Abstract
The inorganic biopolymer polyphosphate (polyP) occurs in all domains of life and affects myriad cellular processes. A longstanding observation is polyP’s frequent proximity to chromatin, and, in many bacteria, its occurrence as magnesium (Mg2+)-enriched condensates embedded in the nucleoid region, particularly in response to stress. The physical basis of the interaction between polyP, DNA and Mg2+, and the resulting effects on the organization of the nucleoid and polyP condensates, remain poorly understood. Here, using a minimal system of polyP, Mg2+, and DNA, we find that DNA can form shells around polyP-Mg2+ condensates. These shells show reentrant behavior, that is, they form within a window of Mg2+ concentrations, representing a tunable architecture with potential relevance in other multicomponent condensates. This surface association tunes condensate size and DNA morphology in a manner dependent on DNA length and concentration, even at DNA concentrations orders of magnitude lower than found in the cell. Our work also highlights the remarkable capacity of two primordial inorganic species to organize DNA.
Subject terms: Biopolymers in vivo, DNA, Chromatin structure, Biophysical chemistry
Polyphosphate affects many cellular processes and is often found near cellular DNA. Here, the authors show that DNA forms shells around polyP-Mg2+ condensates in vitro, controlling condensate size and DNA morphology in a manner dependent on DNA properties.
Introduction
Polyphosphate (polyP) is a structurally simple, inorganic polymer consisting of a few to many hundreds of orthophosphate units linked by phosphoanhydride bonds. Biosynthesis of polyP is found in all three domains of life and affects myriad cellular processes. In bacteria, polyP has been implicated in promoting cellular fitness with pleiotropic effects on biofilm formation, motility, cell cycle, and oxidative stress resistance1–4. In eukaryotic organisms, including humans, polyP has been linked with a wide variety of cellular processes from blood clotting and innate immunity to mitochondrial bioenergetics and cancer signaling5,6. How synthesis of this simple polyanion exerts a broad range of effects on cellular physiology has remained enigmatic.
A unifying organizational feature of polyP across evolution is that this polymer is frequently observed in close proximity to chromatin. In eukaryotes, from yeast to protists to metazoans, including human cells, polyP has been found in the nucleus, and in some cases in the nucleolus7–14. In bacteria, polyP has also been observed in the nucleoid region, forming spatially and temporally organized membraneless condensates15–19. Embedding of polyphosphate granules within the nucleoid of diverse bacterial taxa has been observed at least since the 1960s15,17,18,20–22. The organization of polyP granules within the nucleoid region appears to be both species and condition-specific: in the opportunistic human pathogen Pseudomonas aeruginosa, polyP granules are transiently evenly spaced on the long axis of the cell in the nucleoid region during nitrogen starvation18. In Caulobacter crescentus, polyP granules form at the ¼ and ¾ positions in the nucleoid region, and disruption of chromosome segregation can alter the granule organization, suggesting a functional association15. Further support of a functional association between polyP and the nucleoid in bacteria includes observed effects of polyP on cell cycle progression in diverse taxa and recruitment of DNA binding proteins to polyP condensates18,23–26. Collectively, these observations of conservation, organization, and function suggest that polyP condensates could be a fundamentally important feature of bacterial chromatin.
Therefore, we must first understand how polyP and DNA interact. Despite the known structural and functional association between these two polyanions in vivo, the mechanistic basis of interaction between polyP and DNA remains poorly understood. An interesting and relevant starting point for mediating this interaction is divalent cations, particularly Mg2+. In addition to Mg2+’s relative abundance as a divalent cation and known association with nucleic acids, numerous studies in diverse bacteria have used elemental analysis to show that polyP granules are enriched in divalent cations, including Mg2+16,18,27,28. Coupled with the observation that depletion of Mg2+ from the minimal medium prevents Aerobacter aerogenes from making polyP granules29, Mg2+ appears to be highly relevant to a simplified model system. Moreover, as Mg2+ has been well known to induce homotypic phase separation with polyanionic polymers30–32, including RNA, developing an understanding of the ways DNA, polyP, and Mg2+ interact could expand our understanding of multicomponent condensate systems and the properties that modulate their morphology and organization30,33–37. New insight into the physical chemistry of this system will also be valuable in future industrial and biomedical applications, as proposed for example with multi-layer polyP-Ca2+ nanoparticles as a drug delivery vehicle38.
Thus, from evolutionary and biophysical perspectives, characterizing the emergent properties of this multicomponent system of polyP-Mg2+-DNA is a key starting point to understanding fundamental principles of polyP interaction with DNA, with potential relevance for further understanding polyP granules and their relationship with chromatin structure and function. The polyP-DNA-Mg2+ interaction system we focus on here can be expanded on in future studies to include the effects of proteins and other cellular partners that may vary across biological systems.
In this work, we probe the condensation properties of this polyP-Mg2+-DNA system using multiple biophysical and imaging methods to ask: what are the properties of the polyP-Mg2+-DNA interface? How does the formation of the multicomponent system affect the organization of DNA? And how does DNA tune the organization and dynamics of polyP condensates? Our studies reveal an unexpected architecture in this multicomponent system, with polyP condensates and DNA both providing control elements that can synergistically tune each other’s properties.
Results
Long polyP undergoes Mg2+-driven reentrant phase transitions
As a starting point for our multicomponent system, we first tested the ability of Mg2+ to drive phase separation of polydisperse polyP in a length regime that would be expected to be found in bacteria, which can make chains in the 100–1000 s of orthophosphates in length1,4,12. Based on our and other previous work on divalent cation-driven RNA/polyanion phase separation30–32,39,40, we were interested in understanding the Mg2+ concentration dependence and possible non-monotonic characteristics of this process. As a model in vitro system, we therefore charted the Mg2+ induced phase separation of long-chain polyP (P700- mean: 113 kDa, mode(nP): 1000–1300, range: 10–208 kDa, according to the manufacturer) at pH 7.5. We used 1 mg/mL P700, corresponding to around 9.8 mM polyP in terms of Pi units. Studies in E. coli, P. aeruginosa, and other species show that cells accumulate >100 nmol polyP per mg protein, which corresponds to 20–50 mM polyP in terms of Pi units41–44. For these studies, we employed absorption spectroscopy measurements, which can be used to quantify light scattering induced by phase separation, a method that has been previously used for such studies32,45. Additionally, confocal fluorescence microscopy with ~10% AlexaFluor647-labeled polyP was used to visualize the morphologies of condensates.
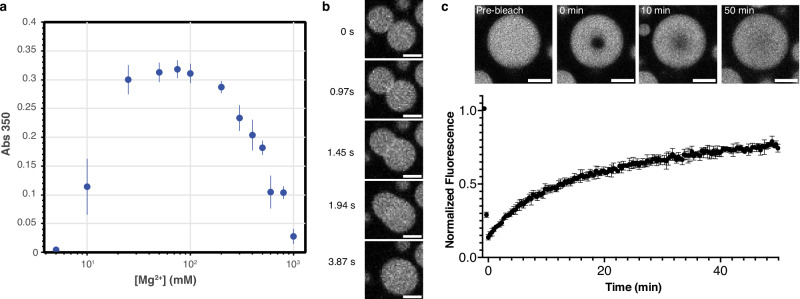
The absorbance data indicate an onset of phase separation around 10 mM Mg2+ (Fig. 1a). Imaging studies confirm that the absorbance increase corresponds to the formation of spherical droplets that show facile fusion on the few-second timescale, consistent with liquid-like behavior (Fig. 1b, Supplementary Movie 1). Bleached regions in condensates reached around 75% recovery within 50 min in fluorescence recovery after photobleaching (FRAP) experiments (Fig. 1c, Supplementary Fig. 1a). These observations are similar for condensates formed across multiple Mg2+ concentrations, with slightly slower recovery occurring as Mg2+ concentrations increased to the right of the peak absorbance (Supplementary Methods, Supplementary Fig. 1b). Compared to some other protein-RNA systems which can recover within seconds to a few minutes for a similar size of bleached region46–48, polyP recovery in polyP-Mg2+ condensates is relatively slow.
Fig. 1. PolyP-Mg2+ coacervates exhibit reentrant phase transition and are dynamic.
a Phase boundary curve for polyP-Mg2+ coacervates as determined by the solution turbidity ([polyP] = 1 mg/mL, 50 mM HEPES-NaOH, pH 7.5). Individual points represent the mean of three replicates, while error bars represent the standard deviation. b Representative confocal fluorescence microscopy images of polyP-Mg2+ mixtures that correspond to 100 mM MgCl2 of the phase diagram 8 min after droplet induction. Images represent fusion of polyP-Mg2+ coacervates ([polyP] = 1 mg/mL, polyP-AF647 = 10% polyP, [Mg2+] = 100 mM, 50 mM HEPES-NaOH, pH 7.5; scale bar = 2 µm). A movie showing a larger field of view of droplet fusion is available (Supplementary Movie 1). c PolyP-Mg2+ coacervates reached around 75% recovery within 50 min in Fluorescence Recovery After Photobleaching (FRAP) experiments (dbleached ROI = 1.7 µm, ddroplets = 8.4–8.5 µm, n = 4). Points represent the time-binned averages of four independent runs and error bars represent the SD. Representative images from a single run showing recovery at select time points are inset (scale bar = 5 µm).
As would be predicted based on prior work, the absorbance data also reveal reentrant behavior with a reduction in scattering observed for Mg2+ concentrations above 100 mM. This rollover is similar to the behavior demonstrated previously for RNA-protein and other condensates35,45,49–51. This effect can be attributed to droplet dissolution past the charge-balance region around 100 mM Mg2+, where the surface interaction valences of smaller polyP species (single molecules or clusters) are quenched by excess Mg2+, thus terminating the network and preventing larger condensate formation. It is noteworthy that complete dissolution is observed at high Mg2+ concentration, indicating a lack of residual networking interactions in this reentrant region as observed in some other reentrant systems such as RNA polyA-Mg2+31,32. Furthermore, time-series imaging reveals the formation of dynamic vacuolar species during dissolution (Supplementary Fig. 1c, d, Supplementary Movie 2), similar to reported non-equilibrium dynamics of RNA-peptide complex coacervate systems45,52.
Overall, these experiments establish the fundamental characteristics of the polyP-Mg2+ system for this biologically relevant polyP size range. We show that even in the absence of other cellular factors likely to further modulate phase behavior, low mM Mg2+ concentrations are sufficient in vitro to drive the formation of condensates which demonstrate reentrant behavior at higher Mg2+ concentrations.
DNA interacts with polyP-Mg2+ droplets, forming shells that display reentrant behavior
We next studied the effects of the inclusion of circular double-stranded DNA in the system. Since the observed polyP granule embedding within the bacterial nucleoid may also involve other cellular factors, we aimed to test the intrinsic morphology and physical principles for this simplified DNA-polyP-Mg2+ system. Based on prior cellular and in vitro work30,50,53–57, we could envision several scenarios. These would include the partitioning of DNA into the polyP-Mg2+ droplets or the formation of a core-shell architecture, with Mg2+, in either case, potentially mediating the interaction of the two polyanions. Other possibilities include DNA exclusion from polyP-Mg2+ condensates or the formation of separate or partially interacting DNA-Mg2+ condensates. These latter scenarios where DNA forms condensate in the presence of Mg2+ are less likely given the lack of reported evidence of DNA condensation by divalent cations except in a limited set of conditions36,37,58,59.
As a starting point for understanding how DNA might behave in the presence of polyP-Mg2+ condensates, we used pUC19, a standard, circular 2.7 kb plasmid DNA, at a final concentration of 10 µg/mL, labeled with the intercalating dye YOYO-1. To facilitate droplet visualization and quantification, we first probed the system near the peak concentration of 100 mM Mg2+ in the polyP-Mg2+ phase transition curve. Experiments were carried out by pre-mixing the two polyanions followed by induction of phase separation by the addition of Mg2+.
