Abstract
In contrast to humans, several primate species are believed to have harbored simian immunodeficiency viruses (SIVs) since ancient times. In particular, the geographically dispersed species of African green monkeys (AGMs) are all infected with highly diversified SIVagm viruses at high prevalences (greater than 50% of sexually mature individuals) without evident diseases, implying that the progenitor monkeys were infected prior to their dispersal. If this is correct, AGMs would be expected to have accumulated frequent resistance-conferring polymorphisms in host genes that are important for SIV replication. Accordingly, we analyzed the coding sequences of the CCR5 coreceptors from 26 AGMs (52 alleles) in distinct populations of the four species. These samples contained 29 nonsynonymous coding changes and only 15 synonymous nucleotide substitutions, implying intense functional selection. Moreover, 24 of the resulting amino acid substitutions were tightly clustered in the CCR5 amino terminus (D13N in the vervets and Y14N in the tantalus species) or in the first extracellular loop (Q93R and Q93K in all species). The Y14N substitution was extremely frequent in the 12 wild-born African tantalus, with 7 monkeys being homozygous for this substitution and 4 being heterozygous. Although two of these heterozygotes and the only wild-type homozygote were naturally infected with SIVagm, none of the Y14N homozygotes were naturally infected. A focal infectivity assay for SIVagm indicated that all five tested SIVagms efficiently use CCR5 as a coreceptor and that they also use CXCR6 (STRL33/Bonzo) and GPR15 (BOB) with lower efficiencies but not CXCR4. Interestingly, the D13N, Y14N, Q93R, and Q93K substitutions in AGM CCR5 all strongly inhibited infections by the SIVagm isolates in vitro. The Y14N substitution eliminates a tyrosine sulfation site that is important for infections and results in partial N-linked glycosylation (i.e., 60% efficiency) at this position. Nevertheless, the CCR5(Y14N) component that lacks an N-linked oligosaccharide binds the chemokine MIP-lβ with a normal affinity and is fully active in signal transduction. Similarly, D13N and Q93R substitutions did not interfere with signal transduction. Thus, the common substitution polymorphisms in AGM CCR5 strongly inhibit SIVagm infections while substantially preserving chemokine signaling. In contrast, polymorphisms of human CCR5 are relatively infrequent, and the amino acid substitutions are randomly situated and generally without effects on coreceptor function. These results support an ancient coevolution of AGMs and SIVagm viruses and establish AGMs as a highly informative model for learning about host proteins that play critical roles in immunodeficiency virus infections.
Although human immunodeficiency virus type 1 (HIV-1) and HIV-2 have become prevalent only during the last few decades and probably arose by zoonoses from chimpanzees (11, 20) and sooty mangabeys (21, 28), respectively, there is evidence that several primate species have been infected since ancient times. For example, African green monkeys (AGMs) consist of four major species, i.e., vervets (Chlorocebus pygerythrus), tantalus (C. tantalus), grivets (C. aethiops), and sabaeus (C. sabaeus), that are geographically segregated and dispersed throughout Africa (2, 5, 50, 56). All tested populations are heavily infected (greater than 50% of sexually mature adults) by distantly related strains of the simian immunodeficiency virus SIVagm (2, 5, 26, 30–33, 41, 50). These results have implied that the AGM progenitor population was infected prior to its speciation and geographic dispersal (2, 5, 30, 31). Moreover, the highly evolved endemic infections of AGMs and sooty mangabeys have not been associated with diseases, whereas the emergent infections of humans and the experimental transfers of SIVs into naive primate species have resulted in immunodeficiencies (25, 27, 38, 56).
The latter observations are consistent with the hypothesis that prolonged coevolution of AGMs with SIVagm viruses may have resulted in selection for mutations in host genes that control susceptibility to virus-induced immunodeficiency. Indeed, it is well known that attenuations of infectious diseases often result from selection for resistance alleles of host genes, with classic examples being the resistances to malaria conferred by high-frequency polymorphisms for sickle cell hemoglobin and thalassemia (55, 64). Accordingly, genes that mediate host resistances, including those involved in immune responses, are generally extraordinarily polymorphic (55). The resulting diversity of the host species may also select for a corresponding diversity of the infectious agent, which can further decrease the likelihood that any single infection is highly pathogenic.
There has been great interest in identifying human genes that cause resistance to HIV-1 infections and to AIDS progression. A major focus has concerned alleles of the HIV-1 coreceptor CCR5 and other coreceptors or chemokines that may modulate HIV-1 pathogenesis (24, 29, 35, 42, 45, 47–49, 53, 61, 63, 65). However, since HIV infections are relatively recent and occur at low frequencies in most populations, the resistance alleles that have been found evolved independently of HIV and may have been selected by other infections. For example, the Δ32 CCR5 human resistance allele occurs in approximately 13% of Caucasians and may have been selected for resistance to poxvirus (3, 39, 42, 53, 61, 63) or to rheumatoid arthritis (22). It is a null allele, and the encoded protein is absent from cell surfaces (6, 42, 59, 61). Similarly, amino acid substitutions in human CCR5 occur at low frequencies and at random positions in the protein, and they have generally not been shown to cause resistances to HIV-1 infections (3, 6, 10, 66). In contrast, we would anticipate that AGM CCR5 polymorphisms might occur at relatively high frequencies and that the prevalent amino acid substitutions would cluster at critical sites that are required for infections by immunodeficiency viruses but are of lesser importance for chemokine reception.
To address this, we previously analyzed CCR5-coding sequences derived from five AGMs (37). Four of these AGMs were heterozygous for nonsynonymous CCR5 mutations that caused amino acid substitutions D13N, Y14N, Q93R, and Q93K, and one was homozygous for the wild-type allele. These substitutions all inhibited AGM CCR5 function as a coreceptor for different R5 strains of HIV-1. We also found several substitution mutations in a study of four other AGM CCR5 alleles (51). We have now extended these analyses by studying the CCR5-coding sequences of 26 AGM DNAs (52 alleles), including samples from different species living in Africa. The latter monkeys had known histories, and some were naturally or experimentally infected with SIVagm. Moreover, we developed a focal infectivity assay for quantitatively determining titers of SIVagm infections, and we used it to study the coreceptor requirements and effects of AGM CCR5 polymorphisms on these infections. In addition, we quantitatively analyzed signal transduction by the wild-type and mutant CCR5 proteins. Our results support the hypothesis that CCR5 substitution polymorphisms are prevalent in AGMs and that they have resulted from prolonged selection pressures caused by the large reservoirs of SIVagm and by the countervailing pressure to preserve chemokine-dependent CCR5 signaling activity. An important corollary is that AGMs are likely to contain a repository of mechanistically informative polymorphisms in different genes that are critical for infections and pathogenesis of immunodeficiency viruses.
MATERIALS AND METHODS
Cells and viruses.
AGM cell lines CV-1, BS-C-1, and Vero were from the American Type Culture Collection (Manassas, Va.). BS-C-1 and Vero cells were maintained in minimum essential medium (MEM) with 10% fetal bovine serum (FBS) and 0.1 mM MEM nonessential amino acids (Life Technologies, Inc., Grand Island, N.Y.). HeLa-CD4 (clone HI-J) cells were previously described (33). CV-1-CD4 (clone CB.14) cells were generated by transduction with the SFF-CD4 retroviral vector and limiting-dilution cloning as previously described for HeLa-CD4 cells (33). HeLa and CV-1 cells were grown in Dulbecco MEM (DMEM) supplemented with 10% FBS (Life Technologies). Molt4-8 cells infected with SIVagm strains from vervets (strains 12, Cpa266, and Cpa27) (12, 34) were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) (contributed by Ronald Desrosiers). Molt4-8 cells were grown in RPMI 1640 medium supplemented with 10% FBS (Life Technologies). Supernatants were harvested every 3 or 4 days over a period of 2 weeks in culture and filtered through a 0.45-μm-pore-size filter. The grivet SIVagm strain gri-1 (19) and the sabaeus SIVagm strain sab-2 (2) were obtained as cell-free-virus-containing tissue culture supernatants (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH [contributed by Jonathan Allan]), which were incubated with approximately 2 × 106 uninfected Molt 4-8 cells (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH [contributed by Ronald Desrosiers]) for 2 h at 37°C, washed, and placed into culture. Supernatants from the infected Molt4-8 cells were then harvested as described above.
AGM genomic DNA.
We analyzed DNAs from peripheral blood mononuclear cells (PBMC) of the four AGM species and from the three AGM cell lines (CV-1 [ATCC CCL-70], BS-C-1 [ATCC CCL-26], and Vero [ATCC CCL-81]). The genomic DNA samples from sabaeus monkeys 9308, 9310, 9312, and 9314 and vervet monkeys 6243, 9648, and 9649 were kindly provided by Jonathan Allan (Department of Virology and Immunology, Southwest Foundation for Biomedical Research, San Antonio, Tex.). PBMC were isolated from our other sabaeus monkeys that were wild born in Senegal (Table 1). One was uninfected, two were naturally infected, and one was experimentally infected by wild-type virus SIVagm.sab92018 according to a previously described protocol (14). PBMC were also isolated from 12 tantalus AGMs wild born in distinct geographic regions of the Central African Republic. One was uninfected, three were naturally infected by SIVagm.tan, and eight were experimentally infected (Table 1). Experimental infection of tantalus B87 by intravenous inoculation of SIVagm.tanB14 (50) was previously described (52). The other seven tantalus monkeys were similarly infected either by SIVagm.tanB14 (tantalus B85, B126, B127, and B143) or by three other wild-type SIVagm viruses (tantalus B111, B133, and B142). Genomic DNAs were prepared by standard methods (4).
