Abstract
Acute myeloid leukemia (AML) is a deadly hematopoietic malignancy. Although many patients achieve complete remission with standard induction therapy, a combination of cytarabine and anthracycline, ~40% of patients have induction failure. These refractory patients pose a treatment challenge, as they do not respond to salvage therapy or allogeneic stem cell transplant. Herein, we show that AML patients who experience induction failure have elevated expression of the NF-κB target gene tumor necrosis factor alpha-induced protein-3 (TNFAIP3/A20) and impaired necroptotic cell death. A20High AML are resistant to anthracyclines, while A20Low AML are sensitive. Loss of A20 in AML restores sensitivity to anthracycline treatment by inducing necroptosis. Moreover, A20 prevents necroptosis in AML by targeting the necroptosis effector RIPK1, and anthracycline-induced necroptosis is abrogated in A20High AML. These findings suggest that NF-κB-driven A20 overexpression plays a role in failed chemotherapy induction and highlights the potential of targeting an alternative cell death pathway in AML.
Subject terms: Acute myeloid leukaemia, Cancer therapeutic resistance
Patients with Acute myeloid leukemia (AML) who do not respond to standard chemotherapy have limited treatment options. Here, the authors show that high expression of the gene TNFAIP3/A20 in AML cells contributes to chemotherapy resistance by blocking a type of cell death called necroptosis.
Introduction
Acute myeloid leukemia (AML) is a type of blood cancer that affects people of all ages. If left untreated, AML is always fatal. The primary treatment for most AML subtypes involves either chemotherapy or targeted therapy (www.cancer.org). Fortunately, AML patients who sustain a complete remission (CR) with chemotherapy can have positive outcomes and can be cured of the disease1. On the other hand, patients who do not respond to initial treatment or develop refractory disease experience very poor outcomes. Young and fit older patients may undergo standard induction chemotherapy, which involves using the nucleoside analog, cytarabine (Ara-C), along with an anthracycline drug such as daunorubicin or idarubicin1. This treatment regimen, commonly known as “7+3,” together with consolidation or allogeneic stem cell transplantation is the standard of care for AML and, to date, is the only treatment that has the potential to be curative2. Despite the positive outcomes seen in patients who respond to induction chemotherapy, relapse and refractory disease is the most common causes of death. Prognostic classification, which combines cytogenetic and mutational status, can estimate overall and event-free survival but is not effective in predicting induction failure3. Various research groups have developed prediction models that consider clinical characteristics, laboratory variables, cytogenetics, and mutational status to better predict induction failure, although their success rates vary4. Despite decades of treatment with 7+3 chemotherapy and extensive research in this area, the reason why some patients initially respond while others are refractory to chemotherapy remains unresolved. The outlook is particularly dismal for AML patients who are initially refractory to induction chemotherapy, referred to as primary induction failure, as long-term survival is typically no higher than 10%5. Understanding the mechanisms behind treatment response in AML patients undergoing standard induction therapy will assist in making risk-adapted treatment decisions and identifying new potential therapeutic targets. Several mechanisms have been implicated in primary chemotherapy resistance in AML, including expression of detoxifying enzymes and drug efflux pumps, mutations in topoisomerase, increased expression of pro-survival genes, and metabolic adaptations6,7. For example, SAMHD1, a deoxynucleoside triphosphate triphosphohydrolase, has been shown to reduce the efficacy of cytarabine (Ara-C), a key chemotherapeutic agent used in AML treatment8. SAMHD1 hydrolyzes Ara-CTP, the active metabolite of Ara-C, leading to decreased drug efficacy. There is also ongoing research to improve the prediction of treatment failure in AML. Since it is widely viewed that chemotherapy failure is attributed to cell-intrinsic dysregulation of cell survival mechanisms, gene expression, and signaling patterns in leukemic cells at the time of diagnosis have been utilized to categorize patients according to their response to chemotherapy9–14. These lines of inquiry have identified that increased Nuclear factor-kappa B (NF-κB) signaling at diagnosis correlates with chemotherapy resistance15,16, however, it is not well understood mechanistically how NF-κB signaling contributes to poor outcomes.
NF-κB is a family of conserved transcription factors consisting of NF-κB1 (p50), NF-κB2 (p52), RelA (p65), RelB, and c-Rel homo or heterodimers that respond to signals by inducing the expression of numerous genes17,18. Under physiological conditions, NF-κB is in an inactive state in the cytoplasm through its interaction with NF-κB inhibitory proteins (IκBs). The NF-κB pathway can be activated through extracellular stimuli and/or intracellular signals that activate the IκB kinase (IKK) complex. Next, IKK promotes IκB phosphorylation and degradation, which results in nuclear translation of NF-κB. NF-κB has garnered significant attention as a therapeutic target in cancer as it regulates critical biological functions related to cancer cell survival, proliferation, and therapy resistance mechanisms19. Elevated activation NF-κB at diagnosis or in response to chemotherapy leads to dysregulated apoptotic responses, which is thought to mediate chemoresistance19. For example, NF-κB induces the expression of anti-apoptotic genes, such as BCL2 family members, under specific cellular conditions. Notably, NF-κB can regulate both pro- and anti-survival pathway, which is likely a reason for the limited success of NF-κB specific therapies in cancer15. NF-κB signaling has been implicated in chemotherapy resistance in AML15,16. In these studies, much attention has been focused on the expression of anti-apoptotic genes, such as BCL2, BCLXL, and MCL1, as drivers of NF-κB-mediated chemoresistance20,21. However, these studies have not confirmed that expression of BCL2-related genes are the mechanistic reason for how NF-κB signaling contributes to chemotherapy responses. In recent studies, cytarabine was shown to induce a senescence-like phenotype that enabled survival of AML cells22. Cytarabine treatment also resulted in expression of inflammatory genes, many of which are a result of NF-κB signaling. Notwithstanding, the precise mechanism by which NF-κB activation contributes to chemotherapy failure and resistance in AML remains unresolved. Given the pleiotropic role of NF-κB, we posited that increased NF-κB signaling, through specific target genes, results in diminished chemotherapy responses in AML.
In this study, we examine the mechanisms by which NF-κB activation contributes to the failure of induction chemotherapy in AML patients by identifying the critical NF-κB effector genes and putting forth a new therapeutic approach that targets alternative programmed cell death pathways. We show that AML patients who undergo induction failure exhibit increased expression of the NF-κB target gene TNFAIP3 (A20) and impaired necroptotic cell death, leading to a worse prognosis with chemotherapy. A20High AML samples are resistant to anthracyclines, while A20Low AML samples are sensitive. Loss of A20 in AML cells restores sensitivity to anthracycline treatment by inducing necroptosis. Moreover, our studies revealed that A20 plays a critical role in AML development as deletion or knockdown of A20 effectively suppresses leukemic cells by mediating spontaneous necroptosis. A20 prevents necroptosis in AML by targeting the necroptosis effector RIPK1, and anthracycline-induced necroptosis is abrogated in A20High AML cells. These findings reveal that A20 overexpression via NF-κB plays a role in failed chemotherapy induction and highlights the potential of targeting an alternative cell death pathway in AML.
Results
Select NF-κB target genes are associated with induction failure in AML
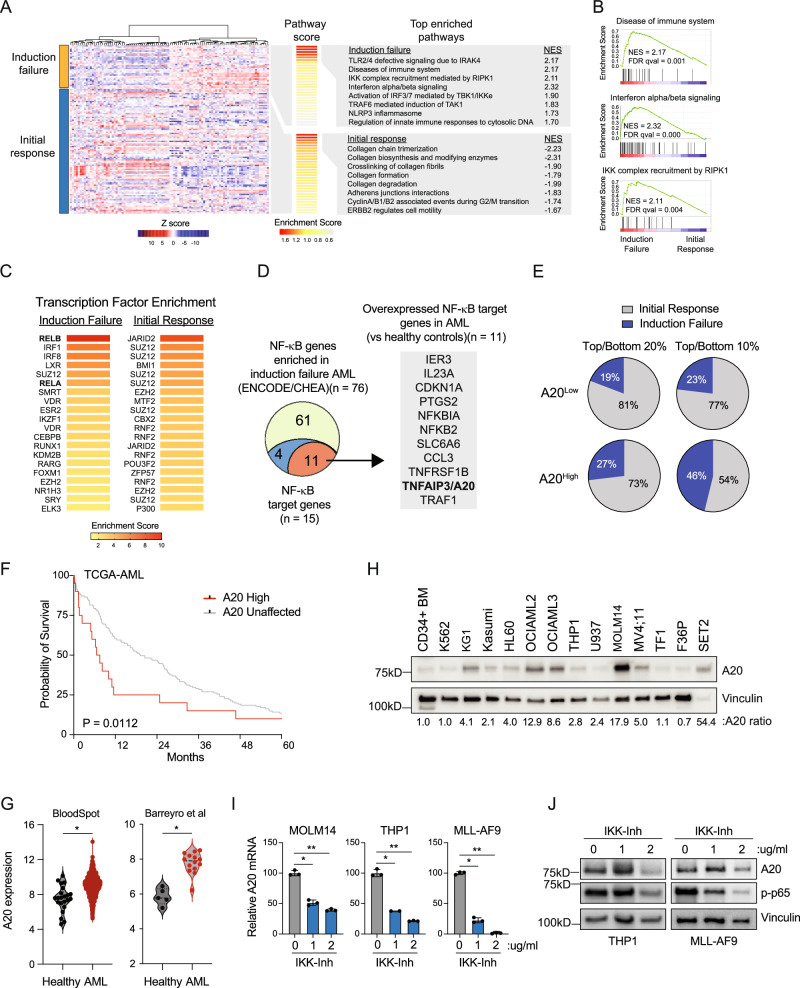
To identify gene expression and signaling networks in AML patients at diagnosis that correlate with induction chemotherapy responses, we performed an unsupervised hierarchical clustering analysis of RNA sequencing data from TCGA AML (www.cancer.gov/tcga). We first stratified AML patients based on their initial response to chemotherapy. (Fig. 1A, Supplementary Data 1). Group 1 (n = 35) consists of patients that initially failed to achieve a complete response (CR) following induction chemotherapy (Induction failure), whereas Group 2 (n = 92) represents patients that initially achieved a CR after the first round of induction chemotherapy (Initial response) (Fig. 1A). As expected, AML patients who failed induction chemotherapy were enriched for mutations in TP53, NPM1, and NRAS (Supplementary Fig. 1A). We next focused on gene expression signatures correlated with induction failure. Among the top enriched pathways, expression of inflammatory gene programs, interferon signaling, and IKK/RIPK1 activation were uniquely enriched in AML patients that initially failed induction therapy (Fig. 1B). The differentially expressed genes observed in AML patients that initially failed induction therapy are significantly enriched for DNA binding motifs of NF-κB (RELB and RELA, adjusted P = 1.4 × 10−11 and 7.8 × 10−11) (Fig. 1C, Supplementary Data 2). In contrast, AML patients that initially responded to induction therapy expressed genes enriched for the polycomb repressive complex (JARID2, SUZ12, and BMI), but not for NF-κB binding sites (Fig. 1C, Supplementary Data 3). Prior to treatment, a mid/high NF-κB signature, defined by all identified NF-κB target genes, was associated with a higher incidence of induction failure compared with patients with a low NF-κB signature (Supplementary Fig. 1B). These findings confirm that NF-κB signaling programs define patients with a greater likelihood of induction chemotherapy failure.
Fig. 1. The NF-κB target gene TNFAIP3/A20 is associated with induction failure in AML.
A Differentially expressed genes in TCGA AML patients with chemotherapy induction failure (n = 35) or initial response (n = 92) and top enriched pathways from REACTOME. Normalized enrichment score (NES). B GSEA plots comparing TCGA AML patients with induction failure or initial response in select immune-related enriched pathways. Normalized enrichment score (NES), false discovery rate (FDR). C Transcription factor enrichment analysis was based on the top and bottom differentially expressed genes from (A)(cut-offs ≥0.025 and <−0.025). D NF-κB genes from ENCODE/ChEA that are enriched in induction failure AML (n = 76), including NF-κB target genes (blue/red, n = 15), and NF-κB target genes that are overexpressed in AML compared to healthy controls (red, n = 11). E Proportion of TCGA AML patients with low expression of A20 (bottom 20% n = 26; bottom 10% n = 13) or high expression of A20 (bottom 20% n = 26; bottom 10% n = 13) that experienced induction failure or had an initial response to 7 + 3 chemotherapy. F Kaplan–Meier survival analysis of TCGA AML patients with high TNFAIP3/A20 expression (Z-score >2.0) versus all other AML patients (A20 unaffected). Gehan-Breslow-Wilcoxon test was used to determine significance. G A20 expression in AML patient blasts (n = 1730) versus healthy BM cells (n = 22)(BloodSpot56), or in AML GMPs (n = 5) versus healthy control GMPs (n = 14)(GSE35008 and GSE35010). Student’s t test (unpaired, two-tailed) was used to determine significance. H Immunoblot of healthy human cells and human AML cell lines (n = 3 independent experiments). I Quantitative PCR (qPCR) analysis of A20 mRNA expression in human AML cell lines (MOLM14, THP1), and mouse AML cells (MLL-AF9) cultured with IL-1B and treated IKK VII of for 20 min (n = 2–3 technical replicates in 3 independent experiments). Student’s t test (unpaired, two-tailed) was used to determine significance. J Immunoblot of A20 protein abundance in human AML cell lines (MOLM14, THP1), and mouse AML cells (MLL-AF9) cultured with IL-1β and treated with IKK VII for 20 min (n = 2 independent immunoblots). Error bars represent the standard error of the mean. *P < 0.05; **P < 0.01. Source data are provided as a source data file.
