Abstract
This report presents the design and synthesis of quinazolinone-based derivatives as promising SARS-CoV-2 3CL protease inhibitors. Two novel series, namely, febrifugine analogues 4a–i and quinazolinone conjugated benzimidazoles 9a–c, were successfully synthesized starting from isatoic anhydride. The synthesized quinazolinone derivatives were evaluated for their cytotoxicity against cancer cell lines and SARS-CoV-2 3CL inhibitory activity. The results showed that the synthesized compounds did not have significant toxicity for the non-cancer HEK293 cell line and MCF-7, MDA-MB-231, HEPG2 and HEPG2.2.15 cancer cell lines. Notably, compound 9b exhibited anti-3CL enzymatic activity in a dose-dependent manner, with the calculated IC50 value of 10.73 ± 1.17 μM. Docking results highlighted the interaction between 9b and 3CL protease through hydrogen bonding with key amino acids, including His41, Met49, Cys145, Met165, Arg188, His164, and Glu166, at the active site of the protease. Pharmacokinetic studies and ADME analyses provide valuable insights into the potential of compound 9b as a drug candidate. These findings support the new scaffold as a candidate for 3CLpro inhibition and advanced anti-coronavirus drug research.
This report presents the design and synthesis of quinazolinone-based derivatives as promising SARS-CoV-2 3CL protease inhibitors.
1. Introduction
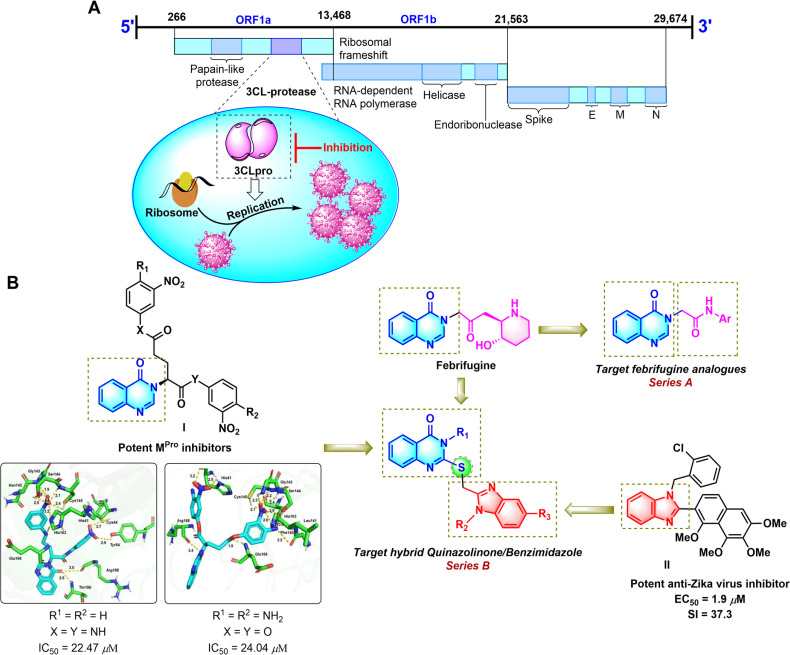
Coronaviruses (CoVs), which are known to cause respiratory and gastrointestinal infections in humans and animals, belong to the Coronaviridae family and are positive-sense RNA, single-stranded viruses.1–3 SARS-CoV and MERS-CoV are the two novel βCoVs that display high mortality rates compared to other CoVs. SARS-CoV-2 is a novel CoV that causes COVID-19 disease by entering the host cells and binding to angiotensin-converting enzyme 2 (ACE 2) leading to respiratory and gastrointestinal sickness.4–6 Current therapeutic methods for COVID-19 mainly rely on the conserved enzymes and proteins such as nucleoside-triphosphate hydrolase, RNA-dependent RNA polymerase (RdRP, nsp12), helicase (NTPase/helicase, nsp13), the spike protein (S), papain-like protease (PLpro) and main proteases (Mpro or 3CLpro, nsp5). Among these, the main protease (Mpro or 3CLpro), which plays a key role in the replication cycle of CoVs has emerged as an attractive target for the treatment of infectious diseases caused by CoVs, including COVID-19 (Fig. 1A).7,8 Because there have not been protease inhibitors for the treatment of COVID-19 approved by the FDA, continuous efforts have been made to discover 3CLpro inhibitory agents with the desired pharmacokinetic nature.
Fig. 1. The 3C-like proteinase (3CLpro) of cleavage site numbers of the polypeptide chain (A). Rationally designed template for targeted molecules (B).
Quinazolinone is one of the most important nitrogen-containing heterocycles in medicinal chemistry.9–11 This privileged scaffold is widely found in natural alkaloids, such as luotonin F and febrifugine. Moreover, some studies have indicated that synthetic derivatives containing the quinazolinone moiety exhibit a broad range of biological activities,11,12 especially anticancer13 and antiviral activities.14 Besides, benzimidazole is an important class of fused nitrogen-containing heterocycles with diverse biological activities, including anti-bacterial,15 anti-fungal,16 antihistaminic,17 anti-inflammatory,18 anti-HIV,19 anti-hepatitis C virus (HCV),20 and anti-Zika virus activity.21
Nowadays, molecular hybridization has become an increasingly attractive method in drug design and development by combining two or more known active pharmacophores to produce a new hybrid molecule with enhanced biological activity and increased interaction with more than one biological target.22 The activity of hybrid structures combining the quinazolinone core structure with other biologically active pharmacophores has been reported in the literature.23,24 Some novel hybrid quinazoline-based pyrazolinone and imidazolinone derivatives have recently been found as potent cytotoxic agents.25,26 Furthermore, cytotoxicity and dihydrofolate reductase inhibition activity of quinazolinone conjugating with benzimidazole have already been reported.27,28 Yet, the antiviral activity, especially the SARS-CoV-2 3CL protease inhibition activity, of quinazolinone/benzimidazole hybrids, to the best of our knowledge, has not been reported. In an effort to develop heterocyclic compounds with potential anticancer and antiviral activities,21,29–31 we herein report the design and synthesis of two series of quinazolinone-based SARS-CoV-2 3CLpro inhibitors including the febrifugine analogs N-substituted acetamide 4(3H)-quinazolinone (series A, Fig. 1B) and the benzimidazole conjugated 4(3H)-quinazolinones (series B). For rationalization of the antiviral activity, a molecular docking study with ligand binding free energy estimation and pharmacokinetic studies were performed.
