Abstract
The L1 and L2 capsid genes of human papillomavirus type 31 (HPV-31) are expressed upon keratinocyte differentiation from a promoter located in the E7 open reading frame (ORF) of the early region. Late transcripts must therefore pass through and ignore the early polyadenylation sequences to use the downstream late AAUAAA element located at the end of the L1 ORF. To identify sequences which modulate downstream capsid gene expression, a variety of substitution mutations were introduced into the early polyadenylation signal and studied first in the context of polycistronic luciferase reporter constructs. Removal of the G/U-rich cleavage stimulation factor (CstF) binding sites and the degenerate cleavage and polyadenylation specificity factor binding sites, UAUAUA, had minimal effect on downstream expression as defined by luciferase activities. This is in contrast to the deletion of the HPV-31 early AAUAAA element, which resulted in a dramatic increase in downstream expression. Additional sequences within the first 800 bp of the L2 ORF were also found to negatively regulate capsid expression in luciferase assays. To determine how these mutations influence gene expression in the context of the complete HPV-31 genome, recombinant genomes were constructed that contained a substitution in the AAUAAA sequence, an inserted strong CstF binding site, an inserted simian virus 40 (SV40) late poly(A) signal, or a substitution of the 5′-most 800 nucleotides of the L2 ORF. Reductions in both transient and stable replication were observed with the recombinant genomes containing the strong CstF site or the late SV40 signal, suggesting that alterations in the strength of the upstream poly(A) signal influence expression of viral replication factors. Similarly, disruption of the L2 ORF resulted in a significant reduction in genome replication and an inability to be maintained stably. In contrast, genomes containing a substitution of the AAUAAA sequence had increased levels of transient and stable replication. Quantitation of late transcripts following keratinocyte differentiation in methylcellulose also showed a reduction in downstream capsid gene expression in lines containing genomes with the strong CstF site or the late SV40 signal mutations, while a significant increase in expression was detected in the lines with genomes lacking the AAUAAA sequence. These studies demonstrate that capsid gene expression in HPV-31 requires an inefficient early poly(A) signal which is defined primarily by the AAUAAA element as well as a major negative regulatory element located within the L2 ORF.
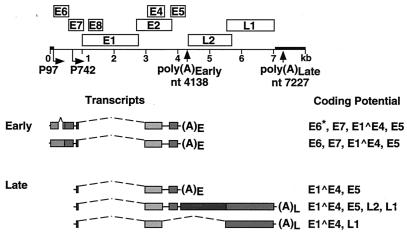
Human papillomaviruses (HPVs) are small double-stranded DNA viruses that target cutaneous or mucosal epithelium for infection. The productive HPV life cycle is dependent upon epithelial differentiation (19, 30). Upon entry into the basal cells, viruses establish and maintain their viral DNA as autonomously replicating nuclear plasmids at a low copy number per cell. HPV genomes are transcribed into polycistronic messages through the use of multiple promoters, two polyadenylation signals, and extensive splicing (Fig. 1) (21). In the lower portion of infected epithelia, HPV-16 and -31 messages initiate primarily at the early promoter P97 and use a polyadenylation signal, AAUAAA, located downstream of the E5 ORF (21, 35, 36, 39, 49). In suprabasal cells, late messages initiate at the differentiation-dependent promoter located within the E7 open reading frame (ORF) and use a polyadenylation site at the end of either the E5 gene or the L1 gene (17, 21, 22, 34). The nature and the location of these polyadenylation elements are conserved among most papillomaviruses (3, 24).
FIG. 1.
Map of the HPV-31 genome showing the major early and late transcripts. Late messages terminate either near pAEarly or at a downstream consensus hexanucleotide element, pALate.
Papillomavirus capsid expression is restricted to the most differentiated layers of the epithelium (2). Transcripts encoding capsid genes initiate at the late promoter, P742, and must bypass the early polyadenylation signal to preferentially use the downstream poly(A) site (21, 22, 35). Differentiation promotes an increase in readthrough of the early signal which is in part due to changes in the activities and levels of polyadenylation factors (46). Downstream capsid gene expression is also influenced by numerous inhibitory elements located within the late coding region. cis-Acting negative regulatory elements have been identified through reporter assays in the 3′ untranslated region (UTR) of bovine papillomavirus type 1 (BPV-1) and HPV-16 late mRNAs (12, 13, 25, 28). In BPV-1, this element inhibits late polyadenylation site usage through the binding of U1 small nuclear ribonucleoprotein to an unutilized 5′ splice site (15). Additional inhibitory elements have been found in the HPV-16 L1 and L2 ORFs (42). It is, however, unclear what functions, if any, these elements have in the productive viral life cycle.
Due to the complexity of the vegetative life cycle of papillomaviruses, the requirements for a productive infection have been largely studied through the use of heterologous systems. With recent advances in cell culture techniques allowing the use of cell lines containing recombinant viral genomes, the requirements for a productive HPV infection are beginning to be elucidated. The in vitro synthesis of HPV can be accomplished through cotransfection of recircularized viral genomes with a drug resistance marker into normal human foreskin keratinocytes and has been successfully demonstrated for HPV-16, -18, and -31 (8, 10, 11, 32). In HPV-31, these methods have led to the identification of cis elements and trans-acting factors required for the productive viral life cycle (20, 27, 43–45, 47). Studies using recombinant HPV genomes have determined that late-gene expression requires episome maintenance, DNA amplification, and induction of the late promoter P742 (11). However, the cis elements and trans-acting factors involved in the differentiation-dependent induction of capsid gene expression remain largely unknown. The studies presented here examine the role of the HPV-31 early polyadenylation signal in regulating downstream capsid gene expression in recombinant HPV-31 genomes. In addition, the role of the early polyadenylation signal in the establishment and maintenance of the viral genome within the host keratinocyte was investigated.
MATERIALS AND METHODS
Cell lines and cell culture.
Normal human keratinocytes (NHKs) were derived from neonatal human foreskin epithelium as previously described and maintained in serum-free Keratinocyte Growth Medium (Clonetics) (16). The keratinocyte line LKP-31 maintains the HPV-31 genome in an episomal form and has been described previously (41). The CIN-612 line was derived from a cervical biopsy (5). SCC13 cells are a human squamous cell carcinoma line (38). LKP-31, CIN-612, and SCC13 cells were maintained in E medium with mitomycin C (Boehringer Mannheim)-treated fibroblast feeders (31). To induce differentiation, cells were suspended in semisolid medium containing 1.6% methylcellulose as previously described (41). For transient-expression assays, CIN-612 or LKP-31 cells were transfected with 0.5 μg of each reporter construct and 2.5 μg of pSP72 (Promega) using Lipofectamine (Gibco-BRL) in Opti-MEM (Gibco-BRL) according to the manufacturer's instructions. At 24 h after transfection, fibroblast feeders were removed with phosphate-buffered saline containing 0.5 mM EDTA, and keratinocytes were split either into semisolid medium or onto freshly treated fibroblasts. After 24 h, luciferase activities were determined through the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions.
Transfection of NHKs.
Transfection of viral DNA into NHKs has been described by Frattini et al. (11). Briefly, to remove the bacterial vector sequences, 10.0 μg of the p599-HPV31 plasmid and mutants were digested with HindIII overnight, followed by heat inactivation. Plasmids were ligated at a concentration of 10 ng/μl using T4 DNA ligase (10 U/900 μl) at 16°C overnight (Gibco-BRL). The DNA was precipitated with isopropyl alcohol and resuspended in TE buffer (10 mM Tris-Cl [pH 7.4rsqb], 1.0 mM EDTA). The entire precipitated ligation was cotransfected with 2.0 μg of pSV2Neo into NHKs with Lipofectace (Gibco-BRL) according to the manufacturer's instructions. Transfected cells were plated onto mitomycin-treated J2 fibroblast feeders in E medium 1 day after transfection. Selection began 2 days after transfection with G418 (200 μg/ml) (Gibco-BRL) every 2 days for a total of 4 days, and then G418 at 100 μg/ml every 2 days for 4 more days. After selection, pooled populations were expanded for analysis.
Recombinant plasmids.
