Abstract
Primary human keratinocytes with ectopic expression of high-risk human papillomavirus (HPV) E6 and E7 oncoproteins display abnormal centrosome numbers, multipolar mitoses, and aneusomy. However, it has not been explored whether these abnormalities can occur in cells containing HPV episomes where E6 and E7 expression is under viral transcriptional control. Here, we demonstrate that centrosome abnormalities and genomic instability occur in organotypic raft cultures of human keratinocytes with episomal HPV-16 even at low copy numbers. We conclude that HPV-16 DNA, when maintained as an episome, can disturb centrosome homeostasis and subvert genomic integrity of the host cell during early stages of the viral infection.
Cervical cancer is tightly associated with infection by certain high-risk human papillomavirus (HPV) types such as HPV-16 or HPV-18, whereas low-risk types like HPV-6 are associated with benign genital warts (21). High-risk HPVs encode two oncoproteins, E6 and E7, that subvert the crucial cell cycle regulatory molecules p53 (15) and pRB (2, 7), respectively. Genomic instability as manifested by numeric chromosome imbalances (i.e., aneuploidy) is an early step during carcinogenic progression (11, 20) and can be detected already in preinvasive high-risk HPV-positive cervical malignancies (10, 12). We have previously shown that abnormal multipolar mitoses, which are histopathological hallmarks of these lesions (3), are associated with abnormal centrosome numbers and that overexpression of HPV-16 E6 and E7 oncoproteins can cause such alterations (4). Centrosomes serve as major microtubule organizing centers in interphase and mitotic cells and ensure accurate chromosome segregation and symmetry of the cell division process (19).
Previously, we have analyzed centrosome abnormalities in primary keratinocytes with ectopic expression of high-risk HPV E6 and/or E7 oncoproteins. This was to model the situation in the majority of cervical cancers, wherein E6 and E7 expression is dysregulated due to chromosomal integration of HPV DNA and disruption of viral E2 gene regulation (21). However, in precursor lesions, the HPV-16 genome is maintained as an episome, and expression of E6 and E7 is tightly regulated and restrained by the viral E2 transcriptional repressor. Given that aneuploidy is detected already in preinvasive cervical lesions, we sought to determine whether centrosome abnormalities and signs of genomic instability can be detected in cells where E6 and E7 are expressed from the HPV-16 episome. To address this question, we analyzed organotypic raft cultures of keratinocytes that maintain HPV-16 episomes (8). Here, we report that centrosome abnormalities and chromosomal imbalances are present in these raft cultures. Moreover, such abnormalities already occur in basal cells, which contain HPV-16 at a low copy number. Thus, episomal expression of HPV-16 DNA during productive viral infection is sufficient to induce these alterations, and these processes do not require HPV genome amplification or viral integration.
Detection of HPV-16 DNA in organotypic keratinocyte raft cultures by fluorescence in situ hybridization (FISH).
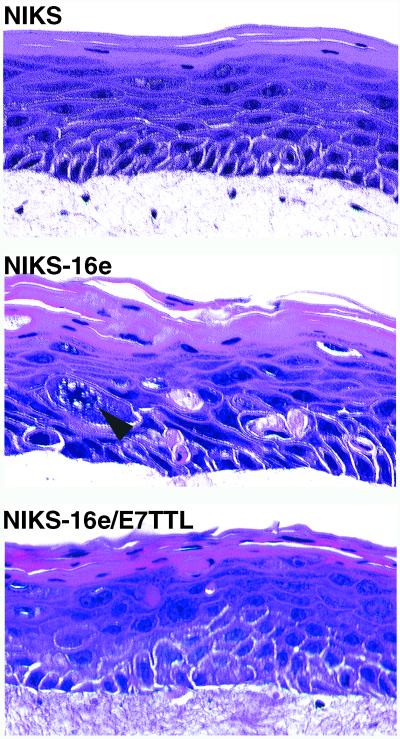
Rafts were generated from a spontaneously immortalized keratinocyte cell line (NIKS [1]) that supports the viral life cycle of HPV-16, as reported previously (8). These cells were chosen because, unlike normal human keratinocytes (17), they support episomal maintenance of HPV genomes in the absence of E7 function. This cell line also retains normal differentiation properties (Fig. 1) (1). Rafts were also prepared from NIKS harboring either wild-type episomal HPV-16 (NIKS-16e–BC-1-EP/SL HPV16 [8]) or E7-deficient HPV-16 episomes with a translational termination linker (TTL) inserted at nucleotide 711 of the E7 gene (NIKS-16/E7TTL–BC-1-EP/SL HPV-16/E7TTL [8]). These lines have been previously analyzed for the episomal maintenance of HPV-16 genomes by Southern blotting (8, 9). The lines contain 5 to 50 HPV-16 episomes per cell (9), and there is no evidence for viral integration (8, 9). Morphological analysis of raft cultures using hematoxylin and eosin staining revealed a relatively normal tissue architecture in NIKS rafts. In contrast, the disturbed tissue stratification in NIKS-16e and NIKS-16e/E7TTL rafts, together with nuclear abnormalities, was reminiscent of HPV-associated preinvasive genital squamous lesions (Fig. 1).
