Abstract
Arabidopsis (Arabidopsis thaliana) contains about 130 ATP-binding cassette (ABC) proteins, which are likely to contribute to the transport of diverse materials, including toxic substances. However, the substrates of ABC transporters remain unknown in most cases. We tested which ABC transporter is involved in detoxification of lead [Pb(II)]. Among the many tested, we found that the message level of only AtPDR12 increased in both shoots and roots of Pb(II)-treated Arabidopsis, suggesting that it may be involved in the detoxification of Pb(II). AtPDR12-knockout plants (atpdr12) were used to further test this possibility. In Pb(II)-containing medium, atpdr12 plants grew less well and had higher Pb contents than those of wild-type plants. In contrast, AtPDR12-overexpressing Arabidopsis plants were more resistant to Pb(II) and had lower Pb contents than wild-type plants. The mutant phenotypes and their Pb contents, as well as the localization of the GFP:AtPDR12 fusion protein at the plasma membrane, suggest that AtPDR12 functions as a pump to exclude Pb(II) and/or Pb(II)-containing toxic compounds from the cytoplasm. Inhibition of glutathione synthesis by addition of buthionine sulfoximine to the growth medium exacerbated the Pb(II)-sensitive phenotype of atpdr12 plants, consistent with a glutathione-dependent detoxification mechanism operating in parallel with an AtPDR12-dependent mechanism. Thus, we propose that AtPDR12 is an ABC transporter that contributes to Pb(II) resistance in Arabidopsis.
Heavy metals contaminate soils as a result of industrial processes and agricultural practices (Ross, 1994) and are a dangerous stress to plants that grow in the contaminated soils. Plants grown in contaminated agricultural fields contain the heavy metals and thus endanger animals who feed on them. Among the heavy metals, lead (Pb) is particularly important, since it is toxic to many organisms, widespread in the human environment, and causes multiple serious health problems in growing children and adults (as stated in a U.S. Environmental Protection Agency fact sheet at http://www.epa.gov/iaq/lead.html).
Many transporters have been identified that detoxify heavy metals in diverse organisms. Since cells have to remove heavy metals from the cytoplasm to avoid toxicity, some transporters extrude them across the plasma membrane and others sequester them into inactive organelles. Transporters that have been shown to extrude heavy metals across the plasma membrane are P-type pumps, including ZntA from Escherichia coli, a Pb(II)/Cd(II)/Zn(II)-transporting ATPase that mediates resistance to toxic concentrations of Pb(II), Cd(II), and Zn(II) by pumping these metals out of cells (Rensing et al., 1997; Sharma et al., 2000). CAD2 from yeast (Saccharomyces cerevisiae) has a similar role (Shiraishi et al., 2000). Many different types of transporters sequester heavy metals into inactive organelles. ScYCF1 is an ATP-binding cassette (ABC) transporter of yeast that contributes to Cd(II) resistance by pumping Cd(II) conjugated to glutathione into the vacuole (Szczypka et al., 1994; Li et al., 1997). ScYCF1 has been found to confer Pb(II) resistance as well, possibly by a similar mechanism (Song et al., 2003). In Schizosaccharomyces pombe, a half-size ABC transporter has been shown to transport phytochelatin-Cd complexes (Ortiz et al., 1995). In plants, CAX, a Ca2+/H+ exchanger, has been shown to exchange Cd(II) with protons at the vacuolar membrane (Hirschi et al., 2000), thereby sequestering Cd into the vacuole. An Arabidopsis (Arabidopsis thaliana) P-type ATPase, AtHMA3, improves Cd(II) and Pb(II) resistance when expressed in Cd(II)-sensitive ycf1 yeast and is localized at the vacuolar membrane; it has therefore been suggested to function as a vacuolar sequester of toxic heavy metals (Gravot et al., 2004).
ABC transporters have been reported to transport many cytotoxic compounds, such as drugs, in addition to heavy metals (Rogers et al., 2001; Sanchez-Fernandez et al., 2001; Martinoia et al., 2002). Arabidopsis has about 130 ABC proteins, but the functions of most of them are unknown (Sanchez-Fernandez et al., 2001). Many investigators have searched for a plant ortholog of ScYCF1, but it has not yet been found. The only ABC transporter that has been suggested to transport Cd(II) is AtMRP3, which partially restores Cd(II) resistance when expressed in a Δycf1 mutant (Tommasini et al., 1998). ABC proteins of Arabidopsis play important roles in normal developmental processes as well. For example, AtMDR1 and AtPGP1 have roles in polar-auxin transport across the plasma membrane (Noh et al., 2001). The double mutant atmdr1atpgp1 shows auxin-signaling or distribution-defective phenotypes, such as dwarfism, and reduced apical dominance in adult plants (Noh et al., 2001). AtATM1, a mitochondrial inner membrane ABC protein, transports Fe/S cluster precursor from mitochondrial matrix to the cytosol (Kispal et al., 1999; Kushnir et al., 2001). AtPMP2 is localized to the peroxisome and transports acyl-CoA derivatives from the cytosol into the peroxisomal matrix (Zolman et al., 2001). Soluble ABC protein AtNAP1 is required for transport of the chlorophyll precursor protoporphyrin IX from the chloroplast envelope into the stroma (Moller et al., 2001). An AtMRP5 knockout plant, atmrp5-1, shows abnormal stomatal movement in light and, when treated with calcium or ABA, and displays reduced water use, indicating a role for AtMRP5 in guard cell functioning (Klein et al., 2003).
In this paper, we present evidence that a member of the pleiotropic drug resistance (PDR) subfamily of ABC transporters in Arabidopsis has a role in Pb(II) detoxification. We show that AtPDR12 expression is strongly induced by Pb(II) treatment and that AtPDR12-knockout plants are more sensitive to Pb(II) than wild-type plants. In addition, AtPDR12-knockout plants contain more Pb than wild-type plants and AtPDR12-overexpressing plants are more resistant to Pb(II) than wild-type plants. These results, as well as localization of AtPDR12 to the plasma membrane, suggest that AtPDR12 confers Pb(II) resistance by pumping Pb(II) and/or Pb(II)-containing toxic compounds out of the cell from the cytoplasm.
