Abstract
Infections by low-risk papillomavirus types, such as human papillomavirus (HPV) type 6 (HPV-6) and HPV-11, induce benign genital warts that rarely progress to malignancy. In contrast, lesions induced by high-risk HPV types have the potential to progress to cancer. Considerable information is available concerning the pathogenesis of high-risk HPV types, but little is known about the life cycle of low-risk HPV types. Although functionally distinct, both high- and low-risk virus types infect keratinocytes and induce virion production upon differentiation. This information suggests that they may share common mechanisms for regulating their productive life cycles. Using tissue culture methods developed to study high-risk HPV types, we examined the ability of HPV-11 to be stably maintained as episomes following transfection of normal human keratinocytes with cloned viral DNA. HPV-11 genomes were found to be maintained in keratinocytes for extended passages in cultures in 14 independent experiments involving transfection of cloned HPV-11 DNA. Interestingly, the HPV-11-positive cells exhibited an extended life span that averaged approximately twofold longer than that of control neomycin-transfected cells. In organotypic cultures, HPV-11-positive cells exhibited altered differentiation patterns, but the extent of disruption was less severe than that seen with high-risk HPV types. In addition, the amplification of HPV-11 DNA, as well as the induction of several viral messages, was observed following differentiation of transfected cells in semisolid media. To determine whether global changes in cellular gene expression induced by HPV-11 were similar to those observed with high-risk HPV-31 (Y. E. Chang and L. A. Laimins, J. Virol. 74:4174–4182, 2000), microarray analysis of 7,075 expressed sequences was performed. A spectrum of cellular genes different from that previously reported for HPV-31 was found to be activated or repressed by HPV-11. The expression of only a small set of genes was similarly altered by both high- and low-risk HPV types. This result suggests that different classes of HPVs have distinct effects on global cellular transcription patterns during infection. The methods described allow for a genetic analysis of HPV-11 in the context of its differentiation-dependent life cycle.
Both high- and low-risk human papillomaviruses (HPVs) infect keratinocytes in the genital tract. The high-risk HPV types, such as HPV type 16 (HPV-16), HPV-18, and HPV-31, are the etiologic agents of cervical cancers; their oncoproteins are able to efficiently immortalize keratinocytes in tissue cultures (11, 20). The low-risk types, such as HPV-6 and HPV-11, induce benign genital warts but are unable to immortalize cells in vitro. Although functionally distinct, both virus types are able to induce virion production upon differentiation, suggesting that they share common mechanisms for modulating cell cycle control (7).
The productive life cycles of all HPV types are closely associated with the differentiation program of epithelial cells (13). Consequent to infection, viral genomes are established and replicate as episomes in basal cells coincident with cellular replication. Following replication in basal cells, HPV-positive daughter cells migrate away from the basal layer and begin to differentiate. In the suprabasal layers, HPV-positive cells are induced to enter S phase, resulting in amplification of the viral genomes, expression of late transcripts, production of capsid proteins, and assembly of progeny virions (12). Similar events are thought to occur in the productive life cycles of all papillomaviruses, although details of these processes have been studied only for the high-risk types.
Tissue culture methods have been developed to propagate high-risk types of HPVs with keratinocytes derived from biopsies or transfected with cloned HPV DNAs (2–4, 16). Keratinocytes isolated from biopsies of low-grade cervical lesions can be propagated as monolayer cultures and often become immortal. These cell lines maintain viral DNA as episomes and, upon differentiation either in organotypic cultures or by suspension in semisolid media, induce late viral functions, such as amplification of viral DNA, activation of late transcripts, and synthesis of virions (14, 21). Similar effects are seen with cell lines that maintain viral episomes generated by transfection of normal human keratinocytes with cloned high-risk HPV DNA (3, 4, 16, 21). In contrast, cells that contain only integrated copies of viral DNA fail to amplify genomes and are unable to activate late gene expression upon differentiation (3). Two major promoters for high-risk HPV-31 have been identified by these methods (8, 19). In undifferentiated cells, the majority of transcripts initiate upstream of the E6 open reading frame and terminate downstream of E5. These transcripts are polycistronic and express a variety of HPV genes as a result of alternative splicing. Upon differentiation, transcripts that encode the capsid genes are induced from a promoter in the E7 open reading frame (8, 19). The use of a promoter in the early region to activate capsid gene expression requires an additional level of regulation through the differential use of tandem polyadenylation sites (25).
The ability to duplicate the productive HPV life cycle using transfected cloned DNAs has allowed for a genetic analysis of viral functions in a physiologically relevant context (10, 23, 24, 26). These studies have been facilitated by the ability of high-risk HPV genomes to efficiently immortalize normal human keratinocytes. Despite the inability of low-risk HPV types to immortalize normal keratinocytes, we investigated whether similar methods could be used to study the life cycle of HPV-11 in a tissue culture model. We observed that cloned HPV-11 genomes could be readily established as episomes in normal keratinocytes following transfection. These HPV-11-positive keratinocyes exhibited an extended life span in monolayer cultures as well as altered patterns of differentiation in organotypic rafts. Microarray analysis of HPV-11-positive cells identified cellular genes that were activated or repressed, and many were distinct from those previously seen with high-risk HPV-31 (1). This study establishes a system for the genetic manipulation of the low-risk genome that can elucidate the differences between low- and high-risk virus types.
MATERIALS AND METHODS
Cell cultures and plasmids.