2. Results
2.1. Chemical design and synthesis
Based on the fact that febrifugine showed interesting in silico energetic and interactive activities with the COVID-19 ACE2 receptor,32 we designed a series of febrifugine analogues targeting the SARS-CoV-2 3CLpro (series A, Fig. 1B). On the other hand, in our previous findings, we discovered that the quinazolinone-containing compounds I were potent SARS-CoV-2 3CL protease inhibitors with the IC50 values of 22.47 μM and 24.04 μM (Fig. 1B).30 Furthermore, benzimidazole containing derivative II inhibited an African ZIKV strain with a selectivity index (SI = CC50/EC50) of 37.3.21 Therefore, it was expected that the combination of such inherent antiviral scaffolds as quinazolinone and benzimidazole would enhance the antiviral activity of the resulting structures. Accordingly, the unique quinazolinone-benzimidazole hybrids (series B) were designed, in which the thioether-linked benzimidazole moiety was connected to the quinazolinone ring at the N-3 position. The presence of the thioether linkage is expected to enhance the activity by improving the liposolubility of the hybrids leading to good tissue permeability.33
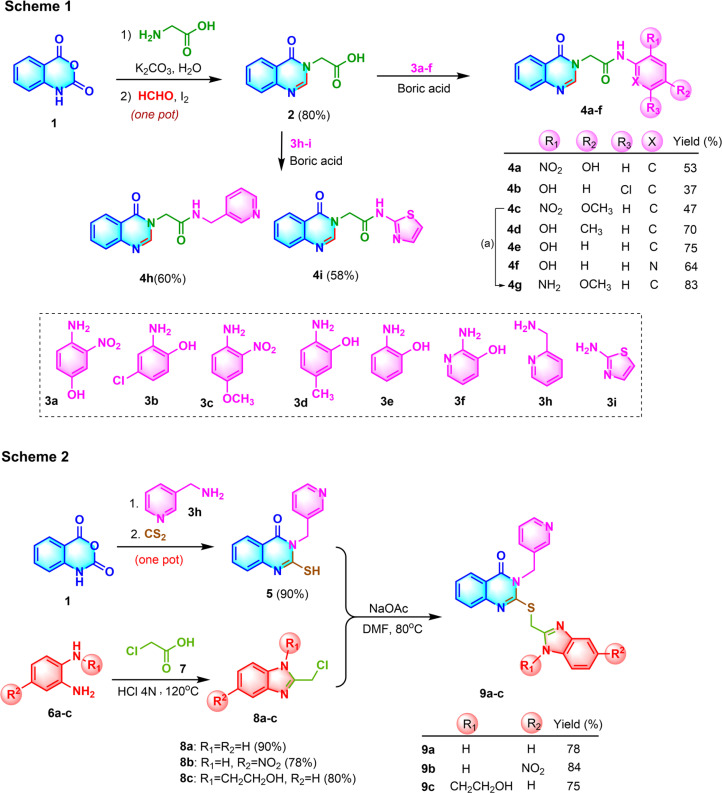
The synthetic pathway toward the febrifugine analogues (series A) is depicted in Scheme 1 (Fig. 2). The quinazolinone core structure 2 was obtained in good yield based on a one-pot tandem ring opening of isatoic anhydride 1 by glycine followed by iodine-catalyzed oxidative cyclization with formaldehyde using water as the only solvent under mild basic conditions.29 The carboxyl moiety of 2 was then coupled with the corresponding aromatic amines 3a–f using boric acid as the coupling agent to afford the desired febrifugine analogues 2-quinazolinyl-N-arylacetamides 4a–f, 4h–i. For the synthesis of 4g, the NO2 group of 4c was reduced to the corresponding amino group using zinc as the reductant under mild reaction conditions.
Fig. 2. Schematic of the synthesis of the quinazolinone based SARS-CoV-2 3CL protease inhibitor. Synthesis of febrifugine analogues (Scheme 1) (a: Zn, CH3COONH4, MeOH, 83%) and quinazolinone conjugated benzimidazole (Scheme 2).
For the synthesis of quinazolinone-conjugated benzimidazole derivatives (series B), two fragments including the 2-mercapto-3-(pyridin-3-ylmethyl)quinazolin-4(3H)-one 5 and 1,5-disubstituted 2-chloromethylbenzimidazoles 8a–c were prepared (Scheme 2, Fig. 2).
Starting from isatoic anhydride 1, the tandem ring opening by pyridin-3-ylmethanamine 3h followed by base-promoted cyclization with carbon disulfide accomplished the quinazolinone core structure 5 bearing the thiol moiety at the C-2 position of the ring, which could function as an anchor to connect to the benzimidazole moiety. The benzimidazole-containing fragments 8a–c were prepared by the condensation of o-phenylenediamine derivatives 6a–c with 2-chloroacetic acid 7 under acidic conditions.31 Finally, the two fragments 5 and 8a–c were connected via a nucleophilic substitution reaction to afford three desired quinazolinone-conjugated benzimidazoles 9a–c in rather good yields. The structures of all the synthesized quinazolinone derivatives were characterized by 1H-NMR, 13C-NMR, FT-IR and HR-MS, and the detailed data are included in the ESI file.†
2.2. Cytotoxicity of quinazolinone derivatives
Cytotoxicities of 13 quinazolinone derivatives on two breast cancer cell lines, the hepatocellular carcinoma (HCC), parent HepG2 and its hepatitis B virus (HBV) transformed HepG2.2.15 and the non-cancer HEK293 cell lines were evaluated. The results presented in Table 1 indicate that all quinazolinone compounds had slight or no toxicity against all cancer cell lines as well as the non-cancer HEK293 cell.
Cytotoxicity of the quinazolinone-based derivatives against the healthy cell line and different cancer cell lines.
| Compd | IC50a (μM) | ||||
|---|---|---|---|---|---|
| Non-cancer cellsb | Cancer cellsc | ||||
| HEK293 | MCF-7 | MDA-MB-231 | HEPG2 | HEPG2.2.15 | |
| 4a | >100 | >100 | >100 | >100 | >100 |
| 4b | >100 | >100 | >100 | >100 | >100 |
| 4c | >100 | >100 | >100 | >100 | >100 |
| 4d | >100 | >100 | >100 | >100 | >100 |
| 4e | >100 | >100 | >100 | 54.89 ± 15.23 | >100 |
| 4f | >100 | >100 | >100 | >100 | >100 |
| 4g | >100 | >100 | >100 | >100 | >100 |
| 4h | >100 | 58.16 ± 8.04 | 34.39 ± 1.53 | 51.36 ± 10.22 | 51.98 ± 28.42 |
| 4i | >100 | >100 | >100 | 47.67 ± 16.16 | 79.47 ± 10.14 |
| 5 | >100 | >100 | >100 | >100 | 46.35 ± 4.75 |
| 9a | 50.19 ± 4.10 | >100 | >100 | >100 | >100 |
| 9b | >100 | >100 | >100 | >100 | >100 |
| 9c | >100 | >100 | >100 | >100 | >100 |
IC50: 50% inhibitory concentration. The data are shown in mean ± SD.
HEK293 (human embryonic kidney).
MCF-7 and MDA-MB-231 (human breast cancer), HepG2 and HepG2.2.15 (human hepatocellular carcinoma).
2.3. Screening for SARS-CoV-2 3CL enzyme inhibitory activity
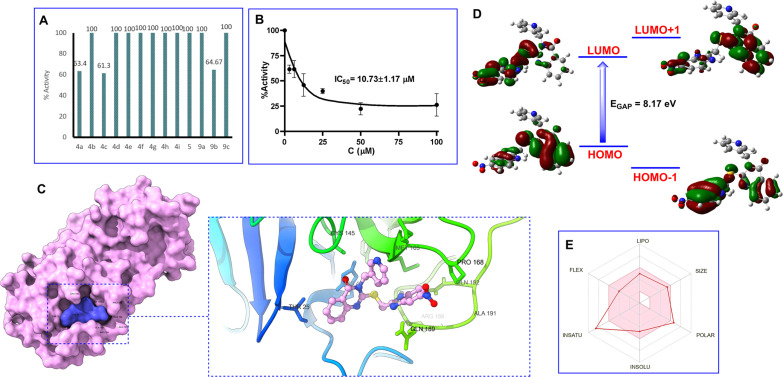
The synthesized compounds 4a–i, 5 and 9a–c were initially screened at a concentration of 50 μM in an assay buffer with a 3CL concentration of 50 nM. The results revealed that three compounds, namely 4a, 4c, and 9b, demonstrated inhibitory activity against 3CL protease exceeding 50% (Fig. 3A). Further investigation of the IC50 values of 4a, 4c, and 9b was conducted. However, due to the solubility of the tested compounds in the assay buffer, the exploration of the IC50 value was initially conducted for compound 9b at various concentrations of 100, 50, 25, 12.5, 6.25 and 3.125 μM in 3CL (50 nM). The results shown in Fig. 3 indicated that the IC50 of compound 9b for 3CL in the presence of 9% EtOH was found as 10.73 ± 1.17 μM (Fig. 3B). A negative control in the presence of 9% EtOH and without 9b showed no inhibition of 3CL activity. Studies have been continuously conducted in order to determine the IC50 values of compounds 4a and 4c.
Fig. 3. Percent activity on SARS-CoV-2 3CLpro at 50 μM concentration of quinazolinone compounds (A). Dose–response curve analysis of 9b for inhibitory activity against SARS-CoV-2 3CLpro (B). Three-dimensional protein–ligand interactions and optimal docking pose between 9b and the SARS-CoV-2 3CL protease active site. (C) HOMO, LUMO, and charge density difference (D). The bioavailability radar criteria based on lipophilicity (LIPO), molecular weight (SIZE), polarity (POLAR), solubility (INSOLU), saturation (INSATU), and flexibility (FLEX) using SwissADME tools (E).