Plasmid p599-31WT is a derivative of pBR322-HPV31 containing a modified version of pBR322 (11, 14). pBR322 was digested with ClaI and Eco47III, filled in with Klenow fragment, and ligated. The resulting minimal pBR322 was digested with EcoRI and a custom EcoRI and HindIII adapter linker (5′-AATTTTAAGCTTAA) was inserted. The HPV-31 genome was obtained from pBR322-HPV31 by digestion with EcoRI, ligated, and inserted into the HindIII site of the modified pBR322 plasmid. p599-31CP (CP) was created by PCR mutagenesis, replacing the early AAUAAA element with the sequence GGATCC. The 5′ product was obtained by PCR amplification using an upstream primer at nucleotide (nt) 3361 of HPV-31 (5′-cgggtaccgagctcGAATTCC) containing an EcoRI site and a downstream primer at nt 4170 (5′-GGTAATAATAAAAAAAAAGTAAAAAAGggatccATACCAATACCA), where capital letters indicate HPV-31 sequence and lowercase letters denote restriction enzyme sites or random sequence. The 3′ product was obtained by PCR amplification using an upstream primer at nt 4121 (5′-GGTATTGGTATTGGTATggatccCTTTACTTTTTTTTTATTATTACC) and a downstream primer at nt 4697 (5′-ggagatctGCAGGTGTAGGAGGCTGC). The products were combined and amplified using the original primers at nt 3361 and 4697, with the final species being inserted into the EcoRI and PpuMI sites of p599-HPV31. p599-31CP also lacks one of six repeated sequences, GGTATT, immediately upstream of the AAUAAA substitution which does not appear to influence readthrough activity when multiple repeats were substituted in pPolyA Luc-C15.2. Plasmid p599-31LD (LD) was made through the same approach, making a 5′ product using a downstream primer at nt 4195 (5′-caacatacacaacacacaccCGCATGGTAATAATAAAA) and a 3′ product using an upstream primer at nt 4177 (5′-gtgtgtgttgtgtatgttgGCACTAACGTGCGTC). Plasmid p599-31SV (SV) was constructed by cloning the simian virus 40 (SV40) late polyadenylation signal from pGL3-Basic (Promega) into p599-31WT using AvrII at nt 4071 and BglII at nt 4165. The AvrII and BglII sites in HPV-31WT were created by PCR mutagenesis. Plasmid p599-31BL (BL) was constructed by PCR mutagenesis, replacing nt 4219 to 5022 of p599-31WT with sequence from the β-galactosidase gene. The 5′ product was obtained from PCR amplification using p599-31WT, the upstream primer at nt 3361, and a downstream primer at nt 4217 (5′-cactccaggatccGTAGCAGACGCACGTTTAGTG). The 3′ product was produced as described below in the construction of the luciferase reporter plasmid pPolyA Luc-BL8. The products were combined, amplified using the outer primers, and inserted into the EcoRI and StuI sites of p599-31WT. All sequences produced through PCR amplification for recombinant genomes were sequenced in their entirety (Northwestern University Biotechnology Center). Plasmid p599-BL also contains an inadvertent substitution at nt 3448 which is silent in E2 but alters the E4 coding sequence. Studies have shown that mutations in E4 have no effect on transient replication (D. J. Klumpp and L. A. Laimins, unpublished data). The HPV-31 E1 and E2 expression vectors pSG-E1 and -E2, respectively, are based on pSG5 (Stratagene) and have been described by Frattini and Laimins (9). Plasmid pSL1–2 contains the E1∧E4,L1 cDNA from CIN-612 cells between nt 877 and 5760 of HPV-31 (21).
Luciferase reporter plasmids. (i) Poly(A) site mutations.
Plasmid pPolyA Luc-Control has been described previously (46). Plasmid pPolyA Luc-1500 was created by PCR amplification of p599-31WT using an upstream primer at nt 3797 containing an EcoRI site (5′-atagaattcATATGACTATTTAGCCTAATG) and a downstream primer at nt 5679 containing a BglII site (5′ ggagatctAGCAGCCTAGCACTGCCTG). The amplicon was cloned into the EcoRI and BamHI sites of pPolyA Luc-Control. Plasmids containing mutations in the early polyadenylation signal and L2 ORF were constructed using the pPolyA Luc-1500 reporter. pPolyA Luc-P15.1 was created by PCR amplification of p599-31CP, which lacks the AAUAAA element at nt 4138. The reporter pPolyA Luc-P15.2 was produced through PCR mutagenesis using p599-31WT and contains substitutions in both UAUAUA elements to CGGCCG at nt 3999 and CCCGGG at nt 4014. pPolyA Luc-C15.1 was constructed by replacing the sequence GGTATTGGTATTGGTATTGGT between nt 4103 and 4123 with ACGTAACCGTACACCTACAGCA. The plasmid pPolyA Luc-C15.2 was constructed by replacing ACTTTTTTTTT at nt 4151 to 4161 with CGAACACCCAT. pPolyA Luc-C15.3 was made by substituting AACGTGCGTCTGCT with GTCAAACCACAACC at nt 4202 to 4215. pPolyA Luc-L15 and pPolyA Luc-SV15 contain the late CstF binding site and SV40 late polyadenylation signal, respectively, as previously described, produced by PCR amplification and cloned into pPolyA Luc-1500.
(ii) Luciferase reporter plasmids with L2 substitution mutations.
The plasmid pPolyA Luc-E15 was created by PCR amplification of the HPV-31 E1 ORF between nt 901 and 2350 using primers containing BamHI and EagI sites. The PCR product was cloned into the BamHI and EagI sites of pPolyA Luc-1500. The BamHI site was created in pPolyA Luc-1500 by PCR mutagenesis at nt 4218. pPolyA Luc-EL8 contains sequence from nt 901 to 1695 of E1 ORF between the BamHI and StuI sites of pPolyA Luc-1500. The reporter pPolyA Luc-LE8 was made by PCR amplification of E1 nt 1724 to 2350, containing StuI and EagI sites, and cloned into the StuI and EagI sites of pPolyA Luc-1500. pPolyA Luc-EL4 was made by a two-step PCR amplification reaction. The E1 sequence was obtained by PCR amplification of nt 901 to 1296 (downstream primer, 5′-CATGTGTGCTACTGTACCATCTGCTGC). The L2 sequence was made by PCR amplification of nt 4620 to 5128 (upstream primer, 5′-ATGGTACAGTAGCACACATGAAAATCCTAC). The PCR products were then amplified using primers in E1 at nt 901 and in L2 at nt 5128 which contained BamHI and StuI sites and created an E1-L2 hybrid sequence. The sequence was cloned into the BamHI and StuI sites of pPolyA Luc-1500. Plasmid pPolyA Luc-LE4 was created by PCR amplification of L2 at nt 4018 to 4620 (downstream primer, 5′-GTTGTTTGTTGCTCCTCTAGAACACTTGTTACATCTAAAAGT) and of E1 at nt 1298 to 1695 (upstream primer, 5′-TAGAGGAGCAACAAACAAC). PCR products were combined and amplified using primers in L2 at nt 4018 containing a BamHI site and in E1 at nt 1695 containing a StuI site. The L2-E1 hybrid PCR product was cloned into the BamHI and StuI sites of pPolyA Luc-1500. The reporter pPolyA Luc-BL8 was constructed by PCR amplification of the β-galactosidase gene at nt 906 to 1701 from pSV-β-galactosidase (Promega) using primers that contained BamHI and StuI sites. The product was cloned into the BamHI and StuI sites of pPolyA Luc-1500. Plasmid pPolyA Luc-CPB was created by a two-step PCR amplification reaction. The 5′ product was obtained by PCR amplification of nt 3797 to 4223 from pPolyA Luc-P15.1, while the 3′ product was obtained by PCR amplification of nt 4211 to 5022 from pPolyA Luc-BL8. The products were PCR amplified with primers at nt 3797 and nt 5022, containing BamHI and StuI sites, respectively, and inserted into the BamHI and StuI sites of pPolyA Luc-1500.
Transient-replication assays.
Transient-replication assays were completed as previously described (20). Briefly, viral DNA was digested with HindIII and unimolecularly ligated. Samples were combined with carrier DNA and equimolar amounts of E1 and E2 expression plasmids (pSG-E1 and pSG-E2). SCC13 cells were transfected by electroporation at 250 V, 960 μF (Bio-Rad GenePulser) and plated onto mitomycin C-treated fibroblast feeders. At 5 days posttransfection, low-molecular-weight DNA was isolated through Hirt extraction (18). Samples were digested with DpnI to remove residual methylated DNA and with BanII to linearize viral genomes. After agarose gel electrophoresis and blotting to a nylon membrane (Magna; Micron Separations), DNA was detected with a radiolabeled HPV-31 DNA probe (HpaI-EcoRI fragment) and examined by autoradiography.
Southern blot analysis.
