Abstract
The steady-state level and metabolic half-life of retinoblastoma tumor suppressor protein pRB are decreased in cells that express high-risk human papillomavirus (HPV) E7 proteins. Here we show that pRB degradation is a direct activity of E7 and does not reflect a property of cell lines acquired during the selection process for E7 expression. An amino-terminal domain of E7 that does not directly contribute to pRB binding but is required for transformation is also necessary for E7-mediated pRB degradation. Treatment with inhibitors of the 26S proteasome not only blocks E7-mediated pRB degradation but also causes the stabilization of E7. Mutagenic analyses, however, reveal that the processes of proteasomal degradation of E7 and pRB are not linked processes. HPV type 16 E7 also targets the pRB-related proteins p107 and p130 for destabilization by a proteasome-dependent mechanism. Using the SAOS2 flat-cell assay as a biological indicator for pRB function, we demonstrate that pRB degradation, not solely binding, is important for the E7-induced inactivation of pRB.
Human papillomaviruses (HPVs) are DNA viruses with small circular genomes that cause epithelial hyperplasias ranging from benign papillomas (warts) to premalignant lesions that can progress to squamous cell carcinomas (reviewed in reference 25). There are over 100 different HPV types, approximately 30 of which specifically infect anogenital tract mucosa. These HPVs are classified as low risk or high risk, depending on the clinical prognosis of the lesions they cause. Low-risk HPVs, such as HPV type 6 (HPV-6) and HPV-11, cause condylomata acuminata (genital warts), while high-risk HPVs, including HPV-16 and HPV-18, are associated with squamous intraepithelial lesions that can progress to cervical carcinomas. Integration of the HPV genome into a host cell chromosome is a frequent event during malignant progression and results in the consistent but dysregulated expression of the HPV E6 and E7 proteins (reviewed in reference 55).
High-risk HPV E6 proteins target tumor suppressor protein p53 for ubiquitin-mediated proteasomal degradation by interacting with and reprogramming the E6-AP ubiquitin ligase (38, 39, 51). E6-mediated degradation of p53 compromises the ability of the host cell to engage cell cycle checkpoints and apoptotic responses (33). High-risk HPV E7 oncoproteins target retinoblastoma tumor suppressor protein pRB (19). High-risk HPV E7 proteins interact with pRB at a higher efficiency than do low-risk HPV E7 proteins (22, 34). Interaction of E7 with hypophosphorylated pRB causes the disruption of growth-suppressive pRB-E2F complexes (10), promoting the G1-S cell cycle transition. E7-mediated cellular transformation correlates with pRB binding (22); however, there are mutations in E7 that impair cellular transformation and immortalization without affecting pRB binding (4, 8, 20, 28, 35). Furthermore, the E7 protein of HPV-1a, a low-risk cutaneous HPV, can interact with pRB and transactivate E2F-dependent promoters with the same efficiency as can high-risk HPV-16 E7, yet it scores negative in standard transformation assays (13, 40). These reports strongly suggest that high-risk HPV E7 proteins also target as-yet-unidentified cellular processes that contribute to transformation.
The steady-state level and metabolic half-life of pRB are decreased in HPV-16 E7-expressing cells, suggesting that E7 can induce the degradation of pRB (6, 7, 30). The ubiquitin-proteasome system has been implicated as inhibitors of the 26S proteasome increase pRB levels in E7-expressing cells (6, 7). Moreover, HPV-16 E7 is unable to induce pRB destabilization in a cell line expressing a temperature-sensitive E1 enzyme of the ubiquitin system (7). HPV-16 E7 can interact with the S4 subunit of the 26S proteasome. The S4 subunit has been proposed to function as a bridge between pRB and the 26S proteasome, allowing E7 to direct the proteasomal degradation of pRB without the need for ubiquitination (5). The HPV-16 E7 oncoprotein is itself a target of ubiquitination at the amino-terminal residue, followed by degradation through the 26S proteasome (37). However, it is not known whether the degradation of E7 is linked to its ability to target pRB for degradation.
We have established a rapid cell-based degradation assay using the pRB-deficient osteosarcoma cell line SAOS2. We show that this system accurately recapitulates the pRB-destabilizing activity of E7 that has been previously described for stable cell lines (31). Our experiments also demonstrate that pRB degradation is a direct activity of E7 and does not reflect a property that E7-expressing cell lines acquire during the selection process. HPV-16 E7 also targets the pRB-related proteins p107 and p130 for proteasome-dependent degradation. Inhibition of the 26S proteasome not only blocks E7-mediated pRB degradation but also causes a dramatic stabilization of E7, supporting recently published results showing that E7 is normally turned over by the proteasome (37). Mutational analyses of HPV-16 E7 indicate that degradation of E7 and E7-mediated pRB degradation are not directly connected. Using a biological indicator for pRB function in SAOS2 cells, we demonstrate that pRB degradation is important for E7-induced inactivation of pRB.
MATERIALS AND METHODS
Plasmids.
Wild-type HPV-16 E7 and the 16E7 ΔD21-C24, 16E7 ΔP6-E10, and 16E7 C91S mutants (35) were subcloned into the BamHI site of the CMV-BamHI-Neo plasmid (3). HPV-16 E7, 16E7 ΔD21-C24, 16E7 C24G, 16E7 E26G, 16E7 ΔP6-E10, and HPV-1a E7 with a hemagglutinin (HA) epitope tag at their carboxyl termini were generated by PCR. PCR products were then subcloned into the BamHI site of the CMV-BamHI-Neo plasmid. Both amino- and carboxyl-terminal FLAG-epitope-tagged 16E7 constructs were generated by PCR. The FLAG-16E7 PCR product was subcloned into the NotI site of the pcDNA3 expression plasmid. The 16E7-FLAG PCR product was subcloned into the HindIII and NotI sites of the pcDNA3 expression plasmid (Invitrogen). The 16E7 K60,97R mutant was generated using a PCR-based QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). Plasmids were sequenced to confirm the presence of corresponding mutations. All primer sequences used in subcloning and site-directed mutagenesis are available upon request.
CMV-pRB, CMV-HA-p107, and pcDNA3-HA-p53 expression plasmids were kind gifts from P. Hinds (Harvard Medical School, Boston, Mass.). Expression plasmids for pRB truncation mutants were kindly provided by W. Sellers (Dana-Farber Cancer Institute, Boston, Mass.) (41), and the pRB double point mutant pRB Y756F/N757A (16) was provided by F. Dick and N. Dyson (MGH Cancer Center, Charlestown, Mass.). The CMV-p130 expression plasmid was obtained from D. Cobrinik (Columbia University, New York, N.Y.), and the pcDNA3-HPV-16 E6 expression plasmid was a generous gift from P. Howley (Harvard Medical School). A pBABE-HPV-1a E7 expression plasmid, from which pCMV HPV-1a-HA-E7 was constructed, was obtained from M. Tommasino (Deutsches Krebsforschungszentrum, Heidelberg, Germany). The green fluorescent protein (GFP) expression plasmid, EGFP-C1, was purchased from Clontech. LXSN-based vectors were kindly provided by D. A. Galloway (Fred Hutchinson Cancer Research Center, Seattle, Wash.).
Cell lines and cell culturing.