Human foreskin keratinocytes (HFKs) were derived from neonatal human foreskin epithelia as previously described (6) and were maintained in serum-free keratinocyte growth medium (Clonetics, San Diego, Calif.). HPV-11 genome transfectants and control HFKs were grown in serum-containing medium (E medium) supplemented with 5 ng of mouse epidermal growth factor (Collaborative Biomedical Products, Bedford, Mass.)/ml in the presence of mitomycin C-treated J2 3T3 fibroblast feeders kindly provided by the Howard Green laboratory. All cells were treated with 0.5 mM EDTA in phosphate-buffered saline [PBS]) to remove fibroblast feeders prior to harvesting. Plasmid pBR322.HPV11 contains the HPV-11 genome inserted into the BamHI site of pBR322, and pSV2neo encodes the neomycin drug resistance gene.
Transfection of HFKs.
Ten micrograms of the pBR322.HPV11 construct was digested with BamHI to release viral genomes. The restriction enzyme was heat inactivated, and genomes were unimolecularly ligated in the same buffer with T4 DNA ligase (10 U/900 μl). The DNA was then precipitated with isopropyl alcohol and resuspended in 10 mM Tris–1 mM EDTA (pH 7.5). The religated DNA was cotransfected with 2 μg of the selectable marker, pSV2neo, into HFKs with LipofectAce (Gibco BRL, Grand Island, N.Y.) as described by the manufacturer. At 1 day posttransfection, cells were plated onto mitomycin C-treated fibroblast feeders in E medium. Selection began with G418 (Gibco BRL) on day 2 posttransfection as follows: 200 mg of G418/ml every 2 days for a total of 4 days and then 100 mg of G418/ml every 2 days for an additional 4 days. After selection, pooled populations were expanded for analyses.
Differentiation of keratinocytes in raft cultures.
HFKs and HPV-11 transfectants were induced to differentiate in raft cultures as previously described (15). Briefly, cells were plated on a solidified collagen matrix containing J2 3T3 fibroblasts, allowed to grow to confluence, and then transferred to a metal grid, which provided an air-liquid interface for differentiation. Cultures were harvested at 14 days, fixed in 4% paraformaldehyde, paraffin embedded, sectioned, and stained with hematoxylin and eosin for visualization of differentiated raft tissue.
Differentiation of keratinocytes in semisolid media.
HFKs and HPV-11 transfectants were suspended in 1.6% methylcellulose to induce differentiation. The methylcellulose solution was prepared by adding half of the final volume of E medium containing 5% fetal bovine serum to autoclaved dry methylcellulose (4,000 cps; Sigma, St. Louis, Mo.) and heating the mixture in a 60°C water bath for 20 min. The remaining E medium containing 10% fetal bovine serum was added, and the mixture was stirred at 4°C overnight until clear. Approximately 1 × 106 to 2 × 106 keratinocytes were harvested by trypsinization, resuspended in 1 ml of E medium, and added dropwise to a 6-cm petri dish containing 15 ml of 1.6% methylcellulose. Cells were stirred with a pipette and incubated at 37°C in a humidified CO2 incubator for various times. Cells in methylcellulose were harvested by scraping into two 50-ml conical tubes, washing with PBS (50 ml) three times, combining into a 15-ml conical tube for a final PBS wash, and pelleting by centrifugation. Samples were then subjected to Southern analyses to detect HPV-11 genomic DNA and Northern analyses to examine transcripts.
Southern blot analyses.
Total genomic DNA from HPV-11 transfectants was prepared by resuspension of the cell pellet in lysis buffer (400 mM NaCl, 10 mM Tris-HCl [pH 7.4], 10 mM EDTA); the addition of RNase A (50 μg/ml), proteinase K (50 μg/ml), and sodium dodecyl sulfate (SDS) (0.2%); and incubation at 37°C overnight. DNA was sheared by passage through an 18-gauge needle approximately 10 times, extracted with phenol-chloroform, and precipitated with ethanol. Five micrograms of total genomic DNA was digested with DpnI to remove any residual input DNA. Digested DNA was separated on an 0.8% agarose gel, treated, and alkaline transferred to a DuPont GeneScreen Plus nylon membrane (NEN Research Products, Boston, Mass.) as described by the manufacturer. The membrane was prehybridized in 50% formamide–4× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])–5× Denhardt's solution–1% SDS–10% dextran sulfate–0.1 mg of denatured herring sperm DNA/ml for 1 h at 42°C. The HPV-11 probe was prepared by gel purification of the entire HPV-11 genome from pBR322.HPV11 digested with BamHI and labeling with the Ready-to-go DNA labeling kit (Amersham Pharmacia, Piscataway, N.J.). The labeled probe was purified with ProbeQuant G-50 Micro Columns (Amersham Pharmacia), denatured, added to fresh hybridization solution, and incubated with the membrane at 42°C overnight. The membrane was washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS for 15 min at room temperature, twice with 0.5× SSC–0.1% SDS for 15 min at room temperature, twice with 0.1× SSC–0.1% SDS for 15 min at room temperature, and once with 0.1× SSC–1% SDS for 30 min at 50°C. Hybridizing species were visualized by autoradiography.
Analyses of HPV-11 late transcripts.