Total genomic DNA of transfectants was isolated by resuspending cell pellets in lysis buffer (400 mM NaCl, 10 mM Tris-Cl [pH 7.4], 10 mM EDTA). Samples were incubated overnight at 37°C with 50 μg of proteinase K per ml and 0.2% sodium dodecyl sulfate (SDS). DNA was sheared by passage through an 18-gauge needle 10× and extracted with phenol-chloroform. Samples were treated with RNase A (50 μg/ml) at 37°C for 1 h, followed by phenol-chloroform extraction and ethanol precipitation. Southern blots were completed using 10.0 μg of total DNA digested with DpnI and/or BlpI. Digested samples were run on an 0.8% agarose gel overnight and alkaline transferred to GeneScreen Plus nylon membranes (NEN). Membranes were prehybridized in 50% (vol/vol) formamide–4× standard saline phosphate (0.18 M NaCl, 10 mM phosphate [pH 7.4], 1.0 mM EDTA)–5× Denhardt's solution (0.02% polyvinylpyrollidone, 0.02% Ficoll, 0.02% bovine serum albumin)–1.0% SDS–10% (vol/vol) dextran sulfate–0.1 mg of denatured herring sperm DNA per ml for 1 h at 42°C. The HPV-31 probe was prepared by gel purification of an XbaI-HindIII fragment and labeled with the Ready-to-go DNA labeling kit (Amersham Pharmacia). Labeled probe was purified with Probe Quant G-50 microcolumns (Amersham Pharmacia), denatured, and added to fresh hybridization solution, which was incubated overnight at 42°C. The membrane was washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0015 M sodium citrate)–0.1% SDS for 15 min at room temperature, twice with 0.5× SSC–0.1% SDS for 15 min at room temperature, and once with 0.1× SSC–1.0% SDS for 30 min at 50°C. Membranes were visualized by autoradiography and quantitated through phosphorimaging.
Real-time RT-PCR analysis.
Quantitative reverse transcription (RT)-PCR was completed using the LightCycler Instrument (Roche) and the RNA amplification kit SYBR Green I (Roche) according to the manufacturer's instructions. The RNA standards were made by in vitro transcription reactions using the linearized plasmid pSL1–2 and the Riboprobe Combination System T3/T7 (Promega) according to the manufacturer's instructions. In vitro transcription reactions were treated with DNase I to remove template DNA, phenol-chloroform extracted, and ethanol precipitated prior to use. Transcripts containing the E1∧E4 splice site were amplified by using an upstream primer at nt 770 (5′-AGCACACAAGTAGATATTCGC) and a downstream primer at nt 3487 (5′-GTCGCCTCGCAACAACTTG), amplifying a 301-nt product from spliced RNA. Transcripts containing the E4∧L1 splice site were amplified by an upstream primer at nt 3408 (5′-CGACGACGTCTACTAAGCG) and a downstream primer at nt 5696 (5′-ATGGATGGCCTACTGTAAGC), amplifying a 328-nt product from spliced RNA. Template RNA was prepared as described above. RT-PCR was completed using 2.0 μg of total cellular RNA unless otherwise noted. The cycle profile was as follows, using a slope of 20°C/s: reverse transcription, 55°C for 10 min, denaturation at 95°C for 30 s, and amplification at 95°C for 1 s, 58°C for 10 s, and 72°C for 14 s (slope, 2°C/s) for 45 cycles; and melting curve, 97°C for 0 s, 65°C for 20 s, and 99°C for 0 s (slope, 0.1°C/s). Acquisition of fluorescence was completed as a single reading following each amplification cycle and as a continuous reading during the melting curve. Product specificity was determined by both melting-peak analysis and agarose gel electrophoresis (data not shown).
RESULTS
Readthrough activity is regulated by the early AAUAAA element.
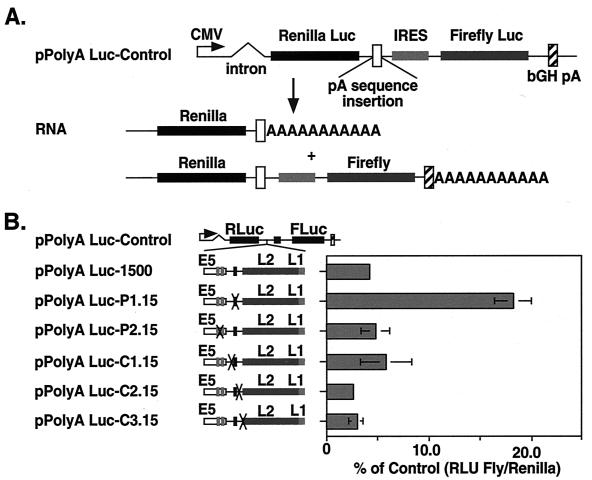
For the majority of RNA polymerase II transcripts, efficient polyadenylation requires a canonical AAUAAA element and a downstream GU- or U-rich (G/U-rich) sequence that binds the cleavage stimulatory factor (CstF) (7). Analysis of HPV-31 sequence around the site of early polyadenylation identified a single early AAUAAA element and two degenerative upstream polyadenylation elements, UAUAUA, in addition to several G/U-rich elements (46). In order to examine the regulatory elements within the early polyadenylation signal of HPV-31 that influence readthrough, we first took advantage of a dual luciferase reporter plasmid, pPolyA Luc-Control, that was used previously to show that the early polyadenylation signal allows a significant amount of readthrough to downstream expression (46). The reporter plasmid contains a cytomegalovirus promoter driving expression of the Renilla luciferase gene as well as the firefly luciferase gene, separated by the encephalomyocarditis virus internal ribosome entry site (IRES) (Fig. 2A). This reporter assay examines steady-state RNA levels and therefore indirectly measures polyadenylation site usage and downstream expression.
FIG. 2.
Identification of HPV-31 early polyadenylation cis elements which regulate downstream gene expression. (A) Schematic of pPolyA Luc-Control reporter construct containing a cytomegalovirus (CMV) promoter driving expression of both the Renilla (Rluc) and firefly (Fluc) luciferase genes. The genes are separated by an IRES element from encephalomyocarditis virus and contain a downstream polyadenylation signal from bovine growth hormone (bGH pA). (B) The reporter pPolyA Luc-1500 contains sequences from upstream of the E5 ORF to 1,500 nt of the late coding region. The HPV-31 sequence includes the early polyadenylation signal AAUAAA (solid vertical box) and degenerative signal UAUAUA (shaded vertical box). pPolyA Luc-P1.15 and -P2.15 contain substitutions in the AAUAAA and UAUAUA elements, respectively. pPolyA Luc-C1.15, -C2.15, and -C3.15 contain substitutions which reduce the G/U content within previously defined CstF binding sites. Plasmids were transfected into LKP-31 cells as described in Materials and Methods. Luciferase activities were determined and are illustrated graphically as the ratio of relative light units (RLU) from the downstream firefly luciferase to the upstream Renilla luciferase as a percentage of that with pPolyA Luc-Control. The ratios are presented as the standard deviations from three experiments.
We first investigated the role of the hexanucleotide element AAUAAA in modulating downstream expression. Plasmid pPolyA Luc-1500, which contains wild-type HPV-31 sequences from upstream of the E5 ORF to 1,500 nt into the L2 ORF, was constructed (Fig. 2B). We next constructed a derivative plasmid, pPolyA Luc-P1.15, by PCR mutagenesis, in which the HPV-31 AAUAAA element was removed and substituted with random sequences (Fig. 2B). An additional plasmid, pPolyA Luc-P2.15, was constructed, in which the two degenerative hexanucleotide sequences, UAUAUA, were removed from pPolyA Luc-1500. These reporter plasmids were transfected into LKP-31 cells, which stably maintain the HPV-31 genome as episomal DNA. With pPolyA Luc-P1.15, we observed a 4.3-fold increase in downstream expression compared to pPolyA Luc-1500. This is in contrast to pPolyA Luc-P2.15, where removal of the degenerative UAUAUA sequences had little effect on expression. This indicated that a major regulator of readthrough was the consensus hexanucleotide sequence and that the degenerative sequences had little effect.
We previously identified three weak G/U-rich CstF binding sites near the HPV-31 early polyadenylation signal through RNA-binding studies (46). To determine the role of the early CstF binding sites in regulating downstream expression, mutations were introduced into pPolyA Luc-1500 which reduced the G/U content of each element to generate plasmids pPolyA Luc-C1.15, -C2.15, and -C3.15 (Fig. 2B). Upon transfection into LKP-31 cells, only minor differences in readthrough activities were detected with these mutants compared to pPolyA Luc-1500 and the above hexanucleotide substitution mutant. These data suggest that the G/U-rich regions have little influence on readthrough and downstream expression. This is consistent with our previous observations that these elements are weak CstF sites (46).
Elements within the 5′-most 800 nt of L2 influence readthrough activity.
