Abstract
Class II histone deacetylases (HDACs) repress transcription by associating with a variety of transcription factors and corepressors. Phosphorylation of a set of conserved serine residues in the N-terminal extensions of class II HDACs creates binding sites for 14-3-3 chaperone proteins, which trigger nuclear export of these HDACs, thereby derepressing specific target genes in a signal-dependent manner. To identify intracellular signaling pathways that control phosphorylation of HDAC5, a class II HDAC, we designed a eukaryotic cDNA expression screen in which a GAL4-dependent luciferase reporter was expressed with the DNA-binding domain of GAL4 fused to the N-terminal extension of HDAC5 and the VP16 transcription activation domain fused to 14-3-3. The transfection of COS cells with cDNA expression libraries results in activation of luciferase expression by cDNAs encoding HDAC5 kinases or modulators of such kinases that enable phosphorylated GAL4–HDAC5 to recruit 14-3-3-VP16 with consequent reconstitution of a functional transcriptional complex. Our results reveal a remarkable variety of signaling pathways that converge on the signal-responsive phosphorylation sites in HDAC5, thereby enabling HDAC5 to connect extracellular signals to the genome.
Keywords: endothelial differentiation genes, lysophosphatidic acid receptors, Rho signaling, sphingosine-1 phosphate
Changes in histone acetylation represent a key mechanism for the modulation of gene transcription (1). Acetylation of nucleosomal histones by histone acetyltransferases enhances transcription by relaxing the condensed structure of the nucleosome, whereas deacetylation of histones by histone deacetylase (HDAC) activity reverses this process and promotes chromatin condensation and transcriptional repression. The recruitment of histone acetyltransferases and HDACs by specific transcription factors enables these chromatin-modifying enzymes to regulate specific sets of target genes.
Three different classes of HDACs can be distinguished by their structures, expression patterns, and catalytic mechanisms (2). The four class II HDACs (HDAC4, HDAC5, HDAC7, and HDAC9) contain a bipartite structure with an N-terminal extension of ≈600 aa followed by a catalytic domain (3). The N-terminal regions of class II HDACs interact with a variety of transcription factors and corepressors and contain a set of conserved phosphorylation sites that control their distribution between the nucleus and the cytoplasm (4–10). The signal-responsive serines in the N-terminal regions of class II HDACs are phosphorylated by calcium/calmodulin-dependent protein kinase (CaMK) and protein kinase D (PKD) (5–9, 11, 12). When phosphorylated, these sites recruit the chaperone protein 14-3-3, which masks the nuclear localization sequence in class II HDACs with consequent export of the HDAC/14-3-3 complex from the nucleus to the cytoplasm and derepression of specific genes (4, 6, 13–16). Thus, phosphorylation of class II HDACs provides a mechanism for coupling external signals to the genome.
Class II HDACs have been implicated in a variety of developmental and signal-dependent processes. HDAC5 and HDAC9 suppress hypertrophic growth of cardiomyocytes, such that knockout mice lacking these HDACs develop grossly enlarged hearts in response to cardiac stress (8, 17). HDAC9 also has been shown to repress the responsiveness of skeletal muscle genes to motor innervation (18). The actions of these HDACs in cardiac and skeletal myocytes correlate with their ability to associate with and repress the activity of myocyte enhancer factor 2 (MEF2). HDAC4 acts as a repressor of chondrocyte hypertrophy by means of its association with the Runx2 transcription factor, a master regulator of chondrocyte hypertrophy; chondrocytes from HDAC4 knockout mice undergo precocious and ectopic hypertrophy, resulting in lethal ossification of endochondral cartilage (19). HDAC7 has been implicated in negative selection and apoptosis of T cells as a result of its association with the orphan nuclear receptor Nur77 (11).
In an effort to identify kinases and other signaling molecules capable of regulating the phosphorylation of HDAC5, we designed a eukaryotic cDNA expression screen for cDNAs whose products could induce the interaction of GAL4-HDAC5 and 14-3-3-VP16 chimeric proteins and consequent activation of a GAL4-dependent luciferase reporter in transfected COS cells. Here, we describe a collection of regulators of HDAC5 phosphorylation uncovered in this screen. Our results reveal a remarkable number of signaling pathways that culminate with the phosphorylation of HDAC5 and suggest that HDAC5 integrates diverse signaling pathways and transduces their effects to “downstream” target genes as a result of its signal-dependent nuclear export.
Materials and Methods
Constructs and cDNA Expression Library. HDAC5 derivatives in which residues 2–664 were fused to the DNA-binding domain of GAL4 (GAL4BD-HDAC5), and 14-3-3 was fused to the VP16 transcription activation domain have been described (9). Human fetal heart and mouse embryonic day 10.5 cDNA expression libraries were purchased from Invitrogen. cDNA pools were prepared by using the PerfectPrep Plasmid 96 Vac Direct Bind kit (Eppendorf). Each cDNA pool used in the screening contained 50–100 single cDNA clones.
