Abstract
The species diversity of early-diverging fungi has long lagged behind that of higher fungi, posing a significant obstacle to our comprehensive understanding of the fungal kingdom. Our ongoing research endeavors aim to address this gap by exploring the species diversity of early-diverging fungi in China. In this study, we describe three novel species within the Backusella, namely B.ellipticasp. nov., B.fujianensissp. nov., and B.variisporasp. nov., based on phylogenetic and morphological analyses. In the phylogenetic analysis of the ITS (internal transcribed spacer), LSU (large subunit of ribosomal RNA gene), and RPB1 (RNA polymerase II largest subunit gene) regions, the B.elliptica and B.fujianensis cluster closely with B.gigacellularis, B.ovalispora, and B.solicola, and the B.variispora is closely related to B.locustae and B.pernambucensis. Morphologically, B.elliptica is distinguished by elliptical sporangiospores, as well as cylindrical and hemispherical columellae. The B.fujianensis is characterized by elliptical sporangiospores, and various types of columellae such as hemispherical, subglobose, depressed globose and conical. The B.variispora is characterized by subglobose to globose sporangiospores, as well as hemispherical, subglobose to globose columellae. Additionally, the sporangiophores are long and monopodially branched in B.elliptica and B.fujianensis, while short and simple or sympodially branched in B.variispora. Physiologically, the maximum growth temperatures of B.elliptica (32 °C), B.fujianensis (35 °C), and B.variispora were (35 °C) were determined. With the inclusion of these newly described taxa, the total number of Backusella species known from China now stands at 12. Finally, we provide a key to facilitate the morphological identification of Backusella species from Asia.
Key words: Fungal diversity, morphology, Mucorales, phylogeny, physiology
Introduction
Currently, there is a remarkable increase in the number of documented fungal species owing to advances in molecular evidence. For instance, the 10th edition of the Dictionary of the Fungi in 2008 recorded approximately 100,000 species (Kirk et al. 2008), but now the Fungal Names database reported 156,781 species (assessed on March 7, 2024; Wang et al. 2023a).
Early-diverging fungi, also known as basal or lower fungi, are important in biotechnological areas, such as production of enzymes, lipids and antifungal proteins, and anaerobic members colonizing the digestive tracts of herbivorous vertebrates play a significant role in the breakdown of lignocellulosic feed (Flad et al. 2020). This group of fungi is well-known as pathogens for human, livestock and amphibians, causing diseases such as mucormycosis and chytridiomycosis (Voigt et al. 2021). They encompass a diverse array of evolutionary lineages, morphological characteristics, and ecological distributions, with 17 phyla currently recognized (Galindo et al. 2021; Voigt et al. 2021; Wijayawardene et al. 2022). However, compared to higher fungi (Ascomycota and Basidiomycota, 153,609 species; https://nmdc.cn/fungalnames/; assessed on March 7, 2024), there were significantly limited studies on the evolutionary relationship and species diversity of the early-diverging fungal lineages (Benny et al. 2016; Spatafora et al. 2016; Galindo et al. 2021; Voigt et al. 2021; Zhao et al. 2023a), with only 3,172 species documented (https://nmdc.cn/fungalnames/; assessed on March 7, 2024; Wang et al. 2023a).
In China, studies of early-diverging fungi mainly focused on Entomophthoromycota, Glomeromycota, Kickxellomycota, Mucoromycota, and Mortierellomycota. Notably, from 1980s to 2010s, R.Y. Zheng (Chinese Academy of Sciences), Z.Z. Li (Anhui Agricultural University), S.M. Ho (National Taipei University of Education) and their colleagues have been engaged in these groups of fungi for nearly half a century (Zheng and Chen 1986, 1998; Ho and Chen 1990; Ho 1995a, 1995b, 1996, 2000, 2001, 2002a, 2002b, 2003, 2004, 2006a, 2006b; Li et al. 1999; Li 2000; Liu et al. 2001, 2008; Ho and Chang 2003; Ho et al. 2004, 2007, 2008; Liu 2004; Ho and Hsu 2005; Ho and Benny 2007, 2008; Zheng et al. 2007; Ho and Kirk 2009; Ho and Chuang 2010; Wang et al. 2013, 2014; Zheng and Liu 2014; Liu and Zheng 2015), and in 2018, Zheng and Liu made a summary of 452 species of chytrid, zygomycotan, and glomeromycotan fungi in China (Zheng and Liu 2018). Since then, H. Zhao and his colleagues have contributed 109 new species and new records in Mucoromycota from China, including new species and new records of Absidia (Zhao et al. 2021, 2022a, 2022b, 2023a; Zong et al. 2021), Backusella, Circinella (Zhao et al. 2023a), Cunninghamella (Zhao et al. 2021, 2323a; Wang et al. 2022a), Gongronella (Wang et al. 2023b; Zhao et al. 2023a), Lichtheimia, Mucor, Syncephalastrum (Zhao et al. 2023a), and Umbelopsis (Wang et al. 2022b; Zhao et al. 2023a). During the same period, Y. Nie and his colleagues also described 15 new species and five new records of Entomophthoromycota from China (Nie et al. 2024), covering genera Azygosporus (Cai et al. 2021), Capillidium (Wang et al. 2010a; Nie et al. 2020a, 2022a), Conidiobolus s.s. (Wang et al. 2010a, b; Nie et al. 2017, 2020b, 2023), and Neoconidiobolus (Nie et al. 2012, 2016, 2018, 2021, 2022b). Up to now, early diverging fungi in China accommodated a total of 581 chytrid, zygomycotan, and glomeromycotan species. We expect to conduct a series of studies on the species diversity of early-diverging fungi, and this is the first article in the series, reporting new species of the genus Backusella.
Backusella was proposed by C. Hesseltine and J. Ellis in 1969, characterized by transitorily recurved sporangiophores, and classified within Backusellaceae, Mucorales, Mucoromycetes, and Mucoromycota (Ellis and Hesseltine 1969; Walther et al. 2013; Urquhart et al. 2021; Wijayawardene et al. 2022; Zhao et al. 2023a). Members of Backusella are widely distributed on various substrates, such as soil, litter, toads, wood, invertebrates, and herbivore dung (Santos et al. 2023; Zhao et al. 2023a). In the 20th century, only three species were described in Backusella. From the beginning of this century, the species of Backusella rapidly increased with a total of 38 species being reported (www. indexfungorum.org; accessed on March 8, 2024; de Souza et al. 2014; Lima et al. 2016; Nguyen et al. 2021; Urquhart et al. 2021; de Lima et al. 2022; Hurdeal et al. 2022; Cordeiro et al. 2023; Santos et al. 2023; Zhao et al. 2023a). However, research in China was relatively limited, with only nine Backusella species reported, accounting for 23.68% (9/38; Zheng et al. 2013; Zhao et al. 2023a), and only 15 Chinese occurrences out of the worldwide 3,030 (less than 1%) in the Global Biodiversity Information Facility database (GBIF 2024). In this study, soil samples were collected from Fujian and Hainan Provinces, China, and subjected to the isolation and identification of early-diverging fungi. Subsequently, three novel species within the Backusella were delineated through comprehensive approaches involving morphology, molecular phylogeny, and maximum growth temperatures.
Materials and methods
Samples and strains
During the field trips in Fujian and Hainan Provinces, China, soil samples were collected for the isolation of early-diverging fungi strains. Fujian Province is located along the southeast coast of China and has the subtropical monsoon climate. The air temperature is significantly affected by the monsoon in Fujian Province, with warm winters and an average annual temperature ranging from 15.7 °C to 23.7 °C. The annual precipitation is relatively abundant in Fujian Province, generally between 1400 and 2000 millimeters, decreasing from southeast to northwest. Hainan Province is located at the southern of China, with a tropical monsoon maritime climate. The annual average temperature ranges from 22.5 °C to 25.6 °C, and the annual precipitation is 1500–2500 millimeters.
