Abstract
Tat-specific cytotoxic T cells have previously been shown to exert positive Darwinian selection favoring amino acid replacements of an epitope of simian immunodeficiency virus (SIV). The region of the tat gene encoding this epitope falls within a region of overlap between the tat and vpr reading frames, and nonsynonymous nucleotide substitutions in the tat reading frame were found to occur disproportionately in such a way as to cause synonymous changes in the vpr reading frame. Comparison of published complete SIV genomes showed Tat to be the least conserved at the amino acid level of nine proteins encoded by the virus, while Vpr was one of the most conserved. Numerous parallel amino acid changes occurred within the Tat epitope independently in different monkeys, and purifying selection on the vpr reading frame, by limiting acceptable nonsynonymous substitutions in the tat reading frame, evidently has enhanced the probability of parallel evolution.
The phenomenon of viral proteins encoded by overlapping reading frames has attracted the attention of evolutionary biologists since its discovery (4, 9, 11, 12, 15). One question of evolutionary interest raised by this phenomenon is how natural selection can act simultaneously on two different protein products encoded in different reading frames by the same DNA sequence. Recently, we reported evidence of positive Darwinian selection exerted by the host immune system on a portion of the Tat protein of simian immunodeficiency virus (SIV) (1). The portion of the Tat protein which is subject to positive selection is encoded by a reading frame that overlaps that encoding the Vpr protein (Fig. 1). In the present paper, we examine in further detail natural selection on the Tat and Vpr proteins in order to understand how natural selection in one reading frame affects the evolution of a protein encoded by an overlapping reading frame.
FIG. 1.
Schematic map of the SIV genome, showing the nine proteins encoded.
The region of the Tat protein subject to positive selection is an 8-amino-acid peptide epitope presented to cytotoxic T cells (CTL) by rhesus monkeys (Macaca mulatta) possessing a class I major histocompatibility complex molecule known as Mamu-A*01. Allen et al. (1) studied the evolution of this region in an experimental system involving Mamu-A*01-positive (A*01+) animals and Mamu-A*01-negative (A*01−) controls. Because both groups of monkeys were infected with the same viral inoculum, it was possible to compare the evolution of the Tat epitope in the two groups. In virus from A*01+ monkeys, there was an enhanced rate of nonsynonymous nucleotide substitution, leading to variant forms of the epitope that were experimentally shown not to be bound by the Mamu-A*01 molecule (1). No such evidence was found in the case of A*01− controls (1). The results of this study provide perhaps the most convincing evidence to date of CTL-driven selection on a virus. Because this is a particularly well understood example of positive selection at the molecular level, it provides an excellent opportunity for studying the evolution of an overlapping reading frame in the presence of such selection.
Holmes and colleagues (5) presented evidence that natural selection, presumably exerted by the host immune system, can lead to convergent or parallel amino acid substitutions in the hypervariable V3 loop of the envelope glycoprotein gp120 of human immunodeficiency virus type 1 (HIV-1). However, this conclusion was based on phylogenetic analyses of sequences collected from a single patient; and because only a short gene segment was sequenced, the reliability of phylogenetic inferences in this case is unclear. There is considerably stronger evidence of parallel evolution of HIV-1 in response to pharmacological agents; for example, parallel amino acid changes in the HIV-1 protease have been documented for different patients treated with the same protease inhibitors (2). In general, the existence of convergent or parallel evolution at the amino acid sequence level has been controversial (3), although in recent years a number of purported cases have been described in the literature (for a review, see reference 6). Because this study involved the evolution of SIV in separate, noninteracting monkey hosts, it provides an opportunity for an unequivocal demonstration of parallel evolution in a protein under positive selection.
MATERIALS AND METHODS
A total of 18 rhesus monkeys, 10 A*01+ (here designated A to H and J and K) and 8 A*01−, were infected with a molecularly cloned virus, SIVMAC239 (16). After 8 weeks of infection, a 98-codon segment of the SIV tat gene was amplified and sequenced; this segment overlaps a 50-codon segment of the vpr reading frame. The 98 codons of tat include the codons encoding the peptide STPESANL known to be bound and presented by the Mamu-A*01 molecule to CTL (1). A total of 159 such sequences from infected monkeys were compared with 19 sequences from the inoculum. For further details of sequencing and immunological methods, see the work of Allen et al. (1).
