmadpour et al. [ 23] concluded that Iranians carrying the CC genotype at position -607 have greater resistance to VL infection. Furthermore, Kumar et al. [ 21], Ahmadpour et al. [ 23], and Moravej et al. [ 22], when analyzing the -137G/C region of the IL-18 gene, found no association with the development of the disease.
rs2227473 and rs1026786 polymorphisms
Babaloo et al. [ 24], when studying the SNP rs2227473 A/G of the IL-22 gene in an Iranian population, found no significant association with the development of VL. However, Kalani et al. [ 25] concluded that the A allele and the AA genotype of rs1026786 may be protective factors against the disease.
-307 and rs1800629 polymorphisms
Karplus et al. [ 26] when evaluating the genetic variability in the TNF locus and the development of VL in a Brazilian population, concluded that there is a strong association between the -307 promoter region and asymptomatic infection. When analyzing the SNP rs1800629, Ejghal et al. [ 27] and Ortiz-Flores et al. [ 28] observed that there is no association with disease progression.
rs2430561 polymorphism
Maghsood et al. [ 29], when studying the +874 A/T region of IFN-γ in an Iranian population, reported that there were no significant associations between resistance and susceptibility. However, the highest frequency of the wild-type genotype T was found in healthy VL seropositive individuals. As in a previous study, Kalani et al. [ 18] demonstrated that carriers of the TT genotype have a greater amount of circulating IFN-γ. Thus, these individuals may be more resistant to VL than to AT and AA. In addition, the presence of the AT genotype was considered a predictive factor for the susceptibility of individuals from the endemic area of Iran. However, Al-Bashier [ 30], when evaluating the same region in Iraqi individuals, concluded that carriers of the mutant allele A had a 2.54-fold increase in the risk of developing the disease compared with those with the wild-type T allele. Moreover, the +874A allele was considered a risk factor for the development of VL.
rs2243250, rs2070874, rs79071878, IL4RP2 and IL4RP1 polymorphisms
Mishra et al. [ 31], when studying an Indian population, did not observe an association between the SNPs rs2243250, rs2070874, and rs79071878 and the development of the disease. However, Mohamed et al. [ 32], when conducting a study in Sudan, showed a positive association between the SNPs IL4RP2 and IL4RP1 and the development of VL. In a study in Brazil, Jerônimo et al. [ 33] noted an association between IL-4 34C/T (rs2070874) and delayed-type hypersensitivity (DTH).
rs1800871, rs1800896 and rs3024498 polymorphisms
When evaluating the SNP -819 (rs1800871) of the IL-10 gene, Hajilooi et al. [ 34] observed a significant association between the polymorphism and VL. The CT genotype was more common in the group of patients than in the other groups (asymptomatic and healthy). This may suggest that the presence of this genetic characteristic is a possible risk factor for the development of the disease. When evaluating the same region, Kalani et al. [ 18] and Ahmed et al. [ 35] did not find a significant association.
Hajilooi et al. [ 36] reported that the AG genotype of region -1082 (rs1800896), which could influence the expression of this interleukin, was also considered a risk factor for the disease. Other studies, such as Ahmed et al. [ 35], Al-Bashier [ 30], and Kalani et al. [ 18], on the same region of the IL-10 gene, did not find an association between the SNPs and the development of the disease.
Moreover, Mishra et al. [ 37], when studying Indian individuals, concluded that the rs3024498 polymorphism (5311A > G, 3'UTR) is associated with VL. The presence of the risk-reducing G allele and the AG and GG genotypes are related to protection against the disease.
rs1800469 polymorphism
Frade et al. [ 17], when analyzing the -509 CT gene region of TGF-β1 in a Brazilian population, concluded that this SNP was associated with general susceptibility to infection and severity of the clinical disease. The presence of the T allele results in a 1.9-fold greater risk of developing the disease in individuals with VL and asymptomatic ones than in individuals without the presence of infection. In addition, the presence of this allele was also significantly related to the bleeding conditions of individuals with VL in this population. Hamidi et al. [ 38], when evaluating the same region, obtained similar results regarding the presence of the genotype and susceptibility to the disease.
rs3819024, rs3819025, rs8193038, rs3748067, rs8193036 and rs2275913 polymorphisms
Rasouli et al. [ 39], when evaluating the IL-17 gene, concluded that the AA genotype of the SNPs rs3819024, rs3819025, and rs8193038 are considered resistance markers and that the A allele and AA genotype of rs3748067 are susceptibility factors for the development of VL in an Iranian population. Khatonier et al. [ 40] evaluated an Indian population and reported that individuals carrying the rs8193036TT genotype and the T allele are more susceptible to the disease. The rs2275913A allele is considered a susceptibility factor. Another study carried out by Sadighi et al. [ 41], with other SNPs and again in an Iranian population, did not obtain significant results.
Meta-analysis results
rs2430561 polymorphism
For the IFN-γ SNP, 3 studies, including 275 cases and 324 controls, were counted in the final analysis [ 18, 29, 30]. There was a significant association between the dominant model (OR 1.64, 95% CI 1.13-2.38, I2 = 0%, p < 0.01) and the heterozygous model (OR 1.72, 95% CI 1.15-2.57, I2 = 0%, p < 0.01) and disease progression. For the other models, there were no associations: recessive (OR 1.01, 95% CI 0.68-1.50, I2 = 50%, p = 0.96), allelic (OR 1.34, 95% CI 0.83-2.17, I2 = 62%, p = 0.23) and, homozygous (OR 1.38, 95% CI 0.86-2.22, I2 = 35%, p = 0.18) ( Figure 2). Furthermore, a control group was not in the HWE ( Table 2).
Figure 2. Odds ratios and 95% confidence intervals for the associations between the rs2430561 polymorphism and the progression of leishmaniasis in the models: (A) dominant (AA+AT vs. TT), (B) recessive (AA vs. TA+TT), (C) allelic (A vs. T), (D) homozygous (AA vs. TT), and (E) heterozygous (AT vs. TT).