2.4. Theoretical studies
The docking simulations underscored the robust binding affinity of 9b for the protease (−8.1 kcal mol−1) (Fig. 3C). The quinazolinone ring was found to form hydrogen bonds with His41 (4.93 Å), Met49 (5.37 Å), and Cys145 (5.55 Å) while the benzimidazole ring exhibited significant interactions with Met165 (4.59 Å) and Arg188 (6.31 Å). Additionally, the pi-donor H-bond interactions between 9b and two amino acids, His164 (6.61 Å) and Glu166 (5.99 Å) were also observed.
Examination of the HOMO and the LUMO of 9b inhibitor revealed the energy of the HOMO and LUMO as −9.59 eV and −1.42 eV, respectively, corresponding to the energy gap (Egap) value of 8.17 eV (Fig. 3D). Studies on the drug-likeness and bioavailability indicated that compound 9b had a topological polar surface area (TPSA) value less than 150 Å and a number of rotatable bonds (RB) of 6 (see ESI†). Moreover, compound 9b possessed hydrogen bond acceptor (HBA) and donor (HBD) features, with the number being less than 10 and 5, respectively. The compound exhibited a lack of blood–brain barrier (BBB) penetration and poor gastrointestinal (GI) absorption, as depicted by the A boiled-egg model (see ESI†). In addition, compound 9b adhered to the three rules (Lipinski, Ghose, and Muegge) and violated two rules (Veber and Egan) as they failed to meet the threshold for the TPSA value.
3. Discussion
Febrifugine has been used for a long time as medicine to treat the symptoms of fevers caused by malaria parasites. Febrifugine also takes part in changing the nitric oxide production and tumor necrosis factor-α in mouse macrophages.34 It has been reported that structural modification of febrifugine leads to decreased toxicity while maintaining its antiparasitic activity.35,36 Notably, an in silico analysis indicated that febrifugine displayed remarkably energetic and interactive properties with the COVID-19 ACE2 receptor.32 Based on these findings, nine febrifugine analogues 4a–i targeting the SARS-CoV-2 3CLpro were rationally designed by substitution of the 3-hydroxypiperidinylmethyl ketone moiety of febrifugine with various N-substituted arenyl/thiazolyl amide or N-benzylamide fragments (Scheme 1, Fig. 2). On the other hand, a number of studies have shown that synthetic quinazolinone-containing derivatives possess a wide spectrum of biological properties,11,12 especially anticancer13 and antiviral activities.14 Liu et al. reported the design and synthesis of a series of quinazolinone-containing myricetin derivatives, which showed potential antiviral activity better than ningnanmycin.14 Furthermore, in our earlier research, by combining a peptidomimetic backbone derived from glutamine or glutamate and a quinazolinone core structure, two compounds of structure I were synthesized and found as potent SARS-CoV-2 3CLpro inhibitors, with the IC50 values of 22.47 μM and 24.04 μM (Fig. 1B).30 On the other hand, we have also previously discovered that structure II containing the benzimidazole moiety exhibited promising anti-Zika virus activity with the EC50 value of 1.9 μM.21 Structure–activity relationship analysis indicated that substituents at the C-2, N-1, and C-5 positions of the benzimidazole ring were important for antiviral activity. Therefore, we hypothesized that a combination of the inherent antiviral skeletons such as quinazolinone and benzimidazole would produce new hybrid structures with enhanced antiviral activity. Accordingly, three quinazolinone-conjugated benzimidazoles 9a–c have been designed and successfully synthesized starting from isatoic anhydride (Scheme 2, Fig. 2). The synthetic procedure can be conveniently applied to synthesize library of quinazolinone/benzimidazole hybrids bearing various substituents at the N-3 position in the quinazolinone and the N-1 and C-5 positions in the benzimidazole moieties by choosing different substituted benzylamines 3 as well as different N-substituted o-phenylenediamine 6.31
The cytotoxic assays indicated that only compound 4h showed meaningful cytotoxic activity on the MCF-7 and MDA-MB-231 cells with IC50 = 58.16 and 34.39 μM, respectively. The cytotoxic activity of compound 4h might be due to its modification to bridge the 4-quinazolinone core and benzene or heterocyclic rings with an additional carbon bond. For the non-cancer HEK293 cell, all quinazolinone compounds did not have significant toxicity except for compound 9a. In the cytotoxic study of quinazolinone derivatives on HCC, two compounds 4h and 4i, indicated weak cytotoxicities on HepG2 cells and HepG2.2.15 cells at the IC50: 4h (51.36 μM and 51.98 μM), 4i (47.67 μM and 79.47 μM). Interestingly, 4e and 5 were excluded from each other, which had IC50 = 54.89 μM in HepG2 and IC50 = 46.35 μM in HepG2.2.15, however, had slight or no toxicity to another cell line.
Screening results for SARS-CoV-2 3CL enzyme inhibitory activity of compounds 4a–i, 5 and 9a–c at the concentration of 50 μM showed that three compounds (4a, 4c, and 9b) exhibited more than 50% inhibitory activity against 3CL protease (Fig. 3A). Compounds 4a and 4c share the common structural features of an electron-withdrawing NO2 group at the C-2 and an electron-releasing OH or OCH3 groups at the C-4 positions in the benzene ring. The presence of the NO2 group seemed to play a critical role in inducing the activity of the compound. Reduction of the NO2 group to the corresponding NH2 group led to a complete loss of the activity (compared compound 4a, 4c with compound 4g). Similarly, the activity of compound 9b was likely attributed to the presence of the NO2 group in the benzene ring of the benzimidazole moiety. Notably, the IC50 value of 9b for 3CLpro was found as 10.73 μM (Fig. 3B), which showed around 2-fold higher activity than compounds I (IC50 = 22.47 and 24.04 μM) (Fig. 1B).30 These results indicated the potential of the benzimidazole-conjugated quinazolinone scaffold as a SARS-CoV-2 3CLpro inhibitor.
To elucidate the structure–function relationship of 3CLpro inhibition with compound 9b, docking simulations of 9b were performed focusing on its tight binding with crucial amino acids at the active site of the 3C-like proteinase.30 Accordingly, the compound was found to form hydrogen bonds with His41 (4.93 Å), Met49 (5.37 Å), and Cys145 (5.55 Å) while the benzimidazole ring exhibited significant interactions with Met165 (4.59 Å) and Arg188 (6.31 Å). Additionally, the pi-donor H-bond interactions between 9b and two amino acids, His164 (6.61 Å) and Glu166 (5.99 Å) were also observed. These results provided valuable insights into the molecular mechanism of the 3CLpro inhibition activity of compound 9b.
The properties of HOMOs and LUMOs provide significant information for chemists. It has been known that a compound with a large HOMO–LUMO gap tends to be more stable against oxidation or reduction processes, serving as a useful guideline in drug design and development. Examination of the HOMO and the LUMO of the 9b inhibitor revealed the Egap value of 8.17 eV (Fig. 3D). This suggested that the inhibitor had considerable stability and resistance to oxidation or reduction processes, a desirable trait for pharmaceutical agents.
Studies on the drug-likeness of compound 9b showed that the molecule exhibited the potential to establish flexible interactions with the target protease based on a TPSA and RB. Moreover, the molecular weight below 500 g mol−1 suggests the potential to penetrate the cell membrane. Besides, the preliminary pharmacokinetic and ADME analyses provide valuable insights into the potential of the 9b compound as a drug candidate (Fig. 3E), offering a comprehensive understanding of pharmacological properties and safety profiles, and laying a solid foundation for further optimization and development in the future.
4. Conclusions
This paper describes the design and synthesis of quinazolinone derivatives as promising SARS-CoV-2 3CL inhibitors and advocates 9b as a potent candidate for further optimization studies. These discoveries bolster the new scaffold as a potential candidate that might contribute to the development of structurally interesting anti-coronavirus agents.
