Abstract
Nanofiber structures of some peptides and proteins as biological materials have been studied extensively, but their molecular mechanism of self-assembly and reassembly still remains unclear. We report here the reassembly of an ionic self-complementary peptide RADARADARADARADA (RADA16-I) that forms a well defined nanofiber scaffold. The 16-residue peptide forms stable β-sheet structure and undergoes molecular self-assembly into nanofibers and eventually a scaffold hydrogel consisting of >99.5% water. In this study, the nanofiber scaffold was sonicated into smaller fragments. Circular dichroism, atomic force microscopy, and rheology were used to follow the kinetics of the reassembly. These sonicated fragments not only quickly reassemble into nanofibers that were indistinguishable from the original material, but their reassembly also correlated with the rheological analyses showing an increase of scaffold rigidity as a function of nanofiber length. The disassembly and reassembly processes were repeated four times and, each time, the reassembly reached the original length. We proposed a plausible sliding diffusion model to interpret the reassembly involving complementary nanofiber cohesive ends. This reassembly process is important for fabrication of new scaffolds for 3D cell culture, tissue repair, and regenerative medicine.
Keywords: atomic force microscopy, circular dichroism, dynamic behaviors, ionic self-complementary peptides, nanofiber hydrogels
Molecular design, development, and fabrication of biological materials are a prerequisite for the advancement of medical technologies. These include scaffolds for fostering tissue regeneration, tissue engineering in regenerative medicine, and controlled drug release (1–7). Synthetic polymers and biodegradable biomaterials have had a significant impact in medicine over the last two decades (8–10). However, the continuous discovery and design of materials of biological origins are of great interest to multiple and diverse scientific and medical communities. The fabrication of materials at the molecular scale from “the bottom up,” one molecule at a time through synthesis and one unit at a time through self-assembly, has many advantages (11, 12). This approach is not only flexible and simple, but these materials can be tailor-made, thus facilitating the incorporation of many biochemically and medically desired features.
We previously reported the discovery and development of a class of self-assembling peptide scaffold materials to culture cells in three dimensions (13–17). These short, 8- to 16-residue (≈2.5–5 nm in length) peptides are chemically synthesized and form extremely stable β-sheet structures in water (13, 14). They not only self-assemble to form stable nanofibers, but also form higher-order nanofiber scaffolds, namely, hydrogels with extremely high water content [>99.5 (wt/vol)% water] (15–17). The gelation process is accelerated either by changing to neutral pH or adding physiological concentrations of salt solutions (13–15, 18–21). However, although it has high water content and is relatively soft, once the scaffold is formed, it limits the movement of biomolecules and other nanoscale and microscale entities (T. Savin, P. Doyle, R. Ellis-Behnke, and L. Spirio, personal communications).
To obtain a homogenous distribution of biological molecules in the peptide scaffolds for 3D tissue cultures (22, 23), controlled drug release, and other uses, it is important to evenly mix the substances to initiate the culture. We asked whether the self-assembled nanofibers could undergo reassembly after mechanical breakage through sonication. Sonication usually breaks the weak chemical bonds including hydrogen, ionic bonds, and hydrophobic interactions, but not the covalent peptide bonds. Thus, this process may allow reassembly of the mechanically dissembled individual peptides. The fragmented assemblies may contain compatible and complementary ends through both hydrophobic and ionic interactions. We report here the observation of reproducibly dynamic reassembly of self-assembling peptide that becomes a simple, reliable, and reproducible material process for producing well ordered peptide nanofiber scaffolds. Our findings will not only facilitate further development of well defined and tailor-made 3D cell culture systems for diverse cell systems, but may also have significance for studying and fabricating a broad range of fibrous biological materials.
Materials and Methods
Preparation of Peptide Nanofibers. The peptide RADA16-I, [COCH3]-RADARADARADARADA-[CONH2] (molecular weight 1,712), was commercially synthesized and purified (Syn-Pep, Dublin, CA, or Massachusetts Institute of Technology Biopolymers Laboratory). The solution of RADA16-I was prepared by dissolving the peptide powder with Milli-Q water and Tris·HCl buffer (pH 7.5). Final concentration of the peptide in 20 mM Tris·HCl was 3 mM or 0.5% (5 mg/ml). The peptide solution was sonicated for 30 min with an ultrasonic cleaner (50T, VWR Scientific) before each measurement at the maximal power setting.
CD. The peptides either sonicated or not sonicated were in Tris·HCl buffer (pH 7.5) and used for CD measurement. The samples of the sonicated peptides were removed from time periods of 2, 8, 16, 32, and 64 min after sonication or before sonication as a control.