In order to compare the evolution of these regions in experimentally infected monkeys with that in other SIV populations, we also analyzed six complete SIV genomes available in the GenBank database: three from Cercocebus torquatus hosts (AF077017, L03295, and M31325), two from Macaca nemestrina (U79412 and M83293), and one each from Macaca arctoides and M. mulatta (M83293 and U72748, respectively). These represent all available SIV genomes including complete coding sequences for all nine protein-encoding genes. Coding sequences for the nine genes gag, pol, vif, vpx, vpr, tat, rev, env, and nef (Fig. 1) were aligned at the amino acid level using the CLUSTAL W program (19). In computing pairwise distances among a set of sequences, we did not include any site at which the alignment postulated a gap in any sequence in the set, so that a comparable data set was used for each comparison.
The number of synonymous substitutions per synonymous site (dS) and the number of nonsynonymous substitutions per nonsynonymous site (dN) were estimated by the method of Nei and Gojobori (13). This method is known to overestimate dS and slightly underestimate dN when there is a significant transitional bias at twofold degenerate sites; as a consequence, alternative methods that incorporate an estimate of the transition/transversion ratio (R) at such sites have been proposed elsewhere (10, 20). However, such methods require a reliable estimate of R, which requires data for a large number of sites. An unbiased estimate of R can be obtained by comparing fourfold degenerate sites in phylogenetically independent comparisons (7). Fourfold degenerate sites are preferable for estimating R, since only at these sites are the effects of transitional bias and purifying selection not confounded (7). The sequences from the experimental study were short, while in the case of complete SIV genomes, the number of sequences was small. Furthermore, overlapping reading frames add an additional complication to the estimation of R even at fourfold degenerate sites. Because nonsynonymous transversions typically cause more radical amino acid changes than do nonsynonymous transitions, purifying selection will in many cases act more strongly on the former. Therefore, in the case of overlapping reading frames, the effects of transitional bias and purifying selection are confounded even at fourfold degenerate sites. Because it was impossible to incorporate all these factors into the estimation of dS and dN, we used the unmodified Nei and Gojobori method (13), following the recommendation of Nei and Kumar (14) that a simple method is preferable when complex factors influence the pattern of nucleotide substitution. Such a simple method has the advantage of making fewer assumptions than do alternative methods, and if there is a transitional bias, the unmodified Nei and Gojobori method provides a conservative test of the hypothesis of positive selection (14).
In the experimental study, because sequences from each monkey were independent, mean dS and dN within hosts and between samples and the inoculum were compared by t tests. In comparisons of complete genomes, standard errors of mean dS and dN were computed by the bootstrap method (14).
RESULTS
Selection on tat and vpr.
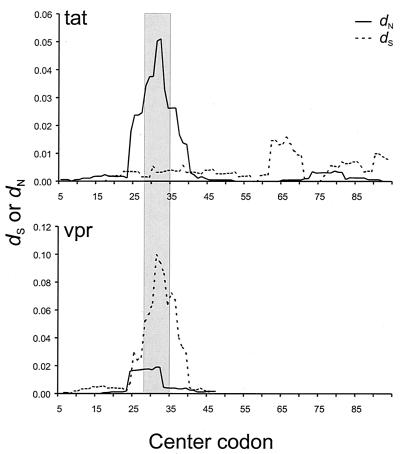
Figure 2 illustrates the mean of dS and dN for comparisons between samples from A*01+ monkeys and the inoculum in a sliding window analysis of tat and vpr reading frames. In the tat reading frame, a strong peak in dN was observed in the region of the STPESANL epitope, while in the vpr reading frame, there was a corresponding peak in dS in the same region (Fig. 2). Table 1 summarizes the means of dS and dN in epitope and nonepitope regions in both tat and vpr reading frames. In comparisons of the tat reading frame of samples from infected monkeys with that of the viral inoculum, the mean dN for A*01+ monkeys significantly exceeded the mean dS in the STPESANL epitope but not elsewhere in the gene (Table 1). No such pattern was seen in the STPESANL epitope in the case of A*01− monkeys (Table 1). Likewise, in comparisons within samples from A*01+ monkeys, mean dN significantly exceeded mean dS (Table 1). Again, no difference was seen in the case of A*01− monkeys (Table 1).
FIG. 2.
