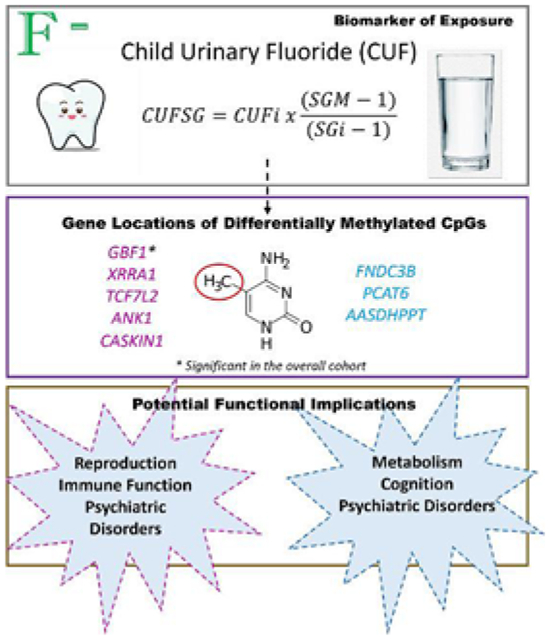
Abstract
Exposure to fluoride in early childhood has been associated with altered cognition, intelligence, attention, and neurobehavior. Fluoride-related neurodevelopment effects have been shown to vary by sex and very little is known about the mechanistic processes involved. There is limited research on how fluoride exposure impacts the epigenome, potentially leading to changes in DNA methylation of specific genes regulating key developmental processes. In the Cincinnati Childhood Allergy and Air Pollution Study (CCAAPS), urine samples were analyzed using a microdiffusion method to determine childhood urinary fluoride adjusted for specific gravity (CUFsg) concentrations. Whole blood DNA methylation was assessed using the Infinium MethylationEPIC BeadChip 850 k Array. In a cross-sectional analysis, we interrogated epigenome-wide DNA methylation at 775,141 CpG loci across the methylome in relation to CUFsg concentrations in 272 early adolescents at age 12 years. Among all participants, higher concentrations of CUF were associated with differential methylation of one CpG (p < 6 × 10−8) located in the gene body of GBF1 (cg25435255). Among females, higher concentrations of CUFsg were associated with differential methylation of 7 CpGs; only three CpGs were differentially methylated among males with no overlap of significant CpGs observed among females. Secondary analyses revealed several differentially methylated regions (DMRs) and CpG loci mapping to genes with key roles in psychiatric outcomes, social interaction, and cognition, as well as immunologic and metabolic phenotypes. While fluoride exposure may impact the epigenome during early adolescence, the functional consequences of these changes are unclear warranting further investigation.
Keywords: Fluoride exposure, Epigenetics, Pediatric environmental health
Graphical Abstract
1. Introduction
Fluoride exposure is nearly ubiquitous due to its presence in groundwater, fluoridation of water and table salt, its presence in food, and its wide use in commercial products to prevent dental caries. (Water Fluoridation Data and Statistics, 2019) Fluorinated water is supplied to nearly 75 % and 39 % of United States (U.S.) and Canadian residents, respectively, through community water systems (Water Fluoridation Data and Statistics, 2019; Public Health Capacity and knowledge Management Unit: The State of community Water Fluoridation (CWF) across Canada, 2022). In Mexico, approximately 39 % of drinking water comes from groundwater sources and for regions where naturally occurring fluoride in water is <0.7 mg/L, sodium fluoride or potassium fluoride is added to salt at levels of 250 mg/Kg (Martinez-Mier et al., 2003). Drinking water fluorosis is endemic across 27 provinces, cities, and autonomous regions in North and South China due to its location in the fluoride belt, a region where excess fluoride has been detected in groundwater (Wang et al., 2023).
Birth cohort studies from Canada, Mexico, India, and China suggest prenatal and early childhood fluoride exposure is associated with impaired cognition (Bashash et al., 2017), decreased intelligence (IQ) (Das and Mondal, 2016; Green et al., 2019; Yu et al., 2018; Till et al., 2020), and attention-deficit hyperactivity disorder (Malin and Till, 2015). In the U.S., fluoride exposure prenatally and during early adolescence has been associated with increases in neurobehavior problems including anxiety and depression symptoms (Adkins et al., 2022; Malin et al., 2024). Observations in human studies are supported by numerous animal studies linking postnatal fluoride exposure to anxiety- and depression-like behavior suggesting an imbalance in excitation and inhibition (Li et al., 2019; Liu et al., 2014; Lu et al., 2019). Furthermore, studies have identified sex-specific fluoride effects on neurodevelopment (Green et al., 2019; Adkins et al., 2022).
Despite the associations between fluoride and neurodevelopment, very little is known about the mechanistic processes by which fluoride impacts neurodevelopment. Epigenetic alterations that can affect gene expression without changing the underlying DNA sequence can respond to environmental exposures in ways that directly affect gene transcription and disease risk (Wu et al., 2023; Martin and Fry, 2018). DNA methylation, the most widely studied epigenetic mechanism, modifies genome function through the addition of methyl groups to cytosine to form 5-methyl-cytosine (5mC). Sexual dimorphisms exist both naturally in how the methylome responds during puberty as well because of environmental insults (Martin and Fry, 2018; Thompson et al., 2018). Specifically, regarding fluoride, very few studies have investigated the association between fluoride exposure and changes to DNA methylation and even fewer considered sex-specific effects. Of the 17 published studies, only 9 were human observational studies (cross-sectional and case-control) and all of them were conducted among Chinese cohorts (Zhang et al., 2017; Wu et al., 2019; Wu et al., 2018; Pan et al., 2020; Sun et al., 2020; Gao et al., 2020; Meng et al., 2021; Wang et al., 2021; He et al., 2022); the majority (n = 8) were conducted in fluorosis-impacted adults and utilized a targeted DNA methylation approach (i.e., specific genes); one considered sex-specific effects (Gao et al., 202C). Targeted DNA methylation analyses conducted in humans and animals suggest fluoride exposure impacts gene imprinting, oocyte maturation, estrogenic processes, bone development/metabolism, and cell cycle progression through DNA methylation modifications.