RESULTS
Expression of AtPDR12 Is Elevated by Pb(II) Treatment
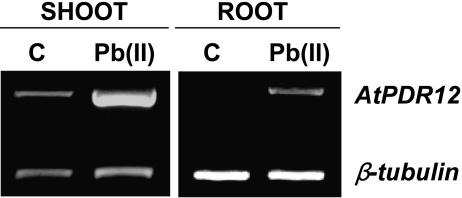
As a first step in the identification of ABC transporters involved in Pb(II) transport, we searched databases (http://www.ncbi.nlm.nih.gov/ and http://www.ddbj.nig.ac.jp/E-mail/clustalw-e.html) to select Arabidopsis ABC transporters with high amino acid sequence homology to ABC transporters of other organisms that are reported to transport toxic substances. We then performed reverse transcription (RT)-PCR experiments to test whether the transcript levels of these candidate ABC transporters are up-regulated when Arabidopsis is treated with Pb(II). The expression of AtMRP4, AtPDR1, AtPDR4, AtPDR6, and AtPDR9 did not change much upon Pb(II) treatment (data not shown), whereas that of AtPDR12 increased both in shoots and in roots (Fig. 1). Without Pb(II) treatment, the AtPDR12 transcript was expressed in shoots but not detected in roots.
Figure 1.
Pb(II)-induced elevation of AtPDR12 expression detected by RT-PCR. Total RNA was extracted from shoots and roots of 2-week-old wild-type Arabidopsis without (C) or with (Pb[II]) 0.5 mm Pb(NO3)2 treatment for 24 h. β-Tubulin was used as a loading control.
AtPDR12-Knockout Plants Are Less Resistant to Pb(II) Than Wild-Type Plants
To test whether AtPDR12 contributes to Pb(II) resistance in plants, we obtained seeds of 2 AtPDR12-knockout alleles from the Salk Institute Genomic Analysis Laboratory. These knockout plants have T-DNA inserted into 2 different sites of the 21st exon of the AtPDR12 gene (Fig. 2A; http://signal.salk.edu/cgi-bin/tdnaexpress, SALK stock nos. SALK_ 013945 and SALK_005635). Homozygous knockout plants (atpdr12-1 and atpdr12-2) were selected by PCR using 1 T-DNA-specific primer and 2 AtPDR12-specific primers with genomic DNA as a template. The PCR using 2 AtPDR12-specific primers did not produce any product (right lanes of Fig. 2B, subsections a and c), but the PCR using a T-DNA-specific primer and 1 AtPDR12-specific primer produced 730- and 760-bp-size bands from genomic DNA of atpdr12-1 and 2, respectively, as expected (right lanes of Fig. 2B, subsections b and d). These PCR products were sequenced using T-DNA-specific primers, and insertion sites were confirmed. To confirm that the homozygous lines do not have the AtPDR12 transcript, RT-PCR was performed using total RNA extracted from the plants. PCR using 2 AtPDR12-specific primers did not produce any product from the homozygous knockout plants, as it did from wild-type plants (Fig. 2C).
Figure 2.
Selection of AtPDR12-knockout plants. A, T-DNA insertion sites in the AtPDR12 gene in 2 alleles of AtPDR12-knockout plants. The dark gray and white boxes represent the SALK_013945 line (atpdr12-1) and SALK_005635 line (atpdr12-2), respectively. B, PCR for genotype analysis using genomic DNA as template and two gene-specific primers (subsections a and c) and a T-DNA-specific and one AtPDR12-specific primer (subsections b and d). C, RT-PCR using total RNA from wild-type and AtPDR12-knockout plants. Note the absence of the AtPDR12 message in atpdr12-1 and atpdr12-2 plants. Total RNA of wild-type and AtPDR12-knockout plants was extracted from 14-d-old Arabidopsis seedlings treated with 0.5 mm Pb(NO3)2 for 24 h.
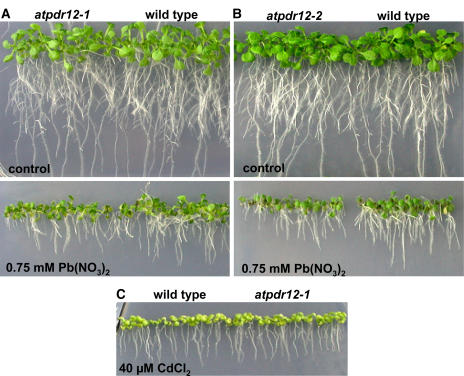
We obtained the next generation of seeds from the selected AtPDR12-homozygous knockout plants and tested their resistance to Pb(II) and Cd(II). In control medium, the growth of AtPDR12-knockout plants (atpdr12-1 and atpdr12-2) was similar to that of wild-type plants, and they reproduced normally (Fig. 3, A and B, top). In Pb(II)-containing medium, the growth of roots and shoots was reduced more in AtPDR12-knockout plants than in wild-type plants (Fig. 3, A and B, bottom). Although addition of Pb(NO3)2 may have caused some precipitation of P and S, the plant symptoms did not resemble those of P and S deficiency, which cause dark-green coloration and chlorosis of young leaves, respectively. Rather, the Pb(II)-treated plants displayed the typical symptom of Pb(II)-treated plants, reduced root growth. Growth of AtPDR12-knockout plants was not different from that of wild-type plants in 30, 40, or 50 μm Cd(II)-containing media (Fig. 3C; data not shown) or in media containing Zn(II), Cu(II), Al(III), or the antifungal drug sclareol (data not shown). These results demonstrate that AtPDR12 specifically confers Pb(II) resistance, at least under our experimental conditions.
Figure 3.
Reduced growth of AtPDR12-knockout plants in medium containing Pb(NO3)2. A to C, Photographs of wild-type and AtPDR12-knockout homozygous plants (atpdr12-1 and -2) germinated and grown on one-half MS agar media without (control) or with Pb(NO3)2 (A and B) or CdCl2 (C) for 3 weeks.
