Summary
Interspecific hybridization can be consequential for insular species. The Critically Endangered Aeolian wall lizard, Podarcis raffonei, severely declined due to interactions with the invasive Italian wall lizard, Podarcis siculus. The largest population of P. raffonei survives on a narrow peninsula (Capo Grosso) that is mildly connected to the island of Vulcano, which has been entirely invaded by P. siculus. Recent observation of individuals with an intermediate phenotype raised concern over the risk that hybridization might swamp this last stronghold. We genetically characterized lizards from Vulcano, considering individuals showing multiple phenotypes (native, invasive, and “intermediate”). Hybridization rate was low (∼3%), with just two F1 hybrids and two backcrosses, suggesting that hybridization does not currently represent a major threat. However, we identified low genetic diversity, a small effective population size, and a low Ne/Nc ratio. Management strategies are urgently needed to control invasive species and maintain the genetic diversity of P. raffonei.
Subject areas: Ecology, Evolutionary biology, Zoology
Graphical abstract
Highlights
-
•
Podarcis siculus is thought to be responsible for the decline of Podarcis raffonei
-
•
Detection of intermediate phenotype individuals raised alarm of hybrid swamping
-
•
Genomic analysis reveals low hybridization rate (∼3%) on Capo Grosso (Vulcano)
-
•
Low genetic diversity and Ne in the largest remaining population of Podarcis raffonei
Ecology; Evolutionary biology; Zoology
Introduction
Climate change, habitat modification, and the intentional or unintentional movement of organisms by humans, are causing species distribution shifts across the planet. One outcome of these disturbances is increased rates of interspecific hybridization, which can be especially problematic for rare or endangered species that encounter more abundant species.1 Hybridization can threaten a species’ genetic integrity and even contribute to species extinction,1,2,3 yet in some cases hybridization can also increase diversity and may aid evolutionary rescue of endangered species.4,5,6 Nonetheless, the effects of interspecific hybridization are highly species- and context-specific, and to predict its potential evolutionary outcomes, a robust quantification of hybridization is required. Such evaluations are especially pressing for rare and endangered species, where an understanding of the rate and mode of interspecific hybridization is required to design the most appropriate conservation and management plans.
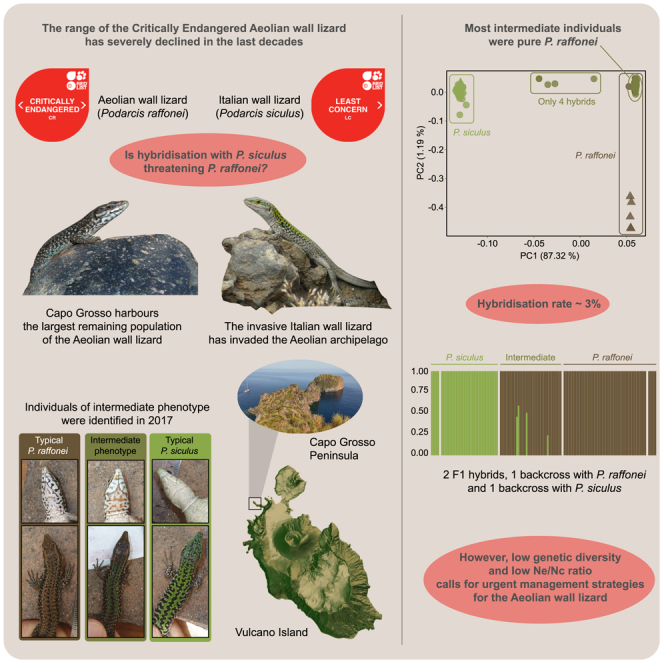
Alien invasive species are a major cause of extinction risk in reptiles, with particularly strong effects in island environments.7,8,9 In this respect, the endemic Aeolian wall lizard, Podarcis raffonei, provides an emblematic case. Once probably widespread across the Aeolian archipelago (Southern Italy), this lizard is now restricted to three tiny islets (Scoglio Faraglione, Strombolicchio, and La Canna) and a narrow peninsula (Capo Grosso; Figure 1). With an estimated range of ∼1.6 ha,10 P. raffonei is the rarest and most threatened reptile in Europe. Listed as Critically Endangered by the IUCN since 2006, the species remains understudied and underprotected.11 The island of Vulcano hosts the only population of P. raffonei that is not microinsular as it occurs on the narrow peninsula of Capo Grosso (surface <0.7 ha) connected to a main island. Until 1999, the species was recorded in multiple locations across Vulcano island (Vulcanello and Gran Cratere),12 but these populations are now considered to be extinct.13,14,15 Interspecific interactions with the Italian wall lizard, P. siculus, have been regarded as a major cause of the near extinction of P. raffonei.12 P. siculus is native to the Italian Peninsula, Sicily, and the northern Adriatic coast, but has been introduced by humans across the world.16,17 P. siculus was likely introduced in Vulcano and other Aeolian islands a few centuries ago18 and is now widespread across the archipelago.12,13,14 The invasion by P. siculus has thus been proposed as a key driver for P. raffonei’s relict distribution.12,19,20
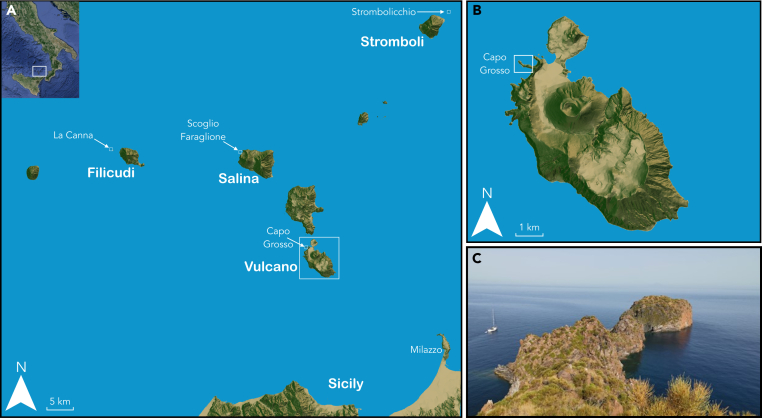
Figure 1.
Geographic locations of the Aeolian wall lizard (Podarcis raffonei)
(A) Map of the Aeolian Islands in southern Italy. Inset shows the location of the Aeolian Islands in Italy. The locations of the Aeolian wall lizard (Podarcis raffonei) are the three islets: La Canna (close to Filicudi), Scoglio Faraglione (close to Salina), Strombolicchio (close to Stromboli) and the peninsula of Capo Grosso on the island of Vulcano. Sampled individuals include: the typical brown phenotype (P. raffonei; n = 50) and putative hybrid lizards with an intermediate phenotype (n = 38) from Capo Grosso; pure P. raffonei individuals from Scoglio Faraglione (n = 5); and pure P. siculus lizards from mainland Vulcano (n = 35) and from mainland northern Sicily, Milazzo (n = 5).
(B) Zoom-in on the island of Vulcano, showing the small peninsula of Capo Grosso.
(C) Photograph of Capo Grosso: P. raffonei is confined to the small distal portion of the peninsula. Orographic maps were created in R21 with rayshader.22.
P. siculus might impact P. raffonei through both competition and hybridization. Competition has been proposed as a reason for decline because P. siculus has a larger body size, higher aggressiveness, excellent thermoregulation, behavioral plasticity, and tolerance to disturbed environments.12,23,24,25,26,27 Moreover, an early allozyme study analyzed 101 lizards from Vulcano, identifying 15 individuals as F1 hybrids between P. raffonei and P. siculus (hybrid ratio: ∼15%19), suggesting that hybridization might further accelerate the decline of P. raffonei.10,12,19 However, these early studies only used a small panel of allozyme markers (only four “diagnostic” loci19). The assessment of hybridization patterns requires large (>100) panels of markers, as a smaller number of markers can yield both severe underestimation and overestimation of hybridization rates.28 Unfortunately, such assessments are so far lacking.
More recently, observations of lizards from Capo Grosso with a green dorsal-coloration phenotype (hybrid-like) resulted in renewed alarm.10 P. raffonei and P. siculus differ only slightly in morphology and color pattern, meaning the two species are difficult to tell apart.19,29 P. raffonei possesses dark markings on the throat and is generally brown on the dorsal surface, whereas P. siculus has a white throat with no dark spots and shows a green dorsal pattern. In a survey of 131 lizards in 2017, a large number (>50%) of individuals from Capo Grosso showed an intermediate phenotype, with green patterns and dark markings on the throat (Figure 2). This was suspected to indicate extensive hybridization10; however, individuals from Capo Grosso kept in captive-breeding programs showed strong plasticity in color pattern with a seasonal shift toward a green phenotype.10 Thus, the true hybrid status of the Capo Grosso population of P. raffonei remains unknown. Given the high-priority conservation status of the Aeolian wall lizard, and the potentially catastrophic effects of widespread hybridization with the Italian wall lizard, there is a pressing need to quantify the actual hybridization rate using a large panel of genomic markers. Such an assessment would also provide measures of genetic diversity and effective population size (NE) for the largest extant population of P. raffonei, with key consequences for the management of this island endemic.
Figure 2.
Color patterns of male lizards captured on Vulcano Island and Capo Grosso
Podarcis raffonei (left) and the Italian wall lizard P. siculus (right), and patterns of individuals with the intermediate phenotype (middle), adapted from Ficetola et al. (2021).10 Typical phenotypes of P. raffonei and intermediate phenotypes were observed on the Capo Grosso peninsula, whereas typical phenotypes of P. siculus were observed across the main island of Vulcano.
In this study, we use single-nucleotide polymorphisms (SNPs) derived from double-digest restriction-site associated DNA sequencing (ddRAD-seq), mitochondrial sequencing, and phenotypic data, to investigate the status of P. raffonei from the Capo Grosso peninsula. First, we quantified the genetic status of lizards from Capo Grosso and mainland Vulcano, to evaluate the extent of hybridization between P. raffonei and P. siculus, and to assess the relationship between genotype, maternal species, and color pattern. Second, after the exclusion of hybrids, we performed an assessment of genetic diversity and effective population size (NE) of P. raffonei from Capo Grosso. These results have key implications for the conservation and management of this Critically Endangered species, and provide general insights on the conservation of small, insular populations under the increasing threat of hybridization with invasive species.
Results
ddRAD data quality control, de novo assembly and reference alignment
After removal of five low-quality samples, we generated over 224 million paired-end reads (average 1,835,851 ± SE 85,979 per individual) for 133 lizards: Capo Grosso brown phenotype (n = 50); Capo Grosso intermediate phenotype (n = 38); Vulcano P. siculus (n = 35); Scoglio Faraglione P. raffonei (n = 5); Milazzo P. siculus (n = 5) (Table S1). De novo assembly of the RAD data separately for P. raffonei and P. siculus showed that the best parameter for merging alleles into loci (-M) was 2 for both species. We merged the loci across individuals from both species (-n) using an optimized value of 2. Average coverage overall was 31X. One individual (CGI38) was retained for analysis, despite low coverage (5.4X) as it showed evidence of being a hybrid in a preliminary assessment of the dataset. In the reference-guided analysis, P. raffonei and P. siculus samples showed different alignment rates to the P. raffonei reference genome, with P. raffonei samples showing an average alignment rate of 92.5%, and P. siculus samples showing an average alignment rate of 72% (Table S1). This alignment bias was alleviated by retaining loci occurring in both species (-p 1 in populations).