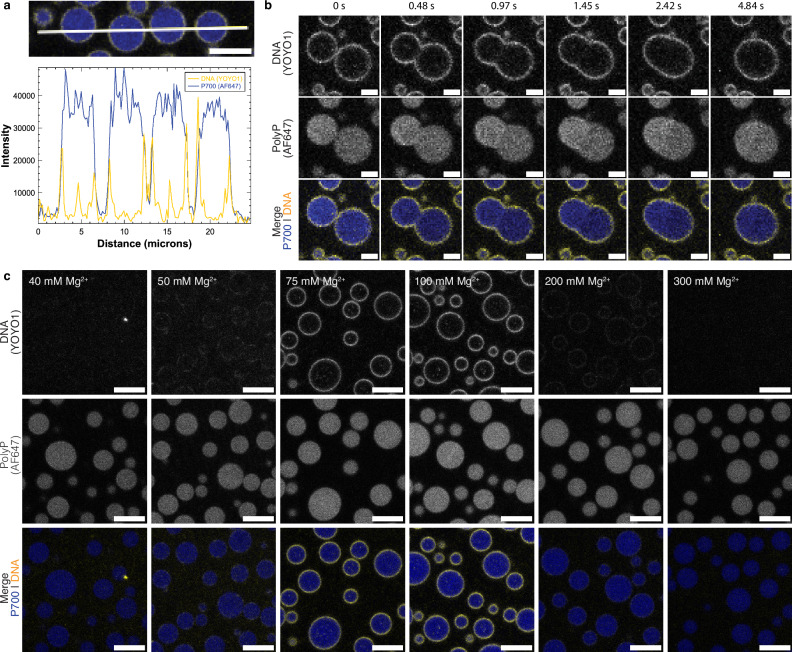
Upon induction of phase separation by the addition of Mg2+, we observed that pUC19 DNA forms a shell (Fig. 2a, yellow) associated with the surface of the polyP-Mg2+ droplets (blue). A 3D construction of confocal fluorescence microscopy images confirms the surface association of DNA across different planes (Supplementary Fig. 2a). To confirm that shell formation is not an artifact of the intercalating cationic dye, we demonstrated shell formation with covalently labeled DNAs. We first used 5′ Cy5 end-labeled pUC19 and were able to see faintly visible shells (Supplementary Fig. 2b, left). To increase the dye density and therefore signal, we also performed experiments with an ATTO488 end-labeled, 400-mer duplex DNA, which displayed shells (Supplementary Fig. 2b, right). These controls in the absence of YOYO-1 and with a significantly lower dye-labeling density indicate that DNA forms shells around the polyP-Mg2+ droplets.
Fig. 2. DNA interacts with the surface of polyP-Mg2+ coacervates and forms shells that exhibit reentrant behavior.
a Intensity profiles across polyP-Mg2+-DNA coacervate confocal image showing the surface localization of DNA [P700] = 1 mg/mL, P700-AF647 = 10% of the total polyP, 50 mM HEPES-NaOH, pH 7.5; scale bar = 5 µm; P700, blue; DNA (YOYO-1, 1 µM, yellow). b Confocal fluorescence microscopy images at different time points of polyP-Mg2+-DNA coacervate fusion (for conditions described in (a), scale bar = 2 µm). See Supplementary Fig. 2c for the full frame fusion and Supplementary Movie 3 for a wider field-of-view video. c Confocal fluorescence microscopy of polyP-Mg2+ coacervates and pUC19 (2.7 kb) plasmid under different MgCl2 conditions. DNA forms a shell on the surface of polyP-Mg2+ coacervates within a Mg2+ concentration range of 50–200 mM. For each Mg2+ concentration, N = 1. Three channels corresponding to Alexa Fluor 647 (P700), YOYO-1 (DNA), and the merge of these two channels are shown (10–12 min, scale bar = 5 μm).
A key question that arises from these observations is whether shell formation restricts the fusion of polyP-Mg2+ droplets. This question is especially relevant given our prior observations that in P. aeruginosa under nitrogen starvation conditions, initial coarsening of smaller granules results in fewer larger polyP granules that are transiently evenly spaced in the nucleoid but do not coalesce to a single larger droplet15,17,18,20–22. An examination of time-lapse images of the system shows rapid fusion of these droplets (Fig. 2b, Supplementary Fig. 2c, Supplementary Movie 3), suggesting that, under these conditions, pUC19 shells do not substantially restrict droplet fusion. Further quantification supports this idea. Looking at various relaxation events suggests that, once fusion is initiated, relaxation occurs on a few-second timescale and is similar for no DNA and pUC19 conditions when adjusting for approximate droplet size (Supplementary Methods, Supplementary Fig. 3, Supplementary Note 1). We also conducted FRAP on whole droplets in the presence and absence of DNA to test whether DNA shells significantly restrict polyP exchange between dense and dilute phases (Supplementary Methods). The recovery curves were similar for both cases, suggesting that DNA shells do not substantially prohibit exchange (Supplementary Fig. 4).
We next asked whether the non-monotonic phase behavior of the polyP-Mg2+ components also resulted in modulation of the DNA shell. Since polyP and DNA do not form droplets without Mg2+ under our conditions, we hypothesized that DNA interacts with positive charges (Mg2+) on the surface of polyP-Mg2+ droplets. Based on prior work on reentrant behavior in intramolecular and intermolecular DNA and RNA condensation processes mediated by polyvalent counterions45,51,60, we anticipated that there would be a charge inversion for the condensates in the region of the peak in polyP-Mg2+ phase separation (Fig. 1a), i.e., the surface of the droplets becomes negatively and positively charged in the regions to the left and right of the peak respectively. Therefore, a prediction from the charge-based DNA:polyP-Mg2+ droplet interaction model is that shell formation could be reduced in the lower Mg2+ concentration region.
To test this model, we carried out a series of imaging experiments, checking for DNA shell formation at different polyP-Mg2+ ratios. We observed that in keeping with the interaction model, shell formation is substantially reduced below 50 mM Mg2+ (Fig. 2c, Supplementary Fig. 2d). Interestingly, shell formation also is not observed above 200 mM Mg2+ (Fig. 2c, Supplementary Fig. 2d). We can rationalize this latter observation using the same mechanism as we discussed for reentrance in the polyP-Mg2+ system. At high Mg2+ concentrations, the charges on DNA molecules are screened by the excess Mg2+, thus reducing the propensity to interact with the droplet surfaces. Although the predominant DNA density appears uniform on the surface, we also observe puncta both on the surface at low Mg2+ where shells are less prominent, and occasionally within the condensates at Mg2+ concentrations where shells form (Fig. 2c).
Our results therefore show that pUC19 DNA forms shells at the surface of polyP-Mg2+ droplets within a concentration range around the maximum in the reentrant curve in Fig. 1a.
The condensate interface exhibits distinct morphologies as a function of DNA concentration and length
To determine the morphological features of the DNA shells on the surface of polyP-Mg2+ condensates, we turned to the higher resolution provided by cryo-electron tomography (cryo-ET). Cryo-ET is a particularly powerful and agnostic approach to determine the structural properties of interfaces at high resolution across a wide range of length scales. We probed how DNA concentration and length, properties that could affect the number of surface contacts, global orientation, and packing dynamics, affect the architecture of DNA at the interface. Because YOYO-1 can both introduce supercoiling61 and, under long incubation times, induce nicking62, the following experiments were performed in the absence of YOYO-1.
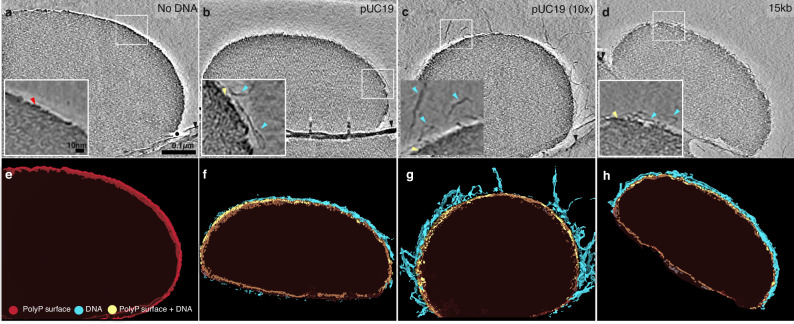
Representative tomographic slices of polyP condensates incubated with different types of DNA are shown in Fig. 3a–d, with the associated 3-dimensional renderings shown in Fig. 3e–h, respectively (refer to Supplementary Fig. 5a–f for corresponding low magnification images of grids). In the absence of DNA, the interface of polyP-Mg2+ condensates exhibits a dense edge (Fig. 3a, red arrow; e). We also observe a dense edge in the presence of DNA, which could be a combination of polyP-Mg2+ and DNA (Fig. 3b–d, yellow arrow). To represent this ambiguity the surface rendering displays this feature in yellow (Fig. 3f–h). A dense edge has been previously noted for polyP granules in vivo in Acetonema longum spores and is also visible in several other systems16,18,63. With 10 µg/mL pUC19 plasmid DNA (2.7 kb), we observe distinct filaments protruding from the surface compared to the no DNA case (Fig. 3a–d, Supplementary Fig. 5a, b). Condensates formed with 10-fold more DNA (100 µg/mL pUC19) exhibit protruding filaments that are both more numerous and extend further from the surface, with some filaments extending more than 100 nm from the surface (Fig. 3d, Supplementary Fig. 6c). In the presence of longer, circular DNA (15 kb) at 10 µg/mL, we observe filaments protruding a similar distance from the surface as with circular pUC19 (Fig. 3b, c, Supplementary Fig. 6d). Alternative views of the 3D renderings highlighting the different surface textures are available in Supplementary Fig. 7 and Supplementary Movies 4–7.
Fig. 3. Cryo-electron tomography shows topologies of different types of DNA on polyP condensates.
a–d Representative tomographic slices of polyP condensates incubated with different types of DNA. Red arrow highlights the dense edge of polyP, cyan arrows highlight DNA, yellow arrows highlight the dense edge+DNA surface, and the black arrow highlights the carbon hole (scale bar = 100 nm, inset scale bar = 10 nm). Representative slices were selected from multiple droplets and tomograms from a single experimental run (see “Methods” section for details). The phenotypes shown are consistently observed in multiple tomograms and droplets (see Supplementary Fig. 6 for additional examples). The entire dataset is deposited on EMDB under the following accession ID: EMPIAR-11701 (e–h) 3-dimensional renderings of tomograms shown in (a–d), respectively. The dense edge of polyP condensate is shown in red, the dense edge+DNA is shown in yellow, and DNAs are shown in cyan.
To determine the effect of DNA concentration and length on the thickness and density of the interface, we performed subtomogram averaging on thousands of randomly selected ~30 nm cubic regions spanning the interface (Supplementary Fig. 8a). We quantified the thickness of the dense edge by drawing eight x–y plane density profiles on the mid-section of the average maps perpendicular to the edge and averaging the thickness values (Supplementary Figs. 8b–g and 9). The measured thickness of the dense edge is 4.6 ± 0.7 nm in the absence of DNA, which is not significantly different upon the addition of 10 µg/mL circular pUC19 (Supplementary Fig. 8). We observe an additional outer layer of intermediate density between the background and the dense edge in the presence of 100 µg/mL pUC19 DNA (Supplementary Fig. 8d, cyan arrow) which we attribute to the protruding filaments. These findings are consistent with DNA adsorbing to the surface of polyP-Mg2+ condensates as a thin shell. The DNA shell’s packing architecture, including the length and density of filaments, depends on DNA concentration.
For reference, we also tested linearized pUC19 and 15 kb and found that for plasmids of both lengths, DNA tends to lie flat along the surface compared to the more ‘hairy’ circular forms and results in a smoother surface texture that is more difficult to decouple from the polyP-Mg2+ condensate surface (Supplementary Fig. 10, and Supplementary Fig. 6e, f, Supplementary Fig. 7m–q). In the case of a linearized 15 kb plasmid, the thickness of the dense edge increases (Supplementary Fig. 8h).