TABLE 1.
CCR5 polymorphisms and natural histories of AGMs used in this investigation
| AGM subspecies | Sample | Origina | Polymorphisms and linkages (where determined)b | Infection status |
|---|---|---|---|---|
| Tantalus | B57 | CAR | A278G (Q93R), G618A | Naturally infected |
| B83 | CAR | T40A (Y14N), T660C | Naturally infected | |
| B85 | CAR | T52A (Y14N), G696A | Experimentally infected | |
| B87 | CAR | T40A (Y14N)∗ | Experimentally infected | |
| B111 | CAR | T40A (Y14N)∗ | Experimentally infected | |
| B126 | CAR | T40A (Y14N) | Experimentally infected | |
| B127 | CAR | T40A (Y14N), G439A (V147M) | Experimentally infected | |
| B129 | CAR | T40A (Y14N)∗ | Uninfected | |
| B132 | CAR | T40A (Y14N)∗ | Experimentally infected | |
| B141 | CAR | T40A (Y14N), C678T | Naturally infected | |
| B142 | CAR | T40A (Y14N)∗ | Experimentally infected | |
| B143 | CAR | T40A (Y14N)∗ | Experimentally infected | |
| Sabaeus | D92017 | Senegal | None | Naturally infected |
| D94005 | Senegal | T84C | Naturally infected | |
| D96010 | Senegal | T84C∗, A160G (N57S), A487G (R163G) | Experimentally infected | |
| D96029 | Senegal | None | Uninfected | |
| 9308 | Colony | T84C | Uninfected | |
| 9310 | Colony | T84C | Uninfected | |
| 9312 | Colony | T84C, A278G (Q93R) | Uninfected | |
| 9314 | Colony | T84C | Uninfected | |
| Vervet | 6243c | Colony | G37A (D13N), T112A (S38T)-C277A (Q93K) | Uninfected |
| 9648 | Colony | None | Uninfected | |
| 9649 | Colony | None | Uninfected | |
| Vero | Unknown | T84C | Uninfected | |
| Grivet | BS-C-1 | Unknown | T84C, C277A (Q93K) | Uninfected |
| Unknown | CV-1c | Unknown | T40A (Y14N)-G1056C (L352F), T300C | Uninfected |
Monkeys from the Central African Republic (CAR) and from Senegal were captured in distinct geographic regions between 1988 and 1995, and they did not derive from a single breeding troop. Several monkey DNAs were obtained from virus-free primate colonies maintained in the United States or the Caribbean. The geographic origins of the monkeys used to derive the BS-C-1 and CV-1 cell lines are unknown.
Nucleotide substitutions were determined as described in Materials and Methods. Each substitution is reported as the wild-type nucleotide followed by the base number starting from the A in the ATG start codon followed by the substituted base. Nonsynonymous substitutions are followed by parentheses showing the predicted amino acid change. Amino acid substitutions are numbered by the codon starting from the ATG start codon. All substitutions were heterozygous (i.e., both the wild-type and mutant bases were seen in electropherograms from that sample), except those marked with an asterisk, which were homozygous. Where linkage was determined, the linked substitutions are connected by a dash.
Linkage of the nucleotide substitutions in these genomic DNA samples was determined by cloning of the entire CCR5-coding sequence from both alleles. In all cases, the nucleotide substitutions identified by direct sequencing of the PCR products were identical to those found in the cloned CCR5-coding sequences. For sample Vervet 6243, the T112A (S38T) and C277A (Q93K) substitutions were found to be on one allele, whereas the G37A (D13N) substitution was found on the other allele. As previously reported (37), there were two alleles identified from the CV-1 cell line, one which contained the T300C substitution relative to the consensus AGM nucleotide sequence and one which contained the T40A (Y14N) and G1056C (L352F) substitutions.
Sequence analysis of CCR5-coding regions.
Two approximately 600-bp PCR products were generated by amplifying the genomic DNA with the Pfu proofreading thermostable DNA polymerase (Stratagene, La Jolla, Calif.). Conditions for the PCRs were as previously described (37). Briefly, Pfu was used according to the manufacturer's recommendations in 100-μl reaction mixtures, with 500 ng of genomic DNA as a template. An initial denaturing step of 95°C for 45 s was followed by 25 cycles of 95°C for 45 s, 55°C for 45 s, and 72°C for 2.5 min; this was followed by a final extension step of 72°C for 10 min. The 5′ half of the CCR5-coding region was amplified with the primers CCR5F (5′ cggcggggatccGGGTGGAACAAGATGGATTATC 3′) and AGMR (5′ ACTGTATGGAAAATGAGAGCTGC 3′). The 3′ half was amplified with the primers AGMF2 (5′ CTCCCAAGAATCATCTTTACCAG 3′) and CCR5R (5′ gccgccctcgagCCACTTGAGTCCGTGTCACAAG 3′), where the lowercase letters indicate 5′ extensions which are not derived from CCR5 sequences. The PCR products were then purified by agarose gel electrophoresis and gel extraction using a Qiaquick gel extraction kit (Qiagen Inc., Valencia, Calif.). Purified PCR products (20 ng) were sequenced in both directions using the same primers used for the PCRs by automated fluorescent dye-terminator sequencing in the Microbiology and Molecular Immunology Core Facility at Oregon Health Sciences University on a 377 DNA sequencer (PE Applied Biosystems, Foster City, Calif.). Because the PCR products overlap, this analysis allowed for the sequence of the entire CCR5-coding region to be determined from both strands for all except the 5′ and 3′ ends of approximately 20 bp, which were determined from only one strand. For DNA samples from CV-1 cells, the cloning of the full-length CCR5-coding sequences was as previously described (37). In this study the full-length coding region was amplified from sample 6243 using the CCR5F and CCR5R primers. These ≈1.1-kb PCR products were subcloned into pBluescript II (KS+) (Stratagene) and sequenced as previously described (37). For both the CV-1 and 6243 DNA samples, the analysis of the full-length alleles confirmed the sequencing of the 600-bp PCR products and allowed for determination of the linkage of the polymorphisms in these monkeys. The PCR and sequencing results reported in this study were unambiguous and reproducible.
Coreceptor expression vectors.
The plasmid encoding CXCR4 cDNA (pcDNA3-LESTR) was provided by Marcal Loetscher (Theodor Kocher Institute, University of Bern, Bern, Switzerland). pcDNA3 expression vectors containing human, NIH/Swiss mouse, wild-type AGM, AGM(D13N), AGM(Q93R), and AGM(Y14N, L352F) CCR5-coding sequences were previously described (37). pcDNA3-AGM(S38T, Q93K) was constructed by subcloning the AGM(S38T, Q93K) (from AGM sample 6243 [see above]) coding sequence from pBluescript II (KS+)-AGM(S38T, Q93R) into pcDNA3 (Invitrogen, Carlsbad, Calif.) using the BamHI and XhoI restriction sites flanking the CCR5-coding region. The AGM(S38T)-coding sequence was generated by splicing the BamHI-to-MscI fragment from pBluescript II (KS+)-AGM(S38T, Q93K) into pBluescript II (KS+)–wild-type AGM digested with the same enzymes. Likewise, the AGM(Q93K)-coding sequence was generated by splicing the MscI-to-BglII fragment from pBluescript II (KS+)-AGM(S38T, Q93K) into pBluescript II (KS+)–wild-type AGM digested with the same enzymes. The resulting AGM(S38T) and AGM(Q93K) CCR5-coding sequences were then subcloned into pcDNA3 using the BamHI and XhoI restriction sites flanking the CCR5-coding region. The pcDNA3-CXCR6 and pcDNA3-BOB plasmids were generated by PCR amplification of the human CXCR6 (46)- and BOB-coding sequences from pBABE-Bonzo and pBABE-BOB vectors (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH [contributed by Dan Littman]) using primers specific to these coreceptors and containing BamHI and XhoI restriction sites flanking the coding region. The resulting PCR products were then subcloned into pcDNA3 as described above.
Coreceptor activity assays.
Coreceptor activity assays using SIVagm virus isolates were performed by the immunoperoxidase focal infectivity method essentially as described previously for HIV-1 (9, 37). Briefly, coreceptors were transiently expressed in HeLa-CD4 (clone HI-J) or CV-1-CD4 (clone CB.14) cells by transfection by the calcium phosphate method as previously described (37). At 48 h after transfection, cells were seeded at 2 × 104 cells/well in 24-well cluster plates. The cells were infected 24 h after seeding by incubating the cells with various dilutions of the SIVagm stocks in DMEM supplemented with 0.1% FBS in the presence of 8 μg of DEAE-dextran per ml for 2 h at 37°C, after which time the cells were fed with DMEM supplemented with 10% FBS. Seventy-two hours later, cells were fixed and stained as previously described for HIV-1 (9), except that a 1:1,000 dilution of the pooled heat-inactivated sera from four SIVagm-seropositive AGMs (kindly provided by Ronald C. Desrosiers, New England Regional Primate Research Center) was used as the primary antibody and a horseradish peroxidase-conjugated goat anti-monkey immunoglobulin G antibody which cross-reacted with AGM immunoglobulin G (Sigma, St. Louis, Mo.) was used as the second antibody. Controls with no SIVagm and with serial dilutions of SIVagm stocks showed that the resulting immunoperoxidase staining was specific to SIVagm infections (results not shown).
Flow cytometry.
