Abstract
The objective of this systematic review (SR) was to select studies on the use of gene editing by CRISPR technology related to plant resistance to biotic stresses. We sought to evaluate articles deposited in six electronic databases, using pre-defined inclusion and exclusion criteria. This SR demonstrates that countries such as China and the United States of America stand out in studies with CRISPR/Cas. Among the most studied crops are rice, tomatoes and the model plant Arabidopsis thaliana. The most cited biotic agents include the genera, Xanthomonas, Manaporthe, Pseudomonas and Phytophthora. This SR also identifies several CRISPR/Cas-edited genes and demonstrates that plant responses to stressors are mediated by many complex signaling pathways. The Cas9 enzyme is used in most articles and Cas12 and 13 are used as additional editing tools. Furthermore, the quality of the articles included in this SR was validated by a risk of bias analysis. The information collected in this SR helps to understand the state of the art of CRISPR/Cas aimed at improving resistance to diseases and pests to understand the mechanisms involved in most host–pathogen relationships. This SR shows that the CRISPR/Cas system provides a straightforward method for rapid gene targeting, providing useful information for plant breeding programs.
Keywords: biotic stress, CRISPR/Cas, plant diseases, phytopathogens, pests, plant genetic improvement
1. Introduction
Biotic stresses caused by pests and pathogens such as viruses, bacteria, fungi, oomycetes, nematodes, and insects are largely responsible for low productivity in various crops [1]. In addition, the continuous increase in several new pest species makes the control of these pathogens challenging [2]. Microorganisms have specific characteristics and are classified into groups. Biotrophic microorganisms depend on the living plant to feed and complete their life cycle; necrotrophs, during their feeding habit, kill the host plant, and hemibiotrophs initially depend on the living plant (behaving like biotrophs) in order to survive and complete their cycle with a necrotrophic phase; where the host is degraded [3,4].
The plant and the pathogen are intertwined in a battle of recognition and evasion where a multilayered defense system, including pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI), has evolved in plants to fight invading pathogens for survival [5]. In general, PTI uses pattern recognition receptors to monitor PAMPs on the cell surface. Meanwhile, ETI relies on leucine-rich repeat receptors with a nucleotide-binding domain to recognize pathogen effectors inside the cell [6,7,8].
Thus, understanding the molecular mechanisms of pathogen–host interactions, especially the identification of key targets related to defense responses in plants, would offer a great opportunity to design broad-spectrum and durable resistance in various crops [5,9,10]. Hence, plant breeding programs are looking for effective and long-lasting techniques to improve crops. However, some challenges, such as the complex inheritance of the vast majority of agronomic traits and the strong genotype–environment interaction, are still challenging [11].
Currently, three types of genome editing tools are widely used by researchers, including zinc finger nuclease (ZFN) [12], transcription activator-like effector nuclease (TALEN) [13], and CRISPR-clustered/associated regularly interspaced short palindromic repeats (CRISPR/Cas) [14]. ZFN and TALEN have not been widely used due to high costs and failures. The CRISPR/Cas system (which includes Cas9, Cas12, and Cas13) from a prokaryotic organism has transformed the field of gene editing with high efficiency and easy handling and application. Compared to the previous two generations of genome editing techniques, the CRISPR/Cas system is flexible, simple, stable, and easy to transform. These resources allowed for ZFN and TALEN to be replaced by CRISPR/Cas, which has become one of the main genome editing techniques.
CRISPR is composed of CRISPR RNA (crRNA) (transcribed from the spacer sequences) and transactivating crRNA, or single chimeric guide RNA (sgRNA) (formed by the fusion of crRNA and tracrRNA) for targeting and the specificity of targeting [14,15]. The Cas9 protein-RNA complex (from Streptococcus pyogenes) is formed by the combinations of the crRNA spacer to a target sequence close to an adjacent motif of the proto-spacer (PAM—3 base pair (bp) motifs essential for spacer acquisition and target cleavage) [15,16,17].
Due to its ease of execution, the CRISPR/Cas system has become the tool of choice for gene editing in any species of interest. By generating a double-strand break (DSB) at the desired site by the Cas-gRNA complex, the host–cell repairs the DNA lesion via the non-homologous end joining (NHEJ) pathway, resulting in short insertions or deletions, consequently leading to gene knockouts. Another form of repair is the homology-directed repair (HDR) pathway, which is more precise and has a lower probability of error [18,19]. In plants, the system has been used to knock out all members or a single member of a multigenic family [20] and even several unrelated genes [21], with the NHEJ pathway being the most reported [22].
Several studies have been published to demonstrate the different genes that positively or negatively regulate resistance to various pests and pathogens in model plants and diverse crops, such as Arabidopsis thaliana, where genes such as ZAR1, UGT71C3, and miR398b have been studied [23,24,25], in rice, SWEET14, eIF4G, and PRAF2 [26,27,28], in maize, ZmACD6, Zmksl2, and JAZ15 [29,30,31], in tomato, SlWRKY16, SlWAT1, and SlDMR6 [32,33,34], and in soybean, Rfg1, Rpp1, and GmLMM1 [35,36,37].
In addition, CRISPR technology has evolved rapidly and has shown great potential for plant biology, especially with regard to CRISPR/Cas9 variants, such as CRISPR/Cas12 and CRISPR/Cas13, which offer better specificity for DNA and RNA, respectively. For precise changes in a single base, without causing double-strand breaks, reducing off-target mutations, base editing methods have been implemented [38,39]. Other improved delivery tools, such as the use of nanoparticles and viral vectors, allow for the efficient introduction of the CRISPR system into plant cells and the delivery of RNP (ribonucleoprotein) complexes, rather than DNA plasmids, is being used to improve efficiency and reduce off-target effects [40]. Other advanced techniques allow for the simultaneous editing of multiple sites in the genome, making it easier to modify multiple traits at the same time, and systems such as Prime Editing and CRISPR 3.0 are emerging, allowing for precise insertions and deletions without the need for DNA breaks [38,39,41].
Thus, off-target effects have been significantly reduced due to improvements in the specificity of the CRISPR/Cas system. In addition, these data facilitate reliability and safety, allowing for regulatory approval, coupled with strategies such as temporary editing, where CRISPR machinery is rapidly degraded after editing, as well aa the ability to edit multiple genes simultaneously, which facilitates the engineering of complex traits in plants, such as disease resistance and nutritional trait improvements [39,40,41,42].
In order to systematically gather and review current research on the use of CRISPR/Cas technology in gene editing for biotic stress tolerance, this study presents a systematic review (SR) of articles published in the last twelve years. It also aims to contribute to the SR previously carried out on the use of CRISPR/Cas technology in gene editing for tolerance to abiotic stresses [22]. Here, we describe how the technique has been applied to pest and pathogen resistance studies and the locations and crops, among other data, for which it is possible to detect the current research trend on the subject and its impact on crops.
2. Materials and Methods
To carry out this SR, the State of the Art through Systematic Review (StArt) software (version 3.0.3 Beta) was used, developed, and made available by the Software Engineering Research Laboratory of the Federal University of São Carlos, at https://www.lapes.ufscar.br/resources/tools-1/start-1, accessed on 15 August 2023.
The review was prepared following the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines [43], structured in a set of evidence-based items that help authors report a wide range of systematic reviews and meta-analyses and can be used in plant, animal, and health intervention areas. A PRISMA checklist was drawn up to minimize bias in this SR, available at https://doi.org/10.5281/zenodo.13869284 (accessed on 29 September 2024). The SR process using StArt occurred in three stages: planning, execution, and summarization.
2.1. Planning
In order to plan the SR, a protocol was developed, available at https://doi.org/10.5281/zenodo.13371943 (accessed on 26 August 2024), which includes a description of the SR, the research objectives, the main/guiding question, the research questions (Table 1), the search string, the source mechanism, the inclusion and exclusion criteria, and the definition of the types of study. The question guiding the SR was based on the Population Intervention Comparison Results strategy [44] (Table 1). Thus, this SR aims to answer the following research question: how has CRISPR/Cas technology been used in gene editing in plants for biotic stress tolerance over the last twelve years?
Table 1.
Description of the PICOS strategy used to develop the RS research questions on the use of CRISPR/Cas technology to edit tolerance genes/resistance to biotic stresses from studies published in the last 12 years.
| Description | Abbreviation | Components of the Question |
|---|---|---|
| Population | P | Agricultural varieties under biotic stresses |
| Interest/Intervention | I | Gene editing in plants using CRISPR/Cas technology for disease resistance |
| Comparison | C | Plant breeding methods |
| Outcome | O | Editing genes that confer resistance to biotic stresses in plants |
| Study type | S | Scientific articles and literature reviews |
After drafting the main research questions, secondary questions were elaborated (Table 2).
Table 2.
Guiding questions for this SR on the use of CRISPR/Cas technology to edit tolerance genes/resistance to biotic stresses from studies published in the last 12 years.
| Research Questions |
|---|
| 1. In which country was the study performed? |
| 2. What culture is the article about? |
| 3. Which biotic agent is addressed in the study? |
| 4. Which genes are reportedly associated with disease and pest resistance in plants? |
| 5. Which nuclease is used in conjunction with the CRISPR tool? |
| 6. What methodology is used to use CRISPR? |
| 7. What method is used to prove the effectiveness of the tool? |
| 8. What techniques/tools are associated with CRISPR/Cas9? |
| 9. What transformation method was used? |
| 10. Which explant was used to transform the plants? |
| 11. What are the main vectors used to express Cas9 and/or gRNA in plants? |
| 12. Were any unusual phenotypic characteristics observed in the plants after genetic transformation? Which ones? |
| 13. What is the characteristic obtained after mutating the plant? |
2.2. Execution
Searches were performed in different electronic databases such as Pub Med Central, Springer, Scopus, Web of Science and sites such as Google Scholar and CAPES Periodicals Portal. For the Google Scholar, Springer, PubMed Central, CAPES Periodicals, Web of Science and Scopus databases, the following keywords were used: (“CRISPR/Cas9” OR “CRISPR-Cas9” OR “CRISPR-Cas in plants”) AND (“plant resistance” OR “plant disease resistance”) AND (“plant disease” OR “biotic factors” OR “disease resistance” OR “plant pathogens” OR “pests” OR “plant parasite”).
For the Web of Science and Scopus databases, another search string was also used, with the following keywords: (“CRISPR” OR “CRISPR/Cas9” OR “CRISPR-Cas9” OR “CRISPR-Cas in plants”) AND (“biotic factors” OR “pathogen resistance” OR “phytopathogen resistance” OR “plant disease resistance” OR “disease resistance” OR “plant resistance” OR “pest resistance” OR “parasite resistance”), seeking to include as many studies as possible.
The Boolean connectives “AND” and “OR” were used to differentiate search terms and group synonymous terms, respectively. The search results in each database were imported into the BIBTEX, MEDLINE, or RIS formats, compatible with the StArt software. The bibliographic survey was performed from January 2013 to July 2024.
To select the articles, the title, abstract, and keywords were analyzed. Articles that met the terms of the search sequence and did not deviate from the proposed theme were accepted and submitted to the extraction stage. At this stage, only articles that answered the research questions (Table 2) previously established in the SR protocol were accepted as an inclusion criterion. Exclusion criteria were also used to extract the following articles: theses, dissertations, manuals, book chapters, review articles, papers not written in English, papers without a clear contribution, papers published prior to 2013, or papers that were off-topic.
2.3. Data Summarization
The data obtained from the scientific articles was summarized in tables, graphs, word clouds, and bibliometric maps. The graphs were constructed using the R version 4.4.1 statistical environment [45], using the ggplot2, reshape2, and ggpubr packages. The bibliometric analyses were performed according to the metadata of the selected articles using the VOSviewer_1.6.17 program [46] to verify the networks of interactions between keywords and between authors and co-citations. Word clouds containing the journals used to publish the articles, genes edited, tools, and software used to support the CRISPR/Cas tool over the last twelve years were generated online and free of charge (https://www.wordclouds.com/, accessed on 18 November 2023), based on the frequency of the data.
2.4. Risk of Bias Analysis
To assess the risk of bias, the adapted Cochrane Collaboration Tool [47] was used. The methodological quality was analyzed by three authors (MSM, FdSN, and AdJR), and the articles selected in the extraction stage were subjected to four questions (Table 3) in order to further reduce data bias. These are essential questions that confirm whether editing using CRISPR/Cas was effective, reaching the target site or not.
Table 3.
Questions to evaluate the methodological quality of the articles included in the SR on the use of CRISPR/Cas technology to edit tolerance genes/resistance to biotic stresses from studies published in the last 12 years.
| Risk of Bias |
|---|
| 1. Has off-target analysis been performed? |
| 2. Has the pathogen been inoculated? |
| 3. Has phenotypic analysis been performed after mutation in the plant? |
| 4. Does the article answer at least 50% of the research questions? |
Systematic errors in scientific studies that cause distortions in the results can happen; it is complex to state whether a study is biased or not, but systematic errors in scientific studies can be estimated and minimized through a careful evaluation of its methodological quality. Rigorous practices such as protocol development, the use of PICOS strategy, PRISMA checklist, and the others described above, significantly reduce the risks of bias.
The risk of bias can be classified as low, moderate, or high when the study presents negative responses (“no”) of up to 25%, between 25 and 75%, and greater than 75%, respectively.
3. Results
3.1. Bibliographical Survey
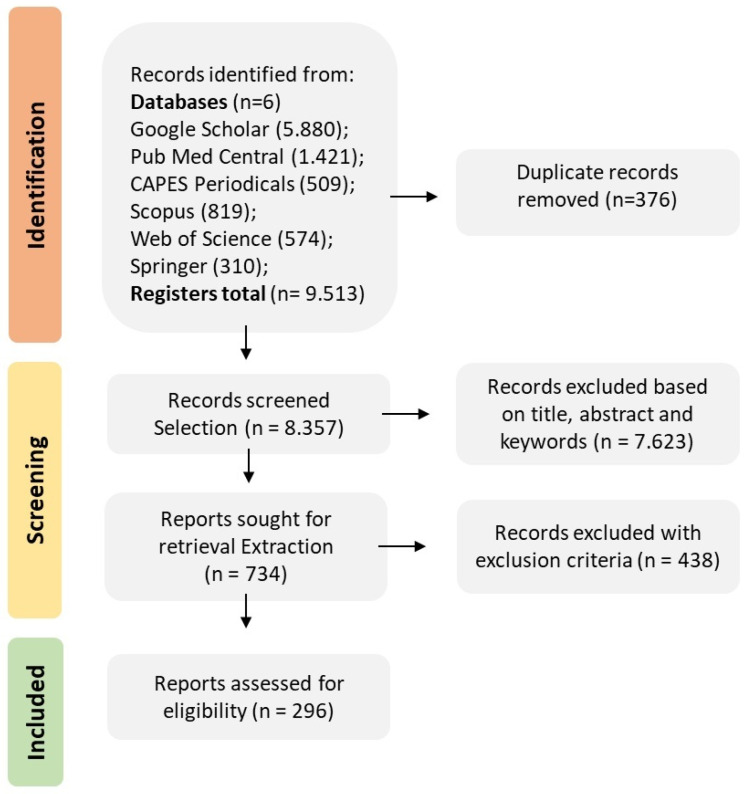
Initially, 9513 studies related to the proposed topic were identified from the search strings in the selected electronic databases, which are widely used in plants. Google Scholar showed 5880 studies, Pub Med Central showed 1421, CAPES Periodicals Portal showed 509, Scopus showed 819, Web of Science showed 574, and Springer, 310. Although the Web of Science and Scopus databases use two search strings, Google Scholar contributed 61.8% of the articles submitted, which is justified by its broad search spectrum. From this total, 376 were detected as duplicates by the StArt software.
After analyzing the title, abstract, and keywords, 7623 studies were rejected and 734 were submitted to the extraction stage. The texts were read in full, resulting in 296 accepted articles (Figure 1). These selected articles met the inclusion/exclusion criteria because they are related to the theme of this SR, which aimed to include as many studies as possible on the use of CRISPR/Cas technology in the editing of tolerance/resistance genes to biotic stresses in the last 12 years; then, the information was deposited in the Supplementary Materials (Table S1) for consultation.
Figure 1.
A PRISMA flow diagram with the respective stages of the process of selecting studies for inclusion/exclusion in the systematic review of the CRISPR/Cas technology used to edit genes for tolerance/resistance to biotic stress in plants according to the databases [43].
The studies evaluated covered the period from January 2013 to July 2024, with 2021 considered the year with the highest number of publications as to CRISPR/Cas technology in the editing of genes related to resistance to biotic stressors, contributing 21.6% of the articles. The other years had 16.6% (2022), 16.2% (2020), 11.5% (2023), 10.5 (2024), 9.8% (2019), 7.4% (2018), 3.7% (2017); for the years 2016, 2015, 2014 and 2013, less than 2% were obtained.
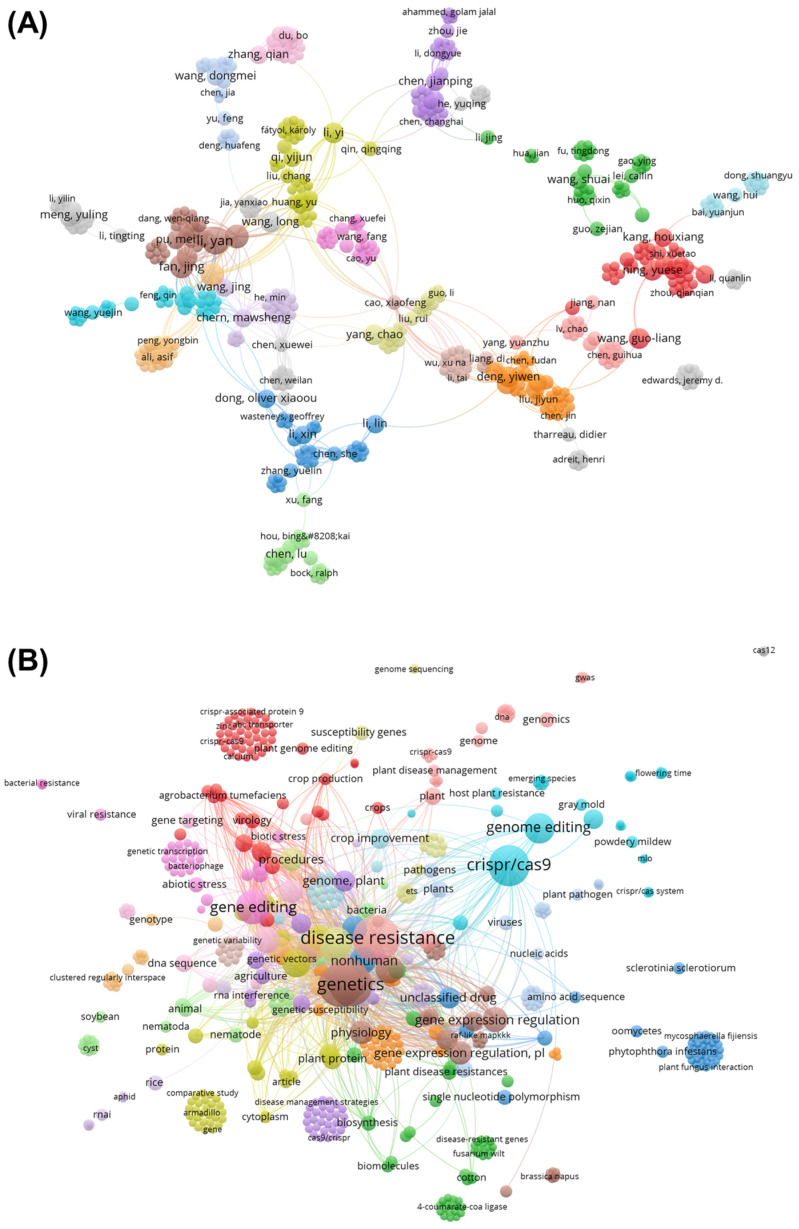
Considering the frequency of authors and all the keywords in the articles selected in the extraction phase, bibliometric maps were developed to represent the co-occurrence of these words (Figure 2). The size of the circles represents the number of times these words were repeated; the larger the circle, the more times the author and journal were cited. Colors indicate different groups of authors and keywords and the thickness of the lines the correlation between these words. The thicker the line, the higher the occurrence of the term.
Figure 2.
Bibliometric indicators of the collaboration network between authors and keywords of the selected articles on CRISPR/Cas technology and biotic factors. (A) Collaborators who have published the most on CRISPR/Cas and biotic stresses in the last 12 years. (B) Keywords of the selected articles on CRISPR/Cas technology used for gene editing of tolerance/resistance to biotic stresses in plants during the extraction phase of this systematic review. Different colors for each circle indicate collaboration between groups.
Twenty clusters were formed and identified using different colors, according to the degree of similarity between the authors’ works (Figure 2A). Authors such as Yan Li, Jing Fan, Long Wang, Yuese Ning, Yi Li, Chao Yang, Liang Guo Wang, Qian Zhang, Jianping Chen, and Jing Wang are responsible for a large bibliographic contribution. These data demonstrate a trend in centralized research related to Chinese authors with not much exchange of information between Chinese researchers and those from the rest of the world. The links or distances infer the correlation between these authors and their collaboration on other works. Some small grouped but isolated nodes can be observed, but they show minimal contribution to the studies performed by the authors included in these groups.
For the keywords, approximately 92 nodes and 12 clusters were observed, which defined the main research themes in this area. The most relevant groups according to the size of each circle refer, in order of relevance, to the following words: disease resistance, genetics, CRISPR/Cas9, gene editing, and genome editing. These words form core groups associated with several other terms of collaboration with a theme that constitutes the smaller groups formed, for example, by the terms vectors, crop breeding and regulation of gene expression. Words such as Cas12, RNAi, bacterial resistance, and genomic sequencing appear in isolation, which indicates lower frequency and low correlation with other studies (Figure 2B).
3.2. Plant of Origin and Plant Cultures Edited Using CRISPR/Cas Technology
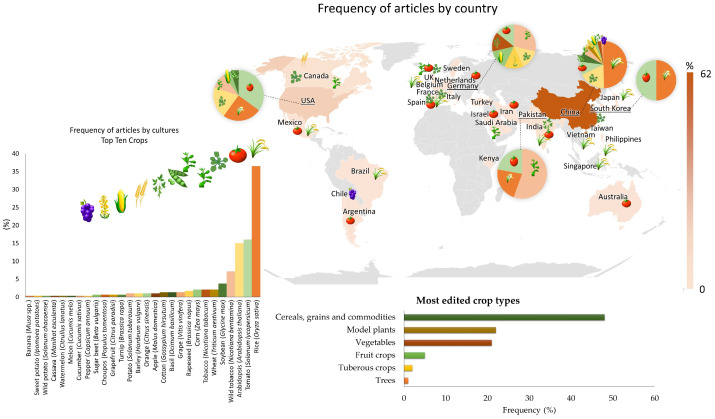
Of the 296 research papers, 158 (59.8%) originated in China, 30 (11.4%) in the United States of America (USA), 12 (4.5%) in Germany, 7 (2.7%) in South Korea, 5 (1.9%) in Canada and Pakistan, 4 (1.5%) in Spain and Saudi Arabia, 3 (1.1%) in India, Israel, Japan, and the Netherlands, and 2 (0.8%) in Australia, the Philippines, Sweden and the United Kingdom. The other countries only had one article submitted, which represents just 0.4% of the publications on the subject (Figure 3).
Figure 3.
Frequency of articles according to country of publication and crop edited by CRISPR/Cas technology for plant disease tolerance/resistance. More than one plant species per article was considered in calculating the frequency.
China and the USA are the countries that produce and disseminate the most scientific knowledge on the subject. However, all continents, except for Antarctica, have contributed to this area of research. The articles identified 28 plant species used for gene editing related to resistance to biotic factors. Overall, the types of crops most edited by the CRISPR technique include cereals, grains and agricultural commodities, with 48% of the studies represented mainly by rice, followed by studies with model plants, represented mainly by Arabidopsis. Other studies have included vegetables (21%), fruits (5%), tubers (2%) and trees (1%) (Figure 3).
Rice (Oryza sativa) was the most studied crop, present in 36.5% (109) of the studies, followed by tomato (Solanum lycopersicum) with 16% (48), Arabidopsis thaliana with 15% (45), wild tobacco (Nicotiana benthamiana) with 7.2% (22), soybean (Glycine max) with 3.8% (12), wheat (Triticum aestivum), tobacco (Nicotiana tabacum), and corn (Zea mays), with 2% (6), rapeseed (Brassica napus) with 1.7% (5), and grape (Vitis vinifera L.), basil (Ocimum basilicum), and cotton (Gossypium hirsutum) with 1.4% (4). The other species had a frequency of less than 1% of the studies (Figure 3).
