Abstract
The master regulator of the DNA damage response, the transcription factor p53, orchestrates multiple downstream responses and coordinates repair processes. In response to double-strand DNA breaks, p53 exhibits pulses of expression, but how it achieves temporal coordination of downstream responses remains unclear. Here, we show that p53’s posttranslational modification state is altered between its first and second pulses of expression. We show that acetylations at two sites, K373 and K382, were reduced in the second pulse, and these acetylations differentially affected p53 target genes, resulting in changes in gene expression programs over time. This interplay between dynamics and modification may offer a strategy for cellular hubs like p53 to temporally organize multiple processes in individual cells.
p53 posttranslational modifications change between its oscillations and affect its transcriptional output after DNA damage.
INTRODUCTION
Dynamic changes in protein levels have been shown in recent years to play important roles in processing and transferring information in many biological systems (1, 2). This mode of signaling encodes information in the frequency, amplitude, duration, or other features of the temporal signal. An additional well-established mechanism for information transfer in cells is through the addition or removal of posttranslational modifications (PTMs), which can affect the function, localization, or stability of the modified protein (3, 4). Ultimately, cellular responses depend on the combined effect of multiple regulatory mechanisms. The interplay between protein dynamics and protein PTMs is especially intriguing when the protein levels oscillate, as each pulse is composed of newly synthesized proteins with no memory of the PTMs in previous pulses. A paradigm for studying the relationship between protein dynamics and PTMs is the tumor suppressor protein p53, which oscillates in response to DNA damage and is also subjected to a large number of PTMs.
The transcription factor p53 is central for the response to cellular stress such as DNA damage. In unstressed conditions, p53 is marked for degradation by one of its target genes, the E3 ubiquitin ligase Mouse Double Minute 2 (MDM2), leading to low levels of p53 (Fig. 1A). Upon cellular stress, p53 degradation is suppressed, and nuclear p53 accumulates and activates multiple cellular programs including DNA repair, metabolic regulation, cell cycle arrest, senescence, and apoptosis (5, 6). The transcriptional activity of p53 is regulated by several PTM enzymes (7), and certain PTMs were shown to favor activation of genes in specific programs. For example, phosphorylations of p53 on serine(S)15 (8, 9) and S46 (10, 11) and acetylations of lysine(K)120 (12, 13) and K373/K382 (14–16) were associated with apoptotic gene induction, whereas acetylation at K320 promoted cell cycle arrest (17). The dynamics of p53 following DNA damage also play a role in its transcriptional activity. Double-strand breaks (DSBs) induced by irradiation lead to oscillations in total p53 levels, whose frequency and amplitude are independent of the radiation dose (Fig. 1A) (18, 19). Altering these dynamics into a sustained p53 response affected the choice of target genes and consequent cellular outcomes (20–22). Most studies of p53 dynamics monitored total p53 levels and did not capture the dynamics of p53 modifications. The combined effect of p53 dynamics and the temporal changes in its PTM on gene expression has therefore not been explored. It is unknown, for example, whether p53 PTMs differ between pulses, which would open the possibility that different p53 pulses activate different downstream programs.
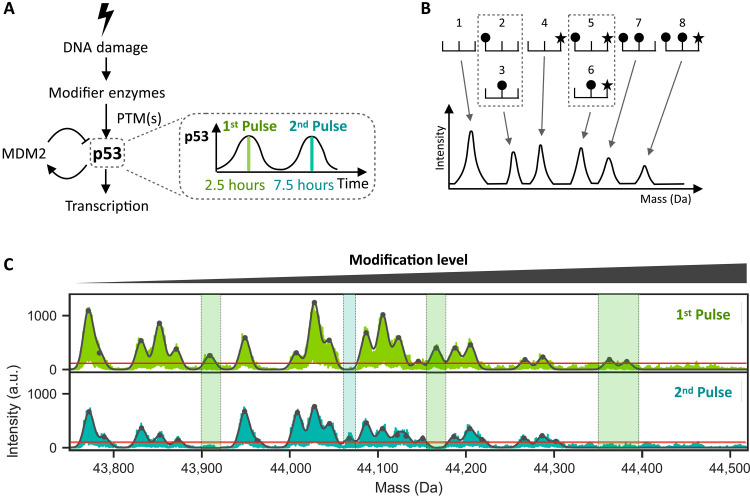
Fig. 1. The overall state of p53 PTMs changes between its first and second pulses following DNA damage.
(A) DNA damage activates enzymes, which activate p53 through PTMs. Activated p53 subsequently transcribes target genes including MDM2, an E3 ubiquitin ligase that leads to p53 degradation. The resulting negative feedback loop leads to oscillatory p53 dynamics with its first peak at 2.5 hours and second peak at 7.5 hours after initial DNA damage. (B) Schematic depicting a protein with 3 sites of modifications (top), the first two sites being subject to one type of modification (circle) and the third site to a different type of modification (star). The eight possible modified forms are shown. An I2MS spectrum (bottom) can distinguish forms that are unique in mass (1, 4, 7, 8) while those sharing the same mass cannot be distinguished (2 and 3, 5 and 6). (C) I2MS mass spectra of p53 proteins isolated at the peaks of the first (top) or second (bottom) pulses as shown in arbitrary units (a.u.). The furthest left peak shows the least modified form of p53, and all peaks to the right represent combinations of p53 PTMs. Black contour line represents fitted Gaussian distributions to each peak in the spectrum, while horizontal red line distinguishes the limit of detection (detailed in Materials and Methods). Shaded green and turquoise areas indicate modification states that differ between the first and second p53 peaks.
RESULTS
p53’s overall modification states differ between its pulses after DNA damage
To identify potential changes in p53 PTMs over time after DNA damage, we immunoprecipitated p53 (detailed in Materials and Methods and fig. S1A) at the first and second pulses following irradiation (Fig. 1A and fig. S1B) and characterized its modification state using individual ion mass spectrometry (I2MS). This recently introduced mass spectrometry method directly measures the masses of intact protein molecules without enzymatic digestion (23). I2MS generates spectra showing series of peaks at different masses, with the lowest-mass peak representing the least modified state of the protein and subsequent peaks representing more highly modified forms (Fig. 1B). While this method enables the simultaneous detection of all modifications present on a given protein, it does not decipher the specific modifications present nor the identities of the modified amino acid residues (Fig. 1B). The lowest mass species of p53 (43773 Da) was consistent with a singly cysteinylated p53, and the distribution of peaks to the right represented the landscape of modification patterns on p53 (Fig. 1C). The number of distinct peaks following irradiation was relatively restricted, considering the large number of previously reported modifications (24, 25) (Fig. 1C). In addition, no p53 molecules above 44,400 Da were detected, suggesting that no p53 molecules were phosphorylated more than eight times on the basis of mass shift, consistent with previous in vitro studies using recombinant p53 (26). Different peak landscapes were observed between the first and second p53 pulses, with several peaks absent in the second pulse compared to the first one (Fig. 1C, shaded green areas). One modification state was exclusively detected in the second pulse (Fig. 1C, shaded turquoise area). These findings demonstrate that distribution of modified p53 species changes between its pulses, prompting us to identify the specific modifications that differ.
Specific lysine acetylations vary between p53’s pulses due to regulation of HDAC enzymes
We next identified the specific PTMs at the first and second p53 pulses using a candidate-based approach. Several modifications were detectable by Western blotting of total cell lysate with modification-specific antibodies, whereas other modifications were detectable only after enrichment of p53 by immunoprecipitation (Fig. 2A). All tested phosphorylations, as well as acetylation of one lysine residue K381, were relatively constant between the two pulses. Notably, acetylations (Ac) of two lysine residues, K373 and K382, were lower in the second pulse than in the first (Fig. 2, A and B). Monitoring these acetylations at higher temporal resolution showed that their levels rose and declined with total p53 levels during the first pulse but remained low thereafter (Fig. 2, C and D). Together, these results show that at least two acetylation sites of p53 change dynamically following irradiation, leading to pulse-dependent modifications and the possibility of differentially active p53 pulses.
