Abstract
INTRODUCTION
The apolipoprotein E (APOE) ε4 allele exerts a significant influence on peripheral inflammation and neuroinflammation, yet the underlying mechanisms remain elusive.
METHODS
The present study enrolled 54 patients diagnosed with late‐onset Alzheimer's disease (AD; including 28 APOE ε4 carriers and 26 non‐carriers). Plasma inflammatory cytokine concentration was assessed, alongside bulk RNA sequencing (RNA‐seq) and single‐cell RNA sequencing (scRNA‐seq) analysis of peripheral blood mononuclear cells (PBMCs).
RESULTS
Plasma tumor necrosis factor α, interferon γ, and interleukin (IL)‐33 levels increased in the APOE ε4 carriers but IL‐7 expression notably decreased. A negative correlation was observed between plasma IL‐7 level and the hippocampal atrophy degree. Additionally, the expression of IL‐7R and CD28 also decreased in PBMCs of APOE ε4 carriers. ScRNA‐seq data results indicated that the changes were mainly related to the CD4+ Tem (effector memory) and CD8+ Tem T cells.
DISCUSSION
These findings shed light on the role of the downregulated IL‐7/IL‐7R pathway associated with the APOE ε4 allele in modulating neuroinflammation and hippocampal atrophy.
Highlights
The apolipoprotein E (APOE) ε4 allele decreases plasma interleukin (IL)‐7 and aggravates hippocampal atrophy in Alzheimer's disease.
Plasma IL‐7 level is negatively associated with the degree of hippocampal atrophy.
The expression of IL‐7R signaling decreased in peripheral blood mononuclear cells of APOE ε4 carriers
Dysregulation of the IL‐7/IL‐7R signal pathways enriches T cells.
Keywords: Alzheimer's disease, apolipoprotein E allele 4, bulk RNA sequencing, hippocampal atrophy, interleukin 7R signaling pathway, peripheral blood mononuclear cells, single‐cell RNA sequencing
1. INTRODUCTION
Mounting evidence has suggested that polymorphisms in the apolipoprotein E (APOE) gene, particularly the APOE ε4 allele, are closely linked to the development of Alzheimer's disease (AD). 1 APOE ε4 is recognized as a primary susceptibility gene for AD, due to its role in accelerating amyloid plaque formation, tau protein phosphorylation, and intensifying the neuroinflammatory process. 2 , 3 APOE ε4 allele carriers exhibit marked disparities in immune cells and inflammatory cytokines compared to non‐carriers, both in the central nervous system (CNS) and peripheral blood. 2 , 4 However, the exact mechanisms of APOE ε4 in affecting neuroinflammation in AD remain elusive.
APOE is a major apolipoprotein with a critical role in lipid metabolism, particularly in the transport and clearance of lipids. 3 It has isoform‐dependent effects on the immune system and inflammatory responses, potentially because of the differences in lipid metabolism. 5 , 6 , 7 , 8 Due to the structural differences in two residues of its domain (Arg158 and Cys112), the three APOE isoforms have distinct lipid‐bound structures (ε2, ε3, and ε4), leading to varying affinities for different lipoprotein receptors. 1 APOE ε4 shows a higher affinity for very‐low‐density lipoprotein (VLDL) particles and cholestenone, while APOE ε2 and APOE ε3 display a greater affinity for high‐density lipoprotein receptors rich in phospholipids. 1 , 9 Such variation in lipid metabolism will result in disparities in immune cell activation and the antigen presentation process. 6 , 10 , 11 In antigen‐presenting cells (APCs), cholesterol‐enriched specialized microdomains known as lipid rafts serve as sites where major histocompatibility complex class II (MHC‐II) molecules concentrate, thereby optimizing T‐cell activation. 12 APOE ε4 carriers show higher levels of low‐density lipoprotein and cholestenone, which may potentially worsen T‐cell activation and inflammation. Moreover, the APOE ε4 isoform correlates with increased levels of activated T cells and heightened susceptibility to apoptosis in CD4(+) T cells, 13 underscoring the intricate impact of APOE ε4 on immune cell dynamics.
Apart from regulating T cells, the APOE genotype also affects cytokine expression levels, and impacts both systemic circulation and the brain. 8 , 14 , 15 , 16 , 17 For instance, lipopolysaccharide (LPS) induction has demonstrated a more pronounced increase in interleukin (IL)‐1β, IL‐6, and tumor necrosis factor (TNF)‐α levels among APOE ε4 carriers. 8 This isoform‐dependent effect extends to the levels of IL‐10, IL‐13, and IL‐8 as well. 16 , 18 , 19 The altered cytokine profiles, stemming from Th1 and Th2 cells, underscore the influence of the APOE ε4 genotype on both innate and adaptive immune responses. 20 Notably, cytokines such as TNF‐α, interferon (IFN)‐γ, IL‐1β, IL‐6, IL‐7, IL‐10, IL‐12, IL‐17, IL‐18, IL‐33, and monocyte chemoattractant protein 1 (MCP‐1) are also implicated in activating the innate immune response of microglial cells. 21 , 22 , 23 , 24 However, the clinical importance of these inflammatory factors remains to be fully elucidated.
Typically, IL‐7 is a 25‐kD protein that binds to a heterodimer receptor, IL‐7R, which consists of the IL‐7R α and universal γ chains. It mainly targets lymphocytes and promotes the activity of B progenitor cells, thymocytes, and peripheral mature T cells from human or mouse bone marrow. 25 Initially characterized as a survival factor for T and B cell precursors in mice, playing a critical role in their T and B cell development, 25 , 26 IL‐7 is now recognized as a key regulator in the lymphatic system. A previous study observed a negative association between plasma IL‐7 and age in APOE ε4–carrying familial Alzheimer's disease (FAD) patients, suggesting its potential involvement in neuroinflammation. 16 Furthermore, IL‐7 has also been linked to hippocampal neuronal death 27 , 28 , 29 and may contribute to structural deterioration in AD, particularly among APOE ε4 carriers. 30 The relationship between hippocampal atrophy and inflammatory factors in APOE ε4 carriers warrants further investigation.
To deepen our understanding of how the APOE ε4 allele affects immune cells and cytokines in AD, we analyzed clinical profiles and cytokine levels using bulk and single‐cell RNA sequencing of peripheral blood mononuclear cells (PBMCs). Our study unveiled aberrant regulation of the IL‐7R signal in T cells, offering novel insights into neuroinflammatory mechanisms.
2. METHODS
2.1. Subjects
Fifty‐eight patients diagnosed with AD were enrolled in this study after providing informed and explicit consent to participate. The study protocol was approved by the Clinical Research Ethics Committee of the First Affiliated Hospital of Shantou University Medical College (approval number: 2020‐115‐XZ2). All participants were informed individually of the detailed research protocol. Written informed consent or assent with proxy consent was obtained from all subjects prior to enrollment, in accordance with the latest Declaration of Helsinki. Diagnosis was established by neuroimaging and assessments conducted by neurology specialists, encompassing a comprehensive analysis of neuropsychological evaluations, magnetic resonance imaging (MRI) data, and clinical manifestations. The inclusion criteria for this study were as follows 31 : (1) participants aged between 50 and 85 years; (2) self‐reported memory loss persisting for at least 1 year; (3) a Mini‐Mental State Examination (MMSE) score of ≤ 26 (for individuals with at least 7 years of education: MMSE score ≤ 26; for those with 1–7 years of education: MMSE score ≤ 24; for individuals with < 1 year of education: MMSE score ≤ 19); (4) MRI scans indicating medial temporal atrophy (MTA), which includes the hippocampus, entorhinal cortex, and amygdala; and (5) a MTA visual evaluation scale score of ≥ 2.
RESEARCH IN CONTEXT
Systematic review: We searched PubMed using the terms “Alzheimer's disease,” “apolipoprotein allele,” “peripheral blood mononuclear cells,” and “IL7R,” since January 1, 1990. However, the effect of the apolipoprotein E (APOE) ε4 allele on the peripheral monocyte pathway has not yet been fully addressed.
Interpretation: This study is the first to validate the importance of the interleukin (IL)‐7R pathway in regulating neuroinflammation at the APOE ε4 allele. We found that the APOE ε4 allele decreased the expression of IL‐7R in peripheral blood mononuclear cells. The changes mainly occurred in the CD4+ Tem and CD8+ Tem T cells. Plasma IL‐7 also decreased in the APOE ε4 carriers, which was related to the degree of hippocampal atrophy and cognitive function. The present study may provide novel insights into the inflammatory mechanisms associated with APOE ε4 alleles.
Future directions: The IL‐7/IL‐7R pathway may play a critical role in regulating peripheral T cells and neuroinflammation, and may become a target for future therapeutic interventions in the treatment of Alzheimer's disease.