The SAOS2 human osteosarcoma cell line was maintained in Dulbecco's modified Eagle's medium supplemented with 15% fetal bovine serum, 50 U of penicillin/ml, and 50 μg of streptomycin/ml. Human foreskin keratinocytes (HFKs) from neonatal human foreskins were prepared as described previously (29) and maintained in keratinocyte serum-free medium (Gibco BRL).
Transfections, retrovirus infections, and immunological methods.
SAOS2 cells were seeded at 3.3 × 105 cells per 60-mm plate and transfected using the calcium phosphate method (11). For protein half-life determinations, SAOS2 cells were transfected as described above. At 24 h posttransfection, the cells were treated with cycloheximide (30 μg/ml; Sigma) for various times. Stable HPV-16 E7-expressing pools of HFKs were prepared after retrovirus transfer with an LXSN-based vector followed by G418 selection (29).
SAOS2 cells and HFKs were lysed in 150 mM NaCl–1% Nonidet P-40–50 mM Tris-HCl (pH 8.0) buffer supplemented with leupeptin (1 μg/ml), aprotinin (1 μg/ml), sodium orthovanadate (45 nM), and phenylmethylsulfonyl fluoride (0.01%). Protein concentrations were determined using the Bradford method (Bio-Rad). Samples (100 μg) of total protein lysate were subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and then transferred to polyvinylidene difluoride membranes (Immobilon P; Millipore).
The following commercial antibodies were used: for pRB, Ab-5 (Oncogene Science); for p53, Ab6 (Oncogene Science); for HPV-16 E7, sc6981 (Santa Cruz Biotechnology, Inc.) and 8C9 (Zymed Laboratories); for GFP, catalog no. 8367-1 (Clontech); for glutathione S-transferase, catalog no. 3818-1 (Clontech); for HA, HA.11 clone 16B12 (Covance); and for p130, RB2 (Transduction Laboratories). Complexes of primary and horseradish peroxidase-linked secondary antibodies (Amersham) were detected using enhanced chemiluminescence (NEN Life Sciences). Blots were acquired using a Bio-Rad Fluor-S Max Multi-imager and quantified using the manufacturer's software. Cotransfected GFP was used as an internal control to normalize for differences in transfection efficiency between individual plates.
Proteasome inhibitor treatments.
SAOS2 cells were transfected as described above. At 40 h posttransfection, cells were treated with either 10 or 40 μM MG132 or Lactacystin (BIOMOL, Plymouth Meeting, Pa.) for 4 h. Cells were then harvested by scraping into boiling lysis buffer containing 100 mM Tris-HCl (pH 6.8), 200 mM dithiothreitol (DTT), 4% SDS, 20% glycerol, 0.2% bromophenol blue, and 10% β-mercaptoethanol. Lysates were boiled for an additional 5 min. Equal volumes of lysates were subjected to SDS-PAGE and immunoblotting.
SAOS2 flat-cell assay.
SAOS2 cells were transfected in duplicate as described above. At 24 h posttransfection, cells were split 1:2 into 100-mm plates, selected for 10 to 14 days in 500-μg/ml puromycin (Sigma), and stained for senescence-associated β-galactosidase activity as described previously (49). Numbers of flat, blue cells were determined by counting 20 random ×200 fields per plate. The reported results represent averages and standard deviations from two independent experiments.
RESULTS
HPV-16 E7-mediated destabilization of pRB upon transient cotransfection in SAOS2 cells.
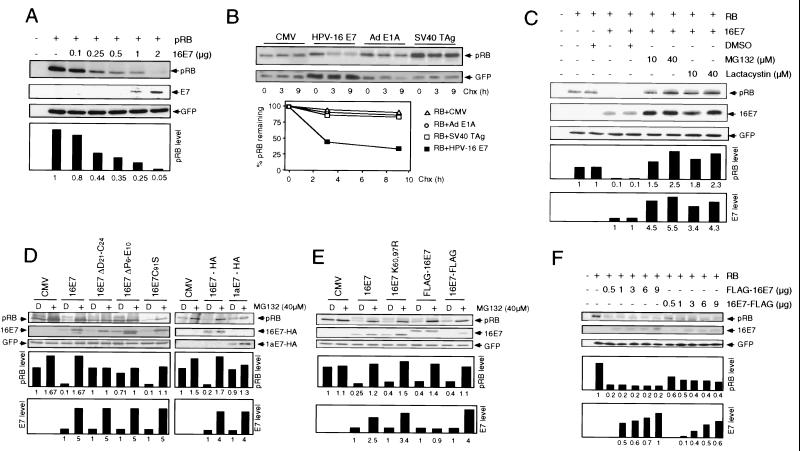
To study HPV-16 E7-mediated destabilization of pRB, we developed a rapid cell-based assay using the human osteosarcoma cell line SAOS2, which contains no full-length nuclear pRB (44). Initially, we ascertained that E7 destabilizes pRB in SAOS2 cells upon transient cotransfection to a similar extent as was previously observed for E7-expressing normal human fibroblasts and keratinocytes. A pRB expression plasmid was transfected into the SAOS2 cell line, alone or in combination with HPV-16 E7. An expression plasmid for GFP was routinely cotransfected to normalize for equal transfection efficiency in each reaction. Cotransfection of increasing amounts of HPV-16 E7 caused a dose-dependent reduction of the steady-state levels of pRB (Fig. 1A).
FIG. 1.
