Abstract
Impaired secretion of an essential blood coagulation factor fibrinogen leads to hepatic fibrinogen storage disease (HFSD), characterized by the presence of fibrinogen-positive inclusion bodies and hypofibrinogenemia. However, the molecular mechanisms underlying the biogenesis of fibrinogen in the endoplasmic reticulum (ER) remain unexplored. Here we uncover a key role of SEL1L-HRD1 complex of ER-associated degradation (ERAD) in the formation of aberrant inclusion bodies, and the biogenesis of nascent fibrinogen protein complex in hepatocytes. Acute or chronic deficiency of SEL1L-HRD1 ERAD in the hepatocytes leads to the formation of hepatocellular inclusion bodies. Proteomics studies followed by biochemical assays reveal fibrinogen as a major component of the inclusion bodies. Mechanistically, we show that the degradation of misfolded endogenous fibrinogen Aα, Bβ, and γ chains by SEL1L-HRD1 ERAD is indispensable for the formation of a functional fibrinogen complex in the ER. Providing clinical relevance of these findings, SEL1L-HRD1 ERAD indeed degrades and thereby attenuates the pathogenicity of two disease-causing fibrinogen γ mutants. Together, this study demonstrates an essential role of SEL1L-HRD1 ERAD in fibrinogen biogenesis and provides insight into the pathogenesis of protein-misfolding diseases.
Subject terms: Endoplasmic reticulum, Protein aggregation, Liver diseases, ER-associated degradation
Fibrinogen, a key blood clotting factor, is produced in the endoplasmic reticulum (ER) of hepatocytes. Here, authors uncover the vital role of ER- associated degradation in preventing fibrinogen aggregation and ensuring its proper biogenesis.
Introduction
Fibrinogen is a highly abundant plasma glycoprotein complex essential for blood clot formation and hemostasis (i.e., the cessation of bleeding)1, as well as wound healing, inflammation, infection, angiogenesis, and tumor growth and metastasis2–4. It is produced by hepatocytes as three polypeptide chains, Aα, Bβ and γ, which undergo extensive folding, assembly, and maturation processes to form a 340 kDa hexameric (AαBβγ)2 complex with 29 disulfide bonds in the endoplasmic reticulum (ER) prior to entering the secretory pathway1,5. Clinically, qualitative or quantitative defects in fibrinogen have been observed in congenital or acquired hypo-, a- and dys-fibrinogenemia6,7. Hepatic fibrinogen storage disease (HFSD) is a specific type of hypofibrinogenemia condition caused by impaired fibrinogen assembly and secretion in hepatocytes8,9. Patients with HFSD exhibit fibrinogen-positive inclusion bodies in the liver with hepatic damage and low circulating fibrinogen levels, yet most cases do not present hemostatic issues8–16. Despite clinical reports of HFSD for over two decades11,17, the molecular events regulating the biogenesis of fibrinogen in the ER and the formation of fibrinogen-positive inclusion bodies remain poorly understood.
The synthesis, folding, and assembly of secretory and membrane proteins take place in the ER18,19. However, a portion of nascent proteins fail to attain their native conformation and are subsequently targeted for proteasomal degradation via an ER quality control mechanism known as ER-associated degradation (ERAD)20–23. The SEL1L-HRD1 protein complex represents the most conserved ERAD machinery24–31. Using conditional and cell type-specific Sel1L-deficient mice, we have demonstrated an indispensable role of SEL1L as an obligatory cofactor for the E3 ligase HRD132,33. Subsequently, we and others have collectively revealed the vital significance of this ERAD protein complex in a range of cell types during many physiological processes, including but not limited to, energy, lipid and iron metabolism, immunity, gut and kidney homeostasis, cartilage development, and the maintenance of hematopoietic stem cell quiescence34–51. Recently, we identified several SEL1L and HRD1 variants in humans with ERAD-associated neurodevelopmental disorders with onset in infancy, in short ENDI syndrome52,53. In the liver, we previously reported that hepatocyte-specific deletion of Sel1L in mice (Sel1Lflox/flox;Albumin-Cre, Sel1LAlb) caused transient growth retardation, altered energy and iron homeostasis, and predisposed animals to chemically induced liver tumors42,54,55. Nonetheless, our understanding of the importance of SEL1L-HRD1 ERAD in hepatic protein biogenesis and its pathological relevance remains very limited.
In this study, we serendipitously uncovered a crucial role of SEL1L-HRD1 ERAD in the formation of hepatic intracytoplasmic inclusion bodies, which surprisingly contained a large amount of fibrinogen proteins, resembling those found in HFSD. Indeed, subsequent mechanistic studies revealed that SEL1L-HRD1-mediated ERAD of all three fibrinogen chains was a prerequisite for the biogenesis of mature fibrinogen protein complex.
Results
Hepatic SEL1L-HRD1 ERAD deficiency leads to the formation of inclusion bodies in the liver
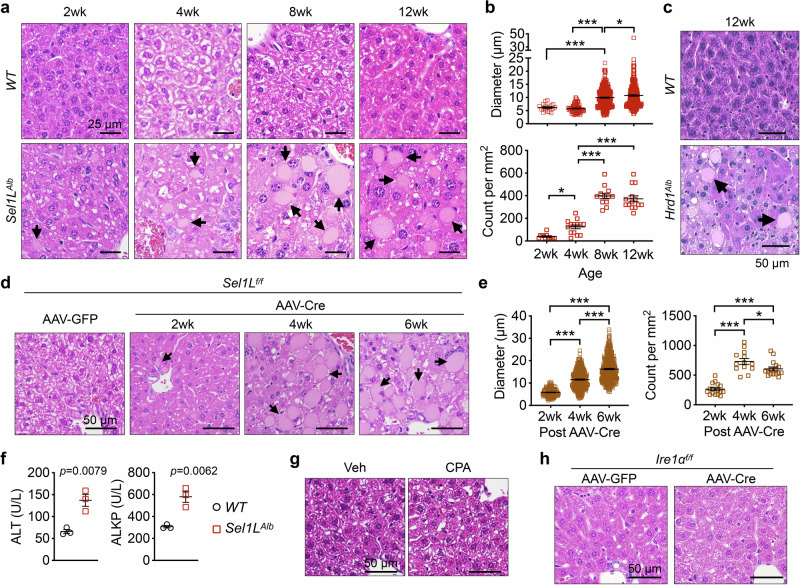
Histological examination of the livers of wild-type (Sel1Lflox/flox, WT) and Sel1LAlb cohorts from 2- to 12-weeks of age revealed the presence of globular inclusion bodies throughout the lobules in Sel1LAlb mice (arrows, Fig. 1a). The inclusion bodies were round, sharply bordered, palely eosinophilic, and ranged between 5 and 40 μm in diameter (Fig. 1a and quantitated in Fig. 1b). The inclusion bodies progressively enlarged in size and increased in numbers with age and were consistently observed in the livers of both male and female Sel1LAlb mice (Fig. 1a, b and S1a, b). Hepatic inclusion bodies were also observed in hepatocyte-specific Hrd1-deficient (Hrd1flox/flox;Albumin-Cre, Hrd1Alb) mice at 12 weeks of age43 (arrows, Fig. 1c), supporting the notion that this phenomenon is caused by defects in SEL1L-HRD1 ERAD function. The formation of inclusion bodies in Sel1LAlb mice was not due to developmental defects of hepatocytes, as acute deletion of SEL1L in hepatocytes of adult mice using liver-tropic adeno-associated virus serotype 8 (AAV8) expressing the Cre recombinase driven by a hepatocyte-specific thyroxine-binding globulin (TBG) promoter (Fig. S1c) also led to the formation of inclusion bodies within 2 weeks (Fig. 1d, e). A similar observation was made in Hrd1flox/flox mice injected with AAV8-Cre (Fig. S1d, e). In addition, circulating levels of liver enzymes, including alanine aminotransferase (ALT) and alkaline phosphatase (ALKP), but not aspartate aminotransferase (AST) or serum total bilirubin (TBIL), were mildly elevated in Sel1LAlb mice compared to those of WT littermates at 6 weeks of age (Fig. 1f and S1f). However, there was no histological evidence of cell apoptosis or fibrosis in Sel1LAlb livers (Fig. S2a, b), pointing to mild liver damage. Taken together, these data demonstrate that chronic or acute deletion of hepatic Sel1L or Hrd1 leads to the formation of inclusion bodies in the liver, which progressively enlarge with age.
Fig. 1. Formation of inclusion bodies in Sel1L-deficient livers.
a–b Representative images of H&E stained liver sections of male WT (Sel1Lf/f) and Sel1LAlb (Sel1Lf/f;Albumin-Cre) littermates at different ages. The size and number of inclusion bodies found in Sel1LAlb livers are quantitated in b. N = 3 per cohort. c Representative images of H&E stained liver sections of 12-week-old WT and Hrd1Alb littermates. N = 3. d–e Sel1Lf/f mice were injected i.v. with AAV8 expressing hepatocyte-specific TBG promoter-driven Cre or GFP. Representative images of H&E-stained liver sections of mice at different time points after the injection are shown d. The size and number of inclusion bodies are quantitated in e. N = 4 for GFP cohort, n = 3 for 2 wk and 4 wk post-AAV-Cre, n = 5 for 6 wk post-AAV-Cre. In (a), (c) and (d), arrows point to inclusion bodies. f Serum from 6-week-old WT and Sel1LAlb littermates were analyzed for alanine aminotransferase (ALT) and alkaline phosphatase (ALKP). N = 3. g–h Representative images of H&E stained liver sections of (g) WT mice 4 weeks after i.p. injections of vehicle (Veh) or cyclopiazonic acid (CPA) at 5 mg/kg body weight every other day, n = 3; and (h) Ire1αf/f mice 6 weeks after an i.v. injection of AAV8 expressing TBG-driven Cre or GFP, n = 5. For inclusion body quantitation, 12 to 16 images taken from indicated number of mice per cohort were analyzed. Values, mean ± SEM. *, p < 0.05; ***, p < 0.001 by one-way ANOVA with Tukey multiple comparison test in (b) and (e), and by two-tailed Student’s t test in f. Source data are provided as a Source Data file.
The induction of CREBH, FGF21, or deficiency of IRE1α is insufficient for the formation of hepatic inclusion bodies
As we previously reported that SEL1L-HRD1 ERAD deficiency in hepatocytes is associated with mild unfolded protein response (UPR) and elevated levels of an ER-resident transcription factor CREBH and stress hormone FGF2141–43,55, we next investigated their role in the formation of inclusion bodies. The challenge of a pharmacological ER stress inducer, cyclopiazonic acid (CPA)55, that depletes ER calcium56, did not induce visible inclusion bodies in the livers of WT mice (Fig. 1g). Similarly, acute deletion of a key UPR sensor IRE1α in hepatocytes via AAV8-Cre injection into Ire1aflox/flox mice failed to induce the formation of hepatic inclusion bodies (Fig. 1h and S2c). Moreover, the overexpression of CREBH or FGF21 in hepatocytes did not induce hepatic inclusion bodies (Fig. S2d, e)55,57,58. Together, these data shows that the induction of CREBH, FGF21, or deficiency of IRE1a in hepatocytes is insufficient to induce the formation of hepatic inclusion bodies.