Mean numbers of synonymous (dS) and nonsynonymous (dN) nucleotide substitutions per site in comparisons of tat and vpr reading frames in a sliding nine-codon window in comparisons between samples from Mamu-A*01+ monkeys and the inoculum. The vertical bar marks the location of the STPESANL epitope in Tat.
TABLE 1.
Mean numbers (± standard errors of the means) of synonymous (dS) and nonsynonymous (dN) numbers of nucleotide substitutions per 100 sites in comparisons of Tat and Vpr proteins regionsa
| Reading frame and animal type | Comparison | Epitope
|
Remainder
|
||
|---|---|---|---|---|---|
| dS | dN | dS | dN | ||
| tat | |||||
| A*01− | Vs inoculum | 0.2 ± 0.2 | 1.1 ± 0.5 | 0.5 ± 0.2 | 0.3 ± 0.1 |
| Within | 0.5 ± 0.5 | 0.9 ± 0.3 | 0.6 ± 0.2 | 0.4 ± 0.1 | |
| A*01+ | Vs inoculum | 0.4 ± 0.4 | 5.6 ± 0.4***††† | 0.4 ± 0.1 | 0.1 ± 0.0 |
| Within | 0.8 ± 0.8 | 7.2 ± 1.1***††† | 0.5 ± 0.2 | 0.2 ± 0.1 | |
| vpr | |||||
| A*01− | Vs inoculum | 0.4 ± 0.3 | 1.5 ± 0.7 | 0.7 ± 0.4 | 0.2 ± 0.1 |
| Within | 0.9 ± 0.6 | 1.2 ± 0.6 | 0.9 ± 0.4 | 0.4 ± 0.2 | |
| A*01+ | Vs inoculum | 9.3 ± 1.6†† | 2.0 ± 0.5** | 0.3 ± 0.1 | 0.1 ± 0.0 |
| Within | 13.9 ± 2.7†† | 2.2 ± 0.6** | 0.6 ± 0.2 | 0.3 ± 0.1 | |
In the tat reading frame, the epitope region encompasses the eight codons aligned with the STPESANL epitope. In the vpr reading frame, it encompasses nine codons overlapping those eight codons in tat. **, P < 0.01; ***, P < 0.001, both by paired sample t tests of the hypothesis that mean dS = mean dN. ††, P < 0.01; †††, P < 0.001, both by t tests of the hypothesis that mean dS or mean dN in Mamu-A*01+ animal data equals the corresponding value in Mamu-A*01− animal data.
In the vpr reading frame, in A*01− monkeys, no significant difference between mean dS and mean dN was seen either in the nine codons overlapping the STPESANL epitope or in the remainder of the gene (Table 1). However, in A*01+ monkeys, mean dS was significantly greater than mean dN in the region corresponding to the STPESANL epitope (Table 1). Thus, positive selection favoring amino acid changes in the STPESANL epitope of the Tat protein in virus infecting A*01+ monkeys evidently resulted in a burst of synonymous changes in the vpr reading frame (Fig. 2; Table 1).
This finding was further analyzed by considering all possible nonsynonymous changes that might occur in the STPESANL epitope. There were 49 such possible changes, of which 32 would also cause a nonsynonymous change in the vpr reading frame, while the remaining 17 would cause a synonymous change in the vpr reading frame. Of the 32 possible nonsynonymous changes in tat that are also nonsynonymous in vpr, only 4 were actually observed in the viral sequences from A*01+ monkeys. On the other hand, 9 of 17 possible nonsynonymous changes in tat that are synonymous in vpr were observed. The difference between observed and expected is highly significant (P = 0.0002; Fisher's exact test). This result shows that positively selected nonsynonymous changes in the tat gene occurred disproportionately in such a way as not to change the amino acid sequence of Vpr.
