Abstract
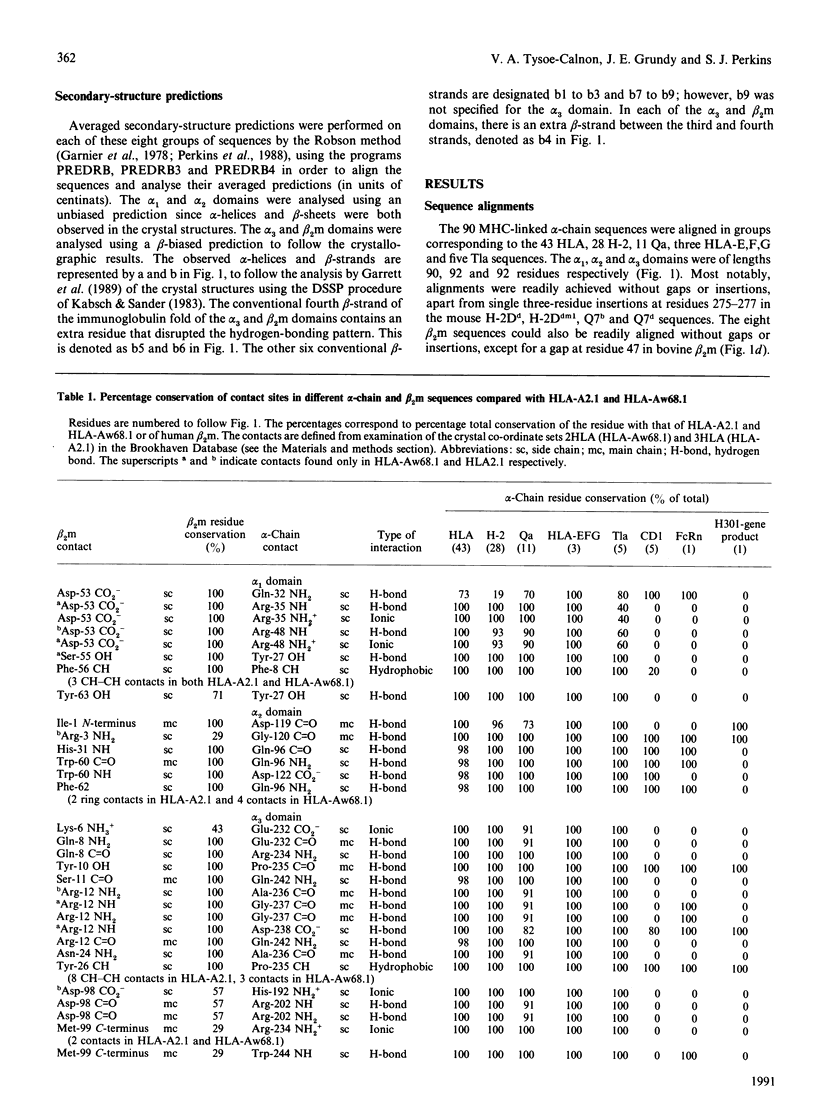
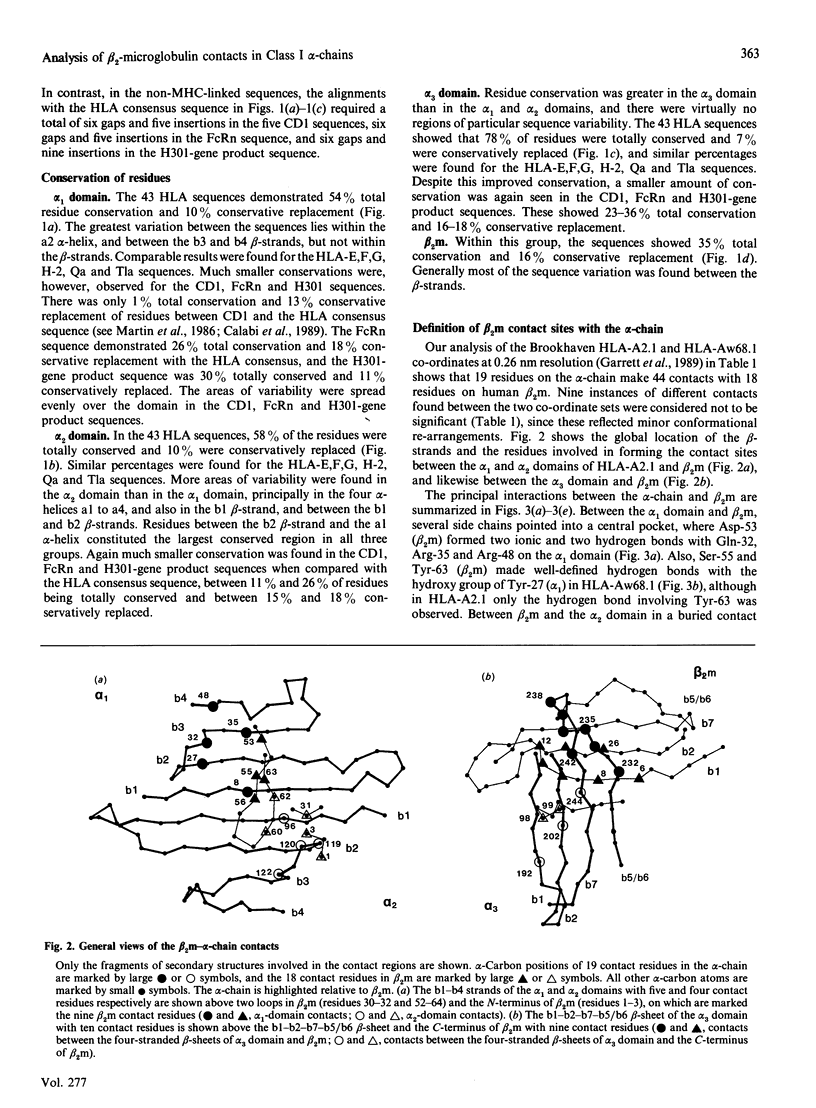
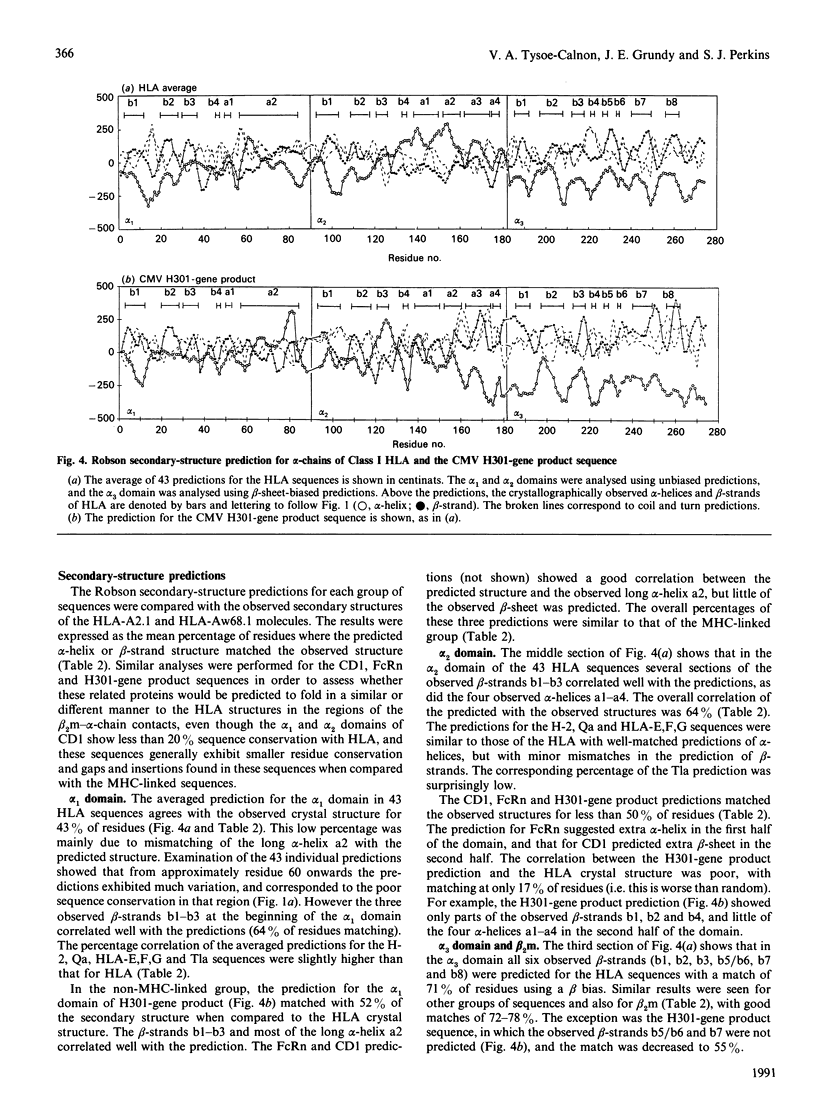
beta 2-Microglobulin (beta 2m) binds non-covalently to the alpha 1, alpha 2 and alpha 3 domains of the alpha-chain of Class I major-histocompatibility-complex (MHC) molecules. On the basis of the crystal structures of human leucocyte antigens HLA-A2.1 and HLA-Aw68.1, we have used molecular-graphics analyses to define 44 contact points between 19 alpha-chain residues and 18 beta 2m residues. In 88 other alpha-chain sequences from the HLA-A, HLA-B, HLA-C, HLA-D, HLA-E, HLA-F and HLA-G locus products in man and the H-2, Qa and Tla loci in mouse, 37 contact sites were conserved to 90% or more, and in beta 2m sequences from seven other species 40% of contact sites were totally conserved. Four distinct regions form the contact points between the alpha-chain and beta 2m, one on each of the alpha 1 and alpha 2 domains and two on the alpha 3 domain. We have further studied the alpha-chain sequences of three non-MHC molecules, human CD1 and rat Fc receptor (FcRn), known to bind to beta 2m, and a third molecule, the putative product of the H301 (UL18) gene of human cytomegalovirus (CMV). CMV has been shown to bind beta 2m, and it has been postulated that the H301-gene product, which has sequence similarity to Class I HLA, is the protein responsible. These sequences exhibited much lower residue conservation with the MHC-linked group, although the alpha 3 domain was the most highly conserved, and gaps and insertions were required for optimal alignments with the 90 alpha-chain sequences. Of the 44 beta 2m-alpha-chain contacts defined for Class I HLA, 24 alpha-chain contact sites were conserved in CD1, 25 in FcRn and 17 in the H301-gene product. For CD1 and FcRn, the majority of the conserved beta 2m contacts were found in the alpha 2 domain and the major contact region in the alpha 3 domain. Together with the use of secondary-structure predictions, it was concluded that the binding of beta 2m in CD1 and FcRn was MHC-like at the alpha 3 domain, and probably also at the alpha 2 domain for FcRn, but non-MHC-like for the alpha 1 domain of both molecules and the alpha 2 domain of CD1. In the H301-gene product sequence, only the beta 2m contacts with the main region of the alpha 3 domain were noticeably conserved.(ABSTRACT TRUNCATED AT 400 WORDS)