NF-κB signaling has been previously implicated in chemotherapy resistance in AML. However, these studies do not provide the precise mechanism by which NF-κB signaling contributes to induction chemotherapy failure and resistance in AML. NF-κB transcription factors exert their effects by inducing the expression of select genes in a cell-type and temporal manner. As such, we sought to define the key NF-κB target genes that mediate induction chemotherapy failure in AML. We selected the enriched consensus NF-κB targets in the induction failure AML based on ENCODE and ChEA datasets (n = 76) and prioritized these genes for ones that have confirmed NF-κB DNA-binding motifs (n = 15) (Supplementary Data 4). We next narrowed these induction failure NF-κB target genes to ones that are also overexpressed in AML as compared to healthy controls (n = 11) (Fig. 1D). Notably, BCL2 family members were not among this set of genes. Instead, we focused on TNFAIP3 (A20) as it is an NF-κB target gene that is associated with numerous human cancers, yet has not been implicated in AML nor in chemotherapy resistance. When we stratify TCGA AML patients by A20 expression, we see that the 20% of patients with the highest expression experience induction failure at a higher rate compared to the 20% of patients with the lowest expression of A20 (Fig. 1E). This trend is even more clear in the 10% of patients with the highest and lowest expression of A20, with nearly half of the patients in the high A20 group experiencing induction failure (46%). Moreover, A20 expression correlates with worse overall survival in AML patients undergoing chemotherapy (Fig. 1F). Collectively, we found that patients who failed to achieve complete response after induction chemotherapy had enriched NF-κB signaling pathways, particularly the gene TNFAIP3 (A20), which was associated with higher induction failure rates and worse overall survival compared to those who initially responded to therapy.
A20 is overexpressed in AML via NF-κB signaling
Inactivating mutations and deletions of A20 are common in lymphoma, but not in AML23. Paradoxically, our analysis indicates that A20 is overexpressed in AML at diagnosis and specifically in patients that fail induction chemotherapy. Analysis of publicly available datasets revealed that A20 expression is high in adult and pediatric AML as compared to other human cancer types (Supplementary Fig. 1C, D). Unlike many other human cancers with elevated A20 expression, inactivating mutations in A20 are not observed in AML (Supplementary Fig. 1C). We next validated in independent data sets that A20 mRNA is higher in AML hematopoietic progenitor cells as compared to healthy progenitor cells (Fig. 1G). Examination of A20 protein levels confirmed that A20 expression is also elevated in the majority of AML cell lines examined as compared to healthy BM cells (Fig. 1H). Immunoblotting of A20 confirmed that full-length WT protein is expressed in the majority of analyzed AML cell lines (Fig. 1H). Although A20 is a validated NF-κB target gene, it has not been confirmed that activation of NF-κB is responsible for elevated A20 expression in AML. The NF-κB inhibitor IKK-Inhibitor suppressed A20 mRNA and protein expression in mouse and human AML cells, indicating that A20 is regulated by NF-κB in AML (Fig. 1I, J). To determine whether A20 overexpression correlates with specific clinical features in AML patients, we stratified AML patients based on high (Z score >1.5) or low A20 expression (Z score <1.5). AML patients with elevated A20 expression were not enriched for any of the common mutations or subtypes at diagnosis (Supplementary Fig. 1E). Upon further examination of less prevalent mutations, we observed that elevated A20 was significantly more prevalent in AML patients with TP53 mutations (n = 14 patients) as compared to wild-type TP53 (n = 159 patients; P = 0.002). This observation is consistent with the induction failure preferentially observed in TP53-mutant AML24.
A20 expression impacts sensitivity of AML cells to doxorubicin
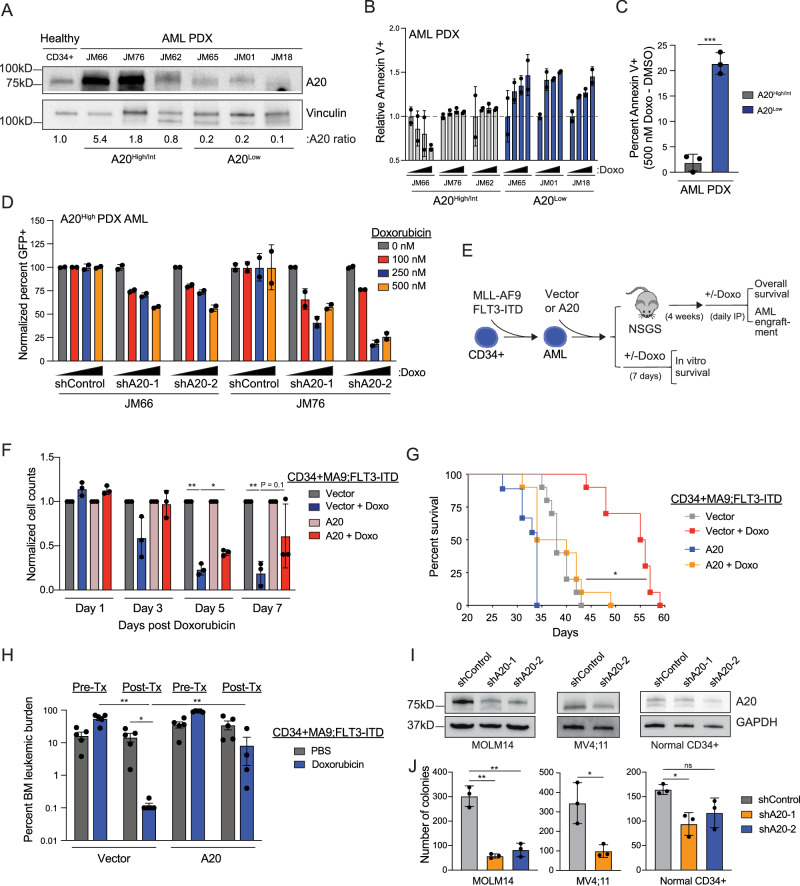
To determine whether A20 expression impacts sensitivity to chemotherapy in AML, a panel of primary AML samples were evaluated following treatment with doxorubicin or cytarabine. We found that AML samples that are resistant to doxorubicin correlated with higher levels of A20 compared with samples that are sensitive to doxorubicin (Supplementary Fig. 2A–D). However, there is no correlation between A20 expression and response to cytarabine (Supplementary Fig. 2B), suggesting that A20 is implicated in resistance to anthracyclines (doxorubicin) and not nucleoside analogs (cytarabine). Moreover, we did not observe a correlation between A20 expression and sensitivity to venetoclax and azacitidine (Supplementary Fig. 2E). We next evaluated the response of an independent panel of primary AML samples with varying levels of A20, ranging from low (A20Low AML) to high (A20High AML) levels of A20 (relative to healthy CD34+ cells) to doxorubicin (Fig. 2A, Supplementary Data 5). To capture multiple forms of cell death without discrimination, we evaluated Annexin V staining alone by flow cytometry25. The concentrations of doxorubicin selected are based on clinically relevant free drug exposure levels26. A20 expression in AML samples positively correlated with doxorubicin resistance. Specifically, A20High AML samples were generally more resistant to doxorubicin treatment, whereas A20Low AML showed increased sensitivity to doxorubicin, particularly at higher doxorubicin concentrations (Fig. 2B, C). Moreover, we asked whether reducing A20 expression in an A20High AML will resensitize the cells to doxorubicin treatment. Knockdown of A20 in two A20High AML samples (JM66 and JM76) decreased cell viability upon doxorubicin treatment as compared to control samples (shControl) (Fig. 2D). Of note, both of these samples were obtained from patients that had high-risk disease (ClinicalTrials.gov; NCT04293562). These findings suggest that elevated A20 expression results in diminished sensitivity of AML cells to doxorubicin.
Fig. 2. A20 expression impacts sensitivity to doxorubicin in AML cells.
A Immunoblot of AML patient-derived xenografts (PDX). B Flow cytometry analysis of Annexin V positive AML PDX cells following treatment with increasing doses of doxorubicin (0, 100, 250, 500 nM) for 24 and 48 h (n = 2 replicates). C Percent of Annexin V positive cells following treatment with doxorubicin in A20High/Int and A20Low AML PDX cells (calculated from panel B). Dots represent individual patients (n = 3 patients per group). Student’s t test (unpaired, two-tailed) was used to determine significance. D Flow cytometry analysis of GFP positive cells in AML PDX cells following transfection with scrambled control (shControl) or two independent A20 short hairpin RNAs (shA20) with or without doxorubicin treatment for 24 h (n = 2 independent technical replicates). E Outline of in vivo xenograft and in vitro study to determine if A20 overexpression modulates sensitivity to Doxorubicin. Human CD34+MLL-AF9;FLT3-ITD cells were transduced with empty vector or A20. F Viability of MLL-AF9;FLT3-ITD cells expressing an empty vector or A20 following treatment with doxorubicin (n = 3 technical replicates in 2 independent experiments). Student’s t test (unpaired, two-tailed) was used to determine significance. G Kaplan Meier survival analysis of xenograft study in non-conditioned NSGS mice (n = 10 per group). Mantel-Cox test was used to determine significance. H BM engraftment in NSGS mice transplanted with CD34 + MLL-AF9;FLT3-ITD overexpressing empty vector or A20 before and after intravenous Doxorubicin treatment or 1X PBS (n = 5 biological replicates). Student’s t test (unpaired, two-tailed) was used to determine significance. I Immunoblot in AML cell lines and normal human CD34+ cells expressing scrambled control (shControl) or two independent A20 short hairpin RNAs (shA20). J Colony forming capacity of AML cell lines and normal human CD34+ cells expressing scrambled control (shControl) or two independent A20 short hairpin RNAs (shA20) was assessed after 7 days (n = 3 technical replicates in 3 independent experiments). Student’s t test (unpaired, two-tailed) was used to determine significance. Error bars represent the standard error of the mean. *P < 0.05; **P < 0.01; ***P < 0.001. Source data are provided as a source data file.
To investigate whether elevated levels of A20 can make AML cells resistant to doxorubicin, we overexpressed A20 in human CD34+MLL-AF9;FLT3-ITD AML cells. These human AML cells were selected as they are inherently sensitive in vitro and in vivo to doxorubicin (Fig. 2E). Overexpression of A20 alone resulted in increased expansion of untreated AML cells, further supporting the role of A20 in AML (Supplementary Fig. 2F). Treatment with doxorubicin led to a reduction in AML cell viability harboring an empty vector, however overexpression of A20 resulted in a negligible effect on cell viability during the course of treatment with doxorubicin (Fig. 2F). Moreover, the AML cells were xenografted into immunocompromised mice and monitored for overall survival and leukemic cell engraftment following doxorubicin treatment (Fig. 2E). As expected, mice xenografted with MLL-AF9;FLT3-ITD AML cells expressing empty vector were sensitive to doxorubicin administration as they exhibited a significantly prolonged overall survival (Fig. 2G) and reduced leukemic engraftment (Fig. 2H) as compared to vehicle-treated mice. In contrast, mice xenografted with MLL-AF9;FLT3-ITD AML cells expressing A20 that were treated with doxorubicin succumbed to disease and exhibited leukemic burden similar to vehicle-treated mice (Fig. 2G, H), indicating that A20 expression diminishes the sensitivity of AML cells to doxorubicin. We also observed that mice xenografted with MLL-AF9;FLT3-ITD AML cells expressing A20 developed disease more rapidly than mice engrafted with vector-expressing AML cells. Collectively, these findings suggest that A20 overexpression contributes to doxorubicin resistance and may also influence the fitness of AML cells.
A20 is required for AML cell survival and leukemic stem/progenitor cell function
Our observations revealed that A20 expression is elevated at diagnosis and contributes to diminished sensitivity of AML cells to chemotherapy. To gain an understanding for the reason for A20 overexpression in AML, we next investigated the requirement of A20 on AML cell function by knocking down A20 in a panel of human AML cell lines and patient-derived samples (Fig. 2I). Knockdown of A20 with independent shRNAs resulted in a significant reduction in leukemic progenitor function of AML cell lines (MOLM14 and MV4;11) as compared to control cells expressing a non-targeting shRNA (shControl) but had a minimal impact on colony formation of normal CD34+ cells (Fig. 2J). These findings revealed that A20 expression is critical for the function of human AML cells.
