Abstract
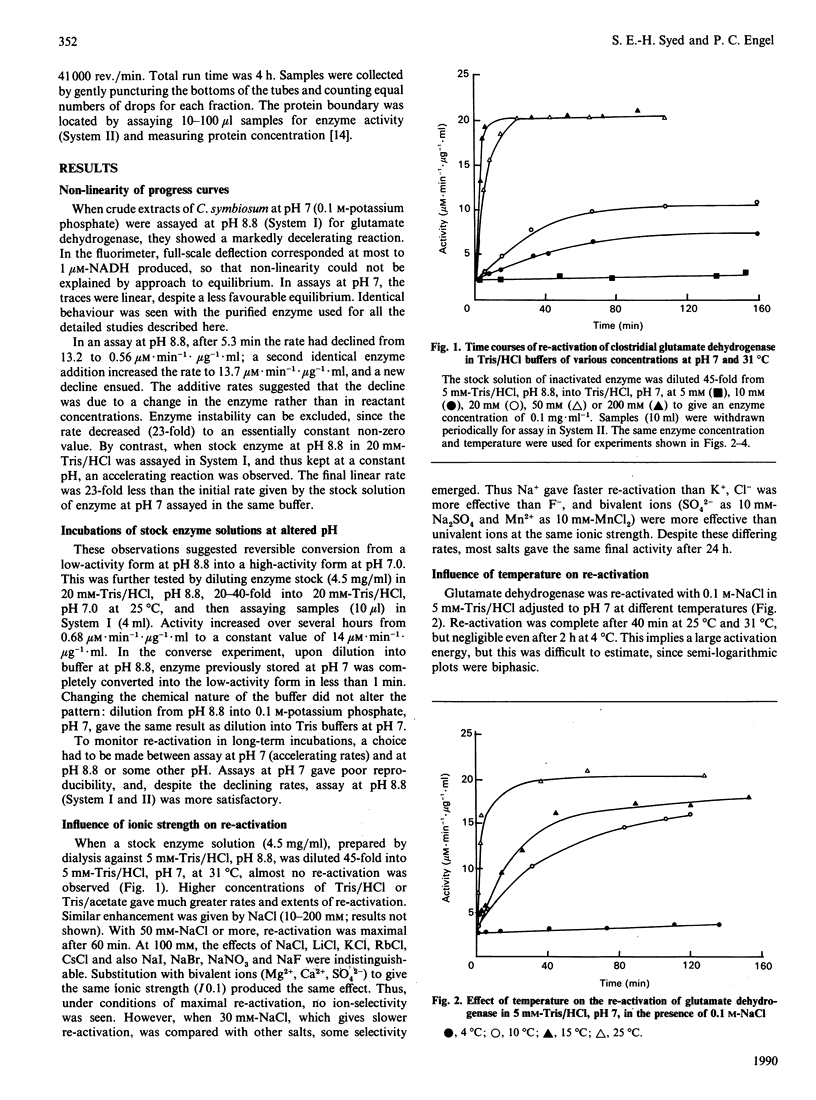
1. On transferring Clostridium symbiosum glutamate dehydrogenase from pH 7 to assay mixtures at pH 8.8, reaction time courses showed a marked deceleration that was not attributable to the approach to equilibrium of the catalysed reaction. The rate became approximately constant after declining to 4-5% of the initial value. Enzyme, stored at pH 8.8 and assayed in the same mixture, gave an accelerating time course with the same final linear rate. The enzyme appears to be reversibly converted from a high-activity form at low pH to a low-activity form at high pH. 2. Re-activation at 31 degrees C upon dilution from pH 8.8 to pH 7 was followed by periodic assay of the diluted enzyme solution. At low ionic strength (5 mM-Tris/HCl), no re-activation occurred, but various salts promoted re-activation to a limiting rate, with full re-activation in 40 min. 3. Re-activation was very temperature-dependent and extremely slow at 4 degrees C, suggesting a large activation energy. 4. 2-Oxoglutarate, glutarate or succinate (10 mM) accelerated re-activation; L-glutamate and L-aspartate were much less effective. 5. The monocarboxylic amino acids alanine and norvaline appear to stabilize the inactive enzyme: 60 mM-alanine does not promote re-activation, and, as substrates at pH 8.8 for enzyme stored at pH 7, alanine and norvaline give progress curves showing rapid complete inactivation. 6. Mono- and di-nucleotides (AMP, ADP, ATP, NAD+, NADH, NADP+, CoA, acetyl-CoA) at low concentrations (10(-4)-10(-3) M) enhance re-activation at pH 7 and also retard inactivation at pH 8.8. 7. The re-activation rate is independent of enzyme concentration: ultracentrifuge experiments show no changes in molecular mass with or without substrates. 8. The activation-inactivation appears to be due to a slow pH-dependent conformational change that is sensitively responsive to the reactants and their analogues.