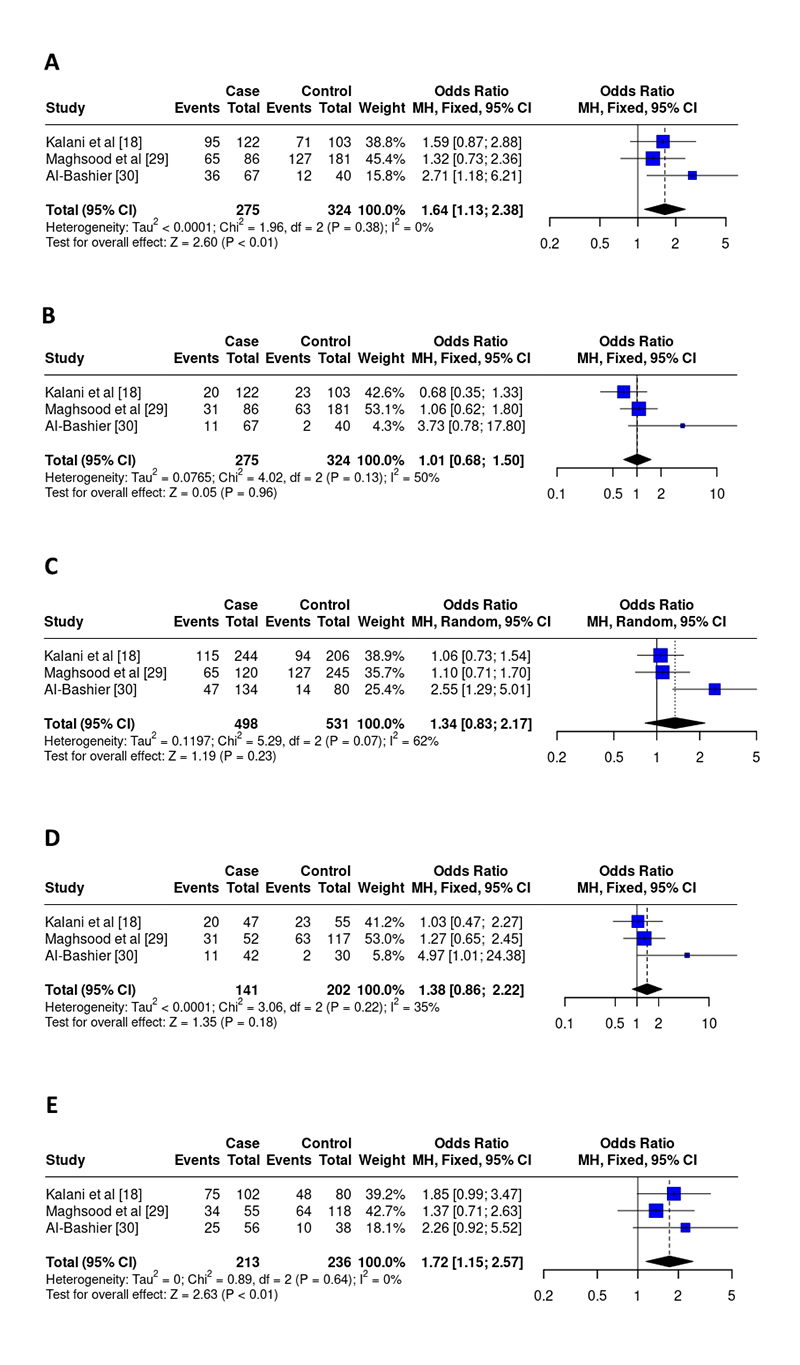
Table 2. Distribution of genotwypes and alleles from the studies included in the meta-analysis and HWE of controls.
| Cases | Control | HWE | MAF | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AT | TT | A | T | AA | AT | TT | A | T | |||
| Study | ||||||||||||
| rs2430561 | ||||||||||||
| Kalani et al. 18 | 20 | 75 | 27 | 115 | 129 | 23 | 48 | 32 | 94 | 112 | 0.55 | 0.46 |
| Maghsood et al [29] | 31 | 34 | 21 | 65 | 55 | 63 | 64 | 54 | 127 | 118 | < 0.001 | 0.52 |
| Al-Bashier [30] | 11 | 25 | 31 | 47 | 87 | 2 | 10 | 28 | 14 | 66 | 0.32 | 0.18 |
| Cases | Control | HWE | MAF | |||||||||
| GG | GC | CC | G | C | GG | GC | CC | G | C | |||
| Study | ||||||||||||
| rs187238 | ||||||||||||
| Kumar et al. [21] | 128 | 63 | 13 | 319 | 89 | 136 | 112 | 19 | 384 | 150 | 0.65 | 0.72 |
| Moravej et al. [22] | 73 | 36 | 9 | 182 | 54 | 93 | 50 | 13 | 236 | 76 | 0.13 | 0.76 |
| Ahmadpour et al. [23] | 59 | 22 | 10 | 81 | 32 | 116 | 52 | 17 | 169 | 69 | 0.006 | 0.67 |
| Cases | Control | HWE | MAF | |||||||||
| GG | GA | AA | G | A | GG | GA | AA | G | A | |||
| Study | ||||||||||||
| rs1800896 | ||||||||||||
| Kalani et al. [18] | 16 | 44 | 60 | 76 | 164 | 19 | 33 | 50 | 71 | 113 | 0.004 | 0.35 |
| Al-Bashier [30] | 17 | 29 | 21 | 63 | 71 | 6 | 22 | 12 | 34 | 46 | 0.53 | 0.43 |
| Ahmed et al. [35] | 4 | 19 | 21 | 27 | 61 | 4 | 21 | 15 | 30 | 50 | 0.51 | 0.36 |
| Hajilooi et al. [36] | 8 | 102 | 0 | 118 | 102 | 35 | 142 | 10 | 212 | 162 | < 0.001 | 0.57 |
| Cases | Control | HWE | MAF | |||||||||
| TT | TC | CC | T | C | TT | TC | CC | T | C | |||
| Study | ||||||||||||
| rs1800871 | ||||||||||||
| Kalani et al. [18] | 11 | 45 | 64 | 67 | 173 | 11 | 32 | 59 | 54 | 150 | 0.07 | 0.26 |
| Hajilooi et al. [34] | 2 | 96 | 2 | 100 | 100 | 28 | 140 | 22 | 184 | 196 | < 0.001 | 0.52 |
| Ahmed et al. [35] | 11 | 14 | 19 | 36 | 52 | 6 | 15 | 19 | 27 | 53 | 0.30 | 0.34 |
HWE: Hardy-Weinberg equilibrium; MAF: minor allele frequency.
