Abstract
MicroRNAs (miRNAs) have been implicated in human disorders, from cancers to infectious diseases. Targeting miRNAs or their target genes with small molecules offers opportunities to modulate dysregulated cellular processes linked to diseases. Yet, predicting small molecules associated with miRNAs remains challenging due to the small size of small molecule-miRNA datasets. Herein, we develop a generalized deep learning framework, sChemNET, for predicting small molecules affecting miRNA bioactivity based on chemical structure and sequence information. sChemNET overcomes the limitation of sparse chemical information by an objective function that allows the neural network to learn chemical space from a large body of chemical structures yet unknown to affect miRNAs. We experimentally validated small molecules predicted to act on miR-451 or its targets and tested their role in erythrocyte maturation during zebrafish embryogenesis. We also tested small molecules targeting the miR-181 network and other miRNAs using in-vitro and in-vivo experiments. We demonstrate that our machine-learning framework can predict bioactive small molecules targeting miRNAs or their targets in humans and other mammalian organisms.
Subject terms: Machine learning, RNA, miRNAs, Drug discovery
Here the authors developed and experimentally validated sChemNET, a deep learning framework to predict small molecules affecting microRNA function based on chemical structure and sequence data. sChemNET predicts bioactive small molecules on the basis of sparse chemical datasets.
Introduction
RNA molecules, essential to cellular function, are required for cellular information transfer, cell structure, and gene regulation and have been implicated in many human diseases1–3. One primary class of small non-coding regulatory RNAs, the microRNAs (miRNAs), are central to post-transcriptional gene regulation, modulating the levels of more than half of the human transcripts4. Dysregulated miRNAs have been implicated in the pathology of metabolic and cardiovascular disorders, cancers, hepatitis, and emergent infectious diseases such as COVID-195–8. Evidence also indicates that highly stable circulating miRNAs can be present in the blood of diseased individuals8–10, which implies that miRNAs could be helpful as biomarkers and therapeutic targets.
miRNAs have also been validated as therapeutic targets11,12. Complementary oligonucleotides are being developed to inhibit miRNAs13. For instance, mimics of miR-34 have been designed to repress oncogene expression and block tumor growth14; oligonucleotides complementary to miR-122 are being developed to treat hepatitis C virus15, and an antisense oligonucleotide to miR-2392 is being explored for treating COVID-198. However, the development of therapeutic oligonucleotides to target miRNA has proven challenging due to the requirements of delivery technology, stability, and potentially high toxicity16.
An attractive alternative is to target miRNAs or their gene targets with small molecules17,18. Numerous studies (see review by Chen et al.19) have demonstrated that miRNA levels or their targets can be modulated by small molecules20, and systematic principles for targeting RNAs with small molecules are under development12,21–23. However, this avenue of research is hindered by a lack of ability to infer which small molecules will be bioactive against which miRNAs19. The major challenge in identifying bioactive small molecules that interact with a given miRNA or its function is the limited predictive understanding of the chemical principles by which molecules are bioactive against miRNAs21.
To assist researchers in developing small molecule modulators of miRNAs or their downstream effectors, we proposed and developed a deep-learning framework called sChemNET. sChemNET predicts scores that a given small molecule can modulate a given miRNA, or the expression of its mRNA targets, based on its chemical features. Previous machine-learning models for predicting small molecules targeting miRNAs could be applied only to a small set of small molecules that had been experimentally identified24–26. In contrast, our approach can be applied to infer novel bioactive chemicals from any chemical library with information about the 2D chemical structure of the compounds.
sChemNET’s ability to integrate chemical structure information of small molecules with previously unknown interactions with miRNAs is critical to achieving good prediction performance for the bioactivities of a broader set of small molecules. Furthermore, sChemNET can be helpful for smaller chemical datasets available for model organisms by integrating cross-species data and modeling miRNAs sequence information.
We used sChemNET’s predictions to create a mapping between pharmacological classes of small molecules and miRNAs and exploit this map to design machine learning-guided in-vitro and in-vivo assays that validate sChemNET-predicted small molecules for miR-451, an erythrocyte-specific miRNA whose demise leads to profound anemia under oxidative stress, and also, to experimentally validate the predicted effects of vitamin D on other miRNAs relevant to breast cancer and the mitochondria.
Results
sChemNET: a deep learning framework for predicting drug targets in the presence of sparse and small-size chemical datasets
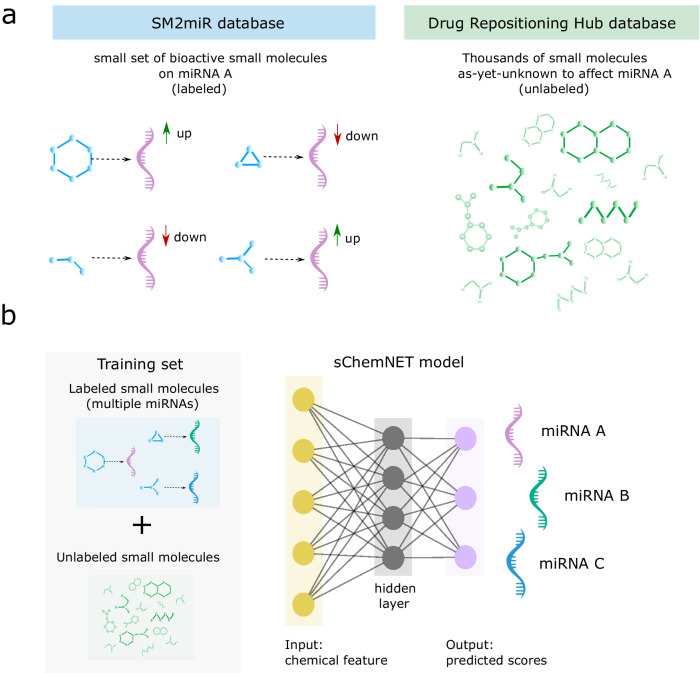
We developed a deep-learning predictive model that incorporates information about small molecules with known and yet-unknown biological activity on miRNAs in a neural network model to predict small molecules targeting miRNAs (or their downstream targets) on the basis of their chemical structure alone. We combined ~2400 such “unlabeled” small molecules with a small number of “labeled” small molecules (i.e., known to affect miRNA expression levels or the expression of its targets) to build a two-layered neural network for small chemical datasets (sChemNET) – see Fig. 1a. In sChemNET, the chemical structure information of the labeled and unlabeled small molecules is fed into the model and distributed over a set of hidden layers of nodes (Fig. 1b). The output layer of the network represents each of the miRNAs, and the model outputs a predicted score for each miRNA for a given small molecule’s chemical feature.
Fig. 1. Overview of our deep learning framework for predicting miRNA targeted by small molecules in the presence of sparse and small-size chemical datasets.
a sChemNET integrates labeled and unlabeled chemical structure information to predict bioactive small molecules against miRNAs or their mRNA targets. (Left) Labeled small molecules (sky-blue) are known to affect the expression level of miRNAs or their mRNA targets, as curated in the SM2miR database. The dotted arrow represents the experimentally verified small molecule-miRNA association and the up and down-arrows (in green and red, respectively) represent whether there is up-or down-regulation of the expression level (Right). The Drug Repurposing Hub database was used to obtain thousands of small molecules yet unknown to affect miRNAs (a.k.a. unlabeled) shown in green. b sChemNET is a two-layered, fully connected neural network model that incorporates unlabeled chemical structure information during training to enhance prediction performance when only a small set of bioactive small molecule-miRNA dataset is available for training. The trained sChemNET model provides predicted scores for each miRNA given a small molecule’s chemical fingerprint (obtained from its 2D chemical structure representation). Nodes represent input chemical features (yellow), hidden units (gray), and miRNAs’ predicted scores represent the output (purple). Solid lines show the connection between the layers. Molecules with known bioactivity, labeled, molecules without a bioactivity designation, unlabeled. Different miRNAs are illustrated with different colors.
sChemNET’s key idea is to train a learning model using a large amount of unlabeled chemical structure information. To learn the probability that the ith small molecule, with chemical feature , affects the uth miRNA, sChemNET aims to minimize the following loss function:
| 1 |
The first summation in our model applies a fitting constraint to the labeled chemical information (small molecule-miRNA associations () with ), designed to learn a high prediction score for known associations between small molecules and miRNAs. To learn a model for each miRNA , sChemNET exploits the labeled information available for all other miRNAs, such that their relative contribution to the learning is weighted based on their sequence similarities to the miRNA target through , where (see “Methods” section). The second summation in Eq. (1) is the fitting constraint on the unlabeled chemical information (small molecule-miRNA associations () with ). Unlabeled small molecules are assigned low prediction scores to each miRNA during learning, and their overall relative importance is controlled with the hyperparameter . Typically, is used. Unlabeled small molecules have unknown biological activity against targeted miRNA , and they are introduced here due to the small-size chemical dataset available for training the model. The goal of the second term is to allow the neural network to learn from a broader range of chemical space that is mapped to a low probability score; our modeling was motivated by our recent work on zero-driven regularization in non-negative matrix decomposition models27,28.
Predicting small molecules targeting miRNAs or their targets in Homo sapiens
We trained and tested sChemNET using small-size chemical datasets with labeled information about the bioactivities of each small molecule on miRNAs. We used the Small Molecule to miRNA (SM2miR)20 database to obtain manually curated information on small molecules that affect the expression levels of either specific miRNAs or their corresponding mRNA targets (see “Methods” section). Our dataset only provides positive label information, that is, true positives, or (Eq. 1). There is not explicit source for negative labels and in Eq. (1) represents unlabeled small molecule-miRNA associations. In SM2miR, we found several small molecule-miRNA associations across 18 species (Supplementary Fig. 1). For Homo sapiens, we used 1102 associations between 131 small molecules and 126 miRNAs (see Supplementary Fig. 2a). The number of bioactive small molecules for each miRNA varies between 5 to 35, and its distribution follows a long-tailed pattern (see Supplementary Fig. 2a). The average number of shared bioactive small molecules for miRNAs is (mean ), indicating that most miRNAs tend to share a small number of bioactive small molecules (see distribution in Supplementary Fig. 3). To obtain a large set of unlabeled small molecules, we used the Drug Repurposing Hub database29, which contains structurally and therapeutically diverse small molecules that have reached clinical trials; including most FDA-approved drugs. We obtained 6,302 unlabeled small molecules with unique PubChem CIDs that we used together with the 131 small molecules from SM2miR to build a set of 6,433 small molecules. Chemical input feature information for each small molecule was obtained from their MACCS chemical fingerprints calculated from their SMILES representation. Sequence similarities between miRNAs were obtained by re-scaling the Needleman-Wunsch score obtained using miRNA mature sequences from the miRBase database30 (see “Methods” section and Supplementary Fig. 4).