Full text
PDF
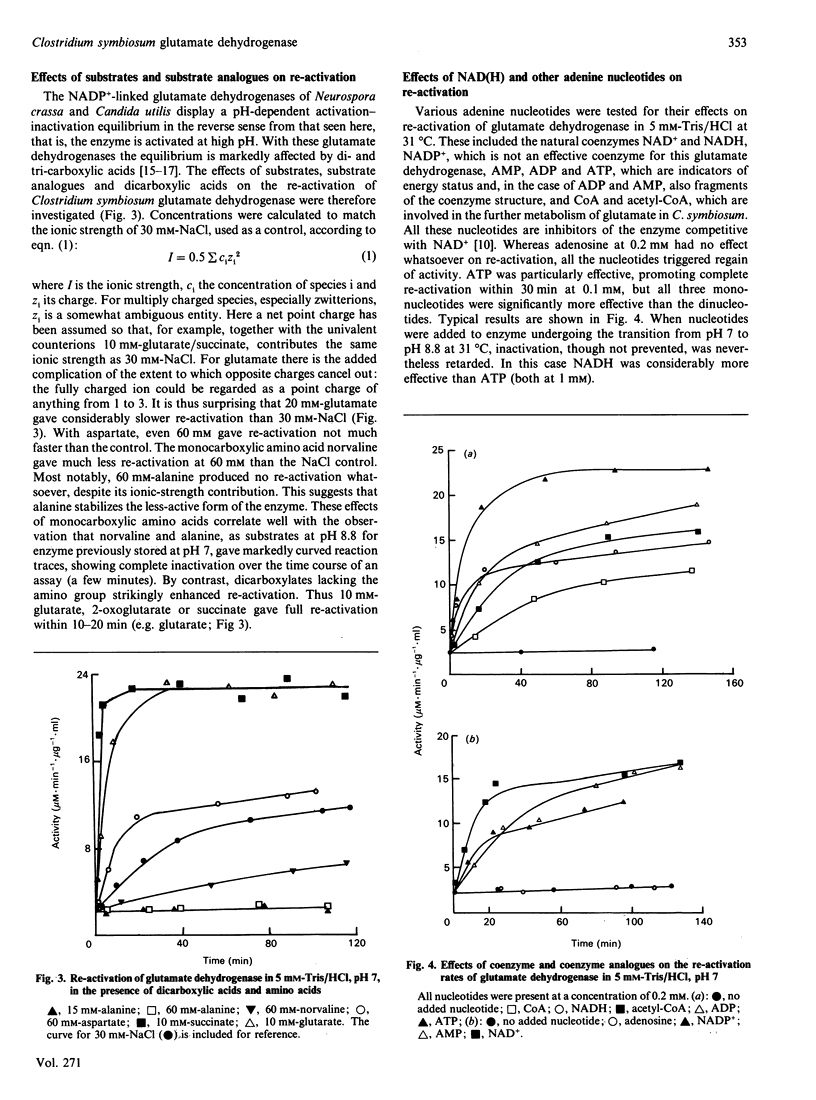


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ashby B., Wootton J. C., Fincham J. R. Slow conformational changes of a Neurospora glutamate dehydrogenase studied by protein fluorescence. Biochem J. 1974 Nov;143(2):317–329. doi: 10.1042/bj1430317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker P. J., Farrants G. W., Rice D. W., Stillman T. J. Recent progress on the structure and function of glutamate dehydrogenase. Biochem Soc Trans. 1987 Aug;15(4):748–751. doi: 10.1042/bst0150748. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- DALZIEL K. Kinetic studies of liver alcohol dehydrogenase. Biochem J. 1962 Aug;84:244–254. doi: 10.1042/bj0840244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRIEDEN C. Glutamic dehydrogenase. I. The effect of coenzyme on the sedimentation velocity and kinetic behavior. J Biol Chem. 1959 Apr;234(4):809–814. [PubMed] [Google Scholar]
- Frieden C. Kinetic aspects of regulation of metabolic processes. The hysteretic enzyme concept. J Biol Chem. 1970 Nov 10;245(21):5788–5799. [PubMed] [Google Scholar]
- Gore M. G., Greenwood C. Studies on the binary and ternary complexes formed by a Neurospora glutamate dehydrogenase and its substrates. Biochem Biophys Res Commun. 1975 Feb 17;62(4):997–1002. doi: 10.1016/0006-291x(75)90421-0. [DOI] [PubMed] [Google Scholar]
- Hershko A., Kindler S. H. Mode of interaction of purine nucleotides and amino acids with glutamate dehydrogenase. Biochem J. 1966 Dec;101(3):661–664. doi: 10.1042/bj1010661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephs R. Electron microscope studies on glutamic dehydrogenase: subunit structure of individual molecules and linear associates. J Mol Biol. 1971 Jan 28;55(2):147–153. doi: 10.1016/0022-2836(71)90188-4. [DOI] [PubMed] [Google Scholar]
- Kinnaird J. H., Fincham J. R. The complete nucleotide sequence of the Neurospora crassa am (NADP-specific glutamate dehydrogenase) gene. Gene. 1983 Dec;26(2-3):253–260. doi: 10.1016/0378-1119(83)90195-6. [DOI] [PubMed] [Google Scholar]
- MARTIN R. G., AMES B. N. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem. 1961 May;236:1372–1379. [PubMed] [Google Scholar]
