Abstract
Plants have mechanisms for repairing and tolerating detrimental effects by various DNA damaging agents. A tolerance pathway that has been predicted to be present in higher plants is translesion synthesis (TLS), which is catalyzed by polymerases. In Arabidopsis (Arabidopsis thaliana), however, the only gene known to be involved in TLS is the Arabidopsis homolog of REV3, AtREV3, which is a putative catalytic subunit of Arabidopsis DNA polymerase ζ. A disrupted mutant of AtREV3, rev3, was previously found to be highly sensitive to ultraviolet-B (UV-B) and various DNA damaging agents. REV1 and REV7 are thought to be components of translesion synthesis in plants. In this study, we identified the Arabidopsis homologs of REV1 and REV7 (AtREV1 and AtREV7). Several mutants carrying disrupted AtREV1 and AtREV7 genes were isolated from Arabidopsis T-DNA-inserted lines. An AtREV1-disrupted mutant, rev1, was found to be moderately sensitive to UV-B and DNA cross-linkers. A rev1rev3 double mutant, like rev3, showed high sensitivity to UV-B, γ-rays, and DNA cross-linkers. An AtREV7-disrupted mutant, rev7, was possibly sensitive to cis-diamminedichloroplatinum(II), a kind of DNA cross-linker, but it was not sensitive to acute UV-B and γ-ray irradiation. On the other hand, the aerial growth of rev7, like the aerial growth of rev1 and rev3, was inhibited by long-term UV-B. These results suggest that a TLS mechanism exists in a higher plant and show that AtREV1 and AtREV7 have important roles in tolerating exposure to DNA-damaging agents.
Plants are continuously exposed to stressful environments due to their sessile lifestyle. UV-B, which accounts for only a small part of the energy in sunlight, causes alterations in physiological processes, growth, and development of plants (Hopkins et al., 2002; Takeuchi et al., 2002). UV-B and certain environmental stresses target DNA and generate various types of DNA lesions. UV-B induces DNA lesions such as cyclobutane pyrimidine dimers (CPDs), pyrimidine (6-4) pyrimidone dimers [(6-4) photoproducts], and some minor lesions (Jagger, 1985; Umlas et al., 1985). Various types of DNA lesion are also induced by reactive oxygen species, which are generated by exposure to UV-B (Dai et al., 1997; Hidge et al., 2002), air pollutants such as ozone (Floyd et al., 1989), ionizing radiation, and many chemicals (Friedberg, 1995). In several organisms, DNA lesions can be repaired by various mechanisms, such as photorepair, excision repair (dark repair), and recombination repair (for review, see Hays, 2002). Most DNA repair mechanisms are thought to be conserved in higher plants (for review, see Britt, 1999).
Another tolerance mechanism is the damage tolerance pathway, which allows a cell to tolerate such damage, thereby allowing DNA replication to be completed. One such tolerance mechanism is translesion synthesis (TLS; Broomfield et al., 2001). In TLS, DNA damage is bypassed and the nascent DNA strand is extended by specialized DNA polymerases. TLS has the ability to bypass DNA lesions, but DNA replication is not always accurate, which can lead to mutations (for review, see Friedberg et al., 2000). Three TLS-type polymerases are conserved among species and have been well characterized in yeast and mammals. These are DNA polymerase η (McDonald et al., 1997; Masutani et al., 1999), Rev1 (Nelson et al., 1996a; Gibbs et al., 2000; Simpson and Sale, 2003), and DNA polymerase ζ (Nelson et al., 1996b; Sonoda et al., 2003). DNA polymerase ζ contains two subunits encoded by the REV3 and REV7 genes. Rev1 and DNA polymerase ζ are thought to be major proteins of the error-prone TLS, which generates mutations as a result of insertion of the wrong bases.
TLS has also been proposed to be present in higher plants based on bioinformatics analyses (Lawrence et al., 2000; Ohmori et al., 2001; Kimura et al., 2002). However, the functions and physiological roles of these genes are unknown. A disrupted mutant of AtREV3, a gene for the putative catalytic subunit of Arabidopsis (Arabidopsis thaliana) DNA polymerase ζ, was found to be sensitive to UV-B and various DNA-damaging agents (Sakamoto et al., 2003). These results suggest that Arabidopsis has a functional TLS DNA polymerase and that AtREV3 plays a role in tolerance to UV-B and other environmental stresses.
Another TLS-type polymerase, REV1, was first identified in yeast (Saccharomyces cerevisiae) through a screening for mutants that were hypomutable in response to UV irradiation (Lemontt, 1971). REV1 is a Y-family DNA polymerase, which is distinct from the existing A-C and X families of DNA polymerases, and includes DNA polymerase IV (DinB) and DNA polymerase V (UmuC) in Escherichia coli, RAD30 in, and polymerase η, ι, and κ in vertebrates (Ohmori et al., 2001). REV7 was also identified in yeast as a gene related to UV-induced reversion and the rev7 mutant showed some sensitivity to UV (Lawrence et al., 1985a). In yeast, Rev1 possesses deoxycytidyl transferase activity and is capable of inserting a C opposite an abasic site in vitro. The resultant mismatch is efficiently extended by DNA polymerase ζ (Nelson et al., 1996b; Lin et al., 1999). Rev7 is the regulatory subunit of DNA polymerase ζ and acts to stabilize and enhance polymerase activity (Nelson et al., 1996b). In higher plants, however, the functions of REV1 and REV7 are unknown.
In this study, to clarify the error-prone TLS pathway in Arabidopsis, we identified the Arabidopsis homologs of REV1 and REV7 (AtREV1 and AtREV7) and characterized the corresponding mutants. Disruption of these genes was found to cause moderate sensitivity to UV-B and other DNA-damaging agents, but less sensitivity than was observed in the AtREV3-disrupted mutant. These results suggest that Arabidopsis has all the components needed for the error-prone TLS and, like other eukaryotes, has a TLS pathway.