Total RNA was isolated from methylcellulose-treated normal HFKs and HPV-11 transfectants with TRIzol reagent (Gibco BRL) as described by the manufacturer and examined by Northern analyses as follows. Ten micrograms of total RNA was separated on a 1.0% agarose–2.2 M formaldehyde gel in 1× morpholinepropanesulfonic acid (MOPS) buffer (10× MOPS buffer is 0.2 M MOPS, 50 mM sodium acetate, and 10 mM EDTA) and transferred to a Zeta-Probe membrane (Bio-Rad) as described by the manufacturer. After cross-linking, the membrane was prehybridized in 1 mM EDTA–0.5 M Na2HPO4–7% SDS for 10 min at 65°C. The HPV-11 probe was prepared by gel purification of the E4-E5 region of HPV-11 from pBR322.HPV11 digested with HindIII and BamHI and labeling with the Ready-to-go DNA labeling kit. The labeled probe was purified with ProbeQuant G-50 Micro Columns, denatured, added to fresh hybridization solution, and incubated with the membrane overnight at 65°C. The membrane was washed twice with 2× SSC–10% SDS for 5 min at room temperature and twice with 0.2× SSC–1% SDS for 30 min at 55°C. Hybridizing species were visualized by autoradiography.
Microarray analyses.
Total RNA was isolated from normal HFKs and HPV-11 transfectants with TRIzol reagent. Fibroblast feeders were removed prior to RNA isolation by treatment with 0.5 mM EDTA in PBS. Poly(A) RNA was further purified using Oligotex columns (Qiagen, Valencia, Calif.). Generation of cDNA, fluorescent labeling with Cy3 and Cy5, and hybridization were performed as previously described (1). A total of 7,075 human genes and expressed sequence tags (ESTs) were examined on the Human UniGem V array (Incyte Pharmaceuticals, Palo Alto, Calif.) and analyzed with the help of GEM tools 2.4 software. A gridding and region detection algorithm was used to determine each element. The area surrounding each element was used to calculate a local background and was subtracted to calculate Cy3/Cy5 ratios. The average of the resulting Cy3 and Cy5 signals gave a ratio that was used to balance or normalize the signals. Confirmation of a representative subset of the microarray findings was carried out by Northern analyses as described above.
RESULTS
Low-risk HPV-11 genomes are stably maintained in HFKs for multiple passages.
We first investigated whether it was possible to isolate keratinocytes that maintain episomal copies of HPV-11 following transfection of cloned viral DNA. For these experiments, we used tissue culture techniques previously developed for the study of HPV-31 and HPV-18 (3, 4). HFKs were transfected, together with plasmid pSV2neo in trans, through the use of liposomes with recircularized HPV-11 DNA restricted from pBR322.HPV11 as described in Materials and Methods. Following selection for neomycin resistance, pooled colonies were expanded; a subset was harvested approximately 4 weeks posttransfection. Typically, 20 to 50 individual colonies were pooled for these analyses.
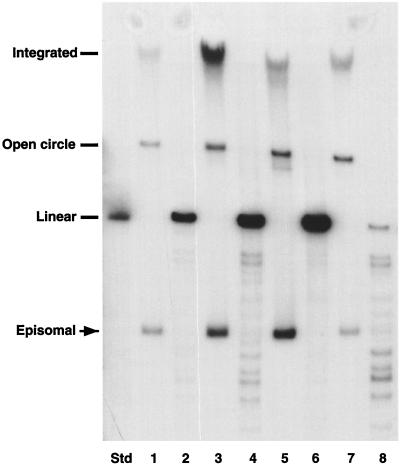
Southern analyses were performed on DpnI-digested total genomic DNA from 14 separate pooled cultures of HFKs isolated from four different donors. The results of four analyses, each representing a different primary HFK isolate, are shown in Fig. 1. In all analyses, supercoiled copies of HPV-11 DNA were observed. By comparison to copy number standards, we observed an average copy number of approximately 50 at early passages, with a two- to threefold variation between pooled transfected colonies from different donors. Southern analyses of transfectants at late passages, as well as frozen and thawed cells, showed retention of HPV-11 episomes, although there was a reduction in copy numbers at later passages, prior to senescence (data not shown).
FIG. 1.
Autoradiogram of Southern analysis of HFKs stably transfected with cloned HPV-11 DNA. Std, linear HPV-11 DNA standard. Each pair of numbered lanes (e.g., 1 and 2) represents an independently derived pooled culture. Equal amounts of total genomic DNA were digested with DpnI to remove residual input DNA. Even-numbered lanes were digested with DpnI and HindIII to linearize the HPV-11 genomes, while odd-numbered lanes contained DNAs which were digested with DpnI alone. Faster-migrating species in odd-numbered lanes represent supercoiled episomes of HPV-11.
The short-term maintenance of transfected HPV-11 genomes as episomes has been previously observed (17), but these cells quickly lost viral genomes and did not exhibit extended life spans. In our studies, episomal copies of the HPV-11 genome were maintained for multiple passages in cultures. Furthermore, HPV-11-positive cells exhibited life spans that, on average, were approximately twice those of cells transfected with a simian virus 40-driven neomycin resistance gene alone (Table 1). One passage consists of approximately a 1-to-5 split every 4 days. Results from multiple transfection experiments with the same donor keratinocytes, as well as transfection experiments with different donor backgrounds, exhibited comparable extensions in life spans. In some instances, HPV-positive keratinocytes could be maintained in cultures for over 3 months without undergoing senescence. One pooled culture, HFK-HPV11-D, was still actively dividing after 54 passages. We cannot exclude the possibility that this significant extension of life span in the HFK-HPV11-D cells was due to a spontaneous mutation of a cellular gene, as we have not seen similar effects with other pooled cultures of HPV-11-positive cells. Overall, these data indicate that keratinocytes transfected with cloned HPV-11 sequences exhibit extended life spans and stably replicate viral genomes as episomes.