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ajitkumar P., Geier S. S., Kesari K. V., Borriello F., Nakagawa M., Bluestone J. A., Saper M. A., Wiley D. C., Nathenson S. G. Evidence that multiple residues on both the alpha-helices of the class I MHC molecule are simultaneously recognized by the T cell receptor. Cell. 1988 Jul 1;54(1):47–56. doi: 10.1016/0092-8674(88)90178-x. [DOI] [PubMed] [Google Scholar]
- Amiot M., Dastot H., Degos L., Dausset J., Bernard A., Boumsell L. HLA class I molecules are associated with CD1a heavy chains on normal human thymus cells. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4451–4454. doi: 10.1073/pnas.85.12.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold B., Burgert H. G., Archibald A. L., Kvist S. Complete nucleotide sequence of the murine H-2Kk gene. Comparison of three H-2K locus alleles. Nucleic Acids Res. 1984 Dec 21;12(24):9473–9487. doi: 10.1093/nar/12.24.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk S. P., Bleicher P. A., Terhorst C. Isolation and characterization of a cDNA and gene coding for a fourth CD1 molecule. Proc Natl Acad Sci U S A. 1989 Jan;86(1):252–256. doi: 10.1073/pnas.86.1.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck S., Barrell B. G. Human cytomegalovirus encodes a glycoprotein homologous to MHC class-I antigens. Nature. 1988 Jan 21;331(6153):269–272. doi: 10.1038/331269a0. [DOI] [PubMed] [Google Scholar]
- Becker J. W., Reeke G. N., Jr Three-dimensional structure of beta 2-microglobulin. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4225–4229. doi: 10.1073/pnas.82.12.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkman P. J., Saper M. A., Samraoui B., Bennett W. S., Strominger J. L., Wiley D. C. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987 Oct 8;329(6139):506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- Bjorkman P. J., Saper M. A., Samraoui B., Bennett W. S., Strominger J. L., Wiley D. C. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature. 1987 Oct 8;329(6139):512–518. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- Bouillot M., Choppin J., Cornille F., Martinon F., Papo T., Gomard E., Fournie-Zaluski M. C., Levy J. P. Physical association between MHC class I molecules and immunogenic peptides. Nature. 1989 Jun 8;339(6224):473–475. doi: 10.1038/339473a0. [DOI] [PubMed] [Google Scholar]
- Brown J. H., Jardetzky T., Saper M. A., Samraoui B., Bjorkman P. J., Wiley D. C. A hypothetical model of the foreign antigen binding site of class II histocompatibility molecules. Nature. 1988 Apr 28;332(6167):845–850. doi: 10.1038/332845a0. [DOI] [PubMed] [Google Scholar]