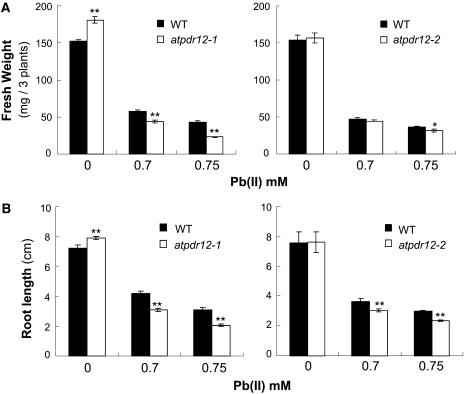
For quantitative analyses of the phenotypes, we tested various concentrations of Pb(NO3)2 and chose two concentrations that showed the most striking differences between the knockout and wild-type plants. Figure 4 shows the fresh weight and root length of AtPDR12-knockout and wild-type plants grown on Pb(II)-containing one-half Murashige and Skoog (MS) agar plates without (control) and with 0.7 mm or 0.75 mm Pb(NO3)2 for 4 weeks. In the control medium without additional Pb(II), atpdr12-1 plants had 1.2-fold higher fresh weight and 1.1-fold higher root length than wild-type plants. In contrast, atpdr12-2 plants did not display any differences in fresh weight and root length compared to wild-type plants (Fig. 4, A and B). However, in Pb(NO3)2-containing media, both fresh weight and root length were more severely reduced in AtPDR12-knockout plants than in wild-type plants; the fresh weights of atpdr12-1 and atpdr12-2 were reduced to 52% and 87% of wild type, and root lengths were decreased to 67% and 79% of wild type in medium containing 0.75 mm Pb(NO3)2 (Fig. 4, A and B).
Figure 4.
Phenotypic analysis of wild-type and AtPDR12-knockout plants (atpdr12-1 and atpdr12-2) grown in Pb(NO3)2-containing media. A, Fresh weight. B, Root length. Wild-type and AtPDR12-knockout seeds were sown and grown on Pb(NO3)2-containing one-half MS plates for 4 weeks. Black and white bars represent wild-type and AtPDR12-knockout plants, respectively. The average of 2 experiments with 4 replicates each is shown (n = 8). Error bars represent se. **P < 0.001 and *P < 0.05 by Student's t test.
AtPDR12-Knockout Plants Contain More Pb
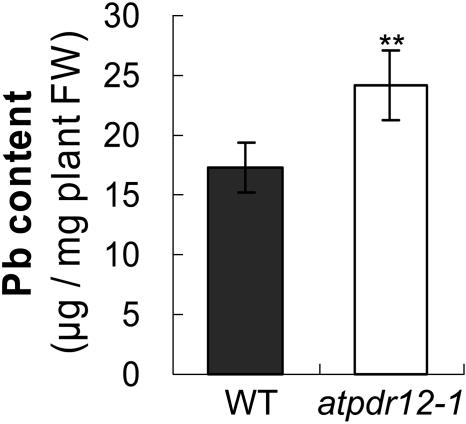
We measured the Pb contents of 2-week-old wild-type and AtPDR12-knockout plants whose roots were soaked in 1 mm Pb(NO3)2 for 2 d. Shoots of atpdr12-1 plants contained 1.4-fold more Pb than those of wild-type plants (P < 0.001; Fig. 5). Although the Pb content of atpdr12-2 plants seemed slightly higher than that of wild-type plants in Pb(NO3)2-containing media, the difference was not statistically significant (data not shown). Additionally, the phenotypes of atpdr12-2 plants were less pronounced than those of atpdr12-1 plants with respect to fresh weight and root length. We have no valid explanation for this difference, except perhaps for the observation that the T-DNA insertion site of atpdr12-2 is closer to the stop codon than that of atpdr12-1. We then examined the possibility that Pb treatment of the knockout plants causes changes in the levels of other metal ions, using an inductively coupled plasma spectrometer. While potassium, copper, magnesium, iron, zinc, and manganese levels remained the same, the Pb level was confirmed to be higher in AtPDR12-knockout plants (atpdr12-1) than in the wild type (data not shown).
Figure 5.
Increased accumulation of Pb in AtPDR12-knockout plants. Wild-type and AtPDR12-knockout plants were grown for 2 weeks on one-half MS agar plates, then their roots were soaked with 1 mm Pb(NO3)2 for 2 d, and shoots were harvested in pools of 3 plants for measurement of the Pb contents using an atomic absorption spectrometer. A representative from 3 independent experiments with similar results is shown (n = 17 in the representative experiment; n = 1 means a pool of 3 plants.) Error bars represent se. **P < 0.001 by Student's t test.