3.3. Biotic Stresses in Plants
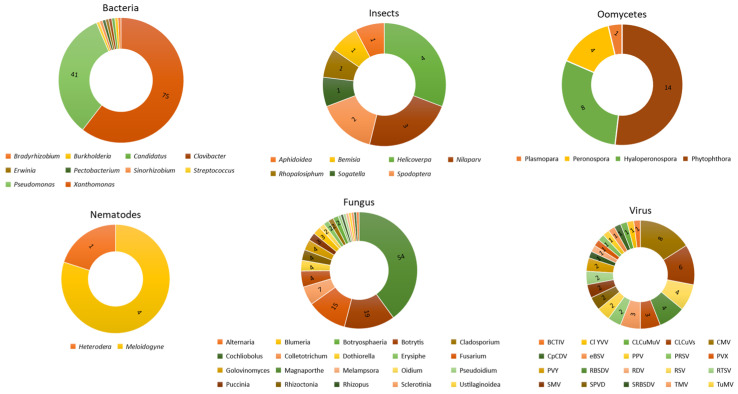
The biotic agents cited in the literature were bacteria, fungi, viruses, oomycetes, insects, and nematodes, accounting for 51.8, 28.3, 10.5, 5.7, 2.7, and 1%, respectively. The genera Xanthomonas (75) and Pseudomonas (41) account for 92% of the studies on bacteria. For fungi, the most studied genera were Magnaporthe (54), Botrytes (19), Fusarium (15), Sclerotinia (7), and Verticillium (6) (Figure 4).
Figure 4.
The most-studied biotic agents (bacteria, insects, oomycetes, nematodes, fungi, and viruses) in the last twelve years for resistance/tolerance to plant diseases using CRISPR/Cas technology. More than one biotic agent per article was considered in calculating frequency.
The most-cited viruses were cucumber mosaic virus (CMV) (8), cotton leaf curl virus (CLCuVs) (6), rice streak virus (RSV), rice black-streaked dwarf virus (RBSDV) (4), tomato yellow leaf curl virus (TYLCV) (3), and tobacco mosaic virus (TMV) (3). The oomycete Phytophthora (14), followed by Hyaloperonospora (8) and Peronospora (4), were the most covered. The insect genera Helicoverpa (4), Nilaparvata (3), Spodoptera (2), Sogatella (1), Rhopalosiphum (1), Bemisia (1), and Aphidoidea (1), and the nematode genera Meloidogyne (4) and Heterodera (1), were also observed in the studies (Figure 4).
As a result, the diseases most frequently covered were bacterial leaf blight (BLB), bacterial leaf streak (BLS) in rice, and bacterial spot in tomatoes. For fungi, brusone in rice, gray mold in tomatoes, and Fusarium wilt in various crops, among other diseases, were the most commonly observed (Figure 4).
3.4. Types of Explants
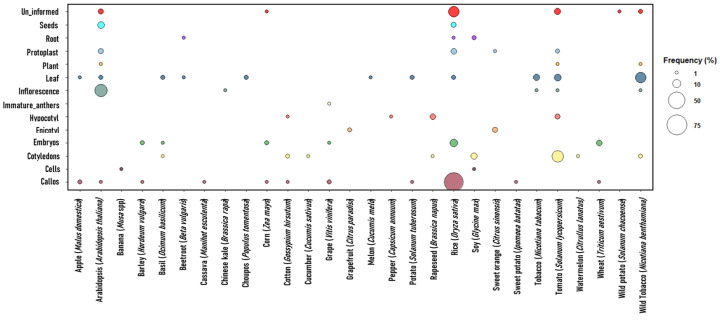
The explants used for plant transformation via CRISPR/Cas varied according to the plant species. Callus, cells, cotyledons, embryos, epicotyl, hypocotyl, anthers, inflorescence, leaf disks/leaves, plants, protoplasts, roots, and seeds were found as transforming materials (Figure 5).
Figure 5.
Explants used for the transformation of the different plant species covered in studies on gene editing via CRISPR/Cas for tolerance/resistance to biotic stress in the last 12 years. The colors of the circles represent each explant and the size of the circumference the frequency of each explant in different crops.
For the rice crop, transformation via CRISPR/Cas was mainly performed using embryogenic calli, with a frequency of over 50%. Embryos, protoplasts, seeds, leaves, and roots were also used as transforming sources, but with a frequency of less than 10%. In tomatoes, the most commonly used explants were cotyledons, with a frequency of more than 10% of the studies performed on this crop, followed by leaves (<10%). In Arabidopsis, the inflorescence (>10%), seeds, protoplasts, leaves, and plant (<10%) were used as explants. In wild tobacco, the leaves (>10%), inflorescence, plant, and cotyledons (<10%) were used as the main transformation explants (Figure 5).
3.5. Plant Disease Resistance/Susceptibility Genes
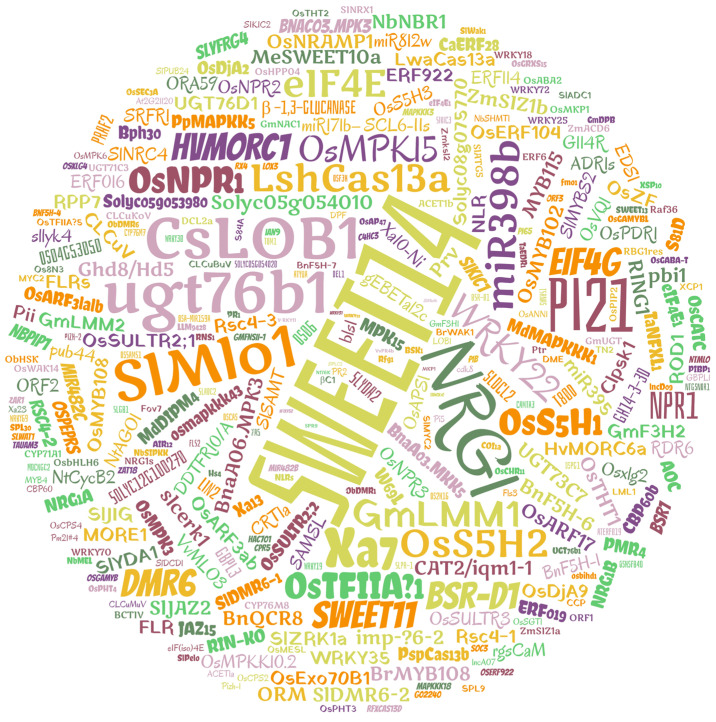
A word cloud designed from the genes cited in the papers as potential targets for resistance/susceptibility to plant diseases. The sucrose efflux transporter gene (SWEET14) appears prominently in the center of the word cloud and was the most cited in the papers, especially those related to resistance to bacteria Xoo in rice, followed by N Requirement Gene 1 (NRG1), Pi21 resistance genes, LateraL Organ Boundaries 1 (CsLOB1), Mildew resistance locus o 1 (SlMlo1), Dependent Glycosyl Transferases (UGT76b1), and the Xa7 resistance gene. The gene families WRKY (14), SWEET/OsSWEET (12), UGT (7), Xa (7), and Solyc (5) have also been extensively studied. In the articles selected for this SR, 337 genes related to tolerance/resistance to biotic factors were covered; some papers used CRISPR/Cas technology to edit more than one gene (Figure 6).
Figure 6.
Word cloud of CRISPR/Cas edited genes in different plant species related to tolerance/resistance/susceptibility to biotic stresses.
3.6. Auxiliary Methods to CRISPR/Cas
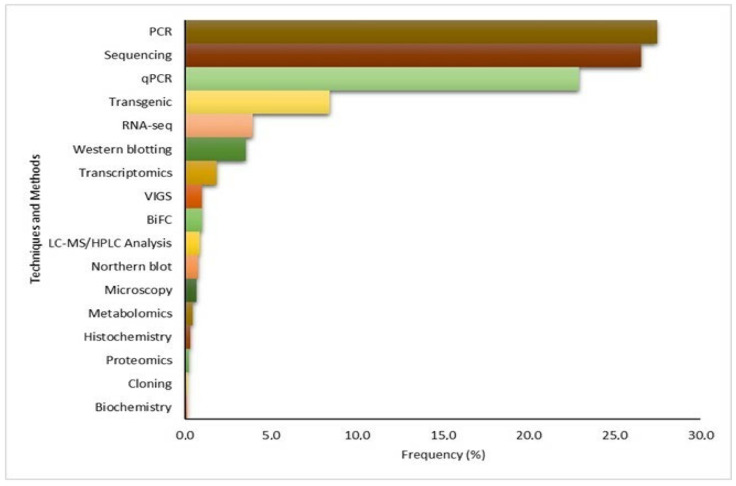
The methodological strategies most used in the studies collected to validate and support the CRISPR/Cas tool were PCR (27.4%), sequencing (26.5%), qPCR (22.9%), transgenics (8.4%), RNA-seq (3.9%), Western blotting (3.5%), transcriptomics (1.8%), virus-induced gene silencing (VIGS) (1.0%), bimolecular fluorescence complementation (BiFC) assay (1.0%), LC-MS/HPLC liquid chromatography analysis (0.9%), Northern blot (0.7%), microscopy (0.6%), metabolomics (0.4%), histochemistry (0.3%), and proteomics (0.2%) (Figure 7). The other methods accounted for less than 0.1% of the studies.
Figure 7.
Auxiliary tools and analyses used with the CRISPR/Cas technique for validation and comparison between knockout with control and/or the overexpression of mutants identified in articles on tolerance/resistance to biotic stresses in the last 12 years.
PCR, sequencing, and qPCR techniques were mainly used to demonstrate the efficacy of the CRISPR/Cas tool and detect on- and off-target mutations.
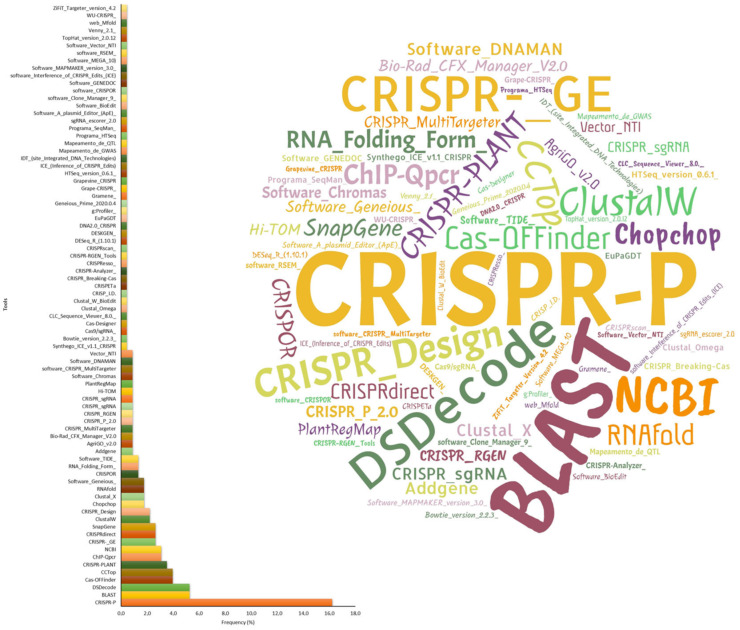
Certain types of software were also used to complement CRISPR/Cas-related analyses. The CRISPR-P version 2.0 software appears in 16.2% of the articles as an auxiliary method to CRISPR/Cas to predict target sites and/or mutations. Other widely used software/programs included BLAST, DSDecode, Cas-OFFinder, CCTop, CRISPR-PLANT, NCBI, CRISPR-GE, CRISPRdirect, SnapGene, ClustalW, CRISPR Design, CHOPCHOP, ClustalX, RNAfold, Geneious, CRISPOR, RNA Folding Form, and TIDE (Figure 8).
Figure 8.
Frequency and word cloud of tools and software that help the CRISPR/Cas technique to search for specific target sites identified in studies on tolerance/resistance to biotic stresses in the last 12 years.
3.7. Use of CRISPR/Cas Technology
Most of the CRISPR/Cas methods used in the 296 studies selected for this SR had already been validated by other authors. The method used by Ma et al. (2015) [48], Xing et al. (2014) [49] and Wang et al. (2015) [50], and showed great reproducibility, being used in 24.7% of the studies (Table S2) to precisely edit plant genomes, deleting regions responsible for unwanted characteristics or inserting gain-of-function mutations.
For the CRISPR tool to be effective as Cas, endonuclease must be used. Of the studies collected, 98.3% (291) used Cas9 as an accessory to this editing system. Other endonucleases such as Cpf1, formerly known as Cas12a (2) and Cas13 (3), were also mentioned, but they were not very common.
Several vectors have been used to express Cas and/or single guide RNA (gRNA), but the most commonly cited is pCAMBIA and pYLCRISPR/Cas. The most widely used delivery method for introducing the gene of interest into plant cells was carried out by Agrobacterium tumefaciens (286) and Agrobacterium rhizogenes (6), occurring mainly via electroporation and heat shock.
3.8. Phenotypic Analysis and Characteristics Obtained after Mutation
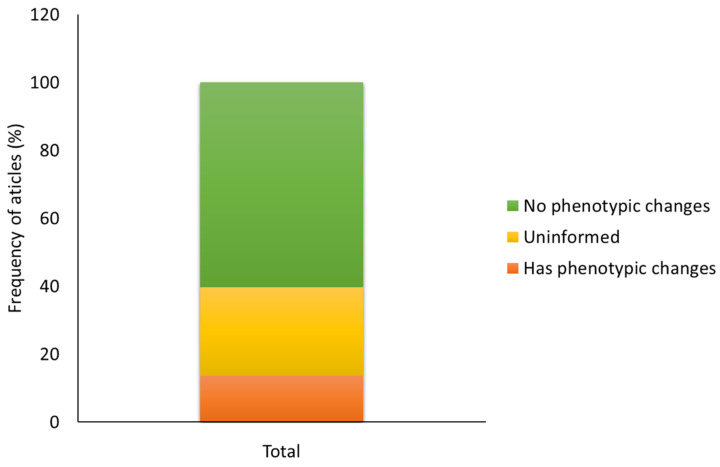
Considering the agronomic characteristics and visible symptoms of the disease after mutation of the plants, 60.2% of the studies indicated that the phenotype was preserved, 13.7% inferred that the plants showed unusual characteristics after mutagenesis, such as dwarfism, albinism, and more aggressive symptoms of the disease, such as wider lesions than would be characteristic, and 26.1% of the articles did not perform this type of analysis or did not record having carried it out (Figure 9).
Figure 9.
Frequency of articles that performed a phenotypic analysis of plants after mutation and the pathogen inoculation test.
Greater resistance to plant diseases was observed in approximately 70% of the studies and higher plant susceptibility after gene mutation was noted in 28% of the studies, indicating that these genes are related to plant defense/immunity response (Table S2).
3.9. Sources of Bias in the Included Studies
To assess risk of bias in individual studies, an adaptation of the Cochrane risk of bias tool protocol was performed, which is composed of domains; according to the reviewers’ judgment, the study/outcome is classified as having a high, low or unclear risk of bias. The domains assigned to this SR are important and necessary questions in studies related to gene editing by CRISPR/Cas technology. Thus, questions such as “Was phenotypic analysis performed after mutation in the plant?”, “Was off-target analysis performed?” were used to classify the methodological quality of the selected articles. And three authors did these analyses independently to avoid potential biases.
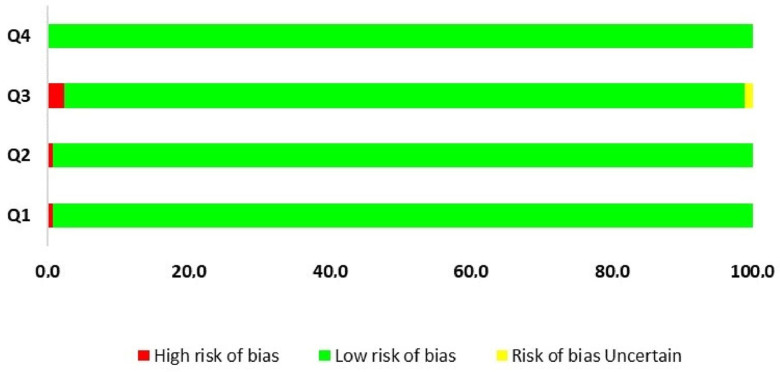
Based on the classification defined for the risk of bias and the questions designed to measure the risk, it can be inferred that 98.6% of the articles presented a low risk of bias (Figure 10). Only six articles did not answer question 3 (“Was phenotypic analysis performed after mutation in the plant?”) and presented a high risk for this question. Three studies had an uncertain answer; however, the other questions were answered, which does not invalidate these studies from contributing to this SR. For question 1 (“Was off-target analysis performed?”), only two studies did not answer. The other questions were answered in full, confirming the good methodological and bibliographical quality of this study (Table S3).
Figure 10.
Risk of bias analysis based on the following questions: “Q1: Was off-target analysis performed? Q2: Was the pathogen inoculated? Q3: Was phenotypic analysis performed after mutation in the plant? Q4: Does the article answer at least 50% of the research questions?”.
4. Discussion
4.1. Bibliographic Survey
This SR presents a compilation of data extracted from articles carefully selected between 2013 and 2024, with the aim of expanding knowledge on the use of CRISPR/Cas technology in plant gene editing for resistance to biotic stresses. The application of the CRISPR/Cas system in plants began in 2013 [51,52,53,54]; however, until 2015, the works consisted mainly of preliminary studies and the validation of techniques and protocols. Literature reviews were rejected to avoid bias, and letters to the editor and non-peer-reviewed articles were also disregarded. For this reason, and to obtain more recent studies on the subject, articles from the last twelve years were considered.
The year 2021 saw the largest bibliographic contribution on the subject, which may be related to the increased demand for food in the world and the negative effects of the COVID-19 pandemic [55], which led to an 18% increase in production in 2021 and an 11% increase in 2022 [56], stimulating agribusiness and studies focused on the genetic improvement of crops in order to minimize food shortages. The amount of data obtained on the subject in recent years reveals its importance and the need for investment in this area of research aiming to provide returns for the population and rural producers. Furthermore, these data reveal that technology is evolving rapidly and could contribute to overcoming food shortages for exponentially growing populations [57].
The biometric analysis demonstrated that the keywords “disease resistance” or “CRISPR/Cas9” present in the search string are also the most cited words in the selected articles and indicate that, over the last twelve years, more than 5000 studies have focused on this topic.
Keywords such as “viral resistance”, “DNA”, “genomics”, “oomycetes”, “soybean”, and “Sclerorotinia sclerotiorum” appear in isolation despite being related to the topic; this is because such words are found mainly in the body of the text and not in the titles, abstracts, and keywords of the selected articles.
The authors who have produced the most studies on the subject are from research institutions located mainly in China and the USA. The Rice Research Institute and Key Lab for Major Crop Diseases located at Sichuan Agricultural University in China is responsible for a major contribution to gene-editing work using CRISPR [58,59,60].
4.2. Study Sites and Edited Crops
Most of the studies included in this SR originated in China (140), which is in line with the data on agricultural production. Despite having less than 10% of the world’s productive land, the country ranks first in the production of cereals, cotton, fruit, vegetables, meat, poultry, and fishery products, as well as accounting for 25% of the world’s grain production [61]. This makes the country a major contributor to crop improvement studies using the CRISPR/Cas tool.
The studies performed in the USA were also representative (27). The country is the third largest food producer in the world and the first when it comes to exporting corn and soybeans, the main agricultural commodities [61]. Countries such as Germany, South Korea, Canada, Pakistan, Spain, and India have also contributed to studies on the subject.
Rice is the second-most-produced food crop in the world and the first-most-cultivated in China, which accounts for 30% of world production [61]. It is a monocot considered a model, because its genome is small and easy to manipulate when compared to other crops, which justifies the large number of studies (107) using CRISPR/Cas technology as a gene-editing tool for improving this crop [62,63,64,65].
In addition to rice, 27 other plant species have been covered in gene-editing studies in this SR. Tomato is the second-most-cited crop (47 articles) and the sixth most important crop economically, with a production of more than 100 million tons per year [61]. The model crops, Arabidopsis thaliana and Nicotiana sp., were also well cited in the selected papers; this may be related to the large amount of information already validated on these species and because their genomes have already been sequenced [66,67,68,69].
4.3. Biotic Stresses
Biotic stressors such as pathogens, insect pests, and weeds reduce the yield and quality of agricultural production. In high-yielding crops such as wheat, rice, corn, potatoes, and soybeans, losses can range from 17.2% in potatoes to 30% in rice [70]. Several diseases affect rice cultivation. Bacterial leaf blight (BLB), caused by Xanthomonas oryzae pv. oryzae (Xoo), is considered one of the most important bacterial diseases of rice. Irrigated or rainfed areas are common for growing this species and favor the development of the disease due to the abundance of water facilitating the dispersion of the pathogen, or through the high availability of nitrogen [62,71,72,73,74,75].
Bacterial leaf streak of rice caused by X. oryzae pv. oryzicola (Xoc) is also another disease that has been widely covered in the studies collected [76,77,78]. The genus Xanthomonas has been the most studied in gene editing via CRISPR/Cas in the last twelve years [75]. The studies seek to understand the mechanisms involved in plant defense against pathogens in order to make them resistant/tolerant to diseases.
Brusone is the main fungal disease of rice, caused by Magnaporthe oryzae, which establishes itself in the plant under favorable environmental conditions and causes damage to grain quality, plant height, and the number of tillers [79]. Rice is grown and consumed worldwide and is a staple food for around 2.5 billion people [61], so it is necessary to understand the biology of these pathogens to develop strategies to control these diseases.
Pseudomonas syringae was another pathogen that was mentioned frequently [80,81,82,83]. This bacterium is found in a wide variety of plants and penetrates host tissues through lesions or structures such as stomata [84]. The species has been widely used to elucidate questions about plant immunity and bacterial pathogenesis. In the selected articles, the bacterium is mainly present in studies with the model plant A. thaliana [66,85].
Tomatoes are an economically essential vegetable worldwide and their production is also threatened by many pathogens [86,87,88]. Gray mold, caused by Botrytis cinerea, rarely occurs in the field; however, in protected environments, humidity becomes a problem, favoring the development of the fungus, which infects the plant through wounds and causes the rapid rotting of the fruit, resulting in harvest losses. Other biotic agents have also been addressed in the studies, such as fungi (such as Fusarium), CMV viruses, CLCuVs, and the Phytophthora oomycete.
4.4. Types of Explants
The genetic transformation of plants is based on the insertion of transgenes into totipotent plant cells, which then regenerate into fertile plants. Small fragments of living tissue isolated from a plant specimen, called explants, are used [89]. The explants used for transformation via Agrobacterium or bioballistics can vary depending on the plant species, including calli, embryos, protoplasts, inflorescence, leaves, hypocotyls, epicotyls, and cotyledons [2,62,90,91,92,93,94,95].
When transformation occurs by electroporation, protoplasts undergo membrane destabilization after being subjected to high voltage, resulting in temporary pores in the cell membrane, allowing for the influx of DNA molecules that will integrate into the genome of the species to be mutated [96]. This type of method requires plants to be obtained entirely from protoplasts, which requires mastery of the production and regeneration of this type of explant, which is still a challenge in tissue culture [97].
The vast majority of the studies collected for this research used embryogenic calli and leaves as explants for various plant crops. In rice, the use of calli as an explant source is predominant [62,74,98,99,100]. Calli are formed practically from any fragment of the plant; in rice, seeds are commonly used to induce calli, which grow slowly as an amorphous cell mass through stimuli supplemented with specific phytohormones [89].
In tomatoes, transformations have been carried out mainly from cotyledon and leaf explants [81,93,101]. Most tissue culture tests in this species have been performed to achieve organogenesis over somatic embryogenesis [102]. In studies performed by Costa et al. (2000) [103], the tomato varieties ‘IPA-5’ and ‘IPA-6’ demonstrated favorable regeneration capacity (97 and 80%, respectively) from cotyledons when inserted into a supplemented Murashige and Skoog medium.
Studies with A. thaliana have mainly used flowers/inflorescences as a source of explants [92,104,105,106]. The floral immersion method is considered simple, fast, and efficient, and consists of immersing developing floral tissues in a solution containing Agrobacterium tumefaciens, sucrose, and detergent to transform the plants [107,108].
In tobacco, leaves were the most commonly used explants [68,109,110]. In direct somatic embryogenesis tests, using leaf tissue explants from different tobacco genotypes, different Agrobacterium strains, and different transformation methods, the transformation and regeneration rates varied [111,112]. The success of the transformation system involves the integration of the DNA into the host genome, the expression, inheritance, and stability of the exogenous DNA, as well as the regeneration of explants that depend largely on the genotype, origin, and age of the explant and plant growth regulators used to supplement culture media [113].
4.5. Plant Disease Resistance/Susceptibility Genes
The gene that stood out in CRISPR/Cas studies for resistance to biotic stresses was SWEET14 (Figure 6). SWEET genes encode sugar transporter proteins and often function as susceptibility (S) genes, the recessive alleles of which provide resistance [114]. This gene has been extensively studied and reviewed in studies involving the bacterial pathogen Xoo, which causes bacterial rust in rice [114,115]. In the context of plant–pathogen interaction, transcription activator-like effectors (TALEs) of the pathogen function in diverting the nutritional resources of rice, inducing the expression of OsSWEET14 and thus causing susceptibility [72,114]. The activation of SWEET14 by the pathogen results in an increase in the amount of sucrose available in the phloem apoplast, providing a source of nutrition for the pathogen promoting its proliferation [116].
