Abstract
Recent evidence suggests that amyloidogenic oligomers may be the toxic species in cell cultures. Thus, it is crucial to understand their structure and oligomerization mechanism in atomistic detail. By employing tens of fast central processing units and an advanced phase-space sampling algorithm, parallel-tempering molecular dynamics, we have explored the energy landscape of amyloidogenic peptide oligomerization in explicit water. A pentapeptide, DFNKF, derived from human calcitonin and its mutant, DFAKF, was simulated with a total simulation time of ≈500 ns. The detailed oligomerization process of a DFNKF parallel β-sheet formation at 300 K has been characterized. The assembly of a parallel β-sheet from the amorphous state mainly occurs via a “bottleneck” channel where the interstrand Asn–Asn stacking is the major interaction. The interactions of Asn–Asn stacking include both backbone and side-chain hydrogen bonds. The Asn–Asn interactions work like “glue” by sticking the DFNKF strands together and assist the “on-pathway” oligomerization. The Asn–Asn stacking observed here is similar to the Asn ladder commonly found in globular β-helix proteins. A control run shows that when Asn is mutated to Ala, the stability and population of the DFAKF parallel β-sheet is decreased. Furthermore, our in vitro mutagenesis experiments show that the ability of DFAKF peptides to form amyloid fibrils is significantly reduced, in agreement with the simulations. Knowledge of the energy landscape of oligomerization may provide hints for rational drug design, preventing amyloid-associated diseases.
Keywords: amyloid oligomerization, bottleneck channel, β-helix, human calcitonin, mutagenesis
Proteins fold in vivo into their unique 3D structures to perform their biological functions. Yet, under certain conditions, proteins can misfold and aggregate, leading to the malfunctioning of the organism (1). Amyloid fibrosis is one such example, involving severe diseases such as Alzheimer's disease, Prion disorder, and Parkinson's disease (1). Although amyloidogenic diseases are characterized by the deposition of insoluble amyloid fibrils, recent evidence suggests that amyloid oligomers and protofibrils may actually be the toxic agents (2, 3). The increasing appreciation of the importance of the toxic oligomers in the pathological process has attracted considerable attention, focusing not only on their structures but also on the details of their oligomerization mechanism (4–6).
X-ray diffraction data indicate that amyloid fibrils from peptides of unrelated origins have a similar cross-β-fibril organization (7). Also, amyloidogenic proteins do not share sequence or structural homology. The noncrystallinity and insolubility of amyloid fibrils complicate the determination of their structures in atomic detail by conventional methods, such as x-ray crystallography and solution NMR. Nevertheless, the mechanisms of formation and the structure of amyloid fibrils are starting to be revealed by a number of experimental (8–10) and molecular-modeling methods (4, 5, 11). Solid-state NMR studies of Aβ-fibrils provide valuable insights toward the understanding of the structures of the amyloid fibrils (8). Molecular dynamics (MD) simulations at various levels of complexity, ranging from the continuum solvent model (5) to all-atom models with explicit solvent (4, 12), have also been used, complementing the experimental macroscopic views of peptide aggregation.
Recently, we searched for potential minimal peptides that are able to form ordered amyloid (13). We found that a pentapeptide, DFNKF, derived from human calcitonin (hCT) assembles into highly ordered fibrils. Experimentally, the DFNKF peptides can form amyloid fibrils similar to the full-length hCT. hCT tends to aggregate, and its amyloid depositions are associated with medullary carcinoma of the thyroid (14). Our group has studied its stability and dynamics by constant-temperature MD (CTMD). The results showed that the most stable organization of the DFNKF protofibril is a parallel β-stranded sheet (15). Even small oligomer-like trimer and tetramer were stable with a parallel organization for a sufficient time in 350-K MD simulations (16). This result indicates that the size of the crucial nucleus seed for amyloid formation could be quite small, and it supported the experimental observations of the rapid amyloid fibril formation. Recently, solid-state NMR data on the DFNKF peptide by isotope labeling of atoms in two Phe positions have been reported (17). The authors have demonstrated that a mixed model of 70% in-register antiparallel structure and 30% other structure (the first Phe forms a hydrogen bond with Asn) provided a best fit to the “limited” solid-state NMR data. However, additional data are essential to understand the DFNKF amyloid structure in atomic detail (A. Naito, personal communication). For the full-length calcitonin peptide, the authors suggested a mixture of antiparallel and parallel β-sheets under acidic conditions. Other studies have indicated that some amyloidogenic peptides can adopt a mixed organization with parallel and antiparallel arrangements (6, 9, 18). Recent experiments have suggested that amyloid fibrils have distinct morphologies and different morphologies have different underlying molecular structures (19).
The main difficulty in computational studies of the amyloid assembly mechanism arises from limited computer power and simulation techniques. Parallel-tempering MD (PTMD) is a powerful approach that can sample the conformational space more effectively (20) than CTMD. At room temperature (RT), proteins are usually trapped at some local energy minima. To fold into their native structure, proteins have to transverse the rugged energy landscape and escape the energy traps. These barrier-crossing processes are believed to be the most time-consuming steps in protein folding (21). Several computational algorithms, such as Monte-Carlo minimization (22), umbrella sampling, multicanonical sampling (23), and parallel-replica dynamics (24), have been developed to speed up protein-folding simulations (25, 26). Designed to overcome the barrier-crossing problems, PTMD runs a series of MD simulations at various temperatures ranging from low to high temperature. It has been applied to study the folding of peptides and miniproteins successfully (27–31).