Formation of single membrane-bound inclusion bodies in Sel1LAlb hepatocytes
To explore the nature of inclusion bodies, we next performed transmission electron microscopy (TEM). In WT hepatocytes, organelles such as the ER, mitochondria, and glycogen granules appeared with normal morphology (Fig. 2a). In contrast, we noted dilated ER (white arrows) and large cytosolic inclusion bodies with homogeneous electron density in Sel1LAlb hepatocytes (Fig. 2b–d). The inclusion bodies were notably bound by single membranes (red arrows, Fig. 2b, c), likely representing the ER membranes (discussed more below). We also noted the likely fusion of a small vesicle with the inclusion body (green arrow, Fig. 2c), providing a plausible explanation for the progressive enlargement of the inclusion bodies as shown in Fig. 1. The electron density of the inclusion bodies was uniformly low, distinct from that of lipid droplets, excluding the possibility of the inclusion bodies being lipid-laden (Fig. 2d). Morphologically, mitochondria appeared normal in Sel1LAlb hepatocytes (Fig. 2a–d). Thus, inclusion bodies in Sel1LAlb hepatocytes are single membrane-bound structures in the cytosol containing non-lipid materials.
Fig. 2. Inclusion bodies are encased in ER membrane with the identification of fibrinogen chains as a possible major component.
a–d TEM images of liver tissues from 6-week-old WT and Sel1LAlb littermates with insets of higher magnification shown (n = 2 mice each genotype). White arrowheads, normal ER; red arrows, inclusion bodies bounded by a single membrane; white arrows, dilated ER; green arrow, a vesicle in the process of fusing to a large inclusion body; N, nucleus; m, mitochondria; IB, inclusion bodies; LD, lipid droplets. Glycogen granules are noted in both WT and Sel1LAlb livers. e Proteomics analysis of purified microsomes from WT and Sel1LAlb livers, showing the fibrinogen chains significantly over-represented in the Sel1LAlb samples. Values represent scaled abundances of proteins from tandem mass tag (TMT) labeling-based mass spectrometry. P values were caculated by two-tailed Student’s t test without adjustments for multiple comparisons.
ERAD deficiency-associated inclusion bodies contain fibrinogen
We next explored the composition of inclusion bodies. Periodic acid-Schiff (PAS) staining for polysaccharides showed that, unlike in glycogen-ladened WT livers, inclusion bodies in Sel1LAlb livers were largely PAS negative (Fig. S3a). Staining of α1-antitrypsin revealed no abnormal protein aggregation in Sel1LAlb livers (Fig. S3b). Furthermore, albumin, a highly abundant protein secreted by hepatocytes, was able to reach to the sinusoidal space of the Sel1LAlb livers (arrowheads, Fig. S3c). Fibronectin and transferrin receptor 1 (TFR1), homodimers with multiple intra- and inter-molecular disulfide bonds from hepatocytes, were both secreted normally, and not present in the inclusion bodies (Fig. S3d, e). These data exclude the possibility of ERAD deficiency-associated inclusion bodies being either glycogen-containing Lafora bodies or α1-antitrypsin inclusions59,60, and more importantly, demonstrate that Sel1L deficiency does not cause a general secretion defect in hepatocytes.
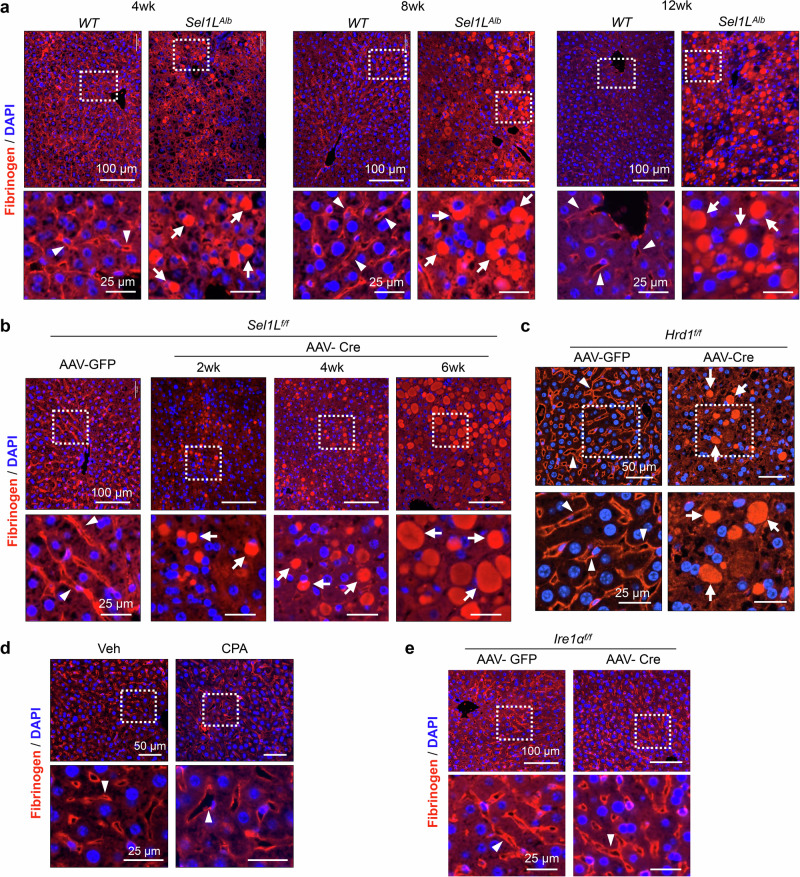
We then employed a non-biased proteomics screen of proteins in the purified microsomes from WT and Sel1LAlb livers (ProteomeXchange Dataset PXD035243)54. Two known SEL1L-HRD1 ERAD substrates, IRE1α and OS944,61, were enriched in Sel1LAlb microsomes (Fig. 2e). Interestingly, we noted that all three fibrinogen chains were significantly over-represented in the microsomal fractions of Sel1LAlb livers compared to those of WT livers (Fig. 2e). Immunofluorescent staining showed that, in WT livers, fibrinogen was detected largely on the apical membrane of hepatocytes or sinusoidal space (arrowheads, Fig. 3a). In contrast, fibrinogen accumulated largely in the inclusion bodies of Sel1LAlb hepatocytes (arrows, Fig. 3a). The abundance of these fibrinogen-positive inclusion bodies increased with age (Fig. 3a). Similar observations were obtained in inducible Sel1L- and Hrd1-deficient mice within 2-4 weeks of SEL1L or HRD1 deletion (Fig. 3b, c). As controls, fibrinogen localization to the apical membrane was unaffected by CPA-induced ER stress, inducible Ire1α deficiency, or FGF21 overexpression (Fig. 3d, e and S3f). Providing clinical relevance to our findings, these fibrinogen-positive inclusion bodies indeed resemble those found in the livers of patients with HFSD, exhibiting notable features such as robust fibrinogen positivity, absence of α1-antitrypsin, albumin, and PAS staining8–15. Taken together, we conclude that SEL1L-HRD1 ERAD deficiency in hepatocytes leads to the formation of fibrinogen-containing inclusion bodies.
Fig. 3. Fibrinogen is largely trapped in the inclusion bodies of SEL1L-HRD1 ERAD-deficient hepatocytes.
Fibrinogen staining using a rabbit anti-serum recognizing all chains of fibrinogen in liver sections from: (a) WT and Sel1LAlb littermates at different ages; (b) Sel1Lf/f mice at different time points after an i.v. injection of AAV8 expressing TBG-driven Cre or GFP; (c) Hrd1f/f mice 4 weeks after an i.v. injection of AAV8-TBG-Cre or GFP; (d) WT mice 4 weeks after i.p. injections of vehicle (Veh) or cyclopiazonic acid (CPA) at 5 mg/kg body weight every other day; (e) Ire1αf/f mice 6 weeks after an i.v. injection of AAV8-TBG-Cre or GFP. Insets of higher magnification shown below. Arrowheads point to apical and sinusoidal localization of fibrinogen; arrows point to inclusion bodies containing fibrinogen. Representative data from n = 3 mice per cohort.
Fibrinogen is complexed with ER chaperones in inclusion bodies
We next explored the molecular mechanism underlying the formation of fibrinogen inclusion bodies. We first performed an exploratory immunoprecipitation-based mass spectrometry, without replication, to analyze fibrinogen interactomes (ProteomeXchange Dataset PXD047658). The fibrinogen antibody used here was highly specific for all three fibrinogen chains (Fig. S4a)62,63. There were a total of 39 candidate proteins with 2-fold or higher of peptide-spectrum matches (PSMs) in Sel1LAlb liver lysates over WT lysates (Supplementary Data 1, Supplementary Data 2, and Fig. S4b). Pathway analyses showed that protein processing and folding were among the top pathways increased in fibrinogen interactomes in Sel1LAlb liver (Fig. S4c). Indeed, 10 top hits were ER chaperones involved in protein folding and ERAD substrate recruitment such as OS9, protein disulfide isomerases (PDI), GRP78/BiP, GRP94, and calreticulin (Fig. S4b). Further co-immunoprecipitation and immunostaining assays confirmed the interaction and colocalization, respectively, of fibrinogen chains with BiP and GRP94 (Fig. 4a and S4d–f). Both PDI and OS9 proteins were enriched in the inclusion bodies as well (Fig. S4f). Furthermore, BiP immunogold-TEM indeed revealed that while BiP was detected in ER sheets in WT hepatocytes, BiP was highly enriched in inclusion bodies of Sel1LAlb hepatocytes (Fig. 4b, c). Biochemically, sucrose gradient fractionation of protein complexes based on the size showed that BiP and PDI co-migrated with fibrinogen into the last (heaviest) fraction as high molecular weight (HMW) aggregates in Sel1LAlb hepatocytes (red arrows, Fig. S5). Together, our data demonstrate that fibrinogen within the inclusion bodies is physically associated with various ER chaperones, pointing to a misfolded or folding intermediates stage.
Fig. 4. Fibrinogen is in a misfolded or folding intermediate state in Sel1L-deficient hepatocytes.
a Primary hepatocytes were isolated from WT and Sel1LAlb littermates and stained with the indicated antibodies. Note the co-localizations of fibrinogen and an ER marker KDEL (recognizing BiP and GRP94) in the inclusion bodies of Sel1LAlb hepatocytes (white arrows). White dotted lines outline individual hepatocytes. Representative data from two independent repeats. b, c TEM couple to Immunogold labeling of BiP in WT and Sel1LAlb livers. White arrows mark the localization of BiP in normal ER in WT hepatocytes. Representitave image of eight hepatocytes from one mouse per genotype shown. Red arrows mark BiP in inclusion bodies in Sel1LAlb hepatocytes. White dotted lines mark the boundary of inclusion bodies. N, nucleus; m, mitochondria.
