Correction to: Scientific Reports10.1038/srep27078, published online 01 June 2016
This Article contains errors.
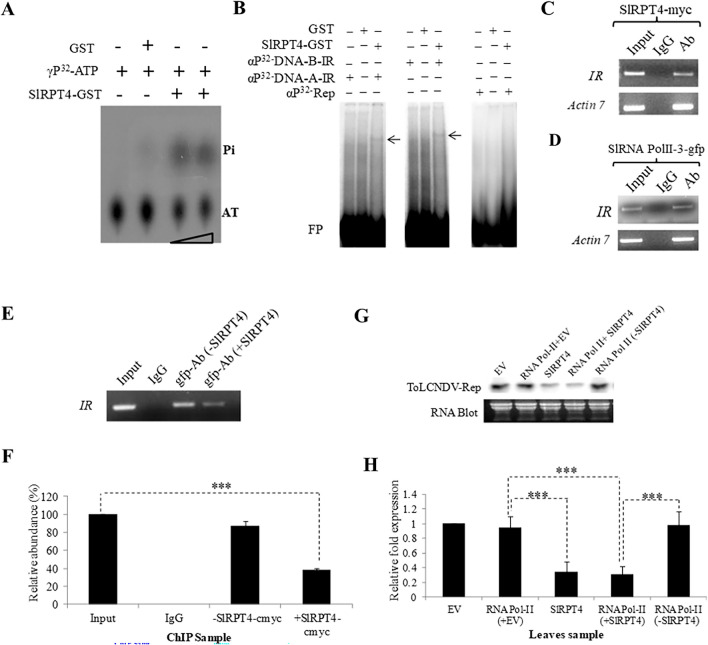
In Fig. 1H, “RNA Pol-II (-SIRPT4)” is incorrectly labelled as “RNA Pol-II”. The correct Fig. 1 and accompanying legend appear below.
Fig. 1.
Molecular characterization of SlRPT4 protein. (A) Thin-layer chromatography (TLC) to evaluate ATPase activity. Figure shows dissociation of Pi from γP32-labelled ATP. Level of Pi was enriched upon increasing the amount of protein. GST protein was used as a negative control of the experiment. (B) DNA binding activity of SlRPT4-GST protein. Binding of SlRPT4 protein onto αP32-dCTP-labeled corresponding fragment of DNA-A-IR, DNA-B-IR and Rep regions are shown by retarded DNA–protein complex through EMSA on 6% native polyacrylamide gel. Signs, + / − represent the presence/absence of components. GST protein was used as a control substrate. In vivo binding assay was performed by transiently overexpressing SlRPT4 (C); SlRNA PolII subunit-3-gfp construct (D); and SlRPT4 and SlRNA PolII subunit-3 co-infiltration (E), in ToLCNDV infected leaves. Figures depict the amplification of IR fragments from the chomatin immuno-precipitated from the sample by using tag-corresponding to c-myc and gfp. (E) Relative abundance of IR specific fragments in the experimental samples. (F) Accumulation of Tomato leaf curl New Delhi virus ToLCNDV specific Rep transcripts in HT (ToLCNDV infected cultivar H-88–78-1) and HSlRPT4+T (SlRPT4 silenced H-88–78-1 infected with ToLCNDV), (G) Northern hybridization showing the accumulation of Rep transcripts, (H) Relative accumulation of Rep transcripts in the leaf samples infiltrated with empty vector (EV), SlRPT4-myc and RNA Pol II-3-gfp construct alone, and co-infiltrated with RNA Pol II-3-gfp and SlRPT4-myc construct. Fragment corresponding to ToLCNDV-Rep gene was used as probe. Total RNA is shown as equivalent loading in the experiment. Data depicts means ± SD of three independent experiments (n = 3); *P < 0.05; **P < 0.01; ***P < 0.001.
In Fig. 2A, upper panel, the representative image corresponding to HTRV:SIRPT4 is inadvertently duplicated for HTRV:00. The representative image of HTRV:00 is also erroneously included as the control. Furthermore, the control condition is not defined in the accompanying figure legend. The correct Fig. 2 and revised legend appear below.
Fig. 2.
Plant phenotyping and molecular analysis of SlRPT4 silenced tomato plants. (A) phenotype of experimental tomato plants. ToLCNDV tolerant cultivar H-88–78-1 plants were infiltrated with Agrobacterium containing the TRV:SlRPT4 construct (HSlRPT4). Subsequent to SlRPT4 silencing process, ToLCNDV infection was performed. The photographs of the plants were taken at 7, 14, 21 and 28 days post inoculation (dpi). Control, cv. H-88–78-1 without virus, mock or silencing treatments, HSlRPT4, SlRPT4 silenced H-88–78-1; HTRV:00, vector infiltrated control plant; HTRV:00+T vector infiltrated control plant infected with ToLCNDV; HSlRPT4+T, SlRPT4 silenced H-88–78-1 infected with ToLCNDV; HT, cultivar H-88–78-1 agroinfiltrated with ToLCNDV. Level of viral DNA in HT (ToLCNDV infected cultivar H-88–78-1) and HTRV:SlRPT4+T (SlRPT4 silenced H-88–78-1 infected with ToLCNDV) at different dpi. (B) Southern blot of tomato genomic DNA from all experimental plants were hybridized with ToLCNDV-coat protein gene specific probe. Replicative forms of ToLCNDV genome are designated as open circular (OC), linear (Lin), supercoiled (SC) and single strand (SS). TRV:00 infiltrated H-88–78-1 was taken as a mock control. Ethidium bromide-stained DNA from each experiment were shown as equivalent loading. (C) Relative accumulation of viral DNA in the samples HT and HSlRPT4+T at different time points. Data depicts means ± SD of three independent experiments (n = 3); *P < 0.05; **P < 0.01; ***P < 0.001.
In Figure S3B, the “Slpds” blot is incorrect and is a duplication of the “ToLCNDV-Rep” blot of Fig. 1G. Additionally, the control condition is not defined in the accompanying figure legend. The correct Figure S3 and revised legend are provided in the Supplementary Information file linked to this notice.
In Figure S5A, the HTRV:SIRPT4+T plant is erroneously labelled as HTRV:SIRPT4. The lower leaf image provided for HTRV:SIRPT4+T is also incorrect and is an inadvertent duplication of the day 14 image of HT shown in the upper panel of Fig. 2A. The correct Figure S5 is included in the linked Supplementary Information file.
These changes do not affect the conclusions of the Article.
Supplementary Information
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-75420-2.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.