Inter- and intra-species genetic structure and identification of genetic hybrids
Principal-components analysis (PCA) showed strong genetic differentiation between P. siculus lizards sampled from Vulcano and brown P. raffonei individuals sampled from Capo Grosso (Figure 3A). PC1 (87% of the variance) clearly separated the two species, and evidenced that 34 of the intermediate phenotype lizards sampled from Capo Grosso clustered with the brown P. raffonei lizards. Four of the intermediate-phenotype lizards were placed in the genetic space between the two species: CGI13, CGI14, CGI38, and CGI36. PC2 (1% of the variance) separated the P. raffonei lizards sampled from Capo Grosso from lizards sampled from Scoglio Faraglione. P. siculus lizards sampled from Milazzo showed very little genetic differentiation from P. siculus from Vulcano, as indicated by PC3 (0.93% of the variance). Genetic differences were well-reflected in the FST estimates among the four sampling locations (excluding intermediate phenotypes). A very high FST was observed between brown P. raffonei lizards from Capo Grosso and P. siculus from Vulcano (FST = 0.92). A moderately high FST was also found between P. raffonei lizards from Capo Grosso and P. raffonei from Scoglio Faraglione (FST = 0.42). A very low FST was observed between P. siculus lizards from Vulcano and Milazzo (FST = 0.07).
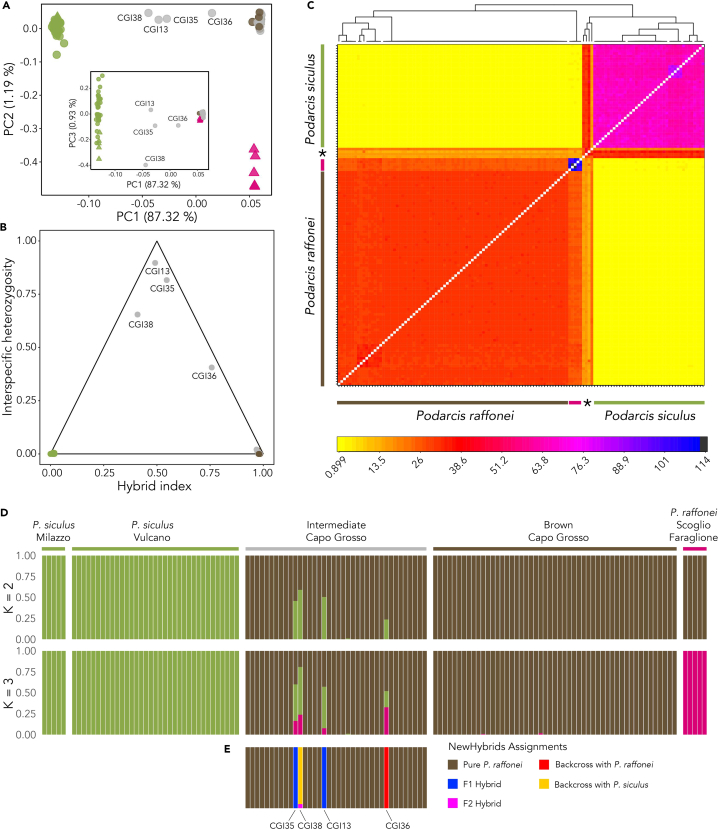
Figure 3.
Genetic structure and identification of genetic hybrids
(A) Principal-components analysis (PCA Green circles represent P. siculus lizards from Vulcano; green triangles represent P. siculus lizards from Milazzo; brown circles represent brown-phenotype lizards from Capo Grosso; pink triangles represent P. raffonei lizards from Scoglio Faraglione; gray circles represent intermediate-phenotype lizards from Capo Grosso. Main plot shows the variance explained by PC1 and PC2; inset shows the variance explained by PC1 and PC3.
(B) Triangle plot depicting the hybrid index, and heterozygosity at species diagnostic SNPs inferred by introgress. Green represents P. siculus lizards; brown represents brown-phenotype lizards, gray represents intermediate-phenotype lizards.
(C) Co-ancestry relationships from fineRADstructure. Two main clusters are observed, representing P. siculus and P. raffonei. All intermediate-phenotype lizards (except the four hybrids, identified by the asterisk) cluster within the P. raffonei cluster. P. siculus from Milazzo are nested within the P. siculus cluster. P. raffonei lizards from Scoglio Faraglione show a distinct cluster compared to the P. raffonei cluster of Capo Grosso. Color legend depicts the proportion of shared ancestry between the haplotypes. The tree atop shows crude relationships between the haplotypes.
(D) Individual ancestries from Admixture showing results for K = 2 and K = 3.
(E) Assignment of the intermediate-phenotype lizards using NewHybrids. The color legend describes the assignment to each of the five genotype classes that were identified.
See also Figures S1, S2, and S3.
Analysis using introgress confirmed that most intermediate-phenotype lizards are in fact P. raffonei, with the analysis indicating just four genetic hybrids based on the hybrid index and interspecific heterozygosity (Figure 3B). CGI13 (male) and CGI35 (female) showed F1 hybrid indices of approximately 50% (CGI13: 0.492 [0.467–0.512]; CGI35: 0.546 [0.523–0569]). CGI36 (male) had a backcross hybrid index: 0.758 (0.737–0.778). The low coverage individual (CGI38; male) showed a hybrid index more closely related to P. siculus, although with larger 95% confidence intervals: 0.409 (0.344–0.476).
The co-ancestry method of fineRADstructure using the full haplotype information also showed two clearly defined clusters representing the two species (Figure 3C). Podarcis raffonei from Scoglio Faraglione clustered within the P. raffonei cluster, and P. siculus from Milazzo clustered within the P. siculus cluster, each with higher co-ancestries to the other samples from these sampling locations. The fineRADstructure analysis further confirmed the identification of the four hybrids. The two suspected F1 hybrids (CGI13 and CGI35) showed equally shared donor and recipient ancestries to both P. siculus and P. raffonei. Sample CGI36 showed higher shared co-ancestry to P. raffonei. Sample CGI38 (low coverage) showed approximately equally shared donor and recipient ancestries to both species, although with some small haplotype blocks showing a stronger signature of donor ancestry from P. siculus.
Cross-validation (CV) error from Admixture showed that K = 3 was the best K for describing the allocation of individual ancestries, but K = 2 also showed a low CV error rate (Figure S1). At K = 2, individuals from the two species clustered into two clearly defined groups (>99% assignment probability (Figure 3D). Most of the intermediate phenotype samples showed the same cluster assignment (>99%) as the brown-phenotype P. raffonei from Capo Grosso and P. raffonei from Scoglio Faraglione. Four putative hybrids were clearly apparent from their shared assignment probabilities to the two species clusters. Two samples showed approximately 50% assignment probability to P. raffonei (CGI13: 50%; CGI35: 54%). Sample CGI36 showed a high assignment probability to P. raffonei (CGI36: 0.76%). CGI38 (low coverage) showed a higher assignment to P. siculus (CGI38; 59%). At K = 3, the P. raffonei samples from Scoglio Faraglione were separated from the Capo Grosso P. raffonei cluster and the four hybrid individuals also showed some shared ancestry with this location. Further assessment of population structure within the groups showed that the best K was 1 for both the Capo Grosso P. raffonei individuals and the Vulcano P. siculus individuals, and there was no effect of sampling year (Figure S2). Hierarchical analysis on all P. siculus samples also showed K = 1 as the best value for K, although examination of the cluster probabilities for K = 2 showed that lizards sampled from Milazzo formed a homogeneous group compared to samples from Vulcano (Figure S3).
In the NewHybrids assignment, all panels of diagnostic markers used for simulation were species-diagnostic (i.e., FST = 1). The critical posterior probability of assignment thresholds to each genotype frequency class was 1 for all marker panels across all runs in the simulated datasets. We combined the empirical data with all three marker panels to assess hybrid classification of the intermediate-phenotype individuals. These results also confirmed that the majority (n = 34) of individuals with the intermediate phenotype are P. raffonei (Figure 3E). NewHybrids assigned two of the intermediate-phenotype samples as F1 hybrids (CGI13 and CGI35), and one sample as a P. raffonei backcross (CGI36) with high posterior probabilities (>0.99 across all runs). The low-coverage sample (CGI38) showed the highest assignment probability of being a siculus backcross. However, the reduced data quality of this sample resulted in a lower assignment probability across all marker panels and simulations (0.941; range: 0.940–0.942), with the remaining assignment probability assigning this individual as an F2 hybrid.
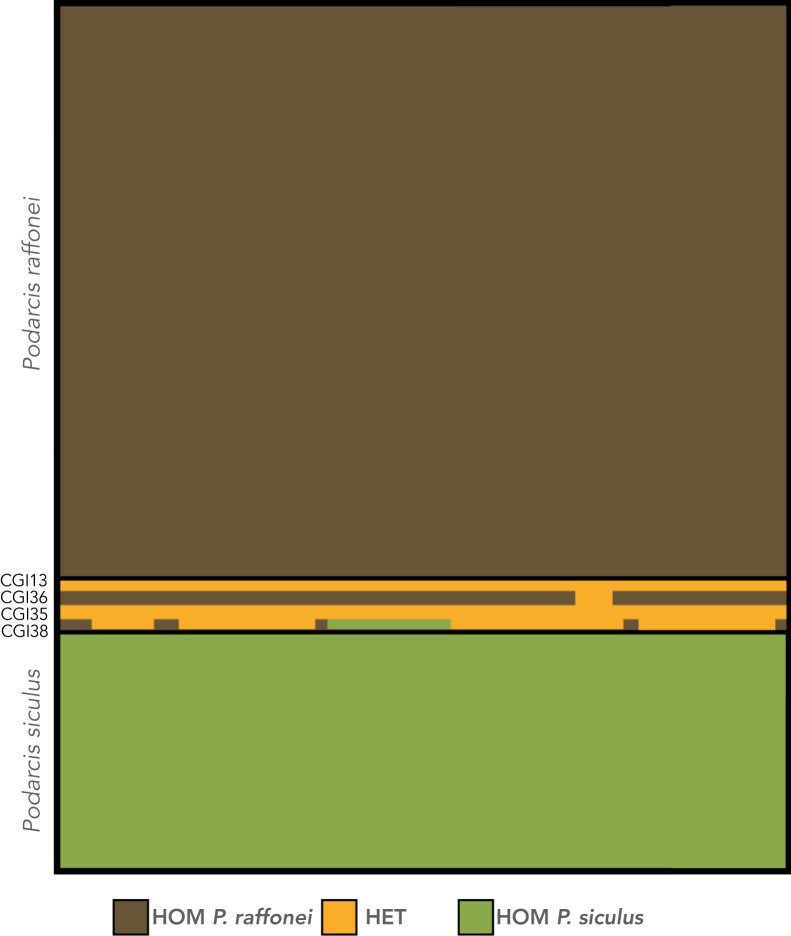
Finally, we used a whitelist of 87 SNPs genotyped across 49 loci to interrogate the individual genotype information of the four identified hybrid individuals (Figure 4). This confirmed the genotype patterns for the four genetic hybrids, with CGI13 and CGI35 showing heterozygous P. raffonei and P. siculus genotypes for the SNPs used in the analysis. CGI36 had a majority of homozygous P. raffonei genotypes, but a stretch of heterozygous genotypes was also seen, as would be expected with a raffonei backcross. In the low-coverage individual (CGI38), a large proportion of the SNPs were heterozygous, but a genotype signature of homozygous P. siculus was also observed.