FIG. 1.
Hematoxylin and eosin staining of raft cultures obtained from a spontaneously immortalized keratinocyte cell line (NIKS), and NIKS stably transfected with wild-type HPV-16 DNA (NIKS-16e) or HPV-16 with disabled E7 oncogene expression (NIKS-16e/E7TTL). The arrowhead indicates an abnormal mitotic figure.
In an HPV-infected epithelium, the HPV DNA is present at a low copy number in basal cells. In suprabasal cells, HPV genome amplification occurs and the viral genome is present at a higher copy number. We used a nonradioactive FISH procedure with HPV-16 DNA as a probe (Fig. 2) to analyze the HPV-16 DNA copy numbers in the different strata of these rafts. HPV-16 DNA isolated from W12E cells (line 20863 [13]) was labeled with digoxigenin (Bioprime DNA Labeling System; Gibco). Sections (4 μm) from formalin-fixed, paraffin-embedded specimens were deparaffinized in xylene and then dehydrated. Slides were then heated to 92°C in 100 mM Tris base–50 mM EDTA (pH 7.0) for 15 min in a microwave oven and digested with Digest-All 3 pepsin solution (Zymed) for 10 s at 37°C. After dehydration in a graded ethanol series, the slides were air dried and 1 μl of digoxigenin-labeled HPV-16 genome probe was added to each slide. Codenaturation of tissue DNA and probe was performed for 3 min at 85°C. Hybridization was then carried out overnight in a humidified chamber at 37°C. After a posthybridization wash, signals were detected with fluorescein isothiocyanate (FITC)–anti-digoxigenin (Boehringer Mannheim) at a dilution of 1:400 for 25 min at 37°C. Nuclei were counterstained with DAPI (4′,6′-diamidino-2-phenylindole; Vector).
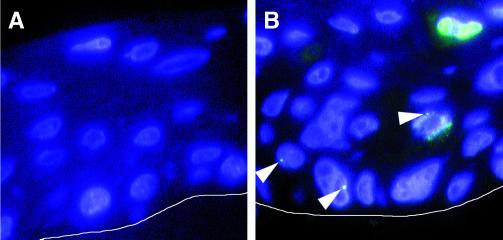
FIG. 2.
(A) FISH analysis of spontaneously immortalized HPV-negative human foreskin keratinocytes (NIKS) grown as organotypic raft cultures. No positive signals for HPV-16 were detected. Nuclei were counterstained with Hoechst 33258. A white line was added to indicate the border between the underlying collagen matrix and the basal layer of the raft. (B) NIKS that harbor HPV-16 episomes (NIKS-16e) were analyzed for the HPV-16 DNA by FISH. Single fluorescence dots indicating low-copy-number amplification were seen in basal cells or occasionally in cells undergoing mitosis (arrowheads). Strong fluorescence signals comprising large parts of the nuclei, indicating high-copy-number viral DNA amplification, were detected in the suprabasal layers of the rafts. Nuclei were counterstained with Hoechst 33258. The white line indicates the border between the underlying collagen matrix and the basal layer of the raft.