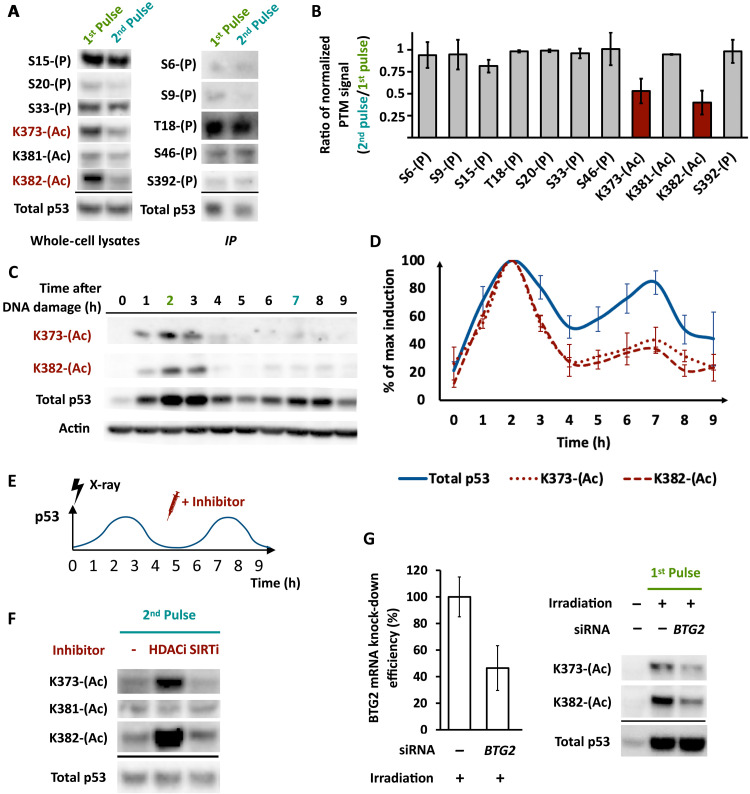
Fig. 2. Specific lysine acetylations vary between p53’s pulses due to regulation of HDAC enzymes.
(A) Phosphorylations or acetylations at the indicated residues of p53 were monitored by Western blots of total cell lysates (left) or of p53 immunoprecipitates (IP) (right) in the first and second pulses. Loading was normalized for total p53. Modifications indicated in red showed differences in levels between the two pulses. (B) Quantification of the Western blot from A, normalized to total p53 levels (n = 3 biological replicates, error bars indicate SD). (C) Western blot of acetylation at lysine-373 (K373-Ac) or lysine-382 (K382-Ac) of p53 at the indicated time points after 10-Gy x-ray irradiation. Actin is shown as a loading control. (D) Quantification of time-course Western blot signal from (B), normalized to the maximum level attained for each species over the time course (n = 3 biological replicates, error bars indicate SD). (E) Schematic of experiment showing addition of inhibitor(s) at the trough between p53 peaks (4.5 hours post irradiation). (F) Western blot of p53 acetylated at K373, K381, or K382 in the presence or absence of HDACi or SIRTi added at the time point shown in A. Total p53 is shown as a loading control. (G) Left: BTG2 mRNA levels following 48-hour knockdown with control or BTG2 short interfering (si)RNA. Right: Western blot of p53 acetylated at K373 or K382 following siRNA silencing of BTG2 (see Materials and Methods) and after the indicated number of hours after irradiation.
Previous work has shown that levels of acetylation on p53 K373 and K382 were affected by histone deacetylase (HDAC) and Sirtuin family deacetylases (14, 15, 27). To investigate which enzymes might regulate K373 and K382 acetylations after irradiation, we treated cells with a pan-HDAC inhibitor romidepsin (HDACi) or a pan-Sirtuin inhibitor Sirtinol (SIRTi) during the second pulse of p53 when acetylation at these sites is normally low (Fig. 2E). Acetylation was not affected by SIRTi treatment. However, HDACi enhanced acetylation at K373 and K382, without affecting K381 acetylation (Fig. 2F), the levels of total p53, or its oscillations (fig. S2A). We concluded that acetylation on p53 K373 and K382 following irradiation is regulated by HDAC enzymes.
We next investigated how HDAC regulation may lead to the change in p53 acetylation between its pulses. Our previous transcriptomics time series of irradiated cells (22) showed all detectable HDACs to be expressed at near-constant levels for 9 hours following irradiation (fig. S2B). We therefore hypothesized that the change in p53 acetylation does not arise because of changes in the levels of HDAC enzymes but rather changes in their activity, which could be determined by interactions with cofactors. Specifically, we aimed to identify HDAC regulators that were differentially expressed between the first and second pulses of p53. We found that the mRNA levels of BTG2, an inhibitor of HDAC1/4/9 (28), were higher in the first pulse of p53 compared to the second pulse (fig. S2C). Knocking down BTG2 by small interfering RNA (siRNA) reduced both K373 and K382 acetylation in the first pulse of p53 (Fig. 2G), suggesting that the reduced levels of BTG2 during p53’s second pulse contribute to the reduction of acetylation at K373 and K382.
Enhancing p53 acetylation differentially affects target gene expression
We next investigated how the reduction of p53 acetylation during its second pulse affected target gene expression. We used HDACi to enhance p53 K373 and K382 acetylation during the second pulse, without affecting total p53 levels (fig. S2A), and measured the effect on the expression of eight canonical p53 target genes, representing major transcriptional programs regulated by p53. Our data revealed varying effects of p53 acetylation on different targets (29). Specifically, the mRNA levels of the proapoptotic gene PUMA and senescence drivers PAI1 and PML significantly increased following irradiation and HDACi treatment compared to irradiation alone, while the mRNA levels of five other targets were not significantly affected by HDACi (Fig. 3A). The HDACi-mediated increases in expression were reduced in a control p53 knockdown cell line (Fig. 3A and fig. S3A). While our results are aligned with previous findings of acetylation of K373 and K382 on p53 being generally more transcriptionally active (14, 30–33), they also demonstrated that the effect of enhancing p53 acetylation is not uniform across all p53 targets, suggesting that the difference in p53 acetylations between its pulses affects the induction of only a fraction of its target genes.
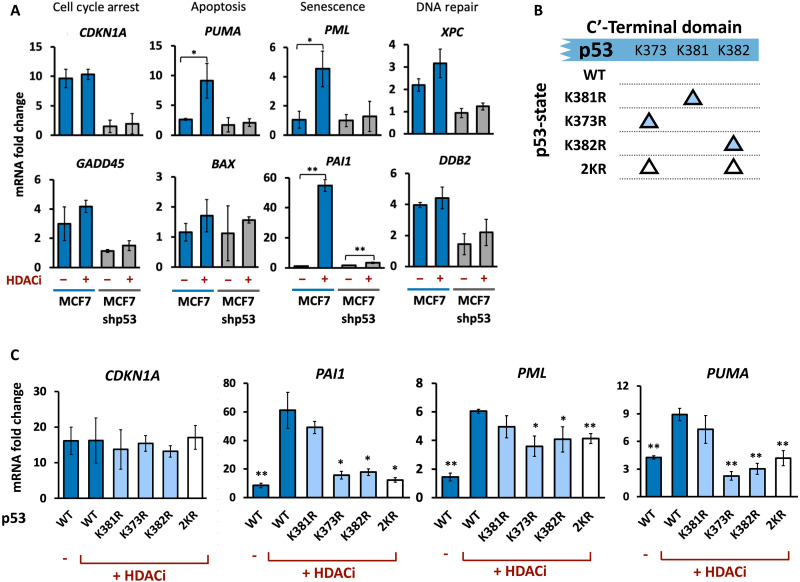
Fig. 3. Enhancing p53 acetylation differentially affects target gene expression.
