Abstract
The complete mitochondrial genome of the shining leaf chafer Mimela junii was sequenced and is herein described. The mitogenome consists of a circular molecule of 16,805 bp, with an overall AT content of 75.7%. It encodes for 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNAs) and contains a non-coding Control Region (CR) characterized by the presence of tandem repeats. The gene order corresponds to the ancestral Pancrustacea model and mitogenome characteristics are congruous with those of hexapods. In the phylogenetic analysis, M. junii is nested within a paraphyletic Anomala with high support, and is herein associated with Anomala corpulenta with medium/low support.
Keywords: Rutelinae, Anomala, mitogenomics, molecular identification, agricultural pest
Graphical Abstract
Introduction
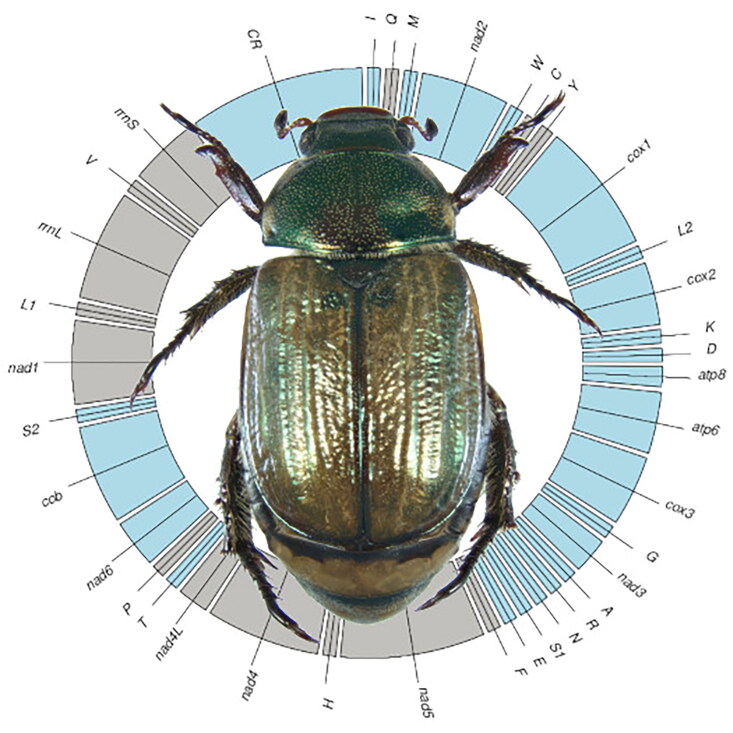
Mimela junii (Duftschmidt, 1805) (Coleoptera: Scarabaeidae; Figure 1), belongs to the diverse sub-family Rutelinae (shining leaf chafers) (Jameson et al. 2003; Ballerio et al. 2014). M. junii is a fairly common species, widely distributed in Central and Southern Europe (Baraud 1992), and is usually associated with sandy soils, both near the coasts and in the interior. The larvae live in the soil and are rhizophagous, while the summer-flying adults feed on leaves of spontaneous shrubs and grasses (Ballerio et al. 2014).
Figure 1.
Mimela junii, adult habitus. Photo by D.B.
Sequencing of the complete mitochondrial genome of M. junii was performed to aid molecular identification within shining leaf chafers, that include notable pests, e.g. Popillia japonica, whose morphological identification is often problematic, especially at the larval stage. This is the first complete mitochondrial genome for the species. A second complete mitochondrial genome is available in the NCBI database for the co-generic species Mimela splendens (MZ064554, unpublished); complete (or semi complete) mitochondrial genomes are available in NCBI for eight additional species from sub-family Rutelinae.
Materials and methods
Larvae of M. junii were manually collected (Siena, Italy: 43.337833 N, 11.336306E) and reared until adult emergence. Species attribution was confirmed by morphological analysis of a set of adult diagnostic characters, i.e. morphology of antenna, clypeus, pronotum and tarsus (Baraud 1992; Ballerio et al. 2014), as well as barcoding (Ratnasingham and Hebert 2007).