Transient Transfections and Luciferase Reporter Assays. In each well of a 24-well plate, 5 × 104 COS cells were cultured in DMEM with 10% FBS. Cells were transfected with 250 ng of cDNA expression library together with 100 ng of upstream activating sequence (UAS)-luciferase reporter plasmid, and 50 ng each of expression plasmids encoding GAL4BD-HDAC5 and VP16-14-3-3 by using 1.4 μl of FuGENE 6 reagent (Roche Molecular Biochemicals). Transfection efficiency was normalized by cotransfection of 10 ng of pCMV-LacZ. At 48 h posttransfection, the cells were harvested in 150 μl of passive lysate buffer (Promega), and 20 μl of cell lysate was used for luciferase or β-galactosidase assays. For sib-selection, positive pools of cDNA expression clones were transformed into Escherichia coli DH5α competent cells and plated on LB agar dishes. For each positive pool, 96 single clones were picked from the dish and grown in LB liquid media. Twelve single clones were combined as subpools, and plasmids were prepared and transfected for the reporter assay as described above. Single clones from the positive subpools were prepared and tested for their ability to promote HDAC5 phosphorylation.
Immunocytochemistry. COS cells plated on glass coverslips were fixed and stained as described in ref. 7. Fluorescent images were collected on a fluorescent microscope (Leica, Deerfield, IL) and were processed with photoshop (Adobe Systems, San Jose, CA). Anti-FLAG (Sigma) and fluorescein-conjugated anti-mouse IgG (Vector Laboratories) were used at a dilution of 1:200.
HDAC Kinase Assay. Kinase assays were performed as described in ref. 8. Briefly, lysates were prepared from COS cells transfected with a microtubule-associating regulatory kinase 2 (Mark2) plasmid. GST–HDAC protein, containing amino acids 208–310 of HDAC4 and 218–328 of HDAC5, was conjugated to glutathione-agarose beads that were washed with PBS and subsequently incubated with COS cell lysate (100 μg) in lysis buffer for 2 h at 4°C. Beads were washed twice with the same buffer, equilibrated with kinase reaction buffer, and then resuspended in kinase reaction buffer (30 μl) containing 12.5 μM ATP and 5 μCi of [γ-32P]-ATP (1 Ci = 37 GBq), and reactions were allowed to proceed for 30 min at room temperature. Reactions were then stopped by boiling with SDS gel-loading buffer, and phosphoproteins were resolved by SDS/PAGE and visualized by autoradiography.
Results
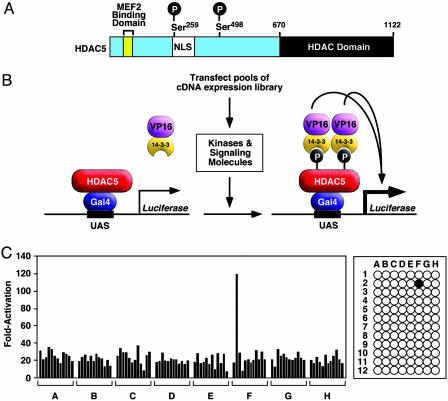
A cDNA Expression Screen for Activators of HDAC5 Phosphorylation. Phosphoserines -259 and -498 in HDAC5 (Fig. 1A) serve as binding sites for 14-3-3 proteins (4–9). In an effort to identify protein kinases capable of phosphorylating the 14-3-3 consensus sites in HDAC5, we designed a eukaryotic expression screen as schematized in Fig. 1B. In this screen, the N-terminal extension of HDAC5 was fused to the GAL4 DNA-binding domain, and 14-3-3 was fused to the VP16 transcription activation domain. In transfected COS cells, HDAC5 is not phosphorylated, so GAL4-HDAC5 cannot interact with 14-3-3-VP16, and a GAL4-dependent luciferase reporter (UAS-luciferase) cannot be activated. Expression plasmids encoding these fusion proteins, together with UAS-luciferase, were transfected into COS cells along with pools of ≈100 cDNA clones each from a fetal human heart or a mouse embryonic day 10.5 cDNA expression library. Any cDNA pool containing a kinase capable of phosphorylating the 14-3-3-binding sites in HDAC5 or an upstream activator of an endogenous kinase in COS cells for these sites will result in the creation of a phospho-14-3-3 recognition motif, which will recruit 14-3-3-VP16 to activate the reporter. Individual clones were isolated from positive pools by sib-selection. Results obtained from a typical screen of cDNA pools in a 96-well plate and the luciferase expression in response to an activating pool containing a cDNA encoding the endothelin 1 (ET-1) receptor type A are shown in Fig. 1C.
Fig. 1.
Schematic diagram of HDAC5 and the cDNA expression screening strategy. (A) Schematic diagram of HDAC5. The positions of the two signal-responsive serines flanking the nuclear localization sequence (NLS) in the N-terminal extension of HDAC5 are shown. Amino acid positions are indicated. (B) The cDNA expression screen. The N-terminal extension of HDAC5 was fused to the DNA-binding domain of GAL4, and 14-3-3 was fused to the activation domain of VP16. A luciferase reporter controlled by the GAL4 DNA-binding site, referred to as the UAS, is expressed at a basal level in control COS cells. Transfection of COS cells with pools of cDNAs results in the activation of UAS-luciferase expression by pools containing kinases or activators of kinases that phosphorylate the 14-3-3-binding sites in HDAC5, resulting in the recruitment of 14-3-3-VP16 and reconstitution of a transcriptional complex. (C) Results from a transfection assay in a representative 96-well plate are shown. Each well received a pool of ≈50–100 cDNAs as described in B. The UAS-luciferase plasmid was specifically activated in well F2. Sib-selection from this pool identified ET-1 receptor A as the activating cDNA.