2.2. MRI scan and hippocampal atrophy degree
We conducted MRI scans using a 3.0 T MRI scanner equipped with a standard 8‐channel head coil. Various imaging sequences, including T1 fluid‐attenuated inversion recovery (FLAIR), T2 FLAIR, T2 weighted image, sagittal T2 fast spin echo, and diffusion‐weighted imaging, were acquired to rule out any irrelevant intracranial lesions. The detailed scanning sequence and methodology are provided in the supplementary materials in supporting information. To assess the degree of hippocampal atrophy, we adopted the medial temporal atrophy scale from a previous publication. 32 Two independent raters, a neurologist and a radiologist, evaluated the MTA scores. Discrepancies were resolved through discussion. Abnormal scores were defined as follows: MTA score of ≥ 1 for patients younger than 65 years old, MTA score of ≥ 2 for those aged 66 to 75 years, and MTA score of ≥ 3 in patients aged ≥ 75 years. Hippocampal atrophy was further quantified by measuring the length of the hippocampal head, hippocampal body, and temporal horn (Figure 1A). A decrease in length corresponded to an increase in the degree of hippocampal atrophy.
FIGURE 1.
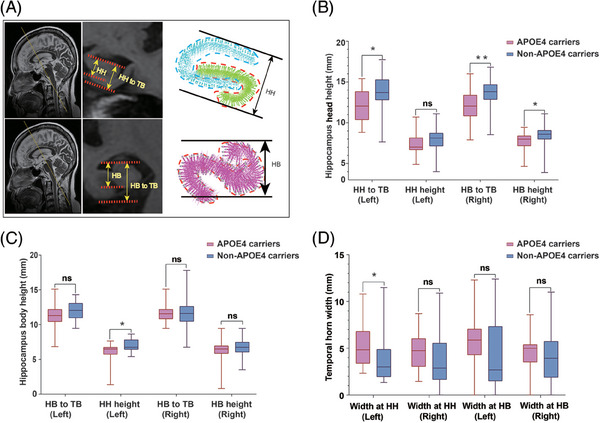
Impact of APOE ε4 allele on hippocampal atrophy. (A) Illustration of the method used for measuring hippocampal dimensions. This measurement involves assessing the distance from the upper region of the hippocampus to the lower region of the hippocampus, or to the TB, as observed in MRI oblique coronal slices of the HH and HB. B, In the case of HH slices, APOE ε4 allele carriers exhibited greater height measurements from the left HH to the TB, as well as the height of the right HH, compared to non‐carriers. This difference in measurements highlights the more pronounced hippocampal atrophy observed in APOE ε4 carriers. APOE, apolipoprotein E; HB, hippocampal body; HH, hippocampal head; MRI, magnetic resonance imaging; TB, temporal bottom
2.3. Blood sampling
We obtained peripheral blood samples and subjected them to centrifugation at 300 g and 4°C for 5 minutes. From the collected blood, 1 mL was used for APOE genotyping, while the remaining 3 mL was designated for plasma and PBMC collection. The resulting supernatant, which contained plasma, was preserved for subsequent examination of inflammatory factor levels. To isolate PBMCs, we used the Histopaque density gradient method for RNA sequencing (RNA‐seq), and detailed procedures are provided in the supplementary materials.
2.4. APOE genotyping
We conducted APOE genotyping using the SNaPshot Multiplex System, following the methodology outlined in a previous publication. The genotyping involved sequencing rs429358 and rs7412 located in exon 4 of the APOE gene. APOE status was categorized based on the presence of one or more copies of ε2, ε3, and ε4. An APOE ε4–positive status was confirmed for individuals with the ε4/ε4, ε3/ε4, or ε2/ε4 genotype, while all other genotypes were classified as non–APOE ε4 carriers. 31
2.5. Inflammatory cytokine measurements using Luminex liquid chip technology
In this study, we used Luminex liquid chip technology to assess the plasma levels of 13 inflammatory cytokines in AD patients. These cytokines included TNF‐α, IFN‐γ, IL‐1β, IL‐5, IL‐6, IL‐7, IL‐8, IL‐10, IL‐13, IL‐17, IL‐18, IL‐33, and MCP‐1. The detailed procedure for this aspect of the study is provided in the supplementary materials.
2.6. Measurement of plasma amyloid beta levels
Additionally, we measured plasma levels of amyloid beta (Aβ)1‐42 and Aβ1‐40 using liquid‐phase flow cytometry, using the Meso Scale Discovery platform (MSD).
2.7. Bulk RNA‐seq and analysis of PBMC
RNA was extracted from the PBMCs of 10 patients with AD (four non‐APOE ε4 carriers and six APOE ε4 carriers) using standard extraction protocols. Subsequently, RNA samples underwent quality control assessment through agarose gel electrophoresis, the Agilent 2100 bioanalyzer, and the NanoPhotometer spectrophotometer. Detailed procedures for bulk RNA‐seq analysis are outlined in the supplementary materials.
2.8. Single‐cell sequencing and analysis of PBMC
To increase sequencing depth and improve cost efficiency, a sample pooling strategy was used by combining data from four non‐carrier and six carriers PBMC samples. The combined dataset was analyzed using the R package Seurat. First, a Seurat object was created for the two sample groups. Stringent thresholds were set for data quality control, requiring cells to have between 200 and 5000 expressed features (nFeatures), between 500 and 15,000 total counts (nCount), and < 20% mitochondrial genes. To account for potential batch effects, the IntegrateData function was used to integrate the two samples. Subsequently, the RunPCA function from the R package Seurat was applied to reduce the dimensionality of the dataset. Clustering analysis was then conducted using the FindClu function, with a resolution parameter set to 1. Single‐cell RNA sequencing (scRNA‐Seq) libraries were prepared using the SeekOne MM Single Cell 3′ library preparation kit. Sequencing was performed on either the Illumina NovaSeq 6000 platform with a paired‐end read length of 150 bp (PE150) or the DNBSEQ‐T7 platform with a read length of PE100.
2.9. Single cell RNA‐seq cell ranger pipeline and cell cluster analysis
The Cell Ranger software (10X Genomics) was used to perform barcode and unique molecular identifier counting, after filtering and alignment to the GRCh37 (hg19) reference genome, to generate the feature‐barcode matrix and determine clusters. Dimensionality reduction was performed using principal component analysis, and the first 10 principal components were used to generate clusters by the k‐means and graph‐based algorithms, respectively. Data analysis was performed by the Loupe Cell Browser software (10X Genomics) on Cloupe files displaying Uniform Manifold Approximation and Projection (UMAP) projections of cell transcriptome.
2.10. Single cell RNA‐seq functional and pathway enrichment analysis
The differentially expressed genes (DEGs) for each cell type were imported into Metascape (http://metascape.org/) for Gene Ontology analysis of biological processes and the Kyoto Encyclopedia of Genes and Genomes (KEGG). The WikiPathways analysis was performed with a false discovery rate (FDR) of < 0.01 as the cut‐off value.
2.11. Pseudo‐temporal analysis, protein–protein interaction network analysis, cell–cell interaction analysis
Subpopulations associated with CD4+ T cells were identified, and quasi‐temporal analysis on all cells was conducted using the Monocle 2 algorithm in the R package (version 2.26.0). Meanwhile, Metascape was used to conduct the protein–protein interaction (PPI) network analysis; whereas the molecular complex detection (MCODE) algorithm was used to identify closely related modules in the network. Additionally, the R package CellChat (v 1.1.3) tool was used to infer cell–cell interaction networks.
2.12. Statistical analysis
The data are presented as mean ± standard deviation. All data were analyzed using the SPSS version 22 (IBM) software, and the comparison between groups was considered statistically significant with a P value of < 0.05. Differences in age, years of schooling, neuropsychological assessment, plasma level of Aβ peptide, plasma level of inflammatory cytokines, and hippocampal height between APOE ε4 carriers and non‐carriers were compared using independent‐samples t test or the Mann–Whitney test. Differences in sex and comorbidities were assessed using the chi‐square test. We used the R package linkET (v. 0.05) to create combined network heat maps. Additionally, we calculated Pearson linear correlation coefficients to analyze the associations between hippocampal height and plasma levels of inflammatory cytokines or Aβ peptides. Linear regression analysis was used to further examine the correlations between hippocampal height and plasma levels of inflammatory cytokines.
3. RESULTS
3.1. Participant characteristics
A total of 54 AD patients were enrolled in this study, with an even distribution between APOE ε4 carriers (n = 28, 78.6% female) and non‐carriers (n = 26, 69.2% female). There were no significant differences between the two groups in terms of age; education level; or diagnoses of hypertension, diabetes, lacunar infarction, arthritis, tumors, or other diseases (Table 1). Cognitive functions were assessed using the Mini‐Mental State Examination (MMSE) and the Montreal Cognitive Assessment (MoCA), revealing no significant differences between the two groups (Table 1). Similarly, visual rating scales for MTA were used to preliminarily evaluate the degree of hippocampal atrophy, with no significant difference observed in the MTA score (t = −1.140; 95% confidence interval [CI], −0.226 to 0.589; P = 0.377). Furthermore, the pathological state was evaluated by measuring plasma levels of Aβ1‐40 and Aβ1‐42, with no significant differences found in these levels between the groups.
TABLE 1.