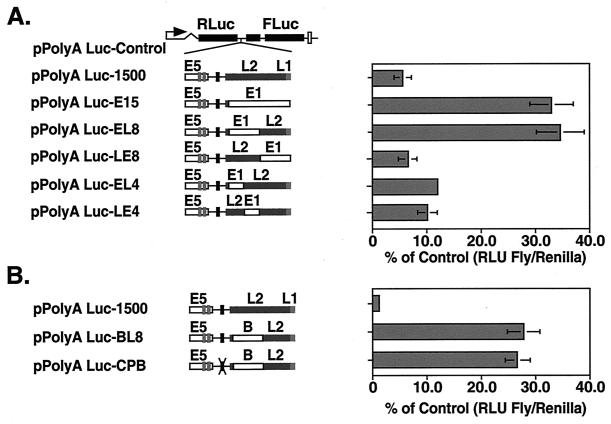
Sequences within the L2 ORFs of both HPV-16 and BPV-1 have been implicated as negative regulators of papillomavirus late gene expression through the use of reporter assays (4, 42). We next investigated if such elements exist in HPV-31 and if these have a role in modulating late gene expression. For these studies, a series of substitutions were introduced into the L2 ORF in pPolyA Luc-1500 (Fig. 3A). The reporter pPolyA Luc-E15 was made by replacing the L2 ORF with sequence from the HPV-31 E1 ORF, which lacks any known cis-acting regulatory elements, including splice sites, and, as such, functions as a neutral spacer sequence (Fig. 3A). The substitutions were made to ensure that the effects we were observing were not due to the changes in the number of nucleotides separating the reporter genes. As shown in Fig. 3A, transfection of pPolyA Luc-E15 into LKP-31 cells resulted in a 6.0-fold increase in readthrough compared to the parental pPolyA Luc-1500 plasmid. These observations confirm the presence of inhibitory elements within the HPV-31 L2 coding region.
FIG. 3.
Identification of sequences within the L2 ORF which influence downstream pPolyA Luc reporter expression. (A) Substitution mutations were introduced into the late coding region of pPolyA Luc-1500, replacing the late coding region with neutral spacer sequence obtained from the HPV-31 E1 ORF (E1). In pPolyA Luc-E15, 1,500 nt of the late coding region was replaced with E1. pPolyA Luc-EL8 and -LE8 contain substitutions in the 5′ and 3′ end of the late region, respectively. The reporters pPolyA Luc-EL4 and -LE4 contain substitutions within the 5′-most 800 nt of L2 with spacer sequence. (B) pPolyA Luc-BL8 contains a substitution within the 5′ end of L2 using spacer sequence obtained from the prokaryotic β-galactosidase gene. In pPolyA Luc-CPB, both AAUAAA and the 5′-most 800 nt of L2 have been replaced by spacer sequence. Reporter plasmids were transfected into LKP-31 cells as described in Materials and Methods. Activities are illustrated graphically as a ratio of RLUs from firefly luciferase to Renilla luciferase and presented as a percentage of the pPolyA Luc-Control value. The ratios are presented as the standard deviations from three experiments with the exception of the pPolyA Luc-EL4 data, which come from a single experiment.
We next sought to localize the inhibitory elements through a series of smaller substitutions. The 800 nt at the 5′ end of the L2 ORF were replaced by sequences from the E1 ORF to generate pPolyA Luc-EL8 (Fig. 3A). Following transfection, downstream luciferase activity was found to increase by 6.3-fold in pPolyA Luc-EL8 compared to pPolyA Luc-1500, suggesting that the inhibitory element was localized to this region. In contrast, when the 3′ half of the L2 ORF was replaced in pPolyA Luc-1500 with a comparably sized region from the E1 ORF (pPolyA Luc-LE8), no difference in downstream expression was observed in transient assays compared to pPolyA Luc-1500. We next made two 400-nt substitutions in the 5′-most 800 nt to generate pPolyA Luc-EL4 and -LE4, but neither of these plasmids exhibited downstream expression levels similar to those seen with pPolyA Luc-EL8. Additional mutations were also made within L2 by deleting small segments of the 5′-most 400 nt in the context of pPolyA Luc-LE4, but these changes failed to restore downstream expression levels to those seen in pPolyA Luc-EL8 (data not shown). These studies demonstrate that the first 800 nt of L2 are responsible for inhibiting readthrough and that sequences throughout this region are required for activity.
The above studies used E1 sequences as a neutral DNA spacer, and it was possible that these sequences may have some uncharacterized effect on gene expression. We therefore constructed a second series of reporter plasmids in which sequences from the prokaryotic β-galactosidase gene were used as spacers. As in pPolyA Luc-EL8, we replaced the complete 800 nt at the 5′ end of the L2 ORF with sequence from the β-galactosidase gene, making pPolyA Luc-BL8. In transient assays, a similar level of downstream expression was observed with pPolyA Luc-BL8 and with the E1 substitution (Fig. 3B).
We next investigated whether altering the AAUAAA element in combination with the 5′-most 800 nt of L2 would further modulate downstream expression. For these studies, substitutions were made in both elements to generate plasmid pPolyA Luc-CPB. Transfection of pPolyA Luc-CPB into LKP-31 cells resulted in an increase in downstream expression similar to that seen with pPolyA Luc-BL8 (Fig. 3B). We conclude that a primary determinant regulating readthrough expression exists within the 5′-most 800 nt of the L2 ORF and that this region negatively regulates downstream capsid gene expression.
Mutations within HPV-31 early polyadenylation region influence genome replication.
It was next important to examine the role of the early polyadenylation signal in regulating downstream capsid gene expression under more physiological conditions during the life cycle of the virus. Since reporter assays identified the AAUAAA element and sequences within the L2 ORF as major regulators of early and late gene expression, we mutated these elements in the context of the complete HPV-31 genome. The AAUAAA element was disrupted in plasmid p599-31CP, and the 5′-most 800 nt of the L2 ORF were replaced with sequence from the β-galactosidase gene in p599-31BL (Fig. 4C). We also constructed genomes into which we inserted a high-affinity CstF binding site as well as the SV40 late polyadenylation signal in place of the HPV-31 early poly(A) signal (Fig. 4C). Previous studies using reporter assays demonstrated that insertion of such strong elements into the early polyadenylation signal significantly reduced readthrough into the late region (46).
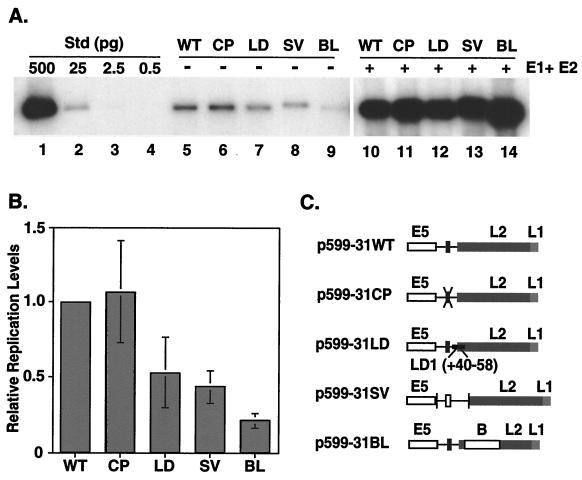
FIG. 4.
Transient-replication assays using HPV-31 genomes containing mutations within the early polyadenylation signal. (A) Transient-replication assays were completed using viral DNA that was released from the bacterial vector, unimolecularly ligated, and transfected into SCC13 cells. After 5 days, low-molecular-mass DNA was isolated through Hirt extraction. DNA was digested with DpnI to remove methylated input DNA, and replication levels were determined through Southern analysis using a 32P-labeled HPV-31 genomic DNA fragment (lanes 5 to 9). Lanes 1 to 4 are DNA standards (Std) consisting of linearized wild-type (WT) HPV-31 DNA representing 500, 25, 2.5, and 0.5 pg, respectively. Replication was restored to wild-type levels upon cotransfection with E1 and E2 expression vectors (lanes 10 to 14). (B) Levels of replication from lanes 5 to 9 have been quantitated through phosphorimage analysis and are presented relative to the wild-type value. The data are presented as the standard deviation from multiple experiments. (C) Mutations were introduced into p599-31WT which contains the HPV-31 genome in pBR322 as described within the Material and Methods. Plasmid p599-31CP contains a substitution in the early AAUAAA element, while p599-31LD contains the late strong CstF binding site 40 nt downstream of AAUAAA. Plasmid p599-31SV contains the complete SV40 late polyadenylation signal in place of the early signal. Plasmid p599-31BL contains a substitution in the 5′-most 800 nt of the L2 ORF using a neutral spacer sequence obtained from the β-galactosidase gene.
It is likely that introducing mutations into the HPV-31 early polyadenylation signal will alter viral early gene expression as well as late gene expression. In fact, changes in the levels of expression of the viral replication proteins E1 and E2 can have profound effects on genome establishment and maintenance within the host keratinocyte (11, 43, 47). We therefore first examined the effects that mutations within the HPV-31 early poly(A) signal have on genome replication using transient assays. For these studies, viral DNAs were released from the bacterial vector, unimolecularly ligated, and transfected into SCC13 keratinocytes by electroporation. After 5 days, low-molecular-weight DNA was isolated and digested with DpnI to remove any residual bacterially methylated input DNA. The viral genomes were linearized to facilitate quantitation, and Southern analysis was then performed (Fig. 4A).