DNA concentration and length modulate the size of polyP-Mg2+ droplets
Our cryo-ET observations provide several key insights into the general structural features of the DNA shells and their dependencies on key DNA parameters, including DNA concentration and length. Given the known ability of adsorbed macromolecules to stabilize emulsions and colloids against fusion/aggregation64,65, we next returned to fluorescence imaging to test whether DNA shells can similarly influence polyP-Mg2+ condensate size distributions. This aspect is especially interesting given the transient organization of multiple non-fusing polyP granules in P. aeruginosa18. As such, here we probed the effects of DNA concentration and length on condensate size distributions. We were also interested in the effects of DNA form (circular vs linear, Supplementary Methods, Supplementary Fig. 11) and GC content (Supplementary Methods, Supplementary Fig. 12), however, in both cases, we were unable to resolve differences under the conditions tested.
We considered several mechanisms that could contribute to the dependence of droplet size on DNA concentration. First, in the case of a thin shell, the total maximum available DNA-polyP interfacial area should be a monotonic function of DNA concentration. Therefore, since the interfacial area of a given volume of polyP-Mg2+ condensate will have an inverse dependence on droplet size, higher DNA concentration should result in smaller droplets given that shell formation must overall be energetically favorable. Furthermore, our cryo-ET results revealed that increasing the concentration of pUC19 results in a brush-like morphology66 of DNA on the droplet surface, which could also result in slowing of droplet fusion and smaller droplets due to the physical/entropic barrier on the droplet surface inhibiting the initiation of fusion. Higher partitioning and packing of surface DNA at higher concentrations could similarly lead to reduced growth. Thus, thermodynamic and kinetic mechanisms could all result in reductions in droplet size as a function of increasing DNA concentration.
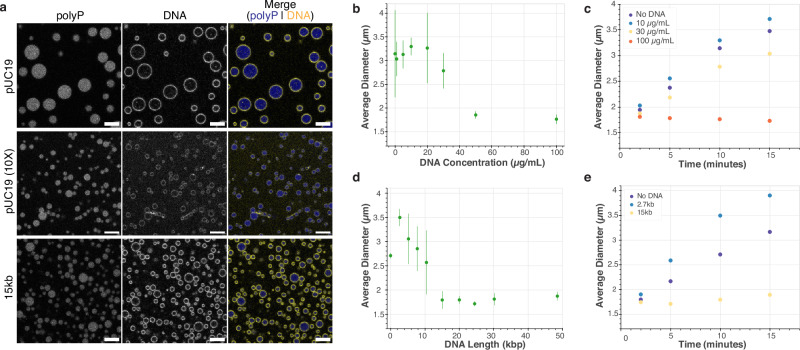
To test this idea, we performed widefield and confocal fluorescence imaging experiments using a series of DNA concentrations ranging from 0 to 100 µg/mL with the same polyP and Mg2+ conditions as previously used (Supplementary Figs. 13 and S14). Droplets decrease in size at higher concentrations of DNA (Fig. 4a (top/middle), 4b), and we also occasionally observe the appearance of rod-like filaments of the micrometer scale, which we did not analyze further (Supplementary Fig. 13 and S14).
Fig. 4. Effect of DNA concentration and length on polyP-Mg2+ size distribution and average droplet size.
a Representative confocal images of polyP-Mg2+ droplets given different DNA concentration (top & middle) and length (top & bottom) ([polyP] = 1 mg/mL with ~10% P700-AF647, [DNA] = 10 µg/mL or 100 µg/mL, [YOYO-1] = 1 µM, 50 mM HEPES, scale bar = 5 µm, n = 1). These representative images are selected from an expanded set of concentrations and lengths, which are available in Supplementary Figs 13, 14, 18, and 19. b Scatter plot showing the average of mean droplet size across three experiments with respect to varied DNA concentrations (error bars = SD of mean diameters of each experiment) ([polyP] = 1 mg/mL with ~10% P700-AF647, [DNA] = as shown, 50 mM HEPES, n = 3). c Scatter plot showing average droplet size as a function of time for three representative DNA concentrations (n = 3). d Scatter plot showing the average of mean droplet size across three experiments with respect to different DNA lengths (error bars = SD of mean diameters of each experiment). ([polyP] = 1 mg/mL with ~10% P700-AF647, [DNA] = 10 µg/mL, [YOYO-1] = 1 µM, 50 mM HEPES, n = 3) e Scatter plot showing average droplet size as a function of time for three representative DNA lengths (n = 3).
To probe the droplet size distribution quantitatively, we employed a MATLAB-based image analysis routine to analyze the widefield images (refer to the “Methods” section for more details, and also to Supplementary Fig. 15 for representative images of the segmentation step). We then plotted the average of the mean droplet size (Fig. 4b) for each replicate distribution as a single statistic to gain insights into our data. The full quantification of the droplet sizes as an empirical cumulative distribution function (ECDF) plot is available in the SI (Supplementary Fig. 16).
Consistent with the mechanisms discussed above, our analyses revealed that increasing the DNA concentration beyond 20 µg/mL indeed leads to a decrease in the droplet size (Fig. 4b) and a corresponding left shift of ECDF curves (Supplementary Fig. 16a). Figure 4c right panel shows the time evolution of the average droplet sizes for three representative DNA concentrations. While the average size of polyP-Mg2+ droplets for DNA concentrations 10 and 30 µg/mL grows with a net positive slope, the average detected droplet size of 100 µg/mL remains low (close to our detection threshold) with near-overlapping ECDF curves at the four time points studied (Fig. 4c and S17) indicating slowing of droplet size growth at high concentrations relative to lower DNA concentrations. Notably, once fusion starts, relaxation of fusing polyP-Mg2+ condensates in the presence of 100 µg/mL pUC19 occurs on a timescale of only a few seconds, similar to that of the no DNA conditions and 10 µg/mL pUC19 conditions (Supplementary Fig. 3). Assuming droplet growth is driven predominantly by fusion, our findings would suggest that DNA affects droplet growth in our in vitro system more through a reduction of fusion events rather than through slowing the coalescence and relaxation process.
We next asked whether DNA length can alter droplet size distributions even if the total base-pair concentration in solution remains constant. It is well known from the polymer physics field that polymer length can intrinsically affect phase separation propensity, often discussed in terms of polyvalency in the condensate literature67–70. Previous studies on DNA condensation as well as phase separation demonstrate DNA-length dependent properties37,71. In the present context, rearrangements of the surface-bound DNA may be increasingly slower as the length increases due to increased avidity, entanglement effects, or through differences in partitioning and surface coverage. Since DNA rearrangement could be important in droplet fusion kinetics, we hypothesized that shell formation with longer DNA could also affect droplet growth by inhibiting the initiation of polyP-Mg2+ condensate fusion.
To test this model, we probed the length-dependence of DNA on droplet formation with polyP-Mg2+, using a range of circular plasmid sizes: 2.7 kb (pUC19 used thus far), 5 kb, 8 kb, 10 kb, 15 kb, 20 kb, 24 kb, 30 kb and commercially available linear phage DNAs Lambda (49 kb) and T4 (166 kb), all at the same weight concentration of 10 μg/mL(refer to Table 1 for exact DNA lengths and additional details). We chose this range of DNA lengths to span a range from below to above the size of bacterial plectonemes (estimated at 10 kb based on EM, simulations, and gene expression microarray in E. coli and 15 kb based on Hi-C and modeling in C. crescentus)72,73. As with the concentration-based experiments, we used widefield fluorescence images coupled with MATLAB to quantify their condensate size distributions (Supplementary Fig. 18) and confocal imaging to confirm the presence of 3D shells (Fig. 4a (top/bottom), Supplementary Figs. 19–S20). The resulting average size and time-dependence data are shown in Fig. 4d, e (also see ECDFs, Supplementary Figs. 21 and S22).
Table 1.
Plasmids used in our study
| Plasmid (nomenclature in paper) | Plasmid name | Source | Size (bp) | Growth strain |
|---|---|---|---|---|
| 2.7 kb | pUC19 | NEB Catalog# N3041S | 2686 | DH5ɑ |
| 5 kb | pETM6 | Addgene #49795135 | 5203 | DH5ɑ |
| 8 kb | pLREX122 | This study | 7768 | DH5ɑ |
| 10 kb | pLREX185 | This study | 10,433 | DH5ɑ |
| 15 kb | pVG1 | Addgene #111444136 | 15,014 | DH5ɑ |
| 20 kb | pEMS1107 | Addgene #29036137 | 20,005 | DH5ɑ |
| 24 kb | pEBTet-SNAP-ALMS1 | Addgene #136828138 | 24,445 | DH5ɑ |
| 30 kb | pLD1 translation factors | Addgene #117760139 | 30,152 | BLR (F- ompT hsdSB(rB- mB-)gal dcm (DE3) Δ(srl-recA)306::Tn10 (TetR)) |
| Lambda DNA | Lambda DNA | NEB Catalog #N3011L | 48,502 | - |
| T4 DNA | T4 GT7 DNA | FUJIFILM Wako Chemicals USA Catalog #318-03971 | 166,000 | - |
| 5.5 kb (44% GC) | pLL346 | Gift from Lasker Lab | 5558 | DH5ɑ |
| 5.5 kb (53% GC) | pLREX240 | This study | 5515 | DH5ɑ |
| 5.5 kb (61% GC) | pOPTO328 | Gift from Lasker Lab | 5515 | DH5ɑ |
Our experiments revealed that increasing the DNA length in the range of 2.7–15 kb shifts the distribution of polyP-Mg2+ droplets to a smaller size (Fig. 4c; left-shifted ECDFs in Supplementary Fig. 21), also reflected in the time-dependence (Fig. 4e). The shift from larger to smaller droplets from 2.7 to 15 kb is also recapitulated in the absence of YOYO-1 (Supplementary Fig. 11). Despite the smaller size of polyP-Mg2+ droplets in the presence of 15 kb DNA, fusion is still observed (Supplementary Movie 8). Similar to what we observe in the higher DNA concentration system, aspect ratio-based fusion quantification demonstrates that condensates with 15 kb plasmids take a similar amount of time to relax to a spherical shape after fusion starts as those in no DNA and 2.7 kb DNA-polyP-Mg2+ systems (Supplementary Fig. 3). The similar relaxation timescale again suggests that the reduction in size is a product of reduced fusion overall rather than a function of the coalescence and relaxation time, provided that fusion is the predominant contributor to coarsening.
However, there were some deviations from this trend of decreasing droplet size with longer DNA. First, condensate size roughly levels off between 15 and 30 kb, which could be due to substantial growth arrest or because the distribution is clustered close to the resolution limit of our analysis. We also curiously observe that T4 DNA exhibits a wider and seemingly anomalous droplet size distribution, which is also reflected in a larger average droplet size and small but positive growth compared to the 15 and 30 kb range (Supplementary Fig. 21); the droplets also tend to cluster together, occasionally moving as a grouped unit (Supplementary Movie 9).
Overall, these length and concentration observations are particularly striking, given the substantial effects observed even at DNA phosphate concentrations 2–3 orders of magnitude lower than the polyP phosphate concentration, especially in the DNA-length regimes relevant to bacterial plectonemes.
As a further control for the potential generality of the DNA shell phenomenon, we tested polyP from a different commercial source which, using previously established gel methods74, are more narrowly dispersed with shorter and longer size distributions compared to our P700 samples, P130, and P300 (Supplementary Methods, Supplementary Fig. 23a)12. We observe DNA shells with both P130 and P300, using YOYO-1-labeled pUC19, and with an end-labeled, 400 bp PCR fragment (Supplementary Methods, Supplementary Fig. 23b). As a point of reference to another recent study of bacterial protein-mediated polyP condensates25, we also explored how P300 droplet size can be modulated by DNA concentration and length. Like that of the P700 system, the droplets with higher concentration pUC19 (50 µg/mL) are reduced in size compared to at 10 µg/mL, and the longer 15 kb plasmid similarly tends to have smaller droplets compared to the shorter pUC19 case (Supplementary Fig. 23c–e). While it is possible that the exact relationship between concentration and length differs in P300 compared to P700, our results demonstrate that these two parameters have the potential to also tune droplet size for polyP of different length distributions.