Given the importance of DNA methylation existing outside of promoter or CpG island regions for regulating phenotypic variability, conducting unbiased epigenome-wide analyses in search of relevant DNA methylation changes resulting from fluoride exposure is paramount. An epigenome-wide association study (EWAS) of urinary fluoride, a contemporary biomarker of fluoride exposure at the population level, was conducted among Chinese children during the early adolescent period (ages 8–12 years) (Wang et al., 2021). The study identified several differentially methylated genes associated with skeletal and neuronal development. It is unclear if similar associations would be found in a population of U.S. children where fluoride exposure levels and patterns are expected to be considerably different due to differences in community water fluoridation, diet, and oral care products.
The goal of this study was to examine the association between fluoride exposure (as indicated by urinary fluoride levels) and epigenome-wide DNA methylation among early adolescents (~ 1 2 years of age) enrolled in a U.S. based cohort. We hypothesize that elevated urinary fluoride levels will be associated with differential DNA methylation at loci implicated in neurodevelopment and the methylation changes will be sex specific.
2. Methods
2.1. Study population
Participants were enrolled in the Cincinnati Children’s Allergy and Air Pollution Study (CCAAPS), a prospective birth cohort of children born between 2001 and 2003 in the Greater Cincinnati Region (Ryan et al., 2005; LeMasters et al., 2006). Enrollment into the CCAAPS study was based on residence at the time of birth and distance to a major highway due to the primary interest in the impact of air pollution on child health. All children reside in communities with water fluoridation. Children and their caregivers completed demographic assessments and were clinically evaluated at the child’s age of 1, 2, 3, 4, 7, and 12 years. Data obtained at the visits included information regarding participant health and general wellbeing, housing characteristics, residential history, and secondhand smoke exposure. The study was approved by the Institutional Review Boards of the University of Cincinnati and Cincinnati Children’s Hospital Medical Center (IRB# 2012–1619). Participants and caregivers provided informed assent and consent, respectively.
2.2. Assessment of fluoride exposure
Spot urine samples were collected under normal (non-fasting) conditions at age 12 years and analyzed at the Oral Health Research Institute of the Indiana University School of Dentistry. Prior to use, childhood urinary fluoride (CUF) samples were stored at −20 °C. In brief, 5 mL of urine was thawed, and ionic fluoride was diffused from the sample using the hexamethyldisiloxane (HDMS)-diffusion method (Martinez-Mier et al., 2011). Urinary specific gravity (SG), used to correct for dilution in urine samples, was measured using a zero-setting calibrated ATAGO® Pen Refractometer under darkened conditions against a standard check (using a fluoride standard traceable to National Institutes of Standards and Technology). Dilution-corrected urinary fluoride was estimated using the following equation: , where CUFSG is the SG-adjusted fluoride concentration (in micrograms of fluoride per milliliter), CUFi is the observed fluoride concentration, SGi is the SG of the individual urine sample, and SGM is the median SG for the cohort (Thomas et al., 2016) (Till et al., 2018). Dilution-corrected CUFsg was used in EWAS analyses.
2.3. Epigenome-Wide Association Studies (EWAS)
2.3.1. Specimen collection and DNA isolation
Whole blood samples were collected for each participant at the age 12-year clinical evaluation. Whole blood was separated into plasma and buffy coat using centrifugation and stored at −80 °C. DNA was extracted and assessed according to the Illumina® HumanMethylationEPIC (EPIC) BeadChip assay at the University of Cincinnati laboratories.
2.3.2. Quality control, preprocessing, and probe filtering
DNA methylation preprocessing and quality control was conducted as previously described by Niu et al. (Niu et al., 2016). In summary, the major steps of this process included filtering out low-quality samples (sex or genetic mismatches), as well as those with poor-performing probes (outliers with detection p-value < 0.01, bead count <3). Quantile normalization was performed to remove any outlier samples in signal intensity due to technical variability. Dye bias correction for Type I and Type II probes was conducted using the RELIC package (Xu et al., 2017). To minimize batch effects, stratified randomization was used to randomize samples to chips and plates. Surrogate variables were calculated using the ComBat function from the sva package in R. (Leek et al., 2012) and included as covariates in downstream analyses as the top five principal components for batch effects. DNA methylation beta values, ranging from 0 (completely unmethylated) to 1 (completely methylated), after trimming for extreme outliers (> 3× interquartile range from the 25th and 75th quartile limits) for EWAS analysis. Cell-type proportions of blood samples was estimated using the Houseman method (Houseman et al., 2012), which provides estimates for the proportions of CD8 + T cells, CD4 + T cells, natural killer (NK) cells, B-cells, monocytes, and nucleated red blood cell (nRBC) estimates. We excluded probes on the sex chromosomes, polymorphic CpGs which overlap with known single-nudeotide-polymorphisms, control probes, and cross-reactive probes (targeting repetitive sequences/co-hybridizing to alternate sequences), which left a total of 773,595 probes for analysis (Chen et al., 2013; McCartney et al., 2016). All pre-processing was done using the ENmix package (version 3.15) in R (Xu et al., 2016).
