Abstract
Background
Despite effective antiretroviral therapy, people with HIV (PWH) experience persistent systemic inflammation and increased morbidity and mortality. Modulating the gut microbiome through fecal microbiota transplantation (FMT) represents a novel therapeutic strategy. We aimed to evaluate proteomic changes in inflammatory pathways following repeated, low-dose FMT versus placebo.
Methods
This double-masked, placebo-controlled pilot study assessed the proteomic impacts of weekly FMT versus placebo treatment over 8 weeks on systemic inflammation in 29 PWH receiving stable antiretroviral therapy (ART). Three stool donors with high Faecalibacterium and butyrate profiles were selected, and their individual stools were used for FMT capsule preparation. Proteomic changes in 345 inflammatory proteins in plasma were quantified using the proximity extension assay, with samples collected at baseline and at weeks 1, 8, and 24. Concurrently, we characterized shifts in the gut microbiota composition and annotated functions through shotgun metagenomics. We fitted generalized additive models to evaluate the dynamics of protein expression. We selected the most relevant proteins to explore their correlations with microbiome composition and functionality over time using linear mixed models.
Results
FMT significantly reduced the plasma levels of 45 inflammatory proteins, including established mortality predictors such as IL6 and TNF-α. We found notable reductions persisting up to 16 weeks after the final FMT procedure, including in the expression of proteins such as CCL20 and CD22. We identified changes in 46 proteins, including decreases in FT3LG, IL6, IL10RB, IL12B, and IL17A, which correlated with multiple bacterial species. We found that specific bacterial species within the Ruminococcaceae, Succinivibrionaceae, Prevotellaceae families, and the Clostridium genus, in addition to their associated genes and functions, were significantly correlated with changes in inflammatory markers.
Conclusions
Targeting the gut microbiome through FMT effectively decreased inflammatory proteins in PWH, with sustained effects. These findings suggest the potential of the microbiome as a therapeutic target to mitigate inflammation-related complications in this population, encouraging further research and development of microbiome-based interventions.
Video Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s40168-024-01919-5.
Keywords: HIV, Systemic inflammation, Fecal microbiota transplant, Proteomics, Shotgun metagenomics, Microbiome
Introduction
Although antiretroviral therapy (ART) has significantly improved outcomes, people with HIV (PWH) still exhibit persistently high levels of inflammatory markers, a factor linked to increased mortality risk [1, 2]. Additionally, PWH exhibit distinct alterations in their gut microbiome composition and function, which seem to sustain immune dysfunction [3–6]. This raises the following question: can we modify the microbiome in PWH to reduce inflammation?
In HIV/AIDS, CD4 + T cells, especially those producing IL17, are significantly depleted in the gut lamina propria. This selective loss of IL17-producing cells, which are crucial for maintaining mucosal barriers, leads to a “leaky gut,” prompting bacterial translocation and systemic inflammation [7]. The gut microbiome plays a key role in this process via various mechanisms. For instance, certain microbes, such as Bifidobacteria, may help prevent mucosal defects, reduce microbial translocation, and support immune recovery. In contrast, others, such as Succinivibrionaceae and Erysipelotrichaceae, may counteract pro-inflammatory molecules and accumulate antiviral compounds [8, 9]. Notably, the Lachnospiraceae and Ruminococcaceae families, which are significant butyrate producers known for maintaining enterocyte barrier integrity and promoting immunotolerance, are often depleted in PWH [10].
However, the microbiome has proven to be a challenging therapeutic target in this population [11]. Previous interventions, including dietary changes [12], prebiotics [13], probiotics [13, 14], and nonabsorbable antibiotics such as rifaximin [15, 16], have shown limited success in modulating the microbiome or reducing inflammation, indicating their resilience to such treatments. Therefore, we conducted a placebo-controlled pilot study with 30 PWH on stable ART. This double-masked study involved randomizing participants to receive either weekly fecal microbiota capsules or a placebo for 8 weeks and selecting stool donors for their butyrate-enriched, anti-inflammatory microbiota profile (a high proportion of Faecalibacterium and a low proportion of Prevotella). The results suggested that fecal microbiota transplantation (FMT) could mitigate HIV-related dysbiosis, increase gut microbiota alpha diversity, and achieve transient donor microbiota integration, particularly in those with recent antibiotic use. FMT notably increased Lachnospiraceae and Ruminococcaceae levels and improved intestinal fatty acid-binding protein levels [17], underscoring its potential benefits for intestinal health and the necessity for further research in this domain.
To extend our understanding of the impact of FMT on inflammation in PWH, we analyzed the effects of FMT on 345 inflammatory proteins in plasma. We also explored host-microbiome interactions, identifying the main correlations between inflammatory proteins and gut microbiota composition and function.
Materials and methods
Study design and setting
This is a post hoc analysis of a randomized, double-masked, placebo-controlled pilot study (REpeated Fecal microbiota REStoration in HIV—REFRESH-), originally designed to test the safety and tolerability of FMT in PWH on ART, with secondary outcomes including changes in CD4 + and CD8 + T cells, CD4/CD8 ratio, gastrointestinal tolerance, and variations in microbiota composition and donor microbiota engraftment in recipients [17]. Participants were recruited from the HIV unit of Hospital Universitario Ramón y Cajal in Madrid, Spain, between January 27 and June 29, 2017. Participants were PWH on stable ART with a plasma HIV RNA concentration < 37 copies/mL for at least 48 weeks and a CD4/CD8 ratio < 1 as an indicator of ongoing immune activation [18]. The exclusion criteria were age < 18 years, pregnancy, planned use of chemotherapy or antibiotics, neutropenia < 500 cells/µL or CD4 counts < 350 cells/µL, active infections, or dysphagia. The original study publication details the data collection, donor screening, FMT preparation, randomization, and sample processing. As previously described, we selected three donors whose stools were in the highest quartile for fecal Bacteroides and Faecalibacterium abundance and butyrate concentrations, and in the lowest quartile for Prevotella abundance, without pooling their samples for capsule preparation [17].
Proteomic profiling of circulating inflammatory proteins
Quantification and quality control
A total of 116 EDTA plasma samples kept at − 80 °C were thawed and vortex-mixed before the plate was loaded for proteomic analysis. We used the Olink inflammation panel to measure 368 inflammatory proteins (Olink Proteomics, Uppsala, Sweden), 115 out of 116 of which passed quality control, and 345 out of the 368 proteins analyzed were detected in more than 50% of the samples. A protein was considered undetected if its expression was below the detection limit in more than 50% of the samples in both the FMT and placebo groups. To mitigate batch effects across the two necessary runs, we ensured an equal representation of samples from both the placebo and FMT groups in each run, and all samples from a single participant were analyzed within the same batch. We measured protein concentrations using proximity extension assay (PEA) technology, and the results are expressed as normalized protein expression (NPX) values, a relative protein quantification unit on a log2 scale [19].
Functional prediction of the proteomic analysis
To infer the most relevant pathways associated with the 46 differentially expressed inflammatory proteins (DEIPs) identified during FMT, we uploaded the list to http://metascape.org [20] using the Express Analysis mode. This tool performs enrichment analysis compared with different RNA-seq and proteomic analysis databases. The METASCAPE network was obtained using Cytoscape. The software compares only previously demonstrated interactions in the protein‒protein interaction network. The Molecular Complex Detection (MCODE) algorithm [21] was used to infer molecular complexes from more extensive protein networks.
Shotgun metagenomics
Sample collection and DNA extraction
Fecal samples were stored in Omnigene Gut Kits (DNA Genotek), which contain a stabilizer solution that preserves the composition of fecal microbial community structure DNA for microbiome analysis better than with the RNAlater and Tris–EDTA kits [22, 23]. Fecal samples were aliquoted and cryopreserved at − 80 °C until use.
Library preparation and sequencing
DNA extraction from fecal samples was performed using the MagNA Pure LC Instrument, a robotic workstation, and the MagNA Pure LC DNA Isolation Kit III (Roche). Subsequent preparation of DNA libraries was performed according to the protocols outlined in the Illumina DNA Prep Reference Guide 1,000,000,025,416–10 utilizing the Illumina DNA Prep Kit (Illumina, reference 20,060,059). The input DNA was standardized to a concentration of 0.2 ng/µl before initiating the library preparation protocol. During the multiplexing stage, Nextera DNA CD Indexes (Illumina, reference 20,018,708) were used. The library size was refined per the Illumina Library Prep protocol stipulations, employing Sample Purification Beads provided within the Prep Kit. The library’s size distribution was confirmed using the Fragment Analyzer 48-Capillary Array alongside the HS NGS Fragment Kit (1–-6000 bp) (Agilent, reference DNF-474–0500). The finalized libraries were sequenced using the NextSeq 2 × 150 bp paired-end reagent kit (NextSeq 500/550 High Output Kit v2.5, reference 20,024,908) on the MiSeq Sequencer. This process was conducted according to the manufacturer’s instructions (Illumina MiSeq reference guide) at the FISABIO Sequencing and Bioinformatics Service in Valencia, Spain.
Preprocessing and quality control
All the sequences used in this analysis passed quality control, during which the length and quality of the reads were filtered using Trimmomatic v0.33 (paired-end method, minimum length of 100, average quality of 30) [24]. We identified outliers using seqkit v0.10.1 [25].
Taxonomic annotation and quantification of bacterial abundance
Shotgun data from 84 samples (54 FMT at weeks 0, 1, 8, and 24, and 30 placebo at weeks 0, 8, and 24) were analyzed using the marker gene taxonomic sequence classifier mOTUs v3.0.1 with default parameters (marker genes cutoff -g 3 and minimum alignment length -l 70 for higher sensitivity) [26].
Assembly and quantification of contig coverage
MetaSPAdes (SPAdes genome assembler v3.15.2 in metaSPAdes mode) was used for the novo assembly of the trimmed reads [27]. Once the contigs were assembled, CoverM v0.6.1 (https://github.com/wwood/CoverM) was used to calculate the abundance of each of these fragments. The coverage of each contig was quantified as RPKM (reads per kilobase of transcript per million reads mapped).