Clinical characteristics and concentration of plasma inflammatory cytokines in APOE ε4–carrying and non‐APOE ε4–carrying Alzheimer's disease.
|
APOE ε4 carriers Mean (SD) |
Non‐APOE ε4 carriers Mean (SD) |
Differences between groups (95% CI) | p‐value | |
|---|---|---|---|---|
| APOE genotype (patients) | 28 | 26 | ||
| Sex [female %] | 22 [78.571] | 18 [69.231] | NA | 0.438 a |
| Age | 72.68 (8.060) | 70.85 (8.048) | –2.569 to 6.234 | 0.407 |
| Education level | 5.821 (4.405) | 6.115 (4.520) | –2.731 to 2.142 | 0.810 |
| Comorbidity | ||||
| Hypertension [%] | 7 [25.000] | 6 [23.767] | NA | 0.870 a |
| Diabetes [%] | 4 [14.286] | 5 [19.231] | NA | 0.629 a |
| Arthritis [%] | 3 [10.714] | 3 [11.538] | NA | 0.924 a |
| Number of lacunar cerebral infarction area | 1.321 (0.819) | 1.423 (0.857) | –0.559 to 0.356 | 0.658 |
| Coronary heart disease [%] | 5 [17.857] | 1 [3.846] | NA | 0.105 a |
| Cancer [%] | 1 [3.571] | 1 [3.846] | NA | 0.958 a |
| Neuropsychological assessments | ||||
| MMSE score | 11.464 (8.230) | 14.961 (8.166) | –7.977 to 0.983 | 0.123 |
| MoCA score | 7.704 (6.317) | 9.960 (6.931) | –5.946 to 1.434 | 0.225 |
| MTA | 2.643 (0.731) | 2.462 (0.761) | –0.226 to 0.589 | 0.377 |
| Plasma Aβ peptides | ||||
| Plasma Aβ1‐40 (pg/mL) | 87.865 (29.050) | 95.154 (37.853) | –30.251 to 15.674 | 0.524 |
| Plasma Aβ1‐42 (pg/mL) | 4.061 (2.319) | 4.372 (2.069) | –1.761 to 1.140 | 0.666 |
| Ratio of plasma Aβ1‐42 and Aβ1‐40 | 0.044 (0.018) | 0.047 (0.021) | –0.016 to 0.010 | 0.648 |
| Plasma levels of inflammatory cytokines | ||||
| TNF‐alpha (pg/mL) | 2.392 (1.445) | 1.803 (1.522) | –0.221 to 1.399 | 0.019 b |
| IFN‐gamma (pg/mL) | 111.391 (16.842) | 95.867 (16.205) | 6.484 to 24.562 | 0.001 |
| IL‐1beta (pg/mL) | 31.069 (6.692) | 30.367 (7.663) | –3.218 to 4.624 | 0.721 |
| IL‐5 (pg/mL) | 10.510 (3.973) | 9.687 (3.695) | –1.276 to 2.923 | 0.435 |
| IL‐6 (pg/mL) | 0.979 (0.760) | 1.218 (1.263) | –0.803 to 0.325 | 0.399 |
| IL‐7 (pg/mL) | 2.708 (2.065) | 4.706 (3.626) | –3.638 to −0.357 | 0.018 |
| IL‐8 (pg/mL) | 4.164 (7.858) | 5.015 (10.101) | –5.773 to 4.071 | 0.730 |
| IL‐10 (pg/mL) | 11.440 (3.786) | 13.054 (4.425) | –3.858 to 0.630 | 0.155 |
| IL‐13 (pg/mL) | 631.916 (238.264) | 610.120 (230.720) | –106.454 to 150.045 | 0.734 |
| IL‐17 (pg/mL) | 11.061 (6.259) | 11.186 (6.861) | –3.707 to 3.458 | 0.945 |
| IL‐18 (pg/mL) | 247.066 (191.891) | 334.452 (252.529) | –209.319 to 34.5477 | 0.156 |
| IL‐33 (pg/mL) | 4.949 (3.004) | 3.094 (1.131) | 0.597 to 3.113 | 0.004 |
| MCP‐1 (pg/mL) | 572.854 (1322.922) | 283.402 (64.963) | –232.104 to 811.008 | 0.258 |
Abbreviations: Aβ, amyloid beta; APOE, apolipoprotein E; CI, confidence interval; IFN, interferon; IL, interleukin; MCP‐1, monocyte chemoattractant protein 1; MMSE, Mini‐Mental State Examination; MoCA, Montreal Cognitive Assessment; MTA, medial temporal atrophy; SD, standard deviation; TNF, tumor necrosis factor.
Indicates analysis with a method of Fisher exact test.
Indicates analysis with a method of Mann–Whitney.
Measurement of plasma inflammatory cytokine levels revealed that APOE ε4 carriers exhibited higher plasma levels of TNF‐α, IFN‐γ, and IL‐33, and lower levels of IL‐7 than non‐carriers.
The plasma levels of 13 inflammatory cytokines potentially linked to AD pathogenesis were measured. While most measured cytokines showed no significant differences between APOE ε4 carriers and non‐carriers (Table 1), a key distinction in the levels of IL‐7, IL‐33, TNF‐α, and IFN‐γ was observed. Specifically, APOE ε4 carriers had significantly lower levels of IL‐7 compared to non‐carriers, recording a 42.5% decrease (t = 2.463; 95% CI, −3.638 to −0.357; P = 0.018). In contrast, plasma levels of TNF‐α, IFN‐γ, and IL‐33 were all significantly higher in the APOE ε4 carriers group compared to the non‐carrier group.
3.2. APOE ε4 carriers exhibited more significant atrophy
Hippocampal atrophy analysis revealed more pronounced structural changes in AD patients who were APOE ε4 carriers compared to non‐carriers (Figure 1B,C,D). Notably, the height of the left hippocampal body displayed a greater degree of atrophy in the APOE ε4 carrier group compared to the non‐carriers (Figure 1C). Furthermore, APOE ε4 carriers exhibited a visible widening of the temporal horn widths within the hippocampal head (Figure 1D), suggesting more extensive atrophy in this brain region.
3.3. Correlation between inflammatory cytokine levels and hippocampal atrophy: plasma IL‐7 correlates with hippocampus atrophy degree
Correlation analysis between plasma levels of inflammatory cytokines and hippocampal height of all patients is illustrated in Figure 2, with additional details provided in Table S1 and Figure S1 in supporting information. Among these inflammatory cytokines, significant correlations with specific aspects of hippocampal height were observed. Specifically, IL‐1β exhibited a significant correlation with the height of the left hippocampal head (P = 0.048, r = 0.270), while IL‐6 displayed a significant correlation with the height of left hippocampal body (P = 0.043, r = −0.276). In the right hippocampal head slices, IL‐5 revealed significant correlations with the right hippocampal body height (P = 0.017, r = 0.323) and the height of the right hippocampal body (P = 0.020, r = 0.316). Additionally, IL‐6 exhibited a significant correlation with the width of the right temporal horn (P = 0.028, r = 0.298). Notably, among all the cytokines analyzed, only IL‐7 consistently demonstrated a significant correlation with the degree of atrophy in the left hippocampal length (Figure 2A,B). This correlation was observed on both the slices of the left hippocampal head (Figure 2C,D), spanning from hippocampal head to temporal bottom (r = 0.298; 95% CI, 0.096 to 0.564; P = 0.029), as well as within the left hippocampal body (Figure 2E,F), ranging from hippocampal body to temporal bottom (r = 0.289; 95% CI, 0.080 to 0.443; P = 0.034) and within the hippocampal body itself (r = 0.274; 95% CI, −0.023 to 0.499; P = 0.045). While IL‐5 exhibited correlations with the right hippocampal body (hippocampal body to temporal bottom: r = 0.323; 95% CI, 0.082 to 0.526; P = 0.017 and hippocampal body: r = 0.312; 95% CI, 0.066 to 0.567; P = 0.020), it did not display any significant correlation with atrophy. Interestingly, all other examined inflammatory cytokines showed no significant associations with the thickness of the hippocampal head, body, or the width of the temporal horn. Table S1 provides additional details from the correlation analysis, including results related to the height of the left hippocampal head.
FIGURE 2.
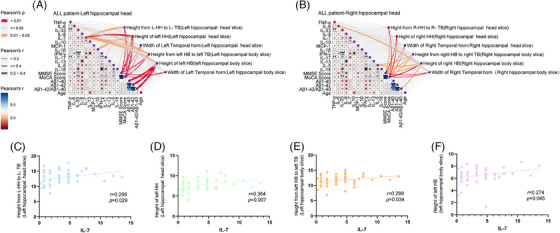
Correlation of IL‐7 with hippocampal atrophy. A, Correlation analysis illustrating the relationship between 13 inflammatory cytokines and the degree of hippocampal atrophy in the left HH slices. B, Correlation analysis shows the association between 13 inflammatory cytokines and hippocampal atrophy in the right HH slices. C, A significant correlation was observed between IL‐7 levels and the height measurement from the L‐HH to the L‐TB in the L‐HH slice. D, A significant correlation was observed between IL‐7 levels and the height measurement of the L‐HH in the L‐HH slice. E, Strong correlation between IL‐7 levels and the height measurement from the L‐HB to the L‐TB in the L‐HB slice. F, Noteworthy correlation between IL‐7 levels and the height measurement of the L‐HB in the L‐HB slice. HB, hippocampal body; HH, hippocampal head; IL, interleukin; L‐HB, left hippocampal body; L‐HH, left hippocampal head; L‐TB, left temporal bottom
Subsequently, a linear correlation analysis was conducted to compare APOE ε4 carriers and non‐carriers using the linear regression method. Among non‐carriers, no significant correlation was identified between the IL‐7 level and the height of both the left and right hippocampus (Figure S1A,B). However, among APOE ε4 carriers, a noteworthy correlation was observed between the IL‐7 level and the height of the left hippocampal head (Figure S1C, hippocampal head to temporal bottom: R 2 = 0.190; 95% CI, 0.072 to 0.729; P = 0.019 and hippocampal head: R 2 = 0.344; 95% CI, 0.149 to 0.532; P = 0.001). Notably, this correlation was not observed in relation to the height of the right hippocampal head (Figure S1D). No significant correlation was found between the width of the bilateral temporal horns and IL‐7 levels in either the APOE ε4 carrier or non‐carrier groups (Figure S1A–D). These results suggest that a clear correlation exists between lower plasma IL‐7 levels and more severe hippocampal atrophy. Notably, this trend was not observed in relation to other inflammatory cytokines.