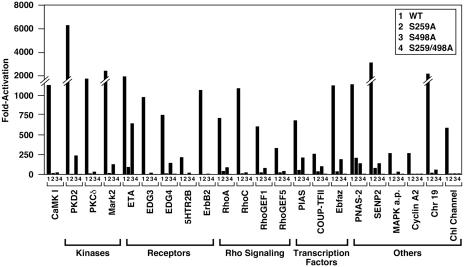
We screened ≈2,000 individual cDNA pools, representing ≈200,000 individual cDNA clones, and identified 36 cDNAs capable of activating the luciferase reporter. As shown in Table 1, the screen resulted in the identification of known and previously uncharacterized HDAC kinases as well as cell-surface receptors and other effector molecules. The specificity of activation of UAS-luciferase by individual clones was tested by mutating serines 259 and 498 of HDAC5 to alanine, which prevents 14-3-3-binding. All clones listed in Table 1 showed a dramatic reduction in their ability to activate UAS-luciferase when either 14-3-3 site was mutated and a complete loss of activity when both 14-3-3 sites were mutated. (Fig. 2 and data not shown). To assess the relative potencies of activating cDNAs, we performed parallel transfection assays with an expression plasmid encoding CaMKI, which is known to phosphorylate serines 259 and 498 of HDAC5 (5). Activating cDNAs evoked increases in expression of UAS-luciferase that were generally of the same magnitude as seen with CaMKI. Characteristics of different activating clones are described below.
Table 1. Positive clones from the screen.
| Activators | No. of clones | cDNA insert |
|---|---|---|
| Kinases | ||
| PKD2 | 1 | 118 amino acid to the end |
| PKCδ | 1 | 237 amino acid to the end |
| Mark2 | 3 | Full length |
| Receptors | ||
| Endothelin receptor type A | 5 | All full length |
| EDG3 | 1 | Full length |
| EDG4 | 3 | All full length |
| EDG7 | 1 | Full length |
| 5HTR2B | 2 | 89 amino acid to the end and full length |
| ErbB-2 | 1 | 487 amino acid to the end |
| Regulators of Rho signaling | ||
| RhoA | 1 | Full length |
| Rho C | 1 | Full length |
| Rho GEF1 | 1 | 383 amino acid to the end |
| Rho GEF5 | 1 | 1002 amino acid to the end |
| Transcriptional regulators | ||
| PIAS 4 | 1 | Full length |
| COUP-TFII | 2 | Both full length |
| Ebfaz | 1 | 6 amino acid to the end |
| Others | ||
| Cyclin A2 | 1 | Full length |
| Chloride channel intracellular protein 1 | 1 | Full length |
| PNAS-2 | 1 | Full length |
| SENP2 | 3 | 349 amino acid to the end and full length |
| Putative MAPK/NFκB activating protein | 1 | Full length |
| Novel | 3 | Unknown |
Proteins encoded by cDNA clones isolated in the expression screen described in Fig. 1A are shown. The number of clones of each type that were isolated and the amino acids encoded by the clones are shown. PKD, protein kinase D; Mark2, microtubule-associating regulatory kinase 2; EDG, endothelial differentiation gene; GEF, guanine nucleotide exchange factor; PIAS, protein inhibitor of activated STAT; Ebfaz, early B cell factor-associated zinc finger gene; SENP2, small ubiquitin-related modifier 1/sentrin/SMT3-specific protease 2.
Fig. 2.
Activation of UAS-luciferase by expression of cDNAs that promote the association of GAL4-HDAC5 and 14-3-3-VP16. COS cells were transfected with UAS-luciferase and expression plasmids encoding GAL4 fused to the wild-type HDAC5 N-terminal extension or mutants of this region in which serine 259 and/or 498 were mutated to alanines, as indicated, along with 14-3-3-VP16 and expression plasmids for individual activating cDNAs. Mutation of single serines severely impaired activation of UAS-luciferase and mutation of both serines abolished activation.
Kinases. As expected, we identified multiple protein kinases in the screen, including clones encoding PKCδ and PKD2, both of which have been shown to stimulate phosphorylation and nuclear export of HDAC5 (9). In addition, three independent cDNAs encoding Mark2 were isolated. Mark2 phosphorylates the sequence KxGS, which is similar to the signal-responsive sites in class II HDACs (20).
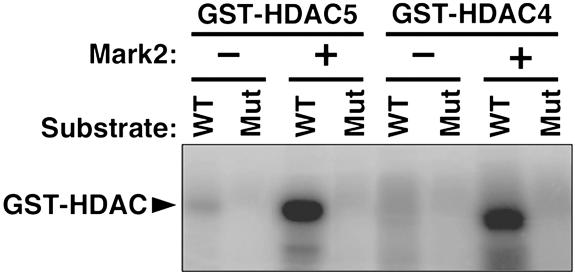
To examine whether Mark2 might act as a direct class II HDAC kinase, we performed an in vitro kinase assay by using extracts from COS cells transfected with the Mark2 expression plasmid and GST-HDAC5 or GST-HDAC4 fusion proteins as substrates. As shown in Fig. 3, extracts from Mark2-expressing cells efficiently supported the phosphorylation of these class II HDACs. Mutation of the signal-responsive serines in the GST-HDAC substrates abolished phosphorylation. We conclude that Mark2 can function as an HDAC kinase.