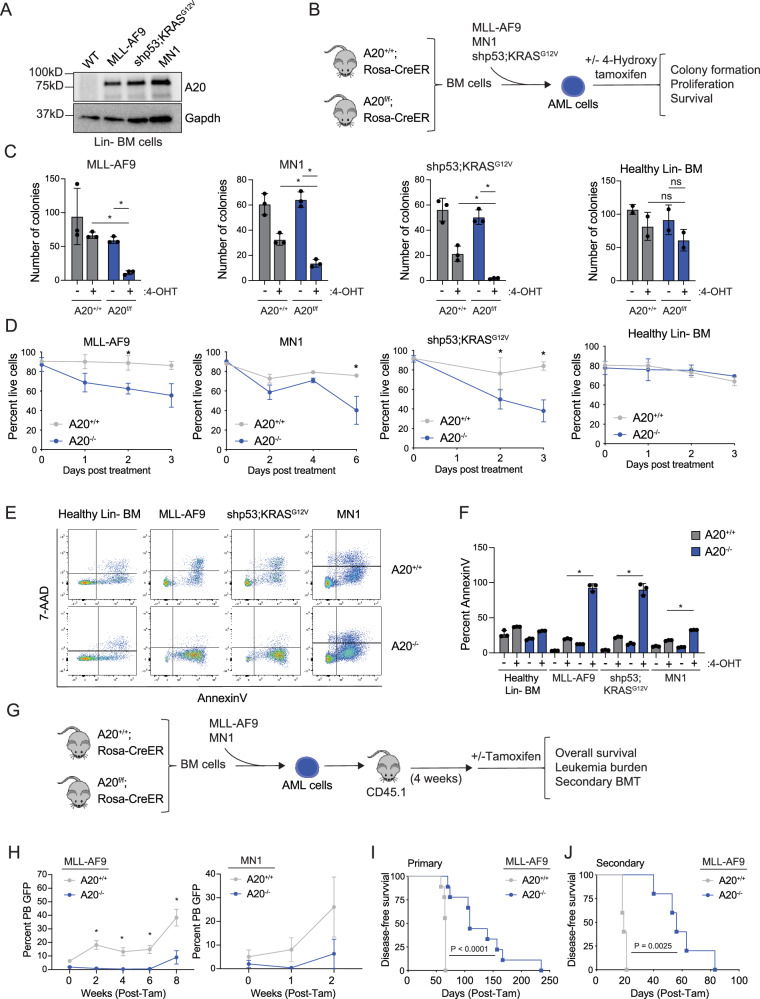
Since A20 is overexpressed in human AML, we wanted to investigate whether A20 protein expression is also elevated in mouse AML models. Retroviral expression of MLL-AF9, MN1, and KRASG12V oncogenes in mouse lineage-negative (Lin-) BM cells resulted in increased A20 protein expression as compared to wild-type Lin- BM cells expressing an empty vector (Fig. 3A). To determine the requirement of A20 in AML, we crossed A20f/f mice to inducible (RosaCreER) recombinase strains that, upon in vivo tamoxifen or in vitro 4-hydroxytamoxifen (4-OHT) treatment, induces deletion of A20, as previously reported27. As above, we modeled AML in Lin- BM cells isolated from A20f/f;RosaCreER mice by retroviral expression of MLL-AF9, MN1, or KRASG12V along with shRNAs targeting p53 (shp53;NRASG12D) (Fig. 3B). In vitro 4-OHT treatment resulted in efficient A20 excision and loss of mRNA and protein expression in the A20fl/fl AML and wild-type Lin- BM cells (Supplementary Fig. 3A–I). Since A20 expression was not significantly impacted in heterozygous A20+/fl AML and wild-type cells, we exclusively characterized the homozygous A20 knockout models. Deletion of A20 in the MLL-AF9, MN1, and shp53;KRASG12V AML cells resulted in a significant loss of leukemic progenitor function in methylcellulose colony assays (Fig. 3C) and reduced proliferation in vitro (Fig. 3D). The deleterious effects following A20 deletion in the AML cells coincided with an increase in pan cell death as indicated by 7-AAD and AnnexinV staining (Fig. 3E, F). In contrast, deletion of A20 did not impact the progenitor function, proliferation, nor viability of wild-type Lin- BM cells (Fig. 3C–F). These findings indicate that A20 expression is necessary for mouse AML cell function.
Fig. 3. A20 is required for AML cell survival and progenitor function.
A Immunoblot of A20 protein in lineage-negative BM from a healthy mouse or leukemic mouse models: MLL-AF9, shp53;KRASG12V, and MN1 (n = 2 independent experiments). B Outline of in vitro experiments. C–F Lineage-negative cells from A20+/+;Rosa-CreER and A20f/f;Rosa CreER mice were transduced with oncogenes MLL-AF9, shp53;KRASG12V, or MN1, sorted, and treated with 4-Hydroxy tamoxifen (4-OHT). Cells were assessed for colony-forming capacity (C), viability by trypan blue exclusion (D), and viability by Annexin V/7AAD staining by flow cytometry (E, F) (n = 3 independent experiments). Student’s t test (unpaired, two-tailed) was used to determine significance. G Overview of transplant studies. After 4 weeks of engraftment, 1 mg of tamoxifen per day for 5 days was injected intraperitoneally, and leukemic burden (H) and overall survival (I) were assessed (n = 9 mice per group). Student’s t test (unpaired, two-tailed) was used to determine significance for (H). Mantel–Cox test was used to determine significance in (I). J Kaplan Meier survival analysis showing survival in secondary recipients (5 mice per group) after receiving BM cells from moribund primary transplant recipients. Mantel–Cox test was used to determine significance. Error bars represent the standard error of the mean. *P < 0.05; **P < 0.01; ***P < 0.001. Source data are provided as a source data file.
We next determined the requirement of A20 on AML development in vivo. Lin- BM cells from A20f/f or wild-type RosaCreER mice (A20+/+) were transduced with MLL-AF9 or MN1 and then transplanted along with helper BM cells into lethally irradiated mice (Fig. 3G). After 4 weeks of engraftment, recipient mice were injected with tamoxifen intraperitoneally to induce deletion of A20 (A20–/–) (Fig. 3G). Mice transplanted with A20+/+ MLL-AF9 or MN1 AML cells exhibited hallmarks of leukemia including splenomegaly and myeloid blasts in the BM, spleen, and peripheral blood (Supplementary Fig. 3J). Mice engrafted with A20–/– MLL-AF9 or MN1 AML cells coincided with reduced circulating AML cells as compared to wild-type AML cells (Fig. 3H). Moreover, recipient mice engrafted with A20–/– MLL-AF9 AML cells had significantly prolonged survival as compared to mice engrafted with A20+/+ MLL-AF9 AML cells (Fig. 3I). To determine whether A20 deletion impacts long-term leukemic cell potential, we performed secondary transplantations using A20+/+ and A20–/– MLL-AF9 AML cells isolated from mice at time of death. Secondary recipient mice engrafted with A20–/– MLL-AF9 AML cells exhibited a significantly extended disease-free survival as compared to mice engrafted with A20+/+ MLL-AF9 AML cells (Fig. 3J). Although A20 is indispensable for AML cells, deletion of A20 in healthy BM did not significantly affect the overall survival of chimeric recipient mice nor blood counts (Supplementary Fig. 4). These results show the dispensable nature of A20 in normal adult hematopoiesis and highlight the requirement of A20 in AML.
A20 protects AML cells from necroptosis
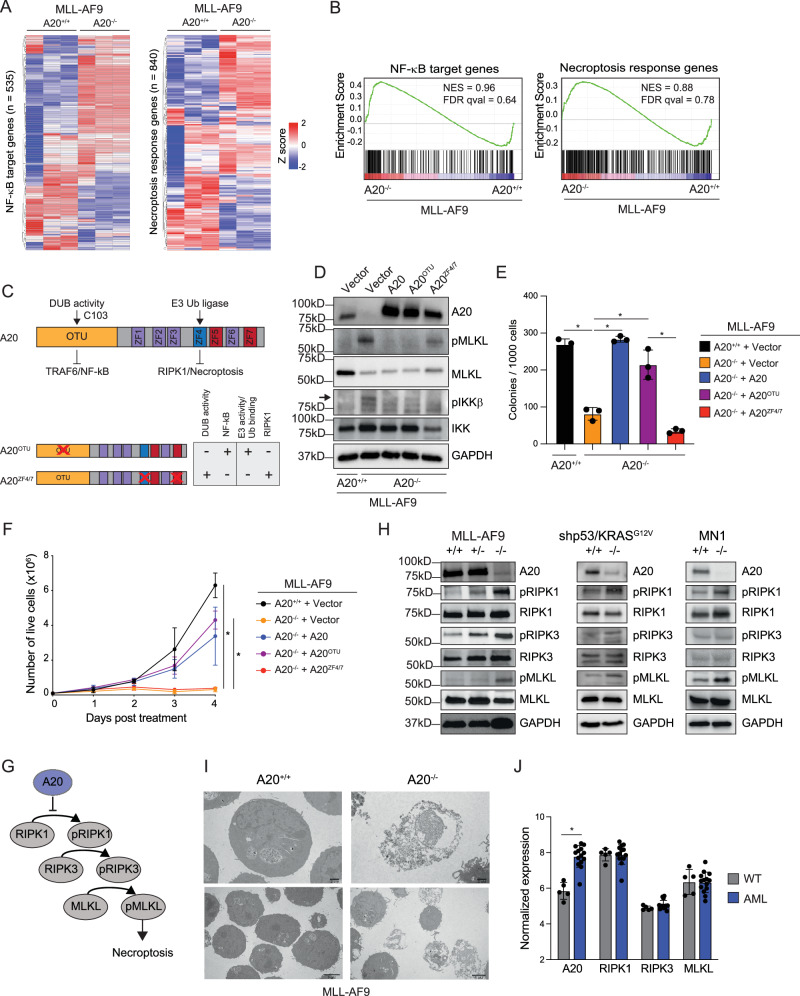
To identify the relevant signaling pathways and gene expression changes following deletion of A20 in AML cells, we performed RNA-sequencing on A20 deficient (A20–/–) and wild-type (A20+/+) MLL-AF9 AML cells. Deletion of A20 resulted in significant differentially expressed genes as compared wild-type MLL-AF9 AML cells. Specifically, 534 genes had a 2-fold decrease in expression and 601 genes had a twofold increase in gene expression (Supplementary Data 6). Since A20 is a negative regulator of NF-κB and necroptosis pathways28,29, we wanted to examine whether these pathways are dysregulated upon A20 deletion in AML cells. A curated list of all NF-κB target genes and a necroptosis response signature genes were evaluated in A20–/– and wild-type MLL-AF9 AML cells (Fig. 4A). Deletion of A20 resulted in a partial enrichment of genes associated with both NF-κB and necroptosis activation as compared to wild-type MLL-AF9 AML cells (Fig. 4A, B).
Fig. 4. A20 protects leukemic cells from necroptosis.
A Differentially expressed genes in A20+/+ and A20–/– MLL-AF9 cells (2-fold, P < 0.05, n = 3). B GSEA plots of NF-κB target genes and necroptosis response genes in A20+/+ and A20–/– MLL-AF9 cells. C Schematic of A20 functional domains. Deubiquitinase activity is conferred by C103 in the OTU domain and is lost in the A20OTU mutant. E3 ubiquitin ligase activity conferred by zinc finger 4 (ZF4), is required for RIPK1 activation and necroptosis and is lost in the A20ZF4/7 mutant. Immunoblot (D), colony forming assay (E), and proliferation assay showing live cells by trypan blue exclusion (F) in A20+/+ MLL-AF9 cells expressing empty vector and A20–/– MLL-AF9 cells expressing either empty vector, A20, A20OTU, or A20ZF4/7 (n = 3 technical replicates in 3 independent experiments). Student’s t test (unpaired, two-tailed) was used to determine significance in (E, F). G Necroptosis activation process. Necrosome effectors, RIPK1, RIPK3, and MLKL, are phosphorylated leading to necroptosis. A20 inhibits necroptosis by causing degradation of RIPK1 via the proteasome. H Immunoblot of necroptosis effectors in A20+/+ and A20–/– cells in MLL-AF9, shp53;KRASG12V, and MN1 leukemia models after 48 h of treatment with 4-OHT. Representative image from at least 2 independent replicates. I Transmission electron microscopy of A20+/+ and A20–/– MLL-AF9 cells after 24 h of treatment with 4-OHT (n = 2 independent experiments, each consisting of 3 technical replicates). J Expression of necroptosis effectors in AML patient GMPs versus healthy control GMPs (GSE35008 and GSE35010) (n = 5 healthy controls, n = 14 AML patients). Student’s t test (unpaired, two-tailed) was used to determine significance. Error bars represent the standard error of the mean. *P < 0.05; **P < 0.01; ***P < 0.001. In each of the immunoblots (D, H), all samples were derived from the same experiment, but different gels for each antibody were processed in parallel. Source data are provided as a source data file.