In addition, the correlations between the levels of 13 inflammatory cytokines and plasma Aβ1‐40, Aβ1‐42, and Aβ1‐42/Aβ were analyzed. As a result, the level of each of the inflammatory cytokines was found to significantly correlate with the levels of Aβ1‐40, Aβ1‐42, and Aβ1‐42/Aβ, as shown in Table S2 in supporting information.
3.4. Plasma IL‐7 levels are associated with the number of APOE ε4 allele carriers and number of lacunar cerebral infarction area
To investigate the factors influencing plasma IL‐7 levels, the Pearson/Spearman correlation method was used to analyze the relationship between plasma IL‐7 and various factors including age, sex, number of APOE ε4 alleles, hypertension, diabetes, arthritis, number of lacunar cerebral infarction areas, coronary heart disease, cancer, MMSE score, MoCA score, Aβ1‐40, Aβ1‐42, and Aβ1‐42/1‐40. The results revealed significant correlations between IL‐7 levels and sex (r = −0.38; P = 0.12), number of APOE ε4 alleles (r = −0.301; P = 0.027), number of lacunar cerebral infarction areas (r = 0.516; P < 0.001), and MTA score (r = −0.306; P = 0.024). Subsequently, linear correlation analysis with IL‐7 as the dependent variable and sex, number of APOE ε4 alleles, number of lacunar cerebral infarction areas, and MTA score as independent variables showed a linear correlation between the number of APOE ε4 alleles (β = −0.309, P = 0.008) and the number of lacunar cerebral infarction areas (β = 0.514, P < 0.001) and IL‐7 levels.
3.5. Bulk RNA‐seq analysis revealed downregulation of IL7R and CD28 genes, which are associated with the biological process of enrichment in lymphocyte activation
In this study, bulk RNA‐seq analysis was conducted to explore the gene expression profiles of PBMCs in AD patients with different APOE genotypes using rigorous criteria (|log2fc| > 1 and FDR < 0.05). Our findings shed light on the alterations in transcriptomic profiles associated with distinct APOE genotypes (Figure 3A). The analysis identified DEGs, among which, FLJ42393, ULBP2, LOC105378721, CNBD2, PALM2AKAP2, and PRSS57 were upregulated, while IL7R, CD28, GCNT4, and USP32P1 were downregulated (Figure 3B,C,D). In our attempt to elucidate the biological relevance of these DEGs, pathway analysis using Metascape unveiled a significant enrichment of immune activation‐related biological processes such as “lymphocyte activation,” “leukocyte activation,” and “cell activation” (Figure 3E). This suggests that the downregulation of IL7R and CD28 genes may play a pivotal role in regulating lymphocyte activation in AD patients with distinct APOE genotypes. To validate this finding, human SY5Y cells expressing APOE ε4 and APOE ε3 were used (Figure S2A in supporting information). The cell experiment corroborated the results obtained by quantitative polymerase chain reaction, demonstrating that APOE ε4 significantly reduced the expression levels of IL7R and CD28 (Figure S2B). This indicates that the downregulation of IL7R and CD28 may be attributed to the presence of the APOE ε4 allele, rather than disease status, thereby explaining differences in APOE ε4 carriers. Further details regarding methods and results are available in the supplementary materials.
FIGURE 3.
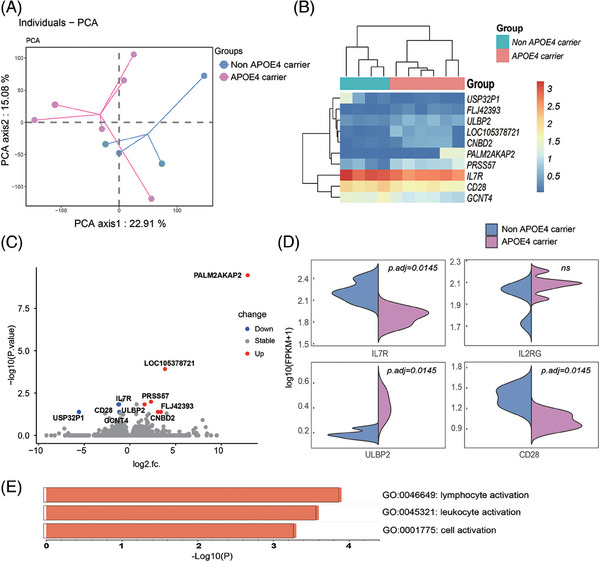
Transcriptomic analysis of PBMCs in AD patients with different APOE genotypes. A, PCA reveals distinct gene expression profiles among PBMCs from AD patients with different APOE genotypes, emphasizing the presence of unique transcriptomic profiles associated with varying APOE genotypes. B, A heatmap visualizes the DEGs that distinguish between different APOE genotypes, providing a comprehensive overview of gene expression variations. C, Volcano plots present the DEGs between different APOE genotypes, highlighting specific genes with significant differential expression. D, Bar plots depict the results of Gene Ontology term enrichment analysis for biological processes associated with the DEGs between different APOE genotypes. Notably, terms related to “lymphocyte activation” are among the enriched processes. AD, Alzheimer's disease; APOE, apolipoprotein E; DEGs, differentially expressed genes; PBMCs, peripheral blood mononuclear cells; PCA, principal component analysis
3.6. Single‐cell RNA‐sequencing showed that the downregulated genes of T cells were enriched in the IL‐7 signaling pathway
In this study, we used scRNA‐seq to explore the gene expression profiles of T cells, specifically focusing on the downregulated genes in association with the IL‐7 signaling pathway. The analysis began with the pooling of 13,850 PBMCs from 10 AD patients, including both non‐APOE ε4 carriers (n = 4) and APOE ε4 carriers (n = 6). The detailed analytical process is outlined in Figure S3A–C in supporting information. To ensure the robustness of the analysis, linear dimensionality reduction was performed on the single‐cell data, systematically evaluating the percentage of information represented by each principal component and identifying the principal components before the inflection point for subsequent analysis (Figure S3D). Subsequently, the top 20 principal components were selected for conducting clustering analysis (Figure S3E). Using the Seurat package, the FindNeighbors and FindClusters functions were used to perform unsupervised clustering analysis on the 13,850 cells, setting a resolution parameter to 1, effectively clustering the cells into 44 distinct categories. The UMAP technique was applied to visualize these clusters, as depicted in Figure S4A,B in supporting information. For further characterization of cell subtypes, marker genes specific to each cell type were identified within our subgroups. Using the CellMarker database, which consolidates extensive information from > 100,000 published papers, encompassing cell markers, tissue types, cell types, cancer data, and their respective sources. Our analysis resulted in the compilation of 13,605 cell markers for 467 cell types across 158 human tissues/subtissues and 9148 cell markers for 389 cell types across 81 mouse tissues/subtissues. These markers were cross‐referenced with relevant literature to generate comprehensive bubble charts (Figure S4C).
In the final phase of our analysis, the identified clusters were annotated based on the results obtained from the bubble chart (Figure S5A‐D in supporting information). This comprehensive annotation revealed the presence of four prominent cell types, namely B cells (n = 620), macrophages (n = 1073), T cells (n = 9412), and natural killer (NK) cells (n = 2745), among others, as illustrated in Figure S5C.
Building upon these annotation results, our next step was to examine the expression pattern of the IL7R gene across the aforementioned cell types, as presented in Figure 4A. Notably, we observed that the expression of IL‐7R in T cells was significantly higher compared to other cell types, indicating the potential significance of T cells in our study. However, upon analyzing the expression distribution of IL‐7R among different types of T cells, no significant difference was discerned between APOE ε4 carriers and non‐carriers, as shown in Figure 4B.
FIGURE 4.