Fig. 3.
Phosphorylation of HDAC4 and HDAC5 by extracts from Mark2-transfected cells. COS cells were transiently transfected with an empty pcDNA3 expression plasmid (-) or a pcDNA3-Mark2 expression plasmid (+). Cell extracts were prepared and used for in vitro kinase assays with GST–HDAC fusion proteins and [γ-32P]ATP. WT, GST fusion proteins with the wild-type amino acid sequence; Mut, GST fusion proteins in which the signal-responsive serines were mutated to alanines.
Cell-Surface Receptors. A number of cell-surface receptors were identified in the expression screen (Table 1 and Fig. 2). Although these receptors require ligand-binding for signaling, we presume that their overexpression leads to signaling in the absence of ligand or that ligand present in culture medium activates the receptors. Consistent with prior studies demonstrating that ET-1 can promote nuclear export of HDAC5 in cardiomyocytes (9), we isolated five independent clones encoding the ET-1 receptor type A. We also identified three clones encoding the cell-surface receptors for lysophosphatidic acid, referred to as endothelial differentiation genes 4 (EDG4) and 7 (EDG7), which belong to a family of G protein-coupled receptors referred to as EDG receptors (21). Another clone encoded the EDG3 receptor, which binds sphingosine-1 phosphate (21). These receptors activate calcium currents and couple to Rho- and PKC-dependent signaling pathways (21, 22).
In addition, two cDNAs encoding the serotonin receptor, 5-hydroxytryptamine receptor 2B (5HTR2B), were found in the screen. The ability of 5HTR2B to promote HDAC5 phosphorylation confirms our prior studies showing that the 4-amino-pyridine derivative, pyridine activator of myocyte hypertrophy, which promotes cardiomyocyte hypertrophy, acts through this receptor to stimulate HDAC5 phosphorylation (23). 5HTR2B triggers intracellular calcium release and PKC activation (24), which is likely to account, at least in part, for the ability of the overexpressed receptor to induce HDAC5 phosphorylation.
Unexpectedly, a clone encoding the tyrosine kinase receptor ErbB-2, which is localized to the plasma membrane, was isolated in the screen. Because ErbB protein kinases are specific for tyrosine phosphorylation (25), the ability of ErbB-2 to activate the UAS-luciferase reporter by inducing the interaction of GAL4-HDAC5 and 14-3-3-VP16 apparently reflects the activation of an endogenous HDAC kinase in COS cells that responds to ErbB-2 signaling.
Regulators of Rho Signaling. Multiple regulators of Rho signaling, including RhoA, RhoC, and the Rho guanine nucleotide exchange factors 1 and 5, were isolated in the screen (Table 1 and Fig. 2). Rho guanine nucleotide exchange factors activate Rho by stimulating the exchange of GDP for GTP, with consequent stimulation of Rho-dependent signaling pathways. RhoA activates two groups of kinases, PKN/PKC-related kinases and Rho kinases (26), raising the possibility that these kinases may phosphorylate HDAC5.
Transcriptional Regulators. Several transcriptional regulators, including protein inhibitor of activated STAT 4 (PIAS), COUP-TFII, and early B-cell factor-associated zinc finger protein (Ebfaz), were as effective as protein kinases and signaling molecules in activation of the UAS-luciferase reporter (Table 1 and Fig. 2). Protein inhibitor of activated STAT proteins function as transcriptional coregulators in various cellular pathways and have been shown to act as E3-like ligases involved in sumoylation (27). Because protein inhibitor of activated STAT can interact with HDAC1 (28) and HDAC1 undergoes small ubiquitin-related modifier modification (29), it could potentially regulate HDAC5 phosphorylation by recruiting HDAC kinases to the complex, or its sumoylation of HDAC5 could make HDAC kinases more accessible to HDAC5.
The orphan nuclear receptor COUP-TFII functions as a transcriptional repressor (30) and is involved in many developmental processes including skeletal muscle differentiation, cardiogenesis, and angiogenesis (31). Early B cell factor-associated zinc finger protein binds to and inhibits the activity of early B cell factor, a basic helix–loop–helix transcription factor required for B cell lineage commitment and development of the olfactory epithelium (32). Because these cDNAs also required serines 259 and 498 for activation, it seems likely that these transcriptional repressors stimulate phosphorylation of these sites through an indirect mechanism.
Clones That Activate HDAC5 Phosphorylation Through Unknown Mechanisms. Finally, there were multiple cDNAs that act through mechanisms that would not be expected to affect phosphorylation of HDAC5, including cyclin A2, chloride intracellular channel 1, the small ubiquitin-related modifier 1/sentrin/SMT3-specific protease 2 (SENP2), and the putative mitogen-activated protein kinase (MAPK)/NFκB activating protein (Table 1 and Fig. 2).