HPV-16 E7 degradation and E7-mediated pRB degradation are 26S proteasome-dependent but separable processes. (A) The human osteosarcoma cell line SAOS2 was transiently transfected with the indicated combinations of plasmids encoding wild-type pRB and wild-type HPV-16 E7 (16E7). Samples (100 μg) were subjected to SDS-PAGE and immunoblot analysis for pRB (upper panel), HPV-16 E7 (middle panel), and cotransfected GFP control (lower panel). Quantitation of pRB levels, normalized for GFP expression, is shown underneath those panels (in arbitrary units based on densitometry). (B) HPV-16 E7, adenovirus E1A, and SV40 T antigen oncoproteins differ in their ability to destabilize pRB. SAOS2 cells were transfected with the indicated combinations of plasmids encoding pRB, HPV-16 E7, adenovirus (Ad) E1A, and SV40 T antigen (TAg) or control plasmid (CMV). At 24 h posttransfection, protein synthesis was blocked by treatment with cycloheximide (Chx). At the indicated times, samples (100 μg) were processed for immunoblot analysis for pRB (upper panel) and cotransfected GFP control (lower panel). Quantitation of pRB (RB) levels, normalized for GFP expression, is shown underneath these panels. (C) Effect of MG132 and Lactacystin on HPV-16 E7 and pRB steady-state levels. SAOS2 cells were transfected with expression plasmids encoding pRB alone or in combination with 16E7. At 40 h posttransfection, cells were treated with 10 or 40 μM MG132 or Lactacystin or mock treated with dimethyl sulfoxide (DMSO) for 4 h. Samples (100 μg) were subjected to SDS-PAGE and immunoblot analysis for pRB (upper panel), HPV-16 E7 (middle panel), and cotransfected GFP control (lower panel). Quantitation of pRB and HPV-16 E7 levels, normalized for GFP expression, is shown underneath these panels. (D) Effect of proteasome inhibition on various HPV E7 proteins. SAOS2 cells were transfected with the indicated combinations of plasmids encoding pRB, HPV-16 E7 (16E7 and 16E7-HA), a pRB-binding- and pRB degradation-deficient 16E7 mutant (16E7 ΔD21-C24), pRB-binding-competent and pRB degradation-deficient versions of E7 (16E7 ΔP6-E10 and 1aE7-HA), and a pRB-binding- and pRB degradation-competent 16E7 mutant (16E7 C91S) or control vector (CMV). Cells were treated with 40 μM MG132 (+) or mock treated with DMSO (D) for 4 h. Samples (100 μg) were subjected to SDS-PAGE and immunoblot analysis for pRB (upper panel), HPV-16 E7 or HPV-16 E7 tagged with HA (middle panel), and cotransfected GFP control or HPV-1a E7 tagged with HA (lower panel). Quantitation of pRB and HPV-16 E7 levels, normalized for GFP expression, is shown underneath these panels. (E) E7 degradation and E7-mediated pRB degradation are not linked. SAOS2 cells were transfected with the indicated combinations of plasmids encoding pRB, HPV-16 E7 (16E7), a lysineless mutant of 16E7 (16E7 K60,97R), an amino-terminal FLAG-tagged version of 16E7 (FLAG-16E7), and a carboxyl-terminal FLAG-tagged version of 16E7 (16E7-FLAG) or control vector (CMV). Cells were treated with 40 μM MG132 (+) or mock treated with DMSO (D) for 4 h. Samples (100 μg) were subjected to SDS-PAGE and immunoblot analysis for pRB (upper panel), HPV-16 E7 (middle panel), and cotransfected GFP control (lower panel). Quantitation of pRB and HPV-16 E7 levels, normalized for GFP expression, is shown underneath these panels. (F) HPV-16 E7 stability correlates with E7-mediated pRB degradation. SAOS2 cells were transiently transfected with the indicated combinations of plasmids encoding pRB and amino- and carboxyl-terminal FLAG-tagged HPV-16 E7. Samples (100 μg) were subjected to SDS-PAGE and immunoblot analysis for pRB (upper panel), HPV-16 E7 (middle panel), and cotransfected GFP control (lower panel). Quantitation of pRB and E7 levels, normalized for GFP expression, is shown underneath these panels.
To confirm that the decrease in pRB levels upon E7 expression reflects altered protein stability, SAOS2 cells were transfected with the pRB expression plasmid alone or in combination with HPV-16 E7 and treated with cycloheximide to inhibit protein synthesis. In the absence of HPV-16 E7, the pRB half-life was greater than 9 h; however, when HPV-16 E7 was coexpressed, the pRB half-life was decreased to approximately 3 h (Fig. 1B). This reduction in pRB stability was specific for HPV-16 E7, as the pRB-binding oncoproteins adenovirus E1A (52) and simian virus 40 (SV40) T antigen (15) did not induce a decrease in the pRB half-life (Fig. 1B).
Because the E6 and E7 oncogenes are consistently coexpressed in HPV-associated cervical carcinomas, we wanted to determine whether the coexpression of p53 and E6 affects pRB destabilization by E7. However, the coexpression of HPV-16 E6 and p53, alone or in combination, had no effect on the E7-induced degradation of pRB in this assay (data not shown).
E7-mediated pRB destabilization and E7 degradation are not linked processes.
Inhibitors of the 26S proteasome can block E7-mediated pRB destabilization (6, 7). Furthermore, HPV-16 E7 is a target for ubiquitination and subsequent degradation by the 26S proteasome (37). To ensure that the proteasome is involved in E7-mediated pRB destabilization as well as E7 degradation in our transient assay system, we treated transfected SAOS2 cells with the proteasome inhibitors MG132 and lactacystin. As shown in Fig. 1C, treatment with MG132 and lactacystin interfered with E7-mediated pRB destabilization and also caused an increase in E7 levels.
To determine whether the degradation of E7 and pRB may be linked, we tested several E7 mutants that differ in their ability to bind and degrade pRB. The steady-state level of each of these E7 mutants increased four- to fivefold upon MG132 treatment (Fig. 1D). In particular, HPV-1a E7 and the 16E7 ΔP6-E10 mutant, which efficiently bind (13, 34) but do not degrade (1, 30) pRB, and the 26S proteasomal S4 subunit-binding-deficient 16E7 C91S mutant were stabilized upon proteasome inhibition (Fig. 1D). Interestingly, the S4 subunit-binding-deficient 16E7 C91S mutant (5) decreased pRB steady-state levels to levels similar to those seen with wild-type 16E7. Moreover, the pRB-binding-deficient 16E7 ΔD21-C24 mutant (35) was also stabilized by proteasome inhibitor treatment (Fig. 1D).
To more clearly separate proteasomal degradation of E7 and E7-mediated pRB destabilization, we generated a lysineless E7 mutant, 16E7 K60,97R, and both amino-and carboxyl-terminal FLAG-tagged versions of 16E7, FLAG-16E7 and 16E7-FLAG, respectively. Consistent with a recent report (37), the removal of both lysines, as in 16E7 K60,97R, and carboxyl-terminal epitope tagging, as in16E7-FLAG, did not alter the sensitivity of E7 to proteasome inhibition. Both 16E7 K60,97R and 16E7-FLAG were stabilized to levels similar to those of wild-type HPV-16 E7 upon MG132 treatment (Fig. 1E). However, levels of the amino-terminal FLAG-tagged version of HPV-16 E7, FLAG-16E7, were unaffected by treatment with MG132 (Fig. 1E). This resistance of FLAG-16E7 to proteasome inhibition suggests amino-terminal conjugation of ubiquitin, as has been reported (37). However, as shown in Fig. 1E, 16E7 K60,97R, FLAG-16E7, and 16E7-FLAG induced pRB degradation to levels similar to those seen with wild-type 16E7. Hence, although E7 degradation and E7-mediated pRB degradation are both proteasome-dependent processes, they occur independently of each other.
To determine whether degradation-resistant FLAG-16E7 was more effective in destabilizing pRB than 16E7-FLAG, we performed a quantitative comparison. Different amounts of FLAG-16E7 and 16E7-FLAG were cotransfected with a constant amount of the pRB expression plasmid in SAOS2 cells, and the steady-state levels of pRB and E7 were determined by immunoblotting. At comparable steady-state levels of the FLAG-tagged E7 proteins, pRB levels were twofold lower when degradation-resistant FLAG-16E7 was coexpressed than when degradation-sensitive 16E7-FLAG was coexpressed, indicating that FLAG-16E7 is more efficient in targeting pRB for degradation (Fig. 1F).
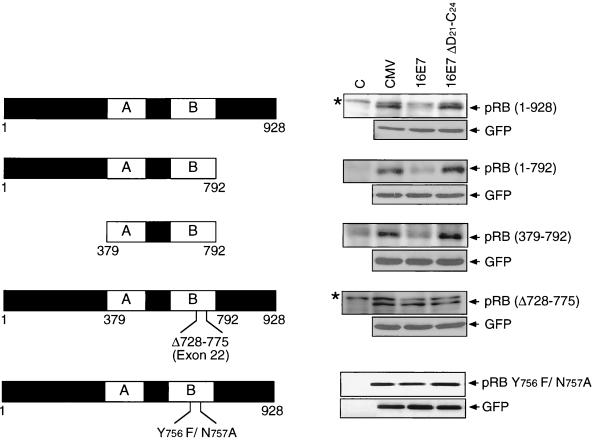
The “small pocket” of pRB is sufficient for E7-mediated pRB destabilization.