RESULTS
Isolation of the Arabidopsis REV1 Homolog Gene and Its Mutant
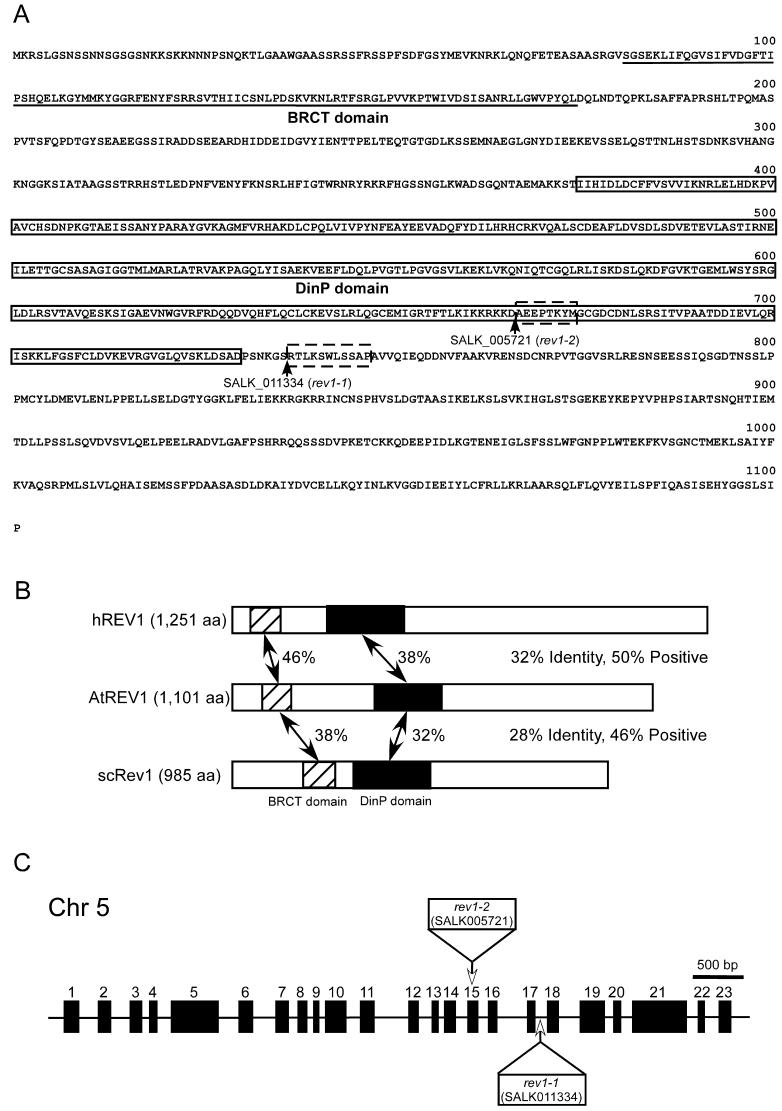
Three genes homologous to Y-family polymerases have been found in the Arabidopsis genome (Ohmori et al., 2001). Based on a BLAST analysis, the three genes (At1g49980, At5g44740, and At5g44750) were most similar to the genes of the DinB, Rad30A, and REV1 proteins, respectively. Thus, the At5g44750 gene was thought to be an Arabidopsis homolog of REV1 gene (AtREV1). The open reading frame (ORF) of the AtREV1 (accession no. AB187523) cDNA has a length of 3,303 bp and encodes a protein of 1,101 amino acids (Fig. 1A). The N terminus of AtREV1 has 46% identity to that of hREV1 and 38% identity to that of scRev1 and contains the BRCA1 terminus (BRCT) domain. The BRCT domain is found in many DNA repair and cell cycle checkpoint proteins and is specific to REV1 proteins of Y-family DNA polymerases (Masuda et al., 2002). The central region has a putative DinP domain (38% and 32% identity to hREV1 and scRev1, respectively; Fig. 1B). The DinP domain is observed within nucleotidyltransferases and DNA polymerases that are involved in DNA repair (accession no. COG0389.1 DinP, National Center for Biotechnology Information [NCBI] Cluster of Orthologous Group domain database).
Figure 1.
Structure of AtREV1 cDNA and T-DNA insertion lines. A, Deduced amino acid sequence of AtREV1 cDNA. Underlined sequence indicates the BRCT domain. Boxed sequence of AtREV1 indicates the DinP domain. Arrowheads show T-DNA insertion sites of SALK_005721 (rev1-2) lines and SALK_011334 (rev1-1) lines. Boxed sequences with broken line are amino acids that are predicted to be deleted due to the T-DNA insertion. B, Schematic representation of the alignment of hREV1, AtREV1, and yeast Rev1p (scREV1). Amino acid sequences were compared by the BLAST search program. Regions with significant identity are hatched or shaded (containing the BRCT and DinP domain, respectively), and percent identity is indicated between AtREV1 and other REV1 proteins. C, Structure of the AtREV1 gene and T-DNA insertion sites. Black rectangles represent 23 exons of the AtREV1 gene. White rectangles and arrows indicate the T-DNA insertion positions in exon 15 (rev1-2) and intron 17 (rev1-1). In rev1-1, a 131-bp deletion of genomic DNA was found around the junction of intron 17 and exon 18. In rev1-2, a 23-bp deletion was found in exon 15. The left border sequences were found at both sides of the inserted T-DNA, and T-DNA was accompanied by the insertion of filler DNA aattaggacg and g on the both sides.
To determine the function of AtREV1, we searched the database of the SALK Institute Genome Analysis Laboratory (http://signal.salk.edu/cgi-bin/tdnaexpress) for AtREV1-disrupted T-DNA insertion lines. Two lines, SALK_011334 and SALK_005721, which we named rev1-1 and rev1-2, were found (Fig. 1C). In both lines, the right border sequence was truncated and the left border sequence was found at both sides of the T-DNA inserted region. rev1-1 has a T-DNA-inserted at the 17th intron and is missing 131 bp, which corresponds to the entire 17th intron and a part of the 18th exon. rev1-2 has a T-DNA inserted at the 15th exon and is missing 23 bp. rev1-2 also has an insertion of filler DNA on both sides of the T-DNA insertion site. AtREV1 transcripts obtained by reverse transcription (RT)-PCR were observed in the wild type but not in rev1-1 and rev1-2, although only the upstream region of the T-DNA insertion site was amplified (data not shown).
Sensitivity of AtREV1-Deficient Mutant to UV-B Irradiation
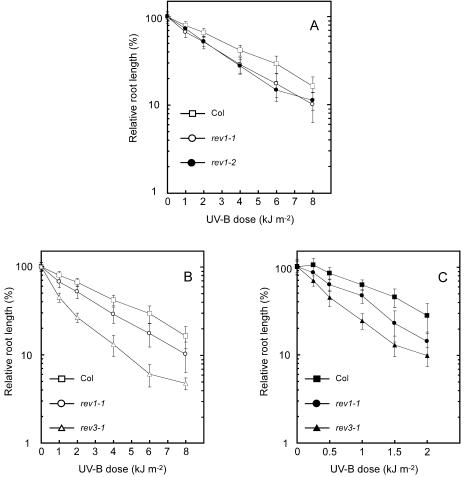
Root growth of rev3-1 was previously shown to be severely inhibited by UV-B irradiation (Sakamoto et al., 2003). To characterize the AtREV1, deficient mutants, rev1-1 and rev1-2 seedlings were irradiated with various doses of UV-B irradiation and grown in the light, and their responses were analyzed by a root-bending assay. Both rev1-1 and rev1-2 plants were found to be more sensitive to UV-B irradiation than the wild type (Fig. 2A). For example, 50% growth inhibition was observed in the rev1-1 plants at a 1.52-fold lower dose of UV-B irradiation than that required to induce a similar inhibition of the wild type.