Cyclin A2 induces the activity of cyclin-dependent kinase 1 during the G1/S transition and activates cyclin-dependent kinase 2 during the G2/M transition (33). Because HDAC5 phosphorylation sites differ from the cyclin-dependent kinase consensus site, and because previous studies have shown that cyclin-dependent kinase inhibitors failed to inhibit HDAC kinase activity (8), we believe that cyclin A2 stimulates HDAC5 phosphorylation by an indirect mechanism.
Chloride channels are frequently involved in calcium fluctuation, and some are activated by calcium. Membrane depolarization caused by chloride conductance or chloride movement also contributes to Ca2+ release from intracellular stores (34). Thus, it is tempting to speculate that chloride intercellular channel 1 regulates HDAC kinase(s) by means of an effect on calcium signaling.
SENP2 reverses the process of sumoylation by removing small ubiquitin-related modifier from modified proteins (35). Recently, it has been proposed that HDACs repress MEF2 activity by potentiating MEF2 sumoylation, and that SENP3, a closely related family member, removes small ubiquitin-related modifier from MEF2 and activates MEF2 (36). Our results suggest that SENP also may activate MEF2 through promoting phosphorylation of HDAC5. It is interesting that two proteins we identified in this screen, protein inhibitor of activated STAT 4 and SENP2, are involved in the same pathway and counteract each other. These findings raise the possibility that sumoylation of HDAC5 is coordinated with phosphorylation. Prior studies also have shown that SENP2 interacts with Axin, a regulator of the Wnt signaling pathway, and therefore has been implicated as a possible regulator of β-catenin degradation (37). Thus, it is conceivable that SENP2 regulates HDAC5 phosphorylation by means of an effect on Wnt signaling.
The putative MAPK/NFκB activating protein was identified originally in a similar expression screen for proteins that can activate MAPK and/or NFκB pathways (38). Because kinases in MAPK pathways do not phosphorylate the 14-3-3 sites in HDAC5, we presume that MAPK/NFκB activating protein may induce HDAC5 phosphorylation either by regulating the NFkB pathway or by other unknown mechanisms.
We also identified three cDNAs encoding previously uncharacterized proteins with unknown function. Clone BC007457 encodes an SNF7 domain-containing protein, clone AK001192 encodes a predicted protein with weak similarity to Splicing factor ARGI-NINE/SERINE-RICH 4, and clone BC006701 encodes a previously uncharacterized ORF on mouse chromosome 19.
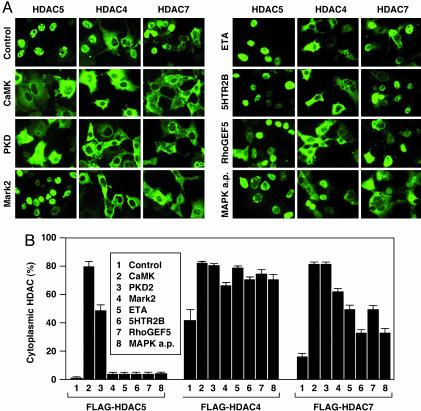
Differential Responses of Class II HDACs to Activators of HDAC5 Phosphorylation. We tested the effects of activating cDNAs on the nuclear export of HDAC4, HDAC5, and HDAC7. In contrast to HDAC5, which is completely nuclear when overexpressed alone in COS cells, HDAC4 and HDAC7 are distributed between the nucleus and the cytoplasm. CaMKI, which we used as a positive control for HDAC5 export, was the most potent export kinase, stimulating ≈100% of cells to translocate HDAC5 from the nucleus to the cytoplasm. PKD2 was about half as potent as CaMK in the nuclear export assay (Fig. 4). Surprisingly, the other activating cDNAs were relatively weak inducers of HDAC5 nuclear export and caused HDAC5 to redistribute to the cytoplasm in only about 5% of cells. HDAC4 was present in the cytoplasm of ≈50% of control COS cells. All of the activating cDNAs stimulated HDAC4 translocation to the cytoplasm, with CaMKI and PKD being most effective. HDAC7 was present in the cytoplasm of ≈20% of COS cells. CaMK and PKD caused a complete relocalization of HDAC7 to the cytoplasm, whereas other activators stimulated HDAC7 nuclear export to intermediate levels.
Fig. 4.
Effects of activating cDNAs on nuclear/cytoplasmic distribution of class II HDACs. (A) COS cells were transiently transfected with expression plasmids encoding FLAG-tagged HDACs, as shown in each column, and activators of HDAC5 phosphorylation, as shown in each row. The subcellular distribution of HDACs was determined by immunostaining as described in Materials and Methods. Representative fields are shown. (B) The percentage of cells containing FLAG-tagged HDACs in the cytoplasm was determined by counting at least 100 transfected cells.
These findings suggest that HDAC4, HDAC5, and HDAC7 are all able to respond to the different types of activators identified in the screen, but the stimulus required for nuclear export of HDAC4 and HDAC7 is less stringent than for HDAC5. Because none of the activators shown in Table 1 were able to stimulate expression of the UAS-luciferase reporter if serines 259 and 498 in HDAC5 (Fig. 2) were mutated to alanines, it is reasonable to conclude that all of these activators enhance the phosphorylation of HDAC5 at one or both of these sites with consequent recruitment of 14-3-3-VP16. Thus, it is puzzling that several of the activating cDNAs have only minimal effects on the nuclear export of HDAC5. One interpretation of this result is that nuclear export requires steps in addition to phosphorylation of the 14-3-3 docking sites in HDAC5. Export kinases such as CaMKI and PKD must be effective in triggering all of the steps required for export, whereas other stimuli might activate only a subset of such steps.