A20 plays a role in regulating diverse signaling pathways which depend upon A20’s ubiquitin editing functions, either via its E3 ligase or deubiquitinase activity. The de-ubiquitin function of A20 via catalytic cysteine (C103) of the OTU domain removes activating ubiquitin chains from TRAF6, which results in diminished NF-κB signaling (Fig. 4C). The E3 ligase function of A20 via zinc finger 4 mediates receptor-interacting serine/threonine-protein kinase 1 (RIPK1) ubiquitination and degradation, which results in suppression of necroptosis-mediated cell death (Fig. 4C). Recent studies suggest that zinc finger 7, which is known to bind M1-linked ubiquitin chains, plays a role in NF-κB activation in a non-catalytic, RIPK1-independent manner30. To distinguish between whether dysregulated NF-κB or necroptosis activation leads to impaired leukemic cell function in A20-deficient AML cells, we expressed A20 mutants that are impaired in primarily suppressing NF-κB (A20OTU) or necroptosis activation (A20ZF4/7) (Fig. 4C). As expected, re-expression of wild-type A20 in A20–/– MLL-AF9 AML prevented necroptosis activation, as indicated by reduced phosphorylated (p) mixed lineage kinase domain-like protein (MLKL), and NF-κB activation, as indicated by reduced pIKKβ (Fig. 4D). However, expression of A20OTU in A20–/– MLL-AF9 AML was unable to inhibit IKKβ but retained its ability to suppress necroptosis (as indicated by reduced pMLKL). In contrast, expression A20ZF4/7 was defective for necroptosis inhibition (sustained pMLKL) but retained its ability to inhibit IKKβ in A20–/– MLL-AF9 AML cells (Fig. 4D). Expression of wild-type A20 or A20OTU restored the leukemic colony-forming ability of A20–/– MLL-AF9 AML cells (Fig. 4E). However, expression of A20ZF4/7 was not able to restore the leukemic colony-forming ability of A20–/– MLL-AF9 AML cells. Further supporting the essential role of A20’s E3 ligase ability in AML, wild-type A20 and A20OTU were able to restore the proliferative capacity of A20-deficient AML cells, whereas A20ZF4/7 was not (Fig. 4F). These findings suggest that A20 is required for restraining necroptosis signaling in AML cells.
Necroptosis is a tightly regulated and multi-step programmed cell death mechanism that is initiated under defined cellular stresses, which include a combination of extracellular stimuli coupled with inactive apoptosis31. Thus, necroptosis is considered as an alternative mode of programmed cell death overcoming apoptosis resistance32. TNFα, a known activator of necroptosis33, positively correlates with A20 expression in AML (Supplementary Fig. 5A), suggesting that the microenvironment in AML is conducive to the induction of necroptosis. Unexpectedly, our findings revealed that MLL-AF9 AML cells are primed and exquisitely sensitive to undergo spontaneous necroptosis. Therefore, we next determined whether A20 restricts spontaneous necroptosis signaling in other subtypes of AML. Necroptosis is executed by a biochemically-defined signaling cascade, which involves the phosphorylation of its main effectors RIPK1, RIPK3, and MLKL (Fig. 4G). Deletion of A20 in the MLL-AF9, MN1, and shp53;KRASG12V AML cells resulted in increased phosphorylated forms of RIPK1, RIPK3, and MLKL, which are critical indicators of necroptosis (Fig. 4H). Knockdown of A20 in human AML cells also resulted in activation of RIPK1 (Supplementary Fig. 5B) and lactate dehydrogenase (LDH) release, an indicator of necrosis, in A20high AML cells (Supplementary Fig. 5C). Notably, the cleavage of Caspase 3, a measure of apoptotic cell death, did not occur in A20-deficient AML cells (Supplementary Fig. 5D). To confirm that MLKL activation leads to necroptosis in A20-deficient AML cells, we used transmission electron microscopy. Indeed, A20–/– MLL-AF9 AML cells exhibited necroptotic cell morphology, which includes cell and organelle swelling, a lack of chromatin condensation, and plasma membrane rupture (Fig. 4I, Supplementary Fig. 5E). Importantly, A20–/– MLL-AF9 AML did not show indicators of apoptotic morphology, such as cell shrinkage and chromatin condensation. These findings indicate that A20 expression is essential for preventing necroptotic-mediated cell death of AML cells, while its ability to regulate NF-κB is dispensable in AML.
The key regulators of the necroptotic pathway are generally downregulated in cancer cells, suggesting that cancer cells evade necroptosis, in addition to apoptosis, to survive. For example, expression of RIPK3 and/or MLKL is frequently downregulated in human cancers, suggesting a selective pressure to minimize necroptosis activation34. In human AML leukemic progenitor cells, expression of RIPK1, RIPK3, and MLKL are nearly identical to healthy control cells, indicating that the canonical regulators of necroptosis are not under selective pressure in AML. In contrast, A20 expression is significantly elevated in the leukemic progenitor cells as compared to healthy cells (Fig. 4J), further reinforcing the requirement of A20 as the central factor in preventing necroptosis in AML.
Necroptosis inhibition restores the survival of A20-deleted AML cells
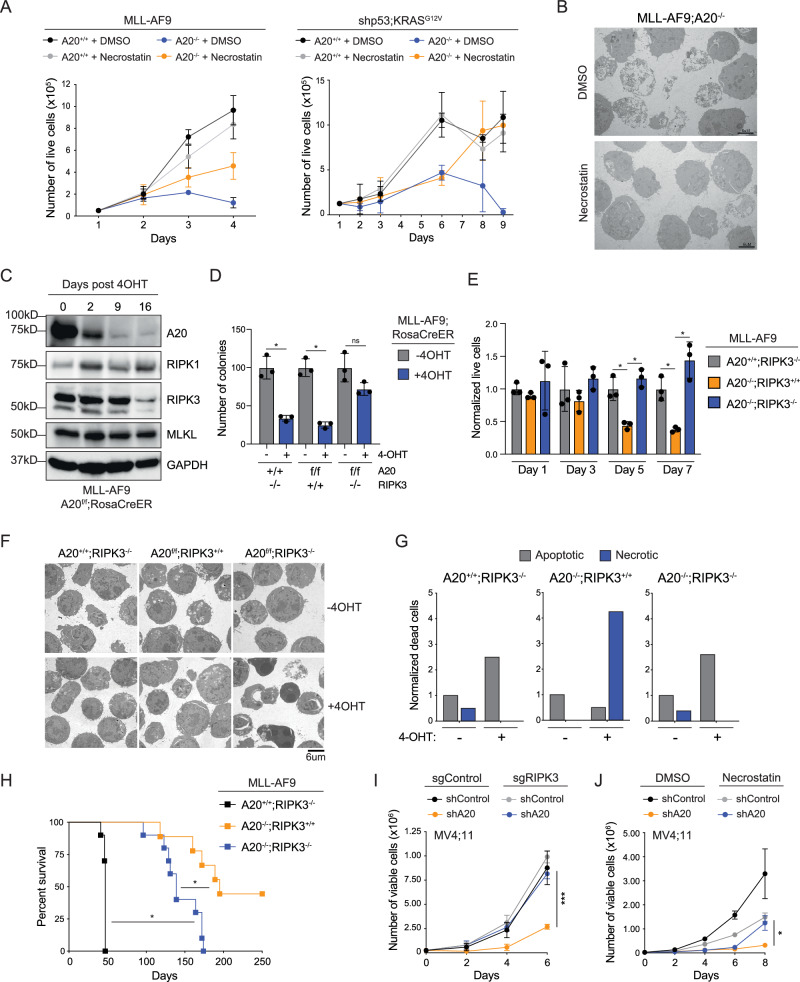
To confirm that link between A20 and necroptosis suppression in AML, we employed pharmacological and genetic approaches. Necroptosis can be inhibited pharmacologically using small molecule inhibitors that interfere with the kinase activity of RIPK1 or RIPK3. Here, we evaluated the RIPK1 kinase inhibitor necrostatin-1 (Nec-1) to prevent necroptosis in A20-deficient AML cells. Nec-1 treatment of A20–/– MLL-AF9 AML or shp53;KRASG12V AML cells rescued the proliferative and survival defect, while not impacting the function of wild-type AML cells (Fig. 5A). Moreover, A20–/– AML cells treated with Nec-1 did not show morphological features of necroptosis (Fig. 5B). In contrast, inhibition of NF-κB activation or apoptosis did not rescue the proliferative defect of A20–/– AML cells (Supplementary Fig. 6A, B). Nec-1 also rescued the leukemic progenitor defect of human AML cells following knockdown of A20 (Supplementary Fig. 6C). These findings confirm that A20 is critical for preventing spontaneous necroptosis in AML.
Fig. 5. Necroptosis inhibition restores A20-deleted leukemic cells.
A Proliferation assay of A20+/+ and A20f/f treated with either vehicle (DMSO) or Necrostatin-1 (2 μM) for 20 min, followed by vehicle or 4-OHT (n = 3 technical replicates in 2 independent experiments). Student’s t test (unpaired, two-tailed) was used to determine significance. B Transmission electron microscopy of A20f/f MLL-AF9 cells treated with vehicle (DMSO) or Necrostatin-1 (2 μM) for 20 min, followed by 4-OHT for 24 h (n = 2 independent experiments, each consisting of 3 technical replicates). Scale bar = 6 μm. C Immunoblot of A20f/f MLL-AF9 cells treated with 4-OHT and collected on the indicated days after growing in liquid culture (n = 2 independent experiments). All samples were derived from the same experiment, but different gels for each antibody were processed in parallel. D Colony forming assay of cells treated with vehicle or 4-OHT for 48 h in liquid culture before plating into methylcellulose with 4-OHT (n = 3 independent technical replicates). Student’s t test (unpaired, two-tailed) was used to determine significance. E Proliferation assay of the indicated MLL-AF9 cells treated with vehicle or 4-OHT (n = 3 technical replicates in 3 independent experiments). Student’s t test (unpaired, two-tailed) was used to determine significance. F Transmission electron microscopy of the indicated MLL-AF9 cells treated with vehicle or 4-OHT (n = 3 technical replicates). Scale bar = 6 μm. G Morphological assessment of dead/dying A20+/+RIPK3–/–, A20f/fRIPK3+/+, and A20f/fRIPK3–/– MLL-AF9 cells treated with vehicle or 4-OHT. H Kaplan–Meier survival analysis of mice transplanted with the indicated MLL-AF9 cells. After 5 weeks of engraftment, tamoxifen per day for 5 days was injected intraperitoneally. Mantel-Cox test was used to determine significance. I Proliferation assay of MV4;11 cells with sgControl or sgRIPK3 expressing shControl or shA20 (n = 3 technical replicates in 2 independent experiments). Student’s t test (unpaired, two-tailed) was used to determine significance. J Proliferation assay of MV4;11 cells expressing shControl or shA20 and treated with vehicle or Necrostatin-1 (2 μM) (n = 3 technical replicates in 2 independent experiments). Student’s t test (unpaired, two-tailed) was used to determine significance. Error bars represent the standard error of the mean. *P < 0.05; **P < 0.01; ***P < 0.001. Source data are provided as a source data file.