Figure 4.
Genotype plot for 87 species-diagnostic SNP markers (i.e., FST = 1) between P. raffonei and P. siculus
Genotypes are colored as being homozygous in P. raffonei (HOM = brown), homozygous in P. siculus (HOM = green) or heterozygous between the two species (HET = yellow). Each block is colored by these genotype calls. Note that the SNPs were called de novo and are therefore not ordered in a positional manner.
Overall, our analyses show that the genetic hybrids comprise two F1s, one raffonei backcross, and one hybrid which is most likely a siculus backcross. If we consider the sampling years, one hybrid (siculus backcross) was detected in 2015 out of 30 captured individuals (only 10 individuals captured in 2015 were sequenced, but none of the non-sequenced individuals showed an intermediate phenotype; this suggests a 2015 hybrid ratio = 0.033), and three hybrids (two F1s, one raffonei backcross) were detected in 2017 among a total of 88 lizards sampled from Capo Grosso (2017 hybrid ratio = 0.034).
Assessment of the maternal species and phenotype of the genetic hybrids
We identified the maternal species of each of the four hybrid individuals by Sanger sequencing the 12S rRNA mtDNA region. Mitochondrial DNA sequences obtained for the hybrids CGI35 and CGI38 were identical to the 12S sequence of a P. siculus individual from Vulcano island (GenBank: KX080573:30); sequences of the hybrids CGI13 and CGI36 show a single substitution (genetic identity of 99.7%) from the sequence found in four P. raffonei individuals from Strombolicchio islet (GenBank: KY562000:31; GenBank: MW619265-267;32). Sequences of P. siculus and P. raffonei show 15 alternatively fixed SNPs at this 12S gene fragment (length: 348 base pairs). This indicated that the two F1 hybrid lizards originated from a P. raffonei female (CGI13), and a P. siculus female (CGI35). The raffonei backcross lizard originated from a P. raffonei female in the backcross or in the F1-producing cross. The low-coverage sample (CGI38) originated from a P. siculus female.
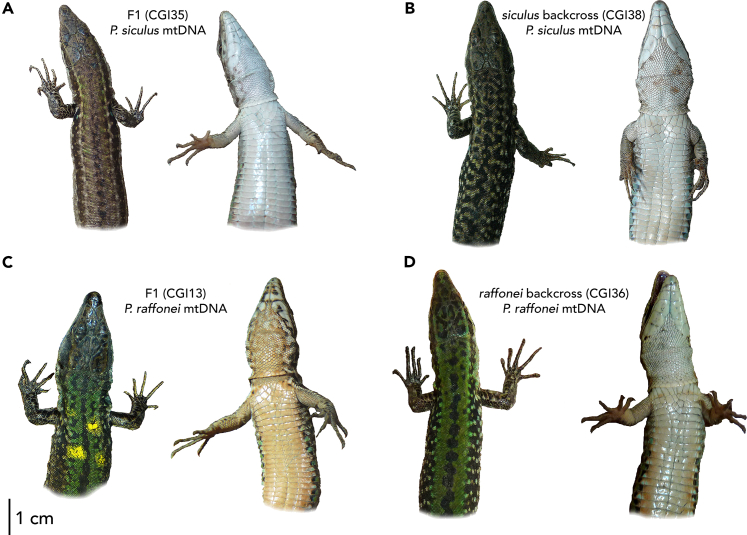
Inspection of the phenotypes suggests hybrids more closely resembled the maternal species in both dorsal and ventral patterns, regardless of hybrid class and sex. CGI13 (F1, P. raffonei maternal haplotype) and CGI36 (raffonei backcross, P. raffonei maternal haplotype) had a phenotype more closely resembling the P. raffonei phenotype with few to many dark markings on the chin shields and throat, orange ventral coloration, a dark vertebral stripe flanked by two brown/green stripes on the dorsal surface (Figure 5A). Conversely, CGI35 (F1, P. siculus maternal haplotype) and CGI38 (siculus backcross, P. siculus mother) resemble a P. siculus phenotype with white belly and throat with pale spots and mostly reticulated dorsal pattern (Figure 5B).
Figure 5.
Dorsal and ventral phenotypes of the four hybrid individuals
Hybrid individuals more closely resemble the maternal species, regardless of hybrid class.
(A) CGI35 (F1, P. siculus maternal haplotype) and (B) CGI38 (siculus backcross, P. siculus maternal haplotype) resemble a P. siculus phenotype with white belly and throat with pale spots and mostly reticulated-blurred dorsal pattern.
(C) CGI13 (F1, P. raffonei maternal haplotype) and (D) CGI36 (raffonei backcross, P. raffonei maternal haplotype) resemble the P. raffonei phenotype with orange ventral coloration, dark markings on the chin shields and throat, a dark vertebral stripe flanked by two green stripes on the dorsal surface.
Genetic diversity and effective population size estimates of P. raffonei
Estimates of genetic diversity were calculated using individuals with the lowest amount of missing data (≤10%) (P. raffonei = 60; P. siculus = 30), using only loci present in all individuals of both species. All estimates of genetic diversity were lower in P. raffonei compared to P. siculus (Table 1). Estimates of the inbreeding coefficient (FIS) were slightly higher in P. siculus, with confidence intervals above zero. Iterative downsampling of the sample size in the P. raffonei dataset did not affect the results, suggesting robustness in the estimations. Estimates of the effective population size (Ne) of P. raffonei from Capo Grosso were 63.8 (95% CI = 54.7–75.5) when including singletons, translating to an Ne/Nc ratio of 0.06. Excluding singletons lead to slightly smaller Ne values but with larger confidence intervals (NE = 49.8; 95% CI = 25.5–133.7).
Table 1.
Genetic diversity estimates for the Aeolian wall lizard (Podarcis raffonei) from Capo Grosso and the Italian wall lizard (Podarcis siculus) from Vulcano
| Species | Dataset | HE | HO | FIS | AR |
|---|---|---|---|---|---|
| Italian wall lizard (Podarcis siculus) | Full dataset; n = 30 | 0.0570 | 0.0547 | 0.041 (0.036–0.046) | 1.25 |
| Aeolian wall lizard (Podarcis raffonei) | Average and range of Full dataset; n = 60 and iterations; n = 30 | 0.022 (0.021–0.023) | 0.022 (0.022–0.023) | −0.005 (95% CI: −0.001–0.002) (−0.014–0.001) |
1.07 (1.06–1.08) |
Estimates of diversity include expected heterozygosity (HE), observed heterozygosity (HO), allelic richness (AR), and the inbreeding coefficient (FIS). Values are shown for both species using the full datasets of individuals with the lowest amount of missing data (P. siculus n = 30 and P. raffonei n = 60) with numbers in brackets representing the values obtained after downsampling iterations for P. raffonei.
Discussion
The promontory of Capo Grosso in the island of Vulcano hosts the largest remaining population of the Critically Endangered Aeolian wall lizard. This population represents the last stronghold for the species on a large island, where P. raffonei has persisted after the historical introduction of P. siculus.18,19,20 Recent observations of lizards with an intermediate phenotype in Capo Grosso resulted in renewed concern, especially given that interspecific hybridization with P. siculus could quickly drive this population to extinction.10 Our high-density panel of SNPs showed limited hybridization (∼3%). Just a handful of intermediated individuals were hybrid, there is no evidence of older hybrid classes, and most individuals sampled from Capo Grosso are in fact pure P. raffonei. Therefore, we cautiously suggest that hybridization does not currently represent the major threat for P. raffonei in Capo Grosso. These findings are in contrast with the speculation that hybridization caused the disappearance of P. raffonei from Capo Grosso (based on phenotypic observations15) and casts doubt on the hypothesis that hybrid swamping was the primary reason for the disappearance of the species from the Aeolian archipelago.
Low contemporary hybridization between the Aeolian wall lizard and the Italian wall lizard
Our genome-wide estimate of hybridization between P. raffonei and P. siculus suggests a low (∼3%) rate of hybridization compared with other congeneric pairs.33 This is in contrast with the previous genetic study on Vulcano, which reported a much higher ratio of F1 hybrids (15%19). This can be explained by the very small panel of loci employed by this early study (four allozymes). Assessments of hybrid zones based on a few (<100) loci can yield inaccurate estimates of introgression rate,34 stressing the need for methods which extensively cover the genome. Despite the low information content in each individual SNP, the large number of markers provided by approaches such as RAD-seq offers a cost-effective method to achieve accurate hybrid classification35 while genotyping a large number of individuals.28 Other studies have also observed discrepancies in the hybridization rate measured using different marker typologies,36,37 suggesting caution in the interpretation of hybridization rates derived by allozyme data and in the comparison of estimates based on different marker types.
Aside from methodological differences, a further potential explanation for the discrepancy between the results here and those of Capula19 is that interspecific interactions on Vulcano Island have probably changed in the last ∼40 years, especially given the dramatic decline of P. raffonei. In 1951, P. raffonei was reported to be the predominant species in the northern areas of the island.38 In the late 1980s, Capula witnessed the replacement of P. raffonei by P. siculus, but still found pure P. raffonei individuals and hybrids in these areas (Vulcanello and at the base of Gran Cratere19; Capula pers. comm.), not far from Capo Grosso (∼1 km). The expansion of invasive lizards (as observed by Capula) probably led to an increased frequency of interspecific encounters and thus more frequent hybridization. On the other hand, the distal portion of the Capo Grosso peninsula, where P. raffonei still occurs, is connected to the main island by a narrow and tortuous rocky isthmus (∼400 m long, minimum width ∼10 m, minimum altitude 1–2 m above-sea-level; Figure 1), which probably represents a strong geographical filter against the invasion of P. siculus. The strength and type of the habitat barrier (in this case the isthmus) can have substantial effects on the flux of alleles in hybrid zones, potentially limiting introgression levels.39 We remain blind about the temporal movement of the hybridization zone, but it can be hypothesized that it has moved over time and that hybridization rate increased at the expansion front of P. siculus, and then decreased in extent as P. siculus outcompeted P. raffonei until near-complete replacement. The P. siculus lizards sampled in this study were mostly from northern Vulcano (Vulcanello), where the historical hybrids were observed, but we found no trace of introgressed alleles from P. raffonei. This suggests that introgression has not been the main driver of the replacement of Aeolian lizards by invasive lizards, and that other processes (e.g., competition;12,14,23) may have taken place and require further investigation.
Demographic and spatial trends of these lizards have been likely determined by their different ecological response to the anthropogenic impact on native habitat. The two species show clear ecological differences, with P. siculus thriving in anthropic environments, including open and urban habitats.14,40 The dispersal of P. siculus is limited by habitat quality and density-dependence, being higher in crowded habitats.41 Opportunistic transects along the Capo Grosso promontory indicate that P. siculus density is very low and decreases along the isthmus (L. Vignoli and B. Gambioli, pers. comm.), further supporting the hypothesis that the isthmus is an efficient barrier to the invasion by P. siculus, possibly preventing extensive hybridization at this site.