The isolation methods followed protocols as in previous studies (Zong et al. 2021; Zhao et al. 2022b, 2023a). In brief, 1 g soil was thoroughly suspended with 9 mL sterilized water. Subsequently, 100 μL of the soil suspension was incubated at 25 °C on plates containing potato dextrose agar (PDA: glucose 20 g/L, potato 200 g/L, agar 20 g/L, and pH 7) medium supplemented with antibiotics (streptomycin sulfate 100 mg/mL, and ampicillin 100 mg/mL). The plates were examined using a stereo microscope (SMZ1500, Nikon Corporation, Japan), and cultures exhibiting morphological characteristics were transferred to new plates containing PDA medium and the same antibiotics. Pure strains were obtained through three generations of subcultures. Finally, all living cultures (strains) were deposited at both Beijing Forestry University and Shandong Normal University, and dried cultures (specimens) were preserved in the Herbarium Mycologicum Academiae Sinicae, Beijing, China (HMAS).
Morphology and maximum growth temperature
The pure cultures were incubated with PDA medium at 25 °C for seven days in darkness, followed by morphological observation and photography under a light microscope (ZEISS, Axioscope 5, Germany). The determination of maximum growth temperature was conducted using established methods (Zheng et al. 2007; Zong et al. 2021; Zhao et al. 2023a). Briefly, pure cultures were inoculated onto the center of the PDA plates and placed in a series of biochemical incubators with a temperature range of 25 °C to 45 °C in 5 °C increments. The cultures were observed every 12 hours. All strains were repeated three times. Once the approximate maximum growth temperature was determined, the temperature was gradually increased until the maximum growth temperature was accurate to within 1 °C.
DNA extraction, PCR amplification, and sequencing
The internal transcribed spacers (ITS), large subunit (LSU) of nuclear ribosomal RNA gene, and largest subunit of RNA polymerase II (RPB1) were used for molecular identification. Firstly, the cultures were grown on PDA plates at 25 °C for one week, followed by extraction of total DNA from mycelia using the GO-GPLF-400 kit (GeneOnBio Corporation, Changchun, China), as per the manufacturer’s instructions. Secondly, the ITS, LSU, and RPB1 regions were amplified using the primer pairs ITS 5 (5′‐GGA AGT AAA AGT CGT AAC AAG G‐3′) and ITS 4 (5′‐TCC TCC GCT TAT TGATAT GC‐3′; White et al. 1990), LR0R (5′‐ACC CGC TGA ACT TAA GC‐ 3′) and LR7 (5′‐TAC TAC CAC CAA GAT CT‐3′; http://www.biology.duke.edu/fungi/mycolab/primers.htm), as well as Af (5′-GAR TGY CCD GGD CAY TTY GG-3′) and Cr (5′-CCN GCD ATN TCR TTR TCC ATR TA-3′), respectively. PCR protocols followed previous studies (Urquhart et al. 2021; Zhao et al. 2022a, 2023b). Thirdly, the PCR products were sequenced by the BGI Tech Solutions Beijing Liuhe Co., Limited (https://www.bgi.com/, Beijing, China). Finally, all sequences generated were checked using Geneious v.9.0.2 (Kearse et al. 2012).
Phylogenetic analyses
ITS, LSU, and PRB1 sequences of Backusella and the outgroup Absidiayunnanensis were obtained from the GenBank database or sequenced in this work (Table 1). Each genetic locus was separately aligned using the MAFFT v.7 (Katoh and Standley 2013), and the poorly-aligned sites were trimmed. The ITS, LSU, and RPB1 regions were concatenated using PhyloSuit v.1.2.3 (Zhang et al. 2020) before phylogenetic analyses. The best optimal model of the concatenated dataset was estimated by ModelTest-NG v.0.1.7 (Darriba et al. 2020).
Table 1.
Taxon information and GenBank accession numbers used in the phylogenetic analyses.
Note: “T”, ‘ET”, and “LT” are represented ex-type, ex-epitype, and ex-lectotype, respectively. “–” is represented absences of sequences.
Maximum Likelihood (ML) and Bayesian Inference (BI) phylogenetic analyses were conducted with RAxML v.8 (Stamatakis 2014) and MrBayes v.3.2.7a (Ronquist et al. 2012), respectively, following the methods described in previous studies (Nie et al. 2020a, 2020b; Zhao et al. 2023a). For ML analysis, 1,000 bootstrap replications were conducted using the best optimal model. For BI analysis, two million generations were run until the standard deviation fell below 0.01, and the first 25% were discarded as burn-in. Meanwhile, ML and BI analyses were carried out using ITS and LSU sequences. Finally, the ML and BI trees were visualized using the Figtree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/). Nodes with ML bootstrap values below 50% and BI posterior probability values of less than 0.9 were not considered.
Results
Phylogeny
The concatenated dataset comprised a total of 2,685 characters derived from 61 strains, including 1,029 characters from ITS sequences, 661 characters from LSU sequences, and 995 characters from RPB1 sequences (Suppl. material 1). A concatenated dataset of ITS and LSU sequences was provided in the supplementary material Suppl. material 2. GTR+I+G model was selected as the most suitable for the analysis. For the BI analysis, the standard deviation was 0.004813 after two million generations were calculated.
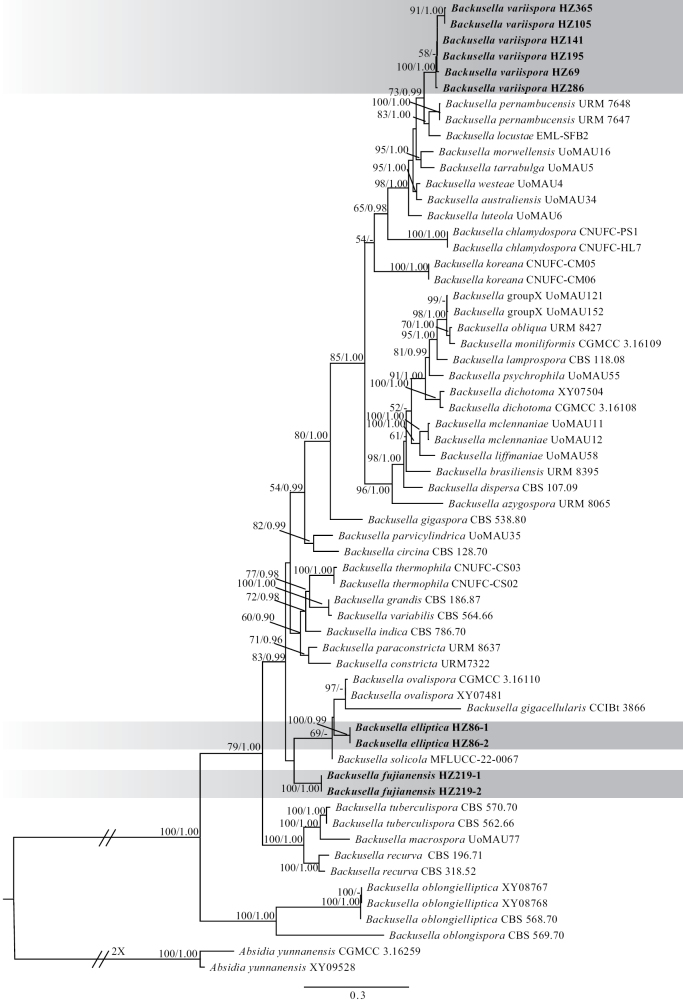
Phylogenetic analyses of the Backusella suggested that three new species, namely B.elliptica, B.fujianensis, and B.variispora, were well supported (Fig. 1, Suppl. material 3). The B.elliptica and B.fujianensis formed a distinct clade with B.gigacellularis, B.ovalispora, and B.solicola. The B.variispora was sister to B.locustae and B.pernambucensis (MLBV 73% / BPP 0.99).
Figure 1.
The Maximum Likelihood phylogenetic tree of the genus Backusella based on ITS, LSU, and RPB1 genetic loci. Two strains of Absidiayunnanensis serve as the outgroups. The new species, Backusellafujianensis, B.elliptica, and B.variispora, are shaded. The Maximum Likelihood bootstrap values (MLBV ≥ 50%) / Bayesian Posterior Probabilities (BPP ≥ 0.90) of each clade are indicated along branches. Some branches are shortened to fit to the page, which are indicated by double slashes and the number of fold times. The scale bar at the bottom left indicates the number of substitutions per site.