We posited that if loss of A20 results in the demise of AML cells via necroptosis then compensatory mechanisms to overcome this death pathway should be observed following prolonged absence of A20. To identify compensatory mechanisms of A20-deficient AML cells, A20–/– MLL-AF9 AML cells were replated in methylcellulose for over 30 days. During this time, the colony-forming potential was gradually restored with each replating (Supplementary Fig. 6D). Although A20 deletion was confirmed by PCR and immunoblotting, RIPK3 protein levels were significantly diminished, suggesting that the emerging A20-deficient clones have downregulated RIPK3 (Fig. 5C). To confirm that RIPK3 is essential for executing necroptosis in A20 deficient AML, we crossed Ripk3 knockout mice to the conditional A20 Rosa-CreER mice and retrovirally infected lineage negative BM cells from these mice with the MLL-AF9 oncogene (Supplementary Fig. 6E). In contrast to RIPK3-proficient MLL-AF9 AML (A20–/–;RIPK3+/+) cells, deletion of A20 in RIPK3-deficient MLL-AF9 AML (A20–/–;RIPK3–/–) cell did not result in loss of leukemic progenitor function in methylcellulose colony assays (Fig. 5D) or reduced proliferation in vitro (Fig. 5E). Moreover, deletion of RIPK3 prevented necroptosis activation, as indicated by the absence of necroptotic cell morphology (Fig. 5F, G). To determine whether the mechanistic basis for suppression of leukemic burden in the absence of A20 is due to RIPK3-dependent necroptosis, we transplanted Lin- BM cells from A20f/f;RIPK3+/+RosaCreER, A20f/f;RIPK3–/–RosaCreER or A20+/+;RIPK3–/–mice expressing MLL-AF9 and then transplanted along with helper BM cells into lethally irradiated mice (Supplementary Fig. 6E). After 4 weeks after engraftment, recipient mice were injected with tamoxifen intraperitoneally to induce deletion of A20 (A20–/–;RIPK3+/+, A20–/–;RIPK3–/–, or A20+/+;RIPK3–/–). Mice transplanted with A20+/+;RIPK3–/–MLL-AF9 AML cells became moribund with leukemia around day 50 (Fig. 5H), while mice transplanted with A20–/–;RIPK3+/+ MLL-AF9 AML cells had significantly prolonged survival in recipient mice and lack of leukemia in approximately half of the mice (Fig. 5H). In contrast, deletion of RIPK3 in A20–/– AML cells (A20–/–;RIPK3–/–) resulted in a fully penetrant disease and accelerated leukemic phenotype (Fig. 5H). The shorter survival of mice engrafted with A20–/–;RIPK3–/– MLL-AF9 AML cells coincided with increased circulating AML cells. Lastly, suppression of human AML cells following knockdown of A20 also is mediated by RIPK3-dependent necroptosis. Deletion of RIPK3 using sgRNAs or treatment with Nec-1 rescued the abrogated cell growth of MV4;11 cells expressing shRNAs targeting A20 (Fig. 5I, Supplementary Fig. 6F). These findings indicate that A20 expression is essential for preventing necroptotic-mediated cell death of AML cells via RIPK3 signaling.
Chemotherapy-induced necroptosis is impeded by A20
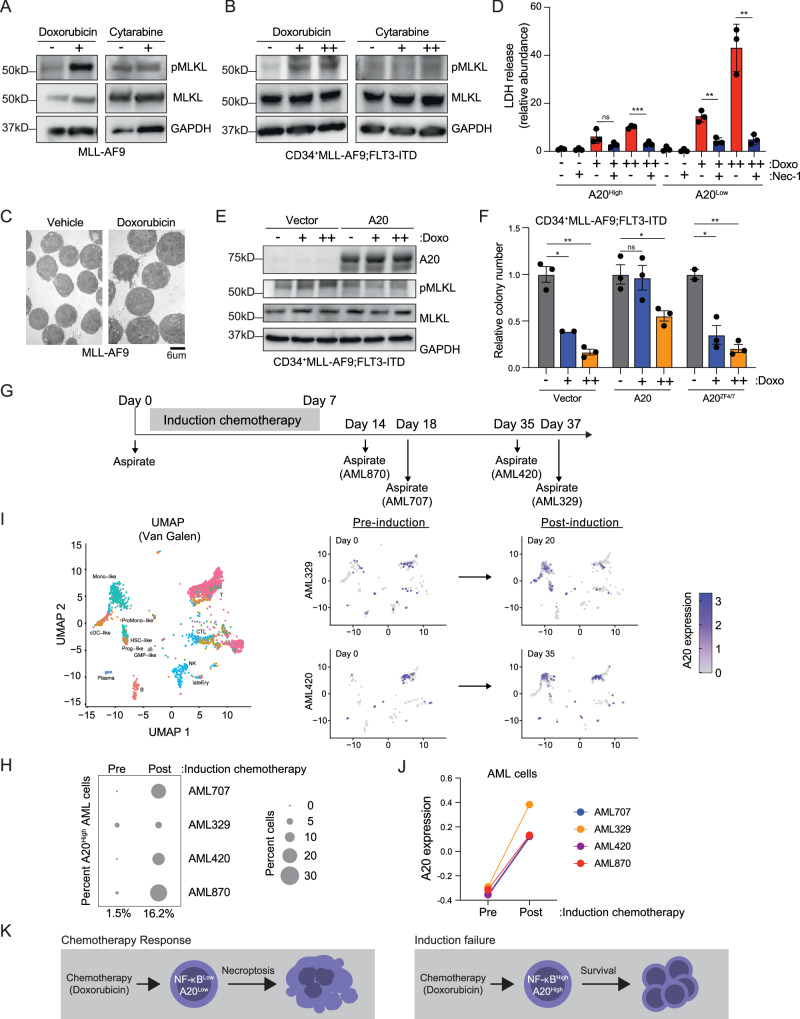
Our data revealed that A20 overexpression restricts sensitivity to doxorubicin chemotherapy in AML and that loss of A20 can initiate necroptosis. Therefore, we next asked whether doxorubicin mediates cell death of AML cells via A20-dependent necroptosis. We first evaluated the effects of doxorubicin and cytarabine on mouse and human AML cells. Mouse MLL-AF9 AML cells or primary human CD34 + MLL-AF9;FLT3-ITD cells treated with doxorubicin resulted in activated MLKL (Fig. 6A, B), which correlated with reduced cell viability (Supplementary Fig. 7A). Moreover, doxorubicin treatment of AML cells for 24 h also resulted in morphological features of necroptosis (Fig. 6C). In contrast, cytarabine at concentrations that impaired cell viability did not induce activated necroptosis (Fig. 6A, B, Supplementary Fig. 7B). To determine whether differences in A20 expression can influence the sensitivity of AML cells to doxorubicin, we evaluated patient-derived AML samples with high or low A20 expression (from Fig. 2A, B). Doxorubicin induced necroptosis in a dose-dependent manner in A20Low AML samples yet had minimal effects on necroptosis induction in A20High AML samples (Fig. 6D). Since human CD34 + MLL-AF9;FLT3-ITD AML cells exhibit low levels of A20 expression and are exquisitely sensitive to doxorubicin, we asked whether overexpression of A20 can lead to doxorubicin resistance. Lentiviral-mediated overexpression of A20 in CD34 + MLL-AF9;FLT3-ITD AML cells prevented induction of necroptosis as evidenced by the absence of pMLKL (Fig. 6E) and increased cell viability (Fig. 6F), as compared to CD34 + MLL-AF9;FLT3-ITD AML cells expressing an empty vector. Even at fivefold higher concentrations of doxorubicin, A20-overexprssing AML cells were less sensitive to doxorubicin-induced cell death as compared to control cells (Supplementary Fig. 7C). Overexpression of A20 did not protect against cytarabine-induced cell death, indicating that doxorubicin and cytarabine mediate AML cell killing through distinct cell death mechanisms (Supplementary Fig. 7D). To confirm that the suppression of necroptosis by A20 is indeed mediated by its E3 ligase function, we compared the viability of CD34 + MLL-AF9;FLT3-ITD AML cells expressing A20ZF4/7 to cells expressing wild-type A20. Although A20 protected the AML cells from doxorubicin-induced necroptotic cell death, AML cells expressing A20ZF4/7 were sensitive to doxorubicin and were not able to restore the leukemic colony forming ability of A20–/– MLL-AF9 AML cells. Further supporting the essential role of A20’s E3 ligase ability in AML, wild-type A20 and A20OTU were able to restore the proliferative capacity of A20-deficient AML cells, whereas A20ZF4/7 was not (Fig. 6F). These findings suggest that A20 protects AML cells from doxorubicin-induced necroptosis. To test this hypothesis at the single cell level, we analyzed a publicly available single-cell RNA-sequencing data set from 4 AML patients prior to treatment and then at the first time point post-induction chemotherapy treatment35. The patients selected had evidence of blasts after induction chemotherapy (1–6%) and a suitable number of malignant cells for analysis based on mutation-specific single-cell genotyping. Although few cells remained after treatment, transcriptomes of samples obtained prior to induction chemotherapy (Day 0) was compared to samples obtained after the last day of initial treatment (Fig. 6G). Prior to induction chemotherapy, A20 expression was observed in a small percentage of AML cells; however, the proportion of AML cells expressing A20 post treatment was higher for all patients (Fig. 6H). In the examination of matched malignant cell types, A20 expression was observed in ~1.5% of malignant cells (referred to as “like”) before treatment (averaged across 4 patients at Day 0). However, in all cases, A20 expression increased to ~16% of the remaining malignant cells after treatment. For example, the proportion of malignant monocytic and progenitor cell clusters from AML329 and AML420 expressing A20 was initially low, but following induction chemotherapy, the proportion of these cells expressing A20 increased (Fig. 6I). A20 expression was also induced in some non-malignant cell populations where NF-κB is operational, such as in clusters containing lymphocytes (see AML420, Fig. 6I). We also observed that the transcript levels of A20 were higher post-chemotherapy as compared to samples prior to therapy (Fig. 6J). Thus, these findings suggest that induction chemotherapy results in the expansion of AML cells expressing A20 and that chemotherapy-exposed AML cells express elevated levels of A20. To determine whether continuous exposure to doxorubicin will select for AML cells with elevated A20, we treated CD34 + MLL-AF9;FLT3-ITD AML cells with increasing concentrations of doxorubicin for 5 days and then examined A20 protein expression. AML cells treated with doxorubicin exhibited increased expression of A20 protein (Supplementary Fig. 7E). The increased expression of A20 was not observed following venetoclax or azacitidine treatment (Supplementary Fig. 7F). These findings suggest that surviving AML cells following chemotherapy exhibit higher levels of A20 expression, which contributes to induction chemotherapy failure by preventing necroptosis (Fig. 6K).
Fig. 6. Chemotherapy-induced necroptosis is impeded by A20 in AML.
A Immunoblot of necroptosis effectors in A20+/+ MLL-AF9 treated with vehicle, Doxorubicin (5 nM), or Cytarabine (500 nM) for 24 h (n = 2 independent experiments). B Immunoblot of CD34 + MLL-AF9;FLT3-ITD cells treated with Doxorubicin (0, 1, 2.5 nM) or Cytarabine (0, 250, 500 nM) for 24 h (n = 2 independent experiments). C Transmission electron microscopy of A20+/+ MLL-AF9 treated with vehicle or Doxorubicin (5 nM) for 24 h (n = 3 technical replicates). Scale bar = 6 μm. D LDH release assay of an A20High (JM62) and A20Low (JM18) PDX AML samples treated with doxorubicin (+, 250 nM; ++, 500 nM) and/or Necrostatin-1 (Nec-1, 30 μM) (n = 3 independent technical replicates). Student’s t test (unpaired, two-tailed) was used to determine significance. E Immunoblot of CD34 + MLL-AF9;FLT3-ITD cells expressing empty vector or wild-type A20 treated with Doxorubicin (0, 1, 2.5 nM) or Cytarabine (0, 250, 500 nM) for 24 h. F Colony numbers of CD34 + MLL-AF9;FLT3-ITD cells expressing empty vector, wild-type A20, or A20ZF4/7 following treatment with Doxorubicin (0, 1, 2.5 nM) (n = 3 independent experiments). Student’s t test (unpaired, two-tailed) was used to determine significance. G Timeline of AML patient sample collection pre- and post-chemotherapy. H Percent of AML cells with A20 overexpression (Z score >0.25) pre- and post-chemotherapy. I UMAP plots of single-cell RNA sequencing data (GSE116256) showing AML cell fate mapping (left), AML cells prior to chemotherapy in patient AML329 and AML420, and AML cells post-chemotherapy (right). J A20 expression in the indicated malignant cells pre- and post-chemotherapy. K Overview of A20-dependent suppression of necroptosis in AML. Error bars represent the standard error of the mean. *P < 0.05; **P < 0.01; ***P < 0.001. For each immunoblot (A, B, E), all samples were derived from the same experiment, but different gels for each antibody were processed in parallel. Source data are provided as a source data file.
Discussion
In summary, this study shows that primary chemotherapy resistance in AML is mediated by defective necroptosis via A20 and is associated with induction therapy failure. Currently, the standard approach for treatment of AML remains “7+3” chemotherapy1. Fortunately, AML can be cured in about ~40% of patients under 60 years old with chemotherapy, but the prognosis for individuals over 60 is worse, with a cure rate below 20%. Moreover, AML patients who fail induction therapy have a significantly worse prognosis, therefore, understanding mechanisms that contribute to induction failure is important for improving response rates and overall survival. Around 40% of AML patients display constitutive NF-κB activation, which supports AML cell survival and resistance to chemotherapy15,16. NF-κB signaling affects AML cell differentiation, growth, survival, and therapy resistance16,36. Despite its critical role, targeting NF-κB has shown limited success in cancer treatment due to its involvement in both pro-survival and anti-survival pathways37. Although NF-κB’s significance in AML is evident, the precise mechanism by which NF-κB activation contributes to chemotherapy failure and resistance in AML remains unclear. This study aimed to identify the essential factors regulated by NF-κB following primary induction failure to 7+3 chemotherapy. The significant findings of this study are that A20, a target gene of NF-κB, is elevated in leukemic cells from AML patients who experience primary induction failure and that A20 expression is critical for preventing necroptotic cell death in AML. We found that doxorubicin primarily induces necroptotic cell death in AML cells, which is consistent with prior studies in other cancers. Thus, elevated expression of A20 prevents necroptosis induced by doxorubicin, the primary cell death mechanism in AML cells.