Discussion
Biomolecular condensates have emerged as a key structural feature of both eukaryotic and, more recently, bacterial chromatin25,75–83. Diverse partners can drive chromatin condensate formation, but the role of polyphosphate, a universal and ancient inorganic polymer, has been largely overlooked in chromatin biology. We hypothesize that polyP condensates are a fundamental feature of bacterial chromatin, and are likely important for chromatin structure and function in all three domains of life.
Empirically, magnesium has been shown to be the dominant cation in bacterial polyP condensates, and divalent cations can drive polyP condensate formation, as they do with RNA. Given the critical role of magnesium in nucleic acid structure and function and the longstanding observation that polyP condensates are embedded in the bacterial nucleoid in diverse species, in this study we have established a fundamental interaction between DNA and polyP mediated by magnesium that determines the properties of these condensates. We discovered that DNA associates with the surface of polyP-Mg2+ coacervates. This surface association both affects the morphology of the DNA and tunes the size of the condensates in a manner dependent on DNA properties.
PolyP-Mg2+ coacervation
In our study, we found that interactions between long-chain polyphosphate, relevant to bacterial physiology, and Mg2+ can result in the formation of coacervates. The formation of coacervates of longer-length polyP in the presence of Mg2+ is consistent with the larger body of polyP-Mg2+ coacervation work and more recently in the context of RNA interactions and condensation39,84. Our observed onset of condensation in this system (~10 mM Mg2+) is substantially lower than reported thresholds of Mg2+-induced phase separation of long polyU RNA and similar to that of short polyA RNA in the absence of crowding agents30–32. Additionally, while relatively rapid fusion resulted in spherical droplets, our FRAP results showed that diffusion and mixing within the resultant droplets is slow, qualitatively similar to previous observations in chromatin85 and the much slower internal rearrangement of polyrA in Mg2+-induced condensation32. Given these observations, it is worth noting that the condensates studied in this work could be considered as network fluids and as such, could exhibit viscoelastic characteristics33,67,70,86–88, an important direction for future work. Given the similarities of our system with other homotypic coacervates of RNA and divalent cations32,40, we predicted that the system would only result in coacervation in a window of relative polyP-Mg2+ concentrations around the charge-balance region. Our demonstration of precisely this type of reentrant behavior highlights the importance of charge-based interactions in mediating networking in these coacervates.
We also observe the presence of a dense edge in our cryo-ET images (Fig. 3a–d), which appears even in the absence of DNA. This is particularly interesting given that a dense edge has also been previously noted for polyP granules in vivo in Acetonema longum spores16. While the dense edge has been hypothesized to be the product of proteins gathering on the surface, it is interesting that a similar feature can be recapitulated in vitro in a system containing only polyP and Mg2+. We speculate that the dense edge may be an outcome of differential hydration of Mg2+ at the surface compared to the droplet interior, and could be similar to differences in hydration, ion concentration, and binding observed in polyP-Ca2+ systems38,89,90.
DNA association with Mg2+-polyP condensate surfaces
Our studies have also revealed that DNAs are preferentially recruited on the condensates’ surface while being relatively depleted from the condensate core. The association of DNA with the polyP-Mg2+ surface presumably arises from favorable interactions between the negatively charged phosphate groups on the backbone of DNA and Mg2+ at the surface of polyP-Mg2+ coacervates. Such a model would also be consistent with the differential hydration of Mg2+ inside and at the surface of polyP coacervates discussed in the previous section and could lead to the emergence of distinct surface properties relative to the internal condensate environment. A charge-based interaction is consistent with our observations of the reentrant nature of the DNA shells which form under a relatively narrow range of Mg2+ concentrations, where we expect both the surface to be positively charged/near-neutral and the divalent cation concentrations to be within a regime that does not screen charge-based interactions. While reentrant behavior has been observed in multiple systems in the biomolecular condensate field, as discussed above, as well as in earlier DNA condensation work, our work now extends the reentrance phenomenon to the phase behavior of charged polymer shells in multicomponent condensates.
While higher-order core-shell architectures have been observed both in cells and recapitulated in vitro30,46,53–56,91–95, there are some notable differences between these multiphase condensate systems and our own. First, in contrast with many previous studies with more comparable concentrations of the different biopolymers, we studied a region of concentration space where DNA phosphate concentrations were generally more than two orders of magnitude lower than those of polyP-Mg2+ (for the majority of experiments, ~15 μM DNA phosphate vs ~10 mM polyP phosphate and >10 mM Mg2+). Our results demonstrate that even much smaller relative concentrations of DNA can exert substantial control on certain properties of polyP-Mg2+ condensates which has potential implications for other cellular condensates where minor or undetected components could be important for biological regulation and function.
Another important contrast with many other described core-shell systems is the lack of DNA condensation in similar Mg2+ concentration regimes in the absence of polyP. Indeed, to the best of our knowledge, divalent cations (like Mg2+ and other alkaline earth metal ions) are not known to condense dsDNA in dilute, bulk solution alone and require additional special conditions like addition of PEG to induce DNA condensation (termed PSI-condensation) or change of solvent conditions (such as changes in dielectric constant)36,58,59. We note yet another contrast: surface confinement of DNA on polyP condensates in our system represents a morphology distinct from that of previously observed DNA condensation induced purely by the addition of polyvalent cations (charge ≧3) that leads to the formation of superstructures such as toroids96. On the other hand, similar to the previously discussed mechanistic understanding for multiphasic core-shell condensates53–56, it is likely that an overall reduction of the interfacial energetic cost is one driving force for DNA shell formation in our polyP-Mg2+-DNA system.
While we cannot rule out the possibility that the multicomponent system here is a form of the multiphase condensates described above, it is tempting to speculate that polyP-Mg2+ induces the adsorption and subsequent condensation of DNA on its surface. We note potentially related observations of adsorption and formation of shell-like structures in Pickering emulsions and some RNA-based condensates97. Such surface-induced adsorption and condensation would be consistent with previous work showing DNA adsorption/condensation on cationic and zwitterionic lipid surfaces58,98–102. Interestingly, for the studied zwitterionic systems, these surface-based interactions appear to be mediated by the divalent counterion Mg2+. Given the thin nature of the DNA shells observed in our work (Figs. 2 and 3), the presumed surface charge dependence of the interaction, and the notable absence of a separate DNA-Mg2+ dense phase, our multicomponent polyP-Mg2+-DNA system thus potentially represents a useful system for studying 2D-DNA condensation, adding to and complementing previously studied DNA-lipid systems.
DNA tuning of droplet growth
We also observed the potential for DNA properties of length and concentration to modulate droplet growth despite relatively low concentrations of DNA. We rationalized differences in droplet size from varied DNA concentration and length to originate from a combination of thermodynamic and kinetic driving forces. While thermodynamic arguments might explain some of the DNA morphology we observe at the interface and the emergence of shells (Figs. 2 and 3), many of our quantitative observations cannot be explained by thermodynamics alone and instead suggest that kinetic factors could play a significant role in controlling droplet growth.
For the DNA concentration dependence, simple consideration of energetic stabilization by shell formation would be consistent with higher DNA concentration correlating with smaller droplets, since the system would try to maximize the DNA-polyP interfacial area. However, that model assumes similar shell morphology for the different DNA concentrations. In contrast, we clearly observe a much more extended brush-like DNA morphology at the 10× DNA concentration, consistent with a physical barrier for fusion and growth. Overall, we therefore conclude that a combination of thermodynamic and kinetic contributions give rise to our observed concentration dependence of droplet size. As a related note, naturally occurring polymer brushes are important in attenuating interactions of large macromolecular assemblies in a variety of biological systems103,104. And polymer brushes have been harnessed in diverse engineering and industrial applications to prevent flocculation of particles105.
Similarly, we attribute trends observed from DNA-length variation to be a consequence of kinetic and thermodynamic contributions. Using the same simple thermodynamic consideration, it could be expected that maintaining the same base-pair concentration of DNA could result in minimal changes to droplet size given the constant potential for contacts with the polyP-Mg2+ condensate surface. However, we observe a clear dependence on DNA length. An interesting consideration is that kinetically driven differences such as ease of rearrangement, entanglement, or jamming67,98,106,107 that scale with DNA length could play a role. Properties that scale non-linearly with DNA length, including unequal numbers of effectively available contacts due to constraints in DNA bending from supercoiling, could also contribute. Moreover, the dependence could also be a product of several thermodynamically driven differences such as partitioning/binding affinity to the surface favoring longer DNA.
Open questions and functional implications of DNA shells
Our work opens questions regarding the material state of DNA shells and their influence on condensate dynamics. As noted above, the mobility and packing of the DNA shells likely impact various condensate dynamics, hence a detailed understanding of this aspect will be an important future direction. In addition, while our data suggest that the exchange of polyP across the condensate interface appears similar without and with pUC19 shells (Supplementary Fig. 4), it is possible that DNA affects exchange to a finer degree than we resolve if we take into account potential uneven shell morphology and limitations of our experimental methods. Therefore, interesting future directions will be a more extensive characterization of this effect across different DNA properties, as well as directly testing the related question of how shells influence Ostwald ripening.
There are also open questions regarding the extent to which other properties of DNA alter its interactions with polyP-Mg2+ condensates. Although we were not able to resolve differences in size distribution with GC content with our current dataset (Supplementary Fig. 12), we cannot rule out the possibility for sequence or GC content to potentially play a finer tuning role than our method allows us to detect. Patterning of high GC regions, and a wider range of GC content, among other sequence-related properties, could all be relevant and will be an interesting avenue for future study. Similarly, while we were not able to resolve differences in the size distribution of linearized and circular plasmids (Supplementary Fig. 11), there are many unexplored facets of DNA topology including positive supercoiling and the distribution of topoisomers. Our cryo-ET suggests that DNA topological state does influence the orientation of DNA on the condensate surface (Supplementary Fig. 10). Even if DNA topological state does not significantly alter droplet size distribution, it could still affect the way DNA interacts with polyP-Mg2+ condensate surfaces, an important topic for further study.
While our work focuses on how DNA shell architecture can affect polyP coacervation, it is also possible that the properties of DNA are altered in functionally important ways as a consequence of this association. Indeed surface association can dramatically alter both packing density and conformation of polymers. The former is well established with polymer brushes, where increased packing density drives polymer extension through repulsive interactions or entropic effects104,105. Interestingly, in the case of polyelectrolyte brushes, multivalent cations can oppose this effect, leading to more collapsed configurations104,105. Magnesium bridging interactions are thought to enable DNA to pack more densely when spatially confined, including confinement to a 2D surface59. Thus packing density and divalent cation partitioning at the interface of polyP condensates may dynamically tune DNA compaction. Given that polyP synthesis is upregulated during growth arrest, condensate formation may be a mechanism to regulate local DNA compaction. Additionally, our observation of reentrance in shell formation suggests a potential avenue for cellular regulation, as has been invoked in the case of RNA-protein reentrant behavior49,50,108. Lastly, since DNA supercoiling can affect many processes, notably transcription, and DNA adsorption of charged surfaces can alter supercoiling109–111, it is intriguing to speculate that in vivo interaction with polyP granules could module DNA supercoiling and associated functions locally and more globally112.