In this study, PTMD as well as mutagenesis were used to explore the energy landscape of the amyloid-forming peptide DFNKF oligomerization. Understanding the mechanism of amyloid oligomerization is crucial, not only because amyloid oligomers are potential toxic species (2, 3) but also because they can act as nucleus seeds for amyloid fibril formation (16). Based on our simulations, we propose a mechanism for the formation of parallel β-sheet of DFNKF trimer. The simulations have pointed toward the significant role of the Asn residue in amyloid formation. A mutated peptide, DFAKF, is also simulated to directly compare the free-energy surface of DFNKF. In addition, in vitro mutational experiments were performed. The experiments further support the simulation results.
Methods
Computational Method and System. The PTMD simulations were performed to study the energy landscape of amyloidogenic peptide oligomerization by an in-house program, mcmd [developed by C.-J.T. and H.-H.(G.)T.]. The following three systems were studied: DFNKF trimer, DFAKF trimer, and DFNKF monomer. The DFNKF pentapeptide is a neutral peptide derived from the hCT. One control run with mutant peptide, DFAKF, was performed to examine the role of Asn in DFNKF amyloid formation. The DFNKF monomer simulation is served as a reference to understand the conformation change during oligomerization. All simulations were performed in explicit water with an all-atom charmm-22 force field (32) under periodic boundary condition. The simulations reached a total simulation time of ≈500 ns. All simulations were performed in the Biowulf PC/Linux cluster at the National Institutes of Health (available at: http://biowulf.nih.gov).
Supporting Information. The details of computational method are described in Supporting Computational Methods, which is published as supporting information on the PNAS web site. The supporting information provided to support the conclusion of this work include the details of computational method and experimental procedures, the energy landscape of DFNKF aggregation along the coordinates of radius of gyration and the total contacts, TEM and CR analysis of DFAKF peptide deposit for a longer incubation time, and one additional simulation of DFNKF aggregation with all initial configurations having a Cα-RMS deviation of >2.0 Å.
Results and Discussion
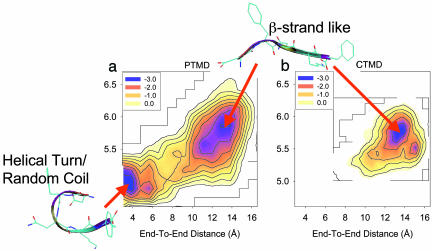
Characterization of DFNKF Monomer Conformations. To understand the energy landscape of the DFNKF oligomerization, the conformations of the DFNKF monomer were characterized first. Characterization of the DFNKF monomer conformations is necessary. It provides a reference to monitor the conformational changes that occur during the course of oligomerization. Fig. 1 shows the energy contours of the DFNKF monomer in the coordinates of end-to-end distance and radius of gyration at 300 K, calculated from the PTMD and CTMD simulations, respectively. The unit used in Fig. 1 is RT. The end-to-end distance was calculated by using the separation between the nitrogen atom of the ammonium group at the N terminus and the carbon atom of the carboxylate group at the C terminus. The radius of gyration was calculated with all of the heavy atoms. For comparison, simulation conditions used for PTMD and CTMD simulations were set to be the same, if possible. The simulations started from an extended structure. The results show that two discrete basins existed in the PTMD simulation, whereas there was only one basin sampled in the CTMD simulation. In the PTMD simulation, the conformations of the DFNKF in its monomeric form were distinguished by two states, the β-strand-like form and the helical-turn/random coil form, as shown in Fig. 1. In contrast, only one conformation, the β-strand-like form, was sampled in the CTMD simulation. It is clear that the CTMD sampled only the conformational space around the initial conformation during the limited simulation time. PTMD sampled both the β-strand-like and the helical-turn/random-coil conformational space. The results described here further validate that the PTMD sampled the conformational space much more effectively than the CTMD at the relevant physiological temperature. To sample the helical-turn/random-coil conformational space by using CTMD, it needs a higher simulation temperature (350 K) and a longer simulation time (12 ns), as demonstrated in the previous study (16).
Fig. 1.
Energy landscapes of the DFNKF monomer at 300 K along two reaction coordinates of the end-to-end distance and the radius of gyration. The unit is in RT. (A) calculated from the PTMD simulation. (B) calculated from the CTMD simulation. The program vmd (50) was used to draw the peptide structures.
In the PTMD simulation, the helical-turn conformation of the DFNKF monomer was found to be 0.21 kcal/mol lower in free energy than the β-strand. There exists a barrier separating the helical-turn/random coil and the β-strand-like conformational space. This energy barrier was characterized to be 2.4 RT (1.43 kcal/mol). Because the barrier is higher than RT, it retards the free interconversion between the β-strand and the helical-turn/random-coil at RT, as shown by the RT CTMD simulation. Amyloid fibrils are believed to have cross β-structures. Thus, the assembly of the cross β-amyloid fibrils from the monomers might happen by means of a conformational change (e.g., helical-turn→β-strand).