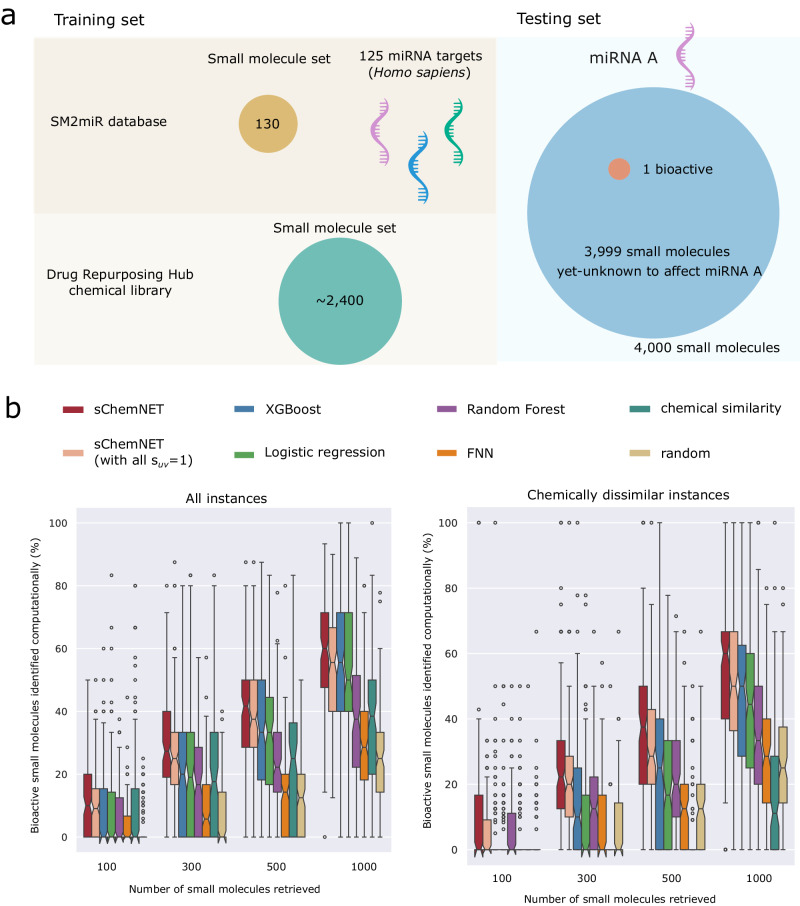
sChemNET’s ability to integrate large amounts of chemical structure information in the presence of small bioactive chemical datasets allows us to simulate a realistic scenario in which a small molecule biologically active against a miRNA is recovered from a large pool of chemicals. To this end, for each known bioactive small molecule-miRNA association, we built a test set containing 4000 small molecules, where only one was experimentally determined to be bioactive, and 3999 were randomly selected small molecules currently unknown to affect miRNAs (see Fig. 2a). We then performed a systematic evaluation using leave-one-out cross-validation (LOOCV) for all the miRNAs (see “Methods” section). For each miRNA, we trained sChemNET with the remaining labeled and unlabeled small molecules and used the trained model to rank all the 4000 small molecules in the test set by their predicted scores. The model’s prediction performance was assessed based on the percentage of known bioactive small molecules that could be retrieved amongst the top 100, 300, 500, or 1000 predicted small molecules.
Fig. 2. sChemNET outperforms other methods at predicting small molecules bioactive against 125 miRNAs in Homo sapiens.
a Training and testing set with labeled and unlabeled chemical structure information built in. Molecules and activity data were selected from the SM2miR and the Drug Repurposing Hub databases. We used these to assess the prediction performance of sChemNET and other computational approaches under a leave-one-out cross validation procedure. (Left) The training set consists of labeled and unlabeled small molecules. The labeled compounds are small molecules known to be bioactive against at least one miRNA from Homo sapiens. The unlabeled set of compounds consists of ~2400 randomly selected small molecules without known activity against the set of miRNAs. (Right) The testing set consists of 4000 small molecules of which only a single small molecule is known to be bioactive against the specific miRNA under evaluation. b Prediction performance of eight computational methods obtained under our leave-one-out cross-validation procedure. Notched boxplots show the distribution around the median of the percentage of recall obtained for 125 miRNAs (y-axis) when retrieving top-K (100, 300, 500, 1000) small molecules from the test set. (Left) When considering all the 1102 instances; (Right) When considering only chemically dissimilar instances between training and testing sets. sChemNET was run with and without () miRNA sequence similarity information in its loss function. FNN stands for Feed-Forward Neural Network and “random” for scores sampled from a uniform distribution. The chemical similarity baseline ranks all the small molecules in the test set based on the max 2D Tanimoto chemical similarity to the bioactive small molecules in the training set. The distribution shows the variation of recall across the 125 miRNAs for all the methods. For the boxplots the center line represents the median and the lines extending from both ends of the box indicate the quartile (Q) variability outside Q1 and Q3 to the minimum and maximum values. The notch represents the 95% confidence interval of the median.
Following Stokes et al.31, to select the model hyperparameters- number of hidden units (), unlabeled regularization parameter (), number of epochs, learning rate, and dropout-, we used a Bayesian optimization approach to hyperparameter search based on a LOOCV of the small molecules known to target miR-224-5p. This miRNA was randomly selected and excluded from further evaluation analysis. sChemNET performed well with , , dropout = 0.174, and learning rate = 0.0346.
Figure 2b (Left) shows sChemNET’s prediction performance at retrieving bioactive small molecules for 125 miRNAs in Homo sapiens. The recall is shown as a percentage (-axis) as a function of the top- number of small molecules retrieved from the test set (-axis). The performance of sChemNET, shown with and without () integrating sequence similarity information is compared with four machine-learning baseline methods that were trained using the same input feature information as sChemNET: XGBoost, Logistic Regression (LR), Random Forest (RF), and a Feed-Forward Neural Network (FNN), and two other baseline approaches that rank each of the 4000 small molecules in the test set based on: (i) the maximum Tanimoto chemical similarity to the set of bioactive small molecules in the training set (chemical similarity, green bars) or (ii) random scores assigned to each small molecule when sampling from a uniform distribution between 0 and 1 (random, brown bars) (see “Methods” section). sChemNET outperforms the baseline methods at different numbers of predictions retrieved by 1–9% for the top 100 small molecules retrieved from the test set, 7–21% (top 300), 5–33% (top 500), and 8-29% in the top 1000. We found that the average improvement in prediction performance of sChemNET over all the competitors is statistically significant (Supplementary Fig. 5). sChemNET achieves good prediction performance even without using sequence similarity information in the loss function (see also Supplementary Fig. 6) but with a slight reduction in prediction performance of ~1.81–3.62% across the different top-K thresholds. In Supplementary Fig. 7, we also show that sChemNET outperforms the competitors in terms of the area under the receiver operating characteristic curve (AUROC) obtained for three miRNAs with the largest number of positive labels.
A key question regarding the utility of our approach in practice concerns its ability to predict bioactive small molecules chemically dissimilar from those available for training the model. Figure 2b (Right) shows the prediction performance of sChemNET when considering instances in which the bioactive small molecules in the test set were chemically dissimilar from those available in the training set (Tanimoto similarity <0.6, see “Methods” section). sChemNET significantly outperforms the baseline methods by 5-9% in the top 100 small molecules retrieved from the test set, 10-24% (top 300), 10-40% (top 500), and 12-34% in the top 1000. Our findings suggest that sChemNET could be helpful in the discovery of novel small molecule modulators of miRNAs or their downstream targets.
The small size of the best-available labeled chemical dataset, which we used for training sChemNET, prompted us to ask whether sChemNET prediction performance varies with the number of bioactive small molecules available for each miRNA target. Supplementary Figure 8 shows the predicted rank of the active small molecules as a function of the number of labels available for training. sChemNET effectively retrieves bioactive small molecules even when as few as four or five bioactive small molecules are available for training the model.
We further performed a prospective evaluation in which we used all our available data from SM2miR 2015 as a training set, and 1180 new associations between 120 small molecules and 123 miRNAs as a test set that we obtained from the RNAInter 2022 database32. This evaluation is a realistic scenario that preserves the chronological order in which the information becomes available. sChemNET outperforms all the baselines methods in the prospective evaluation (Supplementary Fig. 9).
Predicting small molecules targeting miRNAs or their targets in model organisms
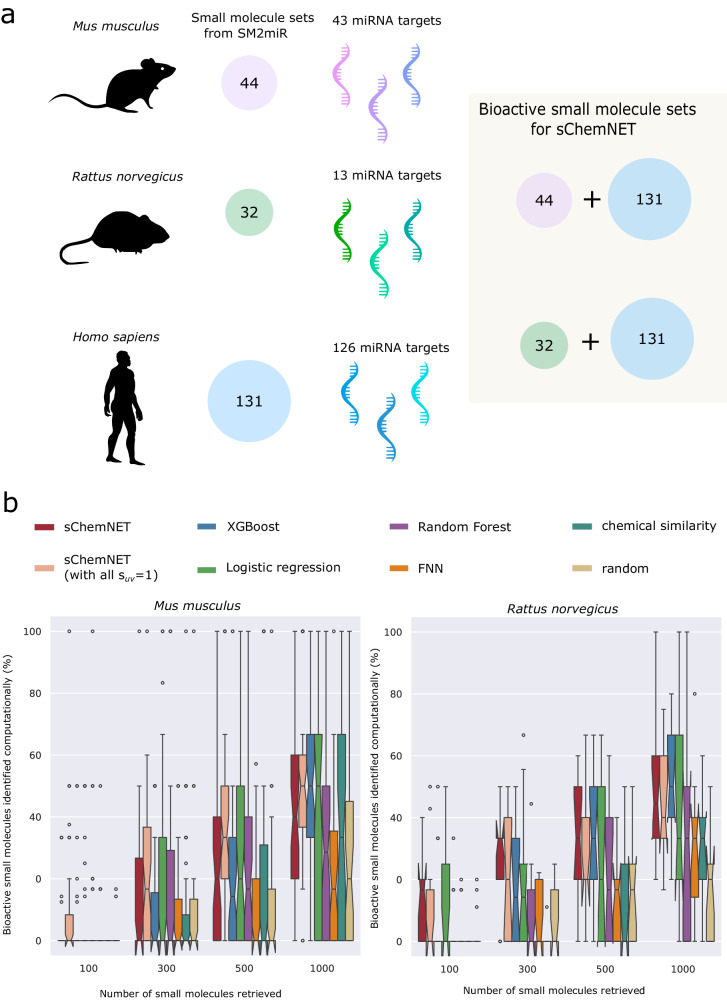
To understand whether sChemNET can be helpful for chemical datasets available for mammalian model organisms, we assessed its prediction performance in small molecule-miRNA datasets available for Mus musculus and Rattus norvergicus. Since fewer miRNA-small molecule associations are known for these models than for Homo sapiens (see the distribution of labeled information in Supplementary Fig. 2b, c), we combined miRNA information from Homo sapiens to train sChemNET for each model organism (see Fig. 3a and “Methods” section). Like our evaluation for Homo sapiens, we combined chemical data from the Drug Repurposing Hub to obtain a broader range of unlabeled chemical structures and performed a LOOCV procedure on the bioactive small molecules against miRNA targets in Mus musculus and Rattus norvergicus, respectively (see “Methods” section). For Mus musculus, we used 272 associations between 44 small molecules known to be bioactive against 43 miRNAs, and for Rattus norvergicus, we used 78 associations between 32 small molecules known to be bioactive against 13 miRNAs.