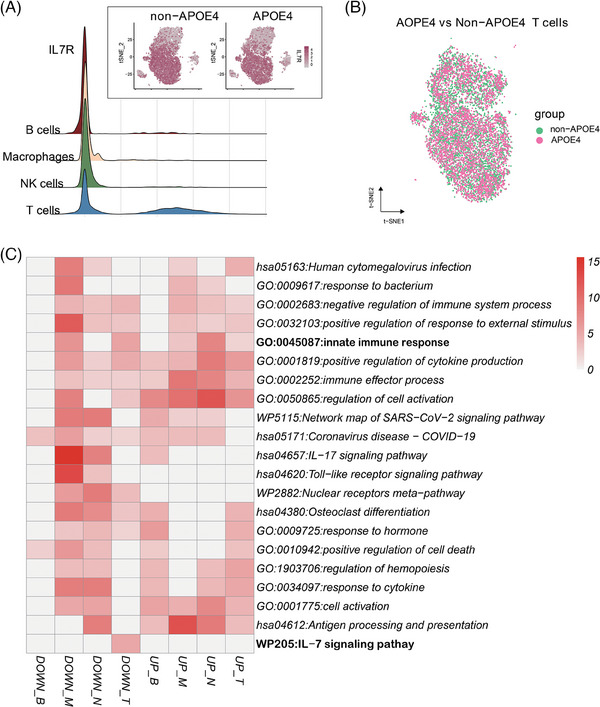
Expression of IL‐7R in various PBMCs and its relationship to APOE genotypes. A, This panel visualizes the expression levels of IL‐7R in different PBMC types, providing an overview of IL‐7R expression across distinct cell populations. B, Distribution analysis reveals the prevalence of T cells among different experimental groups, shedding light on variations in T cell populations associated with APOE genotypes. C, A heatmap displays the DEGs identified between individuals with different APOE genotypes within various PBMC populations. This heatmap helps elucidate the specific gene expression differences associated with APOE genotypes within distinct cell types. APOE, apolipoprotein E; DEGs, differentially expressed genes; IL, interleukin; NK, natural killer; PBMC, peripheral blood mononuclear cell
UMAP analysis was used to visualize the cells in two‐dimensional space and SingleR was used to annotate the cell type. Four cell types were identified: T cell, B cell, NK cell, and macrophages. DEGs for each immune cell type were screened with |log2fc| > 0.25 and FDR < 0.05 between AD patients having different APOE genotypes. Compared to the non‐carriers, the numbers of upregulated genes identified for each cell type of APOE ε4 carriers were: 33 for T cells, 32 for NK cells, 30 for monocytes, and 24 for B cells; whereas the numbers of downregulated genes identified for each cell type were: 40 for T cells, 35 for monocytes, 28 for NK cells, and 12 for B cells. Metascape unveiled a significant enrichment of immune activation–related biological processes such as “innate immune response,” “positive regulation of cytokine production,” “immune effector process,” and “antigen processing and presentation” (Figure 4C). Notably, IL‐7 signaling pathway downregulation was only seen in T cells. These results indicated the direction of the IL‐7 related research would focus on T populations in PBMCs.
3.7. Single cell RNA sequencing analysis of the subsets of T cell subsets in AD
A comprehensive investigation was conducted on T cell populations in AD using scRNA‐seq. To unravel the intricacies within this subset, dimensionality reduction clustering was conducted, with the results elegantly depicted in Figure S6A in supporting information. Our analysis was on delineating the subset of T cells, recognized as pivotal components in this context, encompassing a total of 9412 cells, including 5060 T cells from APOE ε4 carriers and 4352 T cells from non‐carriers (Figure S6B,C). Subsequently, the clustering results were meticulously annotated, drawing insights from a bubble chart (Figure S6D). The identified key subpopulations within T cells comprised CD4+Naive T cells (n = 4591), CD4+Tem cells (n = 1379), CD4+Treg cells (n = 243), CD8+Naive T cells (n = 173), CD4+Tem cells (n = 1070), and CD4+CD8+T cells (n = 1956). Distinguishing among these subpopulations was paramount for unraveling the complexity of T cell responses in AD. APOE ε4 carriers exhibited a lower proportion of CD8+ Tem T cells. This observation suggests a potential link between APOE genotype and specific T cell subpopulations, shedding new light on the pathophysiology of AD.
To gain deeper insights into the molecular landscape of T cell subsets in AD, the differences in gene expression patterns between APOE ε4 carriers and non‐carriers were meticulously scrutinized. The focused analysis centered on genes associated with the IL‐7R‐related pathways, comprising pivotal players such as IL7R, CD28, TREM1, TREM2, AP‐1 (JUN), STAT5A, KLF4, and EGR1. The results, depicted in Figure 5, unearthed intriguing disparities between these two groups. Notably, APOE ε4 carriers exhibited lower expression levels of critical genes within the IL‐7R pathway, including IL7R, CD28, and JUN, in their T cells. This finding pointed to potential regulatory mechanisms specific to APOE ε4 carriers that modulate the expression of these genes. A deeper dive into specific T cell subpopulations revealed distinctive patterns: CD4+ Tem T cells displayed concurrent downregulation of CD28 and JUN, while CD8+ Tem T cells exhibited simultaneous reductions in IL7R and JUN expression. These intricate variations underscore the multifaceted role of T cell subsets in AD and may provide crucial clues to the disease's pathogenesis.
FIGURE 5.
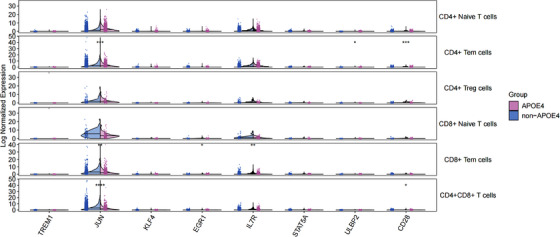
Expression of IL7R and associated pathway genes in key cell subpopulations of APOE ε4 carriers and non‐carriers. This figure provides insight into the expression profiles of IL7R and relevant pathway genes within critical cell subpopulations of both APOE ε4 carriers and non‐carriers. It offers a comparative view of how these genes are expressed in distinct cell clusters associated with different APOE genotypes, highlighting potential differences in gene expression patterns. APOE, apolipoprotein E
Pseudo‐temporal analysis of single cells is a powerful technique used to model gene expression data in a time‐series fashion, shedding light on the dynamic processes of gene expression changes within individual cells. Our pseudo‐temporal analysis unraveled that CD4+ naive cells exhibited a progression toward a CD4+ Tem phenotype, eventually culminating in a CD4+ Treg state (Figure S7A in supporting information). The trend in the differentiation of T cells remained consistent across both APOE ε4 and non‐APOE ε4 groups (Figure S7B). Intriguingly, the expression level of IL‐7R exhibited a gradual increase over time, signifying its role in this differentiation process (Figure S7C). Remarkably, this trend in IL‐7R expression remained consistent across both APOE ε4 and non‐APOE ε4 groups (Figure S7D). This pseudo‐temporal analysis demonstrated the correlation between IL‐7 expression and T cell differentiation.
To elucidate the genetic underpinnings of these PBMC subsets, the DEGs specific to each subset were integrated using a PPI network in Metascape. The metascape analysis also revealed the IL‐7 signaling pathway was enriched in the downregulated genes of T cells (Figure S8 in supporting information). These changes from T cells are mainly enriched in critical biological processes, including “cytokine signaling in the immune system,” “viral myocarditis,” and “antigen processing and presentation” (Figure S9A in supporting information). Within the network, two significant modules comprising 27 key genes that play pivotal roles in T cell differentiation and function were identified. These key genes were derived from CD4+ naive T cells, CD8 effector T cells, and CD8 memory T cells. Functional and pathway enrichment analysis of these key genes highlighted the involvement of “antigen processing and presentation,” “cytokine signaling in the immune system,” and the “IL‐7 signaling pathway” in Module 1 (Figure S9B,C). “IL‐7 signaling pathway,” “IL‐2 signaling pathway” were enriched in the downregulated genes of T cells. The IL‐7−(IL‐7R+IL2RG) interaction played a central role in shaping the IL‐2 family signaling pathway network in both non‐APOE ε4 and APOE ε4 carriers (Figure S9D,E). This comprehensive analysis sheds light on the intricate landscape of T cell subpopulations, providing a foundation for understanding their functional roles and interactions in the context of different APOE genotypes.
4. DISCUSSION
Numerous studies have demonstrated the influence of the APOE ε4 allele on neuroinflammation, particularly regarding proinflammatory and anti‐inflammatory processes. Our study also investigated cytokine profiles, revealing distinct patterns in TNF‐α, IFN‐γ, IL‐33, and IL‐7 expression levels between APOE ε4 carriers and non‐carriers. Correlation analysis with hippocampal atrophy indicated a significantly negative relationship only with IL‐7. Both bulk RNA‐seq profiling and scRNA‐seq of PBMCs also revealed significant downregulation of IL‐7R pathways (IL7R, CD28, JUN1) in PBMCs, particularly associated with CD4+Tem and CD8+Tem T cells. Our research underscores the importance of the IL‐7/IL‐7R pathway in driving inflammation and hippocampal atrophy in individuals with the APOE ε4 allele.