- Browne H., Smith G., Beck S., Minson T. A complex between the MHC class I homologue encoded by human cytomegalovirus and beta 2 microglobulin. Nature. 1990 Oct 25;347(6295):770–772. doi: 10.1038/347770a0. [DOI] [PubMed] [Google Scholar]
- Calabi F., Jarvis J. M., Martin L., Milstein C. Two classes of CD1 genes. Eur J Immunol. 1989 Feb;19(2):285–292. doi: 10.1002/eji.1830190211. [DOI] [PubMed] [Google Scholar]
- Davidson W. F., Kress M., Khoury G., Jay G. Comparison of HLA class I gene sequences. Derivation of locus-specific oligonucleotide probes specific for HLA-A, HLA-B, and HLA-C genes. J Biol Chem. 1985 Nov 5;260(25):13414–13423. [PubMed] [Google Scholar]
- Ellis S. A., Palmer M. S., McMichael A. J. Human trophoblast and the choriocarcinoma cell line BeWo express a truncated HLA Class I molecule. J Immunol. 1990 Jan 15;144(2):731–735. [PubMed] [Google Scholar]
- Ennis P. D., Jackson A. P., Parham P. Molecular cloning of bovine class I MHC cDNA. J Immunol. 1988 Jul 15;141(2):642–651. [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Garrett T. P., Saper M. A., Bjorkman P. J., Strominger J. L., Wiley D. C. Specificity pockets for the side chains of peptide antigens in HLA-Aw68. Nature. 1989 Dec 7;342(6250):692–696. doi: 10.1038/342692a0. [DOI] [PubMed] [Google Scholar]
- Gates F. T., 3rd, Coligan J. E., Kindt T. J. Complete amino acid sequence of murine beta 2-microglobulin: structural evidence for strain-related polymorphism. Proc Natl Acad Sci U S A. 1981 Jan;78(1):554–558. doi: 10.1073/pnas.78.1.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates F. T., 3rd, Coligan J. E., Kindt T. J. Complete amino acid sequence of rabbit beta 2-microglobulin. Biochemistry. 1979 May 29;18(11):2267–2272. doi: 10.1021/bi00578a021. [DOI] [PubMed] [Google Scholar]
- Geraghty D. E., Koller B. H., Orr H. T. A human major histocompatibility complex class I gene that encodes a protein with a shortened cytoplasmic segment. Proc Natl Acad Sci U S A. 1987 Dec;84(24):9145–9149. doi: 10.1073/pnas.84.24.9145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraghty D. E., Wei X. H., Orr H. T., Koller B. H. Human leukocyte antigen F (HLA-F). An expressed HLA gene composed of a class I coding sequence linked to a novel transcribed repetitive element. J Exp Med. 1990 Jan 1;171(1):1–18. doi: 10.1084/jem.171.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey H. M., Buus S., Colon S., Miles C., Sette A. Structural requirements and biological significance of interactions between peptides and the major histocompatibility complex. Philos Trans R Soc Lond B Biol Sci. 1989 Jun 12;323(1217):545–552. doi: 10.1098/rstb.1989.0034. [DOI] [PubMed] [Google Scholar]