Reassembly Assessment with Atomic Force Microscopy (AFM). After sonication of the RADA16-I solution for 30 min, aliquots of 1 μl were removed from the peptide solution and diluted with 19 μl of Milli-Q water (20× dilutions). One microliter sample was immediately deposited onto a freshly cleaved mica surface at varying times as indicated. Each aliquot was left on the mica for ≈15 s, then rinsed with 100 μl of Milli-Q water. The peptide sample on the mica surface was then air-dried, and images were acquired immediately. The images were obtained by scanning the mica surface in air by AFM (Nanoscope IIIa, Digital Instruments, Santa Barbara, CA) operating in tapping mode. Soft silicon cantilevers were chosen (force modulation etched silicon probe, Veeco Probes) with cantilever length of 219 μm, spring constant of 1–5 N/m, and tip radius of curvature of 5–10 nm. AFM scans were taken at 512 × 512-pixels resolution and produced topographic images of the samples in which the brightness of features increases as a function of height. Typical scanning parameters were as follows: tapping frequency ≈60 kHz, RMS amplitude before engage 1–1.2 V, integral and proportional gains 0.2–0.6 and 0.4–1.2, respectively, set point 0.7–1.0 V, and scanning speed 1–1.5 Hz. It must be emphasized that the entire dynamic reassembly experiments were repeated four times for all samples taken from a single tube. It should be pointed out that many experiments using different sample preparations also yield similar results.
Microrheological Analysis. Viscoelastic property was determined with a rheometer (AR1000, TA Instruments, New Castle, DE) in the Massachusetts Institute of Technology Hatsopoulos Microfluids Laboratory at the Department of Mechanical Engineering. After sonicating RADA16-I solution for 30 min, 550 μl of the solution was placed on the plate of a rheometer. A 4-cm-diameter, 2° stainless-steel cone with a truncation at 56 μm was used so that the tip was 56 μm above the plate, and a solvent trap was placed around the cone with vacuum grease applied to obtain a liquid tight seal. Storage modulus G' was measured at a constant frequency of 1 Hz. The sample was tested with 2 μm oscillatory torque at 25°C.
Results and Discussion
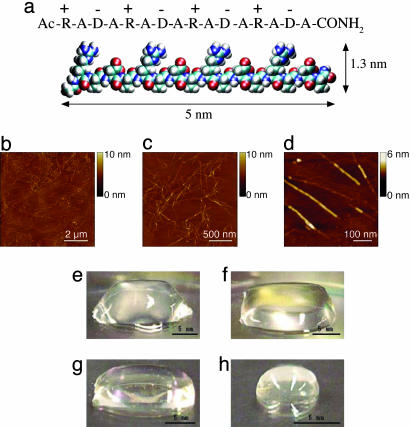
Nanofiber Scaffold Formation. The ionic self-complementary peptide RADA16-I (Fig. 1a) formed nanofibers (Fig. 1 b–d) ranging from a few hundred nanometers to a few microns. Peptide samples in aqueous solution using environmental AFM examination showed similar nanofiber results, suggesting the nanofiber formation is independent of the drying process (see Fig. 6, which is published as supporting information on the PNAS web site). It is interesting to observe that at high resolution the nanofibers appeared to have distinct layers, especially in some segments (Fig. 1d). The difference in height is ≈1.3–1.5 nm, a similar dimension as a single thickness of a peptide. Fig. 1 e–h shows the peptide scaffold hydrogel at various concentrations, 0.6–3 mM (1–5 mg/ml, wt/vol, or 99.5–99.9% water content). The scaffold hydrogel is completely transparent, which is a very important requirement for accurate image collections for uses in 3D tissue cell cultures.
Fig. 1.
Peptide RADA16-I. (a) Amino acid sequence and molecular model of RADA16-I. The dimensions are ≈5 nm long, 1.3 nm wide, and 0.8 nm thick. (b–d) AFM images of RADA16-I nanofiber scaffold, 8 × 8 μm(b), 2 × 2 μm(c), and 0.5 × 0.5 μm(d). Note the different height of the nanofiber, ≈1.3 nm, in d, suggesting a double-layer structure. (e–h) Photographs of RADA16-I hydrogel at various conditions: 0.5 wt% (pH 7.5) (e), 0.1 wt% (pH 7.5, Tris·HCl) (f), 0.1 wt% (pH 7.5, PBS) (g) before sonication, and reassembled RADA16-I hydrogel after four rounds of sonication (h).