Energy Landscape of DFNKF Trimer Oligomerization. The previous comparative MD study, which built different possible models of DFNKF organizations and examined their stabilities by MD simulations, found a stable structure of in-register parallel β-sheet (15). Even small oligomers were also stable for several nanoseconds in 350-K MD simulations (16). However, no stable antiparallel β-sheet structure was found (15). In this PTMD study, although the simulations started from different organizations with partially parallel β-sheet characteristics, the higher-temperature simulations still allowed us to sample the antiparallel β-sheet phase space well. Detailed examination of the trajectories sampled in the lower-temperature replicas showed that only a negligible population of antiparallel β-sheet was sampled. Even though some conformers were associated in an antiparallel arrangement, their population was very low, and their conformations were not in-register antiparallel β-sheet structures. Thus, we focused here on studies of the energy landscape of the parallel β-sheet oligomerization of DFNKF trimer.
To delineate the energy landscape of oligomerization, the ensembles sampled at a given temperature were projected on two reaction coordinates, pQnat and Rg. The pQnat is the fraction of “native” contacts (pQnat) based on the optimized parallel β-sheet of the DFNKF trimer taken from ref. 16. A parallel “native” contact was defined as an interstrand backbone hydrogen bond (33) or a distance between two side-chain geometrical centers of nonadjacent residues <6.7 Å (27), and the Rg is the radius of gyration calculated by using all heavy atoms. The pQnat, with a scale from zero to one, was used to measure the degree of oligomerization, and the Rg was used to monitor the extent of aggregation.
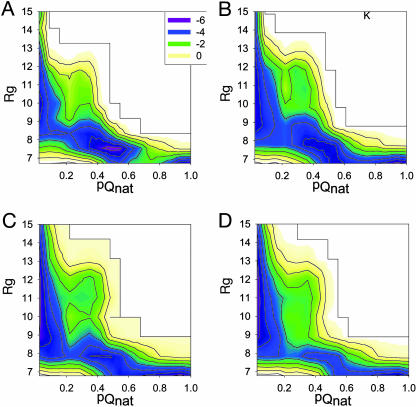
Fig. 2 shows the energy landscapes of DFNKF trimer oligomerization in terms of pQnat and Rg reaction coordinates at the following four different temperatures: 300, 318, 347, and 378 K. For simplicity, to quantify the barriers at different temperatures, the energy scale used in Fig. 2 is given in units of RT instead of kcal/mol. The energy landscapes show the following interesting features relating to the DFNKF trimer oligomerization. (i) The energy landscape is more rugged at low temperature and becomes smoother as the temperature increases. At 300 K, a barrier exists at reaction coordinates pQnat ≈ 0.7 and Rg ≈ 7.0 Å, separating its two neighboring basins. This barrier disappears when the temperature is increased. As a result, the DFNKF trimer sampled all local basins more easily with higher thermal energy than at RT (300 K). (ii) At RT (300 K), the in-register parallel β-sheet of the DFNKF trimer (in the basin with pQnat ≈ 1.0) does not have the lowest free energy. Instead, the basin with pQnat ≈ 0.5 is highly populated. This result indicates that the formation of the in-register parallel β-sheet of the DFNKF trimer could be a kinetic trap. The small oligomer is expected to be less stable. However, the stability of the oligomer will increase with the number of strands, rising dramatically when the critical nucleus seed is formed. At higher temperatures, the population shifted to the lower pQnat side, where the DFNKF trimer gained more entropic energy. (iii) At 300 K, during the early events of aggregation (e.g., Rg > 9.0 Å), the aggregates were amorphous and had a low fraction of parallel “native” contacts (e.g., pQnat < 0.15). Nevertheless, the aggregates with Rg > 9.0 Å had a higher fraction of total contacts (Qtot ≈ 0.40; see Fig. 7, which is published as supporting information on the PNAS web site). This result indicates that the early stage of aggregation was also driven by some “nonnative” interactions. The pQnat increased dramatically when the radius of gyration of the aggregates decreased to ≈8.5 Å. (iv) At 300 K, there was one lower energy barrier at reaction coordinates pQnat ≈ 0.2 and Rg ≈ 8.5 Å. This energy barrier was lower than that with reaction coordinates at pQnat ≈ 0.7 and Rg ≈ 7.0 Å. Nevertheless, its reaction coordinates were narrow, like a “bottleneck,” which retarded the free conversion of structures between two neighboring minima at 300 K. At 318 K, this barrier became broader and was no longer “bottleneck-like.” However, when the temperature continued to increase (347 and 378 K), this barrier became narrower again. This result is because, at higher temperatures, the reaction shifted to the lower pQnat side, and this barrier region was less sampled.
Fig. 2.
Energy landscape of DFNKF trimer oligomerization at four different temperatures [300 (A), 318 (B), 347 (C), and 378 (D) K] by using two reaction coordinates, the fraction of parallel “native” contacts (pQnat) and the radius of gyration (Rg). The units of energy are shown in RT.