To map the structural domains of pRB that are necessary for degradation, we tested truncation mutants of pRB (Fig. 2). Deletion of the amino-terminal and/or the carboxyl-terminal domains of pRB did not interfere with the ability of E7 to target pRB for degradation. Hence, as previously reported (6), the E7-binding domain of pRB, the small pocket (50), is sufficient for degradation by E7. The binding of E7 to pRB is required for degradation, as a pRB mutant lacking amino acid residues 728 to 775, which encompass the core E7-binding site, and an E7-binding-defective pRB double point mutant (pRB Y756F/N757A) are not efficiently degraded by E7 (Fig. 2) (16, 41).
FIG. 2.
The small pocket of pRB is sufficient for E7-mediated degradation. SAOS2 cells were transiently transfected with the indicated combinations of plasmids encoding HA-tagged wild-type or mutant forms of pRB, the E7-binding-defective pRB double point mutant (pRB Y756F/N757A), wild-type HPV-16 E7 (16E7), and a pRB-binding-deficient HPV-16 E7 mutant (16E7 ΔD21-C24) or control vector (CMV). Samples (100 μg) were subjected to SDS-PAGE and immunoblot analysis for the pRB mutants using an HA antibody or a pRB antibody (upper panels) and cotransfected GFP control (lower panels). Untransfected cells were used as controls (C). Background bands are denoted by asterisks.
HPV-16 E7 destabilizes pRB family members p107 and p130.
E7 has also been shown to bind the pRB family members p107 and p130 through its LXCXE domain (18). Previous experiments showed that p107 levels were decreased in E7-expressing cell lines (30). However, other reports have suggested that neither p107 nor p130 levels are affected by E7 (6). Therefore, the effects of HPV-16 E7 on p107 and p130 steady-state levels and half-lives were determined with the SAOS2 cell transient assay.
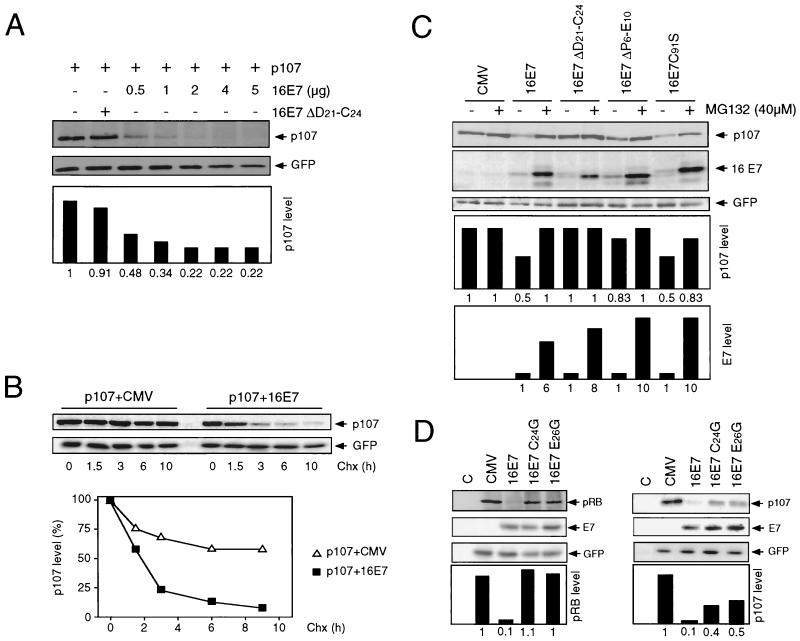
A p107 expression plasmid was transfected alone or in combination with wild-type HPV-16 E7 or the 16E7 ΔD21-C24 mutant. While the 16E7 ΔD21-C24 mutant had no effect on p107 levels, titration of wild-type HPV-16 E7 caused a dramatic dose-dependent decrease in p107 steady-state levels (Fig. 3A). Moreover, the p107 half-life was reduced from over 11 h to 2.5 h when HPV-16 E7 was coexpressed (Fig. 3B). As with pRB, binding of E7 to p107 was necessary but not sufficient for destabilization (Fig. 3C). Furthermore, E7-mediated degradation of p107 could be overcome by treatment with the 26S proteasome inhibitor MG132, suggesting that the proteasome is involved in E7-mediated p107 destabilization (Fig. 3C). To further substantiate that E7 binding to p107 is necessary for p107 degradation, we analyzed two E7 point mutants, 16E7 C24G and 16E7 E26G (14). It has been reported that these mutants are pRB binding deficient but retain binding to p107. Degradation assays showed that, consistent with their binding characteristics, these mutants did not destabilize pRB but retained their ability to target p107 for degradation (Fig. 3D).
FIG. 3.
HPV-16 E7 destabilizes p107 by a proteasome-dependent mechanism. (A) HPV-16 E7 decreases steady-state levels of p107. SAOS2 cells were transiently transfected with the indicated combinations of plasmids encoding HA-tagged p107, wild-type HPV-16 E7 (16E7), or the pRB-binding-deficient mutant 16E7 ΔD21-C24. Samples (100 μg) were subjected to SDS-PAGE and immunoblot analysis for p107 using an HA antibody (upper panel) and cotransfected GFP control (lower panel). Quantitation of p107 levels, normalized for GFP expression, is shown underneath these panels. (B) HPV-16 E7 decreases the half-life of p107. SAOS2 cells were transiently transfected with the indicated combinations of plasmids encoding HA-tagged p107 and HPV-16 E7 (16E7) or control plasmid (CMV). At 24 h posttransfection, protein synthesis was blocked by treatment with cycloheximide (Chx). At the indicated times, samples (100 μg) were harvested and then processed for immunoblot analysis for p107 using an HA antibody (upper panel) or cotransfected GFP control (lower panel). Quantitation of p107 levels, normalized for GFP expression, is shown underneath these panels. (C) Inhibition of the 26S proteasome blocks HPV-16 E7-mediated p107 degradation. SAOS2 cells were transfected with the indicated combinations of plasmids encoding HA-tagged p107, HPV-16 E7 (16E7), and HPV-16 E7 mutants 16E7 ΔD21-C24 (pRB binding and degradation deficient), 16E7 ΔP6-E10 (pRB binding competent and pRB degradation deficient), and 16E7 C91S (pRB binding and degradation competent) or control vector (CMV). Cells were treated with 40 μM MG132 (+) or mock treated with DMSO (−) for 4 h. Samples (100 μg) were subjected to SDS-PAGE and immunoblot analysis for p107 using an HA antibody (upper panel), HPV-16 E7 (middle panel), and cotransfected GFP control (lower panel). Quantitation of p107 and HPV-16 E7 levels, normalized for GFP expression, is shown underneath these panels. (D) Differential destabilization of pRB and p107 by pRB-binding-site E7 point mutants. SAOS2 cells were transfected with the indicated combinations of plasmids encoding pRB or HA-tagged p107, HPV-16 E7 (16E7), and HPV-16 E7 mutants 16E7 C24G and 16E7 E26G (pRB binding and degradation deficient, partially p107 binding competent) or control vector (CMV). C, untransfected controls. Samples (100 μg) were subjected to SDS-PAGE and immunoblot analysis for pRB (left upper panel), p107 using an HA antibody (right upper panel), HPV-16 E7 (middle panels), and cotransfected GFP control (lower panels). Quantitation of pRB and p107 levels, normalized for GFP expression, is shown underneath these panels.