Functional annotation
eggNOG-mapper v2.1.11 was used for functional annotation [28] with the default parameters specified in the eggNOG-mapper web server (http://eggnog-mapper.embl.de/): 0–001 e-value; 60 bit-score; 40 identity percentage; 20% coverage; and 20% subject coverage. The search and annotation steps in eggNOG-mapper were performed using Diamond in blastx mode (-m diamond) [29] on proteins predicted by Prodigal v2.6.3 (–genepred prodigal) [30].
Functional profiling
All scripts for processing eggNOG-mapper results and producing functional profiles and abundance matrices (gene counts and KEGG ortholog terms) were implemented de novo in Python 3.
Taxonomic assignment of KEGG orthologs
To assign taxonomies to the contigs with genes that drive a signal for a KO term, we employed MMseqs2 taxonomyv.0b27c9d7d7757f9530f2efab14d246d268849925 [31]. The contig taxonomy workflow allowed us to perform a search against Genome Taxonomy Database (GTDB) v220 [32] and compute the lowest common ancestor with the 2bLCA algorithm. For each KO term, the assigned taxonomies were counted and ranked according to their frequency of occurrence. This ranking highlights the most prevalent taxa associated with relevant functions within the dataset.
Statistical analysis
Analysis of DEIPs for each week was conducted using the olink_ttest function, which performs individual t-tests for each assay between the FMT and placebo arms. To fully leverage the longitudinal experimental design of our study, we also used generalized additive mixed models to detect DEIPs overall between FMT and Placebo, assuming a Gaussian distribution of the outcome variable, NPX, using the R package mgcv [33]. This approach was chosen to account for the possibility that protein levels might not change at a constant rate (nonlinear) and vary between groups over time. We specified the model as follows:
In this model, “group” is included as a fixed effect, capturing the overall difference between the treatment groups; “s(week, k = 4)” captures the smooth effect of time (week) across all patients, irrespective of the group; “s(week, k = 4, by = group)” allows the smooth effect of time (week) to vary by group, modeling the interaction between group and week and allowing different time trends within each group; and “s(patientid, bs = “re”)” specifies a random effect for individual patients, accounting for repeated measures within each patient. Restricted maximum likelihood (REML) was used as a smoothing parameter estimation method.
To compute correlations between changes in DEIPs and bacterial abundances or bacterial genes and annotated functions over time, we used linear mixed effects models with the lme4 [34] and lmerTest [35] packages, introducing the treatment group and the timepoint as covariates:
This method was chosen because it effectively manages the correlations in our longitudinal data and captures the linear relationships we needed to identify potential bacterial drivers of inflammation. We represented in networks the significant interactions between bacterial abundances or their genes and functions with the DEIPs over time using the igraph package [36]. Analyses were adjusted for false discovery rate (FDR) when indicated using the “p.adjust” function in R with the method set to “BH” (Benjamini-Hochberg), and the FDR threshold was set at 0.05.
Study approval
The research project was authorized by the Ethics Committee (approval number: 165/16), and all participants provided informed consent before the study procedures were initiated. Patients who could not provide informed consent or had oral consent documented with written consent from a representative were not included in the study. Clinical Trials Registry Identification Number (clinicaltrials.gov): NCT03008941.
Results
General characteristics of the study population
We recruited 30 participants who were randomly divided into two groups: the treatment (FMT) group and the placebo group. Samples were collected from both groups at 0 (pre-intervention), 1 (after first FMT), 8 (1 week after last FMT), and 24 (16 weeks after last FMT) weeks. The characteristics of the study population are summarized in Table S1. In summary, 29 participants completed the evaluations. The participants in the study represented a population of middle-aged men who had sex with men and had well-controlled HIV infection. The safety and clinical events during the study are described in a previous publication.
The FMT and placebo groups were balanced regarding ART regimen distribution: 5 vs. 5 participants were on NNRTI-based triple ART, 1 vs. 2 on PI-based triple ART, and 9 vs. 8 on INSTI-based triple ART (p = 0.822). Regarding specific drug use during the study, only two patients (R3 in the placebo group and R24 in the FMT group) were on omeprazole from baseline and continued its use throughout the study. Only one patient (R24 in the FMT group) received statins during the study. Seven subjects received antibiotic treatment in the 14 weeks before the intervention, of whom three were in the FMT arm. However, as shown in our previous study, the use of antibiotics after the baseline did not introduce drastic changes in the microbiota. Furthermore, there were no notable differences in food consumption or intake of energy or nutrients between the groups [17]. The mean alcohol intake was 9.3 vs. 7.6 g/day (p = 0.479).
Fecal microbiota transplantation (FMT) induces changes in the expression pattern of plasma inflammatory proteins
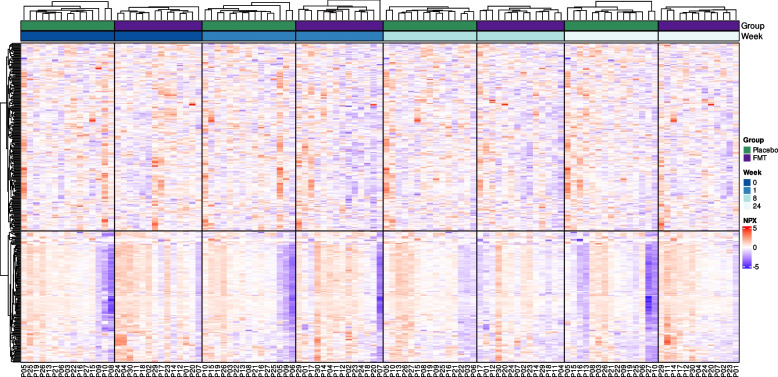
FMT resulted in differences in the expression patterns of plasma inflammatory proteins assessed by PEA. A heatmap visualization (Fig. 1) revealed a complex yet distinct shift from a more inflammatory state at baseline (week 0) toward a less inflammatory profile sustained up to week 24 in the FMT group compared to the placebo group.
Fig. 1.
Heatmap showing the expression levels of plasma inflammatory proteins measured by the proximity extension assay during follow-up. A total of 345 proteins were analyzed in biologically independent samples from 14 individuals in the FMT group and 15 in the placebo group. Unsupervised clustering analysis of the 345 proteins detected is shown in the rows. Supervised clustering analysis of independent observations per patient in the placebo (green) or FMT (purple) arms and at different time points (weeks in blue) are shown in columns. The unit of protein expression is scaled normalized protein expression (NPX)
Unsupervised hierarchical clustering of the 345 proteins assessed (lines in the heatmap of Fig. 1) identified two major clusters of coregulated proteins. To understand the effect of the treatment over time, we clustered the columns in the heatmap by group and week. In the FMT cohort, we found a marked reduction in the expression of inflammatory markers posttreatment (from higher NPX values, in red, to lower values, in blue), with the most pronounced decrease evident at week 1, which persisted after FMT was discontinued. In contrast, the expression of inflammatory proteins in the placebo group did not decrease over time (from lower NPX values in blue to higher values in red). This finding suggested that the FMT intervention had a sustained anti-inflammatory effect.
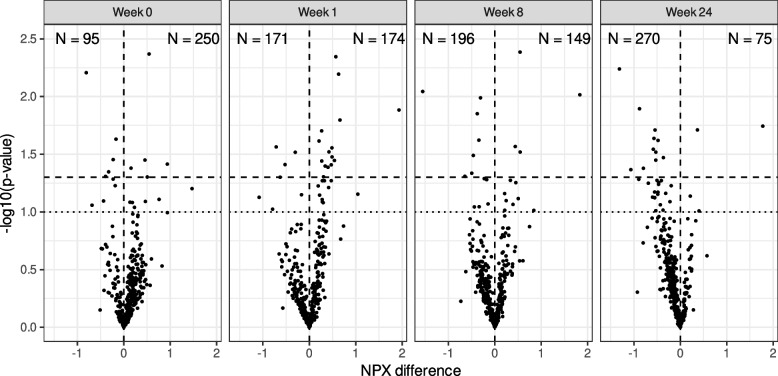
Then, we assessed the impact of FMT on protein expression by comparing the number of DEIPs in the FMT group with that in the placebo group at baseline and after treatment (weeks 1, 8, and 24). Figure 2 shows a decrease in the number of overexpressed inflammatory proteins in the FMT group relative to the placebo group, from 250 proteins at week 0 to 174 at week 1, 149 at week 8, and 75 at week 24. The FMT and placebo groups initially had the same number of differentially expressed proteins. By week 24, however, the FMT group had 12 underexpressed versus two overexpressed proteins compared to the placebo group, suggesting that FMT may progressively reduce inflammation over time.
Fig. 2.
Differential protein expression between the FMT and placebo groups over time. The figure shows the number of proteins overexpressed (positive values) or underexpressed (negative values) in the FMT group compared to the placebo group at different time points (weeks 0, 1, 8, and 24). The text at the top of each graph indicates the number (N) of proteins overexpressed (right) or underexpressed (left). The upper quadrants represent DEIPs with a p value < 0.05 (dashed line) or < 0.1 (dotted line), not adjusted by FDR. n = 29 biologically independent samples from 14 individuals in the FMT group and 15 in the placebo group were measured at four time points
Longitudinal analysis of protein expression revealed 46 proteins differentially expressed after FMT that are commonly implicated in cytotoxicity, cytokine stimulation, and immune cell recruitment to the infection site
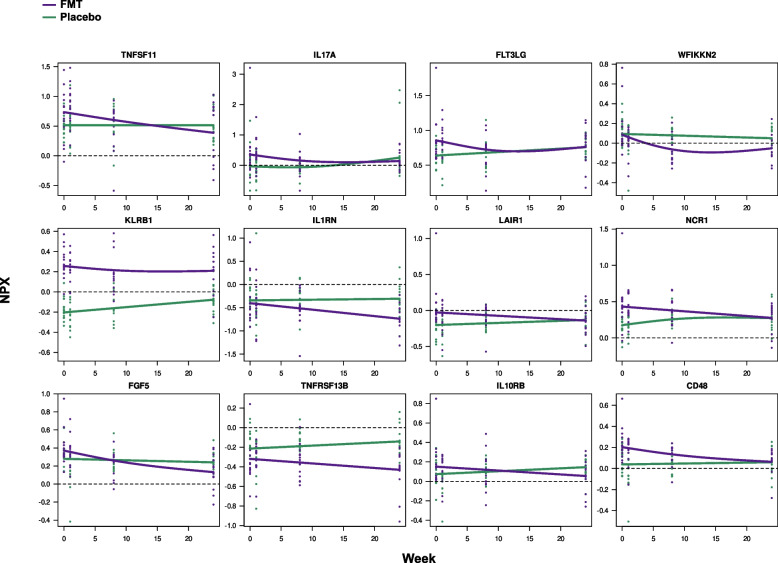
To discern proteins strongly influenced by the intervention, we used generalized additive mixed models (GAM). This approach was chosen because of its flexibility in modeling variables and the nonlinear distribution of protein levels between groups and over time. Notably, no significant alterations in protein expression levels were observed in the placebo arm, except for CTSC. Conversely, in the FMT group, for an exploratory p value threshold < 0.10, the analysis revealed 46 DEIPs, as detailed in Table S2 and Supplemental Figs. 1-3. Those with the strongest statistical significance are illustrated in Fig. 3 to evaluate their dynamics over time. FMT induced a pronounced decrease in the serum levels of 45 out of the 46 investigated proteins, which included pro-inflammatory and regulatory factors. This finding suggested a broad immunomodulatory effect of FMT. Interestingly, Persephin (PSPN), a neurotrophic factor that primarily supports the survival and differentiation of specific neuronal populations, was the sole protein whose expression increased post-FMT. A general description of the main functions of the 46 DEIPs is provided in Table S3.