The main strategy when using CRISPR/Cas9 in relation to the SWEET14 gene is to mutate the coding region of OsSWEET14 to test whether its disruption will result in broad-spectrum resistance to Xoo strains in rice [72,117] or the disruption of the TALE-binding elements of Xoo in rice harboring the recessive resistance allele in order to defuse the arms race between the effectors of the pathogen and their host targets [26,118,119]. Inhibition or SWEET14 editing can reduce the plant’s susceptibility to the pathogen, a potential strategy for the development of resistant cultivars.
Other genes, such as NRG1, Pi21 resistance genes, CsLOB1, SlMlo1, dependent glycosyl transferases (UGT76b1), and the Xa7 resistance gene, were also reported with considerable frequency in the studies (Figure 6). The NRG1 genes are close homologs of the Activated Disease Resistance 1 family of leucine-rich repeat domain proteins (NLRs), the function of which is still unclear, so some studies have reported their functional analysis through CRISPR/Cas9 in Arabidopsis [120,121,122]. The Pi21 gene belongs to the set of R genes that encode NLRs. It is resistant to rice brusone and is, therefore, the target of CRISPR/Cas9 rice-breeding programs to obtain mutant varieties [75,123,124,125].
Plants can prevent pathogen attacks through induced systemic resistance (ISR) and acquired systemic resistance (SAR). What differentiates them are the types of induction in the plant. SAR is activated through disease-causing organisms and relies on salicylic acid (SA) and genes, whereas beneficial microbes induce ISR and are independent of SA [126]. The two forms of resistance are activated from different defense signals when the plant is attacked by pathogens [127].
4.6. CRISPR/Cas Technology for Gene Editing
This SR sought to identify the most commonly used protocols for gene editing via CRISPR over the last twelve years. Among the editing methods used, the protocols proposed by Ma et al. (2015) [48], Xing et al. (2014) [49] and Wang et al. (2015) [50] were the most cited, respectively. The three protocols seek to edit various target genes in dicots and monocots using a multiplexing system, using one to several binary vectors and the Cas9 endonuclease. These results corroborate the findings of [22] in a systematic review of gene editing using CRISPR technology to edit genes tolerant to abiotic stresses.
Different CRISPR/Cas systems have been widely used to generate DSBs at target genomic sites in various plant species. Among the two classes of CRISPR immune systems, Class 2 is simpler than Class 1 and therefore easier to use for the development of genome editing tools [128]. Thus, three Class 2 effectors, Cas9, Cas12, and Cas13, have been extensively used for targeted DNA and RNA cleavage. The Cas9 endonuclease was the most widely used in the articles in this SR (259 studies), followed by Cas13 (3), and Cas12 (2). The effectors Cas9 and Cas12 are DNA-directed endonucleases, while Cas13 is an RNA-directed endonuclease [129].
As evidenced by Jinek et al. (2012) [14], Cas9 nucleases are guided by an RNA hybrid consisting of a crRNA and a tracrRNA. However, most Cas9 genome editing applications use an sgRNA that is designed by fusing crRNA and tracrRNA into a single RNA molecule for Cas9 to cleave DNA [130,131]. Normally, CRISPR/Cas9 requires a target site of 17 to 20 bp directly adjacent to a 5′-NGG PAM sequence (motif adjacent to the protospacer) to be effectively recognized by sgRNA [15,132]. Several authors have used Cas9 [68,133,134,135,136], and although several Cas9 orthologs have been discovered [137], Cas9 from Streptococcus pyogenes (SpCas9) is the nuclease that has been used the most for different genome manipulation experiments due to its high efficiency and simple NGG PAM sequence requirements [129].
The Cas12 endonuclease was identified in this SR with the aim of knocking out Xa13 [138] and PRAF2 [28] to improve resistance to bacterial rust caused by Xanthomonas oryzae pv. Oryzae. Cas12 is a class II type V endonuclease that was developed from Prevotella and Francisella [139]; it cleaves at a distal position of the PAM, generating a staggered break of the DNA double-strand, and recognizes a PAM region rich in T 5′-TTN-3′ [140] and proved to be an efficient alternative in editing these genes. Cas13 cleaves single-stranded RNA [141], and in the studies observed it was used to interfere against RNA viruses in plants, also presenting itself as a viable alternative to the use of Cas9 [142,143,144].
Cas9 and gRNA are regulated by appropriate promoters within a vector. The cauliflower mosaic virus (CaMV35S) is a constitutive promoter widely used for its strong expression in various plant tissues, being effective for mutations throughout the organism. The ubiquitin promoter (UBI), also constitutive and commonly used in monocots, has efficient and stable expression, especially in recalcitrant cultures. In addition, specific tissue promoters can also be used to induce mutations in plants. These allow for more controlled editing, limiting the expression of the system to specific sites, which reduces off-target effects, but can make mutations in the whole organism less efficient. The choice of promoter is crucial for the efficiency of the mutation due to to the objectives of gene editing, such as the need for localization or plant-wide expression, species compatibility, expression and efficiency, and the risks of off-site effects [145,146,147].
Several vectors have been used to express Cas and/or sgRNA, among the most cited being pCAMBIA (46) [63,73,148] a popular vector due to its easy handling, stability, and the existence of a variety of selection and reporter genes [149], and the pYLCRISPR/Cas9 vector (40) [150,151], which is a CRISPR/Cas9 system efficient in multi-locus gene knockout [48]. Other vectors, such as pHEE401E [152], also had considerable frequencies (Table S2). The most widely used delivery method for introducing the gene of interest into plant cells was carried out by Agrobacterium tumefaciens (286) and Agrobacterium rhizogenes (6). This is considered a powerful tool for delivering genes of interest to a host plant due to the efficiency of transformation, the low operating cost, and the simplicity of the transformation and selection protocols [153].
Although Agrobacterium-mediated delivery is very efficient, it also has some disadvantages, such as the need for long periods of tissue culture to recover transgenic plants, the low frequency of stably transformed plants, the narrow range of genotypes within a crop species that can be transformed, and the limitations of the host range of certain Agrobacterium species [154]. The delivery of CRISPR/Cas reagents to plants can be carried out by several methods. The most common in addition to Agrobacterium tumefaciens-mediated transformation include particle bombardment (biobalistics) and protoplast transfection [155,156].
Particle bombardment is useful for recalcitrant plant species, but it can cause physical damage to cells and random DNA integrations. Protoplast transfection, on the other hand, allows for the delivery of ribonucleoproteins (RNPs), reducing the risk of exogenous DNA integration, but the regeneration of complete plants from protoplasts can be challenging in some cultures [155,156]. Additional delivery methods of the CRISPR/Cas system, such as the use of nanoparticles and pollen magnetofection, can be an alternative for more precise and efficient delivery [157].
4.7. Auxiliary Methods to CRISPR/Cas
The main methodological strategies used in the studies collected to validate and support the CRISPR/Cas tool were PCR, sequencing, and qPCR techniques (Figure 7); these were mainly used to prove the efficacy of CRISPR/Cas-mediated editing and detect on- and off-target mutations (Figure 7). The PCR technique is an essential tool in molecular biology that allows for the amplification of nucleic acid sequences (DNA and RNA) through repetitive cycles in vitro, simulating what occurs in vivo during DNA replication [158].
PCR followed by sequencing has been reported in many studies; however, Zischewski et al. [159] highlight that a disadvantage of screening only potential pre-selected off-target sequences is the risk of overlooking mutations at other loci in the plant genome. In contrast, the use of the unbiased whole-genome-sequencing approach is the most common detection method in plants, allowing for the identification of off-target effects in a less restricted way [160].
Different prediction software were also used to detect off-target effects (Figure 8). The CRISPR-P software was reported in 16.2% of the articles as an auxiliary method to CRISPR/Cas, aimed at predicting target sites and/or mutations. Other software/programs, such as BLAST, DSDecode, Cas-OFFinder, CCTop, CRISPR-PLANT, and CHOPCHOP, also had considerable frequencies. A major concern in CRISPR/Cas9 system applications is its off-target effects that occur when Cas9 acts on untargeted genomic sites and creates cleavages that can lead to adverse outcomes [161].
The tools identified in this SR aid in silico prediction and are generally free online software that can be properly accessed via the Internet. The prediction algorithms of these software are mainly based on sgRNA sequences, so the results of these methods are generally biased toward sgRNA-dependent off-target effects. For epigenetics and chromatin organization experiments, off-target prediction by these in silico tools needs additional experimental validation [161].
4.8. Phenotypic Analysis and Characteristics Obtained after Mutation
In 60.2% of the articles, the phenotype was preserved, with no unusual or unexpected characteristics occurring after mutagenesis. Sixty-one percent exhibited greater resistance to plant diseases and 29% greater susceptibility after editing (Figure 9). This is because most studies are focused on knocking out/silencing genes or knocking in/overexpressing a gene to study and demonstrate its functions. Thus, the technique that cuts double-stranded DNA and generates a DSB will be repaired by the NHEJ repair mechanism; this can be carried out for a specific and individual gene without other side effects [162].
In other articles, the CRISPR/Cas technique has been used to knock-in the overexpression of an individual gene. In this sense, it is possible to edit the genome by cutting the DNA sequence at a specific site, and then, through HDR, a foreign DNA sequence (target gene) will be inserted at this cleavage site [162]. In this way, position effects can be avoided because CRISPR/Cas can be used to precisely insert a foreign gene into a specific location within a genome without interrupting other genes.
In this sense, the overexpression of the OsbHLH6 gene in transgenic rice plants caused responsive gene expression to jasmonic acid and increased susceptibility to the pathogen Magnaporthe oryzae [63]. Similarly, the overexpression of the GmLMM1 gene in Nicotiana benthamiana severely suppressed the production of reactive oxygen species triggered by microbe-associated molecules (bacterial flg22) and the pattern-induced cell death of the oomycete Phytophthora sojae [163].
Thus, the use of the CRISPR/Cas technique associated with gene knockout/silencing or gene knock-in/overexpression has contributed to the elucidation of various plant–pathogen interaction pathways in many pathosystems, without causing unwanted phenotypic changes, such as citrus canker caused by Xanthomonas citri subsp. citri in citrus [2], BLS of rice caused by Xoc and Xanthomonas campestris pv. campestris [73,164,165], Phytophthora sojae in soybeans [144], and Botrytis cinerea in tomatoes [81].
4.9. Sources of Bias in the Included Studies
The aim of SRs is to gather and synthesize data on a given topic that meets pre-established eligibility criteria and methods are used to reduce the chances of data bias [166]. The Cochrane Collaboration Tool was developed to assess the risk of bias of the studies to be included in the SRs and is widely used in health studies [47], which is why the method was adapted to the needs of this SR.
The tool aims to make the process clearer and more precise, free from errors that compromise the quality of the research. Therefore, possible limitations of the primary studies must be carefully assessed so that the results and conclusions obtained are reliable. It is not possible to determine the “quality” of a study without any kind of criteria; it is necessary to observe the design, the conducting of the research, and the analysis and presentation of the results so that the studies are not underestimated or overestimated [47,167].
In order to minimize errors in the choice of studies collected for this SR, inclusion/exclusion criteria, the PRISMA checklist, and questions on the topic (Table 3) were used to confirm whether the use of CRISPR/Cas technology was efficient in gene editing through off-target analysis. Inoculation tests of the pathogen and phenotypic analysis were also considered, as well as articles that answered at least 50% of the research questions (Table 1).
Literature reviews were excluded from the research, as many papers are cited repeatedly in the reviews, overestimating the data. Manuscripts that did not answer at least 75% of the risk-of-bias questions were considered high-risk and were not included in this SR. Only nine articles presented a risk equal to 25% for not answering one of the four questions, which is considered a low risk of bias, and two articles presented moderate risk, which means they answered only 50% of the questions. The articles selected for this SR are highly qualified and the methodologies used are reliable.
5. Final Considerations and Future Perspectives
The growing demand for food is a challenge for society in the face of population growth, changes in consumption patterns, environmental changes, and dealing with pathogens that cause plant diseases and pests. Meeting this demand is based on the need to guarantee global food security.
Biotic and abiotic stresses cause major losses in agricultural production, which calls for novel strategies to subsidize plant tolerance, as conventional practices are insufficient to meet the current and future food needs of the population. The use of the CRISPR/Cas tool can accelerate plant breeding by rapidly modifying genomes in a predictable and accurate way. Due to its efficiency, simplicity, and versatility, CRISPR/Cas has become a popular tool for genome editing and has been widely used in improving the resistance of various crops [57]. The development of disease-resistant varieties with good yields and quality is a fundamental strategy to guarantee global food security and generate employment and income for farmers.
This SR included 296 papers in which plant genes were edited via CRISPR/Cas to confer resistance to plant diseases and pests. We identified that Cas9 endonuclease is widely used in studies; however, this is not the only “molecular scissors” that can help the CRISPR editing system; the use of other enzymes such as Cpf1 (Cas12a), and Cas 13 has been reported in CRISPR studies for editing genes related to plant resistance and could be applied more frequently in future studies.
Genes related to tolerance/resistance to biotic stresses were identified in this SR and the CRISPR/Cas system can be used for gene knockout, gene insertion and gene replacement, resulting in the loss of function, knockdown or activation of mutants, which can lead to the generation of tolerant/resistant plants to the various pathogens. However, some issues are still far from being clarified and serve as a starting point for future studies, such as the fact that the main genes that control important traits of crops have not been identified, which limits the application of CRISPR/Cas in plant breeding; and pathogens continue to modify their genome through evolution to break the already available resistance gained by editing the CRISPR/Cas gene. Thus, it is necessary to design new variants in a short period of time and insert them into the plants. In addition, many genes are represented by multigene families, making it difficult to produce a resistance phenotype by eliminating a single gene, and it is necessary to develop more precise CRISPR/Cas tools to perform multiplex genome editing.
Regarding the methods used for editing, gRNAs were designed with different target sequences to direct Cas9 to specific corresponding sites; however, proper care is important when designing gRNAs, as unwanted targets are a major limitation, and to reduce these challenges, tools and software such as CRISPR-P, CRISPR-GE, BLAST, among others, are used. Among the methods used for mutation detection, PCR and sequencing are the most reported methods that can detect unwanted targets. Explant regeneration in most plants is still a challenge because it is labor-intensive and poses a limitation in CRISPR/Cas-based gene editing.
The information provided in this SR was based on articles with methodological quality confirmed by a risk of bias analysis, which determined that most of the included studies were at low risk of bias. Among the most-studied crops, rice, tomatoes, and the model plant Arabidopsis thaliana stand out. Among the most studied genera of biotic agents are Xanthomonas, Magnaporthe, Phytophthora and cucumber mosaic, belonging to the group of bacteria, fungi, oomycete and viruses, respectively.
Although the use of CRISPR/Cas technology has revolutionized plant breeding in recent years, there are still many challenges to be overcome; its off-target alterations are the main bioethical concern, namely whether they can lead to ecological imbalance, genetic drift, fatal diseases, or a chimeric phenotype in animals or even in humans. Another concern is whether GMOs produced by CRISPR/Cas9 can change the natural ecosystem by changing the mating potential of living organisms. Agricultural foods produced by CRISPR also face the same challenges as GMOs and may be prevented from being consumed in some countries. Despite these concerns, plants developed CRISPR/Cas can also become safe and GMO-free by using ribonucleoproteins (RNPs), i.e., without exogenous DNA. This will also help overcome the hurdles scientists face in commercializing biotech crops. To date, around 128 plant cultivars such as corn, soybeans, cotton, wheat, and sugar cane have been genetically edited, mainly for resistance to insects and/or herbicides, and have been approved by the National Technical Biosafety Commission [168].
Studies on gene editing with CRISPR/Cas for resistance to biotic agents are only beginning. The results obtained so far not only show that this technology offers precise modifications to the plant genome and has been successfully used to confer resistance to diseases and pests, but are also essential mainly to understand the function of genes related to various pathways of plant–pathogen interaction.
Acknowledgments
The authors would like to thank the Postgraduate Program in Biotechnology (PPGBiotec) of the State University of Feira de Santana, as well as CNPq (National Council for Scientific and Technological Development) for the E.P.A. and C.F.F. research productivity grants; Coordination for the Improvement of Higher Education Personnel (CAPES) for the DSc. grants for M.S.M. and F.d.S.N.; and Fapesb (Bahia Research Support Foundation) for the DSc. grants for M.d.S.F. and W.D.d.S.O.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cimb46100659/s1, Table S1: The selection of articles on the use of CRISPR/Cas technology associated with biotic stress tolerance in plants in the last twelve years. Table S2: Methods used for genome editing based on CRISPR/Cas technology and characteristics obtained by the plant after mutation in studies on biotic stresses in the last twelve years. Table S3: An assessment of the risk of bias of 296 articles selected for a systematic review on CRISPR/Cas technology used in gene editing in biotic stress tolerance published in the last twelve years. References [169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203,204,205,206,207,208,209,210,211,212,213,214,215,216,217,218,219,220,221,222,223,224,225,226,227,228,229,230,231,232,233,234,235,236,237,238,239,240,241,242,243,244,245,246,247,248,249,250,251,252,253,254,255,256,257,258,259,260,261,262,263,264,265,266,267,268,269,270,271,272,273,274,275,276,277,278,279,280,281,282,283,284,285,286,287,288,289,290,291,292,293,294,295,296,297,298,299,300,301,302,303,304,305,306,307,308,309,310,311,312,313,314,315,316,317,318,319,320,321,322,323,324,325,326,327,328,329,330,331,332,333,334,335,336,337,338,339,340,341,342,343,344,345,346,347,348,349,350,351,352,353,354,355,356,357,358,359,360,361,362,363,364,365,366,367,368,369,370,371,372,373,374,375,376,377,378,379,380,381,382,383,384] are cited in the Supplementary Materials.
Author Contributions
Conceptualization, M.S.M., F.d.S.N. and A.d.J.R.; methodology, M.S.M., F.d.S.N. and A.d.J.R.; C.F.F., T.A.d.O.M. and E.P.A.; software, M.S.M. and A.d.J.R.; validation, E.P.A. and C.F.F.; formal analysis, M.S.M., F.d.S.N., A.d.J.R. and E.P.A.; research, M.S.M., F.d.S.N., A.d.J.R., M.d.S.F., W.D.d.S.O. and L.S.M.L.; resources, E.P.A.; data curation, M.S.M., A.d.J.R. and F.d.S.N.; writing—preparation of original draft, M.S.M.; writing—review and editing, M.S.M., E.P.A. and C.F.F.; visualization, E.P.A. and C.F.F.; supervision, E.P.A., C.F.F., T.A.d.O.M. and J.A.d.S.-S.; project administration, E.P.A.; acquisition of funding, E.P.A. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research was funded by the International Institute of Tropical Agriculture (IITA)/The Bill and Melinda Gates Foundation—Accelerated Breeding of Better Bananas, ID OPP1093845.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Miller R.N.G., Costa Alves G.S., Van Sluys M.A. Plant Immunity: Unravelling the Complexity of Plant Responses to Biotic Stresses. Ann. Bot. 2017;119:681–687. doi: 10.1093/aob/mcw284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jia H., Omar A.A., Orbović V., Wang N. Biallelic Editing of the LOB1 Promoter via CRISPR/Cas9 Creates Canker-Resistant ‘Duncan’ Grapefruit. Phytopathology. 2022;112:308–314. doi: 10.1094/PHYTO-04-21-0144-R. [DOI] [PubMed] [Google Scholar]
