Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) is associated with Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. The latency-associated nuclear antigen (LANA) is highly expressed in these malignancies and has been shown to play an important role in episomal maintenance, presumably by binding to a putative oriP. In addition, LANA modulates cellular and viral gene expression and interacts with the cellular tumor suppressors p53 and retinoblastoma suppressor protein. Many of these features are reminiscent of Epstein-Barr virus nuclear antigens (EBNAs), a family of six proteins expressed during latency. EBNA-1 is required for episome maintenance, binds to oriP, and strongly activates transcription from two promoters, including its own. We have previously shown that LANA can transactivate its own promoter and therefore asked whether LANA, like EBNA-1, activates transcription by direct binding to DNA. By using recombinant LANA expressed from vaccinia virus vectors for electrophoretic mobility shift assays, we found that LANA does not bind to its own promoter. In contrast, LANA binds specifically to sequences containing an imperfect 20-bp palindrome in the terminal repeat (TR) of KSHV. We further show that the C-terminal domain of LANA is sufficient for site-specific DNA binding. Unlike EBNA-1, which activates transcription through binding of oriP, we found that LANA inhibits transcription from a single TR binding site. A multimerized TR as found in the viral genome results in strong transcriptional suppression when linked to a heterologous promoter. These data suggest that LANA, although fulfilling functions similar to those of EBNA-1, does so by very different mechanisms.
Kaposi's sarcoma-associated herpes virus (KSHV), also called human herpesvirus 8, is associated with Kaposi's sarcoma (KS), primary effusion lymphoma (PEL), and a plasmablastic variant of multicentric Castleman's disease (7, 8, 12). In all of these malignancies, the vast majority of tumor cells are latently infected at a stage during which only a small subset of viral genes are expressed (6, 51, 54). Of these genes, the latency-associated nuclear antigen encoded by ORF73 (LANA) is the only protein consistently shown to be highly expressed in all KS-associated malignancies by in situ hybridization and immunohistochemistry (13, 22, 23, 40). By gene expression profiling and transient transfection assays, we have previously shown that LANA modulates viral and cellular gene expression. In addition, it was demonstrated that LANA induces its own synthesis (25, 43). The LANA promoter also regulates the synthesis of v-cyclin and v-FLICE, two gene products with potential roles in cellular growth deregulation (11, 20, 34).
Based on primary sequence, LANA can be divided into three domains: a conserved proline- and serine-rich N-terminal region, a central region which is variable in length and composed of several acidic repeats including a leucine zipper motif, and a conserved C-terminal domain containing a proline-rich region and a region rich in charged and hydrophobic amino acids.
LANA seems to have multiple functions vital to viral latency. It is capable of acting in trans to maintain a plasmid containing three copies of the KSHV terminal repeat (TR) and 300 bp of viral unique sequence (5). In addition, Cotter et al. showed that DNA fragments containing the TR region can be coimmunoprecipitated with LANA-specific antibodies (9). We and others have shown that LANA modulates cellular gene expression positively and negatively and also activates transcription from its own promoter (25, 43). There is evidence that LANA is able to modulate transcription through two distinct mechanisms: interaction with upstream transcriptional regulators and direct binding of DNA. LANA has been shown to interact with p53, thereby down-regulating p53-dependent transcription (16). Recently, Radkov et al. demonstrated direct interaction of LANA with the retinoblastoma suppressor protein (pRb), leading to activation of E2F-dependent transcription (39). LANA's ability to control transcription by direct DNA binding has been shown only indirectly by using Gal4 fusion proteins together with reporters that contained multiple Gal4 binding sites. First, full-length LANA fused to a Gal4 DNA binding domain represses transcription from a Gal4-dependent promoter up to 10-fold (25). In addition, both N-terminal and C-terminal domains have been shown to down-regulate transcription of this reporter (48). In the case of the N-terminal domain, the effect seems to be due to localization of the mSin3 corepressor complex to the bound DNA (25). These two distinct mechanisms of transcriptional modulation, interaction with upstream coregulators and direct DNA binding, are similar to mechanisms employed by two latency-associated proteins of Epstein Barr virus (EBV), the closest human relative of KSHV. EBNA-2 modulates transcription of both cellular and viral promoters through interaction with several transcription factors and coactivators but does not bind directly to DNA (28). EBNA-1 binds to a DNA element called the family of repeats and an upstream dyad symmetry element in the EBV oriP (2, 41). This binding is necessary for episomal replication and maintenance in vivo (37, 50). A second consequence of EBNA-1 binding to oriP is strong transcriptional activation of the LMP-1 and BamHI C promoters (17, 38, 52). Placing these cis-regulatory elements upstream from a heterologous promoter confers strong EBNA-1-dependent transcriptional activation (42). In addition to binding oriP, EBNA-1 activates its own synthesis by binding at two specific regions in the BamHI Q promoter (47).
Here we provide data showing that LANA does not activate its own promoter through direct binding. We map a cis-regulatory LANA binding element within the TR and show that the C-terminal domain of LANA is sufficient for specific DNA binding. We also show that the LANA binding site does not confer LANA-dependent transcriptional activation, but instead confers suppression, to a heterologous promoter. Together, these data suggest that LANA modulates transcription through different mechanisms than the EBNA proteins of EBV.
MATERIALS AND METHODS
Plasmids.
pTR1, pTR2, and pTR3 were gifts from Mike Lagunoff. pTR1 was created by digesting pML1 (26) with NotI/SrfI and ligating it into NotI/EcoRV of pBluescriptII (pBSII) KS. pTR2 was created from an SrfI/AscI fragment of pML1 cloned into EcoRV of pBSII KS. pTR3 was created from an AscI/NotI fragment cloned into EcoRV/NotI of pBSII KS. pCRII/TR contains the NotI fragment of pML1 cloned into the NotI site of pCRII. pAG4 contains an XmaI fragment from the cosmid Z6 derived from the PEL cell line BC-1 (46) cloned into pBSII KS. pAG9 was created ligating the 114-bp-long ApaI fragment of pML1 into pBSII KS.
pCDNA3/orf73 has been described earlier (43). pEETM1/LANA was created by first cloning the 3′ portion of ORF73, NcoI/AccI, from pCDNA3/orf73 into pEETM1 (a gift from D. Templeton, (Case Western Reserve University). The 5′ portion of ORF73 was amplified by PCR using primers which engineered an additional NcoI site into the translation initiation site. To avoid introduction of mutations, PCR amplifications were performed with the Pfx polymerase, which has proofreading activity (Gibco-BRL), and primers 5′-TCAGACCAGATTTCCCGACCATGG-3′ and 5′-GGAATTCATCATCCTTATTGTCATTGTC-3′, which created an NcoI site at the 5′ terminus of ORF73. This PCR fragment was then cut with NcoI and cloned into the vector pEETM1 containing the 3′ portion of ORF73.
ORF73 deletion mutants were created by PCR, and amplification products were ligated into the pCR-Blunt II Topo vector (Invitrogen, Carlsbad, Calif.). From these vectors, fragments were then created by either EcoRI/XhoI or KpnI/XhoI insertion into the T7 promoter-containing vector pCDNA3.1V5HISA or pCDNA3.1V5HISC, which also contained a C-terminal V5 tag. Primer pairs used for each mutant are as follows: pcDNA3.1V5HISA/orf73A, 5′-GGGGTACCAGATTTCCCGAGGATGG-3′ plus 5′-GGAATTCATCATCCTTATTGTCATTGTC-3′; pcDNA3.1V5HISA/orf73AB, 5′-GGGGTACCAGATTTCCCGAGGATGG-3′ plus 5′-CCGCTCGAGTGTCATTTCC-3′; pcDNA3.1V5HISA/orf73B, 5′-CCATGGACAATGACAATAA-3′ plus 5′-CCGCTCGAGTGTCATTTCC-3′; pCDNA3.1V5HISA/orf73BC, 5′-CCATGGACAATGACAATAA-3′ plus 5′-CCGCTGGAGTGTCATTTCCTGTGGAGAGT-3′; pcDNA3.1V5HISA/orf73C, 5′-CCATGGAAGAGCCCATAAT-3′ plus 5′-CCGCTGGAGTGTCATTTCCTGTGGAGAGT-3′; pcDNA3.1V5HISA/orf73AC, 5′-GGGGTACCAGATTTCCCGAGGATGG-3′ plus 5′-GGAATTCATCATCCTTATTGTCATTGTC-3′ and 5′-GGAATTCGAAGAGCCCATAATCTTG-3′ plus 5′-CCGCTGGAGTGTCATTTCCTGTGGAGAGT-3′ (pcDNA3.1V5HISA/orf73AC required two PCRs with an EcoRI linker engineered between the two; this linker inserted a single amino acid, phenylalanine, between domains A [N terminal] and [C terminal]).