HeLa-CD4 (clone HI-J) cells and HEK293T cells were transiently transfected as described above with the pcDNA3-CCR5 expression vectors. Cells were reseeded into new flasks at 24 h posttransfection and were removed 24 h later with 8 mM EDTA–150 mM NaCl (pH 8.0). Approximately 5 × 105 cells were resuspended in 100 μl of DMEM containing 10% FBS and 0.125 μg of phycoerythrin (PE)-conjugated mouse anti-human CCR5 monoclonal antibody (clone 3A9; B-D PharMingen, San Diego, Calif.). Clone 3A9 was used to detect AGM CCR5 because it cross-reacts with rhesus CCR5 (60) and with AGM CCR5. Cells were incubated with the 3A9 conjugate for 30 min at 37°C, washed three times with DMEM containing 10% FBS, and resuspended in the same medium lacking phenol red. The live stained cells were then analyzed for PE fluorescence using a FACScan flow cytometer (BD Immunocytochemistry Systems, San Jose, Calif.). The gating was set so that only 0.4% of the mock-transfected cells were in the positive window, and we recorded the fluorescence intensity of each cell. Based on the results with both cell types, we concluded that within each experiment the transfection efficiencies for all pcDNA3-CCR5 expression vectors were the same within experimental error. Although not a significant issue in this analysis, CCR5 mutants that are expressed relatively poorly on cell surfaces or that bind the 3A9 antibody weakly can appear to have been transfected less efficiently by this analytical method.
Signal transduction assays.
For expression of cRNA in Xenopus laevis oocytes, cRNAs encoding the CCR5 receptor and the Gβγ K-channel Kir 3.1 were transcribed in vitro with T7 RNA polymerase from the oocyte expression vector as previously described (43). Collagenase-treated, defolliculated stage IV or V oocytes were injected with 50 ng of cRNA and were incubated at 18°C with oocyte Ringer solution ND96 (5 mM HEPES [pH 7.5], 96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2). Experiments were performed at 25°C 3 to 4 days after injection. Potassium currents were measured by two-electrode voltage clamping using a Geneclamp 500 amplifier (Axon Instruments, Foster City, Calif.). Current-voltage protocols were performed with the pClamp 6.0 program (Axon Instruments) run on an IBM-compatible PC interfaced with a Digidata 1200 A/D. Microelectrodes were filled with 3 mM KCl (resistances were <2.5 MΩ), and recordings from voltage-clamped oocytes held at −30 mV were performed with constant perfusion of ND96 containing 100 mM KCl in the presence and absence of different concentrations of ligand. Raw current recordings and current-voltage data were prepared and exported by the pClamp 6.0 software (Axon Instruments). Imported data were analyzed on a Macintosh PowerPC using the analysis software. Data are presented as mean values ± standard errors of the means (SEM). Dose-response curves were fitted to the Michaelis-Menten equation by least squares, and 50% effective concentrations (EC50s) were calculated.
RESULTS
Polymorphisms in the CCR5 genes of AGM species.
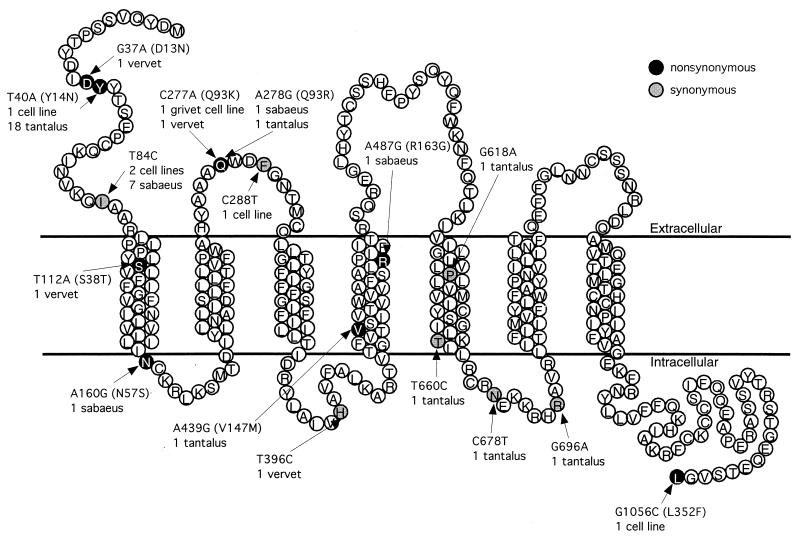
To identify polymorphisms in the CCR5 genes of AGMs and their possible roles in natural transmission of SIVagm viruses, we analyzed the CCR5-coding sequences of 23 AGMs and three cell lines (CV-1, BS-C-1, and Vero) (see Materials and Methods). Based on its high frequency in these samples and its presence in all four AGM species, we defined a consensus CCR5 nucleic acid sequence, which we consider to be the common wild-type sequence of AGMs. It differs from the previously reported CCR5 clone 2 allele of CV-1 cells (37) by only one synonymous nucleotide substitution, T300C. The corresponding wild-type CCR5 protein was encoded by at least 23 of the 52 alleles analyzed in this investigation. Figure 1 shows a synopsis of the nucleotide polymorphisms detected in these samples relative to the consensus nucleotide sequence, mapped onto a topological model of the wild-type AGM CCR5 amino acid sequence. Table 1 summarizes the properties of each monkey, including its species, its geographic origin, its specific polymorphism(s), and its infection status.
FIG. 1.
Summary of nucleotide substitutions in the coding regions of 26 AGM genomic DNA samples. Genomic DNA samples were sequenced as described in Materials and Methods. The origins of the 26 AGM samples were as follows: 3 AGM cell lines and 3 vervet, 8 sabaeus, and 12 tantalus AGMs. The electropherograms from the automated sequencing were examined manually for polymorphic nucleotides. Heterozygous bases were readily apparent by this method. The nucleotide sequence changes relative to the AGM wild-type sequence that were found in these samples are mapped onto a topological model of the amino acid coding sequence of wild-type AGM CCR5. If a codon contained a nonsynonymous change, it is highlighted in black. Synonymous nucleotide substitutions are highlighted in gray. Next to each amino acid for which polymorphic nucleotide substitutions were observed is listed the nucleotide substitution relative to the wild-type sequence (e.g., T40A). For nonsynonymous substitutions, the predicted amino acid change that results is listed in parentheses. Below this is listed the number of alleles from each source of genomic DNA in which this substitution was found.
Overall, these results indicate a very high frequency of CCR5 substitution polymorphisms in these AGM DNAs. Specifically, the 52 CCR5 alleles that were sequenced contained 29 nonsynonymous substitutions and 15 synonymous nucleotide substitutions. Moreover, 24 of the 29 nonsynonymous substitutions caused amino acid sequence changes that were clustered at positions 13 and 14 in the amino-terminal region or position 93 in extracellular loop 1 (ECL1) of the CCR5 protein.
The polymorphism of T for A at nucleotide 40 of the coding sequence (T40A), which causes a Y14N substitution in the CCR5 protein, was the most common substitution observed in this analysis (Fig. 1). This polymorphism occurred exclusively in the 12 tantalus monkeys, which included 7 Y14N homozygotes, 4 heterozygotes, and only 1 wild-type Y14 homozygote. In addition, the CV-1 cell line, which may have derived from a tantalus monkey, was heterozygous for this substitution. These results indicate a substantial dispersal of the Y14N substitution in tantalus monkeys from different locales throughout the Central African Republic.
The tantalus monkey (B57) that was homozygous for the wild-type Y14 sequence was heterozygous for a different nucleotide substitution, A278G, which causes a Q93R amino acid substitution in the ECL1 region of CCR5. This same substitution in the absence of any other coding change was also detected in a heterozygous sabaeus monkey, suggesting that it may be common in different AGM species. Interestingly, an alternative Q93K polymorphism at this same position was present in heterozygous form in the BS-C-1 cell line of grivet origin and in linkage to an S38T substitution in a vervet. Thus, substitutions of Q93 by the basic amino acid R or K were found in all four AGM species.
The vervet mentioned above, which was heterozygous for an allele that encoded Q93K and S38T substitutions, had a D13N coding change in its other CCR5 allele. Although this substitution was found only once in the four vervet DNAs used in our analysis, it was independently reported in DNA from another vervet (16), suggesting that it may be common in this species.
A synonymous nucleotide substitution relative to the AGM CCR5 consensus nucleotide sequence, T84C, was found frequently in CCR5 alleles from sabaeus monkeys. This synonymous substitution also occurred in heterozygous BS-C-1 cells from a grivet and in heterozygous Vero cells from a vervet. Thus, although it is most prevalent in sabaeus monkeys, T84C also occurs in other AGM species. Additional synonymous and nonsynonymous nucleotide substitutions occurred in single AGM DNA alleles and are indicated in Fig. 1 and Table 1. These relatively infrequent changes were randomly located throughout the CCR5-coding sequence.
Coreceptor dependencies of SIVagm infections.
To evaluate the coreceptor requirements of SIVagm isolates, we adapted a focal infectivity assay (9) in which dilutions of the viruses were incubated with CD4-positive adherent cell cultures, followed 72 h later by immunoperoxidase staining using pooled sera from SIVagm-infected AGMs. As shown by the representative data in Table 2, the five tested SIVagm isolates showed strongly stained discrete foci of infection in monolayer cultures of HeLa-CD4 cells that had been previously transiently transfected with the pcDNA3-human CCR5 expression vector. However, in contrast to previous studies using R5 strains of HIV-1, the background levels of infection in the control cultures transfected with vector alone were significant, ranging from approximately 3 to 6% for Cpa27 to approximately 91% for the sab-2 isolate of SIVagm.
TABLE 2.