IL‐7 is primarily produced by bone marrow and thymic stromal cells, 26 , 33 but is influenced by multiple factors. For instance, previous studies have shown that plasma IL‐7 levels may vary based on sex, with different relationships observed between IL‐7 levels and depression scores in female and male AD patients. 34 Age is another factor, with IL‐7 levels decreasing with age in APOE ε4–carrying FAD patients but not in non‐carriers, suggesting a combined effect of age and APOE ε4 on IL‐7 levels. 16 Furthermore, studies have indicated a positive relationship between IL‐7 levels and Toll‐like receptor 4 (TLR4), which is associated with the nuclear factor kappa‐light‐chain‐enhancer of activated B cells intracellular signaling pathway and is severely decreased in AD patients. 35 Our study revealed that plasma IL‐7 levels were influenced by the number of APOE alleles and the number of lacunar cerebral infarction sites, with APOE ε4 alleles exerting a negative effect and lacunar cerebral infarction sites exerting a positive effect. Collectively, these findings indicate that plasma IL‐7 is affected by multiple factors, with the APOE ε4 allele and aging being significant influences.
The role of IL‐7 in inflammation remains a subject of debate. Some studies suggest that it acts as a pro‐inflammatory cytokine in diseases such as viral infections and autoimmune disorders. Elevated IL‐7 levels as associated with increased T cell proliferation and enhanced virus‐clearing abilities, while lower levels may weaken immune function. 36 However, IL‐7/IL‐7R also plays a significant role in the anti‐inflammatory process. 37 , 38 For instance, glucocorticoid (GC) exerts immune‐suppressive effects by binding to GC receptors with GC‐response elements in the N‐terminal region 37 regulating the transcription of target genes. IL‐7 can counteract cell death by GCs, rendering cells more responsive to IL‐7 after exposure to GCs. In addition, IL‐7 has been reported to influence immune balance by promoting the expansion and function of CD4+CD28‐ and CD8+CD28‐ cytotoxic T cells through the Janus kinase pathway. 39 Therefore, IL‐7 likely plays a pivotal role in maintaining immune homeostasis by modulating both pro‐inflammatory and anti‐inflammatory responses.
The effect of the APOE ε4 allele on immune cells may indeed be mediated through the IL‐7/IL‐7R signaling pathway. Studies have reported that individuals expressing the APOE ε4 isoform (ε4/ε3‐ε4/ε4) exhibit increased circulating levels of activated T cells compared to those expressing wild‐type apoE3 (ε3/ε3) or apoE2 isoform (ε2/ε3‐ε2/ε2). 11 However, APOE ε4–carrying AD patients demonstrate higher rates of CD4+ T cell apoptosis and a more rapid decline in CD4+ cell counts compared to non‐carriers. 13 Moreover, when stimulated with various Toll‐like receptor (TLR) ligands, APOE ε4–positive cultures produce significantly higher levels of cytokines involved in the standard TLR‐triggered cascades than apoE3 cultures. 40 In elder controls, plasma IL‐7 levels have a positive correlation with TLR levels, but this correlation is lost in AD patients. 35 IL‐7, essential for lymphocyte development and maintenance, 26 , 41 has been shown to undergo expression changes consistent with CD4+naive T cells transitioning into CD4+ Tem and eventually progressing into CD4+Treg (regulatory T) cells, as evidenced by our scRNA‐seq pseudo‐temporal analysis. The lower levels of plasma IL‐7 observed in APOE ε4–carrying AD patients may hinder CD4+ T cell proliferation. A recent meta‐analysis has reported a decline in CD8+ cells in AD patients compared to healthy individuals. 42 , 43 Similar to CD4 T cells, the changes in CD8 cells are closely related to IL‐7 signaling. The lower expression of IL‐7R in memory CD8+ T cells is also considered a signature of aging cells and is associated with neurocognitive functioning in AD. 44 These findings collectively suggest that the impact of the APOE ε4 allele on neuroinflammation is likely regulated through the IL‐7/IL‐7R signaling pathway.
Our results indicated that, compared to non‐carriers, APOE ε4 has varying effects on the IL‐7R signaling pathway in CD4+ Tem and CD8+ Tem cells. The CD28, JUN1, and ULBP2 expression decreases in CD4+ Tem cells, while IL‐7R and JUN1 levels in CD8+ Tem cells decrease in APOE ε4 carriers. The AP‐1 pathway is crucial for the activation of NK cells, T cells, and phagocytes, particularly in T cells in the process of antigen presentation. 45 The binding of apoE to its receptors triggers dual leucine zipper kinase activation, leading to the activation of extracellular signal‐related kinase 1/2, ultimately increasing Aβ levels through the AP‐1‐mediated transcription of amyloid precursor protein. 5 The activation processes of ε2, ε3, and ε4 isoforms are different in this process, thus exerting different effects on the IL‐7R/AP‐1 pathway. 5 Recent studies have highlighted the association between the survival of memory T cells and the IL‐7–mediated fatty acid esterification and triglyceride (TG) metabolism. Noteworthily, APOE ε4 has been suggested to impair intracellular lipid homeostasis, thereby compromising the regeneration of memory T cells. 9 Additionally, CD28, one of the key costimulatory molecules of T cells, is recruited to the T cell receptor (TCR) lipid raft region for cytokine signal transduction. 46 Lipid metabolism is also vital for the proper functioning of CD28. However, APOE ε4 may disrupt intracellular lipid homeostasis by increasing the unsaturation of fatty acids and the accumulation of intracellular lipid droplets. 9 Furthermore, it is postulated that APOE ε4 may enhance lipid raft formation, 47 primarily because of its increased affinity for the VLDL and TGs. This propensity could potentially render a higher susceptibility to cell death caused by the accumulation of excessive lipids. As a consequence, it is hypothesized that APOE ε4 could accelerate the aging process and contribute to the degeneration of memory T cells. The data from this study unequivocally demonstrate that both the downregulation of IL‐7 signaling and the reduction in CD28 expression are indicative of immune system dysfunction in APOE ε4 carriers. Additionally, the observed downregulations of IL7 and IL7R suggest the age‐related decline and deterioration of the immune function. Therefore, it is postulated that these changes occur during aging, which mirrors other aging processes observed in individuals with AD.
Furthermore, our study found increased expression of PRSS57 and PALM2AKAP2 in APOE ε4 carriers. PRSS57 has been linked to regulatory properties in inflammation and innate immune reactions, 48 with higher levels associated with multiple sclerosis and CD34+ cell apoptosis. 48 The increased expression of PRSS57 may suggest a potential role in APOE ε4's regulation of neuroinflammatory response. PALM2AKAP2, on the other hand, is related to plasma membrane dynamics and has functional links in unusual manners. 49 Its prenylated form has been reported to enhance cancer cell migration by activating ezrin. 50 However, the implications of PRSS57 and PALM2AKAP2 for the pathogenesis of AD and their involvement in the regulation of neuroinflammation by APOE ε4 remain unclear, warranting further research to confirm their functions.
While the relationship between the APOE ε4 allele and plasma IL‐7 levels is evident, its precise role in AD pathogenesis remains elusive. Previous studies have reported lower levels of plasma IL‐7 in dementia patients, with a negative correlation observed with neuropsychiatric symptoms. 15 , 34 , 51 Conversely, other studies have found a negative relationship between plasma IL‐7 levels and daily living abilities. 52 Moreover, the present study did not find a clear correlation between the plasma IL‐7 levels and Aβ1‐42 or Aβ1‐42/Aβ1‐40, which is consistent with previous findings. 14
In our study, a positive correlation was observed between the plasma IL‐7 levels and the degree of left hippocampal atrophy, which has implications for working memory and cognitive functions. We speculate that hippocampal atrophy may result from disorders in neuronal regeneration and maturation due to lower expression of IL‐7/IL‐7R. While IL‐7 is known for its role in immune cell development, its involvement in neuronal regeneration is not well understood and lacks robust research support. However, limited studies suggest that IL‐7 may play a role in the differentiation of neurons and the development of glial cells in neural precursor cells within the brain. 27 , 28 IL‐7 has been detected in both embryonic and adult mouse brains and has been shown to have neuronotrophic effects on primary hippocampal cultures. 53 Additionally, IL‐7, basic fibroblast growth factor (bFGF), and transforming growth factor (TGF) work together to promote neuronal maturation, with Cx33 and Cx40 serving as important markers during this process. IL‐7 alone induces Cx40 expression, while IL‐7 in conjunction with bFGF and TGF induces Cx33 expression in progenitor cells. 29 These findings suggest that IL‐7 may have additional roles beyond immune regulation, such as in neuronal regeneration and maturation, but further experiments are required to confirm this hypothesis.
Many scholars believe that aging of the immune system may accelerate the progression of pathological changes of AD. 54 , 55 This might be related to changes in the IL‐7/IL‐7R pathway. Research on mice has shown that IL‐7 expression declines with age, contributing to thymic atrophy, 56 and affecting TCR cells. 57 Similarly, thymic involution in humans occurs progressively throughout the first three decades of life 58 and is responsible for the aging of immune cells. 58 , 59 , 60 The aging of human immune cells in older adults is also closely related to the expression of IL‐7R. 61 A decline in plasma IL‐7 levels with age has been observed in AD patients, with a more pronounced decline seen in carriers of the APOE ε4 allele compared to non‐carriers. 16 This evidence suggests that the gradual decrease in plasma IL‐7 levels is a consistent and ongoing process, accelerated in APOE ε4 carriers. Therefore, the influence of the APOE ε4 allele on the IL‐7 signaling pathway may be a causal factor, rather than a consequence, of pathological damage in AD.