Fig. 3. sChemNET prediction performance evaluation on mouse and rat miRNAs.
a Small molecule and miRNA set for mammalian model organisms (Mus musculus and Rattus norvergicus) were combined with those available for Homo sapiens for training sChemNET for predicting small molecule-miRNA associations available for these organisms. Silhouettes of model organisms were obtained from https://www.phylopic.org/. b The percentage of bioactive small molecules correctly retrieved from the test set for different numbers of small molecules retrieved by each method under a leave-one-out cross validation procedure. Only chemically dissimilar instances were considered between training and testing sets (Tanimoto chemical similarity <0.6). (Left) Recall obtained for 272 small molecule-miRNA associations for 43 miRNAs from Mus musculus; (Right) Recall obtained for 78 small molecule-miRNA associations for 13 miRNAs from Rattus norvergicus. For the boxplots the center line represents the median and the lines extending from both ends of the box indicate the quartile (Q) variability outside Q1 and Q3 to the minimum and maximum values. The notch represents the 95% confidence interval of the median.
Figure 3b (Left) shows the prediction performance of sChemNET in Mus musculus miRNA data when considering only chemically dissimilar instances in the test set. We observed that sChemNET performs best without using sequence similarity information, and it can retrieve more than 43% of bioactive small molecules within the top 25% of prediction retrieved. Similarly, the prediction performance of the different methods for chemically dissimilar instances of bioactive small molecules for miRNAs in Rattus norvergicus is shown in Fig. 3b (Right). In this dataset, sChemNET outperforms the competitors by 6.18–24.67% in the top 300 (7.5%) of predictions retrieved and by 2.74–20.50% in the top 1000 (12.5%). Logistic regression performs 0.726% better than sChemNET in the top 100 (2.5%). The prediction performance for mammalian organism when using all the small molecule-miRNA instances (i.e. without controlling for chemical similarities) is shown in Supplementary Fig. 11.
Mapping the effects of drugs on miRNAs and experimental validations for miR-451
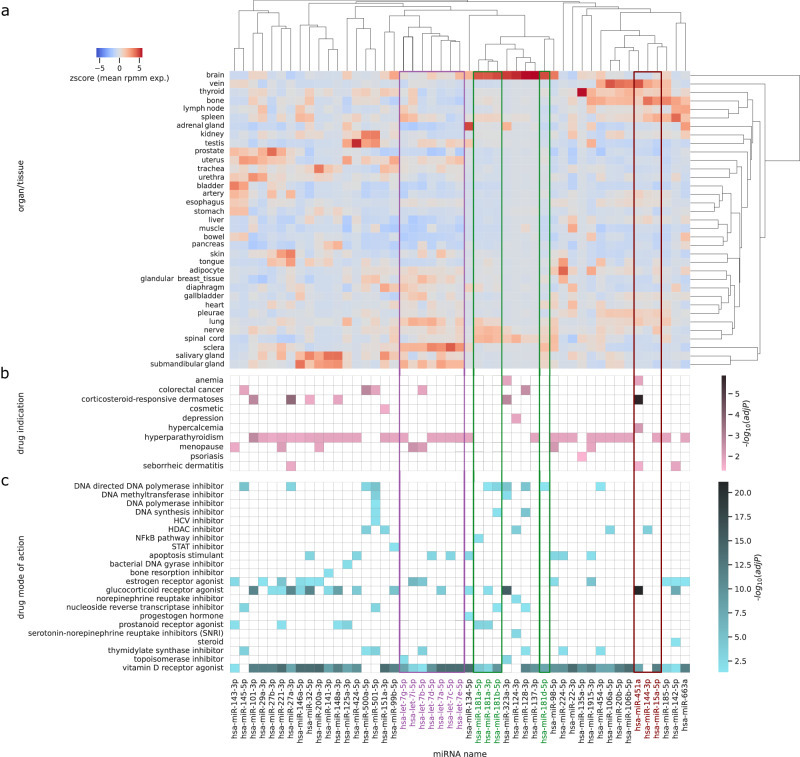
sChemNET’s effectiveness at computationally predicting small molecules bioactive against miRNA activity prompted us to ask whether we could generate a map between miRNAs and small molecules’ pharmacological and chemical classes. To generate the mapping, we calculated the enrichment of the drug mode of action (MoA) and drug indications for ~127 out of ~6300 small molecules predicted in the 98th percentile score for each miRNA belonging to Homo sapiens (see “Methods” section). Figure 4b, c below shows a heatmap for the enrichment obtained for drug MoA and indications for selected miRNAs. In the heatmaps, miRNAs are ordered based on their distance in tissue-specific expression patterns using data from human donors obtained from the miRNA Tissue Atlas database33 (Fig. 4a).
Fig. 4. Heatmaps of miRNA tissue expression, enriched small molecules mode of action and drug indications predicted by sChemNET for selected miRNAs from Homo sapiens.
a miRNAs were ordered using hierarchical clustering based on their z-score expression levels on human tissues from the Tissue Atlas database; Red areas indicate expression and the color is proportional to the z-score. b Enrichment values of drug indications predicted by sChemNET for each miRNA. The color is proportional to the , where is the Benjamini-Hochberg corrected p-value calculated using Fisher’s Exact Test to keep an overall significance below 0.05. White areas indicate non-significant; (c) Enrichment values of drug mode of action predicted by sChemNET for each miRNA. The color is proportional to the, where is the Benjamini–Hochberg corrected p-value calculated using Fisher’s Exact Test to keep an overall significance below 0.05. White areas indicate non-significant.
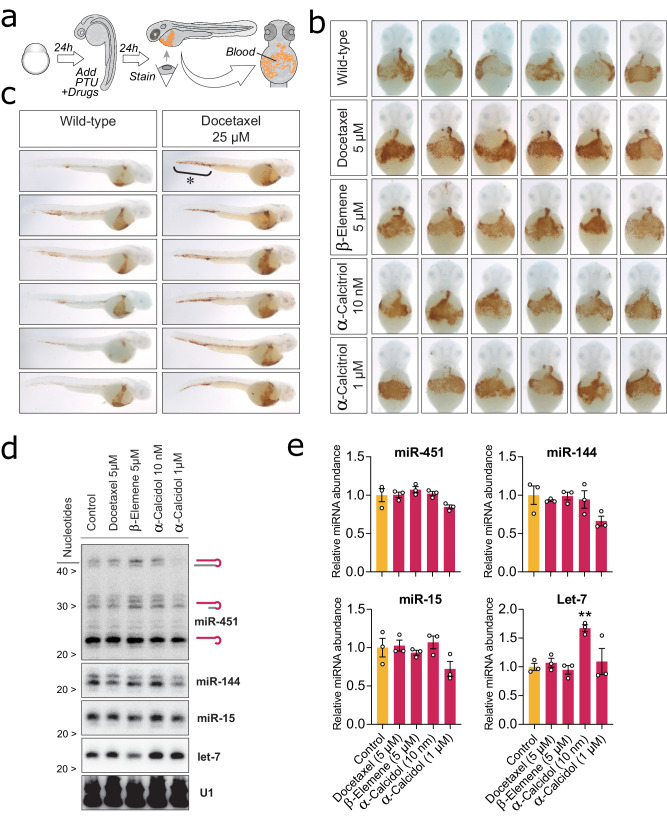
We investigated several compelling associations observed in Fig. 4 in more detail. We first focus our attention to miR-451, an erythrocyte-specific miRNA. To experimentally validate if sChemNET-predicted associations for miR-451 are phenotypically and physiologically relevant, we incubated zebrafish embryos with different drug candidates with the potential to modulate the miR-451 response. Zebrafish embryos are an optimal model for validating small molecules as they are transparent and enable testing the physiological effect of the drugs in the whole organism. Since miR-451 is expressed only in erythrocytes, we focused our analysis on the progress of erythrocyte maturation. 48 hours after fertilization, embryos display robust blood circulation. At this stage, the accumulation of mature erythrocytes can be easily assessed in transparent embryos using O-dianisidine, a hemoglobin-specific stain34. All the drugs were tested in wild-type zebrafish embryos in combination with phenyl-thiourea, a chemical known to induce anemia due to oxidative stress when miR-451 activity is impaired, but not in wild-type embryos35,36. In this PTU-sensitized background, drugs impairing miR-451 activity induce anemia, while miR-451 boosting drugs will increase erythrocyte production (Fig. 5a).