The primary and secondary resistance mechanisms of leukemia clones play a significant role in the development of chemotherapy resistance. This resistance can arise from various factors, including the inherent dormancy of leukemia stem cells, which provides protection against cell cycle-specific treatments. Additionally, there is an increased expression of ATP-binding cassette (ABC) transporters, which effectively pumps cytotoxic drugs out of the cell, contributing to drug resistance. Senescent resistance mechanisms, epigenetic alterations, and metabolic reprogramming enables the leukemic clones to adapt, enhancing their survival against chemotherapy. Furthermore, the BM niches play a crucial role by offering a sanctuary-like environment that shields leukemic clones from the effects of therapeutic drugs. This microenvironmental support contributes to the resilience of leukemic cells and their ability to withstand chemotherapy6. For example, TNFα, which is often elevated in the circulation of AML patients, contributes in the initiation and regulation of necroptosis by inducing RIPK activation33. In AML patients, we observed a positive correlation between A20 and TNF expression, suggesting that the leukemic microenvironment may further promote necroptosis and permitting the expansion of leukemic cells that are intrinsically resistant necroptosis. For example, RIPK3 suppression prevents R-2-hydroxyluterate-induced necroptosis in IDH-mutated AML cells38. The objective of our study was to examine the cell-intrinsic role of necroptosis in AML; however, further studies are necessary to evaluate the microenvironmental contribution to regulating necroptosis in the pathogenesis of AML and following chemotherapy.
Chemotherapy elicits various cellular effects culminating in different forms of cell death. Therefore, common mechanisms of resistance to chemotherapy arise from cancer cells evading these death pathways. For instance, AML cells can develop resistance to BCL2 inhibitors by increasing MCL1 or BCLXL levels, other anti-apoptotic BCL2 family members39. Although much attention has been focused on how chemotherapy induces apoptotic cell death in AML, we found that anthracyclines, the backbone of 7+3 chemotherapy in AML, leads to necroptotic cell death. We find that A20High AML cells are unable to initiate necroptotic cell death. Necroptosis is an evolutionarily conserved and tightly regulated multi-step programmed cell death mechanism triggered by specific cellular stresses. It acts as a backup cell death mode when other cell death pathways, namely apoptosis, are compromised. Unexpectedly, we found that a major subset of AML cells are primed and exquisitely sensitive to undergo spontaneous necroptosis. Unlike other cancers where necroptosis is suppressed through effector downregulation of RIPK1, RIPK3, or MLKL, a subset of AML cells evade necroptosis by expressing A20, which counteracts RIPK1-dependent necroptosis. Indeed, activation of necroptosis with the SMAC mimetic, birinapant, targeted chemotherapy-resistant acute lymphoid leukemia (ALL) cells40,41, suggesting that exploiting necroptosis for therapy-resistant leukemic cells is a viable strategy. At this time, 7+3 chemotherapy is indicated for low- and intermediate-risk AML patients but is less effective for high-risk AML. This is because 7+3 therapy resistance is likely in high-risk patients and an alternative, targeted therapy combination, venetoclax and azacitidine, despite having a similar likelihood of therapy resistance, is more tolerable. In our studies, we show that reducing A20 expression in two high-risk AML patient samples sensitized the leukemic cells to doxorubicin-mediated cell death. This suggests that targeting A20 may enable a greater number of patients to benefit from 7+3 therapy.
Normally, A20 expression remains low unless NF-κB is activated, as seen in AML patients with induction failure. Increased A20 levels result in resistance to anthracycline-induced AML cell death, and reducing A20 expression sensitizes AML cells to anthracyclines, such as doxorubicin. Although A20 typically restrains NF-κB activation as a negative feedback loop28,29, this study reveals that A20 is required for AML cells to inhibit necroptosis, under both steady-state conditions and during anthracycline treatment. Although A20 is considered an anti-inflammatory protein, previous work has shown A20 can also protect certain immune cells and epithelial cells from necroptosis, albeit via distinct mechanisms42. A20-deficient T cells and fibroblasts are susceptible to caspase-independent and kinase RIPK3-dependent necroptosis. For example, A20-deficient T cells exhibited increased formation of RIPK1-RIPK3 complexes. A20 is frequently mutated or deleted in certain cancers, such as in lymphomas, impairing its tumor-suppressive function23. These mutations lead to loss of A20 expression and chronic NF-κB activation. Reduced A20 expression has been implicated in the aging of hematopoietic stem cells27,43,44. In contrast, A20 is elevated and contributes to the expansion of pre-leukemic clones during low-grade inflammation by restricting excessive canonical NF-κB activation45,46. Consistent with these findings, A20 expression is also elevated in AML, even without chemotherapy, but is vital for preventing necroptotic cell death. Interestingly, elevated expression of A20 is observed shortly after induction chemotherapy in AML patients47, which suggests that A20High AML cells have a fitness advantage over A20Low AML cells during treatment and/or that AML cells able to rapidly induce A20 expression are positively selected during treatment. Deleting A20 in AML leads to increased RIPK1 phosphorylation, the initial step in necroptosis activation, which can be rescued through necroptosis inhibition. Contrary to expectations, restricting NF-κB activation does not rescue A20-deficient AML cells, highlighting that A20’s role in preventing necroptosis is distinct.
Necroptosis is a regulated form of necrosis or inflammatory cell death, distinct from apoptosis31. It plays a role in numerous human diseases, particularly in conditions where apoptosis is dysfunctional, such as cancer, neurodegenerative disorders, and inflammatory conditions48. In cancer, necroptosis can serve as an alternative cell death mechanism, especially in tumors resistant to apoptosis. However, its role is complex, as it operates in a context and cellular-dependent manner and can both suppress and promote tumor growth. In neurodegenerative diseases, such as Alzheimer’s and Parkinson’s, necroptosis contributes to neuronal cell death, exacerbating disease progression. Inflammatory diseases like inflammatory bowel disease and systemic inflammatory response syndrome also involve necroptosis, where it can lead to excessive inflammation and tissue damage. Necroptosis has also been studied in myelodysplastic syndrome (MDS). It has been shown that the necroptotic pathway may contribute to the pathogenesis of MDS by promoting excessive cell death of maturing hematopoietic cells and inflammation in the BM49,50. Of note, A20 levels are higher in HSPCs from MDS patients, which may contribute to the resistance of MDS HSPCs to necroptosis and the suppressive effects of chronic inflammation45. We posit that in AML, the leukemic cells have subverted the ability to activate the necroptosis pathways, such as through elevated expression of A20. Thus, targeting necroptosis with small molecule modulators is emerging as a new approach in cancer therapy, offering the advantage of bypassing apoptosis resistance51. Alternatively, our research suggests that targeting A20 may offer a more direct method of inducing necroptosis selectively in cancer cells and avoiding unwanted necroptosis in normal cells. Since inhibitors targeting A20 do not yet exist, future studies are necessary to identify therapeutic strategies that selectively target the ubiquitin binding and E3 ligase function of A20. This study establishes the novel finding that A20 is essential for AML cells in averting necroptosis, uncovering a potential therapeutic vulnerability.
Methods
Institutional Animal Care and Use Committee and Institutional Review Board
The experiments in this study were performed in compliance with the Institutional Animal Care and Use Committee (IACUC) protocols (2023-0049, 2022-0054). AML samples were obtained from bone marrow (BM) or peripheral blood of patients at initial diagnosis with written informed consent and approval of the institutional review board of Cincinnati Children’s Hospital Medical Center and University of Cincinnati, or from the Eastern Cooperative Oncology Group (ECOG). These AML samples had been obtained within the framework of routine diagnostic BM aspirations after written informed consent in accordance with the Declaration of Helsinki. De-identified leukemic cells from peripheral blood and BM of patients were obtained at CCHMC following consent under the IRB approved Studies (2008-0021, 2017-2224, 2023-0031). Participants were not compensated. Sex or gender information was not available.
Compounds and materials
In vitro 4-hydroxytamoxifen (4-OHT; Sigma, Cat No. H7904), dissolved in 100% EtOH, was performed at a dose of 1 μM for 48–96 h. 100% EtOH was used as the vehicle control. In vivo tamoxifen treatment was performed by intraperitoneally injecting mice with 1 mg of tamoxifen (Sigma, Cat No. T-5648) resuspended in corn oil each day for 5 days. Cells were stimulated with 10 ng/mL IL-1 (Peprotech, Cat No. 200-01B) for 30 min. Cells were incubated with necrostatin-1 (Sigma-Aldrich, Cat No. N9037-25MG) at the indicated doses for 20 min prior to stimulation with other agents. Necroptosis was induced by stimulation with 5 μM z-VAD-fmk (Santa Cruz, Cat No. sc3067), 100 nM Birinapant (MedChemExpress, Cat No. HY-16591), and 200 ng/mL TNF-α (Peprotech, Cat No. 300-01A) for 16 h. Doxorubicin was received from the CCHMC pharmacy and was originally from Pfizer (NDC 0069-3032-20). Cytarabine was purchased from Sigma-Aldrich (Cat No. PHR1787).
Mice
All animal studies were approved by the CCHMC IACUC (protocol no. 2019-0072). Mice were housed at the vivarium at Cincinnati Children’s Hospital under a 14 h light/10 h darkness schedule, 30–70% humidity and at 22.2 ± 1.1 °C. An equal mix of male and female Boy J mice (C57BL/6J; CCHMC), or female NSGS (CCHMC) used in this study were 8–10 weeks old and not involved in previous procedures. All mice were bred, housed, and handled in the Association for Assessment and Accreditation of Laboratory Animal Care-accredited animal facility of Cincinnati Children’s Hospital Medical. A20Rosa Cre-ER mice were a gift from A. Ma (UCSF) and Rip3-/- (B6.129-Ripk3tm1.1Fkmc/J) were purchased from The Jackson Laboratory (Cat No. 030284). Male and female recipient and donor mice were used.
BM transplantation experiments
An equal mix of male and female Boy J mice were lethally irradiated and intravenously injected with whole bone marrow containing 500,000 white blood cells from a healthy Boy J mouse (“helper cells”) along with 500,000 MLL-AF9 leukemic cells. An equal mix of male and female Boy J mice were sub-lethally irradiated and intravenously injected with 250 MN1 leukemic cells. For these leukemia studies, mice transplanted with MLL-AF9 AML cells that appeared hunched, scruffy, lethargic, cytopenic, or distressed were euthanized in accordance with the approved IACUC protocol. Specifically, mice that developed severe anemia (Hg <6gms) or showed signs of excessive pain or discomfort (including limited mobility, hindlimb paralysis, etc.) were removed from the study and euthanized. Mice were euthanized with carbon dioxide following the AVMA Guidelines for the Euthanasia of Animals followed by cervical dislocation. Leukemia burden was assessed on a monthly basis by flow cytometric analysis and complete blood counts. Maximal tumor burden was not exceeded in the studies. If mice do not display signs of disease, mice remained on study and were monitored for up to 2 years. Bone marrow cells were immediately extracted by crushing the leg bones (two iliac crests, two femurs, and one tibia) with a mortar and pestle. Spleens were dissociated by crushing through a 0.45μ filter. Red blood cells in peripheral blood, bone marrow, and dissociated spleen were lysed by incubating for 5 min in 1X PharmLyse buffer (BD Biosciences, Cat No. 555899). Cells were washed twice in 1X PBS and then either assessed for leukemic burden and/or used in secondary transplant. Secondary transplant involved intravenously injecting an equal mix of lethally irradiated male and female Boy J mice with 500,000 helper cells along with 500,000 sorted GFP-positive cells. Leukemic burden was assessed by percent GFP-positive cells by flow cytometry, the presence of blasts in bone marrow and spleen cytospins and peripheral blood smears after Wright-Geimsa staining using an automated slide stainer (Siemens Hematek), appearance of the bone marrow in one tibia, spleen, and liver following suspension in 10% formalin, sectioning, and hematoxylin and eosin staining, spleen weight, and complete blood counts of peripheral blood. For survival analyses, time of death was recorded, and Kaplan Meier survival analysis was performed using GraphPad Prism version 9 for Mac (GraphPad Software, www.graphpad.com).
Xenograft experiments
Non-conditioned primary NSGS mice were transplanted with 250,000 cells and given 1.5 mg/kg Doxorubicin or 1X PBS intravenously for three days on days 22, 23, and 24. Engraftment was checked on days 20 and 31 by femoral bone marrow aspirate. Moribund mice were sacrificed and assessed for leukemic burden as above. For survival analyses, time of death was recorded, and Kaplan Meier survival analysis was performed using GraphPad Prism version 9 for Mac (GraphPad Software, www.graphpad.com).