- McPherson M. J., Wootton J. C. Complete nucleotide sequence of the Escherichia coli gdhA gene. Nucleic Acids Res. 1983 Aug 11;11(15):5257–5266. doi: 10.1093/nar/11.15.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P., Markau K., Sund H. Studies of glutamate dehydrogenase. Regulation of glutamate dehydrogenase from Candida utilis by a pH and temperature-dependent conformational transition. Eur J Biochem. 1976 Jun 1;65(2):465–472. doi: 10.1111/j.1432-1033.1976.tb10362.x. [DOI] [PubMed] [Google Scholar]
- OLSON J. A., ANFINSEN C. B. The crystallization and characterization of L-glutamic acid dehydrogenase. J Biol Chem. 1952 May;197(1):67–79. [PubMed] [Google Scholar]
- Rice D. W., Baker P. J., Farrants G. W., Hornby D. P. The crystal structure of glutamate dehydrogenase from Clostridium symbiosum at 0.6 nm resolution. Biochem J. 1987 Mar 15;242(3):789–795. doi: 10.1042/bj2420789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice D. W., Hornby D. P., Engel P. C. Crystallization of an NAD+-dependent glutamate dehydrogenase from Clostridium symbiosum. J Mol Biol. 1985 Jan 5;181(1):147–149. doi: 10.1016/0022-2836(85)90334-1. [DOI] [PubMed] [Google Scholar]
- Syed S. E., Engel P. C. Ox liver glutamate dehydrogenase. The use of chemical modification to study the relationship between catalytic sites for different amino acid substrates and the question of kinetic non-equivalence of the subunits. Biochem J. 1984 Sep 15;222(3):621–626. doi: 10.1042/bj2220621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle F., Becerril B., Chen E., Seeburg P., Heyneker H., Bolivar F. Complete nucleotide sequence of the glutamate dehydrogenase gene from Escherichia coli K-12. Gene. 1984 Feb;27(2):193–199. doi: 10.1016/0378-1119(84)90140-9. [DOI] [PubMed] [Google Scholar]
- West D. J., Tuveson R. W., Barratt R. W., Fincham J. R. Allosteric effects in nicotinamide adenine dinucleotide phosphate-specific glutamate dehydrogenase from Neurospora. J Biol Chem. 1967 May 10;242(9):2134–2138. [PubMed] [Google Scholar]
- Wootton J. C., Chambers G. K., Holder A. A., Baron A. J., Taylor J. G., Fincham J. R., Blumenthal K. M., Moon K., Smith E. L. Amino-acid sequence of NADP-specific glutamate dehydrogenase of neurospora crassa. Proc Natl Acad Sci U S A. 1974 Nov;71(11):4361–4365. doi: 10.1073/pnas.71.11.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YIELDING K. L., TOMKINS G. M. An effect of L-leucine and other essential amino acids on the structure and activity of glutamic dehydrogenase. Proc Natl Acad Sci U S A. 1961 Jul 15;47:983–989. doi: 10.1073/pnas.47.7.983. [DOI] [PMC free article] [PubMed] [Google Scholar]