Fig. 5. Experimental validation on Zebrafish embryos of drugs predicted to modulate the activity of miR-451 or the expression of its targets.
a Depiction of our experimental design. Zebrafish embryos were incubated with different drug candidates predicted by sChemNET in combination with phenyl-thiourea (PTU), a chemical known to induce anemia due to oxidative stress when miR-451 activity is impaired, but not in wild-type embryos. 48 hours after fertilization, embryos display robust blood circulation. At this stage, the accumulation of mature erythrocytes can be easily assessed in transparent embryos using O-dianisidine, a hemoglobin-specific stain. Drugs impairing miR-451 activity induce anemia, while miR-451 boosting drugs will increase erythrocyte production (blood circulation). b Ventral images of 2-day-old embryos stained with O-dianisidine to reveal hemoglobinized cells (brown staining) for wild-type embryos and those treated with docetaxel, -elemene and -calcidol. Blood accumulates in the ventral region (ducts of Cuvier). c Lateral view of another group of embryos to reveal accumulation of the excess of blood in the tail region upon treatment with docetaxel. d Northern blot quantification of miRNA expression in zebrafish embryos treated with drug candidates. Total RNA extracted from 2-day-old embryos was analyzed by Northern blot to reveal the expression of miR-451, miR-144, miR-15, and let-7 under different drug treatments. e Quantification of miRNA expression based on the radioactive signal of miRNA probes after Northern blot assay. Bars represent averaged normalized miRNA expression from three representative experiments (white circles). Error bars indicate the standard error of the mean. **p-value = 0.0091 One-way ANOVA test.
We selected three small molecules for the experimental validation on miR-451 response: (i) the tubulin polymerization inhibitor docetaxel, predicted in sChemNET’s top-3 position and also known to target BCL2, a known gene target of miR-451; (ii) the vitamin D receptor agonist -calcidol, predicted in sChemNET’s top-5 position; and (iii) -elemene, predicted in sChemNET’s top-71 position.
We treated the embryos with docetaxel to experimentally validate our first candidate drug. Figure 5b shows that consistent with the predictions of sChemNET, docetaxel causes a higher accumulation of blood in the ventral region of treated embryos compared to untreated siblings. This finding confirms that docetaxel has a physiological effect on miR-451-induced erythropoiesis. Higher doses of docetaxel (25 µM) further induced erythrocyte production, and erythrocytes started to pool in the tail region (Fig. 5c).
Our second candidate compound was -calcidol, motivated by sChemNET predictions for miR-451 in Fig. 4c, which shows enrichment for vitamin D receptor agonists (adjusted significance ). To test this hypothesis, we treated zebrafish embryos with -calcidol. We observed blood accumulation associated with -calcidol treatment on the ventral region of embryos, even at concentrations as low as 10 nM (see Fig. 5b). Finally, our third candidate compound was -elemene due to its ability to bind to miR-451 targets MMP2 and MMP937,38. Our experiments also confirm that -elemene treatment also induces excess blood in the ventral region of the embryos (Fig. 5b).
To elucidate whether these compounds increased erythrocyte maturation by stimulating miR-451 biogenesis or through modulation of its network of mRNA targets, we analyzed the miR-451 and other miRNA levels by Northern blot (see Fig. 5d, e and Supplementary Fig. 12). Our analyses revealed that miR-451 expression levels did not change upon drug treatment compared to untreated embryos. Consistent with these results, we did not observe changes in miR-144, another erythrocyte-specific miRNA expressed from the same primary transcript as miR-45139. These results suggest that the drugs tested elicit a transcriptional response that mimics the effect of miR-451-mediated regulation.
Only accumulation of let-7 shown in Fig. 5e, expressed in the hematopoietic tissue and elsewhere in the embryo, increased significantly upon treatment with -calcidol, a drug known to increase Dicer expression and hence miRNA processing. Since miR-144 but not miR-451 is also processed by Dicer, we conclude that -calcidol affects Dicer expression outside the hematopoietic tissue.
miRNAs, Vitamin D, and the example of the miRNA-181 isotype family
The most striking association between miRNAs and a drug proved to be vitamin D which was associated with most of the miRNAs examined (last row of Fig. 4c). Initially, this may seem anomalous, until it is recognized that the active form of vitamin D, calcitriol or 1,25-dihydroxy vitamin D (1,25(OH)2D), is active in every tissue and recently been discovered to be central in regulating mitochondrial function which is essential for all tissues.
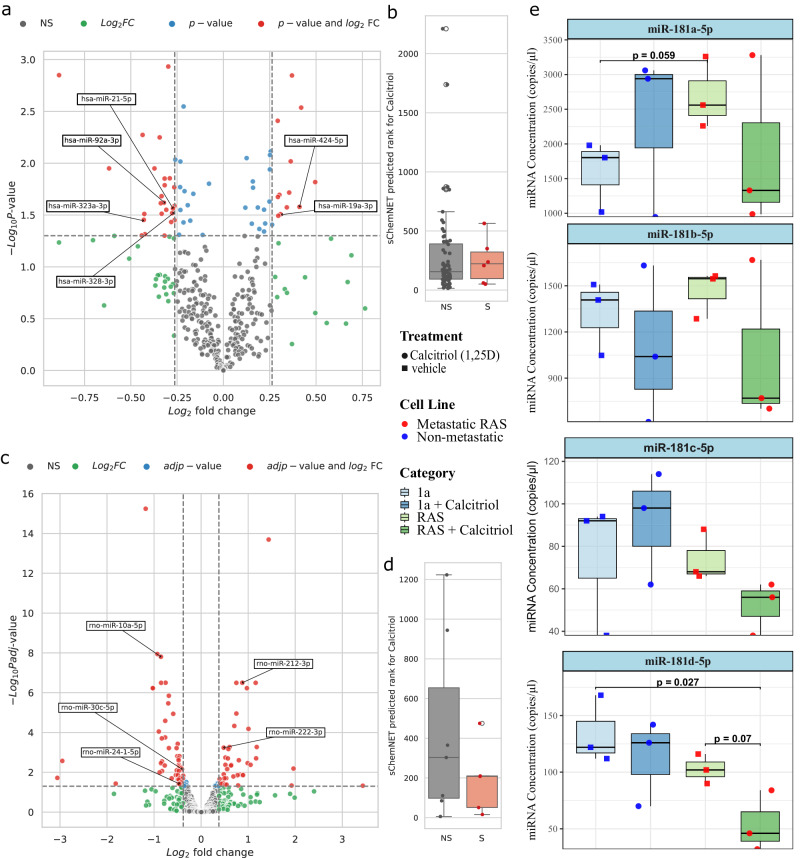
Our experiments in zebrafish embryos indicated that the VDR agonist -calcidol acts directly on miRNA processing. The observed upregulation of mature let-7 most likely occurs by Dicer overexpression. Since Dicer is a central component of the miRNA processing pathway, we analyzed if other miRNAs are associated with other VDR agonists. To experimentally assess the accuracy of miRNAs predicted for calcitriol, we first investigated the correspondence of miRNAs in human neuroblastoma cells (SH-SY5Y) treated with calcitriol using miRNA sequencing. Following 24 h treatment, we observed a small number of miRNAs were differentially expressed in SH-SY5Y cells (Fig. 6a). Two of our predicted miRNAs hsa-miR-424-5p and hsa-miR-19a-3p were upregulated, and four predicted miRNAs were reduced (hsa-miR-21-5p, hsa-miR-92a-3p, hsa-miR-323a-3p, and hsa-miR-328-3p). The mean of sChemNET’s distribution of predicted rank for calcitriol-miRNA interactions was lower for significant miRNAs (mean rank 245) than for the non-significant ones (mean rank 288; see Fig. 6b).
Fig. 6. Experimental validation of miRNAs targeted by the vitamin D receptor agonist calcitriol.
a Enhanced volcano plot of miRNAs differentially expressed in SH-SY5Y cell lines upon drug treatment. P-value was obtained with two-way ANOVA test and contrasts were determined between control and treated using Fisher’s Least Significant Difference (LSD); (b) sChemNET predicted rank for calcitriol for the group of miRNAs found to be non-significant (NS) vs. the group found to be significant in terms of p-value and log2FC (S) in SH-SY5Y cell lines ( for NS and for S). For the boxplots the center line represents the median and the lines extending from both ends of the box indicate the quartile (Q) variability outside Q1 and Q3 to the minimum and maximum values; (c) Enhanced volcano plot of miRNAs differentially expressed under treatment with calcitriol on endothelial progenitor cells and control cells derived from the bone marrow of male Sprague-Dawley rats. P-values were obtained with DESeq2 by the Wald test and adjusted for multiple testing using the Benjamini–Hochberg method. d ChemNET predicted rank for calcitriol for the group of miRNAs found to be non-significant (NS) vs. the group found to be significant in terms of p-value and log2FC (S) in miRNAs from Rattus Norvergicus ( for NS and for S). For the boxplots the center line represents the median and the lines extending from both ends of the box indicate the quartile (Q) variability outside Q1 and Q3 to the minimum and maximum values. e Box plots for the droplet digital PCR (ddPCR) quantification of the copies/ul of miR-181a-5p, miR-181b-5p, miR-181c-5p, and miR-181d-5p in human non-metastatic MCF10CA1a and metastatic MCF10CA1a-ras breast cancer cell line with and without calcitrol treatment determined by ddPCR. The significance between groups is shown by the p-value. There are replicates for each condition. For the boxplots the center line represents the median and the lines extending from both ends of the box indicate the quartile (Q) variability outside Q1 and Q3 to the minimum and maximum values. The p-values were determined by two-side pairwise comparison.
We further tested sChemNET predictions on model organisms. To assess the predictions for miRNAs from Rattus norvergicus, we used previously published data on the expression of miRNAs between calcitriol-treated endothelial progenitor cells and control cells derived from the bone marrow of male Sprague-Dawley rats40 (Fig. 6c and “Methods” section). Figure 6d shows the mean of sChemNET’s distribution of predicted rank for calcitriol-miRNA interactions was lower for significant miRNAs (mean rank 192) than for the non-significant ones (mean rank 434). Our analysis suggests that sChemNET’s predictions can also be helpful for small-sized miRNA chemical datasets available for model organisms.
Vitamin D or its active form calcitriol have long been associated with calcium and phosphate metabolism, which are directly modulated by the mitochondrion, and vitamin D has been associated with regulation of mitochondrial respiration, reactive oxygen specific (ROS) production, cell proliferation, and cell death. Vitamin D acts through the Vitamin D Receptor (VDR) and silencing of the VDR in a variety of cultured human cells not only modulated mitochondrial respiration, ROS production, and apoptosis, it downregulated the protein levels of critical oxidative phosphorylation (OXPHOS) proteins coded in both the mtDNA (COX2 and ATP6) and the nDNA (COX5 and ATP5B)41.