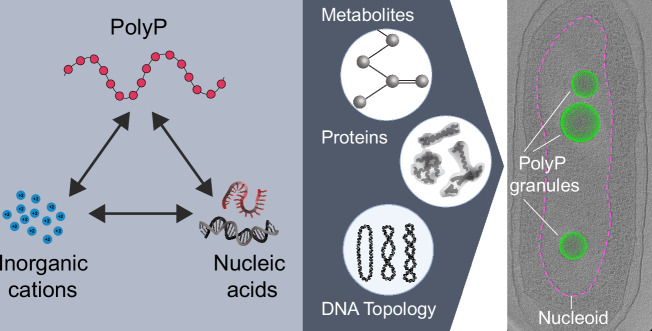
In this work, we have explored the surprisingly complex and tunable Mg2+-mediated condensation behavior of two polyanions with broad relevance in biology and other fields (Fig. 5). How this system plays out in a biological context merits further exploration, particularly balancing considerations of lower free Mg2+ concentrations observed in cells (1–2 mM)113 and the potential impacts of compensatory networking species, including proteins, metabolites, and other metals in vivo. In particular, chromatin binding proteins participate in mediating the interaction between DNA and polyP, as has been shown for the nucleoid-associated protein (NAP), Hfq25. Such interactions may act to bring specific DNA loci to the surface and further tune the conformational state of the DNA. NAPs may also modulate the partition of DNA between the surface and the interior, change the properties of the condensates, and provide additional interactions that substitute for and compete with interactions with cations. For example, the histone H1-like protein AlgP in P. aeruginosa, which has a +55 charge at neutral pH, localizes to the granules and alters their consolidation dynamics26. Thus, from a biophysical perspective, it would be interesting to expand our minimal system to include both chromatin proteins such as AlgP and other well-represented biological polyanions, specifically single-stranded RNA and DNA of varied topological states (Fig. 5). This future direction is particularly relevant given that these polyanions are widely represented in cellular condensates, including ones involved in transcription and RNA regulation97,114,115. Additionally, such single-stranded systems can add a more complex conformational landscape than duplex DNA, another interesting feature with potential broad biological, chemical, and prebiotic relevance.
Fig. 5. A potential framework for polyP-chromatin interactions.
In this study, we have developed a three-component polyP-Mg2+-DNA system (interactions represented by black arrows) which is a fundamental physicochemical interaction unit underlying the functional coupling between polyP granules and chromatin in cells. Our results highlight the tunable nature of a polyP-Mg2+-DNA system, showing that in vitro DNA interacts with and forms reentrant shells around polyP-Mg2+ condensates in the absence of DNA protein binding partners and modulates condensate size in a DNA-length- and concentration-dependent manner. Future in vitro and in vivo studies building on this framework to include cationic organic metabolites, relevant proteins, such as DNA binding proteins known to associate with polyP (Hfq and AlgP, for example), and DNA topology, are needed to understand how polyP granules affect chromatin structure and function in cells (gray arrow). Right: Cryo-ET of nitrogen-starved P. aeruginosa cells with nucleoid region (ribosome depleted) delineated with dashed magenta line, polyphosphate granules shown as green spheres (image: Fig. 5 adapted from Racki et al.18).
Methods
Reagents and stocks
Long-chain polyphosphate P700 was obtained from Kerafast, Inc. (EUI002). This polyphosphate is heterogeneous in size, with approximate polymer lengths ranging from ~200 to 1300 phosphate units; the modal size is about 700 phosphate units. We prepared 100 mg/mL stocks of P700 in water and stored them at −80 °C for long-term storage. The 100 mg/mL P700 stocks were used to further prepare sub-stocks of P700 at 10 mg/mL which were stored at −20 °C. These sub-stocks were used for experiments. Polyphosphate P300 was obtained from RegeneTiss, Inc. (Japan) and suspended in water at a concentration of 1 M polyP (Pi units; 100 mg/mL). We similarly aliquoted 100 mg/mL stocks of P300, stored them at −80 °C for long-term storage, and prepared sub-stocks of P300 at 10 mg/mL stored at −20 °C for experimental use. Polyphosphate P130 was obtained from RegeneTiss, Inc. and suspended in water at a concentration of 0.5 M polyP (Pi units; 50 mg/mL). Magnesium chloride was obtained in dried form (M9272-100) as well as 1 M MgCl2 solution (M1028-100) from Sigma. HEPES solid powder (H3375-100) was obtained from and 1 M stock was prepared in deionized water with the pH adjusted to 7.5 by the addition of 10 N NaOH (306576-100). The stock was stored at 4 °C for long-term storage. Aliquots of DNA labeling dye YOYO-1 (ThermoFisher, Y3601) were stored at −20 °C.
PolyP labeling
We adopted a previously developed polyP end labeling protocol with minor modifications116,117. Briefly, a reaction of P700 with EDAC and AF647 cadaverine (Sigma, A30679) was set up in MOPS buffer, pH 8.0 in the dark at 37 °C in a 1.5 mL Eppendorf tube. The final concentration of P700, EDAC, AF647 cadaverine, and the buffer in the reaction mixture was 100 μM (defined in terms of phosphate ends), 150 mM, 2 mM (20-fold excess), and 100 mM MOPS, pH 8.0 respectively. The Eppendorf tube was agitated occasionally (every 10-15 min). At the end of 1 h incubation at 37 °C, the reaction was stopped by placing the Eppendorf tube on ice and centrifuged briefly to remove any condensation from the top of the tube. Next, excess dye removal was carried out using spin desalting columns. To remove excess dye and buffer exchange (into water), we employed three consecutive 0.5 mL Zeba™ Spin Desalting Columns (7 K MWCO) and followed the manufacturer’s guidelines.
DNA plasmid preparation
To cover a range of DNA sizes, we used plasmids that were in our laboratory as well as commercially available DNA like Lambda-DNA and T4. The in-house plasmid preparation was carried out following the manufacturer’s protocol (Qiagen Midi-kits) and eluted in deionized water. Lambda and T4 DNA were dialyzed from the TE buffer into deionized water using the Pur-A-Lyzer Mini Dialysis Kit (6–8 K MWCO, Sigma PURN60010). The supercoiling state for these plasmids was quantified using gel quantification and can be found in Supplementary Figs. 24–27. The DNA stocks were maintained at −20 °C and thawed on ice prior to the experiments. The plasmids used for Cryo-ET were purified using phenol-chloroform extraction118 and were not rechecked by gel. All plasmids used in this study are available upon request from the corresponding authors. Plasmid information and primer sequences are available in Supplementary Methods, Table 1, and Supplementary Table 1 and 2.
Cy5 end labeling of DNA
Plasmid pUC19 was linearized by using restriction enzymes HindIII (NEB) and XbaI (NEB), purified using the NEB miniprep kit and ligated with a Cy5-labeled oligo duplex. Briefly, a 15-fold excess of Cy5 labeled duplex (pRRC11_56bp_Cy5; /5Cy5/acggccagtgaattcgagctcggtacgatcctctagagtcgacctgcaggcatgca annealed to 5′ phosphorylated primer pRRC10; /5Phos/agcttgcatgcctgcaggtcgactctagaggatcgtaccgagctcgaattcactggccgt) was ligated to linearized pUC19 using T4 ligase in an overnight ligation reaction at room temperature. The excess oligos were removed from ligated DNA using CHROMA SPIN columns and purified DNA was used directly for microscopy experiments.
Absorbance measurements of polyP-Mg2+
Absorbance measurements were carried out with unlabeled polyP. Sample absorbance was measured 15–20 s after droplet induction, with absorbance reported at 350 nm (Nanodrop). To ensure proper mixing, the solution was pipetted up and down 3–4 times before measurement on the Nanodrop 2000/2000c instrument and its software, NanoDrop 200c (1.4.1). Final concentrations: polyP: 1 mg/mL (unlabeled), 50 mM HEPES-NaOH, pH 7.5, MgCl2: 0–1000 mM. The time of induction of droplets by the addition of MgCl2 was used as a reference of t = 0 min for all of our experiments.
Sample preparation
PolyP-Mg2+ condensates with DNA of different lengths: Unlabeled P700 and P700-AF647 were thawed from −80 °C on ice prior to each experiment. A 10× master mix was prepared by adding 100 mg/mL of unlabeled P700 and purified AF647-labeled P700 (termed 10× PolyP Master Mix henceforth). DNA stocks were removed from −20 °C and allowed to thaw on ice at room temperature prior to use in the experiments. Buffer (HEPES-NaOH pH 7.5) was added to DNA in a PCR tube followed by incubation with YOYO-1 dye for 7–8 min. After incubation of DNA with YOYO-1, 10X PolyP Master Mix was added to this solution and droplet induction carried out by mixing an equal volume of appropriate 2× MgCl2 solution. Typically 3–4 fields of view were acquired per time point (t = 2, 5, 10, and 15 min) for three experiments carried out on different days using widefield microscopy. To ensure proper mixing, the solution was pipetted up and down 3–4 times before being introduced to the glass chamber for observation under the microscope (confocal/widefield). Final concentrations: PolyP: 1 mg/mL unlabeled, with ~10% labeled P700-AF647, 50 mM HEPES-NaOH, pH 7.5, MgCl2: 100 mM, DNA concentration: 10 μg/mL, YOYO-1: 1 μM. Note: the control experiment for the ‘No DNA’ case had DNA replaced with water and had a final [YOYO-1] = 1 μM in the solution. All droplets were observed at room temperature. The time at which the MgCl2 solution was added to induce droplet formation was used as the t = 0 min reference in all our studies.
PolyP-Mg2+ condensates with different DNA concentrations: Concentrated pUC19 stock was removed from −20 °C and thawed on ice prior to the experiment. The concentrated stock was then used to prepare dilutions of DNA stocks for each experiment. Buffer (HEPES-NaOH pH 7.5) was added to thawed DNA in a PCR tube followed by incubation for 5–7 min at room temperature. 10× polyP master mix was added to the DNA-buffer solution and droplet induction was carried out by mixing MgCl2 solution as noted previously. Typically 3–4 fields of view were acquired per time point (t = 2, 5, 10, and 15 min) for three experiments carried out on different days using widefield microscopy. To ensure proper mixing, the solution was pipetted up and down 3–4 times before being introduced to the glass chamber for observation under the microscope (confocal/widefield). Final concentrations: polyP: 1 mg/mL unlabeled, with ~10% labeled P700-AF647, 50 mM HEPES-NaOH, pH 7.5, MgCl2: 100 mM, DNA: 0-100 μg/mL. Note: No YOYO-1 was added in these experiments.
Sample chamber surface coating
The surface of Lab-Tek chambers (8-well, ThermoSci 155409) was coated using Tween 20 (Sigma T2700). Each well received 300 μL 10% Tween 20 and incubated for 35–38 min. The Tween 20 was then removed, and each well was washed with 600 μL deionized water. This was repeated for a total of 6 washes. The chamber was allowed to dry completely, and covered, in a vacuum filtration apparatus overnight. These Tween-coated Lab-Tek chambers were used for all widefield and confocal microscopy imaging of polyP condensates.
Confocal microscopy
Confocal images were recorded on a Zeiss LSM 780 laser scanning confocal microscope using the Zeiss software, ZEN 2011 SP7 FP3 (black) (v 14.01.207.201). Samples were imaged at room temperature using a 100× oil immersion objective (Plan-Apochromat 100×/ NA 1.40 Oil DIC M27) at a 16 bit depth with pixel size between 0.17 and 0.08 μm. DNA, through YOYO-1 labeling, was imaged using an Argon laser set at 20% laser power, which excited at 458 nm. The detection range for the YOYO-1 channel was set from 487 to 561 nm. Detector gain was adjusted to 800 and an offset of 450 was applied to reduce undersaturated pixels. PolyP was detected through P700 labeled with Alexa Fluor 647. A HeNe laser set at 40% laser power was applied, exciting at 633 nm. The detection range was set to 637–755 nm with a gain of 800 and an offset of 300. The imaging settings were held constant for all confocal images, except for the polyP-Mg2+-only images and movies used in Fig. 1 and Supplementary Movie 1, where the intensity of the HeNe 633 nm laser at the same detection range was set to 5% and the pinhole for the singular laser was adjusted to 105.5 (or 1 AU).
Z-stacks were collected at 2, 5, 10, and 15 min for samples at different locations for each time point. The frames were separated in z by 0.37 μm, except when otherwise noted. For movies acquired through confocal imaging, frames were collected with no fixed delay resulting in a temporal frame separation of ~484 ms unless otherwise noted.
Images were imported into FIJI/ImageJ(v 2.9.0/1.53t)119 where timestamps and scale bars were added. Some frames were cropped to highlight particular features (e.g., single droplet fusion) or for scaling. No other corrections to the images (e.g. brightness and contrast) were made for all non-FRAP images. Orthoviews and 3D orthosliced views were generated using Imaris Software (RRID:SCR_007370).