W12E cells, which stably maintain HPV-16 episomes (13), were used as controls, and HPV-16 FISH analysis revealed a dot-like fluorescence staining pattern. This indicates that under these conditions HPV episomes tend to form cluster-like structures within the nuclei of infected cells. A similar dot-like staining pattern was detected in NIKS-16e (Fig. 2B) and NIKS-16e/E7TTL, whereas, as expected, no signals were detected in untransfected NIKS (Fig. 2A). The mean signal number per cell, i.e., fluorescent dots per nucleus, was 1.6 in NIKS-16e and 2.0 in NIKS-16e/E7TTL.
Cells with small, well-defined dots were often present in the basal compartment or in the lower third of the suprabasal compartment of the rafts (Fig. 2B). In addition, some cells displayed enlarged or multiple signals (Fig. 2B) or signals that comprised almost the entire nucleus (Fig. 2B). These stronger fluorescence signals were found in 12.4% of the cells that could be evaluated in NIKS-16e rafts, but only in 2.7% of NIKS-16e/E7TTL cells, a result consistent with the observation that the E7 oncogene is required for the full viral life cycle in these cells (9). Cells with enlarged fluorescence signals were frequently found within the upper half of the rafts, and such cells frequently displayed morphological alterations, predominantly binucleation, as exemplified in Fig. 2B. These results agree with previous in situ hybridization studies (8, 9) and demonstrate that, although FISH analysis does not provide exact quantitation of the HPV DNA, it is a valuable tool to study HPV copy number changes related to the viral life cycle in comparison to other cellular alterations.
Episomal expression of HPV-16 DNA is sufficient to induce abnormal centrosome numbers.
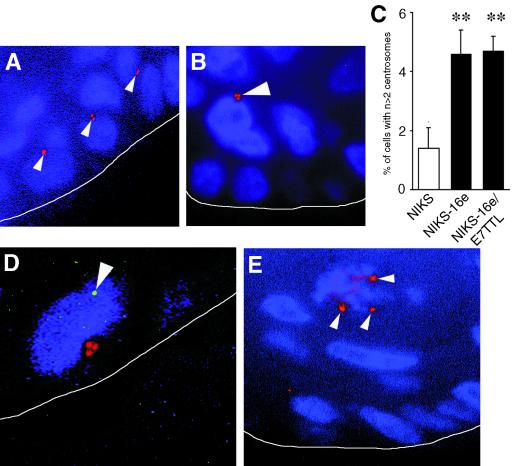
To determine whether episomal expression of HPV DNA in keratinocytes is sufficient to induce centrosome abnormalities, rafts were stained for centrosomes by immunofluorescence for gamma-tubulin (Sigma) as described previously (4). Since normal cells contain one or two centrosomes (Fig. 3A), we considered three or more centrosomes per cell abnormal (Fig. 3B). Numeric centrosome abnormalities were detected in 1.4% of nontransfected NIKS whereas 4.6% of NIKS-16e and 4.7% of NIKS-16e/E7TTL cells showed more than two centrosomes (Fig. 3C). Differences between untransfected NIKS and NIKS-16e and NIKS-16e/E7TTL were statistically significant (Student's t test, P ≤ 0.02). Effectively, these numbers are likely to be higher as in some cases centrosomes could have been lost due to the sectioning of individual cells.
FIG. 3.
(A) Detection of centrosomes in NIKS by immunofluorescence using the pericentriolar marker gamma-tubulin. Normal centrosome numbers are shown, i.e., one or two centrosomes per cell (red, arrowheads). Nuclei counterstained with Hoechst 33258. The white line was added to indicate the border between the underlying collagen matrix and the basal layer of the raft. (B) Abnormal centrosomes (red, arrowhead) in NIKS harboring HPV-16 episomes (NIKS-16e). Nuclei were counterstained with Hoechst 33258. The white line was added to indicate the border between the underlying collagen matrix and the basal layer of the raft. (C) Quantitation of abnormal centrosome numbers in rafts from NIKS and NIKS harboring HPV-16 episomes (NIKS-16e) or NIKS harboring HPV-16 episomes with disrupted E7 expression (NIKS-16e/E7TTL). The means and standard errors of at least triplicate centrosome evaluation are shown (∗∗, P ≤ 0.02 [Student's t test for independent samples]). (D) Abnormal centrosomes (red) in a basal cell with low-copy HPV DNA amplification (arrowhead, green) detected in NIKS harboring HPV-16 episomes (NIKS-16e). Nuclei were counterstained with Hoechst 33258. The white line was added to indicate the border between the underlying collagen matrix and the raft culture. (E) Multipolar mitosis with three participating centrosomes (red, arrowheads) in a raft from NIKS-16e cells. Nuclei were counterstained with Hoechst 33258. The white line was added to indicate the border between the underlying collagen matrix and the raft culture.