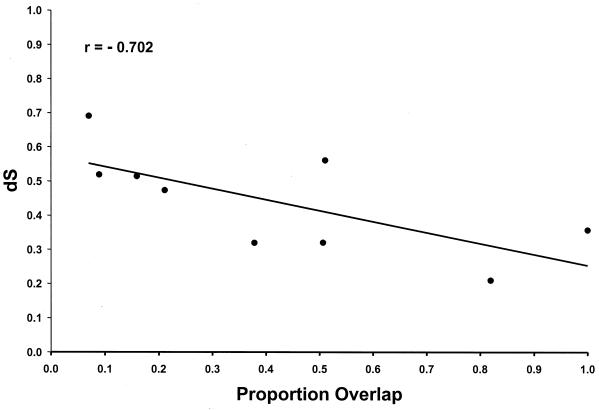
Table 2 shows mean dS and dN for comparisons of nine protein-encoding genes among six complete genomes. The genes were found to differ with respect to both mean dS and mean dN (Table 2). Mean dN was lowest in the pol gene and highest in the tat gene (Table 2). Four genes, one of which was vpr, had mean dN significantly lower than that of tat (Table 2). On the other hand, mean dS was highest in pol and lowest in tat (Table 2). Differences among genes with respect to dN are most plausibly explained by differences in the strength of purifying selection on the protein. Differences among genes with respect to dS can evidently be explained to a considerable extent by differences among genes with respect to overlap with other genes. For the nine genes of SIV, when mean dS was plotted against the proportion of overlap with other genes, there was a significant negative relationship (r = −0.702; R2 = 49.3%; P = 0.035) (Fig. 3). Thus, the genes with the greatest extent of overlap had the lowest mean dS values, presumably as a consequence of purifying selection in the overlapping reading frame, and nearly 50% of the difference among loci with respect to dS was explainable by differences with respect to overlap with other genes.
TABLE 2.
Mean numbers of synonymous (dS) and nonsynonymous (dN) nucleotide substitutions per 100 sites in comparisons of coding regions of SIV genomes
| Gene | % Overlap | dS (±SE)a | dN (±SE)a |
|---|---|---|---|
| gag | 8.9 | 51.9 ± 4.1*** | 4.5 ± 0.5*** |
| pol | 7.0 | 61.9 ± 4.4*** | 3.1 ± 0.3*** |
| vif | 37.8 | 32.0 ± 4.2 | 7.9 ± 1.0 |
| vpx | 51.0 | 56.1 ± 12.1** | 4.8 ± 1.2** |
| vpr | 50.7 | 32.0 ± 6.6 | 5.9 ± 1.4* |
| tat | 81.9 | 20.9 ± 3.8 | 14.3 ± 1.7 |
| rev | 100.0 | 35.7 ± 6.3* | 12.3 ± 2.2 |
| env | 15.9 | 51.4 ± 2.9*** | 9.1 ± 0.6 |
| nef | 21.1 | 47.4 ± 5.0*** | 13.1 ± 1.3 |
*, P < 0.05; **, P < 0.01; ***, P < 0.001, by tests of the hypothesis that dS or dN equals the corresponding value for tat.
FIG. 3.
Plot of mean dS in each SIV gene for comparisons among six SIV genomes versus the proportion of overlap of the coding region with other genes. The line shown is the linear regression line y = 0.574 − 0.321x.
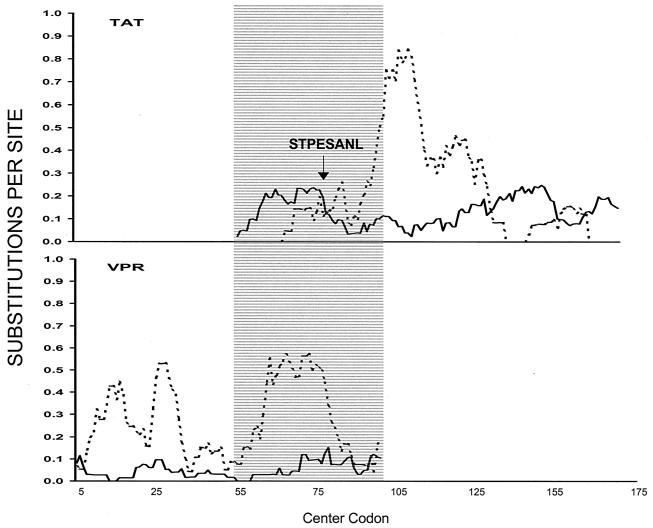
A sliding window analysis of dS and dN over the complete tat and vpr genes from the six complete SIV genomes showed that, in the region of overlap between tat and vpr, dS was relatively low in the tat reading frame, while dN was relatively high (Fig. 4). Conversely, in the vpr reading frame, the region of overlap showed a substantial peak in dS (Fig. 4), presumably a consequence of the high dN in the tat reading frame.
FIG. 4.
Plot of mean dS (dotted line) and mean dN (solid line) in nine-codon sliding window across the vpr and tat genes. The arrow shows the location of the STPESANL epitope, and the shaded area shows the region of overlap between the two reading frames.