Besides the role of habitat changes and species abundance, the low rate of hybridization could also be due to evolutionary mechanisms associated with the genetic differentiation between species and the fitness of hybrid offspring.42 The high genetic differentiation (FST = 0.92) supports a deep divergence between P. raffonei and P. siculus (around 11–18 mya32,43). Nevertheless, Podarcis lizards are typified by pervasive ancestral hybridization and introgression,43,44,45 suggesting that postzygotic reproductive isolation mechanisms are fluid. Hybrid fitness is a further point of consideration when interpreting hybridization rates. In lizards, reduced hybrid fitness can result from changes in intraspecific competition,46 sexual selection,47 and reduced reproductive function, including the production of fewer sperm and fewer eggs.48 The fitness of first-generation hybrids is presumably very low given the observation of atrophied gonads in F1 hybrids.19 The low fitness of later-generation hybrid offspring can be indirectly inferred from the apparent lack of these hybrid classes in our analyses. Negative effects of hybridization are often manifested in later generations, such as F2s and higher order hybrids.49,50 Nevertheless, the detection of a few individuals with backcross genetic signatures indicate that first generation hybrids are not always sterile as previously proposed.19 In a review of hybridization between 94 pairs of genetically distinct lizard species and subspecies, the majority of F1 hybrids were found to be fertile, thus allowing backcrosses with at least one parental species.44 The individual identified as a backcross with P. raffonei is of special concern, as even infrequent F1 hybrids can facilitate backcrossing, leading to the risk of parental genotype displacement.51 Nevertheless, the rarity of backcrosses suggests that they have low fitness. This is further supported by the lack of introgressed alleles observed in P. siculus from Vulcanello (see previous text). Overall, the hybridization between P. raffonei and P. siculus is a spatially complex and highly dynamic process, and strong demographic and ecological factors, including species abundance and habitat disturbance, as well as postzygotic mechanisms, probably determines hybridization rate and extent.
Phenotypic identification of hybrids
The identification of which species acted as the maternal parent of hybrids may assist future studies wishing to address which genetic factors have contributed to the observed fitness outcomes.52 The phenotype of hybrid individuals suggests a maternal effect on the phenotype (Figures 5 53,54), but this remains speculative given the small number of identified hybrids. In any case, the phenotypes of these hybrids are not markedly different from pure individuals, confirming the challenge of morphological identification.10,19 The identification of species identity and hybrids without genetic data15 is thus unreliable, stressing the need for continuous genetic monitoring prior to any management action. Indeed, our genetic analysis corroborates that green coloration is a plastic trait of P. raffonei. Several P. raffonei individuals show a seasonal transition, shifting from the typical brown pattern into a green phenotype during spring.10 Green coloration has not been previously reported for P. raffonei from Vulcano in earlier studies,12,29 but seasonal dorsal color changes have been observed in other Podarcis lizards, including P. siculus, P. waglerianus, P. carbonelli, and P. bocagei.55,56,57,58,59 Further research is needed to understand the ecological and evolutionary drivers of this phenotype, and its fitness implications.
Current genetic status of the Aeolian wall lizard in Capo Grosso
Recent genomic analyses showed that P. raffonei has the lowest genetic diversity of any of the 26 Podarcis species, including other island endemics,43 and the lowest heterozygosity among seven species belonging to distinct squamate families.60 The low genetic diversity of P. raffonei from Capo Grosso corroborates this. Neutral genetic diversity is often used as a surrogate measure of adaptive capacity.61 Nevertheless, low genetic diversity does not always prevent probable adaptation in Podarcis lizards.62 Furthermore, some studies suggest that populations and species can survive for long periods of time with low genetic diversity63 if they can effectively purge genetic load.64 Ongoing research quantifying the genetic diversity and genetic load in P. raffonei will further clarify how at-risk the species is regarding these harmful genetic risk factors.
The effective population size (Ne) is one of the most important indicators of evolutionary potential65 and extinction risk.66 Ne of the Capo Grosso population of P. raffonei was small, ranging from 49.8 (95% CI 25.5–133.7) to 63.8 (95% CI 54.7–75.5), depending on the exclusion or inclusion of singleton markers, respectively. Although comparisons to Ne estimates are challenged by methodology and the marker type used, the Ne estimate for P. raffonei in Capo Grosso is lower compared to other island-endemic reptiles (e.g., Gongylomorphus bojerii: Ne: 99.6–22867; Sceloporus occidentalis becki: Ne: 175–23668) and is similar to islet populations of Podarcis gaigeae: Ne: 39.4–97.7.69 However, because calculating Ne is challenged by biases, the ratio of effective size to census size, Ne/Nc is deemed a more useful indicator of the extent of genetic variation expected in a population.70,71 Our estimate of Ne/Nc for P. raffonei is 0.05–0.06. This is lower than the average Ne/Nc of 0.1, estimated over a range of different animal taxa,70,71,72 and is lower than the most recent review of Ne/Nc calculated for other reptile species (range: 0.08–7373).
The extremely low estimates of genetic diversity and effective population size probably indicate a reduced evolutionary potential of the Capo Grosso population that can determine a limited ability to withstand environmental stressors, thus increasing extinction risk.74 Given recent evidence of a further demographic decline in this population (L. Vignoli and B. Gambioli pers. comm.), we stress the urgency of management plans, combined with robust demographic and genomic monitoring.
Hybridization: Good or bad for small, isolated populations?
Interspecific hybridization is arguably one of the most controversial, and neglected, topics in conservation.75 Hybridization can drive rare species to extinction through genetic swamping, via hybrid replacement of the rare form, or through demographic swamping, where the overall population growth is reduced to the production of maladaptive hybrid individuals.76 Moreover, extinction risk is exacerbated if the hybrids exhibit reduced fitness relative to that of either parental species (i.e., outbreeding depression77). However, for small, isolated populations of rare species which may potentially lack the genetic variation required for adaptation,61 or where variation is required to avoid the negative impacts of realized genetic load,64 natural hybridization may represent a source of novel genetic variation that can increase evolutionary potential.52,78 This is supported by empirical evidence that introgressive hybridization can be central in increasing fitness, driving rapid evolution, and improving environmental stress responses.79,80,81,82 From a conservation perspective, the role of hybridization is controversial due to concerns about the dilution of parental species’ genetic integrity, as well as defining appropriate conservation policies for hybrid populations.1 Nonetheless, correctly defining conservation and management programs for rare species with extremely fragmented populations is arguably more pressing.83 On the basis of empirical studies, the inbreeding depression threat of small populations is more urgent than the potential disadvantages of outbreeding.84 Given the near impossible potential for gene flow between the four geographically isolated populations of P. raffonei, could low natural hybridization with P. siculus represent a mechanism of population rescue for P. raffonei? It might be assumed that the low number of identified hybrids might exclude the possibility of genetic swamping, while representing a suitable scenario for adaptive introgression of positively selected variants that could improve the genetic status of P. raffonei. Empirical evidence suggests that even extremely low fertility or viability of early-generation hybrids does not prevent gene flow and the establishment of novel evolutionary lineages.85,86,87 Our results suggest that perhaps the major concern for P. raffonei is not about hybridization per se but rather the fact that the encounter and natural hybridization with P. siculus comes at the cost of imminent competitive exclusion as observed in mainland Vulcano in the last century.
Implications for the conservation of the Aeolian wall lizard
Genomic data have democratized the field of population genetics and can provide crucial information for conservation and management.88 Our population genomic analysis of the Aeolian wall lizard provides key insights on the causes of its decline, and on possible management scenarios. The interplay between the low fitness of hybrids, alongside demographic and ecological factors, explains the low rate of hybridization, and the lack of introgressed alleles in P. siculus from mainland Vulcano. Together, this suggests that other processes, such as interspecific competition, probably play(ed) a stronger role in the decline of P. raffonei.89 Further studies are needed to disentangle these mechanisms, and whether they are context-dependent, i.e., if P. raffonei can withstand the impact of P. siculus in specific habitat refugia. On the other hand, the very low genetic diversity and evidence of recent population declines highlights the urgency of management actions to avoid the extinction of this important population. Capo Grosso is deemed the largest extant population of P. raffonei, yet the species also survives in three tiny islets. These localities are strongly isolated, but there is very poor information on their genetic features, on their divergence, and on whether they represent Evolutionarily Significant Units (ESUs). The definition of a global management plan for this Critically Endangered species thus requires integrated data, combining extensive, range-wide genetic information with detailed demographic and habitat data on each population. These analyses, together with the results presented here will provide the basis for much needed conservation projects (e.g., the EOLIZARD Life Project; https://cinea.ec.europa.eu/programmes/life_en) that are crucial for ensuring the persistence of this unique component of island biota.
Limitations of the study
Currently, Capo Grosso represents the only known location where P. raffonei is in contact with P. siculus. The previous assessment of hybridization between the two species was performed using lizards collected from two sites on Vulcano island.19 The present study therefore lacks the geographic and ecological scope for disentangling the effect of demographic (species relative abundance), spatial (expanding or introduced populations), and ecological (habitat type) determinants of hybridization rate.3,90 The low hybridization rate observed here might be due to the low hybridization potential between the two species on Capo Grosso, where a few individuals of P. siculus expanding over the promontory became isolated in its distal portion in a population dominated by heterospecific individuals. Indeed, a higher hybridization rate was detected previously on Vulcano in two sites with comparable species relative abundance.19 Moreover, while we found that hybridization remained at similar, low rates in Capo Grosso between 2015 and 2017, data covering a longer time frame are needed to ascertain the temporal dynamics occurring in this area. Future studies are necessary to monitor the demographic and spatial trends of P. siculus on the promontory and its consequences on hybridization rate. Finally, although hybrids are rare, it remains unclear how common mating between P. raffonei and P. siculus is, which would represent wasted reproductive effort and could pose further demographic risk.1 In this respect, data on heterospecific mating, and on hatchling and survival rate of hybrid offspring, are crucial to assess the demographic effect of hybridization on P. raffonei.
Resource availability
Lead contact
Further information and requests for resources should be directed to the lead contact, Daniele Salvi (daniele.salvi2@univaq.it).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
ddRADseq data have been deposited at the European Nucleotide Archive (ENA) under the accession code ENA: PRJEB77477 and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
We thank Delphine Rioux (LECA, UMR UGA-USMB-CNRS, Grenoble, France), Matteo Garzia and Emanuele Berrilli for help with the DNA laboratory work, and Weizhao Yang and Stephanie Sherpa for the help with initial check of the ddRADseq data. We thank Andrea Melotto, Roberto Sacchi, Stefano Scali, and Leonardo Vignoli for their help during fieldwork. This study was funded by the Italian Ministry for Research (PRIN project ‘Hybrind’—2017KLZ3MA to D.S. and G.F.F.), the Mohamed bin Zayed Species Conservation Fund (Project 162514415 to G.F.F.) and the European Commission LIFE Programme (Project 101114121 LIFE22-NAT-IT-LIFE EOLIZARD to D.S.). MAC and IS-R are supported by grant 28014 02/SAICT/2017 and SFRH/BD/95745/2013 from FCT, Portugal. Lizards were captured and handled under permits from the Italian Ministry of Environment (PNM-0004602, PNM-0008287, and MATTM-0037921). JRP is currently supported by funding from the European Union’s Horizon Europe research and innovation programme under the Marie Skłodowska-Curie grant agreement no. 101068395 - “Poly2Adapt”.