(A) qPCR analysis of the mRNA levels of the indicated genes at 8 hours after irradiation treated with or without HDACi, normalized to mRNA levels in unirradiated cells. Error bars represent SD from three biological replicates. *P < 0.05, **P < 0.01 by t test. (B) Schematic of p53 constructs harboring single or double K➔R mutations. (C) qPCR analysis of mRNA levels of CDKN1A, PAI1, PML, and PUMA after the second p53 pulse in cells expressing the indicated p53 variants and treated with or without HDACi. Error bars represent SD from three biological replicates. *P < 0.05, **P < 0.01 by t test (all conditions compared to WT + HDACi).
HDACs play roles in many pathways independent of p53, and their inhibition therefore may alter gene expression regardless of their effects on p53 acetylation. To establish a more direct link between p53 acetylation at K373 and K382 and gene expression, we created lysine-to-arginine substitutions (K➔R) at these sites, which prevent p53 acetylation while retaining the chemical and structural properties of unacetylated lysine (34). We established clonal cell lines expressing p53 fused to mVenus with either single K➔R substitutions at K373 or K382 or a double substitution of K373 and K382 (2KR). We also investigated a K➔R substitution at K381, which does not change in acetylation between the first two pulses and is unaffected by HDACi (Fig. 2F). As a control, we expressed mVenus fused to wild-type (WT) p53. All exogenous p53 variants were expressed from a defined ectopic locus (35) to prevent locus-specific effects (Fig. 3B and fig. S3, B and D). All exogenously expressed p53 vasriants showed the stereotypic oscillatory dynamics in response to irradiation, albeit at lower levels than the endogenous p53 (fig. S3, E and F). Overall, the ratio of mutant to endogenous (WT) p53 was consistently between 1:3 and 1:4. Any potential measured consequences of the mutants, therefore, are expected to be relatively modest. However, because naturally not every molecule of p53 in a cell is expected to be acetylated at any given time, a mixture of WT p53 (which can be acetylated) and mutant p53 (which cannot be acetylated) may represent a more physiologically relevant context.
We next used the p53 mutant lines to examine whether the enhanced gene expression following HDACi treatment (Figs. 2E and 3C) depended on p53 acetylation at these sites. If the effects on gene expression resulted from enhanced p53 acetylation at K373 and K382, we expected that the KR mutants would not show increased target gene expression in the second pulse following HDACi treatment. Single and double KR mutants at 373 and 382 reduced the elevated expression of the three genes (PAI1, PML, and PUMA) induced by HDACi (Fig. 3C), supporting their dependence on p53 acetylation at these sites. In the absence of HDACi, the mRNA levels of these three genes were lower in the second pulse compared to the first. Compared to WT cells, KR mutants generally showed decreased expression of these genes during the first pulse, as expected for genes that are better induced by acetylated p53 (fig. S3G). KR mutants also slightly reduced their expression during the second pulse of p53 (fig. S3G). This may result from further reduction of the small fraction of acetylated p53 during its second pulse or might reflect transcripts expressed during the first p53 pulse that remained in the second pulse because of their stability. To investigate the effects of the mutants while minimizing any confounding effects of endogenous WT p53, we repeated these experiments in cell lines expressing a short hairpin RNA that targeted endogenous p53 but not the ectopically introduced version (fig. S3H). Expression of PAI1, PUMA, and PML, but not CDKN1, was reduced in cells ectopically expressing the 2KR mutant, compared to the WT control (fig. S3I). Furthermore, HDAC inhibition did not enhance gene expression in 2KR cells (fig. S3I), consistent with K373 and K382 being the primary acetylation sites governing expression of these genes.
Single mutants had similar qualitative effects on all three genes, while the double substitution did not alter gene expression additively or synergistically, suggesting functional redundancy between the two acetylation sites. CDKN1A, which was unaffected by HDACi, was also not affected by the KR mutants. K381, which did not show changes in acetylation between p53 pulses (Fig. 2, A, B, and F), did not affect the expression of PAI1, PML, or PUMA when mutated. Together, these results suggest that p53 acetylation at K373 and K382 affects the function of p53 and imply that the first and second pulses of p53 differ not only in their modifications but also in their ability to activate the transcription of specific target genes.
Pulse-dependent modifications of p53 temporally separate its various transcription programs
To assess the effect of the temporal changes in p53 acetylation on gene expression systematically, we performed time-series RNA sequencing at hourly intervals for 9 hours following irradiation in cells expressing either WT or 2KR p53 along with endogenous p53 (fig. S4A). We first verified that time-series RNA sequencing recapitulates the quantitative polymerase chain reaction (qPCR) trends for two target genes (fig. S4B). Direct transcriptional targets of p53 were determined by overlapping a consensus p53 target list (36) with a list of p53-bound sites as determined by chromatin immunoprecipitation sequencing (ChIP-seq) following irradiation (37) (detailed in Materials and Methods). We focused on the resulting 399 positively induced targets of p53 because repressed targets were previously suggested to represent indirect effects of p53 (38). First, to ascertain any differences in gene expression programs between WT and 2KR lines, we used gene set enrichment analysis (GSEA) to calculate the enrichment of a subset of functional gene ontology terms for which at least 20 genes were represented among the set of p53 targets (39, 40). Positive scores represent categories preferentially induced by WT p53, whereas negative scores represent categories preferentially induced by 2KR-expressing cells. Expression of the cell adhesion pathway was most preferentially enriched in WT cells at 3 hours, and the extent of enrichment was reduced by 8 hours (Fig. 4A, top). The DNA repair pathway was equivalently enriched in 2KR cells at 3 and 8 hours (Fig. 4A, bottom). These results suggested that the extent of preferential expression of these pathways in WT or 2KR cells might vary dynamically. Plotting the GSEA scores for these pathways at every time point (Fig. 4B and table S1) revealed that the cell adhesion pathway achieved peak enrichment in WT cells at 3 hours, whereas the DNA repair pathway showed a consistent, steady enrichment in 2KR cells across the time series. Even though WT and 2KR lines were expected to have equivalent, low levels of p53 acetylation during the second pulse, both the cell adhesion and DNA repair pathways showed preferential expression in one line or the other at this time. We expect the continued enrichment reflects long-lived transcripts whose enrichment in the first pulse is maintained at later time points.
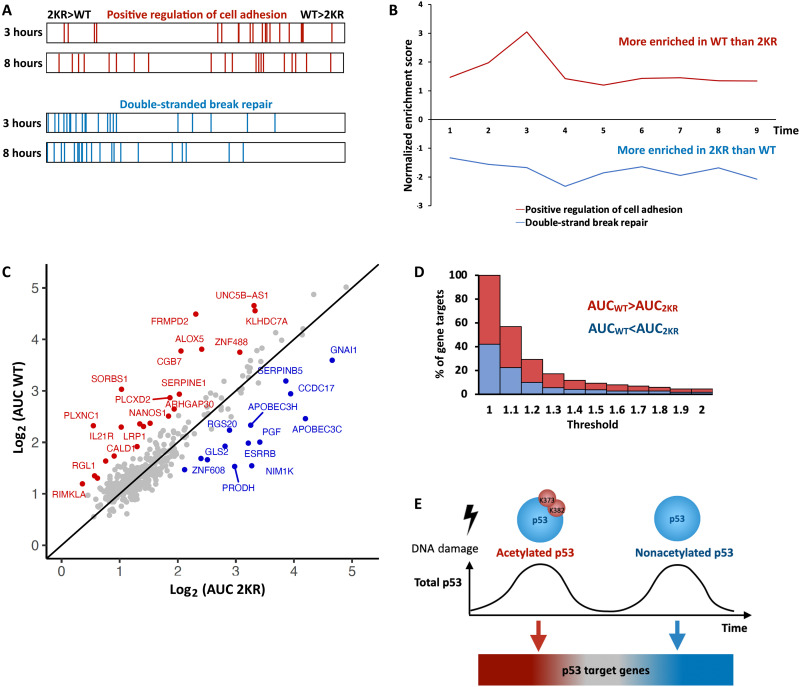
Fig. 4. Pulse-dependent modifications of p53 temporally separate its various transcription programs.