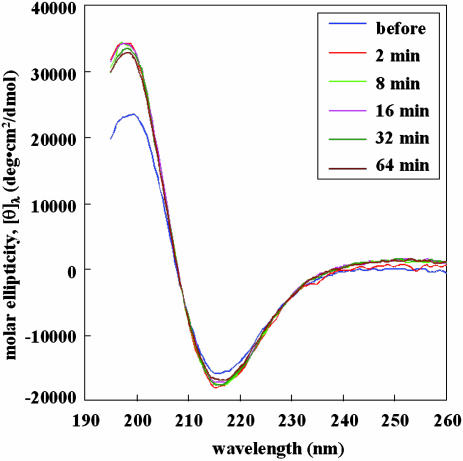
β-Sheet Structure Measurement. It is know that the RADA16-I forms a very stable β-sheet (15), thus its structure can be followed before and after sonication. Since CD only measures the backbone conformation, it cannot precisely determine whether and how the nanofibers reassembled. However, since the structure of β-sheet formation is a prerequisite, CD can determine whether sonication can break the peptide backbones. CD examination of the peptide structures was used to measure the β-sheet at various times before and after sonication (Fig. 2). Before sonication, samples at time points 2, 8, 16, 32, and 64 min were taken after sonication. The typical β-sheet spectra were observed at all time point experiments, indicating the molecular structure and the integrity of the peptides were unchanged before and after sonication. Furthermore, the β-sheet contents (216 nm) remain nearly identical in all time points and slightly higher than the sample before sonication, suggesting conventional β-sheet packing. However, because the degree of β-sheet in the 195-nm region before sonication is different from that after sonication may suggest a different β-sheet twist (ref. 15 and G. Fasman, personal communication in 1994). It is possible that the sonication process resulted in different reassembly packing. This result is not surprising; because reassembly is a dynamic process, variation of packing at molecular backbone is possible. Furthermore, the initial β-sheet packing without sonication may not result in the most well ordered packing. Like packing wood or plastic strips in a box, the initial packing may not result in the tightest packing. A more precise measurement through x-ray fiber diffraction may clarify these subtle differences.
Fig. 2.
CD examination of the peptide structures at various times before and after sonication. Sample time points, 2, 8, 16, 32, and 64 min, were taken after sonication. The typical β-sheet spectra were observed at all time point experiments, indicating the molecular structure and the integrity of the peptides were unchanged before and after sonication. Furthermore, the β-sheet contents (216 nm) remain nearly identical in all time points and slightly higher than the sample before sonication, suggesting tight β-sheet packing. However, the degree of β-sheet twist (195-nm region) before sonication is different, suggesting different β-sheet packing. It is possible that the sonication process removed less packing.
The CD experiments suggest that at the molecular scale the individual peptides did not break into monomers but formed stable β-sheets and were packed together even after repeated sonication times.
Self-Assembling Peptide Reassembly. To unequivocally demonstrate the reassembly process, all samples were taken from a single tube throughout the entire experiment (the experimental design is illustrated in Fig. 7, which is published as supporting information on the PNAS web site). The time points of the peptide samples were obtained from the identical tube after four repeated procedures of sonication and reassembly. As a control, an aliquot of the peptide was removed before the first sonication to determine the nanofiber structure (Figs. 1 and 7).