5. Experimental section
5.1. Chemistry
Reagents sourced commercially were used without additional purification. Reactions were monitored via thin-layer chromatography (TLC) on silica gel plates and visualized under UV light. The synthesized compounds were purified by flash column chromatography and characterized using NMR, FT-IR and HR-MS spectra. Detailed synthetic procedures for febrifugine analogues 4a–i and quinazolinone-conjugated benzimidazoles 9a–c are available in the ESI† file.
5.1.1. N-(4-Hydroxy-2-nitrophenyl)-2-(4-oxoquinazolin-3(4H)-yl)acetamide (4a)
Yellow solid. Rf = 0.30 (Hex : EtOAc = 1 : 2). Mp 297–299 °C. FT-IR (KBr) νmax (cm−1): 3285, 1655, 1683, 1615, 1557, 1540, 1521, 1360, 1292, 1280, 1234, 1217, 1168, 969, 777. HR-ESI-MS found m/z 339.0731 [M − H]− (calcd 339.0729, C16H12N4O5). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 10.37 (s, 1H), 8.32 (s, 1H), 8.15 (dd, J1 = 7.5, J2 = 1.0, 1H), 7.85 (m, 1H), 7.71 (d, J = 8.0, 1H), 7.57 (t, J = 7.7, 1H), 7.43 (d, J = 8.5, 1H), 7.30 (d, J = 3.0, 1H), 7.12 (dd, J1 = 9.0, J2 = 3.0, 1H), 4.87 (s, 2H). 13C-NMR (125 MHz, DMSO-d6, δ ppm): 165.9, 160.1, 155.0, 148.3, 148.0, 134.5, 127.2, 127.1, 126, 121.8, 121.4, 121.1, 110.6, 48.3.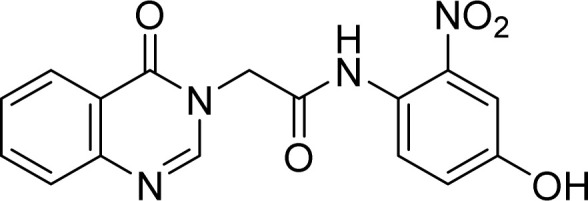
5.1.2. N-(5-Chloro-2-hydroxyphenyl)-2-(4-oxoquinazolin-3(4H)-yl)acetamide (4b)
White solid. Rf = 0.23 (Hex : EtOAc = 1 : 1). Mp 284–286 °C. FT-IR (KBr) νmax (cm−1): 3247, 3222, 3049, 2957, 2923, 2854, 1694, 1673, 1614, 1544, 1477, 1408, 1373, 1293, 1171, 1118, 969, 917, 892, 813, 774. HR-ESI-MS found m/z 330.0643 [M + H]+ (calcd 330.0645, C16H12ClN3O3). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 10.28 (s, 1H), 9.86 (s, 1H), 8.35 (s, 1H), 8.15 (dd, J1 = 8.0, J2 = 1.5, 1H), 7.98 (d, J = 2.5 Hz, 1H), 7.89–7.84 (m, 1H), 7.72 (d, J = 7.5, 1H), 7.59–7.55 (m, 1H), 6.97 (dd, J1 = 8.5, J2 = 2.5, 1H), 6.90 (d, J = 8.5, 1H), 4.97 (s, 2H). 13C-NMR (125 MHz, DMSO-d6, δ ppm): 166.0, 160.22, 148.5, 148.0, 146.2, 134.4, 127.2, 127.0, 125.9, 123.7, 122.0, 121.4, 120.8, 116.2, 48.7.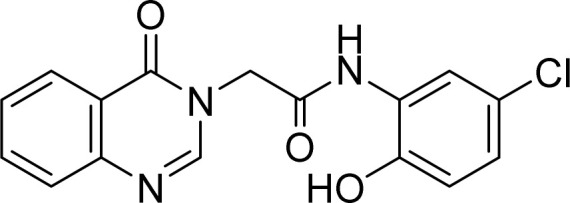
5.1.3. N-(4-methoxy-2-nitrophenyl)-2-(4-oxoquinazolin-3(4H)-yl)acetamide (4c)
Yellow solid. Rf = 0.31 (Hex : EtOAc = 1 : 1). Mp 215–217 °C. FT-IR (KBr) νmax (cm−1): 3313, 3001, 2944, 1678, 1613, 1588, 1466, 1365, 1307, 1280, 1253, 777. HR-ESI-MS found m/z 353.0611 [M − H]− (calcd 353.0886, C17H15N4O5). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 10.43 (s, 1H), 8.33 (s, 1H), 8.16 (dd, J1 = 1.5, J2 = 9.5, 1H), 7.87–7.84 (m, 1H), 7.71 (d, J = 8.5, 1H), 7.58–7.55 (m, 2H), 7.49 (d, J = 3.0, 1H), 7.31 (dd, J1 = 9.0, J2 = 3.0, 1H), 4.89 (s, 2H), 3.84 (s, 3H). 13C-NMR (125 MHz, DMSO-d6, δ ppm): 166.0, 160.1, 156.4, 148.3, 147.9, 143.4, 134.5, 127.5, 127.2, 127.1, 126.0, 123.2, 121.4, 120.2, 109.1, 56.0, 48.3.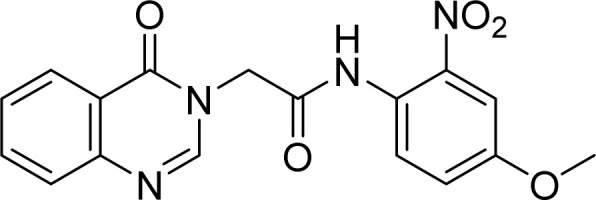
5.1.4. N-(2-Hydroxy-4-methylphenyl)-2-(4-oxoquinazolin-3(4H)-yl)acetamide (4d)
White solid. Rf = 0.36 (Hex : EtOAc = 1 : 2). Mp 242–244 °C. FT-IR (KBr) νmax (cm−1): 3250, 3228, 3054, 1698, 1687, 1670, 1614, 1549, 1519, 1478, 1375, 1322, 1294, 1204, 1172, 1120, 968, 774. HR-ESI-MS found m/z 310.1194 [M − H]− (calcd 310.1192, C17H15N3O3). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 9.64 (s, 1H), 9.57 (s, 1H), 8.35 (s, 1H), 8.15 (dd, J1 = 7.5, J2 = 1.0, 1H), 7.87–7.84 (m, 1H), 7.71 (d, J = 8.0, 1H), 7.64 (s, 1H), 7.58–7.55 (m, 1H), 6.73–6.78 (m, 2H), 4.94 (s, 2H), 3.31 (s, 3H). 13C-NMR (125 MHz, DMSO-d6, δ ppm): 165.5, 160.2, 148.6, 148.1, 145.3, 134.4, 127.4, 127.2, 127.0, 126.0, 125.5, 124.9, 122.5, 121.4, 115.1, 48.7, 20.3.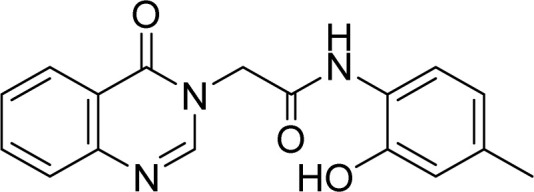
5.1.5. N-(2-Hydroxyphenyl)-2-(4-oxoquinazolin-3(4H)-yl)acetamide (4e)
White solid. Rf = 0.15 (Hex : EtOAc = 1 : 1). FT-IR (KBr) νmax (cm−1): 3264, 3061, 1697, 1677, 1617, 1541, 1476, 1455, 1374, 1300, 1258, 1222, 965, 756. ESI-MS found m/z 295.9 [M + H]+ (calcd 296.1, C16H14N4O2) and m/z 293.8 [M + H]+ (calcd 294.1, C16H13N3O3). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 9.83 (s, 1H), 9.69 (s, 1H), 8.36 (s, 1H), 8.16 (d, J = 8.0, 1H), 7.87–7.80 (m, 2H), 7.71 (d, J = 8.0, 1H), 7.58–7.55 (m, 1H), 6.95–6.92 (m, 1H), 6.90–6.88 (m, 1H), 6.76–6.73 (m, 1H), 4.96 (s, 2H). 13C-NMR (125 MHz, DMSO-d6, δ ppm): 165.6, 160.2, 148.6, 148.1, 147.6, 134.4, 127.2, 127.0, 126.0, 125.8, 124.6, 121.9, 121.4, 118.9, 115.3, 48.7.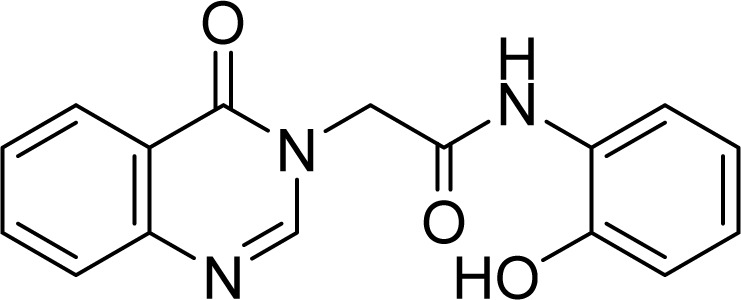