Cell lines.
CV-1 cells, African green monkey fibroblasts, and 293 (human embryonic kidney) cells were obtained from the American Type Culture Collection. Cell monolayers were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin at 37°C under a 5% CO2 atmosphere. BJAB, an EBV-negative Burkitt's lymphoma line (kindly provided by Elliot Kieff, Harvard University), BJAB/73 (43), and BCBL-1 (45) cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum, 0.05 mM β-mercaptoethanol, 1 mM sodium bicarbonate, and 2 mM l-glutamine, penicillin, and streptomycin at 37°C under a 5% CO2 atmosphere.
Expression of recombinant full-length and mutant LANA proteins with the MVA/T7 expression system.
Full-length and mutant proteins were produced by using the modified vaccinia virus T7 (MVA/T7) expression system (53). CV-1 cells were infected with MVA/T7 as previously described (10, 35, 53). Briefly, a confluent 10-cm-diameter plate of CV-1 cells was split 1 to 2 12 h prior to infection with MVA-T7 at an approximate multiplicity of infection of 10. Cells were transfected 1 h postinfection with 1 μg of each plasmid, using Effectine (Qiagen, Valencia, Calif.) as instructed by the manufacturer. Nuclear protein extractions were performed 24 to 36 h posttransfection as previously described (3). Briefly, approximately 107 cells were collected by scraping and lysed in 1 ml of buffer A (20 mM HEPES-KOH [pH 7.9], 25% glycerol, 10 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol [DTT], 0.2 mM phenylmethylsulfonyl fluoride). The nuclei were spun down, and the pellet was lysed in 100 μl of buffer B (20 mM HEPES-KOH [pH 7.9], 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM phenylmethylsulfonyl fluoride). This suspension was then centrifuged, and the supernatant (nuclear extracts) was collected and used in electrophoretic mobility shift assays (EMSA) after protein concentration were determined by Bradford assays (Bio-Rad, Hercules, Calif.).
Western blot analysis.
Protein extracts were electrophoretically separated on sodium dodecyl sulfate–8% polyacrylamide gels. Proteins were transferred to nylon membranes (Amersham, Piscataway, N.J.) by using a Mini-Trans blot unit (Bio-Rad) in transfer buffer containing 25 mM Tris-HCl (pH 8.3), 12 mM glycine, and 10% methanol. Membranes were blocked for 2 h in Tween–Tris-buffered saline (TTBS) containing 5% dry milk. Primary antibodies were diluted in TTBS, and membranes were incubated for 1 h at room temperature. After washing, membranes were incubated with secondary antibody for 1 h. Primary antibodies were either a polyclonal rabbit antiserum specific for the repeat region of LANA (a gift from Andy Polson and Don Ganem) or a monoclonal mouse antibody against the V5 epitope tag (Invitrogen). Peroxidase-conjugated secondary antibodies were either goat anti-rabbit or goat anti-mouse and were diluted 1 to 7,500 prior to incubation. After final washes, Western blots were developed with the Pierce SuperSignal detection system and exposed to Kodak film.
EMSA.
Probes were labeled with Klenow fragment using [α-32P]dCTP (3,000 Ci/mmol; Amersham) as instructed by the manufacturer (Promega, Madison, Wis.). To purify the probe from nonincorporated nucleotides, we used Sephadex 50 spin columns (Boehringer, Indianapolis, Ind.). The fragment for the LANA promoter was generated by digesting pDD83 (11) with PvuII and NheI. The fragments TR1, TR2, and TR3 were generated by digesting the corresponding plasmids pTR1, pTR2, and pTR3 (a kind gift from Michael Lagunoff) with NotI and XhoI. TR4 was generated by digesting pCRII-TR with EagI. TR5 was made by digesting pAG9 with XhoI and KpnI. TR6 was made by digesting pAG4 with XmaI. The 36-bp-long probe TR7 was produced by a NarI/ApaI digest of the PvuII fragment released from pAG9. TR8 was created by annealing two oligonucleotides, 5′-CCCCATGCCCGGGCGGGAGG-3′ and 5′-CCTCCCGCCCGGGCATGGGG-3′. The TR8 fragment was labeled by incubation at 37°C with 5 U of T4 polynucleotide kinase and 100 μCi [γ-32P]ATP. For each lane, about 40,000 cpM of labeled fragment was combined with 0.5 to 1 μg of MVA/T7-infected CV-1 nuclear extract or 5 μg of BCBL-1 or BJAB nuclear extract. The protein extracts were incubated at room temperature for 25 min in a total volume of 20 μl of buffer which contained 10 mM HEPES (pH 7.9), 50 mM KCl, 1 mM EDTA, poly(dI-dC) (0.05 μg/μl), bovine serum albumin (0.5 μg/μl), 10 mM DTT, and 10% glycerol. The samples were then separated by electrophoresis through a native 4% polyacrylamide gel (55 mA in a room kept at 4°C). Gels were dried and either exposed to Kodak film or analyzed with a Storm PhosphorImager.
Transient transfection assays.
293 cells were plated at a density of 4 × 105 cells per well in six-well plates 8 to 12 h prior to transfection. Reporter and effector plasmids were transfected using Fugene (Gibco-BRL) according to the manufacturer's instructions. To monitor transfection efficiency, we transfected pcDNA3/lacZ into parallel wells and stained fixed cells for β-galactosidase activity. Transfection efficiencies were generally between 25 and 35%. We did not use internal standards in each transfection for normalization because the presence of LANA can influence a wide range of reporters, as we have previously shown (43). To normalize, we performed Bradford assays on all lysates and normalized relative light unit values to the protein concentrations as previously reported for other proteins which have the ability to affect a wide range of different reporters in transient transfection assays (49).
RESULTS
Recombinant LANA expression using the MVA/T7 expression system.
Initially, we attempted to study the DNA binding properties of LANA by using BCBL-1 extracts. Using total cellular or nuclear BCBL-1 extracts combined with relatively large DNA probes to screen for LANA binding sites in the LANA promoter resulted in multiple bands of various intensities (data not shown).
Therefore, we chose to utilize a vaccinia virus T7 polymerase expression system to ectopically express LANA. MVA/T7 is a highly attenuated and avian-host-range-restricted vaccinia virus which allows high T7 polymerase-dependent gene expression. Its decreased cytopathic effect in mammalian nonpermissive cells yields more protein and adds additional biosafety to this expression system (53). LANA is posttranslationally modified, which may effect its function (36, 40; A. Polson and D. Ganem, unpublished results). It has previously been shown that vaccinia virus-expressed proteins are posttranslationally modified with respect to N- and O-linked glycosylation, phosphorylation, myristyolation, cleavage, and assembly (14).