Discussion
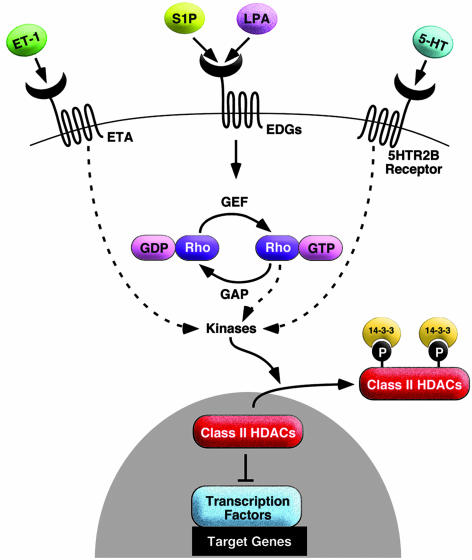
A eukaryotic expression screen to identify HDAC5 kinases and their effectors revealed known and unexpected regulators of HDAC5 phosphorylation. The remarkable number of signaling molecules that induce HDAC5 phosphorylation suggests that the signal-responsive serines in HDAC5 (and in other class II HDACs) serve as a point of convergence of diverse signaling cascades that culminate in the nucleus to control specific programs of gene expression (Fig. 5).
Fig. 5.
Signaling pathways directed at serines 259 and 498 of HDAC5. The ability of class II HDACs to inhibit activity of transcription factors is abolished by phosphorylation at signal-responsive serines, which creates 14-3-3 docking sites and results in the nuclear export of the complex. A variety of inducers of cardiac hypertrophy, including ET-1, sphingosine-1 phosphate, lysophosphatidic acid, serotonin (5-HT), and RhoA signaling, stimulate HDAC kinase activity and consequently derepress HDAC targeted transcription factors. We propose that class II HDACs serve as a nodal point that transmits extracellular and intracellular signals to the genome and controls gene expression.
Phosphorylation of HDAC5 by Mark2. Mark2 was the only unknown HDAC kinase identified from the screen. Mark2, which is expressed in adult human heart, brain, and skeletal muscle, phosphorylates microtubule-associated proteins and triggers microtubule disruption (20). In vitro kinase assays demonstrated that Mark2 could phosphorylate class II HDACs directly at the signal-responsive sites. Mark2-null mice show growth retardation and immune system dysfunction (39). It will be interesting to determine whether any of the actions of Mark2 are mediated by HDAC phosphorylation and whether Mark2 signaling may influence HDAC functions during cardiac development and growth.
Regulation of HDAC5 Phosphorylation by EDG Signaling. EDG receptors 3, 4, and 7 were potent inducers of HDAC5 phosphorylation. These receptors, which bind lysophosphatidic acid and sphingosine-1 phosphate, respectively, have been implicated in cardiac hypertrophy (40, 41) and vascular development (42, 43), but the downstream signals from pathways that link these receptors to the genome have not been identified. Our results suggest that EDG signaling stimulates cardiomyocyte growth, at least in part, by promoting the phosphorylation of class II HDACs.
Regulation of HDAC5 Phosphorylation by Rho Signaling. Activation of Rho signaling by RhoA, RhoC, or Rho guanine nucleotide exchange factor 1 or 5 induced HDAC5 phosphorylation. Rho signaling controls a variety of downstream kinases that are likely to couple Rho activation to HDAC5 phosphorylation. RhoA has been implicated in the signaling pathways whereby ET-1 and phenylephrine induce cardiac hypertrophy (44), but the downstream effectors of RhoA that promote hypertrophy are not fully understood. The ability of RhoA and Rho guanine nucleotide exchange factors to induce the association of GAL4-HDAC5 and 14-3-3-VP16 suggests that these effectors control a protein kinase pathway leading to the phosphorylation of the signal-responsive serines in HDAC5. Given the importance of Rho signaling for differentiation and morphogenesis of cardiac, skeletal, and smooth muscle cells (44, 45) and the role of MEF2 in these processes (46), it is likely that class II HDACs act as intermediaries in these pathways.
Transcriptional Regulators of HDAC5 Phosphorylation. It is curious that we identified several transcriptional regulators that induced the interaction of HDAC5 and 14-3-3 in a manner dependent on the signal-responsive serines in HDAC5. Although we have not directly examined the effects of these activators on HDAC5 phosphorylation, the finding that serine-to-alanine mutations in HDAC5 abolish their stimulatory effects strongly suggests that they act via an HDAC kinase. We currently favor the possibility that these activators induce the expression of one or more HDAC kinases or suppress the expression of phosphatases that act on these sites.