2.4. Covariates
A core set of potential confounders included in all models were selected based on biological relevance and prior literature demonstrating their association with fluoride and their impact on DNA methylation. They included child sex (Martin and Fry, 2018; Thompson et al., 2018), race (Gao et al., 2020), age (Aylward et al., 2015), and total household income (Sanders et al., 2019) - all collected by questionnaire completed by the caregiver. Additional covariates were also included. Serum cotinine is a well-characterized biomarker of tobacco smoke and an environmental exposure strongly associated with changes in DNA methylation and routinely controlled for in EWAS analyses (Joubert et al., 2016; Vives-Usano et al., 2020). Serum cotinine levels for each child were measured using a standardized isotope dilution liquid chromatography/tandem mass spectrometry (IXD/MS/MS) method (Perlman et al., 2016). Maternal depression (as measured by the BDI Score, (Beck et al., 1996)) and child parent relational frustration (measured by Relational Frustration T-Score (Kamphaus and Reynolds, 2008)) were also included as covariates given that both the quality of caregiving and the caregivers psychological status have been shown to contribute to changes in DNA methylation of children (Lester et al., 2013; Provenzi et al., 2020; Mastrotheodoros et al., 2023). Additionally, DNA methylation technical covariates, measurements correcting for systematic sources of variation, included the top five principal components for batch effects and six cell type proportions (Leek et al., 2012).
2.5. Statistical analyses
2.5.1. Epigenome wide association study and Differentially Methylated Regions (DMR) analyses
An EWAS analysis was performed on the total sample (referred to as “Total” group) utilizing robust multiple linear regression (MASS R package) (https://www.stats.ox.ac.uk/pub/MASS4/) to control for potential heteroscedasticity and DNA methylation outliers. The EWAS examined the associations between log-transformed urinary childhood fluoride concentrations (CUFsg) and DNA methylation at 775,141 CpG loci. Probes were annotated to genome build hgl9 according to the package meffil (Min et al., 2018). Genomic inflation factor lambdas (λ) and quantile-quantile plots were generated to assess genomic inflation. For multiple testing correction, we used a Bonferroni correction based on the number of probes being interrogated (p < 6 × 10–08). Sex-specific EWAS analyses were completed for females (“female” group) and males (“male” group) separately to identify potential sex-specific methylation differences. CpGs identified by the EWAS as being significantly differentially methylated in the female or male group were then confirmed via follow up linear regression analyses to formally test for a fluoride by sex interaction.
We also interrogated DMRs. DMRs are defined as differentially methylated genomic regions, or areas of adjacent methylated CpG sites (Rakyan et al., 2011). They can occur in any genomic region and are known to potentially impact gene regulation and expression, regardless of location in the genome. To identify DMRs associated with urinary fluoride concentrations, we used the DMRcate R package (Peters et al., 2015). Settings were adjusted to identify methylated regions within a length of ≤1000base pairs, 2 CpGs present, andp associations with an FDR significance level, as recommended by the developers to minimize Type I errors.
2.5.2. Follow-up analyses
Additional analyses were conducted among the total population, as well as females and males separately. Secondary analyses include an investigation of Blood-Brain Epigenetic Concordance (BECon) (Edgar et al., 2017), Methylation Quantitative Trait Loci (mQTL) (Min et al., 2021), expression Quantitative Trait Methylation (eQTM) (Ruiz-Arenas et al., 2022), and Experimentally Derived Functional Element Overlap Analysis of Regions from EWAS (eFORGE) (Breeze, 2022) as detailed below.
Epigenetic processes are tissue-specific, making the assessment of disease-relevant tissue an important consideration for EWAS. We used two web-based applications to assess correlations between blood and brain DNA methylation. The first is the Blood-Brain Epigenetic Concordance (BECon) application (https://redgar598.shinyapps.io/BECon/), which calculates correlation and variability metrics between blood DNA methylation levels of the significantly differently methylated CpGs and three different brain regions– Brodmann area 7 (parietal cortex), Brodmann area 10 (frontal cortex), and Brodmann area 20 (temporal cortex). The second web-based application is the Hannon Blood Brain Comparison Tool https://epigenetics.essex.ac.uk/bloodbrain/) (Hannon et al., 2018). Using this tool, we examined correlations of DNA methylation at CpG loci identified in the EWAS across four different areas of the brain: prefrontal cortex, entorhinal cortex, superior temporal gyrus and cerebellum.
Given that genetic influence on DNA methylation is common, we also conducted mQTL mapping (cis and trans) of those CpG sites in the main EWAS model conducted among the Total group that were below significance (p < 5.0 × 10−5) to identify possible population-level variations and functional consequences of DNA methylation changes. This was carried out by querying the catalog of cis- and tram- mQTLs from the Genetics of DNA Methylation Consortium (GoDMC) of >30,000 participants of European ancestry (Min et al., 2021) (http://mqtldb.godmc.org.uk/index.php).