In summary, the present study provides compelling evidence that the APOE ε4 allele affects the immune system through the IL‐7 signaling pathway, potentially contributing to the hippocampal atrophy in AD. IL‐7, a key immune cell growth factor, has been increasingly recognized as a valuable tool in immunotherapy. Our research aimed to highlight the importance of IL‐7 in AD and raise awareness among neurologists.
AUTHOR CONTRIBUTIONS
Nai‐Li Wei and Ying‐Jie Zhang: conception, supervision, and design of this article; Ying‐Jie Zhang, writing the manuscript and scRNA‐seq data analysis; Nai‐Li Wei: supervision, funding, and editing the manuscript; Ren‐Hua Wu: supervision and funding in neuroimage; Yan Cheng and Hai‐Liang Tang: MRI scanning, data analysis, and proofreading manuscript; Hai‐Liang Tang, Qi Yue, Xin‐Yi Cai, and Chang‐Hao Lu: PBMC procedure, and bulk RNA‐seq and scRNA‐seq data analysis; Ying‐Jie Zhang, Xin‐Yi Cai, Zhi‐Jie Lu, Ting Hou, An‐Xiang Dai, and Hao‐Xin Liu: clinical data analysis; Sheng‐Liang Xu: clinical evaluation; Kai Huang, Ya‐Qi Wen, Wan‐Yin Ma, Ji‐Tian Guan, Yan Lin, and Wen‐Bin Zheng: MRI scanning; Xin Zeng and Hui Pan: patient diagnosis and evaluation; Yi‐Xuan Hao and Nan Kong: cytological experiments and qPCR. All authors in the article have approved the submitted version of this manuscript.
CONFLICT OF INTEREST STATEMENT
The authors have no relevant financial or non‐financial interests to disclose. Author disclosures are available in the supporting information.
CONSENT STATEMENT
This study was approved by the Clinical Research Ethics Committee of the First Affiliated Hospital of Shantou University Medical College (approval number: 2020‐115‐XZ2). All subjects provided written consent or assent with proxy consent prior to enrollment in accordance with the latest Declaration of Helsinki. This manuscript is in accordance with the authorship statement of ethical standards for manuscripts submitted to this journal.
Supporting information
Supporting Information
Supporting Information
ACKNOWLEDGMENTS
We would like to thank all colleagues of the Department of Neurosurgery in our hospital (Jian‐Guo Xiao, Lie‐Peng Xu, Ji Wu, Xin‐Yuan Peng, Kai‐Xun Lin, Zi‐Dong Zheng, Man‐Qi Chen, Yan‐Rou Li, Rong‐Er Zhou, Xiao‐Fang Ying). They also contributed to the evaluation of the patients. This work was supported by grants from the National Natural Science Foundation of China (82001447; 82020108016), the Natural Science Foundation of Guangdong Province (2214050005144), special funds for Science and Technology of Guangdong Province (210728166901906; STKJ2021075), Guangdong Basic and Applied Basic Research Foundation (2023B1515230008), Dengfeng Project for the construction of high‐level hospitals in Guangdong Province—the First Affiliated Hospital of Shantou University Medical College Supporting Funding (202003‐29), and 2020 Li Ka Shing Foundation Cross‐Disciplinary Research Grant (2020LKSFG11C).
Zhang Y‐J, Cheng Y, Tang H‐L, et al. APOE ε4–associated downregulation of the IL‐7/IL‐7R pathway in effector memory T cells: Implications for Alzheimer's disease. Alzheimer's Dement. 2024;20:6441–6455. 10.1002/alz.14173
Ying‐Jie Zhang, Yan Cheng, Hai‐Liang Tang, Qi Yue, and Xin‐Yi Ca are co‐first authors.
Contributor Information
Ren‐Hua Wu, Email: rhwu@stu.edu.cn.
Nai‐Li Wei, Email: nlwei@stu.edu.cn.
DATA AVAILABILITY STATEMENT
All data presented in this paper are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Koutsodendris N, Nelson MR, Rao A, Huang Y. Apolipoprotein E and Alzheimer's disease: findings, hypotheses, and potential mechanisms. Annu Rev Pathol. 2022;17:73‐99. [DOI] [PubMed] [Google Scholar]
- 2. Crane A, Brubaker WD, Johansson JU, et al. Peripheral complement interactions with amyloid beta peptide in Alzheimer's disease: 2. Relationship to amyloid beta immunotherapy. Alzheimers Dement. 2018;14(2):243‐252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yamazaki Y, Zhao N, Caulfield TR, Liu CC, Bu G. Apolipoprotein E and Alzheimer disease: pathobiology and targeting strategies. Nat Rev Neurol. 2019;15(9):501‐518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xiang Y, Bu XL, Liu YH, et al. Physiological amyloid‐beta clearance in the periphery and its therapeutic potential for Alzheimer's disease. Acta Neuropathol. 2015;130(4):487‐499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang YA, Zhou B, Wernig M, Sudhof TC. ApoE2, ApoE3, and ApoE4 differentially stimulate APP transcription and Aβ secretion. Cell. 2017;168(3):427‐441.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Atehortua L, Morris J, Street SE, Bedel N, Davidson WS, Chougnet CA. Apolipoprotein E‐containing HDL decreases caspase‐dependent apoptosis of memory regulatory T lymphocytes. J Lipid Res. 2023;64(9):100425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ostendorf BN, Bilanovic J, Adaku N, et al. Common germline variants of the human APOE gene modulate melanoma progression and survival. Nat Med. 2020;26(7):1048‐1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iannucci J, Sen A, Grammas P. Isoform‐specific effects of apolipoprotein E on markers of inflammation and toxicity in brain glia and neuronal cells in vitro. Curr Issues Mol Biol. 2021;43(1):215‐225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sienski G, Narayan P, Bonner JM, et al. APOE4 disrupts intracellular lipid homeostasis in human iPSC‐derived glia. Sci Transl Med. 2021;13(583):eaaz4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu C‐C, Wang N, Chen Y, et al. Cell‐autonomous effects of APOE4 in restricting microglial response in brain homeostasis and Alzheimer's disease. Nat Immunol. 2023;24(11):1854‐1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bonacina F, Coe D, Wang G, et al. Myeloid apolipoprotein E controls dendritic cell antigen presentation and T cell activation. Nat Commun. 2018;9(1):3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zilber M‐T, Setterblad N, Vasselon T, et al. MHC class II/CD38/CD9: a lipid‐raft‐dependent signaling complex in human monocytes. Blood. 2005;106(9):3074‐3081. [DOI] [PubMed] [Google Scholar]
- 13. Frey C, Bonert A, Kratzsch T, et al. Apolipoprotein E epsilon 4 is associated with an increased vulnerability to cell death in Alzheimer's disease. J Neural Transm (Vienna). 2006;113(11):1753‐1761. [DOI] [PubMed] [Google Scholar]
- 14. Aksnes M, Aass HCD, Tiiman A, et al. Associations of cerebrospinal fluid amyloidogenic nanoplaques with cytokines in Alzheimer's disease. Transl Neurodegener. 2021;10(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen A, Oakley AE, Monteiro M, et al. Multiplex analyte assays to characterize different dementias: brain inflammatory cytokines in poststroke and other dementias. Neurobiol Aging. 2016;38:56‐67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ringman JM, Elashoff D, Geschwind DH, et al. Plasma signaling proteins in persons at genetic risk for Alzheimer disease: influence of APOE genotype. Arch Neurol. 2012;69(6):757‐764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Albrecht DS, Sagare A, Pachicano M, et al. Early neuroinflammation is associated with lower amyloid and tau levels in cognitively normal older adults. Brain Behav Immun. 2021;94:299‐307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koutsos A, Jackson KG, Lockyer S, Carvalho‐Wells A, Minihane AM, Lovegrove JA. Greater impact of dietary fat manipulation than apolipoprotein E genotype on ex vivo cytokine production—insights from the SATgenε study. Cytokine. 2014;66(2):156‐159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Motta C, Finardi A, Toniolo S, et al. Protective role of cerebrospinal fluid inflammatory cytokines in patients with amnestic mild cognitive impairment and early Alzheimer's disease carrying apolipoprotein E4 genotype. J Alzheimers Dis. 2020;76(2):681‐689. [DOI] [PubMed] [Google Scholar]
- 20. Beyer K, Lao JI, Gómez M, et al. Identification of a protective allele against Alzheimer disease in the APOE gene promoter. Neuroreport. 2002;13(11):1403‐1405. [DOI] [PubMed] [Google Scholar]