Fluorescence recovery after photobleaching (FRAP)
FRAP experiments of polyP-Mg2+ condensates were conducted using the Zeiss LSM 780 laser scanning confocal microscope conditions as noted above. Partial Droplet FRAP (Fig. 1 and Supplementary Fig. 1) is described below. See Supplementary Methods for whole droplet FRAP (Supplementary Fig. 4) methods.
Samples were prepared by adding an equal volume of MgCl2 solution to P700 labeled with ~10% P700-AF647 in HEPES buffer such that final concentrations were 1 mg/mL polyP, 100 mM MgCl2, 50 mM HEPES, pH 7.5. Condensates were allowed to coalesce and fuse for 35–45 min, after which a condensate with a diameter of around 8.5 μm was selected. The offset for the z-plane was calibrated for reflection autofocus.
Each experimental run collected images at three time points before subsequently initiating a bleaching protocol. Bleaching consisted of two rounds of 15 iterative pulses over a circular region at the center of the droplet with a diameter of 1.6 μm at 100% HeNe laser power set to a reduced scan speed (pixel dwell time: 12 μs). Following bleaching, images were collected in 20 s intervals for 52 min with reflection autofocus being applied every 15 scans or roughly every 5 min.
To correct for drift in the xy dimension over the 52 min, images were processed in FIJI where the StackReg plugin120 translation transformation was applied to a cropped frame of the bleached droplet. A circular region equivalent to the bleached ROI size was placed at the bleaching area and measured using FIJI’s measure function. One ROI equivalent in size and shape to the bleached ROI was used as a reference for photobleaching in condensates of around the same size as the bleached condensate and was measured in FIJI. Reference images were then similarly cropped and aligned using the StackReg plugin translation transformation. Time was adjusted to be zero immediately after the bleach by subtracting the time of the fifth scan from all times.
Data from the transformed bleached ROI corresponding to different time points were double normalized following the equation:
| 1 |
where is the average pixel intensity of the bleached ROI at time t, is the average of the three pre-bleach ROI mean pixel intensity, and and are the corresponding averages for the two reference ROIs.
Widefield microscopy
Microscopy images for image analysis were collected using a Nikon Ti2-E inverted microscope with a perfect focusing system (PFS) and a 100× oil immersion objective (Plan-Apochromat phase contrast, N.A. 1.45) at a 16 bit depth with a pixel size of ~0.11 μm. For brightfield, a white LED, and for fluorescence, the Spectra X Light Engine with a 470 nm LED (Lumencor) were used as illumination sources. The camera used for imaging was Prime 95B sCMOS (Photometrics). Image acquisition was controlled using NIS-Elements AR (5.21.01). The following parameters were typically used: For phase contrast: 10% light intensity, 100 ms exposure time, gain = 1.0. For YOYO-1 imaging: 5% light intensity from the 470 nm LED, 30–100 ms exposure time, and a GFP filter cube (466/40 nm excitation filter, 525/50 nm emission filter, 495 nm dichroic mirror, Semrock), gain = 1.0. For AF647 imaging: 10% light intensity from the 640 nm LED, 30–100 ms exposure time, and a quad LED-DA/FI/TR/Cy5-A filter cube(DAPI / FITC / TRITC / Cy5 - Full Multiband Quad Lumencor C19446). For Cy5 imaging: settings similar to AF647 imaging, with the exception of 50% light intensity.
Representative widefield images in the SI were processed using FIJI. Adjustments were made to brightness/contrast by setting the minimum and maximum intensity value to the overall observed min and max values based on the set’s histograms and applying that range equally to all comparable figures. For exact values, please see Supplementary Methods for details.
Size quantification
Images of condensates from different fields of view and experimental conditions at time points corresponding to t = 2, 5, 10, and 15 min were acquired by widefield microscopy as described above. The channel corresponding to 640 nm (P700-AF647) was used for segmentation and droplet size quantification. Custom MATLAB scripts were used for image analysis. Briefly, pre-processing was performed using in-built Matlab function imadjust and imclearborder. The function imadjust maps the intensity values in grayscale images to increase the contrast of the output images and the function imclearborder was used to exclude the droplets at the edge in any given field. The MATLAB function imfindcircles, which employs the circular Hough transform, was used to find circles in the images. Given the limited accuracy of imfindcircles when the value of radius (or rmin) is less than or equal to 5, a default rmin of 6 was used for all of our analysis. Note: the use of rmin sets a minimum radius of droplet detection as 0.66 μm (or diameter 1.32 μm). A default value of parameters rmax = 90 and sensitivity = 0.85 were used for imfindcircles and adjusted as needed for each field of view to capture the most accurate size distribution using manual visual inspection. The codes were able to accurately capture size distribution for larger sizes; we would, however, like to note that the codes were not able to capture droplets with sizes less than rmin 0.66 μm and sets a lower limit for such analysis.
Software
Image processing was carried out using Matlab (R_2023a). Data processing and analysis were performed in Python (CPython 3.10.11, IPython 8.12.0) with NumPy version 1.24.3, Pandas version 1.5.3, Scikit Image v0.23.3121, SciPy v1.11.1122, MatPlotLib v3.7.1123,124, and iqplot v0.3.3125 using Jupyter Notebook (Jupyerlab version 3.6.3). Ortho slice images were generated using Imaris (v10.1), Oxford Instruments Group (Abingdon, UK). Averages in Fig. 4 were calculated from means of three different experiments and the error bar denotes the standard deviation between the experiments using .mean and .std methods of Pandas DataFrame respectively. Data were plotted with Prism version 10.2.3 or Bokeh version 3.1.1126 and the figures were assembled with Adobe Illustrator.
Cryo-ET sample preparation
200 μL of 10 nm gold fiducial beads (Aurion) were centrifuged with a benchtop centrifuge for 20 min at 15,000 RPM and buffer exchanged with HEPES-NaOH buffer, pH 7.5. This procedure was repeated twice, and the beads were resuspended in a final volume of 100 μL of HEPES-NaOH buffer, pH 7.5. Afterward, 2 μL of gold fiducial beads were added to 4 μL of each sample. The droplet samples for Cryo-ET observation were prepared as described above, but with the following differences: DNA was incubated with HEPES and gold beads for 7 min, followed by the addition of unlabeled P700. Droplets were induced by the addition of Mg2+ and spotted on the grids after one minute of droplet induction. Water was used as a control for the no DNA case. Final concentrations: 1 mg/mL P700 (unlabeled), ~50 mM HEPES- NaOH, Mg2+: 100 mM, DNA: 0–100 μg/mL.
Quantifoil R2/1 copper 200-mesh grids were glow-discharged with a Pelco easiGlow using the following parameters: set-15 mA, glow-25 s, and hold-10 s. 4 μL of the samples containing the fiducial beads were deposited onto the grid and plunge-frozen into a propane/ethane mixture using a Vitrobot (Thermo Fisher Scientific) with the following parameters: 2.5 s blot time, 0 s wait time, 0.5 s drain time, 0 blot force, and 1 blot total.
Cryo-ET data collection and reconstruction
Cryo-ET samples were imaged using a 300 keV transmission electron microscope, Titan Krios (Thermo Fisher Scientific), equipped with a Gatan K3 direct electron detector and an energy filter (slit width of 20 eV was used). The data collection package SerialEM (4.0)127 was used to run PACEtomo (1.5)128 for tilt series acquisition. 35 image stacks were collected from −51° to +51° for each tilt series with an increment of 3°, a target defocus of −6 μm, a pixel size of 1.67 Å/pixel, and a total dose of approximately 100 e−/Å2. Each stack contained 10 frames, which were aligned using MotionCor2 (1.6.3)129 and then assembled into the drift-corrected stack using IMOD (4.11). The drift-corrected stacks were aligned using fiducial markers and reconstructed by IMOD130.
Subtomogram averaging
The subtomogram averaging package I3131 (version 0.9.9.3) was used to average the condensate edges. For each tomogram, the coordinate of the center of the condensate and multiple coordinates of the condensate edges were manually selected: polyP (2231 particles), polyP + pUC19 (1847 particles), polyP + pUC19 (10x) (1791 particles), polyP + 15 kb (1726 particles), polyP + linear pUC19 (1769 particles), polyP + linear 15 kb (1191 particles). An in-house script was used to calculate the Euler angles to orient particles in a consistent orientation. Subtomogram averaging was performed using bin4 particles reconstructed in the Weighted Back-Projection (WBP) method. The “graph” function in IMOD was used to generate density profiles.
3D segmentation and visualization of Cryo-ET data
The representative tomograms shown in the figure panels have been denoised by IsoNet (0.2)132. 3D segmentations were generated using Dragonfly (2022.2) Deep Learning software (Comet Technologies Canada Inc, previously Object Research Systems)133. A 2D U-Net model was trained on an individual tomogram using hand-segmented frames of the corresponding tomogram. The model was then applied to the tomogram to generate a full 3D segmentation of the tomogram and then manually corrected. This process was repeated for each tomogram shown in Fig. 3, Supplementary Figs. 7 and 10. The model was trained iteratively to distinguish the polyP interior, the dense edge of the condensate, the extruding DNA, and the background. Due to an inability to fully distinguish the dense edge and tightly wound DNA, the dense edge feature was depicted in yellow as shown in Fig. 3f–h, Supplementary Figs. 7 and 10, and Supplementary Movies 5–7. Videos and 3D rendering images shown in figure panels were generated using UCSF ChimeraX (1.6.1)134.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We gratefully acknowledge support from the US NIH (NIGMS Grant R35 GM130375 to A.A.D., Grant DP2-GM-739-140918 to L.R.R, as well as S10OD032467 to Scripps Research for equipment), Scripps Research start-up funds (to D.P. and L.R.R.), and a Postdoctoral Fellowship from the American Heart Association (Award #903967 to R.C.). D.A.G is supported by the Pew Scholars Program. We thank Anthony Milin and Ya-Ting Chang for their help with preliminary studies, Polina Cobb for making plasmid pLREX185, and Keren Lasker’s lab for the gift of plasmids pLL346 and pOPTO328. We thank Toshikazu Shiba (RegeneTiss, Japan) for the gift of P300. We would also like to thank Megan Bergkessel, Keren Lasker, and Samrat Mukhopadhyay for insightful feedback on this work, and The Scripps Research Institute Core Microscopy Facility for the use of confocal microscopy instrumentation and image analysis software.
Author contributions
Conceived the project and designed research, R.C., J.K.A.T., A.A.D., and L.R.R.; Performed Research, R.C., J.K.A.T., T. Boyd, N.H.T., T. Bai, and D.P.; Contributed reagents and/or Analytical tools, R.C., J.K.A.T., D.A.G., and N.H.T.; Analyzed Data, R.C., J.K.A.T., T. Boyd, N.H.T., T. Bai, and D.P.; Managed the project, A.A.D. and L.R.R.; Wrote the manuscript, R.C., J.K.A.T., A.A.D., and L.R.R.; Edited the manuscript, all authors. Funding acquisition, R.C., D.A.G., D.P., A.A.D., and L.R.R.
Peer review
Peer review information
Nature Communications thanks Anthony Vecchiarelli and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Fluorescence microscopy images used in this are available from the Zenodo repository (Zenodo record 13324261). Representative tomograms and raw cryo-ET data are available at the EMDB (the Electron Microscopy Data Bank), under the following accession IDs: EMD-42152, EMD-42153, EMD-42154, EMD-42155, and EMPIAR-11701. Source data are provided with this paper.
Code availability
Custom codes used for analysis in this study are available on GitHub, as follows. Aspect Ratio code: https://github.com/Deniz-Lab/condensate-fusion-aspect-ratio. FindCircles code: https://github.com/RackiLab/Chawla_and_Tom_2024_etal.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ravi Chawla, Jenna K. A. Tom.
Contributor Information
Ashok A. Deniz, Email: deniz@scripps.edu
Lisa R. Racki, Email: lracki@scripps.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53469-x.