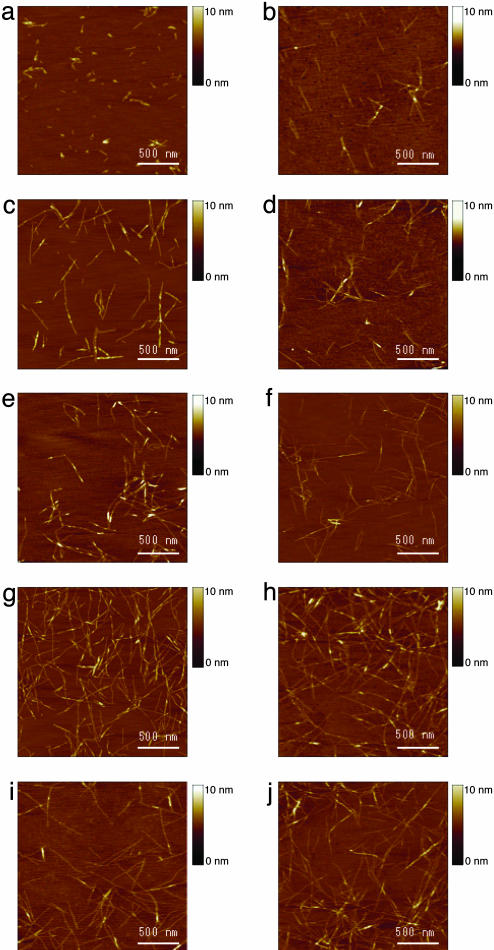
AFM images revealed that the nanofibers range from several hundred nanometers to a few microns in length before sonication. After sonication, the fragments were broken into ≈20–100 nm. The kinetics of the nanofiber reassembly was followed closely at 1, 2, 4, 8, 16, 32, and 64 min and 2, 4, and 24 h (Fig. 3). The nanofiber length reassembly is a function of time: by 2 h, the nanofibers had essentially reassembled to their original length. This remarkable and rapid reassembly is interesting because there may be little nucleation for regrowth of the nanofiber from the addition of monomers that could be produced only during sonication. It is plausible that a large population of the sonicated nanofiber fragments contains many overlapped cohesive ends caused by undisrupted alanine hydrophobic sides that may quickly find each other (Fig. 1d). The situation is analogous and commonly found in sonicated and enzymatic digested DNA fragments.
Fig. 3.
AFM images of RADA16-I nanofiber at various time points after sonication. The observations were made by using AFM immediately after sample preparation: 1 min (a), 2 min (b), 4 min (c), 8 min (d), 16 min (e), 32 min (f), 64 min (g),2h(h),4h(i), and 24 h (j). Note the elongation and reassembly of the peptide nanofibers over time.
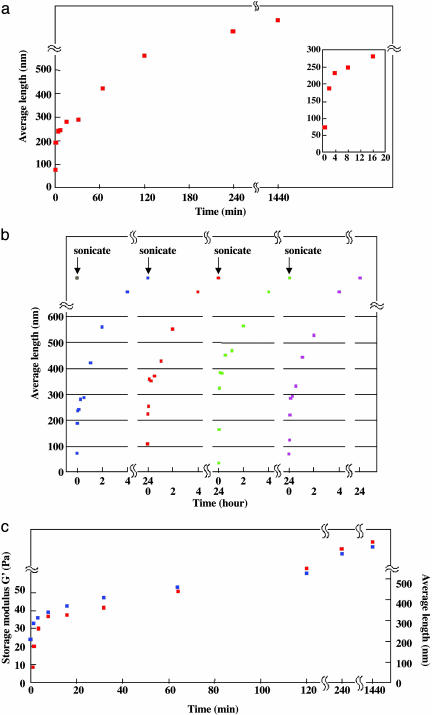
Kinetics of Nanofiber Reassembly. The kinetics of reassembly is shown in Fig. 4. The first reassembly kinetics is shown as a function of time, and the initial 32-min reassembly is shown in detail in Fig. 4a Inset. Perhaps, like DNA reassembly, the reassembly largely depends on the concentrations of the short complementary fragments. In this case, the fragments are the sonicated peptide nanofibers with the possible presence of sonicated monomers.
Fig. 4.
Dynamic reassembly of RADA16-I self-assembling peptide followed by AFM and rheology. (a) Kinetics of the average reassembled length of RADA16-I nanofiber obtained from AFM. The increasing average length of RADA16-I fibers grew as a function of time. (Inset) Initial 20 min, a log-phase reassembly. (b) Four repeated reassembly processes observed by AFM. The average length of RADA16-I nanofiber is shown as a function of time. Note the repeated complete reassembly of the nanofiber length, suggesting robust and reproducible reassembly events. It is likely that this process can be repeated many more times. (c) Increase of elasticity of RADA16-I scaffold hydrogel after sonication. Storage modulus G' (blue square) is shown as a function of time (left y axis). The average length (red square) of RADA16-I fiber from four time-repeated experiments was also plotted in the same way (right y axis).
The reassembled peptide nanofibers were subjected to sonication for three additional times. In each case, it was observed that the final nanofibers reached the original length, from a few hundred nanometers to a few micrometers (Fig. 4b). This reassembly progress correlates well with the rheology experiments where the viscoelastic property increases as a function of time, thus the nanofiber length. The storage modulus G' (Pa) of the scaffold hydrogel became higher and higher, reaching >50 Pa, whereas the nanofibers reassemble into >1,000-nm average fiber length (Fig. 4c). These observations are consistent with each other, namely, the longer the nanofibers, the higher the storage modulus G'.