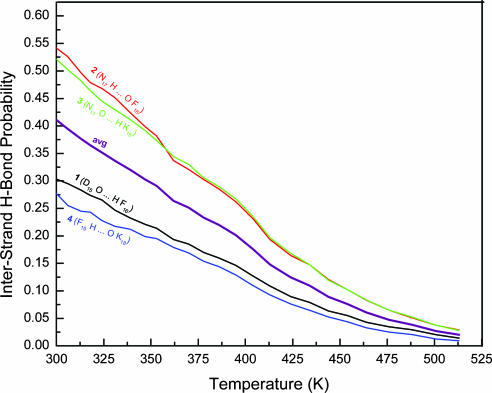
Backbone Hydrogen Bond Probability. To identify which residue of DFNKF has a significant role during the oligomerization and in stabilizing the parallel β-sheet, the probabilities of individual interstrand backbone hydrogen bonds were calculated over a wide range of temperatures. For clarity, each residue is denoted by one subscript number and abbreviated by its one-letter code. Because the DFNKF peptide is derived from residues 15–19 of hCT, the sequence numbers of DFNKF are kept the same as those in the original hCT and shown as subscript numbers to the one-letter code. For example, the residue Asp is abbreviated as D15. The hydrogen bonds were numbered from the N to C termini with a parallel β-sheet organization. For example, the backbone hydrogen bond, D15 O... H F16 nearest to the N termini was numbered as 1, and the backbone hydrogen bond, F19 H... O K18 closest to the C termini was numbered as 4. Fig. 3 shows the probabilities of interstrand backbone hydrogen bonds and their average as a function of temperatures of 300–513 K. The average hydrogen bonds, as well as individual hydrogen bonds, decreased gradually with temperature, indicating a steady decrease of parallel β-sheet characteristics. Hydrogen bonds 2 and 3, which locate at the interior of the β-sheets, were relatively more stable than hydrogen bonds 1 and 4, which are close to the N or C terminus. The latter hydrogen bonds are more likely to be broken when “attacked” by water molecules. Similar results were observed in the β-hairpin simulations (34), in which native hydrogen bonds near the N/C termini were less stable than those close to the turn. The less stable backbone hydrogen bonds (1 and 4) also indicate that the residues near the N/C termini are relatively more flexible. Both more stable hydrogen bonds (2 and 3) are associated with the Asn residues making up the highly populated interstrand Asn–Asn interactions. The interactions with higher populations are expected to have more important roles in stabilizing the parallel β-sheet of the DFNKF trimer and the oligomerization mechanism.
Fig. 3.
Temperature dependence of interstrand backbone hydrogen bond probabilities of the DFNKF trimer.
The stability of the interior backbone hydrogen bonds may arise mainly for two reasons. (i) Because they are located at the interior region of the strands, they are well protected by other residues from being attacked by water molecules; and (ii) the specific side-chain–side-chain interactions, π–π stacking of F16–F16 and the hydrogen bonds between Asn side chains, may have significant roles in stabilizing the interior backbone hydrogen bonds, helping the formation and stabilization of the β-sheet. The significant role of aromatic amino acids in molecular recognition and in the self-assembly of amyloid and amyloid-related structures has been demonstrated in refs. 35–37. Moreover, Asn-rich and Gln-rich peptides were found to tend to aggregate (38, 39). Together, it appears that the existence of a Phe/aromatic ring together with Asn/Gln residues may serve as a common denominator for amyloid formation of various amyloidogenic peptides. Some examples of aromatic–Asn or Asn–aromatic conjugates in amyloidogenic peptides are summarized in Table 1 (13, 40–43).
Table 1. Amyloidogenic peptide sequences highlighting the aromatic–Asn or Asn–aromatic conjugates.
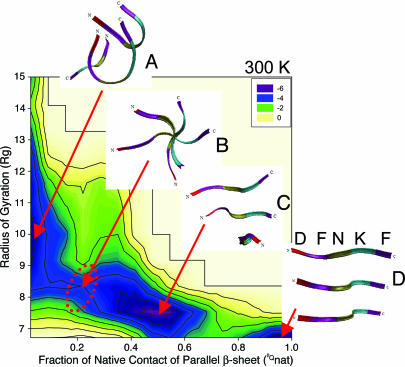
Pathway of DFNKF Trimer Oligomerization. Combining the energy landscapes and backbone hydrogen bond probability discussed above led us to propose a populated oligomerization pathway of the parallel β-sheet DFNKF trimer, characterized at 300 K. In Fig. 4, the structures selected to represent the different energy minima are the structures that are highly populated in the specific basins. Here, we describe one populated pathway; however, other pathways are also possible. The oligomerization reaction occurs along the pQnat coordinate (pQnat increases from zero to one; from basin A to D). At pQnat ≈ 0 (basin A), the conformations of the DFNKF trimer were amorphous, with less structural characteristics of parallel β-sheet. In this basin, strands formed helical-turn, random-coil, or β-strand-like structures. Helical-turn/random-coil structure, the lower energy conformer of the DFNKF monomer, dominated the population of this basin. Basin B (0.1 < pQnat < 0.2 and Rg ≈ 8.5 Å) is a narrow channel interconnecting basins A and C (the highest populated basin). In this region, the three strands were stacked one-by-one, forming a screw axis along the three aligned Asn residues. The structure shown in basin B is shown from the top of the screw axis. When the Asn–Asn stacking is formed, the oligomerization progresses smoothly to basin C. Basin C (pQnat ≈ 0.5 and Rg < 8.5 Å) is a larger basin containing complex structures with ≈50% of the parallel β-sheet characteristics. They generally had the two following characteristics. (i) The Asn–Asn stacking occurred as shown in basin B. (ii) The interior backbone hydrogen bonds were formed; however, the N/C-terminal residues were less in-register to form the β-sheet. The reaction barrier was characterized at pQnat ≈ 0.7, where the formation of the parallel β-sheet of the DFNKF trimer is retarded. This is the time-limiting step, mainly arising from the interstrand packing of the β-sheet at the N/C termini. Last, the well organized parallel β-sheet of the DFNKF trimer (pQnat ≈ 1.0) formed at basin D.