An eQTM analysis was implemented to explore whether there was any association between DNA methylation of specific CpG sites and gene expression. A catalog of 39,749 eQTMs with significant associations to gene expression from children’s blood compiled by the Exposome-omics-Wide Association Study (ExWAS) conducted in the Human Early Life Exposome (HELIX) project was used to identify whether our significantly differentially methylated CpGs were associated with a change in gene expression. The catalog was downloaded from http://www.helixomics.isglobal.org/. We utilized the significantly methylated CpG sites to cross-reference with the catalog. Any matches were considered significant associations to the specified gene.
To assess the relationship of specific CpG sites of interest and disease etiology, we utilized the web-based tool eFORGE (experimentally-derived Functional element Overlap analysis of ReGions from EWAS) (Breeze, 2022). eFORGE interrogates any overlap of CpG sites of interest and regulatory elements for tissues involved in disease processes. The tool selects background probes based on gene-centric categories, such as promoter or gene body regions, and CpG island-centric categories (shelves or islands) with chosen input probes as a guide. It is recommended that genome-wide significance is increased from typical EWAS cutoffs due to differences in tissue-specific DNA methylation. The suggested number of CpG sites of interest input for query is between 100 and 1000 (referred to as “input probes”) to minimize false positive results. Thus, for this analysis we utilized the cutoff of p < 5.0 × 10−5 to increase the number of sites within the recommended range of input probes. Female and male groups were analyzed separately.
The genes for which significant CpG loci reside were queried using the GWAS Atlas (https://atlas.ctglab.nl/) (Watanabe et al., 2019). Results are calculated with the PheWAS function, which plots traits that are significantly correlated to the genetic expression of the gene of interest (p < 0.05). The atlas includes a compilation of 4756 GWAS and 3302 unique traits. Plots are arranged by phenotype domain and p-value. Text labels include phenotypes of similar sample sizes.
3. Results
3.1. Participant characteristics
Participant characteristics are described in Table 1. EWAS analyses were completed for three separate analytic groups (see Methods for additional details on analytic groups): total cohort (n = 272), females only (n = 124), and males only (n = 148). The average age of participants was 12.15 ± 0.77 years with most of the participants identifying as white (76 %). Specific gravity adjusted CUF levels (CUFsg) were similar among males (median 0.81 μg/mL; IQR 0.11) and females (median 0.81 μg/mL; IQR 0.49) and did not vary by income (Kruskal-Wallis χ = 4.38, DF = 4, p = 0.88). There were also no significant differences in race/ethnicity, age, total household income, maternal BDI, or relational frustration T-score between males and females (Table 1). Serum cotinine levels were significantly higher in males versus females (0.43 versus 0.23 ng/mL).
Table 1.
Demographics and Characteristics of CCAAPS Participants at the 12-Year Visit.
| Total samplea | Females | Males | |
|---|---|---|---|
| (n = 272) | (n = 124) | (n = 148) | |
| Characteristic(s) | n (%) or Mean (SD) | n (%) or Mean (SD) | n (%) or Mean (SD) |
| Sex | |||
| Male | 148 (54 %) | – | 148 (100 %) |
| Female | 124 (46 %) | 124 (100 %) | – |
| Race/Ethnidtyb | |||
| White | 207 (76 %) | 95 (77 %) | 112 (76%) |
| Black/More Than One Race | 65 (24 %) | 29 (23 %) | 36 (24 %) |
| Age (years) | 12.12 (0.77) | 12.26 (0.83) | 12.00 (0.70) |
| Serum Cotinine (ng/mL) | 0.34 (1.39) | 0.23 (0.62) | 0.43 (1.79) |
| Total Household | |||
| Incomec | |||
| <$20,000 | 28 (10 %) | 13 (10%) | 15 (10 %) |
| $20,000–<$40,000 | 34 (13 %) | 14 (11 %) | 20 (14 %) |
| $40,000–<$70,000 | 51 (19 %) | 25 (20 %) | 26 (18 %) |
| $70,000–<$90,000 | 37 (14 %) | 13 (10%) | 24 (16 %) |
| >$90,000 | 111 (41 %) | 54 (44 %) | 57 (39 %) |
| Prefer not to answer | 10 (4 %) | 4 (3%) | 6 (4%) |
| Maternal BDI Score | 6.28 (7.27) | 6.62 (8.26) | 6.01 (6.36) |
| Relational Frustration T-Score | 47.79 (9.28) | 47.94 (10.18) | 47.66 (8.48) |
| Fluoride in μg/mL [Median (IQR)] | |||
| Child urinary fluoride (CUF) | 0.82 (0.56–1.18) | 0.71 (0.51–1.08) | 0.92 (0.65–1.27) |
| Specific Gravity (SG) | 1.02 (1.01–1.02) | 1.02 (1.01–1.03) | 1.02 (1.02–1.03) |
| Fluoride SG corrected (CUFsg) | 0.81 (0.62–1.09) | 0.81 (00.59–1.09) | 0.81 (0.64–1.08) |
| Log-transformed | −0.21 | −0.21 | −0.21 |
| CUFsg | (−0.47–0.09) | (−0.51–0.09) | (−0.44–0.08) |
Total participants with complete and usable fluoride and epigenetic data.
Self-reported at visit.
One participant is missing household income data in each group except for “males”.