Accumulation and aggregation of all three fibrinogen chains in Sel1LAlb livers
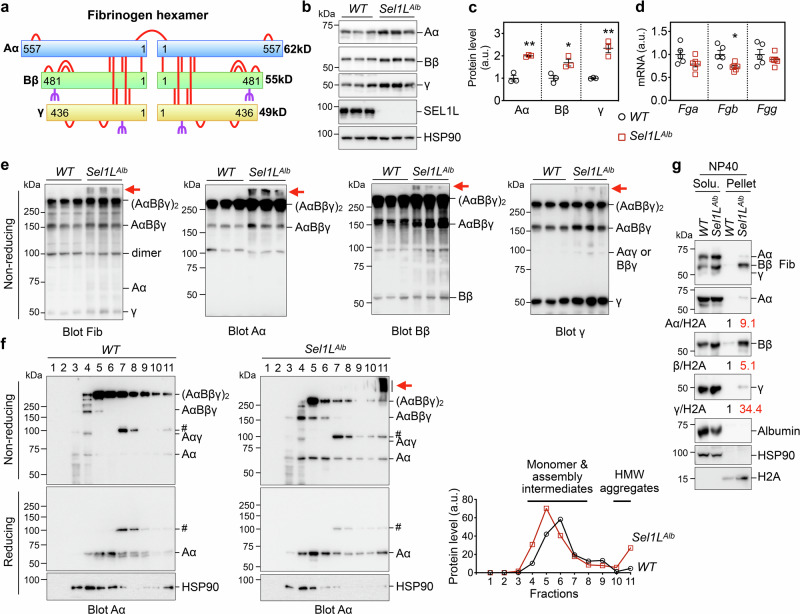
We next defined the biochemical nature of fibrinogen inclusions. Fibrinogen is a 340 kDa hexamer formed by three polypeptide chains, Aα, Bβ and γ, via a total of 29 inter- or intra-molecular disulfide bonds (Fig. 5a)1,5. Bβ and γ are glycosylated, but not Aα (Fig. 5a). Denaturing Western blot analyses of individual chains64,65 revealed that all three chains were increased by ~2 folds in Sel1LAlb livers compared to those of WT livers (Fig. 5b, c), independently of gene transcription (Fig. 5d).
Fig. 5. Fibrinogen chains accumulate and form insoluble aggregates in Sel1L-deficient livers.
a The overall structure of fibrinogen showing a hexamer of Aα, Bβ and γ chains. The 29 highly conserved disulfide bonds are shown as red bars, and the glycosylation sites on Bβ and γ are marked in purple. Aα is not glycosylated. Image created in BioRender. Tushi, N. (2024) BioRender.com/l49l811. (b–e) Liver samples from 6-week-old WT and Sel1LAlb littermates were analyzed for: (b) reducing Western blot analysis with quantitation normalized to HSP90 shown in (c); (d) qPCR analysis; (e) non-reducing SDS-PAGE and Western blot analysis using the same protein lysates shown in (b). In (b) and (e), fibrinogen proteins were analyzed by antibodies specific for each chain or all fibrinogen chains (Fib). Red arrows point to high molecular weight aggregates of fibrinogen. N = 3 for Western blot analyses, n = 5 for WT and n = 6 for Sel1LAlb for qPCR analyses. Values, mean ± SEM. *, p < 0.05; **, p < 0.01 by two-tailed Student’s t test. f Sucrose gradient fractionation (fractions 1 to 11 from top to bottom) of liver samples from WT and Sel1LAlb littermates analyzed by nonreducing or reducing SDS-PAGE using an Aα chain-specific antibody. Red arrow points to fibrinogen aggregates. #, a non-specific band. Quantitation of reduced Aα is shown on the right with fractions with Aα monomers, assembly intermediates and high molecular weight (HMW) aggregates labeled. HSP90, a loading control. g Liver samples from 6-week-old WT and Sel1LAlb littermates were prepared in lysis buffer containing 0.5% Nonidet P-40 (NP40) and analyzed for protein distribution in the NP40 soluble and NP40 insoluble (pellet) fractions by Western blot. The distribution of HSP90 and H2A marks the soluble and insoluble fractions, respectively. Quantitation of fibrinogen chains normalized to H2A in NP40 insoluble fraction is shown below the blots. (f–g), representative data from two independent repeats. Source data are provided as a Source Data file.
In addition to monomeric chains, several bands can be detected around 100, 160 and 340 kDa in non-reducing Western blot, representing fibrinogen oligomeric complexes including dimeric fibrinogen Aαγ or Bβγ, trimeric AαBβγ, and hexameric (AαBβγ)2, respectively (Fig. 5e)5. Of note, we included antibodies specifically recognizing all fibrinogen chains (labeled as Fib)62,63 or individual chains in this experiment (Fig. 5e). These oligomeric forms of fibrinogen were either unchanged or moderately elevated in Sel1LAlb livers compared to WT littermates (Fig. 5e). The most notable difference was the appearance of HMW aggregates with MW of over 400 kDa in Sel1LAlb livers, but not WT livers (red arrows, Fig. 5e). Sucrose gradient fractionation analyses further showed that, unlike in WT livers where fibrinogen Aα formed largely hexamers, it formed significantly more HMW aggregates in Sel1LAlb livers (Fig. 5f).
Fibrinogen Aα HMW aggregates were sensitive to the treatment of a reducing agent β-mercaptoethanol (Fig. 5f), pointing to the involvement of inter-molecular disulfide bonds in the formation of HMW aggregates. Furthermore, all three fibrinogen chains accumulated in the detergent-insoluble pellet fraction of Sel1LAlb livers compared to WT livers (Fig. 5g). Similar observations were consistently obtained in three other ERAD deficient mouse models, inducible Sel1L- (Fig. S6a–c), and two Hrd1-deficient mouse models (Fig. S6d, e), where fibrinogen chains accumulated and aggregated in the absence of SEL1L-HRD1 ERAD, without concomitant changes in their mRNA levels. Taken together, we conclude that SEL1L-HRD1 ERAD is required for fibrinogen complex assembly, and that ERAD deficiency leads to the accumulation and aggregation of all three fibrinogen chains in hepatocytes.
ER retention of fibrinogen chains in Sel1LAlb hepatocytes
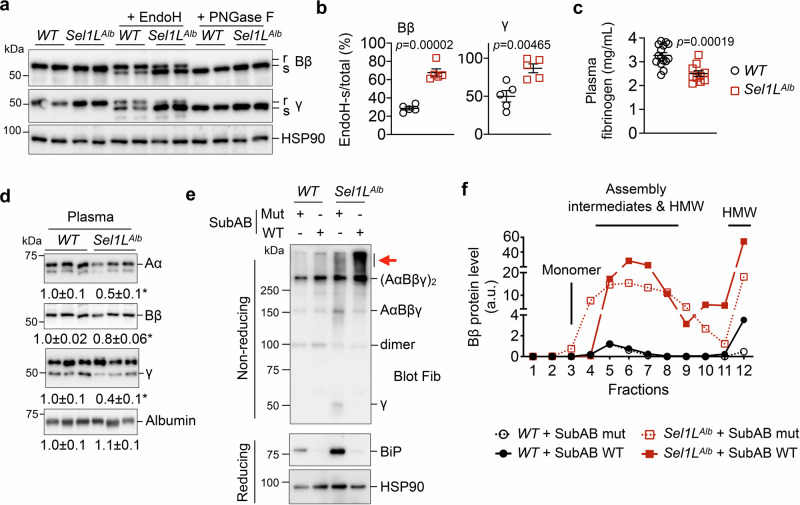
We next examined the impact of ERAD on fibrinogen secretion. We first performed an endoglycosidase H (EndoH) digestion assay to distinguish fibrinogen pools that are in the ER with high mannose glycosylation (EndoH sensitive) vs. those that have matured and exited the ER with complex glycosylation (EndoH resistant). In WT hepatocytes, about 70% of Bβ and 50% of γ were EndoH resistant (Fig. 6a, b), suggesting that they are able to fold and readily exit the ER. In contrast, ~ 65% Bβ and 85% γ chains were EndoH sensitive in the absence of SEL1L, suggesting that they are retained in the ER (Fig. 6a, b). The differential mobility observed in EndoH-treated samples resulted from glycosylation, as the EndoH-resistant form was sensitive to PNGase F treatment, which removes all N-linked glycan from glycoproteins (Fig. 6a). In keeping with these findings, circulating levels of fibrinogen in Sel1LAlb mice were significantly reduced by 20-50% as measured by ELISA and Western blot analyses (Fig. 6c, d). The reduction is specific for fibrinogen, as plasma levels of albumin were unaffected by Sel1L deficiency (Fig. 6d), again excluding the possibility that ER retention of fibrinogen is due to a general secretory defect. Similar observations were obtained in inducible Sel1L-deficient livers (Fig. S7a, b).
Fig. 6. Fibrinogen is retained in the ER of Sel1L-deficient hepatocytes, with BiP attenuating its further aggregation.
a–b Western blot analysis of fibrinogen Bβ and γ in liver lysates treated with EndoH or PNGase F. R and s, EndoH-resistant and sensitive, respectively. The percentages of EndoH-sensitive Bβ and γ chains are quantitated in b. N = 5 per cohort. c–d ELISA (c) and Western blot (d) analyses of inferior vena cava (IVC) plasma from 6-week-old WT and Sel1LAlb littermates. Quantitation of Western blot is shown below the blots. WT, n = 15; Sel1LAlb, n = 10 for ELISA; n = 3 for Western blot. Values, mean ± SEM. *, p < 0.05 or as labeled by two-tailed Student’s t test. (e-f) Primary hepatocytes isolated from WT and Sel1LAlb littermates were treated with 1 µg/mL WT SubAB or mutant SubAA272B (mut) for 12 hours and analyzed by: (e) non-reducing and reducing SDS-PAGE followed by Western blot analyses, with red arrow pointing to fibrinogen aggregates; and (f) sucrose gradient fractionation (fractions 1 to 12 from top to bottom) showing the quantitation of reduced Bβ chain, with monomers, assembly intermediates and HMW aggregates labeled. Western blot images for (f) are shown in Fig. S7c. Representative data from two independent repeats. Source data are provided as a Source Data file.
Depletion of BiP by the treatment of a BiP-specific protease SubAB66 led to a marked fibrinogen aggregation in Sel1LAlb hepatocytes, while exerting a subtle effect in WT hepatocytes, as shown by non-reducing Western blot (red arrows, Fig. 6e) and sucrose gradient fractionation (Fig. S7c and quantitated in Fig. 6f), pointing to an important role of BiP in preventing the aggregation of fibrinogen chains in inclusion bodies. As circulating levels of fibrinogen were modestly reduced, the coagulation function measured by prothrombin time was not altered in Sel1LAlb mice (Fig. S7d), which is consistent with observations made in fibrinogen Aα heterozygous (Fib+/−) mice67. Taken together, we conclude that SEL1L-HRD1 ERAD is required for fibrinogen maturation in the ER and that, without ERAD, a fraction of fibrinogen is trapped as aggregates or BiP-associated folding intermediates, leading to the formation of hepatic inclusion bodies and hypofibrinogenemia.