Focal infectivity assay of SIVagm isolates in human HeLa-CD4 cells transfected with human CCR5
| SIVagm isolatea | Titer (foci/well)b
|
|
|---|---|---|
| Human CCR5 transfected | Mock transfected | |
| gri-1 | 633 | 42 |
| Cpa226 | 111 | 10 |
| Cpa27 | 685 | 19 |
| Strain 12 | 1,170 | 204 |
| sab-2 | 598 | 547 |
SIVagm isolates were from grivet (gri-1), vervet (Cpa226, Cpa27, and strain 12), or sabacus (sab-2) AGM subspecies.
Titers were determined by immunoperoxidase staining and microscopic inspection of foci of infected cells as described in Materials and Methods. Numbers represent foci from one well of a 24-well cluster plate seeded with 2 × 104 HeLa-CD4 (clone HI-J) cells that had been transfected with either pcDNA3-hCCR5 or pcDNA3 (mock transfected) 24 h prior to infection. The results shown are for a single representative experiment. The average backgrounds for the three experiments represented graphically in Fig. 2 were as follows: gri-1, 14%; Cpa226, 10%; Cpa27, 6%; and strain 12, 18%. As expected, the background percentages varied somewhat in different experiments due to differences in transfection efficiencies.
These results raised the possibility that the SIVagm isolates might use not only CCR5 but also, to different extents, alternative coreceptors such as CXCR4 (18) or CXCR6 (13, 46) that are known to be expressed in HeLa cells (13, 18). Similar to the case for HeLa cells, we found substantial background levels of SIVagm infections in U87MG-CD4 cells (results not shown), which also express CXCR6 (13), but not in the AGM cell line CV-1-CD4 (Table 3). Although the latter cells provided a zero background system to identify the coreceptor dependencies of the SIVagm isolates, the foci of infection in these CV-1-CD4 cultures were slightly dispersed and were very difficult to count accurately due to the patterns of cell growth and migration in the monolayers. Consequently, we were able only to semiquantitatively measure viral titers in the control and coreceptor-expressing CV-1-CD4 cultures. As shown by the data in Table 3, this analysis demonstrated that the five tested SIVagm isolates, including sab-2, all use CCR5 efficiently and CXCR6 with lower efficiency and that three of the five isolates also used GPR15 with low efficiency under the conditions of this assay. However, these viruses were completely unable to use CXCR4. Thus, it seems unlikely that the CXCR4 present in HeLa-CD4 cells acts as a coreceptor for SIVagm isolates. A qualification to these conclusions is that the coreceptors analyzed for Tables 2 and 3 were the human proteins.
TABLE 3.
Utilization of different coreceptors by SIVagm isolates in CV-1-CD4 cells
| SIVagm isolatea | Relative infectivity in cells transfected with the following coreceptorb:
|
||||
|---|---|---|---|---|---|
| CCR5 | CXCR6 | GPR15(BOB) | CXCR4 | mock | |
| gri-1 | ++ | + | − | − | − |
| Cpa226 | ++++ | + | − | − | − |
| Cpa27 | ++++ | +++ | + | − | − |
| Strain 12 | ++++ | ++ | + | − | − |
| sab-2 | ++++ | ++ | + | − | − |
SIVagm isolates were from grivet (gri-1), vervet (Cpa226, Cpa27, and strain 12), or sabaeus (sab-2) AGM subspecies.
The assay was performed as described for HeLa-CD4 cells in Table 2. Titers in the CV-1-CD4 (clone CB.14) monolayer cultures were scored approximately, for reasons described in the text. Each + represents approximately a threefold increase in the titer. The background infectivity of these viruses in mock-tranfected CV-1-CD4 cells was zero. The same CXCR4 expression vector was biologically active when assayed with X4 strains of HIV-1.
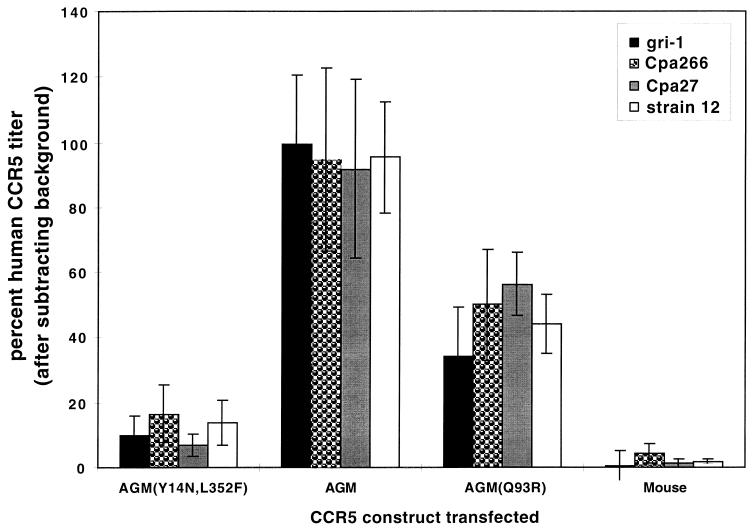
To address the latter issue and to initially evaluate effects of AGM CCR5 polymorphisms on SIVagm infections, we used the quantitative focal infectivity assay in HeLa-CD4 cells to compare human and mouse CCR5s with several of the full-length CCR5 alleles that we had isolated from AGMs. The background titers obtained in the mock-transfected cultures were subtracted from the data for each assay, and the results were normalized relative to the titers obtained using human CCR5 as a standard. In these assays, we used the four SIVagm isolates gri-1, Cpa226, Cpa27, and strain 12 but were unable to use sab-2 because of its very high background level of infectivity in HeLa-CD4 cells (Table 2). As shown by the results in Fig. 2, these SIVagm isolates used wild-type AGM CCR5 with approximately the same efficiency as human CCR5 (P = 0.3 [n = 12] as determined by the paired-comparisons t test). In contrast, we previously showed that HIV-1 isolates use wild-type AGM CCR5 only inefficiently due to the G163R difference between the human and AGM proteins (62). Thus, the SIVagm viruses interact differently with CCR5 than does HIV-1 and are adapted to efficiently employ wild-type AGM CCR5. In addition, these results clearly show that all of the SIVagm isolates were severely inhibited in their utilization of the naturally occurring CCR5 variant alleles AGM(Y14N, L352F) and AGM(Q93R), compared to wild-type AGM or human CCR5s (P ≤ 0.001; n = 12). In other studies we have shown that the rare L352F substitution, which affects the intracellular carboxyl-terminal region of CCR5, has no effect on these infectivity assays (37). Moreover, we have previously shown that coreceptors are overexpressed in these transient-transfection assays and that weak coreceptors become much more efficient when they are overexpressed (36). We conclude that the Y14N and Q93R substitutions in AGM CCR5 severely attenuate infections by all of the SIVagm strains that we employed.
FIG. 2.
SIVagm infections of HeLa-CD4 cells expressing wild-type AGM, AGM(Y14N, L352F), AGM(Q93R), and NIH/Swiss mouse CCR5s. HeLa-CD4 cells (clone HI-J) were transiently transfected with the pcDNA3 expression vectors for the CCR5 proteins indicated, as well as for human CCR5. The number of infected foci above the mock-transfected background number was determined for each CCR5 and for human CCR5. The percent human CCR5 titer for each virus isolate was then calculated as 100 × [(number of foci in each well) − (number of foci in the same experiment in mock-transfected cells)]/[(number of foci in the same experiment in cells transfected with human CCR5) − (number of foci in the same experiment in mock-transfected cells)]. Results are the means from three independent experiments, and the error bars represent the SEM.
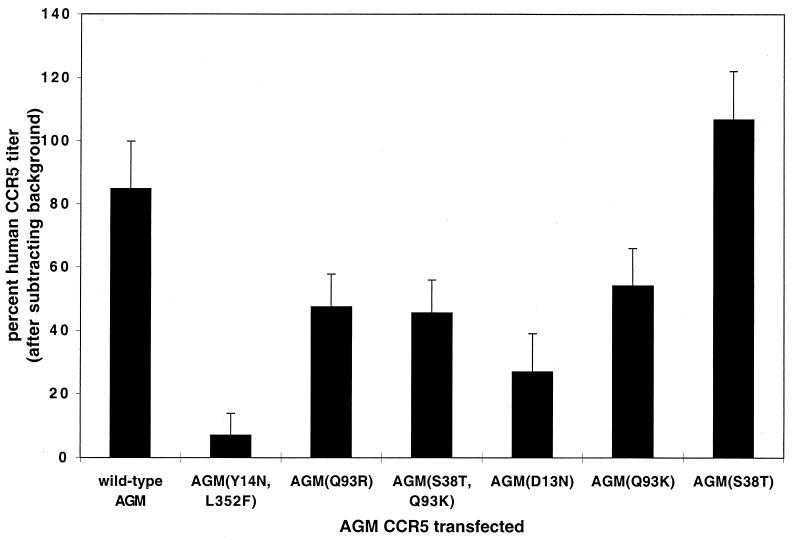
A more comprehensive analysis of the effects of AGM CCR5 polymorphisms on SIVagm infections was subsequently done with the Cpa226 virus. The results in Fig. 3 are the composite data from multiple (n = 6) assays of the AGM CCR5 variants indicated. The results support the conclusions described above and suggest that all of the substitutions in the amino-terminal region of AGM CCR5 (i.e., Y14N and D13N) and in ECL1 (Q93R and Q93K) strongly inhibit SIVagm Cpa226 infections, whereas the rare substitution S38T has no effect. In this context, it is notable that human CCR5 differs from wild-type AGM CCR5 in containing N rather than D at position 13. Moreover, mutagenesis studies have suggested that the D13N substitution in macaque CCR5 severely inhibits CD4-independent binding of SIVmac gp120 (44) and inhibits CD4-independent infection of SIVmac (16). However, the effect of the D13N substitution clearly depends on the overall CCR5 context, because the AGM(D13N) CCR5 mutant is severely attenuated for SIVagm infection, whereas wild-type human CCR5 contains N13 but is a highly efficient coreceptor for tested SIVagm isolates. Similarly, the Q93R mutation has a severely inhibitory effect on R5 HIV-1 infections in the context of AGM CCR5 but not in the context of human CCR5 (reference 37 and data not shown). These results support other evidence that attenuations in viral interactions with one region of CCR5 can be compensated for by enhancements in interactions with other regions (E. Platt, S. Kuhmann, and D. Kabat, unpublished data).