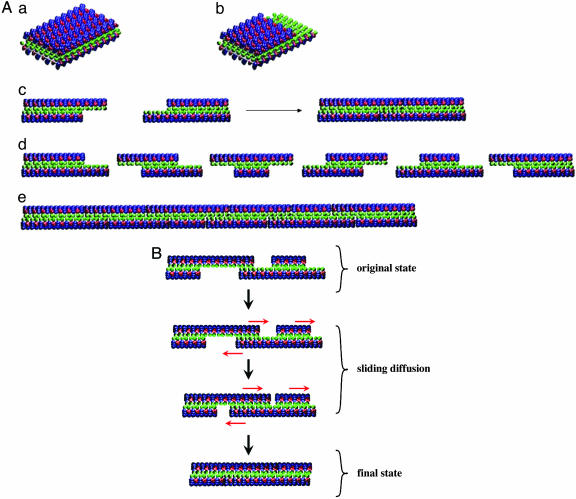
A Suggested Plausible Reassembly Process. To understand dynamic reassembly, we proposed a plausible sliding diffusion molecular model to interpret these observations of reassembly of the self-assembling RADA16-I peptides (Fig. 5). Unlike the left-handed helical structures observed in KFE8 (18), a different self-assembling peptide, no helical structures were observed for RADA16-I with AFM and transmission electron microscopy (16).
Fig. 5.
A proposed molecular sliding diffusion model for dynamic reassembly of self-assembling RADA16-I peptides. When the peptides form stable β-sheets in water, they form intermolecular hydrogen bonds along the peptide backbones. The β-sheets have two distinctive sides, one hydrophobic with an array of alanines and the other with negatively charged aspartic acids and positively charged arginines (see Fig. 1a). These peptides form antiparallel β-sheet structures (gray arrows indicate the directions). The alanines form overlap packed hydrophobic interactions in water; these structures are found in silk fibroin from silkworm and spiders. On the charged sides, both positive and negative charges are packed together through intermolecular ionic interactions in a checkerboard-like manner. (A) These nanofiber fragments can form various assemblies similar to restriction-digested DNA fragments. (a) Blunt ends. (b) Semiprotruding ends (see Fig. 1d). (c) These fragments with protruding ends could reassemble readily through hydrophobic interactions. (d) The fragments with semiprotruding and various protruding ends. (e) These fragments can reassemble readily. (B) The proposed sliding diffusion pathway of a single peptide nanofiber. When the fragments of nanofiber first meet, the hydrophobic sides may not fit perfectly but have gaps. However, the nonspecific hydrophobic interactions permit the nanofiber to slide diffusively along the fiber in either direction, which minimizes the exposure of hydrophobic alanines and eventually fills the gaps. The sliding diffusion phenomenon was also reported for nucleic acids of poly(A) and poly(U) in 1956 (27, 28). Green indicates alanines; red indicates negatively charged aspartic acids; and blue indicates positively charged arginines. For clarity, these β-sheets are not presented as twisted strands.
For molecular modeling clarity, these RADA16-I β-sheets are presented as nontwisted strands. It is known that these peptides form stable β-sheet structure in water, thus they not only form the intermolecular hydrogen bonding on the peptide backbones, but they also have two distinctive sides, one hydrophobic with array of overlapping alanines (Fig. 5, green color sandwiched inside), similar to that found in silk fibroin or spider silk assemblies (24–26). The other side of the backbones has negatively charged (-) aspartic acids, represented as red in Fig. 5, and positively charged (+) arginines, represented as blue in Fig. 5.
The alanines form packed hydrophobic interactions in water; during sonication the hydrophobic interaction could be disrupted mechanically. However, these hydrophobic cohesive ends could find each other quickly in water since the exposure of hydrophobic alanine arrays to water is energetically unfavorable. Because the hydrophobic alanines interaction is nonspecific, they can slide diffusively along the nanofiber, like trains on train tracks. The same sliding diffusion phenomenon was also observed in nucleic acids where poly(A) and poly(U) form complementary base pairings that can slide diffusively along the chains (27, 28). If, however, the bases are heterogonous, containing G, A, T, and C, the bases cannot undergo sliding diffusion. Likewise, if the hydrophobic side of the peptides does not always contain alanine, such as valine and isoleucine, it would become more difficult for sliding diffusion to occur because of structure constraint.