Surprisingly, we observed that removal of the AAUAAA element resulted in a modest increase in genome replication (Fig. 4A, lanes 5 and 6, 4B) rather than a reduction or abrogation of activity. In contrast, genomes containing the high-affinity late CstF binding site replicated at levels that were slightly lower than wild-type levels (Fig. 4A, lanes 5 and 7, and 4B), while the SV40 polyadenylation signal replacement mutant replicated at significantly reduced levels (Fig. 4A, lanes 5 and 8, and 4B). The genomes containing the SV40 late polyadenylation signal are approximately 100 nt larger than either the wild-type HPV-31 genome or the mutated HPV early poly(A) genomes, and this accounts for the slight shift in genome mobility seen in Fig. 4A. Interestingly, replacing the 5′-most 800 nt of the L2 ORF resulted in a dramatic decrease in genome replication (Fig. 4A, lanes 5 and 9, and 4B). Overall, these results were unexpected, as it was anticipated that more efficient early polyadenylation would occur in the late CstF and SV40 mutant genomes, leading to increased levels of transcripts encoding replication protein and higher replication rates. Equally unexpected is the increase in the replication of the AAUAAA deletion genome, which we had expected to replicate less efficiently, if at all.
Altered replication ability may be the result of either disruption of cis elements involved in replication or altered expression levels of viral factors. To test if the HPV poly(A) mutant genomes were indeed diminished in their ability to express replication factors, as we suspected, transient assays were performed with cotransfected E1 and E2 expression vectors. In these assays, an expression defect could be restored, while a cis defect could not. As shown in Fig. 4A, replication levels of the genomes containing the late CstF binding site (lane 12), the SV40 polyadenylation signal (lane 13), and the substitution in the L2 ORF (lane 14) were all restored to wild-type levels (lane 10). Comparable levels of replication were also observed between 31CP and wild-type genomes (Fig. 4A, lanes 10 and 11). These data suggest that the changes that we introduced in the HPV-31 early polyadenylation signal most likely influence the overall expression levels of HPV-31 replication factors.
Recombinant genomes containing altered polyadenylation signals are stably maintained within NHKs.
The productive HPV life cycle requires stable maintenance of the viral genome as extrachromosomal elements within the host keratinocyte. It was therefore important to determine whether genomes containing mutations within the HPV-31 early polyadenylation signal could be stably maintained. Wild-type and mutant genomes were excised from the vector backbone, unimolecularly ligated, and cotransfected with the pSV2neo plasmid into normal NHKs. After selection of the cells with G418, colonies were pooled and expanded. Four to six weeks after transfection, total cellular DNA was extracted from the cells, digested with DpnI to remove any residual input DNA, and analyzed by Southern blotting. As shown in Fig. 5 (lane 9), several prominent HPV-31 species were detected in lanes containing DNA from wild-type genome transfections corresponding to supercoiled, open-circle, and linear forms of DNA. Sample DNA isolated from wild-type, 31CP, 31LD, and 31SV mutant transfections all contained episomal forms of DNA consistent with extrachromosomal maintenance of the viral genome (Fig. 5, lanes 9 to 12). In contrast, the 31BL mutant was unable to be stably maintained within primary keratinocytes (data not shown). A low level of high-molecular-weight HPV DNA corresponding to integrated copies was seen in all transfected cell lines and is most likely due to incomplete religation of viral genomes prior to transfection.
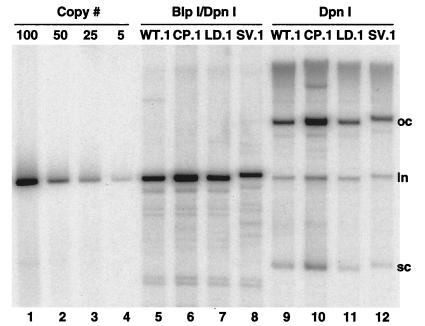
FIG. 5.
Southern blot analysis of NHK lines containing recombinant wild-type and mutant HPV-31 genomes. NHK lines containing recombinant HPV-31 genomes were established as described in Materials and Methods. Southern blot analysis was completed using 10.0 μg of total DNA isolated from low-passage 31WT.1, 31CP.1, 31LD.1, and 31SV.1 cells lines. DNA was digested with either DpnI, to remove residual input DNA (lanes 9 to 12), or DpnI and BlpI, to linearize HPV-31 DNA, allowing quantitation (lanes 5 to 8). Specific DNAs were detected with 32P-labeled HPV-31 genomic DNA fragment. Several species of DNA are observed, including open circle (oc), linear (ln), and supercoiled (sc). Lanes 1 to 4 are genome copy number controls representing 100, 50, 25, and 5 copies per cell, respectively.
Quantitative phosphorimaging analysis revealed genome copy numbers within the wild-type-transfected cells (31WT.1) to be approximately 80 copies per cell (Fig. 5, lane 5). Comparable copy numbers were observed in 31LD.1-transfected cells (Fig. 5, lane 7). However, DNA from the 31CP.1 cells showed a higher copy number at approximately 130 copies per cell (Fig. 5, lane 6), while a slightly lower copy number was observed in 31SV.1-transfected cells at approximately 70 copies per cell (Fig. 5, lane 8). Southern analysis of DNA isolated from later passages and from other independent transfections confirmed that episomes were retained at comparable levels relative to the wild type. The differences in copy numbers in stable assays paralleled the differences observed in transient assays. Primary NHK lines obtained from different foreskin donors have been shown to vary in the average HPV-31 copy number maintained per cell (L. Laimins, unpublished data). However, in repeated transfections of the same donor NHK cells, similar copy numbers for wild-type HPV-31 were obtained. Experiments were also completed in an additional primary NHK isolate obtained from a different foreskin donor and showed similar patterns of maintenance with comparable ratios of copy numbers (data not shown). The overall copy number per cell in these additional isolates was, however, reduced from those shown in Fig. 5. All mutant genomes except those containing the substitution in the L2 ORF and the SV40 polyadenylation signal were consistently found in an episomal state. In the 31SV-transfected cells, episomal maintenance was seen in only one of three experiments. Overall, these data suggest that alterations in the early polyadenylation signal do not prevent the stable maintenance of viral genomes within NHKs, though the average copy number per cell varied depending on the specific mutation. In addition, introduction of a strong polyadenylation signal resulted in a diminished ability to maintain episomes.
Levels of early gene expression are similar between 31WT, 31CP, 31LD, and 31SV cell lines.
Since the mutations within the early poly(A) signal may alter early gene expression in addition to modifying readthrough into the late region, we investigated whether the levels of early gene expression varied significantly between the cell lines through the use of quantitative real-time RT-PCR. Quantitative real-time RT-PCR allows an accurate measure of transcript levels, as opposed to approximations obtained through RNase protection assays. For these studies, total RNA was isolated from the cell lines described above, and RT-PCR was performed using a set of primers which span the splice donor at nt 877 and the acceptor at nt 3295. These primers allow quantitation of E1∧E4-containing transcripts, which are present in most early transcripts (Fig. 6B). RNA standards were synthesized through in vitro transcription reactions using a linearized plasmid template containing the E1∧E4,L1 cDNA. RT-PCR products were quantitated using SYBR green dye, which incorporates into double-stranded DNA. After each round of cycling, the dye binds to the PCR product, which can then be detected by fluorescence. From these experiments, the concentration of E1∧E4-containing transcripts was determined from a standard curve (Fig. 6A). Amplification of the correct mRNA target sequence was then confirmed by melting-curve analysis and visualized by ethidium bromide staining following agarose gel electrophoresis (data not shown).
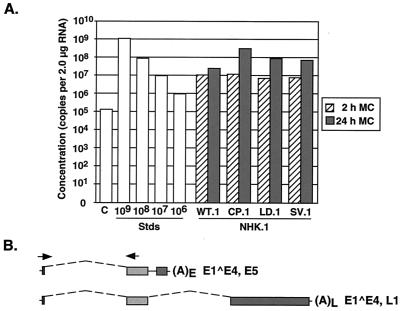
FIG. 6.
Quantitation through real-time RT-PCR of E1∧E4-containing transcript levels within undifferentiated NHK cells containing recombinant HPV-31 mutants. (A) The concentration of E1∧E4 transcripts was determined through real-time RT-PCR using 2.0 μg of total RNA isolated from NHK lines containing recombinant genomes. Total RNA was isolated from lines placed in semisolid medium for 2 and 24 h. Quantitation was completed through SYBR green I dye incorporation into the amplicon, followed by quantitation of fluorescence. The RNA standard was synthesized through in vitro transcription reactions from an E1∧E4,L1 cDNA and amplified using the same primer set. Levels of E1∧E4-containing transcripts were determined from a standard curve (Stds). Similar results were seen in multiple experiments. (B) Quantitation was completed using a primer set which spans E1∧E4 using a donor splice site at nt 877 and an acceptor at nt 3295.