FIG. 3.
SIVagm (Cpa226) infections of HeLa-CD4 cells expressing AGM CCR5s with naturally occurring amino acid substitutions. HeLa-CD4 cells (clone HI-J) were transiently transfected with the pcDNA3 expression vectors for the CCR5 proteins indicated, as well as for human CCR5. The percentage of the human CCR5 titer for each assay was then calculated as described in the legend to Fig. 2. Results are the means from six independent experiments, and the error bars represent the SEM.
In studies using HeLa-CD4 and HEK293T cells, we found that all of the CCR5 expression vectors described above were transfected with equal efficiencies within experimental error, as indicated by the percentages of cells that stained with the 3A9 monoclonal antibody. These percentages were approximately 5 to 6% for the HeLa-CD4 cells and 90 to 95% for the HEK293T cells. In addition, the flow cytometry results indicated that the levels of CCR5 cell surface expression in the populations of transfected cells were also approximately the same for all CCR5s with the exception of the AGM CCR5(S38T) and AGM CCR5(S38T, Q93K) proteins, which appeared to be expressed on cell surfaces at levels that were only 50 to 60% as high as those for the other CCR5s (data not shown). These results suggested that the S38T mutation inhibits processing of CCR5 to cell surfaces or binding of the 3A9 antibody. In either case, these studies clearly established that the effects of the mutations on CCR5 coreceptor activities in Fig. 2 and 3 were not caused by differences in transfection efficiencies or in processing of the CCR5s to cell surfaces. Indeed, the only mutant that appeared to be expressed on cell surfaces with reduced efficiency, AGM CCR5(S38T), was as active as human and wild-type AGM CCR5s in mediating SIVagm infections. Additional evidence concerning expression of these CCR5s on cell surfaces is shown below; also see references 36, 37, and 62.
Effects of AGM CCR5 polymorphisms on natural and experimental SIVagm infections.
Table 1 summarizes our evidence concerning the infection status of the monkeys used in this investigation. Some of the AGMs were naturally infected, and some of the seronegative monkeys were experimentally infected after their capture in different regions of Senegal or the Central African Republic. Interestingly, among the tantalus the naturally infected monkeys included one Q93R heterozygote that lacked any Y14 substitution (Tan B57) and two of the four Y14N heterozygotes (Tan B83 and Tan B141). In contrast, none of the seven Y14N tantalus homozygotes was naturally infected at the time of capture, although several were later experimentally infected. These experimentally infected monkeys are all seropositive and appear to contain continuously replicating SIVagm viruses, suggesting that the viruses may have become adapted to the severe Y14N lesion. This distribution of the natural infections in the 12 tantalus monkeys would be expected to occur at random with a low probability (P ≤ 0.02), implying that monkeys homozygous for CCR5(Y14N) are probably resistant to natural transmission of SIVagm. However, analyses of additional tantalus monkeys caught in the wild will be necessary to definitively evaluate this hypothesis and to determine the degree of resistance to infection and to viral replication.
Biochemical effects of the CCR5(Y14N) mutation.
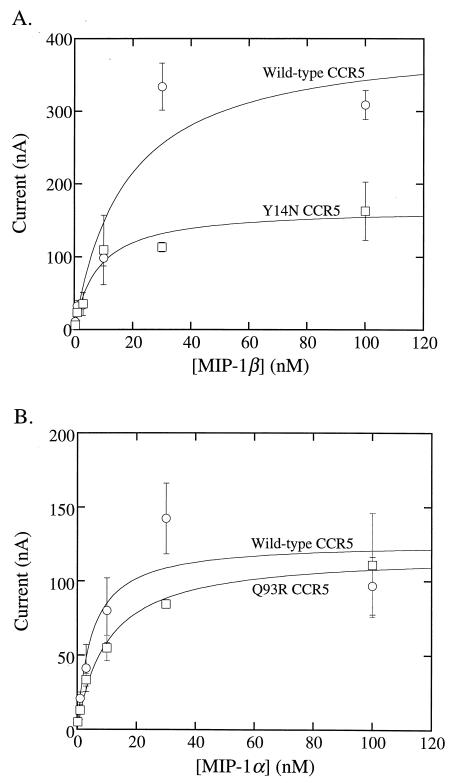
Previous evidence has established that tyrosine sulfation occurs at the Y3, Y10, Y14, and Y15 positions, which are common to human and AGM CCR5s, and that these sulfations are important for HIV-1 infections (17). Thus, Y-to-A substitutions at these sites severely inhibit coreceptor activity (36, 58). Although the Y14A mutation was not individually analyzed, simultaneous Y-to-A mutations at several of these tyrosines also severely attenuated chemokine-dependent signal transduction (17). In addition, as illustrated in Fig. 1, the CCR5(Y14N) mutation converts a YYT sequence common to human and AGM CCR5s into an NYT consensus sequence for potential N-linked glycosylation. Consistent with this, we have previously shown that the CCR5(Y14N) mutation results in incomplete N-linked glycosylation of human CCR5, with approximately 60% of the molecules acquiring this modification (36). Interestingly, however, the remaining 40% of CCR5(Y14N) molecules that lacked this N-linked oligosaccharide bound 125I-labeled MIP-lβ with the same affinity as wild-type CCR5 (36).
To further investigate this issue, we compared chemokine-induced signal transduction by the wild-type and mutant CCR5s. This was done using a sensitive signal transduction assay in which the CCR5s were coexpressed in Xenopus oocytes with the Kir 2.1 Gβγ-coupled potassium channel, which enables signal transduction through heterotrimeric G protein-coupled receptors to be monitored by two-electrode voltage clamp recording (43). As shown in Fig. 4A, the chemokine concentration dependencies (EC50s) for activations of wild-type and Y14N mutant versions of human CCR5s were not significantly different. However, in agreement with our evidence that only 40% of the CCR5(Y14N) is unglycosylated and able to bind 125I-labeled MIP-1β, the quantity of signaling was approximately 40% as high for CCR5(Y14N) as for wild-type CCR5. This evidence suggests that the CCR5(Y14N) molecules that lack an N-linked oligosaccharide are fully active in MIP-1β binding and in signal transduction. Thus, the unglycosylated asparagine side chain at position 14 severely inhibits infections while preserving substantial chemokine receptor function. Similarly, Fig. 4B shows that wild-type and Q93R mutant AGM CCR5s have very similar signal transduction properties. Since wild-type human and AGM CCR5s contain N13 and D13 residues, respectively, and are both active in chemokine-mediated signaling (Fig. 4) (62), this sequence difference also does not significantly perturb signal transduction in this assay system.
FIG. 4.
Effects of substitutions at amino acids 14 and 93 on activation of CCR5 signal transduction by MIP-1α and MIP-1β. X. laevis oocysts were injected with cRNA for CCR5 (see Materials and Methods). Inward currents were measured by two-electrode voltage clamp analyses 3 to 4 days later. (A) Activation of wild-type human CCR5 (circles) by Mip-1β was compared to the corresponding activation of Y14N human CCR5 (squares). (B) Activation of wild-type AGM 2 CCR5 (circles) by Mip1-α was compared to activation of Q93R AGM CCR5 (squares). Inward potassium currents were measured at −80 mV during voltage pulses in three or four oocytes. Error bars represent the SEM. The EC50s of activation were as follows: wild-type human CCR5, 17.3 ±10.9 nM MIP-1β; Y14N human CCR5, 8 ± 2.8 nM MIP-1β; wild-type AGM CCR5, 4.8 ± 3.0 nM MIP-1α; Q93R AGM CCR5, 10.2 ± 1.8 nM MIP-1α.
DISCUSSION
These results substantiate previous preliminary evidence which suggested that nonsynonymous substitution polymorphisms might occur relatively frequently in AGM CCR5 genes and that these polymorphisms might often cause amino acid substitutions at the critical sites required for infections by immunodeficiency viruses (37). Thus, in a previous analysis of DNAs derived from five AGMs, we found heterozygosity for linked Y14N and L352F substitutions in CV-1 cells, heterozygosity for a D13N substitution in vervet 6243, heterozygosity for a Q93R substitution in sabaeus 9312, heterozygosity for a Q93K substitution in BS-C-1 cells derived from a grivet, and homozygosity for the wild-type allele in Vero cells from a vervet (37). Furthermore, the D13N substitution was also detected in another vervet (16). The two AGM monkeys used in our previous study were derived from populations that became established in the Caribbean islands in the 17th to 18th centuries, and it was unclear whether those samples would be representative of outbred African AGMs. Consequently, in the present investigation we supplemented our earlier data by analyzing DNAs from 21 additional AGMs, most of which were tantalus and sabaeus monkeys that were wild born and independently captured between 1988 and 1995 in at least three distinct regions of the Central African Republic or in distinct regions of Senegal (see additional information concerning monkey origins in Table 1). As summarized in Fig. 1 and Table 1, the composite data from the 26 AGM DNAs (52 CCR5 alleles) include 29 nonsynonymous nucleotide substitutions, of which 24 are predicted to cause tightly clustered D13N or Y14N substitutions in the CCR5 amino terminus or Q93R and Q93K substitutions in ECL1. Thus, the overall frequency of nonsynonymous substitutions in these alleles is 0.56 per allele. A high frequency of nonsynonymous substitutions was also detected by other methods in our previous study of four AGM CCR5 alleles (51).