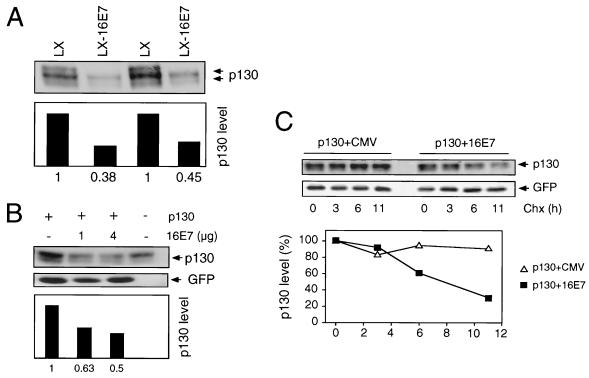
Next, we investigated the effect of HPV-16 E7 on p130 steady-state levels as well as half-life. The steady-state levels of p130 were clearly reduced in HPV-16 E7-expressing populations of HFKs (Fig. 4A). Similar E7-mediated decreases in p130 levels were also detected in the SAOS2 cell transient assay (Fig. 4B). These alterations were due to decreased protein stability, as the half-life of p130 was consistently decreased from over 11 h to approximately 6 h upon coexpression of E7 (Fig. 4C).
FIG. 4.
HPV-16 E7 destabilizes p130. (A) Decreased p130 steady-state levels in HPV-16 E7-expressing pools of HFKs. Two independent, matched populations of HFKs infected with control LXSN (LX) and HPV-16 E7-expressing LXSN (LX-16E7) vectors are shown. Pools of cells were obtained after G418 selection. Extracts (100 μg) were prepared and subjected to SDS-PAGE and p130 immunoblot analysis. Quantitation of p130 levels is shown underneath the blot. (B) HPV-16 E7 decreases p130 steady-state levels in the SAOS2 cell transient assay. SAOS2 cells were transiently transfected with the indicated combinations of plasmids encoding p130 and HPV-16 E7 (16E7). Samples (100 μg) were subjected to SDS-PAGE and immunoblot analysis for p130 (upper panel) and cotransfected GFP control (lower panel). Quantitation of p130 levels, normalized for GFP expression, is shown underneath these panels. Untransfected cells are shown as controls. (C) HPV-16 E7 decreases the half-life of p130. SAOS2 cells were transiently transfected with the indicated combinations of plasmids encoding p130 and HPV-16 E7 (16E7) or control plasmid (CMV). At 24 h posttransfection, protein synthesis was blocked by treatment with cycloheximide (Chx). At the indicated times, samples (100 μg) were harvested and then processed for immunoblot analysis for p130 (upper panel) or cotransfected GFP control (lower panel). Quantitation of p130 levels, normalized for GFP expression, is shown underneath these panels.
E7-mediated pRB degradation is important for functional inactivation of pRB in an SAOS2 flat-cell assay.
The flat-cell assay with SAOS2 cells represents a unique activity assay for pRB (24, 26). The assay is carried out by transfecting SAOS2 cells with a pRB expression plasmid and a selectable marker. Upon reexpression of pRB, SAOS2 cells arrest in G1. The G1 cell cycle arrest and continuous drug selection allow the pRB expression plasmid to be maintained episomally for over 2 weeks (24). During this time of pRB expression, SAOS2 cells undergo morphological changes that are collectively termed the flat-cell phenotype. These pRB-induced morphological changes in the SAOS2 cell line correlate with the expression of senescence-associated β-galactosidase (53) as well as osteogenic differentiation markers (41). Remarkably, induction of the flat-cell phenotype does not depend on the ability of pRB to interact with E2F-1, and it is unique to pRB, as the expression of p107 and p130 induces growth arrest but not the flat-cell phenotype (41). Cotransfection of pRB with inactivators of the tumor suppressor, such as adenovirus E1A or cyclin D, prevents the induction of the flat-cell phenotype (24).
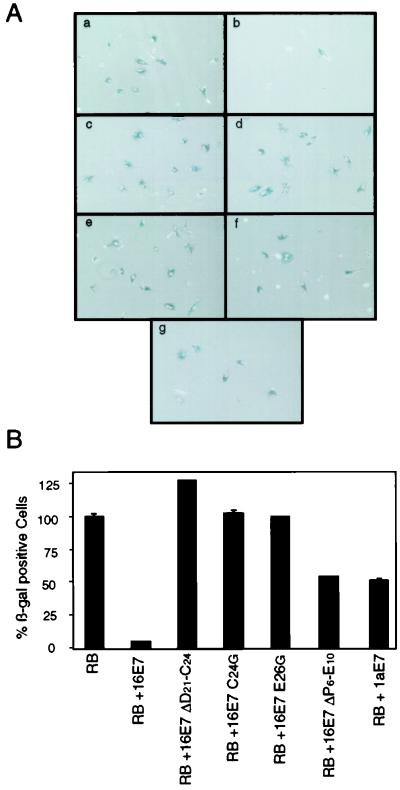
We carried out the SAOS2 flat-cell assay by transfecting pRB alone or in combination with wild-type HPV-16 E7 or E7 proteins that do not bind pRB (16E7 ΔD21-C24, 16E7 C24G, and 16E7 E26G) or that bind but do not degrade pRB (16E7 ΔP6-E10 and HPV-1a E7). Drug-resistant cells were evaluated for flat-cell formation and stained for senescence-associated β-galactosidase activity. Coexpression of HPV-16 E7 with pRB caused a 92% reduction in the average number of senescence-associated β-galactosidase-positive cells relative to the findings obtained with the pRB control (Fig. 5), while the pRB-binding-deficient 16E7 C24G, 16E7 E26G, and 16E7 ΔD21-C24 mutants had no effect on flat-cell induction (Fig. 5). Versions of HPV-16 E7 that bind but do not degrade pRB, such as the 16E7 ΔP6-E10 mutant and HPV-1a E7, caused a 50% reduction in β-galactosidase-positive cells (Fig. 5). This result suggests that the binding of E7 to pRB is only partially effective in inactivating pRB function in this assay.
FIG. 5.
Abrogation of pRB functions by HPV-16 E7-mediated degradation. (A) HPV-16 E7 inactivates pRB in an SAOS2 flat-cell assay. SAOS2 cells were transfected with the indicated combinations of plasmids encoding pRB alone (panel a) or in combination with carboxyl-terminal HA-tagged HPV-16 E7 (16E7) (panel b), HPV-16 E7 mutants 16E7 ΔD21-C24, 16E7 C24G, 16E7 E26G, and 16E7 ΔP6-E10 (panels c, d, e, and f, respectively), and HPV-1a E7 (panel g). At 10 days posttransfection, cells were stained for senescence-associated β-galactosidase (β-gal) activity. (B) Quantitation of the results from panel A. The results represent averages and standard deviations from two independent experiments. RB, pRB.