rs187238 polymorphism
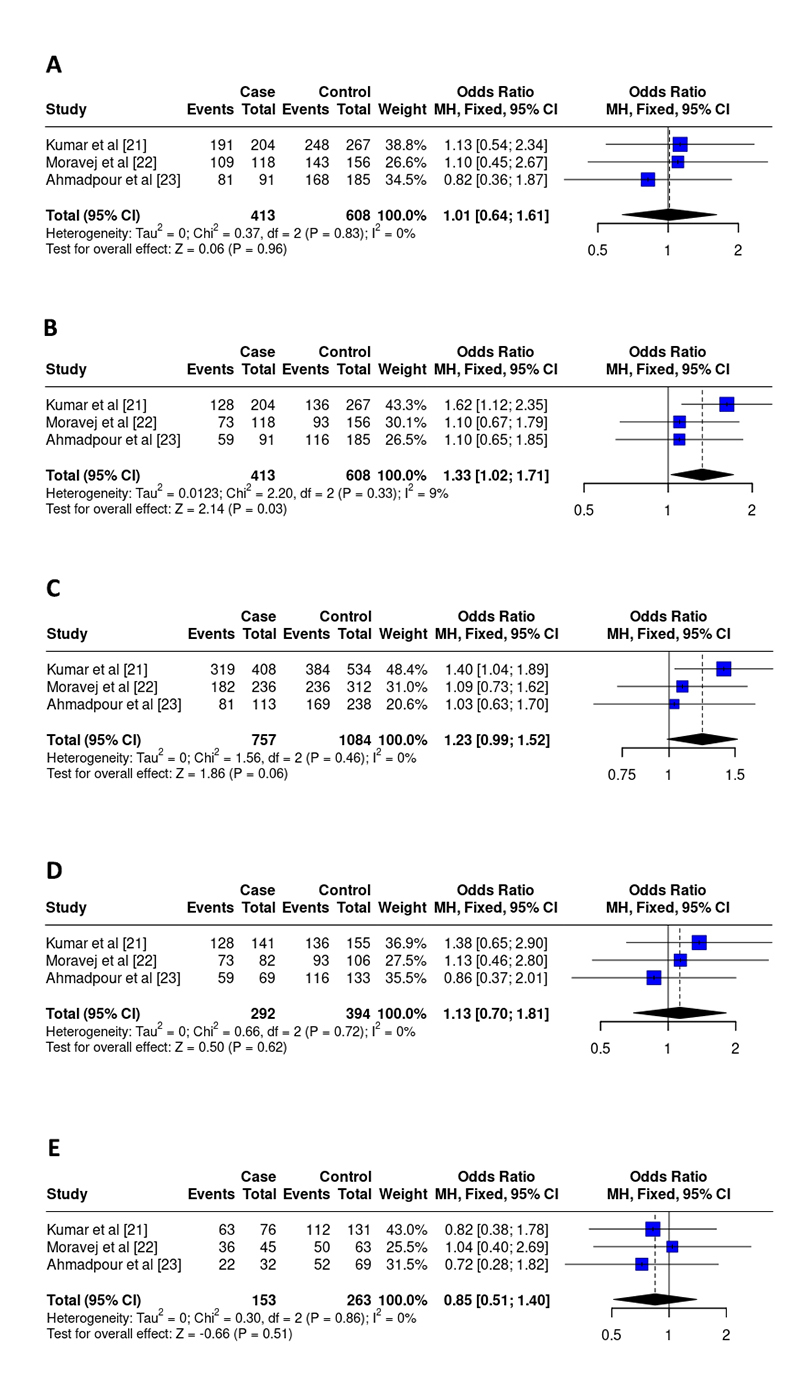
For the IL-18 SNP, 3 studies, including 413 cases and 608 controls, were included in the final analysis [ 21, 22, 23]. There was a significant association between the recessive model (OR 1.33, 95% CI 1.02-1.71, I2 = 9%, p = 0.03) and disease progression. For the other models, there were no associations: dominant (OR 1.01, 95% CI 0.64-1.61, I2 = 0%, p = 0.96), allelic (OR 1.23, 95% CI 0.99-1.52, I2 = 0%, p = 0.06), homozygous (OR 1.13, 95% CI 0.70-1.81, I2 = 0%, p = 0.62), heterozygous (OR 0.85, 95% CI 0.51-1.40, I2 = 0%, p = 0.51) ( Figure 3). Additionally, a control group was not in the HWE ( Table 2).
Figure 3. Odds ratios and 95% confidence intervals for the association between the rs187238 polymorphism and the progression of leishmaniasis in the models: (A) dominant (GG+GC vs. CC), (B) recessive (GG vs GC+CC), (C) allelic (G vs C), (D) homozygous (GG vs CC), and (E) heterozygous (GC vs CC).