Cell lines, heathy control CD34+ cells, and patient-derived AML xenograft samples
Adult peripheral blood-mobilized CD34+ cells (healthy control samples) were purchased from the Yale School of Medicine Cooperative Center of Excellence in Hematology Cell Preparation and Analysis Core. Participants were not compensated. MOLT16, F36P, SET2, OCIAML2, and OCIAML3 cells were purchased from DSMZ. MOLM13 were purchased from AddexBio. TF1, HL60, and THP1 cells were purchased from ATCC. MOLM14 (N Shah Lab, UCSF), MV4;11 (HL Grimes Lab, CCHMC), 293T (S. Wells lab, CCHMC), and CD34 + MLL-AF9;FLT3-ITD (J Mulloy Lab, CCHMC) were gifts from other labs. All cell lines were validated. Previously generated PDX models were obtained from the Humanized Mouse Resource of the Cancer & Blood Diseases Institute (CBDI) at Cincinnati Children’s Hospital Medical Center as previously described52. AML samples were RBC lysed and coated with OKT3 antibody (OKT3, BioXCell). Primary NSGS mice were given a single 30 mg/kg intraperitoneal dose of busulfan 24 h prior to intravenous or intrafemoral injection of the OKT3-coated cell preparations. After ~60 days (median 56 days, average 70 days) in xenografted mice, single cell spleen preparations were isolated and cultured in IMDM, 20% FBS, and 10 ng/mL of cytokines (SCF, TPO, FLT3L, IL3, IL-6). MOLM13, MOLM14, and SET-2 were cultured in RPMI-1640 medium (Fisher, Cat No. MT10040CV) with 20% FBS (Biotechne, Cat No. S11550) and 1% penicillin–streptomycin (Fisher, Cat No. NC2077587). MV4;11, MOLT16, and THP1 were cultured in RPMI-1640 medium with 10% FBS and 1% penicillin–streptomycin. TF1 were cultured in RPMI-1640 medium with 10% FBS and 1% penicillin–streptomycin supplemented with 10 ng/ml recombinant human IL-3 (Peprotech, Cat No. 200-03). OCIAML2 and OCIAML3 were cultured with Modified Eagle Medium a (VWR, Cat No. SH30265.FS) with 20% FBS and 1% penicillin–streptomycin. F36P were cultured in RPMI-1640 medium with 20% FBS and 1% penicillin–streptomycin supplemented with 10 ng/ml recombinant human G-CSF (Peprotech, Cat No. 300-23) and 293 T were cultured in Dulbecco Modified Eagle Medium (Fisher, Cat No. SH30022FS) with 10% FBS and 1% penicillin–streptomycin. For short-term in vitro culture, AML PDX cells were maintained in Iscove’s modified Dulbecco’s medium (Cellgro, Cat No. 10-016-CV) containing 20% FBS, 1% penicillin–streptomycin, and 10 ng/mL human SCF, TPO, FLT3L, IL3, and IL6 (Peprotech, Cat No. 300-07, 300-18, 300-19, 200-03, and 200-06).
Plasmids
pMSCV pGK MLL-AF9 GFP was a gift from A. Kumar lab (CCHMC). pLKO.1 puro shp53 was purchased from Dharmacon (Cat No. RMM3981-201743939), where puro was replaced with GFP from pLKO shControl-GFP (Sigma, Cat No. SHC202) using BamHI and KpnI restriction sites. pLKO.1 shTNFAIP3/A20 puro-1 (TCRN0000050958) and pLKO.1 shTNFAIP3/A20 puro-2 (TCRN0000050959) were purchased from the CCHMC shRNA library. In both plasmids, puro was replaced with GFP from pLKO shControl-GFP (Sigma, Cat No. SHC202) using BamHI and KpnI restriction sites. pLVET-HA-KRAS-G12V-IRES-GFP was a gift from M. Azam (CCHMC). pSF91-MN1-GFP was a gift from F. Kuchenbauer (BC Cancer Research Centre). pSP72-E-hA20 (Cat No. 5931), pSP72-E-hA20-ZF4/ZF7 (Cat No. 5934), and pCAGGS-E-hA20 C103S (Cat No. 5355) were purchased from BCCM/LMBP. hA20 sequences were subconed into pMSCV-IRES-mCherry using XhoI and EcoRI restriction sites.
Immunoblot
Protein extracts were prepared by lysing 1e6 cells in 50 uL sodium dodecyl sulfate (SDS) sample buffer containing benzonase (Millipore Sigma, 70746) and incubating on ice for 20 min. Samples were boiled at 95 °C for 5 min, separated by SDS-polyacrylamide gel electrophoresis (BIO-RAD), and transferred to nitrocellulose membranes (BIO-RAD, 162-0112). Immunoblot analysis was performed with the antibodies listed in the Key Resource Table (Supplementary Data 7). Blots were developed using ECL Western Blotting Substrate (Pierce, Cat No. 32106) or SuperSignal West Femto Substrate (Thermo Scientific, Cat No. 34096) and imaged on a BIO-RAD ChemiDoc Touch Imaging system.
PCR
For quantitative PCR, total RNA was extracted and purified using Quick-RNA MiniPrep (Zymo research, Cat No. R1055) and reverse transcription was carried out using High-Capacity cDNA Reverse Transcription Kit (Thermo-Fisher, Cat No. 4387406). Quantitative RT–PCR was performed using Taqman probes to A20 (human and mouse, Applied Biosystems, Cat No. 4331182) and GAPDH (human, Applied Biosystems, 4351370; mouse, Life Tech, 4331182) along with the Taqman Master Mix (Life Tech, Cat No. 4324020). For genotyping, genomic DNA was extracted by incubating cells with 4 parts 40 mM NaOH at 95 °C for 15 min, followed by addition of 1 part 10 mM Tris-HCl, pH 6.8. Polymerase chain reaction (PCR) was performed on the genomic DNA using primers flanking the DNA sequence containing flox sites and PCR master mix (Life Tech, Cat No. 4359187). Thermocycler conditions used were the PCR master mix manufacturer’s recommendations.
Flow cytometry
Red blood cells in peripheral blood, bone marrow, and dissociated spleen were lysed by incubating for 5 min in 1X PharmLyse buffer (BD Biosciences, Cat No. 555899). After washing twice in 1X PBS, cells were resuspended in 2% BSA/1X PBS and analyzed by flow cytometry on a BD Biosciences FACSCanto. Cells growing in culture were washed twice in 1X PBS, and either resuspended in 2% BSA/1X PBS and analyzed by flow cytometry on a BD Biosciences FACSCanto or resuspended in Sorting Buffer (Ca/Mg2+-free 1X PBS, 2% BSA, 25 mM HEPES pH 7.0, 1 mM EDTA) and sorted for live (7AAD-negative), GFP-positive cells on a BD Biosciences FACSAria. For viability assays, cells were stained with Annexin V and 7AAD using a BD Pharmingen kit (Cat No. 559763) according to the manufacturer’s instructions.
Growth assays
Cells were initially plated at a uniform density in 12-well plates. Cells were counted every other day by mixing 1-to-1 with trypan blue (Fisher, Cat No. ICN1691049) and quantifying trypan blue-negative cells on a Countess II automated cell counter (Life Technologies). Cells were split to maintain appropriate confluency, and dilution factors were recorded to adjust cell counts. In growth assays involving drug treatment, cells were collected and resuspended in fresh media with drug every other day.
Colony forming assays
One thousand mouse BM cells per replicate were plated in methylcellulose (Stemcell Technologies, Cat No. M3434) in triplicate. Colonies propagated in culture were scored at day 7, and for longitudinal studies, were pooled and replated at 1.0 × 103 cells per replicate into the same medium and scored after 7 days. For human cell lines (MOLM14, MV4;11), 250 cells per replicate were plated in methylcellulose (Stemcell Technologies, Cat No. H4434) in triplicate. Colonies were quantified after 7–10 days using the STEMvision counter (StemCell Technologies).
Lactate dehydrogenase (LDH) release assay
Cells were plated at a uniform density and then treated with the indicated inhibitors. The LDH-GloTM cytotoxicity assay (Promega, Cat No. J2380) was performed according to the manufacturer’s protocol.
RNA-sequencing and analysis
Total RNA was extracted from MLL-AF9 cells that were treated with 4-OHT (1 μM) for 48 h in triplicate using a Quick-RNA MiniPrep kit (Zymo Research, Cat No. R1055). Sequencing results were demultiplexed and converted to FASTQ format using Illumina bcl2fastq software. The sequencing reads were aligned to the mouse genome (mm10/GRCm38) using the HISAT2 (v.2.2.1) (http://daehwankimlab.github.io/hisat2/) aligner. The featureCounts (v.2.0.3) (https://subread.sourceforge.net/) was utilized to generate counts for each gene based on how many aligned reads overlap its exons. These counts were then normalized and used to test for differential expression using edgeR package (https://bioconductor.org/packages/release/bioc/html/edgeR.html) RNA libraries were prepared according to the Illumina TruSeq Stranded mRNA (polyA capture) library protocol by the DNA Sequencing and Genotyping Core at CCHMC. The analysis of RNA sequencing was performed with iGEAK53. For differential gene expression analysis using limma-voom, P values were computed using the moderated t-test assuming a normal distribution of transformed/normalized/smoothed gene counts. Gene set enrichment analysis (GSEA) was performed as previously described54. Transcription factor enrichment analysis was performed using Enrichr55. P values were computed from Fisher’s exact test.
Virus production and transduction
To generate virus, transfection of 293T cells (for transducing human cells) or PlatE cells (for transducing mouse cells) with viral packaging vectors and transfer plasmid was performed using TransIT®-LT1 Transfection Reagent (Mirus, Cat No. MIR2305) according to the manufactures’ recommendation. Viral packaging plasmids pMICK (2 μg) and pCLDN (6 μg) for lentivirus production or M57 (10 μg) and RD114 (3 μg) for retrovirus production were used with 12 μg of transfer plasmid. The following day, the media was exchanged. After 24 h, viral supernatant was collected, filtered using a 0.45μ filter, and stored at −80 °C. Cells were transduced by incubation with viral supernatant and 8 μg/mL of polybrene (Millipore, Cat No. TR-1003-G) overnight. GFP-positive cells were either sorted 72 h post-transduction or, in the case of MLL-AF9- and MN1-transduced cells, expanded in M3434 methylcellulose every 7 days for 3 weeks. Methylcellulose-expanded cells were analyzed by flow cytometry to ensure >95% GFP-positive cells.
Generation of CRISPR/Cas9 mutant cells
To generate the RIPK3KO clones, MV4;11 cells were suspended in buffer R with Cas9-NLS and a modified synthetic gRNA targeting exon 2 of RIPK3 (Synthego; CCGCCCCCTTGGTGTCCATCGAGGAACT) and electroporated (1700 mV × 20 ms × 1 pulse) using the Neon Transfection system (Invitrogen). Transfected cells were recovered for 48 h in antibiotic-free RPMI-1640 with 1% FBS. Following recovery, transfected cells were plated in 96 well plates at a target density of 0.3 cells/well to isolate single clones. Clones were expanded and screened for RIPK3 deletion by immunoblotting. Deletion was confirmed by PCR amplification of the PAM site for Sanger sequencing.
Transmission electron microscopy
Two million cells were collected, washed twice in 1X PBS, and resuspended in 3% glutaraldehyde fixative. Cells were sectioned into thin sections by the CCHMC Integrated Pathology Research Facility (RRID:SCR_022637). Cells were imaged on a transmission electron microscope (Hitachi).