Fig. 3.
The first 12 DEIPs from the 46 identified DEIPs were selected based on their greater statistical significance, not adjusted by FDR, for the differences between treatment slopes in the FMT vs. placebo groups. Each scatter plot shows individual expression values for each participant, with smoothed mean values represented by purple (FMT) and green (placebo) lines. The study involved 29 participants (14 FMTs and 15 placebos), with longitudinal measurements taken at weeks 0, 1, 8, and 24. The trajectories of the remaining 36 DEIPs are shown in Figs. S1–S3
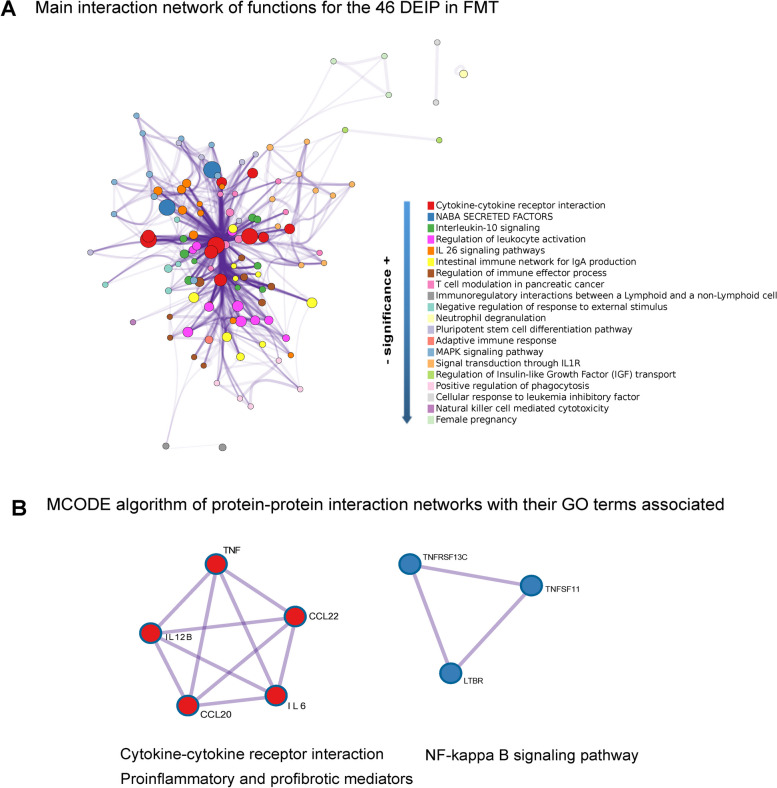
Next, we conducted a network analysis using METASCAPE to explore the biological functions of the 46 DEIPs, as summarized in Fig. 4A. The central nodes within the network highlight proteins involved in similar functions, depicted by different colors, and their connections, depicted by purple edges. The highlighted pathways included cytokine‒cytokine receptor interaction, IL10 signaling, and IL6 signaling pathways. These factors are crucial for immune regulation, suggesting that FMT may influence innate and adaptive immune responses in PWH. The occurrence of pathways such as T-cell modulation in pancreatic cancer and the regulation of tumor necrosis factor production indicate that FMT has a profound effect on systemic immune functions, potentially beyond gastrointestinal physiology. The central, highly interconnected nodes (e.g., cytokine interactions, MAPK signaling) suggest that these pathways are pivotal hubs in the network altered by FMT. Their central roles indicate that FMTs might broadly affect inflammatory and immune responses, possibly explaining the overall decrease in the serum levels of inflammatory proteins.
Fig. 4.
Network analysis of differentially expressed inflammatory proteins in PWH receiving repeated FMT vs. placebo. A This Cytoscape network graph visualizes the biological functions and pathways associated with the 46 DEIPs between PWH who underwent FMT and those who received a placebo. The nodes represent the number of proteins involved in the indicated process and their color functional clusters. The size of the nodes represents the number of proteins associated with that function (the larger the node is, the greater the number of selected proteins involved in that function). The color represents its cluster identity (i.e., nodes of the same color belong to the same cluster). Edges connect the related biological processes, and their width shows the strength of the connection between the proteins and the Gene Ontology (GO) term assigned. Terms with a similarity score > 0.3 are linked by an edge (the thickness of the edge represents the similarity score). The purple intensity denotes the superposition of the edges. B These subnetworks illustrate two distinct molecular complexes identified within the protein-to-protein interaction network of the differentially expressed inflammatory proteins. The tool finds protein‒protein interactions previously demonstrated experimentally. Each node represents a protein, and each edge denotes the physical protein‒protein interaction described between them. The MCODE algorithm groups nodes (elements in the network) together based on the strength and number of interactions they have with each other. Although the MCODE algorithm itself does not directly assign functional meaning to the identified subnetworks, the main functions (GO terms) related to these proteins with a p value < 0.01 are indicated below the subnetworks. Detailed information about the enrichment analysis, statistical values, and specific proteins associated with the functions is provided in Table S4 and its caption
Two functional complexes were found to be significant when we explored clusters of protein‒protein interactions using the MCODE algorithm, which revealed densely connected regions in large protein interaction networks (Fig. 4B): (i) Cytokine‒cytokine receptor interaction complex, which includes critical inflammatory mediators such as TNFɑ and IL6 and chemokines (CCL20 and CCL22). The interactions among these proteins, which drive inflammatory responses, and their modulation by FMT could explain the observed decrease in pro-inflammatory proteins (Fig. 3 and Supplemental Figs. 1-3). (ii) The NF-kappa B signaling pathway cluster, which comprises TNF and its receptors. Given that NF-kappa B is a transcription factor that regulates inflammation-related genes, its prominence post-FMT suggests significant regulation of inflammatory gene expression.
Correlation network between bacterial species abundance and inflammatory protein expression
To investigate the microbial drivers behind the observed shifts in inflammation, we fitted mixed models correlating changes in microbiome composition at the species level with alterations in the 46 previously selected DEIPs in plasma. From the 2074 distinct bacterial species detected, we identified 385 associations between changes in fecal bacterial species and plasma DEIPs at an adjusted p value threshold of 0.05. Several species within the Firmicutes phylum and families Ruminococcaceae, Succinivibrionaceae, Prevotellaceae, and the Clostridium genus showed the strongest associations with DEIPs in plasma (Table S5).
We specifically examined the associations between microbial species and three anti-inflammatory proteins relevant to HIV immunopathogenesis: Galectin-9 (LGALS9), which disrupts gut epithelial tight junctions and correlates with microbial translocation [37]; IL10RB, which limits gut inflammation by inhibiting pro-inflammatory cytokines, down-regulating MHC class II expression, and controlling immune responses to commensal bacteria [38–40]; and IL1RN, which inhibits pro-inflammatory IL1 signaling, maintaining gut homeostasis and limiting inflammation [41, 42].
The Clostridiales order had the most robust impact on inflammation, as almost all species exhibiting changes directly correlated with alterations in at least 15 plasma DEIPs belonged to this group (Table S6). Notably, several species within the genus Clostridium, known for their role in producing SCFAs such as butyrate, which have anti-inflammatory properties, were repeatedly linked to several DEIPs [40]. Specifically, Clostridium species correlated the DEIPs with anti-inflammatory properties IL10RB, LAIR1, IL1RN.
The Succinivibrionaceae and Prevotellaceae families, as well as genera previously reported as functionally relevant, like Faecalibacterium, Erysipelotrichaceae, Roseburia, Lactobacillus and Lactococcus, showed significant associations with various DEIPs.
Additionally, the Ruminococcaceae and Succinivibrionaceae families were recurrently associated with these DEIPs with anti-inflammatory properties (Table S6). The Succinivibrionaceae and Prevotellaceae families and other species previously reported as functionally relevant, included in the Faecalibacterium, Erysipelotrichaceae, Roseburia, Lactobacillus and Lactococcus genera, showed significant associations with various DEIPs.