Fig. 4.
Characterization of the oligomerization pathway of the parallel β-sheet of DFNKF trimer at 300 K. The formation of parallel β-sheet of DFNKF trimer follows the reaction coordinate of the fraction of parallel “native” contacts (pQnat) from 0.0 to 1.0 (from basins A to D). (A) Amorphous state. The structures aggregate with the combination of helical-turn, random-coil, and β-strand-like conformers with less or no β-sheet structural characteristics. (B) Bottleneck channel. The structures in this narrow region contained significant amount of Asn–Asn interactions. The structure shown here is viewed from the top of the screw axis forming along the aligned Asn–Asn contacts. (C) A highly populated basin. This basin contained various complex structures with ≈50% of in-register parallel β-sheet characteristics. (D) In-register parallel β-sheet of DFNKF trimer. For clarity, the color–residue correspondences are also labeled in structure D. The program vmd (50) was used to create images of the peptide structures.
To form the parallel β-sheet of the DFNKF trimer, the amorphous aggregates (basin A) pass through a narrow channel (basin B). This channel is narrow, acting as a bottleneck pathway to form the parallel β-sheet structures. The analysis shows that the structures in this channel contained a significant amount of Asn–Asn stacking between strands. The Asn–Asn stacking interactions mainly include interstrand backbone and side-chain hydrogen bonds of Asn. The Asn–Asn stacking characterized here is similar to the Asn ladder commonly observed in naturally occurring β-helices (44). The Asn–Asn stacking works like “glue” by sticking the DFNKF peptides together and help them get into the “on-pathway” reaction coordinates to form the parallel β-sheet of the DFNKF oligomers. Previous CTMD studies showed that when the Asn was mutated to Ala, the parallel β-sheet DFAKF tetramer quickly lost its β-sheet integrity (16). Our results here suggest that the Asn–Asn stacking is the key interaction in the formation of parallel β-sheet of DFNKF oligomers.
To examine how sensitive are the energy landscape and the results to the initial configurations, one addition simulation of DFNKF peptides was performed, with all initial configurations having a Cα-RMS deviation of >2.0 Å (see Supporting Computational Methods for details). The results show that the basin with parallel in-register β-sheet was not sampled within the same simulation time. This result might be expected. Because all initial configurations are far away from this basin, it will take a longer time for this basin to be sampled. Nevertheless, the featured bottleneck channel is still observed in the 300-K energy surface, which further supports our conclusion.
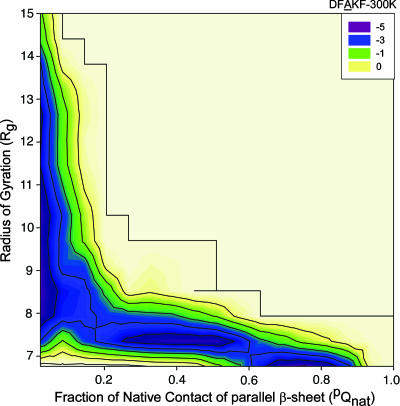
Energy Landscape of DFAKF Trimer Aggregation. One run was performed with the mutant peptide (DFAKF) by using the same simulation protocol as for the DFNKF to further examine the role of Asn in amyloid formation. The energy landscape of DFAKF aggregation (Fig. 5) is generally shifted to the smaller pQnat regions as compared with the energy landscape of the DFNKF oligomerization (Fig. 4). The DFAKF peptides are prone to form amorphous aggregate with less (or no) features of native contacts as compared with DFNKF (basin A in Fig. 4), which might be simply ascribed to the larger hydrophobicity of DFAKF than DFNKF. However, no obvious population with pQnat ≈ 1.0 is observed for DFAKF. The basin with pQnat ≈ 0.75 is the closest one near the in-register parallel β-sheet (pQnat ≈ 1.0). In addition, the largest basin in the DFNKF energy landscape with pQnat ≈ 0.5 (basin C in Fig. 4) is shifted to the basin with pQnat ≈ 0.4 in the DFAKF energy surface. At the same time, the population of this basin in DFAKF is lower than the corresponding one in the DFNKF energy surface. Together, the parallel β-sheet of DFAKF peptides is short-lived as compared with the parallel β-sheets of DFNKF. Namely, the tendency of DFAKF to form stable and quasistable parallel β-sheet trimer is reduced compared with DFNKF. However, whether or not the DFAKF can form amyloid fibril cannot be simply predicted from the simulation.
Fig. 5.
Energy landscape of DFAKF trimer aggregation at 300 K using two reaction coordinates, the fraction of parallel “native” contacts (pQnat) and the radius of gyration (Rg). The units of energy are given in RT.