All DNA from one individual was used for sequencing. A second specimen, as well as the DNA of a third, were deposited in the collection of the Department of Life Sciences (URL: www.dsv.unisi.it, contact: D. Badano, davide.badano@unisi.it) under vouchers MJU6_DB and MJU7_DB_DNA, respectively.
DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega). Sequencing (TruSeq DNA PCR-Free) was performed at Macrogen Europe (The Netherlands) on a Novaseq6000 (Illumina) applying a 150 bp PE layout to produce a total of ∼332 million PE reads. Sequences were trimmed in fastp (ver. 0.23.2; Chen et al. 2018). Reads were assembled de novo in MegaHit (ver. 1.2.9; Li et al. 2015), under default settings, as well as in NovoPlasty (ver. 4.3.4; Dierckxsens et al. 2017), under multiple k-mer lengths (33 to 143) using the cox1 sequence from the MegaHit assembly as the seed. MegaHit produced a contig that was manually circularized based on terminal overlaps. NovoPlasty produced circular genomes (k = 33, 55, 121) or linear contigs that could be readily circularized based on terminal overlaps (k = 77, 99, 111). All assemblies were identical in sequence apart for a short segment of the CR (∼376 bp) containing tandem repeats. The sequence obtained with k = 121 (the longest k that circularized automatically) was selected as the most reliable and is herein described. Coverage was assessed in bbmap ver. 15/9/2022 (Brian Bushnell 2020). Sequencing coverage (Supplementary Figure S1a) was stable throughout the genome at ∼14000x, with a marked spike in the region interested by the repeats, suggesting that the length of the repeated sequence may have been underestimated (e.g. Nardi et al. 2024). Automatic annotation was performed using MITOS2 (ver. 2; Bernt et al. 2013) and manually revised in accordance with the Popillia japonica (OP974626; Nardi et al. 2024) genome. CR repeats were identified using Tandem Repeats Finder (update 2022; Benson 1999). The genome was prepared for submission using Aln2tbl (Pons et al. 2021) and the genome map was drawn using Chloroplot (Zheng et al. 2020).
All available complete mitochondrial genomes of Rutelinae were downloaded from NCBI (Table 1). Following automatic annotation, Melolontha melolontha (Melolonthinae) was included as outgroup. Single PCGs were retroaligned using MUSCLE (ver. 3.8.425; Edgar 2004) in AliView (ver. 1.28; Larsson 2014), end-trimmed and concatenated using EZmito (Cucini et al. 2021). Starting from initial partitions by strand/codon, the evolutionary model was optimized using Partition Finder (Lanfear et al. 2012) and Model Finder (Kalyaanamoorthy et al. 2017) in IQtree2 (ver. 2.0.7; Minh et al. 2020). The Maximum Likelihood tree, including bootstrap support, was identified in IQtree2. The analysis was repeated based on 1st and 2nd codon positions as well as on amino-acid sequences. The optimal Bayesian tree, with posterior probabilities, was identified in BEAST2 (ver. 2.7.7; Bouckaert et al. 2014) applying the same partitions and model.
Table 1.
Species included in the phylogenetic analysis.
| Species | NCBI | Reference | Sub-family | Tribe | Sub-tribe |
|---|---|---|---|---|---|
| Popillia japonica | OP974626 | Nardi et al. 2024 | Rutelinae | Anomalini | Popillina |
| Popillia mutans | MF997049 | Song and Zhang 2018 | Rutelinae | Anomalini | Popillina |
| Anomala aulax | PP350013 | unpublished | Rutelinae | Anomalini | Anomalina |
| Anomala corpulenta | OL449520 | Qu et al. 2023 | Rutelinae | Anomalini | Anomalina |
| Anomala rufiventris | OR208200 | Long et al. 2024 | Rutelinae | Anomalini | Anomalina |
| Anomala russiventris | MW829593 | Li et al. 2022 | Rutelinae | Anomalini | Anomalina |
| Mimela junii | PQ067309 | This study | Rutelinae | Anomalini | Anomalina |
| Mimela splendens * | MZ064554 | unpublished | Rutelinae | Anomalini | Anomalina |
| Callistethus plagiicollis | OR208201 | Long et al. 2024 | Rutelinae | Anomalini | Anomalina |
| Adoretus sp. * * | JX412788 | Timmermans et al. 2015 | Rutelinae | Adoretini | Adoretina |
| Melolontha melolontha | OW285245 | Ashworth 2023 | Melolonthinae | Melolonthini | Melolonthina |
*species attribution tentatively not confirmed. **incomplete genome including compete PCGs.