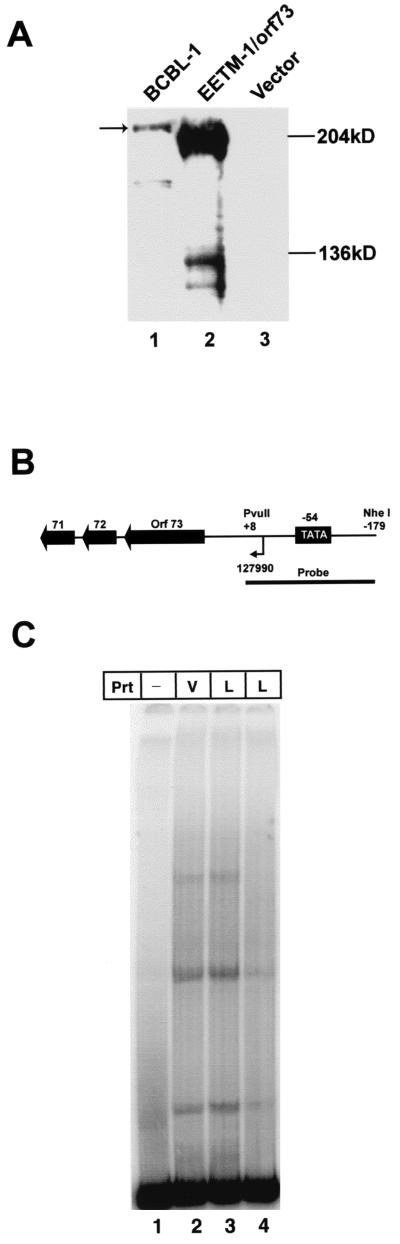
First, we cloned the entire coding region of LANA into pEETM, which contains an N-terminal EE tag and an internal ribosomal entry site, which greatly increases translation efficiency (10). Full-length EE-tagged LANA was produced by transfecting pEETM/LANA into CV-1 cells 1 h after infection with MVA/T7 as described in Materials and Methods (MVA/T7 was a gift from D. Templeton with permission from Bernard Moss, National Institutes of Health). Nuclear extracts were prepared 48 h after infection or transfection, and LANA expression was monitored by Western blot analysis in comparison to BCBL-1 cells, which are latently infected with KSHV and express LANA (21, 45). As shown in Fig. 1A, we obtained high amounts of LANA expression from the MVA/T7 expression system in comparison to BCBL-1 (0.8 μg of nuclear extract from CV-1 cells and 4.5 μg from BCBL-1 cells were loaded in lanes 1 and 2). Estimating the signal of EE-tagged LANA to be about 25-fold higher than that of BCBL-1 (compare lanes 1 and 2) suggests that LANA expression in MVA/T7-infected CV-1 cells is about 100-fold higher than in BCBL-1 cells. The EE-tagged LANA migrates significantly faster than LANA from BCBL-1 cells. Sequencing of our LANA clone, which was originally derived from a lung KS tumor (54), revealed that the central domain of LANA is 480 bp shorter than the sequence isolated from BC-1 cells (46) (GenBank accession no. AF360120). Therefore, the protein that we expressed in MVA/T7-infected and BJAB/orf73 cells is only 1,003 amino acids (aa) in length, not 1,163 aa as previously reported. Expressing the same ORF73 clone from the eukaryotic expression vector pcDNA3 in 293 and BJAB cells resulted in a protein that migrated identically to the EE-tagged LANA (data not shown). LANA from different PEL tumors vary in molecular weight due to length polymorphism in the central acidic domain. LANA from BCBL-1 cells was reported to be comparable in length to LANA from BC-1 cells (1,163 aa), which explains the difference in size between BCBL-1 protein and our recombinant EE-tagged LANA (18).
FIG. 1.
LANA expressed by the MVA/T7 system does not bind to sequences within the LANA promoter. (A) Western blot analysis of LANA expression in vaccinia virus-infected CV-1 and BCBL-1 cells. Nuclear extracts prepared as described in Materials and Methods were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 8% gels, transferred, and probed for the presence of LANA by using a monoclonal antibody specific for LANA (24). Lane 1 contains 4.5 μg of nuclear extract from BCBL-1 cells, while lanes 2 and 3 contain 0.8 μg of extracts from MVA/T7-infected CV-1 cells. Lane 2 contains approximately 25-fold more LANA than lane 1, suggesting a 100-fold enrichment over BCBL-1-derived LANA. The mobility of the recombinant protein is higher because the central domain of the LANA clone that we used is 477 nt shorter than reported for BC-1 cells. (B) Diagram of the LANA promoter depicting sequences previously shown to confer LANA responsiveness and the probe used for EMSAs assays (20, 43). Previously published transcriptional start site (+1) and TATA box are indicated (11) (all nucleotide sequence annotations are those of Russo et al. [46]) (C) EMSAs with the LANA promoter using vaccinia virus-expressed LANA protein do not show any specific binding. Lane 1 contains probe alone. Lanes 2 to 4 contain 0.4 μg of protein extract (Prt) from MVA/T7-infected CV-1 cells transfected with either empty vector (pEETM; V) or vector containing the full-length LANA sequence (L). Lane 4 contains a 50-fold excess of cold competitor (LANA promoter fragment unlabeled). None of the visible bands are specific for extracts containing LANA protein.
LANA does not bind to its own promoter.
Using transient transfection assays, LANA has been shown to transactivate its own promoter, which also drives the transcription of two other genes, for v-cylin and v-FLICE, encoded by ORF71 and ORF72 (Fig. 1B) (20, 25, 43). This is functionally similar to EBNA-1 of EBV, which strongly transactivates its own promoter by direct DNA binding (47). We therefore asked whether LANA binds a cis-regulatory element in its own promoter. A region of the LANA promoter previously shown to be activated by LANA nucleotides (nt) +8 to −179 (43) (+1 being the transcription initiation site), was used to prepare a radiolabeled probe for EMSA (Fig. 1B). Radiolabeled probe was incubated with nuclear extracts containing either EE-tagged LANA or control extracts and separated on native polyacrylamide gels as described in Materials and Methods. No LANA-specific complexes were detected in these assays. All detectable bands were present in lanes which contained LANA and lanes which contained control extract (Fig. 1C, compare lanes 2 and 3). These data suggested that LANA acts to modulate transcription of its promoter through interaction with upstream factors and not by direct binding to a DNA element. This would be comparable to EBNA-2 of EBV, which modulates transcription of viral and cellular genes through interaction with RBP-Jκ (19, 29, 30).
LANA specifically binds to a fragment within the TR.
Previously published data suggested that LANA binds to DNA. It was demonstrated that LANA is sufficient for episomal maintenance and that a fragment containing three TRs plus 600 bp of unique sequence at the left end of the KSHV genome does have oriP activity in BJAB cells stably expressing LANA (5). Further, it was shown that radiolabeled cosmids spanning either the left or right region of the KSHV genome could be immunoprecipitated with a LANA-specific antibody (9). These data suggested that LANA might bind specifically to sequences within TRs. We therefore tested the ability of EE-tagged LANA expressed in MVA/T7-infected cells to bind the TR by EMSAs.