5.1.6. N-(3-Hydroxypyridin-2-yl)-2-(4-oxoquinazolin-3(4H)-yl)acetamide (4f)
White solid. Rf = 0.24 (Hex : EtOAc = 3 : 1). Mp 215–217 °C. HR-ESI-MS found m/z 297.0983 [M + H]+ (calcd 297.0988, C15H12N4O3). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 8.36 (s, 1H), 8.15 (dd, J1 = 8.0, J2 = 1.5, 1H), 7.90 (dd, J1 = 4.5, J2 = 1.5, 1H), 7.87–7.84 (m, 1H), 7.41 (d, J = 7.5, 1H), 7.58–7.55 (m, 1H),7.28 (dd, J1 = 7.5, J2 = 1.5, 1H), 7.14 (dd, J1 = 8.0, J2 = 4.5, 1H), 5.03 (s, 2H). 13C-NMR (125 MHz, DMSO-d6, δ ppm): 167.1, 160.2, 148.5, 148.0, 145.1, 145.0, 139.8, 138.4, 134.6, 127.2, 127.1, 126.0, 124.7, 122.2, 121.4, 48.5.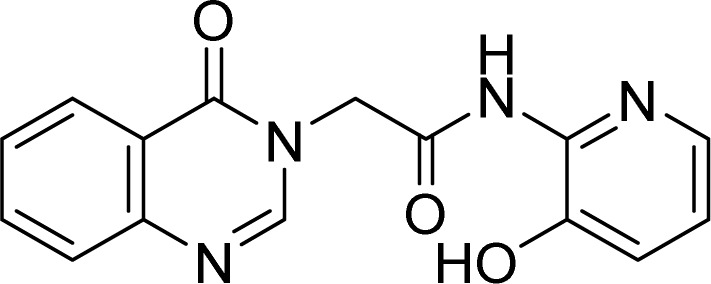
5.1.7. N-(2-Amino-4-methoxyphenyl)-2-(4-oxoquinazolin-3(4H)-yl)acetamide (4g)
White solid. Rf = 0.42 (Hex : EtOAc = 1 : 4). Mp 219–221 °C. FT-IR (KBr) νmax (cm−1): 3455, 3359, 3254, 2923, 2853, 1660, 1611, 1542, 1514, 1468, 1378, 1325, 1294, 1262, 1212, 1173, 1105, 1033, 777, 700. HR-ESI-MS found m/z 323.1141 [M–H]− (calcd 323.1144, C17H16N4O3). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 9.43 (s, 1H), 8.35 (s, 1H), 8.22–8.16 (m, 1H), 7.87–7.84 (m, 1H), 7.71 (d, J = 8.0, 1H). 7.57 (t, J = 7.5, 1H), 6.96 (d, J = 8.5, 1H), 6.29 (d, J = 2.5, 1H), 6.12 (dd, J1 = 11.0, J2 = 2.5, 1H), 4.90 (s, 2H), 4.84 (s, 2H), 3.66 (s, 3H). 13C-NMR (125 MHz, DMSO-d6, δ ppm): 165.7, 160.4, 158.2, 148.6, 148.1, 144.1, 134.4, 127.2, 127.1, 126.0, 121.5, 115.5, 101.7, 100.3, 54.8, 48.6.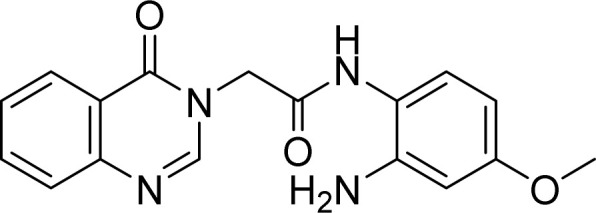
5.1.8. 2-(4-Oxoquinazolin-3(4H)-yl)-N-(pyridin-3-ylmethyl)acetamide (4h)
White solid. Rf = 0.13 (EtOAc : MeOH = 4 : 1). Mp 215–217 °C. FT-IR (KBr) νmax (cm−1): 3279, 3070, 2991, 2956, 1685, 1658, 1608, 1559, 1470, 1367, 1259, 783, 755. ESI-MS found m/z 294.9 [M + H]+ (calcd 295.1, C16H14N4O2) and m/z 292.8 [M + H]+ (calcd 293.1, C16H14N4O2). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 8.88 (t, J = 6.0, 1H), 8.51 (d, J = 2.0, 1H), 8.47 (dd, J1 = 4.5, J2 = 1.5, 1H), 8.33 (s, 1H), 8.15 (dd, J1 = 8.0, J2 = 1.5, 1H), 7.86–7.83 (m, 1H), 7.71–7.68 (m, 2H), 7.58–7.54 (m, 1H), 7.36 (dd, J1 = 8.0, J2 = 5.0, 1H), 4.72 (s, 2H), 4.36 (d, J = 6.0, 2H). 13C-NMR (125 MHz, DMSO-d6, δ ppm): 166.9, 160.3, 148.7, 148.5, 148.1, 148.1, 135.0, 134.5, 134.4, 127.2, 127.0, 126.0, 123.4, 121.5, 48.2.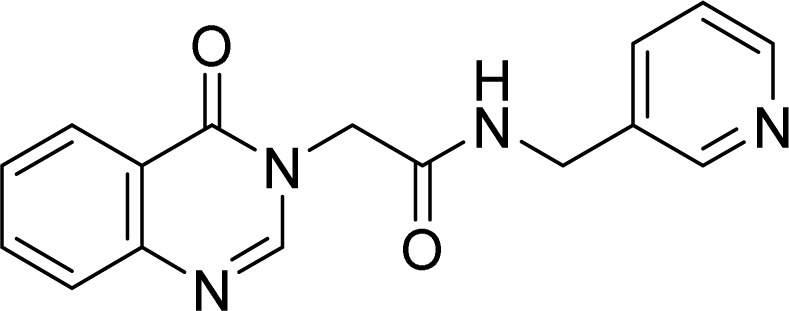
5.1.9. 2-(4-Oxoquinazolin-3(4H)-yl)-N-(thiazol-2-yl)acetamide (4i)
White solid. Rf = 0.33 (Hex : EtOAc = 1 : 1). Mp 215–217 °C. HR-ESI-MS found m/z 287.0599 [M + H]+ (calcd 287.0603, C13H10N4O2S). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 12.61 (s, 1H), 8.37 (s, 1H), 8.15 (dd, J1 = 8.0, J2 = 1.0, 1H), 7.89–7.86 (m, 1H), 7.73 (d, J = 7.5, 1H), 7.60–7.56 (m, 1H), 7.50 (d, J = 3.5 Hz, 1H), 7.25 (d, J = 3.5 Hz, 1H), 4.98 (s, 2H). 13C-NMR (125 MHz, DMSO-d6, δ ppm): 165.7, 160.2, 157.5, 148.3, 148.0, 137.8, 134.6, 127.2, 125.9, 121.3, 113.9, 48.2.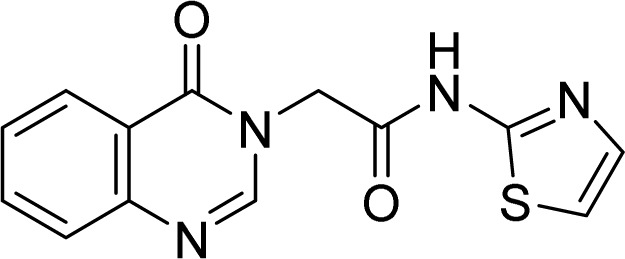
5.1.10. 2-Mercapto-3-(pyridin-3-ylmethyl)quinazolin-4(3H)-one (5)
White solid. 1H-NMR (500 MHz, DMSO-d6, δ ppm): 8.62 (s, 1H), 8.45 (d, J = 4.0, 1H), 7.97 (dd, J1 = 8.0, J2 = 1.0, 1H), 7.78–7.75 (m, 1H), 7.43 (d, J = 8.0, 1H), 7.35 (td, J1 = 7.5, J2 = 1.0, 1H), 7.32 (dd, J1 = 8.0, J2 = 5.0, 1H), 5.68 (s, 2H)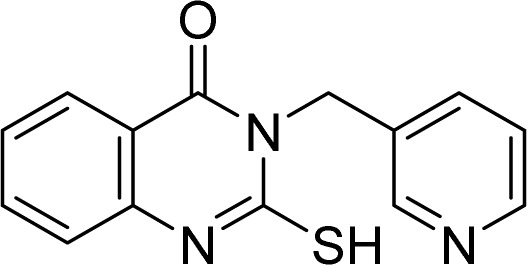
5.1.11. 3-(((1H-Benzo[d]imidazole-2-yl)methyl)thio)-2-(pyridin-3-ylmethyl)isoquinolin-1(2H)-one (9a)
White solid. FT-IR (KBr) νmax (cm−1): 3633, 3199, 1684, 1553, 1431, 1159, 745. HR-ESI-MS found m/z 400.1232 [M + H]+ (calcd 400.1232, C22H17N5OS). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 8.61 (d, J = 2.0, 1H), 8.49 (dd, J1 = 4.5, J2 = 1.5, 1H), 8.12 (dd, J1 = 8.0, J2 = 1.0, 1H), 7.85–7.82 (m, 1H), 7.72 (dt, J1 = 8.0, J2 = 2.0, 1H), 7.66 (d, J = 8.0, 1H), 7.52–7.48 (m, 3H), 7.35 (ddd, J1 = 8.0, J2 = 5.0, J3 = 1.0, 1H), 7.18–7.15 (m, 1H), 5.28 (s, 2H), 4.82 (s, 2H). 13C-NMR (125 MHz, DMSO-d6, δ ppm): 160.9, 155.6, 149.6, 148.7, 148.6, 146.7, 135.0, 134.8, 131.4, 126.5, 126.3, 123.6, 121.9, 118.8, 44.9, 29.5.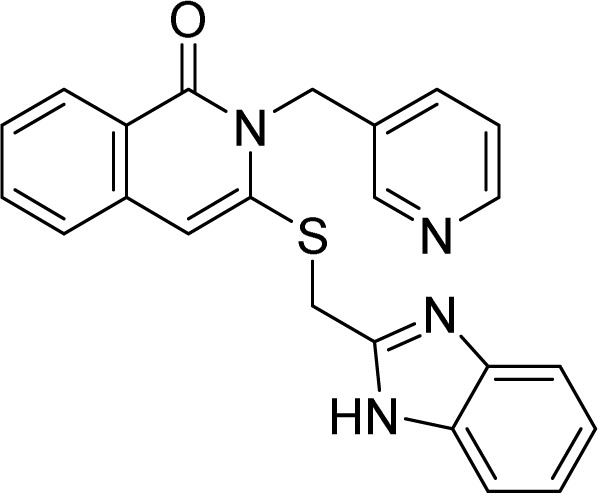