- Groves M. L., Greenberg R. Complete amino acid sequence of bovine beta 2-microglobulin. J Biol Chem. 1982 Mar 10;257(5):2619–2626. [PubMed] [Google Scholar]
- Grundy J. E., McKeating J. A., Griffiths P. D. Cytomegalovirus strain AD169 binds beta 2 microglobulin in vitro after release from cells. J Gen Virol. 1987 Mar;68(Pt 3):777–784. doi: 10.1099/0022-1317-68-3-777. [DOI] [PubMed] [Google Scholar]
- Grundy J. E., McKeating J. A., Ward P. J., Sanderson A. R., Griffiths P. D. Beta 2 microglobulin enhances the infectivity of cytomegalovirus and when bound to the virus enables class I HLA molecules to be used as a virus receptor. J Gen Virol. 1987 Mar;68(Pt 3):793–803. doi: 10.1099/0022-1317-68-3-793. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Koller B. H., Geraghty D. E., Shimizu Y., DeMars R., Orr H. T. HLA-E. A novel HLA class I gene expressed in resting T lymphocytes. J Immunol. 1988 Aug 1;141(3):897–904. [PubMed] [Google Scholar]
- Lancet D., Parham P., Strominger J. L. Heavy chain of HLA-A and HLA-B antigens is conformationally labile: a possible role for beta 2-microglobulin. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3844–3848. doi: 10.1073/pnas.76.8.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liabeuf A., le Borgne de Kaouel C., Kourilsky F. M., Malissen B., Manuel Y., Sanderson A. R. An antigenic determinant of human beta 2-microglobulin masked by the association with HLA heavy chains at the cell surface: analysis using monoclonal antibodies. J Immunol. 1981 Oct;127(4):1542–1548. [PubMed] [Google Scholar]
- Ljunggren H. G., Stam N. J., Ohlén C., Neefjes J. J., Höglund P., Heemels M. T., Bastin J., Schumacher T. N., Townsend A., Kärre K. Empty MHC class I molecules come out in the cold. Nature. 1990 Aug 2;346(6283):476–480. doi: 10.1038/346476a0. [DOI] [PubMed] [Google Scholar]
- Long E. O., Jacobson S. Pathways of viral antigen processing and presentation to CTL: defined by the mode of virus entry? Immunol Today. 1989 Feb;10(2):45–48. doi: 10.1016/0167-5699(89)90303-4. [DOI] [PubMed] [Google Scholar]
- Martin L. H., Calabi F., Lefebvre F. A., Bilsland C. A., Milstein C. Structure and expression of the human thymocyte antigens CD1a, CD1b, and CD1c. Proc Natl Acad Sci U S A. 1987 Dec;84(24):9189–9193. doi: 10.1073/pnas.84.24.9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L. H., Calabi F., Milstein C. Isolation of CD1 genes: a family of major histocompatibility complex-related differentiation antigens. Proc Natl Acad Sci U S A. 1986 Dec;83(23):9154–9158. doi: 10.1073/pnas.83.23.9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryanski J. L., Abastado J. P., MacDonald H. R., Kourilsky P. Intradomain H-2Kd/Dd recombinants define the same regions as crucial for recognition by alloreactive or major histocompatibility complex-restricted cytolytic T cells. Eur J Immunol. 