Taxonomy
. Backusella elliptica
H. Zhao & X.Y. Liu sp. nov.
CD916D96-7E33-5503-929B-5ACE0755E359
Fungal Names: FN 571901
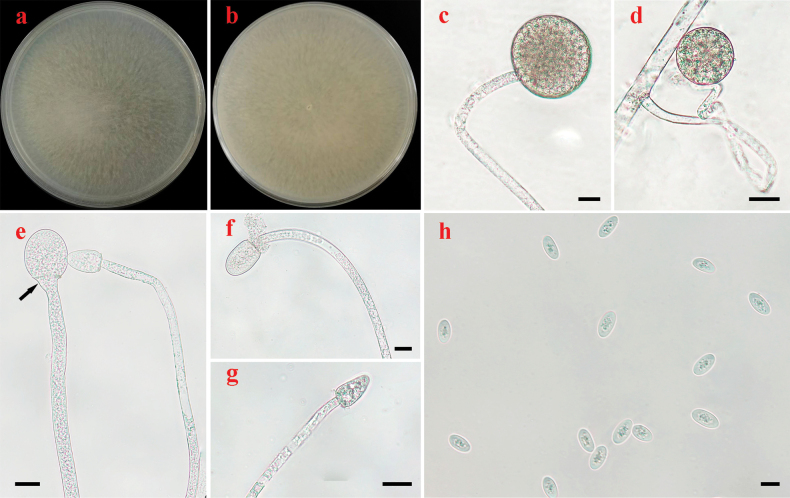
Figure 2.
Morphologies of Backusellaelliptica ex-holotype HZ86-1 a, b colonies on PDA (a obverse b reverse) c long sporangiophores with multi-spored sporangia d short sporangiophores with multi-spored sporangia e–g sporangiophores with columellae h sporangiospores. Scale bars: 20 μm (c–g); 10 μm (h).
Etymology.
elliptica (Lat.) refers to the species having elliptical sporangiospores.
Holotype.
HMAS 352890.
Colonies on PDA at 25 °C for 4 days, reaching 90 mm in diameter, more than 15 mm high, flat, granulate, initially white, soon becoming pale mouse-grey, reverse straw-yellow stramineus. Hyphae aseptate at first, septate with age, hyaline, 5.0–18.5 μm in diameter. Rhizoids absent. Stolons absent. Long sporangiophores arising directly from substrate mycelia or aerial mycelia, transitorily curved, monopodially branched, usually with large terminal sporangia, erect, bent or rarely curved. Sporangia globose, hyaline to brownish, rough-walled, multi-spored, with more than 50 sporangiospores per sporangium, deliquescent-walled, 75.0–95.0 μm in diameter. Short sporangiophores unbranched, curved, ending with a multi-spored sproangiolum. Multi-spored sporangiola globose, hyaline, containing more than 10 sporangiospores, 30.0–50.0 μm in diameter, persistent-walled. Uni-spored sporangiola unknown. Apophyses rarely present. Collars, if present, small. Columellae usually cylindrical and rarely hemispherical, hyaline, with small droplets, 27.0–54.5 × 20.0–43.5 μm on the top of long sporangiophores, and usually conical, hyaline, with small droplets, 20.0–30.0 × 10.0–20.0 μm on the short sporangiophores. Sporangiospores elliptical, hyaline, with small droplets, 11.0–16.5 × 6.5–8.5 μm wide. Azygosporangia absent. Chlamydospores absent. Zygospores absent.
Materials examined.
China • Hainan Province, Ledong Li Autonomous Country, 18°42'35"N, 108°52'36"E, from forest soil sample, 11 April 2023, Heng Zhao (holotype HMAS 352890, living ex-holotype culture HZ86-1, and living culture HZ86-2).
GenBank accession numbers.
ITS, PP477393 and PP477394; LSU, PP477403 and PP477404, RPB1, PP709513 and PP709514.
Maximum growth temperature.
32 °C.
. Backusella fujianensis
H. Zhao & X.Y. Liu sp. nov.
0A30502B-18E4-5F5B-B2CC-EDE3C0761A86
Fungal Names: FN 571900
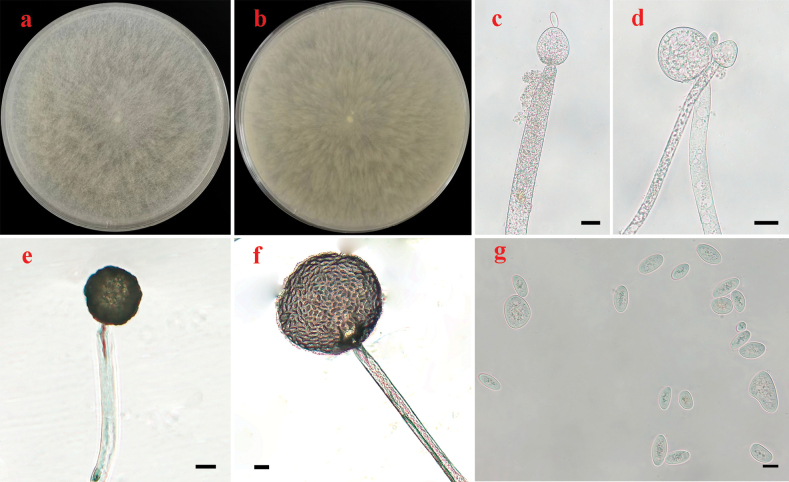
Figure 3.
Morphologies of Backusellafujianensis ex-holotype HZ219-1 a, b colonies on PDA (a obverse b reverse) c, d sporangiophores with columellae e short sporophore with multi-spored sporangia f tall sporophore with multi-spored sporangia g sporangiospores. Scale bars: 20 μm (c–f); 10 μm (g).
Etymology.
fujianensis (Lat.) refers to Fujian province where the type was collected.
Holotype.
HMAS 352889.
Colonies on PDA at 25 °C for 4 days, reaching 90 mm in diameter, more than 15 mm high, granulate, lobed and scaly, initially white, soon becoming pale mouse-grey, reverse straw-yellow stramineus. Hyphae aseptate at first, septate with age, hyaline, 5.5–25.5 μm in diameter. Rhizoids absent. Stolons absent. Long sporangiophores arising directly from substrate or aerial mycelia, transitorily curved, monopodially branched, usually with large terminal sporangia, erect, bent or curved. Sporangia subglobose to globose, hyaline to brownish, rough-walled, multi-spored, with more than 50 sporangiospores per sporangium, persistent-walled, 70.0–160.0 μm in diameter. Short sporangiophores unbranched, ending with a multi-spored sporangiolum. Multi-spored sporangiola subglobose to globose, hyaline, containing more than 20 sporangiospores, 45.0–65.0 μm in diameter, persistent-walled. Uni-sporangiola unknown. Apophyses absent. Collars if present, small. Columellae hemispherical, depressed globose to subglobose, hyaline to light brown, 36.0–64.5 × 33.0–63.5 μm in long sporangiophores, and conical and hemispherical, hyaline, 13.0–21.0 × 12.0–20.0 μm in short sporangiophores. Sporangiospores elliptical, rarely irregular, hyaline, with droplets, 12.0–21.5 × 6.0–10.5 μm. Azygosporangia absent. Chlamydospores absent. Zygospores absent.
Materials examined.
China • Fujian Province, Wuyishan City, 27°48'59"N, 117°42'46"E, from forest soil sample, 15 October 2022, Heng Zhao (holotype HMAS 352889, living ex-holotype culture HZ219-1, and living culture HZ219-2).
GenBank accession numbers.
ITS, PP477391 and PP477392; LSU, PP477401 and PP477402, RPB1, PP709511 and PP709512.
Maximum growth temperature.
35 °C.