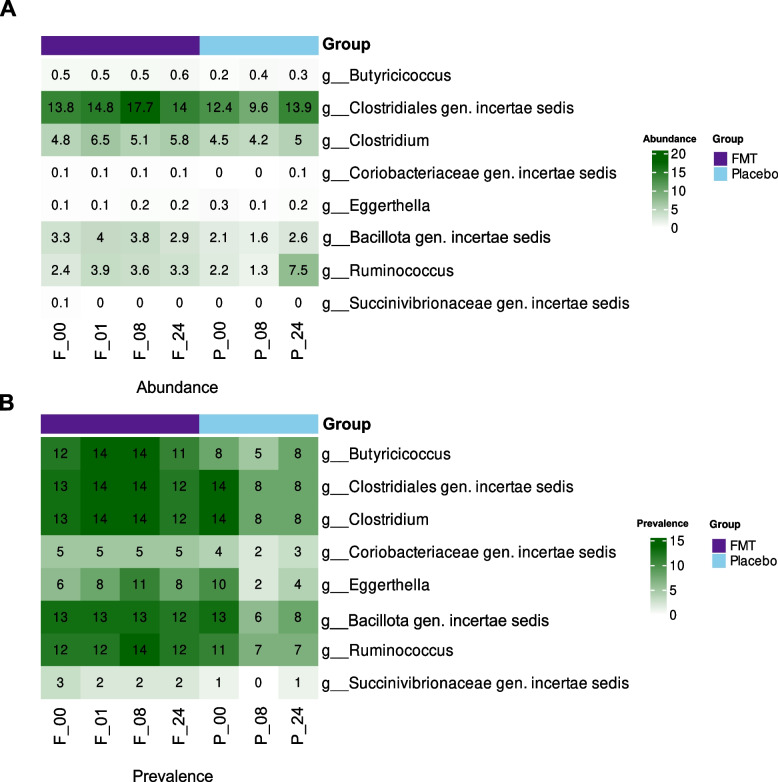
To further refine the biological relevance and mitigate the risk of false positive associations, we assessed the occurrence of the genus whose species showed the strongest associations with changes in DEIPs among participants across study timepoints. We did this by calculating the prevalence of bacteria and summarizing their abundance across visits (Fig. 5 and Table S7), which helps determine the presence of bacteria within the study participants. Specifically, we observed the consistent presence of genera such as Butyricicoccus, Clostridiales gen. incertae sedis, Clostridium, Bacillota gen. incertae sedis, and Ruminococcus in the FMT group.
Fig. 5.
Distribution and prevalence of key bacterial genera identified in the study. A Relative abundance (%) and B number of participants with the presence of key bacterial species significantly correlated with five or more DEIPs in plasma, aggregated at the genus level. In the X-axis, the F or P prefixes denote the study group (FMT or placebo, respectively), and the numeric suffixes indicate the weeks from baseline
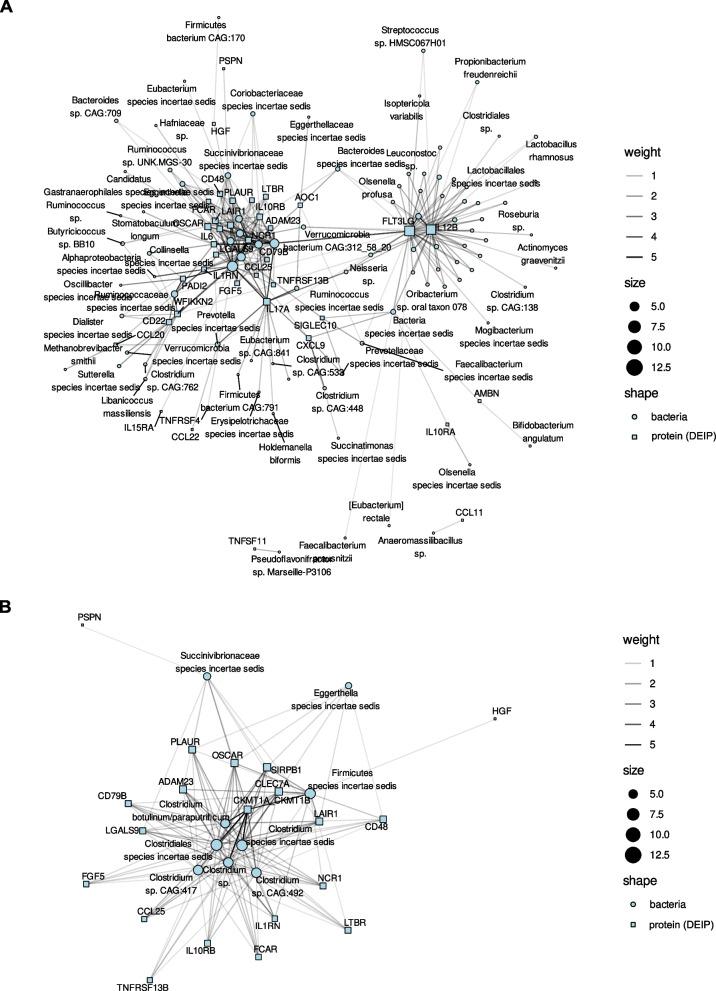
In parallel, certain proteins were recurrently linked to bacterial species. For instance, changes in FLT3LG, IL12B, and IL17A correlated with at least 20 distinct bacterial species. This subset of proteins might serve as biomarkers for microbial-driven inflammatory responses in the host (Table S8). The relationships between the fecal bacterial species and the 46 DEIPs in plasma are illustrated in the network analysis in Fig. 6.
Fig. 6.
Correlation networks between bacterial species abundance and inflammatory protein expression A The network illustrates significant correlations between the abundance of fecal bacterial species and the expression levels of DEIPs in plasma, including species showing at least one significant association with a DEIP after FDR correction. B The network focuses on bacterial species correlated with at least five DEIPs. Circles represent bacterial species, and squares represent DEIPs. The edge intensity on a grayscale indicates the strength of the correlation, with darker edges denoting stronger associations. The node size reflects the number of significant correlations. Clostridium species, particularly within the Clostridiales order, exhibited numerous correlations with DEIPs, such as OSCAR, CLEC7A, SIRPB1, ADAM23, and IL1RN, highlighting their role in modulating inflammation through the production of anti-inflammatory SCFAs, such as butyrate. Other significant associations included genera such as Ruminococcus and Butyricicoccus and members of the Succinivibrionaceae family. The genera classified as incertae sedis within Clostridiales and Firmicutes (Table S8) also exhibited substantial correlations, suggesting important roles in inflammation
Changes in the bacterial gene counts correlate with changes in the expression of inflammatory proteins in plasma following FMT
Next, we aimed to elucidate the bacterial genes potentially associated with the observed shifts in the expression of inflammatory proteins following FMT. We fitted mixed models to establish correlations between the relative abundance of bacterial genes and alterations in the 46 significant DEIPs. With an adjusted p value threshold of 0.05, we identified 585 significant correlations between changes in bacterial gene abundances and DEIPs (Table S9).
A subset of the bacterial genes, identified by eggNOG-mapper as yhcR, ectB, grdI, wzm, XK27_00670, and yulB, proved to be particularly significant, as their alterations were strongly correlated with changes in more than 20 of the 46 selected DEIPs (Table S10). Among the DEIPs correlated with at least 15 bacterial species detailed in Table S8, FLT3LG, IL12B, IL17A, and OSCAR were also correlated with at least 15 bacterial genes, emphasizing the potential influence of the gut microbiome on these proteins (Table S11).
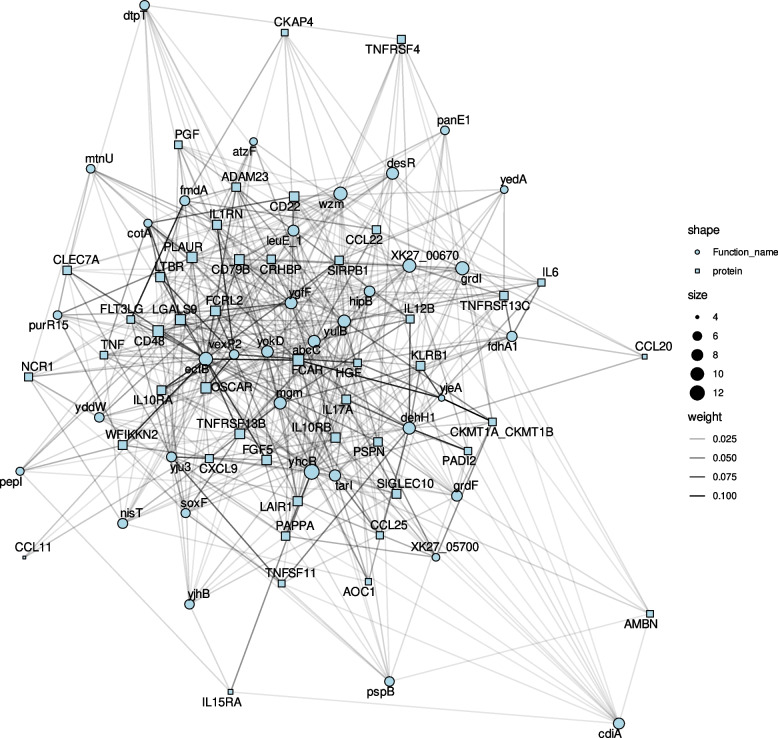
We also examined the bacterial genes associated with the DEIP with anti-inflammatory properties that were consistently associated with Clostridium sp. (Table S6), including IL1RN, IL10RB, LAIR1, and LGALS9. The genes identified by eggNOG-mapper as abcC, wzm, yjhB, XK27_05700, desR, cdiA, ectB, leuE_1, mgm, dehH1, hipB, napF, vexP2, yokD, and yulB were associated with at least two of these proteins. Their functions and potential mechanisms influencing inflammation following FMT are summarized in Table S12. The relationships between changes in bacterial gene counts and the 46 DEIP proteins are illustrated in the network analysis in Fig. 7.
Fig. 7.