3.2. EWAS results
Overall, higher CUFsg concentrations were associated with increases in methylation at cg25435255 located in an open sea region of chromosome 10 mapping to GBF1. There was no evidence of genomic inflation (λ = 1.0) (Fig. 1A and B, Table 2). Among females, CUFsg was significantly associated with DNA methylation at 7 CpG loci with no evidence of genomic inflation (λ = 1.1) (Fig. 2A and B). Similar to the total EWAS, the strongest relationship was observed between increasing CUFsg concentrations and increasing methylation at cg25435255. Among the 7 total CpG sites, five demonstrated an increase in methylation with increasing fluoride concentration (cg25435255, cg14665389, cg21470142, cg12434747, cg02602408); the remaining two exhibited a decrease in methylation (cg13828087, cg12773035) (Table 2). Among males, CUFsg was significantly associated with DNA methylation at three CpG loci with no evidence of genomic inflation (λ = 1.0) or overlap with significantly differentially methylated CpGs identified in the female only EWAS (Fig. 2C and D, Table 2). The methylation status of all three CpG loci were positively associated with CUFsg concentrations (Table 2). All three loci annotated to gene bodies: cg14324167 in FNDC3B on chromosome 3, cg13993945 in PCAT6 on chromosome 1, and cg19292625 in AASDHPPT on chromosome 11. Sex-specific effects were confirmed by interaction terms (PCUF * SEX < 0.05) for 4 (cgl3828087, cg14665389, cg12434747, cg02602408) out of 7 CpGs identified in the female EWAS and 2 (cgl3993945, cgl9292625) out of 3 CpGs identified in the male EWAS (Supplemental Fig. 1).
Fig. 1.
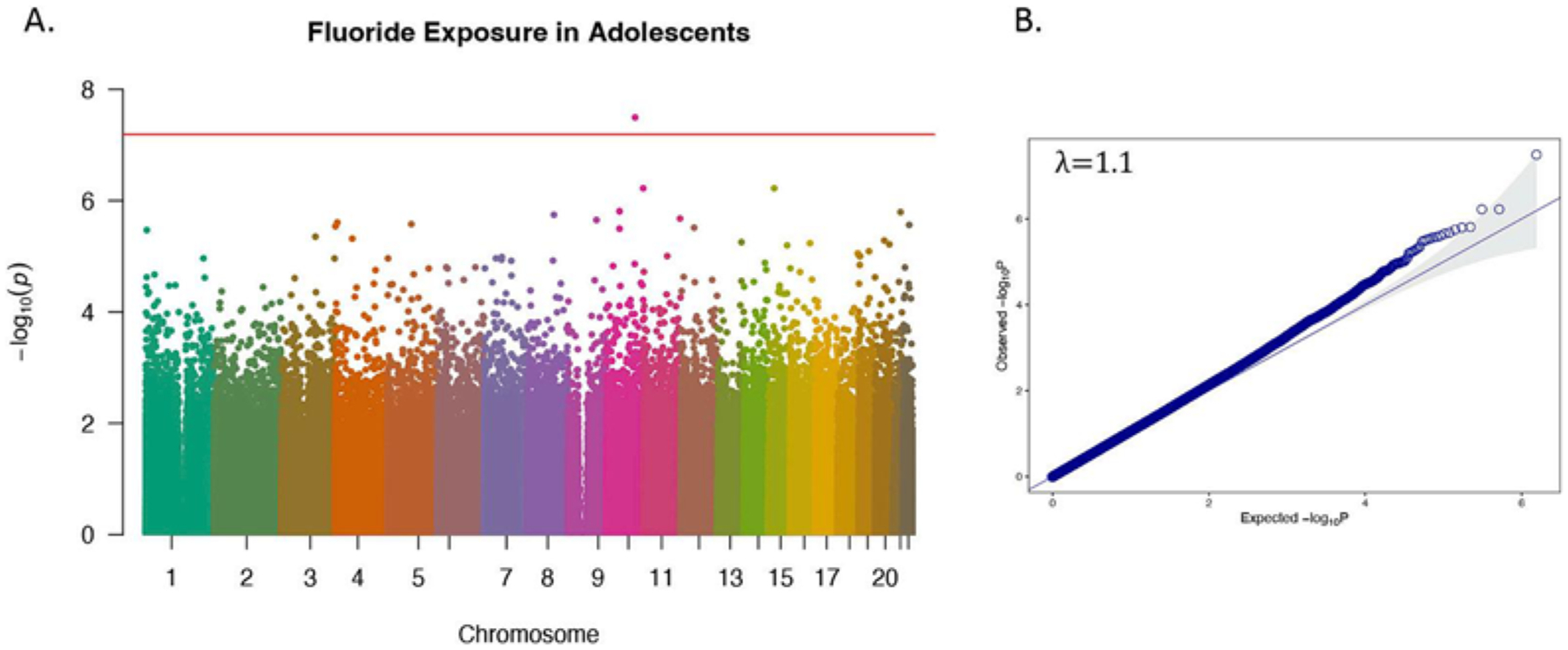
Manhattan plot (A) and related quantile-quantile plot (B) showing sites of DNA methylation in whole blood associated with log-transformed childhood urinary fluoride (CUFsg) concentration in the CCAAPS total cohort. Red line indicates epigenome-wide significance based on Bonferroni, (p < 6 × 10−8). Methylation of CpG sites that are significantly associated with log-transformed CUFsg are depicted by dots above the red line.
Table 2.