TABLE 1.
Life spans of HFKs transfected with HPV-11 genomes
| HFK populationa | No. of passages
|
No. of experiments | |
|---|---|---|---|
| Transfected | Control | ||
| A | 10 | 5 | 4 |
| B | 14 | 5 | 4 |
| C | 24 | 11 | 3 |
| D | >54b | 10 | 3 |
A, B, C, and D denote primary keratinocytes isolated from different donors. All were transfected with HPV-11.
Cells were stopped and frozen at passage 54.
HPV-11 transfectants exhibit altered differentiation in raft cultures.
It was important to examine whether the morphological differentiation of HPV-11-positive keratinocytes was altered in organotypic raft cultures, as was previously observed for cells transfected with high-risk HPV-31 or HPV-18 (3, 4, 16). Normal keratinocytes rapidly lose nuclei upon differentiation, while cells that express high-risk HPVs maintain nuclear staining throughout the suprabasal layers. To investigate whether HPV-11-transfected keratinocytes exhibited altered differentiation programs similar to those seen with HPV-31 transfectants, we performed organotypic raft culture analyses.
As shown in Fig. 2A and B, respectively, untransfected cells and neomycin-treated control transfectants demonstrated normal differentiation patterns in the raft cultures, with nuclear staining predominantly localized to cells in the basal layers. In contrast, HPV-31-positive cells showed a dramatically altered differentiation pattern, with a thickening of the basal layer and nuclear staining throughout all layers (Fig. 2C). While organotypic rafts of HPV-11 transfectants demonstrated an altered differentiation pattern compared to that of neomycin-treated controls (Fig. 2D to F), the changes appeared to be less severe than those seen with HPV-31-positive cells. Similar results were observed for raft cultures of six different HPV-11 transfectants in two independent experiments. Although nuclei were generally maintained throughout all differentiated layers, some regions of the HPV-11 rafts exhibited less pronounced retention of nuclei. We cannot exclude the possibility that this observation was due to the generation during transfection of neomycin-resistant keratinocytes that lacked HPV-11 DNA. In addition, we observed a spectrum of changes in the degree to which nuclei were retained in suprabasal cells in different pooled cultures, and this finding correlated with the average HPV-11 copy number in these cells.
FIG. 2.
Stained sections of organotypic raft cultures. Normal and transfected HFKs were induced to differentiate in raft cultures as described in Materials and Methods. Paraffin-embedded tissues were sectioned and stained with hematoxylin and eosin for visualization of differentiation. (A) Normal HFKs. (B) Neomycin-treated control transfected HFKs. (C) HPV-31-transfected HFKs. (D to F) Three independent HPV-11-transfected HFKs. The asterisk in panel A indicates the basal layer, while the bar identifies suprabasal cells. The arrows in panels C and F identify nuclei in suprabasal layers.
HPV-11 genomes are amplified in HFKs upon differentiation.
To determine whether the altered differentiation of HPV-11-transfected cells correlated with the induction of late viral functions, we examined the ability of these stably transfected cells to induce differentiation-dependent amplification of viral DNA. A simple method to induce differentiation is the suspension of keratinocytes in semisolid media. Previous studies using HPV-31-positive keratinocytes demonstrated that suspension in methylcellulose leads to viral DNA amplification that can be detected by Southern analyses (21). HPV-11-positive cells were suspended in methylcellulose as described in Materials and Methods, and total genomic DNA was isolated at various times for Southern analyses.
As shown in Fig. 3, amplification of HPV-11 DNA was detected following suspension in methylcellulose, with some transfectants exhibiting an increased signal by 24 h (Fig. 3A) while others exhibited maximum amplification at 48 or 72 h (Fig. 3B and C, respectively). In addition, approximately one-half of the pooled cultures did not exhibit amplification of viral DNA (Fig. 3D). It appeared that pooled cultures with high average copy numbers of HPV-11 episomes were more likely to show amplification, while cultures with low average copy numbers were less likely to do so. In addition, pooled cultures that amplified viral DNA were able to do so reproducibly in multiple experiments.
FIG. 3.
Autoradiograms of four independent Southern analyses showing differentiation-dependent amplification of HPV-11 DNA following suspension in methylcellulose. The results for three different pooled HPV-11 cultures (HPV-11-1, HPV-11-2, and HPV-11-3) are shown. In addition, two separate experiments using one pooled culture (HPV-11-1) are shown. HPV-11-positive cells were suspended in semisolid media at the indicated times to induce differentiation. Equal amounts of total genomic DNA were digested with HindIII to linearize the HPV-11 genomes and analyzed by Southern analyses as described in Materials and Methods. (A) HPV-11-1 cells with amplification following suspension in methylcellulose. (B) Amplification of DNA from HPV-11-1 cells in a second experiment. (C) Amplification of HPV-11-3 DNA at 72 h in methylcellulose. (D) Lack of significant amplification of HPV-11 DNA from HPV-11-2 cells.
HPV-11 differentiation-dependent transcripts are expressed upon suspension in methylcellulose.