Regardless of the developmental target of a miRNA, it would be essential that the miRNA also modulate mitochondrial bioenergetics to have an integrated effect on the cellular and developmental function. This is powerfully demonstrated by miR-2392 which not only enters the mitochondrion to bind to the mtDNA but also has “seed” binding sites in in 362 nDNA coded mRNAs8,42. miRNA-181 provides an example of the critical importance for a miRNA to regulate both developmental as well as mitochondrial functions. miRNA-181 is developmentally regulated, predominantly expressed in the multiple areas of the brain (Fig. 4a), though it is also active in immune, neuronal, and heart tissues43–45. As predicted, miRNA-181 has been found to be a powerful negative effector of mitochondrial biogenesis, mitophagy, and apoptosis45,46.
There are four mature forms of miR-181 (miR-181a-5p, miR-181b-5p, miR-181c-5p, miR-181d-5p). These are transcribed from three chromosomal clusters: miR-181-a1 and miR-181-b1 on chromosome 1, miR-181-a2 and miR-181-b2 on chromosome 9, and miR-181c and miR-181d on chromosome 1945. In neuronal cells, miR-181a/b act within the cytosol to reduce OXPHOS in favor of glycolysis through inhibition of the mRNAs for the master mitochondrial biogenesis transcription factor, peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) gene PPARGC1A, the mitochondrial nuclear regulator factor 1 gene (NRF1), as well as the structural gene mRNAs COX11, COQ10B, and PRDX344,45,47. In the heart, miR-181c enters the mitochondrion and binds to the mtDNA COXI transcript resulting in suppression of OXPHOS48.
Given that the four different isotypes of miR-181 all affect mitochondrial function, but have subtly different mRNA targets, if follows that the clinical effects of vitamin D would differentially overlap with the functional profiles of the different miR-181 isotypes. Since it is established that miR-181 directly regulates mitochondrial biogenesis and bioenergetics, and the requirement for mitochondrial function is ubiquitous, it follows that predictions made from the function of a wide range of miRNAs would also modulate mitochondrial function and thus be related to vitamin D metabolism.
miR-181s have been shown to be overexpressed in several cancer types49–51 including breast cancer, and has been demonstrated to be involved with greater proliferation, invasiveness, and metastasis when overexpressed52. There has been evidence of dysregulation of the miR-181 family in a number of cancer types, including colorectal, breast, lung, and prostate cancers45,53. Depending on the target genes involved, studies have demonstrated that miR-181s can function as tumor suppressors54. In addition, it has been reported that VDR agonists have the ability to alter the expression of miR-181 in cancer cells55. VDR agonists with the capacity to control miRNA expression has been identified as possible cancer treatment drugs56.
To experimentally determine the impact of vitamin D receptor agonist on miR-181 family (miR-181a, b, c and d) and breast cancer, we utilized a non-metastatic MCF10CA1a and metastatic MCF10CA-ras breast cancer cell line with and without calcitriol treatment (Fig. 6e). We then quantified miRNA concentration for each condition using droplet digital PCR (ddPCR; see “Methods” section). As expected, the miR-181a-5p and miR-181c-5p increased in copies/ when comparing the metastatic to the non-metastatic cell line. On average, the calcitriol treatment reduced the amounts of the miR-181 family for the metastatic cell line. For the non-metastatic cell line, calcitriol caused an increase in the amount of miR-181a-5p, with no difference in miR-181b-5p and a decrease in miR-181c-5p and miR-181d-5p (Fig. 5e).
Discussion
Proteins remain the predominant class of pharmaceutical drug targets21. Yet, many disease-related proteins remain undruggable to date, thus hindering any possibilities for the development of treatments for rare and/or complex human diseases. Targeting RNA molecules, such as microRNAs or their downstream targets, have been proposed as alternative treatment strategies due to the ability of miRNAs to regulate pathway networks related to disease8,57. Despite our growing knowledge of miRNA-disease associations, we lack advances in miRNA-based therapeutics, as there are currently no clinically approved miRNA-based drugs available for treatment. To harness the potential of altering miRNA function to improve human health, systematic principles and computational methods are needed to support the development of RNA-based therapeutics. In this paper, we introduced a deep learning approach, sChemNET, for predicting small molecules that might affect miRNA function. Our model learns non-linear relationships between chemical features of small molecules and miRNAs by learning from small bioactive miRNA-chemical datasets and large corpus of chemical structures as-yet-unknown to affect miRNAs. We show that sChemNET can be useful for predicting small molecule-miRNA associations obtained from Homo sapiens and other mammalian model organisms. We also show that sChemNET provides predictive understanding of the chemical principles by which small molecules are bioactive against a particular miRNA function (see Fig. 4), and how this knowledge can be used as hypothesis generator for the experimental design. To facilitate exploration of the predictions, we provide them in Supplementary Datasets 1.
In our study, we used the Drug Repositioning Hub database29 to obtain an unlabeled set of chemical structures. This chemical library is known to contain therapeutically and chemically diverse compounds, which includes most of the FDA-approved drugs. Hence, it is likely to include already potential RNA binders58 which might also affect RNA function. To understand whether our unlabeled set of small molecules were already bias towards our labeled set of small molecules, we calculated 46 different physicochemical properties of each small molecule using SwissADME59 and compared the distribution of mean between the Euclidean distances among the labeled small molecules (intra-group) and between the labeled and unlabeled small molecules (inter-group). We found that the mean of the distribution of physicochemical distances underlying the inter-group of small molecules is significantly greater than the mean of the distribution of the intra-group (One-sided Welch’s t-test Significance p-value < 1.10e-30). This suggests that, in terms of physicochemical properties distances, there are statistically significant differences between the labeled and unlabeled small molecules, which indicates that our unlabeled set is not biased towards our labeled set.
sChemNET has several limitations. First, it can only predict whether a small molecule might affect the transcriptional program of a miRNA, but about the exact molecular mechanism of action is unknown. Our experimental validations in zebrafish embryos and human cells demonstrate that the small molecules predicted by sChemNET can act either directly on the miRNAs, affecting their processing or expression, or modulating the expression of genes in the miRNA-target network. Either mechanism of action is valid, as both pathways will allow the desired output, which complements miRNA activity. One case is -calcidol, which does not affect the levels of miR-451 - or its cluster partner miR-144 -, but still boosts blood production. The reason that -calcidol does not affect the levels of the erythrocyte-specific miR-144 is because Dicer and miR-144 are engaged in a negative feedback loop in erythrocytes. Dicer processes miR-144, but at the same time is a target of miR-14436, effectively canceling a potential drug-induced increase in miR-144 output.
A second limitation of sChemNET is that the small molecule-miRNA interaction data comes from different experimental conditions and cell lines, so it could happen that sChemNET predictions generalize better to those cell lines, as miRNA expression is known to be tissue-specific33 (see also Fig. 4). To assist researchers with this last limitation, we manually curated the cell lines/tissues for each miRNA from the dataset used for training (see Supplementary Datasets 2).
Figure 4 represents the final mapping generated with sChemNET that connects drug molecular effects, miRNAs, drug indications, and miRNA tissue expression. Strikingly, Fig. 4c reveals that most miRNAs were significantly associated with vitamin D receptor agonists. This can be attributed to the fact that vitamin D and its metabolites can exert a pleiotropic effect over miRNA expression, as already described in different physiological contexts, mediated by direct promoter activation of miRNA genes60,61. Many miRs, including miR-106a-5p, miR-106b, miR-134, miR-135a, miR-141, miR-146a, miR-181, miR-1915, miR-20b, miR-22, miR-224, miR-27b, miR-29a, miR-98 miR-99b, and let-7a/b/d/e/f are regulated by calcitriol or calcifediol in vitro60. Several miRNAs, including miR-106b, miR-141, miR-146a, miR-221, miR-32, miR-424, miR-99b-5p, and let-7a/b/d/f, are associated with serum or tissue levels of vitamin D3 metabolites in patients60. The untranslated region of the vitamin D receptor mRNA contains binding sites for miR-27 and miR-12562. Other studies refer to the dysregulation of miR-125 in the context of vitamin D62. miR-134, miR-663, and miR-125 were dysregulated in relation to calcifediol status in adult acute myeloid leukemia patients, although none of them remained significant after multiple test corrections62. α-calcidiol is also known to be converted to calcitriol, and thus functions equivalently to calcitriol in the dysregulation of miRNAs63.
In addition to demonstrating the impact of targeting miR-181s from our predictions for breast cancer (Fig. 6e), the predictions produced from this work can assist for future work with determining alternative treatment strategies for many diseases such as different cancer types (Fig. 4). For example, sChemNET also predicted that miR-501-5p is significantly associated with colorectal cancer and drug mode of actions related to anticancer drugs (Fig. 4c). In a study by Ma Xiang et al. it was observed that miR-501-5p promotes gastric cancer cell proliferation and migration by targeting and downregulating LPAR164. In another study, Zhang et al. discovered miR-501–3p restricts prostate cancer cell growth by targeting CREPT to inhibit the expression of cyclin D165. In their work, Zhao et al. note that miRNA-501-5p expression in prostate cancer cells was elevated while PINX1 expression was decreased when compared to the normal prostate epithelial cells. PINX1 was a target of miR-501-5p, which was downregulated to encourage prostate cancer cell invasion, migration, and proliferation66. This is another example of how sChemNET’s predictions can further be utilized to generate novel hypotheses to develop therapies and treatment for patients in the clinic. We believe that once the correct miRNAs are determined associated with the diseases, sChemNET can be an essential tool for rapid treatment response and potentially can also assist with future pandemics, when it is important for rapid development of repurposing existing small molecule drugs for treatment.
On the basis of the results of this pilot study, we propose that a next step would be the generation of an expanded experimental mapping based on sChemNET’s predictions between chemical families of small molecules and miRNA bioactivity in-vitro and in-vivo, that is, whether they up-or down-regulate predicted miRNAs and/or their corresponding mRNA targets, including their effects on model organisms related to specific human diseases. An initial step could be to profile all FDA-approved drugs on miRNAs and their downstream targets or to assess sChemNET in other chemical libraries to discover novel compounds. These efforts will also allow us to expand sChemNET’s predictions capabilities to many other human miRNAs and chemical families. In the meantime, even an incomplete mapping generated by sChemNET in Fig. 4 will accelerate progress in characterizing miRNAs targeted by small molecules, finding new uses for existing drugs, and in understanding the miRNA alteration mechanisms of diseases.