In NIKS-16e cells approximately 50% of cells displaying abnormal centrosomes numbers were basal cells. A similar distribution was observed in rafts from untransfected NIKS, although the overall incidence of centrosome abnormalities was lower in these cells (Fig. 3C). However, in NIKS-16e/E7TTL cells, the proportion of basal cells with centrosome abnormalities was lower and represented only approximately 20% of the cells with abnormal centrosome numbers.
To investigate directly whether centrosome abnormalities occur in basal cells that contain HPV-16 genomes at a low copy number, we performed a representative costaining experiment for HPV-16 DNA by FISH and centrosomes using immunofluorescence for gamma-tubulin on rafts obtained from NIKS-16e cells. As shown in Fig. 3D, centrosome abnormalities can occur in basal cells that contain HPV-16 genomes at a low copy number. This finding further supports the notion that high-risk HPV-induced centrosome abnormalities occur in cells that maintain HPV-16 genomes at a low copy number prior to viral DNA amplification or HPV genome integration.
Genomic Instability in organotypic rafts expressing HPV-16 DNA episomally.
Next, we analyzed the raft cultures for the occurrence of abnormal multipolar mitoses associated with abnormal centrosome numbers. We found sporadic multipolar mitoses in rafts from NIKS-16e (Fig. 3E) and NIKS-16e/E7TTL cells, indicating that the abnormal centrosomes detected in the rafts cultures are functional and can induce centrosome-related mitotic defects.
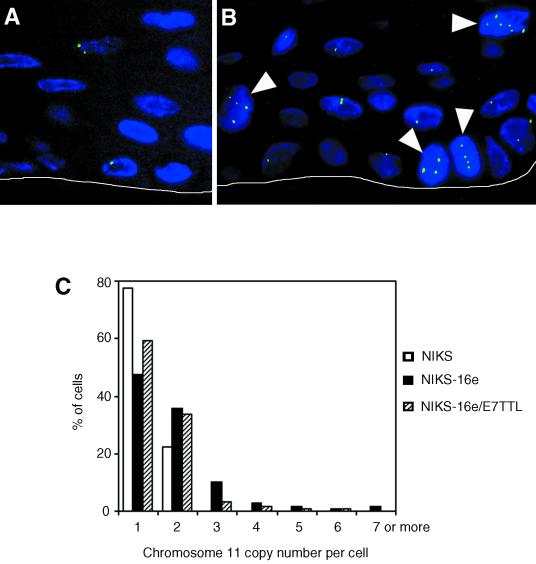
Given the finding of centrosome abnormalities and multipolar mitoses in the raft cultures, we next determined whether these rafts also show numeric chromosome imbalances as a reflection of aneuploidy. NIKS are genetically stable and near-diploid (1). FISH analysis using a Spectrum Green-labeled α-satellite probe for chromosome 11 (D11Z1; Vysis) was employed as previously described (4) for enumeration of the chromosome 11 copy numbers (Fig. 4). Sectioning of the paraffin-embedded tissue can result in loss of signals; hence, only cells with at least one fluorescence signal were analyzed.
FIG. 4.
(A) Normal copy number of chromosome 11 (green) in NIKS. Nuclei were counterstained with DAPI. The white line was added to indicate the border between the underlying collagen matrix and the basal layer of the raft. (B) Genomic instability as manifested by aneusomy of chromosome 11 (arrowheads indicate cells with three or more copies of chromosome 11, respectively) in a raft generated from NIKS harboring HPV-16 (NIKS-16e). Nuclei were counterstained with DAPI. The white line was added to indicate the border between the underlying collagen matrix and the basal layer of the raft. (C) Quantitation of chromosome 11 copy number variability in rafts generated from NIKS, NIKS harboring HPV-16 episomes (NIKS-16e), or NIKS harboring episomes in which expression of the E7 oncogene is disrupted (NIKS-16e/E7TTL). At least 100 nuclei were assessed for each cell line studied.