Author contributions
Conceptualization, D.S. and G.F.F.; methodology, J.R.P., J.F.O., and D.S.; investigation, J.R.P., G.F.F., I.S-.R., M.A.C., and D.S.; writing—original draft, J.R.P. and D.S.; writing—review and editing, J.R.P., G.F.F., and D.S.; visualization, J.R.P., J.F.O., and D.S.; funding acquisition, G.F.F. and D.S.; supervision, D.S.; project administration, D.S.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Aeolian wall lizard (Podarcis raffonei) | Field collection | see Table S1 for more details |
| Italian wall lizard (Podarcis siculus) | Field collection | see Table S1 for more details |
| Chemicals, peptides, and recombinant proteins | ||
| SbfI restriction enzyme | New England Biolabs Inc. | NEB #R3642 |
| MspI restriction enzyme | New England Biolabs Inc. | NEB #R0106 |
| Illumina adapters | Illumina | N/A |
| T4 DNA ligase | New England Biolabs Inc. | NEB #M0202 |
| Taq-Phusion High-Fidelity | New England Biolabs Inc. | NEB #M0530 |
| Critical commercial assays | ||
| QIAgen DNeasy Blood & Tissue Kit | Qiagen | Cat #69506 |
| AMPure XP beads | Beckman Coulter | Cat #A63882 |
| QIAquick Gel Extraction Kit | Qiagen | Cat #28706X4 |
| QIAgen MinElute PCR Purification Kit | Qiagen | Cat # 28006 |
| Illumina HiSeq 2500 | Illumina | N/A |
| Deposited data | ||
| ddRADseq data from 133 lizards: Podarcis raffonei (NCBI Taxonomy ID: 65483), Podarcis siculus (NCBI Taxonomy ID: 65484) and the four identified hybrids (NCBI Taxonomy ID: 3239811) | This study | ENA BioProject Number: PRJEB77477 |
| Oligonucleotides | ||
| 12S mtDNA gene, primer forward: 12Sa | Kocher et al.91 | CTGGGATTAGATACCCCACTAT |
| 12S mtDNA gene, primer reverse: 12Sb | Kocher et al.91 | GAGGGTGACGGGGCGGTGTGT |
| Software and algorithms | ||
| fastp v0.23.2 | Chen et al.92 | https://github.com/OpenGene/fastp |
| Stacks v2.60 | Rochette et al.93 | https://catchenlab.life.illinois.edu/stacks/ |
| bwa mem v0.7.17 | Li94 | https://github.com/lh3/bwa |
| vcftools v0.1.17 | Danecek et al.95 | https://vcftools.github.io/index.html |
| R v4.0.2 | R Core Team21 | https://www.r-project.org/ |
| ggplot2 v3.4.3 | Wickham et al.96 | https://ggplot2.tidyverse.org/ |
| tidyverse v2 | Wickham et al.97 | https://www.tidyverse.org/ |
| Plink v1.9 | Purcell et al.98 | https://www.cog-genomics.org/plink/ |
| Hierfstat v0.5.11 | Goudet99 | https://github.com/jgx65/hierfstat |
| fineRADstructure v0.3.2 | Malinsky et al.100 | https://github.com/millanek/fineRADstructure |
| introgress v1.2.3 | Gompert and Alex Buerkle101 | https://github.com/zgompert/introgress |
| Admixture v1.3 | Alexander et al.102 | https://dalexander.github.io/admixture/ |
| NewHybrids v2.0 | Anderson and Thompson103 | https://github.com/eriqande/newhybrids |
| parallelenewhybrid v1.0.1 | Wringe et al.104 | https://github.com/bwringe/parallelnewhybrid |
| hybriddetective | Wringe et al.105 | https://github.com/bwringe/hybriddetective |
| Genotype Plot v0.2.1 | Whiting106 | https://github.com/JimWhiting91/genotype_plot |
| NeEstimator v2.1 | Do et al.107 | https://www.molecularfisherieslaboratory.com.au/neestimator-software/ |
| rayshader v0.38.1 | Morgan-Wall22 | https://www.rayshader.com/ |
Experimental model and study participant details
Study sites, species and sampling
Sampling occurred during the spring and summer of 2015 and 2017 and comprised a total of 138 lizards (all information related to the animals used in this study can be found in Table S1). Adult lizards were captured with a noose. For each lizard, we recorded the sex and took standard photographs of dorsal and ventral patterns. Sex was determined in the field using sexual secondary characters and hemipenis eversion, and was confirmed bioinformatically by calculating the proportion of reads aligning to the Z/W chromosomes. A 2 cm tail clip was obtained and stored in 95–100% EtOH for genetic analysis. In Capo Grosso, we sampled 54 lizards with the brown phenotype typical of P. raffonei (M:F ratio 2.33:1) and 38 lizards with a green dorsal-colouration phenotype (“intermediate”) (M:F ratio 2.45:1), matching the description of hybrids reported in previous studies.19,29 Furthermore, we sampled 36 lizards from the main island of Vulcano (M:F ratio 4.8:1), matching the typical phenotype of P. siculus (Figures 1 and 2). To ensure the correct identification of each species group from the Capo Grosso/Vulcano system, we also sampled five pure male P. raffonei and five pure male P. siculus from two locations where the two species do not overlap: P. raffonei from the islet of Scoglio Faraglione and P. siculus from Milazzo (mainland Sicily). We have no reason to believe that the male bias in our sample groups affected our inference of hybridization rates. Lizards were captured and handled under permits from the Italian Ministry of Environment (PNM-0004602, PNM-0008287, and MATTM-0037921).
Method details
DNA extraction and ddRAD library preparation
Genomic DNA was extracted using the DNeasy Blood & Tissue kit (Qiagen, Germany), following manufacturer guidelines. Double-digested RAD (Restriction site Associated DNA) was conducted using a modified version of the protocol described in108 (see18). ddRAD library preparation included an initial digestion of 300 ng of DNA in a 34 μL reaction (2 h at 37°C; 10 U each of SbfI and MspI, New England Biolabs Inc.). Standard Illumina adapters were ligated using 60 cycles of digestion at 37°C (2 min) and ligation at 16°C (4 min) with 400 U of T4 ligase (New England Biolabs Inc.), followed by heat-inactivation at 65°C (10 min). Digested-ligated products were purified using 1.5:1 ratio of AMPure XP beads (Beckman Coulter, USA). Size selection was performed using a BluePippin to retain total fragment sizes of 250-500bp and were purified using the QIAquick Gel Extraction Kit (Qiagen, Germany). Libraries were obtained by pooling 10 x 20 μL of the each PCR reaction per library, each consisting of 2.5 μL of DNA, 0.2 mM of dNTPs, 0.15 μM of primer, 3% dimethyl sulfoxide (DMSO) and 0.4 U of Taq-Phusion High-Fidelity (New England Biolabs Inc.). PCR conditions were as follows: initial denaturation at 98°C (10 min), 12 cycles of 98°C (10 s), 66°C (30 s), 72°C (1 min), final extension period at 72°C (10 min). Libraries were purified with QIAgen MinElute PCR Purification Kit (Qiagen, Germany) and were sequenced on an Illumina HiSeq 2500 (2 x 125 bp).
Data processing
Reads shorter than 125 bp in length were removed using fastp v0.23.2.92 Stacks v2.6093 was used to assemble loci. Trimmed data were cleaned and demultiplexed using the module process_radtags. For hybrid detection, we used a de novo approach to avoid biassed estimations of allele frequencies by aligning to a single species’ genome. P. raffonei and P. siculus were first assembled separately to remove low quality samples109 and to perform parameter optimization.110 Due to the high divergence time between the species (11–18 million years32,43), parameters were optimized for each species separately in ustacks (M parameter). We then optimized species-specific loci into a catalog by assessing the n parameter across species. The catalog for de novo optimization consisted of loci assembled from 20 individuals with the highest coverage from each of pure P. raffonei and pure P. siculus. For estimates of genetic diversity and effective population size, we used RAD loci aligned to the reference genome of P. raffonei (GCF_027172205.160). Cleaned reads were aligned to the genome using bwa mem v0.7.17,94 marking shorter split hits as secondary reads (-M option). Secondary reads were removed from the alignment files prior to processing with ref_map. As different analyses require differently filtered datasets, we created several subsets from our data using the populations module. Further details of these filters can be found below in each relevant section.
Quantification and statistical analyses
Genetic structure and hybrid identification
The de novo assembled loci were used for analyses of genetic structure and hybrid identification. We created a whitelist of loci present only in P. siculus green and P. raffonei brown individuals (no intermediate phenotypes), including the pure known individuals from Scoglio Faraglione (P. raffonei) and Milazzo (P. siculus). Loci had to be present in both groups (-p 2), and in 50% of individuals from each of these groups (-r 0.5), including alleles present at a minor allele count of 2 (--min-mac 2). This whitelist was used to sample loci across all individuals, inclusive of the intermediate-phenotype samples. The dataset was then assessed for depth in vcftools v0.1.1795 and was filtered accordingly (--minDP 3 and --max-meanDP 80), keeping only biallelic sites at a genotype quality of 30, resulting in a linkage-pruned dataset (--write-single-SNP) comprised of 2,623 variant sites. The same procedure was used to generate data for the full haplotype (all SNPs for each RAD locus), resulting in a dataset comprising 17,254 variant sites. Data manipulation and plotting was performed in R v4.0.221 with ggplot296 and tidyverse.97
A Principal Components Analysis (PCA) was performed using the linkage-pruned dataset in Plink v1.9.98 Pairwise-FST between the two species was calculated using the linkage-pruned dataset in Hierfstat v0.5.11.99 The full haplotype information was used for high-resolution inference of the recent shared ancestry between the samples using fineRADstructure v0.3.2,100 running the model for 100,000 MCMC iterations with a thinning interval of 1000, discarding the first 10,000 iterations as burn-in, and building a tree with 10,000 hill-climbing iterations. We categorized fixed loci from individuals showing the highest differences in PCA space (36 P siculus samples and 55 brown P. raffonei individuals), resulting in 1081 markers. Note that samples with an intermediate phenotype were not used for marker selection. These fixed markers were used to estimate hybrid indices in introgress v1.2.3.101 Hybrid indices and their confidence limits were estimated using the est.h function using 1000 bootstraps.
Admixture v1.3102 was used for maximum-likelihood (ML) estimates of individual ancestries using the linkage-pruned dataset. To assess variability in ML estimates, the algorithm was run ten times, with the number of groups (K) varying between 1 and 7 with a 10-fold cross-validation (CV = 10). After identification of the optimal K, we performed a hierarchical analysis to identify further sub-structuring within the groups using Admixture. We also performed admixture analysis to assess whether there were any biases introduced into the data by sampling year (2015 versus 2017).