(A) GSEA barcode of “positive regulation of cell adhesion” pathway genes (top) or “DSB DNA repair” pathway genes (bottom) in 2KR compared to WT cells at 3 or 8 hours post irradiation. (B) Normalized Enrichment Score (NES) for the pathways shown in (A) at the indicated time points as calculated by GSEA. Positive values represent enrichment in WT p53, while negative values represent enrichment in 2KR. (C) Scatter plot of the fold-change AUC from 1 to 3 hours for p53 target genes in WT versus 2KR cells. Genes enriched at least 1.5-fold in WT or 2KR are colored red or blue, respectively. (D) Stacked bar-graph showing the percentage of gene targets having a WT:2KR (for WT > 2KR) or 2KR:WT (for WT < 2KR) AUC ratio at or above the indicated threshold. Red bars represent genes enriched in WT cells, and blue bars represent genes enriched in 2KR cells. (E) Model depicting how changes in acetylation on distinct pulses of p53 lead to different transcriptional activation of target genes. Red genes are better induced under K373/K382 acetylated p53. Blue genes are better induced under nonacetylated p53. The shift is gradual with grey genes in the middle having no preference.
To capture the enrichment of individual genes in WT or 2KR cells, we calculated the integrated fold change in expression [area under the curve (AUC)] from 1 to 3 hours for each p53 target gene in each cell line (fig. S4C). This period captures the first p53 peak when acetylation levels are highest in WT cells and minimizes the effects of RNA stability that dominate gene expression at later time points (37, 41). The AUC for each transcript in WT and 2KR cells was plotted (Fig. 4C). We identified 22 and 14 genes enriched at least 1.5-fold in WT or 2KR cells, respectively. Plotting the percentage of genes exceeding various enrichment thresholds (Fig. 4D) revealed that approximately half of p53 targets were similarly expressed in WT and 2KR cells, as they were not captured when a threshold of 1.1-fold enrichment was applied. The percentage of genes captured at increasingly higher thresholds decreased gradually, suggesting that many genes have slight preferences, but not absolute requirements, for the acetylation state of p53. The fact that not all genes were regulated similarly in WT and 2KR lines suggested that acetylated p53 did not simply enhance transcription activity in general, but rather, acetylation at K373 and K382 affected the ability of p53 to activate specific genes. We therefore sought to determine whether reduced acetylations at K373 and K382 have functional consequences. We tested two main functions related to DNA damage response, DNA repair and cell viability. First, we examined the intensity of γ-H2AX, a marker of DNA breaks, in the top 10% of cells expressing WT or 2KR p53. This analysis revealed that the rates of DNA repair were accelerated in the 2KR mutant (fig. S4D), as expected on the basis of its preferential activation of DNA repair pathways (Fig. 4B). Cell viability assay showed similar levels of nonviable cells in WT and 2KR cells under untreated conditions or following our standard sublethal irradiation dose alone (fig. S4E). Inducing both p53 expression and acetylation with irradiation plus HDACi suppressed cell death in nonacetylatable 2KR cells in comparison to WT cells (fig. S4E). Whereas the cell death induced by HDACi may result from its effects either on p53 or on other targets such as histones, the difference in viability between WT and 2KR cells is consistent with enhanced expression of proapoptotic genes in WT cells (Figs. 3C and 4C and fig. S3G). Note that the presence of endogenous WT p53 in the background may also dampen the effects of the 2KR mutant on viability. Overall, we suggest that the dynamic variations in p53 acetylation status between its pulses, combined with the distinct preferences of different target genes for the PTM state of p53, could allow different gene expression programs to be expressed at different times during the DNA damage response (Fig. 4E).
DISCUSSION
We have identified dynamical changes in the landscape of p53 modifications and found two acetylations that differ between the first and second pulses of p53 oscillations. p53 pulses can persist in individual cells for several days following DNA damage (42), and subsequent pulses may show different modification patterns. However, extending the analyses of p53 PTM landscapes to later times after DNA damage using bulk assays such as I2MS or Western blots is challenging because p53 oscillations become desynchronized in cell populations. New technologies to measure PTM in single cells will be required to identify such potential changes. It is intriguing to speculate about the mechanism responsible for the changes in p53 PTMs between its first and second pulses. Because p53 returns to baseline levels between peaks of oscillations, alterations to the PTM landscape likely reflect changes in how newly synthesized molecules are modified rather than addition or removal of PTMs on existing molecules. A switch from acetylation to methylation between the first and second pulses has been proposed as a mechanism of regulating target genes with oscillatory expression (33). Our previous work (22), combined with our identification of BTG2 as a pulse-specific HDAC regulator, suggest that regulation of the p53 PTMs over time occur via changes in the enzymes’ subcellular localization or binding partners rather than changes in their levels.
Our finding that p53 PTMs change between its pulses and that these changes can alter transcriptional programs raises the interesting possibility that PTM dynamics may allow cellular responses to evolve over time following a single stimulus. While we focused here on two dynamically changing p53 acetylations (K373 and K382), we cannot rule out the effects of other known acetylations, for example K120 and K164 in p53’s DNA binding domain (43). Nevertheless, our results indicate that preferential activation of some genes by K373/K382- acetylated p53 may be a mechanism to ensure they are expressed transiently and immediately following DNA damage. For example, LRP1, which promotes cell survival, is activated by p53 and repressed by p53-dependent microRNAs 103 and 107, resulting in LRP1 expression only under low doses of DNA damage (44). Its requirement for K373/K382-acetylated p53 may provide another layer of regulation to ensure that its expression is restricted to the early phases of the DNA damage response, thus allowing cell death programs to predominate at later stages. In addition, many genes with a preference for K373/K382-acetylated p53 appear to be involved in extracellular communication (Fig. 4C), encoding secreted proteins (SERPINE1 and CGB7) (45, 46), or UNC5B-AS1, an antisense RNA regulator of the secreted WNT agonist RSPO4 (47, 48), transmembrane proteins (IL21R and PLXNC1) (49, 50), LRP1 (44), KLHDC7A (51), or peripheral membrane proteins that transmit extracellular cues SORBS1 (52), FRMPD2 (53), and RGL1 (54). It will be interesting to determine the significance of an early burst of p53-mediated extracellular communication following DNA damage.
More generally, the resetting of PTMs in each p53 pulse may allow it to continuously monitor the status of DNA repair and activate appropriate programs, e.g. apoptosis if damage persists. Such integration of dynamics and PTMs may also occur for nuclear factor κB (NF-κB), a transcription factor that oscillates between nuclear and cytoplasmic compartments and its dynamics was shown to affect gene expression and chromatin modifications (55). Nuclear export of NF-κB requires deacetylation, raising the possibility that every cycle of localization refreshes NF-κB’s PTMs (56). Although there are only a small number of examples to date, it is likely that integration of oscillations and PTMs may be a widely used strategy by transcription factors to continuously monitor the internal and external environments to execute appropriate cellular responses over time.