Results
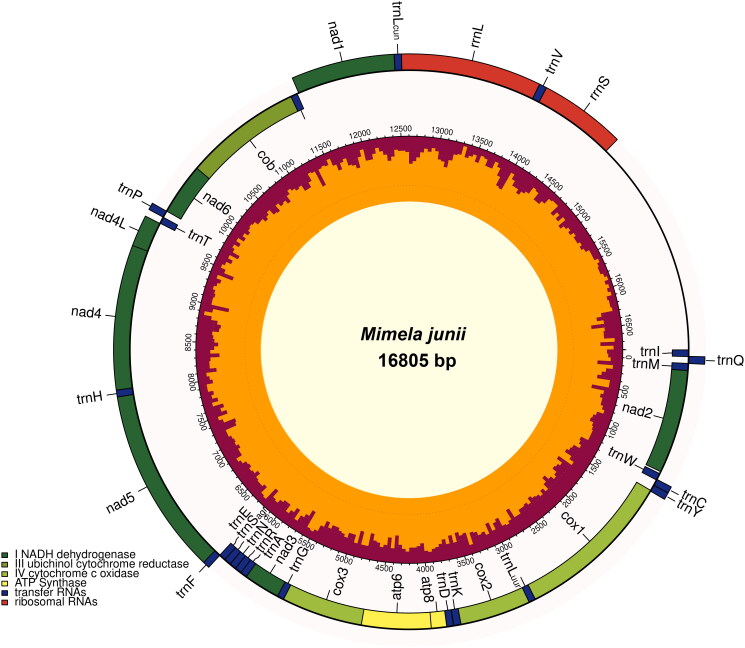
The mitochondrial genome of M. junii is a circular molecule 16,805 bp long and conforms, in structure and gene content, to the model generally observed in Metazoa (Figure 2). It encodes for the canonical 13 protein subunits (cox1/2/3, atp8/6, nad2/3/6, cob on the plus strand; nad1/4/4L/5 on the minus strand), two ribosomal RNAs (rrnS, rrnL, on the minus strand), a complete set of 22 tRNAs (trnI, trnM, trnW, trnLuur, trnK, trnD, trnG, trnA, trnR, trnN, trnSagn, trnE, trnT, trnSucn on the plus strand; trnQ, trnC, trnY, trnF, trnH, trnP, trnLcun, trnV on the minus strand) as well as a long non coding region between rrnS and trnI (CR: 2114 bp). An array of repeats is observed in the CR (Supplementary Figure S1b). The genome organization is compact, with 11 gene overlaps (1 to 8 bp, total 40 bp) and six small spacers (1 to 20 bp, total 42 bp). All PCGs start with a canonical initiation codon. Seven PCGs end with a canonical termination codon and five with an incomplete T signal. The gene order conforms to the Pancrustacea model.
Figure 2.
Mimela junii mitochondrial genome map. Genes encoded on the plus strand are indicated in the inner circle, genes encoded in the minus strand are indicated in the outer circle. GC content is shown within the map.
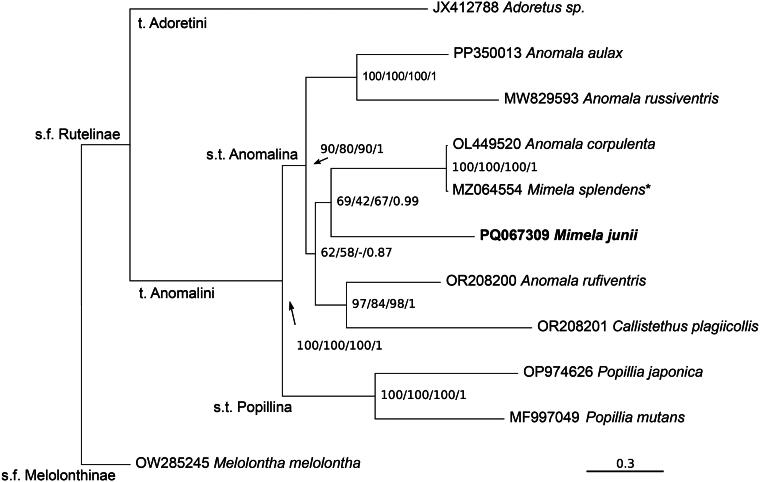
All phylogenetic analyses converged to the same topology (Figure 3), apart from a weakly supported node in the amino-acid dataset. M. junii appears to be nested within genus Anomala with high support and related to A. corpulenta, as well as the unconfirmed M. splendens, with medium/low support.