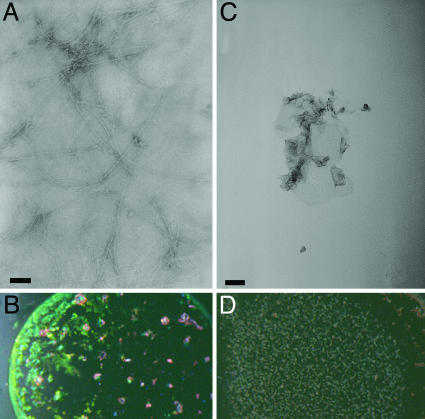
In Vitro Mutant Experiments. Compelling evidence (45, 46) suggests that Asn/Gln-rich domains have a high propensity to form self-propagation of amyloid fibrils. Of particular interest, Wickner and coworkers (46) recently generated five Ure2p variants (46% N/Qs) in which the amino acid sequences were randomly scrambled, but the amino acid composition was kept. All of the five scrambled Ure2p variants still form prion in vivo and amyloid fibers in vitro. Because all Ure2p variants still contain 46% N/Qs and all formed amyloids, the results indicated that the formation of the Ure2p amyloid is mainly driven by Q/N runs. Our work also indicated the significant roles of Asn in DFNKF amyloid formation. To further validate the role of Asn, in vitro mutant experiments were performed by mutating the Asn to Ala. The detailed description of the experimental procedure is given in supporting information. We first studied the ability of the DFAKF peptide to form amyloid fibrils under the same conditions as described for the DFNKF wild-type fragment (13). The incubation of 10 mg/ml DFNKF peptide for 2 days resulted in massive formation of typical amyloid fibrils as observed by transmission electron microscopy (TEM) analysis (Fig. 6A). The peptide deposits showed typical and very strong green-gold birefringence under polarized light upon staining with Congo red (Fig. 6B). In contrast, no amyloid fibrils were observed when the DFAKF peptide was incubated under the same experimental conditions. Extensive analysis of a large area by using electron microscopy did not reveal any typical amyloid fibrils. Only occasional amorphous aggregates (Fig. 6C) were observed with low occurrence. Also no birefringence could be observed with the DFAKF peptide under the same conditions (Fig. 6D).
Fig. 6.
Analysis of the DFNKF and DFAKF peptides. (A) Fibrillar structures formed by the DFNKF pentapeptide after 2 days of incubation at concentration of 10 mg/ml as observed by TEM. (B) CR analysis of DFNKF protein deposits shows typical amyloid birefringence. (C) Amorphous aggregates formed by the DFAKF peptide after 2 days of incubation at concentration of 10 mg/ml as observed at low frequency by TEM. (D) CR analysis of the mutant peptide DFAKF. (Scale bars, 100 nm.)
To study whether the undetectable fibrilar structures is a result of a slow kinetics of formation, a solution of the peptide under the same experimental conditions was monitored for 7 weeks. Only after 2 weeks of incubation, some fibrillar structures were observed (Fig. 8A, which is published as supporting information on PNAS web site). Nevertheless, the amount of fibrillar structures was very small and the fibrils did not show the typical entangled morphology as commonly observed with potent amyloidogenic peptides. Moreover, the peptide solution did not show typical birefringence (Fig. 8B). After 7 weeks of incubation no significant change in the amount or order of the fibrillar structures could be observed. Together, it appears that although the substitution of the Asn residue did not abolish the ability of the peptide to form amyloid fibrils, it reduced it very significantly.
Because the formation of amyloid fibrils is a nucleation-dependent process and, thus, is highly concentration-dependent, we extended our studies to examine the ability of the DFAKF analogue to form amyloid fibrils at a 2-fold concentration of 20 mg/ml. However, no typical amyloid fibrils or CR birefringence was observed with the peptide at this concentration (data not shown). The increased concentration only resulted in the increase of amorphous aggregation. These in vitro experiments further validate the significant role of Asn in amyloid fibril formation identified in our simulations and support the proposed oligomerization mechanism derived from the simulations.
Significant Features of Parallel β-Sheet Amyloid Model. The parallel β-sheet model presents features that contribute to more favorable interactions than those observed in the antiparallel β-sheet model. Within the in-register parallel β-sheet packing, identical residues stack over each other along the fibril axis. Similar phenomena were observed in the naturally occurring β-helix where the chemically similar residues are consistently aligned in successive helical tiers yielding aliphatic, aromatic, and polar stacks (44). Our analysis of the β-helices has led to similar results (data not shown). Further, additional evidence has suggested that amyloids and β-helices may share a similar folding motif (47–49). In the case of the DFNKF, the in-register parallel β-sheet model has two aromatic interactions between β-strands in the same sheet from F16–F16 and F19–F19 stacking. In contrast, the in-register antiparallel β-sheet model has only one aromatic interaction from F16–F19 stacking (17). Moreover, when the F19 was trimmed, the tetrapeptide DFNK still can form amyloid, even though less ordered (13). In this case, no obvious aromatic stacking exists in the in-register antiparallel β-sheet DFNK model. However, the F16–F16 stacking still remains in the inregister parallel β-sheet DFNK model. In addition, our findings toward the significant role of Asn in amyloid formation. In the in-register parallel β-sheet topology of Asn/Gln-rich peptides, each Asn and Gln side chain can have hydrogen bonds with its two neighboring strands forming a side-chain hydrogen-bonding network. In contrast, in the antiparallel model, each side chain can form hydrogen bond with only one other side chain (38), where the Asn side-chain–side-chain hydrogen bonds are discontinuous. The successive Asn side-chain–side-chain hydrogen-bonding network is frequently found in globular β-helix proteins and is commonly called “Asn ladder.” The directional Asn side-chain–side-chain hydrogen bonds, as well as the backbone hydrogen bonds, can have an important role in assisting amyloid fibril elongation.