The posterior probability of each intermediate-phenotype individual belonging to one of each discrete genotype classes (pure P. siculus, pure P. raffonei, F1 hybrid, F2 hybrid, siculus backcross and raffonei backcross) was estimated using NewHybrids v2.0.103 Data were analyzed in parallel using parallelenewhybrid v1.0.1104 as implemented in hybriddetective.105 All analyses were performed with 200,000 MCMC sweeps, discarding the first 50,000 sweeps as burn-in, using Jeffreys-like priors for both the allele frequency (θ) and mixing proportion (π). To evaluate marker efficiency, we explored three random subsets of the 50 most-informative markers (high FST, linkage-pruned). Convergence of the simulations was checked by assessment of the critical posterior probability of assignment thresholds to each genotype frequency class.
To confirm the accuracy of the classification of each individual into a particular genotype class, we simulated data using a panel of loci derived from brown phenotype P. raffonei from Capo Grosso, P. raffonei from Scoglio Faraglione, P. siculus from Vulcano and P. siculus from Milazzo. Based on our empirical data, we simulated n = 50 of each pure class (P. siculus or P. raffonei), n = 5 F1s, n = 3 F2s, n = 2 siculus backcross and n = 2 raffonei backcross using the HybridLab algorithm.111 We simulated three independent datasets, each of which was analyzed three times. Pure individuals from the simulated data were then combined with the empirical data of the intermediate phenotype individuals, using the z option (assignment of the known genotype frequency category) for the known non-hybrid individuals.
Finally, we created a whitelist containing RAD loci genotyped across all individuals (i.e., no missing data) which were alternatively fixed between pure P. siculus and pure P. raffonei (FST = 1) to explore the signature of individual genotypes. This analysis was conducted in attempts to better clarify the hybrid status of the low coverage individual (CGI38). The stringent filtering resulted in 87 SNPs genotyped across 49 marker loci. Genotypes were plotted using Genotype Plot v0.2.1.106
Characterizing the maternal species and phenotypes of the genetic hybrids
As mitochondria are inherited in a matrilinear way, a hybrid lizard contains the mitochondrial DNA (mtDNA) of its ‘mother species’. The maternal species of each of the identified hybrid individuals was therefore inferred by sequencing a fragment of the 12S rRNA mtDNA region91 using PCR primers and protocols described previously.30 We characterized the overall phenotype of hybrids by inspecting the distribution of characteristics typical of the two parental species as reported in literature (see29) for P. raffonei: color of back generally brown (greener in spring) with small dark dots often aligned in a vertebral stripe, the presence of evident dark markings on the throat (usually absent in P. siculus); and for P. siculus: extensive green in the back with a striped or reticulated pattern, a uniform white throat and belly.
Estimating the genetic diversity and effective population size of P. raffonei in Capo Grosso
For estimates of genetic diversity, we included only individuals sampled in 2017 to avoid any temporal biases. Reference-aligned loci were filtered so that they had to be present in both P. raffonei and P. siculus (-p 2) and present in all individuals (-r 1) at a minor-allele count of 2 (--min-mac 2), and loci mapping to the Z and W chromosome were excluded (total loci = 26,831). To account for any potential artifacts due to missing data, we estimated genetic diversity based only on individuals with the lowest (≤10%) amount of missing data (P. raffonei = n60; P. siculus = n30). As the P. siculus dataset comprised half the number of individuals, we evaluated any potential sample size bias by randomly downsampling the P. raffonei dataset to the same number of individuals as the P. siculus dataset (n = 30). Observed heterozygosity (H0), expected heterozygosity (HE), the inbreeding coefficient (FIS) and allelic richness (AR) were calculated in Hierfstat v 0.5.11.99 The FIS values were bootstrapped over loci 1000 times to obtain 95% confidence intervals. Allelic richness estimates were rarefied against a sample size of 20 diploids.
Estimates of the effective population size (Ne) were calculated using the linkage disequilibrium (LD) method implemented in NeEstimator v2.1.107 Estimates were only calculated for the identified true P. raffonei lizards from Capo Grosso (n = 74) with no missing data (6,772 loci). The P. siculus dataset comprised too few individuals to infer accurate estimates. We excluded the Z and W chromosomes from analysis. The Ne was estimated both with and without singletons and 95% confidence limits were calculated using jack-knifing, using a Pcrit value = 0.05. Resulting effective population size estimates were used to calculate the Ne/Nc ratio by using N-mixture model census size (Nc) estimates for the Capo Grosso population: 1050 (847–1280).10
Published: October 5, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.111097.
Contributor Information
Gentile Francesco Ficetola, Email: francesco.ficetola@unimi.it.
Daniele Salvi, Email: daniele.salvi2@univaq.it.
Supplemental information
References
- 1.Allendorf F.W., Leary R.F., Spruell P., Wenburg J.K. The problems with hybrids: setting conservation guidelines. Trends Ecol. Evol. 2001;16:613–622. doi: 10.1016/S0169-5347(01)02290-X. [DOI] [Google Scholar]
- 2.Levin D.A., Francisco-Ortega J., Jansen R.K. Hybridization and the extinction of rare plant species. Conserv. Biol. 1996;10:10–16. https://www.jstor.org/stable/2386938 [Google Scholar]
- 3.Rhymer J.M., Simberloff D. Extinction by hybridization and introgression. Annu. Rev. Ecol. Syst. 1996;27:83–109. doi: 10.1146/annurev.ecolsys.27.1.83. [DOI] [Google Scholar]
- 4.Stelkens R.B., Brockhurst M.A., Hurst G.D.D., Greig D. Hybridization facilitates evolutionary rescue. Evol. Appl. 2014;7:1209–1217. doi: 10.1111/eva.12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann A., Griffin P., Dillon S., Catullo R., Rane R., Byrne M., Jordan R., Oakeshott J., Weeks A., Joseph L., et al. A framework for incorporating evolutionary genomics into biodiversity conservation and management. Clim. Chang. Responses. 2015;2:1. doi: 10.1186/s40665-014-0009-x. [DOI] [Google Scholar]
- 6.Vedder D., Lens L., Martin C.A., Pellikka P., Adhikari H., Heiskanen J., Engler J.O., Sarmento Cabral J. Hybridization may aid evolutionary rescue of an endangered East African passerine. Evol. Appl. 2022;15:1177–1188. doi: 10.1111/eva.13440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Böhm M., Collen B., Baillie J.E.M., Bowles P., Chanson J., Cox N., Hammerson G., Hoffmann M., Livingstone S.R., Ram M., et al. The conservation status of the world’s reptiles. Biol. Conserv. 2013;157:372–385. doi: 10.1016/j.biocon.2012.07.015. [DOI] [Google Scholar]
- 8.Spatz D.R., Zilliacus K.M., Holmes N.D., Butchart S.H.M., Genovesi P., Ceballos G., Tershy B.R., Croll D.A. Globally threatened vertebrates on islands with invasive species. Sci. Adv. 2017;3 doi: 10.1126/sciadv.1603080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox N., Young B.E., Bowles P., Fernandez M., Marin J., Rapacciuolo G., Böhm M., Brooks T.M., Hedges S.B., Hilton-Taylor C., et al. A global reptile assessment highlights shared conservation needs of tetrapods. Nature. 2022;605:285–290. doi: 10.1038/s41586-022-04664-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ficetola G.F., Silva-Rocha I., Carretero M.A., Vignoli L., Sacchi R., Melotto A., Scali S., Salvi D. Status of the largest extant population of the Critically Endangered Aeolian lizard Podarcis raffonei (Capo Grosso, Vulcano island) PLoS One. 2021;16 doi: 10.1371/journal.pone.0253631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gippoliti S., Capula M., Ficetola G.F., Salvi D., Andreone F. Threatened by legislative conservationism? The case of the Critically Endangered Aeolian lizard. Front. Ecol. Evol. 2017;5:1. doi: 10.3389/fevo.2017.00130. [DOI] [Google Scholar]
- 12.Capula M., Luiselli L., Bologna M.A., Ceccarelli A. The decline of the Aeolian wall lizard, Podarcis raffonei: causes and conservation proposals. Oryx. 2002;36:66–72. doi: 10.1017/S0030605302000108. [DOI] [Google Scholar]
- 13.Lo Cascio P. Attuali conoscenze e misure di conservazione per le popolazioni relitte dell’endemica lucertola delle Eolie, Podarcis raffonei (Squamata Sauria) Naturalista siciliano. 2010;4:295–317. [Google Scholar]
- 14.D’Amico M., Bastianelli G., Lo F., Francesco P., Lo Valvo M. The spreading of the invasive Italian wall lizard on Vulcano, the last island inhabited by the Critically Endangered Aeolian wall lizard. Herpetol. Conserv. Biol. 2018;13:146–157. [Google Scholar]
- 15.Lo Cascio P., Sciberras A. In: Life on islands. 1. Biodiversity in Sicily and surrounding islands. La Mantia T., Badalamenti E., Carapezza A., Lo Cascio P., Troia A., editors. Edizioni Danaus; 2020. “Cold-blooded” Travellers Around Sicily: How Introductions and Extinctions Have Shaped the Recent Herpetofauna of Circum-Sicilian and Maltese Islands; pp. 355–390. [Google Scholar]
- 16.Silva-Rocha I., Salvi D., Harris D.J., Freitas S. Molecular assessment of Podarcis sicula populations in Britain, Greece and Turkey reinforces a multiple-origin invasion pattern in this species. Acta Herpetol. 2014;9:253–258. doi: 10.13128/Acta_Herpetol-14968. [DOI] [Google Scholar]
- 17.Bonardi A., Ficetola G.F., Razzetti E., Canedoli C., Falaschi M., Lo Parrino E., Rota N., Padoa-Schioppa E., Sindaco R. ReptIslands: Mediterranean islands and the distribution of their reptile fauna. Glob. Ecol. Biogeogr. 2022;31:840–847. doi: 10.1111/geb.13490. [DOI] [Google Scholar]
- 18.Sherpa S., Salvi D., Silva-Rocha I., Capblancq T., Paris J.R., Carretero M.A., Ficetola G.F. Reconstructing the complex colonisation histories of lizards across Mediterranean archipelagos. J. Biogeogr. 2024;51:157–172. doi: 10.1111/jbi.14739. [DOI] [Google Scholar]
- 19.Capula M. Natural hybridization in Podarcis sicula and P. wagleriana (Reptilia: Lacertidae) Biochem. Syst. Ecol. 1993;21:373–380. doi: 10.1016/0305-1978(93)90028-P. [DOI] [Google Scholar]