Figure 3.
Phylogenetic tree of Mimela junii (in bold) in the context of Sub-family Rutelinae. Nodes corresponding to named taxa are thus labeled. s.f. sub-family, t. tribe, s.t. sub-tribe. Branch lengths are from the 1+2+3 codon position analysis; root node not to scale. Support at nodes is indicated as: boostrap 1+2+3 codon position/bootstrap 1+2 codon position/bootstrap aminoacids/posterior probability 1+2+3 codon position. *species attribution tentatively not confirmed. See Table 1 for full species information.
Discussion and conclusion
The uncertainty in determining the number of CR repeats may be due to a methodological bias, as Illumina short reads are not appropriate to resolve complex repeated structures (e.g. Nardi et al. 2024), or have a biological basis, as heteroplasmy in the number of repeats (Nardi et al. 2001, 2012) may be at play. This suggest caution in the interpretation of CR assemblies obtained using short reads and underlines the relevance of the few complete mitochondrial genomes produced based on a combination of long and short reads (e.g. Gastineau et al. 2024; Nardi et al. 2024).
Sequence MZ064554, identified in NCBI as Mimela splendens, was tentatively considered as of unconfirmed attribution because: a) it is almost identical (99.4%) to sequence OL449520 (identified as A. corpulenta in NCBI); and b) both are identified as A. corpulenta based on their barcode in BOLD.
Based on our phylogenetic reconstruction (Figure 3), and in line with the accepted classification of the group (Bouchard et al. 2011), sub-family (s.f.) Rutelinae includes tribe (t.) Adoretini and t. Anomalini, and the latter sub-tribe (s.t.) Popillina and s.t. Anomalina. Within Anomalina, the genus Anomala is recovered as paraphyletic with respect to Mimela and Callisthetus, with M. junii nested within genus Anomala and associated with A. corpulenta and (the unconfirmed) M. splendens. Mitochondrial genomes of Rutelinae have been included into multiple phylogenetic analyses (Timmermans et al. 2015; Song and Zhang 2018; Li et al. 2022; Qu et al. 2023; Long et al. 2024). Limited to shared sequences, our results are identical to previously published phylogenetic trees.
The availability of the mitochondrial genome of M. junii and other Rutelinae is liable to foster molecular identification, including preimaginal stages. Future work along this line should consider sequencing additional mitochondrial genomes from Rutelinae. We nevertheless wish to underline that molecular identification is only as good as the underlying taxonomic identification of the reference material. As such the source material should be confirmed by positive, multi-disciplinary expertise, especially in species-rich and economically relevant taxa such as shining leaf chafers.
The complete sequence of the mitochondrial genome of the shining leaf chafer Mimela junii herein described will promote molecular identification of leaf chafers, a group that include relevant pest, alongside many economically irrelevant, species.
Supplementary Material
Funding Statement
This work was supported by the NBFC to University of Siena/Department of Life Sciences, funded by the Italian Ministry of University and Research, PNRR, Missione 4 Componente 2, ‘Dalla ricerca all’impresa’, Investimento 1.4, Project CN00000033.
Authors contributions statement
FF conceived the study; FF and DB provided samples; FN, CC, RF and AC conducted experiments and analyzed the data; FN drafted the manuscript. All authors reviewed and edited the manuscript.
Ethical statement
The research was exempt from ethical approval or permissions because it does not involve regulated species.
Disclosure statement
No potential competing interest was reported by the authors.