In summary, here we explore the energy landscape of an amyloid-forming pentapeptide, DFNKF. Our study suggests the significant role of Asn in amyloid formation. The Asn–Asn stacking between strands constitutes a bottleneck channel in the formation of the parallel β-sheet of DFNKF trimer. When the amyloid elongates, the Asn–Asn stacking can be extended to a larger Asn ladder, as commonly present in globular β-helices. Moreover, our in vitro mutant experiment further supports the simulation results. Additional simulations and experiments would elucidate the oligomerization mechanism and structures of amyloid-forming peptides with various sequences. In particular, we focus on the role of Asn/Gln in amyloid formation.
Supplementary Material
Acknowledgments
We thank Dr. A. Naito (Yokohama National University, Yokohama, Japan) for very insightful discussions. This study used the high-performance computational capabilities of the Biowulf PC/Linux cluster at the National Institutes of Health (Bethesda). This work was supported by National Cancer Institute Contract NO1-CO-12400. R.N. was supported in part by the Israel Science Foundation Center of Excellence in Geometric Computing and its Applications.
Author contributions: H.-H.(G.)T., C.-J.T., and R.N. designed research; H.-H.(G.)T. and M.R. performed research; H.-H.(G.)T., K.G., and E.G. contributed new reagents/analytic tools; H.-H.(G.)T., M.R., C.-J.T., K.G., and E.G. analyzed data; and H.-H.(G.)T. and R.N. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MD, molecular dynamics; PTMD, parallel-tempering MD; CTMD, constant-temperature MD; hCT, human calcitonin; RT, room temperature.
References
- 1.Dobson, C. M. (1999) Trends Biochem. Sci. 24, 329-332. [DOI] [PubMed] [Google Scholar]
- 2.Hardy, J. & Selkoe, D. J. (2002) Science 297, 353-356. [DOI] [PubMed] [Google Scholar]
- 3.Kayed, R., Head, E., Thompson, J. L., McIntire, T. M., Milton, S. C., Cotman, C. W. & Glabe, C. G. (2003) Science 300, 486-489. [DOI] [PubMed] [Google Scholar]
- 4.Klimov, D. K. & Thirumalai, D. (2003) Structure (London) 11, 295-307. [DOI] [PubMed] [Google Scholar]
- 5.Gsponer, J., Haberthur, U. & Caflisch, A. (2003) Proc. Natl. Acad. Sci. USA 100, 5154-5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santini, S., Wei, G. H., Mousseau, N. & Derreumaux, P. (2004) Structure (London) 12, 1245-1255. [DOI] [PubMed] [Google Scholar]
- 7.Serpell, L. C. (2000) Biochim. Biophys. Acta 1502, 16-30. [DOI] [PubMed] [Google Scholar]
- 8.Petkova, A. T., Ishii, Y., Balbach, J. J., Antzutkin, O. N., Leapman, R. D., Delaglio, F. & Tycko, R. (2002) Proc. Natl. Acad. Sci. USA 99, 16742-16747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tycko, R. (2004) Curr. Opin. Struct. Biol. 14, 96-103. [DOI] [PubMed] [Google Scholar]
- 10.Jaroniec, C. P., MacPhee, C. E., Bajaj, V. S., McMahon, M. T., Dobson, C. M. & Griffin, R. G. (2004) Proc. Natl. Acad. Sci. USA 101, 711-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma, B. & Nussinov, R. (2002) Proc. Natl. Acad. Sci. USA 99, 14126-14131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klimov, D. K., Straub, J. E. & Thirumalai, D. (2004) Proc. Natl. Acad. Sci. USA 101, 14760-14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reches, M., Porat, Y. & Gazit, E. (2002) J. Biol. Chem. 277, 35475-35480. [DOI] [PubMed] [Google Scholar]
- 14.Arvinte, T., Cudd, A. & Drake, A. F. (1993) J. Biol. Chem. 268, 6415-6422. [PubMed] [Google Scholar]
- 15.Haspel, N., Zanuy, D., Ma, B. Y., Wolfson, H. & Nussinov, R. (2005) J. Mol. Biol. 345, 1213-1227. [DOI] [PubMed] [Google Scholar]
- 16.Tsai, H. H., Zanuy, D., Haspel, N., Gunasekaran, K., Ma, B., Tsai, C. J. & Nussinov, R. (2004) Biophys. J. 87, 146-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naito, A., Kamihira, M., Inoue, R. & Saito, H. (2004) Magn. Reson. Chem. 42, 247-257. [DOI] [PubMed] [Google Scholar]
- 18.Hwang, W. M., Marini, D. M., Kamm, R. D. & Zhang, S. Q. (2003) J. Chem. Phys. 118, 389-397. [Google Scholar]