Correlation network of bacterial genes correlated with DEIPs. The network illustrates significant correlations between bacterial genes and DEIPs in plasma, focusing on genes significantly associated with at least 5 DEIPs. Circles represent bacterial genes, and squares represent DEIPs. The edge intensity on a grayscale indicates the strength of the correlation, with darker edges denoting stronger associations. The node size reflects the number of significant correlations. Several bacterial genes exhibited multiple significant correlations with DEIPs, highlighting their potential role in modulating inflammation
Connections between changes in the annotated bacterial functions and the expression of inflammatory proteins in plasma following FMT
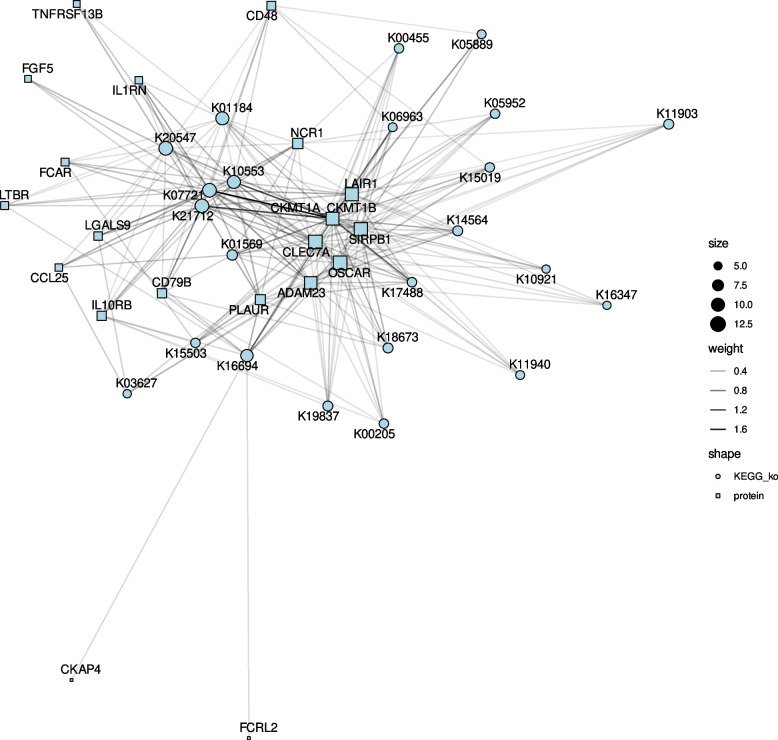
Finally, we fitted mixed models in which we correlated the changes in the functional annotations (KOs) from the bacterial genomes with the dynamics of the 46 previously selected plasma DEIPs. For an adjusted p value < 0.05, we identified 730 significant associations between changes in functional annotation numbers and DEIPs, while 272 associations remained when a more stringent threshold was applied (adjusted p value < 0.01) (Table S13). These associations are mainly related with metabolic pathways, biosynthesis of secondary metabolites, ABC transporters, and the two-component system (KEGG mapper output of Table S13 with more than 10 associations).
We selected the most relevant functions based on several criteria. First, functions associated with changes in a high number of DEIPs include the following: polygalacturonase, which correlated with changes in 16 DEIPs and it was detected in 5 samples; type VI secretion system secreted protein Hcp, which correlated with 8 DEIPs and it was present in 15 samples; and 4Fe-4S ferredoxin, which correlated with 7 DEIPs and it was present in 8 samples. Second, functions present in at least 30 samples and linked with at least 2 DEIPs were divided into three subcategories: (i) those correlated with pro-inflammatory proteins (IL6, IL17A, FLT3LG, CCL20, IL1RN) including ATP-dependent ion protease, epsilon-lactone hydrolase, and several others; (ii) those correlated with anti-inflammatory proteins (LAIR1, TNFRSF13C, IL10RA, TNFRSF13C, IL10RB) including demethylmenaquinone methyltransferase / 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase, [glutamine synthetase] adenylyltransferase / [glutamine synthetase]-adenylyl-L-tyrosine phosphorylase, and others; (iii) those correlated with proteins with context-dependent effects (LGALS9, TNFRSF4, LTBR) including CRISPR-associated protein Csy3 and (heptosyl)LPS beta-1,4-glucosyltransferase. Collectively, these functions could impact systemic inflammation by influencing gut barrier integrity, microbiome composition, and host immune responses (Table S14). These relationships between changes in bacterial annotated functions and the 46 plasma DEIPs proteins are illustrated in the network analysis in Fig. 8.
Fig. 8.
Correlation network of KEGG orthologs functions correlated with DEIPs. The network highlights significant correlations between bacterial annotated functions (KEGG orthologs) and DEIPs, focusing on functions significantly associated with at least 5 DEIPs. Circles represent KEGG orthologs, and squares represent DEIPs. The edge intensity on a grayscale indicates the strength of the correlation, with darker edges indicating stronger associations. The node size reflects the number of significant correlations. The network underscores the functional pathways involved in inflammation, with several KEGG orthologs showing substantial associations with DEIPs, suggesting their role in inflammatory processes
The DEIPs most connected with changes in annotated bacterial functions were the pro-inflammatory IL12B, linked to 167 functions, and FLT3LG, linked to 148 functions (Table S15) of the 730 significant DEIP-function associations detailed in Table S13. Mixed models indicated that all the estimates between IL12B and the annotated functions were negative (Table S13), suggesting that microbiome changes following FMT may inhibit this pro-inflammatory cytokine, which remains elevated despite ART [43] and is associated with cardiovascular risk [44]. Conversely, nearly all associations between FLT3LG and the annotated functions were positive, suggesting that microbiome changes following FMT may elevate this anti-inflammatory molecule. When FLT3LG was administered to humanized mice, it showed to sustain high levels of plasmacytoid dendritic cells, key producers of type I interferons [45]. Remarkably, 107 KEGG ortholog terms simultaneously correlated with both IL12B and FLT3LG.
The functions linked with changes in both IL12B and FLT3LG were predominantly assigned to Streptococcus genus, including S. thermophilus (with known probiotic properties) [46], which appears frequently across multiple functions: DNA-3-methyladenine glycosylase I, penicillin-binding protein 2A, hydroxymethylglutaryl-CoA reductase, porphyrinogen peroxidase, adenine deaminase, and the NarL family two-component system. Other notable taxa include Bifidobacterium spp. and Acidaminococcus intestini for DNA-3-methyladenine glycosylase I, Mitsuokella jalaludinii for putative transposase, and Faecalibacterium prausnitzii for NarL family two-component system. A description of the functions correlated with both IL12B and FLT3LG simultaneously (Table S14), their roles, and the potential mechanisms by which they could affect gut-associated inflammation are summarized in Table S16.
Discussion
In this pilot randomized controlled study investigating the effects of repeated oral FMTs on systemic inflammation in PWH receiving ART, we observed significant reductions in the expression of a broad array of inflammatory proteins. Notably, these effects persisted until the final visit, 16 weeks postintervention, suggesting the sustainable modulation of systemic inflammation.
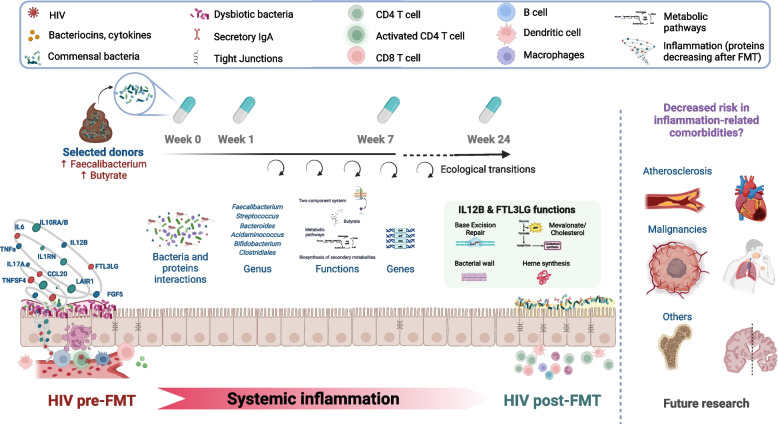
Unlike previous interventions that targeted the gut microbiome with prebiotics, probiotics, and synbiotics (reviewed in [11]), our study directly measured changes in inflammation by assessing a comprehensive panel of inflammatory proteins. Additionally, whereas previous pilot studies noted only limited engraftment of donor microbiota following three different modalities of FMT [17, 47, 48], our current research identified potential key microbial species whose changes correlated significantly with long-lasting variations in inflammatory marker levels, thereby highlighting their potential for targeted interventions in the field of microbiome therapeutics. We hypothesize that the long-lasting effects may be due to the selection of key bacterial strains for the proliferation of other beneficial microbes, events of secondary succession (a cascade of ecological changes), increase in the production of anti-inflammatory metabolites, and/or expansion of relevant immune cells. The key findings of our study, along with potential mechanisms underlying shifts in inflammatory proteins, are conceptualized in Fig. 9.
Fig. 9.
Conceptual model of changes in gut-associated inflammation in PWH following FMT from selected. This figure illustrates the systemic inflammation observed in PWH before FMT and the ecological transitions following FMT, which lead to shifts in inflammatory protein expression. Selected donor microbiota, rich in Faecalibacterium and butyrate producers, was administered weekly from week 0 to 7. The interactions between bacterial genera, genes, and metabolic functions are highlighted, with particular focus on the roles of IL12B and FLT3LG. These proteins are linked to key functions such as base excision repair, bacterial wall synthesis, and dendritic cell differentiation, which may contribute to the observed reduction in systemic inflammation and a potential decreased risk in inflammation-related comorbidities like atherosclerosis and malignancies. Future research is needed to explore these connections further
To date, no interventions targeting the microbiota of PWH on ART, including prebiotics [49, 50], probiotics [14, 51], synbiotics [13], or rifaximin [15], have convincingly proven to ameliorate inflammation or enhance immune recovery. Although some studies have reported mixed effects of diverse inflammatory cytokines, the evaluation of inflammation has typically been restricted to a limited set of molecules [17, 47]. Our previously reported study indicated that repeated oral FMT induces modest yet enduring changes in the gut microbiota structure. This was particularly notable in the Ruminococcaceae and Lachnospiraceae families, which are commonly depleted in PWH [10] and are major butyrate producers. Concurrently, there was a reduction in IFABP, a biomarker of intestinal barrier integrity and an independent predictor of mortality in PWH [17]. Here, we further elucidated the effects of FMT on inflammation, achieving broader resolution.
Among the 46 DEIPs analyzed following FMT, 45 exhibited down-regulation across pro-inflammatory and regulatory domains. Notably, enhanced signaling via cytokine‒cytokine receptor interactions and the IL10 pathway suggested a shift toward an anti-inflammatory profile in patients undergoing FMT. However, this extensive regulation suggests a broad systemic impact beyond mere anti-inflammatory effects, challenging simplistic interpretations of immune responses. A considerable number of proteins with an inflammatory function display dual roles, exerting a distinct influence on the immune response depending on the biological context, the presence of other signaling molecules, and the specific environmental conditions. Such widespread down-regulation might reflect a resetting of the immune system, which often remains in a state of heightened activation in chronic HIV infection [1, 2, 18] despite effective viral suppression through ART.