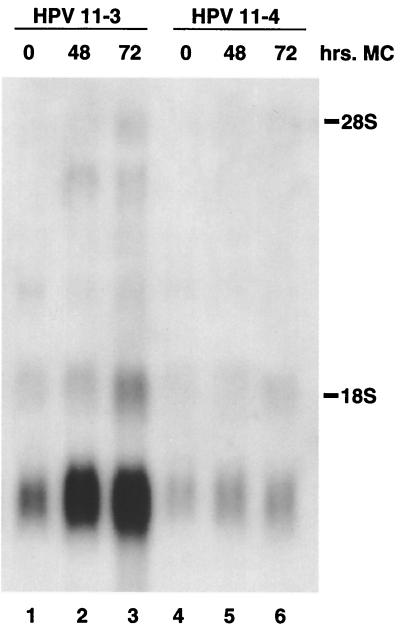
We next examined the patterns of viral expression in several pooled HPV-11 transfectants which stably maintained viral episomes. We chose to examine cultures that were able to amplify viral DNA upon differentiation. The results of Northern analyses for two independent pooled transfection cultures are shown in Fig. 4. In undifferentiated keratinocytes, two major sets of transcripts were observed. The major viral transcript detected was approximately 1.5 kb, while a less prominent transcript of approximately 2.0 kb was also seen. These could potentially initiate upstream of E6 or upstream of E7 and encode E1, E4, or E5 and would be consistent with the two early promoters previously described for low-risk HPV types (22). Following differentiation in methylcellulose, three additional transcripts appeared or showed more pronounced intensity. The larger transcripts have the potential to encode messages for L1 or L2, but a more detailed analysis would be required for definitive identification. For HPV-31-positive cells, the intensity of transcripts encoding E1 E4 has been shown to increase upon differentiation (8, 19). The increased intensity of the band at about 1.4 kb would be consistent with such transcripts (Fig. 4). The pattern of hybridization seen in Fig. 4 is very similar to that observed with RNA extracted from an HPV-11-positive condyloma acuminatum lesion (18). In that analysis, a major hybridizing species at 1.4 kb was also observed, with less intense bands at 1.8, 4.4, and 4.7 kb. This result suggests that the keratinocytes which stably maintain HPV-11 episomes and which were derived following transfection of cloned DNA have transcription patterns similar to those of cells isolated from HPV-11-positive lesions.
FIG. 4.
Northern analysis of HPV-11-positive cells following suspension in methylcellulose (MC). Total RNA was isolated at various times from two pooled cultures (HPV-11-3 and HPV-11-4) that had demonstrated amplification following suspension in methylcellulose. An increase in a 1.2-kb band is seen upon differentiation in both cultures, and additional, larger transcripts are also induced most prominently at 72 h. 28S and 18S rRNA markers correspond to molecular sizes of approximately 4.7 and 1.8 kb, respectively. The Northern blot was hybridized with a probe from the E4-E5 region of HPV-11.
Microarray analyses.
Previous studies have used microarray methods to examine global changes in cellular gene expression induced by HPV-31 in keratinocytes that stably maintain viral episomes. Accordingly, we examined HPV-11-positive keratinocytes for global alterations in cellular gene expression to determine whether changes induced by low-risk types were similar to those observed for high-risk types. For this analysis, mRNA was isolated from a proliferating monolayer culture of pooled keratinocytes that maintained transfected HPV-11 genomes extrachromosomally and that demonstrated amplification upon differentiation. For comparison, mRNA was isolated from an untransfected monolayer culture of cells from the same HFK donor as that used for the generation of the HPV-11-positive keratinocytes at a comparable passage number. A total of 7,075 expressed sequences were analyzed by microarray analysis. Only 5 genes were found to be repressed by HPV-11 gene products by more than 2.0-fold, while 41 were repressed by 1.6-fold or more (Table 2). We also observed that only one interferon-inducible gene, that for interferon-induced protein 1-8U, was even moderately repressed (−1.2-fold). This result is in contrast to the significant number of interferon-inducible genes that were found to be repressed in the HPV-31 microarray analysis (1).
TABLE 2.