Figure 2.
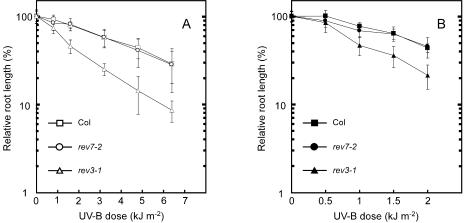
UV-B light sensitivities of the rev1-1 and rev3-1 mutants. A, UV-B dose-response curve for root growth of the wild-type (Col, Columbia), rev1-1, and rev1-2 seedlings in the light condition. Three-day-old wild-type (rectangles), rev1-1 (white circles), and rev1-2 (black circles) seedlings were exposed to UV-B light and then incubated under continuous white light for 3 d. B and C, UV-B dose-response curves for root growth in the wild type (rectangles), rev1-1 (circles), and rev3-1 (triangles) under light (B, white symbols) and dark (C, black symbols) conditions. For all sections, root growth rate was measured using Scion Image software. Each value represents an average of 15 to 36 measurements, and error bars indicate sds.
rev3-1 was highly sensitive to UV-B irradiation under both light and dark conditions. rev1-1 was sensitive to UV-B but less sensitive than rev3-1 under the light condition, while under the dark condition growth was slightly affected by UV-B doses examined (Fig. 2, B and C). Fifty percent growth inhibition was observed in the rev1-1 plants at a 1.49-fold lower dose of UV-B irradiation under dark condition than that required to induce a similar inhibition of the wild type.
Hypersensitivity of rev1rev3 Double Mutant
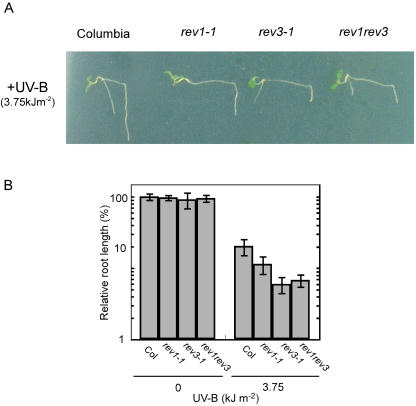
In yeast and mammals, REV1 protein and DNA polymerase ζ, which consists of the REV3 and REV7 proteins, play major roles in the error-prone TLS (Nelson et al., 1996b). REV1 appears to cooperate with DNA polymerase ζ in a common pathway of DNA lesion bypass (Lawrence et al., 2000; Guo et al., 2001, 2004; Haracska et al., 2001). To investigate the possibility that REV1 and REV3 have related functions in Arabidopsis, we generated a rev1-1 rev3-1 double mutant. RT-PCR showed that both AtREV1 and AtREV3 transcripts were suppressed in the rev1rev3 plants. There was no difference in root growth between the wild-type and mutant plants grown in the absence of UV-B (Fig. 3B). Exposure of the rev1rev3 double mutant to 3.75 kJ m−2 of UV-B severely inhibited its root growth (Fig. 3, A and B). The inhibition of root growth was similar to that of rev3-1 and more than that of rev1-1. This suggests that AtREV1 and AtREV3 play roles in a common pathway of UV tolerance.
Figure 3.
UV-B light sensitivity of rev1rev3 double mutant A, Root growth of wild-type, rev1-1, rev3-1, and rev1rev3 seedlings in the light condition. Seedlings were exposed to 3.75 kJ m−2 UV-B and then incubated under continuous white light as described in Figure 2 and “Materials and Methods.” B, Relative root length of plants grown for 3 d after 3.75 kJ m−2 UV-B exposures. Col indicates Columbia (wild type). Root length was measured using Scion Image software. Each value represents an average of 15 to 20 measurements. Error bars indicate sds.
Isolation of the Arabidopsis REV7 Mutant
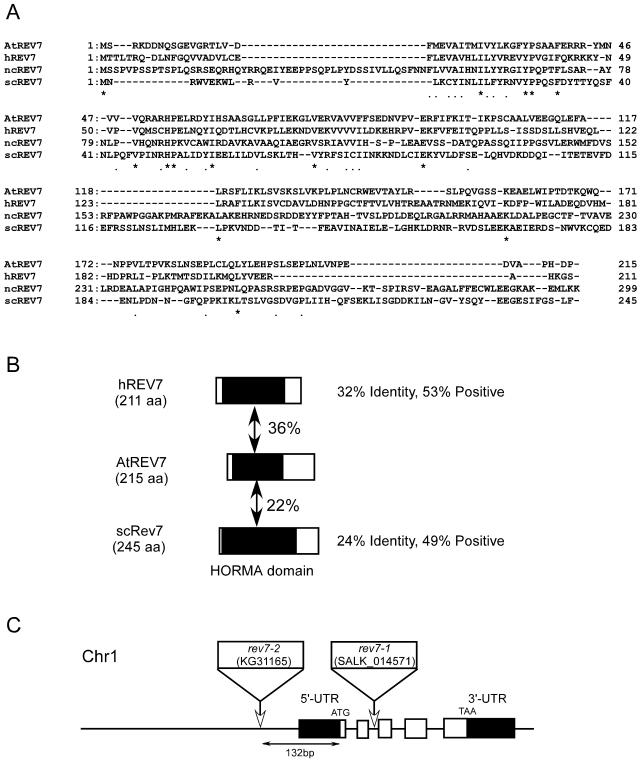
AtREV7 (At1g16590) showed significant homology to REV7 genes of other organisms at the amino acid level (Fig. 4). The protein encoded by AtREV7 is predicted to contain 215 amino acids. The N-terminal region of AtREV7 showed weak homology to the N-terminal regions of the human and yeast counterparts of AtREV7 (36% and 22% identity, respectively; Fig. 4B). In the human and yeast genes, these regions contain a HORMA domain (Aravind and Koonin, 1998).
Figure 4.
Alignment of REV7 proteins. A, Alignment of Rev7 proteins from Arabidopsis (AtREV7), human (hREV7), N. crassa (ncREV7), and yeast (scREV7). Amino acid sequences were aligned using Genetyx software (Software Development, Tokyo). Residues that are conserved among all organisms are shown as asterisks (*) and residues that are conserved among more than three kinds of organisms are shown as dots (.). B, Schematic representation of the alignment of hREV7, AtREV7, and yeast REV7 (scREV7). Regions with significant identity are shaded (putative HORMA domain), and percent identity is indicated between AtREV7 and other REV7 proteins. C, Structure of AtREV7 gene and T-DNA insertion site. White boxes represent five exons of the AtREV7 gene and black boxes represent untranslated regions that were supported by the sequence of the expressed sequence tag clone. 5′-UTR and 3′-UTR indicate upstream and downstream, respectively, of the ORF of AtREV7. White rectangles and arrows indicate the T-DNA insertion positions in the upstream region of the ORF (rev7-2) and intron 2 (rev7-1). In rev7-1, left border sequences were found at both sides of the inserted T-DNA. A substitution of three bases was found at the junction of downstream left border and wild-type genome sequence (TGA in Col to AAT in rev7-1). In rev7-2, the T-DNA was accompanied by the insertion of a filler DNA, tcagcgcattttct.