MATERIALS AND METHODS
Cell culture
MCF7 were grown in RPMI 1640 (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (Gemini Bio catalog no. 100-106, lot no. A54H74L). MCF7-shp53 was a gift from Agami Lab in which a constitutive short hairpin RNA for TP53 is expressed constitutively. For fluorescence microscopy, standard RPMI was replaced with RPMI 1640 without phenol red (Thermo Fisher Scientific). The identity of MCF7 was confirmed by DNA fingerprinting with small tandem repeat profiling and tested negatively for mycoplasma contamination. MCF7 cells were irradiated with 10 Gy using a RS-2000 X-Ray irradiator. Romidepsin/Depsipeptide (HDACi) (Adooq Bioscience, FK228) and Sirtinol (SIRTi) (Sigma-Aldrich, 566320) were added at 10 nM, and 50 μM, respectively.
Cell line generation
Site-directed mutagenesis of p53 was formed using a Q5 Site-directed mutagenesis kit (NEB) and cloned into AAVS1-homology plasmid downstream of the EF1α promoter and fused to mVenus at the C terminus using Gibson assembly with NEBuilding HiFi DNA Assembly (NEB). Hind III and Nde I restriction enzyme sites were introduced into the noncoding sequence upstream of the p53 start codon and p53-mVenus linker region, respectively, and used subsequently to replace WT p53 with mutant p53 sequences by ligation. CRISPR-Cas9–mediated homology-based recombination was performed using TransIT-LT1 transfection reagent (Mirus) with a combination of plasmids containing AAVS-EF1α-p53-mVenus, plasmid expressing Cas9, and guide RNA containing plasmid AAVS-sgRNA(T1) (Addgene, Plasmid no. 41817), at 3:2:1 ratio. Following 48 hours of transfection, successful integrants were selected using G418 (800 μg/ml) for 7 days. Cells were then clonally expanded and confirmed for AAVS1 locus integration by PCR. Primers used in fig. S3C. were as follows: Forward (black) 5′ GCCGCTTTCTGTCTGCAGC, reverse (orange) 5′ CCAGTCATAGCCGAATAGCC and (green) 5′ GGAGACTAGGAAGGAGGAGG.
Live cell fluorescence microscopy and single-cell tracking and quantification
Cells were grown in poly-d-lysine–coated glass-bottom plates (MatTek) and imaged using a Nikon Eclipse TE-2000 inverted microscope with a 20× Plan-Apo objective and a Hammamatsu Orca ER camera, equipped with environmental chamber maintaining temperature, 5% CO2, and humidity. Images were acquired every 15 min for 10 hours using the MetaMorph Software. Cell tracking was done using a semiautomated method developed in the lab (41), which enables (i) automatic identification of single-cell centroids based on intensity and shape information, (ii) centroid linkage and track propagation using nearest-neighbor criteria, and (iii) real-time user correction of tracking. Tracking data were then used to quantify the intensity of fluorescent reporter from background-subtracted images. Only cells that remained within the field of view throughout the entire duration of the experiment were analyzed.
Cell viability measurement
A total of 1 × 105 MCF7 cells were plated in a 6-cm dish. Forty-eight hours after, cells were subjected to treatment as indicated. Cell viability assays were performed 24 hours after treatment following trypsinization of cells followed by 0.4% Trypan Blue (Bio-Rad no. 1450013) staining and quantified using TC20 Cell Counter (Bio-Rad no. 1450102).
Protein extraction and immunoprecipitation
Pellets of (approximately 100 million) treated MCF7 cells were thawed on ice and resuspended in lysis buffer [2× tris-buffered saline (TBS), 1% Triton X-100, 1× HALT protease, and phosphatase inhibitor cocktail]. Cells were disrupted and DNA sheared with a probe sonicator (Thermo Fisher Scientific, Model FB120) equipped with 3.175-mm probe on ice (45% power, 45 s total, 2 s on, 3 s off). Protein concentrations were quantified using the BCA Protein Assay Kit (Thermo Fisher Scientific) following the manufacturer’s protocol.
To prepare beads cross-linked to antibody, 750 ml of Pierce Protein A/G magnetic beads (Thermo Fisher Scientific no. 88803) was added to a 15-ml conical tube, washed with 1× TBS (5 ml), and collected with a magnet. Beads were resuspended in 1× TBS (6 ml), 200 μg of anti-p53 (Thermo Fisher Scientific no. 13-4000, clone PAB1801) was added, and mixture was incubated at 4°C with end-over-end mixing on a Tube Revolver (Thermo Fisher Scientific catalog no. 88861051) for 16 to 24 hours. Buffers for cross-linking were prepared fresh. Beads were washed twice with 6 ml of Wash Buffer (500 mM triethanolamine, pH 8.9) and suspended in Cross-linking Buffer [dimethyl pimelimidate (13 mg/ml) and 500 mM triethanolamine, pH 8.9]. Mixture was incubated at ambient temperature with end-over-end mixing for 1 hour. Beads were collected with a magnet, washed once with 5 ml of Quenching Buffer (500 mM ethanolamine, pH 8.9), and resuspended in 6 ml of Quenching Buffer. Mixture was incubated at ambient temperature with end-over-end mixing for 15 min. Beads were washed once with 6 ml of Wash Buffer, once with 6 ml of 100 mM glycine (pH 2.5), once with 6 ml of Wash Buffer, and twice with 6 ml of 1× TBS. Beads were resuspended in 4.5 ml of TBS and added 500 μl of bovine serum albumin (2 mg/ml). Beads were aliquoted 1 ml each in 1.5-ml LoBind tubes and stored at 4°C.
To immunoprecipitate p53, cell lysates were diluted with 6.5 ml of Lysis Buffer. DNA was completely digested with the addition of 30 ml of 1 M MgCl2 and 3 ml of benzonase (750 units, MilliporeSigma, E1014) and incubation at ambient temperature for 15 min. Lysate was clarified by centrifugation (3220g, 30 min, 4°C) and transferred to a new tube. To each lysate, magnetic beads crosslinked with anti-p53 antibody were added (500 μl per lysate) and mixture was incubated at 4°C with end-over-end mixing for 16 hours. Beads were washed once with 5 ml of Lysis Buffer, resuspended in 5 ml of Lysis Buffer, and transferred to a new 15-ml conical tube. Beads were further washed once with 5 ml of Lysis Buffer, once with 6 ml of High Salt Buffer (3× TBS, 1 mM EDTA, and 1% Triton X-100), and twice with 6 ml of 1× TBS. Beads were resuspended with 1 ml of 1× TBS and transferred to a 1.5 ml of LoBind tube. Beads were washed twice with 1 ml of TBS and once with 500 ml of water. Proteins were eluted from beads by incubating beads in 150 ml of 0.5% formic acid at 37°C for 10 min with 1000-rpm shaking. Beads were collected by a magnet, and eluate was transferred to another 1.5-ml LoBind tube. Trace beads were removed by centrifugation (20,000g, 10 min, 4°C), and supernatant was transferred to a sample vial (Microsolv, catalog no. 9532C-0CV-RS).
Individual ion mass spectrometry
Protein samples were handled by a PAL3 robot (CTC Analytics) integrated with a SampleStream device (Integrate Protein Technologies) as described previously (23, 39). The SampleStream device includes a PEEK channel containing an Ultracel 5-kDa PLCC composite regenerated cellulose ultrafiltration membrane (MilliporeSigma, catalog no. PLCCC05205). Samples (70 ml) were injected into a sample loop and then washed with Spray Buffer (70% water, 29.9% acetonitrile, and 0.1% acetic acid) during two 250-ml focusing steps. After focusing, the sample was eluted from the channel back into the LCP tool of the PAL3 robot (80 ml), and the LCP tool deposited the cleaned sample into an empty LC sample vial. All samples were stored at 4°C when not actively manipulated by the robot. A total of 70 ml of the sample was sprayed through an Ion Max Source (Thermo Fisher Scientific) fitted with a HESI II probe and 34-gauge needle insert at a flow rate of 1 ml/min delivered by a PAL3 robot (CTC Analytics) and analyzed by a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) with the Analyzer CE-HV supply board modified to maintain the Orbitrap center electrode at 1 kV. Instrument conditions included eFT off, trapping gas pressure setting of 0.5, 2.8 kV spray voltage, 0 L/min sheath gas, 10 V in-source collision-induced-dissociation, 320°C source temperature. Injection times ranged from 1 to 400 ms and were optimized for each sample to collect hundreds to thousands of individual ions (depending on spectral complexity). Data were acquired from 600- to 2000–mass/charge ratio (m/z) range for 70 min using 140,000 FT resolving power (at 200 m/z), which corresponded to 2 s transients. Data were collected as STORI files, which contain time-domain signals at specific frequencies where individual ions were detected. STORI files were processed with STORIboard (Proteinaceous, build 1.0.21257.1), which uses a voting algorithm to assign charge to each ion and assemble a mass spectrum by kernel density estimation. Mass spectra were exported as mzML files. Spectra were visualized and annotated with mMass (version 5.5.0). Visualization of I2MS spectra were derived from MSModDetector (57).