On the charged side, both positive and negative charges are packed together through intermolecular ionic interactions in a checkerboard manner (looking from the top). Likewise, the collectively complementary + and - ionic interactions may also facilitate the reassembly. Like restriction-digested DNA fragments, these nanofiber fragments could form various assemblies: blunt, semiprotruding, and protruding ends. The fragments with semiprotruding and various protruding ends and blunt ends can reassemble readily through hydrophobic and ionic interactions.
Other Implications. Nanofibers are ubiquitous and have been found in many self-assembled biological and nonbiological materials, including short peptides, as short as two to five residues (29), medium-sized peptides (30–33), natural occurring proteins (34–42), selected proteins from random peptide/protein libraries (43), artificially designed proteins (44–47), lipids (48, 49), and polysaccharides (50), and synthetic carbon nanofibers (Electrovac, Klosterneuburg, Austria, www.electrovac.com/de/cnt_cnt.htm and Synthecon, Houston, www.synthecon.com).
Unlike processed polymer fibers in which the fragments of polymers cannot readily undergo reassembly without addition of catalysts or through material processing, the supramolecular self-assembly and reassembly event we uncovered here is likely to be widespread in many unrelated fibrous biological materials. Self-assembly and reassembly are very important properties for fabricating novel materials, and it is necessary to fully understand their detailed processes to design better biological materials.
It should also be pointed out that self-assembling peptides have some structural features similar to other protein fibrils commonly found in amyloids. It has been reported that several proteins undergo conformational changes to form stable β-sheets that further form fibrils. Addition of monomer of the same proteins results in fibril growth (51–55).
Furthermore, this finding may have significant implications beyond supramolecular chemistry and biological materials self-assembly. For instance, nanofibers are also found in protein amyloid (33–42, 51–55). It is possible that some of the self-assembled amyloid nanofibers will undergo reassembly to resist drug and other treatments. Understanding such a dynamic amyloid nanofiber formation and finding a way to combat it remains a formidable challenge (56–58).
Supplementary Material
Acknowledgments
We thank Alexander Rich for stimulating discussions and suggesting the sliding diffusion model of the nanofiber assembly that he and Davies found in nucleic acid poly(A) and poly(U) (27, 28); Wonmuk Hwang for making molecular models of the RADA16-I peptide and the various assemblies; and Xiaojun Zhao, Steve Yang, Andrea Lomander, and Jessica Dai for helpful discussions. This work was supported in part by Defense Advanced Research Project Agency/BioComp grants, Defense Advanced Research Project Agency/Air Force Office of Scientific Research grants, and National Science Foundation Grant CCR-0122419 to the Center for Bits and Atoms of the Media Laboratory at the Massachusetts Institute of Technology. We also thank Menicon, Ltd. Nagoya, Japan, for generously supporting H.Y. and our laboratory at the Massachusetts Institute of Technology and Intel Corporation for its academic donation of a computer cluster to our laboratory at the Massachusetts Institute of Technology that was used for the molecular modeling.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: AFM, atomic force microscopy.
References
- 1.Wang, Y., Ameer, G. A., Sheppard, B. J. & Langer, R. (2002) Nat. Biotechnol. 20, 602-606. [DOI] [PubMed] [Google Scholar]