Parallel changes in the Tat epitope.
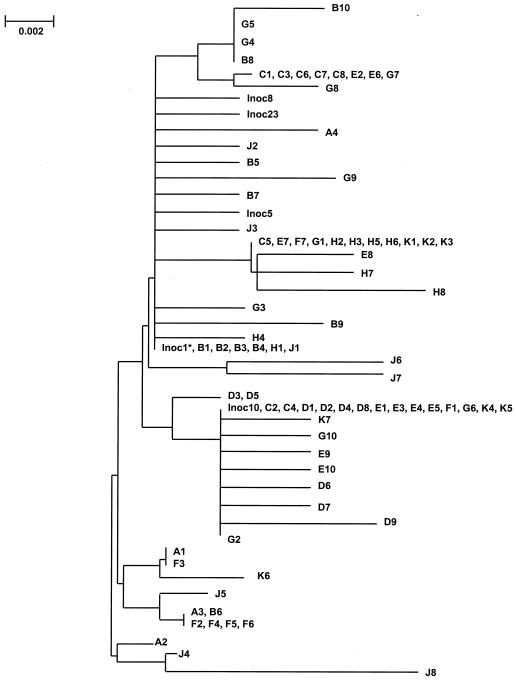
Figure 5 shows a phylogenetic tree of sequences from the inoculum and from viral samples taken from A*01+ monkeys. This phylogenetic tree showed very poor resolution, and clusters within the tree frequently included sequences derived from different monkeys. Thus, it seems that in this case phylogenetic analysis did not accurately reflect the evolutionary relationships among SIV sequences. As illustrated in Table 3, the same amino acid replacements occurred independently in the epitope region in different monkeys. It was evidently this independent or parallel occurrence of the same amino acid replacements in the epitope that caused misleading clustering pattern in the phylogenetic tree.
FIG. 5.
Phylogenetic tree of sequences from Mamu-A*01+ monkeys (individual monkeys are designated A to H and J and K) and inoculum (Inoc) sequences. The asterisk indicates a number of additional identical sequences. The phylogeny was constructed by the neighbor-joining method (17) on the basis of the number of nucleotide substitutions per site estimated by the method of Jukes and Cantor (8).
TABLE 3.
Amino acid replacements in Tat epitopea
| Inoculum(a) and/or isolate(s) | Change to SIVMAC239 Tat epitope positions 28 to 35
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| S | T | P | E | S | A | N | L | ||
| Inocula 1*, 5, 8, 23, and B1 to B5; B9; G9; H1; H4; J1; J3; J6 and J7 | |||||||||
| Inocula 10 and C2; C4; D1 and D2; D4; D6 to D9; E1; E3 to E5; E9 and E10; F1; G2; G6; G10; K4 and K5; K7 | P | ||||||||
| D3; D5 | A | ||||||||
| J4 | F | ||||||||
| B7 | A | ||||||||
| C1; C3; C6 to C8; E2; E6; G7 | I | ||||||||
| G8 | I | D | |||||||
| A4 | G | ||||||||
| C5; E7 and E8; F7; G1; H2 and H3; H5 to H8; K1 to K3 | L | ||||||||
| J2 | P | ||||||||
| B8; B10; G4 and G5 | D | ||||||||
| G3 | S | ||||||||
| A1; F3 | R | ||||||||
| A2 | Q | ||||||||
| A3; B6; F2; F4 to F6 | P | ||||||||
| K6 | K | R | |||||||
| J5 | P | P | |||||||
| J8 | F | Q | |||||||
Isolates from infected monkeys are identified by a letter (for the monkey) and a number.
Amino acid replacements in Tat were classified as parallel if they occurred independently in the viral populations within at least two A*01+ monkey hosts; the remainder were classified as nonparallel. A minimum of 27 amino acid replacements occurred in the Tat epitope in all A*01+ monkeys; of these, 19 (70.4%) were parallel between populations with at least one other host. By contrast, of 11 amino acid replacements observed in tat outside the epitope, none were parallel. The difference between epitope and nonepitope regions with respect to the proportions of parallel and nonparallel amino acid replacements was highly significant (P = 0.007; Fisher's exact test). Thus, parallel evolution of amino acid replacements occurs disproportionately in the portion of Tat that is under positive selection.