References
- 1.Rao, N. N., Gómez-García, M. R. & Kornberg, A. Inorganic polyphosphate: essential for growth and survival. Annu. Rev. Biochem.78, 605–647 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Gray, M. J. & Jakob, U. Oxidative stress protection by polyphosphate-new roles for an old player. Curr. Opin. Microbiol.24, 1–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowlin, M. Q. & Gray, M. J. Inorganic polyphosphate in host and microbe biology. Trends Microbiol.29, 1013–1023 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denoncourt, A. & Downey, M. Model systems for studying polyphosphate biology: a focus on microorganisms. Curr. Genet.67, 331–346 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Baker, C. J., Smith, S. A. & Morrissey, J. H. Polyphosphate in thrombosis, hemostasis, and inflammation. Res Pract. Thromb. Haemost.3, 18–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desfougères, Y., Saiardi, A. & Azevedo, C. Inorganic polyphosphate in mammals: where’s Wally? Biochem. Soc. Trans.48, 95–101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lichko, L. P., Kulakovskaya, T. V. & Kulaev, I. S. Inorganic polyphosphate and exopolyphosphatase in the nuclei of Saccharomyces cerevisiae: dependence on the growth phase and inactivation of the PPX1 and PPN1 genes. Yeast23, 735–740 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Azevedo, C., Livermore, T. & Saiardi, A. Protein polyphosphorylation of lysine residues by inorganic polyphosphate. Mol. Cell58, 71–82 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Bru, S. et al. Polyphosphate is involved in cell cycle progression and genomic stability in Saccharomyces cerevisiae. Mol. Microbiol.101, 367–380 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Pilatus, U., Mayer, A. & Hildebrandt, A. Nuclear polyphosphate as a possible source of energy during the sporulation of Physarum polycephalum. Arch. Biochem. Biophys.275, 215–223 (1989). [DOI] [PubMed] [Google Scholar]
- 11.Negreiros, R. S. et al. Inorganic polyphosphate interacts with nucleolar and glycosomal proteins in trypanosomatids. Mol. Microbiol.110, 973–994 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bondy-Chorney, E. et al. A broad response to intracellular long-chain polyphosphate in human cells. Cell Rep.33, 108318 (2020). [DOI] [PubMed]
- 13.Jimenez-Nuñez, M. D. et al. Myeloma cells contain high levels of inorganic polyphosphate which is associated with nucleolar transcription. Haematologica97, 1264–1271 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie, L. et al. Accumulation of nucleolar inorganic polyphosphate is a cellular response to cisplatin-induced apoptosis. Front. Oncol.9, 1410 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henry, J. T. & Crosson, S. Chromosome replication and segregation govern the biogenesis and inheritance of inorganic polyphosphate granules. Mol. Biol. Cell24, 3177–3186 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tocheva, E. I. et al. Polyphosphate storage during sporulation in the gram-negative bacterium Acetonema longum. J. Bacteriol.195, 3940–3946 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raschdorf, O., Plitzko, J. M., Schüler, D. & Müller, F. D. A tailored galK counterselection system for efficient markerless gene deletion and chromosomal tagging in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol.80, 4323–4330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Racki, L. R. et al. Polyphosphate granule biogenesis is temporally and functionally tied to cell cycle exit during starvation in Pseudomonas aeruginosa. PNAS114, E2440–E2449 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frank, C., Pfeiffer, D., Aktas, M. & Jendrossek, D. Migration of polyphosphate granules in Agrobacterium tumefaciens. Microb. Physiol.32, 71–82 (2022). [DOI] [PubMed] [Google Scholar]
- 20.Voelz, H., Voelz, U. & Ortigoza, R. O. The polyphosphate overplus phenomenon in Myxococcus xanthus and its influence on the architecture of the cell. Arch. Mikrobiol.53, 371–388 (1966). [DOI] [PubMed] [Google Scholar]
- 21.Murata, K., Hagiwara, S., Kimori, Y. & Kaneko, Y. Ultrastructure of compacted DNA in cyanobacteria by high-voltage cryo-electron tomography. Sci. Rep.6, 34934 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosigkeit, H., Kneißle, L., Obruča, S. & Jendrossek, D. The multiple roles of polyphosphate in Ralstonia eutropha and other bacteria. Micro. Physiol.31, 163–177 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Boutte, C. C., Henry, J. T. & Crosson, S. ppGpp and polyphosphate modulate cell cycle progression in Caulobacter crescentus. J. Bacteriol.194, 28–35 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gross, M. H. & Konieczny, I. Polyphosphate induces the proteolysis of ADP-bound fraction of initiator to inhibit DNA replication initiation upon stress in Escherichia coli. Nucleic Acids Res.48, 5457–5466 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beaufay, F. et al. Polyphosphate drives bacterial heterochromatin formation. Sci. Adv.7, eabk0233 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chawla, R., Klupt, S., Patsalo, V., Williamson, J. R. & Racki, L. R. The histone H1-like protein AlgP facilitates even spacing of polyphosphate granules in Pseudomonas aeruginosa. mBio13, e0246321 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward, S. K., Heintz, J. A., Albrecht, R. M. & Talaat, A. M. Single-cell elemental analysis of bacteria: quantitative analysis of polyphosphates in Mycobacterium tuberculosis. Front. Cell Infect. Microbiol2, 63 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Y. et al. The composition and implications of polyphosphate-metal in enhanced biological phosphorus removal systems. Environ. Sci. Technol.53, 1536–1544 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Smith, I. W., Wilkinson, J. F. & Duguid, J. P. Volutin production in Aerobacter aerogenes due to nutrient imbalance. J. Bacteriol.68, 450–463 (1954). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boeynaems, S. et al. Spontaneous driving forces give rise to protein−RNA condensates with coexisting phases and complex material properties. Proc. Natl Acad. Sci. USA116, 7889–7898 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onuchic, P. L., Milin, A. N., Alshareedah, I., Deniz, A. A. & Banerjee, P. R. Divalent cations can control a switch-like behavior in heterotypic and homotypic RNA coacervates. Sci. Rep.9, 12161 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tom, J. K. A., Onuchic, P. L. & Deniz, A. A. Short PolyA RNA homopolymers undergo Mg2+-mediated kinetically arrested condensation. J. Phys. Chem. B126, 9715–9725 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alshareedah, I., Moosa, M. M., Raju, M., Potoyan, D. A. & Banerjee, P. R. Phase transition of RNA−protein complexes into ordered hollow condensates. Proc. Natl Acad. Sci. USA117, 15650–15658 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, X. et al. An inorganic biopolymer polyphosphate controls positively charged protein phase transitions. Angew. Chem. Int. Ed.59, 2679–2683 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Poirier, M. G., Monhait, T. & Marko, J. F. Reversible hypercondensation and decondensation of mitotic chromosomes studied using combined chemical–micromechanical techniques. J. Cell. Biochem.85, 422–434 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Bloomfield, V. A. DNA condensation by multivalent cations. Biopolymers44, 269–282 (1997). [DOI] [PubMed] [Google Scholar]
- 37.Muzzopappa, F., Hertzog, M. & Erdel, F. DNA length tunes the fluidity of DNA-based condensates. Biophys. J.120, 1288–1300 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schröder, H. C. et al. Inorganic polyphosphate: coacervate formation and functional significance in nanomedical applications. IJN17, 5825–5850 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wadsworth, G. M. et al. RNAs undergo phase transitions with lower critical solution temperatures. Nat. Chem.15, 1693–1704 (2023). [DOI] [PMC free article] [PubMed]
- 40.Pullara, P., Alshareedah, I. & Banerjee, P. R. Temperature-dependent reentrant phase transition of RNA–polycation mixtures. Soft Matter18, 1342–1349 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gray, M. J. et al. Polyphosphate is a primordial chaperone. Mol. Cell53, 689–699 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ault-Riché, D., Fraley, C. D., Tzeng, C. M. & Kornberg, A. Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J. Bacteriol.180, 1841–1847 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim, H. Y. et al. Alginate, inorganic polyphosphate, GTP and ppGpp synthesis co-regulated in Pseudomonas aeruginosa: implications for stationary phase survival and synthesis of RNA/DNA precursors. Mol. Microbiol.27, 717–725 (1998). [DOI] [PubMed] [Google Scholar]
- 44.Rao, N. N., Liu, S. & Kornberg, A. Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. J. Bacteriol.180, 2186–2193 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banerjee, P. R., Milin, A. N., Moosa, M. M., Onuchic, P. L. & Deniz, A. A. Reentrant phase transition drives dynamic substructure formation in ribonucleoprotein droplets. Angew. Chem. Int. Ed.56, 11354–11359 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fisher, R. S. & Elbaum-Garfinkle, S. Tunable multiphase dynamics of arginine and lysine liquid condensates. Nat. Commun.11, 4628 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor, N. O., Wei, M.-T., Stone, H. A. & Brangwynne, C. P. Quantifying dynamics in phase-separated condensates using fluorescence recovery after photobleaching. Biophys. J.117, 1285–1300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fang, J. et al. Spatial and functional arrangement of Ebola virus polymerase inside phase-separated viral factories. Nat. Commun.14, 4159 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Milin, A. N. & Deniz, A. A. Reentrant phase transitions and non-equilibrium dynamics in membraneless Organelles. Biochemistry57, 2470–2477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tom, J. K. A. & Deniz, A. A. Complex dynamics of multicomponent biological coacervates. Curr. Opin. Colloid Interface Sci.56, 101488 (2021). [Google Scholar]
- 51.Teif, V. B. & Bohinc, K. Condensed DNA: condensing the concepts. Prog. Biophys, Mol. Biol.105, 208–222 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Erkamp, N. A. et al. Spatially non-uniform condensates emerge from dynamically arrested phase separation. Nat. Commun.14, 684 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feric, M. et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell165, 1686–1697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mountain, G. A. & Keating, C. D. Formation of multiphase complex coacervates and partitioning of biomolecules within them. Biomacromolecules21, 630–640 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Lu, T. & Spruijt, E. Multiphase complex coacervate droplets. J. Am. Chem. Soc.142, 2905–2914 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaur, T. et al. Sequence-encoded and composition-dependent protein-RNA interactions control multiphasic condensate morphologies. Nat. Commun.12, 872 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niewidok, B. et al. Single-molecule imaging reveals dynamic biphasic partition of RNA-binding proteins in stress granules. J. Cell Biol.217, 1303–1318 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koltover, I., Wagner, K. & Safinya, C. R. DNA condensation in two dimensions. Proc. Natl Acad. Sci. USA97, 14046–14051 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nguyen, T. T. Grand-canonical simulation of DNA condensation with two salts, effect of divalent counterion size. J. Chem. Phys.144, 065102 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Besteman, K., Van Eijk, K. & Lemay, S. G. Charge inversion accompanies DNA condensation by multivalent ions. Nat. Phys.3, 641–644 (2007). [Google Scholar]
- 61.Kundukad, B., Yan, J. & Doyle, P. S. Effect of YOYO-1 on the mechanical properties of DNA. Soft Matter10, 9721–9728 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Åkerman, B. & Tuite, E. Single- and double-strand photocleavage of DNA by YO, YOYO and TOTO. Nucleic Acids Res.24, 1080–1090 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oikonomou, C. M. & Jensen, G. J. The atlas of bacterial & archaeal cell structure: an interactive open-access microbiology textbook. J. Microbiol Biol. Educ.22, e00128–21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pawar, A. B., Caggioni, M., Ergun, R., Hartel, R. W. & Spicer, P. T. Arrested coalescence in Pickering emulsions. Soft Matter7, 7710–7716 (2011). [Google Scholar]
- 65.Folkmann, A. W., Putnam, A., Lee, C. F. & Seydoux, G. Regulation of biomolecular condensates by interfacial protein clusters. Science373, 1218–1224 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Milner, S. T. Polymer brushes. Science251, 905–914 (1991). [DOI] [PubMed] [Google Scholar]
- 67.De Gennes, P.-G. Scaling Concepts in Polymer Physics (Cornell University Press, Ithaca, 1979).