Significantly differentially methylated CpG sites associated with child urinary fluoride.
| EWAS Model | CpG Site | P-Value | Beta | Std Err | N of Individuals | Chr | Position | Gene | Relation to Island |
|---|---|---|---|---|---|---|---|---|---|
| Totala | cg25435255 | 3.21E-08 | 0.0028 | 0.0005 | 264 | 10 | 104086857 | GBF1 | Open Sea |
| Femaleb | cg25435255 | 2.51E-11 | 0.0038 | 0.0006 | 119 | 10 | 104086857 | GBF1 | Open Sea |
| cg13828087 | 3.56E-10 | −0.0057 | 0.0009 | 119 | 11 | 74556295 | XRRA1 | Open Sea | |
| cg14665389 | 2.44E-09 | 0.0055 | 0.0009 | 117 | 10 | 114710815 | TCF7L2 | North Shore | |
| cg21470142 | 5.47E-09 | 0.0047 | 0.0008 | 119 | 8 | 41732895 | ANK1 | North Shore | |
| cg12434747 | 6.73E-09 | 0.0131 | 0.0023 | 118 | 15 | 70740470 | - | Open Sea | |
| cg12773035 | 5.61E-08 | −0.0093 | 0.0017 | 118 | 16 | 2239574 | CASKIN1 | South Shelf | |
| cg02602408 | 5.42E-08 | 0.0243 | 0.0045 | 117 | 11 | 26100166 | - | Open Sea | |
| cg14324167 | 5.00E-08 | 0.0056 | 0.001 | 146 | 3 | 171851893 | FNDC3B | Open Sea | |
| Malec | cg13993945 | 2.92E-08 | 0.0255 | 0.0046 | 145 | 1 | 202779497 | PCAT6 | North Shore |
| cg19292625 | 5.49E-08 | 0.0051 | 0.0009 | 146 | 11 | 105951315 | AASDHPPT | South Shelf |
Sites of CpG methylation significantly associated with CUF concentrations in the total cohort.
Sites of CpG methylation significantly associated with log transformed CUFsg concentrations in females.
Sites of CpG methylation significantly associated with log transformed CUFsg concentrations in males.
Fig. 2.
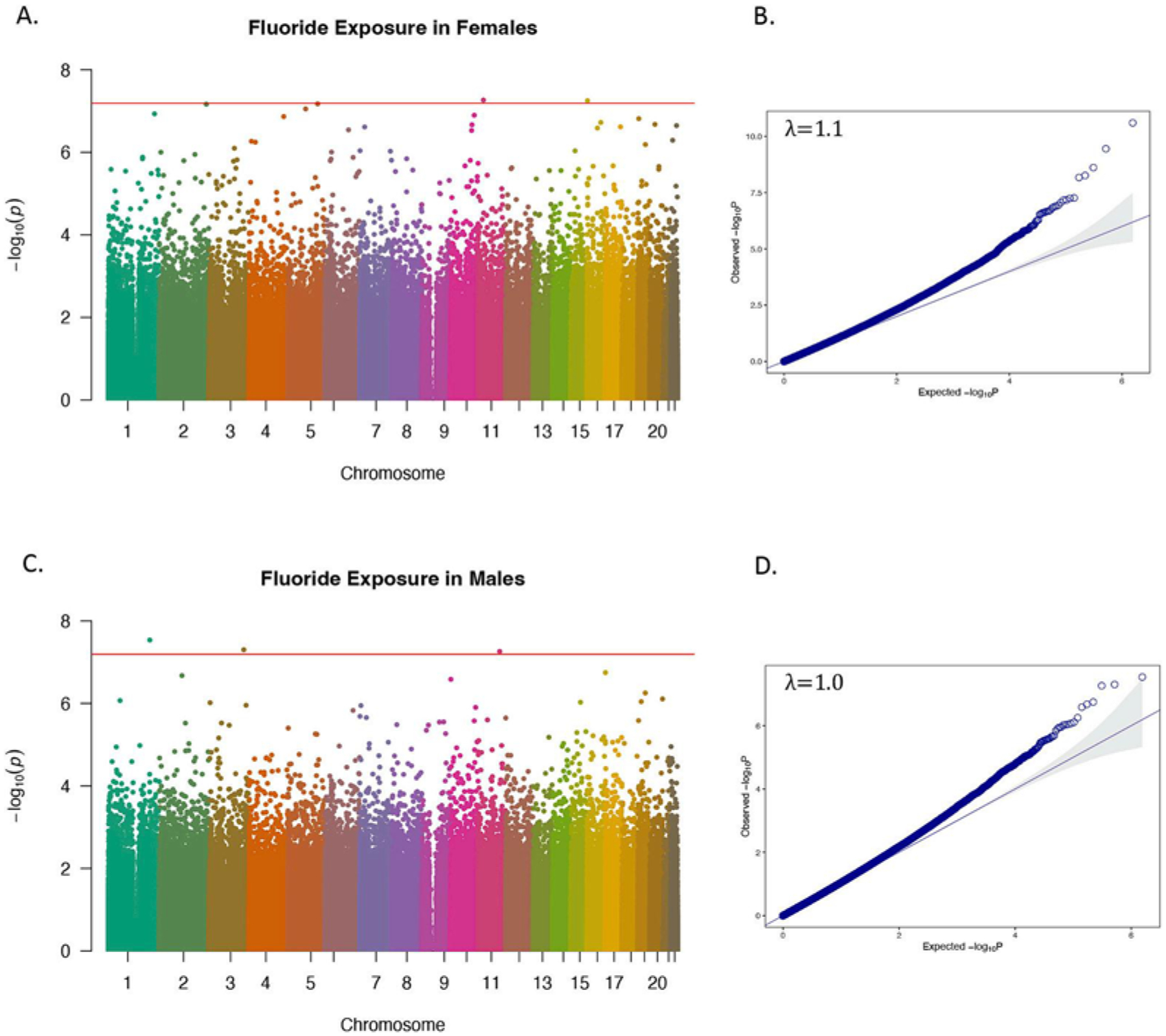
Manhattan plots (A, C) and related quantile-quantile plots (B, D) showing sites of DNA methylation in whole blood associated with log-transformed childhood urinary fluoride (CUFsg) concentration in the CCAAPS among females (A, B) and males (C, D). Red line indicates epigenome-wide significance based on Bonferroni, (p < 6 × 10−8). Methylation of CpG sites that are significantly associated with log-transformed CUFsg are depicted by dots above the red line.