- 3.Velásquez A.C., Castroverde C.D.M., He S.Y. Plant–Pathogen Warfare under Changing Climate Conditions. Curr. Biol. 2018;28:R619–R634. doi: 10.1016/j.cub.2018.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horbach R., Navarro-Quesada A.R., Knogge W., Deising H.B. When and How to Kill a Plant Cell: Infection Strategies of Plant Pathogenic Fungi. J. Plant Physiol. 2011;168:51–62. doi: 10.1016/j.jplph.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 5.Gosavi G., Yan F., Ren B., Kuang Y., Yan D., Zhou X., Zhou H. Applications of CRISPR Technology in Studying Plant-Pathogen Interactions: Overview and Perspective. Phytopathol. Res. 2020;2:21. doi: 10.1186/s42483-020-00060-z. [DOI] [Google Scholar]
- 6.Laflamme B., Dillon M.M., Martel A., Almeida R.N.D., Desveaux D., Guttman D.S. The Pan-Genome Effector-Triggered Immunity Landscape of a Host-Pathogen Interaction. Science. 2020;367:763–768. doi: 10.1126/science.aax4079. [DOI] [PubMed] [Google Scholar]
- 7.Pruitt R.N., Gust A.A., Nürnberger T. Plant Immunity Unified. Nat. Plants. 2021;7:382–383. doi: 10.1038/s41477-021-00903-3. [DOI] [PubMed] [Google Scholar]
- 8.Ninkuu V., Yan J., Fu Z., Yang T., Ziemah J., Ullrich M.S., Kuhnert N., Zeng H. Lignin and Its Pathway-Associated Phytoalexins Modulate Plant Defense against Fungi. J. Fungi. 2022;9:52. doi: 10.3390/jof9010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang J., Gu L., Zhang Y., Yan T., Kong G., Kong L., Guo B., Qiu M., Wang Y., Jing M., et al. An Oomycete Plant Pathogen Reprograms Host Pre-mRNA Splicing to Subvert Immunity. Nat. Commun. 2017;8:2051. doi: 10.1038/s41467-017-02233-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tripathi J.N., Ntui V.O., Tripathi L. Precision Genetics Tools for Genetic Improvement of Banana. Plant Genome. 2023;17:e20416. doi: 10.1002/tpg2.20416. [DOI] [PubMed] [Google Scholar]
- 11.Bhat J.A., Ali S., Salgotra R.K., Mir Z.A., Dutta S., Jadon V., Tyagi A., Mushtaq M., Jain N., Singh P.K., et al. Genomic Selection in the Era of Next Generation Sequencing for Complex Traits in Plant Breeding. Front. Genet. 2016;7:221. doi: 10.3389/fgene.2016.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim Y.G., Cha J., Chandrasegaran S. Hybrid restriction enzymes: Zinc finger fusions to fok I cleavage domain. Proc. Natl. Acad. Sci. USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christian M., Cermak T., Doyle E.L., Schmidt C., Zhang F., Hummel A., Hummel A., Bogdanove A.J., Voytas D.F. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh S., Ramakrishna W. Application of CRISPR–Cas9 in Plant–Plant Growth-Promoting Rhizobacteria Interactions for next Green Revolution. 3 Biotech. 2021;11:492. doi: 10.1007/s13205-021-03041-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah S.A., Erdmann S., Mojica F.J.M., Garrett R.A. Protospacer Recognition Motifs: Mixed Identities and Functional Diversity. RNA Biol. 2013;10:891–899. doi: 10.4161/rna.23764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khatodia S., Bhatotia K., Passricha N., Khurana S.M.P., Tuteja N. The CRISPR/Cas Genome-Editing Tool: Application in Improvement of Crops. Front. Plant Sci. 2016;7:506. doi: 10.3389/fpls.2016.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barakate A., Stephens J. An Overview of CRISPR-Based Tools and Their Improvements: New Opportunities in Understanding Plant–Pathogen Interactions for Better Crop Protection. Front. Plant Sci. 2016;7:765. doi: 10.3389/fpls.2016.00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung Y.-J., Nogoy F.M., Lee S.-K., Cho Y.-G., Kang K.-K. Application of ZFN for Site Directed Mutagenesis of Rice SSIVa Gene. Biotechnol. Bioproc E. 2018;23:108–115. doi: 10.1007/s12257-017-0420-9. [DOI] [Google Scholar]
- 20.Endo M., Mikami M., Toki S. Multigene Knockout Utilizing Off-Target Mutations of the CRISPR/Cas9 System in Rice. Plant Cell Physiol. 2015;56:41–47. doi: 10.1093/pcp/pcu154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowder L.G., Zhang D., Baltes N.J., Paul J.W., Tang X., Zheng X., Voytas D.F., Hsieh T.-F., Zhang Y., Qi Y. A CRISPR/Cas9 Toolbox for Multiplexed Plant Genome Editing and Transcriptional Regulation. Plant Physiol. 2015;169:971–985. doi: 10.1104/pp.15.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nascimento F.D.S., Rocha A.D.J., Soares J.M.D.S., Mascarenhas M.S., Ferreira M.D.S., Morais Lino L.S., Ramos A.P.D.S., Diniz L.E.C., Mendes T.A.D.O., Ferreira C.F., et al. Gene Editing for Plant Resistance to Abiotic Factors: A Systematic Review. Plants. 2023;12:305. doi: 10.3390/plants12020305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L., Wang W.-S., Wang T., Meng X.-F., Chen T., Huang X.-X., Li Y., Hou B.-K. Methyl Salicylate Glucosylation Regulates Plant Defense Signaling and Systemic Acquired Resistance. Plant Physiol. 2019;180:2167–2181. doi: 10.1104/pp.19.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martel A., Laflamme B., Seto D., Bastedo D.P., Dillon M.M., Almeida R.N.D., Guttman D.S., Desveaux D. Immunodiversity of the Arabidopsis ZAR1 NLR Is Conveyed by Receptor-Like Cytoplasmic Kinase Sensors. Front. Plant Sci. 2020;11:1290. doi: 10.3389/fpls.2020.01290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gou X., Zhong C., Zhang P., Mi L., Li Y., Lu W., Zheng J., Xu J., Meng Y., Shan W. miR398b and AtC2GnT Form a Negative Feedback Loop to Regulate Arabidopsis thaliana Resistance against Phytophthora parasitica. Plant J. 2022;111:360–373. doi: 10.1111/tpj.15792. [DOI] [PubMed] [Google Scholar]
- 26.Arulganesh T., Kumam Y., Kumar K.K., Arul L., Kokiladevi E., Nakeeran S., Varanavasiappan S., Manonmani S., Sudhakar D. Genome Editing of Elite Rice Cultivar, CO51 for Bacterial Leaf Blight Resistance. Electron. J. Plant Breed. 2021;12:1060–1068. doi: 10.37992/2021.1204.147. [DOI] [Google Scholar]
- 27.Wang W., Ma S., Hu P., Ji Y., Sun F. Genome Editing of Rice eIF4G Loci Confers Partial Resistance to Rice Black-Streaked Dwarf Virus. Viruses. 2021;13:2100. doi: 10.3390/v13102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu J., Li Q., Wang C., Wang M., Zeng D., Zhang F., Zhai W., Zhou Y. Identification of Quantitative Trait Loci Associated with Resistance to Xanthomonas oryzae pv. oryzae pathotypes Prevalent in South China. Crop J. 2022;10:498–507. doi: 10.1016/j.cj.2021.05.009. [DOI] [Google Scholar]
- 29.Zhang Z., Guo J., Zhao Y., Chen J. Identification and Characterization of Maize ACD6-like Gene Reveal ZmACD6 as the Maize Orthologue Conferring Resistance to Ustilago maydis. Plant Signal. Behav. 2019;14:e1651604. doi: 10.1080/15592324.2019.1651604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding Y., Murphy K.M., Poretsky E., Mafu S., Yang B., Char S.N., Christensen S.A., Saldivar E., Wu M., Wang Q., et al. Multiple Genes Recruited from Hormone Pathways Partition Maize Diterpenoid Defences. Nat. Plants. 2019;5:1043–1056. doi: 10.1038/s41477-019-0509-6. [DOI] [PubMed] [Google Scholar]
- 31.Ma L., Sun Y., Ruan X., Huang P.-C., Wang S., Li S., Zhou Y., Wang F., Cao Y., Wang Q., et al. Genome-Wide Characterization of Jasmonates Signaling Components Reveals the Essential Role of ZmCOI1a-ZmJAZ15 Action Module in Regulating Maize Immunity to Gibberella Stalk Rot. Int. J. Mol. Sci. 2021;22:870. doi: 10.3390/ijms22020870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanika K., Schipper D., Chinnappa S., Oortwijn M., Schouten H.J., Thomma B.P.H.J., Bai Y. Impairment of Tomato WAT1 Enhances Resistance to Vascular Wilt Fungi Despite Severe Growth Defects. Front. Plant Sci. 2021;12:721674. doi: 10.3389/fpls.2021.721674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomazella D.P.D.T., Seong K., Mackelprang R., Dahlbeck D., Geng Y., Gill U.S., Qi T., Pham J., Giuseppe P., Lee C.Y., et al. Loss of Function of a DMR6 Ortholog in Tomato Confers Broad-Spectrum Disease Resistance. Proc. Natl. Acad. Sci. USA. 2021;118:e2026152118. doi: 10.1073/pnas.2026152118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jhu M.-Y., Farhi M., Wang L., Philbrook R.N., Belcher M.S., Nakayama H., Zumstein K.S., Rowland S.D., Ron M., Shih P.M., et al. Heinz-Resistant Tomato Cultivars Exhibit a Lignin-Based Resistance to Field Dodder (Cuscuta campestris) Parasitism. Plant Physiol. 2022;189:129–151. doi: 10.1093/plphys/kiac024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan Y., Liu J., Lyu S., Wang Q., Yang S., Zhu H. The Soybean Rfg1 Gene Restricts Nodulation by Sinorhizobium Fredii USDA193. Front. Plant Sci. 2017;8:1548. doi: 10.3389/fpls.2017.01548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagy E.D., Stevens J.L., Yu N., Hubmeier C.S., LaFaver N., Gillespie M., Gardunia B., Cheng Q., Johnson S., Vaughn A.L., et al. Novel Disease Resistance Gene Paralogs Created by CRISPR/Cas9 in Soy. Plant Cell Rep. 2021;40:1047–1058. doi: 10.1007/s00299-021-02678-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X., Wang D., Chen J., Wu D., Feng X., Yu F. Nematode RALF-Like 1 Targets Soybean Malectin-Like Receptor Kinase to Facilitate Parasitism. Front. Plant Sci. 2021;12:775508. doi: 10.3389/fpls.2021.775508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rasheed A., Gill R.A., Hassan M.U., Mahmood A., Qari S., Zaman Q.U., LLyas M., Aamer M., Batool M., Li H., et al. A Critical Review: Recent Advancements in the Use of CRISPR/Cas9 Technology to Enhance Crops and Alleviate Global Food Crises. Curr. Issues Mol. Biol. 2021;43:1950–1976. doi: 10.3390/cimb43030135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ijaz M., Khan F., Zaki H.E.M., Khan M.M., Radwan K.S.A., Jiang Y., Qian J., Ahmed T., Shahid M.S., Luo J., et al. Recent Trends and Advancements in CRISPR-Based Tools for Enhancing Resistance against Plant Pathogens. Plants. 2023;12:1911. doi: 10.3390/plants12091911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vats S., Kumawat S., Brar J., Kaur S., Yadav K., Magar S.G., Jadhav P.V., Salvi P., Sonah H., Sharma S., et al. Opportunity and challenges for nanotechnology application for genome editing in plants. Plant Nano Biol. 2022;1:100001. doi: 10.1016/j.plana.2022.100001. [DOI] [Google Scholar]
- 41.Erdoğan İ., Cevher-Keskin B., Bilir Ö., Hong Y., Tör M. Recent Developments in CRISPR/Cas9 Genome-Editing Technology Related to Plant Disease Resistance and Abiotic Stress Tolerance. Biology. 2023;12:1037. doi: 10.3390/biology12071037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vidya N., Arun M. Updates and Applications of CRISPR/Cas Technology in Plants. J. Plant Biol. 2023;66:499–518. doi: 10.1007/s12374-023-09383-8. [DOI] [Google Scholar]
- 43.Page M.J., McKenzie J.E., Bossuyt P.M., Boutron I., Hoffmann T.C., Mulrow C.D., Shamseer L., Tetzlaff J.M., Akl E.A., Brennan S.E., et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. Int. J. Surg. 2021;88:105906. doi: 10.1016/j.ijsu.2021.105906. [DOI] [PubMed] [Google Scholar]
- 44.Santos C.M.D.C., Pimenta C.A.D.M., Nobre M.R.C. The PICO Strategy for the Research Question Construction and Evidence Search. Rev. Latino-Am. Enferm. 2007;15:508–511. doi: 10.1590/S0104-11692007000300023. [DOI] [PubMed] [Google Scholar]
- 45.R Core Team, R . A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2016. [(accessed on 20 August 2023)]. Available online: https://www.R-project.org. [Google Scholar]
- 46.Van Eck N.J., Waltman L. Visualizing Bibliometric Networks. In: Ding Y., Rousseau R., Wolfram D., editors. Measuring Scholarly Impact. Springer International Publishing; Cham, Switzerland: 2014. pp. 285–320. [Google Scholar]
- 47.Higgins J.P.T., Altman D.G., Gotzsche P.C., Juni P., Moher D., Oxman A.D., Savovic J., Schulz K.F., Weeks L., Sterne J.A.C., et al. The Cochrane Collaboration’s Tool for Assessing Risk of Bias in Randomised Trials. BMJ. 2011;343:d5928. doi: 10.1136/bmj.d5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma X., Zhang Q., Zhu Q., Liu W., Chen Y., Qiu R., Wang B., Yang Z., Li H., Lin Y., et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant. 2015;8:1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 49.Xing H.-L., Dong L., Wang Z.-P., Zhang H.-Y., Han C.-Y., Liu B., Wang X.-C., Chen Q.-J. A CRISPR/Cas9 Toolkit for Multiplex Genome Editing in Plants. BMC Plant Biol. 2014;14:327. doi: 10.1186/s12870-014-0327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z.-P., Xing H.-L., Dong L., Zhang H.-Y., Han C.-Y., Wang X.-C., Chen Q.-J. Egg Cell-Specific Promoter-Controlled CRISPR/Cas9 Efficiently Generates Homozygous Mutants for Multiple Target Genes in Arabidopsis in a Single Generation. Genome Biol. 2015;16:144. doi: 10.1186/s13059-015-0715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miao J., Guo D., Zhang J., Huang Q., Qin G., Zhang X., Wan J., Gu H., Qu L.-J. Targeted Mutagenesis in Rice Using CRISPR-Cas System. Cell Res. 2013;23:1233–1236. doi: 10.1038/cr.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng Z., Zhang B., Ding W., Liu X., Yang D.-L., Wei P., Cao F., Zhu S., Zhang F., Mao Y., et al. Efficient Genome Editing in Plants Using a CRISPR/Cas System. Cell Res. 2013;23:1229–1232. doi: 10.1038/cr.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie K., Yang Y. RNA-Guided Genome Editing in Plants Using a CRISPR–Cas System. Mol. Plant. 2013;6:1975–1983. doi: 10.1093/mp/sst119. [DOI] [PubMed] [Google Scholar]
- 54.Feng Z., Mao Y., Xu N., Zhang B., Wei P., Yang D.-L., Wang Z., Zhang Z., Zheng R., Yang L., et al. Multigeneration Analysis Reveals the Inheritance, Specificity, and Patterns of CRISPR/Cas-Induced Gene Modifications in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2014;111:4632–4637. doi: 10.1073/pnas.1400822111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.UN . Policy Brief: The Impact of COVID-19 on Food Security and Nutrition. United Nations; Rome, Italy: 2020. [(accessed on 16 August 2023)]. pp. 1–22. Available online: https://unsdg.un.org/resources/policy-brief-impact-covid-19-food-security-and-nutrition. [Google Scholar]
- 56.FAO . The Future of Food and Agriculture: Trends and Challenges. FAO; Rome, Italy: 2017. [(accessed on 6 February 2024)]. Available online: https://www.fao.org/3/i6583e/i6583e.pdf. [Google Scholar]
- 57.Nidhi S., Anand U., Oleksak P., Tripathi P., Lal J.A., Thomas G., Kuca K., Tripathi V. Novel CRISPR-Cas Systems: An Updated Review of the Current Achievements, Applications, and Future Research Perspectives. Int. J. Mol. Sci. 2021;22:3327. doi: 10.3390/ijms22073327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y., Cao X., Zhu Y., Yang X., Zhang K., Xiao Z., Wang H., Zhao J., Zhang L., Li G., et al. Osa-miR398b Boosts H2O2 Production and Rice Blast Disease-resistance via Multiple Superoxide Dismutases. New Phytol. 2019;222:1507–1522. doi: 10.1111/nph.15678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y., Tong Y., He X., Zhu Y., Li T., Lin X., Mao W., Ghulam Nabi Gishkori Z., Zhao Z., Zhang J., et al. The Rice miR171b–SCL6-IIs Module Controls Blast Resistance, Grain Yield, and Flowering. Crop J. 2022;10:117–127. doi: 10.1016/j.cj.2021.05.004. [DOI] [Google Scholar]
- 60.Chen J.-F., Zhao Z.-X., Li Y., Li T.-T., Zhu Y., Yang X.-M., Zhou S.-X., Wang H., Zhao J.-Q., Pu M., et al. Fine-Tuning Roles of Osa-miR159a in Rice Immunity Against Magnaporthe oryzae and Development. Rice. 2021;14:26. doi: 10.1186/s12284-021-00469-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.FAOSTAT Food and Agriculture Organization of the United Nations. [(accessed on 6 February 2024)]. Available online: http://www.fao.org/faostat/en/#home.
- 62.Hou Y., Wang Y., Tang L., Tong X., Wang L., Liu L., Huang S., Zhang J. SAPK10-Mediated Phosphorylation on WRKY72 Releases Its Suppression on Jasmonic Acid Biosynthesis and Bacterial Blight Resistance. iScience. 2019;16:499–510. doi: 10.1016/j.isci.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meng F., Yang C., Cao J., Chen H., Pang J., Zhao Q., Wang Z., Qing Fu Z., Liu J. A bHLH Transcription Activator Regulates Defense Signaling by Nucleo-cytosolic Trafficking in Rice. J. Integr. Plant Biol. 2020;62:1552–1573. doi: 10.1111/jipb.12922. [DOI] [PubMed] [Google Scholar]
- 64.Shen W., Zhang X., Liu J., Tao K., Li C., Xiao S., Zhang W., Li J. Plant Elicitor Peptide Signalling Confers Rice Resistance to Piercing-sucking Insect Herbivores and Pathogens. Plant Biotechnol. J. 2022;20:991–1005. doi: 10.1111/pbi.13781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hou J., Xiao H., Yao P., Ma X., Shi Q., Yang J., Hou H., Li L. Unveiling the Mechanism of Broad-spectrum Blast Resistance in Rice: The Collaborative Role of Transcription Factor OsGRAS30 and Histone Deacetylase OsHDAC1. Plant Biotechnol. J. 2024;22:1740–1756. doi: 10.1111/pbi.14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang F., Kimberlin A.N., Elowsky C.G., Liu Y., Gonzalez-Solis A., Cahoon E.B., Alfano J.R. A Plant Immune Receptor Degraded by Selective Autophagy. Mol. Plant. 2019;12:113–123. doi: 10.1016/j.molp.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 67.Huang J., Sun Y., Orduna A.R., Jetter R., Li X. The Mediator Kinase Module Serves as a Positive Regulator of Salicylic Acid Accumulation and Systemic Acquired Resistance. Plant J. 2019;98:842–852. doi: 10.1111/tpj.14278. [DOI] [PubMed] [Google Scholar]
- 68.Wang Y., Gong Q., Wu Y., Huang F., Ismayil A., Zhang D., Li H., Gu H., Ludman M., Fátyol K., et al. A Calmodulin-Binding Transcription Factor Links Calcium Signaling to Antiviral RNAi Defense in Plants. Cell Host Microbe. 2021;29:1393–1406.e7. doi: 10.1016/j.chom.2021.07.003. [DOI] [PubMed] [Google Scholar]
- 69.Gu H., Lian B., Yuan Y., Kong C., Li Y., Liu C., Qi Y. A 5′ tRNA-Ala-Derived Small RNA Regulates Anti-Fungal Defense in Plants. Sci. China Life Sci. 2022;65:1–15. doi: 10.1007/s11427-021-2017-1. [DOI] [PubMed] [Google Scholar]
- 70.Savary S., Willocquet L., Pethybridge S.J., Esker P., McRoberts N., Nelson A. The Global Burden of Pathogens and Pests on Major Food Crops. Nat. Ecol. Evol. 2019;3:430–439. doi: 10.1038/s41559-018-0793-y. [DOI] [PubMed] [Google Scholar]
- 71.Kim Y.-A., Moon H., Park C.-J. CRISPR/Cas9-Targeted Mutagenesis of Os8N3 in Rice to Confer Resistance to Xanthomonas oryzae pv. oryzae. Rice. 2019;12:67. doi: 10.1186/s12284-019-0325-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zeng X., Luo Y., Vu N.T.Q., Shen S., Xia K., Zhang M. CRISPR/Cas9-Mediated Mutation of OsSWEET14 in Rice Cv. Zhonghua11 Confers Resistance to Xanthomonas oryzae pv. oryzae without Yield Penalty. BMC Plant Biol. 2020;20:313. doi: 10.1186/s12870-020-02524-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang M., Shi H., Li N., Wei N., Tian Y., Peng J., Chen X., Zhang L., Zhang M., Dong H. Aquaporin OsPIP2;2 Links the H2O2 Signal and a Membrane-Anchored Transcription Factor to Promote Plant Defense. Plant Physiol. 2022;188:2325–2341. doi: 10.1093/plphys/kiab604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu X., Yu Y., Yao W., Yin Z., Wang Y., Huang Z., Zhou J., Liu J., Lu X., Wang F., et al. CRISPR/Cas9-mediated Simultaneous Mutation of Three Salicylic Acid. 5-hydroxylase (OsS5H) Genes Confers Broad-spectrum Disease Resistance in Rice. Plant Biotechnol. J. 2023;21:1873–1886. doi: 10.1111/pbi.14099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang J., Fang Y., Wu H., Zhao N., Guo X., Mackon E., Peng H., Huang S., He Y., Qin B., et al. Improvement of Resistance to Rice Blast and Bacterial Leaf Streak by CRISPR/Cas9-Mediated Mutagenesis of Pi21 and OsSULTR3;6 in Rice (Oryza sativa L.) Front. Plant Sci. 2023;14:1209384. doi: 10.3389/fpls.2023.1209384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang W., Ju Y., Zuo L., Shang L., Li X., Li X., Feng S., Ding X., Chu Z. OsHsfB4d Binds the Promoter and Regulates the Expression of OsHsp18.0-CI to Resistant against Xanthomonas oryzae. Rice. 2020;13:28. doi: 10.1186/s12284-020-00388-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang L., Chen J., Zhao Y., Wang S., Yuan M. OsMAPK6 Phosphorylates a Zinc Finger Protein OsLIC to Promote Downstream OsWRKY30 for Rice Resistance to Bacterial Blight and Leaf Streak. J. Integr. Plant Biol. 2022;64:1116–1130. doi: 10.1111/jipb.13249. [DOI] [PubMed] [Google Scholar]
- 78.Wu T., Zhang H., Yuan B., Liu H., Kong L., Chu Z., Ding X. Tal2b Targets and Activates the Expression of OsF3H03g to Hijack OsUGT74H4 and Synergistically Interfere with Rice Immunity. New Phytol. 2022;233:1864–1880. doi: 10.1111/nph.17877. [DOI] [PubMed] [Google Scholar]
- 79.Prabhu A.S., de Faria J.C., de Carvalho J.R.P. Effect of blast on dry matter, grain production and its components in upland rice. Braz. Agric. Res. 1996;21:495–500. [Google Scholar]
- 80.Wang Y., Li Y., Rosas-Diaz T., Caceres-Moreno C., Lozano-Duran R., Macho A.P. The Immune-associated nucleotide-binding 9 Protein is a Regulator of Basal Immunity in Arabidopsis thaliana. Mol. Plant-Microbe Interact. 2019;32:65–75. doi: 10.1094/MPMI-03-18-0062-R. [DOI] [PubMed] [Google Scholar]
- 81.Jeon J.E., Kim J.-G., Fischer C.R., Mehta N., Dufour-Schroif C., Wemmer K., Mudgett M.B., Sattely E. A Pathogen-Responsive Gene Cluster for Highly Modified Fatty Acids in Tomato. Cell. 2020;180:176–187.e19. doi: 10.1016/j.cell.2019.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang S., Zhu S., Kumar P., MacMicking J.D. A Phase-Separated Nuclear GBPL Circuit Controls Immunity in Plants. Nature. 2021;594:424–429. doi: 10.1038/s41586-021-03572-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen Y.-L., Lin F.-W., Cheng K.-T., Chang C.-H., Hung S.-C., Efferth T., Chen Y.-R. XCP1 Cleaves Pathogenesis-Related Protein 1 into CAPE9 for Systemic Immunity in Arabidopsis. Nat. Commun. 2023;14:4697. doi: 10.1038/s41467-023-40406-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hirano S.S., Upper C.D. Bacteria in the Leaf Ecosystem with Emphasis on Pseudomonas syringae—A Pathogen, Ice Nucleus, and Epiphyte. Microbiol. Mol. Biol. Rev. 2000;64:624–653. doi: 10.1128/MMBR.64.3.624-653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang X., Wang Y., Lin J., Chen L., Li Y., Liu Q., Wang G., Xu F., Liu L., Hou B. The Novel Pathogen-responsive Glycosyltransferase UGT73C7 Mediates the Redirection of Phenylpropanoid Metabolism and Promotes SNC1-dependent Arabidopsis Immunity. Plant J. 2021;107:149–165. doi: 10.1111/tpj.15280. [DOI] [PubMed] [Google Scholar]