Although data concerning polymorphisms in human CCR5-coding sequences have been somewhat variable in different studies (3, 10, 66), a relatively comprehensive analysis of the CCR5-coding regions of 50 DNA samples from each of five ethnic groups implied that the frequency of nonsynonymous substitutions is in the range of approximately 0.07 to 0.16 per allele in these groups (3). In Caucasians the Δ32 CCR5 deletion allele has a null phenotype and occurs at a frequency of approximately 0.05 to 0.13 (3, 42, 53, 59, 61, 63). Moreover, the ratio of nonsynonymous to synonymous nucleotide sequence changes has been approximately 1.0 throughout evolution of the CCR5-coding region of primates (54), whereas it is approximately 1.9 for the AGM sequences in our study. Considered together, this evidence supports the hypothesis that the AGM CCR5-coding sequences have evolved under relatively intense selection pressure caused by the high-frequency SIVagm infections (greater than 50% of sexually mature adults [26, 31, 41, 50]) that occur in the AGM subspecies.
A corollary of this hypothesis that is supported by this investigation is that the AGM substitution polymorphisms would be expected to frequently modify critical sites required for SIVagm infections. Accordingly, the D13N, Y14N, Q93R, and Q93K substitutions in AGM CCR5 all strongly inhibit infections by SIVagm isolates within the context of AGM CCR5 as indicated by our focal infectivity assay for these viruses (Fig. 2 and 3). As determined using the 3A9 monoclonal antibody, none of these mutations altered cell surface expression of the CCR5 proteins in human cells. Moreover, these mutations did not substantially perturb cell surface expression or chemokine-stimulated signaling by these CCR5s in Xenopus oocytes (Fig. 4). Consequently, we conclude that these mutations specifically inhibit coreceptor activities of AGM CCR5s. In contrast, the nonsynonymous substitutions found in human CCR5 are randomly situated throughout the protein (3, 6, 10, 66). Moreover, extensive alanine scanning mutagenesis studies of human CCR5 have demonstrated that very few substitutions cause significant losses of coreceptor function (15, 23, 58). Thus, the AGM CCR5 substitutions not only are comparatively frequent but also are exceptionally informative concerning the critical sites required for immunodeficiency virus infections.
Further studies will be required to determine whether these alleles influence transmission or outcome of infections by the SIVagm viruses found in the same AGM populations as the mutations. In particular, it is possible that during their coevolution specific SIVagm strains have become adapted to specific mutations in CCR5, either by altered interactions with CCR5 or by utilizing another coreceptor, and these adaptations could conceivably change the pattern of virus transmission or replication in these monkey populations. In addition, it will be interesting to learn whether viruses that are present in experimentally infected monkeys carrying these mutant CCR5 alleles have adapted to the mutations. Presumably, mutations that attenuate virus interactions with CCR5 would selectively eliminate infections of cells that express low levels of CD4 or CCR5 on their surfaces (36, 57).
We emphasize that the active sites identified in this study are relevant to the AGM CCR5 protein and that somewhat different results can occur when these substitutions are incorporated into human CCR5. For example, AGM(D13N) CCR5 is attenuated as a coreceptor for SIVagm, whereas these same viruses efficiently use human CCR5, which also contains N at position 13 (Fig. 2 and 3). Similarly, Q93R and Q93K substitutions strongly inhibit SIVagm (Fig. 2 and 3) or R5 HIV-1 infections (37) in the context of AGM CCR5 but have reduced effects on HIV-1 infections in the context of human CCR5 (data not shown). These results are consistent with other evidence that weakened interactions of immunodeficiency viruses with one region of CCR5 can be compensated for by strong interactions with other sites in the coreceptor (E. Platt, S. E. Kuhmann, and D. Kabat, unpublished data). Specifically, we found that forced passaging of the R5 HIV-1 isolate JR-CSF in HeLa-CD4/CCR5(Y14N) cultures resulted in outgrowth of adapted viruses that are less dependent on the amino-terminal region of CCR5 and more reliant on increased interactions with the ECL2 region.
A limitation in our present investigation derives from the small number of SIVagm isolates that we were able to analyze. This is a significant issue in all studies, in part because SIVagm viruses are ancient and separated into four genetic subtypes, each specific for its natural AGM host (2, 5, 26, 30, 31, 41, 50), and in part because the available SIVagm strains have all been isolated following their replication in human Molt4-8 cells (1, 12, 19, 34). These cells, which have been uniquely useful for growing stocks of SIVagm (1, 12, 19, 34), contain a low concentration of CCR5 (40), and it is conceivable that the viruses have become altered by this passage (8). However, we believe that such adaptations must have been limited, because the SIVagm strains all efficiently utilize wild-type AGM CCR5 (Fig. 2 and 3), which is a poor coreceptor for HIV-1 (37, 62). A thorough analysis of this issue would require studies of numerous SIVagm samples derived directly from naturally infected AGMs of distinct species.
Additional studies will also be required to test our preliminary evidence that tantalus monkeys homozygous for the severe Y14N CCR5 substitution may be resistant to natural infections by SIVagm variants that are endemic in their environment. Although our evidence supports this hypothesis and is consistent with the prevalence of SIVagm infections determined in a survey of tantalus monkeys of all ages (50), we cannot exclude the possibility that naturally occurring SIVagm strains in tantalus monkey populations might have become adapted to use CCR5(Y14N) or another cell surface component as a coreceptor. The possibility of viral adaptation is supported by the fact that several of these Y14N homozygous tantalus monkeys were experimentally infected with SIVagm after their capture (see Table 1) and have maintained low viral loads (reference 52 and unpublished results). Similarly, a SIVrcm from a red-capped mangabey homozygous for a partially deleted CCR5 null allele adapted to this mutation of its host and exclusively uses the alternative coreceptor CCR2b (7).
Despite its severely debilitating effect on CCR5 coreceptor activity, the Y14N mutation has only a minor quantitative effect on its function in chemokine binding (36) and in signal transduction (Fig. 4). This mutation eliminates a tyrosine sulfation site that is critical for coreceptor activity (17) and creates an NYT sequence that is partially (ca. 60%) modified by N-linked glycosylation (36). Although these N-glycosylated CCR5 molecules are inactive in both signaling and coreceptor function, our results imply that the ungylcosylated CCR5(Y14N) component (i.e., 40% of the total) is fully active in chemokine binding (62) and in signal transduction (Fig. 4). Thus, the Y14N substitution appears to cause an ideal combination of resistance to SIVagm with retention of substantially normal chemokine signaling. Similarly, the Q93R substitution has no effect on chemokine signaling by CCR5 (Fig. 4), and the D13 and N13 residues are also both fully compatible with CCR5 chemokine signaling (Fig. 4) (62). These results support the hypothesis that the AGM CCR5 polymorphisms were selected for their resistance to SIVagm infections and for their preservation of CCR5 signaling.
In summary, our results support previous evidence that AGMs have been infected at high frequencies by SIVagm viruses since ancient times (2, 5, 26, 30, 31, 41, 50). As a consequence of this prolonged virus-host coevolution, the AGM CCR5-coding sequence has become highly polymorphic, with relatively frequent nonsynonymous nucleotide substitutions that strongly inhibit coreceptor activity but have only minor effects on chemokine binding or signaling.
Our study thus provides strong initial evidence for a true coevolution of SIVs with their natural hosts. The evident consequence of this coevolution is a balance between viruses that persist at high frequency and hosts that do not progress to disease. An important corollary of this interpretation is that AGMs are likely to contain a repository of abundant polymorphisms in other genes that have important roles in controlling infections and in limiting pathogenesis of immunodeficiency viruses. As in the example of CCR5, these polymorphisms would be expected to specifically alter the sites that are critical for the resistance properties of the encoded proteins. From this perspective, further analyses of AGMs and other endemically infected primates may be exceptionally helpful for identifying protein targets relevant to disease reduction and for developing treatments to counteract immunodeficiency viruses, including HIV-1.
ACKNOWLEDGMENTS
This research was supported by NIH grant CA67358 and by the French Agency for AIDS Research (ANRS).
We thank Jonathan Allen for donation of DNA samples from several AGMs derived from colony monkey populations and Ronald Desrosiers for generously providing us with pooled sera from SIVagm-infected AGMs. The SIVagm strains 12, Cpa266, and Cpa27 used in this study were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH (contributed by Ronald Desrosiers), as was the Molt4-8 cell line. The SIVagm strains gri-1 and sab-2 were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH (contributed by Jonathan Allan). The CXCR6- and GPR15 (BOB)-coding regions were amplified from plasmids obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH (contributed by Dan Littman).