Statistics and reproducibility
The number of animals, cells, and experimental/biological/technical replicates can be found in the figure legends. Based on our extensive experience, mouse experiments were performed using >5 recipients per condition to detect 65% relative treatment differences with 80% power at a significance level of 0.05. Differences among multiple groups were assessed by one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison posttest for all possible combinations. Comparison of two group was performed using a Student’s t test (unpaired, two-tailed). Unless otherwise specified, results are depicted as the mean ± standard deviation. For correlation analysis, Pearson correlation coefficient (r) was calculated. D’Agostino and Pearson and Shapiro–Wilk tests were performed to assess data distributions. For Kaplan–Meier analysis, Mantel–Cox test was used. GraphPad Prism (version 10.1.1) was used for statistical analysis. For correlative analyses, Spearman rank test was used. The number of animals, cells, and experimental/biological/technical replicates can be found in the figure legends. Differences among multiple groups were assessed by one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison posttest for all possible combinations. Comparison of two group was performed using the Mann–Whitney test or the Student’s t test (unpaired, two tailed) when sample size allowed. Unless otherwise specified, results are depicted as the mean ± standard deviation or standard error of the mean. A normal distribution of data was assessed for data sets >30. For correlation analysis, Pearson correlation coefficient (r) was calculated. For Kaplan–Meier analysis, Mantel–Cox test was used. All graphs and analyses were generated using GraphPad Prism software. No data were excluded from the analyses. The experiments were not randomized, and the investigators were not blinded during experiments or outcome assessments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Information
Source data
Acknowledgements
We thank J Bailey and V Summey for assistance with transplantations (CCHMC Comprehensive Rodent and Radiation Facility; RRID:SCR_022624). We thank the Viral Vector Facility (RRID:SCR_022641), the Research Flow Cytometry Facility (RRID:SCR_022635) and DNA sequencing and Genotyping Core at CCHMC. We thank Deedra Nicolet and Ann-Kathrin Eisfeld (The Ohio State University) for providing the link and assistance with published data from the Alliance for Clinical Trials in Oncology. We thank members of the Starczynowski lab, T Chlon lab, L Jones lab, and L Lee lab and Dr. Douglas Green for helpful discussion and suggestions. This work was supported in parts by the National Institute of Health (U54DK126108, R35HL135787, R01CA275007) to DTS, Cincinnati Children’s Hospital Research Foundation to DTS, and Cancer Free Kids grants to DTS and AEC.
Author contributions
A.E.C. and A.H. performed the experiments. K.H. oversaw the mouse colonies and performed procedures on live mice (injections, bleeds, health monitoring). K.C. analyzed TCGA-AML, BEAT AML, and RNA sequencing data. A.M. generated the A20 Rosa Cre-ER mouse model. B.V., E.O., M.W., and J.P.P. developed the AML PDX samples and performed drug sensitivity assays and RNA sequencing. A.E.C. and D.T.S. analyzed the data, interpreted the results, and wrote the manuscript. D.T.S. supervised the project and conceived of the original idea. All authors discussed the results and had the opportunity to contribute to the manuscript.
Peer review
Peer review information
Nature Communications thanks Anna Eiring, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Source data are provided with this paper. The RNA sequencing data generated in this study has been deposited at NCBI’s GEO repository with accession number GSE250372. Publicly available RNA-sequencing data of AML patients were downloaded from The Cancer Genome Atlas via cBioPortal (cbioportal.org) (GSE68833) and BEAT AML (Vizome, http://www.vizome.org/aml/). Publicly available microarray data of patients with AML along with healthy controls were downloaded from GSE35008 and GSE35010. Alliance AML patient response to treatment and TNFAIP3 expression were obtained from the Publicly available Alliance Statistics and Data Management Center (https://www.allianceforclinicaltrialsinoncology.org)(GSE137851). Source data are provided with this paper.
Competing interests
DTS serves on the scientific advisory board at Kurome Therapeutics and is a consultant for and/or received funding from Kurome Therapeutics, Captor Therapeutics, Treeline Biosciences, and Tolero Therapeutics. DTS has equity in Kurome Therapeutics. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53629-z.
References
- 1.Rowe, J. M. The “7+3” regimen in acute myeloid leukemia. Haematologica107, 3 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thol, F., Schlenk, R. F., Heuser, M. & Ganser, A. How I treat refractory and early relapsed acute myeloid leukemia. Blood126, 319–327 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Papaemmanuil, E. et al. Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med.374, 2209–2221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thol, F. Can we forecast induction failure in acute myeloid leukemia? Haematologica103, 375–377 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burnett, A., Wetzler, M. & Lowenberg, B. Therapeutic advances in acute myeloid leukemia. J. Clin. Oncol.29, 487–494 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Niu, J., Peng, D. & Liu, L. Drug resistance mechanisms of acute myeloid leukemia stem cells. Front. Oncol.12, 896426 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McNeer, N. A. et al. Genetic mechanisms of primary chemotherapy resistance in pediatric acute myeloid leukemia. Leukemia33, 1934–1943 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider, C. et al. SAMHD1 is a biomarker for cytarabine response and a therapeutic target in acute myeloid leukemia. Nat. Med.23, 250–255 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Mrozek, K., Radmacher, M. D., Bloomfield, C. D. & Marcucci, G. Molecular signatures in acute myeloid leukemia. Curr. Opin. Hematol.16, 64–69 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng, S. W. et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature540, 433–437 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Theilgaard-Monch, K. et al. Gene expression profiling in MDS and AML: potential and future avenues. Leukemia25, 909–920 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Valk, P. J. et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N. Engl. J. Med.350, 1617–1628 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Walker, C. J. et al. Gene expression signature predicts relapse in adult patients with cytogenetically normal acute myeloid leukemia. Blood Adv.5, 1474–1482 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wouters, B. J., Lowenberg, B. & Delwel, R. A decade of genome-wide gene expression profiling in acute myeloid leukemia: flashback and prospects. Blood113, 291–298 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Francesco, B., et al. NF-kappaB: a druggable target in acute myeloid leukemia. Cancers14, 3557 (2022). [DOI] [PMC free article] [PubMed]
- 16.Zhou, J., Ching, Y. Q. & Chng, W. J. Aberrant nuclear factor-kappa B activity in acute myeloid leukemia: from molecular pathogenesis to therapeutic target. Oncotarget6, 5490–5500 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilmore, T., Gapuzan, M. E., Kalaitzidis, D. & Starczynowski, D. Rel/NF-kappa B/I kappa B signal transduction in the generation and treatment of human cancer. Cancer Lett.181, 1–9 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Taniguchi, K. & Karin, M. NF-kappaB, inflammation, immunity and cancer: coming of age. Nat. Rev. Immunol.18, 309–324 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Godwin, P. et al. Targeting nuclear factor-kappa B to overcome resistance to chemotherapy. Front. Oncol.3, 120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosman, M. C., Schuringa, J. J., Quax, W. J. & Vellenga, E. Bortezomib sensitivity of acute myeloid leukemia CD34(+) cells can be enhanced by targeting the persisting activity of NF-kappaB and the accumulation of MCL-1. Exp. Hematol.41, 530–538 e531 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Paz-Priel, I., Ghosal, A. K., Kowalski, J. & Friedman, A. D. C/EBPalpha or C/EBPalpha oncoproteins regulate the intrinsic and extrinsic apoptotic pathways by direct interaction with NF-kappaB p50 bound to the bcl-2 and FLIP gene promoters. Leukemia23, 365–374 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duy, C. et al. Chemotherapy induces senescence-like resilient cells capable of initiating AML recurrence. Cancer Discov.11, 1542–1561 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato, M. et al. Frequent inactivation of A20 in B-cell lymphomas. Nature459, 712–716 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Zhao, D. et al. TP53 mutations in AML patients are associated with dismal clinical outcome irrespective of frontline induction regimen and allogeneic hematopoietic cell transplantation. Cancers15, 3210 (2023). [DOI] [PMC free article] [PubMed]
- 25.Crowley, L. C., Marfell, B. J., Scott, A. P. & Waterhouse, N. J. Quantitation of apoptosis and necrosis by annexin V binding, propidium iodide uptake, and flow cytometry. Cold Spring Harb. Protoc.2016, pdb-prot087288 (2016). [DOI] [PubMed]
- 26.Liston, D. R. & Davis, M. Clinically relevant concentrations of anticancer drugs: a guide for nonclinical studies. Clin. Cancer Res.23, 3489–3498 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith, M. A. et al. TNFAIP3 plays a role in aging of the hematopoietic system. Front. Immunol.11, 536442 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malynn, B. A. & Ma, A. A20: A multifunctional tool for regulating immunity and preventing disease. Cell Immunol.340, 103914 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martens, A. & van Loo, G. A20 at the crossroads of cell death, inflammation, and autoimmunity. Cold Spring Harb. Perspect. Biol.12, a036418 (2020). [DOI] [PMC free article] [PubMed]
- 30.Razani, B. et al. Non-catalytic ubiquitin binding by A20 prevents psoriatic arthritis-like disease and inflammation. Nat. Immunol.21, 422–433 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinlich, R., Oberst, A., Beere, H. M. & Green, D. R. Necroptosis in development, inflammation and disease. Nat. Rev. Mol. Cell Biol.18, 127–136 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Hadian, K. & Stockwell, B. R. The therapeutic potential of targeting regulated non-apoptotic cell death. Nat. Rev. Drug Discov.22, 723–742 (2023). [DOI] [PubMed] [Google Scholar]
- 33.Grootjans, S., Vanden Berghe, T. & Vandenabeele, P. Initiation and execution mechanisms of necroptosis: an overview. Cell Death Differ.24, 1184–1195 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koo, G. B. et al. Methylation-dependent loss of RIP3 expression in cancer represses programmed necrosis in response to chemotherapeutics. Cell Res.25, 707–725 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Galen, P. et al. Single-Cell RNA-Seq reveals AML hierarchies relevant to disease progression and immunity. Cell176, 1265–1281.e1224 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guzman, M. L. et al. Nuclear factor-kappaB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood98, 2301–2307 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Ramadass, V., Vaiyapuri, T. & Tergaonkar, V. Small molecule NF-kappaB pathway inhibitors in clinic. Int. J. Mol. Sci.21, 5164 (2020). [DOI] [PMC free article] [PubMed]
- 38.Zhu, S. et al. RIPK3 deficiency blocks R-2-hydroxyglutarate-induced necroptosis in IDH-mutated AML cells. Sci. Adv.10, eadi1782 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei, Y. et al. Targeting Bcl-2 proteins in acute myeloid leukemia. Front Oncol.10, 584974 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McComb, S. et al. Activation of concurrent apoptosis and necroptosis by SMAC mimetics for the treatment of refractory and relapsed ALL. Sci. Transl. Med.8, 339ra370 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Mezzatesta, C. & Bornhauser, B. C. Exploiting necroptosis for therapy of acute lymphoblastic leukemia. Front Cell Dev. Biol.7, 40 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Onizawa, M. et al. The ubiquitin-modifying enzyme A20 restricts ubiquitination of the kinase RIPK3 and protects cells from necroptosis. Nat. Immunol.16, 618–627 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakagawa, M. M. & Rathinam, C. V. A20 deficiency in hematopoietic stem cells causes lymphopenia and myeloproliferation due to elevated Interferon-gamma signals. Sci. Rep.9, 12658 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakagawa, M. M., Davis, H. & Rathinam, C. V. A20 deficiency in multipotent progenitors perturbs quiescence of hematopoietic stem cells. Stem Cell Res.33, 199–205 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muto, T. et al. Adaptive response to inflammation contributes to sustained myelopoiesis and confers a competitive advantage in myelodysplastic syndrome HSCs. Nat. Immunol.21, 535–545 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jakobsen, N. A. et al. Selective advantage of mutant stem cells in human clonal hematopoiesis is associated with attenuated response to inflammation and aging. Cell Stem Cell31, 1127–1144.e17 (2024). [DOI] [PubMed]
- 47.Jakobsen, I., Sundkvist, M., Bjorn, N., Green, H. & Lotfi, K. Early changes in gene expression profiles in AML patients during induction chemotherapy. BMC Genomics23, 752 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ye, K., Chen, Z. & Xu, Y. The double-edged functions of necroptosis. Cell Death Dis.14, 163 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner, P. N. et al. Increased Ripk1-mediated bone marrow necroptosis leads to myelodysplasia and bone marrow failure in mice. Blood133, 107–120 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Croker, B. A. & Kelliher, M. A. BID-ding on necroptosis in MDS. Blood133, 103–104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, Y., Dong, G. & Sheng, C. Targeting necroptosis in anticancer therapy: mechanisms and modulators. Acta Pharm. Sin. B10, 1601–1618 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barreyro, L. et al. Blocking UBE2N abrogates oncogenic immune signaling in acute myeloid leukemia. Sci. Transl. Med.14, eabb7695 (2022). [DOI] [PubMed] [Google Scholar]
- 53.Choi, K. & Ratner, N. iGEAK: an interactive gene expression analysis kit for seamless workflow using the R/shiny platform. BMC Genomics20, 177 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform.14, 128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gislason, M. H. et al. BloodSpot 3.0: a database of gene and protein expression data in normal and malignant haematopoiesis. Nucleic Acids Res.52, D1138–D1142 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Information
Data Availability Statement
Source data are provided with this paper. The RNA sequencing data generated in this study has been deposited at NCBI’s GEO repository with accession number GSE250372. Publicly available RNA-sequencing data of AML patients were downloaded from The Cancer Genome Atlas via cBioPortal (cbioportal.org) (GSE68833) and BEAT AML (Vizome, http://www.vizome.org/aml/). Publicly available microarray data of patients with AML along with healthy controls were downloaded from GSE35008 and GSE35010. Alliance AML patient response to treatment and TNFAIP3 expression were obtained from the Publicly available Alliance Statistics and Data Management Center (https://www.allianceforclinicaltrialsinoncology.org)(GSE137851). Source data are provided with this paper.