Data availability statement
The genome sequence is publicly available in Genbank of NCBI (https://www.ncbi.nlm.nih.gov/) under accession number PQ067309. The associated BioProject, SRA, and Bio-Sample codes are PRJNA1139055, SRR29924876, and SAMN42749332, respectively.
References
- Ashworth M. 2023. The genome sequence of a cockchafer, Melolontha melolontha (Linnaeus, 1758). Wellcome Open Res. 8:222. PMID: 38618196; PMCID: PMC11015117. doi: 10.12688/wellcomeopenres.19434.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballerio A, Rey A, Uliana M, Rastelli M, Rastelli S, Romano M, Colacurcio, L.. Coleotteri Scarabeoidei d’Italia. 2014. [accessed 2024 Aug 1]. https://www.societaentomologicaitaliana.it/Coleotteri%20Scarabeoidea%20d’Italia%202014/scarabeidi/home.htm.
- Baraud JFdF. 1992. 78. Coléoptères Scarabaeoidea d’Europe. Bulletin Mensuel de la Société Linnéene de Lyon. 78(Suppl):1–856. [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573–580. PMID: 9862982; PMCID: PMC148217. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF.. 2013. Nov MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. Epub 2012 Sep 7. PMID: 22982435. doi: 10.1016/j.ympev.2012.08.023. [DOI] [PubMed] [Google Scholar]
- Bouchard P, Bousquet Y, Davies AE, Alonso-Zarazaga MA, Lawrence JF, Lyal CH, Newton AF, Reid CA, Schmitt M, Slipiński SA, et al. 2011. Family-group names in Coleoptera (Insecta). Zookeys. 88(88):1–972. PMID: 21594053; PMCID: PMC3088472. doi: 10.3897/zookeys.88.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ.. 2014. Apr 10 BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 10(4):e1003537. PMID: 24722319; PMCID: PMC3985171. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brian Bushnell . 2020. BBMap homepage on sourceforge, http://sourceforge.net/projects/bbmap/.
- Chen S, Zhou Y, Chen Y, Gu J.. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890. PMID: 30423086; PMCID: PMC6129281. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucini C, Leo C, Iannotti N, Boschi S, Brunetti C, Pons J, Fanciulli PP, Frati F, Carapelli A, Nardi F.. 2021. EZmito: a simple and fast tool for multiple mitogenome analyses. Mitochondrial DNA B Resour. 6(3):1101–1109. PMID: 33796755; PMCID: PMC7995877. doi: 10.1080/23802359.2021.1899865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. PMID: 28204566; PMCID: PMC5389512. doi: 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. PMID: 15034147; PMCID: PMC390337. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastineau R, Lemieux C, Turmel M, Otis C, Boyle B, Coulis M, Gouraud C, Boag B, Murchie AK, Winsor L, et al. 2024. The invasive land flatworm Arthurdendyus triangulatus has repeated sequences in the mitogenome, extra-long cox2 gene and paralogous nuclear rRNA clusters. Sci Rep. 14(1):7840. PMID: 38570596; PMCID: PMC10991399. doi: 10.1038/s41598-024-58600-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson ML, Paucar-Cabrera A, Solís A.. 2003. Synopsis of the new world genera of Anomalini (Coleoptera: Scarabaeidae: Rutelinae) and description of a new genus from Costa Rica and Nicaragua. an. 96(4):415–432. doi: 10.1603/0013-8746(2003)096[0415:SOTNWG]2.0.CO;2. [DOI] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. Epub 2017 May 8. PMID: 28481363; PMCID: PMC5453245. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SY, Guindon S.. 2012. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29(6):1695–1701. Epub 2012 Jan 20. PMID: 22319168. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 30(22):3276–3278. Epub 2014 Aug 5. PMID: 25095880; PMCID: PMC4221126. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu CM, Luo R, Sadakane K, Lam TW.. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 31(10):1674–1676. Epub 2015 Jan 20. PMID: 25609793. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- Li Y, Nie RE, Lu Y, Lee S, Zhao Z, Wu L, Sun H, Bai M.. 2022. Seven new mitochondrial genomes of phytophagous scarab beetles (Coleoptera: Scarabaeidae) and phylogenetic implications. Zootaxa. 5138(3):324–338. PMID: 36095833. doi: 10.11646/zootaxa.5138.3.6. [DOI] [PubMed] [Google Scholar]