- 86.Shu P., Li Z., Min D., Zhang X., Ai W., Li J., Zhou J., Li Z., Li F., Li X. CRISPR/Cas9-Mediated SlMYC2 Mutagenesis Adverse to Tomato Plant Growth and MeJA-Induced Fruit Resistance to Botrytis cinerea. J. Agric. Food Chem. 2020;68:5529–5538. doi: 10.1021/acs.jafc.9b08069. [DOI] [PubMed] [Google Scholar]
- 87.Son G.H., Moon J., Shelake R.M., Vuong U.T., Ingle R.A., Gassmann W., Kim J.-Y., Kim S.H. Conserved Opposite Functions in Plant Resistance to Biotrophic and Necrotrophic Pathogens of the Immune Regulator SRFR1. Int. J. Mol. Sci. 2021;22:6427. doi: 10.3390/ijms22126427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zheng H., Jin R., Liu Z., Sun C., Shi Y., Grierson D., Zhu C., Li S., Ferguson I., Chen K. Role of the Tomato Fruit Ripening Regulator MADS-RIN in Resistance to Botrytis cinerea Infection. Food Qual. Saf. 2021;5:fyab028. doi: 10.1093/fqsafe/fyab028. [DOI] [Google Scholar]
- 89.Torres A.C., Ferreira A.T., de SÀ F.G., Buso J.A., Caldas L.S., Nascimento A.S., Romano E. Glossário de Biotecnologia Vegetal. Ministério da Agricultura e do Abastecimento; Brasília, Brazil: 2000. p. 128. [Google Scholar]
- 90.Galli M., Martiny E., Imani J., Kumar N., Koch A., Steinbrenner J., Kogel K. CRISPR/Sp. Cas9-mediated Double Knockout of Barley Microrchidia MORC1 and MORC6a Reveals Their Strong Involvement in Plant Immunity, Transcriptional Gene Silencing and Plant Growth. Plant Biotechnol. J. 2022;20:89–102. doi: 10.1111/pbi.13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang X., Wang Y., Wang N. Highly Efficient Generation of Canker-Resistant Sweet Orange Enabled by an Improved CRISPR/Cas9 System. Front. Plant Sci. 2022;12:769907. doi: 10.3389/fpls.2021.769907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li Z., Zhang Y., Ren J., Jia F., Zeng H., Li G., Yang X. Ethylene-responsive Factor ERF114 Mediates Fungal Pathogen Effector PevD1-induced Disease Resistance in Arabidopsis thaliana. Mol. Plant Pathol. 2022;23:819–831. doi: 10.1111/mpp.13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li Y., Shu P., Xiang L., Sheng J., Shen L. CRISPR/Cas9-Mediated SlATG5 Mutagenesis Reduces the Resistance of Tomato Fruit to Botrytis Cinerea. Foods. 2023;12:2750. doi: 10.3390/foods12142750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang J., Li J., Saeed S., Batchelor W.D., Alariqi M., Meng Q., Zhu F., Zou J., Xu Z., Si H., et al. Identification and Functional Analysis of lncRNA by CRISPR/Cas9 During the Cotton Response to Sap-Sucking Insect Infestation. Front. Plant Sci. 2022;13:784511. doi: 10.3389/fpls.2022.784511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yin J., Wang L., Jin T., Nie Y., Liu H., Qiu Y., Yang Y., Li B., Zhang J., Wang D., et al. A Cell Wall-Localized NLR Confers Resistance to Soybean Mosaic Virus by Recognizing Viral-Encoded Cylindrical Inclusion Protein. Mol. Plant. 2021;14:1881–1900. doi: 10.1016/j.molp.2021.07.013. [DOI] [PubMed] [Google Scholar]
- 96.Ozyigit I.I. Gene Transfer to Plants by Electroporation: Methods and Applications. Mol. Biol. Rep. 2020;47:3195–3210. doi: 10.1007/s11033-020-05343-4. [DOI] [PubMed] [Google Scholar]
- 97.Grosser J.W., Ollitrault P., Olivares-Fuster O. Somatic Hybridization in Citrus: An Effective Tool to Facilitate Variety Improvement. In Vitro Cell. Dev. Biol. Plant. 2000;36:434–449. doi: 10.1007/s11627-000-0080-9. [DOI] [Google Scholar]
- 98.Xie Z., Yan B., Shou J., Tang J., Wang X., Zhai K., Liu J., Li Q., Luo M., Deng Y., et al. A Nucleotide-Binding Site-Leucine-Rich Repeat Receptor Pair Confers Broad-Spectrum Disease Resistance through Physical Association in Rice. Phil. Trans. R. Soc. B. 2019;374:20180308. doi: 10.1098/rstb.2018.0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Qiu J., Lu F., Xiong M., Meng S., Shen X., Kou Y. Comparative Transcriptomic Analysis Reveals the Mechanistic Basis of Pib-Mediated Broad Spectrum Resistance against Magnaporthe oryzae. Funct. Integr. Genom. 2020;20:787–799. doi: 10.1007/s10142-020-00752-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fang H., Shen S., Wang D., Zhang F., Zhang C., Wang Z., Zhou Q., Wang R., Tao H., He F., et al. A Monocot-Specific Hydroxycinnamoylputrescine Gene Cluster Contributes to Immunity and Cell Death in Rice. Sci. Bull. 2021;66:2381–2393. doi: 10.1016/j.scib.2021.06.014. [DOI] [PubMed] [Google Scholar]
- 101.Prihatna C., Barbetti M.J., Barker S.J. A Novel Tomato Fusarium Wilt Tolerance Gene. Front. Microbiol. 2018;9:1226. doi: 10.3389/fmicb.2018.01226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Souza G.F.M.V., Luz J.M.Q., Arruda A.S., Santana D.G., Teixeira M.S.S.C., Londe L., Silva A.S., Figueira E.R. Capacidade de regeneração in vitro de tomateiro Santa Clara. Plant Cell Cult. Micropropag. 2006;2:88–93. [Google Scholar]
- 103.Costa M., Nogueira F., Figueira M., Otoni W.C., Brommonschenkel S.H., Cecon P.R. Influence of the antibiotic timentin on plant regeneration of tomato (Lycopersicon esculentum Mill.) cultivars. Plant Cell Rep. 2000;19:327–332. doi: 10.1007/s002990050021. [DOI] [PubMed] [Google Scholar]
- 104.Zhang Y., Guo W., Chen L., Shen X., Yang H., Fang Y., Ouyang W., Mai S., Chen H., Chen S., et al. CRISPR/Cas9-Mediated Targeted Mutagenesis of GmUGT Enhanced Soybean Resistance against Leaf-Chewing Insects through Flavonoids Biosynthesis. Front. Plant Sci. 2022;13:802716. doi: 10.3389/fpls.2022.802716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lu W., Deng F., Jia J., Chen X., Li J., Wen Q., Li T., Meng Y., Shan W. The Arabidopsis thaliana Gene AtERF019 Negatively Regulates Plant Resistance to Phytophthora parasitica by Suppressing PAMP-triggered Immunity. Mol. Plant Pathol. 2020;21:1179–1193. doi: 10.1111/mpp.12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Su T., Wang W., Wang Z., Li P., Xin X., Yu Y., Zhang D., Zhao X., Wang J., Sun L., et al. BrMYB108 Confers Resistance to Verticillium Wilt by Activating ROS Generation in Brassica Rapa. Cell Rep. 2023;42:112938. doi: 10.1016/j.celrep.2023.112938. [DOI] [PubMed] [Google Scholar]
- 107.Clough S.J., Bent A.F. Floral Dip: A Simplified Method for Agrobacterium-mediated Transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 108.Rod-in W., Sujipuli K., Ratanasut K. The floral-dip method for rice (Oryza sativa) transformation. J. Agric. Technol. 2014;10:467–474. [Google Scholar]
- 109.Li Z.-C., Ren Q.-W., Guo Y., Ran J., Ren X.-T., Wu N.-N., Xu H.-Y., Liu X., Liu J.-Z. Dual Roles of GSNOR1 in Cell Death and Immunity in Tetraploid Nicotiana Tabacum. Front. Plant Sci. 2021;12:596234. doi: 10.3389/fpls.2021.596234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jogam P., Sandhya D., Alok A., Peddaboina V., Singh S.P., Abbagani S., Zhang B., Allini V.R. Editing of TOM1 Gene in Tobacco Using CRISPR/Cas9 Confers Resistance to Tobacco Mosaic Virus. Mol. Biol. Rep. 2023;50:5165–5176. doi: 10.1007/s11033-023-08440-2. [DOI] [PubMed] [Google Scholar]
- 111.Bakhsh A., Anayol E., Ozcan S. Comparison of Transformation Efficiency of Five Agrobacterium tumefaciens Strains in Nicotiana Tabacum L. Emir. J. Food Agric. 2014;26:259. doi: 10.9755/ejfa.v26i3.16437. [DOI] [Google Scholar]
- 112.Japelaghi R.H., Haddad R., Valizadeh M., Dorani E.U., Javaran M.J. High-Efficiency Improvement of Agrobacterium-Mediated Transformation of Tobacco (Nicotiana tabacum) J. Plant Mol. Breed. 2018;6:38–50. doi: 10.22058/jpmb.2019.92266.1170. [DOI] [Google Scholar]
- 113.Caldas L.S., Padmaja H., Ferreira M.E. Meios Nutritivos. In: Torres A.C., Caldas L.S., Buso J.A., editors. Cultura de Tecidos e Transformação Genética de Plantas. Embrapa-CNPH; Brasília, Brazil: 1998. pp. 87–132. [Google Scholar]
- 114.Gupta P.K., Balyan H.S., Gautam T. Sweet Genes and TAL Effectors for Disease Resistance in Plants: Present Status and Future Prospects. Mol. Plant Pathol. 2021;22:1014–1026. doi: 10.1111/mpp.13075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jiang N., Yan J., Liang Y., Shi Y., He Z., Wu Y., Zeng Q., Liu X., Peng J. Resistance Genes and Their Interactions with Bacterial Blight/Leaf Streak Pathogens (Xanthomonas oryzae) in Rice (Oryza sativa L.)—An Updated Review. Rice. 2020;13:3. doi: 10.1186/s12284-019-0358-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ji J., Yang L., Fang Z., Zhang Y., Zhuang M., Lv H., Wang Y. Plant SWEET family of sugar transporters: Structure, evolution and biological functions. Biomolecules. 2022;12:205. doi: 10.3390/biom12020205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zafar K., Khan M.Z., Amin I., Mukhtar Z., Yasmin S., Arif M., Ejaz K., Mansoor S. Precise CRISPR-Cas9 Mediated Genome Editing in Super Basmati Rice for Resistance against Bacterial Blight by Targeting the Major Susceptibility Gene. Front. Plant Sci. 2020;11:575. doi: 10.3389/fpls.2020.00575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Duy P.N., Lan D.T., Pham Thu H., Thi Thu H.P., Nguyen Thanh H., Pham N.P., Auguy F., Bui Thi Thu H., Manh T.B., Cunnac S., et al. Improved Bacterial Leaf Blight Disease Resistance in the Major Elite Vietnamese Rice Cultivar TBR225 via Editing of the OsSWEET14 Promoter. PLoS ONE. 2021;16:e0255470. doi: 10.1371/journal.pone.0255470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xu Z., Xu X., Gong Q., Li Z., Li Y., Wang S., Yang Y., Ma W., Liu L., Zhu B., et al. Engineering Broad-Spectrum Bacterial Blight Resistance by Simultaneously Disrupting Variable TALE-Binding Elements of Multiple Susceptibility Genes in Rice. Mol. Plant. 2019;12:1434–1446. doi: 10.1016/j.molp.2019.08.006. [DOI] [PubMed] [Google Scholar]
- 120.Castel B., Ngou P., Cevik V., Redkar A., Kim D., Yang Y., Ding P., Jones J.D.G. Diverse NLR Immune Receptors Activate Defence via the RPW 8-NLR NRG 1. New Phytol. 2019;222:966–980. doi: 10.1111/nph.15659. [DOI] [PubMed] [Google Scholar]
- 121.Wu Z., Li M., Dong O.X., Xia S., Liang W., Bao Y., Wasteneys G., Li X. Differential Regulation of TNL-mediated Immune Signaling by Redundant Helper CNLs. New Phytol. 2019;222:938–953. doi: 10.1111/nph.15665. [DOI] [PubMed] [Google Scholar]
- 122.Wang W., Liu N., Gao C., Rui L., Jiang Q., Chen S., Zhang Q., Zhong G., Tang D. The Truncated TNL Receptor TN2-mediated Immune Responses Require ADR1 Function. Plant J. 2021;108:672–689. doi: 10.1111/tpj.15463. [DOI] [PubMed] [Google Scholar]
- 123.Nawaz G., Usman B., Peng H., Zhao N., Yuan R., Liu Y., Li R. Knockout of Pi21 by CRISPR/Cas9 and iTRAQ-Based Proteomic Analysis of Mutants Revealed New Insights into M. oryzae Resistance in Elite Rice Line. Genes. 2020;11:735. doi: 10.3390/genes11070735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tao H., Shi X., He F., Wang D., Xiao N., Fang H., Wang R., Zhang F., Wang M., Li A., et al. Engineering Broad-spectrum Disease-resistant Rice by Editing Multiple Susceptibility Genes. J. Integr. Plant Biol. 2021;63:1639–1648. doi: 10.1111/jipb.13145. [DOI] [PubMed] [Google Scholar]
- 125.Zhou Y., Xu S., Jiang N., Zhao X., Bai Z., Liu J., Yao W., Tang Q., Xiao G., Lv C., et al. Engineering of Rice Varieties with Enhanced Resistances to Both Blast and Bacterial Blight Diseases via CRISPR/Cas9. Plant Biotechnol. J. 2022;20:876–885. doi: 10.1111/pbi.13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vlot A.C., Vendas J.H., Lenk M., Bauer K., Brambilla A., Sommer A., Chen Y., Wenig M., Shahran N. Systemic propagation of immunity in plants. New Phytol. 2020;229:1234–1250. doi: 10.1111/nph.16953. [DOI] [PubMed] [Google Scholar]
- 127.Boller T., Félix J.A. Renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Ver. Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 128.Koonin E.V., Makarova K.S. Evolutionary Plasticity and Functional Versatility of CRISPR Systems. PLoS Biol. 2022;20:e3001481. doi: 10.1371/journal.pbio.3001481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Karmakar S., Das P., Panda D., Xie K., Baig M.J., Molla K.A. A Detailed Landscape of CRISPR-Cas-Mediated Plant Disease and Pest Management. Plant Sci. 2022;323:111376. doi: 10.1016/j.plantsci.2022.111376. [DOI] [PubMed] [Google Scholar]
- 130.Wada N., Ueta R., Osakabe Y., Osakabe K. Precision Genome Editing in Plants: State-of-the-Art in CRISPR/Cas9-Based Genome Engineering. BMC Plant Biol. 2020;20:234. doi: 10.1186/s12870-020-02385-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ghosh S., Dey G. Biotic and Abiotic Stress Tolerance through CRISPR-Cas Mediated Genome Editing. J. Plant Biochem. Biotechnol. 2022;31:227–238. doi: 10.1007/s13562-021-00746-1. [DOI] [Google Scholar]
- 132.Talakayala A., Ankanagari S., Garladinne M. CRISPR-Cas Genome Editing System: A Versatile Tool for Developing Disease Resistant Crops. Plant Stress. 2022;3:100056. doi: 10.1016/j.stress.2022.100056. [DOI] [Google Scholar]
- 133.Khan S., Mahmood M.S., Rahman S.U., Rizvi F., Ahmad A. Evaluation of the CRISPR/Cas9 System for the Development of Resistance against Cotton Leaf Curl Virus in Model Plants. Plant Prot. Sci. 2020;56:154–162. doi: 10.17221/105/2019-PPS. [DOI] [Google Scholar]
- 134.Wang X., Tu M., Wang D., Liu J., Li Y., Li Z., Wang Y., Wang X. CRISPR/Cas9-mediated Efficient Targeted Mutagenesis in Grape in the First Generation. Plant Biotechnol. J. 2018;16:844–855. doi: 10.1111/pbi.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu C., Zhang Y., Tan Y., Zhao T., Xu X., Yang H., Li J. CRISPR/Cas9-Mediated SlMYBS2 Mutagenesis Reduces Tomato Resistance to Phytophthora infestans. Int. J. Mol. Sci. 2021;22:11423. doi: 10.3390/ijms222111423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Távora F.T.P.K., Meunier A.C., Vernet A., Portefaix M., Milazzo J., Adreit H., Tharreau D., Franco O.L., Mehta A. CRISPR/Cas9-Targeted Knockout of Rice Susceptibility Genes OsDjA2 and OsERF104 Reveals Alternative Sources of Resistance to Pyricularia oryzae. Rice Sci. 2022;29:535–544. doi: 10.1016/j.rsci.2022.04.001. [DOI] [Google Scholar]
- 137.Molla K.A., Karmakar S., Islam M.T. Wide Horizons of CRISPR-Cas-Derived Technologies for Basic Biology, Agriculture, and Medicine. In: Islam M.T., Bhowmik P.K., Molla K.A., editors. CRISPR-Cas Methods. Springer; New York, NY, USA: 2020. pp. 1–23. (Springer Protocols Handbooks). [Google Scholar]
- 138.Yu K., Liu Z., Gui H., Geng L., Wei J., Liang D., Lv J., Xu J., Chen X. Highly Efficient Generation of Bacterial Leaf Blight-Resistant and Transgene-Free Rice Using a Genome Editing and Multiplexed Selection System. BMC Plant Biol. 2021;21:197. doi: 10.1186/s12870-021-02979-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Anzalone A.V., Koblan L.W., Liu D.R. Genome Editing with CRISPR–Cas 124. Nucleases, Base Editors, Transposases and Prime Editors. Nat. Biotechnol. 2020;38:824–844. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- 140.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A., et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B.T., Shmakov S., Makarova K.S., Semenova E., Minakhin L., et al. C2c2 Is a Single-Component Programmable RNA-Guided RNA-Targeting CRISPR Effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhan X., Zhang F., Zhong Z., Chen R., Wang Y., Chang L., Bock R., Nie B., Zhang J. Generation of Virus-resistant Potato Plants by RNA Genome Targeting. Plant Biotechnol. J. 2019;17:1814–1822. doi: 10.1111/pbi.13102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Aman R., Ali Z., Butt H., Mahas A., Aljedaani F., Khan M.Z., Ding S., Mahfouz M. RNA Virus Interference via CRISPR/Cas13a System in Plants. Genome Biol. 2018;19:1. doi: 10.1186/s13059-017-1381-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yu Y., Pan Z., Wang X., Bian X., Wang W., Liang Q., Kou M., Ji H., Li Y., Ma D., et al. Targeting of SPCSV-RNase3 via CRISPR-Cas13 Confers Resistance against Sweet Potato Virus Disease. Mol. Plant Pathol. 2022;23:104–117. doi: 10.1111/mpp.13146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kummari D., Palakolanu S.R., Kishor P.B.K., Bhatnagar-Mathur P., Singam P., Vadez V., Sharma K.K. An Update and Perspectives on the Use of Promoters in Plant Genetic Engineering. J. Biosci. 2020;45:119. doi: 10.1007/s12038-020-00087-6. [DOI] [PubMed] [Google Scholar]
- 146.Porto M.S., Pinheiro M.P.N., Batista V.G.L., Dos Santos R.C., De Albuquerque Melo Filho P., De Lima L.M. Plant Promoters: An Approach of Structure and Function. Mol. Biotechnol. 2014;56:38–49. doi: 10.1007/s12033-013-9713-1. [DOI] [PubMed] [Google Scholar]
- 147.Singha D.L., Das D., Sarki Y.N., Chowdhury N., Sharma M., Maharana J., Chikkaputtaiah C. Harnessing Tissue-Specific Genome Editing in Plants through CRISPR/Cas System: Current State and Future Prospects. Planta. 2022;255:28. doi: 10.1007/s00425-021-03811-0. [DOI] [PubMed] [Google Scholar]
- 148.Chen X., Liu P., Mei L., He X., Chen L., Liu H., Shen S., Ji Z., Zheng X., Zhang Y., et al. Xa7, a New Executor R Gene That Confers Durable and Broad-Spectrum Resistance to Bacterial Blight Disease in Rice. Plant Commun. 2021;2:100143. doi: 10.1016/j.xplc.2021.100143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Leclercq J., Szabolcs T., Martin F., Montoro P. Development of a New pCAMBIA Binary Vector Using Gateway® Technology. Plasmid. 2015;81:50–54. doi: 10.1016/j.plasmid.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 150.Zhang Y., Lin X.-F., Li L., Piao R.-H., Wu S., Song A., Gao M., Jin Y.-M. CRISPR/Cas9-mediated knockout of Bsr-d1 enhances the blast resistance of rice in Northeast China. Plant Cell Rep. 2024;43:100. doi: 10.1007/s00299-024-03192-0. [DOI] [PubMed] [Google Scholar]
- 151.Wang C., Chen S., Feng A., Su J., Wang W., Feng J., Chen B., Zhang M., Yang J., Zeng L., et al. Xa7, a Small Orphan Gene Harboring Promoter Trap for AvrXa7, Leads to the Durable Resistance to Xanthomonas oryzae pv. oryzae. Rice. 2021;14:48. doi: 10.1186/s12284-021-00490-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li L.-S., Ying J., Li E., Ma T., Li M., Gong L.-M., Wei G., Zhang Y., Li S. Arabidopsis CBP60b Is a Central Transcriptional Activator of Immunity. Plant Physiol. 2021;186:1645–1659. doi: 10.1093/plphys/kiab164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hwang H.-H., Yu M., Lai E.-M. Agrobacterium-Mediated Plant Transformation: Biology and Applications. Arab. Book. 2017;15:e0186. doi: 10.1199/tab.0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Baltes N.J., Gil-Humanes J., Voytas D.F. Progress in Molecular Biology and Translational Science. Volume 149. Elsevier; Amsterdam, The Netherlands: 2017. Genome Engineering and Agriculture: Opportunities and Challenges; pp. 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chen K., Wang Y., Zhang R., Zhang H., Gao C. CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture. Annu. Rev. Plant Biol. 2019;70:667–697. doi: 10.1146/annurev-arplant-050718-100049. [DOI] [PubMed] [Google Scholar]
- 156.Saini H., Thakur R., Gill R., Tyagi K., Goswami M. CRISPR/Cas9-Gene Editing Approaches in Plant Breeding. GM Crops Food. 2023;14:1–17. doi: 10.1080/21645698.2023.2256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sandhya D., Jogam P., Allini V.R., Abbagani S., Alok A. The Present and Potential Future Methods for Delivering CRISPR/Cas9 Components in Plants. J. Genet. Eng. Biotechnol. 2020;18:25. doi: 10.1186/s43141-020-00036-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ehtisham M., Wani F., Wani I., Kaur P., Nissar S. Polymerase Chain Reaction (PCR): Back to Basics. Indian J. Contemp. Dent. 2016;4:30. doi: 10.5958/2320-5962.2016.00030.9. [DOI] [Google Scholar]
- 159.Zischewski J., Fischer R., Bortesi L. Detection of On-Target and off-Target Mutations Generated by CRISPR/Cas9 and Other Sequence-Specific Nucleases. Biotechnol. Adv. 2017;35:95–104. doi: 10.1016/j.biotechadv.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 160.Modrzejewski D., Hartung F., Sprink T., Krause D., Kohl C., Wilhelm R. What Is the Available Evidence for the Range of Applications of Genome-Editing as a New Tool for Plant Trait Modification and the Potential Occurrence of Associated off-Target Effects: A Systematic Map. Environ. Evid. 2019;8:27. doi: 10.1186/s13750-019-0171-5. [DOI] [Google Scholar]
- 161.Guo C., Ma X., Gao F., Guo Y. Off-Target Effects in CRISPR/Cas9 Gene Editing. Front. Bioeng. Biotechnol. 2023;11:1143157. doi: 10.3389/fbioe.2023.1143157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhang D., Zhang Z., Unver T., Zhang B. CRISPR/Cas: A Powerful Tool for Gene Function Study and Crop Improvement. J. Adv. Res. 2021;29:207–221. doi: 10.1016/j.jare.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang D., Liang X., Bao Y., Yang S., Zhang X., Yu H., Zhang Q., Xu G., Feng X., Dou D. A Malectin-like Receptor Kinase Regulates Cell Death and Pattern-triggered Immunity in Soybean. EMBO Rep. 2020;21:e50442. doi: 10.15252/embr.202050442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lin H., Wang M., Chen Y., Nomura K., Hui S., Gui J., Zhang X., Wu Y., Liu J., Li Q., et al. An MKP-MAPK Protein Phosphorylation Cascade Controls Vascular Immunity in Plants. Sci. Adv. 2022;8:eabg8723. doi: 10.1126/sciadv.abg8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ma H., Li J., Ma L., Wang P., Xue Y., Yin P., Xiao J., Wang S. Pathogen-Inducible OsMPKK10.2-OsMPK6 Cascade Phosphorylates the Raf-like Kinase OsEDR1 and Inhibits Its Scaffold Function to Promote Rice Disease Resistance. Mol. Plant. 2021;14:620–632. doi: 10.1016/j.molp.2021.01.008. [DOI] [PubMed] [Google Scholar]
- 166.Egger M., Smith D.G., Altman D.G. Systematic Reviews in Health: Meta-Analysis in Context. John Wiley & Sons; Hoboken, NJ, USA: 2001. [Google Scholar]
- 167.Higgins J.P., Savović J., Page M.J., Elbers R.G., Sterne J.A. Cochrane Handbook for Systematic Reviews of Interventions. John Wiley & Sons; Chichester, UK: 2019. Assessing Risk of Bias in a Randomized Trial; pp. 205–228. [DOI] [Google Scholar]
- 168.Comissão Técnica Nacional de Biossegurança Ministério da Ciência, Tecnologia e Inovação. (CTNBio). Tabela de Plantas—Uso Comercial: Plantas Geneticamente Modificadas Aprovadas Para Comercialização. [(accessed on 8 November 2023)];2018 Available online: http://ctnbio.mctic.gov.br/liberacao-comercial/-/document_library_display/SqhWdohU4BvU/view/1684467.