rs1800871 polymorphism
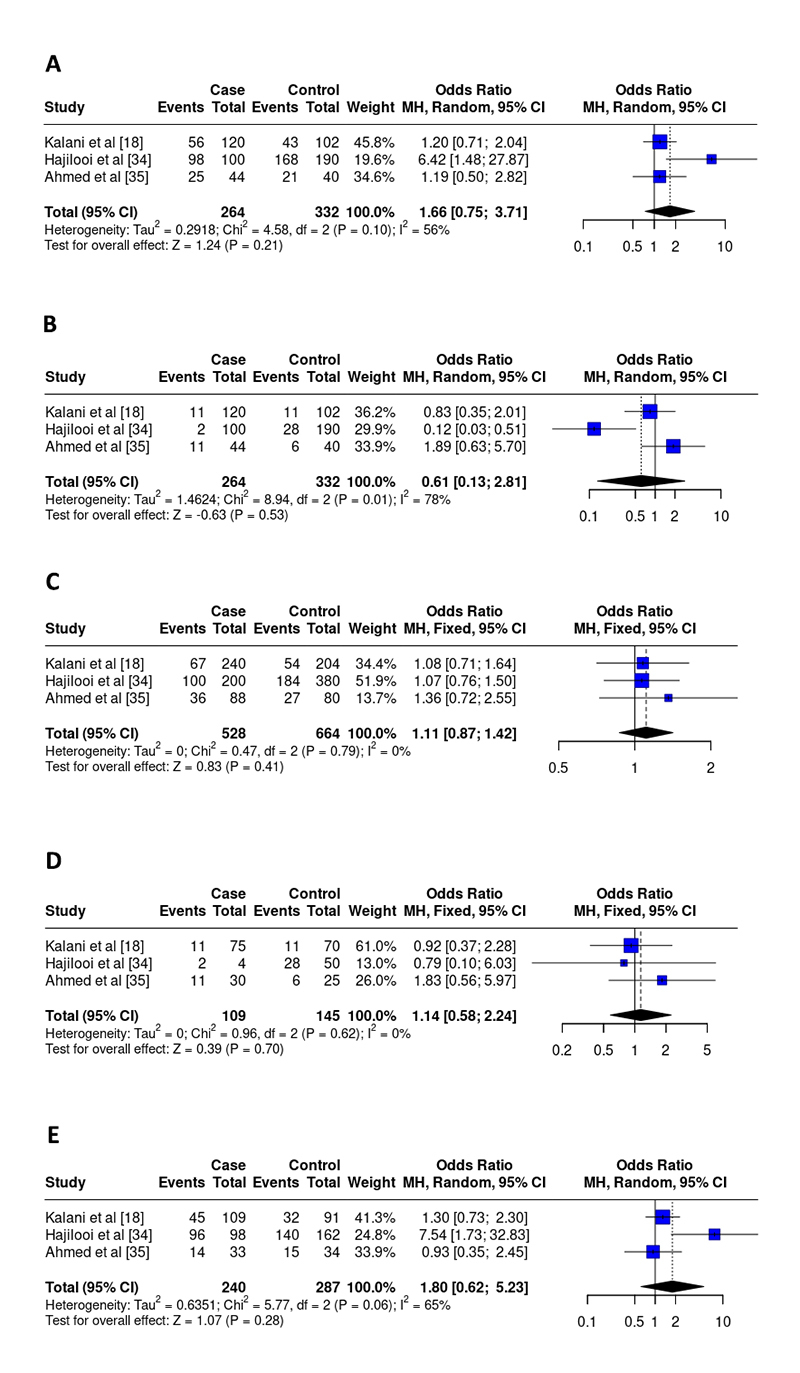
For the IL-10 SNP -819, 3 studies [ 18, 34, 35] encompassing 264 patients and 332 controls were included in the final analysis. No associations were detected between the development of VL and the rs1800871 SNP in any of the genetic models: dominant (OR 1.66, 95% CI 0.75-3.71, I2 = 56%, p = 0.21), recessive (OR 0.61, 95% CI 0.13 -2.81, I2 = 78%, p = 0.53), allelic (OR 1.11, 95% CI 0.87-1.42, I2 = 0%, p = 0.41), homozygous (OR 1.14, 95% CI 0.58-2.24, I2 = 0%, p = 0.70), heterozygous (OR 1.80, 95% CI 0.62-5.23, I2 = 65%, p = 0.28) ( Figure 4). Furthermore, a control group was not in HWE ( Table 2).
Figure 4. Odds ratios and 95% confidence intervals for the association between the rs1800871 polymorphism and the progression of leishmaniasis in the models: (A) dominant (TT+TC vs. CC), (B) recessive (TT vs. TC+CC), (C) allelic (T vs. C), (D) homozygous (TT vs. CC), and (E) heterozygous (TC vs. CC).
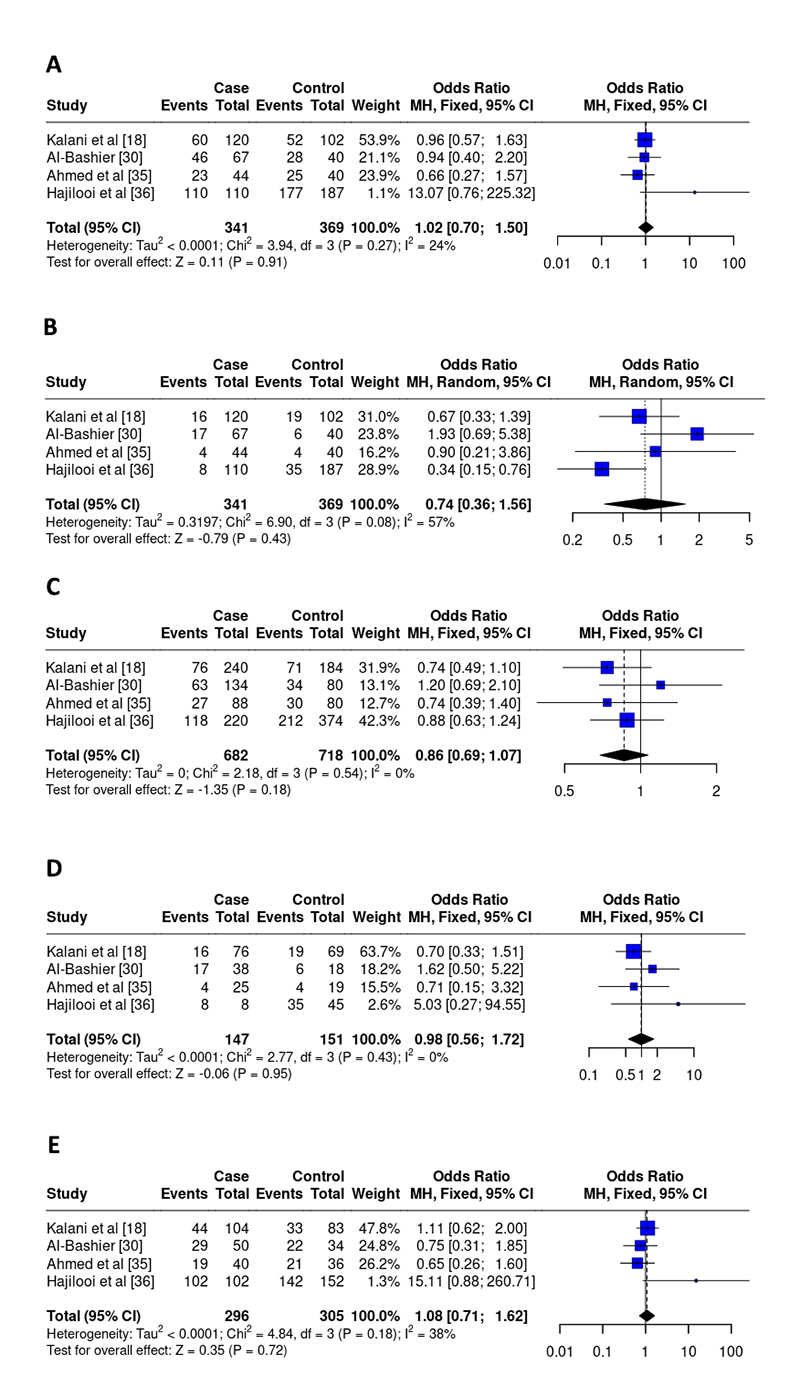
rs1800896 polymorphism
For the IL-10 SNP-1082, 4 studies were included in the final analysis [ 18, 30, 35, 36], including 341 cases and 369 controls. OR values showed no associations between disease development and genetic models: dominant (OR 1.02, 95% CI 0.70-1.50, I2 = 24%, p = 0.91), recessive (OR 0.74, 95% CI 0.36 -1.56, I2 = 57%, p = 0.43), allelic (OR 0.86, 95% CI 0.69-1.07, I2 = 0%, p = 0.18), homozygous (OR 0.98, 95% CI 0.56-1.72, I2 = 0%, p = 0.95), and heterozygous (OR 1.08, 95% CI 0.71-1.62, I2 = 38%, p = 0.72) ( Figure 5). Two control groups in this meta-analysis were not within HWE ( Table 2).