AtPDR12-Overexpressing Arabidopsis Plants Are More Resistant to Pb(II)
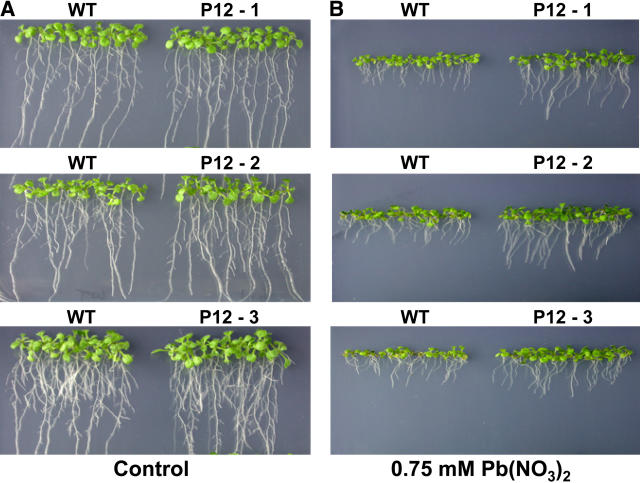
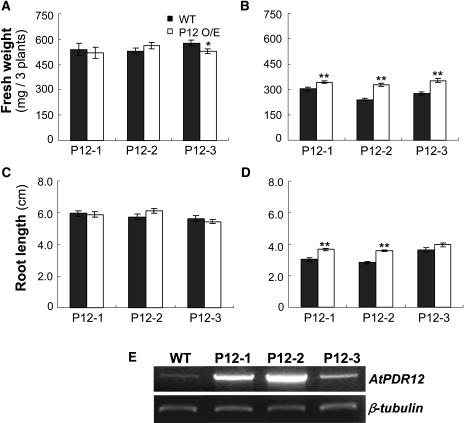
If AtPDR12 contributes to Pb(II) resistance, AtPDR12-overexpressing plants may be more resistant to Pb(II) than wild-type plants. The AtPDR12 gene was inserted into the pBI121 vector with a 35S promoter (pBI121:AtPDR12) and the construct was introduced into wild-type Arabidopsis plants. We selected 3 T2 lines that had only 1 copy of pBI121:AtPDR12, and T3 homozygous lines were chosen from the next generation and tested for their resistance to Pb(II). In one-half MS control medium without Pb(NO3)2, growth of AtPDR12-overexpressing and wild-type plants was similar (Fig. 6A). However, in medium containing 0.75 mm Pb(NO3)2, the shoots and roots of AtPDR12-overexpressing plants grew slightly better than those of wild-type plants (Fig. 6B). Quantitative analyses showed that the fresh weights and root lengths of AtPDR12-overexpressing and wild-type plants were similar in control medium except for one AtPDR12-overexpressing line, which had a slightly reduced fresh weight (Fig. 7, A and C). In Pb(NO3)2-containing medium, fresh weight and root length were consistently higher in the 3 AtPDR12-overexpressing plants than in the wild-type plant; the fresh weight of AtPDR12-overexpressing lines were 1.14-, 1.37-, and 1.28-fold higher than that of the wild type (Fig. 7B), and root lengths of 2 AtPDR12-overexpressing lines were 1.23- and 1.29-fold longer than that of the wild-type plant (Fig. 7D). To investigate the AtPDR12 transcript level in AtPDR12-overexpressing plants, total RNA was extracted from Pb(II)-treated AtPDR12-overexpressing and wild-type plants. The 3 AtPDR12-overexpressing plants had a higher AtPDR12 transcript level compared to wild-type plants (Fig. 7E).
Figure 6.
Enhanced Pb(II) resistance of AtPDR12-overexpressing Arabidopsis. A, Growth in control medium. B, Growth in medium containing 0.75 mm Pb(NO3)2. T3 homozygous lines of AtPDR12-overexpressing plants and wild-type plants were grown vertically on one-half MS agar media with 1.5% Suc with or without 0.75 mm Pb(NO3)2. Photographs were taken 2 weeks after sowing. P12-1, -2, and -3, Three independent lines of AtPDR12-overexpressing T3 homozygous plants.
Figure 7.
Quantitative analyses of the enhanced Pb(II) resistance of AtPDR12-overexpressing Arabidopsis. A and B, Fresh weight in one-half MS agar plates (A) and in Pb(NO3)2-containing media (B). C and D, Root length in one-half MS agar plates (C) and in Pb(NO3)2-containing media (D). Seeds of wild-type and AtPDR12-overexpressing lines were sown and grown on one-half MS plates with and without 0.75 mm Pb(NO3)2 for 2 weeks. A representative result of 2 independent experiments that gave similar results is shown (fresh weight, n = 10; root length, n = 60 in each experiment). Error bars represent se. *P < 0.05 and **P < 0.001 by Student's t test. E, AtPDR12 transcript levels. Total RNA of wild-type and AtPDR12-overexpressing plants was extracted from shoots and roots of 14-d-old Arabidopsis seedlings treated with 0.5 mm Pb(NO3)2 for 24 h. P12-1, -2, and -3, Three independent lines of AtPDR12-overexpressing T3 homozygous plants.
To test whether AtPDR12-overexpressing plants had reduced Pb contents compared to wild-type plants, AtPDR12-overexpressing and wild-type plants were grown on 0.7 mm Pb(NO3)2-containing medium for 2 weeks and then their shoots were prepared. Pb contents of 3 independent lines of AtPDR12-overexpressing plants (P12-1, P12-2, and P12-2) had Pb contents that were 73%, 68%, and 72% of that of wild-type plants (P < 0.05 for all 3 lines; Fig. 8).
Figure 8.
Reduced accumulation of Pb in AtPDR12-overexpressing plants. Wild-type and 3 independent lines of AtPDR12-overexpressing plants (P12-1, -2, and -3) were grown for 2 weeks on 0.7 mm Pb(NO3)2-containing medium. Their shoots were then harvested for measurement of Pb content using an atomic absorption spectrometer. Black and white bars represent wild-type and AtPDR12-overexpressing lines, respectively. The average of 2 independent experiments, each constituted of 8 independent replicates, is shown (n = 16). Error bars represent se. *P < 0.05 by Student's t test.
Pb(II) Resistance Conferred by AtPDR12 Is Independent of Glutathione
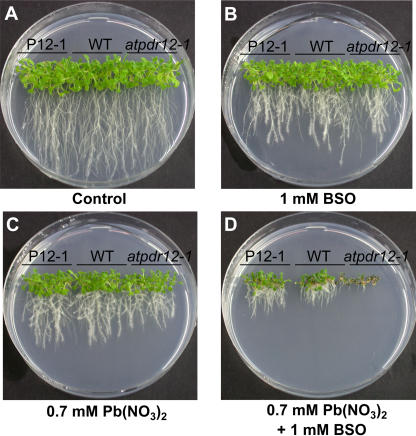
Because Pb(II) can form complexes with thiol compounds, we investigated whether the Pb(II) detoxification mechanism of AtPDR12 is related to glutathione. To test this possibility, we compared the growth of wild-type and AtPDR12-transgenic plants in medium containing buthionine sulfoximine (BSO), an inhibitor of glutathione synthesis. In control or 1 mm BSO-containing media, the growth rates of AtPDR12-overexpressing (P12-1), -knockout (atpdr12-1), and wild-type plants were similar (Fig. 9, A and B). In 0.7 mm Pb(NO3)2-containing medium, AtPDR12-overexpressing and -knockout plants showed slightly enhanced and slightly reduced growth, respectively, compared to the wild-type plants (Fig. 9C), as found in previous experiments. In medium containing both 1 mm BSO and 0.7 mm Pb(NO3)2, this growth difference was even more striking than in medium containing Pb(NO3)2 alone (Fig. 9D). This amplified phenotypes under low-glutathione condition indicates that AtPDR12 does not use glutathione for Pb(II) detoxification.