Although the generalized reduction in inflammatory marker levels indicates that FMT can efficiently shape immune responses—potentially reducing the risk of inflammation-related comorbidities—a reduction in both pro-inflammatory and regulatory proteins may reflect a move toward homeostasis in an activated immune system. In contrast to this general trend, the Persephin (PSPN) level increased post-FMT. Given the critical role of PSPN in neuronal survival and differentiation, its up-regulation post-FMT raises intriguing possibilities regarding the impact of the gut microbiota on the gut-brain axis, hinting at specific pathways involved in gut-immune-neural axis restoration or a unique compensatory response to microbiome modification [52].
To further investigate the mechanism of these proteins, we performed an enrichment analysis of the 46 DEIPs. Most of the identified proteins are related to functions that include pro-inflammatory cytokines and chemokines, such as TNF, a central mediator of acute inflammation; IL1B and IL6, cytokines involved in fever and acute phase reactions; and CCL20 and CCL22, chemokines responsible for immune cell chemotaxis. In addition, some proteins were grouped in a second cluster, suggesting their role in the modulation of adaptive immune responses, as they are involved in lymphoid tissue organization and B-cell function. For example, TNFRSF13C is essential for B-cell development, TNFSF11 (RANKL) is involved in T-cell and dendritic cell regulation and bone metabolism, and LTBR is crucial for lymphoid tissue development. Thus, these 46 DEIPs play critical roles in the immune response and clinical progression of HIV by affecting both pro-inflammatory and anti-inflammatory pathways. For instance, IL6 is frequently reported to be elevated in PWH on ART and serves as an independent predictor of mortality in this population [1]. TNF-α is associated with inflammation and HIV persistence during ART, partly through signaling [53, 54]. The expression of macrophage inflammatory protein-3 alpha (CCL20), a protein involved in recruiting cells to sites of inflammation, is typically elevated in HIV patients [55]. Conversely, key proteins in the IL1 pathway, such as IL10RA, the IL10 receptor, and IL1RN, which are IL1 receptor antagonists, could be particularly relevant in the pathogenesis of inflammation-related cardiovascular events [56, 57]. Interestingly, microbiome-derived signals can influence the expression of these proteins in the epithelium [58–60].
Network analysis revealed a nuanced landscape of protein expression changes following FMT, suggesting that not all DEIPs are uniformly influenced by alterations in the microbiome. We identified a subset of proteins, including pro-inflammatory (IL6, IL17A, CCL20, IL1RN), anti-inflammatory (FLT3LG, LAIR1, IL10RA, IL10RB, and TNFRSF13C), and context-dependent proteins (LGALS9, TNFRSF4, LTBR), that exhibited significant correlations with multiple bacterial taxa and were prevalent across samples. This finding suggested targeted modulation by the microbiome, likely through specific microbial metabolic activities or immunomodulatory mechanisms. For example, in PWH, gut dendritic cells are activated by Prevotella sp. [61], which was positively correlated with changes in IL17-A in our study. Prevotella has been found to promote the maturation and activation of dendritic cells, thereby enhancing their ability to present antigens and activate T cells [61, 62].
FLT3LG plays a crucial role in developing and maturing dendritic cells [63] correlated in our study with changes in Faecalibacterium prausnitzii, a dominant commensal of the human gut and a major butyrate producer [64]. Conversely, changes in pro-inflammatory IL12B, typically increased in PWH despite ART [46] and associated with cardiovascular risk [47], were negatively linked to 167 bacterial functions. Other DEIPs highlight the potential role of FMT in shaping pathogen-specific defense mechanisms. These include CCL20, which is essential for mucosal immunity and is directly regulated by certain bacteria, such as Prevotella sp. [65]; IL17A, whose expression modulates the microbiome composition [66]; and CLEC7A, which is relevant for antifungal immunity and innate immune responses [67]. Collectively, all these changes in bacterial functions suggest that FMT can reduce systemic inflammation in PWH by modulating gut barrier integrity, microbiome composition, and host immune responses, thereby lowering pro-inflammatory signals like IL12B and enhancing anti-inflammatory responses like FLT3LG. The association of these proteins with a broad array of bacterial species highlights their potential as biomarkers for evaluating the efficacy and understanding the biological regulatory effects of FMT.
We identified specific bacterial species significantly associated with changes in plasma DEIPs following FMT. Species within the Clostridiales order, including Clostridium sp. and Ruminococcus sp., were frequently correlated with proteins such as OSCAR, CLEC7A, IL17A, and FLT3LG. These findings align with our selection of stool donors based on high butyrate and Faecalibacterium abundances, emphasizing the role of butyrate-producing bacteria in modulating immune responses. Our previous analysis revealed pronounced engraftment in the Lachnospiraceae and Ruminococcaceae families, with their abundance remaining elevated after 48 weeks [17]. Butyrate can inhibit NF-κB signaling [68], partly by inhibiting human histone deacetylases [28, 49]. This inhibition facilitates the transcription of genes involved in regulatory T-cell function, such as Foxp3 [69]. As a result, butyrate induces a tolerogenic response in human dendritic cells. Our network analysis of protein‒protein interactions revealed that the NF-kB signaling pathway was significantly regulated post-FMT, suggesting that this pathway is critical for microbiome changes post-FMT.
Among the list of DEIPs associated with at least 20 bacterial species, FLT3LG, IL12B, and IL17A were also correlated with at least 15 bacterial genes. Consequently, these proteins could serve as indicators of microbial-driven inflammatory responses. Additionally, we investigated the subset of genes more strongly associated with changes in DEIPs with anti-inflammatory properties (Table S12). EctB, which is involved in ectoine biosynthesis, may have protective effects on human cells, thereby mitigating inflammatory responses, while grdl and wzm, which encode a glycosyltransferase and an ABC transporter, respectively, potentially modulate immune recognition and the antigenic load, respectively, contributing to an anti-inflammatory environment. Additionally, the roles of XK27_00670 in metabolic processes and yubL in sporulation could influence microbial community dynamics and stability, further impacting host inflammatory pathways and improving gut barrier function [70].
Lastly, we selected the most relevant annotated bacterial functions, revealing associations with changes in DEIPs. Key functions identified included polygalacturonase, which was correlated with changes in 16 DEIPs and detected in 5 samples; type VI secretion system secreted protein Hcp, 4Fe-4S ferredoxin, ATP-dependent Lon protease, epsilon-lactone hydrolase, demethylmenaquinone methyltransferase, [glutamine synthetase] adenylyltransferase; and CRISPR-associated protein Csy3, (heptosyl) LPS beta-1,4-glucosyltransferase. We focused on the pro-inflammatory IL12B and the anti-inflammatory FLT3LG, due to their high number of associations with multiple bacterial functions. The functions associated with changes in IL12B and FLT3LG were primarily linked to the Streptococcus genus, including S. thermophilus, and involved various enzymes and proteins. Other notable bacteria like Bifidobacterium spp., Acidaminococcus intestini, and Faecalibacterium prausnitzii were also highlighted for their roles in specific functions, with these associations and their potential impact on gut inflammation detailed Table S16.
Several factors must be taken into consideration when interpreting our results. First, our previous analysis utilized 16S rRNA sequencing across 11 study visits, providing higher temporal resolution but limited taxonomic detail [17]. In contrast, the current analysis employs shotgun metagenomics over four study visits, allowing us to achieve species-level resolution, albeit with less frequent sampling. This difference in methodology provides more detailed taxonomic insights in the current study but less temporal resolution. Here, we used mOTUs3 to profile the microbiome composition at the species level. Given our limited sample size, we chose this method to ensure greater accuracy in role assignment, thereby mitigating the risk of false discoveries despite its lower sensitivity than other tools [71]. In this study, we measured microbiome function indirectly by assigning significant bacterial genes to their functions or proteins. However, a direct assessment would have required analyzing higher functional levels of the microbiome, such as its transcriptome, proteome, or metabolome, which should be considered in future studies.
The strengths of our pilot study include (i) the randomized controlled trial design, which allows us to attribute observed changes directly to the FMT intervention rather than to natural microbial variations; (ii) the use of a novel proteomic assay, which allows for a more detailed, efficient and precise measurement of inflammatory biomarkers than did previous studies [17, 47, 48]; (iii) the application of species-level resolution in microbial analysis; (iv) the lack of significant differences in dietary intake between the groups, minimizing confounding variables; (v) the comprehensive longitudinal analysis, which helps in understanding the changes over time; and (vi) the careful selection of donors with microbiota profiles high in Faecalibacterium spp. and butyrate, which target anti-inflammatory properties.
While randomized controlled trials are essential for establishing causal relationships, our longitudinal correlation analysis between fecal bacteria and plasma proteins should be considered preliminary. The immune system is intricately regulated and often follows a nonlinear response pattern to interventions. This motivates further mechanistic studies to elucidate how FMT affects inflammation. Additionally, future research should directly measure microbiome functions through metatranscriptomics, metaproteomics, or metabolomics. Factors that may enhance the effects of FMT, including donor selection, baseline microbiome composition, inflammatory profiles, specific concomitant antiretroviral drugs, and the potential need for antibiotic preconditioning regimens, need to be explored.
In the context of a rapidly advancing field featuring live biotherapeutic products and synthetic bacterial communities currently undergoing phase 2 trials, our results could inform targeted subsequent investigations. In PWH, individuals diagnosed at advanced stages of the disease—typically showing increased inflammation and higher risk of great outcomes—represent a potential target population. Furthermore, the effects of down-regulation of inflammatory proteins following FMT in PWH treatment show promise in other clinical settings, such as enhancing the efficacy of PD-1 inhibitors [72]. Our group and others are currently evaluating whether repeated FMT has an effect in cancer progression. In fact, we are currently analyzing results from a pilot trial evaluating repeated oral FMT as a strategy to enhance immunotherapy in lung cancer (https://clinicaltrials.gov/study/NCT04924374).