Genes and ESTs activated or repressed by HPV-11
| GenBank accession no. | Gene name | Difference in expression (fold) |
|---|---|---|
| V01512 | v-fos homolog | −3.5 |
| AI272010 | Early growth response 1 | −3.3 |
| AI817864 | Centromere protein | −3.3 |
| M60047 | Heparin-binding protein | −2.2 |
| U49260 | Mevalonate decarboxylase | −2.2 |
| J04076 | Early growth response 2 | −1.9 |
| AF006043 | 3-Phosphoglycerate dehydrogenase | −1.9 |
| X64330 | ATP citrate lyase | −1.9 |
| M83665 | High-mobility-group protein 2 | −1.8 |
| U02019 | Heteroribonucleoprotein | −1.8 |
| L12711 | Transketolase | −1.8 |
| X53416 | Filamin A | −1.8 |
| Y11484 | Phosphoenolpyruvate carboxykinase 2 | −1.8 |
| D13633 | KIAA008 protein | −1.8 |
| U25725 | Centromere protein F | −1.8 |
| AI684439 | EST | −1.7 |
| X12597 | High-mobility-group protein 1 | −1.7 |
| U23143 | Serine hydroxymethyltransferase | −1.7 |
| X16396 | Methylene tetrahydrofolate dehydrogenase | −1.7 |
| AI631255 | Proliferating cell nuclear antigen | −1.7 |
| M60278 | Diphtheria toxin receptor | −1.7 |
| AC004770 | hFEN1 | −1.7 |
| AI417997 | HOX 2 | −1.7 |
| U82984 | Brain 1NB | −1.7 |
| AI707962 | Small cytokine member 19 | −1.6 |
| AA315491 | Nuclear helicase-DEAD box family | −1.6 |
| AF060866 | Zinc finger protein 200 | −1.6 |
| AF01808 | Mediterranean fever | −1.6 |
| U50327 | Protein kinase C substrate 80K | −1.6 |
| F26137 | Metallothionein II | −1.6 |
| AI800815 | Minichromosome maintenance-deficient protein 2 | −1.6 |
| L1683 | Forkhead | −1.6 |
| AA987219 | PHAPI2b | −1.6 |
| M98326 | Valyl-tRNA synthetase | −1.6 |
| S78825 | ID-binding protein | −1.6 |
| U70323 | Spinocerebellar ataxia 2 | −1.6 |
| AI743113 | Hairy homolog | −1.6 |
| X54942 | CDC28 | −1.6 |
| AF09565 | U5 snRNP | −1.6 |
| M80244 | Solute carrier family 7 transporter | −1.6 |
| AF064093 | C42C1.9 | −1.6 |
| AA302123 | Interferon p27 | 4.4 |
| X03557 | p56 interferon inducible | 3.1 |
| AA307912 | EST | 2.8 |
| AI802950 | Microfibril glycoprotein | 2.8 |
| M77349 | Transforming growth factor beta-induced 68K | 2.8 |
| AA937212 | EST | 2.7 |
| AA552360 | Translation initiation factor kinase | 2.6 |
| Y15060 | Galactosyltransferase | 2.6 |
| AI86325 | EST | 2.5 |
| AA58374 | S100 calcium-binding protein | 2.5 |
| AA336592 | Spermidine acetyltransferase | 2.5 |
| U51903 | GTPase-activating protein | 2.5 |
| M31551 | Plasminogen activator inhibitor type II | 2.5 |
| L341551 | Laminin | 2.5 |
| AA675919 | Testis 5 cDNA | 2.5 |
| U50931 | Defensin | 2.4 |
| AF007153 | EST | 2.4 |
| AA460433 | Leukocyte protease inhibitor | 2.4 |
| AF010309 | Quinone oxidoreductase | 2.4 |
| N92840 | EST | 2.4 |
| H03260 | EST | 2.4 |
| AI245471 | Ras homolog | 2.3 |
| AI763171 | Kid12 | 2.3 |
| AI336522 | EST | 2.3 |
| AI341820 | EST | 2.3 |
| AI306588 | EST | 2.3 |
| AI288887 | EST | 2.3 |
| D79783 | EST | 2.3 |
| AB097935 | KIAA0554 protein | 2.3 |
| AF097935 | Desmoglein | 2.3 |
| AI243872 | EST | 2.2 |
| AA243868 | EST | 2.2 |
| U20860 | Angiotensin receptor 2 | 2.2 |
| AI610676 | EST | 2.2 |
| AA889281 | Parathyroid tumor clone | 2.2 |
| AF059195 | v-maf homolog | 2.2 |
| AI192628 | RNase 6 precursor | 2.2 |
| AA488617 | Adenylyl cyclase-associated protein | 2.2 |
| X14787 | Thrombospondin 1 | 2.2 |
| AI087319 | EST | 2.2 |
| X78686 | Neutrophil-activating peptide | 2.2 |
| AA403225 | Ring finger protein 3 | 2.2 |
| W84433 | LTR4 element | 2.2 |
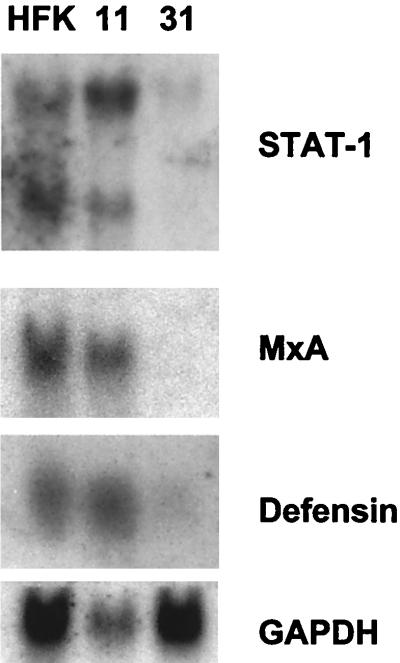
Figure 5 shows a direct comparison of the expression of Stat-1, MxA, and defensin for normal human keratinocytes, HPV-11-positive cells, and HPV-31-positive cells. The expression of these genes in HPV-11-positive cells was slightly elevated or comparable to that seen in normal keratinocytes. In contrast, the levels of these transcripts were substantially reduced in HPV-31-positive cells. Interestingly, the expression of several keratinocyte-specific genes, such as those for desmoplakin and desmocollin, was not significantly altered by HPV-11 (data not shown).
FIG. 5.
Northern analysis of normal human keratinocytes and HPV-11- and HPV-31-positive cells for the expression of Stat-1, MxA, and defensin. RNAs isolated from proliferating monolayer cultures were analyzed. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control and indicates that the RNA in the HPV-11 lane was reduced compared to that in either the normal HFK or the HPV-31 lane.