Figure 9.
Glutathione-independent Pb(II)-resistance mechanism of AtPDR12. A, Growth of AtPDR12 mutants in control medium. B, Growth in 1 mm BSO-containing medium. C, Growth in medium containing 0.7 mm Pb(NO3)2. D, Growth in both 1 mm BSO and 0.7 mm Pb(NO3)2-containing medium. AtPDR12-overexpressing (P12-1), -knockout (atpdr12-1), and wild-type (WT) plants were grown vertically on one-half MS agar media without or with Pb(NO3)2 or BSO. Photographs were taken 3 weeks after sowing.
AtPDR12 Is Localized at the Plasma Membrane and Expressed in Many Organs of Arabidopsis
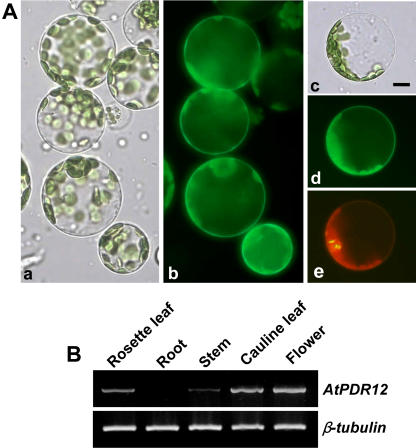
To investigate the localization of AtPDR12, we isolated protoplasts from Arabidopsis whole seedlings and introduced a green fluorescent protein (GFP):AtPDR12 construct (GFP gene fused to the 5′ end of AtPDR12 gene) into them using the polyethylene glycol transformation method (Jin et al., 2001). After 30 h of incubation, the fluorescence of GFP:AtPDR12 was localized at the boundary of the cell, most likely the plasma membrane (Fig. 10A, subsections a and b). To confirm the plasma membrane localization, we used AtAHA2:red fluorescent protein (RFP) as a control. AtAHA2 is an Arabidopsis H+-ATPase that is known to localize to the plasma membrane. When GFP:AtPDR12 and AtAHA2:RFP were transformed into the Arabidopsis protoplasts at the same time, AtAHA2 and AtPDR12 were colocalized at the boundary of the cell (Fig. 10A, subsections c–e), confirming the plasma membrane localization of GFP-AtPDR12. Plasma membrane localization of AtPDR12 was confirmed by transforming the protoplasts with a second construct of the fused genes, AtPDR12:GFP (GFP gene fused to the 3′ end of AtPDR12 gene; data not shown).
Figure 10.
Intracellular localization of the GFP:AtPDR12 fusion protein and tissue-specific expression in Arabidopsis. A, Subsections a and b, Localization of GFP:AtPDR12. Subsection a, Bright field image of mesophyll cell protoplasts transformed with GFP:AtPDR12. Subsection b, Fluorescent image of the cells shown in subsection a. Subsections c to e, Colocalization of GFP:AtPDR12 and AtAHA2:RFP. Subsection c, Bright field image of a mesophyll cell protoplast transformed with GFP:AtPDR12 and AtAHA2:RFP. Subsection d, Fluorescence image of GFP:AtPDR12 in the cell shown in subsection c. Subsection e, Fluorescence image of AtAHA2:RFP in the cell shown in subsection c. Bar = 5 μm. B, RT-PCR analysis of the AtPDR12 expression pattern. Total RNA was extracted from 2-week-old plants grown on one-half MS agar plate (rosette leaf and root) and 5-week-old plant grown in soil (cauline leaf, stem, and flower).
To investigate the tissue-specific expression pattern of AtPDR12 in wild-type plants, we extracted total RNA from each tissue and performed RT-PCR. AtPDR12 transcripts were detected in rosette leaves, stems, cauline leaves, and flowers, but not in roots (Fig. 10B).
DISCUSSION
AtPDR12 has been studied before (Van den Brule and Smart, 2002; Campbell et al., 2003), but information about it has been limited to its induction by defense-related molecules and the susceptibility of its knockout plants to sclareol. In this study, we cloned the AtPDR12 gene and studied its function in Pb(II) resistance. Results we obtained are highly suggestive of a role for AtPDR12 in Pb(II) resistance. First, we showed that the expression level of AtPDR12 was strongly induced by Pb(II) treatment (Fig. 1). Second, we showed that the shoots and roots of AtPDR12-knockout plants grow less on Pb(II)-containing medium than those of wild-type plants (Figs. 3 and 4). Third, we showed that AtPDR12-overexpressing plants, when grown on Pb(II)-containing medium, had improved growth of shoots and roots compared to wild-type plants (Figs. 6 and 7). We propose that the improved resistance of the AtPDR12-overexpressing plants is due to the exclusion of Pb(II), because GFP:AtPDR12 was localized to the plasma membrane (Fig. 10), and the AtPDR12-knockout and -overexpressing plants had increased or reduced levels of Pb, respectively (Figs. 5 and 8). It remains unknown whether AtPDR12 transports Pb(II) and/or Pb-containing compounds directly or indirectly. It may also transport other substrates generated by Pb(II) toxicity, or it may even function as a regulator of an unknown Pb(II) transporter. This problem will be difficult to resolve until a flux assay for Pb(II) becomes available.
To test whether AtPDR12 responds specifically to Pb(II), AtPDR12-knockout plants were tested for growth on media containing various metal ions such as Zn(II), Cd(II), Cu(II), and Al(III). However, their growth rates were not different from those of wild-type plants (data not shown). AtPDR12 is unique in the sense that it confers resistance to Pb(II) but not to Cd(II), whereas the heavy metal transporters of other organisms, including ZntA and Ycf1, confer resistance to both Pb(II) and Cd(II). Thus, AtPDR12 appears to be the first ABC transporter gene that contributes specifically to Pb(II) resistance.