Two T-DNA insertion lines of AtREV7, SALK_014571 and KG31165, referred to here as rev7-1 and rev7-2, respectively, were found. AtREV7 transcripts were detected in wild-type plants while they were not detected in either rev7-1 or rev7-2 by RT-PCR (data not shown).
Wild-type, rev7-2, and rev3-1 seedlings were irradiated with UV-B and then grown under light and dark conditions to examine their recovery (Fig. 5). During recovery under both light and dark conditions, rev7-2 showed little sensitivity to UV-B, while rev3-1 showed a strong sensitivity to UV-B. The UV-B sensitivity of rev7-1 was similar to that of to rev7-2 (data not shown). This suggests that the AtREV7 gene is dispensable in the tolerance of Arabidopsis seedlings to short-term UV-B irradiation.
Figure 5.
UV-B light sensitivities of the rev7-2 and rev3-1 mutants. UV light dose-response curve for root growth in the wild type (rectangle), rev7-2 (circles), and rev3-1 (triangles) under light (A, white symbols) and dark (B, black symbols) conditions. Root growth rate was measured using Scion Image. Each value represents an average of 15 to 18 measurements. Error bars indicate sds.
Sensitivity of rev1 and rev7 to Other DNA-Damaging Agents
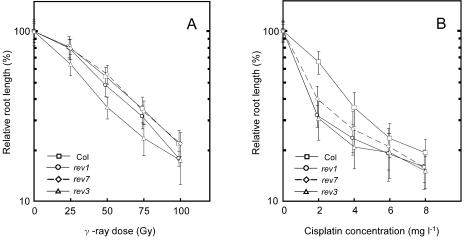
In other organisms, REV1 is involved in the replication of not only UV-caused DNA damage but also other types of DNA damage (Sakai et al., 2003; Simpson and Sale, 2003). A REV7-disrupted mutant in yeast was also found to be sensitive to methyl methane sulfate (MMS) and ethyl methane sulfonate (Lawrence et al., 1985b). The Arabidopsis rev3 mutant was not sensitive to MMS, but it showed a strong sensitivity to γ-rays and mitomycin C (MMC). The root lengths of rev1 and rev7 were hardly sensitive to γ-rays and showed only slightly shorter than the root length of the wild type. Fifty percent growth inhibition was observed in rev1 and rev7 at 1.14-fold and 1-fold lower dose, respectively, of γ-rays than that required to induce a similar inhibition of wild type (Fig. 6A). On the other hand, the growth of rev1, like that of rev3, was moderately inhibited by MMC (data not shown) and cis-diamminedichloroplatinum(II) (cisplatin; Fig. 6B), which are thought to cause intrastrand or interstrand cross-links on double-stranded DNA. Unlike UV-B, cisplatin caused an inhibitory phenotype in rev7 (Fig. 6). The root lengths of rev1 and rev7 were inhibited significantly only at the lowest cisplatin concentration used. Fifty percent growth inhibition was observed in the rev1 and rev7 at 2.4-fold and 2.02-fold lower dose, respectively, of cisplatin concentration than that required to induce a similar inhibition of wild type. The rev1rev3 double mutant, like rev3, was sensitive to γ-rays and cisplatin (data not shown). Following exposure to 50 gray γ-rays, root growths in rev1rev3 and rev3, (33.21% ± 3.19% and 31.97% ± 4.45%, respectively) were less than those in rev1 and the wild type (40.96% ± 6.08%, and 54.33% ± 6.99%, respectively). In the presence of 2 mg L−1 of cisplatin, root growths in rev1rev3, rev1 and rev3 (29.90% ± 4.98%, 34.37% ± 4.89%, and 30.92% ± 7.71%, respectively) were less than root growth in the wild type (65.62% ± 10.04%). This result suggests that AtREV1 and AtREV7 are also required to bypass various kinds of DNA lesion.
Figure 6.
Sensitivities of rev1 and rev7 to DNA-damaging agents. Three-day-old seedlings were irradiated with γ-rays (A) or explanted to cisplatin-containing (B) agar-plates and incubated for another 3 d. Root growth rate was measured using Scion Image software. Each value represents an average of 15 to 36 measurements. Error bars indicate sds.
AtREV1 and AtREV7 Have Important Roles under Long-Term UV-B Irradiation
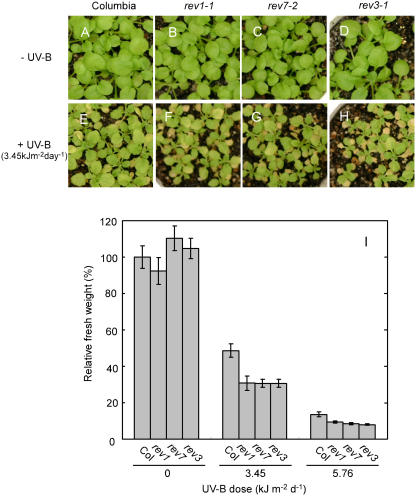
The growth of REV mutants were not significantly different from the growth of wild-type plants under non-UV-B irradiation conditions (Fig. 7, A–D), whereas the growth of the wild type decreased in a UV-B dose-dependent manner (Fig. 7I). After the plants were exposed to UV-B for 11 d, growth was more inhibited in the mutants than in the wild type (Fig. 7, E–H). Growth of the mutants was inhibited even at a low dose of UV-B irradiation (3.45 kJ m−2 s−1 d−1). Surprisingly, under long-term UV-B irradiation, the fresh weight of rev7 was as low as the fresh weights of rev1 and rev3. However, under short term UV-B irradiation, the root growth of rev7 was not significantly different from that of the wild type. The data shown in Figure 7I suggest that the AtREV7 gene is necessary for normal growth under UV-B irradiation.
Figure 7.
Effects of long-term UV-B exposure on growth of the rev1, rev7, and rev3 mutants. The wild-type (Columbia) and mutant (rev3, rev1, and rev7) plants were grown under white light until they were 11-d-old, and then the plants were grown with or without UV-B irradiation for 11 d. A to H, Photographs of 22-d-old plants grown under no UV-B (− UV-B, A–D) or 3.45 kJ m−2 d−1 UV-B irradiation (+ UV-B, E–H) for 11 d. I, Effect of different UV-B doses on relative fresh weight of aerial parts of 22-d-old wild-type (Col), rev3, rev1, and rev7 seedlings. Each value represents an average of 16 to 20 measurements. Error bars indicate ses.