. Backusella variispora
H. Zhao & X.Y. Liu sp. nov.
4A18494A-04C8-579F-A256-B08D2353EC95
Fungal Names: FN 571902
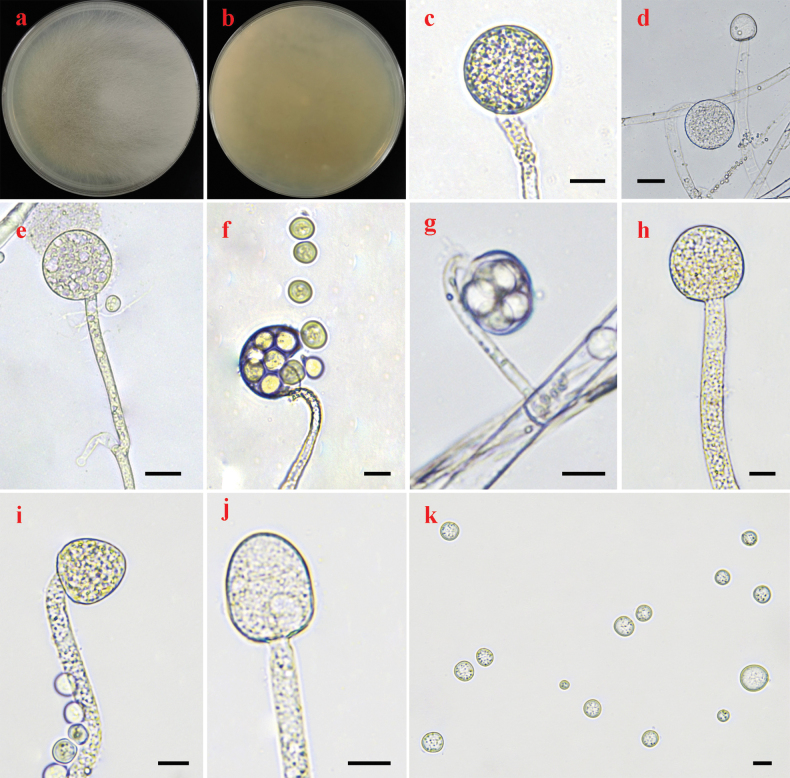
Figure 4.
Morphologies of Backusellavariispora ex-holotype HZ69 a, b colonies on PDA (a obverse b reverse) c–e long sporangiophores with multi-spored sporangia f, g short sporangiophores with multi-spored sporangia h–j sporophore with columellae k sporangiospores. Scale bars: 10 μm (c, f–k); 20 μm (d, e).
Etymology.
variispora (Lat.) refers to the species having an uneven size of sporangiospores.
Holotype.
HMAS 352891.
Colonies on PDA at 25 °C for 4 days, reaching 90 mm in diameter, more than 15 mm high, flat, granulate, initially white, soon becoming pale mouse-grey, irregular at margin. Hyphae aseptate at first, septate with age, hyaline, 4.5–11.5 μm in diameter. Rhizoids absent. Stolons absent. Long sporangiophores arising directly from substrate mycelia, transitorily curved, monopodially branched, with large terminal sporangia, erect, bent or curved. Sporangia globose, hyaline to brownish, wall rough with spines, deliquescent, rough, multi-spored, with more than 20 sporangiospores per sporangium, 30.5–60.0 μm in diameter. Short sporangiophores simple or sympodial, ending with a multi-spored. Multi-spored sporangiola subglobose to globose, with numerous spines, hyaline, containing 5–10 sporangiospores, persistent-walled, 14.5–26.0 μm in diameter. Apophyses absent. Collars absent. Columellae hemispherical, subglobose to globose, hyaline, 21.0–32.5 × 20.0–33.0 μm in long sporangiophores, conical and hemispherical, hyaline, 14.5–18.5 × 14.0–18.0 μm in short sporangiophores. Sporangiospores subglobose to globose, hyaline, with droplets, 5.0–16.0 μm in diameter. Azygosporangia absent. Chlamydospores absent. Zygospores absent.
Materials examined.
China • Hainan Province, Ledong Li Autonomous County, 18°42'35"N, 108°52'36"E, from soil sample, 11 April 2023, (holotype HMAS 352891, living ex-holotype culture HZ69) • Changjiang Li Autonomous County, 19°7'18"N, 109°7'7"E, from soil sample, 12 April 2023, Heng Zhao (living cultures HZ105, HZ195, and HZ365) • Lingshui Li Autonomous County, 18°42'8"N, 109°50'13"E, from forest soil sample, 9 April 2023, Heng Zhao (living cultures HZ141 and HZ286).
GenBank accession numbers.
ITS, PP477395–PP477400; LSU, PP477405–PP477410, RPB1, PP709511 and PP709515.
Maximum growth temperature.
35 °C.
Discussion
In this study, three novel species, Backusellafujianensis, B.elliptica, and B.variispora were proposed based on phylogenetic relationships, morphological characteristics, and maximum growth temperatures. Phylogenetic analyses showed that the B.elliptica and B.fujianensis are closely related to B.gigacellularis, B.ovalispora, and B.solicola, and the B.variispora is closely related to B.locustae and B.pernambucensis.
These three new species are morphologically distinguished from their closely-related species. In detail, the B.gigacellularis differs from B.elliptica by fewer sporangiospores in multi-spored sporangiola (3–4 vs. more than 10), the absence of collars, the presence of giant cells, and the irregular sporangiospores (de Souza et al. 2014). The B.ovalispora differs from B.fujianensis by a faster growth speed (3d vs. 4d reaching 90 mm on PDA), the presence of uni-spored sporangiola, less sporangiospores in multi-spored sporangiola (3–4 vs. more than 10), and globose or subglobose columellae (Zhao et al. 2023a). The B.solicola differs from B.elliptica by subglobose columellae, fewer sporangiospores of multi-spored sporangiola (4–8 vs. more than 10), and the presence of the uni-spored sporangiola, chlamydospores, and rhizoids (Hurdeal et al. 2022).
The B.gigacellularis differs from B.fujianensis by the fewer multi-spored sporangiola (up to 23 μm in diameter vs. 43–64 μm in diameter) and fewer sporangiospores (3–4 vs. more than 20), the absence of collar, and the presence of giant cells (de Souza et al. 2014). The B.ovalispora differs from B.fujianensis by faster growth speed (3d vs. 4d reaching 90 mm on PDA), the presence of uni-spored sporangiola, less sporangiospores in multi-spored sporangiola (3–4 vs. more than 20; Zhao et al. 2023a). The B.solicola differs from B.fujianensis by forming oblong to cylindrical columellae, fewer sporangiospores in multi-spored sporangiola (4–8 vs. more than 20), and the presence of the unispored sporangiola, chlamydospores, and rhizoids (Hurdeal et al. 2022). In addition, the B.elliptica differs from B.fujianensis by the absence of depressed globose to subglobose columellae, the presence of apophyses, and the lower maximum growth temperature (32 °C vs. 35 °C). The B.locustae differs from B.variispora by the larger sporangiospores (9–23.5 × 10.5–25.5 μm vs. 14.5–26.0 μm in diameter) and multi-spored sporangiola (31–59 × 33.5–61.5 μm vs. 5.0–16.0 μm in diameter; Wanasinghe et al. 2018). The B.pernambucensis differs from B.variispora by the presence of rhizoids and giant cells, and more sporangiospores in multi-spored sporangiola (up to 15 vs. 5–10; Cordeiro et al. 2023).
Recent studies have highlighted the significance of maximum growth temperature as a distinguishing characteristic among Backusella species. These studies have categorized maximum growth temperatures into three groups: no higher than 33 °C; between 33 °C and 35 °C; 36 °C or higher (Cordeiro et al. 2023; Santos et al. 2023). In this study, maximum growth temperatures of B.fujianensis, B.elliptica, and B.variispora were 35 °C, 32 °C, and 35 °C, respectively. However, it’s worth noting that the grouping based on maximum growth temperature is not entirely consistent with the results of the phylogenetic analyses (Cordeiro et al. 2023).