Fibrinogen chains are bona fide endogenous ERAD substrates
To delineate how ERAD controls the maturation of nascent fibrinogen, we next asked whether fibrinogen is a bona fide endogenous substrate of SEL1L-HRD1 ERAD. While a previous study showed that fibrinogen is degraded by the proteasome in vitro68, the molecular nature of this process, such as the machinery mediating this degradation event, remains unknown. All three fibrinogen chains interacted with HRD1 and were polyubiquitinated in HRD1-transfected HEK293T cells in an E3-ligase dependent manner as the polyubiquitination was blocked by a ligase-dead HRD1 C2A mutant44,69 (Fig. 7a). Similarly, loss of HRD1 in cells treated with a proteasome inhibitor MG132 attenuated the polyubiquitination of three fibrinogen chains (Fig. S8). Moreover, the loss of SEL1L in primary hepatocytes caused a marked stabilization of all three endogenous fibrinogen chains following the treatment of a translation inhibitor cycloheximide (CHX) (Fig. 7b). Deletion of HRD1 or SEL1L in human Huh7 hepatocytes also stabilized endogenous fibrinogen chains (Fig. 7c), pointing to the conservation of this degradation event. Hence, we conclude that nascent fibrinogen chains are highly misfolding prone, ubiquitinated and degraded by SEL1L-HRD1 ERAD.
Fig. 7. Three fibrinogen chains are bona fide endogenous ERAD substrates.
a Western blot analysis of immunoprecipitates of HA-agarose in HEK293T cells transfected with indicated plasmids, showing that HRD1 is sufficient for the ubiquitination of fibrinogen chains. HRD1-C2A, an E3 ligase dead mutant. Cells were treated with a proteasome inhibitor MG132 for the last 3 hr prior to immunoprecipitation. Representative data from two independent repeats. b–c Western blot analysis of endogenous fibrinogen Aα, Bβ and γ decay after a translation inhibitor cycloheximide (CHX) treatment in (b) primary hepatocytes isolated from WT and Sel1LAlb littermates, and (c) WT, SEL1L−/− and HRD1−/− Huh7 hepatocytes. The degradation is quantitated relative to the percentage of control. In (b), n = 4 for Aα and Bβ; n = 3 for γ each genotype. In c, n = 5 for WT and HRD1−/−; n = 3 for SEL1L−/−. N indicates independent samples. Values, mean ± SEM. *, p < 0.05; **, p < 0.01; ***, p < 0.001 comparing knockout to WT cells by two-way ANOVA with Sidak multiple comparison test. Source data are provided as a Source Data file.
The assembly of fibrinogen chains protects them from SEL1L-HRD1 ERAD
A previous study showed that genetic deletion of fibrinogen Aα chain abolishes the secretion of fibrinogen Bβ and γ, without causing their accumulation in the ER67; however, the underlying mechanism remained unclear. In light of our findings, we next tested whether unassembled fibrinogen Bβ and γ chains are degraded by SEL1L-HRD1 ERAD. To this end, we employed lipid nanoparticle (LNP)-encapsulated small interfering RNA (siRNA) to knockdown fibrinogen Aα mRNA (siFga) in the liver, without affecting the mRNA levels of the other two chains (Fig. S9a)70,71. An siRNA sequence targeting luciferase (siLuc) was used as a negative control. Consistent with previous reports70,71, the LNP-siFga treatment effectively reduced the protein levels of fibrinogen chains in WT livers and led to the depletion of plasma fibrinogen in WT mice (Fig. 8a and S9b). Strikingly, this reduction was reversed by ERAD deficiency, as protein levels of fibrinogen chains were significantly elevated in the livers of Sel1LAlb mice receiving LNP-siFga compared to control Sel1LAlb mice (Fig. 8a). This elevation was not due to gene transcription (Fig. S9a), pointing to the model that orphan Bβ and γ chains in the absence of Aα chain are targeted for proteasomal degradation by SEL1L-HRD1 ERAD. This finding is in line with the ubiquitination of individual fibrinogen chains in transfected HEK293T cells (Fig. 7a and S8). Furthermore, Fga knockdown in Sel1LAlb livers caused marked aggregation of Bβ and γ chains (Fig. 8b). The accumulated fibrinogen Bβ and γ chains were nearly 100% EndoH sensitive, i.e. ER retained (Fig. 8c), leading to further enlarged hepatic inclusion bodies (Fig. 8d) and abolished fibrinogen secretion (Fig. S9b). Together, these data suggest that the assembly of fibrinogen chains may suppress their degradation by ERAD and be a prerequisite for ER export.
Fig. 8. Assembly of fibrinogen complex prevents SEL1L-HRD1 ERAD of individual chains.
8-week-old WT and Sel1LAlb littermates were injected intravenously with lipid nanoparticle (LNP)-encapsulated small interfering RNA against fibrinogen Aα chain (siFga) or luciferase (siLuc) for control. 11 days after injection, mice were examined for: (a) reducing Western blot analysis of liver samples with quantitation normalized to HSP90 shown on the right; (b) non-reducing SDS-PAGE and Western blot analysis using the same liver lysates shown in a; (c) Western blot analysis of liver lysates treated with EndoH, with the percentages of EndoH-sensitive Bβ and γ chains shown on the right; and (d) H&E and fibrinogen staining of liver sections, with the size of inclusion bodies in H&E-stained Sel1LAlb samples quantitated on the right. N = 3. In (d), 12 to 16 images taken from 3 mice per cohort were analyzed. Values, mean ± SEM. *, p < 0.05; ***, p < 0.001 by two-way ANOVA with Tukey multiple comparison test in (a) and (c), and by student’s t test in d. Source data are provided as a Source Data file.
SEL1L-HRD1 ERAD degrades fibrinogen γ disease mutants and attenuates its pathogenicity towards wildtype Aα and Bβ chains
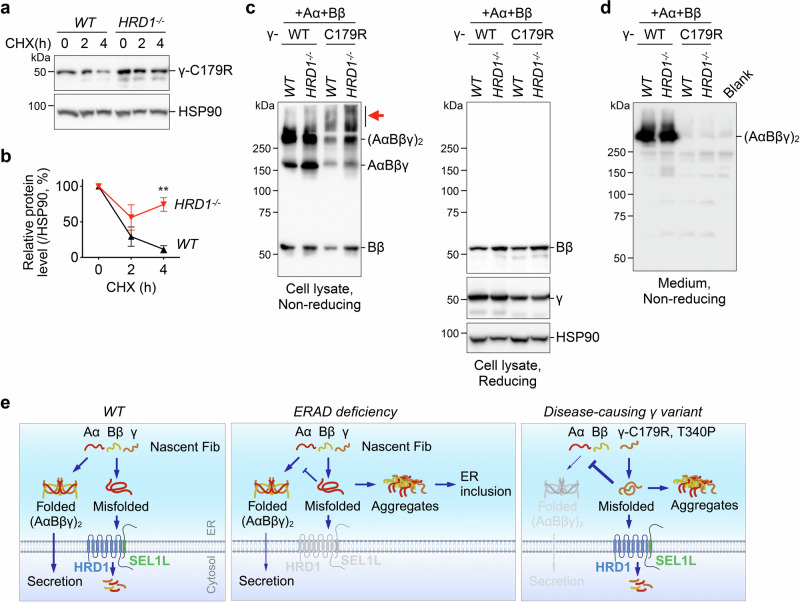
To investigate the pathological importance of SEL1L-HRD1 ERAD, we explored its role in the pathogenicity of two disease-causing fibrinogen γ mutants, C179R and T340P, reported to cause hypofibrinogenemia and HFSD, respectively14,17, in HEK293T cells that do not express endogenous fibrinogen. SEL1L-HRD1 ERAD was indeed responsible for the clearance of both mutants from the ER as they were stabilized in HRD1−/− HEK293T cells compared to that in WT cells (Fig. 9a, b and Fig. S10a, b). We next asked whether the expression of γ-C179R and γ-T340P may alter the maturation and assembly of fibrinogen hexamers. We co-expressed a combination of fibrinogen Aα-WT and Bβ-WT with either WT- or mutant γ chains in HEK293T cells followed by non-reducing and reducing SDS-PAGE and Western blot. In keeping with the observations described above, fibrinogen γ-WT formed trimer and hexamers with Aα and Bβ in WT and more so in HRD1−/− cells (Fig. 9c). In contrast, γ-C179R mutant failed to form a large amount of trimer and hexamers with Aα and Bβ, but rather induced the formation of HMW aggregates of Bβ chain in WT HEK293T cells and much more so in HRD1−/− cells (Fig. 9c). This HMW aggregation was accompanied by the complete absence of fibrinogen hexamers in the medium (Fig. 9d). The HMW complexes were formed via aberrant disulfide bonds as they were sensitive to the treatment of β-mercaptoethanol (Fig. 9c). Similar observations were made for γ-T340P (Fig. S10c-d). Taken together, we conclude that SEL1L-HRD1 ERAD negatively controls the pathogenicity of fibrinogen γ mutants.
Fig. 9. SEL1L-HRD1 ERAD degrades a disease-causing fibrinogen γ variant and mitigates its pathogenicity.
a–b Western blot analysis of fibrinogen γ-C179R decay after a translation inhibitor CHX treatment in WT and HRD1−/− HEK293T cells transfected with a plasmid expressing γ-C179R. The degradation is quantitated relative to the percentage of control. N = 3, indicating independent samples. Values, mean ± SEM. **, p < 0.01 by two-way ANOVA with Sidak multiple comparison test. c–d WT and HRD1−/− HEK293T cells were transfected with Aα-WT, Bβ-WT, and either γ-WT or γ-C179R. Cell lysates (c) and cell culture medium (d) were analyzed by non-reducing or reducing SDS-PAGE as indicated. Red arrow points to fibrinogen Bβ aggregates. Culture medium from non-transfected cells was included as a blank control in d. Representative data from two independent repeats. e Model showing a critical role of SEL1L-HRD1 ERAD in the biogenesis of mature fibrinogen protein complex in the ER. SEL1L-HRD1 ERAD in hepatocytes mediates the degradation of endogenous fibrinogen chains, thereby ensuring their proper assembly and subsequent secretion (left). In the absence of ERAD, fibrinogen chains accumulate and aggregate in ER-derived hepatic inclusion bodies, interfering the folding and secretion of nascent fibrinogen protein (middle). SEL1L-HRD1 ERAD also degrades a disease-causing fibrinogen γ mutant, thereby attenuating its pathogenicity (right). Image created in BioRender. Tushi, N. (2024) BioRender.com/k40b037. Source data are provided as a Source Data file.