1989 Jan;19(1):193–196. doi: 10.1002/eji.1830190131. [DOI] [PubMed] [Google Scholar]
- Mattson D. H., Shimojo N., Cowan E. P., Baskin J. J., Turner R. V., Shvetsky B. D., Coligan J. E., Maloy W. L., Biddison W. E. Differential effects of amino acid substitutions in the beta-sheet floor and alpha-2 helix of HLA-A2 on recognition by alloreactive viral peptide-specific cytotoxic T lymphocytes. J Immunol. 1989 Aug 15;143(4):1101–1107. [PubMed] [Google Scholar]
- Mauxion F., Kress M. Nucleotide sequence of rat beta 2-microglobulin cDNA. Nucleic Acids Res. 1987 Sep 25;15(18):7638–7638. doi: 10.1093/nar/15.18.7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer W. E., Jonker M., Klein D., Ivanyi P., van Seventer G., Klein J. Nucleotide sequences of chimpanzee MHC class I alleles: evidence for trans-species mode of evolution. EMBO J. 1988 Sep;7(9):2765–2774. doi: 10.1002/j.1460-2075.1988.tb03131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maziarz R. T., Burakoff S. J., Bluestone J. A. The alpha-3 domain of class I MHC proteins influences cytotoxic T lymphocyte recognition of antigenic determinants located within the alpha-1 and alpha-2 domains. J Immunol. 1988 Jun 15;140(12):4372–4377. [PubMed] [Google Scholar]
- McKeating J. A., Griffiths P. D., Grundy J. E. Cytomegalovirus in urine specimens has host beta 2 microglobulin bound to the viral envelope: a mechanism of evading the host immune response? J Gen Virol. 1987 Mar;68(Pt 3):785–792. doi: 10.1099/0022-1317-68-3-785. [DOI] [PubMed] [Google Scholar]
- Morita T., Delarbre C., Kress M., Kourilsky P., Gachelin G. An H-2K gene of the tw32 mutant at the T/t complex is a close parent of an H-2Kq gene. Immunogenetics. 1985;21(4):367–383. doi: 10.1007/BF00430802. [DOI] [PubMed] [Google Scholar]
- Novotný J., Auffray C. A program for prediction of protein secondary structure from nucleotide sequence data: application to histocompatibility antigens. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):243–255. doi: 10.1093/nar/12.1part1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen M. J., Kissonerghis A. M., Lodish H. F. Biosynthesis of HLA-A and HLA-B antigens in vivo. J Biol Chem. 1980 Oct 25;255(20):9678–9684. [PubMed] [Google Scholar]