It would be interesting to explore other modern machine-learning approaches with low-dimensionality chemical fingerprint as input for sChemNET. We have benchmarked the RDKit and ECFP4 and ECFP6 chemical fingerprints and found that MACCS outperforms them (Supplementary Fig. 10). Although ECFP-based offers better chemical representation, it is likely that MACCS outperform it due to overfitting in the presence of our small and sparse labeled dataset. It would be also interesting to build datasets that contain true negative small molecule-miRNA associations and to model its confidence with sChemNET with an additional term in the loss function. Finally, future modeling research also requires combining datasets about direct binding58 and regulation between small molecules and miRNAs.
Methods
Chemical datasets
We used the SM2miR database20 version 27 April 2015, to obtain manually curated associations between small molecules and miRNAs. In the database, each small molecule was mapped to its PubChem identifier (CID), and each miRNA was mapped to its miRbase identifier. In total, we found 4244 small molecule-miRNA associations across 18 species. For each organism under study, we only kept miRNAs with at least five small molecule associations. For Homo sapiens, we used 1102 associations between 131 small molecules and 126 miRNA targets. For Mus musculus, we used 272 associations between 44 small molecules and 43 miRNAs. For Rattus norvergicus, we used 78 associations between 32 small molecules and 13 miRNAs.
To obtain a prospective set, we mined the RNAInter database32 and found 1180 new associations for Homo sapiens between 123 miRNAs and 120 small molecules. These associations were not present in our set from SM2miR.
We used the Drug Repurposing Hub29 to obtain a chemical library of small molecules without known activity against miRNAs. The Drug Repurposing Hub contains structurally and therapeutically diverse small molecules that have reached clinical trials for diverse indications. For each organism under study, we kept only the small molecules that were not in our sets from SM2miR. In total, our final set of small molecules for Homo sapiens, Mus musculus, and Rattus norvergicus, was 6302, 6281, and 6294, respectively.
Canonical Simplified Molecular-Input Line-Entry System (SMILES) of each small molecule was obtained from PubChem using CIDs identifiers. To query PubChem, we used the https://pubchempy.readthedocs.io/en/latest/ library in Python.
Chemical structure representation
Each small molecule was represented by its MACCS fingerprint67, a 127 binary feature vector in which each element contains a value of ‘1’ if the chemical substructure is present in the small molecule or a value of ‘0’ otherwise. The MACCS chemical fingerprint was computed from the SMILES chemical structure information using RDKit68.
miRNA sequence similarity and linear re-scaling for sChemNET loss function
miRNA mature sequences were obtained from the miRBase database30 using the miRNA identifiers. Following previous work69,70, we computed miRNA sequence similarities using the mature miRNA sequences using global alignment Needleman-Wunsch algorithm in BioPython v1.76 (with a match score of 1, and mismatch and gap scores of zero). To obtain the that we used in our loss function in Eq.(1), raw sequence similarity scores from the Needleman-Wunsch algorithm were linearly rescaled between and 1, as follows:
Where is the alignment score obtained between miRNA and all other miRNAs, is the slope and is the minimum value of . For Homo sapiens, we found that works best, and for model organisms we used .
The sChemNET model training
We modeled the small molecule-miRNA interaction using a multi-task two-layered feed-forward neural network that learns a score mapping between the chemical features of a small molecule and a set of miRNAs. sChemNET is trained for each miRNA by integrating labeled and unlabeled chemical structure information, and it can be trained with or without integrating miRNA sequence information. When sChemNET is trained without sequence information, it amounts to setting for any miRNA pair () in Eq. (1). The sChemNET model architecture consisted of a set of input chemical features, a set of hidden units fully connected to the input features (with a dropout parameter , batch normalization, and Relu activation function), followed by a set of output units, representing each of the miRNAs, fully connected to the hidden units (with a dropout parameter and sigmoid activation function). To train the sChemNET model, the loss function in Eq. (1) was minimized using an ADAM optimizer with default parameters in Tensorflow/Keras v2.8.0 (beta1 = 0.9, beta2 = 0.999, epsilon=1e-7).
Notice that although sChemNET uses labeled information from other miRNAs during the learning, sChemNET is not a global learning model, that can be learned only once. The sChemNET learning model is optimized for each miRNA of interest. A global model can be obtained only for the case in which we ignore the sequence similarity information, that is, for all the cases.
Hyperparameter search using Bayesian optimization
In the sChemNET architecture, the following hyperparameters were optimized: number of hidden units (), unlabeled regularization parameter (), number of epochs (), learning rate (), and dropout (). We used a Bayesian optimization procedure (https://github.com/fmfn/BayesianOptimization) for hyperparameter search in the following bounds , Number of hidden units were tested for a discrete set of . For every organism, the hyperparameter search was performed on a randomly selected miRNA that was then removed from subsequent performance evaluation. The optimal set of hyperparameters was selected based on the minimum mean rank of the predicted bioactive small molecules that was held out.
In-silico LOOCV evaluation procedure
We used Leave-One-Out Cross Validation (LOOCV) procedure to assess the prediction performance of the model. In this procedure, for each miRNA , we removed one of its bioactive small molecules and placed it on a test set. In total, the test set consisted of 4000 small molecules where only one was known to be bioactive against miRNA target , and the remaining 3999 were randomly selected from the set of small molecules without known activity against miRNA target . We used the remaining labeled and unlabeled small molecules to train sChemNET for miRNA target . The model then assigned a score to each of the 4000 small molecules in the test set that we stored. We repeated the procedure for each association known for miRNA . The prediction performance was then obtained as the recall of active small molecules amongst the top- small molecules retrieved from the test set, for . We repeated the whole procedure for each miRNA target in each organism separately. The recall for a given miRNA at the top- was computed as follows:
Prospective evaluation
We used the RNAInter database to obtain a prospective evaluation set. We found 1180 prospective associations between 123 miRNAs and 120 small molecules. Using this prospective evaluation set, for each miRNA, we used our 2015 dataset (SM2miR database) to train the models and the 2022 dataset (RNAInter database) as a test set. To avoid information leakage from similar chemical structures, we only kept in the test set chemically dissimilar compounds from those in training (Tanimoto similarity < 0.6). We only considered cases in which we had at least five associations in the test set. In the test set, we also incorporated 4000 randomly selected small molecules that were unknown to be bioactive against the miRNA under evaluation. The remaining unlabeled small molecules were used for training. We then framed a binary classification performance and used the area under the receiver operating characteristic curve (AUROC) to calculate the model’s prediction performance for each miRNA.
Baseline methods
The following methods were used to score each of the small molecules in the test set:
Chemical similarity baseline. Each small molecule in the test set was scored based on the maximum chemical similarity to an active small molecule in the training set. Chemical similarities were computed using the 2D Tanimoto chemical similarity based on the binary fingerprints.
Random baseline. Each small molecule in the test set was assigned a random score sampled from a uniform distribution between 0 and 1.
Machine Learning baselines. We also implemented machine-learning baselines using sklearn that work with the same dataset as sChemNET. These includes Logistic Regression, Random Forest (best hyperparameter set, ‘n_estimators’: 2, ‘min_samples_split’:10, ‘min_sample_leaf’: 3, ‘max_features’:2, ‘max_depth’: 50, ‘bootstrap’: True), XGBoost (best hyperparameter set, ‘subsample’: 0.5, ‘n_estimators’: 1000, ‘min_samples_split’: 5, ‘min_samples_leaf’: 5, ‘max_depth’: 3, ‘learning_rate’: 0.02).
Enrichment analysis of drug mode of action and indication
We trained sChemNET for each miRNA using all the available labeled small molecules and a randomly selected set of 2,400 unlabeled small molecules. Then, sChemNET was used to rank the remaining set of unlabeled small molecules based on the average prediction score of 20 random independent repetitions. Small molecules amongst the 98th percentile score was then kept as predictions for the miRNA. We then retrieved the mode of action and indication information of small molecules from the Drug Repurposing Hub database. The enrichment score was calculated based on -values calculated using Fisher’s Exact Test and adjusted with Benjamin-Hochberg correction for multiple testing.
Samples collected on cancer cell lines treated with calcitriol
Human Harvey-ras transformed non-metastatic and metastatic MCF10CA1a were cultured as previously described71,72. Cells were treated for 72 h with 10 nM 1α,25-dihydroxyvitamin D (1,25(OH)2D, Biomol, Plymouth Meeting, PA) in 100% ethanol vehicle (final concentration <0.1%), with media changed every 24 h and harvested at 70-80% confluence for each dish.
miRNA concentration quantification on cancer cell lines with droplet digital PCR
MiRNA extractions from frozen cell pellets were carried out using the QIAGEN miRNeasy serum/plasma kit (#217184). Quantitation of miRNA samples was done using a NanoDrop 2000 Spectrophotometer (ThermoFisher Scientific). cDNA was synthesized from miRNA samples using the QIAGEN miRCURY LNA RT Kit (Cat. 339340) using a concentration of 5 ng/ml for the miRNA per sample. Next, samples were mixed with a 1:20 dilution of the generated cDNA with the BioRad QX200 ddPCR Evagreen Supermix (Cat. 1864034) and the appropriate miRNA primers from miRCURY LNA miRNA PCR Assays (QIAGEN. BioRad QX200 Automated Droplet Generator (Cat. 1864101) was used to create emulsion droplets. With the C1000 Touch Thermal Cycler with 96–Deep Well Reaction Module (Bio-Rad) the following PCR reaction was used for all the primers: 1 cycle 95 °C for 5 min, 40 cycles of 95 °C for 30 s, and 53 °C for 1 min (the annealing temperature can change depending on the primer), 1 cycle of 4 °C for 5 min, and 1 cycle of 90 °C for 5 min. We have optimized the annealing temperature for miR-181a-5p, miR-181b-5p, miR-181c-5p, and miR-181d-5p to be 53 °C. Finally, the QX200 Droplet Digital PCR System (Bio-Rad) quantified the amount of miRNA for each primer set per sample. QuantaSoft software (Bio-Rad) generated the data for each primer set and sample. The same threshold setting was used for all samples per primer set. These values were used for all miRNA analysis. The data was plotted using ggplot2 in R and Student’s t tests were performed to determine the overall significance between the groups.