REFERENCES
- 1.Allan J S, Kanda P, Kennedy R C, Cobb E K, Anthony M, Eichberg J W. Isolation and characterization of simian immunodeficiency viruses from two subspecies of African green monkeys. AIDS Res Hum Retroviruses. 1990;6:275–285. doi: 10.1089/aid.1990.6.275. [DOI] [PubMed] [Google Scholar]
- 2.Allan J S, Short M, Taylor M E, Su S, Hirsch V M, Johnson P R, Shaw G M, Hahn B H. Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J Virol. 1991;65:2816–2828. doi: 10.1128/jvi.65.6.2816-2828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansari-Lari M A, Liu X-M, Metzker M L, Rut A R, Gibbs R A. The extent of genetic variation in the CCR5 gene. Nat Genet. 1997;16:221–222. doi: 10.1038/ng0797-221. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 5.Bibollet-Ruche F, Brengues C, Galat-Luong A, Galat G, Pourrut X, Vidal N, Veas F, Durand J P, Cuny G. Genetic diversity of simian immunodeficiency viruses from West African green monkeys: evidence of multiple genotypes within populations from the same geographical locale. J Virol. 1997;71:307–313. doi: 10.1128/jvi.71.1.307-313.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanpain C, Lee B, Tackoen M, Puffer B, Boom A, Libert F, Sharron M, Wittamer V, Vassart G, Doms R W, Parmentier M. Multiple nonfunctional alleles of CCR5 are frequent in various human populations. Blood. 2000;96:1638–1645. [PubMed] [Google Scholar]
- 7.Chen Z, Kwon D, Jin Z, Monard S, Telfer P, Jones M S, Lu C Y, Aguilar R F, Ho D D, Marx P A. Natural infection of a homozygous delta24 CCR5 red-capped mangabey with an R2b-tropic simian immunodeficiency virus. J Exp Med. 1998;188:2057–2065. doi: 10.1084/jem.188.11.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng-Mayer C, Seto D, Levy J A. Altered host range of HIV-1 after passage through various human cell types. Virology. 1991;181:288–294. doi: 10.1016/0042-6822(91)90494-v. [DOI] [PubMed] [Google Scholar]
- 9.Chesebro B, Wehrly K. Development of a sensitive quantitative focal assay for human immunodeficiency virus infectivity. J Virol. 1988;62:3779–3788. doi: 10.1128/jvi.62.10.3779-3788.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen O J, Paolucci S, Bende S M, Daucher M, Moriuchi H, Moriuchi M, Cicala C, Davey R T, Jr, Baird B, Fauci A S. CXCR4 and CCR5 genetic polymorphisms in long-term nonprogressive human immunodeficiency virus infection: lack of association with mutations other than CCR5-Delta32. J Virol. 1998;72:6215–6217. doi: 10.1128/jvi.72.7.6215-6217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corbet S, Muller-Trutwin M C, Versmisse P, Delarue S, Ayouba A, Lewis J, Brunak S, Martin P, Brun-Vezinet F, Simon F, Barre-Sinoussi F, Mauclere P. env sequences of simian immunodeficiency viruses from chimpanzees in Cameroon are strongly related to those of human immunodeficiency virus group N from the same geographic area. J Virol. 2000;74:529–534. doi: 10.1128/jvi.74.1.529-534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniel M D, Li Y, Naidu Y M, Durda P J, Schmidt D K, Troup C D, Silva D P, MacKey J J, Kestler III H W, Sehgal P K, et al. Simian immunodeficiency virus from African green monkeys. J Virol. 1988;62:4123–4128. doi: 10.1128/jvi.62.11.4123-4128.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng H K, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 14.Diop O M, Gueye A, Dias-Tavares M, Kornfeld C, Faye A, Ave P, Huerre M, Corbet S, Barre-Sinoussi F, Muller-Trutwin M C. High levels of viral replication during primary simian immunodeficiency virus SIVagm infection are rapidly and strongly controlled in African green monkeys. J Virol. 2000;74:7538–7547. doi: 10.1128/jvi.74.16.7538-7547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dragic T, Trkola A, Lin S W, Nagashima K A, Kajumo F, Zhao L, Olson W C, Wu L, Mackay C R, Allaway G P, Sakmar T P, Moore J P, Maddon P J. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J Virol. 1998;72:279–285. doi: 10.1128/jvi.72.1.279-285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edinger A L, Blanpain C, Kunstman K J, Wolinsky S M, Parmentier M, Doms R W. Functional dissection of CCR5 coreceptor function through the use of CD4-independent simian immunodeficiency virus strains. J Virol. 1999;73:4062–4073. doi: 10.1128/jvi.73.5.4062-4073.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard N P, Gerard C, Sodroski J, Choe H. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 18.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 19.Fomsgaard A, Hirsch V M, Allan J S, Johnson P R. A highly divergent proviral DNA clone of SIV from a distinct species of African green monkey. Virology. 1991;182:397–402. doi: 10.1016/0042-6822(91)90689-9. [DOI] [PubMed] [Google Scholar]
- 20.Gao F, Bailes E, Robertson D L, Chen Y, Rodenburg C M, Michael S F, Cummins L B, Arthur L O, Peeters M, Shaw G M, Sharp P M, Hahn B H. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 21.Gao F, Yue L, White A T, Pappas P G, Barchue J, Hanson A P, Greene B M, Sharp P M, Shaw G M, Hahn B H. Human infection by genetically diverse SIVSM-related HIV-2 in west Africa. Nature. 1992;358:495–499. doi: 10.1038/358495a0. [DOI] [PubMed] [Google Scholar]
- 22.Garred P, Madsen H O, Petersen J, Marquart H, Hansen T M, Freiesleben Sorensen S, Volck B, Svejgaard A, Andersen V. CC chemokine receptor 5 polymorphism in rheumatoid arthritis. J Rheumatol. 1998;25:1462–1465. [PubMed] [Google Scholar]
- 23.Genoud S, Kajumo F, Guo Y, Thompson D, Dragic T. CCR5-mediated human immunodeficiency virus entry depends on an amino-terminal gp120-binding site and on the conformational integrity of all four extracellular domains. J Virol. 1999;73:1645–1648. doi: 10.1128/jvi.73.2.1645-1648.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez E, Bamshad M, Sato N, Mummidi S, Dhanda R, Catano G, Cabrera S, McBride M, Cao X H, Merrill G, O'Connell P, Bowden D W, Freedman B I, Anderson S A, Walter E A, Evans J S, Stephan K T, Clark R A, Tyagi S, Ahuja S S, Dolan M J, Ahuja S K. Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes. Proc Natl Acad Sci USA. 1999;96:12004–12009. doi: 10.1073/pnas.96.21.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartung S, Boller K, Cichutek K, Norley S G, Kurth R. Quantitation of a lentivirus in its natural host: simian immunodeficiency virus in African green monkeys. J Virol. 1992;66:2143–2149. doi: 10.1128/jvi.66.4.2143-2149.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayami M, Ido E, Miura T. Survey of simian immunodeficiency virus among nonhuman primate populations. Curr Top Microbiol Immunol. 1994;188:1–20. doi: 10.1007/978-3-642-78536-8_1. [DOI] [PubMed] [Google Scholar]
- 27.Hirsch V M, Dapolito G, Johnson P R, Elkins W R, London W T, Montali R J, Goldstein S, Brown C. Induction of AIDS by simian immunodeficiency virus from an African green monkey: species-specific variation in pathogenicity correlates with the extent of in vivo replication. J Virol. 1995;69:955–967. doi: 10.1128/jvi.69.2.955-967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirsch V M, Olmsted R A, Murphey-Corb M, Purcell R H, Johnson P R. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature. 1989;339:389–392. doi: 10.1038/339389a0. [DOI] [PubMed] [Google Scholar]
- 29.Ioannidis J P, O'Brien T R, Rosenberg P S, Contopoulos-Ioannidis D G, Goedert J J. Genetic effects on HIV disease progression. Nat Med. 1998;4:536. doi: 10.1038/nm0598-536. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 30.Jin M J, Hui H, Robertson D L, Müller M C, Barré-Sinoussi F, Hirsch V M, Allan J S, Shaw G M, Sharp P M, Hahn B H. Mosaic genome structure of simian immunodeficiency virus from west African green monkeys. EMBO J. 1994;13:2935–2947. doi: 10.1002/j.1460-2075.1994.tb06588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson P R, Fomsgaard A, Allan J, Gravell M, London W T, Olmsted R A, Hirsch V M. Simian immunodeficiency viruses from African green monkeys display unusual genetic diversity. J Virol. 1990;64:1086–1092. doi: 10.1128/jvi.64.3.1086-1092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jolly C, Phillips-Conroy J E, Turner T R, Broussard S, Allan J S. SIVagm incidence over two decades in a natural population of Ethiopian grivet monkeys (Cercopithecus aethiops aethiops) J Med Primatol. 1996;25:78–83. doi: 10.1111/j.1600-0684.1996.tb00198.x. [DOI] [PubMed] [Google Scholar]