DISCUSSION
Retinoblastoma tumor suppressor protein pRB and the related “pocket proteins” p107 and p130 play important roles in regulating G1-S cell cycle checkpoint control. To establish and maintain a replication-competent host cellular milieu, the large tumor antigens of polyomaviruses, the adenovirus E1A proteins, and the E7 proteins of many HPVs each target pRB. Each of these viral oncoproteins contains a conserved sequence motif, LXCXE, that represents the core pRB-binding site. Despite this striking sequence conservation, the strategies that these viral oncoproteins have developed for pRB inactivation are surprisingly diverse. The adenovirus E1A proteins contain an additional low-affinity pRB-binding site in amino-terminal conserved region 1 (17). In polyomavirus tumor antigens, an amino-terminal J domain, the signature sequence of the DnaJ or Hsp40 chaperone proteins (32) that is dispensable for interaction with pRB, is required for functional pRB inactivation (43, 54). SV40 T-antigen-mediated disruption of pRB-E2F complexes is an ATP-dependent process, suggesting that SV40 T-antigen-mediated disruption of pRB function has characteristics of an enzymatic reaction (reviewed in reference 36).
Similarly, the notion that the high-risk HPV E7 proteins may target pRB by a stoichiometric protein-protein interaction has been challenged by reports that HPV-16 E7 can destabilize pRB (6, 7, 30). While there are pRB-binding-competent E7 mutants that are transformation or immortalization defective (4, 8, 20, 28, 35), the ability of E7 to target pRB for degradation correlates more closely with viral replication (48) and cellular transformation (31).
We have used a rapid SAOS2 cell-based assay to analyze the biochemical parameters of E7-mediated pRB degradation. In a previously reported assay using transient transfection with the SAOS2 cell line, HPV-16 E7 was able to induce pRB degradation; however, unlike the results for stable E7-expressing cells, p107 and p130 were not destabilized (6). In contrast, our assay with SAOS2 cells accurately parallels the activity of E7 in stable fibroblast and keratinocyte populations (31) (Fig. 1). E7-induced pRB degradation is a rapid and direct effect of E7 expression that is unaffected by the presence of p53 and/or the HPV-16 E6 oncoprotein (data not shown). In addition, E7 expression decreases the steady-state levels and half-lives of p107 (Fig. 3) and p130 (Fig. 4).
Proteasome inhibitor treatment caused not only an increase in pRB levels but also a dramatic augmentation of E7 levels. The HPV-16 E7 protein has a short half-life (45) and, as has been recently reported, is normally turned over through proteasomal degradation (37). Our results show that although E7 degradation and E7-mediated pRB degradation are both proteasome dependent, they are separable. The amino-terminal tagged version of E7 which is resistant to proteasomal degradation, presumably because it is not ubiquitinated (37), efficiently degrades pRB in the SAOS2 cell assay (Fig. 1E). Moreover, transformation-incompetent 16E7 ΔP6-E10 and HPV-1a E7, versions of E7 that bind pRB with an efficiency similar to that of HPV-16 E7 and are equally susceptible to proteasomal degradation, are unable to induce the degradation of pRB (Fig. 1D). Similarly, the pRB-binding-deficient HPV-16 E7 mutant 16E7 ΔD21-C24 is also stabilized under these conditions. Moreover, the proteasomal S4 subunit-binding-deficient mutant 16E7 C91S (5) is also stabilized, strongly suggesting that an interaction with the S4 subunit does not fully account for the rapid turnover of HPV-16 E7 (Fig. 1D). Taken together, our results suggest that E7-mediated pRB degradation is not directly related to E7 turnover.
The biological activities of pRB are primarily regulated by phosphorylation and by phosphorylation-dependent binding to regulatory proteins. However, pRB is also regulated at the level of protein turnover. pRB is cleaved by caspases during apoptosis (2, 12, 27, 47). However, a general inhibitor of caspases does not markedly interfere with E7-induced pRB degradation (30) (data not shown), and versions of pRB lacking the major caspase cleavage sites in the carboxyl terminus are all efficiently degraded by E7 (Fig. 2). Most strikingly, pRB is targeted for proteasomal degradation by gankyrin, an ankyrin repeat protein (23). Overexpression of gankyrin and decreased pRB levels have been observed for hepatomas, correlating pRB degradation with transformation (23).
What is the mechanism of E7-induced pRB degradation? At this point we have not yet identified the factor(s) that is involved, but our studies have started to illuminate this process. One possible model is that E7 induces pRB degradation through an additional cellular factor, similar to p53 degradation through a complex of high-risk HPV E6 proteins with the E6-AP ubiquitin ligase (38). This model is supported by the finding that coexpression of the pRB-binding-deficient HPV-16 E7 mutant 16E7 ΔD21-C24 causes a consistent increase in pRB-induced flat-cell formation in SAOS2 cells (Fig. 5). Based on the inability of the 16E7 ΔP6-E10 mutant to degrade pRB, we propose that the amino terminus of HPV-16 E7 may be one of the determinants for E7-mediated pRB degradation. Alternatively, the binding of HPV-16 E7 may displace a stabilizing cellular factor bound to pRB. Our results do not conclusively rule out this model, but we consider it less likely, given that HPV-1a E7, adenovirus E1A, and SV40 T antigen do not interfere with pRB stability (Fig. 1).
In these and other experiments, we have been unable to unambiguously demonstrate that E7 induces multiubiquitination of pRB. Interestingly, pRB ubiquitination has not been observed in gankyrin-mediated proteasomal degradation of pRB (23). Hence, it is possible that E7-mediated pRB degradation by the proteasome may not require ubiquitination, adding pRB to a growing list of proteins that are turned over by the proteasome independent of multiubiquitination (42).
Our studies also demonstrate that HPV-16 E7 can target the pRB family members p107 and p130 for destabilization. By use of point mutations within the pRB-binding site of E7 that abrogate pRB binding but not p107 binding (14), resulting in mutants 16E7 C24G and 16E7 E26G, p107 can be specifically targeted for degradation by E7 (Fig. 3D). Interestingly, although 16E7 C24G and 16E7 E26G are impaired for transformation, both mutants retain activities in human cell immortalization assays comparable to that of HPV-16 E7 (14); this result suggests that E7-mediated p107 destabilization may contribute to such activity. The pRB-related pocket proteins p107 and p130 function as efficient growth suppressors and target E2F transcription factor complexes, but they have not been conclusively connected with tumor suppressor activity. However, recent studies have shown that inactivation of pRB as well as p107 and p130 is necessary for SV40 T-antigen-mediated transformation (46). Furthermore, p16INK4a-induced cell cycle arrest requires p107 and/or p130, even in the presence of functional pRB (9). Some reports have also implicated p130 in regulating cellular differentiation (reviewed in reference 21). HPV-16 E7 has also been linked to inhibition of differentiation, and it is possible that E7-induced p130 degradation contributes to the ability of E7 to delay keratinocyte differentiation (29).