Not much is known about the Pb(II) resistance mechanisms of plants. We previously showed that oxalate secretion is important for Pb resistance in certain varieties of rice (Yang et al., 2000). Glutathione is also known to be important for Pb(II) resistance. Glutathione is required for glutathione-conjugated vacuolar sequestration, an important pathway of heavy metal detoxification in plants (Grill et al., 1989; Li et al., 1997). Glutathione is a precursor of phytochelatin, which binds to Pb(II), and phytochelatin synthesis increases in plants exposed to Pb(II) (Mehra et al., 1995). In addition, we showed that expression of YCF1 enhanced plant resistance to Pb(II) via a glutathione-dependent pathway (Song et al., 2003). In this paper, we have confirmed the importance of glutathione in Pb(II) resistance, since addition of BSO further reduced the growth of all plant genotypes tested, including the wild-type, AtPDR12-knockout, and AtPDR12-overexpressing plants, all of which already displayed growth in the presence of Pb(II) (Fig. 9, compare C and D). When the glutathione-dependent mechanisms of Pb(II) resistance were blocked by BSO, the contribution of AtPDR12 to Pb(II) resistance became more apparent (Fig. 9D). Thus, plants seem to have at least two distinct mechanisms for Pb(II) resistance, a glutathione-dependent mechanism and a glutathione-independent mechanism that depends on AtPDR12.
AtPDR12 may function in plant defense responses, since previous reports showed that the AtPDR12 transcript was highly elevated under sclareol treatment (Van den Brule and Smart., 2002) and germination of AtPDR12-knockout seeds was inhibited by sclareol (Campbell et al., 2003). Supporting this possibility, the AtPDR12 transcript is induced by fungal-pathogen inoculation or treatment with defense-related signal molecules, including salicylic acid, jasmonic acid, methyl jasmonate, and ethylene (Campbell et al., 2003). Although we used the same line of AtPDR12-knockout plants used by Campbell et al. (2003), atpdr12-2, we were not able to observe any remarkable phenotypes on sclareol-containing media (data not shown). However, our results do not exclude the possibility that AtPDR12 is associated with other plant defense responses.
AtPDR12 may have functions unrelated to Pb detoxification as well, because AtPDR12 is expressed in shoots and flowers under normal conditions (Fig. 9). It is commonly found in many organisms that one protein has multiple functions that are not related to each other (Lee et al., 2002; Kim et al., 2003). Moreover, ABC transporters of many organisms are known to transport more than one substrate. For example, ScPDR5 exports cycloheximide, oligomycin, steroids, and antifungal drugs (Kolaczkowski et al., 1996; Mahe et al., 1996). Therefore, AtPDR12 may transport materials important for the normal growth and development of plants. Further studies are necessary to understand the substrates of AtPDR12 under normal conditions.
In summary, our results strongly support the possibility that AtPDR12 contributes to the resistance of Arabidopsis to Pb(II) by pumping or regulating transport of Pb(II) or Pb(II)-related toxic compounds to the exterior of the cell.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds of Arabidopsis (Arabidopsis thaliana) wild-type ecotype Colombia-0, AtPDR12-overexpressing, and AtPDR12-knockout plants were surface sterilized, placed in the dark at 4°C for 2 d, and then sown on agar plates of one-half MS with 1.5% Suc (Murashige and Skoog, 1962) or in soil. For phenotype tests, heavy metals and drugs were added to one-half MS medium.
Cloning of AtPDR12 and Plasmid Construction
The full AtPDR12 sequence was isolated from a cDNA by PCR. The cDNA was synthesized using total RNA extracted from Pb(II)-treated roots of Arabidopsis wild-type plants. The primers used were AtPDR12-TFF (5′-CCCGGGGGGGATCCATGGAGGGAACTAGTTT TCACCAAGCGAGTA-3′) and AtPDR12-TFR (5′-GGATCCGCGGCCGCCTATCGTTTTTGGAAATTGAAACTCTTGATTC-3′). The primers contained two restriction enzyme sites for easy cloning. The PCR product was ligated into the pGEM-T easy vector (Promega, Madison, WI) using T4 DNA ligase. The fidelity of the AtPDR12 sequence (GenBank accession no. NM_101421) was confirmed by automated DNA sequencing (ABI 3100, Perkin-Elmer Applied Biosystems, Foster City, CA).
Assay of ABC Protein Transcript Levels after Pb(II) Treatment of Arabidopsis
To test expression levels of ABC proteins under Pb(II) stress, total RNA was extracted from Pb(II)-treated and water-treated roots of Arabidopsis. RT-PCR was performed using an RT-PCR kit (Invitrogen, Carlsbad, CA) with each ABC protein-specific primer set. The primer sequences employed are listed in Table I. As a control for RT-PCR, Tub1 and Tub2 primers were used.
Table I.
Primer sequences used for RT-PCR that tested transcript levels of some ABC proteins of Arabidopsis
| Primer Name | Primer Sequence (5′ to 3′) |
|---|---|
| AtMRP4-RF | CCACTTGTGCTCAAGGGGATCACTCTTGAC |
| AtMRP4-RR | TCCAGCAAGCGAGCCGGGCTATCGAACTCT |
| AtPDR1-RF | GGATCCCCCGGGATGAGGACAGTGAGGAATACA |
| AtPDR1-RR | AAGCCCACTAACACGTAGCAATCGG |
| AtPDR4-RF | GCGTTCGAATGGTCCGCGGTGAAGTTC |
| AtPDR4-RR | CAGTAATGACAGAGGAAGAATAGGCAGTC |
| AtPDR6-RF | ACCGGGATCGAATGATCTCTACTTCC |
| AtPDR6-RR | AGGCGATGTGGACAACGGCTACAACT |
| AtPDR9-RF | CGGGAAAGATTCGCAGGGATGTACTCAGCG |
| AtPDR9-RR | GGCAATGGGAAAAGCGATTTGAACCGC |
| AtPDR12-RF | GAAGCGGCTTTAGGAGTCGATTTCGC |
| AtPDR12-RR | CGTCCACTCGAATCCTATCATAGCG |
| Tub1 | CTCACAGTCCCGGAGC TGACAC |
| Tub2 | GCTTCAGTGAACTCCATCTCGT |
Selection of AtPDR12-Knockout Arabidopsis
Two alleles of AtPDR12-knockout Arabidopsis seeds, SALK_013945 and SALK_005635, were obtained from the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu/cgi-bin/tdnaexpress). The seeds were sown and grown on soil for 4 weeks, after which genomic DNA was extracted from the plants. Homozygous AtPDR12-knockout plants were selected by PCR using 3 primers. In the case of the SALK_013945 line, a T-DNA-specific primer (pROKLBb1, 5′-GCGTGGACCGCTTGCTGCAACT-3′) and 2 AtPDR12-specific primers (AtPDR12-TDNA-F, 5′-ATCAACACGGTCCCAGGAATGC-3′ and AtPDR12-TDNA-R, 5′-CCATACGTTCTCGTGCAAGCGA-3′) were used. In the case of the SALK_005635 line, a T-DNA-specific primer and 2 gene-specific primers (AtP12K/O-005635-LP, 5′-CTAATCCAGTGTTGTTGATCATCCT-3′ and AtP12K/O-005635-RP, 5′-GAGCTAATCAAGGAGCTAAGCCAGC-3′) were used.