3.3. Identification of differentially methylated regions (DMRs)
DMR analyses were performed to look for regions of differential methylation significantly associated with CUFsg concentrations. In the results of the total group, one significant (p < 6.80 × 10−22) DMR located on chromosome 6 at position 32064212–32064497 was identified and contained six total CpG sites (Supplemental Table 1). Among females, there were ten regions that exhibited differential methylation associated with CUFsg concentrations (Supplemental Table 2). The most significant association (p < 1.65 × 10−12) was in a region on chromosome 6 (position 29520527–29521803). There are thirty-five differentially methylated CpG sites in this region, none of which are in gene-containing regions. The remaining significantly associated methylated regions contained between three and ten CpGs and p values ranged from p < 1.56 × 10−10 to p < 3.25 × 10−07 (Supplemental Table 2). None of these regions overlapped with the male group regional analysis, which had only one significantly methylated region on chromosome 6 (position 32,064,497–32,064,613) containing six CpG sites (Supplemental Table 3), none of which are in gene-containing regions. One of the six CpG sites, cg03556669, is also included in the DMR significantly associated with CUFsg concentrations in the total group. There was no overlap between the significantly differentially methylated CpGs identified in the EWAS and DMR analyses.
3.4. Follow-up analyses (mQTL, eQTM, BECon, Hannon blood brain comparison, eFORGE, GWAS adas/PhEWAS plot)
We used various web-based tools to further explore the potential functional consequences of significant increases or decreases in DNA methylation associated with CUFsg concentrations in both female and male adolescents.
To consider whether any genetic influence could be contributing to phenotypic effect, we analyzed the CpG sites with the strongest associations in the female group using the GoDMC website. One CpG loci demonstrated potential associations with genetic influences on methylation (cgl2434747, Supplemental Table 4) but was not detected as a cis-eQTM from the HELIX Exposome-omics-Wide Association Analysis database (Ruiz-Arenas et al., 2022; Maitre et al., 2022). No CpG sites of interest in the total or male group indicated there was any significant genetic influence contributing to phenotypic effects.
To evaluate whether there was any tissue correlation between blood and brain, we utilized BECon (Edgar et al., 2017). Of the 7 significant CpGs identified in the female only EWAS (which includes the single CpG site from the total EWAS), four CpGs are reported to have negative moderate correlations (correlation >0.40) between blood and brain DNA methylation levels in BA10 (cg25435255), BA20 (cg25435255, cg12434747), and BA7 (cg25435255, cg21470142) (Fig. 3). Of the three significant CpGs identified in the male only EWAS, none of them were correlated to brain DNA methylation levels. However, the methylation status of cgl9292625 was significantly correlated (p = 0.027) with the cerebellum in the Hannon Blood Brain analysis (Supplemental Fig. 2).
Fig. 3.
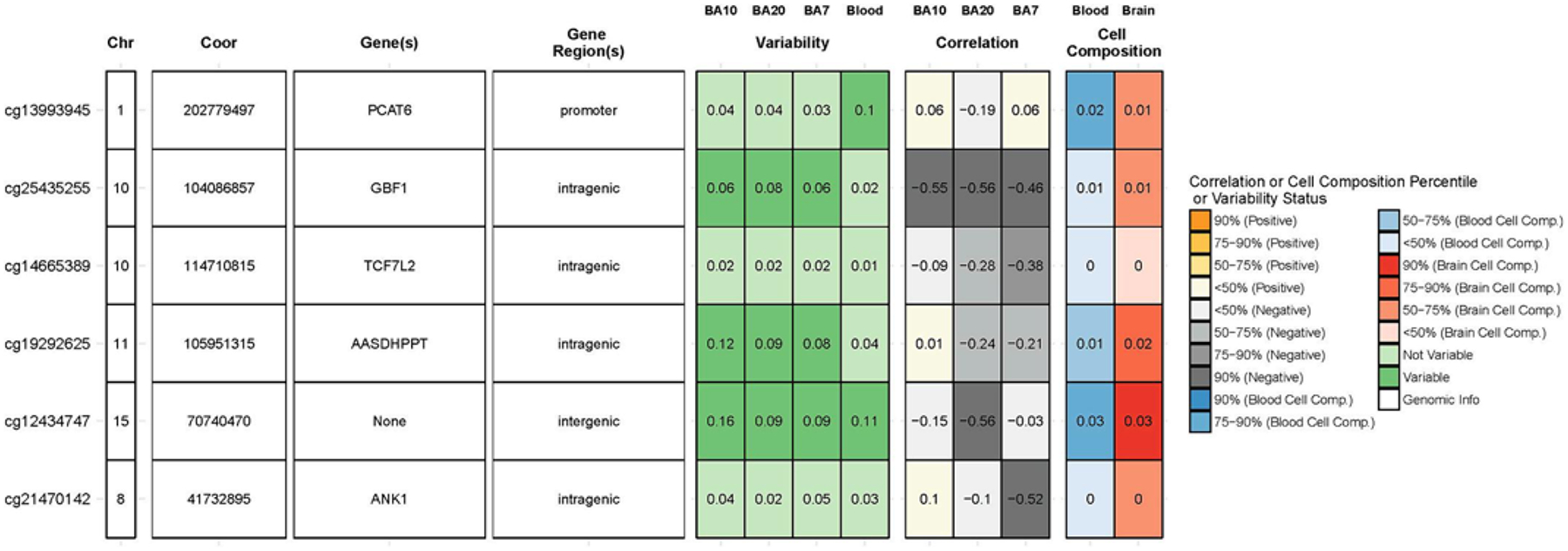
BECon analysis for blood/brain correlations. Six (4 female, 2 male) significant CpG sites were used to assess blood/brain correlation. BA10: Brodmann area 10 (prefrontal cortex); BA20: Brodmann area 20 (temporal cortex); BA7: Brodmann area 7 (parietal cortex).