Backusella species are distributed around the world, such as in Brazil (13 species; Cordeiro et al. 2023; Santos et al. 2023), Australia (10 species; Urquhart et al. 2021), South Korea (seven species; Wanasinghe et al. 2018; Nguyen et al. 2021), and Thailand (one species; Hurdeal et al. 2022). Although the study of Backusella species diversity was carried out relatively late in China (Zheng et al. 2013; Zhao et al. 2023a), 12 species have been discovered, including the three novel species in this study. A total of 21 Backusella species were reported from Asia (Walther et al. 2013; Zheng et al. 2013; Wanasinghe et al. 2018; Nguyen et al. 2021; Hurdeal et al. 2022; Zhao et al. 2023a). Since the characters of Backusellagranulispora were unavailable, we provide herein a synoptic key to the other 20 Asian Backusella species.
Key to species of Backusella from Asia
| 1 | Sporangiospores mainly subglobose to globose, ovoid, or irregularly polyhedral | 2 |
| – | Sporangiospores mainly ellipsoidal | 11 |
| 2 | Sporangiospores mainly ovoid or irregularly polyhedral | 3 |
| – | Sporangiospores mainly subglobose to globose | 4 |
| 3 | Sporangiospores mainly ovoid | B.ovalispora |
| – | Sporangiospores mainly irregularly polyhedral | B.tuberculispora |
| 4 | Azygosporangia subglobose to globose | B.dichotoma |
| – | Azygosporangia absent | 5 |
| 5 | Chlamydospores abundant in substrate hyphae, in chains | 6 |
| – | Chlamydospores absent | 7 |
| 6 | Short sporangiophores simple or rebranched; uni-spored 13.5–23.0 μm; columellae variable in shape, including subglobose, conical, ellipsoidal, cylindrical, hemispherical, near pyriform, or sometimes bell-shaped, long conical | B.chlamydospora |
| – | Short sporangiophores simple or simple or sympodial; uni-spored 23.5–40.0 μm; columellae hemispherical or conical | B.moniliformis |
| 7 | Uni-spored present, subglobose to globose | 8 |
| – | Uni-spored absent | 10 |
| 8 | Giant cells present, globose to oval | B.koreana |
| – | Giant cells absent | 9 |
| 9 | Uni-spored sporangiola are quite common, 18−24 μm in diameter; multi-spored sporangiola 13−33 μm in diameter | B.circina |
| – | Uni-spored sporangiola are rare, 9−14 μm in diameter; multi-spored sporangiola 14−41 μm in diameter | B.lamprospora |
| 10 | Multi-spored sporangiola contain roughly 4–25 sporangiospores, 31.0–59.0 × 33.5–61.5 μm | B.locustae |
| – | Multi-spored sporangiola contain roughly 5–10 sporangiospores, 14.5–26.0 μm in diameter | B.variispora |
| 11 | Chlamydospores abundant | B.solicola |
| – | Chlamydospores absent | 12 |
| 12 | Giant cells present | 13 |
| – | Giant cells absent | 15 |
| 13 | Presence of cylindrical columellae, 62 × 58 µm | B.indica |
| – | Absences of cylindrical columellae | 14 |
| 14 | Sporangiospores globose to broadly ellipsoid, 8–12 × 7–10 µm | B.dispersa |
| – | Sporangiospores oblongly ellipsoidal, in young cultures rather uniform, 39.2–40.5 × 14.9–15.5 µm, in ageing cultures smaller spores, 14 × 5 µm and up | B.oblongielliptica |
| 15 | Uni-spored rare, globose, up to 15 μm diameter | B.thermophila |
| – | Uni-spored absent | 16 |
| 16 | Columellae no more than 70 µm | 17 |
| – | Columellae up to 70 µm | 18 |
| 17 | Columellae depressed globose to subglobose, apophysate, maximum growth temperature 35 °C | B.fujianensis |
| – | Columellae usually cylindrical, nonapophysate, maximum growth temperature 32 °C | B.elliptica |
| 18 | Presence of pyriform columellae, up to 110 × 75 µm | B.oblongispora |
| – | Absences of pyriform columellae | 19 |
| 19 | Sporangia up to 250(-300) µm in diameter, columella conical to cylindrical-ellipsoidal, 115–200 × 100–180 µm | B.grandis |
| – | Sporangia up to 100(-150) µm in diameter, columella applanate conical or cylindrical, 70 × 75 (85 × 100) µm | B.variabilis |
Supplementary Material
Acknowledgements
We thank Zhao-Xue Zhang, Xin-Yi Wang, and Shu-Bin Liu (Shandong Normal University) for soil collection.
Citation
Zhao H, Nie Y, Huang B, Liu X-Y (2024) Unveiling species diversity within early-diverging fungi from China I: three new species of Backusella (Backusellaceae, Mucoromycota). MycoKeys 109: 285–304. https://doi.org/10.3897/mycokeys.109.126029
Funding Statement
The research was supported by the National Natural Science Foundation of China (Nos. 32170012 and 32370007).
Contributor Information
Bo Huang, Email: bhuang@ahau.edu.cn.
Xiao-Yong Liu, Email: liuxy@sdnu.edu.cn.
Additional information
Conflict of interest
The authors have declared that no competing interests exist.
Ethical statement
No ethical statement was reported.
Funding
The research was supported by the National Natural Science Foundation of China (Nos. 32170012 and 32370007).
Author contributions
H. Zhao took charge of the drawings, DNA sequencing, data analyses, and drafted the paper; Y. Nie collected specimens and revised the paper; B. Huang and X.Y. Liu revised the paper and provided funding.
Author ORCIDs
Heng Zhao https://orcid.org/0000-0003-2938-5613
Yong Nie https://orcid.org/0000-0001-8964-1661
Bo Huang https://orcid.org/0000-0001-6032-7396
Xiao-Yong Liu https://orcid.org/0000-0002-8808-010X
Data availability
The sequences were deposited in the GenBank database (Table 1).