DISCUSSION
Our results show that, in a region of the SIV tat gene subject to positive selection driven by host CTL recognition, nonsynonymous nucleotide changes occurred in such a way as to cause predominately synonymous changes in the overlapping vpr reading frame. Positively selected amino acid changes known to eliminate binding by the host class I major histocompatibility complex (1) were able to occur in the tat reading frame with minimal change in the protein encoded by the overlapping vpr reading frame. In comparisons among SIV genomes, the vpr gene showed evidence of stronger purifying selection than did the tat gene, as evidenced by lower mean dN in vpr than in tat (Table 2). The region overlap between the two genes was characterized by a higher mean dN in tat than in vpr and by a high mean dS in vpr (Fig. 4). The peak of synonymous substitution in this region of vpr evidently was at least in part a consequence of nonsynonymous substitution in the same region of tat. Thus, this example shows that natural selection is able to favor amino acid residue replacements in one protein while simultaneously maintaining conserved and presumably functionally important residues in another protein encoded by an overlapping reading frame. This portion of the Vpr protein corresponds to the N-terminal portion of a relatively unstructured C-terminal domain directly following an amphipathic α-helix believed to be involved in Vpr dimerization (18). Although certain experimentally induced mutations in this region of HIV-1 did not affect dimerization (18), the relatively low dN values in this region of SIV (Fig. 2 and 4) suggest that it is subject to functional constraint in SIV.
One of the most useful techniques for studying the effects of natural selection at the molecular level is the comparison of the numbers of synonymous (dS) and nonsynonymous (dN) nucleotide substitutions per site (6). In most organisms, differences among genes with respect to dN are much more pronounced than differences with respect to dS. This occurs because differences in dN reflect differences with respect to the strength of purifying selection, which may vary substantially among different proteins. Differences in dS reflect mainly differences in mutation rate among genes, and these differences are usually not very substantial. In the case of SIV, however, the extent of variation in dS among genes is nearly as great as the extent of variation in dN. For the values in Table 2, the ratio of the highest to lowest dN values is 4.6, while the ratio of the highest to lowest dS values is 3.3. The coefficient of variation in dN values is 49.3%, while that in dS values is 34.0%. This is a very high level of variation in dS among genes. Furthermore, a high proportion of this variation (nearly 50%) is explainable by the extent of overlap with other genes (Fig. 3). Thus, even in the absence of positive selection, the existence of substantial overlap among genes has a major effect on the evolution of SIV.
In spite of the reduction of the observed rate of synonymous substitution due to overlapping reading frames, comparison of dS and dN showed robust evidence of positive selection on the STPESANL epitope of Tat. The mean value of dN in the epitope region of tat (Table 1) was 9 times as great as mean dS in within-sample comparisons and 14 times as great as mean dS in comparisons with the inoculum (Table 1). Thus, even if dS were increased by a factor of 3.3, representing the ratio of dS in pol to that in tat (Table 2), mean dN would still substantially exceed mean dS in the epitope region.
In the literature of molecular evolution, it is customary to distinguish between, on the one hand, convergent or parallel evolution and, on the other hand, chance occurrence of the same substitution in two or more independent lineages (14). It is assumed that true convergent or parallel evolution implies natural selection (3). In practice, however, it may be very difficult to distinguish between selectively driven and chance events of parallel amino acid replacement. The present case is exceptional in that there is independent evidence, derived from both the pattern of nucleotide substitution and immunological evidence of changes in peptide binding as a result of mutations (1), that natural selection has operated on the tat gene of SIV infecting A*01+ monkeys. In addition, the same amino acid replacements were seen to occur independently in viral populations inhabiting separate hosts (Table 3). Furthermore, several of these replacements are relatively radical amino acid changes from the point of view of the chemical properties of amino acids, particularly S→L, A→D, and L→R (Table 3). The present study thus provides a particularly well documented example of parallel evolution at the amino acid level under positive Darwinian selection.
Indeed, it seems likely that the existence of overlapping reading frames has enhanced the occurrence of parallel amino acid changes in this case. Although amino acid changes in the Tat epitope are selectively favored in virus infecting A*01+ monkeys, only a subset of possible changes were actually observed. The fact that nonsynonymous changes in tat which cause synonymous changes in vpr are observed disproportionately suggests that only a limited number of such changes will be permitted by purifying selection on the vpr gene. If the number of acceptable amino acid replacements in Tat is limited, the probability of parallel evolution will in turn be greater than that under standard amino acid substitution models.