Conclusions
In this pilot study, we explored the potential of oral FMT to reduce inflammation in PWH. FMT lowered plasma inflammatory protein levels compared to those in patients treated with a placebo, and FMT was found to be an established independent predictor of mortality, as were IL6 and TNF [1, 69]. The shift in inflammation persisted up to 16 weeks after the final FMT procedure. We identified changes in FT3LG, IL6, IL10RB, IL12B, and IL17A, which correlated with multiple bacterial species and functions. This subset of proteins might serve as biomarkers for microbial-driven inflammatory responses in the host.
Furthermore, we found specific bacterial species within the Ruminococcaceae, Succinivibrionaceae, Prevotellaceae, and Clostridium genera, as well as their associated genes and functions, that were significantly correlated with changes in inflammatory markers. These results support the notion that the gut microbiome could be a therapeutic target for mitigating inflammation in PWH. Further research is warranted to explore the potential of FMT and other microbiome-based interventions.
Supplementary Information
Acknowledgements
We thank all the patients and healthcare workers who participated in the study.
Abbreviations
- PWH
People with HIV
- FMT
Fecal microbiota transplant
- ART
Antiretroviral therapy
- GAMs
Generalized additive models
- DEIPs
Differentially expressed inflammatory proteins
- PEA
Proximity extension assay
- NPX
Normalized protein expression level
- MCODE
Molecular Complex Detection
- GO
Gene Ontology
Authors’ contributions
Conceptualizations: SSV; Methodology: SSV, EM, CDG, AT, MJG, SGBP, JHC; Patient recruitment: SSV, JPM, FD, MJV, SM; Laboratory measurements: EM, LMP, LL, MJG; Shotgun metagenomic analysis: LMF, SGB, JHC. Bioinformatic analysis: CD, AT; Supervision of bioinformatic analysis: SSV, EM. Funding acquisition: SSV; Project supervision: SSV, EM; Writing – original draft: SSV, EM, CDG; Writing – review and editing: all authors.
Funding
This work was funded by the Instituto de Salud Carlos III and Fondos FEDER, Acción Estratégica en Salud (PI18/00154, ICI20/00058 and PI21/00041), and co-financed by the European Development Regional Fund “A way to achieve Europe” (ERDF), a Gilead Fellowship (GLD16-00030), a crowdfunding project from the precipita platform of the Fundación Española para la Ciencia y la Tecnología (FECYT) and a restricted grant from Finch Therapeutics. The SEIMC-GESIDA Foundation supported this study through safety and data monitoring (GESIDA 9116). The funders of the study had no role in the study design, data collection, data analysis, data interpretation, or writing of the report. Claudio Díaz-García was supported by a “Contrato predoctoral de formación en investigación en salud” (FI22/00111) funded by the Instituto de Salud Carlos III (ISCIII) and the European Union (NextGenerationEU).
Data Availability
The data used for these analyses are available as supporting materials, and the code necessary to reproduce the results can be found in our GitHub repository (https://github.com/einlabryc/REFRESH_proteomics). The sequences are available at the European Nucleotide Archive database under accession number PRJEB75958.
Declarations
Ethics approval and consent to participate
The study was approved by the Ethics Committee (approval number: 165/16), and all participants signed an informed consent before the initiation of the study. Clinical Trials Registry Identification Number (clinicaltrials.gov): NCT03008941.
Consent for publication
Figure 9 was created with biorender.com, which confirmed the publication and license rights.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Claudio Díaz-García and Elena Moreno are co-first authors.
Santiago Moreno and Sergio Serrano-Villar are co-senior authors.
Contributor Information
Elena Moreno, Email: emolmo@salud.madrid.org.
Sergio Serrano-Villar, Email: sergio.serrano@salud.madrid.org.
References
- 1.Tenorio AR, Zheng Y, Bosch RJ, Krishnan S, Rodriguez B, Hunt PW, et al. Soluble markers of inflammation and coagulation but not T-cell activation predict non-AIDS-defining morbid events during suppressive antiretroviral treatment. J Infect Dis. 2014;210:1248–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunt PW, Lee SA, Siedner MJ. Immunologic biomarkers, morbidity, and mortality in treated HIV infection. J Infect Dis. 2016;214:S44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vujkovic-Cvijin I, Dunham RM, Iwai S, Maher MC, Albright RG, Broadhurst MJ, et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med. 2013;5:193ra91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vázquez-Castellanos JF, Serrano-Villar S, Jiménez-Hernández N, Soto Del Rio MD, Gayo S, Rojo D, et al. Interplay between gut microbiota metabolism and inflammation in HIV infection. ISME J. 2018;12:1964–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li SX, Sen S, Schneider JM, Xiong K-N, Nusbacher NM, Moreno-Huizar N, et al. Gut microbiota from high-risk men who have sex with men drive immune activation in gnotobiotic mice and in vitro HIV infection. Silvestri G, editor. PLOS Pathog. 2019;15:e1007611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gelpi M, Vestad B, Raju SC, Hansen SH, Høgh J, Midttun Ø, et al. Association of the kynurenine pathway of tryptophan metabolism with human immunodeficiency virus-related gut microbiota alterations and visceral adipose tissue accumulation. J Infect Dis. 2022;225:1948–54. [DOI] [PubMed] [Google Scholar]
- 7.Schuetz A, Deleage C, Sereti I, Rerknimitr R, Phanuphak N, Phuang-Ngern Y, et al. Initiation of ART during early acute HIV infection preserves mucosal Th17 function and reverses HIV-related immune activation Desrosiers. RC, editor. PLoS Pathog. 2014;10:e1004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serrano-Villar S, Rojo D, Martínez-Martínez M, Deusch S, Vázquez-Castellanos JF, Bargiela R, et al. Gut bacteria metabolism impacts immune recovery in HIV-infected individuals. EBioMedicine. 2016;8:203–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vujkovic-Cvijin I, Swainson LA, Chu SN, Ortiz AM, Santee CA, Petriello A, et al. Gut-resident Lactobacillus abundance associates with IDO1 inhibition and Th17 dynamics in SIV-infected macaques. Cell Rep. 2015;13:1589–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vujkovic-Cvijin I, Somsouk M. HIV and the gut microbiota: composition, consequences, and avenues for amelioration. Curr HIV/AIDS Rep. 2019;16:204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brenchley J, Serrano-Villar S. From dysbiosis to defense: harnessing the gut microbiome in HIV/SIV therapy. Microbiome. 2024;Accepted on April 26th. [DOI] [PMC free article] [PubMed]
- 12.Pastor-Ibáñez R, Blanco-Heredia J, Etcheverry F, Sánchez-Palomino S, Díez-Fuertes F, Casas R, et al. Adherence to a supplemented Mediterranean diet drives changes in the gut microbiota of HIV-1-infected individuals. Nutrients. 2021;13:1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serrano-Villar S, De Lagarde M, Vázquez-Castellanos J, Vallejo A, Bernadino JI, Madrid N, et al. Effects of immunonutrition in advanced human immunodeficiency virus disease: a randomized placebo-controlled clinical trial (Promaltia Study). Clin Infect Dis. 2019;68:20–30. [DOI] [PubMed]
- 14.Presti RM, Yeh E, Williams B, Landay A, Jacobson JM, Wilson C, et al. A randomized, placebo-controlled trial assessing the effect of VISBIOME ES probiotic in people with HIV on antiretroviral therapy. Open Forum Infect Dis. 2021;8:ofab550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tenorio AR, Chan ES, Bosch RJ, Macatangay BJC, Read SW, Yesmin S, et al. Rifaximin has a marginal impact on microbial translocation, T-cell activation and inflammation in HIV-positive immune non-responders to antiretroviral therapy – ACTG A5286. J Infect Dis. 2015;211:780–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams BB, Green SJ, Bosch RJ, Chan ES, Jacobson JM, Margolis DM, et al. Four weeks of treatment with rifaximin fails to significantly alter microbial diversity in rectal samples of HIV-infected immune non-responders (ACTG A5286) which may be attributed to rectal swab use. Pathog Immun. 2019;4:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serrano-Villar S, Talavera-Rodríguez A, Gosalbes MJ, Madrid N, Pérez-Molina JA, Elliott RJ, et al. Fecal microbiota transplantation in HIV: a pilot placebo-controlled study. Nat Commun. 2021;12:1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serrano-Villar S, Sainz T, Lee SA, Hunt PW, Sinclair E, Shacklett BL, et al. HIV-infected individuals with low CD4/CD8 ratio despite effective antiretroviral therapy exhibit altered T cell subsets, heightened CD8+ T cell activation, and increased risk of non-AIDS morbidity and mortality. Silvestri G, editor. PLoS Pathog. 2014;10:e1004078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wik L, Nordberg N, Broberg J, Björkesten J, Assarsson E, Henriksson S, et al. Proximity extension assay in combination with next-generation sequencing for high-throughput proteome-wide analysis. Mol Cell Proteomics. 2021;20:100168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown Ashlee, , Denise Lynch, Anne Bouevitch, Evgueni Doukhanine. OMNIgene®•GUT provides easy self-collection and stabilization of liquid fecal samples for microbiome profiling. White Pap DNA Genotek. 2018; Available from: https://www.dnagenotek.com/ROW/pdf/PD-WP-00056.pdf
- 23.Choo JM, Leong LE, Rogers GB. Sample storage conditions significantly influence faecal microbiome profiles. Sci Rep. 2015;5:16350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen W, Le S, Li Y, Hu F. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation Zou. Q, editor. PLOS ONE. 2016;11:e0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milanese A, Mende DR, Paoli L, Salazar G, Ruscheweyh H-J, Cuenca M, et al. Microbial abundance, activity and population genomic profiling with mOTUs2. Nat Commun. 2019;10:1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. Using SPAdes de novo assembler. Curr Protoc Bioinforma. 2020;70: e102. [DOI] [PubMed] [Google Scholar]
- 28.Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J. eggNOG-mapper v2: functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale Tamura. K, editor. Mol Biol Evol. 2021;38:5825–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buchfink B, Reuter K, Drost H-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods. 2021;18:366–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirdita M, Steinegger M, Breitwieser F, Söding J, Levy Karin E. Fast and sensitive taxonomic assignment to metagenomic contigs Kelso. J, editor. Bioinformatics. 2021;37:3029–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parks DH, Chuvochina M, Rinke C, Mussig AJ, Chaumeil P-A, Hugenholtz P. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 2022;50:D785–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wood SN, Pya N, Säfken B. Smoothing parameter and model selection for general smooth models. J Am Stat Assoc. 2016;111:1548–63. [Google Scholar]
- 34.Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67. Available from: http://www.jstatsoft.org/v67/i01/.