Figure 5. Odds ratios and 95% confidence intervals for the association between the rs1800896 polymorphism and the progression of leishmaniasis in the models: (A) dominant (GG+GA vs. AA), (B) recessive (GG vs. GA+AA), (C) allelic (G vs. A), (D) homozygous (GG vs. AA), and (E) heterozygous (GA vs. AA).
Quality evaluation criteria
Scores ranged from 59% to 90% ( Table 3). Studies incomplete obtained scores between 59 to 68% [ 23, 24, 29, 32, 41]. Articles with slightly more detailed information obtained a percentage of around 77% [ 28, 30, 31, 33, 35, 38, 39]. Scores between 82 and 91% [ 14, 15, 16, 17, 18, 19, 20, 21, 22, 25, 26, 27, 34, 36, 37, 40] were given to those with great clarity. Therefore, the quality of the studies included in this review was considered satisfactory.
Table 3. Quality evaluation of the included studies.
| Criteria | [14] | [15] | [16] | [17] | [18] | [19] | [20] | [21] | [22] | [23] | [24] | [25] | [26] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Question/objective sufficiently described? | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Study design evident and appropriate? | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Method of subject/comparison group selection or source of information/input variables described and appropriate? | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 |
| Subject (and comparison group, if applicable) characteristics sufficiently described? | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 0 | 1 | 1 |
| If interventional and random allocation was possible, was it described? | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| If interventional and blinding of investigators was possible, was it reported? | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| If interventional and blinding of subjects was possible, was it reported? | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Criteria | [14] | [15] | [16] | [17] | [18] | [19] | [20] | [21] | [22] | [23] | [24] | [25] | [26] |
| Outcome and (if applicable) exposure measure(s) well defined and robust to measurement/misclassification bias? Means of assessment reported? | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 |
| Sample size appropriate? | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Analytic methods described/justified and appropriate? | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 |
| Some estimate of variance is reported for the main results? | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 1 | 1 | 2 |
| Controlled for confounding? | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Results reported in sufficient detail? | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 |
| Conclusions supported by the results? | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 |
| Maximum points | 22 | 22 | 22 | 22 | 22 | 22 | 22 | 22 | 22 | 22 | 22 | 22 | 22 |
| Total points | 18 | 18 | 19 | 19 | 19 | 18 | 19 | 20 | 19 | 14 | 13 | 18 | 19 |
| Summary score (%) | 82 | 82 | 86 | 89 | 86 | 82 | 86 | 91 | 86 | 64 | 59 | 82 | 86 |
| Criteria | [27] | [28] | [29] | [30] | [31] | [32] | [33] | [34] | [35] | [36] | [37] | [38] | |
| Question/objective sufficiently described? | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | |
| Study design evident and appropriate? | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| Method of subject/comparison group selection or source of information/input variables described and appropriate? | 2 | 2 | 1 | 1 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | |
| Subject (and comparison group, if applicable) characteristics sufficiently described? | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | |
| If interventional and random allocation was possible, was it described? | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| If interventional and blinding of investigators was possible, was it reported? | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| If interventional and blinding of subjects was possible, was it reported? | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| Outcome and (if applicable) exposure measure(s) well defined and robust to measurement/misclassification bias? Means of assessment reported? | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| Sample size appropriate? | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 2 | 1 | 2 | 2 | 2 | |
| Analytic methods described/justified and appropriate? | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| Criteria | [27] | [28] | [29] | [30] | [31] | [32] | [33] | [34] | [35] | [36] | [37] | [38] | |
| Some estimate of variance is reported for the main results? | 2 | 2 | 0 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 0 | |
| Controlled for confounding? | 0 | 0 | 0 | 0 | 0 | 0 0 | 0 | 0 | 0 | 0 | 0 0 | 0 | |
| Results reported in sufficient detail? | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| Conclusions supported by the results? | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| Maximum points | 22 | 22 | 22 | 22 | 22 | 22 | 22 | 22 | 22 | 22 | 22 | 22 | |
| Total points | 19 | 17 | 13 | 17 | 17 | 15 | 17 | 19 | 17 | 19 | 20 | 17 | |
| Summary score (%) | 86 | 77 | 59 | 77 | 77 | 68 | 77 | 86 | 77 | 86 | 91 | 77 | |
| Criteria | [39] | [40] | [41] | ||||||||||
| Question/objective sufficiently described? | 2 | 2 | |||||||||||
| Study design evident and appropriate? | 2 | 2 | |||||||||||
| Method of subject/comparison group selection or source of information/input variables described and appropriate? | 2 | 2 | 2 | ||||||||||
| Subject (and comparison group, if applicable) characteristics sufficiently described? | 0 | 1 | 0 | ||||||||||
| If interventional and random allocation was possible, was it described? | N/A | N/A | N/A | ||||||||||
| If interventional and blinding of investigators was possible, was it reported? | N/A | N/A | N/A | ||||||||||
| If interventional and blinding of subjects was possible, was it reported? | N/A | N/A | N/A | ||||||||||
| Outcome and (if applicable) exposure measure(s) well defined and robust to measurement/misclassification bias? Means of assessment reported? | 2 | 2 | 1 | ||||||||||
| Sample size appropriate? | 1 | 2 | 2 | ||||||||||
| Analytic methods described/justified and appropriate? | 2 | 2 | 2 | ||||||||||
| Some estimate of variance is reported for the main results? | 2 | 2 | 1 | ||||||||||
| Controlled for confounding? | 0 | 0 | 0 | ||||||||||
| Results reported in sufficient detail? | 2 | 2 | 1 | ||||||||||
| Conclusions supported by the results? | 2 | 2 | 1 | ||||||||||
| Maximum points | 22 | 22 | 22 | ||||||||||
| Total points | 17 | 19 | 13 | ||||||||||
| Summary score (%) | 77 | 86 | 59 | ||||||||||
0 if the response is ‘no’; 1 if the response is ‘partial’; 2 if the response is ‘yes’ followed by N/A if not applicable.