- 33.Kabat D, Kozak S L, Wehrly K, Chesebro B. Differences in CD4 dependence for infectivity of laboratory-adapted and primary patient isolates of human immunodeficiency virus type 1. J Virol. 1994;68:2570–2577. doi: 10.1128/jvi.68.4.2570-2577.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kikukawa R, Koyanagi Y, Harada S, Kobayashi N, Hatanaka M, Yamamoto N. Differential susceptibility to the acquired immunodeficiency syndrome retrovirus in cloned cells of human leukemic T-cell line Molt-4. J Virol. 1986;57:1159–1162. doi: 10.1128/jvi.57.3.1159-1162.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kostrikis L G, Neumann A U, Thomson B, Korber B T, McHardy P, Karanicolas R, Deutsch L, Huang Y, Lew J F, McIntosh K, Pollack H, Borkowsky W, Spiegel H M, Palumbo P, Oleske J, Bardeguez A, Luzuriaga K, Sullivan J, Wolinsky S M, Koup R A, Ho D D, Moore J P. A polymorphism in the regulatory region of the CC-chemokine receptor 5 gene influences perinatal transmission of human immunodeficiency virus type 1 to African-American infants. J Virol. 1999;73:10264–10271. doi: 10.1128/jvi.73.12.10264-10271.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuhmann S E, Platt E J, Kozak S L, Kabat D. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J Virol. 2000;74:7005–7015. doi: 10.1128/jvi.74.15.7005-7015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuhmann S E, Platt E J, Kozak S L, Kabat D. Polymorphisms in the CCR5 genes of African green monkeys and mice implicate specific amino acids in infections by simian and human immunodeficiency viruses. J Virol. 1997;71:8642–8656. doi: 10.1128/jvi.71.11.8642-8656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurth R, Norley S. Simian immunodeficiency viruses of African green monkeys. Curr Top Microbiol Immunol. 1994;188:21–33. doi: 10.1007/978-3-642-78536-8_2. [DOI] [PubMed] [Google Scholar]
- 39.Lalani A S, Masters J, Zeng W, Barrett J, Pannu R, Everett H, Arendt C W, McFadden G. Use of chemokine receptors by poxviruses. Science. 1999;286:1968–1971. doi: 10.1126/science.286.5446.1968. [DOI] [PubMed] [Google Scholar]
- 40.Lee B, Sharron M, Montaner L J, Weissman D, Doms R W. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci USA. 1999;96:5215–5220. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Naidu Y M, Daniel M D, Desrosiers R C. Extensive genetic variability of simian immunodeficiency virus from African green monkeys. J Virol. 1989;63:1800–1802. doi: 10.1128/jvi.63.4.1800-1802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 43.Madani N, Kozak S L, Kavanaugh M P, Kabat D. gp120 envelope glycoproteins of human immunodeficiency viruses competitively antagonize signaling by coreceptors CXCR4 and CCR5. Proc Natl Acad Sci USA. 1998;95:8005–8010. doi: 10.1073/pnas.95.14.8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin K A, Wyatt R, Farzan M, Choe H, Marcon L, Desjardins E, Robinson J, Sodroski J, Gerard C, Gerard N P. CD4-independent binding of SIV gp120 to rhesus CCR5. Science. 1997;278:1470–1473. doi: 10.1126/science.278.5342.1470. [DOI] [PubMed] [Google Scholar]
- 45.Martin M P, Dean M, Smith M W, Winkler C, Gerrard B, Michael N L, Lee B, Doms R W, Margolick J, Buchbinder S, Goedert J J, O'Brien T R, Hilgartner M W, Vlahov D, O'Brien S J, Carrington M. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science. 1998;282:1907–1911. doi: 10.1126/science.282.5395.1907. [DOI] [PubMed] [Google Scholar]
- 46.Matloubian M, David A, Engel S, Ryan J E, Cyster J G. A transmembrane CXC chemokine is a ligand for HIV-coreceptor Bonzo. Nat Immunol. 2000;1:298–304. doi: 10.1038/79738. [DOI] [PubMed] [Google Scholar]
- 47.McDermott D H, Zimmerman P A, Guignard F, Kleeberger C A, Leitman S F, Murphy P M. CCR5 promoter polymorphism and HIV-1 disease progression. Multicenter AIDS Cohort Study (MACS) Lancet. 1998;352:866–870. doi: 10.1016/s0140-6736(98)04158-0. [DOI] [PubMed] [Google Scholar]
- 48.Michael N L, Chang G, Louie L G, Mascola J R, Dondero D, Birx D L, Sheppard H W. The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nat Med. 1997;3:338–340. doi: 10.1038/nm0397-338. [DOI] [PubMed] [Google Scholar]
- 49.Michael N L, Louie L G, Rohrbaugh A L, Schultz K A, Dayhoff D E, Wang C E, Sheppard H W. The role of CCR5 and CCR2 polymorphisms in HIV-1 transmission and disease progression. Nat Med. 1997;3:1160–1162. doi: 10.1038/nm1097-1160. [DOI] [PubMed] [Google Scholar]
- 50.Müller M C, Saksena N K, Nerrienet E, Chappey C, Hervé V M, Durand J-P, Legal-Campodonico P, Lang M C, Digoutte J P, Georges A J, Georges-Courbot M-C, Sonigo P, Barré-Sinoussi F. Simian immunodeficiency viruses from central and western Africa: evidence for a new species-specific lentivirus in tantalus monkeys. J Virol. 1993;67:1227–1235. doi: 10.1128/jvi.67.3.1227-1235.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muller-Trutwin M C, Corbet S, Hansen J, Georges-Courbot M C, Diop O, Rigoulet J, Barre-Sinoussi F, Fomsgaard A. Mutations in CCR5-coding sequences are not associated with SIV carrier status in African nonhuman primates. AIDS Res Hum Retroviruses. 1999;15:931–939. doi: 10.1089/088922299310647. [DOI] [PubMed] [Google Scholar]
- 52.Muller-Trutwin M C, Corbet S, Tavares M D, Herve V M, Nerrienet E, Georges-Courbot M C, Saurin W, Sonigo P, Barre-Sinoussi F. The evolutionary rate of nonpathogenic simian immunodeficiency virus (SIVagm) is in agreement with a rapid and continuous replication in vivo. Virology. 1996;223:89–102. doi: 10.1006/viro.1996.0458. [DOI] [PubMed] [Google Scholar]
- 53.Mummidi S, Ahuja S S, Gonzalez E, Anderson S A, Santiago E N, Stephan K T, Craig F E, O'Connell P, Tryon V, Clark R A, Dolan M J, Ahuja S K. Genealogy of the CCR5 locus and chemokine system gene variants associated with altered rates of HIV-1 disease progression. Nat Med. 1998;4:786–793. doi: 10.1038/nm0798-786. [DOI] [PubMed] [Google Scholar]
- 54.Mummidi S, Bamshad M, Ahuja S S, Gonzalez E, Feuillet P M, Begum K, Galvis M C, Kostecki V, Valente A J, Murthy K K, Haro L, Dolan M J, Allan J S, Ahuja S K. Evolution of human and non-human primate CC chemokine receptor 5 gene and mRNA. Potential roles for haplotype and mRNA diversity, differential haplotype-specific transcriptional activity, and altered transcription factor binding to polymorphic nucleotides in the pathogenesis of HIV-1 and simian immunodeficiency virus. J Biol Chem. 2000;275:18946–18961. doi: 10.1074/jbc.M000169200. [DOI] [PubMed] [Google Scholar]
- 55.Murphy P M. Molecular mimicry and the generation of host defense protein diversity. Cell. 1993;72:823–826. doi: 10.1016/0092-8674(93)90571-7. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 56.Norley S G. SIVagm infection of its natural African green monkey host. Immunol Lett. 1996;51:53–58. doi: 10.1016/0165-2478(96)02555-2. [DOI] [PubMed] [Google Scholar]
- 57.Platt E J, Wehrly K, Kuhmann S E, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rabut G E, Konner J A, Kajumo F, Moore J P, Dragic T. Alanine substitutions of polar and nonpolar residues in the amino-terminal domain of CCR5 differently impair entry of macrophage- and dualtropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:3464–3468. doi: 10.1128/jvi.72.4.3464-3468.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rana S, Besson G, Cook D G, Rucker J, Smyth R J, Yi Y, Turner J D, Guo H H, Du J G, Peiper S C, Lavi E, Samson M, Libert F, Liesnard C, Vassart G, Doms R W, Parmentier M, Collman R G. Role of CCR5 in infection of primary macrophages and lymphocytes by macrophage-tropic strains of human immunodeficiency virus: resistance to patient-derived and prototype isolates resulting from the Δccr5 mutation. J Virol. 1997;71:3219–3227. doi: 10.1128/jvi.71.4.3219-3227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rottman J B, Ganley K P, Williams K, Wu L, Mackay C R, Ringler D J. Cellular localization of the chemokine receptor CCR5. Correlation to cellular targets of HIV-1 infection. Am J Pathol. 1997;151:1341–1351. [PMC free article] [PubMed] [Google Scholar]
- 61.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 62.Siciliano S J, Kuhmann S E, Weng Y, Madani N, Springer M S, Lineberger J E, Danzeisen R, Miller M D, Kavanaugh M P, DeMartino J A, Kabat D. A critical site in the core of the CCR5 chemokine receptor required for binding and infectivity of human immunodeficiency virus type 1. J Biol Chem. 1999;274:1905–1913. doi: 10.1074/jbc.274.4.1905. [DOI] [PubMed] [Google Scholar]
- 63.Smith M W, Dean M, Carrington M, Winkler C, Huttley G A, Lomb D A, Goedert J J, O'Brien T R, Jacobson L P, Kaslow R, Buchbinder S, Vittinghoff E, Vlahov D, Hoots K, Hilgartner M W, O'Brien S J. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science. 1997;277:959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- 64.Weatherall D, Clegg J, Kwiatkowski D. The role of genomics in studying genetic susceptibility to infectious disease. Genome Res. 1997;7:967–973. doi: 10.1101/gr.7.10.967. [DOI] [PubMed] [Google Scholar]
- 65.Winkler C, Modi W, Smith M W, Nelson G W, Wu X, Carrington M, Dean M, Honjo T, Tashiro K, Yabe D, Buchbinder S, Vittinghoff E, Goedert J J, O'Brien T R, Jacobson L P, Detels R, Donfield S, Willoughby A, Gomperts E, Vlahov D, Phair J, O'Brien S J. Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. ALIVE Study, Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC) Science. 1998;279:389–393. doi: 10.1126/science.279.5349.389. [DOI] [PubMed] [Google Scholar]
- 66.Zhang L, Carruthers C D, He T, Huang Y, Cao Y, Wang G, Hahn B, Ho D D. HIV type 1 subtypes, coreceptor usage, and CCR5 polymorphism. AIDS Res Hum Retroviruses. 1997;13:1357–1366. doi: 10.1089/aid.1997.13.1357. [DOI] [PubMed] [Google Scholar]