5.1.12. 3-(((5-Nitro-1H-benzo[d]imidazole-2-yl)methyl)thio)-2-(pyridin-3-ylmethyl)isoquinolin-1(2H)-one (9b)
White solid. FT-IR (KBr) νmax (cm−1): 3438, 2994, 1695, 1686, 1548, 1472, 1336, 1167, 670. HR-ESI-MS found m/z 445.1081 [M + H]+ (calcd 445.1083, C22H16N6O3S). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 13.08 (s, 1H), 8.62 (d, J = 1.5, 1H), 8.49 (dd, J1 = 4.5, J2 = 1.5, 1H), 8.42 (d, J = 1.5, 1H), 8.11 (dd, J2 = 8.0, J2 = 1.5, 1H), 8.08 (dd, J1 = 9.0, J2 = 2.0, 1H), 7.84–7.81 (m, 1H), 7.72 (dt, J1 = 8.0, J2 = 2.0, 1H), 7.67 (d, J = 9.0, 1H), 7.63 (d, J = 8.0, 1H), 7.49 (td, J1 = 7.5, J2 = 1.0, 1H), 7.36 (ddd, J1 = 8.0, J2 = 5.0, J3 = 1.0, 1H), 5.39 (s, 2H), 5.87 (s, 2H). 13C-NMR (125 MHz, DMSO-d6, δ ppm): 160.9, 155.4, 148.8, 148.6, 146.6, 142.5, 134.9, 134.7, 131.3, 126.5, 126.3, 126.1, 123.6, 118.8, 44.9, 29.4.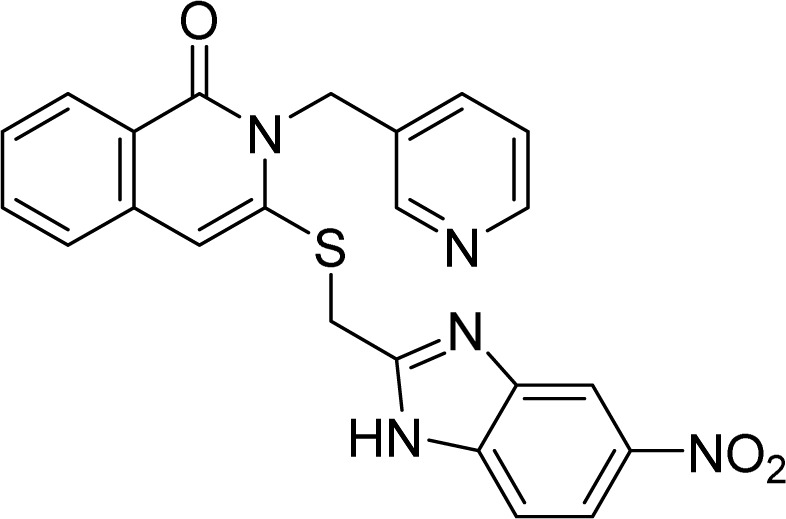
5.1.13. 3-(((1-(2-Hydroxyethyl)-5-nitro-1H-benzo[d]imidazole-2-yl)methyl)thio)-2-(pyridin-3-ylmethyl)isoquinolin-1(2H)-one (9c)
White solid. FT-IR (KBr) νmax (cm−1): 3449, 2947, 1675, 1551, 1414, 721. HR-ESI-MS found m/z 444.1493 [M + H]+ (calcd 444.1494, C24H21N5O2S). 1H-NMR (500 MHz, DMSO-d6, δ ppm): 8.61 (d, J = 2.0, 1H), 8.49 (dd, J1 = 4.5, J2 = 1.5, 1H), 8.11 (dd, J1 = 8.0, J2 = 1.5, 1H), 7.85–7.82 (m, 1H), 7.73 (dt, J1 = 8.0, J2 = 2.0, 1H), 7.63 (d, J = 8.0, 1H), 7.57 (d, J = 9.0, 1H), 7.49 (td, J1 = 8.0, J2 = 1.0, 1H), 7.35 (ddd, J1 = 7.5, J2 = 4.5, J3 = 0.5, 1H), 7.21 (td, J1 = 7.5, J2 = 1.0, 1H), 7.16 (td, J1 = 7.5, J2 = 1.0, 1H), 5.38 (s, 2H), 4.92 (s, 2H), 4.47 (t, J = 5.5, 2H), 3.75 (t, J = 5.5, 2H). 13C-NMR (125 MHz, DMSO-d6, δ ppm): 170.4, 160.9, 155.8, 150.2, 148.7, 148.6, 146.7, 142.1, 135.2, 135.0, 134.7, 131.4, 126.6, 126.3, 126.0, 123.6, 122.1, 121.6, 118.7, 118.6, 110.5, 59.7, 44.9, 28.5, 25.3.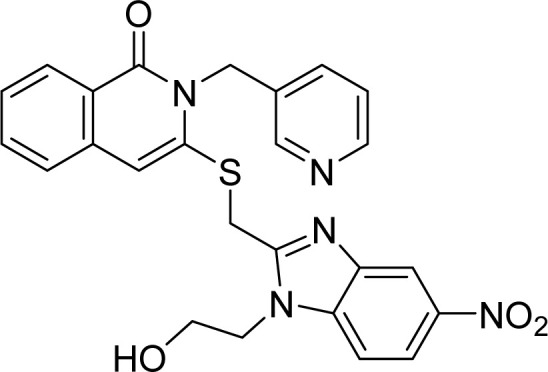
5.2. Cytotoxicity assays
The cytotoxicity of the synthetic quinazolinone derivatives was conducted at two human breast cancer cell lines (MCF-7 and MDA-MB-231), and two human hepatocellular carcinoma cell lines, (HepG2 and HepG2.2.15), along with the control non-cancerous HEK293 cell line. All cell lines were purchased from the Bioresource Collection and Research Center (BCRC) of the Food Industry Research and Development Institute (Hsinchu, Taiwan). MCF-7 cells were cultured at RPMI-1640 supplemented with 10% FBS; MDA-MB-231 cells were cultured at DMEM/F12 (1 : 1) supplemented with 10% FBS. HepG2, HepG2.2.15 and HEK 293 cells were cultured at DMEM supplemented with 10% FBS. All cell lines were maintained at 37 °C, 5% CO2 incubator with humidity, and were routinely sub-cultured every two days to ensure the experiment quality. 24 hours before the experiment, the cells were detached from the culture dishes using 0.05% trypsin to become the cell suspension and a small aliquot (100 μL) from the suspended cells was stained by 0.4% trypan blue (ratio = 1 : 1) to count the cell numbers and exclude the dead cells. 2.5 × 103 cells from each cell line were seeded at each well of the 96-well plates and the plates were stood in the incubator overnight. On the experiment day, the full culture medium was removed from the wells and replaced by serial-diluted quinazolinone compounds (at the concentrations of 100, 50, 25, 12.5, 6.25, and 0 μM) in the reduced culture medium containing 1% FBS quadruplicately. The treated cells were cultured for another 48 hours and then stained with 0.02% crystal violet solution followed by washing out the detached dead cells and extra dye with water. The adherent dyed cells were recognized as live cells, and they were re-dissolved in 1% SDS to measure the absorbance at the wavelength of 595 nm. For estimation of the half-maximal inhibitory concentration (IC50), a straight line (linear regression) of drug concentrations (x-axis) against cell inhibition rates (y-axis) was plotted and the data were fitted with the equation.