Supplementary materials
The concatenated sequences of ITS, LSU, and RPB1 regions
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Heng Zhao, Yong Nie, Bo Huang, Xiao-Yong Liu
Data type
phy
The concatenated sequences of ITS and LSU regions
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Heng Zhao, Yong Nie, Bo Huang, Xiao-Yong Liu
Data type
phy
The Maximum Likelihood phylogenetic tree of the genus Backusella based on ITS and LSU genetic loci
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Heng Zhao, Yong Nie, Bo Huang, Xiao-Yong Liu
Data type
References
- Benny GL, Benjamin R. (1975) Observations on Thamnidiaceae (Mucorales). New taxa, new combinations, and notes on selected species. Aliso 8(3): 301–351. 10.5642/aliso.19750803.10 [DOI] [Google Scholar]
- Benny GL, Smith ME, Kirk PM, Tretter ED, White MM. (2016) Challenges and future perspectives in the systematics of Kickxellomycotina, Mortierellomycotina, Mucoromycotina, and Zoopagomycotina. Biology of Microfungi: 65–126. 10.1007/978-3-319-29137-6_5 [DOI]
- Cai Y, Nie Y, Zhao H, Wang Z, Zhou Z, Liu XY, Huang B. (2021) Azygosporus gen. nov., a synapmorphic clade in the family Ancylistaceae. MycoKeys 85: 161–172. 10.3897/mycokeys.85.73405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro TRL, Walther G, Lee HB, Nguyen TTT, de Souza CAF, Lima DX, de Oliveira RJV, Góes-Neto A, Tomé LMR, Kurzai O, Voigt K, Santiago ALCMA. (2023) A polyphasic approach to the taxonomy of Backusella reveals two new species. Mycological Progress 22(2): 16. 10.1007/s11557-023-01864-x [DOI] [Google Scholar]
- Crous PW, Carnegie AJ, Wingfield MJ, Sharma R, Mughini G, Noordeloos ME, Santini A, Shouche YS, Bezerra JDP, Dima B, Guarnaccia V, Imrefi I, Jurjević Ž, Knapp DG, Kovács GM, Magistà D, Perrone G, Rämä T, Rebriev YA, Shivas RG, Singh SM, Souza-Motta CM, Thangavel R, Adhapure NN, Alexandrova AV, Alfenas AC, Alfenas RF, Alvarado P, Alves AL, Andrade DA, Andrade JP, Barbosa RN, Barili A, Barnes CW, Baseia IG, Bellanger J-M, Berlanas C, Bessette AE, Bessette AR, Biketova AYu, Bomfim FS, Brandrud TE, Bransgrove K, Brito ACQ, Cano-Lira JF, Cantillo T, Cavalcanti AD, Cheewangkoon R, Chikowski RS, Conforto C, Cordeiro TRL, Craine JD, Cruz R, Damm U, de Oliveira RJV, de Souza JT, de Souza HG, Dearnaley JDW, Dimitrov RA, Dovana F, Erhard A, Esteve-Raventós F, Félix CR, Ferisin G, Fernandes RA, Ferreira RJ, Ferro LO, Figueiredo CN, Frank JL, Freire KTLS, García D, Gené J, Gęsiorska A, Gibertoni TB, Gondra RAG, Gouliamova DE, Gramaje D, Guard F, Gusmão LFP, Haitook S, Hirooka Y, Houbraken J, Hubka V, Inamdar A, Iturriaga T, Iturrieta-González I, Jadan M, Jiang N, Justo A, Kachalkin AV, Kapitonov VI, Karadelev M, Karakehian J, Kasuya T, Kautmanová I, Kruse J, Kušan I, Kuznetsova TA, Landell MF, Larsson K-H, Lee HB, Lima DX, Lira CRS, Machado AR, Madrid H, Magalhães OMC, Majerova H, Malysheva EF, Mapperson RR, Marbach PAS, Martín MP, Martín-Sanz A, Matočec N, McTaggart AR, Mello JF, Melo RFR, Mešić A, Michereff SJ, Miller AN, Minoshima A, Molinero-Ruiz L, Morozova OV, Mosoh D, Nabe M, Naik R, Nara K, Nascimento SS, Neves RP, Olariaga I, Oliveira RL, Oliveira TGL, Ono T, Ordoñez ME, de M. Ottoni A, Paiva LM, Pancorbo F, Pant B, Pawłowska J, Peterson SW, Raudabaugh DB, Rodríguez-Andrade E, Rubio E, Rusevska K, Santiago ALCMA, Santos ACS, Santos C, Sazanova NA, Shah S, Sharma J, Silva BDB, Siquier JL, Sonawane MS, Stchigel AM, Svetasheva T, Tamakeaw N, Telleria MT, Tiago PV, Tian CM, Tkalčec Z, Tomashevskaya MA, Truong HH, Vecherskii MV, Visagie CM, Vizzini A, Yilmaz N, Zmitrovich IV, Zvyagina EA, Boekhout T, Kehlet T, Læssøe T, Groenewald JZ. (2019) Fungal Planet description sheets: 868–950. Persoonia 42: 291–473. 10.3767/persoonia.2019.42.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T. (2020) ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Molecular Biology and Evolution 37(1): 291–294. 10.1093/molbev/msz189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima CLF, Lundgren JDAL, Nguyen TTT, Cordeiro TRL, Lima DX, Gurgel LMS, da Costa DP, Lee HB, Santiago ALCMA. (2022) Two new species of Backusella (Mucorales, Mucoromycota) from soil in an upland forest in northeastern Brazil with an identification key of Backusella from the Americas. Journal of Fungi 8(10): 1038. 10.3390/jof8101038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza JI, Marano AV, Pires-Zottarelli CLA, Chambergo FS, Harakava R. (2014) A new species of Backusella (Mucorales) from a Cerrado reserve in Southeast Brazil. Mycological Progress 13(4): 975–980. 10.1007/s11557-014-0981-3 [DOI] [Google Scholar]
- Ellis J, Hesseltine C. (1969) Two new members of the Mucorales. Mycologia 61(5): 863–872. 10.1080/00275514.1969.12018810 [DOI] [Google Scholar]
- Flad V, Young D, Seppälä S, Hooker C, Youssef N, Podmirseg SM, Nagler M, Reilly M, Li Y, Fliegerová K. (2020) Chapter 17: the biotechnological potential of anaerobic gut fungi, genetics and biotechnology. Springer, New York, 413–437. 10.1007/978-3-030-49924-2_17 [DOI]
- Galindo LJ, López-García P, Torruella G, Karpov S, Moreira D. (2021) Phylogenomics of a new fungal phylum reveals multiple waves of reductive evolution across Holomycota. Nature Communications 12(1): 4973. 10.1038/s41467-021-25308-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- GBIF (2024) GBIF Backbone Taxonomy. Checklist dataset. https://www.gbif.org/ [accessed on 22 March 2024]
- Ho HM. (1995a) Notes on two coprophilous species of the genus Circinella (Mucorales) from Taiwan. Fungal Science 10(01–04): 23–27. [Google Scholar]
- Ho HM. (1995b) Zygospore wall in early stage of Rhizopussexualis (Mucoraceae) smooth or warted. Fungal Science 10(01–04): 29–31. [Google Scholar]
- Ho HM. (1996) The outdoor fungal airspora in Hualien (I) - the agar plate method. Taiwania 41(1): 67–80. [Google Scholar]
- Ho HM. (2000) Notes on Zygomycetes of Taiwan (I). Fungal Science 15(01–02): 65–68. [Google Scholar]
- Ho HM. (2001) The Merosporangiferous fungi from Taiwan (I): Two new records of Syncephalis. Taiwania 46(4): 318–324. [Google Scholar]
- Ho HM. (2002a) Notes on Zygomycetes of Taiwan (II): Two Thamnidiaceae (Mucorales) fungi. Fungal Science 17(3–4): 87–92. [Google Scholar]
- Ho HM. (2002b) The Merosporangiferous fungi from Taiwan (II): Two new records of Syncephalis. Taiwania 47(1): 37–42. [Google Scholar]
- Ho HM. (2003) The Merosporangiferous fungi from Taiwan (III): Three new records of Piptocephalidaceae (Zoopagales, Zygomycetes). Taiwania 48(1): 53–59. [Google Scholar]
- Ho HM. (2004) The Merosporangiferous Fungi from Taiwan (IV): Two new records of Piptocephalis (Piptocephalidaceae, Zoopagales). Taiwania 49(3): 188–193. [Google Scholar]
- Ho HM. (2006a) The Merosporangiferous Fungi from Taiwan (VI): Two New Records of Piptocephalis (Piptocephalidaceae, Zoopagales, Zygomycetes). Taiwania 51(3): 210–213. [Google Scholar]
- Ho HM. (2006b) A new species of Piptocephalis from Taiwan. Botanical Studies (47): 453–456.