Most importantly, our studies demonstrate that E7-mediated pRB degradation is necessary for efficient inactivation of pRB function (Fig. 5). The SAOS2 flat-cell assay that we used to address this issue probes several independent biological activities of pRB. In addition to E2F-mediated G1 growth arrest, this assay also detects a second function that maps to a carboxyl-terminal domain of pRB (41). Although the molecules that functionally interface with this region of pRB are unknown, they regulate cellular senescence and/or differentiation (41, 53) and comprise an important facet of the function of pRB as a tumor suppressor (41). Hence, the ability of E7 to target pRB for proteolytic degradation provides an effective mechanism to simultaneously subvert the multiple biological activities of this tumor suppressor protein.
ACKNOWLEDGMENTS
We thank D. Cobrinik, N. Dyson, D. A. Galloway, P. Howley, W. Sellers, M. Tommasino, and J. Wang for providing expression plasmids and the members of the laboratories of K. Münger and P. Hinds, especially Lily Yeh-Lee, Siribang-on Piboonniyom, and David Thomas, for helpful advice and technical assistance. We are grateful to Phil Hinds, Grace Gill, and Valerie Zacny for critical comments on the manuscript.
This work was supported by NIH grant CA66980 (to K.M.). S.L.G. was supported in part by the Training Program in Environmental Health Sciences, Program in Biological Sciences in Public Health, Harvard School of Public Health, grant NIH 2T32ES07155, and a dissertation fellowship from the Ford Foundation. K.M. is a Ludwig Scholar.
REFERENCES
- 1.Alunni-Fabbroni M, Littlewood T, Deleu L, Caldeira S, Giarre M, Dell' Orco M, Tommasino M. Induction of S phase and apoptosis by the human papillomavirus type 16 E7 protein are separable events in immortalized rodent fibroblasts. Oncogene. 2000;19:2277–2285. doi: 10.1038/sj.onc.1203570. [DOI] [PubMed] [Google Scholar]
- 2.An B, Dou Q P. Cleavage of retinoblastoma protein during apoptosis: an interleukin 1 beta-converting enzyme-like protease as candidate. Cancer Res. 1996;56:438–442. [PubMed] [Google Scholar]
- 3.Baker S J, Markowitz S, Fearon E R, Willson J K V, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 4.Banks L, Edmonds C, Vousden K. Ability of the HPV16 E7 protein to bind RB and induce DNA synthesis is not sufficient for efficient transforming activity in NIH 3T3 cells. Oncogene. 1990;5:1383–1389. [PubMed] [Google Scholar]
- 5.Berezutskaya E, Bagchi S. The human papillomavirus E7 oncoprotein functionally interacts with the S4 subunit of the 26S proteasome. J Biol Chem. 1997;272:30135–30140. doi: 10.1074/jbc.272.48.30135. [DOI] [PubMed] [Google Scholar]
- 6.Berezutskaya E, Yu B, Morozov A, Raychaudhuri P, Bagchi S. Differential regulation of the pocket domains of the retinoblastoma family proteins by the HPV16 E7 oncoprotein. Cell Growth Differ. 1997;8:1277–1286. [PubMed] [Google Scholar]
- 7.Boyer S N, Wazer D E, Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- 8.Brokaw J L, Yee C L, Münger K. A mutational analysis of the amino terminal domain of the human papillomavirus type 16 E7 oncoprotein. Virology. 1994;205:603–607. doi: 10.1006/viro.1994.1688. [DOI] [PubMed] [Google Scholar]
- 9.Bruce J L, Hurford R K, Classon M, Koh J, Dyson N J. Requirements for cell cycle arrest by p16ink4a. Mol Cell. 2000;6:737–742. doi: 10.1016/s1097-2765(00)00072-1. [DOI] [PubMed] [Google Scholar]
- 10.Chellappan S, Kraus V B, Kroger B, Münger K, Howley P M, Phelps W C, Nevins J R. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between the transcription factor E2F and the retinoblastoma gene product. Proc Natl Acad Sci USA. 1992;89:4549–4553. doi: 10.1073/pnas.89.10.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen W D, Otterson G A, Lipkowitz S, Khleif S N, Coxon A B, Kaye F J. Apoptosis is associated with cleavage of a 5 kDa fragment from RB which mimics dephosphorylation and modulates E2F binding. Oncogene. 1997;14:1243–1248. doi: 10.1038/sj.onc.1201096. [DOI] [PubMed] [Google Scholar]
- 13.Ciccolini F, Dipasquale G, Carlotti F, Crawford L, Tommasino M. Functional studies of E7 proteins from different HPV types. Oncogene. 1994;9:2633–2638. [PubMed] [Google Scholar]
- 14.Davies R, Hicks R, Crook T, Morris J, Vousden K. Human papillomavirus type 16 E7 associates with a histone H1 kinase and with p107 through sequences necessary for transformation. J Virol. 1993;67:2521–2528. doi: 10.1128/jvi.67.5.2521-2528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeCaprio J A, Ludlow J W, Figge J, Shew J Y, Huang C M, Lee W H, Marsilio E, Paucha E, Livingston D M. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 16.Dick F A, Sailhamer E, Dyson N J. Mutagenesis of the pRB pocket reveals that cell cycle arrest functions are separable from binding to viral oncoproteins. Mol Cell Biol. 2000;20:3715–3727. doi: 10.1128/mcb.20.10.3715-3727.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dyson N, Guida P, McCall C, Harlow E. Adenovirus E1A makes two distinct contacts with the retinoblastoma protein. J Virol. 1992;66:4606–4611. doi: 10.1128/jvi.66.7.4606-4611.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dyson N, Guida P, Münger K, Harlow E. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J Virol. 1992;66:6893–6902. doi: 10.1128/jvi.66.12.6893-6902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dyson N, Howley P M, Münger K, Harlow E. The human papillomavirus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 20.Edmonds C, Vousden K H. A point mutational analysis of human papillomavirus type 16 E7 protein. J Virol. 1989;63:2650–2656. doi: 10.1128/jvi.63.6.2650-2656.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grana X, Garriga J, Mayol X. Role of the retinoblastoma protein family, pRB, p107 and p130 in the negative control of cell growth. Oncogene. 1998;17:3365–3383. doi: 10.1038/sj.onc.1202575. [DOI] [PubMed] [Google Scholar]
- 22.Heck D V, Yee C L, Howley P M, Münger K. Efficiency of binding to the retinoblastoma protein correlates with the transforming capacity of the E7 oncoproteins of the human papillomaviruses. Proc Natl Acad Sci USA. 1992;89:4442–4446. doi: 10.1073/pnas.89.10.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higashitsuji H, Itoh K, Nagao T, Dawson S, Nonoguchi K, Kido T, Mayer R J, Arii S, Fujita J. Reduced stability of retinoblastoma protein by gankyrin, an oncogenic ankyrin-repeat protein overexpressed in hepatomas. Nat Med. 2000;6:96–99. doi: 10.1038/71600. [DOI] [PubMed] [Google Scholar]
- 24.Hinds P W, Mittnacht S, Dulic V, Arnold A, Reed S I, Weinberg R A. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 25.Howley P M. Papillomavirinae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 947–978. [Google Scholar]