Discussion
In this study, we reported that the SEL1L-HRD1 ERAD complex prevents the formation of ER-derived inclusion bodies in hepatocytes. In studying the nature of inclusions, we found that SEL1L-HRD1 ERAD regulates the biogenesis of an essential blood coagulation factor, fibrinogen, in the ER of hepatocytes. SEL1L-HRD1 ERAD mediates the degradation of endogenous unassembled, likely misfolded, fibrinogen chains, a key step in the maturation of the nascent fibrinogen protein complex. In the absence of SEL1L-HRD1 ERAD, fibrinogen accumulates and aggregates in inclusion bodies together with ER chaperones such as BiP, leading to hypofibrinogenemia and mild liver damage. We further demonstrated that SEL1L-HRD1 ERAD attenuates the pathogenicity of two disease-causing fibrinogen γ mutants. This study demonstrates an essential role of SEL1L-HRD1 ERAD in fibrinogen biogenesis and provides insights into the pathogenesis of protein-misfolding diseases (Fig. 9e).
In this study, we found that fibrinogen is retained in the inclusion bodies of Sel1L-deficient hepatocytes, leading to its reduced secretion. In contrast, ceruloplasmin, another SEL1L-HRD1 ERAD substrate, was found to be increasingly secreted by Sel1L-deficient hepatocytes55. Indeed, the secretion of other proteins such as albumin and fibronectin was unaffected by SEL1L-HRD1 ERAD deficiency. Together these observations highlight the specificity of SEL1L-HRD1 ERAD in mediating the degradation and quality control of fibrinogen, and a selective retention of misfolded fibrinogen in specialized ER-derived inclusions. This conclusion is supported by the recent proteomic screens of endogenous SEL1L-HRD1 ERAD substrates, which identified approximately 100 endogenous proteins in different cell and tissue types72. In addition to fibrinogen, several ER chaperones were identified in the inclusion bodies, including but not limited to, BiP, PDI, and OS9. As previous studies have demonstrated the role of these chaperones in regulating protein folding and degradation5,20,73, we speculate that fibrinogen in the inclusion bodies may be in the state of folding intermediates, misfolded protein, or protein aggregates. It is unclear whether properly folded or assembled fibrinogen is also present in the inclusion bodies. Whether other secretory proteins are retained in the inclusion bodies, and what feature(s) in the proteins dictates their incorporation into the inclusions, are open and exciting questions.
Previous studies have shown that SEL1L-HRD1 ERAD deficiency induces a very low level of ER stress, without triggering cell death or inflammation in many cell types42,44,74. How cells adapt to ERAD deficiency remains an open question, which may involve the induction of ER chaperones and/or activation of ER-phagy51,75. We propose that inclusion bodies may serve as another adaptive mechanism to sequester misfolded ERAD substrates when SEL1L-HRD1 ERAD function is impaired. Relevant to this study, recent reports on α1-antitrypsin Z mutant-induced inclusions showed that COPII and SNARE-mediated vesicle trafficking mediates the transport of Z-α1-antitrypsin between the ER and inclusion bodies, and that Z-α1-antitrypsin undergoes phase transition to a solid state in inclusion bodies76,77. Moreover, adipocytes deficient in both SEL1L and autophagy were found to form the coalescence of ER fragments (CERFs) containing misfolded lipoprotein lipase that is able to undergo phase separation in vitro51. It will be of great interest to examine whether similar trafficking and phase transition mechanisms are applicable in the formation of fibrinogen inclusion bodies.
Hepatic inclusion bodies containing fibrinogen have been reported clinically in HFSD patients8,9,78–80. This abnormality can be caused either genetically by dominant-negative mutations of fibrinogen Aα and γ chains11–15,81–83, or acquired under pathological conditions such as hepatocellular carcinoma, chronic hepatitis B virus infection, and COVID-19 infection9,84,85. In both congenital and acquired HFSD, hepatic inclusion bodies exhibit distinctive protein profiles, featuring the presence of fibrinogen but the absence of other proteins like α1-antitrypsin, albumin or serum amyloid A9,11,85. The direct link between SEL1-HRD1 ERAD activity and HFSD is yet to be demonstrated in vivo, requiring the generation of mouse models carrying specific fibrinogen mutations or diseases. In addition to ERAD, recent reports implicated autophagy and lysosome-mediated degradation in the clearance of WT and HFSD-causing mutant fibrinogen chains in vitro68,86,87. Intriguingly, an autophagy-enhancing drug, carbamazepine, was shown to alleviate liver damage in HFSD patients, further supporting a possible role of autophagy in HFSD and hepatic fibrinogen inclusions88,89. While the relationship between ERAD and autophagy in both normal liver physiology and disease states remains largely unclear, we speculate that impaired ERAD activity may contribute to the pathogenesis of various HFSD conditions and that enhancing ERAD function and optimizing the substrate targeting to ERAD machinery may hold therapeutic promise for protein-misfolding diseases associated with defects in ER protein folding and maturation.
Methods
Study approval
All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of University of Virginia, Wayne State University, and University of North Carolina at Chapel Hill.
Mouse studies
Hepatocyte-specific Sel1L deficient (Sel1Lflox/flox;Albumin-Cre, i.e. Sel1LAlb) mice and wildtype (Sel1Lflox/flox, WT) littermates, hepatocyte-specific Hrd1 deficient (Hrd1flox/flox;Albumin-Cre, i.e. Hrd1Alb) mice and wildtype (Hrd1flox/flox, WT) littermates, Ire1αflox/flox mice, as well as hepatocyte-specific ApoE promoter-driven FGF21 transgenic or WT littermates were maintained on a C57BL/6 J background and described previously42,43,58,90. Adenovirus-mediated overexpression of CREBH or GFP was performed via an intravenous injection of recombinant adenovirus as previously described57. For CPA treatment, 8-week-old mice were injected intraperitoneally with CPA (Sigma 239805) at 5 mg/kg body weight every other day for 4 weeks55. Mice were fed a standard rodent chow diet (Inotiv, no.7912) and housed in a temperature-controlled environment with 12 hour light/dark cycles with free access to food and water. For inferior vena cava (IVC) plasma collection, mice were anesthetized with isoflurane and 0.5 mL blood was drawn from IVC into a syringe with 0.05 mL of 3.2% sodium citrate. Tissues were collected and immediately either fixed in 10% neutral buffered formalin or frozen in liquid nitrogen upon collection.
AAV- and LNP-mediated gene delivery
To induce an acute deletion of SEL1L, HRD1 or IRE1α in the liver, 9-week-old Sel1Lflox/flox, Hrd1flox/flox, or Ire1αflox/flox mice were intravenously injected with AAV8 expressing hepatocyte-specific TBG promoter-driven Cre or GFP at a dose of 1×1012 viral genome copies per mouse and euthanized at the indicated time. AAV8 was purchased at the University of Michigan Vector Core (Ann Arbor, MI). To knockdown fibrinogen Aα, 8-week-old WT and Sel1LAlb littermates were intravenously injected with 2 mg/kg LNP-encapsulated siRNA against Fga (siFga) or luciferase (siLuc) and euthanized 11 days later. LNPs were prepared as previous described70.
Fibrinogen ELISA, blood coagulation, and blood chemistry measurement
Fibrinogen concentration in citrated IVC plasma was measured using a fibrinogen assay kit (ICL-Lab E90FIB) according to the manufacturer’s instructions. Prothrombin time (PT) was measured by adding thromboplastin (Pacific Hemostasis) to citrated IVC plasma. Clotting time was assessed when mechanical translocation of a bead was inhibited using the STart machine (Stago). For blood chemistry of the liver panel, blood serum was analyzed by the Liasys analyzer (AMS Alliance) at the University of Michigan In-Vivo Animal Core (Ann Arbor, MI) on a fee-for-service basis.
Isolation of primary hepatocytes
Mice were anesthetized with isoflurane inhalation, and the liver was perfused first with a warm perfusion buffer (Invitrogen 17701), and then with warm HBSS (Gibco 14025-076) containing 10 mM HEPES, 4 mM NaOH, and 1 mg/ml collagenase (Worthington LS004196). Digested livers were carefully removed, dispersed in a cold wash medium (DMEM with 1% sodium pyruvate, 1% penicillin-streptomycin, 10% FBS), and passed through a 100 μm cell strainer. The cells were then washed three times by centrifugation at 50 x g for 5 minutes at 4 °C. The resulting hepatocyte pellets were resuspended in a hepatocyte culture medium (DMEM with 1% penicillin-streptomycin and 10% FBS) and seeded in collagen-coated plates or chamber slides. 4 hours after plating, cells were carefully washed with 1x PBS and cultured in a fresh hepatocyte culture medium overnight. To examine the degradation of endogenous fibrinogen, cells were pre-treated with 1 μg/mL brefeldin A (BFA, Cayman Chemical 11861) for 30 minutes, then treated with vehicle or 150 μM cycloheximide (CHX, Sigma 01810) for 2 or 4 hours. For SubAB treatment, cells were treated with 1 µg/mL wildtype SubAB or mutant SubAA272B for 12 hours before analyses.
Cell culture
HEK293T cells were originally obtained from ATCC. Huh7 cells were originally obtained from JCRB Cell Bank. WT and HRD1−/− HEK293T cells were generated as previously described35,44. WT, HRD1−/−, and SEL1L−/− Huh7 cells were generated using lentivirus-carried CRISPR/Cas9 expressing a nontargeting control guide (5’-GATCGTTTCCGCTTAACGGC-3’), guides targeting human HRD1 (5’-ACACCAGTTCTACCCCACTG-3’), or SEL1L (5’-ACTGCAGGCAGAGTAGTTGC-3’). Huh7 and HEK293T cells were cultured in DMEM (Gibco) containing 10% FBS and 1% penicillin-streptomycin. For transfection, HEK293T cells were transfected with various plasmids using 80 μg/mL polyethylenimine (PEI, Sigma 408727). Plasmids used for transfection are: pcDNA3-mFga-HA, pcDNA3-mFgb-HA, pcDNA3-mFgg-HA, pcDNA3-Hrd1-WT-Myc, pcDNA3-Hrd1-C2A-Myc, and pcDNA3-6His-Ub. Fibrinogen γ point mutations C179R and T340P were generated using site-directed mutagenesis from pcDNA3-mFgg-HA. To examine protein ubiquitination or degradation, cells were treated with 10 μM MG132 (Cayman Chemical 10012628) for 3 hours, or 1 μg/mL brefeldin A (BFA, Cayman Chemical 11861) for 30 minutes followed by 150 μM cycloheximide (CHX, Sigma 01810) for 2 or 4 hours before collection as indicated in the figure legends.