- 20.Capula M. Genetic variation and differentiation in the lizard, Podarcis wagleriana (Reptilia: Lacertidae) Biol. J. Linn. Soc. Lond. 1994;52:177–196. doi: 10.1111/j.1095-8312.1994.tb00986.x. [DOI] [Google Scholar]
- 21.R Core Team . R Foundation for Statistical Computing; 2021. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 22.Morgan-Wall T. rayshader: Create Maps and Visualize Data in 2D and 3D. R package version 0.38.1. 2024 https://github.com/tylermorganwall/rayshader https://www.rayshader.com [Google Scholar]
- 23.Capula M. Proc. Sixth Ord. Gen. Meeting Societas Europaea Herpetologica. 1992. Competitive Exclusion between Podarcis Lizards from Tyrrhenian Islands: Inference from Comparative Species Distributions; pp. 89–93. [Google Scholar]
- 24.Downes S., Bauwens D. An experimental demonstration of direct behavioural interference in two Mediterranean lacertid lizard species. Anim. Behav. 2002;63:1037–1046. doi: 10.1006/anbe.2002.3022. [DOI] [Google Scholar]
- 25.Damas-Moreira I., Oliveira D., Santos J.L., Riley J.L., Harris D.J., Whiting M.J. Learning from others: an invasive lizard uses social information from both conspecifics and heterospecifics. Biol. Lett. 2018;14 doi: 10.1098/rsbl.2018.0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Damas-Moreira I., Riley J.L., Harris D.J., Whiting M.J. Can behaviour explain invasion success? A comparison between sympatric invasive and native lizards. Anim. Behav. 2019;151:195–202. doi: 10.1016/j.anbehav.2019.03.008. [DOI] [Google Scholar]
- 27.Caruso Y. Thermoregulation comparisons between a threatened native and an invasive lizard species. Herpetol. J. 2021;31:70–76. doi: 10.33256/hj31.2.7076. [DOI] [Google Scholar]
- 28.Ravagni S., Sanchez-Donoso I., Vilà C. Biased assessment of ongoing admixture using STRUCTURE in the absence of reference samples. Mol. Ecol. Resour. 2021;21:677–689. doi: 10.1111/1755-0998.13286. [DOI] [PubMed] [Google Scholar]
- 29.Capula M., Lo Cascio P. In: Fauna d’Italia, Reptilia. Corti C., Capula M., Luiselli L., Razzetti E., Sindaco R., editors. Edizioni Calderini de Il Sole 24 ORE; Bologna: 2011. Podarcis Raffonei (Mertens, 1952) pp. 401–407. [Google Scholar]
- 30.Mendes J., Harris D.J., Carranza S., Salvi D. Evaluating the phylogenetic signal limit from mitogenomes, slow evolving nuclear genes, and the concatenation approach. New insights into the Lacertini radiation using fast evolving nuclear genes and species trees. Mol. Phylogenet. Evol. 2016;100:254–267. doi: 10.1016/j.ympev.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 31.Salvi D., Pinho C., Harris D.J. Digging up the roots of an insular hotspot of genetic diversity: decoupled mito-nuclear histories in the evolution of the Corsican-Sardinian endemic lizard Podarcis tiliguerta. BMC Evol. Biol. 2017;17:63. doi: 10.1186/s12862-017-0899-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salvi D., Pinho C., Mendes J., Harris D.J. Fossil-calibrated time tree of Podarcis wall lizards provides limited support for biogeographic calibration models. Mol. Phylogenet. Evol. 2021;161 doi: 10.1016/j.ympev.2021.107169. [DOI] [PubMed] [Google Scholar]
- 33.Caeiro-Dias G., Brelsford A., Kaliontzopoulou A., Meneses-Ribeiro M., Crochet P.-A., Pinho C. Variable levels of introgression between the endangered Podarcis carbonelli and highly divergent congeneric species. Heredity. 2021;126:463–476. doi: 10.1038/s41437-020-00386-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boecklen W.J., Howard D.J. Genetic analysis of hybrid zones: numbers of markers and power of resolution. Ecology. 1997;78:2611–2616. doi: 10.2307/2265918. [DOI] [Google Scholar]
- 35.Twyford A.D., Ennos R.A. Next-generation hybridization and introgression. Heredity. 2012;108:179–189. doi: 10.1038/hdy.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dupuis J.R., Sperling F.A.H. Hybrid dynamics in a species group of swallowtail butterflies. J. Evol. Biol. 2016;29:1932–1951. doi: 10.1111/jeb.12931. [DOI] [PubMed] [Google Scholar]
- 37.Miralles A., Secondi J., Pabijan M., Babik W., Lemaire C., Crochet P.-A. Inconsistent estimates of hybridization frequency in newts revealed by SNPs and microsatellites. Conserv. Genet. 2023;25:215–225. doi: 10.1007/s10592-023-01556-9. [DOI] [Google Scholar]
- 38.Mertens R. Die Mauereidechsen der Liparischen Inseln, gesammelt von Dr. Antonino Trischitta. Senckenbergiana biologica, Frankfurt/Main. 1955;36:25–40. [Google Scholar]
- 39.Barton N.H. The dynamics of hybrid zones. Heredity. 1979;43:341–359. doi: 10.1038/hdy.1979.87. [DOI] [Google Scholar]
- 40.Corti C., Biaggini M., Berti R. Different habitats, different pressures? Analysis of escape behaviour and ectoparasite load in Podarcis sicula (Lacertidae) populations in different agricultural habitats. Amphib. Reptil. 2009;30:453–461. doi: 10.1163/156853809789647068. [DOI] [Google Scholar]
- 41.Vignoli L., Vuerich V., Bologna M.A. Experimental study of dispersal behaviour in a wall lizard species (Podarcis sicula) (Sauria Lacertidae) Ethol. Ecol. Evol. 2012;24:244–256. doi: 10.1080/03949370.2011.643922. [DOI] [Google Scholar]
- 42.Barton N.H., Hewitt G.M. Analysis of Hybrid Zones. Annu. Rev. Ecol. Syst. 1985;16:113–148. doi: 10.1146/annurev.es.16.110185.000553. [DOI] [Google Scholar]
- 43.Yang W., Feiner N., Pinho C., While G.M., Kaliontzopoulou A., Harris D.J., Salvi D., Uller T. Extensive introgression and mosaic genomes of Mediterranean endemic lizards. Nat. Commun. 2021;12:2762. doi: 10.1038/s41467-021-22949-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jančúchová-Lásková J., Landová E., Frynta D. Are genetically distinct lizard species able to hybridize? A review. Curr. Zool. 2015;61:155–180. doi: 10.1093/czoolo/61.1.155. [DOI] [Google Scholar]
- 45.Yang W., Feiner N., Salvi D., Laakkonen H., Jablonski D., Pinho C., Carretero M.A., Sacchi R., Zuffi M.A.L., Scali S., et al. Population genomics of wall lizards reflects the dynamic history of the Mediterranean basin. Mol. Biol. Evol. 2022;39 doi: 10.1093/molbev/msab311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.MacGregor H.E.A., While G.M., Barrett J., Pérez i de Lanuza G., Carazo P., Michaelides S., Uller T. Experimental contact zones reveal causes and targets of sexual selection in hybridizing lizards. Funct. Ecol. 2017;31:742–752. doi: 10.1111/1365-2435.12767. [DOI] [Google Scholar]
- 47.While G.M., Michaelides S., Heathcote R.J.P., MacGregor H.E.A., Zajac N., Beninde J., Carazo P., Pérez I de Lanuza G., Sacchi R., Zuffi M.A.L., et al. Sexual selection drives asymmetric introgression in wall lizards. Ecol. Lett. 2015;18:1366–1375. doi: 10.1111/ele.12531. [DOI] [PubMed] [Google Scholar]
- 48.Gorman G.C., Licht P., Dessauer H.C., Boos J.O. Reproductive failure among the hybridizing Anolis lizards of Trinidad. Syst. Biol. 1971;20:1–18. doi: 10.1093/sysbio/20.1.1. [DOI] [Google Scholar]
- 49.Edmands S. Heterosis and outbreeding depression in interpopulation crosses spanning a wide range of divergence. Evolution. 1999;53:1757–1768. doi: 10.1111/j.1558-5646.1999.tb04560.x. [DOI] [PubMed] [Google Scholar]
- 50.Muraro M., Falaschi M., Ficetola G.F. Patterns of performance variation between animal hybrids and their parents: a meta-analysis. Evol. Biol. 2022;49:482–496. doi: 10.1007/s11692-022-09585-x. [DOI] [Google Scholar]
- 51.Rhode J.M., Cruzan M.B. Contributions of heterosis and epistasis to hybrid fitness. Am. Nat. 2005;166:E124–E139. doi: 10.1086/491798. [DOI] [PubMed] [Google Scholar]
- 52.Chan W.Y., Hoffmann A.A., van Oppen M.J.H. Hybridization as a conservation management tool. Conserv. Lett. 2019;12 doi: 10.1111/conl.12652. [DOI] [Google Scholar]
- 53.Soletchnik P., Huvet A., Le Moine O., Razet D., Geairon P., Faury N., Goulletquer P., Boudry P. A comparative field study of growth, survival and reproduction of Crassostrea gigas, C. angulata and their hybrids. Aquat. Living Resour. 2002;15:243–250. doi: 10.1016/S0990-7440(02)01175-0. [DOI] [Google Scholar]
- 54.Kirk H., Vrieling K., Klinkhamer P.G.L. Maternal effects and heterosis influence the fitness of plant hybrids. New Phytol. 2005;166:685–694. doi: 10.1111/j.1469-8137.2005.01370.x. [DOI] [PubMed] [Google Scholar]
- 55.Galán P. Cambios estacionales de coloración y comportamiento agonístico, de cortejo y de apareamiento en el lacértido Podarcis bocagei. Rev. Esp. Herpetol. 1995;9:57. [Google Scholar]
- 56.Faraone F., Lo Valvo M., Others . Riassunti del 6 Congresso Nazionale Societas Herpetologica Italica (SHI), (ITA) 2006. Seasonal Variation in Color of the Sicilian Wal Lizard Podarcis Wagleriana; p. 25. [Google Scholar]
- 57.Sá-Sousa P. Lagartija de Carbonell - Podarcis carbonelli Pérez-Mellado, 1981. Salvador Milla; 2015. [DOI] [Google Scholar]
- 58.Pellitteri-Rosa D., Gazzola A., Todisco S., Mastropasqua F., Liuzzi C. Lizard colour plasticity tracks background seasonal changes. Biol. Open. 2020;9 doi: 10.1242/bio.052415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Storniolo F., Zuffi M.A.L., Coladonato A.J., Di Vozzo L., Giglio G., Gini A.E., Leonetti F.L., Luccini S., Mangiacotti M., Scali S., et al. Patterns of variations in dorsal colouration of the Italian wall lizard Podarcis siculus. Biol. Open. 2021;10 doi: 10.1242/bio.058793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gabrielli M., Benazzo A., Biello R., Ancona L., Fuselli S., Iannucci A., Balacco J., Mountcastle J., Tracey A., Ficetola G.F., et al. A high-quality reference genome for the Critically Endangered Aeolian wall lizard. J. Hered. 2023;114:279–285. doi: 10.1093/jhered/esad014. [DOI] [PubMed] [Google Scholar]
- 61.Kardos M., Armstrong E.E., Fitzpatrick S.W., Hauser S., Hedrick P.W., Miller J.M., Tallmon D.A., Funk W.C. The crucial role of genome-wide genetic variation in conservation. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2104642118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sherpa S., Paris J.R., Silva-Rocha I., Di Canio V., Carretero M.A., Ficetola G.F., Salvi D. Genetic depletion does not prevent rapid evolution in island-introduced lizards. Ecol. Evol. 2023;13 doi: 10.1002/ece3.10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pečnerová P., Lord E., Garcia-Erill G., Hanghøj K., Rasmussen M.S., Meisner J., Liu X., van der Valk T., Santander C.G., Quinn L., et al. Population genomics of the muskox’ resilience in the near absence of genetic variation. Mol. Ecol. 2024;33 doi: 10.1111/mec.17205. [DOI] [PubMed] [Google Scholar]