DISCUSSION
In recent years, DNA repair systems and genome maintenance systems have been found in higher plants (for review, see Hays, 2002). The TLS pathway is also an important mechanism for genome maintenance in several eukaryotes (Lawrence, 2002). In higher plants, Sakamoto et al. (2003) first found and characterized AtREV3, the putative catalytic subunit of Arabidopsis DNA polymerase ζ, indicating the existence of a damage-induced TLS pathway in plants. In this study, we identified the REV1 and REV7 homologs of Arabidopsis, which are considered to cooperate in the TLS pathway. The sensitivities of the Arabidopsis rev mutants to UV-B, cisplatin, and γ-rays were different (Table I). These data suggest that the overall sensitivities are in the order AtREV3 mutant > AtREV1 mutant > AtREV7 mutant. All three rev mutants were moderately sensitive to DNA cross-linkers. rev3 was sensitive to γ-rays, while rev1 and rev7 were only slightly sensitive to γ-rays. These results suggest that the Arabidopsis REV genes have roles in tolerating DNA damaging agents, but their function might be redundant.
Table I.
Sensitivities of Arabidopsis REV homolog mutants to UV-B, cisplatin, and γ-rays
++, More sensitive; +, sensitive; ±, possibly sensitive; −, same as the wild type; n.t., not tested.
| DNA-Damaging Agent
|
Sensitivity
|
DNA Damage
|
||||
|---|---|---|---|---|---|---|
| rev1 | rev7 | rev1rev3 | rev3a | |||
| Short-term UV-B | Light | + | − | ++ | ++ | |
| Dark | ± | − | + | + | CPD, 6-4PP, etc. | |
| Long-term UV-B | + | + | n.t. | + | ||
| Cisplatin | ± | ± | + | + | DNA crosslink, alkylation, etc. | |
| γ-Rays | − | − | + | + | DSB, base damage, etc. | |
Sakamoto et al. (2003) and this study.
In yeast, three kinds of REV genes, REV1, REV3 and REV7, were found. Deletions of these genes cause a decrease in survival and a decrease in reversion under UV-irradiation or some mutagens and then suppress the induction of mutation due to misincorporation of bases opposite the site of bypassed damages (Lemontt, 1971; McKee and Lawrence, 1979; Lawrence et al., 1985a, 1985b). In this study, rev1 and rev7 were sensitive to UV-B and DNA cross-linkers to some extent. Concerning the measurement of a reversion frequency of intact plant, there is only a suitable method, which detects the mutation as reversions of specific GUS (β-glucuronidase) reporter transgene alleles, for intact higher plants (Kovalchuk et al., 2000). In the further study, the reversion frequency of these mutants will be able to measure by using this method.
Both root and aerial tissue growth of rev1 were inhibited by UV-B irradiation (Fig. 2 and 7). rev1 was less sensitive to UV-B than rev3. In other organisms, a deficiency of REV1 protein results in a moderate or lower sensitivity to UV-irradiation (Lemontt, 1971; Gibbs et al., 2000; Sakai et al., 2003). On the other hand, our results also show that the root growth of the rev1rev3 double mutant, like that of rev3-1, was sensitive to UV-B, while the root growth of rev1-1 was only moderately sensitive to UV-B (Fig. 3). REV1 cooperates or interacts functionally with REV3 that mediates DNA polymerase ζ in the error-prone TLS (Nelson et al., 1999; Rajpal et al., 2000). This result suggests that AtREV1 also cooperates with AtREV3. In several organisms, TLS has been described by a two-step, two-polymerase model. REV1 is the first enzyme and aids in the incorporation of one or two nucleotides opposite the lesion, while DNA polymerase ζ is the second polymerase that extends the primer beyond the lesion site to positions downstream of the damage. When DNA is exposed to UV-B, many kinds of DNA lesions are generated (Friedberg, 1995). The major products of the lesions are CPDs, and (6-4) photoproducts, and the minor products include 8-oxoguanine, pyrimidine hydrate, and thymine glycols. Other minor effects are DNA-DNA cross-links and strand breaks. AtREV1 may respond to some of these types of DNA damage, and this could explain why rev1 was less sensitive to UV-B than rev3.
If insertion type polymerases other than AtREV1 are the first insertion type polymerases in TLS, they might incorporate bases opposite the DNA lesions that AtREV1 is essentially unresponsive to. In several organisms, some polymerases, such as DNA polymerases η and ι, were also able to insert bases opposite a damaged template and cooperate with DNA polymerases ζ and κ to catalyze the translesion synthesis across from the lesion (Johnson et al., 2000; Haracska et al., 2001; Prakash and Prakash, 2002; Guo et al., 2003, 2004). Two Arabidopsis homologs of Y-family DNA polymerases other than REV1 (At5g44750) have been identified, DNA polymerase η (At5g44740) and DNA polymerase κ (At1g49980). Recently, an Arabidopsis DNA polymerase κ, named AtPOLK, was characterized by using an in vitro assay of a recombinant protein (García-Ortiz et al., 2004). In our preliminary tests, however, T-DNA insertion lines of Arabidopsis DNA polymerase η and κ showed sensitivities to UV-B similar to those shown by the wild-type plants (data not shown). This suggests that Arabidopsis Y-family DNA polymerases other than AtREV1 scarcely contribute to the TLS system. The difference in the sensitivities of rev1 and rev3 to UV-B may mean that DNA polymerases other than a Y-family DNA polymerase are required for the TLS pathway. In Arabidopsis, it is not clear how a particular mechanism for TLS for a particular type of DNA damage is selected. Multiple DNA polymerase may be responsible for repairing UV-B-induced damage, which may explain the difference of sensitivity to UV-B between rev1 and rev3.
REV7 is thought to be a regulatory protein that enhances the activity of DNA polymerase ζ (Baynton et al., 1999; Murakumo et al., 2000; Masuda et al., 2003). AtREV7 is similar to the REV7 proteins of human and other organisms at the amino acid level (Fig. 4). The HORMA domain is a domain that is conserved among several REV7proteins and some cell-cycle-related proteins (MAD2 and Hop1). This domain might recognize chromatin states that result from DNA damage and might act as an adaptor that recruits other proteins involved in repair (Aravind and Koonin, 1998). Therefore, we predict that AtREV7 has similar recognition functions. In Neurospora crassa, the UV-B sensitivity of a REV7-deficient mutant was similar to that of a REV3-deficient mutant (Sakai et al., 2003). However, in this study, the root growth of an AtREV7-disrupted mutant, rev7, showed little sensitivity to UV-B (Fig. 5), while its aerial growth, like the aerial growth of rev1 and rev3, was inhibited by long-term UV-B irradiation (Fig. 7). These results show that, in rev7, DNA damage accumulated by long-term UV-B inhibits growth, but that DNA damage caused by short-term UV-B does not affect the recovery of root growth.