Selective Responsiveness of Class II HDACs to Different Signaling Pathways. It is remarkable that such a broad range of signaling pathways and transcriptional regulators modulate phosphorylation of the signal-responsive serines in HDAC5. These findings point to HDAC5 as a nodal point in intracellular signaling pathways that integrates diverse upstream signals and transmits them to the transcriptional machinery. Although we have focused primarily on the regulation of HDAC5 phosphorylation, our results indicate that other class II HDACs display unique responses to the various signaling molecules that we have identified. Such specificity allows different sets of target genes to be modulated by these signaling pathways depending on which class II HDACs are expressed in a particular cell type. Additional specificity of action can be achieved by the interaction of class II HDACs with different transcriptional activators and corepressors that may be cell type-specific. Modification of this expression screen to employ different class II HDACs, and cDNA expression libraries from different cell types should allow the identification of a large spectrum of molecules regulating a particular HDAC in tissues of interest.
Acknowledgments
We thank Xiumin Li for technical support, Kunhua Song for advice, A. Diehl and A. Tizenor for graphic work, and J. Page for editorial work. This work was supported by grants from the Donald W. Reynolds Center for Clinical Cardiovascular Research, the National Institutes of Health, the Muscular Dystrophy Association, the Texas Advanced Technology Program, and the Robert A. Welch Foundation.
Author contributions: S.C. and E.N.O. designed research; S.C., S.B., S.L., and E.N.O. performed research; S.C., S.B., and E.N.O. analyzed data; and S.C. and E.N.O. wrote the paper.
Abbreviations: HDAC, histone deacetylase; CaMK, calcium/calmodulin-dependent protein kinase; PKD, protein kinase D; MEF2, myocyte enhancer factor 2; ET-1, endothelin 1; Mark2, microtubule-associating regulatory kinase 2; EDG, endothelial differentiation gene; SENP2, small ubiquitin-related modifier 1/sentrin/SMT3-specific protease 2; UAS, upstream activating sequence.
References
- 1.Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074-1080. [DOI] [PubMed] [Google Scholar]
- 2.Grozinger, C. M. & Schreiber, S. L. (2002) Chem. Biol. 9, 3-16. [DOI] [PubMed] [Google Scholar]
- 3.Verdin, E., Dequiedt, F. & Kasler, H. G. (2003) Trends Genet. 19, 286-293. [DOI] [PubMed] [Google Scholar]
- 4.Grozinger, C. M. & Schreiber, S. L. (2000) Proc. Natl. Acad. Sci. USA 97, 7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKinsey, T. A., Zhang, C. L., Lu, J. & Olson, E. N. (2000) Nature 408, 106-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKinsey, T. A., Zhang, C. L. & Olson, E. N. (2000) Proc. Natl. Acad. Sci. USA 97, 14400-14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang, C. L., McKinsey, T. A. & Olson, E. N. (2001) Proc. Natl. Acad. Sci. USA 98, 7354-7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang, C. L., McKinsey, T. A., Chang, S., Antos, C. L., Hill, J. A. & Olson, E. N. (2002) Cell 110, 479-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vega, R. B., Harrison, B. C., Meadows, E., Roberts, C. R., Papst, P. J., Olson, E. N. & McKinsey, T. A. (2004) Mol. Cell. Biol. 24, 8374-8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKinsey, T. A. & Olson, E. N. (2004) Trends Genet. 20, 206-213. [DOI] [PubMed] [Google Scholar]
- 11.Dequiedt, F., Kasler, H., Fischle, W., Kiermer, V., Weinstein, M., Herndier, B. G. & Verdin, E. (2003) Immunity 18, 687-698. [DOI] [PubMed] [Google Scholar]
- 12.Parra, M., Kasler, H., McKinsey, T. A., Olson, E. N. & Verdin, E. (2005) J. Biol. Chem. 280, 13762-13770. [DOI] [PubMed] [Google Scholar]
- 13.Li, X., Song, S., Liu, Y., Ko, S. H. & Kao, H. Y. (2004) J. Biol. Chem. 279, 34201-34208. [DOI] [PubMed] [Google Scholar]
- 14.Kao, H. Y., Verdel, A., Tsai, C. C., Simon, C., Juguilon, H. & Khochbin, S. (2001) J. Biol. Chem. 276, 47496-47507. [DOI] [PubMed] [Google Scholar]