In parental NIKS (Fig. 4A), all evaluable cells showed one or two signals per cell. Abnormal chromosome 11 copy numbers (i.e., more than two per cell; Fig. 4B) were observed in 16.8% of NIKS-16e cells but only in 6.8% of NIKS-16e/E7TTL rafts. These results support our previous findings and show that expression of HPV-16 E7 enhances the process of genomic destabilization (4, 6).
Cells with aneusomy for chromosome 11 were found in the basal layer in 29.6% of cells in the NIKS-16e rafts and in 23.1% of cells in the NIKS-16e/E7TTL rafts (Fig. 4B). This indicates that low-copy-number episomal expression of HPV-16 is sufficient to induce genomic instability as manifested by aneusomy for chromosome 11. Hence, genomic instability can develop in basal cells that contain episomal HPV-16 at a low copy number.
We have previously shown that HPV-16 E6 and E7 oncoproteins cooperate to induce abnormal centrosome numbers, multipolar mitoses, and numerical chromosome imbalances in primary human keratinocytes (4, 6). However, we discovered striking mechanistic differences between high-risk HPV E6 and E7 with respect to centrosome abnormalities (reviewed in reference 5). Overexpression of either E6 or E7 resulted in abnormal centrosome numbers in stable cell lines, whereas only HPV-16 E7 had an immediate effect on centrosome numbers when expressed transiently (4). Since the raft cultures used in the present study are generated from stable cell lines, as expected, both NIKS-16e cells and NIKS-16e/E7TTL cells in which expression of E7 is disrupted show centrosome abnormalities and aneuploidy. Interestingly, however, only 20% of centrosome abnormalities were in basal cells in the NIKS-16e/E7TTL rafts compared to 50% in NIKS-16e rafts. These results support the model that HPV E7 can rapidly uncouple centrosome synthesis from the cell division cycle (4, 6). Expression of high-risk HPV E6 cooperates with E7 to induce these changes most likely by relaxing mitotic checkpoint functions (16, 18).
This is the first report to show that episomal expression of HPV-16 DNA is sufficient to induce centrosome abnormalities. These findings suggest that centrosome-related mitotic dysfunction and increased genomic plasticity can already occur in cells that contain high-risk HPV at a low copy number. While most of these cells may not be endowed with an inherent growth advantage and in fact may represent abortive events (6), this increases the overall risk that some of these cells may acquire mutations that contribute to malignant progression. Interestingly, it has been suggested that progressive genomic destabilization may facilitate the stages of carcinogenic progression, including integration of episomal DNA (14). Viral integration associated with disruption of E2 expression during malignant progression leads to dysregulated expression of E6 and E7, which may cause an additional increase in genomic instability. Further mechanistic understanding of these processes may help to develop better strategies to suppress the development of genomic instability at early stages of malignant progression.
Acknowledgments
The first two authors contributed equally to this work.
This study was supported by NIH grant CA66980 to K.M. and American Cancer Society grant RSG-96-043-06 and NIH grants CA22443 and CA07175 to P.F.L. S.D. is supported by a fellowship from the Deutsche Forschungsgemeinschaft (Du 343/1-1). Anette Duensing is a postdoctoral fellow of the Dr. Mildred Scheel Stiftung. E.R.F. is a postdoctoral fellow of the Leukemia and Lymphoma Society of America. K.M. is a Ludwig Scholar.