5.3. Molecular cloning and protein expression of Mpro
The SARS-CoV-2 Mpro gene was obtained from Addgene and cloned into the pGEX-5X vector, which has a glutathione S-transferase (GST) tag attached to the N-terminus of the recombinant Mpro protein as previously described.30 After the recombinant plasmid was transformed into E. coli BL21 (DE3) competent cells, isopropyl thio-β-d-thiogalactopyranoside (IPTG) (0.5 mM) was added to induce recombinant Mpro protein expression for 10 hours at 16 °C and 180 rpm. The expressed proteins were purified using Glutathione Sepharose 4 Fast Flow resin, followed by cleavage with Factor Xa and analysis of protein purity by SDS-PAGE. The protein appears as a homogeneous band on SDS-PAGE. Protein concentration was determined by Bradford assay, aliquoted into vials, and stored at −80 °C until use. To confirm enzyme activity and stability, GC376 (a preclinical cysteine protease inhibitor that binds to Mpro of feline coronavirus (FCoV)) was used as a positive control in each experiment to verify the activity of the enzyme in that assay.37,38 Purified proteins are stable at −80 °C for at least six months.
5.4. In vitro inhibitory activity assay of 3CL protease
Protease activity assays were performed in a 384-well black flat-bottomed microtiter plate (Thermo Scientific™ Nunc™). In a final volume of 25 μL, the recombinant SARS-CoV-2 3CL protease was added at a final concentration of 50 nM and mixed with different concentrations of compounds in assay buffer (final concentration: 20 mM Tris pH 7.3, 100 mM NaCl, 1% DTT, 1% EDTA, 1% DMSO) and pre-incubated at 37 °C for 30 min. The FRET substrate, DABCYL-KTSAVLQSGFRKME-EDANS, was then added to a final concentration of 50 μM. The completed reaction was incubated at 37 °C for 1 h. First, the activity of the purified recombinant protein was tested against the positive control substrate GC-376 and an IC50 similar to that reported in the literature to confirm that the activity of the enzyme was close to that reported in the literature.37,38 This enzyme activity was assumed to be 100% for subsequent calculation of inhibitory activity. Blank wells contained the same compound concentration as substrate but without 3CL protease. Inhibition rates were then calculated by comparison with control wells without added inhibitors. The fluorescence signal (excitation/emission, 355 nm/460 nm) of the released EDANS was measured using a fluorometer (Fluoroskan Ascent FL). IC50 values were determined by nonlinear regression (GraphPad Prism 8.0.1).
5.5. Theoretical studies
The 3D structure of the target 3CL main protease was retrieved from the Protein Data Bank (ID PDB 6W63, https://www.rcsb.org/structure/6W63). The protein structure was prepared by removing water molecules, heteroatoms, and any co-crystallized ligands. Ligand structures were prepared by optimizing their geometry by DFT (6-31G). The study was conducted following the descriptions of our previous molecular docking studies.39–42 Specifically, for a 3D grid encompassing the active site of the protein, the grid dimensions were defined based on the size and shape of the active site. The prepared ligand structures were docked into the active site of the protein using Autodock Vina. In order to gain deeper insights into the pharmacokinetic properties and drug likeliness, the analysis was conducted by utilizing the online tool SwissADME.43
Data availability
The data supporting this article have been included as part of the ESI.†
Author contributions
B. T. B. H.: conceptualization, visualization, supervision, writing – original draft, writing – review & editing. D. L. D. T., H. N. H. G., V. T. H., and L. H. P. M. performed the chemical synthesis. H. N. H. G., F.-P. C., H.-C. L., and T.-K. W. performed the bioassay. N. C. Q. and T. Q. D. analyzed molecular docking and theoretical studies. N. C. Q. designed, drew and presented images. All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
We thank Can Tho University and National Yang Ming Chiao Tung University for providing laboratory facilities and equipment support.