- 169.Hu H., Zhang Y., Yu F. A CRISPR/Cas9-based vector system enables the fast breeding of selection-marker-free canola with Rcr1-rendered clubroot resistance. J. Exp. Bot. 2023;75:1347–1363. doi: 10.1093/jxb/erad471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Xu G., Zhong X., Shi Y., Liu Z., Jiang N., Liu J., Ding B., Li Z., Kang H., Ning Y., et al. A Fungal Effector Targets a Heat Shock–Dynamin Protein Complex to Modulate Mitochondrial Dynamics and Reduce Plant Immunity. Sci. Adv. 2020;6:eabb7719. doi: 10.1126/sciadv.abb7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Xu S., Wei X., Yang Q., Hu D., Zhang Y., Yuan X., Kang F., Wu Z., Yan Z., Luo X., et al. A KNOX II Transcription Factor Suppresses the NLR Immune Receptor BRG8-Mediated Immunity in Rice. Plant Commun. 2024;5:101001. doi: 10.1016/j.xplc.2024.101001. [DOI] [PubMed] [Google Scholar]
- 172.Mizobuchi R., Sugimoto K., Tsushima S., Fukuoka S., Tsuiki C., Endo M., Mikami M., Saika H., Sato H. A MAPKKK Gene from Rice, RBG1res, Confers Resistance to Burkholderia glumae through Negative Regulation of ABA. Sci. Rep. 2023;13:3947. doi: 10.1038/s41598-023-30471-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang H., Bi Y., Yan Y., Yuan X., Gao Y., Noman M., Li D., Song F. A NAC Transcription Factor MNAC3-centered Regulatory Network Negatively Modulates Rice Immunity against Blast Disease. J. Integr. Plant Biol. 2024;66:2017–2041. doi: 10.1111/jipb.13727. [DOI] [PubMed] [Google Scholar]
- 174.Wang Z., Hardcastle T.J., Canto Pastor A., Yip W.H., Tang S., Baulcombe D.C. A Novel DCL2-Dependent miRNA Pathway in Tomato Affects Susceptibility to RNA Viruses. Genes Dev. 2018;32:1155–1160. doi: 10.1101/gad.313601.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Campo S., Sánchez-Sanuy F., Camargo-Ramírez R., Gómez-Ariza J., Baldrich P., Campos-Soriano L., Soto-Suárez M., San Segundo B. A Novel Transposable Element-derived microRNA Participates in Plant Immunity to Rice Blast Disease. Plant Biotechnol. J. 2021;19:1798–1811. doi: 10.1111/pbi.13592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wu D., Von Roepenack-Lahaye E., Buntru M., De Lange O., Schandry N., Pérez-Quintero A.L., Weinberg Z., Lowe-Power T.M., Szurek B., Michael A.J., et al. A Plant Pathogen Type III Effector Protein Subverts Translational Regulation to Boost Host Polyamine Levels. Cell Host Microbe. 2019;26:638–649.e5. doi: 10.1016/j.chom.2019.09.014. [DOI] [PubMed] [Google Scholar]
- 177.Chai L.-X., Dong K., Liu S.-Y., Zhang Z., Zhang X.-P., Tong X., Zhu F.-F., Zou J.-Z., Wang X.-B. A Putative Nuclear Copper Chaperone Promotes Plant Immunity in Arabidopsis. J. Exp. Bot. 2020;71:6684–6696. doi: 10.1093/jxb/eraa401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kumar A., Harloff H., Melzer S., Leineweber J., Defant B., Jung C. A Rhomboid-like Protease Gene from an Interspecies Translocation Confers Resistance to Cyst Nematodes. New Phytol. 2021;231:801–813. doi: 10.1111/nph.17394. [DOI] [PubMed] [Google Scholar]
- 179.Kim C., Park J., Choi G., Kim S., Vo K.T.X., Jeon J., Kang S., Lee Y. A Rice Gene Encoding Glycosyl Hydrolase Plays Contrasting Roles in Immunity Depending on the Type of Pathogens. Mol. Plant Pathol. 2022;23:400–416. doi: 10.1111/mpp.13167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mishra R., Mohanty J.N., Mahanty B., Joshi R.K. A Single Transcript CRISPR/Cas9 Mediated Mutagenesis of CaERF28 Confers Anthracnose Resistance in Chilli Pepper (Capsicum annuum L.) Planta. 2021;254:5. doi: 10.1007/s00425-021-03660-x. [DOI] [PubMed] [Google Scholar]
- 181.Liu S., Zhang X., Xiao S., Ma J., Shi W., Qin T., Xi H., Nie X., You C., Xu Z., et al. A Single-Nucleotide Mutation in a Glutamate Receptor-Like Gene Confers Resistance to Fusarium Wilt in Gossypium hirsutum. Adv. Sci. 2021;8:2002723. doi: 10.1002/advs.202002723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yu G., Zou J., Wang J., Zhu R., Qi Z., Jiang H., Hu Z., Yang M., Zhao Y., Wu X., et al. A Soybean NAC Homolog Contributes to Resistance to Phytophthora sojae Mediated by Dirigent Proteins. Crop J. 2022;10:332–341. doi: 10.1016/j.cj.2021.08.009. [DOI] [Google Scholar]
- 183.Sun Z., Zang Y., Zhou L., Song Y., Chen D., Zhang Q., Liu C., Yi Y., Zhu B., Fu D., et al. A Tomato Receptor-like Cytoplasmic Kinase, SlZRK1, Acts as a Negative Regulator in Wound-Induced Jasmonic Acid Accumulation and Insect Resistance. J. Exp. Bot. 2021;72:7285–7300. doi: 10.1093/jxb/erab350. [DOI] [PubMed] [Google Scholar]
- 184.Zhao S., Hong W., Wu J., Wang Y., Ji S., Zhu S., Wei C., Zhang J., Li Y. A Viral Protein Promotes Host SAMS1 Activity and Ethylene Production for the Benefit of Virus Infection. eLife. 2017;6:e27529. doi: 10.7554/eLife.27529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Li C., Wang K., Huang Y., Lei C., Cao S., Qiu L., Xu F., Jiang Y., Zou Y., Zheng Y. Activation of the BABA-induced Priming Defence through Redox Homeostasis and the Modules of TGA1 and MAPKK5 in Postharvest Peach Fruit. Mol. Plant Pathol. 2021;22:1624–1640. doi: 10.1111/mpp.13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Monino-Lopez D., Nijenhuis M., Kodde L., Kamoun S., Salehian H., Schentsnyi K., Stam R., Lokossou A., Abd-El-Haliem A., Visser R.G.F., et al. Allelic Variants of the NLR Protein Rpi-chc1 Differentially Recognize Members of the Phytophthora infestans PexRD12/31 Effector Superfamily through the Leucine-rich Repeat Domain. Plant J. 2021;107:182–197. doi: 10.1111/tpj.15284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Liu J., Chen X., Liang X., Zhou X., Yang F., Liu Jia He S.Y., Guo Z. Alternative splicing of rice WRKY62 and WRKY76 transcription factor genes in pathogen defense. Plant Physiol. 2016;171:1427–1442. doi: 10.1104/pp.15.01921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Guo X., Chen J., Gao M., Li D. An Aminobutyric Acid Transaminase in Zea mays Interacts with Rhizoctonia solani Cellulase to Participate in Disease Resistance. Front. Plant Sci. 2022;13:860170. doi: 10.3389/fpls.2022.860170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wang Z., Zhou L., Lan Y., Li X., Wang J., Dong J., Guo W., Jing D., Liu Q., Zhang S., et al. An Aspartic Protease 47 Causes Quantitative Recessive Resistance to Rice Black-streaked Dwarf Virus Disease and Southern Rice Black-streaked Dwarf Virus Disease. New Phytol. 2022;233:2520–2533. doi: 10.1111/nph.17961. [DOI] [PubMed] [Google Scholar]
- 190.Fu S., Wang K., Ma T., Liang Y., Ma Z., Wu J., Xu Y., Zhou X. An Evolutionarily Conserved C4HC3-Type E3 Ligase Regulates Plant Broad-Spectrum Resistance against Pathogens. Plant Cell. 2022;34:1822–1843. doi: 10.1093/plcell/koac055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Shen S., Peng M., Fang H., Wang Z., Zhou S., Jing X., Zhang M., Yang C., Guo H., Li Y., et al. An Oryza-Specific Hydroxycinnamoyl Tyramine Gene Cluster Contributes to Enhanced Disease Resistance. Sci. Bull. 2021;66:2369–2380. doi: 10.1016/j.scib.2021.03.015. [DOI] [PubMed] [Google Scholar]
- 192.Gao H., Yang M., Yang H., Qin Y., Zhu B., Xu G., Xie C., Wu D., Zhang X., Li W., et al. Arabidopsis ENOR3 Regulates RNAi-Mediated Antiviral Defense. J. Genet. Genom. 2018;45:33–40. doi: 10.1016/j.jgg.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 193.Atarashi H., Jayasinghe W.H., Kwon J., Kim H., Taninaka Y., Igarashi M., Ito K., Yamada T., Masuta C., Nakahara K.S. Artificially Edited Alleles of the Eukaryotic Translation Initiation Factor 4E1 Gene Differentially Reduce Susceptibility to Cucumber Mosaic Virus and Potato Virus Y in Tomato. Front. Microbiol. 2020;11:564310. doi: 10.3389/fmicb.2020.564310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ma Z., Qin G., Zhang Y., Liu C., Wei M., Cen Z., Yan Y., Luo T., Li Z., Liang H., et al. Bacterial Leaf Streak 1 Encoding a Mitogen-activated Protein Kinase Confers Resistance to Bacterial Leaf Streak in Rice. Plant J. 2021;107:1084–1101. doi: 10.1111/tpj.15368. [DOI] [PubMed] [Google Scholar]
- 195.Zhang K., Zhuo C., Wang Z., Liu F., Wen J., Yi B., Shen J., Ma C., Fu T., Tu J. BnaA03.MKK5-BnaA06.MPK3/BnaC03.MPK3 Module Positively Contributes to Sclerotinia sclerotiorum Resistance in Brassica napus. Plants. 2022;11:609. doi: 10.3390/plants11050609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Shi S., Wang H., Nie L., Tan D., Zhou C., Zhang Q., Li Y., Du B., Guo J., Huang J., et al. Bph30 Confers Resistance to Brown Planthopper by Fortifying Sclerenchyma in Rice Leaf Sheaths. Mol. Plant. 2021;14:1714–1732. doi: 10.1016/j.molp.2021.07.004. [DOI] [PubMed] [Google Scholar]
- 197.Shi H., Li Q., Luo M., Yan H., Xie B., Li X., Zhong G., Chen D., Tang D. BRASSINOSTEROID-SIGNALING KINASE1 Modulates MAP KINASE15 Phosphorylation to Confer Powdery Mildew Resistance in Arabidopsis. Plant Cell. 2022;34:1768–1783. doi: 10.1093/plcell/koac027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Oliva R., Ji C., Atienza-Grande G., Huguet-Tapia J.C., Perez-Quintero A., Li T., Eom J.-S., Li C., Nguyen H., Liu B., et al. Broad-Spectrum Resistance to Bacterial Blight in Rice Using Genome Editing. Nat. Biotechnol. 2019;37:1344–1350. doi: 10.1038/s41587-019-0267-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Gao M., He Y., Yin X., Zhong X., Yan B., Wu Y., Chen J., Li X., Zhai K., Huang Y., et al. Ca2+ Sensor-Mediated ROS Scavenging Suppresses Rice Immunity and Is Exploited by a Fungal Effector. Cell. 2021;184:5391–5404.e17. doi: 10.1016/j.cell.2021.09.009. [DOI] [PubMed] [Google Scholar]
- 200.Zhao Q., Liu R., Zhou Q., Ye J., Meng F., Liu J., Yang C. Calcium-Binding Protein OsANN1 Regulates Rice Blast Disease Resistance by Inactivating Jasmonic Acid Signaling. Plant Physiol. 2023;192:1621–1637. doi: 10.1093/plphys/kiad174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Liu H., Li J., Wang S., Hua J., Zou B. CHROMATIN REMODELING 11-Dependent Nucleosome Occupancy Affects Disease Resistance in Rice. Plant Physiol. 2023;193:1635–1651. doi: 10.1093/plphys/kiad381. [DOI] [PubMed] [Google Scholar]
- 202.Wang L., Ma Z., Kang H., Gu S., Mukhina Z., Wang C., Wang H., Bai Y., Sui G., Zheng W., et al. Cloning and Functional Analysis of the Novel Rice Blast Resistance Gene Pi65 in Japonica Rice. Theor. Appl. Genet. 2022;135:173–183. doi: 10.1007/s00122-021-03957-1. [DOI] [PubMed] [Google Scholar]
- 203.Peng S., Guo D., Guo Y., Zhao H., Mei J., Han Y., Guan R., Wang T., Song T., Sun K., et al. CONSTITUTIVE EXPRESSER OF PATHOGENESIS-RELATED GENES 5 Is an RNA-Binding Protein Controlling Plant Immunity via an RNA Processing Complex. Plant Cell. 2022;34:1724–1744. doi: 10.1093/plcell/koac037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Sun L., Alariqi M., Wang Y., Wang Q., Xu Z., Zafar M.N., Yang G., Jia R., Hussain A., Chen Y., et al. Construction of Host Plant Insect-Resistance Mutant Library by High-Throughput CRISPR/Cas9 System and Identification of A Broad-Spectrum Insect Resistance Gene. Adv. Sci. 2024;11:2306157. doi: 10.1002/advs.202306157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ichimaru K., Yamaguchi K., Harada K., Nishio Y., Hori M., Ishikawa K., Inoue H., Shigeta S., Inoue K., Shimada K., et al. Cooperative Regulation of PBI1 and MAPKs Controls WRKY45 Transcription Factor in Rice Immunity. Nat. Commun. 2022;13:2397. doi: 10.1038/s41467-022-30131-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Norouzi M., Nazarain-Firouzabadi F., Ismaili A., Ahmadvand R., Poormazaheri H. CRISPR/Cas StNRL1 Gene Knockout Increases Resistance to Late Blight and Susceptibility to Early Blight in Potato. Front. Plant Sci. 2024;14:1278127. doi: 10.3389/fpls.2023.1278127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Tripathi J.N., Ntui V.O., Ron M., Muiruri S.K., Britt A., Tripathi L. CRISPR/Cas9 Editing of Endogenous Banana Streak Virus in the B Genome of Musa Spp. Overcomes a Major Challenge in Banana Breeding. Commun. Biol. 2019;2:46. doi: 10.1038/s42003-019-0288-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Olivares F., Loyola R., Olmedo B., Miccono M.D.L.Á., Aguirre C., Vergara R., Riquelme D., Madrid G., Plantat P., Mora R., et al. CRISPR/Cas9 Targeted Editing of Genes Associated with Fungal Susceptibility in Vitis vinifera L. Cv. Thompson Seedless Using Geminivirus-Derived Replicons. Front. Plant Sci. 2021;12:791030. doi: 10.3389/fpls.2021.791030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Elliott K., Veley K.M., Jensen G., Gilbert K.B., Norton J., Kambic L., Yoder M., Weil A., Motomura-Wages S., Bart R.S. CRISPR/Cas9-Generated Mutations in a Sugar Transporter Gene Reduce Cassava Susceptibility to Bacterial Blight. Plant Physiol. 2024;195:2566–2578. doi: 10.1093/plphys/kiae243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Dutta T.K., Rupinikrishna K., Akhil V.S., Vashisth N., Phani V., Pankaj, Sirohi A., Chinnusamy V. CRISPR/Cas9-Induced Knockout of an Amino Acid Permease Gene (AAP6) Reduced Arabidopsis thaliana Susceptibility to Meloidogyne incognita. BMC Plant Biol. 2024;24:515. doi: 10.1186/s12870-024-05175-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wang L., Chen S., Peng A., Xie Z., He Y., Zou X. CRISPR/Cas9-Mediated Editing of CsWRKY22 Reduces Susceptibility to Xanthomonas citri subsp. Citri in Wanjincheng Orange (Citrus sinensis (L.) Osbeck) Plant Biotechnol. Rep. 2019;13:501–510. doi: 10.1007/s11816-019-00556-x. [DOI] [Google Scholar]
- 212.Pramanik D., Shelake R.M., Park J., Kim M.J., Hwang I., Park Y., Kim J.-Y. CRISPR/Cas9-Mediated Generation of Pathogen-Resistant Tomato against Tomato Yellow Leaf Curl Virus and Powdery Mildew. Int. J. Mol. Sci. 2021;22:1878. doi: 10.3390/ijms22041878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Xiao Z., Yang W., Yang A., Deng L., Geng R., Xiang H., Kong W., Jiang C., Li X., Chen Z., et al. CRISPR/Cas9-Mediated Knockout of NtMYC2a Gene Involved in Resistance to Bacterial Wilt in Tobacco. Gene. 2024;927:148622. doi: 10.1016/j.gene.2024.148622. [DOI] [PubMed] [Google Scholar]
- 214.Sun Q., Lin L., Liu D., Wu D., Fang Y., Wu J., Wang Y. CRISPR/Cas9-Mediated Multiplex Genome Editing of the BnWRKY11 and BnWRKY70 Genes in Brassica napus L. Int. J. Mol. Sci. 2018;19:2716. doi: 10.3390/ijms19092716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Zhang M., Liu Q., Yang X., Xu J., Liu G., Yao X., Ren R., Xu J., Lou L. CRISPR/Cas9-Mediated Mutagenesis of Clpsk1 in Watermelon to Confer Resistance to Fusarium oxysporum f.Sp. niveum. Plant Cell Rep. 2020;39:589–595. doi: 10.1007/s00299-020-02516-0. [DOI] [PubMed] [Google Scholar]
- 216.Zhou H., Bai S., Wang N., Sun X., Zhang Y., Zhu J., Dong C. CRISPR/Cas9-Mediated Mutagenesis of MdCNGC2 in Apple Callus and VIGS-Mediated Silencing of MdCNGC2 in Fruits Improve Resistance to Botryosphaeria dothidea. Front. Plant Sci. 2020;11:575477. doi: 10.3389/fpls.2020.575477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Hasley J.A.R., Navet N., Tian M. CRISPR/Cas9-Mediated Mutagenesis of Sweet Basil Candidate Susceptibility Gene ObDMR6 Enhances Downy Mildew Resistance. PLoS ONE. 2021;16:e0253245. doi: 10.1371/journal.pone.0253245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Huang Q., Lin B., Cao Y., Zhang Y., Song H., Huang C., Sun T., Long C., Liao J., Zhuo K. CRISPR/Cas9-Mediated Mutagenesis of the Susceptibility Gene OsHPP04 in Rice Confers Enhanced Resistance to Rice Root-Knot Nematode. Front. Plant Sci. 2023;14:1134653. doi: 10.3389/fpls.2023.1134653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Wan D.-Y., Guo Y., Cheng Y., Hu Y., Xiao S., Wang Y., Wen Y.-Q. CRISPR/Cas9-Mediated Mutagenesis of VvMLO3 Results in Enhanced Resistance to Powdery Mildew in Grapevine (Vitis vinifera) Hortic. Res. 2020;7:116. doi: 10.1038/s41438-020-0339-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Debbarma J., Saikia B., Singha D., Das D., Keot A., Maharana J., Velmurugan N., Arunkumar K., Reddy P., Chikkaputtaiah C. CRISPR/Cas9-Mediated Mutation in XSP10 and SlSAMT Genes Impart Genetic Tolerance to Fusarium Wilt Disease of Tomato (Solanum lycopersicum L.) Genes. 2023;14:488. doi: 10.3390/genes14020488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Perk E.A., Arruebarrena Di Palma A., Colman S., Mariani O., Cerrudo I., D’Ambrosio J.M., Robuschi L., Pombo M.A., Rosli H.G., Villareal F., et al. CRISPR/Cas9-Mediated Phospholipase C 2 Knock-out Tomato Plants Are More Resistant to Botrytis Cinerea. Planta. 2023;257:117. doi: 10.1007/s00425-023-04147-7. [DOI] [PubMed] [Google Scholar]
- 222.Miyoshi S., Unung O.O., Kaya H., Yaeno T., Kobayashi K. CRISPR/Cas9-Mediated Resurrection of Tobacco NB-LRR Class Virus Resistance Gene from a Susceptible Allele with Partial Duplication. J. Gen. Plant Pathol. 2024:1–11. doi: 10.1007/s10327-024-01189-x. [DOI] [Google Scholar]
- 223.Wang X., Li D., Tan X., Cai C., Zhang X., Shen Z., Yang A., Fu X., Liu D. CRISPR/Cas9-Mediated Targeted Mutagenesis of Two Homoeoalleles in Tobacco Confers Resistance to Powdery Mildew. Euphytica. 2022;219:67. doi: 10.21203/rs.3.rs-1892545/v1. [DOI] [Google Scholar]
- 224.Li M.-Y., Jiao Y.-T., Wang Y.-T., Zhang N., Wang B.-B., Liu R.-Q., Yin X., Xu Y., Liu G.-T. CRISPR/Cas9-Mediated VvPR4b Editing Decreases Downy Mildew Resistance in Grapevine (Vitis vinifera L.) Hortic. Res. 2020;7:149. doi: 10.1038/s41438-020-00371-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Santillán Martínez M.I., Bracuto V., Koseoglou E., Appiano M., Jacobsen E., Visser R.G.F., Wolters A.-M.A., Bai Y. CRISPR/Cas9-Targeted Mutagenesis of the Tomato Susceptibility Gene PMR4 for Resistance against Powdery Mildew. BMC Plant Biol. 2020;20:284. doi: 10.1186/s12870-020-02497-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.García-Murillo L., Valencia-Lozano E., Priego-Ranero N.A., Cabrera-Ponce J.L., Duarte-Aké F.P., Vizuet-de-Rueda J.C., Rivera-Toro D.M., Herrera-Ubaldo H., De Folter S., Alvarez-Venegas R. CRISPRa-Mediated Transcriptional Activation of the SlPR-1 Gene in Edited Tomato Plants. Plant Sci. 2023;329:111617. doi: 10.1016/j.plantsci.2023.111617. [DOI] [PubMed] [Google Scholar]
- 227.Mubarik M.S., Khan S.H., Sadia B., Ahmad A. CRISPR-Cas9 based suppression of cotton leaf curl virus in Nicotiana benthamina. Int. J. Agric. Biol. 2019;22:517–522. [Google Scholar]
- 228.Zhang X., Low Y.C., Lawton M.A., Simon J.E., Di R. CRISPR-Editing of Sweet Basil (Ocimum basilicum L.) Homoserine Kinase Gene for Improved Downy Mildew Disease Resistance. Front. Genome Ed. 2021;3:629769. doi: 10.3389/fgeed.2021.629769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Liu S., Zhang F., Su J., Fang A., Tian B., Yu Y., Bi C., Ma D., Xiao S., Yang Y. CRISPR-targeted Mutagenesis of Mitogen-activated Protein Kinase Phosphatase 1 Improves Both Immunity and Yield in Wheat. Plant Biotechnol. J. 2024;22:1929–1941. doi: 10.1111/pbi.14312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Dong S., Liu X., Han J., Miao H., Beckles D.M., Bai Y., Liu X., Guan J., Yang R., Gu X., et al. CsMLO8/11 Are Required for Full Susceptibility of Cucumber Stem to Powdery Mildew and Interact with CsCRK2 and CsRbohD. Hortic. Res. 2024;11:uhad295. doi: 10.1093/hr/uhad295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Zhao Y., Hu K., Yao G., Wang S., Peng X., Zhang C., Zeng D., Zong K., Lyu Y., Zhang H. D-Cysteine Desulfhydrase DCD1 Participates in Tomato Resistance against Botrytis cinerea by Modulating ROS Homeostasis. Veg. Res. 2023;3:21. doi: 10.48130/VR-2023-0021. [DOI] [Google Scholar]
- 232.Jiang W., Zhou H., Bi H., Fromm M., Yang B., Weeks D.P. Demonstration of CRISPR/Cas9/sgRNA-Mediated Targeted Gene Modification in Arabidopsis, Tobacco, Sorghum and Rice. Nucleic Acids Res. 2013;41:e188. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ortigosa A., Gimenez-Ibanez S., Leonhardt N., Solano R. Design of a Bacterial Speck Resistant Tomato by CRISPR/Cas9-mediated Editing of SlJAZ 2. Plant Biotechnol. J. 2019;17:665–673. doi: 10.1111/pbi.13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Chandrasekaran J., Brumin M., Wolf D., Leibman D., Klap C., Pearlsman M., Sherman A., Arazi T., Gal-On A. Development of Broad Virus Resistance in Non-transgenic Cucumber Using CRISPR/Cas9 Technology. Mol. Plant Pathol. 2016;17:1140–1153. doi: 10.1111/mpp.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Yıldırım K., Kavas M., Küçük İ.S., Seçgin Z., Saraç Ç.G. Development of Highly Efficient Resistance to Beet Curly Top Iran Virus (Becurtovirus) in Sugar Beet (B. Vulgaris) via CRISPR/Cas9 System. Int. J. Mol. Sci. 2023;24:6515. doi: 10.3390/ijms24076515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Zhang H., Wang F., Song W., Yang Z., Li L., Ma Q., Tan X., Wei Z., Li Y., Li J., et al. Different Viral Effectors Suppress Hormone-Mediated Antiviral Immunity of Rice Coordinated by OsNPR1. Nat. Commun. 2023;14:3011. doi: 10.1038/s41467-023-38805-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Ding Y., Dommel M.R., Wang C., Li Q., Zhao Q., Zhang X., Dai S., Mou Z. Differential Quantitative Requirements for NPR1 between Basal Immunity and Systemic Acquired Resistance in Arabidopsis thaliana. Front. Plant Sci. 2020;11:570422. doi: 10.3389/fpls.2020.570422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Liao Y., Ali A., Xue Z., Zhou X., Ye W., Guo D., Liao Y., Jiang P., Wu T., Zhang H., et al. Disruption of LLM9428/OsCATC Represses Starch Metabolism and Confers Enhanced Blast Resistance in Rice. Int. J. Mol. Sci. 2022;23:3827. doi: 10.3390/ijms23073827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ma J., Chen J., Wang M., Ren Y., Wang S., Lei C., Cheng Z., Sodmergen Disruption of OsSEC3A Increases the Content of Salicylic Acid and Induces Plant Defense Responses in Rice. J. Exp. Bot. 2018;69:1051–1064. doi: 10.1093/jxb/erx458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zhang Y., Yu Q., Gao S., Yu N., Zhao L., Wang J., Zhao J., Huang P., Yao L., Wang M., et al. Disruption of the Primary Salicylic Acid Hydroxylases in Rice Enhances Broad-spectrum Resistance against Pathogens. Plant Cell Environ. 2022;45:2211–2225. doi: 10.1111/pce.14328. [DOI] [PubMed] [Google Scholar]