Production of AtPDR12-Overexpressing Arabidopsis
To make AtPDR12-overexpressing plants, AtPDR12 was fused to pBI121 using the BamHI site of the pBI121 vector (Jefferson et al., 1987), and the construct was introduced into the Agrobacterium tumefaciens GV3101 strain, which was used to transform Arabidopsis ecotype Columbia by the floral-dipping method (Clough and Bent, 1998). The transformed seeds were selected on 30 μg/mL kanamycin-containing one-half MS agar plates. Transcripts of AtPDR12-overexpressing lines were detected by RT-PCR using primers described in Table I and total RNA extracted from Pb(NO3)2-treated 2-week-old seedlings.
Measurements of Fresh Weight, Root Length, and Metal Contents in Arabidopsis
Arabidopsis plants were grown on one-half MS-agar medium supplemented with 1.5% (w/v) Suc without (control) or with 0.7 mm or 0.75 mm Pb(NO3)2 for 4 weeks or 2 weeks. Plants were collected and weighed and their root length was measured. The shoots of plants were digested with 11 n HNO3 at 200°C overnight. Digested samples were diluted with 0.5 n HNO3 and analyzed using an atomic absorption spectrometer (SpectrAA-800, Varian, Palo Alto, CA) or inductively coupled plasma spectrometer (IRIS/AP, Thermo Jarrell Ash, Franklin, MA).
Intracellular Localization of AtPDR12 in Arabidopsis
To investigate AtPDR12 localization, we made 2 different AtPDR12-GFP fusion constructs, an N-terminal and a C-terminal GFP construct, which we denoted GFP:AtPDR12 and AtPDR12:GFP, respectively. The former construct was made by inserting AtPDR12 into the 326GFP-3G vector (CLONTECH Laboratories, Palo Alto, CA) using the SmaI and SalI sites, and the latter construct was made by inserting AtPDR12 into the 326GFP vector (CLONTECH) using the BamHI site. AtAHA2:RFP was used as a plasma membrane marker (Kim et al., 2001). The two constructs were introduced into Arabidopsis protoplasts by the polyethylene glycol method (Abel and Theologis, 1998; Jin et al., 2001). Expression of constructs was monitored using a Zeiss Axioskop2 fluorescence microscope (Zeiss, Oberkochen, Germany). The filter sets used were Filter set 44 (excitation, BP 455–495; beamsplitter, FT500; emission, BP 505–555) for GFP and Filter set 20 (excitation, BP 540–552; beamsplitter, FT560; emission, BP 575–640) for RFP.
Tissue-Specific Expression of AtPDR12
To investigate the tissue-specific RNA level of AtPDR12, RT-PCR was performed using total RNA extracted from each organ. Rosette leaves and roots were harvested from wild-type plants grown on one-half MS plate for 2 weeks. Flowers, stems, and cauline leaves were harvested from wild-type plants grown in soil for 5 weeks.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number NM_101421.
Acknowledgments
We thank the Salk Institute Genomic Analysis Laboratory for the AtPDR12-knockout seeds and Mrs. Jung and Han for technical assistance.
This work was supported by Pohang Steel Company (grants to Y.L.) and by the National Research Laboratory program of the Ministry of Science and Technology of Korea.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.058107.
References
- Abel S, Theologis A (1998) Transient gene expression in protoplasts of Arabidopsis thaliana. Methods Mol Biol 82: 209–217 [DOI] [PubMed] [Google Scholar]
- Campbell EJ, Schenk PM, Kazan K, Penninckx IA, Anderson JP, Maclean DJ, Cammue BP, Ebert PR, Manners JM (2003) Pathogen-responsive expression of a putative ATP-binding cassette transporter gene conferring resistance to the diterpenoid sclareol is regulated by multiple defense signaling pathways in Arabidopsis. Plant Physiol 133: 1272–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Gravot A, Lieutaud A, Verret F, Auroy P, Vavasseur A, Richaud P (2004) AtHMA3, a plant P1B-ATPase, functions as a Cd/Pb transporter in yeast. FEBS Lett 561: 22–28 [DOI] [PubMed] [Google Scholar]
- Grill E, Löffler S, Winnacker E-L, Zenk MH (1989) Phytochelatins, the heavy-metal-binding peptides of plants, are synthesized from glutathione by a specific γ-glutamylcysteine dipeptidyl transpeptidase (phytochelatin synthase). Proc Natl Acad Sci USA 86: 6838–6842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschi KD, Korenkov VD, Wilganowski NL, Wagner GJ (2000) Expression of Arabidopsis CAX2 in tobacco: altered metal accumulation and increased manganese tolerance. Plant Physiol 124: 125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JB, Kim YA, Kim SJ, Lee SH, Kim DH, Cheong G-W, Hwang I (2001) A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13: 1511–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Eu YJ, Yoo CM, Kim YW, Pih KT, Jin JB, Kim SJ, Stenmark H, Hwang I (2001) Trafficking of phosphatidylinositol 3-phosphate from the trans-golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13: 287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee S, Park JB, Lee SD, Kim JH, Ha SH, Hasumi K, Endo A, Suh P-G, Ryu SH (2003) Hydrogen peroxide induces association between glyceraldehyde 3-phosphate dehydrogenase and phospholipase D2 to facilitate phospholipase D2 activation in PC12 cells. J Neurochem 85: 1228–1236 [DOI] [PubMed] [Google Scholar]