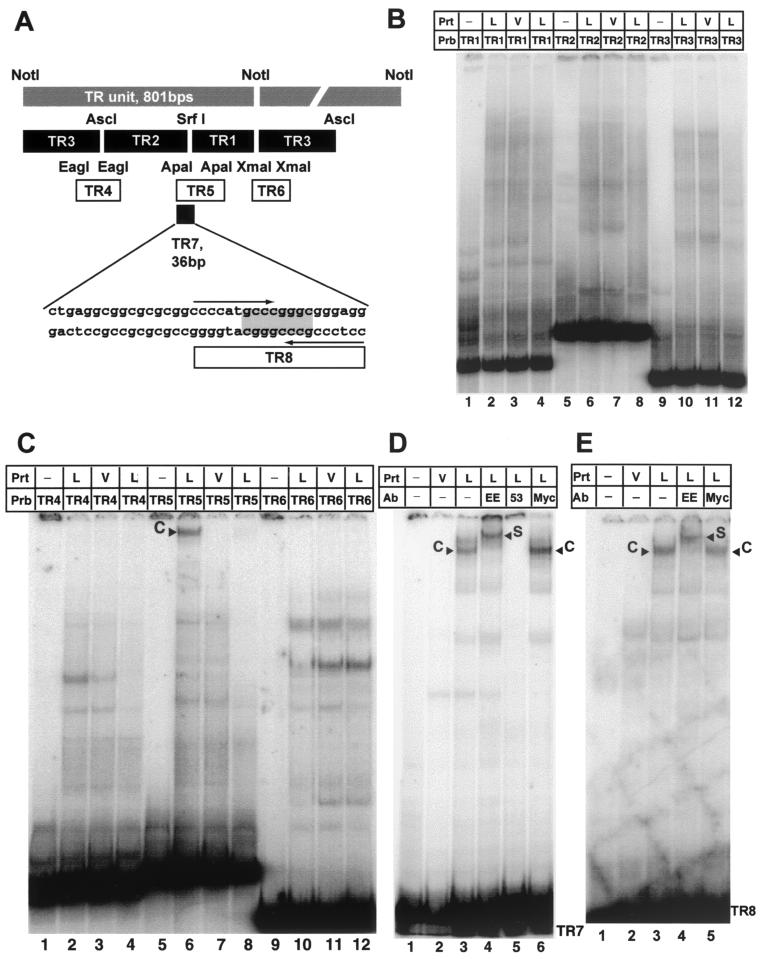
The 801-bp TR unit of KSHV, defined by a unique NotI site, is highly repetitive and GC rich. We therefore prepared all probes by conventional cloning without the use of PCR amplification steps. Plasmids pML1, containing one copy of the NotI TR, and three subclones termed pTR1, pTR2, and pTR3 were a gift from Mike Lagunoff and Don Ganem. Figure 2A gives an overview of all TR-specific probes used in this analysis. First, TR1, TR2, and TR3 were tested for binding to EE-tagged LANA as described above. None of these fragments showed a LANA-specific complex compared to control nuclear extract (Fig. 2B). We next generated fragments spanning the overlapping regions of our original three probes (Fig. 2A). TR4 and TR6 did not produce LANA-specific bands. In contrast, TR5, a 114-bp-long ApaI fragment from nt 556 to 670 (nucleotide positions refer to those of Lagunoff and Ganem [26]) formed a high-molecular-weight complex with the LANA nuclear extract but not with control extract (Fig. 2C, lanes 6 and 7). This complex was efficiently competed away by the addition of a 50-fold excess of the nonlabeled TR5 fragment (Fig. 2C, lane 8). These data show that EE-tagged LANA specifically binds to a cis element within the TR. Because neither the TR1 nor the TR2 fragment was able to bind LANA, we concluded that the SrfI site used to generate these fragments must lie within the LANA binding site. EBNA-1 binds a 16-bp imperfect palindrome (1, 4, 41). Examination of the sequence surrounding the SrfI site revealed that it was the center of a 20-bp imperfect palindrome. To further narrow the LANA binding site, we used a NarI restriction site to isolate a 36-bp fragment including the 20-bp palindrome and 16 bp of sequence to its left (Fig. 2A). EMSA of this fragment, termed TR7, showed a specific complex formed with the LANA extract but not with control extract. To prove that this complex indeed was specific for LANA, we performed EMSAs using specific antibodies. The complex formed on TR7 was shown to supershift with an antibody against the N-terminal EE tag of LANA. In addition, LN53 (24), a monoclonal antibody against the EQEQE repeat in LANA's leucine zipper region, inhibited complex formation (Fig. 2D, lanes 3 to 5). These data show that LANA binds specifically to a 36-bp region containing a 20-bp imperfect palindrome. We used T4 kinase to label a 20-bp synthetic oligonucleotide, creating probe TR8 (Fig. 2A). EMSA analysis revealed that TR8 displayed the same binding specificity as TR7 (Fig. 2E), indicating that this palindrome is sufficient for LANA binding. A smaller fragment containing the 8-bp SrfI site was not sufficient to bind LANA (data not shown).
FIG. 2.
LANA binds specifically to a 20-bp imperfect palindrome within the TR. (A) Diagram of the TR unit as defined by the NotI restriction site (26). Shown are all probes used to identify the LANA binding site within TR7. Plasmids pTR1, pTR2, and pTR3 (kindly provided by M. Lagunoff) were digested with XhoI and NotI to generate the probes TR1, TR2 and TR3, which divide the TR unit into three pieces small enough to be analyzed by EMSA. EagI, ApaI, and XmaI were used to generate fragments TR4, TR5, and TR6, which span the overlapping regions of the first three probes. TR7, 36 bp in length, was created by digestion with ApaI and NarI. The palindromic SrfI site is highlighted; the larger 20-bp imperfect palindrome is indicated by arrows. (B) LANA does not bind to TR1, TR2, and TR3. Probes were radiolabeled with Klenow polymerase and incubated with 4 μg of nuclear extract prepared from MVA/T7-infected CV-1 cells transfected with empty vector (V) or LANA expression vector (L), as indicated above the lanes. Lanes 1, 5, and 9 contain probes (Prb) TR1, TR2, and TR3 in the absence of protein extracts (Prt); lanes 2, 6, and 10 contain protein nuclear extracts of MVA/T7-infected CV-1 cells expressing LANA; lanes 3, 7, and 11 contain extracts from vector-transfected cells. As additional control, lanes 4, 8, and 12 contain LANA and a 50-fold excess of the appropriate unlabeled probe. (C) LANA binds specifically to TR5. Lanes 1, 5, and 9 contain the corresponding probes without protein extracts; lanes 2, 6, and 10 contain protein nuclear extracts CV-1 cells expressing LANA (L); lanes 3, 7, and 11 contain extracts from cells transfected with pEETM-1 vector (V). As a control for specificity, lanes 4, 8, and 12 contain in addition to LANA extract and radiolabeled probe a 50-fold excess of the appropriate unlabeled probe as competitor. Probe TR5 forms a high-molecular-weight complex with LANA extracts (lane 6, labeled C) but not with control extract (lane 7). The observed complex is competed efficiently by the addition of a 50-fold excess of unlabeled probe TR5 as competitor (lane 8). (D) LANA binds specifically to probe TR7 (36 bp in length). Lanes 1 and 2 contain probe TR7 only and control extract from vector-transfected cells (V); lanes 3 to 6 contain protein extract from vaccinia virus-infected CV-1 cells expressing LANA (L). To show that the observed complex is LANA specific, extracts were incubated with three different antibodies (Ab). Lanes 4 and 5 contain EE tag-specific antibody (LANA is tagged at the N terminus [Fig. 1]) or LN53, a LANA-specific monoclonal antibody. Lane 6 contains an unrelated control antibody against a Myc epitope not present in LANA. TR7 forms a complex (labeled C) with LANA-containing extract (lane 3); no complex is seen in lane 2, containing control extract. Lane 4, which contains an antibody to the EE tag, supershifts the complex (labeled S). Lane 5 contains LANA monoclonal antibody LN53, which prevents complex formation. As additional confirmation, unrelated anti-Myc antibody in lane 6 has no effect on complex formation. (E) LANA binds specifically to TR8. Lanes 1 and 2 contain probe TR8 only and control extract from vector-transfected cells (V); lanes 3 to 5 contain protein extract from vaccinia virus-infected CV-1 cells expressing LANA (L); lane 4 contains an antibody specific to the EE tag; lane 5 contains an unrelated control antibody against a Myc epitope not present in LANA. TR8 forms a complex (labeled C) with LANA-containing extract (lane 3); no complex is seen in lane 2, containing control extract. Lane 4, which contains an antibody to the EE tag, supershifts the complex (labeled S).
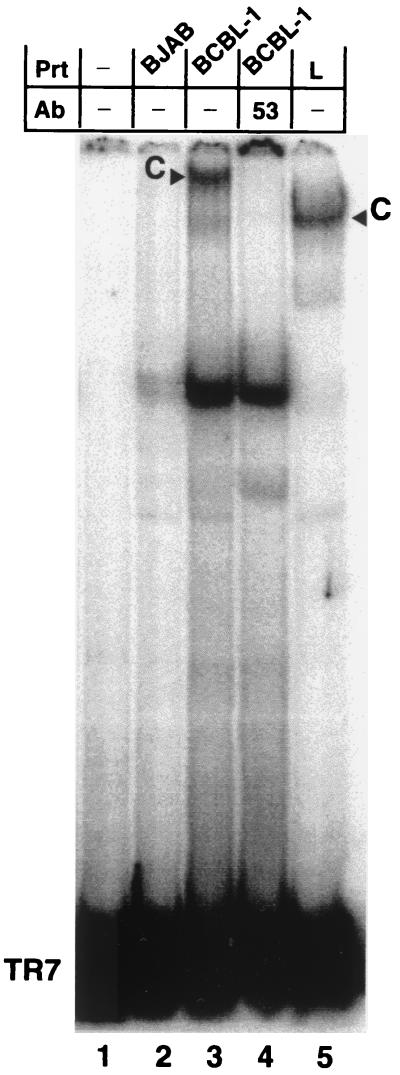
To show that this binding activity was relevant to physiological LANA concentrations in KSHV-infected cells, we isolated nuclear protein extracts from latently infected BCBL-1 cells and analyzed binding to the TR7 fragment (Fig. 3). In this assay, a large complex appears with the BCBL-1 nuclear extract but not with nuclear extract derived from an uninfected B-cell line, BJAB, used as negative control. The large-complex formation is inhibited by the monoclonal antibody LN53, but a smaller, nonspecific complex present at low levels in the BJAB lane is not inhibited, indicating that the LN53 antibody specifically inhibits LANA from binding to the TR7 fragment (Fig. 3, lanes 2 to 4). The BCBL-1 complex migrates in the same area as but slower than the complex formed by EE-tagged LANA. This observation is consistent with the size difference between the LANA expressed in BCBL-1 cells and that expressed in MVA/T7-infected CV-1 cells (Fig. 1A). In summary, these data clearly show that LANA expressed ectopically, or from BCBL-1 cells, has specific DNA binding activity.