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra03267e
References
- Zumla A. Chan J. F. Azhar E. I. et al. . Nat. Rev. Drug Discovery. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis M. Vandeweerd V. Verbeke R. et al. . Transdisciplinary Insights. 2020;4:1–173. [Google Scholar]
- Woo P. C. Lau S. K. Huang Y. et al. . Exp. Biol. Med. 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- Maier H. J. Bickerton E. Britton P. Methods Mol. Biol. 2015:1–23. doi: 10.1007/978-1-4939-2438-7. [DOI] [PubMed] [Google Scholar]
- Ge X. Y. Li J. L. Yang X. L. et al. . Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R. Zhao X. Li J. et al. . Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannalire R. Cerchia C. Beccari A. R. et al. . J. Med. Chem. 2022;65:2716–2746. doi: 10.1021/acs.jmedchem.0c01140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q. Xiong Y. Zhu G. H. et al. . MedComm. 2022;3:e151. doi: 10.1002/mco2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary A. P. Shukla A. K. Pandey J. Kant P. Chem. Biol. Interact. 2018;8:62–83. [Google Scholar]
- Rajput R. Mishra A. P. Int. J. Pharm. Pharm. Sci. 2012;4:66–70. [Google Scholar]
- Sharma P. C. Kaur G. Pahwa R. et al. . Curr. Med. Chem. 2011;18:4786–4812. doi: 10.2174/092986711797535326. [DOI] [PubMed] [Google Scholar]
- Khan I. Ibrar A. Abbas N. Saeed A. Eur. J. Med. Chem. 2014;76:193. doi: 10.1016/j.ejmech.2014.02.005. [DOI] [PubMed] [Google Scholar]
- (a) Zhan X. Xu Y. Qi Q. et al. . Chem. Biodiversity. 2018;15:e1700513. doi: 10.1002/cbdv.201700513. [DOI] [PubMed] [Google Scholar]; (b) El-Bordany E. A. Ali R. S. J. Heterocycl. Chem. 2018;55:1223–1231. [Google Scholar]; (c) -Ze Li H. He H.-Y. Han Y.-Y. et al. . Molecules. 2010;15:9473. doi: 10.3390/molecules15129473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Liu T. Peng F. Cao X. et al. . ACS Omega. 2021;6:30826–30833. doi: 10.1021/acsomega.1c05256. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wang Z. Wang M. Yao X. et al. . Eur. J. Med. Chem. 2012;53:275. doi: 10.1016/j.ejmech.2012.04.010. [DOI] [PubMed] [Google Scholar]; (c) Krishnan S. K. Ganguly S. Veerasamy R. Jan B. Eur. Rev. Med. Pharmacol. Sci. 2011;15:673–681. [PubMed] [Google Scholar]
- Yoon Y. K. Ali M. A. Wei A. C. et al. . Eur. J. Med. Chem. 2015;93:614–624. doi: 10.1016/j.ejmech.2013.06.025. [DOI] [PubMed] [Google Scholar]
- Chandrika N. T. Shrestha S. K. Ngo H. X. Garneau-Tsodikova S. Bioorg. Med. Chem. 2016;24:3680–3686. doi: 10.1016/j.bmc.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iemura R. Kawashima T. Fukuda T. et al. . J. Med. Chem. 1986;29:1178–1183. doi: 10.1021/jm00157a010. [DOI] [PubMed] [Google Scholar]
- Mader M. Dios A. D. Shih C. et al. . Bioorg. Med. Chem. Lett. 2008;18:179–183. doi: 10.1016/j.bmcl.2007.10.106. [DOI] [PubMed] [Google Scholar]
- Rao A. Chimirri A. Clercq E. D. et al. . IL Farmaco. 2002;57:819–823. doi: 10.1016/s0014-827x(02)01300-9. [DOI] [PubMed] [Google Scholar]
- (a) Delang L. Froeyen M. Herdewijn P. Neyts J. Antiviral Res. 2012;93:30–38. doi: 10.1016/j.antiviral.2011.10.012. [DOI] [PubMed] [Google Scholar]; (b) E Diwani H. I. Abdel-Mohsen H. T. Salama I. et al. . Chem. Pharm. Bull. 2014;62:856–866. doi: 10.1248/cpb.c13-01009. [DOI] [PubMed] [Google Scholar]; (c) Ding K. Wang A. Boerneke M. A. et al. . Bioorg. Med. Chem. Lett. 2014;24:3113–3117. doi: 10.1016/j.bmcl.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hue B. T. B. Nguyen P. H. De T. Q. et al. . ChemMedChem. 2020;15:1–12. [Google Scholar]
- Kerru N. Singh P. Koorbanally N. et al. . Eur. J. Med. Chem. 2017;142:179–212. doi: 10.1016/j.ejmech.2017.07.033. [DOI] [PubMed] [Google Scholar]
- Hassanzadeh F. Sadeghi-Aliabadi H. Jafari E. et al. . Res. Pharm. Sci. 2019;14:408–413. doi: 10.4103/1735-5362.268201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanzadeh F. Sadeghi-Aliabadi H. Nikooei S. et al. . Res. Pharm. Sci. 2019;14:130–137. doi: 10.4103/1735-5362.253360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malekzadeh M. Dadkhah S. Khodarahmi G. A. et al. . Res. Pharm. Sci. 2022;17:22–34. doi: 10.4103/1735-5362.329923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadi P. Khodarahmi G. Jahanian-Najafabadi A. et al. . Chem. Biodiversity. 2017;14:e1600411. doi: 10.1002/cbdv.201600411. [DOI] [PubMed] [Google Scholar]
- (a) Singla P. Luxami V. Paul K. J. Photochem. Photobiol., B. 2017;168:156–164. doi: 10.1016/j.jphotobiol.2017.02.009. [DOI] [PubMed] [Google Scholar]; (b) Auti P. S. George G. Paul A. T. RSC Adv. 2020;10:41353. doi: 10.1039/d0ra06642g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taherian E. Khodarahmi G. Khajouei M. R. et al. . Results Pharma Sci. 2019;14:247–254. [Google Scholar]
- Bui H. T. B. Do K. M. Nguyen H. T. D. et al. . Tetrahedron. 2021;98:132426. [Google Scholar]
- Giang H. N. H. Chou F. P. Chen C. Y. et al. . Viruses. 2023;15:287. doi: 10.3390/v15020287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quy N. P. Hue B. T. B. Kiep M. D. et al. . Chem. Pharm. Bull. 2022;70:448–453. doi: 10.1248/cpb.c22-00162. [DOI] [PubMed] [Google Scholar]
- Benslama O. Mansouri N. Arhab R. Not. Sci. Biol. 2021;13:10835. [Google Scholar]
- Hosamani K. M. Reddy D. S. Devarajegowda H. C. Kurjogic M. M. RSC Adv. 2015;5:64566. [Google Scholar]
- Murata K. Takano F. Fushiya S. Oshima Y. J. Nat. Prod. 1998;61:729–733. doi: 10.1021/np970433o. [DOI] [PubMed] [Google Scholar]
- Kikuchi H. Tasaka H. Hirai S. et al. . J. Med. Chem. 2002;6:2563–2570. doi: 10.1021/jm010448q. [DOI] [PubMed] [Google Scholar]
- Takaya Y. Tasaka H. Chiba T. et al. . J. Med. Chem. 1999;42:3163–3166. doi: 10.1021/jm990131e. [DOI] [PubMed] [Google Scholar]
- Fu L. Ye F. Feng Y. Yu F. Wang Q. Wu Y. Zhao C. Sun H. Huang B. Niu P. et al. . Nat. Commun. 2020;11:4417. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong W. Khan M. B. Fischer C. Arutyunova E. Lamer T. Nat. Commun. 2020;11:4282. doi: 10.1038/s41467-020-18096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D Tran Q. Nguyen C. Q. Dang Q. L. et al. . ACS Omega. 2023;8:48994–49008. doi: 10.1021/acsomega.3c06612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Ngoc H. Nguyen C. Q. Vo K. A. T. et al. . Physiol. Mol. Plant Pathol. 2023;127:102123. [Google Scholar]
- Nguyen H. P. Tran Q. D. Nguyen C. Q. et al. . RSC Adv. 2022;12:22108–22118. doi: 10.1039/d2ra01969h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H. P. Hue B. T. B. Pham M. Q. et al. . New J. Chem. 2023;47:7622–7631. [Google Scholar]
- Daina A. Michielin O. Zoete V. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting this article have been included as part of the ESI.†