In contrast to the small number of genes found to be repressed by HPV-11 gene products, we observed 76 expressed sequences to be activated by twofold or more. The 43 genes activated by more than 2.2-fold are shown in Table 2. The genes activated to the highest degree were members of the interferon-inducible family—those for p27 (+4.4-fold) and the 56-kDa protein (3.1-fold). No other readily discernible families of genes could be identified. Other notable genes activated by HPV-11 included those for the transforming growth factor β-induced 86-kDa protein (2.8-fold), defensin (2.4-fold), desmoglein 1 (2.3-fold), Stat-1 (1.7-fold), and COX2 (1.7-fold). The expression of six representative genes (those for Stat-1, MxA, IFI-56, HBD-1, SKALP, and PAI-2) in the microarray analysis was also confirmed by Northern analysis (data not shown). This analysis indicates that high- and low-risk HPV types target distinct sets of cellular genes during their productive life cycles and that only a small number of genes are similarly affected.
DISCUSSION
An understanding of the basic mechanisms that regulate the productive life cycle of HPVs requires knowledge of the differences, as well as similarities, in the actions of low- and high-risk viruses. Using methods developed for the study of the productive life cycles of high-risk HPV-31, HPV-18, and HPV-16 (2–4, 16), we were able to efficiently isolate transfected normal keratinocytes that stably maintained HPV-11 episomes. While transfected HPV-11 sequences were not able to immortalize normal keratinocytes, we consistently observed an extended life span for the HPV-11-positive cells in cultures. In some instances, these cells were maintained in continuous cultures for over 3 months without undergoing senescence. We suspect that the expression of E6, E7, or E5 is responsible for this extended life span, based on our knowledge of the actions of bovine papillomavirus type 1 and high-risk HPV gene products (7).
Because our system allows for genetic analyses of HPV-11, the identification of the responsible protein(s) can be undertaken. While the low-risk E6 protein has no identified binding partners (7), HPV-11 E7 has been shown to associate with members of the retinoblastoma family, albeit with a lower affinity than high-risk E7 (5, 9). In addition, it has been shown that HPV-31 E6 and E7 are required for the stable maintenance or segregation of episomes, an activity that is independent of their roles in immortalization (26). It is important to investigate whether similar activities hold true for the corresponding low-risk proteins.
The life cycles of both high- and low-risk HPV types are linked to epithelial cell differentiation. The amplification of viral genomes in suprabasal cells requires that HPV proteins block the normal process of cell cycle exit upon differentiation. This process appears to be controlled at least in part by the retinoblastoma family of proteins; the ability of low-risk E7 proteins to alter the regulatory function of these factors may explain the altered differentiation of HPV-11-positive cells (9). In a preliminary analysis of cyclin proteins, including cyclins A, E, and B, no differences in levels were observed between normal and HPV-11-positive cells (J. T. Thomas, unpublished data). Similarly, we did not observe dramatic changes in the expression of cell cycle regulatory factors, such as p21, p27, and p57, in HPV-11-positive cells. In cells containing high-risk HPV types, the levels of cyclins A, E, and B were increased, while no change was seen in the levels of the inhibitors p57 and p27 (J. T. Thomas, unpublished).
Interestingly, the changes in cellular gene expression induced by low-risk HPVs were in large part different from those seen with high-risk HPV-31. The most notable of these was the lack of repression of interferon-inducible genes, as was seen in cells containing high-risk HPV types (1). Many viruses have evolved mechanisms by which they interfere with the interferon response. It would be surprising if HPV-11 did not block this response by some mechanism, but our studies suggest that this process is likely to occur in a manner distinct from that used by high-risk HPV-31. Using microarray analyses, we observed that the expression of only a small subset of genes was altered to similar extents by both high- and low-risk HPVs. Because our studies were performed with proliferating monolayer cultures, it is possible that additional changes in cellular gene expression occurred upon differentiation. Furthermore, our microarray analyses were limited to only 7,075 genes, and additional changes in cellular gene expression may be detected when all human expressed sequences can be screened. Microarray analyses can exhibit an intrinsic degree of variability in the expression of individual genes between experiments. We suspect that there may be some variability in the expression of individual genes in multiple microarray analyses, but we have focused our comparative analysis on whether the expression of families of genes is altered in cells that maintain high-risk versus low-risk HPV genomes. From our studies, we conclude that there are differences in the spectra of genes whose expression is modulated by different HPV types.
The generation of keratinocytes that maintain HPV-11 genomes as episomes provides an important tool for the future analysis of low-risk infections. Moreover, the ability to perform a genetic analysis of HPV-11 will allow for the determination of the functions of low-risk viral proteins. It is possible that the seemingly subtle functions of low-risk proteins will be detectable in this system, allowing for a greater understanding of these clinically significant infections.
ACKNOWLEDGMENTS
This work was supported by grants from the National Cancer Center (CA74202) and the National Institute of Allergy and Infectious Diseases (U19AI31494).
We thank Ann Roman for helpful advice on this project.