- 241.Li R., Zhang J., Li Z., Peters R.J., Yang B. Dissecting the Labdane-related Diterpenoid Biosynthetic Gene Clusters in Rice Reveals Directional Cross-cluster Phytotoxicity. New Phytol. 2022;233:878–889. doi: 10.1111/nph.17806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Zhang H., Li L., He Y., Qin Q., Chen C., Wei Z., Tan X., Xie K., Zhang R., Hong G., et al. Distinct Modes of Manipulation of Rice Auxin Response Factor OsARF17 by Different Plant RNA Viruses for Infection. Proc. Natl. Acad. Sci. USA. 2020;117:9112–9121. doi: 10.1073/pnas.1918254117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Gao G., Zhou L., Liu J., Wang P., Gong P., Tian S., Qin G., Wang W., Wang Y. E3 Ligase SlCOP1-1 Stabilizes Transcription Factor SlOpaque2 and Enhances Fruit Resistance to Botrytis cinerea in Tomato. Plant Physiol. 2024;196:1196–1213. doi: 10.1093/plphys/kiae404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Zhang X., Cheng J., Lin Y., Fu Y., Xie J., Li B., Bian X., Feng Y., Liang W., Tang Q., et al. Editing Homologous Copies of an Essential Gene Affords Crop Resistance against Two Cosmopolitan Necrotrophic Pathogens. Plant Biotechnol. J. 2021;19:2349–2361. doi: 10.1111/pbi.13667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Hong Y., Meng J., He X., Zhang Y., Liu Y., Zhang C., Qi H., Luan Y. Editing miR482b and miR482c Simultaneously by CRISPR/Cas9 Enhanced Tomato Resistance to Phytophthora infestans. Phytopathology®. 2021;111:1008–1016. doi: 10.1094/PHYTO-08-20-0360-R. [DOI] [PubMed] [Google Scholar]
- 246.Navet N., Tian M. Efficient Targeted Mutagenesis in Allotetraploid Sweet Basil by CRISPR/Cas9. Plant Direct. 2020;4:e00233. doi: 10.1002/pld3.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Mubarik M.S., Wang X., Khan S.H., Ahmad A., Khan Z., Amjid M.W., Razzaq M.K., Ali Z., Azhar M.T. Engineering Broad-Spectrum Resistance to Cotton Leaf Curl Disease by CRISPR-Cas9 Based Multiplex Editing in Plants. GM Crops Food. 2021;12:647–658. doi: 10.1080/21645698.2021.1938488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Peng A., Chen S., Lei T., Xu L., He Y., Wu L., Yao L., Zou X. Engineering Canker-resistant Plants through CRISPR/Cas9-targeted Editing of the Susceptibility Gene CsLOB1 Promoter in Citrus. Plant Biotechnol. J. 2017;15:1509–1519. doi: 10.1111/pbi.12733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Pyott D.E., Sheehan E., Molnar A. Engineering of CRISPR/Cas9-mediated Potyvirus Resistance in Transgene-free Arabidopsis Plants. Mol. Plant Pathol. 2016;17:1276–1288. doi: 10.1111/mpp.12417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Tashkandi M., Ali Z., Aljedaani F., Shami A., Mahfouz M.M. Engineering Resistance against Tomato Yellow Leaf Curl Virus via the CRISPR/Cas9 System in Tomato. Plant Signal. Behav. 2018;13:e1525996. doi: 10.1080/15592324.2018.1525996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Pathi K.M., Rink P., Budhagatapalli N., Betz R., Saado I., Hiekel S., Becker M., Djamei A., Kumlehn J. Engineering Smut Resistance in Maize by Site-Directed Mutagenesis of LIPOXYGENASE 3. Front. Plant Sci. 2020;11:543895. doi: 10.3389/fpls.2020.543895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Wang F., Wang C., Liu P., Lei C., Hao W., Gao Y., Liu Y.-G., Zhao K. Enhanced Rice Blast Resistance by CRISPR/Cas9-Targeted Mutagenesis of the ERF Transcription Factor Gene OsERF922. PLoS ONE. 2016;11:e0154027. doi: 10.1371/journal.pone.0154027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Bui T.P., Le H., Ta D.T., Nguyen C.X., Le N.T., Tran T.T., Van Nguyen P., Stacey G., Stacey M.G., Pham N.B., et al. Enhancing Powdery Mildew Resistance in Soybean by Targeted Mutation of MLO Genes Using the CRISPR/Cas9 System. BMC Plant Biol. 2023;23:533. doi: 10.1186/s12870-023-04549-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Kourelis J., Malik S., Mattinson O., Krauter S., Kahlon P.S., Paulus J.K., Van Der Hoorn R.A.L. Evolution of a Guarded Decoy Protease and Its Receptor in Solanaceous Plants. Nat. Commun. 2020;11:4393. doi: 10.1038/s41467-020-18069-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Jiang T., Jiao T., Hu Y., Li T., Liu C., Liu Y., Jiang X., Xia T., Gao L.-P. Evolutionarily Conserved 12-Oxophytodienoate Reductase Trans-lncRNA Pair Affects Disease Resistance in Tea (Camellia sinensis) via the Jasmonic Acid Signaling Pathway. Hortic. Res. 2024;11:uhae129. doi: 10.1093/hr/uhae129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Ji H., Mao H., Li S., Feng T., Zhang Z., Cheng L., Luo S., Borkovich K.A., Ouyang S. Fol-milR1, a Pathogenicity Factor of Fusarium oxysporum, Confers Tomato Wilt Disease Resistance by Impairing Host Immune Responses. New Phytol. 2021;232:705–718. doi: 10.1111/nph.17436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Lüdke D., Roth C., Kamrad S.A., Messerschmidt J., Hartken D., Appel J., Hörnich B.F., Yan Q., Kusch S., Klenke M., et al. Functional Requirement of the Arabidopsis Importin-α Nuclear Transport Receptor Family in Autoimmunity Mediated by the NLR Protein SNC1. Plant J. 2021;105:994–1009. doi: 10.1111/tpj.15082. [DOI] [PubMed] [Google Scholar]
- 258.Kumar N., Galli M., Ordon J., Stuttmann J., Kogel K., Imani J. Further Analysis of Barley MORC 1 Using a Highly Efficient RNA-guided Cas9 Gene-editing System. Plant Biotechnol. J. 2018;16:1892–1903. doi: 10.1111/pbi.12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Zhao N., Guo A., Wang W., Li B., Wang M., Zhou Z., Jiang K., Aierxi A., Wang B., Adjibolosoo D., et al. GbPP2C80 Interacts with GbWAKL14 to Negatively Co-Regulate Resistance to Fusarium and Verticillium wilt via MPK3 and ROS Signaling in Sea Island Cotton. Adv. Sci. 2024;11:2309785. doi: 10.1002/advs.202309785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Zhou X., Jiang G., Yang L., Qiu L., He P., Nong C., Wang Y., He Y., Xing Y. Gene Diagnosis and Targeted Breeding for Blast-Resistant Kongyu 131 without Changing Regional Adaptability. J. Genet. Genom. 2018;45:539–547. doi: 10.1016/j.jgg.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 261.Veley K.M., Okwuonu I., Jensen G., Yoder M., Taylor N.J., Meyers B.C., Bart R.S. Gene Tagging via CRISPR-Mediated Homology-Directed Repair in Cassava. G3. 2021;11:jkab028. doi: 10.1093/g3journal/jkab028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Zhou J., Peng Z., Long J., Sosso D., Liu B., Eom J., Huang S., Liu S., Vera Cruz C., Frommer W.B., et al. Gene Targeting by the TAL Effector PthXo2 Reveals Cryptic Resistance Gene for Bacterial Blight of Rice. Plant J. 2015;82:632–643. doi: 10.1111/tpj.12838. [DOI] [PubMed] [Google Scholar]
- 263.Ma J., Yang S., Wang D., Tang K., Feng X.X., Feng X.Z. Genetic Mapping of a Light-Dependent Lesion Mimic Mutant Reveals the Function of Coproporphyrinogen III Oxidase Homolog in Soybean. Front. Plant Sci. 2020;11:557. doi: 10.3389/fpls.2020.00557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Brauer E.K., Balcerzak M., Rocheleau H., Leung W., Schernthaner J., Subramaniam R., Ouellet T. Genome Editing of a Deoxynivalenol-Induced Transcription Factor Confers Resistance to Fusarium graminearum in Wheat. Mol. Plant-Microbe Interact. 2020;33:553–560. doi: 10.1094/MPMI-11-19-0332-R. [DOI] [PubMed] [Google Scholar]
- 265.Yoon Y.-J., Venkatesh J., Lee J.-H., Kim J., Lee H.-E., Kim D.-S., Kang B.-C. Genome Editing of eIF4E1 in Tomato Confers Resistance to Pepper Mottle Virus. Front. Plant Sci. 2020;11:1098. doi: 10.3389/fpls.2020.01098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Jia H., Zhang Y., Orbović V., Xu J., White F.F., Jones J.B., Wang N. Genome Editing of the Disease Susceptibility Gene CsLOB1 in Citrus Confers Resistance to Citrus Canker. Plant Biotechnol. J. 2017;15:817–823. doi: 10.1111/pbi.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Li S., Lin D., Zhang Y., Deng M., Chen Y., Lv B., Li B., Lei Y., Wang Y., Zhao L., et al. Genome-Edited Powdery Mildew Resistance in Wheat without Growth Penalties. Nature. 2022;602:455–460. doi: 10.1038/s41586-022-04395-9. [DOI] [PubMed] [Google Scholar]
- 268.Liu M., Kang H., Xu Y., Peng Y., Wang D., Gao L., Wang X., Ning Y., Wu J., Liu W., et al. Genome-wide Association Study Identifies an NLR Gene That Confers Partial Resistance to Magnaporthe oryzae in Rice. Plant Biotechnol. J. 2020;18:1376–1383. doi: 10.1111/pbi.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Laura M., Forti C., Barberini S., Ciorba R., Mascarello C., Giovannini A., Pistelli L., Pieracci Y., Lanteri A.P., Ronca A., et al. Highly Efficient CRISPR/Cas9 Mediated Gene Editing in Ocimum basilicum ‘FT Italiko’ to Induce Resistance to Peronospora belbahrii. Plants. 2023;12:2395. doi: 10.3390/plants12132395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Huang Y.-Y., Liu X.-X., Xie Y., Lin X.-Y., Hu Z.-J., Wang H., Wang L.-F., Dang W.-Q., Zhang L.-L., Zhu Y., et al. Identification of FERONIA-like Receptor Genes Involved in Rice-Magnaporthe oryzae Interaction. Phytopathol. Res. 2020;2:14. doi: 10.1186/s42483-020-00052-z. [DOI] [Google Scholar]
- 271.Dong O.X., Ao K., Xu F., Johnson K.C.M., Wu Y., Li L., Xia S., Liu Y., Huang Y., Rodriguez E., et al. Individual Components of Paired Typical NLR Immune Receptors Are Regulated by Distinct E3 Ligases. Nat. Plants. 2018;4:699–710. doi: 10.1038/s41477-018-0216-8. [DOI] [PubMed] [Google Scholar]
- 272.Wang J., Tian D., Gu K., Yang X., Wang L., Zeng X., Yin Z. Induction of Xa10-like Genes in Rice Cultivar Nipponbare Confers Disease Resistance to Rice Bacterial Blight. Mol. Plant-Microbe Interact. 2017;30:466–477. doi: 10.1094/MPMI-11-16-0229-R. [DOI] [PubMed] [Google Scholar]
- 273.Yang Z., Huang Y., Yang J., Yao S., Zhao K., Wang D., Qin Q., Bian Z., Li Y., Lan Y., et al. Jasmonate Signaling Enhances RNA Silencing and Antiviral Defense in Rice. Cell Host Microbe. 2020;28:89–103.e8. doi: 10.1016/j.chom.2020.05.001. [DOI] [PubMed] [Google Scholar]
- 274.Yang W., Liu C., Fu Q., Jia X., Deng L., Feng C., Wang Y., Yang Z., Yang H., Xu X. Knockout of SlbZIP68 Reduces Late Blight Resistance in Tomato. Plant Sci. 2023;336:111861. doi: 10.1016/j.plantsci.2023.111861. [DOI] [PubMed] [Google Scholar]
- 275.Cao Y., Yan X., Ran S., Ralph J., Smith R.A., Chen X., Qu C., Li J., Liu L. Knockout of the Lignin Pathway Gene BnF5H Decreases the S/G Lignin Compositional Ratio and Improves Sclerotinia sclerotiorum Resistance in Brassica napus. Plant Cell Environ. 2022;45:248–261. doi: 10.1111/pce.14208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Zhou H., Liu B., Weeks D.P., Spalding M.H., Yang B. Large Chromosomal Deletions and Heritable Small Genetic Changes Induced by CRISPR/Cas9 in Rice. Nucleic Acids Res. 2014;42:10903–10914. doi: 10.1093/nar/gku806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Wang L., Zhao L., Zhang X., Zhang Q., Jia Y., Wang G., Li S., Tian D., Li W.-H., Yang S. Large-Scale Identification and Functional Analysis of NLR Genes in Blast Resistance in the Tetep Rice Genome Sequence. Proc. Natl. Acad. Sci. USA. 2019;116:18479–18487. doi: 10.1073/pnas.1910229116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Li R., Cui L., Martina M., Bracuto V., Meijer-Dekens F., Wolters A.-M.A., Moglia A., Bai Y., Acquadro A. Less Is More: CRISPR/Cas9-Based Mutations in DND1 Gene Enhance Tomato Resistance to Powdery Mildew with Low Fitness Costs. BMC Plant Biol. 2024;24:763. doi: 10.1186/s12870-024-05428-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Qin P., Fan S., Deng L., Zhong G., Zhang S., Li M., Chen W., Wang G., Tu B., Wang Y., et al. LML1, Encoding a Conserved Eukaryotic Release Factor 1 Protein, Regulates Cell Death and Pathogen Resistance by Forming a Conserved Complex with SPL33 in Rice. Plant Cell Physiol. 2018;59:887–902. doi: 10.1093/pcp/pcy056. [DOI] [PubMed] [Google Scholar]
- 280.Pröbsting M., Schenke D., Hossain R., Häder C., Thurau T., Wighardt L., Schuster A., Zhou Z., Ye W., Rietz S., et al. Loss of Function of CRT1a (Calreticulin) Reduces Plant Susceptibility to Verticillium longisporum in Both Arabidopsis thaliana and Oilseed Rape (Brassica napus) Plant Biotechnol. J. 2020;18:2328–2344. doi: 10.1111/pbi.13394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Ramos R.N., Zhang N., Lauff D.B., Valenzuela-Riffo F., Figueroa C.R., Martin G.B., Pombo M.A., Rosli H.G. Loss-of-Function Mutations in WRKY22 and WRKY25 Impair Stomatal-Mediated Immunity and PTI and ETI Responses against Pseudomonas syringae pv. Tomato. Plant Mol. Biol. 2023;112:161–177. doi: 10.1007/s11103-023-01358-0. [DOI] [PubMed] [Google Scholar]
- 282.Yang T., Song L., Hu J., Qiao L., Yu Q., Wang Z., Chen X., Lu G. Magnaporthe oryzae Effector AvrPik-D Targets a Transcription Factor WG7 to Suppress Rice Immunity. Rice. 2024;17:14. doi: 10.1186/s12284-024-00693-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Meng G., Xiao Y., Li A., Qian Z., Xie Y., Yang L., Lin H., Yang W. Mapping and Characterization of the Rx3 Gene for Resistance to Xanthomonas euvesicatoria pv. euvesicatoria Race T1 in Tomato. Theor. Appl. Genet. 2022;135:1637–1656. doi: 10.1007/s00122-022-04059-2. [DOI] [PubMed] [Google Scholar]
- 284.Wang N., Liu Y., Dong C., Zhang Y., Bai S. MdMAPKKK1 Regulates Apple Resistance to Botryosphaeria dothidea by Interacting with MdBSK1. Int. J. Mol. Sci. 2022;23:4415. doi: 10.3390/ijms23084415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Bastet A., Zafirov D., Giovinazzo N., Guyon-Debast A., Nogué F., Robaglia C., Gallois J. Mimicking Natural Polymorphism in eIF4E by CRISPR-Cas9 Base Editing Is Associated with Resistance to Potyviruses. Plant Biotechnol. J. 2019;17:1736–1750. doi: 10.1111/pbi.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Zhu X., Kuang Y., Chen Y., Shi J., Cao Y., Hu J., Yu C., Yang F., Tian F., Chen H. miR2118 Negatively Regulates Bacterial Blight Resistance through Targeting Several Disease Resistance Genes in Rice. Plants. 2023;12:3815. doi: 10.3390/plants12223815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Yang Z., Hui S., Lv Y., Zhang M., Chen D., Tian J., Zhang H., Liu H., Cao J., Xie W., et al. miR395-Regulated Sulfate Metabolism Exploits Pathogen Sensitivity to Sulfate to Boost Immunity in Rice. Mol. Plant. 2022;15:671–688. doi: 10.1016/j.molp.2021.12.013. [DOI] [PubMed] [Google Scholar]
- 288.Huang X., Zhu G., Liu Q., Chen L., Li Y., Hou B. Modulation of Plant Salicylic Acid-Associated Immune Responses via Glycosylation of Dihydroxybenzoic Acids. Plant Physiol. 2018;176:3103–3119. doi: 10.1104/pp.17.01530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Roberts R., Liu A.E., Wan L., Geiger A.M., Hind S.R., Rosli H.G., Martin G.B. Molecular Characterization of Differences between the Tomato Immune Receptors Flagellin Sensing 3 and Flagellin Sensing 2. Plant Physiol. 2020;183:1825–1837. doi: 10.1104/pp.20.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Xu J., Wang X., Zu H., Zeng X., Baldwin I.T., Lou Y., Li R. Molecular Dissection of Rice Phytohormone Signaling Involved in Resistance to a Piercing-sucking Herbivore. New Phytol. 2021;230:1639–1652. doi: 10.1111/nph.17251. [DOI] [PubMed] [Google Scholar]
- 291.Zhou X., Zhong T., Wu M., Li Q., Yu W., Gan L., Xiang X., Zhang Y., Shi Y., Zhou Y., et al. Multiomics Analysis of a Resistant European Turnip ECD04 during Clubroot Infection Reveals Key Hub Genes Underlying Resistance Mechanism. Front. Plant Sci. 2024;15:1396602. doi: 10.3389/fpls.2024.1396602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Zhang P., Du H., Wang J., Pu Y., Yang C., Yan R., Yang H., Cheng H., Yu D. Multiplex CRISPR/Cas9-mediated Metabolic Engineering Increases Soya Bean Isoflavone Content and Resistance to Soya Bean Mosaic Virus. Plant Biotechnol. J. 2020;18:1384–1395. doi: 10.1111/pbi.13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Nizan S., Amitzur A., Dahan-Meir T., Benichou J.I.C., Bar-Ziv A., Perl-Treves R. Mutagenesis of the Melon Prv Gene by CRISPR/Cas9 Breaks Papaya Ringspot Virus Resistance and Generates an Autoimmune Allele with Constitutive Defense Responses. J. Exp. Bot. 2023;74:4579–4596. doi: 10.1093/jxb/erad156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Liao Y., Bai Q., Xu P., Wu T., Guo D., Peng Y., Zhang H., Deng X., Chen X., Luo M., et al. Mutation in Rice Abscisic Acid2 Results in Cell Death, Enhanced Disease-Resistance, Altered Seed Dormancy and Development. Front. Plant Sci. 2018;9:405. doi: 10.3389/fpls.2018.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Ijaz S., Haq I.U., Razzaq H.A. Mutation Introduced in DDTFR10/A Gene of Ethylene Response Element-Binding Protein (EREBP) Family through CRISPR/Cas9 Genome Editing Confers Increased Fusarium Wilt Tolerance in Tomato. Physiol. Mol. Biol. Plants. 2023;29:1–10. doi: 10.1007/s12298-022-01273-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Kim S.-Y., Bengtsson T., Olsson N., Hot V., Zhu L.-H., Åhman I. Mutations in Two Aphid-Regulated β-1,3-Glucanase Genes by CRISPR/Cas9 Do Not Increase Barley Resistance to Rhopalosiphum padi L. Front. Plant Sci. 2020;11:1043. doi: 10.3389/fpls.2020.01043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Kieu N.P., Lenman M., Wang E.S., Petersen B.L., Andreasson E. Mutations Introduced in Susceptibility Genes through CRISPR/Cas9 Genome Editing Confer Increased Late Blight Resistance in Potatoes. Sci. Rep. 2021;11:4487. doi: 10.1038/s41598-021-83972-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Yang Z., Xing J., Wang L., Liu Y., Qu J., Tan Y., Fu X., Lin Q., Deng H., Yu F. Mutations of Two FERONIA-like Receptor Genes Enhance Rice Blast Resistance without Growth Penalty. J. Exp. Bot. 2020;71:2112–2126. doi: 10.1093/jxb/erz541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Lin L., Zhang X., Fan J., Li J., Ren S., Gu X., Li P., Xu M., Xu J., Lei W., et al. Natural Variation in BnaA07.MKK9 Confers Resistance to Sclerotinia Stem Rot in Oilseed Rape. Nat. Commun. 2024;15:5059. doi: 10.1038/s41467-024-49504-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Liu H., Dong S., Gu F., Liu W., Yang G., Huang M., Xiao W., Liu Y., Guo T., Wang H., et al. NBS-LRR Protein Pik-H4 Interacts with OsBIHD1 to Balance Rice Blast Resistance and Growth by Coordinating Ethylene-Brassinosteroid Pathway. Front. Plant Sci. 2017;8:127. doi: 10.3389/fpls.2017.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Wu D., Guo J., Zhang Q., Shi S., Guan W., Zhou C., Chen R., Du B., Zhu L., He G. Necessity of Rice Resistance to Planthoppers for OsEXO70H3 Regulating SAMSL Excretion and Lignin Deposition in Cell Walls. New Phytol. 2022;234:1031–1046. doi: 10.1111/nph.18012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Zhu Z., Yin J., Chern M., Zhu X., Yang C., He K., Liu Y., He M., Wang J., Song L., et al. New Insights into Bsr-d1-mediated Broad-spectrum Resistance to Rice Blast. Mol. Plant Pathol. 2020;21:951–960. doi: 10.1111/mpp.12941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Atanasov K.E., Liu C., Erban A., Kopka J., Parker J.E., Alcázar R. NLR Mutations Suppressing Immune Hybrid Incompatibility and Their Effects on Disease Resistance. Plant Physiol. 2018;177:1152–1169. doi: 10.1104/pp.18.00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Macovei A., Sevilla N.R., Cantos C., Jonson G.B., Slamet-Loedin I., Čermák T., Voytas D.F., Choi I., Chadha-Mohanty P. Novel Alleles of Rice eIF4G Generated by CRISPR/Cas9-targeted Mutagenesis Confer Resistance to Rice tungro spherical virus. Plant Biotechnol. J. 2018;16:1918–1927. doi: 10.1111/pbi.12927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Biswal A.K., Wu T.-Y., Urano D., Pelissier R., Morel J.-B., Jones A.M., Biswal A.K. Novel Mutant Alleles Reveal a Role of the Extra-Large G Protein in Rice Grain Filling, Panicle Architecture, Plant Growth, and Disease Resistance. Front. Plant Sci. 2022;12:782960. doi: 10.3389/fpls.2021.782960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Han H., Wang Y., Zheng T., Peng Q., Qiu L., Hu X., Lin H., Xi D. NtAGO1 Positively Regulates the Generation and Viral Resistance of Dark Green Islands in Nicotiana Tabacum. Plant Physiol. Biochem. 2022;174:1–10. doi: 10.1016/j.plaphy.2022.01.028. [DOI] [PubMed] [Google Scholar]
- 307.Wang Z., Yan X., Zhang H., Meng Y., Pan Y., Cui H. NtCycB2 Negatively Regulates Tobacco Glandular Trichome Formation, Exudate Accumulation, and Aphid Resistance. Plant Mol. Biol. 2022;108:65–76. doi: 10.1007/s11103-021-01222-z. [DOI] [PubMed] [Google Scholar]
- 308.Cha J.Y., Uddin S., Macoy D.M., Shin G.-I., Jeong S.Y., Ali I., Hwang J.-W., Ji M.G., Lee S.C., Park J.H., et al. Nucleoredoxin Gene SINRX1 Negatively Regulates Tomato Immunity by Activating SA Signaling Pathway. Plant Physiol. Biochem. 2023;200:107804. doi: 10.1016/j.plaphy.2023.107804. [DOI] [PubMed] [Google Scholar]
- 309.Ruan B., Hua Z., Zhao J., Zhang B., Ren D., Liu C., Yang S., Zhang A., Jiang H., Yu H., et al. Os ACL-A2 Negatively Regulates Cell Death and Disease Resistance in Rice. Plant Biotechnol. J. 2019;17:1344–1356. doi: 10.1111/pbi.13058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Wang X., Wang Z., Lu Y., Huang J., Hu Z., Lou J., Fan X., Gu Z., Liu P., Ma B., et al. OsACA9, an Autoinhibited Ca2+-ATPase, Synergically Regulates Disease Resistance and Leaf Senescence in Rice. Int. J. Mol. Sci. 2024;25:1874. doi: 10.3390/ijms25031874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Liu J., Shen Y., Cao H., He K., Chu Z., Li N. OsbHLH057 Targets the AATCA cis-Element to Regulate Disease Resistance and Drought Tolerance in Rice. Plant Cell Rep. 2022;41:1285–1299. doi: 10.1007/s00299-022-02859-w. [DOI] [PubMed] [Google Scholar]