- Kispal G, Csere P, Prohl C, Lill R (1999) The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J 18: 3981–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M, Perfus-Barbeoch L, Frelet A, Gaedeke N, Reinhardt D, Mueller-Roeber B, Martinoia E, Forestier C (2003) The plant multidrug resistance ABC transporter AtMRP5 is involved in guard cell hormonal signalling and water use. Plant J 33: 119–129 [DOI] [PubMed] [Google Scholar]
- Kolaczkowski M, van der Rest M, Cybularz-Kolaczkowska A, Soumillion JP, Konings WN, Goffeau A (1996) Anticancer drugs, ionophoric peptides, and steroids as substrates of the yeast multidrug transporter Pdr5p. J Biol Chem 271: 31543–31548 [DOI] [PubMed] [Google Scholar]
- Kushnir S, Babiychuk E, Storozhenko S, Davey MW, Papenbrock J, De Rycke R, Engler G, Stephan UW, Lange H, Kispal G, et al (2001) A mutation of the mitochondrial ABC transporter Sta1 leads to dwarfism and chlorosis in the Arabidopsis mutant starik. Plant Cell 13: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Guo Y, Ohta M, Xiong L, Stevenson B, Zhu JK (2002) LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase. EMBO J 21: 2692–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZS, Lu YP, Zhen RG, Szczypka M, Thiele DJ, Rea PA (1997) A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato)cadmium. Proc Natl Acad Sci USA 94: 42–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahe Y, Lemoine Y, Kuchler K (1996) The ATP binding cassette transporters Pdr5 and Snq2 of Saccharomyces cerevisiae can mediate transport of steroids in vivo. J Biol Chem 271: 25167–25172 [DOI] [PubMed] [Google Scholar]
- Martinoia E, Klein M, Geisler M, Bovet L, Forestier C, Kolukisaoglu U, Muller-Rober B, Schulz B (2002) Multifunctionality of plant ABC transporters: more than just detoxifiers. Planta 214: 345–355 [DOI] [PubMed] [Google Scholar]
- Mehra RK, Kodati VR, Abdullah R (1995) Chain length-dependent Pb(II)-coordination in phytochelatins. Biochem Biophys Res Commun 215: 730–736 [DOI] [PubMed] [Google Scholar]
- Moller SG, Kunkel T, Chua NH (2001) A plastidic ABC protein involved in intercompartmental communication of light signaling. Genes Dev 15: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Noh B, Murphy AS, Spalding EP (2001) Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13: 2441–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz DF, Kreppel L, Spaser DM (1995) Transport of metal-binding peptides by HMT1, a fission yeast ABC-type vacuolar membrane protein. J Biol Chem 270: 4721–4727 [DOI] [PubMed] [Google Scholar]
- Rensing C, Mitra B, Rosen BP (1997) The ZntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc Natl Acad Sci USA 94: 14326–14331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers B, Decottignies A, Kolaczkowski M, Carvajal E, Balzi E, Goffeau A (2001) The pleiotropic drug ABC transporters from Saccharomyces cerevisiae. J Mol Microbiol Biotechnol 3: 207–214 [PubMed] [Google Scholar]
- Ross SM (1994) Sources and forms of potentially toxic metal in soil-plant system. In SM Ross, ed, Toxic Metals in Soil-Plant Systems. John Wiley & Sons, New York, pp 3–25
- Sanchez-Fernandez R, Davies TG, Coleman JO, Rea PA (2001) The Arabidopsis thaliana ABC protein superfamily: a complete inventory. J Biol Chem 276: 30231–30244 [DOI] [PubMed] [Google Scholar]
- Sharma R, Rensing C, Rosen BP, Mitra B (2000) The ATP hydrolytic activity of purified ZntA, a Pb(II)/Cd(II)/Zn(II)-translocating ATPase from Escherichia coli. J Biol Chem 275: 3873–3878 [DOI] [PubMed] [Google Scholar]
- Shiraishi E, Inouhe M, Joho M, Tohoyama H (2000) The cadmium-resistant gene, CAD2, which is a mutated putative copper-transporter gene (PCA1), controls the intracellular cadmium-level in the yeast S. cerevisiae. Curr Genet 37: 79–86 [DOI] [PubMed] [Google Scholar]
- Song W-Y, Sohn EJ, Martinoia E, Lee YJ, Yang Y-Y, Jasinski M, Forestier C, Hwang I, Lee Y (2003) Engineering tolerance and accumulation of lead and cadmium in transgenic plants. Nat Biotechnol 21: 914–919 [DOI] [PubMed] [Google Scholar]
- Szczypka MS, Wemmie JA, Moye-Rowley WS, Thiele DJ (1994) Yeast metal resistance protein similar to human cystic fibrosis transmembrane conductance regulator (CFTR) and multidrug resistance-associated protein. J Biol Chem 269: 22853–22857 [PubMed] [Google Scholar]
- Tommasini R, Vogt E, Fromenteau M, Hortensteiner S, Matile P, Amrhein N, Martinoia E (1998) An ABC-transporter of Arabidopsis thaliana has both glutathione-conjugate and chlorophyll catabolite transport activity. Plant J 13: 773–780 [DOI] [PubMed] [Google Scholar]
- Van den Brule S, Smart CC (2002) The plant PDR family of ABC transporters. Planta 216: 95–106 [DOI] [PubMed] [Google Scholar]
- Yang Young-Yell, Jung Ji-Young, Song Won-Young, Suh Hak-Soo, Lee Youngsook (2000) Identification of rice varieties with high tolerance or sensitivity to lead, and characterization of the mechanism of tolerance. Plant Physiol 124: 1019–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Silva ID, Bartel B (2001) The Arabidopsis pxa1 mutant is defective in an ATP- binding cassette transporter-like protein required for peroxisomal fatty acid beta-oxidation. Plant Physiol 127: 1266–1278 [PMC free article] [PubMed] [Google Scholar]