- 21. Adrian M, Weber M, Tsai M‐C, et al. Polarized microtubule remodeling transforms the morphology of reactive microglia and drives cytokine release. Nat Commun. 2023;14(1):6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Werneburg S, Feinberg PA, Johnson KM, Schafer DP. A microglia‐cytokine axis to modulate synaptic connectivity and function. Curr Opin Neurobiol. 2017;47:138‐145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Trombetta BA, Carlyle BC, Koenig AM, et al. The technical reliability and biotemporal stability of cerebrospinal fluid biomarkers for profiling multiple pathophysiologies in Alzheimer's disease. PloS One. 2018;13(3):e0193707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lau S‐F, Chen C, Fu W‐Y, et al. IL‐33‐PU.1 transcriptome reprogramming drives functional state transition and clearance activity of microglia in Alzheimer's disease. Cell Reports. 2020;31(3):107530. [DOI] [PubMed] [Google Scholar]
- 25. Barata JT, Durum SK, Seddon B. Flip the coin: IL‐7 and IL‐7R in health and disease. Nat Immunol. 2019;20(12):1584‐1593. [DOI] [PubMed] [Google Scholar]
- 26. Bradley LM, Haynes L, Swain SL. IL‐7: maintaining T‐cell memory and achieving homeostasis. Trends Immunol. 2005;26(3):172‐176. [DOI] [PubMed] [Google Scholar]
- 27. Moors M, Vudattu NK, Abel J, et al. Interleukin‐7 (IL‐7) and IL‐7 splice variants affect differentiation of human neural progenitor cells. Genes Immun. 2010;11(1):11‐20. [DOI] [PubMed] [Google Scholar]
- 28. Mehler MF, Rozental R, Dougherty M, Spray DC, Kessler JA. Cytokine regulation of neuronal differentiation of hippocampal progenitor cells. Nature. 1993;362(6415):62‐65. [DOI] [PubMed] [Google Scholar]
- 29. Rozental R, Morales M, Mehler MF, et al. Changes in the properties of gap junctions during neuronal differentiation of hippocampal progenitor cells. J Neurosci. 1998;18(5):1753‐1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pievani M, Galluzzi S, Thompson PM, Rasser PE, Bonetti M, Frisoni GB. APOE4 is associated with greater atrophy of the hippocampal formation in Alzheimer's disease. NeuroImage. 2011;55(3):909‐919. [DOI] [PubMed] [Google Scholar]
- 31. Wei N, Chen J. Repetitive transcranial magnetic stimulation for Alzheimer's disease based on apolipoprotein E genotyping: protocol for a randomized controlled study. Front Aging Neurosci. 2021;13:758765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scheltens P, Leys D, Barkhof F, et al. Atrophy of medial temporal lobes on MRI in “probable” Alzheimer's disease and normal ageing: diagnostic value and neuropsychological correlates. J Neurol Neurosurg Psychiatry. 1992;55(10):967‐972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang H‐Y, Luther SA. Expression and function of interleukin‐7 in secondary and tertiary lymphoid organs. Semin Immunol. 2012;24(3):175‐189. [DOI] [PubMed] [Google Scholar]
- 34. Hall JR, Wiechmann A, Edwards M, Johnson LA, O'Bryant SE. IL‐7 and depression: the importance of gender and blood fraction. Behav Brain Res. 2016;315:147‐149. [DOI] [PubMed] [Google Scholar]
- 35. Kilic U, Elibol B, Uysal O, et al. Specific alterations in the circulating levels of the SIRT1, TLR4, and IL7 proteins in patients with dementia. Exp Gerontol. 2018;111:203‐209. [DOI] [PubMed] [Google Scholar]
- 36. Lawson BR, Gonzalez‐Quintial R, Eleftheriadis T, et al. Interleukin‐7 is required for CD4(+) T cell activation and autoimmune neuroinflammation. Clin Immunol. 2015;161(2):260‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shimba A, Ikuta K. Glucocorticoids regulate circadian rhythm of innate and adaptive immunity. Front Immunol. 2020;11:2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Franchimont D, Galon J, Vacchio MS, et al. Positive effects of glucocorticoids on T cell function by up‐regulation of IL‐7 receptor α. J Immunol. 2002;168(5):2212‐2218. [DOI] [PubMed] [Google Scholar]
- 39. Xia CS, Long Y, Liu Y, Alifu A, Zeng X, Liu C. IL‐7 promotes the expansion of circulating CD28‐ cytotoxic T lymphocytes in patients with IgG4‐related disease via the JAK signaling. Front Immunol. 2022;13:922307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gale SC, Gao L, Mikacenic C, et al. APOε4 is associated with enhanced in vivo innate immune responses in human subjects. J Allergy Clin Immunol. 2014;134(1):127‐1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Unsinger J, McGlynn M, Kasten KR, et al. IL‐7 promotes T cell viability, trafficking, and functionality and improves survival in sepsis. J Immunol. 2010;184(7):3768‐3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hu B, Jadhav RR, Gustafson CE, et al. Distinct age‐related epigenetic signatures in CD4 and CD8 T cells. Front Immunol. 2020;11:585168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang L‐T, Zhang C‐P, Wang Y‐B, Wang J‐H. Association of peripheral blood cell profile with Alzheimer's disease: a meta‐analysis. Front Aging Neurosci. 2022;14:888946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Young JJ, Park H‐J, Kim M, et al. Aging gene signature of memory CD8+ T cells is associated with neurocognitive functioning in Alzheimer's disease. Immun Ageing. 2023;20(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hofmeister RKA, Benbernou N, Rajnavolgyi E, Muegge K, Durum SK. Interleukin‐7: physiological roles and mechanisms of action. Cytokine Growth Factor Rev. 1999;10(1):41‐60. [DOI] [PubMed] [Google Scholar]
- 46. Tavano R, Contento RL, Baranda SJ, et al. CD28 interaction with filamin‐A controls lipid raft accumulation at the T‐cell immunological synapse. Nat Cell Biol. 2006;8(11):1270‐1276. [DOI] [PubMed] [Google Scholar]
- 47. Lee SI, Jeong W, Lim H, et al. APOE4‐carrying human astrocytes oversupply cholesterol to promote neuronal lipid raft expansion and Aβ generation. Stem Cell Reports. 2021;16(9):2128‐2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. AhYoung AP, Eckard SC, Gogineni A, et al. Neutrophil serine protease 4 is required for mast cell‐dependent vascular leakage. Commun Biol. 2020;3(1):687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hu B, Copeland NG, Gilbert DJ, Jenkins NA, Kilimann MW. The paralemmin protein family: identification of paralemmin‐2, an isoform differentially spliced to AKAP2/AKAP‐KL, and of palmdelphin, a more distant cytosolic relative. Biochem Biophys Res Commun. 2001;285(5):1369‐1376. [DOI] [PubMed] [Google Scholar]
- 50. Vishnubalaji R, Alajez NM. Epigenetic regulation of triple negative breast cancer (TNBC) by TGF‐β signaling. Sci Rep. 2021;11(1):15410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hall JR, Wiechmann AR, Johnson LA, et al. Biomarkers of vascular risk, systemic inflammation, and microvascular pathology and neuropsychiatric symptoms in Alzheimer's disease. J Alzheimers Dis. 2013;35(2):363‐371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hall JR, Johnson LA, Barber RC, et al. Biomarkers of basic activities of daily living in Alzheimer's disease. J Alzheimers Dis. 2012;31(2):429‐437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Araujo DM, Cotman CW. Trophic effects of interleukin‐4, ‐7 and ‐8 on hippocampal neuronal cultures: potential involvement of glial‐derived factors. Brain Res. 1993;600(1):49‐55. [DOI] [PubMed] [Google Scholar]
- 54. Shue F, White LJ, Hendrix R, et al. CSF biomarkers of immune activation and Alzheimer's disease for predicting cognitive impairment risk in the elderly. Sci Adv. 2024;10(14):eadk3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Guo L, Li X, Gould T, Wang ZY, et al. T cell aging and Alzheimer's disease. Front Immunol. 2023;14:1154699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Aspinall R, Andrew D. Age‐associated thymic atrophy is linked to a decline in IL‐7 production. Exp Gerontol. 2002;37(2‐3):455‐463. [DOI] [PubMed] [Google Scholar]
- 57. Laky K, Lewis JM, Tigelaar RE, Puddington L. Distinct requirements for IL‐7 in development of TCR gamma delta cells during fetal and adult life. J Immunol. 2003;170(8):4087‐4094. [DOI] [PubMed] [Google Scholar]
- 58. Asnafi V, Beldjord K, Libura M, et al. Age‐related phenotypic and oncogenic differences in T‐cell acute lymphoblastic leukemias may reflect thymic atrophy. Blood. 2004;104(13):4173‐4180. [DOI] [PubMed] [Google Scholar]
- 59. Thomas R, Wang W, Su DM. Contributions of age‐related thymic involution to immunosenescence and inflammaging. Immun Ageing. 2020;17:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Elyahu Y, Monsonego A. Thymus involution sets the clock of the aging T‐cell landscape: implications for declined immunity and tissue repair. Ageing Res Rev. 2021;65:101231. [DOI] [PubMed] [Google Scholar]
- 61. Kim H‐R, Hong MS, Dan JM, Kang I. Altered IL‐7Ralpha expression with aging and the potential implications of IL‐7 therapy on CD8+ T‐cell immune responses. Blood. 2006;107(7):2855‐2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Data Availability Statement
All data presented in this paper are available from the corresponding author upon reasonable request.