Immunoblot quantification and immunofluorescence
Total proteins (50 μg) were loaded into each lane of a 4 to 12% Bis-Tris gradient gel (NuPAGE), and transferred to 0.45-μm polyvinylidene difluoride membranes (GE Healthcare). Membranes were blocked with 5% BSA, incubated with primary antibody overnight, washed and incubated with secondary HRP conjugate antibodies, followed by a last wash and developed using chemiluminescence substrate (Thermo Fisher Scientific no. 34580). Membranes were exposed using Bio-rad Chemidoc. Primary antibodies used (all 1:1000 unless otherwise stated): β-ACT (Sigma-Aldrich no. A5316, 1:10,000 dilutions), total p53 (Santa Cruz Biotechnology no. sc-126, 1:5,000 dilutions), phospho-p53 Ser6 (CST no. 9285), Ser9 (CST no. 9288), Ser15 (CST no. 9284), Thr18 (Invitrogen no. PA5-12660), Ser20 (CST no. 9287), Ser46 (CST no. 2521), Ser392 (CST no. 9281); acetyl-p53 Lys373 (Abcam no. ab62376), Lys381 (Abcam no. ab61241), and Lys382 (CST no. 2525). Secondary antibodies used: anti-mouse immunoglobulin G (IgG) horseradish peroxidase (HRP)–linked (Invitrogen no. 62-6520, 1:10,000 dilutions), anti-rabbit IgG HRP-linked (CST no. 7074, 1:10,000 dilutions). Quantifications of band intensity were performed using densitometry analysis with background subtraction by Image Lab integrated software (BioRad). For immunofluorescence cells were fixed for 15 min using 0.4% Paraformaldehyde. Membranes were permeabilized using 5% BSA with 0.1% Triton X-100 for 1 hour. Primary antibody for γH2A.X (Millipore no. 05-636) was used at 1:250 dilution with fluorescent secondary Alexa Fluor 647 antibody (Invitrogen no. A21141) at 1:1,000 dilutions. Cells were imaged using a Nikon eclipse TE-2000 inverted microscope with a 20× Plan Apo objective and a Hammamatsu Orca ER camera. Nuclear signal was quantified using CellProfiler (58), and signal was normalized automatically using built-in CellProfiler settings. Individual nuclei were identified on the basis of a size filter of 10 to 25 μm. The top 10% of Venus expressing cells were correlated to its respective γH2A.X intensity.
siRNA-mediated knockdown
A total of 1 × 105 MCF7 cells were plated in a 6-cm dish. After 24 hours, 10 μl of 5 μM siRNA (Horizon ON-TARGETplus SMARTpool Human BTG2 siRNA no. 012308-00-0005 and Non-Targeting Control (Scrambled) siRNA no. 001810-10-05) were used along with 5 μl of DharmaFECT transfection reagent 1 (Horizon no. 2001-03) with Opti-MEM I Medium (Thermo Fisher Scientific) without serum, combined and incubated at room temperature for 20 min before adding to full serum media to cells, all accordingly to the manufacturer’s protocol. Forty-eight hours after the transfection, cells were harvested for RNA extraction.
RNA extraction
A total of 2.5 × 105 cells per condition were subjected to cell lysis and RNA extraction including deoxyribonuclease I treatment according to the manufacturer’s protocol (Qiagen RNeasy). RNA concentrations were determined using a Nanodrop (Thermo Fisher Scientific).
Quantitative reverse transcription PCR
One microgram of extracted RNA was used to generate complementary DNA (cDNA) using the high-capacity cDNA reverse transcription protocol (Applied Biosystems). qPCRs were then performed using 1/40 of the total of cDNA, 100 nM primer, and SYBR Green reagent following the manufacturer’s protocol (Applied Biosystems). Reactions were normalized to ACTB1 as a loading control.
Quantitative reverse transcription PCR primers used (F = forward, R = reverse, all written as 5′➔3′):
ACTB1 (F: ACCTTCTACAATGAGCTGCG, R: CCTGGATAGCAACGTACATGG)
BAX (F: CTGACGGCAACTTCAACTGG, R: GATCAGTTCCGGCACCTTGG)
CDKN1A (F: TGTCACTGTCTTGTACCCTTG, R: GGCGTTTGGAGTGGTAGAA)
DDB2 (F: TCATTGTTGTGGGCCGATAC, R: TGGCTCCAGATGAGAATGT).
GADD45 (F: GCAATATGACTTTGGAGGAATTCTC, R: TGACTCAGGGCTTTGCTG)
PAI1 (F: GTGGACTTTTCAGAGGTGGAG, R: GAAGTAGAGGGCATTCACCAG).
PML (F: AGACTCAGATGCCGAAAACTC, R: GGTCAGCAAGGTTCTCGTC).
PUMA (F: CGACCTCAACGCACAGTACG, R: GGGTGCAGGCACCTAATTGG).
XPC (F: GTCTCTACAGCCAATTCCTCTG, R: CCTTTGCTGGTCTTTGGTTTG).
RNA sequencing and library preparation
Libraries were prepared using a SciClone G3 NGSx workstation (Perkin Elmer) using the Kapa mRNA HyperPrep kit (Roche Applied Science). Polyadenylated mRNAs were captured using oligo-dT-conjugated magnetic beads (Kapa mRNA HyperPrep kit, Roche Sequencing, KR1352–v4.17) from 500 ng of total RNA on a Perkin Elmer SciClone G3 NGSx automated workstation. Poly-adenylated mRNA samples were immediately fragmented to 300 to 400 bp using heat and magnesium. First-strand synthesis was completed using random priming followed by second-strand synthesis and A-tailing. dUTP was incorporated into the second strand to allow strand-specific sequencing of the library. Libraries were enriched and indexed using 9 cycles of amplification (Kapa mRNA HyperPrep kit, Roche Sequencing) with PCR primers, which included dual 8 bp index sequences to allow for multiplexing (IDT for Illumina unique dual 8 bp indexes). Excess PCR reagents were removed through magnetic bead-based cleanup using Kapa Pure magnetic beads on a SciClone G3 NGSx workstation (Perkin Elmer). The resulting libraries were assessed using a 4200 TapeStation (Agilent Technologies) and quantified by qPCR (Roche Sequencing). Libraries were pooled and sequenced on one lane of an Illumina NovaSeq S4 flow cell using paired-end, 100 bp reads.