Previous reports have shown that a deficiency of the REV gene leads to high sensitivity to several mutagens. In Arabidopsis, rev3 was sensitive to MMC but insensitive to MMS (Sakamoto et al., 2003). In our study, rev1 and rev7 were moderately sensitive to DNA cross-linkers (Fig. 6). In other studies, exposure to DNA cross-linkers damaged some bases and generated DNA inter-strand cross-links (ICLs), which led to mutations, chromosomal rearrangements, and cell death (Friedberg, 1995; Dronkert and Kanaar, 2001). Although the repair mechanism for ICLs in yeast and mammals is unclear, in one model, nucleotide excision repair (NER) and recombination repair cut out and unwind the ICLs, and TLS simultaneously synthesizes a new DNA strand opposite the site formed by the cross-link (Dronkert and Kanaar, 2001). Sakamoto et al. (2003) and this study showed that all the REV gene-deficient mutants were sensitive to DNA cross-linkers to some extent. Two other reports showed that Rev3- or Rev1-deficient DT40 cells were also sensitive to cisplatin (Sonoda et al., 2003; Simpson and Sale, 2003). Simpson and Sale (2003) suggested that Rev1 and TLS are critical for efficient tolerance or repair of ICLs and that it is likely involved in other pathways. In higher plants, many genes involved in NER and recombinant repair have been isolated, and some of them were found to be involved in tolerance to DNA cross-linkers (Xu et al., 1998; Li et al., 2002). Therefore, it is possible that TLS works synergistically with NER and/or recombinant repair in plants. Further studies are needed to confirm these hypotheses.
In conclusion, we isolated two new genes, AtREV1 and AtREV7, which appear to be involved in TLS in plants. These mutants are moderately sensitive to UV-B and some DNA-damaging agents such as MMC and cisplatin, although the sensitivities differed among the mutants. Our results indicate that plants have a TLS mechanism similar to TLS mechanisms in other eukaryotes and that plant REV genes help plants to survive under UV-B and in stressful environments.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis ecotype Columbia was used for the wild-type plant in this study. The rev1-1 line (SALK_011334), rev1-2 line (SALK_005721), and rev7-1 line (SALK_014571) were provided by the SALK Institute Genomic Analysis Laboratory (Alonso et al., 2003). To construct the double mutant rev1rev3, a rev3-1 plant (Sakamoto et al., 2003) was crossed with a rev1-1 plant, and the resulting F1 plant was self-pollinated. Among the UV-B sensitive plants in these F2 progeny, the rev1-1 rev3-1 plants were selected by PCR. Plants were grown at 23°C in a growth chamber (LH200RD, NK System, Osaka) under continuous white light from fluorescent lamps (approximately 40 μmol m−2 s−1, FL40SS-W/ 37, Toshiba Lighting and Technology, Tokyo; Sakamoto et al., 2003).
Isolation of AtREV1 and AtREV7 cDNAs
The AtREV1 and AtREV7 genes were predicted to be the At5g44750.1 and At1g16590 genes, which were annotated by The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org/). The 3.3-kb AtREV1 (accession no. AB187523) cDNA was prepared by amplifying the total cDNA with the primers REV1-5TER (5′-ATGAAGCGTAGCTTGGGTTCAAATTCTTC-3′) and REV1-3TER (5′-GACTCGTGTCATGGGAAGAAATGATTATC-3′). The AtREV7 cDNA was prepared similarly with the primers REV7-5TER (5′-TGATCAAGGAATCCTTACCTCC-3′) and REV7-3TER (5′-ACATCCTCGGGGTTGACAAGGT-3′). The PCR products for AtREV1 and AtREV7 were cloned into pGEM-T Easy (Promega, Madison, WI) for sequencing. AtREV1 and AtREV7 were analyzed by the NCBI Conserved Domain Search program (http://www.ncbi.nim.nih.gov/BLAST), which searches for related protein families and putative functional domains.
Screening of an AtREV7 T-DNA Insertion Line
To find an AtREV7 T-DNA insertion line, we searched the T-DNA insertion sequence databases of the SALK Institute Genomic Analysis Laboratory. We found one line, SALK_014571, which we named rev7-1. rev7-1 had a T-DNA inserted at the first intron, and the right border sequence was truncated and left border sequence was found at both sides of the T-DNA inserted region. To search for the other rev7 allele, one of the lines was screened from the T-DNA insertion line collection of the Kazusa DNA Research Institute. By screening for AtREV7-knockout lines by PCR, we isolated one line (KG31165) possessing a T-DNA insertion at 132 bp upstream of the ORF. We named this line rev7-2. The rev7-2 line was isolated from the T-DNA insertion line stocks prepared by the Kazusa DNA Research Institute with a primer specific for T-DNA (pGTAC-LUS_LBP, 5′-AAGAAAATGCCGATACTTCATTGGC-3′) and a primer specific for AtREV7 (REV7-3, 5′-ACATCCTCGGGGTTGACAAGGT-3′). A set of DNA pool prepared from many insertion lines were amplified. To identify the T-DNA insertion line, the amplified products were separated by agarose-gel electrophoresis, and AtREV7-positive signals for PCR products were detected by Southern-blot analysis with the AtREV7 cDNA fragment as a probe.
Identification of the T-DNA Insertion Sites
The left border-flanking sequence was amplified with the primer LBa1 (5′-GCGTGGACCGAGTATTTTTCAACTTT-3′) and one of the following gene-specific primers: REV1-7 (5′-ACATCCTCGGGGTTGACAAGGT-3′), REV1-4 (5′-AGAAGGATCTGCACTGTCAAGCT-3′), or REV7-2 (5′-CAGTCAGGTAAGTTCCTGATTC-3′). To determine the other border sequence of rev1-1, thermal asymmetric interlaced-PCR was employed, as described previously (Liu et al., 1995). The specific primers were REV1-1_TR1 (5′-CGAGTGACATCATTCATCAC-3′), REV1-1_TR2 (5′-CTTGCGCTCTGTTGTTACTGCAG-3′), and REV1-1_TR3 (5′-GTATATGGGTTGTGGAGACTGTGAC-3′). To identify the other border sequence of rev1-2 and rev7-1, the genomic DNA from these lines was amplified with the LBa1 primer and the specific primer REV1-22 (5′-CTAGACCTACGGTCAGTTACAGACAG-3′) or REV7-6 (5′-TGAGTGGTTCAGACAAACTTGGG-3′). For the rev7-2 mutant, the left border flanking sequence was amplified with pGTAC-LUS_LBP and REV7-6 and then sequenced. The rev7-2 mutant also has only a left border sequence and we could not find a right border sequence.