- Long T, Zhu W, Yang L, Long J, Chang Z, Chen X.. 2024. First report of the complete mitochondrial genome of 3 beetles (Coleoptera: Scarabaeidae) harming Gastrodia elata (Asparagales: Orchidaceae). J Insect Sci. 24(1):12. PMID: 38387434; PMCID: PMC10883712. doi: 10.1093/jisesa/ieae009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R.. 2020. May 1 IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. Erratum in: Mol Biol Evol. 2020 Aug 1;37(8):2461. doi: 10.1093/molbev/msaa131. PMID: 32011700; PMCID: PMC7182206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardi F, Boschi S, Funari R, Cucini C, Cardaioli E, Potter D, Asano SI, Toubarro D, Meier M, Paoli F, et al. 2024. The direction, timing and demography of Popillia japonica (Coleoptera) invasion reconstructed using complete mitochondrial genomes. Sci Rep. 14(1):7120. PMID: 38531924; PMCID: PMC10965970. doi: 10.1038/s41598-024-57667-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardi F, Carapelli A, Fanciulli PP, Dallai R, Frati F.. 2001. Jul The complete mitochondrial DNA sequence of the basal hexapod Tetrodontophora bielanensis: evidence for heteroplasmy and tRNA translocations. Mol Biol Evol. 18(7):1293–1304. doi: 10.1093/oxfordjournals.molbev.a003914. [DOI] [PubMed] [Google Scholar]
- Nardi F, Carapelli A, Frati F.. 2012. Repeated regions in mitochondrial genomes: distribution, origin and evolutionary significance. Mitochondrion. 12(5):483–491. Epub 2012 Jul 20. PMID: 22820120. doi: 10.1016/j.mito.2012.07.105. [DOI] [PubMed] [Google Scholar]
- Pons J, Ensenyat JJ, Bover P, Serra M, Nardi F.. 2021. Aln2tbl: building a mitochondrial features table from a assembly alignment in fasta format. Mitochondrial DNA B Resour. 6(9):2732–2735. PMID: 34447886; PMCID: PMC8386716. doi: 10.1080/23802359.2021.1966334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu C-j, Zhu Y, Jiang C, Qu M-j, Wang X-y, Li X.. 2023. Whole mitochondrial genome and phylogeny analysis of Anomala corpulenta. Biotechnology Bulletin. 39(2):263–273. doi: 10.13560/j.cnki.biotech.bull.1985.2022-0689. [DOI] [Google Scholar]
- Ratnasingham S, Hebert PD.. 2007. bold: the Barcode of Life Data System (http://www.barcodinglife.org). Mol Ecol Notes. 7(3):355–364. PMID: 18784790; PMCID: PMC1890991. doi: 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song N, Zhang H.. 2018. The mitochondrial genomes of phytophagous scarab beetles and systematic implications. J Insect Sci. 18(6):11. PMID: 30508200; PMCID: PMC6275328. doi: 10.1093/jisesa/iey076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans MJ, Barton C, Haran J, Ahrens D, Culverwell CL, Ollikainen A, Dodsworth S, Foster PG, Bocak L, Vogler AP.. 2015. Family-level sampling of mitochondrial genomes in Coleoptera: compositional heterogeneity and phylogenetics. Genome Biol Evol. 8(1):161–175. PMID: 26645679; PMCID: PMC4758238. doi: 10.1093/gbe/evv241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Poczai P, Hyvönen J, Tang J, Amiryousefi A.. 2020. Chloroplot: an online program for the versatile plotting of organelle genomes. Front Genet. 11:576124. PMID: 33101394; PMCID: PMC7545089. doi: 10.3389/fgene.2020.576124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequence is publicly available in Genbank of NCBI (https://www.ncbi.nlm.nih.gov/) under accession number PQ067309. The associated BioProject, SRA, and Bio-Sample codes are PRJNA1139055, SRR29924876, and SAMN42749332, respectively.