Transcriptomic analyses and GSEA
Raw RNA-sequencing data were processed using the bcbio-nextgen toolkit (https://github.com/bcbio/bcbio-nextgen). The bcbio-nextgen pipeline includes aligning reads to the human reference genome hg38 with STAR (59) and quantifying transcripts with Salmon (60). Genes with a mean average count <10 across all samples or those with 0 counts in >90% of samples were removed from downstream analysis. For the remaining occasional missing values, a pseudocount was added and then the counts were normalized using DESeq2 (61). p53 gene targets were selected on the basis of a previously developed method (22) that accounts for p53 binding to DNA from previous ChIP-seq experiments (37). The fold changes were calculated on the basis of comparison to expression at time point 0 hours, and the significance was determined for targets showing greater than 1.5-fold change for one or more time points, with FDR <0.1 (Benjamini–Hochberg corrected), and Pearson’s correlation > 0.5 between the two biological replicates. Clustering was done on z-scores using Fuzzy c-means clustering. GSEA was performed for every time point using the conventional method and database (62). As a ranking metric, we selected the ratio between the fold changes between WT and mutant condition. For full raw results, see GEO Database Bank Accession Number: GSE256300.
Acknowledgments
We thank all members of the Gunawardena, Kelleher, and Lahav Labs for discussion and critical comments on experiments and the manuscript especially L. Broger, P. Balbi, D. Sun, and N. (S.) Nandagopal. We also thank H. Wesseling, R. Fellers, R. Eisert, M. Kalocsay, and H. Zhou for advice on mass spectrometry. RNA-sequencing acquisitions were performed by the Bauer Sequencing Facilities, and analyses were reviewed and aided through the support of V. Barrera Burgos and the Harvard School of Public Health Bioinformatic Core.
Funding: This work was supported by grants from the National Institutes of Health GM139572 (G.L.), GM105375 (J.G.), GM108569 (N.K.) and F32CA246894 (B.D.); the German Research Foundation 445690853 (M.F.); and Ludwig Center at Harvard (G.L.).
Author contributions: D.L., N.K., A.J., J.G., and G.L. conceived experiments and analyses. Mass spectrometry experiments were performed and analyzed by B.D., M.F., and D.L. RNA-sequencing experiment biostatistical analyses were scripted by M.F. and J.F. All other experiments were designed and performed by D.L. with help from A.S. and G.B. D.L., A.J., and G.L. wrote the paper supported by editorial and methodological inputs from B.D., M.F., A.S., N.K., and J.G.
Competing interests: N.K. is involved with commercialization in the area of top-down proteomics and consults for Thermo Fisher Scientific as it moves to commercialize individual ion MS technology. All authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
The PDF file includes:
Figs. S1 to S4
Legend for table S1
References
Other Supplementary Material for this manuscript includes the following:
Table S1
REFERENCES AND NOTES
- 1.Behar M., Hoffmann A., Understanding the temporal codes of intra-cellular signals. Curr. Opin. Genet. Dev. 20, 684–693 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Purvis J. E., Lahav G., Encoding and decoding cellular information through signaling dynamics. Cell 152, 945–956 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beltrao P., Bork P., Krogan N. J., van Noort V., Evolution and functional cross-talk of protein post-translational modifications. Mol. Syst. Biol. 9, 714 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prabakaran S., Lippens G., Steen H., Gunawardena J., Post-translational modification: Nature’s escape from genetic imprisonment and the basis for dynamic information encoding. Wiley Interdiscip. Rev. Syst. Biol. Med. 4, 565–583 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieging K. T., Attardi L. D., Deconstructing p53 transcriptional networks in tumor suppression. Trends Cell Biol. 22, 97–106 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaiser A. M., Attardi L. D., Deconstructing networks of p53-mediated tumor suppression in vivo. Cell Death Differ. 25, 93–103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kruse J.-P., Gu W., Modes of p53 regulation. Cell 137, 609–622 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li D. W.-C., Liu J.-P., Schmid P. C., Schlosser R., Feng H., Liu W.-B., Yan Q., Gong L., Sun S.-M., Deng M., Liu Y., Protein serine/threonine phosphatase-1 dephosphorylates p53 at Ser-15 and Ser-37 to modulate its transcriptional and apoptotic activities. Oncogene 25, 3006–3022 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Kumari R., Kohli S., Das S., p53 Regulation upon genotoxic stress: Intricacies and complexities. Mol. Cell. Oncol. 1, e969653 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oda K., Arakawa H., Tanaka T., Matsuda K., Tanikawa C., Mori T., Nishimori H., Tamai K., Tokino T., Nakamura Y., Taya Y., p53AIP1, A potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102, 849–862 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Rinaldo C., Prodosmo A., Mancini F., Iacovelli S., Sacchi A., Moretti F., Soddu S., MDM2-regulated degradation of HIPK2 prevents p53Ser46 phosphorylation and DNA damage-induced apoptosis. Mol. Cell 25, 739–750 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Tang Y., Luo J., Zhang W., Gu W., Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell 24, 827–839 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Sykes S. M., Mellert H. S., Holbert M. A., Li K., Marmorstein R., Lane W. S., McMahon S. B., Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell 24, 841–851 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roy S., Tenniswood M., Site-specific acetylation of p53 directs selective transcription complex assembly. J. Biol. Chem. 282, 4765–4771 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Knights C. D., Catania J., Giovanni S. D., Muratoglu S., Perez R., Swartzbeck A., Quong A. A., Zhang X., Beerman T., Pestell R. G., Avantaggiati M. L., Distinct p53 acetylation cassettes differentially influence gene-expression patterns and cell fate. J. Cell Biol. 173, 533–544 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reed S. M., Quelle D. E., p53 Acetylation: Regulation and consequences. Cancer 7, 30–69 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chao C., Wu Z., Mazur S. J., Borges H., Rossi M., Lin T., Wang J. Y. J., Anderson C. W., Appella E., Xu Y., Acetylation of mouse p53 at lysine 317 negatively regulates p53 apoptotic activities after DNA damage. Mol. Cell. Biol. 26, 6859–6869 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lahav G., Rosenfeld N., Sigal A., Geva-Zatorsky N., Levine A. J., Elowitz M. B., Alon U., Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat. Genet. 36, 147–150 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Geva-Zatorsky N., Rosenfeld N., Itzkovitz S., Milo R., Sigal A., Dekel E., Yarnitzky T., Liron Y., Polak P., Lahav G., Alon U., Oscillations and variability in the p53 system. Mol. Syst. Biol. 2, 2006.0033 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purvis J. E., Karhohs K. W., Mock C., Batchelor E., Loewer A., Lahav G., p53 Dynamics control cell fate. Science 336, 1440–1444 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stewart-Ornstein J., Lahav G., p53 Dynamics in response to DNA damage vary across cell lines and are shaped by efficiency of DNA repair and activity of the kinase ATM. Sci. Signal. 10, eaah6671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiménez A., Lu D., Kalocsay M., Berberich M. J., Balbi P., Jambhekar A., Lahav G., Time-series transcriptomics and proteomics reveal alternative modes to decode p53 oscillations. Mol. Syst. Biol. 18, e10588 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kafader J. O., Melani R. D., Senko M. W., Makarov A. A., Kelleher N. L., Compton P. D., Measurement of individual ions sharply increases the resolution of orbitrap mass spectra of proteins. Anal. Chem. 91, 2776–2783 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Gu B., Zhu W.-G., Surf the post-translational modification network of p53 regulation. Int. J. Biol. Sci. 8, 672–684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeHart C. J., Chahal J. S., Flint S. J., Perlman D. H., Extensive post-translational modification of active and inactivated forms of endogenous p53. Mol. Cell. Proteomics 13, 1–17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.C. J. DeHart, L. Fornelli, L. C. Anderson, R. T. Fellers, D. Lu, C. L. Hendrickson, G. Lahav, J. Gunawardena, N. L. Kelleher, A multi-modal proteomics strategy for characterizing posttranslational modifications of tumor suppressor p53 reveals many sites but few modified forms. bioRxiv 455527 [Preprint] (2018). 10.1101/455527. [DOI]