Discussion
VL is a complex multifactorial disease whose exact mechanisms leading to its development are not fully known. Factors such as host-parasite interactions, Leishmania species, environmental conditions, and immunological and genetic factors can contribute to the progression of the disease [ 3- 4]. Disease progression can be associated with the individual immunological profile. The resistance or susceptibility of clinical manifestations are associated with responses that are not yet fully understood, but single-base polymorphisms in immunological coding genes may be responsible for the change in this profile [ 3, 6, 42].
The association between the IFN-γ +874T/A polymorphism and VL progression has been studied in several studies [ 18, 29, 30], but they presented different results. Maghsood et al. [ 29] demonstrated that the rs2430561 polymorphism was not associated with the clinical features of the disease, but healthy seropositive individuals with the TT genotype showed greater secretion of this cytokine. Kalani et al. [ 18] concluded that the AT genotype is associated with susceptibility and that the TT genotype is associated with resistance in Iranians. However, Al-Bashier [ 30] concluded that the A allele was considered a risk factor for the development of the disease. Due to the importance of this cytokine in the immune responses associated with the development of VL and to better understand the role of this SNP, we performed this meta-analysis. Our results indicated that there was a significant association between rs2430561 and disease progression in the dominant model (OR 1.64, 95% CI 1.13-2.38; I2 = 0%, p = <0.01) and heterozygous model (OR 1.72, 95% CI 1.15-2.57; I2 = 0%, p = <0.01). These results indicate that the IFN-γ +874T/A polymorphism may play an important role in disease progression. According to the analysis of the dominant model, the +874 AA genotype was associated with a 1.64-fold increased risk of developing the disease. This result corroborates previous studies by Kalani et al. [ 18] and Al-Bashier [ 30]. This may be because the AA genotype leads to low production of IFN-γ, which may increase the chance of developing VL. However, these results must be interpreted with caution, as the low number of studies included in the analysis may have influenced the outcome.
Regarding rs187238 of the promoter region of the IL-18 gene, previous studies carried out with Indian and Iranian individuals did not show an association between this SNP and the progression of kala-azar [ 21, 22, 23]. In contrast, our recessive model demonstrated that the GG genotype was associated with a 1.33-fold increased risk of developing the disease. These differences in results may be associated with the limited number of patients and controls when carrying out an individual analysis of each study [ 22]. When carrying out a group analysis, these results may change due to the influence of the weight of all research.
Previous studies between SNPs in the promoter region of the IL-10 gene have demonstrated an association between them and the development of VL. Hajilooi et al. [ 34], when evaluating rs1800871, demonstrated that the CT genotype was a risk factor for mortality in Iranians. In another study, Hajilooi et al. [ 36] demonstrated that the AG genotype of the region -1082 was also considered a risk factor for the disease. However, the results are contradictory, and other studies did not observe a significant association [ 18, 30, 35]. Our results indicate that there was no association between the presence of polymorphisms and resistance or susceptibility. This can be explained by the fact that vulnerability to VL may not be related only to these SNPs but also to several other characteristics, such as ethnicity, Leishmania species, and host conditions, among others, thus demonstrating that the mechanisms responsible for the disease they are caused by a combination of multigenetic and environmental factors [ 3, 34]. Furthermore, other components of innate and acquired immunity and other cytokines are also involved in the clinical progression of this disease. Besides, it may be related to the combined inheritance of SNP and polymorphic haplotypes that are inherited together by linkage blocks and may influence disease predisposition.
Finally, the limitations of the present study should be noted. In the final analyses, there are a few studies published and incorporated into the meta-analysis for all SNPs, which may have somehow altered the results. Furthermore, there were methodological differences between the studies, such as variation in the types of control groups, ethnicity, lack of a single standard test for detecting VL, and others. These characteristics may have affected the analyses, the forest plot, and the heterogeneity values.
In conclusion, the main clinical outcomes found were: susceptibility -511CC genotype of IL-1β, rs2227513 (AG genotype) of IL-22 in Iranians. Resistance: -511 TT of IL-1β, T allele of the region -656G/T, CC genotype at position -607 of IL-18, TT genotype of +874 A/T of IFN-γ in Iranians. Risk factor: TT genotype in the -251 T/A of IL-8, AG genotype of the region -1082 (rs1800896) in Iranians, T allele of region -509 of TGF-β1 in Brazilians. Protective factor: G allele of the region -656G/T of IL-18 in Indians, A allele, and the AA genotype of rs1026786 of IL-22 in Iranians.
In addition, this meta-analysis demonstrated that the +874 AA polymorphisms of IFN-γ and -137GG of the IL-18 gene may be associated with the risk of developing VL; however, these data should be interpreted with caution due to the small sample size. Therefore, future research on this topic with greater methodological rigor is necessary to better understand the relationship between SNPs and the progression of VL. Understanding molecular conditions may aid in the discovery of new therapeutic targets for future pharmacological interventions.
Abbreviations
ARMS-PCR: amplification refractory mutation system-polymerase chain reaction; AS-PCR: allele-specific polymerase chain reaction; IL: interleukin; PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism; SNP: single nucleotide polymorphism: Th1: T helper 1; Th17: T helper 17; Th2: T helper 2; VL: visceral leishmaniasis; WHO: World Health Organization.
Acknowledgments
The authors would like to thank Catarine Aragone de Albuquerque Mello for her contribution to the English language edition.
Supplementary material.
The following online material is available for this article:
Funding Statement
This study was partially supported by the Brazilian Coordination for the Improvement of Higher Education Personnel (CAPES), Finance Code 001, and by the National Council for Scientific and Technological Development (CNPq), process 310426/2022-7.
Footnotes
Funding: This study was partially supported by the Brazilian Coordination for the Improvement of Higher Education Personnel (CAPES), Finance Code 001, and by the National Council for Scientific and Technological Development (CNPq), process 310426/2022-7.
Ethics approval: Not applicable.
Consent for publication: Not applicable.
Availability of data and materials.