- Ho H, Benny GL. (2007) Two new species of Syncephalis from Taiwan, with a key to the Syncephalis species found in Taiwan. Botanical Studies 48(3): 319–324. [Google Scholar]
- Ho HM, Benny GL. (2008) A new species of Syncephalis from Taiwan. Botanical Studies 49: 45–48. [Google Scholar]
- Ho HM, Chang LL. (2003) Notes on Zygomycetes of Taiwan (III): Two Blakeslea species (Choanephoraceae) new to Taiwan. Taiwania 48(4): 232–238. [Google Scholar]
- Ho HM, Chen ZC. (1990) Morphological study of Gongronellabutleri (Mucorales) from Taiwan. Taiwania 35(4): 259–263. [Google Scholar]
- Ho HM, Chuang SC. (2010) Notes on Zygomycetes of Taiwan (IX): Two new records of Dispira (Dimargaritales, Zygomycetes) in Taiwan. Fungal Science 25(1): 13–18. [Google Scholar]
- Ho HM, Hsu CH. (2005) The Merosporangiferous Fungi from Taiwan (V): Two new records of Coemansia (Kickxellaceae, Kickxellales, Zygomycetes). Taiwania 50(1): 22–28. [Google Scholar]
- Ho HM, Kirk PM. (2009) Piptocephalisformosana, a new species from Taiwan. Botanical Studies 50(1): 69–72. [Google Scholar]
- Ho HM, Chuang SC, Chen SJ. (2004) Notes on Zygomycetes of Taiwan (IV): Three Absidia species (Mucoraceae). Fungal Science 19(3–4): 125–131. [Google Scholar]
- Ho HM, Chien CY, Chuang SC. (2007) Notes on Zygomycetes of Taiwan (V): Linderinapennispora new to Taiwan. Fungal Science 22(01–02): 35–38. [Google Scholar]
- Ho HM, Chuang SC, Hsien CY. (2008) Notes on Zygomycetes of Taiwan (VI): Chaetocladiumbrefeldii new to Taiwan. Fungal Science 23: 21–25. [Google Scholar]
- Hurdeal VG, Jones EBG, Santiago A, Hyde KD, Gentekaki E. (2022) Expanding the diversity of mucoralean fungi from northern Thailand: Novel Backusella species from soil. Phytotaxa 559(3): 275–284. 10.11646/phytotaxa.559.3.5 [DOI] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution 30(4): 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. (2012) Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12): 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk P, Cannon P, Minter D, Stalpers J. (2008) Ainsworth & Bisby’s dictionary of the fungi 10th. CAB International. 10.1017/S0269915X03001204 [DOI]
- Li ZZ. (2000) Flora Fungorum Sinicorum. Vol. 13. Entomophthorales. SciencePress, Beijing, 163 pp. [Google Scholar]
- Li ZZ, Fan MZ, Wang B, Huang B. (1999) Entomophthoralean Fungi in China. Journal of Anhui Agricultural University 26(3): 286–291. [Google Scholar]
- Lima DX, Voigt K, Souza C, Oliveira R, Souza-Motta CM, Santiago ALCMA. (2016) Description of Backusellaconstricta sp. nov. Mucorales, ex Zygomycota from the Brazilian Atlantic Rainforest, including a key to species of Backusella. Phytotaxa 289(1): 59–68. 10.11646/phytotaxa.289.1.4 [DOI] [Google Scholar]
- Liu XY. (2004) Pirella (Thamnidiaceae, Mucorales), a new record genus for China. Mycosystema 23(2): 301–302. [Google Scholar]
- Liu XY, Zheng RY. (2015) New taxa of Ambomucor (Mucorales, Mucoromycotina) from China. Mycotaxon 130(1): 165–171. 10.5248/130.165 [DOI] [Google Scholar]
- Liu XY, Huang H, Zheng RY. (2001) Relationships within Cunninghamella based on sequence analysis of ITS rDNA. Mycotaxon 80: 77–95. [Google Scholar]
- Liu XY, Huang H, Zheng RY. (2008) Delimitation of Rhizopus varieties based on IGS rDNA sequences. Sydowia 60(1): 93–112. [Google Scholar]
- Nguyen TT, Voigt K, Santiago ALCMA, Kirk PM, Lee HB. (2021) Discovery of novel Backusella (Backusellaceae, Mucorales) isolated from invertebrates and toads in Cheongyang, Korea. Journal of Fungi 7(7): 513. 10.3390/jof7070513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y, Yu CZ, Liu XY, Huang B. (2012) A new species of Conidiobolus (Ancylistaceae) from Anhui, China. Mycotaxon 120(1): 427–435. 10.5248/120.427 [DOI] [Google Scholar]
- Nie Y, Tang XX, Liu XY, Huang B. (2016) Conidiobolusstilbeus, a new species with mycelial strand and two types of primary conidiophores. Mycosphere 7(6): 801–809. 10.5943/mycosphere/7/6/11 [DOI] [Google Scholar]
- Nie Y, Tang X, Liu X, Huang B. (2017) A new species of Conidiobolus with chlamydospores from Dabie Mountains, eastern China. Mycosphere : Journal of Fungal Biology 8(7): 809–816. 10.5943/mycosphere/8/7/1 [DOI] [Google Scholar]
- Nie Y, Qin L, Yu DS, Liu XY, Huang B. (2018) Two new species of Conidiobolus occurring in Anhui, China. Mycological Progress 17(10): 1203–1211. 10.1007/s11557-018-1436-z [DOI] [Google Scholar]
- Nie Y, Cai Y, Gao Y, Yu DS, Wang ZM, Liu XY, Huang B. (2020a) Three new species of Conidiobolus sensu stricto from plant debris in eastern China. MycoKeys 73: 133–149. 10.3897/mycokeys.73.56905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y, Yu DS, Wang CF, Liu XY, Huang B. (2020b) A taxonomic revision of the genus Conidiobolus (Ancylistaceae, Entomophthorales): Four clades including three new genera. MycoKeys 66: 55–81. 10.3897/mycokeys.66.46575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y, Wang ZM, Liu XY, Huang B. (2021) A morphological and molecular survey of Neoconidiobolus reveals a new species and two new combinations. Mycological Progress 20(10): 1233–1241. 10.1007/s11557-021-01720-w [DOI] [Google Scholar]
- Nie Y, Zhao H, Wang Z, Zhou Z, Liu XY, Huang B. (2022a) Two new species in Capillidium (Ancylistaceae, Entomophthorales) from China, with a proposal for a new combination. MycoKeys 89: 139–153. 10.3897/mycokeys.89.79537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y, Wang ZM, Zhou ZY, Zhao H, Liu XY, Huang B. (2022b) A new species of Neoconidiobolus (Entomophthorales, Ancylistaceae) from China based on morphological, molecular and physiological evidences. Phytotaxa 574(2): 149–157. 10.11646/phytotaxa.574.2.3 [DOI] [Google Scholar]
- Nie Y, Cai Y, Zhao H, Zhou ZY, Zhao CW, Liu XY, Huang B. (2023) Morphological and phylogenetic analyses reveal two new species in Conidiobolus s.s. (Conidiobolaceae, Entomophthorales) from China. MycoKeys 98: 221–232. 10.3897/mycokeys.98.103603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y, Zhao H, Liu X, Huang B. (2024) Taxonomic outline of entomophthoroid fungi. Mycosystema 43(4): 230301. 10.13346/j.mycosystema.230301 [DOI] [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos FRS, Cordeiro TRL, Lima CLF, Santos MAB, Freitas LWS, Gentekaki E, Gurgel LMS, Souza-Motta CM, Lee HB, Santiago ALCMA. (2023) Discovery of Backusellaparaconstricta sp. nov. (Mucorales, Mucoromycota) in an upland forest in northeastern Brazil with an identification key for Backusella from the Americas. Acta Botanica Brasílica 37: e20230061. 10.1590/1677-941x-abb-2023-0061 [DOI]