ACKNOWLEDGMENTS
This research was supported by NIH grants GM34940 to A.L.H. and RR00167 and AI36466 to D.I.W. David I. Watkins is an Elizabeth Glaser Scientist.
REFERENCES
- 1.Allen T M, O'Connor D H, Jing P, Dzuris J L, Mothé B R, Vogel T U, Dunphy E, Leibl M E, Emerson C, Wilson N, Kunstman K J, Wang X, Allison D B, Hughes A L, Desrosiers R C, Altman J D, Wolinsky S M, Sette A, Watkins D I. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–390. doi: 10.1038/35030124. [DOI] [PubMed] [Google Scholar]
- 2.Crandall K A, Kelsey C R, Imanichi H, Lane H C, Salzman N P. Parallel evolution of drug resistance in HIV: failure of nonsynonymous/synonymous substitution rate to detect selection. Mol Biol Evol. 1999;16:372–382. doi: 10.1093/oxfordjournals.molbev.a026118. [DOI] [PubMed] [Google Scholar]
- 3.Doolittle R F. Convergent evolution: the need to be explicit. Trends Biochem Sci. 1994;19:15–18. doi: 10.1016/0968-0004(94)90167-8. [DOI] [PubMed] [Google Scholar]
- 4.Hein J, Støvlbæk J. A maximum-likelihood approach to analyzing non-overlapping and overlapping reading frames. J Mol Evol. 1995;40:181–189. doi: 10.1007/BF00167112. [DOI] [PubMed] [Google Scholar]
- 5.Holmes E C, Zhang L Q, Simmonds P, Ludlam C A, Leigh Brown A J. Convergent and divergent sequence evolution in the surface envelope glycoprotein of human immunodeficiency virus type 1 within a single infected patient. Proc Natl Acad Sci USA. 1992;89:4835–4839. doi: 10.1073/pnas.89.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hughes A L. Adaptive evolution of genes and genomes. New York, N.Y: Oxford University Press; 1999. [Google Scholar]
- 7.Hughes A L, Green J A, Garbayo J M, Roberts R M. Adaptive diversification within a large family of recently duplicated, placentally expressed genes. Proc Natl Acad Sci USA. 2000;97:3319–3323. doi: 10.1073/pnas.050002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 9.Krakauer D C. Stability and evolution of overlapping genes. Evolution. 2000;54:734–739. doi: 10.1111/j.0014-3820.2000.tb00075.x. [DOI] [PubMed] [Google Scholar]
- 10.Li W-H. Unbiased estimation of the rates of synonymous and non-synonymous substitution. J Mol Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- 11.Miyata T, Yasunaga T. Evolution of overlapping genes. Nature. 1978;272:532–535. doi: 10.1038/272532a0. [DOI] [PubMed] [Google Scholar]
- 12.Mizokami M, Orito E, Ochba K, Ikeo K, Lau J Y N, Gojobori T. Constrained evolution with respect to gene overlap of hepatitis B virus. J Mol Evol. 1997;44(Suppl.):S83–S90. doi: 10.1007/pl00000061. [DOI] [PubMed] [Google Scholar]
- 13.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 14.Nei M, Kumar S. Molecular evolution and phylogenetics. New York, N.Y: Oxford University Press; 2000. [Google Scholar]
- 15.Pavesi A, De Iaco B, Ilde Granero M, Porati A. On the informational content of overlapping genes in prokaryotic and eukaryotic viruses. J Mol Evol. 1997;44:625–631. doi: 10.1007/PL00006185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Regier D, Desrosiers R C. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS Res Hum Retrovir. 1990;6:1221–1231. doi: 10.1089/aid.1990.6.1221. [DOI] [PubMed] [Google Scholar]
- 17.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 18.Schüler W, Wecker K, de Rocquigny H, Baudat Y, Sire J, Roques B P. NMR structure of the (52-96) C-terminal domain of the HIV-1 regulatory protein Vpr: molecular insights into its biological functions. J Mol Biol. 1999;285:2105–2117. doi: 10.1006/jmbi.1998.2381. [DOI] [PubMed] [Google Scholar]
- 19.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Rosenberg H F, Nei M. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc Natl Acad Sci USA. 1998;95:3708–3713. doi: 10.1073/pnas.95.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]