All data generated or analyzed during this study are included in this article.
References
- Okwor I, Uzonna J. Social and Economic Burden of Human Leishmaniasis. Am J Trop Med Hyg. 2016;94(3):489–493. doi: 10.4269/ajtmh.15-0408.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization Leishmaniasis. 2023. [2023 Jan 05]. internet. Available from: https://www.who.int/news-room/fact-sheets/detail/leishmaniasis .
- Bharati K. Human genetic polymorphism and Leishmaniasis. Infect Genet Evol. 2022;98:105203. doi: 10.1016/j.meegid.2021.105203.. [DOI] [PubMed] [Google Scholar]
- Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry ST, Mullikin JC, Mortimore BJ, Willey D. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409(6822):928–933. doi: 10.1038/35057149.. [DOI] [PubMed] [Google Scholar]
- da Silva RR, Vasconcelos FSF, Tavares DDS, Dos Santos PL. Association between interleukin 10 (IL-10) polymorphisms and leishmaniasis progression: a systematic review and meta-analysis. Sci Rep. 2022;12(1):11136. doi: 10.1038/s41598-022-15377-2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anversa L, Tiburcio MGS, Richini-Pereira VB, Ramirez LE. Human leishmaniasis in Brazil: A general review. Rev Assoc Med Bras. 2018;64(3):281–289. doi: 10.1590/1806-9282.64.03.281.. [DOI] [PubMed] [Google Scholar]
- Page MJ, McKenzie JE, Bossuyt PM, Boutron I, C Hoffmann TC, D Mulrow CD, Shamseer L, Tetzlaff JM, Akl EA, Brennan SE, Chou R, Glanville J, Grimshaw JM, Hróbjartsson A, Lalu MM, Li T, Loder EW, Mayo-Wilson E, McDonald S, McGuinness LA, Stewart LA, Thomas J, C Tricco AC, Welch VA, Whiting P, Moher D. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ. 2021;372(71) doi: 10.1136/bmj.n71.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Prospective Register of Systematic Reviews [2023 Jan 09];PROSPERO. 2022 2022:1–4. Available from: https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42022350889 . [Google Scholar]
- Ouzzani M, Hammady H, Fedorowicz Z, Elmagarmid A. Rayyan - a web and mobile app for systematic reviews. Systematic Reviews. 2016;5:210. doi: 10.1186/s13643-016-0384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmet LM, Lee RC, Cook LS. Standard quality assessment criteria for evaluating primary research papers from a variety of fields. Edmonton: Alberta Heritage Foundation for Medical Research. 2004:1–22. https://www.ihe.ca/advanced-search/standard-quality-assessment-criteria-for-evaluating-primary-research-papers-from-a-variety-of-fields [Google Scholar]
- Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2.. [DOI] [PubMed] [Google Scholar]
- RStudio Team . RStudio: Integrated Development for R. RStudio, PBC; Boston, MA: 2020. http://www.rstudio.com [Google Scholar]
- Moravej A, Rasouli M, Kalani M, Asaei S, Kiany S, Najafipour S, Koohpayeh A, Abdollahi A. IL-1β (-511T/C) gene polymorphism not IL-1β (+3953T/C) and LT-α (+252A/G) gene variants confers susceptibility to visceral leishmaniasis. Mol Biol Rep. 2012;39(6):6907–6914. doi: 10.1007/s11033-012-1517-z.. [DOI] [PubMed] [Google Scholar]
- Ahmadi A, Hajilooi M, Solgi G, Abasi M, Bazmani A, Khalilian A, Matini M, Sardarian K. Evaluation of interleukin 8 +2767 A/T polymorphism in visceral leishmaniasis. Asian Pac J Trop Med. 2016;9(11):1075–1077. doi: 10.1016/j.apjtm.2016.09.010.. [DOI] [PubMed] [Google Scholar]
- Hajilooi M, Abasi M, Bazmani A, Ahmadi A, Matini M, Solgi G, Sardarian K. Evaluation of interleukin-8 -251 T/A polymorphisms in visceral leishmaniasis. J Res Health Sci. 2015;15(1):59–61. [PubMed] [Google Scholar]
- Frade AF, Oliveira LC, Costa DL, Costa CH, Aquino D, Van Weyenbergh J, Barral-Netto M, Barral A, Kalil J, Goldberg AC. TGFB1 and IL8 gene polymorphisms and susceptibility to visceral leishmaniasis. Infect Genet Evol. 2011;11(5):912–916. doi: 10.1016/j.meegid.2011.02.014.. [DOI] [PubMed] [Google Scholar]
- Kalani M, Choopanizadeh M, Rasouli M. Influence of genetic variants of gamma interferon, interleukins 10 and 12 on Visceral Leishmaniasis in an endemic area. Iran. Pathog Glob Health. 2019;113(1):14–19. doi: 10.1080/20477724.2019.1568034.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahra’a A, Ad’hiah AH, Idan EM. Interleukin-12B gene polymorphism and visceral Leishmaniasis in Iraqi patients. J Biol Agri Healthcare. 2015;5:16 [Google Scholar]
- Rasouli M, Kalani M, Kiany S. The role of IL15 gene variants in visceral leishmaniasis among Iranian patients. Mol Biol Rep. 2013;40(8):5151–5157. doi: 10.1007/s11033-013-2617-0.. [DOI] [PubMed] [Google Scholar]
- Kumar D, Tiwary P, Chakravarty J, Sundar S. Association of interleukin-18 gene polymorphism with susceptibility to visceral leishmaniasis in endemic area of Bihar, an Indian population. Sci World J. 2014;2014:852104. doi: 10.1155/2014/852104.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravej A, Rasouli M, Asaei S, Kalani M, Mansoori Y. Association of interleukin-18 gene variants with susceptibility to visceral leishmaniasis in Iranian population. Mol Biol Rep. 2013;40(6):4009–4014. doi: 10.1007/s11033-012-2479-x.. [DOI] [PubMed] [Google Scholar]