UV-B Light Source
UV-B light was supplied by a UV-lamp (CSL-15B, COSMO BIO, Tokyo) that radiated at wavelengths of >280 nm with a peak intensity at 312 nm. UV-B light intensity was measured with a UV-B light radiometer (UVX31 sensor and UVX radiometer, UVP).
Root-Bending Assay and Analysis of the Root Growth Rate
The root-bending assay was performed as described previously (Sakamoto et al., 2003). Seedlings were grown vertically on nutritive agar plates (1.5% agar, 2% Suc, and 0.1% [v/v] commercial nutrient; Hyponex, Osaka) under continuous white light (approximately 40 μmol m−2 s−1) for 3 d. For analysis of UV-B sensitivity, seedlings were exposed to 0.25 to 2 kJ m−2 (for the dark condition) or 1.25 to 5 kJ m−2 (for the light condition) UV-B light and then incubated in the dark or under continuous white light for 3 d. The length of root growth after UV-B irradiation was measured using Scion Image software (Scion Corporation, Frederick, MD) and was expressed as a percentage of the average length of nonirradiated wild-type roots.
Long-Term UV-B Irradiation
Plants were grown in a pot containing soil (Metro-Mix 350, Scotts-Sierra Horticultural Products, Marysville, OH) under 16-/8-h photoperiods at 40 μmol m−2 s−1 in a photochamber (BIOTRON, NK System; Osaka). Twelve-day-old seedlings were irradiated with UV-B light at 3.45 or 5.76 kJ m−2 for 16 h d−1 under white light. The UV-B light dose was adjusted by varying the distance from the UV-B lamp to the plants. After 11 d of UV-B light treatment, the fresh weight of each plant was measured and expressed as a percentage of the average fresh weight of nonirradiated wild-type aerial parts. More than 18 plants were used for each data point.
Measurement of Plant Sensitivities to γ-Rays and Cisplatin
Seeds were set on nutritive agar plates and grown vertically under continuous white light for 3 d. The measurements of sensitivities to γ-rays and cisplatin were performed by a root-bending assay as described previously (Sakamoto et al., 2003). To test sensitivity to γ-rays, plants were irradiated with γ-rays from a 60Co irradiation facility (JAERI, Takasaki, Japan). To test sensitivity to cisplatin, the plants were transplanted to the surface of nutritive agar plates supplemented with 2 to 8 mg L−1 cisplatin. The plants were placed vertically so that the new root would grow at a right angle to the previous root. After a 3-d incubation under continuous white light, new root growth was measured as described above.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AB187523.
Acknowledgments
We thank Chihiro Suzuki and Yoshihiro Hase for their technical assistance, and Yutaka Oono, Naoya Shikazono, Satoshi Kitamura, Yuichiro Yokota, Vo Thi Thuong Lan, and Abidur Rahman for their helpful comments. We thank the ABRC and the Salk Institute Genome Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants. We are grateful to James Raymond for careful review of the manuscript.
This work was supported by the Ministry of Education, Science, Sports and Culture of Japan (grant-in-aid 15201010 for scientific research).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.060236.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Steveson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV (1998) The HORMA domain: a common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem Sci 23: 284–286 [DOI] [PubMed] [Google Scholar]
- Baynton K, Bresson-Roy A, Fuchs RPP (1999) Distinct roles for Rev1p and Rev7p during translesion synthesis in Saccharomyces cerevisiae. Mol Microbiol 34: 124–133 [DOI] [PubMed] [Google Scholar]
- Britt AB (1999) Molecular genetics of DNA repair in higher plants. Trends Plant Sci 4: 20–25 [DOI] [PubMed] [Google Scholar]
- Broomfield S, Hryciw T, Xiao W (2001) DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat Res 486: 167–184 [DOI] [PubMed] [Google Scholar]
- Dai Q, Yan B, Hung S, Liu X, Peng S, Miranda MLL, Chavez AQ, Vergara BS, Olszyk DM (1997) Response of oxidative stress defense systems in rice (Oryza sativa) leaves with supplemental UV-B radiation. Physiol Plant 101: 301–308 [Google Scholar]
- Dronkert ML, Kanaar R (2001) Repair of DNA interstrand cross-links. Mutat Res 486: 217–247 [DOI] [PubMed] [Google Scholar]
- Floyd RA, West MS, Hogsett WE, Tingey DT (1989) Increased 8-hydroxyguanine content of chloroplast DNA from ozone-treated plants. Plant Physiol 91: 644–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC (1995) Environmental Damage to DNA. In EC Friedberg, GC Walker, W Siede, eds, DNA Repair and Mutagenesis. American Society of Microbiology Press, Washington DC, pp 19–42
- Friedberg EC, Feaver WJ, Gerlach VL (2000) The many faces of DNA polymerases: strategies for mutagenesis and for mutational avoidance. Proc Natl Acad Sci USA 97: 5681–5683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Ortiz MV, Ariza RR, Hoffman PD, Hays JB, Roldán-Arjona TR (2004) Arabidopsis thaliana AtPOLK encodes a DinB-like DNA polymerase that extends mispaired primer termini and is highly expressed in a variety of tissues. Plant J 39: 84–97 [DOI] [PubMed] [Google Scholar]
- Gibbs PEM, Wang X-D, Li Z, McManus TP, McGregor WG, Lawrence CW, Maher VM (2000) The function of the human homolog of Saccharomyces cerevisiae REV1 is required for mutagenesis induced by UV light. Proc Natl Acad Sci USA 97: 4186–4191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Wu X, Rajpal DK, Taylor J-S, Wang Z (2001) Translesion synthesis by yeast DNA polymerase ζ from templates containing lesions of ultraviolet radiation and acetylaminofluorene. Nucleic Acids Res 29: 2875–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Fischhaber L, Luk-Paszyc MJ, Masuda Y, Jiang Z, Kamiya K, Kisker C, Friedberg EC (2003) Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J 22: 6621–6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Xie Z, Shen H, Xhao B, Wang Z (2004) Translesion synthesis of acetylaminofluorene-dG adducts by DNA polymerase ζ is stimulated by yeast Rev1 protein. Nucleic Acids Res 32: 1122–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Unk I, Johnson RE, Johansson E, Burgers PMJ, Prakash S, Prakash L (2001) Roles of yeast DNA polymerases δ and ζand of Rev1 in the bypass of abasic sites. Genes Dev 15: 945–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays JB (2002) Arabidopsis thaliana, a versatile model system for study of eukaryotic genome-maintenance functions. DNA Repair (Amst) 1: 579–600 [DOI] [PubMed] [Google Scholar]