FIG. 3.
Nuclear extracts from KSHV-infected BCBL-1 cells confirm specific binding to TR7. Lane 1 contains probe TR7 without extract; lane 2 contains nuclear extract from a noninfected Burkitt's lymphoma line, BJAB; lanes 3 and 4 contain 6.75 μg of nuclear extract from BCBL-1 cells latently infected with KSHV; lane 5 contains protein extract from vaccinia virus-infected CV-1 cells expressing LANA. A complex is seen in lane 3, containing BCBL-1 extracts, but not in lane 2, containing the control BJAB extracts. In lane 4, the addition of monoclonal antibody LN53 inhibits this specific complex but not nonspecific complexes seen in the BJAB lane. The difference in size between the complex formed with BCBL-1 extracts and MVA/T7-expressed LANA reflects differences in molecular weight due to heterogeneity in the central domain of the central domain of LANA (Fig. 1) (18).
Mapping of the LANA DNA binding domain.
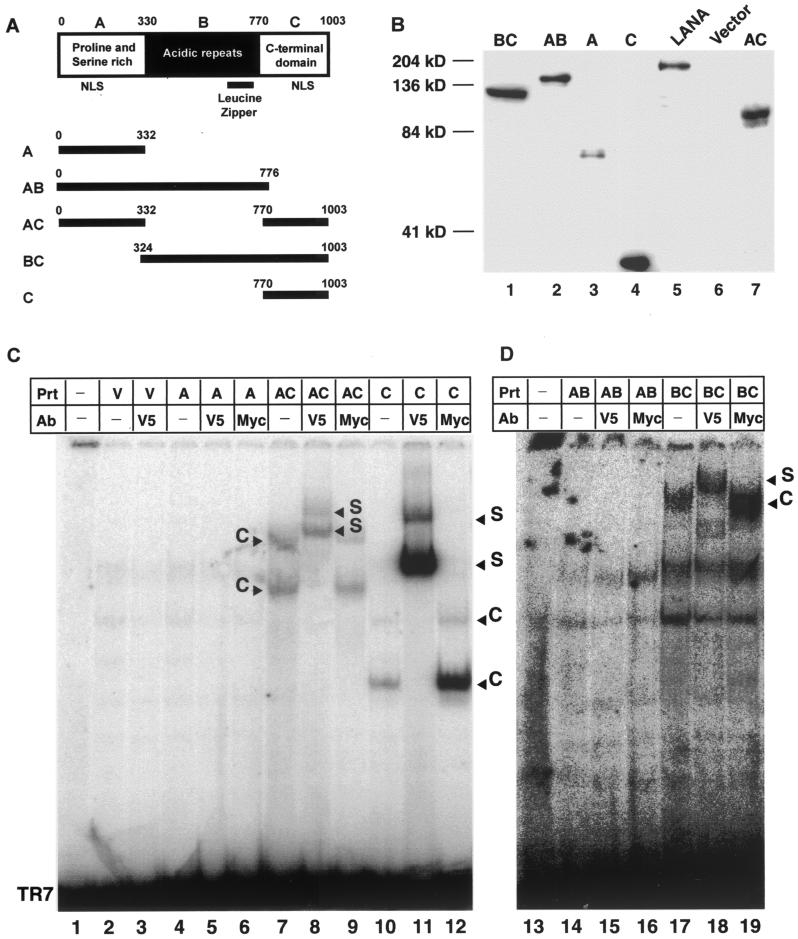
To determine which domain of LANA is sufficient to specifically bind DNA, we prepared a panel of LANA deletion mutants and expressed all proteins by using the MVA/T7 expression system. Based on its primary amino acid sequence, LANA can be divided into three major domains. The highly conserved N-terminal (or A) domain is composed of 330 aa rich in prolines and serines. The C-terminal (or C) domain, also highly conserved, is composed of 229 aa rich in charged and hydrophobic residues. Both N- and C-termini have putative nuclear localization signals (NLSs). The central (B) domain is variable in length (440 aa in EE-tagged LANA) and is composed of several different imperfect acidic repeats followed by a leucine zipper motif. To map the LANA DNA binding domain, we created constructs encoding either full-length LANA or all combinations of either one or two domains (Fig. 4A). All mutant proteins were constructed in pCDNA3.1V5HIS, which provides a C-terminal V5 epitope for immune detection and a His tag for affinity chromatography. This vector facilitates eukaryotic cytomegalovirus promoter-driven expression (see below) and also contains a T7 promoter which can be used to express proteins in MVA/T7-infected cells upon transfection. We then expressed all mutant proteins by using the vaccinia virus system as described above. Before EMSAs were performed, we used immunoblots against the V5 epitope tag of each protein to demonstrate protein expression in the nuclear extracts (Fig. 4B). To adjust for variable expression levels of the different mutants, we loaded different amounts of nuclear extracts, allowing us to detect all LANA mutants by Western blotting. Nuclear extracts loaded in each lane ranged from 0.8 μg for domain C to 29.3 μg for mutant BC (Fig. 4). Despite repeated attempts to express the B domain alone, we could never detect protein expression in either nuclear or cytoplasmic extracts.
FIG. 4.
Mapping of LANA DNA binding domain. (A) Putative domain structure of LANA based on primary sequence features. The N-terminal domain is rich in prolines and serines and contains a putative NLS. The central region of LANA (domain B) is compromised of several repeats and is very acidic. In addition, this region shows length polymorphism in different virus isolates. The C-terminal domain also contains a putative NLS. The C-terminal portion of the B domain contains a leucine zipper motif. All mutant proteins and their coordinates are depicted below the domain model of LANA. All proteins are tagged with a V5 epitope at their N termini. (B) Western blot analysis of LANA deletion mutants. All proteins were expressed in MVA/T7-infected CV-1 cells transfected with the appropriate expression vector. Details of construction, expression, and extraction are described in Materials and Methods. To normalize for expression differences between mutants, we loaded different amounts of total ptotein (A, 1.3 μg; AB, 1.6 μg; AC, 6.1 μg; BC, 29 μg; C, 0.8 μg; LANA, 6.3 μg; and vector control, 5 μg). After electrophoretic separation and transfer, proteins were detected by using a V5-specific antibody. (C) Domain C of LANA is sufficient for specific DNA binding. Lanes 1 and 13 contain probe TR7 in the absence of protein extracts (Prt); lanes 2 to 12 and 14 to 17 contain protein extract from vaccinia virus-infected CV-1 cells transfected with either empty vector pEETM-1 (labeled V) or vector containing one of the mutant LANA proteins (A, AC, C, AB, or BC), as noted above the lanes. Lanes 3, 5, 8, 11, 15, and 18 contain an antibody (Ab) against the V5 tag attached to the C terminus of each protein; lanes 6, 9, 12, 16, and 19 contain an unrelated control antibody specific for the Myc epitope; lanes 7, 10, and 17 containing mutants AC, C, and BC, respectively, which each form specific complexes (labeled C). These complexes are supershifted (S) in lanes 8, 11, and 18 with the antibody against the V5 epitope. The unrelated control antibody against the Myc epitope in lanes 9, 12, and 19 does not a effect the migration of the complexes seen in lanes 7, 10, and 17. No complex formation was detected in lanes 4 to 6 or 14 to 16, which contained mutants A and AB.