SH-SY5Y cell line experiments with calcitriol
The human neuroblastoma cell line SH-SY5Y was obtained from ATCC (CRL-2266) and was cultured in Basic Growth Media (BGM) contain; EMEM (Quality Biological) with 1% v/v GlutaMAX (Gibco), 1% v/v penicillin–streptomycin (Gibco), and 15% v/v hiFBS (Gibco). Cells were subcultured for 2-3 days, and then treated with Calcitrol 10 μM (Sigma) for 24 h. Cells were washed with ice cold PBS. RNA was extracted using the mirNEasy extraction kit (Qiagen) and then libraries were assembled using the QiaSeq protocol for Ion Torrent (Qigen). Library barcodes were called from raw data using cutadapt and uBam data uploaded to Qigen GeneGlobe miRNA Software for processing (https://geneglobe.qiagen.com/us/analyze). Resultant data sheets from 2 runs were merged and processed in Partek Genomics studio. Differential expression was determined using a linear model, with drug treatment and batch ID as contrasts. Data from the results were input into an enhanced volcano for visualization.
Zebrafish strains
Zebrafish strains were bred, handled, and maintained according to the standard laboratory conditions under IACUC protocol PROTO201800373 at Boston University. Experiments were performed in hybrid wild-type strain crosses obtained from AB/TU and TL/NIHGRI breeders.
Drug treatment
Dechorionated wild-type zebrafish embryos at 24 h post-fertilization (hpf) examined under the brightfield microscope and collected for drug treatment. Groups of 15 embryos were distributed in each well of twelve-well plates after coating the wells in agarose. Next, each drug was added to the water containing the embryos at concentrations listed below, and the embryos were incubated with the drugs for 24 h at 28°C. To induce oxidative stress, the embryos were kept in water containing 0.003% phenylthiourea (PTU) (Sigma-Aldrich) from 8 hpf until the end of the experiment. The control group was treated with ethanol or DMSO. All the experiments were done in triplicate.
docetaxel (5 µM, 25 µM)
-elemene (5 µM)
-calcidol (10 nM,1 µM)
Small RNA northern blot
The endogenous miRNAs are detected by processing groups of 30 embryos, treated with drugs and collected at the 48 hpf. Using Trizol (Invitrogen), the total RNA was extracted and resuspended in formamide and 2X loading buffer (8 M urea, 50 mM EDTA, 0.2 mg/ml bromophenol blue, and 0.2 mg/ml xylene cyanol). The extracted total RNAs were separated in 15% denaturing urea polyacrylamide gel in 1X TBE and transferred to a positively charged Zeta-Probe blotting membrane (Bio-Rad) using a semi-dry Trans-Blot SD (Bio-Rad) for 35 min at 20 V (0.68 A). Membranes were UV cross-linked and pre-hybridized with ExpressHyb Hybridization Solution (Clontech) for 1 h at 50 °C. Membranes were blotted with 32P-radiolabelled DNA oligonucleotide probes (StarFire probes, IDT) at 30oC overnight. The oligonucleotide DNA probes hybridized membranes were washed twice with 2X SSC/0.1% SDS followed by 1X SSC/0.1% SDS for 15 min at room temperature. The blots were exposed to a phosphor imaging screen for 1 day and the signal intensity was detected using the Typhoon FLA 7000 phosphor imager (GE Healthcare Life technologies) and analyzed using the ImageQuant TL software (GE Healthcare).
Quantification of miRNA by northern blot
Employing northern blot, the estimate amount of endogenous miR-144, miR-451, miR-15 and let-7 was quantified from 2-day-old zebrafish embryos. The value of each miRNA was normalized with U1 snRNA. The experiments were performed in triplicate. Graphs were generated and statistical analysis was performed using GraphPad Prism.
Preparation of radiolabeled probes
Following StarFire method the radiolabeled DNA probes were prepared. The oligos specific to the targeted miRNA were annealed with universal oligo (5′- TTTTTTTTTT666G6(ddC)-3′ from IDT, where “6” corresponds to a propyne dC modification) via complementary hexamer sequence. Annealed duplexes are then labeled with a-32P-dATP (6 µL of 10 mCi/mL stock) using the 3′−5′ exo- Klenow fragment of DNA polymerase. To stop the reaction 40 µL of 10 mM EDTA solution was added to 10 µL of reaction. The labeled oligos were purified using Micro-Spin G25 columns (GE Health Care) at 3000×g for 3 min. The membranes were probed with 3,000,000 cpm of the P32 labeled Star-Fire probes. Probe sequences used in this study are listed in Supplementary Table 1.
O-dianisidine staining
Hemoglobin content in zebrafish embryos at 48 hpf was performed as previously described by Kretov et al.36. In briefly, embryos were stained in O-dianisidine staining solution (2 mL of water, 2 mL of 0.7 mg/mL O-dianisidine (Sigma-Aldrich), dissolved in 96% ethanol and protected from light, 0.5 mL of 100 mM sodium acetate (pH 4.5), 100 mL of 30% hydrogen peroxide) for 15 minutes at room temperature. After that the embryos were washed 2 times with 1X PBS (pH 7.4) then transferred into 1X PBS. To induce oxidative stress, the embryos were kept in water containing 0.003% phenylthiourea (PTU) (Sigma-Aldrich) from 8 hpf and collected at 48 hpf. The control group was treated with ethanol or DMSO. All the experiments were done in triplicate. Images were captured using a Zeiss Discovery v.12 microscope. Images were processed with ZEN software (Zeiss) and Adobe Photoshop and Illustrator 2021.
Rattus norvegicus miRNA-Seq
Raw miRNA-Seq reads from study PRJNA545400 were downloaded from the sequencing read archive and were trimmed for quality and adapters via pyrpipe73. miRDeep274 was used to process and quantity the reads using the mature and precursor miRNA sequences for Rattus norvegicus from miRBase30. Mature miRNA’s with more than 1 precursor were average across precursors. The subsequent counts were rounded to the nearest integer and ran through DESeq275 for differential expression analysis. This pipeline can be found https://github.com/jahaltom/miRNA-Seq/tree/main.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
D.G. was supported by 2022 KBR SMS SDI Lymphoma grant and the US Air Force grant contract no. FA8075-16-D-0010, task order FA8075-18-F-1690 Explainable Artificial Intelligent Applications within Integrated Dynamic Visualization Environment and the Facultad de Ingeniería (FIUNA). D.T. was supported by NIH grants R01CA232589 and R01CA271597. F.S. was supported by NIH grant R35 CA232105. D.C.Wallace was supported by the Bill & Melinda Gates Foundation grant # INV-046722. D.C. and I. were supported by NIH grant R01GM130935. A.Y. and R.M. were supported by NIH grant RM1HG012334. D.C. and I. were supported by National Institute of Health (US) grant R01GM130935-03.
Author contributions
D.G. conceived of the project. D.G. developed and implemented the ML sChemNET model. Data analysis was performed by D.G., A.B., J.H., F.J.E. D.C. and R.M. Figures generation by D.G., A.B., and D.C. SH-SY5Y experiments and related RNA-seq was done by A.Y. and R.M. Experiments with breast cancer cell lines treated with calcitriol were performed by C.A. and D.T. ddPCR quantification of miRNAs was done by A.B. miRNA-seq analysis was done by J.H. and D.G. mRNA RNASeq-analysis was done by F.J.E. Zebrafish experiments were done by I. and D.C. D.G., A.B.,, D.C.W., V.Z., S.D., S.B., F.J.S.., E.S.W., and R.B. wrote the manuscript. All authors provided additional edits and guidance for the manuscript. D.G. and A.B. supervised and directed the project.
Peer review
Peer review information
Nature Communications thanks Aaron Mackey, Tudor Oprea and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Data availability
The SH-SY5Y data generated in this study have been deposited in the Bioproject NCBI database under accession code PRJNA1115227 [http://www.ncbi.nlm.nih.gov/bioproject/1115227]. The northern blot data and miR-181 quantification data generated in this study are provided in the Supplementary Information/Source Data file. The miRNA-Seq vitamin D treatment data used in this study are available in the Bioproject database under accession code PRJNA545400. Source data are provided with this paper.
Code availability
The sChemNET code is available on https://github.com/diegogalpy/sChemNET/.
Competing interests
D.C.W. is at the Scientific Advisory Boards of Medical Capital Excellence and Pano Therapeutic. Other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-49813-w.