TEM and immunogold labeling
Mice were anesthetized and perfused with fixation buffer containing 2.5% glutaraldehyde, 4% formaldehyde in 0.1 M sodium cacodylate buffer (Electron Microscopy Sciences). Liver tissues were then incubated in the fixation buffer overnight at 4 °C, cut into small pieces (about 1-2 mm cubes) and submitted to the University of Michigan Microscopy Core for washing, embedding and sectioning at 70 nm. The grid containing tissue sections was imaged by a JEOL 1400-plus electron microscope (JEOL) at the University of Michigan Microscopy Core. For immunogold labeling, livers were fixed in a buffer containing 3% paraformaldehyde, 0.2 M sucrose in 0.1 M Sorensen’s phosphate buffer (PH = 7.2) and cut into small pieces and processed by the University of Michigan Microscopy Core. The ultra-thin sections (80 nm) were cut, placed on bare nickel grid and stored at 4 °C. For labeling, the grids were quenched in 80 mM glycine and incubated in blocking solution (EMS 25599) containing 0.2% Tween-20 for 1 hour at room temperature followed by overnight incubation with anti-BiP antibody (1:50, Abcam Ab21685) at 4 °C in a humidifying chamber. The grids were next incubated with colloidal gold antibodies (1:25, Jackson ImmunoResearch) for 1 hour at room temperature followed by post-fixation in 1% glutaraldehyde and contrast staining using 0.5% uranyl acetate. After carbon evaporation, the grids were imaged by JEOL electron microscope at the University of Michigan Microscopy Core.
Sucrose gradient fractionation
Sucrose gradient fractionation was performed as reported previously44. Briefly, primary hepatocytes or liver tissues were lysed by a dounce homogenizer in 1% NP40 lysis buffer (150 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl pH 7.5, 1% NP-40, protease inhibitors and 20 mM N-ethylmaleimide). Extracts were centrifuged through 20%-40% sucrose gradients (in 150 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl pH 7.5 and protease inhibitor) prepared freshly by progressively layering higher to lower density sucrose fractions in 5% increments. Extracts were centrifuged at 243,000x g. for 17 hours at 4 °C using a SW50.1 Swinging-Bucket Rotor (Beckman Ultracentrifuge L8M). Each 3.5 mL gradient was divided evenly into fractions 1-10 for livers or 1-11 for hepatocytes from top to bottom and the pellet was resuspended in 300 μL 1% NP40 lysis buffer as the last fraction. Aliquots of fractions were subjected to Western blot analyses under reducing or non-reducing conditions as described below.
NP-40 solubility assay
Frozen liver tissues were weighed and homogenized in 0.5% NP-40 lysis buffer (50 mM Tris-HCl pH 8.0, 0.5% NP-40, 150 mM NaCl, 5 mM MgCl2 and protease inhibitor) as previously described32. The lysate volume was normalized by tissue weight, centrifuged at 12,000 x g for 10 minutes and the supernatant was collected as the NP-40 soluble fraction. The pellet was then resuspended in 1× SDS sample buffer (50 mM Tris-Cl, 2% SDS, 0.29 M 2-mecaptoethanol, 10% glycerol, 0.01% bromophenyl blue) with the volume normalized to initial tissue weight, heated at 95 °C for 30 minutes, and collected as the NP-40 pellet fraction.
Immunoprecipitation and mass spectrometry
To analyze the ubiquitination of HA-tagged fibrinogen chains, cells were lysed in a reducing lysis buffer (150 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl pH 7.5, 1% NP-40, 1% SDS, 5 mM DTT, and protease inhibitors), denatured at 95 °C for 5 minutes and diluted 10-fold with 1% NP40 lysis buffer before immunoprecipitation. Lysates were then pre-cleared with protein A agarose at 4 °C for 1 hour and incubated with anti-HA agarose (Sigma A2095) at 4 °C overnight with gentle rocking. To perform immunoprecipitation for protein interactions, cells or tissues were lysed by sonication in the 1% NP40 lysis buffer (150 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl pH 7.5, 1% NP-40, protease inhibitors and 5 mM N-ethylmaleimide). To immunoprecipitate BiP/GRP94 or fibrinogen, a total of 2 mg tissue lysate was pre-cleared with protein A agarose at 4 °C for 1 hours, then incubated with 2 μg anti-KDEL antibody (Novus NBP1-97469) or anti-fibrinogen serum (a gift from Dr. Matthew James Flick)62,63 at 4 °C overnight, followed by an incubation in protein A agarose at 4 °C for 2 hours. Immuno-complexes were washed in the 1% NP40 lysis buffer four times and eluted by boiling at 95 °C for 5 minutes in SDS sample buffer. For Immunoprecipitation followed by mass spectrometry analysis of fibrinogen interactomes, ~60 mg liver tissues (one per genotype) were lysed in 1% NP40 lysis buffer by sonication, incubated on ice for 30 minutes, and centrifuged at 16,000 x g for 10 minutes. 5 mL supernatants with 10 mg protein were pre-cleared with protein A agarose at 4 °C for 2 hours, then incubated with 20 μL anti-fibrinogen serum or normal rabbit IgG at 4 °C overnight, followed by an incubation in protein A agarose at 4 °C for 4 hours. Immuno-complexes were washed in the 1% NP40 lysis buffer six times, rinsed by PBS three times, and submitted for mass spectrometry analysis at University of Michigan Proteomics core on a fee-for-service basis as recently described51. Proteins were identified by searching the MS/MS data using Proteome Discoverer (v2.4, Thermo Scientific). False discovery rate (FDR) was determined using Percolator and proteins/peptides with a FDR of ≤1% were retained for further analysis. Pathway analysis was performed by searching the 32 enriched proteins with KEGG pathway annotation against the remaining unenriched protens. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE91 partner repository with the dataset identifier PXD047658.
Endoglycosidase H (EndoH) and PNGase treatment
EndoH and PNGase F (New England BioLabs, P0702L and P0704L) treatment was performed according to the manufacture’s protocol and as previously described92. Briefly, tissue lysates were denatured at 95 °C for 10 minutes with the glycoprotein denaturing buffer, and then digested with EndoH or PNGase F at 37 °C for 1 hour. The reaction was stopped by the addition of 5X denaturing sample buffer and boiled at 95 °C for 5 minutes prior to be loaded onto SDS-PAGE.
Western blot
Preparation of cell and tissue lysates and western blotting was performed as previously described44. For non-reducing SDS-PAGE, lysates were prepared in 5X non-denaturing sample buffer (250 mM Tris-HCl pH 6.8, 1% SDS, 50% glycerol and 0.05% bromophenyl blue) without boiling before being separated on an SDS-PAGE gel. The following antibodies were used in this study: fibrinogen (polyclonal fibrinogen antiserum targeting the entire fibrinogen complex, gift from Dr. Matthew James Flick, 1:10000)62,63, fibrinogen Aα (Proteintech 20645-1-AP, 1:1000), fibrinogen Bβ (Proteintech 16747-1-AP, 1:10000), fibrinogen γ (Proteintech 15841-1-AP, 1:2000), albumin (Proteintech 16475-1-AP, 1:6000), BiP (CST C50B12, 1:3000), SEL1L (Abcam ab78298, 1:2000), HRD1 (Proteintech 13473-1-AP, 1:5000), IRE1α (CST 14C10, 1:2000), KDEL (Novus NBP1-97469, 1:2000), PDI (Enzo ADI-SPA-890, 1:2000), ubiquitin (CST 3933, 1:2000), Myc (CST 2278, 1:2000), HA (Sigma H3663, 1:2000), H2A (CST 12349S, 1:2000), βActin (Proteintech 66009, 1:2000) and HSP90 (Santa Cruz sc7947, 1:4000). Secondary antibodies, goat anti-rabbit or mouse IgG HRP (Bio-rad), were used at 1:6000 dilution. For fibrinogen after immunoprecipitation, Trueblot anti-rabbit IgG HRP (Rockland) was used as secondary antibody at 1:1000 dilution. Western blot membranes were developed using the Clarity Western ECL Substrate (Bio-rad) and signal was detected with a ChemiDOC imager (Bio-rad). Quantification was performed using ImageLab software (Bio-rad).
Histology, immunofluorescence and immunocytochemistry
Tissues were fixed in 10% neutralized formalin and processed for embedding, sectioning and H&E or PAS staining by the University of Michigan In-Vivo Animal Core (Ann Arbor, MI) on a fee-for-service basis. The H&E slides were imaged using an Olympus BX41 miscroscope or Aperio Scanscope (Leica). Inclusion body size and number were quantitated with images taken under 600X magnification using Image J software. At least 12 images randomly taken from liver sections of three or more mice per cohort were used for quantitation. The minimum diameter of each inclusion body was used as a measurement of size. The inclusion body number was quantitated by normalizing to the liver tissue area. Sinus red staining (Electron Microscopy Sciences 26357) was performed according to the manufacture’s protocol. For immunofluorescent staining, paraffin-embedded liver sections of 5 µm were deparaffinized in xylene and re-hydrated using graded ethanol series and distilled water. Antigen retrieval was performed by boiling the slides in 10 mM citric acid buffer (pH 6.0) for 15 minutes, followed by blocking (5% normal donkey serum, 0.5% Tween-20 in PBS) at room temperature for 1 hour. Liver cryosections were used for fibronectin and TFR1 immunofluorescent staining. Briefly, 5 µm cryosections were fixed with 4% paraformaldehyde and permeabilized in 0.5% Triton X-100 for 10 minutes, followed by blocking (5% normal donkey serum, 0.5% Tween-20 in PBS) at room temperature for 1 hour. For staining, slides were incubated with primary antibodies at 4 °C overnight, followed by secondary antibodies at room temperature for 1 hour using Alexa fluor 488 or 594 affinipure donkey anti-mouse or rabbit IgG (H + L) (Jackson ImmunoResearch). Both primary and secondary antibodies were diluted in PBS containing 5% normal donkey serum and 0.5% Tween-20. The nuclei and slides were stained and mounted by prolong gold antifade reagent with DAPI. Fluorescence images were captured under a ZeissLSM710 confocal microscope, Zeiss Imager.Z1 fluorescence microscope, Leica DMi8 Thunder Microscope, or a Cytation C10 Confocal Imaging Reader (BioTek). For immunocytochemistry, primary hepatocytes seeded on chamber slides were fixed with 4% paraformaldehyde, then permeabilized with permeabilization buffer (0.5% Triton X-100, 0.05% SDS in PBS) for 10 minutes, followed by blocking, staining and imaging as described above. The following primary antibodies were used for staining: fibrinogen (from Dr. Matthew James Flick, 1:1000)62,63, fibrinogen Aα (Proteintech 20645-1-AP, 1:100), fibrinogen Bβ (Proteintech 16747-1-AP, 1:100), fibrinogen γ (Proteintech 15841-1-AP, 1:100), KDEL (Abcam ab12223, 1:500), BiP (CST C50B12, 1:200), PDI (Enzo ADI-SPA-890, 1:100), OS9 (Abcam ab109510, 1:100), α1-antitrypsin (Proteintech 16382-1-AP, 1:100), albumin (Bethyl Lab A90-234A, 1:200), fibronectin (Abcam ab2413, 1:200), and TFR1 (Invitrogen 13-6800, 1:100).