- 64.Bertorelle G., Raffini F., Bosse M., Bortoluzzi C., Iannucci A., Trucchi E., Morales H.E., van Oosterhout C. Genetic load: genomic estimates and applications in non-model animals. Nat. Rev. Genet. 2022;23:492–503. doi: 10.1038/s41576-022-00448-x. [DOI] [PubMed] [Google Scholar]
- 65.Waples R.S. What Is Ne. J. Hered. 2022;113:371–379. doi: 10.1093/jhered/esac023. [DOI] [PubMed] [Google Scholar]
- 66.Antao T., Pérez-Figueroa A., Luikart G. Early detection of population declines: high power of genetic monitoring using effective population size estimators. Evol. Appl. 2011;4:144–154. doi: 10.1111/j.1752-4571.2010.00150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Michaelides S., Cole N., Funk S.M. Translocation retains genetic diversity of a threatened endemic reptile in Mauritius. Conserv. Genet. 2015;16:661–672. doi: 10.1007/s10592-014-0691-z. [DOI] [Google Scholar]
- 68.Trumbo D.R., Funk W.C., Pauly G.B., Robertson J.M. Conservation genetics of an island-endemic lizard: low Ne and the critical role of intermediate temperatures for genetic connectivity. Conserv. Genet. 2021;22:783–797. doi: 10.1007/s10592-021-01362-1. [DOI] [Google Scholar]
- 69.Runemark A., Hansson B., Pafilis P., Valakos E.D., Svensson E.I. Island biology and morphological divergence of the Skyros wall lizard Podarcis gaigeae: a combined role for local selection and genetic drift on color morph frequency divergence? BMC Evol. Biol. 2010;10:269. doi: 10.1186/1471-2148-10-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Frankham R. Effective population size/adult population size ratios in wildlife: a review. Genet. Res. 1995;66:95–107. doi: 10.1017/S0016672300034455. [DOI] [PubMed] [Google Scholar]
- 71.Palstra F.P., Fraser D.J. Effective/census population size ratio estimation: a compendium and appraisal. Ecol. Evol. 2012;2:2357–2365. doi: 10.1002/ece3.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Palstra F.P., Ruzzante D.E. Genetic estimates of contemporary effective population size: what can they tell us about the importance of genetic stochasticity for wild population persistence? Mol. Ecol. 2008;17:3428–3447. doi: 10.1111/j.1365-294X.2008.03842.x. [DOI] [PubMed] [Google Scholar]
- 73.Hoban S., Bruford M., D’Urban Jackson J., Lopes-Fernandes M., Heuertz M., Hohenlohe P.A., Paz-Vinas I., Sjögren-Gulve P., Segelbacher G., Vernesi C., et al. Genetic diversity targets and indicators in the CBD post-2020 Global Biodiversity Framework must be improved. Biol. Conserv. 2020;248 doi: 10.1016/j.biocon.2020.108654. [DOI] [Google Scholar]
- 74.Frankham R. Inbreeding and extinction: Island populations. Conserv. Biol. 2008;12:665–675. doi: 10.1111/j.1523-1739.1998.96456.x. [DOI] [Google Scholar]
- 75.Draper D., Laguna E., Marques I. Demystifying negative connotations of hybridization for less biased conservation policies. Front. Ecol. Evol. 2021;9:1. doi: 10.3389/fevo.2021.637100. [DOI] [Google Scholar]
- 76.Todesco M., Pascual M.A., Owens G.L., Ostevik K.L., Moyers B.T., Hübner S., Heredia S.M., Hahn M.A., Caseys C., Bock D.G., Rieseberg L.H. Hybridization and extinction. Evol. Appl. 2016;9:892–908. doi: 10.1111/eva.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wolf D.E., Takebayashi N., Rieseberg L.H. Predicting the risk of extinction through hybridization. Conserv. Biol. 2001;15:1039–1053. doi: 10.1046/j.1523-1739.2001.0150041039.x. [DOI] [Google Scholar]
- 78.Taylor S.A., Larson E.L. Insights from genomes into the evolutionary importance and prevalence of hybridization in nature. Nat. Ecol. Evol. 2019;3:170–177. doi: 10.1038/s41559-018-0777-y. [DOI] [PubMed] [Google Scholar]
- 79.Arnold M.L., Kentner E.K., Johnston J.A., Cornman S., Bouck A.C. Natural hybridisation and fitness. Taxon. 2001;50:93–104. doi: 10.2307/1224513. [DOI] [Google Scholar]
- 80.Grant P.R., Grant B.R., Petren K. Hybridization in the recent past. Am. Nat. 2005;166:56–67. doi: 10.1086/430331. [DOI] [PubMed] [Google Scholar]
- 81.Richards Z.T., Hobbs J.-P.A. Hybridisation on coral reefs and the conservation of evolutionary novelty. Curr. Zool. 2015;61:132–145. doi: 10.1093/czoolo/61.1.132. [DOI] [Google Scholar]
- 82.Zhang Z., Bendixsen D.P., Janzen T., Nolte A.W., Greig D., Stelkens R. Recombining your way out of trouble: the genetic architecture of hybrid fitness under environmental stress. Mol. Biol. Evol. 2020;37:167–182. doi: 10.1093/molbev/msz211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ralls K., Ballou J.D., Dudash M.R., Eldridge M.D.B., Fenster C.B., Lacy R.C., Sunnucks P., Frankham R. Call for a paradigm shift in the genetic management of fragmented populations. Conserv. Lett. 2018;11 doi: 10.1111/conl.12412. [DOI] [Google Scholar]
- 84.Edmands S. Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol. Ecol. 2007;16:463–475. doi: 10.1111/j.1365-294X.2006.03148.x. [DOI] [PubMed] [Google Scholar]
- 85.Arnold M.L., Bulger M.R., Burke J.M., Hempel A.L., Williams J.H. Natural hybridization: how low can you go and still be important? Ecology. 1999;80:371–381. doi: 10.1890/0012-9658(1999)080[0371:NHHLCY]2.0.CO;2. [DOI] [Google Scholar]
- 86.Abbott R.J., Barton N.H., Good J.M. Genomics of hybridization and its evolutionary consequences. Mol. Ecol. 2016;25:2325–2332. doi: 10.1111/mec.13685. [DOI] [PubMed] [Google Scholar]
- 87.Brown M.R., Abbott R.J., Twyford A.D. The emerging importance of cross-ploidy hybridisation and introgression. Mol. Ecol. 2024;33 doi: 10.1111/mec.17315. [DOI] [PubMed] [Google Scholar]
- 88.Hohenlohe P.A., Funk W.C., Rajora O.P. Population genomics for wildlife conservation and management. Mol. Ecol. 2021;30:62–82. doi: 10.1111/mec.15720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ficetola G.F., Melotto A., Scali S., Sacchi R., Salvi D. Interference competition with an invasive species as potential driver of rapid extinction in an island-endemic lizard. Glob. Ecol. Conserv. 2024 doi: 10.1016/j.gecco.2024.e03251. [DOI] [Google Scholar]
- 90.Klein E.K., Lagache-Navarro L., Petit R.J. Demographic and spatial determinants of hybridization rate. J. Ecol. 2017;105:29–38. doi: 10.1111/1365-2745.12674. [DOI] [Google Scholar]
- 91.Kocher T.D., Thomas W.K., Meyer A., Edwards S.V., Pääbo S., Villablanca F.X., Wilson A.C. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc. Natl. Acad. Sci. USA. 1989;86:6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rochette N.C., Rivera-Colón A.G., Catchen J.M. Stacks 2: Analytical methods for paired-end sequencing improve RADseq-based population genomics. Mol. Ecol. 2019;28:4737–4754. doi: 10.1111/mec.15253. [DOI] [PubMed] [Google Scholar]
- 94.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 doi: 10.48550/arXiv.1303.3997. Preprint at. [DOI] [Google Scholar]
- 95.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wickham H. Springer; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 97.Wickham H., Averick M., Bryan J., Chang W., McGowan L., François R., Grolemund G., Hayes A., Henry L., Hester J., et al. Welcome to the Tidyverse. J. Open Source Softw. 2019;4:1686. [Google Scholar]
- 98.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Goudet J. hierfstat, a package for r to compute and test hierarchical F-statistics. Mol. Ecol. Notes. 2005;5:184–186. doi: 10.1111/j.1471-8286.2004.00828.x. [DOI] [Google Scholar]
- 100.Malinsky M., Trucchi E., Lawson D.J., Falush D. RADpainter and fineRADstructure: Population Inference from RADseq Data. Mol. Biol. Evol. 2018;35:1284–1290. doi: 10.1093/molbev/msy023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gompert Z., Alex Buerkle C. introgress: a software package for mapping components of isolation in hybrids. Mol. Ecol. Resour. 2010;10:378–384. doi: 10.1111/j.1755-0998.2009.02733.x. [DOI] [PubMed] [Google Scholar]
- 102.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Anderson E.C., Thompson E.A. A model-based method for identifying species hybrids using multilocus genetic data. Genetics. 2002;160:1217–1229. doi: 10.1093/genetics/160.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wringe B.F., Stanley R.R.E., Jeffery N.W., Anderson E.C., Bradbury I.R. parallelnewhybrid: an R package for the parallelization of hybrid detection using newhybrids. Mol. Ecol. Resour. 2017;17:91–95. doi: 10.1111/1755-0998.12597. [DOI] [PubMed] [Google Scholar]
- 105.Wringe B.F., Stanley R.R.E., Jeffery N.W., Anderson E.C., Bradbury I.R. hybriddetective: A workflow and package to facilitate the detection of hybridization using genomic data in R. Mol. Ecol. Resour. 2017;17:e275–e284. doi: 10.1111/1755-0998.12704. [DOI] [PubMed] [Google Scholar]
- 106.Whiting J.R. JimWhiting91/genotype_plot: Genotype Plot. Zenodo; 2022. [DOI] [Google Scholar]
- 107.Do C., Waples R.S., Peel D., Macbeth G.M., Tillett B.J., Ovenden J.R. NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne ) from genetic data. Mol. Ecol. Resour. 2014;14:209–214. doi: 10.1111/1755-0998.12157. [DOI] [PubMed] [Google Scholar]
- 108.Peterson B.K., Weber J.N., Kay E.H., Fisher H.S., Hoekstra H.E. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cerca J., Maurstad M.F., Rochette N.C., Rivera-Colón A.G., Rayamajhi N., Catchen J.M., Struck T.H. Removing the bad apples: A simple bioinformatic method to improve loci-recovery in de novo RADseq data for non-model organisms. Methods Ecol. Evol. 2021;12:805–817. doi: 10.1111/2041-210X.13562. [DOI] [Google Scholar]
- 110.Paris J.R., Stevens J.R., Catchen J.M. Lost in parameter space: a road map for stacks. Methods Ecol. Evol. 2017;8:1360–1373. doi: 10.1111/2041-210X.12775. [DOI] [Google Scholar]
- 111.Nielsen E.E., Bach L.A., Kotlicki P. hybridlab (version 1.0): a program for generating simulated hybrids from population samples. Mol. Ecol. Notes. 2006;6:971–973. doi: 10.1111/j.1471-8286.2006.01433.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
ddRADseq data have been deposited at the European Nucleotide Archive (ENA) under the accession code ENA: PRJEB77477 and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.