References
- 1.Cooper, T. A., Wan, L. & Dreyfuss, G. RNA and disease. Cell136, 777–793 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Esteller, Manel “Non-coding RNAs in human disease.”. Nat. Rev. Genet.12, 861–874 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Friedman, R. C., Farh, K. K.-H., Burge, C. B. & Bartel, D. P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res.19, 92–105 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iorio, MarilenaV. & Croce, CarloM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med.4, 143–159 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gurha, P. MicroRNAs in cardiovascular disease. Curr. Opin. Cardiol.31, 249–254 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Rupaimoole, R., Calin, G. A., Lopez-Berestein, G. & Sood, A. K. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov.6, 235–246 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDonald, J. T. et al. Role of miR-2392 in driving SARS-CoV-2 infection. Cell Rep.37, 109839 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beheshti, A. et al. Identification of circulating serum multi-microRNA signatures in human DLBCL models. Sci. Rep.9, 17161 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva, S. S., Lopes, C., Teixeira, A. L., Carneiro de Sousa, M. J. & Medeiros, R. Forensic miRNA: potential biomarker for body fluids? Forensic Sci. Int. Genet.14, 1–10 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Rupaimoole, R. & Slack, F. J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov.16, 203–222 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Winkle, M., El-Daly, S. M., Fabbri, M. & Calin, G. A. Noncoding RNA therapeutics — challenges and potential solutions. Nat. Rev. Drug Discov.20, 629–651 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsui, M. & Corey, D. R. Non-coding RNAs as drug targets. Nat. Rev. Drug Discov.16, 167–179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agostini, M. & Knight, R. A. miR-34: from bench to bedside. Oncotarget5, 872 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thakral, S. & Ghoshal, K. miR-122 is a unique molecule with great potential in diagnosis, prognosis of liver disease, and therapy both as miRNA mimic and antimir. Curr. Gene Ther.15, 142–150 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alhamadani, F. et al. Adverse drug reactions and toxicity of the food and drug administration-approved antisense oligonucleotide drugs. Drug Metab. Dispos.50, 879–887 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheridan, C. First small-molecule drug targeting RNA gains momentum. Nat. Biotechnol.39, 6–8 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Fan, R. et al. Small molecules with big roles in microRNA chemical biology and microRNA-targeted therapeutics. RNA Biol.16, 707 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen, X., Guan, N.-N., Sun, Y.-Z., Li, J.-Q. & Qu, J. MicroRNA-small molecule association identification: from experimental results to computational models. Brief Bioinform.10.1093/bib/bby098 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Liu, X. et al. SM2miR: a database of the experimentally validated small molecules’ effects on microRNA expression. Bioinformatics29, 409–411 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Warner, K. D., Hajdin, C. E. & Weeks, K. M. Principles for targeting RNA with drug-like small molecules. Nat. Rev. Drug Discov.17, 547–558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costales, M. G., Childs-Disney, J. L., Haniff, H. S. & Disney, M. D. How we think about targeting RNA with small molecules. J. Med. Chem.63, 8880–8900 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Velagapudi, S. P., Gallo, S. M. & Disney, M. D. Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat. Chem. Biol.10, 291–297 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guan, N.-N., Sun, Y.-Z., Ming, Z., Li, J.-Q. & Chen, X. Prediction of potential small molecule-associated microRNAs using graphlet interaction. Front. Pharmacol.9, 1152 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen, Xing et al. Predicting potential small molecule–miRNA associations based on bounded nuclear norm regularization. Brief Bioinform.22, bbab328 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Qu, J., Chen, X., Sun, Y.-Z., Li, J.-Q. & Ming, Z. Inferring potential small molecule–miRNA association based on triple layer heterogeneous network. J. Cheminf.10, 30 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galeano, Diego et al. Predicting the frequencies of drug side effects. Nat. Commun.11, 4575 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santos, S. de S. et al. Machine learning and network medicine approaches for drug repositioning for COVID-19. Patterns10.1016/j.patter.2021.100396 (2021). [DOI] [PMC free article] [PubMed]
- 29.Corsello, S. M. et al. The Drug Repurposing Hub: a next-generation drug library and information resource. Nat. Med.23, 405–408 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozomara, A., Birgaoanu, M. & Griffiths-Jones, S. miRBase: from microRNA sequences to function. Nucleic Acids Res.47, D155–D162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stokes, J. M. et al. A deep learning approach to antibiotic discovery. Cell180, 688–702.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang, Juanjuan et al. “RNAInter v4. 0: RNA interactome repository with redefined confidence scoring system and improved accessibility. Nucleic Acids Res.50, D326–D332 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ludwig, Nicole et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res.44, 3865–3877 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kretov, D. A., Shafik, A. M. & Cifuentes, D. Assessing miR-451 activity and its role in erythropoiesis. Methods Mol. Biol.1680, 179–190 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Cifuentes, D. et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science328, 1694–1698 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kretov, D. A. et al. Ago2-dependent processing allows miR-451 to evade the global microRNA turnover elicited during erythropoiesis. Mol. Cell78, 317–328.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scrimgeour, N. R., Wrobel, A., Pinho, M. J. & Høydal, M. A. microRNA-451a prevents activation of matrix metalloproteinases 2 and 9 in human cardiomyocytes during pathological stress stimulation. Am. J. Physiol. Cell Physiol.318, C94–C102 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Liang, Y. & Li, S. β-elemene suppresses migration of esophageal squamous cell carcinoma by modulating expression of MMP9 through the PI3K/Akt/NF-κB pathway. Comb. Chem. High. Throughput Screen.26, 2304–2320 (2023). [DOI] [PubMed] [Google Scholar]
- 39.Dore, L. C. et al. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc. Natl Acad. Sci. USA105, 3333–3338 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu, P. et al. Vitamin D (1,25-(OH)2D3) regulates the gene expression through competing endogenous RNAs networks in high glucose-treated endothelial progenitor cells. J. Steroid Biochem. Mol. Biol.193, 105425 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Ricca, C. et al. Vitamin D receptor is necessary for mitochondrial function and cell health. Int. J. Mol. Sci.19, 1672 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guarnieri, J. W. et al. Targeted down regulation of core mitochondrial genes during SARS-CoV-2 infection. bioRxiv10.1101/2022.02.19.481089 (2022).
- 43.Ebert, M. S. & Sharp, P. A. Roles for microRNAs in conferring robustness to biological processes. Cell149, 515–524 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henao-Mejia, J. et al. The microRNA miR-181 is a critical cellular metabolic rheostat essential for NKT cell ontogenesis and lymphocyte development and homeostasis. Immunity38, 984–997 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Indrieri, A., Carrella, S., Carotenuto, P., Banfi, S. & Franco, B. The pervasive role of the miR-181 family in development, neurodegeneration, and cancer. Int. J. Mol. Sci.21, 2092 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ouyang, Y.-B., Lu, Y., Yue, S. & Giffard, R. G. miR-181 targets multiple Bcl-2 family members and influences apoptosis and mitochondrial function in astrocytes. Mitochondrion12, 213–219 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Indrieri, A. et al. miR‐181a/b downregulation exerts a protective action on mitochondrial disease models. EMBO Mol. Med.11, e8734 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Das, Samarjit et al. Divergent effects of miR‐181 family members on myocardial function through protective cytosolic and detrimental mitochondrial microRNA targets. J. Am. Heart Assoc.6, e004694 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ji, D. et al. MicroRNA-181a promotes tumor growth and liver metastasis in colorectal cancer by targeting the tumor suppressor WIF-1. Mol. Cancer13, 86 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, C., Passos Gibson, V. & Hardy, P. The role of MiR-181 family members in endothelial cell dysfunction and tumor angiogenesis. Cells11, 2022 (1670). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neel, J.-C. & Lebrun, J.-J. Activin and TGFβ regulate expression of the microRNA-181 family to promote cell migration and invasion in breast cancer cells. Cell Signal.25, 1556–1566 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Zhai, Z. et al. MiR-181a-5p facilitates proliferation, invasion, and glycolysis of breast cancer through NDRG2-mediated activation of PTEN/AKT pathway. Bioengineered13, 83–95 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Croce, C. M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet.10, 704–714 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marisetty, A. et al. MiR-181 family modulates osteopontin in glioblastoma multiforme. Cancers12, 3813 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh, T. & Adams, B. D. The regulatory role of miRNAs on VDR in breast cancer. Transcription8, 232–241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Campbell, M. J. & Trump, D. L. Vitamin D receptor signaling and cancer. Endocrinol. Metab. Clin. North Am.46, 1009–1038 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hanna, J., Hossain, G. S. & Kocerha, J. The potential for microRNA therapeutics and clinical research. Front. Genet. 10, 478 (2019). [DOI] [PMC free article] [PubMed]
- 58.Zhang, P. et al. Reprogramming of protein-targeted small-molecule medicines to RNA by ribonuclease recruitment. J. Am. Chem. Soc.143, 13044–13055 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Daina, Antoine, Michielin, Olivier & Zoete, Vincent SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep.7, 42717 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giangreco, A. A. & Nonn, L. The sum of many small changes: microRNAs are specifically and potentially globally altered by vitamin D3 metabolites. J. Steroid Biochem. Mol. Biol.136, 86–93 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fernandez, G. J., Ramírez-Mejía, J. M. & Urcuqui-Inchima, S. Vitamin D boosts immune response of macrophages through a regulatory network of microRNAs and mRNAs. J. Nutr. Biochem.109, 109105 (2022). [DOI] [PubMed] [Google Scholar]
- 62.Zhang, Z., Moon, R., Thorne, J. L. & Moore, J. B. NAFLD and vitamin D: evidence for intersection of microRNA-regulated pathways. Nutr. Res. Rev.36, 120–139 (2023). [DOI] [PubMed] [Google Scholar]
- 63.Ryan, Z. C. et al. 1α,25-Dihydroxyvitamin D3 regulates mitochondrial oxygen consumption and dynamics in human skeletal muscle cells. J. Biol. Chem.291, 1514–1528 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma, X. et al. microRNA-501-5p promotes cell proliferation and migration in gastric cancer by downregulating LPAR1. J. Cell. Biochem.121, 1911–1922 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Zhang, Z., Shao, L., Wang, Y. & Luo, X. MicroRNA-501-3p restricts prostate cancer growth through regulating cell cycle-related and expression-elevated protein in tumor/cyclin D1 signaling. Biochem. Biophys. Res. Commun.509, 746–752 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Zhao, Yueguang et al. MicroRNA-501-5p targets PINX1 gene to regulate the proliferation, migration, and invasion of prostatic carcinoma cells. J. Biomater. Tissue Eng.11, 471–477 (2021). [Google Scholar]
- 67.Cereto-Massagué, A. et al. Molecular fingerprint similarity search in virtual screening. Methods71, 58–63 (2015). [DOI] [PubMed] [Google Scholar]
- 68.Landrum, G. RDKit: A Software Suite for Cheminformatics, Computational Chemistry, and Predictive Modeling (Academic Press, 2013).
- 69.Jiang, L., Ding, Y., Tang, J. & Guo, F. MDA-SKF: similarity kernel fusion for accurately discovering miRNA-disease association. Front. Genet.9, 618 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li, L. et al. SCMFMDA: predicting microRNA-disease associations based on similarity constrained matrix factorization. PLoS Comput. Biol.17, e1009165 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sheeley, M. P. et al. 1α,25-dihydroxyvitamin D reduction of MCF10A-ras cell viability in extracellular matrix detached conditions is dependent on regulation of pyruvate carboxylase. J. Nutr. Biochem.109, 109116 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zembroski, AlyssaS. et al. Proteomic characterization of cytoplasmic lipid droplets in human metastatic breast cancer cells. Front. Oncol.11, 576326 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Singh, U., Li, J., Seetharam, A. & Wurtele, E. S. pyrpipe: a Python package for RNA-Seq workflows. NAR Genom. Bioinform.3, lqab049 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Friedländer, MarcR. et al. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res.40, 37–52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The SH-SY5Y data generated in this study have been deposited in the Bioproject NCBI database under accession code PRJNA1115227 [http://www.ncbi.nlm.nih.gov/bioproject/1115227]. The northern blot data and miR-181 quantification data generated in this study are provided in the Supplementary Information/Source Data file. The miRNA-Seq vitamin D treatment data used in this study are available in the Bioproject database under accession code PRJNA545400. Source data are provided with this paper.
The sChemNET code is available on https://github.com/diegogalpy/sChemNET/.