- 27.Zhao Y., Lu S., Wu L., Chai G., Wang H., Chen Y., Sun J., Yu Y., Zhou W., Zheng Q., Wu M., Otterson G. A., Zhu W.-G., Acetylation of p53 at lysine 373/382 by the histone deacetylase inhibitor depsipeptide induces expression of p21Waf1/Cip1. Mol. Cell. Biol. 26, 2782–2790 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Micheli L., D’Andrea G., Leonardi L., Tirone F., HDAC1, HDAC4, and HDAC9 bind to PC3/Tis21/Btg2 and are required for its inhibition of cell cycle progression and cyclin D1 expression. J. Cell. Physiol. 232, 1696–1707 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Bieging K. T., Mello S. S., Attardi L. D., Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer 14, 359–370 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loffreda A., Jacchetti E., Antunes S., Rainone P., Daniele T., Morisaki T., Bianchi M. E., Tacchetti C., Mazza D., Live-cell p53 single-molecule binding is modulated by C-terminal acetylation and correlates with transcriptional activity. Nat. Commun. 8, 313 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofmann T. G., Möller A., Sirma H., Zentgraf H., Taya Y., Dröge W., Will H., Schmitz M. L., Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 4, 1–10 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Wang Y., Chen Y., Chen Q., Zhang X., Wang H., Wang Z., Wang J., Tian C., The role of acetylation sites in the regulation of p53 activity. Mol. Biol. Rep. 47, 381–391 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Friedrich D., Friedel L., Finzel A., Herrmann A., Preibisch S., Loewer A., Stochastic transcription in the p53-mediated response to DNA damage is modulated by burst frequency. Mol. Syst. Biol. 15, e9068 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panda A. K., Nandi S. K., Chakraborty A., Nagaraj R. H., Biswas A., Differential role of arginine mutations on the structure and functions of α-crystallin. Biochim. Biophys. Acta 1860, 199–210 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin S., Kim S. H., Shin S. W., Grav L. M., Pedersen L. E., Lee J. S., Lee G. M., Comprehensive analysis of genomic safe harbors as target sites for stable expression of the heterologous gene in HEK293 cells. ACS Synth Biol 9, 1263–1269 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Fischer M., Census and evaluation of p53 target genes. Oncogene 36, 3943–3956 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hafner A., Stewart-Ornstein J., Purvis J. E., Forrester W. C., Bulyk M. L., Lahav G., p53 pulses lead to distinct patterns of gene expression albeit similar DNA-binding dynamics. Nat. Struct. Mol. Biol. 24, 840–847 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fischer M., Steiner L., Engeland K., The transcription factor p53: Not a repressor, solely an activator. Cell Cycle 13, 3037–3058 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kozyrska K., Pilia G., Vishwakarma M., Wagstaff L., Goschorska M., Cirillo S., Mohamad S., Gallacher K., Carazo Salas R. E., Piddini E., p53 Directs leader cell behavior, migration, and clearance during epithelial repair. Science 375, eabl8876 (2022). [DOI] [PubMed] [Google Scholar]
- 40.Vaddavalli P. L., Schumacher B., The p53 network: Cellular and systemic DNA damage responses in cancer and aging. Trends Genet. 38, 598–612 (2022). [DOI] [PubMed] [Google Scholar]
- 41.Porter J. R., Fisher B. E., Batchelor E., p53 Pulses diversify target gene expression dynamics in an mRNA half-life-dependent manner and delineate co-regulated target gene subnetworks. Cell Syst. 2, 272–282 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reyes J., Chen J.-Y., Stewart-Ornstein J., Karhohs K. W., Mock C. S., Lahav G., Fluctuations in p53 signaling allow escape from cell-cycle arrest. Mol. Cell 71, 581–591.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang Y., Zhao W., Chen Y., Zhao Y., Gu W., Acetylation is indispensable for p53 activation. Cell 133, 612–626 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leslie P. L., Franklin D. A., Liu Y., Zhang Y., p53 Regulates the expression of LRP1 and apoptosis through a stress intensity-dependent microRNA feedback loop. Cell Rep. 24, 1484–1495 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zimmermann G., Ackermann W., Alexander H., Expression and production of human chorionic gonadotropin (hCG) in the normal secretory endometrium: Evidence of CGB7 and/or CGB6 beta hCG subunit gene expression. Biol. Reprod. 86, 87 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Mu X., Higgins P. J., Differential growth state-dependent regulation of plasminogen activator inhibitor type-1 expression in senescent IMR-90 human diploid fibroblasts. J. Cell. Physiol. 165, 647–657 (1995). [DOI] [PubMed] [Google Scholar]
- 47.Fu J., Zhang Y., Wang M., Hu J., Fang Y., Inhibition of the long non-coding RNA UNC5B-AS1/miR-4455/RSPO4 axis reduces cervical cancer growth in vitro and in vivo. J. Gene Med. 23, e3382 (2021). [DOI] [PubMed] [Google Scholar]
- 48.Kim K.-A., Wagle M., Tran K., Zhan X., Dixon M. A., Liu S., Gros D., Korver W., Yonkovich S., Tomasevic N., Binnerts M., Abo A., R-Spondin family members regulate the Wnt pathway by a common mechanism. Mol. Biol. Cell 19, 2588–2596 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu H., Juo Z. S., Shim A. H.-R., Focia P. J., Chen X., Garcia K. C., He X., Structural basis of semaphorin-plexin recognition and viral mimicry from Sema7A and A39R complexes with plexinC1. Cell 142, 749–761 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rochman Y., Spolski R., Leonard W. J., New insights into the regulation of T cells by gamma(c) family cytokines. Nat. Rev. Immunol. 9, 480–490 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gupta V. A., Beggs A. H., Kelch proteins: Emerging roles in skeletal muscle development and diseases. Skelet. Muscle 4, 11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin W.-H., Huang C.-J., Liu M.-W., Chang H.-M., Chen Y.-J., Tai T.-Y., Chuang L.-M., Cloning, mapping, and characterization of the human sorbin and SH3 domain containing 1 (SORBS1) gene: A protein associated with c-Abl during insulin signaling in the hepatoma cell line Hep3B. Genomics 74, 12–20 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Stenzel N., Fetzer C. P., Heumann R., Erdmann K. S., PDZ-domain-directed basolateral targeting of the peripheral membrane protein FRMPD2 in epithelial cells. J. Cell Sci. 122, 3374–3384 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Zago G., Veith I., Singh M. K., Fuhrmann L., de Beco S., Remorino A., Takaoka S., Palmeri M., Berger F., Brandon N., el Marjou A., Vincent-Salomon A., Camonis J., Coppey M., Parrini M. C., RalB directly triggers invasion downstream Ras by mobilizing the Wave complex. eLife 7, e40474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng Q. J., Ohta S., Sheu K. M., Spreafico R., Adelaja A., Taylor B., Hoffmann A., NF-κB dynamics determine the stimulus specificity of epigenomic reprogramming in macrophages. Science 372, 1349–1353 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen L. F., Fischle W., Verdin E., Greene W. C., Duration of nuclear NF- κB action regulated by reversible acetylation. Science 293, 1653–1657 (2001). [DOI] [PubMed] [Google Scholar]
- 57.M. Faizi, R. T. Fellers, D. Lu, B. S. Drown, A. Jambhekar, G. Lahav, N. L. Kelleher, J. Gunawardena, MSModDetector: A tool for detecting mass shifts and post-translational modifications in individual ion mass spectrometry data. bioRxiv 543961 [Preprint] (2023). 10.1101/2023.06.06.543961. [DOI] [PMC free article] [PubMed]
- 58.Stirling D. R., Swain-Bowden M. J., Lucas A. M., Carpenter A. E., Cimini B. A., Goodman A., CellProfiler 4: Improvements in speed, utility and usability. BMC Bioinform. 22, 433 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4
Legend for table S1
References
Table S1