- 2.Lavik, E., Teng, Y. D., Snyder, E. & Langer, R. (2002) Methods Mol. Biol. 198, 89-97. [DOI] [PubMed] [Google Scholar]
- 3.Teng, Y. D., Lavik, E. B., Qu, X., Park, K. I., Ourednik, J., Zurakowski, D., Langer, R. & Snyder, E. Y. (2002) Proc. Natl. Acad. Sci. USA 99, 3024-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee, H., Cusick, R. A., Browne, F., Ho Kim, T., Ma, P. X., Utsunomiya, H., Langer, R. & Vacanti, J. P. (2002) Transplantation 73, 1589-1593. [DOI] [PubMed] [Google Scholar]
- 5.Girotto, D., Urbani, S., Brun, P., Renier, D., Barbucci, R. & Abatangelo, G. (2003) Biomaterials 24, 3265-3275. [DOI] [PubMed] [Google Scholar]
- 6.Perets, A., Baruch, Y., Weisbuch, F., Shoshany, G., Neufeld, G. & Cohen, S. (2003) J. Biomed. Mater. Res. 65, 489-497. [DOI] [PubMed] [Google Scholar]
- 7.Bensaid, W., Triffitt, J. T., Blanchat, C., Oudina, K., Sedel, L. & Petite, H. (2003) Biomaterials 24, 2497-2502. [DOI] [PubMed] [Google Scholar]
- 8.Langer, R. & Vacanti, J. P. (1993) Science 260, 920-926. [DOI] [PubMed] [Google Scholar]
- 9.Langer, R. (1999) J. Control Release 62, 7-11. [DOI] [PubMed] [Google Scholar]
- 10.Hubbell, J. A. (1999) Curr. Opin. Biotechnol. 10, 123-129. [DOI] [PubMed] [Google Scholar]
- 11.Zhang, S. (2003) Mater. Today 6, 20-27. [Google Scholar]
- 12.Zhang, S. (2003) Nat. Biotechnol. 21, 1171-1178. [DOI] [PubMed] [Google Scholar]
- 13.Zhang, S., Holmes, T., Lockshin, C. & Rich, A. (1993) Proc. Natl. Acad. Sci. USA 90, 3334-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang, S., Lockshin, C., Cook, R. & Rich, A. (1994) Biopolymers 34, 663-672. [DOI] [PubMed] [Google Scholar]
- 15.Zhang, S., Holmes, T., DiPersio, M., Hynes, R. O., Su, X. & Rich, A. (1995) Biomaterials 16, 1385-1393. [DOI] [PubMed] [Google Scholar]
- 16.Holmes, T., Delacalle, S., Su, X., Rich, A. & Zhang, S. (2000) Proc. Natl. Acad. Sci. USA 97, 6728-6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kisiday, J., Jin, M., Kurz, B., Hung, H., Semino, C., Zhang, S. & Grodzinsky, A. J. (2002) Proc. Natl. Acad. Sci. USA 99, 9996-10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marini, D., Hwang, W., Lauffenburger, D. A., Zhang, S. & Kamm, R. (2002) NanoLetters 2, 295-299. [Google Scholar]
- 19.Caplan, M., Moore, P., Zhang, S., Kamm, R. & Lauffenburger, D. (2000) Biomacromolecules 1, 627-631. [DOI] [PubMed] [Google Scholar]
- 20.Caplan, M., Schwartzfarb, E., Zhang, S., Kamm, R. & Lauffenburger, D. A. (2002) Biomaterials 23, 219-227. [DOI] [PubMed] [Google Scholar]
- 21.Caplan, M. R., Schwartzfarb, E. M., Zhang, S., Kamm, R. D. & Lauffenburger, D. A. (2002) J. Biomater. Sci. 13, 225-236. [DOI] [PubMed] [Google Scholar]
- 22.Zhang, S. (2002) Biotechnol. Adv. 20, 321-339. [DOI] [PubMed] [Google Scholar]
- 23.Zhang, S. (2004) Nat. Biotechnol. 22, 151-152. [DOI] [PubMed] [Google Scholar]
- 24.Pauling, L. (1961) The Nature of Chemical Bond (Cornell Univ. Press, Ithaca, NY), 3rd Ed.
- 25.Asakura, T., Sugino, R., Okumura, T. & Nakazawa, Y. (2002) Protein Sci. 11, 1873-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayashi, C. Y. & Lewis, R. V. (2000) Science 287, 1477-1479. [DOI] [PubMed] [Google Scholar]
- 27.Rich, A. & Davies, D. R. (1956) J. Am. Chem. Soc. 78, 3548-3549. [Google Scholar]
- 28.Felsenfeld, G., Davies, D. R. & Rich, A. (1957) J. Am. Chem. Soc. 79, 2023-2024. [Google Scholar]
- 29.Reches, M. & Gazit, E. (2003) Science 300, 625-627. [DOI] [PubMed] [Google Scholar]
- 30.Ryadnov, M. G. & Woolfson, D. N. (2003) Nat. Mater. 2, 329-332. [DOI] [PubMed] [Google Scholar]
- 31.Gay, N. J., Packman, L. C., Weldon, M. A. & Barna, J. C. (1991) FEBS Lett. 291, 87-91. [DOI] [PubMed] [Google Scholar]