REFERENCES
- 1.Chang Y E, Laimins L A. Microarray analysis identifies interferon-inducible genes and Stat-1 as major transcriptional targets of human papillomavirus type 31. J Virol. 2000;74:4174–4182. doi: 10.1128/jvi.74.9.4174-4182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flores E, Allen-Hoffman L, Lambert P F. The human papillomavirus type 16 E7 oncogene is required for the productive stage of the life cycle. J Virol. 2000;74:6622–6633. doi: 10.1128/jvi.74.14.6622-6631.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frattini M G, Lim H B, Laimins L A. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc Natl Acad Sci USA. 1996;93:3062–3067. doi: 10.1073/pnas.93.7.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frattini M G, Lim H B, Doorbar J, Laimins L A. Induction of human papillomavirus type 18 late gene expression and genomic amplification in organotypic cultures from transfected DNA templates. J Virol. 1997;71:7068–7072. doi: 10.1128/jvi.71.9.7068-7072.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gage J, Meyers C, Wettstein F. The E7 proteins of the nononcogenic human papillomavirus type 6b (HPV-6b) and of the oncogenic HPV-16 differ in retinoblastoma binding and other properties. J Virol. 1990;64:723–730. doi: 10.1128/jvi.64.2.723-730.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halbert C, Demers G, Galloway D. The E6 and E7 genes of human papillomavirus type 6 have weak immortalizing activity in human epithelial cells. J Virol. 1992;66:2125–2134. doi: 10.1128/jvi.66.4.2125-2134.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howley P M. Papillomavirinae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 947–978. [Google Scholar]
- 8.Hummel M, Hudson J B, Laimins L A. Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. J Virol. 1992;66:6070–6080. doi: 10.1128/jvi.66.10.6070-6080.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones D L, Munger K. Interactions of the human papillomavirus E7 protein with cell cycle regulators. Semin Cancer Biol. 1996;7:327–337. doi: 10.1006/scbi.1996.0042. [DOI] [PubMed] [Google Scholar]
- 10.Klumpp D J, Stubenrauch F, Laimins L A. Differential effects of the splice acceptor at nucleotide 3295 of human papillomavirus type 31 on stable and transient viral replication. J Virol. 1997;71:8186–8194. doi: 10.1128/jvi.71.11.8186-8194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kubbutat M H G, Vousden K H. Role of E6 and E7 oncoproteins in HPV-induced anogenital malignancies. Semin Virol. 1996;7:295–304. [Google Scholar]
- 12.Laimins L A. Human papillomaviruses target differentiating epithelia for virion production and malignant conversion. Semin Virol. 1996;7:305–313. [Google Scholar]
- 13.Laimins L A. Regulation of transcription and replication by human papillomaviruses. In: McCance D J, editor. Human tumor viruses. Washington, D.C.: American Society for Microbiology; 1998. pp. 201–223. [Google Scholar]
- 14.Meyers C, Frattini M, Hudson J, Laimins L A. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science. 1992;257:971–973. doi: 10.1126/science.1323879. [DOI] [PubMed] [Google Scholar]
- 15.Meyers C, Frattini M G, Laimins L A. Tissue culture techniques for the study of human papillomaviruses in stratified epithelia. In: Celis J E, editor. Cell biology: a laboratory handbook. Vol. 1. San Diego, Calif: Academic Press; 1994. pp. 491–499. [Google Scholar]
- 16.Meyers C, Mayer T J, Ozbun M A. Synthesis of infectious human papillomavirus type 18 in differentiating epithelia transfected with viral DNA. J Virol. 1997;71:7381–7386. doi: 10.1128/jvi.71.10.7381-7386.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mungal S, Steinberg B M, Taichman L B. Replication of plasmid-derived human papillomavirus type 11 DNA in cultured keratinocytes. J Virol. 1992;66:3220–3224. doi: 10.1128/jvi.66.5.3220-3224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nassrei M, Hirochika R, Broker T R, Chow L T. A human papillomavirus type 11 transcript encoding an E1 E4 protein. Virology. 1987;159:433–439. doi: 10.1016/0042-6822(87)90482-x. [DOI] [PubMed] [Google Scholar]
- 19.Ozbun M, Meyers C. Characterization of late gene transcripts expressed during vegetative replication of human papillomavirus type 31b. J Virol. 1997;71:5161–5172. doi: 10.1128/jvi.71.7.5161-5172.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfister H. The role of human papillomavirus in anogenital cancer. Obstet Gynecol Clin North Am. 1996;23:579–595. [PubMed] [Google Scholar]
- 21.Ruesch M N, Stubenrauch F, Laimins L A. Activation of papillomavirus late gene transcription and genome amplification upon differentiation in semisolid medium is coincident with expression of involucrin and transglutaminase but not keratin-10. J Virol. 1998;72:5016–5024. doi: 10.1128/jvi.72.6.5016-5024.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smotkin D, Prokoph H, Wettstein F. Oncogenic and nononcogenic human genital papillomaviruses generate the E7 mRNA by different mechanisms. J Virol. 1989;63:1441–1447. doi: 10.1128/jvi.63.3.1441-1447.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stubenrauch F, Colbert A M, Laimins L A. Transactivation by the E2 protein of oncogenic human papillomavirus type 31 is not essential for early and late viral functions. J Virol. 1998;72:8115–8123. doi: 10.1128/jvi.72.10.8115-8123.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stubenrauch F, Lim H B, Laimins L A. Differential requirements for conserved E2 binding sites in the life cycle of oncogenic human papillomavirus type 31. J Virol. 1998;72:1071–1077. doi: 10.1128/jvi.72.2.1071-1077.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terhune S S, Milcarek C, Laimins L A. Regulation of human papillomavirus type 31 polyadenylation during the differentiation-dependent life cycle. J Virol. 1999;73:7185–7192. doi: 10.1128/jvi.73.9.7185-7192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas J T, Hubert W G, Ruesch M N, Laimins L A. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of epiosmes during the viral life cycle in normal human keratinocytes. Proc Natl Acad Sci USA. 1999;96:8449–8454. doi: 10.1073/pnas.96.15.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]