Nuclear extracts containing mutant LANA proteins were then tested in EMSAs for the ability to bind TR7 (Fig. 4C and D). Assays were performed as previously described, and protein concentrations were adjusted based on the Western blot data shown in Fig. 5B. In summary, all mutant LANA proteins containing the C domain (AC, BC, and C) showed distinct shifts and supershifts using the V5 epitope-specific antibody (Fig. 4C and D, lanes 7, 8, 11, 12, 17, and 18). Proteins without the C domain (A and AB) did not show a specific shift (Fig. 4C, lanes 4, 5, 14, and 15). Two additional nonspecific bands are apparent in Fig. 4C. These bands are present in all extracts, including the control extract expressing empty vector. The background is more prominent in these assays because of the low expression levels of mutant proteins AB and BC. For mutants AC, BC, and C, two specific complexes are visible. This pattern is similar to that observed in EMSAs of EBNA-1 (31). The formation of two bands suggests that LANA binds the palindrome as a homodimer; therefore, the lower band might represent a LANA monomer bound to the probe, while the upper band represents LANA dimers bound to the probe. This argument is strengthened by the observation that both complexes supershift with the V5 epitope-specific antibody. In addition, it has previously been shown that the C domain of LANA is capable of forming homodimers (48). In summary, these data demonstrate that the C domain of LANA is sufficient for specific DNA binding.
FIG. 5.
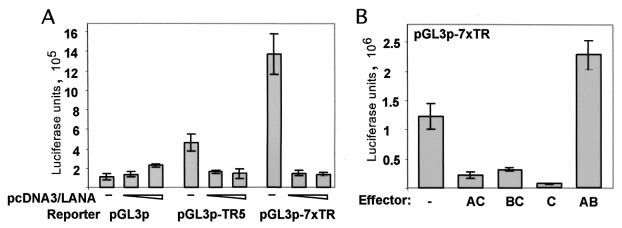
LANA binding to the TR confers transcriptional suppression to a heterologous promoter. Transient transfection assays were performed in 293 cells. TR5 was cloned upstream the SV40 early promoter within the luciferase reporter plasmid pGL3-promoter (pGL3p). This construct was termed pGL3-TR5. Seven copies of the entire TR sequence were inserted into pGL3p, resulting in plasmid pGL3/7XTR. Reporter plasmids (100 ng) were cotransfected with either 500 ng or 2 μg of effector plasmids by lipofection. DNA concentration was equalized in all transfections, using pBS as filler DNA. Shown are mean values ± standard deviations of two independent experiments each performed in duplicate. (A) LANA does inhibit transcription from TR-containing reporters. Transcription from the TR5-containing reporter (pGL3-TR5) is about 4-fold down-regulated in the presence of LANA, while multimerization of the TR results in a more than 10-fold inhibition. (B) Transcriptional suppression is dependent on DNA binding. While AC, BC, and C show transcriptional down-regulation up to 10-fold of the TR containing reporter, no inhibition was detected when mutant AB was used.
LANA binding to the TR confers transcriptional suppression to a heterologous promoter.
LANA has been shown to interact with a variety of proteins involved in transcriptional regulation such as p53, pRb, RING3, and the mSin3 suppressor complex (16, 25, 36, 39). By gene expression profiling, we have shown that LANA can positively or negatively modulate transcription of viral and cellular genes (43). Based on these observations and in analogy to EBNA-1, whose specific DNA binding leads to strong transcriptional activation, we asked whether binding of LANA to TR modulates transcription. We prepared two different reporter constructs and performed transient transfection assays in 293 cells, which have been shown to be semipermissive to KSHV infection (15, 44). First, we cloned TR5 into the vector pGL3-promoter upstream of the simian virus 40 (SV40) early promoter driving luciferase. In addition, we cloned seven TR units from cosmid Z6 derived from BC-1 cells (a kind gift of Yuan Chang, Columbia University) into pGL3-promoter downstream from the luciferase gene. Using lipofection, we cotransfected a constant amount of reporter plasmid together with increasing amounts of the LANA expression vector pcDNA3/LANA into 293 cells. Total DNA concentration was kept constant, and cells were harvested 48 h posttransfection, when total cellular extracts were analyzed for luciferase activity. All obtained raw data were normalized to total protein concentration by Bradford assays. Transfection efficiency was determined by transfecting control wells with a LacZ reporter and ranged from 25 to 35%. Transcription from pGL3-promoter was not affected at low concentrations of LANA expression vector and increased nearly twofold at high concentrations. In contrast, transcription from pGL3-TR5 containing a single LANA binding site was suppressed up to fourfold by the presence of LANA. Furthermore, transcription from pGL3-7xTR was suppressed 10-fold in the presence of LANA (Fig. 5A). These data show that LANA down-regulates transcription from the TR binding site. This is in contrast with EBNA-1, whose binding to DNA results in transcriptional up-regulation. We also observed that TR5, consisting of 114 bp within TR, and more potently the multimerized TR showed enhancer activity up to 10-fold in the absence of LANA (Fig. 5A). This enhancer effect is not due to a possible promoter activity within the TR because the TR confers activity when inserted downstream of the reporter gene; additionally, the TR does not confer transcriptional activity to pGL3-basic, which lacks a polymerase II promoter (data not shown).
We next asked which LANA domains are responsible for the observed transcriptional suppression and whether DNA binding is required for this phenotype. The LANA mutants AC, BC, and C, which were previously shown to bind to DNA (Fig. 4C), suppressed transcription from the pGL3-7xTR reporter (Fig. 5B). In contrast, AB resulted in a less than twofold activation of transcription. Hence, the C domain of LANA is necessary and sufficient for both DNA binding and LANA-dependent transcriptional down-regulation of a TR-containing reporter.
DISCUSSION
By performing EMSAs using vaccinia virus-expressed proteins, we demonstrate that LANA binds to DNA in a sequence-specific manner and that the C-terminal domain of LANA is sufficient for this binding activity.
We recently demonstrated that LANA can modulate cellular and viral gene expression. In analogy to EBNA-1 of EBV, we found that LANA transactivates its own synthesis (43). To elucidate the mechanism of this activation, we examined whether LANA binds to its promoter. In contrast to EBNA-1, which binds sequences within its own promoter, our EMSA data did not show any LANA-specific binding to sequences previously been shown to confer LANA responsiveness in transient transfection assays in cell lines of different origins (20, 25, 43). Therefore, LANA's ability to regulate its own synthesis must be mediated through interactions with transcriptional coactivators. Several reports describe direct interactions of LANA with proteins involved in the regulation of transcription. While LANA interaction with p53 and mSin3 has been shown to confer negative transcriptional effects, the interaction with pRb has been shown to stimulate E2F-dependent transcription (16, 25, 39). However, because there are no E2F binding sites within the LANA promoter, the LANA-pRb interaction does not explain LANA's autoregulation, which also drives the synthesis of v-cyclin and v-FLICE, two proteins with potential roles in deregulation of cellular growth (11, 32). Presumably, LANA might interact with additional unidentified coactivators in order to activate transcription, a mode of action more related to EBNA-2 of EBV, which does not bind to DNA directly but instead activates transcription through interaction coactivators such as RBP-Jκ (19, 29, 30). In contrast, EBNA-1, which is sufficient for episomal maintenance in dividing cells, requires binding to a cis-regulatory element within oriP. Binding to this element also causes strong transcriptional activation of two viral promoters. Mutations in EBNA that abrogate its transcriptional phenotype also neutralize its replication/segregation function (27). In analogy to EBNA-1, it was recently shown that LANA is sufficient for episome maintenance in dividing cells (5). We therefore asked whether LANA also would function as transcription factor when bound to its putative oriP. Two lines of data suggested that oriP of KSHV is located within TR. First, a plasmid containing three copies of TR plus some additional unique sequence was stably maintained in LANA-expressing B cells (5). Second, in the presence of LANA, a specific monoclonal antibody immunoprecipitates radiolabeled KSHV DNA fragments containing the TR region (9).