- 312.Yu S., Li S., Wang W., Tang D. OsCAMTA3 Negatively Regulates Disease Resistance to Magnaporthe oryzae by Associating with OsCAMTAPL in Rice. Int. J. Mol. Sci. 2024;25:5049. doi: 10.3390/ijms25095049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Gao Z., Liu Q., Zhang Y., Chen D., Zhan X., Deng C., Cheng S., Cao L. OsCUL3a-Associated Molecular Switches Have Functions in Cell Metabolism, Cell Death, and Disease Resistance. J. Agric. Food Chem. 2020;68:5471–5482. doi: 10.1021/acs.jafc.9b07426. [DOI] [PubMed] [Google Scholar]
- 314.Hou H., Fang J., Liang J., Diao Z., Wang W., Yang D., Li S., Tang D. OsExo70B1 Positively Regulates Disease Resistance to Magnaporthe oryzae in Rice. Int. J. Mol. Sci. 2020;21:7049. doi: 10.3390/ijms21197049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Cao Y., Zhang Y., Chen Y., Yu N., Liaqat S., Wu W., Chen D., Cheng S., Wei X., Cao L., et al. OsPG1 Encodes a Polygalacturonase That Determines Cell Wall Architecture and Affects Resistance to Bacterial Blight Pathogen in Rice. Rice. 2021;14:36. doi: 10.1186/s12284-021-00478-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Li D., Zhou J., Zheng C., Zheng E., Liang W., Tan X., Xu R., Yan C., Yang Y., Yi K., et al. OsTGAL1 Suppresses the Resistance of Rice to Bacterial Blight Disease by Regulating the Expression of Salicylic Acid Glucosyltransferase OsSGT1. Plant Cell Environ. 2022;45:1584–1602. doi: 10.1111/pce.14288. [DOI] [PubMed] [Google Scholar]
- 317.Wang P., Li J., Zhang Z., Zhang Q., Li X., Xiao J., Ma H., Wang S. OsVQ1 Links Rice Immunity and Flowering via Interaction with a Mitogen-Activated Protein Kinase OsMPK6. Plant Cell Rep. 2021;40:1989–1999. doi: 10.1007/s00299-021-02766-6. [DOI] [PubMed] [Google Scholar]
- 318.Li Y., Liao S., Mei P., Pan Y., Zhang Y., Zheng X., Xie Y., Miao Y. OsWRKY93 Dually Functions Between Leaf Senescence and in Response to Biotic Stress in Rice. Front. Plant Sci. 2021;12:643011. doi: 10.3389/fpls.2021.643011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Wang S., Han S., Zhou X., Zhao C., Guo L., Zhang J., Liu F., Huo Q., Zhao W., Guo Z., et al. Phosphorylation and Ubiquitination of OsWRKY31 Are Integral to OsMKK10-2-Mediated Defense Responses in Rice. Plant Cell. 2023;35:2391–2412. doi: 10.1093/plcell/koad064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Zhu X., Guo L., Zhu R., Zhou X., Zhang J., Li D., He S., Qiao Y. Phytophthora sojae Effector PsAvh113 Associates with the Soybean Transcription Factor GmDPB to Inhibit Catalase-mediated Immunity. Plant Biotechnol. J. 2023;21:1393–1407. doi: 10.1111/pbi.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Vo K.T.X., Lee S.K., Halane M.K., Song M.Y., Hoang T.V., Kim C.Y., Park S.-Y., Jeon J., Kim S.T., Sohn K.H., et al. Pi5 and Pii Paired NLRs Are Functionally Exchangeable and Confer Similar Disease Resistance Specificity. Mol. Cells. 2019;42:637–645. doi: 10.14348/molcells.2019.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Wang P., Wang Y., Hu Y., Chen Z., Han L., Zhu W., Tian B., Fang A., Yang Y., Bi C., et al. Plant hypersensitive induced reaction protein facilitates cell death induced by secreted xylanase associated with the pathogenicity of Sclerotinia sclerotiorum. Plant J. 2024;118:90–105. doi: 10.1111/tpj.16593. [DOI] [PubMed] [Google Scholar]
- 323.Gao Y., Li Z., Yang C., Li G., Zeng H., Li Z., Zhang Y., Yang X. Pseudomonas syringae Activates ZAT18 to Inhibit Salicylic Acid Accumulation by Repressing EDS1 Transcription for Bacterial Infection. New Phytol. 2022;233:1274–1288. doi: 10.1111/nph.17870. [DOI] [PubMed] [Google Scholar]
- 324.Nekrasov V., Wang C., Win J., Lanz C., Weigel D., Kamoun S. Rapid Generation of a Transgene-Free Powdery Mildew Resistant Tomato by Genome Deletion. Sci. Rep. 2017;7:482. doi: 10.1038/s41598-017-00578-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Bi G., Zhou Z., Wang W., Li L., Rao S., Wu Y., Zhang X., Menke F.L.H., Chen S., Zhou J.-M. Receptor-Like Cytoplasmic Kinases Directly Link Diverse Pattern Recognition Receptors to the Activation of Mitogen-Activated Protein Kinase Cascades in Arabidopsis. Plant Cell. 2018;30:1543–1561. doi: 10.1105/tpc.17.00981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Pompili V., Dalla Costa L., Piazza S., Pindo M., Malnoy M. Reduced Fire Blight Susceptibility in Apple Cultivars Using a High-efficiency CRISPR/Cas9-FLP/FRT-based Gene Editing System. Plant Biotechnol. J. 2020;18:845–858. doi: 10.1111/pbi.13253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Hu B., Zhou Y., Zhou Z., Sun B., Zhou F., Yin C., Ma W., Chen H., Lin Y. Repressed OsMESL Expression Triggers Reactive Oxygen Species-mediated Broad-spectrum Disease Resistance in Rice. Plant Biotechnol. J. 2021;19:1511–1522. doi: 10.1111/pbi.13566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Lu H., Luo T., Fu H., Wang L., Tan Y., Huang J., Wang Q., Ye G., Gatehouse A.M.R., Lou Y., et al. Resistance of Rice to Insect Pests Mediated by Suppression of Serotonin Biosynthesis. Nat. Plants. 2018;4:338–344. doi: 10.1038/s41477-018-0152-7. [DOI] [PubMed] [Google Scholar]
- 329.Desmedt W., Kudjordjie E.N., Chavan S.N., Zhang J., Li R., Yang B., Nicolaisen M., Mori M., Peters R.J., Vanholme B., et al. Rice Diterpenoid Phytoalexins Are Involved in Defence against Parasitic Nematodes and Shape Rhizosphere Nematode Communities. New Phytol. 2022;235:1231–1245. doi: 10.1111/nph.18152. [DOI] [PubMed] [Google Scholar]
- 330.Son S., Kim H., Lee K.S., Kim S., Park S.R. Rice Glutaredoxin GRXS15 Confers Broad-Spectrum Resistance to Xanthomonas oryzae pv. oryzae and Fusarium fujikuroi. Biochem. Biophys. Res. Commun. 2020;533:1385–1392. doi: 10.1016/j.bbrc.2020.10.027. [DOI] [PubMed] [Google Scholar]
- 331.Zeng W., Huang H., Lin X., Zhu C., Kosami K., Huang C., Zhang H., Duan C., Zhu J., Miki D. Roles of DEMETER in Regulating DNA Methylation in Vegetative Tissues and Pathogen Resistance. J. Integr. Plant Biol. 2021;63:691–706. doi: 10.1111/jipb.13037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Schultink A., Qi T., Lee A., Steinbrenner A.D., Staskawicz B. Roq1 Mediates Recognition of the Xanthomonas and Pseudomonas Effector Proteins XopQ and HopQ1. Plant J. 2017;92:787–795. doi: 10.1111/tpj.13715. [DOI] [PubMed] [Google Scholar]
- 333.Zhao Z., Feng Q., Liu P., He X., Zhao J., Xu Y., Zhang L., Huang Y., Zhao J., Fan J., et al. RPW8.1 Enhances the Ethylene-signaling Pathway to Feedback-attenuate Its Mediated Cell Death and Disease Resistance in Arabidopsis. New Phytol. 2021;229:516–531. doi: 10.1111/nph.16857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Zhai K., Deng Y., Liang D., Tang J., Liu J., Yan B., Yin X., Lin H., Chen F., Yang D., et al. RRM Transcription Factors Interact with NLRs and Regulate Broad-Spectrum Blast Resistance in Rice. Mol. Cell. 2019;74:996–1009.e7. doi: 10.1016/j.molcel.2019.03.013. [DOI] [PubMed] [Google Scholar]
- 335.Li S., Jia Z., Wang K., Du L., Li H., Lin Z., Ye X. Screening and Functional Characterization of Candidate Resistance Genes to Powdery Mildew from Dasypyrum villosum#4 in a Wheat Line Pm97033. Theor. Appl. Genet. 2020;133:3067–3083. doi: 10.1007/s00122-020-03655-4. [DOI] [PubMed] [Google Scholar]
- 336.Zhou T., Zhang M., Gong P., Li F., Zhou X. Selective Autophagic Receptor NbNBR1 Prevents NbRFP1-Mediated UPS-Dependent Degradation of βC1 to Promote Geminivirus Infection. PLoS Pathog. 2021;17:e1009956. doi: 10.1371/journal.ppat.1009956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Liu F., Chern M., Jain R., Martin J.A., Schakwitz W.S., Ronald P.C. Silencing of Dicer-like Protein 2a Restores the Resistance Phenotype in the Rice Mutant, sxi4 (suppressor of Xa21-mediated Immunity 4) Plant J. 2022;110:646–657. doi: 10.1111/tpj.15692. [DOI] [PubMed] [Google Scholar]
- 338.Zhang Z., Ge X., Luo X., Wang P., Fan Q., Hu G., Xiao J., Li F., Wu J. Simultaneous Editing of Two Copies of Gh14-3-3d Confers Enhanced Transgene-Clean Plant Defense against Verticillium dahliae in Allotetraploid Upland Cotton. Front. Plant Sci. 2018;9:842. doi: 10.3389/fpls.2018.00842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Zhang Y., Bai Y., Wu G., Zou S., Chen Y., Gao C., Tang D. Simultaneous Modification of Three Homoeologs of Ta EDR 1 by Genome Editing Enhances Powdery Mildew Resistance in Wheat. Plant J. 2017;91:714–724. doi: 10.1111/tpj.13599. [DOI] [PubMed] [Google Scholar]
- 340.He N., Huang F., Lu L., Wang X., Li Q.Q., Yang D. SPR9 Encodes a 60 S Ribosomal Protein That Modulates Panicle Spreading and Affects Resistance to False Smut in Rice (Oryza sativa. L) BMC Plant Biol. 2023;23:205. doi: 10.1186/s12870-023-04172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Hu H., Yu F. Studies on the Temporal, Structural, and Interacting Features of the Clubroot Resistance Gene Rcr1 Using CRISPR/Cas9-Based Systems. Hortic. Plant J. 2024;10:1035–1048. doi: 10.1016/j.hpj.2024.04.001. [DOI] [Google Scholar]
- 342.Wu T., Zhang H., Bi Y., Yu Y., Liu H., Yang H., Yuan B., Ding X., Chu Z. Tal2c Activates the Expression of OsF3H04g to Promote Infection as a Redundant TALE of Tal2b in Xanthomonas oryzae pv. oryzicola. Int. J. Mol. Sci. 2021;22:13628. doi: 10.3390/ijms222413628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Xu X., Xu Z., Ma W., Haq F., Li Y., Shah S.M.A., Zhu B., Zhu C., Zou L., Chen G. TALE-Triggered and iTALE-Suppressed Xa1-Mediated Resistance to Bacterial Blight Is Independent of Rice Transcription Factor Subunits OsTFIIAγ1 or OsTFIIAγ5. J. Exp. Bot. 2021;72:3249–3262. doi: 10.1093/jxb/erab054. [DOI] [PubMed] [Google Scholar]
- 344.Ntui V.O., Tripathi J.N., Shah T., Tripathi L. Targeted Knockout of Early Nodulin-like 3 (MusaENODL3) Gene in Banana Reveals Its Function in Resistance to Xanthomonas Wilt Disease. Plant Biotechnol. J. 2024;22:1101–1112. doi: 10.1111/pbi.14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Huang X., Bai X., Qian C., Liu S., Goher F., He F., Zhao G., Pei G., Zhao H., Wang J., et al. TaUAM3, a UDP-Ara Mutases Protein, Positively Regulates Wheat Resistance to the Stripe Rust Fungus. Food Energy Secur. 2023;12:e456. doi: 10.1002/fes3.456. [DOI] [Google Scholar]
- 346.Zhang H., Jing W., Zheng J., Jin Y., Wu D., Cao C., Dong Y., Shi X., Zhang W. The ATP-Binding Cassette Transporter OsPDR1 Regulates Plant Growth and Pathogen Resistance by Affecting Jasmonates Biosynthesis in Rice. Plant Sci. 2020;298:110582. doi: 10.1016/j.plantsci.2020.110582. [DOI] [PubMed] [Google Scholar]
- 347.Lv T., Li X., Fan T., Luo H., Xie C., Zhou Y., Tian C. The Calmodulin-Binding Protein IQM1 Interacts with CATALASE2 to Affect Pathogen Defense. Plant Physiol. 2019;181:1314–1327. doi: 10.1104/pp.19.01060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Guang H., Xiaoyang G., Zhian W., Ye W., Peng W., Linfang S., Bingting W., Anhong Z., Fuguang L., Jiahe W. The Cotton MYB33 Gene Is a Hub Gene Regulating the Trade-off between Plant Growth and Defense in Verticillium dahliae Infection. J. Adv. Res. 2024;61:1–17. doi: 10.1016/j.jare.2023.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Zou J.-P., Zhao Q.-F., Yang T., Shang Y.-F., Ahammed G.J., Zhou J. The E3 Ubiquitin Ligase RING1 Interacts with COP9 Signalosome Subunit 4 to Positively Regulate Resistance to Root-Knot Nematodes in Solanum lycopersicum L. Plant Sci. 2022;322:111344. doi: 10.1016/j.plantsci.2022.111344. [DOI] [PubMed] [Google Scholar]
- 350.Mohnike L., Rekhter D., Huang W., Feussner K., Tian H., Herrfurth C., Zhang Y., Feussner I. The Glycosyltransferase UGT76B1 Modulates N -Hydroxy-Pipecolic Acid Homeostasis and Plant Immunity. The Plant Cell. 2021;33:735–749. doi: 10.1093/plcell/koaa045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Leibman-Markus M., Pizarro L., Schuster S., Lin Z.J.D., Gershony O., Bar M., Coaker G., Avni A. The Intracellular Nucleotide-binding Leucine-rich Repeat Receptor (SlNRC4a) Enhances Immune Signalling Elicited by Extracellular Perception. Plant Cell Environ. 2018;41:2313–2327. doi: 10.1111/pce.13347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Cao Y., Liu L., Ma K., Wang W., Lv H., Gao M., Wang X., Zhang X., Ren S., Zhang N., et al. The Jasmonate-induced bHLH Gene SlJIG Functions in Terpene Biosynthesis and Resistance to Insects and Fungus. J. Integr. Plant Biol. 2022;64:1102–1115. doi: 10.1111/jipb.13248. [DOI] [PubMed] [Google Scholar]
- 353.Zhang J., Wei H., Hong Y., Yang R., Meng J., Luan Y. The lncRNA20718-miR6022-RLPs Module Regulates Tomato Resistance to Phytophthora infestans. Plant Cell Rep. 2024;43:57. doi: 10.1007/s00299-024-03161-7. [DOI] [PubMed] [Google Scholar]
- 354.Hong Y., Liu Q., Cao Y., Zhang Y., Chen D., Lou X., Cheng S., Cao L. The OsMPK15 Negatively Regulates Magnaporthe oryza and Xoo Disease Resistance via SA and JA Signaling Pathway in Rice. Front. Plant Sci. 2019;10:752. doi: 10.3389/fpls.2019.00752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Gu X., Si F., Feng Z., Li S., Liang D., Yang P., Yang C., Yan B., Tang J., Yang Y., et al. The OsSGS3-tasiRNA-OsARF3 Module Orchestrates Abiotic-Biotic Stress Response Trade-off in Rice. Nat. Commun. 2023;14:4441. doi: 10.1038/s41467-023-40176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Li J., Deng F., Wang H., Qiang X., Meng Y., Shan W. The Raf-like Kinase Raf36 Negatively Regulates Plant Resistance against the Oomycete Pathogen Phytophthora parasitica by Targeting MKK2. Mol. Plant Pathol. 2022;23:530–542. doi: 10.1111/mpp.13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Kanda Y., Yokotani N., Maeda S., Nishizawa Y., Kamakura T., Mori M. The Receptor-like Cytoplasmic Kinase BSR1 Mediates Chitin-Induced Defense Signaling in Rice Cells. Biosci. Biotechnol. Biochem. 2017;81:1497–1502. doi: 10.1080/09168451.2017.1325710. [DOI] [PubMed] [Google Scholar]
- 358.Zhao H., Wang X., Jia Y., Minkenberg B., Wheatley M., Fan J., Jia M.H., Famoso A., Edwards J.D., Wamishe Y., et al. The Rice Blast Resistance Gene Ptr Encodes an Atypical Protein Required for Broad-Spectrum Disease Resistance. Nat. Commun. 2018;9:2039. doi: 10.1038/s41467-018-04369-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Chu C., Huang R., Liu L., Tang G., Xiao J., Yoo H., Yuan M. The rice heavy-metal transporter OsNRAMP1 regulates disease resistance by modulating ROS homoeostasis. Plant Cell Environ. 2022;45:1109–1126. doi: 10.1111/pce.14263. [DOI] [PubMed] [Google Scholar]
- 360.Chen J., Wang L., Yang Z., Liu H., Chu C., Zhang Z., Zhang Q., Li X., Xiao J., Wang S., et al. The Rice Raf-like MAPKKK OsILA1 Confers Broad-spectrum Resistance to Bacterial Blight by Suppressing the OsMAPKK4–OsMAPK6 Cascade. J. Integr. Plant Biol. 2021;63:1815–1842. doi: 10.1111/jipb.13150. [DOI] [PubMed] [Google Scholar]
- 361.Wang L., Ran L., Hou Y., Tian Q., Li C., Liu R., Fan D., Luo K. The Transcription Factor MYB115 Contributes to the Regulation of Proanthocyanidin Biosynthesis and Enhances Fungal Resistance in Poplar. New Phytol. 2017;215:351–367. doi: 10.1111/nph.14569. [DOI] [PubMed] [Google Scholar]
- 362.Chen P., Jiang L., Zhang L., Sun B., Lv S., Zhang J., Yu H., Mao X., Fan Z., Li C., et al. The UDP-Glycosyltransferase Gene OsUGT706E2 Negatively Regulates Rice Tolerance to Blast Disease and Abiotic Stresses. Environ. Exp. Bot. 2024;226:105889. doi: 10.1016/j.envexpbot.2024.105889. [DOI] [Google Scholar]
- 363.Xiao G., Laksanavilat N., Cesari S., Lambou K., Baudin M., Jalilian A., Telebanco-Yanoria M.J., Chalvon V., Meusnier I., Fournier E., et al. The Unconventional Resistance Protein PTR Recognizes the Magnaporthe oryzae Effector AVR-Pita in an Allele-Specific Manner. Nat. Plants. 2024;10:994–1004. doi: 10.1038/s41477-024-01694-z. [DOI] [PubMed] [Google Scholar]
- 364.Jiang Y., Guo L., Ma X., Zhao X., Jiao B., Li C., Luo K. The WRKY Transcription Factors PtrWRKY18 and PtrWRKY35 Promote Melampsora Resistance in Populus. Tree Physiol. 2017;37:665–675. doi: 10.1093/treephys/tpx008. [DOI] [PubMed] [Google Scholar]
- 365.Luo D., Huguet-Tapia J.C., Raborn R.T., White F.F., Brendel V.P., Yang B. The Xa7 Resistance Gene Guards the Rice Susceptibility Gene SWEET14 against Exploitation by the Bacterial Blight Pathogen. Plant Commun. 2021;2:100164. doi: 10.1016/j.xplc.2021.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Ninh T.T., Gao W., Trusov Y., Zhao J., Long L., Song C., Botella J.R. Tomato and Cotton G Protein Beta Subunit Mutants Display Constitutive Autoimmune Responses. Plant Direct. 2021;5:e359. doi: 10.1002/pld3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Ai Y., Li Q., Li C., Wang R., Sun X., Chen S., Cai X.-Z., Qi X., Liang Y. Tomato LysM Receptor Kinase 4 Mediates Chitin-Elicited Fungal Resistance in Both Leaves and Fruit. Hortic. Res. 2023;10:uhad082. doi: 10.1093/hr/uhad082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Zhang X., Li N., Liu X., Wang J., Zhang Y., Liu D., Wang Y., Cao H., Zhao B., Yang W. Tomato Protein Rx4 Mediates the Hypersensitive Response to Xanthomonas euvesicatoria pv. perforans Race T3. Plant J. 2021;105:1630–1644. doi: 10.1111/tpj.15138. [DOI] [PubMed] [Google Scholar]
- 369.Liu X., Meng G., Wang M., Qian Z., Zhang Y., Yang W. Tomato SlPUB24 Enhances Resistance to Xanthomonas euvesicatoria pv. perforans Race T3. Hortic. Res. 2021;8:30. doi: 10.1038/s41438-021-00468-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Zhang N., Pombo M.A., Rosli H.G., Martin G.B. Tomato Wall-Associated Kinase SlWak1 Depends on Fls2/Fls3 to Promote Apoplastic Immune Responses to Pseudomonas syringae. Plant Physiol. 2020;183:1869–1882. doi: 10.1104/pp.20.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Malik M.A.M., Haider M.S., Zhai Y., Khan M.A.U., Pappu H.R. Towards Developing Resistance to Chickpea Chlorotic Dwarf Virus through CRISPR/Cas9-Mediated Gene Editing Using Multiplexed gRNAs. J. Plant Dis. Prot. 2023;130:23–33. doi: 10.1007/s41348-022-00677-6. [DOI] [Google Scholar]
- 372.Yao S., Yang Z., Yang R., Huang Y., Guo G., Kong X., Lan Y., Zhou T., Wang H., Wang W., et al. Transcriptional Regulation of miR528 by OsSPL9 Orchestrates Antiviral Response in Rice. Mol. Plant. 2019;12:1114–1122. doi: 10.1016/j.molp.2019.04.010. [DOI] [PubMed] [Google Scholar]
- 373.Wang N., Fan X., He M., Hu Z., Tang C., Zhang S., Lin D., Gan P., Wang J., Huang X., et al. Transcriptional Repression of TaNOX10 by TaWRKY19 Compromises ROS Generation and Enhances Wheat Susceptibility to Stripe Rust. Plant Cell. 2022;34:1784–1803. doi: 10.1093/plcell/koac001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Gravot A., Liégard B., Quadrana L., Veillet F., Aigu Y., Bargain T., Bénéjam J., Lariagon C., Lemoine J., Colot V., et al. Two Adjacent NLR Genes Conferring Quantitative Resistance to Clubroot Disease in Arabidopsis Are Regulated by a Stably Inherited Epiallelic Variation. Plant Commun. 2024;5:100824. doi: 10.1016/j.xplc.2024.100824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Saile S.C., Jacob P., Castel B., Jubic L.M., Salas-Gonzáles I., Bäcker M., Jones J.D.G., Dangl J.L., El Kasmi F. Two Unequally Redundant “Helper” Immune Receptor Families Mediate Arabidopsis thaliana Intracellular “Sensor” Immune Receptor Functions. PLoS Biol. 2020;18:e3000783. doi: 10.1371/journal.pbio.3000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Binyameen B., Khan Z., Khan S.H., Ahmad A., Munawar N., Mubarik M.S., Riaz H., Ali Z., Khan A.A., Qusmani A.T., et al. Using Multiplexed CRISPR/Cas9 for Suppression of Cotton Leaf Curl Virus. Int. J. Mol. Sci. 2021;22:12543. doi: 10.3390/ijms222212543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Chen X., Liu C., Wang H., Liu Q., Yue Y., Duan Y., Wang Z., Zheng L., Chen X., Wang Y., et al. Ustilaginoidea virens-secreted Effector Uv1809 Suppresses Rice Immunity by Enhancing Os SRT 2-mediated Histone Deacetylation. Plant Biotechnol. J. 2024;22:148–164. doi: 10.1111/pbi.14174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Low Y.C., Lawton M.A., Di R. Validation of Barley 2OGO Gene as a Functional Orthologue of Arabidopsis DMR6 Gene in Fusarium Head Blight Susceptibility. Sci. Rep. 2020;10:9935. doi: 10.1038/s41598-020-67006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Ghorbani Faal P., Farsi M., Seifi A., Mirshamsi Kakhki A. Virus-Induced CRISPR-Cas9 System Improved Resistance against Tomato Yellow Leaf Curl Virus. Mol. Biol. Rep. 2020;47:3369–3376. doi: 10.1007/s11033-020-05409-3. [DOI] [PubMed] [Google Scholar]
- 380.Zhang B., Su T., Xin X., Li P., Wang J., Wang W., Yu Y., Zhao X., Zhang D., Li D., et al. Wall-associated Kinase BrWAK1 Confers Resistance to Downy Mildew in Brassica rapa. Plant Biotechnol. J. 2023;21:2125–2139. doi: 10.1111/pbi.14118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Ma W., Zou L., Zhiyuan J., Xiameng X., Zhengyin X., Yang Y., Alfano J.R., Chen G. Xanthomonas oryzae pv. oryzae TALE Proteins Recruit OsTFIIAγ1 to Compensate for the Absence of OsTFIIAγ5 in Bacterial Blight in Rice. Mol. Plant Pathol. 2018;19:2248–2262. doi: 10.1111/mpp.12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Téllez J., Muñoz-Barrios A., Sopeña-Torres S., Martín-Forero A.F., Ortega A., Pérez R., Sanz Y., Borja M., De Marcos A., Nicolas M., et al. YODA Kinase Controls a Novel Immune Pathway of Tomato Conferring Enhanced Disease Resistance to the Bacterium Pseudomonas syringae. Front. Plant Sci. 2020;11:584471. doi: 10.3389/fpls.2020.584471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Liao X., Sun J., Li Q., Ding W., Zhao B., Wang B., Zhou S., Wang H. ZmSIZ1a and ZmSIZ1b Play an Indispensable Role in Resistance against Fusarium Ear Rot in Maize. Mol. Plant Pathol. 2023;24:711–724. doi: 10.1111/mpp.13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Hu T., Huang C., He Y., Castillo-González C., Gui X., Wang Y., Zhang X., Zhou X. βC1 Protein Encoded in Geminivirus Satellite Concertedly Targets MKK2 and MPK4 to Counter Host Defense. PLoS Pathog. 2019;15:e1007728. doi: 10.1371/journal.ppat.1007728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.