- 15.Wang, A. H. & Yang, X. J. (2001) Mol. Cell. Biol. 21, 5992-6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKinsey, T. A., Zhang, C. L. & Olson, E. N. (2001) Mol. Cell. Biol. 21, 6312-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang, S., McKinsey, T. A., Zhang, C. L., Richardson, J. A., Hill, J. A. & Olson, E. N. (2004) Mol. Cell. Biol. 24, 8467-8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mejat, A., Ramond, F., Bassel-Duby, R., Khochbin, S., Olson, E. N. & Schaeffer, L. (2005) Nat. Neurosci. 8, 313-321. [DOI] [PubMed] [Google Scholar]
- 19.Vega, R. B., Matsuda, K., Oh, J., Barbosa, A. C., Yang, X., Meadows, E., McAnally, J., Pomajzl, C., Shelton, J. M., Richardson, J. A., et al. (2004) Cell 119, 555-566. [DOI] [PubMed] [Google Scholar]
- 20.Drewes, G., Ebneth, A., Preuss, U., Mandelkow, E. M. & Mandelkow, E. (1997) Cell 89, 297-308. [DOI] [PubMed] [Google Scholar]
- 21.Panetti, T. S. (2002) Biochim. Biophys. Acta 1582, 190-196. [DOI] [PubMed] [Google Scholar]
- 22.Zhou, H. & Murthy, K. S. (2004) Am. J. Physiol. 286, C1130-C1138. [DOI] [PubMed] [Google Scholar]
- 23.Bush, E., Fielitz, J., Melvin, L., Martinez-Arnold, M., McKinsey, T. A., Plichta, R. & Olson, E. N. (2004) Proc. Natl. Acad. Sci. USA 101, 2870-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox, D. A. & Cohen, M. L. (1995) J. Pharmacol. Exp. Ther. 272, 143-150. [PubMed] [Google Scholar]
- 25.Casalini, P., Iorio, M. V., Galmozzi, E. & Menard, S. (2004) J. Cell. Physiol. 200, 343-350. [DOI] [PubMed] [Google Scholar]
- 26.Riento, K. & Ridley, A. J. (2003) Nat. Rev. Mol. Cell Biol. 4, 446-456. [DOI] [PubMed] [Google Scholar]
- 27.Aravind, L. & Koonin, E. V. (2000) Trends Biochem. Sci. 25, 112-114. [DOI] [PubMed] [Google Scholar]
- 28.Gross, M., Yang, R., Top, I., Gasper, C. & Shuai, K. (2004) Oncogene 23, 3059-3066. [DOI] [PubMed] [Google Scholar]
- 29.David, G., Neptune, M. A. & DePinho, R. A. (2002) J. Biol. Chem. 277, 23658-23663. [DOI] [PubMed] [Google Scholar]
- 30.Achatz, G., Holzl, B., Speckmayer, R., Hauser, C., Sandhofer, F. & Paulweber, B. (1997) Mol. Cell. Biol. 17, 4914-4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou, C., Tsai, S. Y. & Tsai, M. (2000) Biochim. Biophys. Acta 1470, M63-M68. [DOI] [PubMed] [Google Scholar]
- 32.Warming, S., Suzuki, T., Yamaguchi, T. P., Jenkins, N. A. & Copeland, N. G. (2004) Oncogene 23, 2727-2731. [DOI] [PubMed] [Google Scholar]
- 33.Murphy, M., Stinnakre, M. G., Senamaud-Beaufort, C., Winston, N. J., Sweeney, C., Kubelka, M., Carrington, M., Brechot, C. & Sobczak-Thepot, J. (1997) Nat. Genet. 15, 83-86. [DOI] [PubMed] [Google Scholar]
- 34.Kitamura, K. & Yamazaki, J. (2001) Jpn. J. Pharmacol. 85, 351-357. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, H., Saitoh, H. & Matunis, M. J. (2002) Mol. Cell. Biol. 22, 6498-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gregoire, S. & Yang, X. J. (2005) Mol. Cell. Biol. 25, 2273-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishida, T., Kaneko, F., Kitagawa, M. & Yasuda, H. (2001) J. Biol. Chem. 276, 39060-39066. [DOI] [PubMed] [Google Scholar]
- 38.Matsuda, A., Suzuki, Y., Honda, G., Muramatsu, S., Matsuzaki, O., Nagano, Y., Doi, T., Shimotohno, K., Harada, T., Nishida, E., et al. (2003) Oncogene 22, 3307-3318. [DOI] [PubMed] [Google Scholar]
- 39.Hurov, J. B., Stappenbeck, T. S., Zmasek, C. M., White, L. S., Ranganath, S. H., Russell, J. H., Chan, A. C., Murphy, K. M. & Piwnica-Worms, H. (2001) Mol. Cell. Biol. 21, 3206-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hilal-Dandan, R., Means, C. K., Gustafsson, A. B., Morissette, M. R., Adams, J. W., Brunton, L. L. & Heller Brown, J. (2004) J. Mol. Cell. Cardiol. 36, 481-493. [DOI] [PubMed] [Google Scholar]
- 41.Robert, P., Tsui, P., Laville, M. P., Livi, G. P., Sarau, H. M., Bril, A. & Berrebi-Bertrand, I. (2001) J. Mol. Cell. Cardiol. 33, 1589-1606. [DOI] [PubMed] [Google Scholar]
- 42.Liu, Y., Wada, R., Yamashita, T., Mi, Y., Deng, C. X., Hobson, J. P., Rosenfeldt, H. M., Nava, V. E., Chae, S. S., Lee, M. J., et al. (2000) J. Clin. Invest. 106, 951-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kono, M., Mi, Y., Liu, Y., Sasaki, T., Allende, M. L., Wu, Y. P., Yamashita, T. & Proia, R. L. (2004) J. Biol. Chem. 279, 29367-29373. [DOI] [PubMed] [Google Scholar]
- 44.Clerk, A. & Sugden, P. H. (2000) Circ. Res. 86, 1019-1023. [DOI] [PubMed] [Google Scholar]
- 45.Sordella, R., Jiang, W., Chen, G. C., Curto, M. & Settleman, J. (2003) Cell 113, 147-158. [DOI] [PubMed] [Google Scholar]
- 46.Black, B. L. & Olson, E. N. (1998) Annu. Rev. Cell Dev. Biol. 14, 167-196. [DOI] [PubMed] [Google Scholar]