- Spatafora JW, Chang Y, Benny GL, Lazarus K, Smith ME, Berbee ML, Bonito G, Corradi N, Grigoriev I, Gryganskyi A, James TY, O’Donnell K, Roberson RW, Taylor TN, Uehling J, Vilgalys R, White MM, Stajich JE. (2016) A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108(5): 1028–1046. 10.3852/16-042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9): 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquhart A, Douch J, Heafield T, Buddie A, Idnurm A. (2021) Diversity of Backusella (Mucoromycotina) in south-eastern Australia revealed through polyphasic taxonomy. Persoonia 46: 1–45. 10.3767/persoonia.2021.46.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt K, James TY, Kirk PM, Santiago ALCMA, Waldman B, Griffith GW, Fu M, Radek R, Strassert JFH, Wurzbacher C, Jerônimo GH, Simmons DR, Seto K, Gentekaki E, Hurdeal VG, Hyde KD, Nguyen TTT, Lee HB. (2021) Early-diverging fungal phyla: Taxonomy, species concept, ecology, distribution, anthropogenic impact, and novel phylogenetic proposals. Fungal Diversity 109(3): 59–98. 10.1007/s13225-021-00480-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther G, Pawłowska J, Alastruey-Izquierdo A, Wrzosek M, Rodriguez-Tudela J, Dolatabadi S, Chakrabarti A, de Hoog G. (2013) DNA barcoding in Mucorales: An inventory of biodiversity. Persoonia 30(1): 11–47. 10.3767/003158513X665070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanasinghe DN, Phukhamsakda C, Hyde KD, Jeewon R, Lee HB, Jones EG, Tibpromma S, Tennakoon DS, Dissanayake AJ, Jayasiri SC, Gafforov Y, Camporesi E, Bulgakov TS, Ekanayake AH, Perera RH, Samarakoon MC, Goonasekara ID, Mapook A, Li W-J, Senanayake IC, Li J, Norphanphoun C, Doilom M, Bahkali AH, Xu J, Mortimer PE, Tibell L, Tibell S, Karunarathna SC. (2018) Fungal diversity notes 709–839: Taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Diversity 89(1): 1–236. 10.1007/s13225-018-0395-7 [DOI] [Google Scholar]
- Wang CF, Li KP, Liu YJ, Li ZZ, Huang B. (2010a) Three new Chinese records of Conidiobolus. Mycosystema 4: 595–599. [Google Scholar]
- Wang CF, Li KP, Huang B. (2010b) A new record to China – Conidiobolusiuxtagenitus. Journal of Fungal Research 1: 12–14. [Google Scholar]
- Wang YN, Liu XY, Zheng RY. (2013) Four new species records of Umbelopsis (Mucoromycotina) from China. Journal of Mycology 970216: 1–6. 10.1155/2013/970216 [DOI] [Google Scholar]
- Wang YN, Liu XY, Zheng RY. (2014) Umbelopsischangbaiensis sp. nov. from China and the typification of Mortierellavinacea. Mycological Progress 13(3): 657–669. 10.1007/s11557-013-0948-9 [DOI] [Google Scholar]
- Wang YJ, Zhao T, Wu WY, Wang M, Liu XY. (2022a) Cunninghamellaverrucosa sp. nov. (Mucorales, Mucoromycota) from Guangdong Province in China. Phytotaxa 560(3): 274–284. 10.11646/phytotaxa.560.3.2 [DOI] [Google Scholar]
- Wang YN, Liu XY, Zheng RY. (2022b) The Umbelopsisramanniana sensu lato consists of five cryptic species. Journal of Fungi 8(9): 895. 10.3390/jof8090895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Wang K, Cai L, Zhao M, Kirk PM, Fan G, Sun Q, Li B, Wang S, Yu Z, Han D, Ma J, Wu L, Yao Y. (2023a) Fungal names: A comprehensive nomenclatural repository and knowledge base for fungal taxonomy. Nucleic Acids Research 51(D1): D708–D716. 10.1093/nar/gkac926 [DOI] [PMC free article] [PubMed]
- Wang YX, Zhao H, Ding ZY, Ji XY, Zhang ZX, Wang S, Zhang XG, Liu XY. (2023b) Three new species of Gongronella (Cunninghamellaceae, Mucorales) from soil in Hainan, China based on morphology and molecular phylogeny. Journal of Fungi 9(12): 1182. 10.3390/jof9121182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18(1): 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Wijayawardene NN, Hyde KD, Dai DQ, Sánchez-García M, Goto BT, Saxena RK, Erdoğdu M, Selçuk F, Rajeshkumar KC, Aptroot A, Błaszkowski J, Boonyuen N, da Silva GA, de Souza FA, Dong W, Ertz D, Haelewaters D, Jones EBG, Karunarathna SC, Kirk PM, Kukwa M, Kumla J, Leontyev DV, Lumbsch HT, Maharachchikumbura SSN, Marguno F, Martínez-Rodríguez P, Mešić A, Monteiro JS, Oehl F, Pawłowska J, Pem D, Pfliegler WP, Phillips AJL, Pošta A, He MQ, Li JX, Raza M, Sruthi OP, Suetrong S, Suwannarach N, Tedersoo L, Thiyagaraja V, Tibpromma S, Tkalčec Z, Tokarev YS, Wanasinghe DN, Wijesundara DSA, Wimalaseana SDMK, Madrid H, Zhang GQ, Gao Y, Sánchez-Castro I, Tang LZ, Stadler M, Yurkov A, Thines M. (2022) Outline of Fungi and fungus-like taxa – 2021. Mycosphere 13(1): 53–453. 10.5943/mycosphere/13/1/2 [DOI] [Google Scholar]
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. (2020) PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources 20(1): 348–355. 10.1111/1755-0998.13096 [DOI] [PubMed] [Google Scholar]
- Zhao H, Zhu J, Zong TK, Liu XL, Ren LY, Lin Q, Qiao M, Nie Y, Zhang ZD, Liu XY. (2021) Two new species in the family Cunninghamellaceae from China. Mycobiology 49(2): 142–150. 10.1080/12298093.2021.1904555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Nie Y, Zong TK, Dai YC, Liu XY. (2022a) Three new species of Absidia (Mucoromycota) from China based on phylogeny, morphology and physiology. Diversity 14(2): 132. 10.3390/d14020132 [DOI] [Google Scholar]
- Zhao H, Nie Y, Zong TK, Wang YJ, Wang M, Dai YC, Liu XY. (2022b) Species diversity and ecological habitat of Absidia (Cunninghamellaceae, Mucorales) with emphasis on five new species from forest and grassland soil in China. Journal of Fungi 8(5): 471. 10.3390/jof8050471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Nie Y, Zong TK, Wang K, Lv ML, Cui YJ, Tohtirjap A, Chen JJ, Zhao CL, Wu F, Cui BK, Yuan Y, Dai YC, Liu XY. (2023a) Species diversity, updated classification and divergence times of the phylum Mucoromycota. Fungal Diversity 123(1): 49–157. 10.1007/s13225-023-00525-4 [DOI] [Google Scholar]
- Zhao H, Vlasák J, Yuan Y. (2023b) Outline, phylogenetic and divergence times analyses of the genus Haploporus (Polyporales, Basidiomycota): Two new species are proposed. MycoKeys 98: 233–252. 10.3897/mycokeys.98.105684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng RY, Chen GQ. (1986) Blakesleasinensis sp. nov., a further proof for retaining the genus Blakeslea. Acta Mycologica Sinica (Suppl I): 40–55.
- Zheng RY, Chen GQ. (1998) Cunninghamellaclavata sp. nov., a fungus with an unusual type of branching of sporophore. Mycotaxon 69: 187–198. [Google Scholar]
- Zheng RY, Liu XY. (2014) Ambomucor gen. & spp. nov. from China. Mycotaxon 126(1): 97–108. 10.5248/126.97 [DOI] [Google Scholar]
- Zheng RY, Liu XY. (2018) Species Catalogue of China, Vol. 3. Fungi: Chtrid, Zygomycotan, Glomeromycotan Fungi. Science Press, Beijing, 57 pp. [Google Scholar]
- Zheng RY, Chen GQ, Huang H, Liu XY. (2007) A monograph of Rhizopus. Sydowia 59(2): 273–372. [Google Scholar]
- Zheng RY, Liu XY, Wang YN. (2013) Two taxa of the new record genus Backusella from China. Mycosystema 32(3): 330–341. [Google Scholar]
- Zong TK, Zhao H, Liu XL, Ren LY, Zhao CL, Liu XY. (2021) Taxonomy and phylogeny of four new species in Absidia (Cunninghamellaceae, Mucorales) from China. Frontiers in Microbiology 12: 2181. 10.3389/fmicb.2021.677836 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The concatenated sequences of ITS, LSU, and RPB1 regions
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Heng Zhao, Yong Nie, Bo Huang, Xiao-Yong Liu
Data type
phy
The concatenated sequences of ITS and LSU regions
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Heng Zhao, Yong Nie, Bo Huang, Xiao-Yong Liu
Data type
phy
The Maximum Likelihood phylogenetic tree of the genus Backusella based on ITS and LSU genetic loci
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Heng Zhao, Yong Nie, Bo Huang, Xiao-Yong Liu
Data type
Data Availability Statement
The sequences were deposited in the GenBank database (Table 1).