RNA extraction, RT-PCR and quantitative real-time PCR
Liver RNA was isolated as previously described93. Briefly, total RNA from liver tissues was extracted using RNA-Stat 60 (IsoTex Diagnostics), chloroform, and precipitated by isopropanol. RNA quality was determined by measuring the OD260/280 and visualized on an agarose gel. cDNA was generated using the Superscript III reverse transcriptase (Thermofisher). Quantitative PCR (qPCR) was performed using 2X Universal SYBR Green Fast qPCR Mix (Abclonal). 18S was used as the reference. QPCR primer sequences are listed below:
Mouse Fga (GTCCGGGCTCAGTTGATAGA, TAGCGATGACCTGTTGAAGC),
Mouse Fgb (TCTTCAGCACGTACGACAGG, TGTAAAGGCCACCCCAGTAG),
Mouse Fgg (TGGGACAACGACAACGATAA, CCGTCGTCGAAACCATTAGT),
Mouse 18S (ACCGCAGCTAGGAATAATGGA, GCCTCAGTTCCGAAAACCA).
Statistics
Unpaired two-tailed Student’s t test was used for two-group analyses. Two-way ANOVA analysis with Tukey or Sidak multiple comparison test or one-way ANOVA with Tukey multiple comparison test was used for multi-group analyses (GraphPad Prism). Data were presented as the mean ± SEM; p < 0.05 was considered significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We thank Drs. Yuan Zhang, David J. Mangelsdorf and Steven A. Kliewer for sharing reagents; Drs. James P. Luyendyk, Alisa S. Wolberg, Lih Jiin Juang, Ling Qi, and members of the Sun laboratories for their insightful discussions. We also thank the University of Virginia Advanced Microscopy Facility, Wayne State University Microscopy, Imaging and Cytometry Resources Core, Michigan Diabetes Research Center, University of Michigan In-vivo Animal Core, and Vector Core for their support. This work was supported by the funding from NIH (R01DK132068 and R01DK128077 to SS; R01HL160046 to MJF; R01HL166382 to CJK; R01DK120330 and R01DK126908 to DF; DK090313 and R01DK126908 to KZ; R01HL163516 to ZZ; and R21HD104904 to JW), American Society of Nephrology (XW), and National Health and Medical Research Council Investigator Grant 1174876 (JCP).
Author contributions
Z.S. and P.T. performed most in vivo and in vitro studies. W.S.H., M.T., S.A.W., X.W., and N.J.T. assisted with some in vivo tests, biochemical studies and data analyses. S.J. helped with histology analysis. J.W., F.F., A.W.P., J.C.P., Z.Z., K.Z., D.F., C.J.K., and M.J.F. provided critical reagents, insights and discussions. S.S. conceived the study, designed experiments, and wrote the manuscript. All authors commented on and approved the manuscript. First-authorship order was determined based on the extent of involvement during the completion of the study.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
All study data are included in the article and/or SI Appendix. Source data are provided with this paper. The proteomics data of fibrinogen interactomes have been deposited to the ProteomeXchange Consortium with the dataset identifier PXD047658. The previously published proteomics data of purified microsomes can be found at ProteomeXchange Consortium with dataset identifier PXD035243. Source data are provided with this paper.
Competing interests
The authors declared no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhenfeng Song, Pattaraporn Thepsuwan.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53639-x.
References
- 1.Pieters, M. & Wolberg, A. S. Fibrinogen and fibrin: An illustrated review. Res Pr. Thromb. Haemost.3, 161–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luyendyk, J. P., Schoenecker, J. G. & Flick, M. J. The multifaceted role of fibrinogen in tissue injury and inflammation. Blood133, 511–520 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu, X. & Shi, B. Progress in research on the role of fibrinogen in lung cancer. Open Life Sci.15, 326–330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casini A., Neerman-Arbez M., & de Moerloose P. Heterogeneity of congenital afibrinogenemia, from epidemiology to clinical consequences and management. Blood Rev. 100793 (2020). [DOI] [PubMed]
- 5.Tamura, T., Arai, S., Nagaya, H., Mizuguchi, J. & Wada, I. Stepwise assembly of fibrinogen is assisted by the endoplasmic reticulum lectin-chaperone system in HepG2 cells. PLoS One8, e74580 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maghzal, G. J., Brennan, S. O., Homer, V. M. & George, P. M. The molecular mechanisms of congenital hypofibrinogenaemia. Cell Mol. Life Sci.61, 1427–1438 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ariens, R. A. Fibrin(ogen) and thrombotic disease. J. Thromb. Haemost.11, 294–305 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Callea, F., Brisigotti, M., Fabbretti, G., Bonino, F. & Desmet, V. J. Hepatic endoplasmic reticulum storage diseases. Liver12, 357–362 (1992). [DOI] [PubMed] [Google Scholar]
- 9.Zen, Y. & Nishigami, T. Rethinking fibrinogen storage disease of the liver: ground glass and globular inclusions do not represent a congenital metabolic disorder but acquired collective retention of proteins. Hum. Pathol.100, 1–9 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Vu, D. & Neerman-Arbez, M. Molecular mechanisms accounting for fibrinogen deficiency: from large deletions to intracellular retention of misfolded proteins. J. Thromb. Haemost.5, 125–131 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Brennan, S. O., Wyatt, J., Medicina, D., Callea, F. & George, P. M. Fibrinogen brescia: hepatic endoplasmic reticulum storage and hypofibrinogenemia because of a gamma284 Gly–>Arg mutation. Am. J. Pathol.157, 189–196 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asselta, R. et al. Hepatic fibrinogen storage disease: identification of two novel mutations (p.Asp316Asn, fibrinogen Pisa and p.Gly366Ser, fibrinogen Beograd) impacting on the fibrinogen gamma-module. J. Thromb. Haemost.13, 1459–1467 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Brennan, S. O. et al. Novel fibrinogen gamma375 Arg–>Trp mutation (fibrinogen aguadilla) causes hepatic endoplasmic reticulum storage and hypofibrinogenemia. Hepatology36, 652–658 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Brennan, S. O., Davis, R. L., Conard, K., Savo, A. & Furuya, K. N. Novel fibrinogen mutation gamma314Thr–>Pro (fibrinogen AI duPont) associated with hepatic fibrinogen storage disease and hypofibrinogenaemia. Liver Int.30, 1541–1547 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Rubbia-Brandt, L., Neerman-Arbez, M., Rougemont, A. L., Male, P. J. & Spahr, L. Fibrinogen gamma375 arg–>trp mutation (fibrinogen aguadilla) causes hereditary hypofibrinogenemia, hepatic endoplasmic reticulum storage disease and cirrhosis. Am. J. Surg. Pathol.30, 906–911 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Callea F., Francalanci P., and Giovannoni I. Hepatic and Extrahepatic Sources and Manifestations in Endoplasmic Reticulum Storage Diseases. Int J Mol Sci. 22 (2021). [DOI] [PMC free article] [PubMed]
- 17.Terasawa, F. et al. Hypofibrinogenemia associated with a heterozygous missense mutation gamma153Cys to arg (Matsumoto IV): in vitro expression demonstrates defective secretion of the variant fibrinogen. Blood94, 4122–4131 (1999). [PubMed] [Google Scholar]
- 18.Schwarz, D. S. & Blower, M. D. The endoplasmic reticulum: structure, function and response to cellular signaling. Cell. Mol. life Sci.: Cmls.73, 79–94 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chino, H. & Mizushima, N. ER-Phagy: Quality Control and Turnover of Endoplasmic Reticulum. Trends Cell Biol.30, 384–398 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Needham P. G., Guerriero C. J., & Brodsky J. L. Chaperoning Endoplasmic Reticulum-Associated Degradation (ERAD) and Protein Conformational Diseases. Cold Spring Harbor Perspect. Biol. 11 (2019). [DOI] [PMC free article] [PubMed]
- 21.Sun, Z. & Brodsky, J. L. Protein quality control in the secretory pathway. J. Cell Biol.218, 3171–3187 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi, L., Tsai, B. & Arvan, P. New Insights into the Physiological Role of Endoplasmic Reticulum-Associated Degradation. Trends Cell Biol.27, 430–440 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guerriero, C. J. & Brodsky, J. L. The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol. Rev.92, 537–576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hampton, R. Y., Gardner, R. G. & Rine, J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell.7, 2029–2044 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christianson, J. C. et al. Defining human ERAD networks through an integrative mapping strategy. Nat. Cell Biol.14, 93–105 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mueller, B., Lilley, B. N. & Ploegh, H. L. SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER. J. Cell Biol.175, 261–270 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardner, R. G. et al. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J. Cell Biol.151, 69–82 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plemper, R. K. et al. Genetic interactions of Hrd3p and Der3p/Hrd1p with Sec61p suggest a retro-translocation complex mediating protein transport for ER degradation. J. Cell Sci.112, 4123–4134 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Carvalho, P., Goder, V. & Rapoport, T. A. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell126, 361–373 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Mueller, B., Klemm, E. J., Spooner, E., Claessen, J. H. & Ploegh, H. L. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc. Natl Acad. Sci. USA.105, 12325–12330 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sundaram, M. & Greenwald, I. Suppressors of a lin-12 hypomorph define genes that interact with both lin-12 and glp-1 in Caenorhabditis elegans. Genetics135, 765–783 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun, S. et al. Sel1L is indispensable for mammalian endoplasmic reticulum-associated degradation, endoplasmic reticulum homeostasis, and survival. Proc. Natl Acad. Sci. USA.111, E582–E591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin, L. L. et al. SEL1L-HRD1 interaction is required to form a functional HRD1 ERAD complex. Nat. Commun.15, 1440 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhattacharya A., & Qi L. ER-associated degradation in health and disease - from substrate to organism. J. Cell Sci. 132 (2019). [DOI] [PMC free article] [PubMed]
- 35.Yoshida S., et al. Endoplasmic reticulum-associated degradation is required for nephrin maturation and kidney glomerular filtration function. J Clin Invest. 131 (2021). [DOI] [PMC free article] [PubMed]
- 36.Shrestha, N. et al. Sel1L-Hrd1 ER-associated degradation maintains beta cell identity via TGF-beta signaling. J. Clin. Invest.130, 3499–3510 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou, Z. et al. Endoplasmic reticulum-associated degradation regulates mitochondrial dynamics in brown adipocytes. Science368, 54–60 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, L. et al. ER-associated degradation preserves hematopoietic stem cell quiescence and self-renewal by restricting mTOR activity. Blood136, 2975–2986 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu, L. et al. Protein quality control through endoplasmic reticulum-associated degradation maintains haematopoietic stem cell identity and niche interactions. Nat. Cell Biol.22, 1162–1169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei, J. et al. HRD1-mediated METTL14 degradation regulates m(6)A mRNA modification to suppress ER proteotoxic liver disease. Mol. Cell.81, 5052–65.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim, H. et al. Regulation of hepatic circadian metabolism by the E3 ubiquitin ligase HRD1-controlled CREBH/PPARalpha transcriptional program. Mol. Metab.49, 101192 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhattacharya A. et al. Hepatic Sel1L-Hrd1 ER-associated degradation (ERAD) manages FGF21 levels and systemic metabolism via CREBH. EMBO J. 37 (2018). [DOI] [PMC free article] [PubMed]