- Hidge E, Barta C, Kalai T, Vass I, Hideg K, Asada K (2002) Detection of singlet oxygen and superoxide with fluorescent sensors in leaves under stress by photoinhibition or UV radiation. Plant Cell Physiol 43: 1154–1164 [DOI] [PubMed] [Google Scholar]
- Hopkins L, Bond MA, Tobin AK (2002) Ultraviolet-B radiation reduces the rates of cell division and elongation in the primary leaf of wheat (Triticum aestivum L. cv Maris Huntsman). Plant Cell Environ 25: 617–624 [Google Scholar]
- Jagger J (1985) Far-UV Killing, Mutation, and Repair. In J Jagger, ed, Solar-UV Actions on Living Cells. Praeger Publishers, New York, pp 32–58
- Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L (2000) Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature 406: 1015–1019 [DOI] [PubMed] [Google Scholar]
- Kimura S, Uchiyama Y, Kasai K, Namekawa A, Saotome A, Ueda T, Ando T, Ishibashi T, Masahiko M, Furukawa T, et al (2002) A novel DNA polymerase homologous to Escherichia coli DNA polymerase I from a higher plant, rice (Oryza sativa L.). Nucleic Acids Res 30: 1585–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk I, Kovalchuk JH, Hohn B (2000) Genome-wide variation of the somatic mutation frequency in transgenic plants. EMBO J 19: 4431–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CW (2002) Cellular roles of DNA polymerase zeta and Rev1 protein. DNA Repair (Amst) 1: 425–435 [DOI] [PubMed] [Google Scholar]
- Lawrence CW, Gibbs PEM, Murante RS, Wang X-D, Li Z, McManus TP, McGregor WG, Nelson JR, Hinkle DC, Maher VM (2000) Roles of DNA polymerase ζ and Rev1 protein in eukaryotic mutagenesis and translesion replication. In Proceedings of the Cold Spring Harbor Symposia on Quantitative Biology, Vol 65. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 61–69 [DOI] [PubMed]
- Lawrence CW, Nisson PE, Christensen RB (1985. a) REV7, a new gene concerned with UV mutagenesis in yeast. Mol Gen Genet 200: 80–85 [DOI] [PubMed] [Google Scholar]
- Lawrence CW, Nisson PE, Christensen RB (1985. b) UV and mutagenesis rev7 mutants of yeast. Mol Gen Genet 200: 86–91 [DOI] [PubMed] [Google Scholar]
- Lemontt JF (1971) Mutants of yeast defective in mutation induced by ultraviolet light. Genetics 68: 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Schuermann D, Gallego F, Kovalchuk I, Tinland B (2002) Repair of damaged DNA by Arabidopsis cell extract. Plant Cell 14: 263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Xu H, Zhang Y, Wu X, Yuan F, Wang Z (1999) The human REV1 gene codes for a DNA template-dependent dCMP transferase. Nucleic Acids Res 27: 4468–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-G, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insertion junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Masuda Y, Ohmae M, Masuda K, Kamiya K (2003) Structure and enzymatic properties of a stable complex of the human REV1 and REV7 proteins. J Biol Chem 278: 12356–12360 [DOI] [PubMed] [Google Scholar]
- Masuda Y, Takahashi M, Fukuda S, Sumii M, Kamiya K (2002) Mechanisms of dCMP Transferase reactions catalyzed by mouse Rev1 protein. J Biol Chem 277: 3040–3046 [DOI] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399: 700–704 [DOI] [PubMed] [Google Scholar]
- McDonald JP, Levine AS, Woodgate R (1997) The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics 147: 1557–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee RH, Lawrence CW (1979) Genetic analysis of gamma-ray mutagenesis in yeast. I. Reversion in radiation: sensitive strains. Genetics 93: 361–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakumo Y, Roth T, Ihii H, Rasio R, Numata S, Croce CM, Fishel R (2000) A Human REV7 homolog that interacts with the polymerase ζ catalytic subunit hREV3 and the spindle assembly checkpoint protein hMAD2. J Biol Chem 275: 4391–4397 [DOI] [PubMed] [Google Scholar]
- Nelson JR, Gibbs PEM, Nowicka AM, Hinkle DC, Lawrence CW (1999) Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol Microbiol 37: 549–554 [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC (1996. a) Deoxicytidil transferase activity of yeast REV1 protein. Nature 382: 729–731 [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC (1996. b) Thymine-Thymine dimer bypass by yeast DNA polymerase ζ. Science 272: 1646–1649 [DOI] [PubMed] [Google Scholar]
- Ohmori H, Friedberg EC, Fuchs RPP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, et al (2001) The Y-family of DNA polymerases. Mol Cell 8: 7–8 [DOI] [PubMed] [Google Scholar]
- Prakash S, Prakash L (2002) Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev 16: 1872–1883 [DOI] [PubMed] [Google Scholar]
- Rajpal DK, Wu X, Wang Z (2000) Alteration of ultraviolet-induced mutagenesis in yeast through molecular modulation of the REV3 and REV7 gene expression. Mutat Res 461: 133–143 [DOI] [PubMed] [Google Scholar]
- Sakai W, Wada Y, Naoi Y, Ishii C, Inoue H (2003) Isolation and genetic characterization of the Neurospora crassa REV1 and REV7 homologs: evidence for involvement in damage-induced mutagenesis. DNA Repair (Amst) 2: 337–346 [DOI] [PubMed] [Google Scholar]
- Sakamoto A, Lan VTT, Hase Y, Shikazono N, Matsunaga T, Tanaka A (2003) Disruption of the AtREV3 gene causes hypersensitivity to ultraviolet B light and γ-rays in Arabidopsis: implication of the presence of a translesion synthesis mechanism in plants. Plant Cell 15: 2042–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson LJ, Sale JE (2003) Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J 22: 1654–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E, Okada T, Zhao GY, Tateishi S, Araki K, Yamazumi M, Yagi T, Verkaik NS, Gent DC, Takata M, et al (2003) Multiple roles of Rev3, the catalytic subunit of polζ in maintaining genome stability in vertebrates. EMBO J 22: 3188–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi A, Yamaguchi T, Hidema J, Strid A, Kumagai T (2002) Changes in synthesis and degradation of Rubisco and LHCII with leaf age in rice (Oryza sativa L.) growing under supplementary UV-B radiation. Plant Cell Environ 25: 695–706 [Google Scholar]
- Umlas ME, Franklin WA, Chan GL, Haseltine WA (1985) Ultraviolet light irradiation of defined-sequence DNA under conditions of chemical photosensitization. Photochem Photobiol 42: 265–273 [DOI] [PubMed] [Google Scholar]
- Xu H, Swoboda I, Bhalla PL, Sijbers AM, Zhao C, Ong EK, Hoeijmakers JH, Singh MB (1998) Plant homologue of human excision repair gene ERCC1 points to conservation of DNA repair mechanisms. Plant J 13: 823–829 [DOI] [PubMed] [Google Scholar]