REFERENCES
- 1.Allen-Hoffmann B L, Schlosser S J, Ivarie C A R, Sattler C A, Meisner L F, O'Connor S L. Normal growth and differentiation in a spontaneously immortalized near-diploid human keratinocyte cell line, NIKS. J Investig Dermatol. 2000;114:444–455. doi: 10.1046/j.1523-1747.2000.00869.x. [DOI] [PubMed] [Google Scholar]
- 2.Boyer S N, Wazer D E, Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- 3.Crum C P, Ikenberg H, Richart R M, Gissman L. Human papillomavirus type 16 and early cervical neoplasia. N Engl J Med. 1984;310:880–883. doi: 10.1056/NEJM198404053101403. [DOI] [PubMed] [Google Scholar]
- 4.Duensing S, Lee L Y, Duensing A, Basile J, Piboonniyom S, Gonzalez S, Crum C P, Münger K. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc Natl Acad Sci USA. 2000;97:10002–10007. doi: 10.1073/pnas.170093297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duensing S, Münger K. Centrosome abnormalities, genomic instability and carcinogenic progression. Biochim Biophys Acta. 2001;1471:M81–M88. doi: 10.1016/s0304-419x(00)00025-1. [DOI] [PubMed] [Google Scholar]
- 6.Duensing S, Duensing A, Crum C P, Münger K. Human papillomavirus type 16 E7 oncoprotein-induced abnormal centrosome synthesis in an early event in the evolving malignant phenotype. Cancer Res. 2001;61:2356–2360. [PubMed] [Google Scholar]
- 7.Dyson N, Howley P M, Münger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 8.Flores E R, Allen-Hoffmann L, Lee D, Sattler C A, Lambert P F. Establishment of the human papillomavirus type 16 (HPV-16) life cycle in an immortalized human foreskin keratinocyte cell line. Virology. 1999;262:344–354. doi: 10.1006/viro.1999.9868. [DOI] [PubMed] [Google Scholar]
- 9.Flores E R, Allen-Hoffmann L, Lee D, Lambert P F. The human papillomavirus type 16 E7 oncogene is required for the productive stage of the viral life cycle. J Virol. 2000;74:6622–6631. doi: 10.1128/jvi.74.14.6622-6631.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Y S, Reagan J W, Richart R M. Definition of precursors. Gynecol Oncol. 1981;12:S220–S231. doi: 10.1016/0090-8258(81)90076-7. [DOI] [PubMed] [Google Scholar]
- 11.Hashida T, Yasumoto S. Induction of chromosome abnormalities in mouse and human epidermal keratinocytes by the human papillomavirus type 16 E7 oncogene. J Gen Virol. 1991;72:1569–1577. doi: 10.1099/0022-1317-72-7-1569. [DOI] [PubMed] [Google Scholar]
- 12.Heselmeyer K, Schrock E, Du Manoir S, Blegen H, Shah K, Steinbeck R, Auer G, Ried T. Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc Natl Acad Sci USA. 1996;93:479–484. doi: 10.1073/pnas.93.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeon S, Allen-Hoffmann B L, Lambert P F. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J Virol. 1995;69:2989–2997. doi: 10.1128/jvi.69.5.2989-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kessis T D, Connolly D C, Hedrick L, Cho K R. Expression of HPV16 E6 or E7 increases integration of foreign DNA. Oncogene. 1996;13:427–431. [PubMed] [Google Scholar]
- 15.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 16.Thomas J T, Laimins L A. Human papillomavirus oncoproteins E6 and E7 independently abrogate the mitotic spindle checkpoint. J Virol. 1998;72:1131–1137. doi: 10.1128/jvi.72.2.1131-1137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas J T, Hubert W G, Ruesch M N, Laimins L A. Human papillomavirus type 31 oncoprotein E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc Natl Acad Sci USA. 1999;96:8449–8454. doi: 10.1073/pnas.96.15.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson D A, Belinsky G, Chang T H, Schegel R, Münger K. The human papillomavirus-16 E6 oncoprotein decreases the vigilance of mitotic checkpoints. Oncogene. 1997;15:3025–3035. doi: 10.1038/sj.onc.1201495. [DOI] [PubMed] [Google Scholar]
- 19.Urbani L, Stearns T. The centrosome. Curr Biol. 1999;9:R315–R317. doi: 10.1016/s0960-9822(99)80201-2. [DOI] [PubMed] [Google Scholar]
- 20.White A E, Livanos E M, Tlsty T D. Differential disruption of genomic integrity and cell cycle regulation in normal human fibroblasts by the HPV oncoproteins. Genes Dev. 1994;8:666–677. doi: 10.1101/gad.8.6.666. [DOI] [PubMed] [Google Scholar]
- 21.zur Hausen H. Viruses in human cancers. Science. 1991;254:1167–1173. doi: 10.1126/science.1659743. [DOI] [PubMed] [Google Scholar]