- 43.Wei J. et al. HRD1-ERAD controls production of the hepatokine FGF21 through CREBH polyubiquitination. EMBO J. 37 (2018). [DOI] [PMC free article] [PubMed]
- 44.Sun, S. et al. IRE1alpha is an endogenous substrate of endoplasmic-reticulum-associated degradation. Nat. Cell Biol.17, 1546–1555 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, T. et al. Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes Dev.28, 708–722 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi, G. et al. ER-associated degradation is required for vasopressin prohormone processing and systemic water homeostasis. J. Clin. Invest.127, 3897–3912 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim, G. H. et al. Hypothalamic ER-associated degradation regulates POMC maturation, feeding, and age-associated obesity. J. Clin. Invest.128, 1125–1140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sim, H. J. et al. Augmented ERAD (ER-associated degradation) activity in chondrocytes is necessary for cartilage development and maintenance. Sci. Adv.8, eabl4222 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abdon B., et al. Muscle-specific ER-associated degradation maintains postnatal muscle hypertrophy and systemic energy metabolism. JCI insight. 8 (2023). [DOI] [PMC free article] [PubMed]
- 50.Ji, Y. et al. SEL1L-HRD1 endoplasmic reticulum-associated degradation controls STING-mediated innate immunity by limiting the size of the activable STING pool. Nat. Cell Biol.25, 726–739 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, S. A. et al. The mechanisms to dispose of misfolded proteins in the endoplasmic reticulum of adipocytes. Nat. Commun.14, 3132 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang H. H., et al. Hypomorphic variants of SEL1L-HRD1 ER-associated degradation are associated with neurodevelopmental disorders. J. Clin. Invest. 134 (2024). [DOI] [PMC free article] [PubMed]
- 53.Weis D. et al. Biallelic Cys141Tyr variant of SEL1L is associated with neurodevelopmental disorders, agammaglobulinemia, and premature death. J. Clin. Invest.134 (2024). [DOI] [PMC free article] [PubMed]
- 54.Bhattacharya, A. et al. SEL1L-HRD1 ER-associated degradation suppresses hepatocyte hyperproliferation and liver cancer. iScience25, 105183 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thepsuwan, P. et al. Hepatic SEL1L-HRD1 ER-associated degradation regulates systemic iron homeostasis via ceruloplasmin. Proc. Natl Acad. Sci. USA.120, e2212644120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cnop, M. et al. Selective inhibition of eukaryotic translation initiation factor 2 alpha dephosphorylation potentiates fatty acid-induced endoplasmic reticulum stress and causes pancreatic beta-cell dysfunction and apoptosis. J. Biol. Chem.282, 3989–3997 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Kim H., Zheng Z., Walker P. D., Kapatos G., & Zhang K. CREBH Maintains Circadian Glucose Homeostasis by Regulating Hepatic Glycogenolysis and Gluconeogenesis. Mol Cell Biol. 37 (2017). [DOI] [PMC free article] [PubMed]
- 58.Zhang, Y. et al. The starvation hormone, fibroblast growth factor-21, extends lifespan in mice. eLife1, e00065 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hidvegi, T. et al. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science329, 229–232 (2010). [DOI] [PubMed] [Google Scholar]
- 60.Inayat, F. et al. Lafora Disease Masquerading as Hepatic Dysfunction. Cureus10, e3197 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ye, Y., Baek, S. H., Ye, Y. & Zhang, T. Proteomic characterization of endogenous substrates of mammalian ubiquitin ligase Hrd1. Cell Biosci.8, 46 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Flick, M. J. et al. Leukocyte engagement of fibrin(ogen) via the integrin receptor alphaMbeta2/Mac-1 is critical for host inflammatory response in vivo. J. Clin. Invest.113, 1596–1606 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Holmback, K., Danton, M. J., Suh, T. T., Daugherty, C. C. & Degen, J. L. Impaired platelet aggregation and sustained bleeding in mice lacking the fibrinogen motif bound by integrin alpha IIb beta 3. EMBO J.15, 5760–5771 (1996). [PMC free article] [PubMed] [Google Scholar]
- 64.Golanov, E. V. et al. Fibrinogen Chains Intrinsic to the Brain. Front Neurosci.13, 541 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poole, L. G. et al. Chronic liver injury drives non-traditional intrahepatic fibrin(ogen) crosslinking via tissue transglutaminase. J. Thromb. Haemost.17, 113–125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paton, A. W. et al. AB5 subtilase cytotoxin inactivates the endoplasmic reticulum chaperone BiP. Nature443, 548–552 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Suh, T. T. et al. Resolution of spontaneous bleeding events but failure of pregnancy in fibrinogen-deficient mice. Genes Dev.9, 2020–2033 (1995). [DOI] [PubMed] [Google Scholar]
- 68.Xia, H. & Redman, C. The degradation of nascent fibrinogen chains is mediated by the ubiquitin proteasome pathway. Biochem Biophys. Res Commun.261, 590–597 (1999). [DOI] [PubMed] [Google Scholar]
- 69.Kikkert, M. et al. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J. Biol. Chem.279, 3525–3534 (2004). [DOI] [PubMed] [Google Scholar]
- 70.Juang, L. J. et al. Suppression of fibrin(ogen)-driven pathologies in disease models through controlled knockdown by lipid nanoparticle delivery of siRNA. Blood139, 1302–1311 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hur, W. S. et al. Hypofibrinogenemia with preserved hemostasis and protection from thrombosis in mice with an Fga truncation mutation. Blood139, 1374–1388 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wei, X. et al. Proteomic screens of SEL1L-HRD1 ER-associated degradation substrates reveal its role in glycosylphosphatidylinositol-anchored protein biogenesis. Nat. Commun.15, 659 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roy, S., Sun, A. & Redman, C. In vitro assembly of the component chains of fibrinogen requires endoplasmic reticulum factors. J. Biol. Chem.271, 24544–24550 (1996). [DOI] [PubMed] [Google Scholar]
- 74.Sun S., et al. Epithelial Sel1L is required for the maintenance of intestinal homeostasis. Mol Biol Cell. 2015. [DOI] [PMC free article] [PubMed]
- 75.Shrestha N., et al. Integration of ER protein quality control mechanisms defines beta cell function and ER architecture. J. Clin. Invest. 133 (2023). [DOI] [PMC free article] [PubMed]
- 76.Dickens, J. A. et al. The endoplasmic reticulum remains functionally connected by vesicular transport after its fragmentation in cells expressing Z-alpha1-antitrypsin. FASEB J.30, 4083–4097 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chambers, J. E. et al. Z-alpha(1)-antitrypsin polymers impose molecular filtration in the endoplasmic reticulum after undergoing phase transition to a solid state. Sci. Adv.8, eabm2094 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim, P. S. & Arvan, P. Endocrinopathies in the family of endoplasmic reticulum (ER) storage diseases: disorders of protein trafficking and the role of ER molecular chaperones. Endocr. Rev.19, 173–202 (1998). [DOI] [PubMed] [Google Scholar]
- 79.Rutishauser, J. & Spiess, M. Endoplasmic reticulum storage diseases. Swiss Med Wkly.132, 211–222 (2002). [DOI] [PubMed] [Google Scholar]
- 80.Li H., and Sun S. Protein Aggregation in the ER: Calm behind the Storm. Cells. 10 (2021). [DOI] [PMC free article] [PubMed]
- 81.Callea, F. et al. A novel fibrinogen gamma chain mutation (c.1096C>G; p.His340Asp), fibrinogen Ankara, causing hypofibrinogenaemia and hepatic storage. Pathology49, 534–537 (2017). [DOI] [PubMed] [Google Scholar]
- 82.Dib, N. et al. Fibrinogen angers with a new deletion (gamma GVYYQ 346-350) causes hypofibrinogenemia with hepatic storage. J. Thromb. Haemost.5, 1999–2005 (2007). [DOI] [PubMed] [Google Scholar]
- 83.Lee, M. J., Venick, R., Bhuta, S., Li, X. & Wang, H. L. Hepatic Fibrinogen Storage Disease in a Patient with Hypofibrinogenemia: Report of a Case with a Missense Mutation of the FGA Gene. Semin Liver Dis.35, 439–443 (2015). [DOI] [PubMed] [Google Scholar]
- 84.Fraga, M. et al. Hepatocellular type II fibrinogen inclusions in a patient with severe COVID-19 and hepatitis. J. Hepatol.73, 967–970 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moon, W. S., Yu, H. C., Chung, M. J., Kang, M. J. & Lee, D. G. Pale bodies in hepatocellular carcinoma. J. Korean Med Sci.15, 516–520 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Le Fourn, V. et al. Large protein complexes retained in the ER are dislocated by non-COPII vesicles and degraded by selective autophagy. Cell Mol. Life Sci.70, 1985–2002 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kruse, K. B. et al. Mutant fibrinogen cleared from the endoplasmic reticulum via endoplasmic reticulum-associated protein degradation and autophagy: an explanation for liver disease. Am. J. Pathol.168, 1299–1308 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Puls, F. et al. Autophagy-enhancing drug carbamazepine diminishes hepatocellular death in fibrinogen storage disease. J. Hepatol.59, 626–630 (2013). [DOI] [PubMed] [Google Scholar]
- 89.Zhang, M. H., Knisely, A. S., Wang, N. L., Gong, J. Y. & Wang, J. S. Fibrinogen storage disease in a Chinese boy with de novo fibrinogen Aguadilla mutation: Incomplete response to carbamazepine and ursodeoxycholic acid. BMC Gastroenterol.16, 92 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Iwawaki, T., Akai, R., Yamanaka, S. & Kohno, K. Function of IRE1 alpha in the placenta is essential for placental development and embryonic viability. Proc. Natl Acad. Sci. Usa.106, 16657–16662 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res.50, D543–D552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sha H., et al. The ER-associated degradation adaptor protein Sel1L regulates LPL secretion and lipid metabolism. Cell Metab. 2014:pii: S1550–4131(14)00307-6. [DOI] [PMC free article] [PubMed]
- 93.Coate, K. C. et al. FGF21 Is an Exocrine Pancreas Secretagogue. Cell Metab.25, 472–480 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All study data are included in the article and/or SI Appendix. Source data are provided with this paper. The proteomics data of fibrinogen interactomes have been deposited to the ProteomeXchange Consortium with the dataset identifier PXD047658. The previously published proteomics data of purified microsomes can be found at ProteomeXchange Consortium with dataset identifier PXD035243. Source data are provided with this paper.