- Ahmadpour E, Bazmani A, Kohansal MH, Kazemi A, Babaloo Z. IL-18 gene polymorphism in patients with visceral leishmaniasis in East Azarbaijan. Iran J Parasit Dis. 2016;40(3):981–985. doi: 10.1007/s12639-014-0619-z.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babaloo Z, Bazmani A, Ahmadi H, Sedighi S. Evaluation of IL-22 polymorphism in patients with visceral leishmaniasis. Asian Pac J Trop Med. 2014;4:S545–S548. doi: 10.1016/S2222-1808(14)60674-5. [DOI] [Google Scholar]
- Kalani M, Shams SR, Namdarnia S, Choopanizadeh M, Jamshidi J, Moravej A. Interleukine-22 gene variants are associated with susceptibility to visceral leishmaniasis. Exp Parasitol. 2021;226-7:108122. doi: 10.1016/j.exppara.2021.108122. [DOI] [PubMed] [Google Scholar]
- Karplus TM, Jeronimo SM, Chang H, Helms BK, Burns TL, Murray JC, Mitchell AA, Pugh EW, Braz RF, Bezerra FL, Wilson ME. Association between the tumor necrosis factor locus and the clinical outcome of Leishmania chagasi infection. Infect Immun. 2002;70(12):6919–6925. doi: 10.1128/IAI.70.12.6919-6925.2002.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejghal R, Hamdi S, Idrissi M, Mostapha H, Aboubaker EH, Meryem L. Polymorphisms in tumor necrosis factor genes and susceptibility to visceral leishmaniasis in Moroccan children. Asian Pac J Trop Med. 2015;5:380–384. doi: 10.1016/S2222-1808(14)60801-X.. [DOI] [Google Scholar]
- Ortiz-Flores A, De la Rosa-López G, Zavaleta-Villa B, Chávez-López S, Pastor-Santiago J, Guzmán-Bracho C, Romero-Valdovinos M, Martínez-Hernández F, Olivo-Díaz A. Association of leishmaniasis with TNF alpha promoter and SLC11A1 gene polymorphisms in patients of two endemic areas in Mexico. Microbes Infect. 2015;17(5):387–394. doi: 10.1016/j.micinf.2015.01.001.. [DOI] [PubMed] [Google Scholar]
- Maghsood AH, Jadideslam G, Fallah M, Bazmani A. Evaluation of IFN-γ polymorphism in visceral leishmaniasis. J Res Health Sci. 2014;14(2):136–139. [PubMed] [Google Scholar]
- Al-Bashier NM. Impact of IFN-(+ 874T/A) and IL-10 (−1082G/A) on the susceptibility to Visceral Leishmaniasis. Int J Curr Microbiol App Sci. 2014;3(3):662–667. [Google Scholar]
- Mishra A, Jha AN, van Tong H, Singh VK, Gomes CE, Singh L, Velavan TP, Thangaraj K. Analysis of genetic variants in the IL4 promoter and VNTR loci in Indian patients with Visceral Leishmaniasis. Hum Immunol. 2014;75(12):1177–1181. doi: 10.1016/j.humimm.2014.10.007.. [DOI] [PubMed] [Google Scholar]
- Mohamed HS, Ibrahim ME, Miller EN, Peacock CS, Khalil EA, Cordell HJ, Howson JM, El Hassan AM, Bereir RE, Blackwell JM. Genetic susceptibility to visceral leishmaniasis in The Sudan: linkage and association with IL4 and IFNGR1. Genes Immun. 2003;4(5):351–355. doi: 10.1038/sj.gene.6363977.. [DOI] [PubMed] [Google Scholar]
- Jeronimo SM, Holst AK, Jamieson SE, Francis R, Martins DR, Bezerra FL, Ettinger NA, Nascimento ET, Monteiro GR, Lacerda HG, Miller EN, Cordell HJ, Duggal P, Beaty TH, Blackwell JM, Wilson ME. Genes at human chromosome 5q31.1 regulate delayed-type hypersensitivity responses associated with Leishmania chagasi infection. Genes Immun. 2007;8(7):539–551. doi: 10.1038/sj.gene.6364422.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajilooi M, Sardarian K, Dadmanesh M, Matini M, Lotfi P, Bazmani A, Tabatabaiefar MA, Arababadi MK, Momeni M. Is the IL-10 -819 polymorphism associated with visceral leishmaniasis? Inflammation. 2013;36(6):1513–1518. doi: 10.1007/s10753-013-9693-0.. [DOI] [PubMed] [Google Scholar]
- Ahmed ZA, Ad’hiah AH, Idan EM. The association of interleukin-10 gene polymorphisms with visceral leishmaniasis in a sample of Iraqi patients. Int J Curr Res. 2015;7(8):19301–19305. [Google Scholar]
- Hajilooi M, Ahmadi A, Lotfi P, Matini M, Jafari D, Bazmani A, Momeni M. Is the polymorphism at position -1082 of IL-10 gene associated with visceral leishmaniasis? Iran J Public Health. 2014;43(8):1107–1112. [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Nizamuddin S, Arekatla G, Prakash S, Dewangan H, Dominic A, Mishra A, Sudhakar DV, Parine NR, Tupperwar NC, Thangaraj K. IL10 Variant g.5311A Is Associated with Visceral Leishmaniasis in Indian Population. PLoS One. 2015;10(5):e0124559. doi: 10.1371/journal.pone.0124559.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidi M, Hajilooei M, Bazmani A. Association of transforming growth factor-β1 gene polymorphism with Visceral leishmaniasis in an Iranian population. Shiraz E Med J. 2013;14(2):123–129. [Google Scholar]
- Rasouli M, Moazamian E, Nasiri M, Keshavarz M, Asaei S. Interleukin-17A Genetic Polymorphisms as a Prognostic Markers for Resistance to Visceral Leishmaniasis in the Iranian Population. Arch Clin Infect Dis. 2019;14(2):e57163. doi: 10.5812/archcid.57163.. [DOI] [Google Scholar]
- Khatonier R, Khan AM, Sarmah P, Ahmed GU. Role of IL-17 gene polymorphism in Indian kala-azar. J Vector Borne Dis. 2020;57(1):23–30. doi: 10.4103/0972-9062.308795.. [DOI] [PubMed] [Google Scholar]
- Sadighi S, Bazmany A, Babaloo Z, Amadi H. Evaluation of interleukin 17: A polymorphism in patients with visceral. J Anal Res Clin Med. 2014;2(2):77–82. [Google Scholar]
- Mera-Ramírez A, Castillo A, Orobio Y, Gómez MA, Gallego-Marin C. Screening of TNFα, IL-10 and TLR4 single nucleotide polymorphisms in individuals with asymptomatic and chronic cutaneous leishmaniasis in Colombia: a pilot study. BMC Infect Dis. 2017;17(1):177. doi: 10.1186/s12879-017-2281-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article.