- Parham P., Lomen C. E., Lawlor D. A., Ways J. P., Holmes N., Coppin H. L., Salter R. D., Wan A. M., Ennis P. D. Nature of polymorphism in HLA-A, -B, and -C molecules. Proc Natl Acad Sci U S A. 1988 Jun;85(11):4005–4009. doi: 10.1073/pnas.85.11.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnes J. R., Seidman J. G. Structure of wild-type and mutant mouse beta 2-microglobulin genes. Cell. 1982 Jun;29(2):661–669. doi: 10.1016/0092-8674(82)90182-9. [DOI] [PubMed] [Google Scholar]
- Perkins S. J., Haris P. I., Sim R. B., Chapman D. A study of the structure of human complement component factor H by Fourier transform infrared spectroscopy and secondary structure averaging methods. Biochemistry. 1988 May 31;27(11):4004–4012. doi: 10.1021/bi00411a017. [DOI] [PubMed] [Google Scholar]
- Sege K., Rask L., Peterson P. A. Role of beta2-microglobulin in the intracellular processing of HLA antigens. Biochemistry. 1981 Aug 4;20(16):4523–4530. doi: 10.1021/bi00519a003. [DOI] [PubMed] [Google Scholar]
- Shimizu Y., Geraghty D. E., Koller B. H., Orr H. T., DeMars R. Transfer and expression of three cloned human non-HLA-A,B,C class I major histocompatibility complex genes in mutant lymphoblastoid cells. Proc Natl Acad Sci U S A. 1988 Jan;85(1):227–231. doi: 10.1073/pnas.85.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simister N. E., Mostov K. E. An Fc receptor structurally related to MHC class I antigens. Nature. 1989 Jan 12;337(6203):184–187. doi: 10.1038/337184a0. [DOI] [PubMed] [Google Scholar]
- Srivastava R., Duceman B. W., Biro P. A., Sood A. K., Weissman S. M. Molecular organization of the class I genes of human major histocompatibility complex. Immunol Rev. 1985 Jul;84:93–121. doi: 10.1111/j.1600-065x.1985.tb01127.x. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Hood L. Genes of the major histocompatibility complex in mouse and man. Science. 1983 Nov 18;222(4625):727–733. doi: 10.1126/science.6356354. [DOI] [PubMed] [Google Scholar]
- Suggs S. V., Wallace R. B., Hirose T., Kawashima E. H., Itakura K. Use of synthetic oligonucleotides as hybridization probes: isolation of cloned cDNA sequences for human beta 2-microglobulin. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6613–6617. doi: 10.1073/pnas.78.11.6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor W. R. Identification of protein sequence homology by consensus template alignment. J Mol Biol. 1986 Mar 20;188(2):233–258. doi: 10.1016/0022-2836(86)90308-6. [DOI] [PubMed] [Google Scholar]