- 68.Brangwynne, C. P., Tompa, P. & Pappu, R. V. Polymer physics of intracellular phase transitions. Nat. Phys.11, 899–904 (2015). [Google Scholar]
- 69.Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol.18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mittag, T. & Pappu, R. V. A conceptual framework for understanding phase separation and addressing open questions and challenges. Mol. Cell82, 2201–2214 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Keenen, M. M. et al. HP1 proteins compact DNA into mechanically and positionally stable phase separated domains. eLife10, e64563 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Postow, L., Hardy, C. D., Arsuaga, J. & Cozzarelli, N. R. Topological domain structure of the Escherichia coli chromosome. Genes Dev.18, 1766–1779 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo, M. S., Kawamura, R., Littlehale, M. L., Marko, J. F. & Laub, M. T. High-resolution, genome-wide mapping of positive supercoiling in chromosomes. Elife10, e67236 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith, S. A., Wang, Y. & Morrissey, J. H. DNA ladders can be used to size polyphosphate resolved by polyacrylamide gel electrophoresis. Electrophoresis39, 2454–2459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Narlikar, G. J. Phase-separation in chromatin organization. J. Biosci.45, 5 (2020). [PMC free article] [PubMed] [Google Scholar]
- 76.Azaldegui, C. A., Vecchiarelli, A. G. & Biteen, J. S. The emergence of phase separation as an organizing principle in bacteria. Biophys. J.120, 1123–1138 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Janissen, R. et al. Global DNA compaction in stationary-phase bacteria does not affect transcription. Cell174, 1188–1199.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gupta, A., Joshi, A., Arora, K., Mukhopadhyay, S. & Guptasarma, P. The bacterial nucleoid-associated proteins, HU and Dps, condense DNA into context-dependent biphasic or multiphasic complex coacervates. J. Biol. Chem.299, 104637 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krypotou, E. et al. Bacteria require phase separation for fitness in the mammalian gut. Science379, 1149–1156 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Harami, G. M. et al. Phase separation by ssDNA binding protein controlled via protein−protein and protein−DNA interactions. Proc. Natl Acad. Sci. USA117, 26206–26217 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ladouceur, A.-M. et al. Clusters of bacterial RNA polymerase are biomolecular condensates that assemble through liquid-liquid phase separation. Proc. Natl Acad. Sci. USA117, 18540–18549 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guilhas, B. et al. ATP-driven separation of liquid phase condensates in bacteria. Mol. Cell79, 293–303.e4 (2020). [DOI] [PubMed] [Google Scholar]
- 83.Debaugny, R. E. et al. A conserved mechanism drives partition complex assembly on bacterial chromosomes and plasmids. Mol. Syst. Biol.14, e8516 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Umegaki, T. & Kanazawa, T. Viscosity behavior of coacervates of magnesium and calcium highpolyphosphates. Bull. Chem. Soc. Jpn.48, 1452–1454 (1975). [Google Scholar]
- 85.Gibson, B. A. et al. Organization of chromatin by intrinsic and regulated phase separation. Cell179, 470–484.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tanaka, F. Polymer Physics: Applications to Molecular Association and Thermoreversible Gelation (Cambridge Univ. Press, Cambridge, 2011).
- 87.Jawerth, L. et al. Protein condensates as aging Maxwell fluids. Science370, 1317–1323 (2020). [DOI] [PubMed] [Google Scholar]
- 88.Zhou, H.-X. Viscoelasticity of biomolecular condensates conforms to the Jeffreys model. J. Chem. Phys.154, 041103 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Müller, W. E. G. et al. Functional importance of coacervation to convert calcium polyphosphate nanoparticles into the physiologically active state. Mater. Today Bio16, 100404 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Park, Y., Malliakas, C. D., Zhou, Q., Gu, A. Z. & Aristilde, L. Molecular coordination, structure, and stability of metal-polyphosphate complexes resolved by molecular modeling and X-ray scattering: structural insights on the biological fate of polyphosphate. Environ. Sci. Technol.55, 14185–14193 (2021). [DOI] [PubMed] [Google Scholar]
- 91.Jain, S. et al. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell164, 487–498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Protter, D. S. W. & Parker, R. Principles and properties of stress granules. Trends Cell Biol.26, 668–679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Caragine, C. M., Haley, S. C. & Zidovska, A. Nucleolar dynamics and interactions with nucleoplasm in living cells. eLife8, e47533 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lafontaine, D. L. J., Riback, J. A., Bascetin, R. & Brangwynne, C. P. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol.22, 165–182 (2021). [DOI] [PubMed] [Google Scholar]
- 95.Yu, H. et al. HSP70 chaperones RNA-free TDP-43 into anisotropic intranuclear liquid spherical shells. Science371, eabb4309 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lambert, O., Letellier, L., Gelbart, W. M. & Rigaud, J.-L. DNA delivery by phage as a strategy for encapsulating toroidal condensates of arbitrary size into liposomes. Proc. Natl Acad. Sci. USA97, 7248–7253 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tauber, D. et al. Modulation of RNA condensation by the DEAD-Box protein eIF4A. Cell180, 411–426.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maier, B. & Rädler, J. O. Conformation and self-diffusion of single DNA molecules confined to two dimensions. Phys. Rev. Lett.82, 1911–1914 (1999). [Google Scholar]
- 99.Cherstvy, G. A. & Petrov, E. P. Modeling DNA condensation on freestanding cationic lipid membranes. Phys. Chem. Chem. Phys.16, 2020–2037 (2014). [DOI] [PubMed] [Google Scholar]
- 100.Morzy, D. et al. Cations regulate membrane attachment and functionality of DNA nanostructures. J. Am. Chem. Soc.143, 7358–7367 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kato, A., Shindo, E., Sakaue, T., Tsuji, A. & Yoshikawa, K. Conformational transition of giant DNA in a confined space surrounded by a phospholipid membrane. Biophys. J.97, 1678–1686 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Herold, C., Schwille, P. & Petrov, E. P. DNA condensation at freestanding cationic lipid bilayers. Phys. Rev. Lett.104, 148102 (2010). [DOI] [PubMed] [Google Scholar]
- 103.Kumar, S., Yin, X., Trapp, B. D., Hoh, J. H. & Paulaitis, M. E. Relating interactions between neurofilaments to the structure of axonal neurofilament distributions through polymer brush models. Biophys. J.82, 2360–2372 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cuylen, S. et al. Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature535, 308–312 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhao, B. & Brittain, W. J. Polymer brushes: surface-immobilized macromolecules. Prog. Polym. Sci.25, 677–710 (2000). [Google Scholar]
- 106.Aarts, D. G. A. L., Lekkerkerker, H. N. W., Guo, H., Wegdam, G. H. & Bonn, D. Hydrodynamics of droplet coalescence. Phys. Rev. Lett.95, 164503 (2005). [DOI] [PubMed] [Google Scholar]
- 107.Cui, M., Emrick, T. & Russell, T. P. Stabilizing liquid drops in nonequilibrium shapes by the interfacial jamming of nanoparticles. Science342, 460–463 (2013). [DOI] [PubMed] [Google Scholar]
- 108.Henninger, J. E. et al. RNA-mediated feedback control of transcriptional condensates. Cell184, 207–225.e24 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lyubchenko, Y. L. & Shlyakhtenko, L. S. Visualization of supercoiled DNA with atomic force microscopy in situ. Proc. Natl Acad. Sci. USA94, 496–501 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schmatko, T., Muller, P. & Maaloum, M. Surface charge effects on the 2D conformation of supercoiled DNA. Soft Matter10, 2520 (2014). [DOI] [PubMed] [Google Scholar]
- 111.Brouns, T. et al. Free energy landscape and dynamics of supercoiled DNA by high-speed atomic force microscopy. ACS Nano12, 11907–11916 (2018). [DOI] [PubMed] [Google Scholar]
- 112.Higgins, N. P. & Vologodskii, A. V. Topological behavior of plasmid DNA. Microbiol. Spectr.10.1128/microbiolspec.plas-0036-2014 (2015). [DOI] [PMC free article] [PubMed]
- 113.Tyrrell, J., McGinnis, J. L., Weeks, K. M. & Pielak, G. J. The cellular environment stabilizes adenine riboswitch RNA structure. Biochemistry52, 8777–8785 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rhine, K., Vidaurre, V. & Myong, S. RNA droplets. Annu. Rev. Biophys.49, 247–265 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sharp, P. A., Chakraborty, A. K., Henninger, J. E. & Young, R. A. RNA in formation and regulation of transcriptional condensates. RNA28, 52–57 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Baker, C. J., Smith, S. A. & Morrissey, J. H. Diversification of polyphosphate end-labeling via bridging molecules. PLoS ONE15, e0237849 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Choi, S. H. et al. Phosphoramidate end labeling of inorganic polyphosphates: facile manipulation of polyphosphate for investigating and modulating its biological activities. Biochemistry49, 9935–9941 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sambrook, J. & Russell, D. W. Purification of nucleic acids by extraction with phenol:chloroform. Cold Spring Harb. Protoc.2006, pdb.prot4455 (2006). [DOI] [PubMed] [Google Scholar]
- 119.Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Thévenaz, P., Ruttimann, U. E. & Unser, M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process.7, 27–41 (1998). [DOI] [PubMed] [Google Scholar]
- 121.Walt et al. scikit-image: image processing in Python. PeerJ2, e453 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods17, 261–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hunter, J. D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng.9, 90–95 (2007). [Google Scholar]
- 124.Caswell, T. A. et al. matplotlib/matplotlib: REL: v3.7.1. Zenodo10.5281/zenodo.7697899 (2023).
- 125.Bois, J. S. justinbois/iqplot: version 0.3.2. 10.22002/D1.20286 (2022).
- 126.Bokeh Development Team. Bokeh: Python library for interactive visualization http://www.bokeh.pydata.org (2018).
- 127.Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol.152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 128.Eisenstein, F. et al. Parallel cryo electron tomography on in situ lamellae. Nat. Methods20, 131–138 (2023). [DOI] [PubMed] [Google Scholar]
- 129.Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol.116, 71–76 (1996). [DOI] [PubMed] [Google Scholar]
- 131.Winkler, H. 3D reconstruction and processing of volumetric data in cryo-electron tomography. J. Struct. Biol.157, 126–137 (2007). [DOI] [PubMed] [Google Scholar]
- 132.Liu, Y.-T. et al. Isotropic reconstruction for electron tomography with deep learning. Nat. Commun.13, 6482 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dragonfly 2022.2 https://www.theobjects.com/dragonfly (Comet Technologies Canada Inc., 2022).
- 134.Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci.27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xu, P., Vansiri, A., Bhan, N. & Koffas, M. A. G. ePathBrick: a synthetic biology platform for engineering metabolic pathways in E. coli. ACS Synth. Biol.1, 256–266 (2012). [DOI] [PubMed] [Google Scholar]
- 136.Vyas, V. K. et al. New CRISPR mutagenesis strategies reveal variation in repair mechanisms among fungi. mSphere3, e00154–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Portales-Casamar, E. et al. A regulatory toolbox of MiniPromoters to drive selective expression in the brain. Proc. Natl Acad. Sci. USA107, 16589–16594 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lukinavičius, G. et al. Selective chemical crosslinking reveals a Cep57-Cep63-Cep152 centrosomal complex. Curr. Biol.23, 265–270 (2013). [DOI] [PubMed] [Google Scholar]
- 139.Shepherd, T. R. et al. De novo design and synthesis of a 30-cistron translation-factor module. Nucleic Acids Res.45, 10895–10905 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Fluorescence microscopy images used in this are available from the Zenodo repository (Zenodo record 13324261). Representative tomograms and raw cryo-ET data are available at the EMDB (the Electron Microscopy Data Bank), under the following accession IDs: EMD-42152, EMD-42153, EMD-42154, EMD-42155, and EMPIAR-11701. Source data are provided with this paper.
Custom codes used for analysis in this study are available on GitHub, as follows. Aspect Ratio code: https://github.com/Deniz-Lab/condensate-fusion-aspect-ratio. FindCircles code: https://github.com/RackiLab/Chawla_and_Tom_2024_etal.