Real-time RT-PCR analysis demonstrated that the wild-type 31WT.1 line contained approximately 11.2 × 106 copies of E1∧E4-containing transcripts in 2.0 μg of total RNA (Fig. 6A). Similar concentrations were observed for the 31CP.1, 31LD.1, and 31SV.1 cell lines, with approximately 11.7 × 106, 6.7 × 106, and 7.7 × 106 copies per 2.0 μg of RNA, respectively (Fig. 6A). The results are summarized in Table 1. An additional cell line, LKP-31, was included as a control; it contains the wild-type HPV-31 genome. The LKP-31 cell line maintains a high episome copy number with few integrated species. The expression levels in LKP-31 cells were slightly elevated, with approximately 46.6 × 106 copies of E1∧E4-containing transcripts in 2.0 μg of total RNA (Table 1). Analysis of transfectants obtained using a different primary NHK background cell line showed levels of E1∧E4-containing transcripts of approximately 12.7 × 105, 6.8 × 105, and 24.5 × 105 copies per 2.0 μg RNA for 31WT.2, 31CP.2, and 31LD.2-transfected lines, respectively (Table 1). These cell lines maintained fewer episomal copies per cell than the cell lines described above. However, the relative ratio of early transcripts to wild-type genomes was similar to that seen in the previous set of cell lines. We conclude that the HPV-31 early poly(A) mutant genomes express similar levels of viral early transcripts relative to wild-type genomes.
TABLE 1.
Quantitation of late transcripts from NHK lines containing recombinant HPV-31 genomesa
| Cells and genome | No. of transcripts/2 μg of RNA
|
|||
|---|---|---|---|---|
| E1∧E4
|
E4∧L1, 24 h | E4∧L1/E1∧E4 | ||
| 2 h | 24 h | |||
| NHK-1 | ||||
| 31WT | 1.1 × 107 | 2.5 × 107 | 9.8 × 103 | 3.9 × 10−4 |
| 31CP | 1.2 × 107 | 3.2 × 108 | 4.1 × 106 | 1.3 × 10−2 |
| 31LD | 6.7 × 106 | 8.6 × 107 | 1.1 × 104 | 1.3 × 10−4 |
| 31SV | 7.7 × 106 | 6.2 × 107 | 9.6 × 102 | 2.0 × 10−5 |
| NHK, 2b | ||||
| 31WT | 1.3 × 106 | 7.1 × 106 | 7.0 × 104 | 9.8 × 10−3 |
| 31CP | 6.8 × 105 | 6.0 × 106 | 4.1 × 105 | 6.8 × 10−2 |
| 31LD | 2.4 × 106 | 1.1 × 107 | 1.6 × 104 | 1.4 × 10−3 |
| 31SV | —c | — | — | — |
| LKP-31 | ||||
| 31WT | 4.7 × 107 | 5.3 × 108 | 4.2 × 105 | 7.9 × 10−4 |
Quantitative RT-PCR was completed using 2.0 μg of RNA isolated from cells placed in methylcellulose for 2 or 24 h to induce keratinocyte differentiation.
Quantitation of E4∧L1 was completed using 3.0 μg of RNA.
—, cell line contained integrated viral DNA.
Activation of differentiation-dependent promoter P742 occurs in polyadenylation mutant cell lines.
The induction of the late promoter P742 results in an increase in late transcripts encoding E1∧E4,E5 and E1∧E4,L1 (22). These two transcripts utilize alternative polyadenylation signals at the end of E5 and L1, respectively. To determine the levels of transcripts which contain the E1∧E4 splice site upon differentiation, quantitative RT-PCR was performed using total RNA isolated from undifferentiated and differentiated keratinocytes. Epithelial differentiation and induction of late viral functions occur upon suspension of HPV-positive keratinocytes in semisolid medium (26, 40, 41). Primers which span the splice donor at nt 877 and splice acceptor at nt 3295 were used in this reaction. The results are summarized in Table 1. Real-time RT-PCR analysis demonstrated an approximately 11.3-fold induction of E1∧E4-containing transcripts following 24 h in methylcellulose in a cell line containing wild-type genomes (LKP-31). When the keratinocytes harboring mutant genomes were examined, comparable levels of induction were observed for the 31LD.1 and 31SV.1 cell lines at 12.8- and 8.0-fold, respectively (Fig. 6). Interestingly, the 31CP.1 line showed a 26.7-fold induction of E1∧E4 transcripts, which is greater than that observed in the other mutant or wild-type lines. Similar trends were observed using cells from a second NHK donor (Table 1). The level of induction observed in the 31WT.1 line was reduced, but we believe that this is not representative and was not seen in two other wild-type lines (Table 1). We also observed that the levels of DNA amplification were similar in both wild-type and mutant cell lines (data not shown). From these experiments, we conclude that the levels of differentiation-dependent induction of the P742 promoter were similar in cells with wild-type, LD, and SV genomes. The cells containing the genomes with substitutions of the AAUAAA sequence generally induced to a slightly higher level.
Mutations within HPV-31 early polyadenylation signal alter downstream capsid expression levels.
The experiments shown in Fig. 2 using a heterologous reporter system indicated that HPV-31 capsid gene expression requires an inefficient early polyadenylation signal. We next investigated whether alterations within the HPV-31 early polyadenylation signal influence capsid gene expression in cell lines containing recombinant genomes. For these studies, the levels of E4∧L1-containing transcripts were determined by real-time RT-PCR using a primer set that spans the E4∧L1 splice site (Fig. 7B). This primer set will detect the major late transcript, E1∧E4,L1 (Fig. 1) (21). Total RNA was isolated from cell lines placed in semisolid medium for 24 h to induce keratinocyte differentiation. A standard curve was made using RNA produced through an in vitro transcription reaction using a DNA template containing the E1∧E4,L1 cDNA (Fig. 7A).
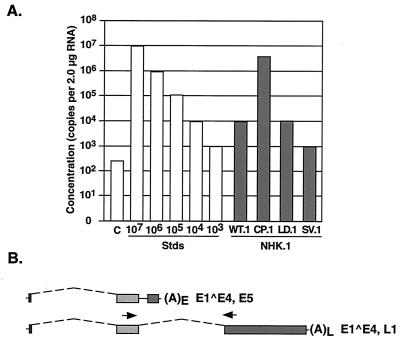
FIG. 7.
Quantitation through real-time RT-PCR of E4∧L1-containing transcript levels from differentiated NHK lines containing recombinant HPV-31 genomes. (A) Real-time RT-PCR was completed using 2.0 μg of total RNA isolated from cells placed in semisolid medium for 24 h. The RNA standard was synthesized through an in vitro transcription reaction using the E1∧E4,L1 cDNA. The concentration of E4∧L1 transcripts was determined through a standard curve (Stds). Similar results were seen in multiple experiments. (B) Quantitation was completed with a primer set that spans E4∧L1 using a splice donor at nt 3590 and a splice acceptor at nt 5552.
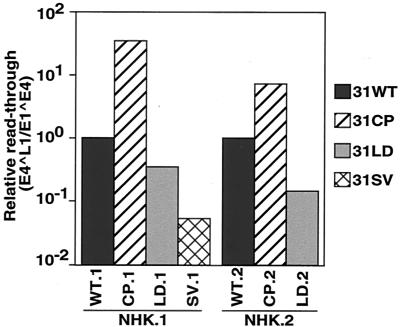
Quantitative RT-PCR demonstrated significant differences in downstream capsid gene expression between cell lines containing wild-type and mutant genomes. The relative differences in expression between E4∧L1-containing transcripts and E1∧E4-containing transcripts, or readthrough activity, between cell lines are presented in Table 1. Quantitation of late transcripts from LKP-31 cells determined the ratio of E4∧L1- to E1∧E4-containing transcripts to be 7.9 × 10−4. Similar results were observed in the 31WT.1 cell line, with a ratio of expression of 3.9 × 10−4. The 31CP.1 cells demonstrated a 32.9-fold increase in downstream expression compared to 31WT.1 cells (Fig. 8). This is in contrast to the dramatic decreases in capsid gene expression levels seen in 31LD.1 and 31SV.1 lines, where downstream expression decreased by approximately 67 and 95%, respectively (Fig. 8). In undifferentiated cells, E4∧L1-containing transcripts were not detectable, and the levels of late transcripts expressed upon differentiation are consistent with those seen in previous studies (21). A similar pattern of capsid gene expression was observed in cell lines using NHK cells from a different foreskin donor, though the overall levels were reduced (Table 1, Fig. 8).
FIG. 8.
Relative readthrough activity of HPV-31 wild-type and mutant polyadenylation signals in the context of the complete genome within differentiated NHKs. The ratio of E4∧L1- to E1∧E4-containing transcripts, or readthrough activity, is presented relative to the wild-type lines. The generation of stable lines was repeated twice using two different foreskin donors, NHK.1 and NHK.2, to ensure reproducibility.