- 32.Aggeli, A., Bell, M., Boden, N., Keen, J. N., Knowles, P. F., McLeish, T. C., Pitkeathly, M. & Radford, S. E. (1997) Nature 386, 259-262. [DOI] [PubMed] [Google Scholar]
- 33.Matsumura, S., Uemura, S. & Mihara, H. (2004) Chemistry 10, 2789-2794. [DOI] [PubMed] [Google Scholar]
- 34.van Raaij, M. J., Mitraki, A., Lavigne, G. & Cusack, S. (1999) Nature 401, 935-938. [DOI] [PubMed] [Google Scholar]
- 35.Kelly, J. W. & Balch, W. E. (2003) J. Cell Biol. 161, 461-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dobson, C. M. (1999) Trends Biochem. Sci. 24, 329-332. [DOI] [PubMed] [Google Scholar]
- 37.Jimenez, J. L., Guijarro, J. I., Orlova, E., Zurdo, J., Dobson, C. M., Sunde, M. & Saibil, H. R. (1999) EMBO J. 18, 815-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krebs, M. R., Wilkins, D. K., Chung, E. W., Pitkeathly, M. C., Chamberlain, A. K., Zurdo, J., Robinson, C. V. & Dobson, C. M. (2000) J. Mol. Biol. 300, 541-549. [DOI] [PubMed] [Google Scholar]
- 39.Rochet, J. C. & Lansbury, P. T., Jr. (2000) Curr. Opin. Struct. Biol. 10, 60-68. [DOI] [PubMed] [Google Scholar]
- 40.Polymeropoulos, M. H. (1998) Ann. Neurol. 44, Suppl. 1, S63-S64. [DOI] [PubMed] [Google Scholar]
- 41.Lee, S. & Eisenberg, D. (2003) Nat. Struct. Biol. 10, 725-730. [DOI] [PubMed] [Google Scholar]
- 42.Scheibel, T., Parthasarathy, R., Sawicki, G., Lin, X. M., Jaeger, H. & Lindquist, S. L. (2003) Proc. Natl. Acad. Sci. USA 100, 4527-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.West, M. W., Wang, W., Patterson, J., Mancias, J. D., Beasley, J. R. & Hecht, M. H. (1999) Proc. Natl. Acad. Sci. USA 96, 11211-11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopez De La Paz, M., Goldie, K., Zurdo, J., Lacroix, E., Dobson, C. M., Hoenger, A. & Serrano, L. (2002) Proc. Natl. Acad. Sci. USA 99, 16052-16057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petka, W. A., Harden, J. L., McGrath, K. P., Wirtz, D. & Tirrell, D. A. (1998) Science 281, 389-392. [DOI] [PubMed] [Google Scholar]
- 46.Nowak, A. P., Breedveld, V., Pakstis, L., Ozbas, B., Pine, D. J., Pochan, D. & Deming, T. J. (2002) Nature 417, 424-428. [DOI] [PubMed] [Google Scholar]
- 47.Schneider, J. P., Pochan, D. J., Ozbas, B., Rajagopal, K., Pakstis, L. & Kretsinger, J. (2002) J. Am. Chem. Soc. 124, 15030-15037. [DOI] [PubMed] [Google Scholar]
- 48.Schnur, J. M. (1993) Science 262, 1669-1676. [DOI] [PubMed] [Google Scholar]
- 49.Spector, M. S., Easwaran, K. R., Jyothi, G., Selinger, J. V., Singh, A. & Schnur, J. M. (1996) Proc. Natl. Acad. Sci. USA 93, 12943-12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Astley, O. M. & Donald, A. M. (2001) Biomacromolecules 2, 672-680. [DOI] [PubMed] [Google Scholar]
- 51.Jarrett, J. T. & Lansbury, P. T., Jr. (1993) Cell 73, 1055-1058. [DOI] [PubMed] [Google Scholar]
- 52.Krebs, M. R., Morozova-Roche, L. A., Daniel, K., Robinson, C. V. & Dobson, C. M. (2004) Protein Sci. 13, 1933-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Inoue, Y., Kishimoto, A., Hirao, J., Yoshida, M. & Taguchi, H. (2001) J. Biol. Chem. 276, 35227-35230. [DOI] [PubMed] [Google Scholar]
- 54.Naiki, H., Higuchi, K., Nakakuki, K. & Takeda, T. (1991) Lab. Invest. 65, 104-110. [PubMed] [Google Scholar]
- 55.Scheibel, T., Kowal, A. S., Bloom, J. D. & Lindquist, S. L. (2001) Curr. Biol. 11, 366-369. [DOI] [PubMed] [Google Scholar]
- 56.Brown, P., Rau, E. H., Johnson, B. K., Bacote, A. E., Gibbs, C. J., Jr., & Gajdusek, D. C. (2000) Proc. Natl. Acad. Sci. USA 97, 3418-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gajdusek, D. C. (1994) Mol. Neurobiol. 8, 1-13. [DOI] [PubMed] [Google Scholar]
- 58.Caughey, B. & Lansbury, P. T., Jr. (2003) Annu. Rev. Neurosci. 26, 267-298. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.