- 19.Petkova, A. T., Leapman, R. D., Guo, Z. H., Yau, W. M., Mattson, M. P. & Tycko, R. (2005) Science 307, 262-265. [DOI] [PubMed] [Google Scholar]
- 20.Sugita, Y. & Okamoto, Y. (2000) Chem. Phys. Lett. 329, 261-270. [Google Scholar]
- 21.Pande, V. S., Baker, I., Chapman, J., Elmer, S. P., Khaliq, S., Larson, S. M., Rhee, Y. M., Shirts, M. R., Snow, C. D., Sorin, E. J. & Zagrovic, B. (2003) Biopolymers 68, 91-109. [DOI] [PubMed] [Google Scholar]
- 22.Li, Z. Q. & Scheraga, H. A. (1987) Proc. Natl. Acad. Sci. USA 84, 6611-6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terada, T., Matsuo, Y. & Kidera, A. (2003) J. Chem. Phys. 118, 4306-4311. [Google Scholar]
- 24.Shirts, M. R. & Pande, V. S. (2001) Phys. Rev. Lett. 86, 4983-4987. [DOI] [PubMed] [Google Scholar]
- 25.Gnanakaran, S., Nymeyer, H., Portman, J., Sanbonmatsu, K. Y. & Garcia, A. E. (2003) Curr. Opin. Struct. Biol. 13, 168-174. [DOI] [PubMed] [Google Scholar]
- 26.Berne, B. J. & Straub, J. E. (1997) Curr. Opin. Struct. Biol. 7, 181-189. [DOI] [PubMed] [Google Scholar]
- 27.Rao, F. & Caflisch, A. (2003) J. Chem. Phys. 119, 4035-4042. [Google Scholar]
- 28.Garcia, A. E. & Onuchic, J. N. (2003) Proc. Natl. Acad. Sci. USA 100, 13898-13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou, R. H. (2003) Proc. Natl. Acad. Sci. USA 100, 13280-13285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pitera, J. W. & Swope, W. (2003) Proc. Natl. Acad. Sci. USA 100, 7587-7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Felts, A. K., Harano, Y., Gallicchio, E. & Levy, R. M. (2004) Prot. Struct. Funct. Bioinform. 56, 310-321. [DOI] [PubMed] [Google Scholar]
- 32.Karplus et al. (1998) J. Phys. Chem. B 102, 3586-3616. [DOI] [PubMed] [Google Scholar]
- 33.Frishman, D. & Argos, P. (1995) Proteins 23, 566-579. [DOI] [PubMed] [Google Scholar]
- 34.Zhou, R. H., Berne, B. J. & Germain, R. (2001) Proc. Natl. Acad. Sci. USA 98, 14931-14936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazor, Y., Gilead, S., Benhar, I. & Gazit, E. (2002) J. Mol. Biol. 322, 1013-1024. [DOI] [PubMed] [Google Scholar]
- 36.Reches, M. & Gazit, E. (2003) Science 300, 625-627. [DOI] [PubMed] [Google Scholar]
- 37.Makin, O. S., Atkins, E., Sikorski, P., Johansson, J. & Serpell, L. C. (2005) Proc. Natl. Acad. Sci. USA 102, 315-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bevivino, A. E. & Loll, P. J. (2001) Proc. Natl. Acad. Sci. USA 98, 11955-11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perutz, M. F., Finch, J. T., Berriman, J. & Lesk, A. (2002) Proc. Natl. Acad. Sci. USA 99, 5591-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tenidis, K., Waldner, M., Bernhagen, J., Fischle, W., Bergmann, M., Weber, M., Merkle, M. L., Voelter, W., Brunner, H. & Kapurniotu, A. (2000) J. Mol. Biol. 295, 1055-1071. [DOI] [PubMed] [Google Scholar]
- 41.Balbirnie, M., Grothe, R. & Eisenberg, D. S. (2001) Proc. Natl. Acad. Sci. USA 98, 2375-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reches, M. & Gazit, E. (2004) Amyloid 11, 81-89. [DOI] [PubMed] [Google Scholar]
- 43.Maury, C. P. J. & Nurmiaholassila, E. L. (1992) Biochem. Biophys. Res. Commun. 183, 227-231. [DOI] [PubMed] [Google Scholar]
- 44.Jenkins, J. & Pickersgill, R. (2001) Prog. Biophys. Mol. Biol. 77, 111-175. [DOI] [PubMed] [Google Scholar]
- 45.Michelitsch, M. D. & Weissman, J. S. (2000) Proc. Natl. Acad. Sci. USA 97, 11910-11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ross, E. D., Baxa, U. & Wickner, R. B. (2004) Mol. Cell. Biol. 24, 7206-7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wetzel, R. (2002) Structure (London) 10, 1031-1036. [DOI] [PubMed] [Google Scholar]
- 48.Guo, J.-T., Wetzel, R. & Xu, Y. (2004) Proteins 57, 357-364. [DOI] [PubMed] [Google Scholar]
- 49.Govaerts, C., Wille, H., Prusiner, S. B. & Cohen, F. E. (2004) Proc. Natl. Acad. Sci. USA 101, 8342-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Humphrey, W., Dalke, A. & Schulten, K. (1996) J. Mol. Graphics 14, 33-38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.