DISCUSSION
In this study, we determined that the early polyadenylation sequence AAUAAA as well as sequences within the first 800 nt of L2 are major regulators of both early and late gene expression in HPV-31. Since late HPV transcripts initiate in the early region, mechanisms must exist to prevent inappropriate readthrough into the late region during the early phases of the viral life cycle but allow it to occur following differentiation. Disruption of the early AAUAAA element resulted in significant increases in readthrough activity and downstream gene expression in the context of both the reporter system and recombinant HPV-31 genomes. Our observations are consistent with studies demonstrating the tight association between readthrough and polyadenylation at the early AAUAAA site (37). Additional regulatory elements such as the degenerative polyadenylation signal UAUAUA as well as putative G/U-rich CstF binding sites had only a modest effect on downstream expression. Studies in BPV-1 identified the degenerative signal UAUAUA, which functions in early polyadenylation, as disruption of this element altered the site of poly(A) addition (1). Our studies show that these degenerative sites have little effect in the context of a wild-type hexanucleotide sequence.
The ultimate test of the importance of these polyadenylation elements is to examine their roles in regulating capsid gene expression during the vegetative life cycle. Quantitation of E1∧E4- and E4∧L1-containing transcripts isolated from differentiated keratinocytes containing wild-type genomes demonstrated a significant amount of readthrough activity from the early to late region. In contrast, introduction of the strong late HPV-31 CstF binding site or the SV40 late polyadenylation signal into the early polyadenylation region resulted in dramatic decreases in capsid gene expression upon keratinocyte differentiation. Such an effect would be anticipated if a weak polyadenylation signal is required for efficient late gene expression. Surprisingly, introduction of this strong polyadenylation signal into the complete HPV-31 genome also resulted in decreased levels of replication in both transient and stable replication studies. Wild-type levels of replication were restored for both mutant genomes upon cotransfection with expression vectors for the replication factors E1 and E2. These data suggest that introducing the late CstF binding site and SV40 late signal into the HPV-31 early polyadenylation signal reduced the levels of expression of viral replication proteins. It had been anticipated that introduction of signals for efficient early polyadenylation would result in increased levels of transcripts encoding the replication factors E1 and E2 since any readthrough transcripts would now be expected to terminate at the early site. Studies using HPV-31 genomes containing mutations within splice donor and acceptor sites demonstrated the importance of specific splicing patterns in early expression through the loss of genome replication and stable maintenance (27, 44, 47). We suspect that making the early polyadenylation signal more efficient may result in altered splicing patterns, leading to a reduction in transcripts expressing replication factors, and studies examining this point have been initiated.
Our studies also demonstrated that removal of the HPV-31 early AAUAAA element from the early polyadenylation signal resulted in an increase in genome copy number in both transient replication and stable maintenance assays. While our previous experiments suggested that efficient HPV-31 replication requires an inefficient early polyadenylation signal, we had anticipated that further mutation of the polyadenylation signal would lead to reduced replication. In fact, we observed higher levels of replication with the more inefficient signal. It is possible that an alternative noncanonical poly(A) signal is used in these mutants, as seen previously in BPV-1 (1). Studies in HPV-16 have demonstrated that altering early polyadenylation through viral genome integration resulted in the loss of an instability element within the early 3′ UTR and an increase in early viral gene expression (23). The early AAUAAA element is conserved in most papillomavirus types with the exception of HPV-6 and -11, which contain degenerative signals, AGUAAA (6). These HPVs are the predominant HPV types in an infected population and are efficient replicators (29). The lack of a consensus AAUAAA polyadenylation signal may provide an advantage for the low-risk viral types by possibly allowing a higher copy number per cell as well as increased levels of expression of viral capsid proteins. The question arises as to why the high-risk types have evolved to include elements that result in reduced viral expression and copy number. It is possible that this reduced level of expression is more beneficial for the long-term persistence of high-risk HPV types. Alternatively, the presence of a consensus hexanucleotide sequence may modulate splicing patterns in a manner that provides an advantage in vivo but is not scored in our tissue culture assays.
In addition to the hexanucleotide sequence, we also identified sequences within 800 nt of the L2 ORF that significantly influence downstream expression. In fact, we believe that this L2 element may be a primary determinant for regulating early as well as late expression. As attempts at identifying smaller elements within this region were unsuccessful, we believe that this inhibitory region functions either as a single large element or cooperatively through multiple redundant elements. In the context of the complete HPV-31 genome, disruption of these sequences resulted in a significant reduction in genome replication and loss of stable maintenance in primary foreskin keratinocytes. Our observations are consistent with studies in BPV-1, which have identified transcription termination sites within the L2 gene (4). A close association between polyadenylation and transcription termination has been demonstrated in other systems, and we suspect that both processes may play important roles in HPV-31 pathogenesis (33). Reporter-based assays examining HPV-16 identified a similar inhibitory element within the 5′ end of the L2 ORF which influenced mRNA stability (42). It remains unclear whether RNA stability, termination, or a combination play a role in the action of the HPV-31 L2 element.
In BPV-1 and HPV-16, a second negative regulatory element has been identified at the end of the L1 gene and has been postulated to make transcripts that traverse L1 unstable in undifferentiated cells (12, 25, 28). It is possible that such an element exists in HPV-31 and that it acts to augment the action of the L2 element. The L1 element could act by destabilizing any transcripts that escape the action of the L2 sequences. However, our studies suggest that the majority of transcripts that pass through the early polyadenylation sequence terminate or are rendered unstable by sequences within the L2 element. Upon differentiation, the negative regulatory effect of the L2 element is abrogated. This could be accomplished through the action of a virally encoded factor or a cellular protein, and efforts to identify these regulators have been initiated. In summary, our studies suggest that the differentiation-specific regulation of capsid gene expression in HPV-31 requires the coordinated action of a least four processes: (i) initiation of transcription at the late P742 promoter, (ii) abrogation of the negative posttranscriptional regulatory effects of the L2 element, (iii) polyadenylation of early transcripts directed by the AAUAAA sequence, and (iv) alternative splicing to generate the E4∧L1 transcripts.
ACKNOWLEDGMENTS
We thank members of the Seifert Lab for technical advice on use of the LightCycler and quantitative RT-PCR. In addition, we thank Richard Longnecker, Pat Hindmarsh, and Stephen Oh for critical reading of the manuscript.
This work was supported by a grant from the National Cancer Institute to L.A.L.