- 35.Kuznetsova A, Brockhoff PB, Christensen RHB. lmerTest package: tests in linear mixed effects models. J Stat Softw. 2017;82. Available from: http://www.jstatsoft.org/v82/i13/. Cited 2024 May 24.
- 36.Csárdi G, Nepusz T, Müller K, Horvát S, Traag V, Zanini F, et al. igraph for R: R interface of the igraph library for graph theory and network analysis. Zenodo; 2024. Available from: https://zenodo.org/doi/10.5281/zenodo.7682609.
- 37.Moar P, Kaur U, Bhagchandani T, Tandon R, Ndhlovu LC. Galectin-9 mediates gut-associated inflammation and disruption of tight junctions. J Immunol. 2023;210:234.22. [Google Scholar]
- 38.Garg A, Spector SA. HIV Type 1 gp120–induced expansion of myeloid derived suppressor cells is dependent on interleukin 6 and suppresses immunity. J Infect Dis. 2014;209:441–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang X, Zhang S, Peng Q, Ling L, Shi H, Liu Y, et al. Sustained IFN-I stimulation impairs MAIT cell responses to bacteria by inducing IL-10 during chronic HIV-1 infection. Sci Adv. 2020;6:eaaz0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papoutsopoulou S, Pollock L, Walker C, Tench W, Samad SS, Bergey F, et al. Impact of interleukin 10 deficiency on intestinal epithelium responses to inflammatory signals. Front Immunol. 2021;12. [DOI] [PMC free article] [PubMed]
- 41.Jeong J-Y, Kim J, Kim B, Kim J, Shin Y, Kim J, et al. IL-1ra secreted by ATP-induced P2Y2 negatively regulates MUC5AC overproduction via PLCβ3 during airway inflammation. Mediators Inflamm. 2016;2016:7984853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mantovani A, Dinarello CA, Molgora M, Garlanda C. Interleukin-1 and related cytokines in the regulation of inflammation and immunity. Immunity. 2019;50:778–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Byrnes AA, Harris DM, Atabani SF, Sabundayo BP, Langan SJ, Margolick JB, et al. Immune activation and IL-12 production during acute/early HIV infection in the absence and presence of highly active, antiretroviral therapy. J Leukoc Biol. 2008;84:1447–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ehinger E, Ghosheh Y, Pramod AB, Lin J, Hanna DB, Mueller K, et al. Classical monocyte transcriptomes reveal significant anti-inflammatory statin effect in women with chronic HIV. Cardiovasc Res. 2021;117:1166–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhardwaj N, Friedlander PA, Pavlick AC, Ernstoff MS, Gastman BR, Hanks BA, et al. Flt3 ligand augments immune responses to anti-DEC-205-NY-ESO-1 vaccine through expansion of dendritic cell subsets. Nat Cancer. 2020;1:1204–17. [DOI] [PubMed] [Google Scholar]
- 46.Metabolic properties, functional characteristics, and practical application of Streptococcus thermophilus: food reviews international: Vol 40, No 2. Available from: 10.1080/87559129.2023.2202406. Cited 2024 Aug 9. [DOI]
- 47.Utay NS, Monczor AN, Somasunderam A, Lupo S, Jiang Z-D, Alexander AS, et al. Evaluation of six weekly oral fecal microbiota transplants in people with HIV. Pathog Immun. 2020;5:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vujkovic-Cvijin I, Rutishauser RL, Pao M, Hunt PW, Lynch SV, McCune JM, et al. Limited engraftment of donor microbiome via one-time fecal microbial transplantation in treated HIV-infected individuals. Gut Microbes. 2017;8:440–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gori A, Rizzardini G, Van’T Land B, Amor KB, Van Schaik J, Torti C, et al. Specific prebiotics modulate gut microbiota and immune activation in HAART-naive HIV-infected adults: results of the “COPA” pilot randomized trial. Mucosal Immunol. 2011;4:554–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serrano-Villar S, Vázquez-Castellanos JF, Vallejo A, Latorre A, Sainz T, Ferrando-Martínez S, et al. The effects of prebiotics on microbial dysbiosis, butyrate production and immunity in HIV-infected subjects. Mucosal Immunol. 2017;10:1279–93. [DOI] [PubMed] [Google Scholar]
- 51.Stiksrud B, Nowak P, Nwosu FC, Kvale D, Thalme A, Sonnerborg A, et al. Reduced levels of D-dimer and changes in gut microbiota composition after probiotic intervention in HIV-infected individuals on stable ART. JAIDS J Acquir Immune Defic Syndr. 2015;70:329–37. [DOI] [PubMed] [Google Scholar]
- 52.Razavi S, Nazem G, Mardani M, Esfandiari E, Salehi H, Esfahani SHZ. Neurotrophic factors and their effects in the treatment of multiple sclerosis. Adv Biomed Res. 2015;4:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu T, Zhang L, Joo D, Sun S-C. NF-κB signaling in inflammation. Signal Transduct Target Ther. 2017;2:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong LM, Jiang G. NF-κB sub-pathways and HIV cure: a revisit. eBioMedicine. 2021;63:103159. [DOI] [PMC free article] [PubMed]
- 55.Packard TA, Schwarzer R, Herzig E, Rao D, Luo X, Egedal JH, et al. CCL2: a chemokine potentially promoting early seeding of the latent HIV reservoir. mBio. 13:e01891–22. [DOI] [PMC free article] [PubMed]
- 56.Hoel H, Ueland T, Knudsen A, Kjær A, Michelsen AE, Sagen EL, et al. Soluble markers of interleukin 1 activation as predictors of first-time myocardial infarction in HIV-infected individuals. J Infect Dis. 2020;221:506–9. [DOI] [PubMed] [Google Scholar]
- 57.Caricchio R, Abbate A, Gordeev I, Meng J, Hsue PY, Neogi T, et al. Effect of canakinumab vs placebo on survival without invasive mechanical ventilation in patients hospitalized with severe COVID-19: a randomized clinical trial. JAMA. 2021;326:230–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doerflinger SY, Throop AL, Herbst-Kralovetz MM. Bacteria in the vaginal microbiome alter the innate immune response and barrier properties of the human vaginal epithelia in a species-specific manner. J Infect Dis. 2014;209:1989–99. [DOI] [PubMed] [Google Scholar]
- 59.Jones-Hall YL, Nakatsu CH. The intersection of TNF. IBD and the microbiome Gut Microbes. 2016;7:58–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matarazzo L, Hernandez Santana YE, Walsh PT, Fallon PG. The IL-1 cytokine family as custodians of barrier immunity. Cytokine. 2022;154;155890. [DOI] [PubMed] [Google Scholar]
- 61.Dillon SM, Lee EJ, Kotter CV, Austin GL, Gianella S, Siewe B, et al. Gut dendritic cell activation links an altered colonic microbiome to mucosal and systemic T-cell activation in untreated HIV-1 infection. Mucosal Immunol. 2016;9:24–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dhodapkar KM, Barbuto S, Matthews P, Kukreja A, Mazumder A, Vesole D, et al. Dendritic cells mediate the induction of polyfunctional human IL17-producing cells (Th17-1 cells) enriched in the bone marrow of patients with myeloma. Blood. 2008;112:2878–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cueto FJ, Sancho D. The Flt3L/Flt3 axis in dendritic cell biology and cancer immunotherapy. Cancers. 2021;13:1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275–83. [DOI] [PubMed] [Google Scholar]
- 65.Xu C, Fan L, Lin Y, Shen W, Qi Y, Zhang Y, et al. Fusobacterium nucleatum promotes colorectal cancer metastasis through miR-1322/CCL20 axis and M2 polarization. Gut Microbes. 2021;13:1980347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Okamura Y, Morimoto N, Ikeda D, Mizusawa N, Watabe S, Miyanishi H, et al. Interleukin-17A/F1 deficiency reduces antimicrobial gene expression and contributes to microbiome alterations in intestines of Japanese medaka (Oryzias latipes). Front Immunol. 2020;11. Available from: https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2020.00425/full. Cited 2024 May 16. [DOI] [PMC free article] [PubMed]
- 67.Ma J, Zhou M, Song Z, Deng Y, Xia S, Li Y, et al. Clec7a drives gut fungus-mediated host lipid deposition. Microbiome. 2023;11:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rodríguez Del Río Á, Giner-Lamia J, Cantalapiedra CP, Botas J, Deng Z, Hernández-Plaza A, et al. Functional and evolutionary significance of unknown genes from uncultivated taxa. Nature. 2024;626:377–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hunt PW, Sinclair E, Rodriguez B, Shive C, Clagett B, Funderburg N, et al. Gut Epithelial barrier dysfunction and innate immune activation predict mortality in treated HIV infection. J Infect Dis. 2014;210:1228–38. [DOI] [PMC free article] [PubMed]
- 70.Apweiler R, Martin MJ, O’Donovan C, Magrane M, Alam-Faruque Y, Antunes R, et al. Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 2012;40:D71–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rodríguez del Río Á, Giner-Lamia J, Cantalapiedra CP, Botas J, Deng Z, Hernández-Plaza A, et al. Functional and evolutionary significance of unknown genes from uncultivated taxa. Nature. 2024;626:377–84. [DOI] [PMC free article] [PubMed]
- 72.Routy B, Lenehan JG, Miller WH, Jamal R, Messaoudene M, Daisley BA, et al. Fecal microbiota transplantation plus anti-PD-1 immunotherapy in advanced melanoma: a phase I trial. Nat Med [Internet]. 2023; Available from: https://www.nature.com/articles/s41591-023-02453-x. Cited 2023 Jul 14. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used for these analyses are available as supporting materials, and the code necessary to reproduce the results can be found in our GitHub repository (https://github.com/einlabryc/REFRESH_proteomics). The sequences are available at the European Nucleotide Archive database under accession number PRJEB75958.