- Townsend A., Elliott T., Cerundolo V., Foster L., Barber B., Tse A. Assembly of MHC class I molecules analyzed in vitro. Cell. 1990 Jul 27;62(2):285–295. doi: 10.1016/0092-8674(90)90366-m. [DOI] [PubMed] [Google Scholar]
- Townsend A., Ohlén C., Bastin J., Ljunggren H. G., Foster L., Kärre K. Association of class I major histocompatibility heavy and light chains induced by viral peptides. Nature. 1989 Aug 10;340(6233):443–448. doi: 10.1038/340443a0. [DOI] [PubMed] [Google Scholar]
- Tysoe-Calnon V. A., Grundy J. E., Nealis A. S., Perkins S. J. High conservation of beta 2-microglobulin contact residues among 82 class I major histocompatability complex alpha-chain sequences. Biochem Soc Trans. 1990 Oct;18(5):930–931. doi: 10.1042/bst0180930. [DOI] [PubMed] [Google Scholar]
- Tüchsen E., Woodward C. Hydrogen exchange of primary amide protons in basic pancreatic trypsin inhibitor: evidence for NH2 group rotation in buried asparagine side chains. Biochemistry. 1987 Dec 15;26(25):8073–8078. doi: 10.1021/bi00399a008. [DOI] [PubMed] [Google Scholar]
- Vega M. A., Bragado R., Ezquerra A., López de Castro J. A. Variability and conformation of HLA class I antigens: a predictive approach to the spatial arrangement of polymorphic regions. Biochemistry. 1984 Feb 28;23(5):823–831. doi: 10.1021/bi00300a007. [DOI] [PubMed] [Google Scholar]
- Watts S., Wheeler C., Morse R., Goodenow R. S. Amino acid comparison of the class I antigens of mouse major histocompatibility complex. Immunogenetics. 1989;30(5):390–392. doi: 10.1007/BF02425281. [DOI] [PubMed] [Google Scholar]
- Welinder K. G., Jespersen H. M., Walther-Rasmussen J., Skjødt K. Amino acid sequences and structures of chicken and turkey beta 2-microglobulin. Mol Immunol. 1991 Jan-Feb;28(1-2):177–182. doi: 10.1016/0161-5890(91)90102-p. [DOI] [PubMed] [Google Scholar]
- Wiley D. MHC gene in cytomegalovirus. Nature. 1988 Jan 21;331(6153):209–210. doi: 10.1038/331209a0. [DOI] [PubMed] [Google Scholar]
- Wolfe P. B., Cebra J. J. The primary structure of guinea pig beta 2-microglobulin. Mol Immunol. 1980 Dec;17(12):1493–1505. doi: 10.1016/0161-5890(80)90175-3. [DOI] [PubMed] [Google Scholar]
- Zijlstra M., Bix M., Simister N. E., Loring J. M., Raulet D. H., Jaenisch R. Beta 2-microglobulin deficient mice lack CD4-8+ cytolytic T cells. Nature. 1990 Apr 19;344(6268):742–746. doi: 10.1038/344742a0. [DOI] [PubMed] [Google Scholar]
- de Préval C., Mach B. The absence of beta 2-microglobulin in Daudi cells: active gene but inactive messenger RNA. Immunogenetics. 1983;17(2):133–140. doi: 10.1007/BF00364753. [DOI] [PubMed] [Google Scholar]