REFERENCES
- 1.Andrews E M, DiMaio D. Hierarchy of polyadenylation site usage by bovine papillomavirus in transformed mouse cells. J Virol. 1993;67:7705–7710. doi: 10.1128/jvi.67.12.7705-7710.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker C C. Post-transcriptional regulation of papillomavirus gene expression., p. III-11–III-16. In: Myers G, Baker C, Munger K, Sverdrup F, McBride A, Bernard H-U, Meissner J, editors. Human papillomaviruses. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1997. [Google Scholar]
- 3.Baker C C, Howley P M. Differential promoter utilization by the bovine papillomavirus in transformed cells and productively infected wart tissues. EMBO J. 1987;6:1027–1035. doi: 10.1002/j.1460-2075.1987.tb04855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker C C, Noe J S. Transcriptional termination between bovine papillomavirus type 1 (BPV-1) early and late polyadenylation sites blocks late transcription in BPV-1-transformed cells. J Virol. 1989;63:3529–3534. doi: 10.1128/jvi.63.8.3529-3534.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bedell M A, Hudson J B, Bolub T R, Turek M E, Hoskin M, Wilbanks G D, Laimins L A. Amplification of human papillomavirus genomes is dependent on epithelial differentiation. J Virol. 1991;63:1247–1255. doi: 10.1128/jvi.65.5.2254-2260.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow L T, Nasseri M, Wolinsky S M, Broker T R. Human papillomavirus types 6 and 11 mRNAs from genital condylomata acuminata. J Virol. 1987;61:2581–2588. doi: 10.1128/jvi.61.8.2581-2588.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colgan D F, Manley J L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 8.Flores E R, Allen-Hoffmann B L, Lee D, Sattler C A, Lambert P F. Establishment of the human papillomavirus type 16 (HPV-16) life cycle in an immortalized human foreskin keratinocyte cell line. Virology. 1999;262:344–354. doi: 10.1006/viro.1999.9868. [DOI] [PubMed] [Google Scholar]
- 9.Frattini M G, Laimins L A. The role of the E1 and E2 proteins in the replication of human papillomavirus type 31b. Virology. 1994;204:799–804. doi: 10.1006/viro.1994.1596. [DOI] [PubMed] [Google Scholar]
- 10.Frattini M G, Lim H B, Doorbar J, Laimins L A. Induction of human papillomavirus type 18 late gene expression and genomic amplification in organotypic cultures from transfected DNA templates. J Virol. 1997;71:7068–7072. doi: 10.1128/jvi.71.9.7068-7072.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frattini M G, Lim H B, Laimins L A. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc Natl Acad Sci USA. 1996;93:3062–3067. doi: 10.1073/pnas.93.7.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furth P A, Baker C C. An element in the bovine papillomavirus late 3′ untranslated region reduces polyadenylated cytoplasmic RNA levels. J Virol. 1991;65:5806–5812. doi: 10.1128/jvi.65.11.5806-5812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furth P A, Choe W-T, Rex J H, Byrne J C, Baker C C. Sequences homologous to 5′ splice sites are required for the inhibitory activity of papillomavirus late 3′ untranslated regions. Mol Cell Biol. 1994;14:5278–5289. doi: 10.1128/mcb.14.8.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldsborough M D, DiSilvestre D, Temple G F, Lorincz A T. Nucleotide sequence of human papillomavirus type 31: a cervical neoplasia-associated virus. Virology. 1989;171:306–311. doi: 10.1016/0042-6822(89)90545-x. [DOI] [PubMed] [Google Scholar]
- 15.Gunderson S I, Polycarpou-Schwarz M, Mattaj I W. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- 16.Halbert C L, Demers G W, Galloway D A. The E6 and E7 genes of human papillomavirus type 6 have weak immortalizing activity in human epithelial cells. J Virol. 1992;66:2125–2134. doi: 10.1128/jvi.66.4.2125-2134.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higgins G D, Uzelin D M, Phillips G E, McEvoy P, Burrel C J. Transcription patterns of human papillomavirus type 16 in genital intraepithelial neoplasia: evidence for promoter usage within the E7 open reading frame during epithelial differentiation. J Gen Virol. 1992;73:2047–2057. doi: 10.1099/0022-1317-73-8-2047. [DOI] [PubMed] [Google Scholar]
- 18.Hirt B. Selective extraction of polyoma DNA from infected mouse cell culture. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 19.Howley P M. Papillomavirinae and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fundamental virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 947–978. [Google Scholar]
- 20.Hubert W G, Kanaya T, Laimins L A. DNA replication of human papillomavirus type 31 is modulated by elements of the upstream regulatory region that lie 5′ of the minimal origin. J Virol. 1999;73:1835–1845. doi: 10.1128/jvi.73.3.1835-1845.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hummel M, Hudson J B, Laimins L A. Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. J Virol. 1992;66:6070–6080. doi: 10.1128/jvi.66.10.6070-6080.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hummel M, Lim H B, Laimins L A. Human papillomavirus type 31b late gene expression is regulated through protein kinase C-mediated changes in RNA processing. J Virol. 1995;69:3381–3388. doi: 10.1128/jvi.69.6.3381-3388.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeon S, Lambert P F. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: Implications for cervical carcinogenesis. Proc Natl Acad Sci USA. 1995;92:1654–1658. doi: 10.1073/pnas.92.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennedy I M, Haddow J K, Clements J B. Analysis of human papillomavirus type 16 late mRNA 3′ processing signals in vitro and in vivo. J Virol. 1990;64:1825–1829. doi: 10.1128/jvi.64.4.1825-1829.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kennedy I M, Haddow J K, Clements J B. A negative regulatory element in the human papillomavirus type 16 genome acts at the level of late mRNA stability. J Virol. 1991;65:2093–2097. doi: 10.1128/jvi.65.4.2093-2097.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klumpp D J, Laimins L A. Differentiation-induced changes in promoter usage for transcripts encoding the human papillomavirus type 31 replication protein E1. Virology. 1999;257:239–246. doi: 10.1006/viro.1999.9636. [DOI] [PubMed] [Google Scholar]
- 27.Klumpp D J, Stubenrauch F, Laimins L A. Differential effects of the splice acceptor at nucleotide3295. of human papillomavirus type 31 on stable and transient viral replication. J Virol. 1997;71:8186–8194. doi: 10.1128/jvi.71.11.8186-8194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koffa M D, Graham S V, Takagaki Y, Manley J L, Clements J B. The human papillomavirus type 16 negative regulatory RNA element interacts with three proteins that act at different posttranscription levels. Proc Natl Acad Sci USA. 2000;97:4677–4682. doi: 10.1073/pnas.070049097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koutsky L. Epidemiology of genital human papillomavirus infection. Am J Med. 1997;102:3–8. doi: 10.1016/s0002-9343(97)00177-0. [DOI] [PubMed] [Google Scholar]
- 30.Laimins L A. The biology of human papillomaviruses: from warts to cancer. Infect Agents Dis. 1993;2:74–86. [PubMed] [Google Scholar]
- 31.Meyers C, Fratini M, Hudson J, Laimins L A. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science. 1992;257:971–973. doi: 10.1126/science.1323879. [DOI] [PubMed] [Google Scholar]
- 32.Meyers C, Mayer T J, Ozbun M A. Synthesis of infectious human papillomavirus type 18 in differentiating epithelium transfected with viral DNA. J Virol. 1997;71:7381–7386. doi: 10.1128/jvi.71.10.7381-7386.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osheim Y N, Proudfoot N J, Beyer A L. EM visualization of transcription by RNA polymerase II: downstream termination requires apoly(A)) signal but not transcript cleavage. Mol Cell. 1999;3:379–387. doi: 10.1016/s1097-2765(00)80465-7. [DOI] [PubMed] [Google Scholar]
- 34.Ozbun M A, Meyers C. Characterization of late gene transcripts expressed during vegetative replication of human papillomavirus type 31b. J Virol. 1997;71:5161–5172. doi: 10.1128/jvi.71.7.5161-5172.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozbun M A, Meyers C. Temporal usage of multiple promoters during the life cycle of human papillomavirus type 31b. J Virol. 1998;72:2715–2722. doi: 10.1128/jvi.72.4.2715-2722.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozbun M A, Meyers C. Two novel promoters in the upstream regulatory region of human papillomavirus type 31b are negatively regulated by epithelial differentiation. J Virol. 1999;73:3505–3510. doi: 10.1128/jvi.73.4.3505-3510.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Proudfoot N. Connecting transcription to messenger RNA processing. Trends Biochem Sci. 2000;25:290–293. doi: 10.1016/s0968-0004(00)01591-7. [DOI] [PubMed] [Google Scholar]
- 38.Rheinwald J G, Beckett M A. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultures from human squamous cell carcinomas. Cancer Res. 1981;41:1657–1663. [PubMed] [Google Scholar]
- 39.Rohlfs M, Winkenback S, Meyer S, Rupp T, Durst M. Viral Transcription in human keratinocyte cell lines immortalized by human papillomavirus type-16. Virology. 1991;183:331–342. doi: 10.1016/0042-6822(91)90146-3. [DOI] [PubMed] [Google Scholar]
- 40.Ruesch M N, Laimins L A. Human papillomavirus oncoproteins alter differentiation-dependent cell cycle exit on suspension in semisolid medium. Virology. 1998;250:19–29. doi: 10.1006/viro.1998.9359. [DOI] [PubMed] [Google Scholar]
- 41.Ruesch M N, Stubenrauch F, Laimins L A. Activation of papillomavirus late gene transcription and genome amplification upon differentiation in semisolid medium is coincident with expression of involucrin and transglutaminase but not keratin-10. J Virol. 1998;72:5016–5024. doi: 10.1128/jvi.72.6.5016-5024.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sokolowski M, Tan W, Jellne M, Schwartz S. mRNA instability elements in the human papillomavirus type 16 L2 coding region. J Virol. 1998;72:1504–1515. doi: 10.1128/jvi.72.2.1504-1515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stubenrauch F, Colbert A M, Laimins L A. Transactivation by the E2 protein of oncogenic human papillomavirus type 31 is not essential for early and late viral functions. J Virol. 1998;72:8115–8123. doi: 10.1128/jvi.72.10.8115-8123.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stubenrauch F, Hummel M, Ifner T, Laimins L. The E8∧E2C protein, a negative regulator of viral transcription and replication, is required for extrachromosomal maintenance of human papillomavirus type 31 in keratinocytes. J Virol. 2000;74:1178–1186. doi: 10.1128/jvi.74.3.1178-1186.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stubenrauch F, Lim H B, Laimins L A. Differential requirements for conserved E2 binding sites in the life cycle of oncogenic human papillomavirus type 31. J Virol. 1998;72:1071–1077. doi: 10.1128/jvi.72.2.1071-1077.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terhune S S, Milcarek C, Laimins L A. Regulation of human papillomavirus type 31 polyadenylation during the differentiation-dependent life cycle. J Virol. 1999;73:7185–7912. doi: 10.1128/jvi.73.9.7185-7192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas J T, Hubert W G, Ruesch M N, Laimins L A. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc Natl Acad Sci USA. 1999;96:8449–8454. doi: 10.1073/pnas.96.15.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y-C, Okayama H, Howley P M. Bovine papillomavirus contains multiple transforming genes. Proc Natl Acad Sci USA. 1985;82:1030–1034. doi: 10.1073/pnas.82.4.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]