We therefore concentrated on a putative LANA binding site within TR. Using EE-tagged LANA expressed in MVA/T7-infected cells, we showed by EMSA that LANA binds specifically to a 20-bp imperfect palindrome called TR8. The importance of this palindrome is further confirmed by the observation that TR8 overlaps with probes TR1 and TR2, neither of which binds LANA (Fig. 2B). TR5, which extends rightward from the palindrome, does not show a higher affinity to LANA, suggesting that these sequences are not required for binding. These data together with our observation that a single SrfI site (the core of the palindrome) does not confer LANA binding suggest that the 20-bp palindrome represents a LANA binding site. The putative LANA binding site is specific for the TRs of KSHV and is not present in the unique long region of KSHV. Examination of LANA's promoter sequence reveals that it contains a short palindrome (nt 127904 to 127913), similar to the SrfI site found in the LANA binding site. However, a fragment containing this sequence was not capable of binding LANA (Fig. 1C). There is no primary sequence homology between this site and the EBNA-1 binding site. However, the EBNA-1 site consists of a 16-bp imperfect inverted repeat to which EBNA-1 binds as a homodimer. To further map the minimal LANA binding motif and to determine the required residues, we are currently performing DNA footprinting analysis utilizing tagged affinity-purified LANA proteins.
Our EMSA analysis using V5-tagged LANA deletion mutants clearly showed that the C-terminal domain of LANA is required for DNA binding. All mutant proteins lacking the C domain did not bind, while mutant proteins containing the C domain did (Fig. 5). It is interesting to note that the C terminus of LANA is also required for the formation of homodimers (48), which suggests that LANA binds to its TR binding site as a dimer, an inference supported by the presence of two LANA-specific bands in the EMSA. Structural analysis of transcription factors, including EBNA-1, which bind as dimers to palindromic target sequences indicate that these factors bind in a highly symmetrical fashion. Therefore, our observation that only LANA mutants which can form dimers bind to TR7 further suggest that the 20-bp imperfect palindrome represents the LANA binding site.
We next analyzed the effect of LANA binding on transcriptional regulation. A single LANA binding site (TR5) inserted upstream of the SV40 early promoter was transcriptionally down-regulated up to fourfold in the presence of LANA. This effect was stronger when a fragment containing seven complete TRs was inserted downstream of the luciferase gene in this reporter plasmid (Fig. 5A). These data show that LANA inhibits transcription when bound to its binding site within the TR. This is in contrast with EBNA-1, which strongly activates transcription when bound to oriP. However, a similar situation exists in human papillomavirus, in which E2 protein binding to the origin causes transcriptional suppression of downstream promoters (32). In addition to the inhibiting effect of LANA, these experiments also revealed a cis-regulatory effect of the TR sequences on transcription. TR5 and the multimerized TR unit functioned as enhancers in transient transfection assays by increasing transcriptional activity up to 10-fold. Using the LANA mutants described above for DNA binding experiments, we examined whether there is a correlation between DNA binding and transcriptional down-regulation. The obtained results were identical to results described for DNA binding. Mutant proteins containing the C terminus of LANA did down-regulate transcription from this reporter, while mutant AB led to a slight increase in transcription (Fig. 5B). These data clearly show that the observed transcriptional inhibition of a TR-containing reporter is dependent on specific DNA binding. This observation is in contrast to data observed by using fusion proteins between LANA and the Gal4 DNA binding domain to direct LANA binding to a multimerized Gal4 binding site-containing reporter (5xGal4SV40). In these assays, both N and C termini fused to Gal4 did result in strong inhibition of a 5xGal4SV40 luciferase reporter in 293T cells (48). Using a similar assay system in HeLa cells, it was shown that only the N terminus, which was shown to interact with the mSin3 complex, but not the C terminus inhibits a 5xGal4/tkCat reporter (25). The different outcomes of these experiments might be attributable to the use of fusion proteins to artificially direct LANA binding. In contrast, our data describe for the first time transcriptional effects of LANA bound to its specific binding site within the TR, presumably the oriP of KSHV. In analogy to EBNA-1, we have shown that mutations in LANA affecting DNA binding also affect its ability to modulate transcription. LANA has previously been shown to interact with proteins involved in transcriptional regulation (i.e. pRb, mSin3, and ATF4/CREB2). The LANA domains responsible for these interactions have been mapped to either the N terminus or the leucine zipper (8a, 25, 39). The fact that neither of these elements is present in the C domain, which is sufficient to confer transcriptional repression, suggests a mechanism independent of these transcriptional regulators. In light of the fact that LANA strongly inhibits transcription when bound to TR, a model can be envisioned by which LANA contributes to the maintenance of latency by silencing genes in the vicinity of the TRs. We are currently performing experiments to address the question whether LANA silences viral genes in the context of the viral genome.
ACKNOWLEDGMENTS
We thank Yuan Chang, Don Ganem, Andy Polson, Mike Lagunoff, and Dennis Templeton for providing reagents. In addition, we thank Don Ganem, Dennis Templeton, and Janet Cross for helpful discussion and critical reading of the manuscript.
R.R. is a Mount Sinai Healthcare Foundation scholar. This work was supported by grants from the American Cancer Society and the National Institutes of Health (CA CA88763-01) to R.R.
ADDENDUM
After submission of this article Ballestas and Kaye (5a) published an article containing results mapping a DNA binding site for LANA within the TR similar to the site shown by our results.
REFERENCES
- 1.Ambinder R F. Epstein-Barr virus infections. In: Thomas E D, Blume K G, Forman S J, editors. Hematopoietic cell transplantation. 2nd ed. Malden, Mass: Blackwell Science Inc.; 1999. pp. 607–615. [Google Scholar]
- 2.Ambinder R F, Shah W A, Rawlins D R, Hayward G S, Hayward S D. Definition of the sequence requirements for binding of the EBNA-1 protein to its palindromic target sites in Epstein-Barr virus DNA. J Virol. 1990;64:2369–2379. doi: 10.1128/jvi.64.5.2369-2379.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews N C, Faller D V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C, et al. DNA sequence and expression of the B95–8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 5.Ballestas M E, Chatis P A, Kaye K M. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 5a.Ballestas M E, Kaye K M. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (tr) sequence and specifically binds DNA. J Virol. 2001;75:3250–3258. doi: 10.1128/JVI.75.7.3250-3258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boshoff C, Whitby D, Hatziioannou T, Fisher C, van der Walt J, Hatzakis A, Weiss R, Schulz T. Kaposi's-sarcoma-associated herpesvirus in HIV-negative Kaposi's sarcoma. Lancet. 1995;345:1043–1044. doi: 10.1016/s0140-6736(95)90780-7. [DOI] [PubMed] [Google Scholar]
- 7.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 8.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 8a.Chunghun L, Hekwang S, Gwack Y, Choe J. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) binds ATF4/Creb2 and inhibits its transcriptional activation activity. J Gen Virol. 2000;81:2645–2652. doi: 10.1099/0022-1317-81-11-2645. [DOI] [PubMed] [Google Scholar]
- 9.Cotter M A, II, Robertson E S. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology. 1999;264:254–264. doi: 10.1006/viro.1999.9999. [DOI] [PubMed] [Google Scholar]
- 10.Deak J C, Cross J V, Lewis M, Qian Y, Parrott L A, Distelhorst C W, Templeton D J. Fas-induced proteolytic activation and intracellular redistribution of the stress-signaling kinase MEKK1. Proc Natl Acad Sci USA. 1998;95:5595–5600. doi: 10.1073/pnas.95.10.5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J Virol. 1998;72:8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupin N, Diss T L, Kellam P, Tulliez M, Du M Q, Sicard D, Weiss R A, Isaacson P G, Boshoff C. HHV-8 is associated with a plasmablastic variant of Castleman disease that is linked to HHV-8-positive plasmablastic lymphoma. Blood. 2000;95:1406–1412. [PubMed] [Google Scholar]