- 26.Huang H-J S, Yee J-K, Shew J-Y, Chen P-L, Bookstein R, Friedmann T, Lee E Y-H P. Suppression of the neoplastic phenotype by replacement of the RB gene in human cancer cells. Science. 1988;242:1563–1566. doi: 10.1126/science.3201247. [DOI] [PubMed] [Google Scholar]
- 27.Janicke R U, Walker P A, Lin X Y, Porter A G. Specific cleavage of the retinoblastoma protein by an ICE-like protease in apoptosis. EMBO J. 1996;15:6969–6978. [PMC free article] [PubMed] [Google Scholar]
- 28.Jewers R J, Hildebrandt P, Ludlow J W, Kell B, McCance D J. Regions of human papillomavirus type 16 E7 oncoprotein required for immortalization of human keratinocytes. J Virol. 1992;66:1329–1335. doi: 10.1128/jvi.66.3.1329-1335.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones D L, Alani R M, Münger K. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 1997;11:2101–2111. doi: 10.1101/gad.11.16.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones D L, Münger K. Analysis of the p53-mediated G1 growth arrest pathway in cells expressing the human papillomavirus type 16 E7 oncoprotein. J Virol. 1997;71:2905–2912. doi: 10.1128/jvi.71.4.2905-2912.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones D L, Thompson D A, Münger K. Destabilization of the RB tumor suppressor and stabilization of p53 contribute to HPV type 16 E7-induced apoptosis. Virology. 1997;239:97–107. doi: 10.1006/viro.1997.8851. [DOI] [PubMed] [Google Scholar]
- 32.Kelley W L, Georgopoulos C. The T/t common exon of simian virus 40, JC, and BK polyomavirus T antigens can functionally replace the J-domain of the Escherichia coli DnaJ molecular chaperone. Proc Natl Acad Sci USA. 1997;94:3679–3684. doi: 10.1073/pnas.94.8.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kessis T D, Slebos R J, Nelson W G, Kastan M B, Plunkett B S, Han S M, Lorincz A T, Hedrick L, Cho K R. Human papillomavirus 16 E6 expression disrupts the p53-mediated cellular response to DNA damage. Proc Natl Acad Sci USA. 1993;90:3988–3992. doi: 10.1073/pnas.90.9.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Münger K, Werness B A, Dyson N, Phelps W C, Harlow E, Howley P M. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phelps W C, Münger K, Yee C L, Barnes J A, Howley P M. Structure-function analysis of the human papillomavirus E7 oncoprotein. J Virol. 1992;66:2418–2427. doi: 10.1128/jvi.66.4.2418-2427.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pipas J M. Molecular chaperone function of the SV40 large T antigen. Dev Biol Stand. 1998;94:313–319. [PubMed] [Google Scholar]
- 37.Reinstein E, Scheffner M, Oren M, Ciechanover A, Schwartz A. Degradation of the E7 human papillomavirus oncoprotein by the ubiquitin-proteasome system: targeting via ubiquitination of the N-terminal residue. Oncogene. 2000;19:5944–5950. doi: 10.1038/sj.onc.1203989. [DOI] [PubMed] [Google Scholar]
- 38.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 39.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 40.Schmitt A, Harry J B, Rapp B, Wettstein F O, Iftner T. Comparison of the properties of the E6 and E7 genes of low- and high-risk cutaneous papillomaviruses reveals strongly transforming and high RB-binding activity for the E7 protein of the low-risk human papillomavirus type 1. J Virol. 1994;68:7051–7059. doi: 10.1128/jvi.68.11.7051-7059.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sellers W R, Novitch B G, Miyake S, Heith A, Otterson G A, Kaye F J, Lassar A B, Kaelin W G., Jr Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheaff R J, Singer J D, Swanger J, Smitherman M, Roberts J M, Clurman B E. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol Cell. 2000;5:403–410. doi: 10.1016/s1097-2765(00)80435-9. [DOI] [PubMed] [Google Scholar]
- 43.Sheng Q, Denis D, Ratnofsky M, Roberts T M, DeCaprio J A, Schaffhausen B. The DnaJ domain of polyomavirus large T antigen is required to regulate Rb family tumor suppressor function. J Virol. 1997;71:9410–9416. doi: 10.1128/jvi.71.12.9410-9416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shew J Y, Lin B T, Chen P L, Tseng B Y, Yang-Feng T L, Lee W H. C-terminal truncation of the retinoblastoma gene product leads to functional inactivation. Proc Natl Acad Sci USA. 1990;87:6–10. doi: 10.1073/pnas.87.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smotkin D, Wettstein F O. The major human papillomavirus protein in cervical cancers is a cytoplasmic phosphoprotein. J Virol. 1987;61:1686–1689. doi: 10.1128/jvi.61.5.1686-1689.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stubdal H, Zalvide J, Campbell K S, Schweitzer C, Roberts T M, DeCaprio J A. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol Cell Biol. 1997;17:4979–4990. doi: 10.1128/mcb.17.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan X, Martin S J, Green D R, Wang J Y J. Degradation of retinoblastoma protein in tumor necrosis factor- and CD95-induced cell death. J Biol Chem. 1997;272:9613–9616. doi: 10.1074/jbc.272.15.9613. [DOI] [PubMed] [Google Scholar]
- 48.Thomas J T, Hubert W G, Ruesch M N, Laimins L A. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc Natl Acad Sci USA. 1999;96:8449–8454. doi: 10.1073/pnas.96.15.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Timmermann S, Hinds P W, Munger K. Re-expression of endogenous p16ink4a in oral squamous cell carcinoma lines by 5-aza-2′-deoxycytidine treatment induces a senescence-like state. Oncogene. 1998;17:3445–3453. doi: 10.1038/sj.onc.1202244. [DOI] [PubMed] [Google Scholar]
- 50.Welch P J, Wang J Y J. A C-terminal protein-binding domain in the retinoblastoma protein regulates nuclear c-abl tyrosine kinase in the cell cycle. Cell. 1993;75:779–790. doi: 10.1016/0092-8674(93)90497-e. [DOI] [PubMed] [Google Scholar]
- 51.Werness B A, Levine A J, Howley P M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 52.Whyte P, Buchkovich K J, Horowitz J M, Friend S H, Raybuck M, Weinberg R A, Harlow E. Association between an oncogene and an antioncogene: the adenovirus E1a proteins bind to the retinoblastoma gene product. Nature. 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- 53.Xu H J, Zhou Y, Ji W, Perng G S, Kruzelock R, Kong C T, Bast R C, Mills G B, Li J, Hu S X. Reexpression of the retinoblastoma protein in tumor cells induces senescence and telomerase inhibition. Oncogene. 1997;15:2589–2596. doi: 10.1038/sj.onc.1201446. [DOI] [PubMed] [Google Scholar]
- 54.Zalvide J, Stubdal H, DeCaprio J A. The J domain of simian virus 40 large T antigen is required to functionally inactivate RB family proteins. Mol Cell Biol. 1998;18:1408–1415. doi: 10.1128/mcb.18.3.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.zur Hausen H. Papillomavirus infections—a major cause of human cancers. Biochim Biophys Acta. 1996;1288:F55–F78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]