- 13.Dupin N, Fisher C, Kellam P, Ariad S, Tulliez M, Franck N, van Marck E, Salmon D, Gorin I, Escande J P, Weiss R A, Alitalo K, Boshoff C. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc Natl Acad Sci USA. 1999;96:4546–4551. doi: 10.1073/pnas.96.8.4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elroy-Stein O, Moss B. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates; 1992. pp. 16.19.1–16.9.12. [Google Scholar]
- 15.Foreman K E, Friborg J, Jr, Kong W P, Woffendin C, Polverini P J, Nickoloff B J, Nabel G J. Propagation of a human herpesvirus from AIDS-associated Kaposi's sarcoma. N Engl J Med. 1997;336:163–171. doi: 10.1056/NEJM199701163360302. [DOI] [PubMed] [Google Scholar]
- 16.Friborg J, Kong W, Hottiger M O, Nabel G J. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 2000;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- 17.Gahn T A, Sugden B. An EBNA-1-dependent enhancer acts from a distance of 10 kilobase pairs to increase expression of the Epstein-Barr virus LMP gene. J Virol. 1995;69:2633–2636. doi: 10.1128/jvi.69.4.2633-2636.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao S J, Zhang Y J, Deng J H, Rabkin C S, Flore O, Jenson H B. Molecular polymorphism of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent nuclear antigen: evidence for a large repertoire of viral genotypes and dual infection with different viral genotypes. J Infect Dis. 1999;180:1466–1476. doi: 10.1086/315098. [DOI] [PubMed] [Google Scholar]
- 19.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J. kappa. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 20.Jeong J, Papin J, Dittmer D. Differential regulation of the overlapping Kaposi's sarcoma-associated herpesvirus vGCR (orf74) and LANA (orf73) promoters. J Virol. 2001;75:1798–1807. doi: 10.1128/JVI.75.4.1798-1807.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kedes D H, Lagunoff M, Renne R, Ganem D. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J Clin Investig. 1997;100:2606–2610. doi: 10.1172/JCI119804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kedes D H, Operskalski E, Busch M, Kohn R, Flood J, Ganem D. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat Med. 1996;2:918–924. doi: 10.1038/nm0896-918. . (Erratum, 2:1041.) [DOI] [PubMed] [Google Scholar]
- 23.Kellam P, Boshoff C, Whitby D, Matthews S, Weiss R A, Talbot S J. Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J Hum Virol. 1997;1:19–29. [PubMed] [Google Scholar]
- 24.Kellam P, Bourboulia D, Dupin N, Shotton C, Fisher C, Talbot S, Boshoff C, Weiss R A. Characterization of monoclonal antibodies raised against the latent nuclear antigen of human herpesvirus 8. J Virol. 1999;73:5149–5155. doi: 10.1128/jvi.73.6.5149-5155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krithivas A, Young D B, Liao G, Greene D, Hayward S D. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J Virol. 2000;74:9637–9645. doi: 10.1128/jvi.74.20.9637-9645.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lagunoff M, Ganem D. The structure and coding organization of the genomic termini of Kaposi's sarcoma-associated herpesvirus. Virology. 1997;236:147–154. doi: 10.1006/viro.1997.8713. [DOI] [PubMed] [Google Scholar]
- 27.Leight E R, Sugden B. EBNA-1: a protein pivotal to latent infection by Epstein-Barr virus. Rev Med Virol. 2000;10:83–100. doi: 10.1002/(sici)1099-1654(200003/04)10:2<83::aid-rmv262>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 28.Liebowitz D, Kieff E. Epstein-Barr virus. In: Roizman B, Whitley R J, Lopez C, editors. The human herpesviruses. New York, N.Y: Raven Press; 1993. pp. 107–172. [Google Scholar]
- 29.Ling P D, Hsieh J J, Ruf I K, Rawlins D R, Hayward S D. EBNA-2 upregulation of Epstein-Barr virus latency promoters and the cellular CD23 promoter utilizes a common targeting intermediate, CBF1. J Virol. 1994;68:5375–5383. doi: 10.1128/jvi.68.9.5375-5383.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ling P D, Rawlins D R, Hayward S D. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc Natl Acad Sci USA. 1993;90:9237–9241. doi: 10.1073/pnas.90.20.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mackey D, Middleton T, Sugden B. Multiple regions within EBNA1 can link DNAs. J Virol. 1995;69:6199–6208. doi: 10.1128/jvi.69.10.6199-6208.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBride A. A., H. Romanczuk, and P. M. Howley. 1991. The papillomavirus E2 regulatory proteins. 266:18411–18414. [PubMed]
- 33.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 34.Moore P S, Kingsley L A, Holmberg S D, Spira T, Gupta P, Hoover D R, Parry J P, Conley L J, Jaffe H W, Chang Y. Kaposi's sarcoma-associated herpesvirus infection prior to onset of Kaposi's sarcoma. AIDS. 1996;10:175–180. doi: 10.1097/00002030-199602000-00007. [DOI] [PubMed] [Google Scholar]
- 35.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. Product review. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 36.Platt G M, Simpson G R, Mittnacht S, Schulz T F. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J Virol. 1999;73:9789–9795. doi: 10.1128/jvi.73.12.9789-9795.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polvino-Bodnar M, Schaffer P A. DNA binding activity is required for EBNA 1-dependent transcriptional activation and DNA replication. Virology. 1992;187:591–603. doi: 10.1016/0042-6822(92)90461-w. [DOI] [PubMed] [Google Scholar]
- 38.Puglielli M T, Woisetschlaeger M, Speck S H. oriP is essential for EBNA gene promoter activity in Epstein-Barr virus-immortalized lymphoblastoid cell lines. J Virol. 1996;70:5758–5768. doi: 10.1128/jvi.70.9.5758-5768.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radkov S A, Kellam P, Boshoff C. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat Med. 2000;6:1121–1127. doi: 10.1038/80459. [DOI] [PubMed] [Google Scholar]
- 40.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S J, Stoiber H, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rawlins D R, Milman G, Hayward S D, Hayward G S. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985;42:859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- 42.Reisman D, Sugden B. trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol Cell Biol. 1986;6:3838–3846. doi: 10.1128/mcb.6.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Renne R, Barry C, Dittmer D, Compitello N, Brown P, Ganem D. Modulation of cellular and viral transcription by the latency-associated nuclear antigen (LANA/ORF73) of Kaposi's sarcoma-associated herpesvirus. J Virol. 2001;75:458–468. doi: 10.1128/JVI.75.1.458-468.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Renne R, Blackbourn D, Whitby D, Levy J, Ganem D. Limited transmission of Kaposi's sarcoma-associated herpesvirus in cultured cells. J Virol. 1998;72:5182–5188. doi: 10.1128/jvi.72.6.5182-5188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 46.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaefer B C, Strominger J L, Speck S H. Redefining the Epstein-Barr virus-encoded nuclear antigen EBNA-1 gene promoter and transcription initiation site in group I Burkitt lymphoma cell lines. Proc Natl Acad Sci USA. 1995;92:10565–10569. doi: 10.1073/pnas.92.23.10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwam D R, Luciano R L, Mahajan S S, Wong L, Wilson A C. Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J Virol. 2000;74:8532–8540. doi: 10.1128/jvi.74.18.8532-8540.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen Y, Shenk T. Relief of p53-mediated transcriptional repression by the adenovirus E1B 19-kDa protein or the cellular Bcl-2 protein. Proc Natl Acad Sci USA. 1994;91:8940–8944. doi: 10.1073/pnas.91.19.8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shirakata M, Hirai K. Identification of minimal oriP of Epstein-Barr virus required for DNA replication. J Biochem (Tokyo) 1998;123:175–181. doi: 10.1093/oxfordjournals.jbchem.a021907. [DOI] [PubMed] [Google Scholar]
- 51.Staskus K A, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson D J, Ganem D, Haase A T. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugden B, Warren N. A promoter of Epstein-Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J Virol. 1989;63:2644–2649. doi: 10.1128/jvi.63.6.2644-2649.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wyatt L S, Moss B, Rozenblatt S. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology. 1995;210:202–205. doi: 10.1006/viro.1995.1332. [DOI] [PubMed] [Google Scholar]
- 54.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]