We also assessed overlap with functional elements among tissues other than brain utilizing the webtool eFORGE, with females and males being analyzed separately. After applying a less restrictive genomic cutoff of p < 5.0 × 10−5, 289 CpG sites and 180 CpG sites were identified and used as the input probes for females and males, respectively. For females, the findings suggest significant enrichment (p < 0.05) in embryonic stem cells, fetal brain, fetal large intestine, fetal kidney, fetal stomach, fetal thymus, induced pluripotent stem cells, pancreas, and placenta. Highly significant enrichment (p < 0.01) was demonstrated for embryonic stem cells, fetal adrenal glands, and gastric tissue (Fig. 4). Among males, the set of 180 differentially methylated CpG sites were not enriched in any of the tissues represented in eFORGE (Supplemental Fig. 3).
Fig. 4.
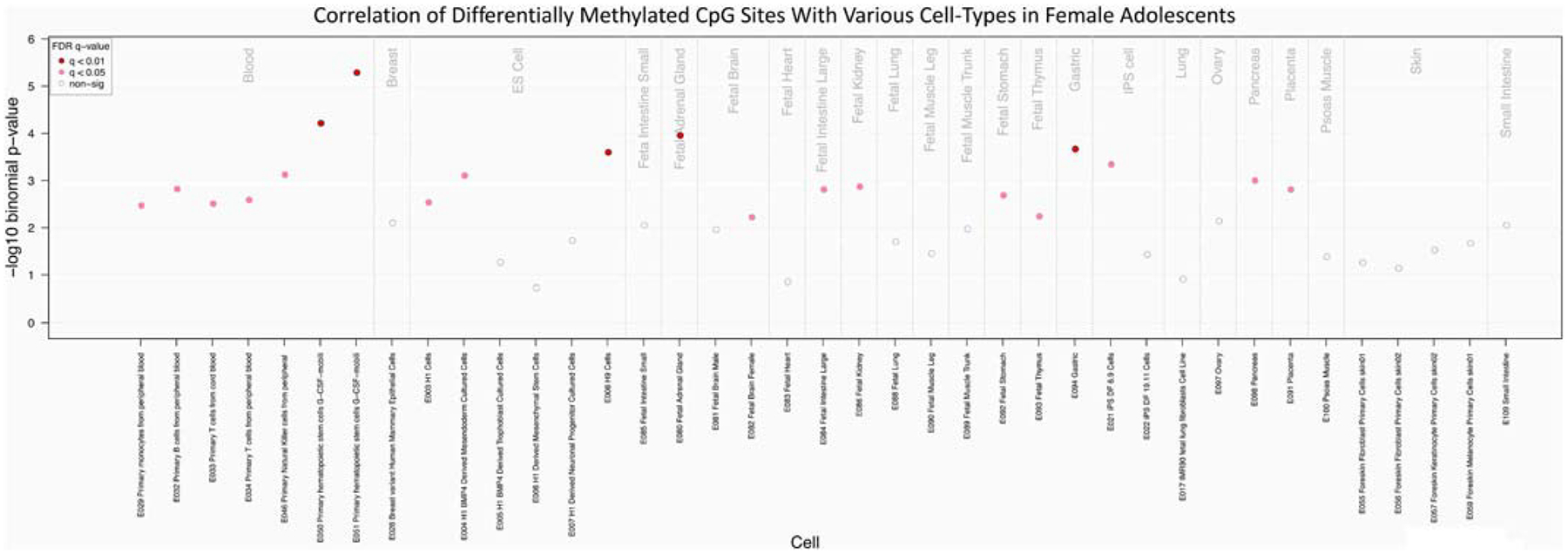
eFORGE (experimentally-derived Functional element Overlap analysis of ReGions from EWAS) Analysis of Females. Various cells from 23 different tissue types were analyzed for tissue specific signal and correlation with 289 CpG sites. FDR q-value are adjusted p-values.
In order to interrogate which traits are significantly correlated to the genetic expression of the genes of interest for which our significant CpG sites are located, we used the PhEWAS function of the GWAS Atlas (Watanabe et al., 2019). GBF1, where cg254352554 is located and was differentially methylated among the total and female groups, is associated with several cognitive phenotypes, such as educational attainment, cognitive performance, and intelligence (Fig. 5). AASDHPPT, cgl9292625 is located and was differentially methylated among males, is most significantly associated with depressive episodes (Fig. 6).
Fig. 5.
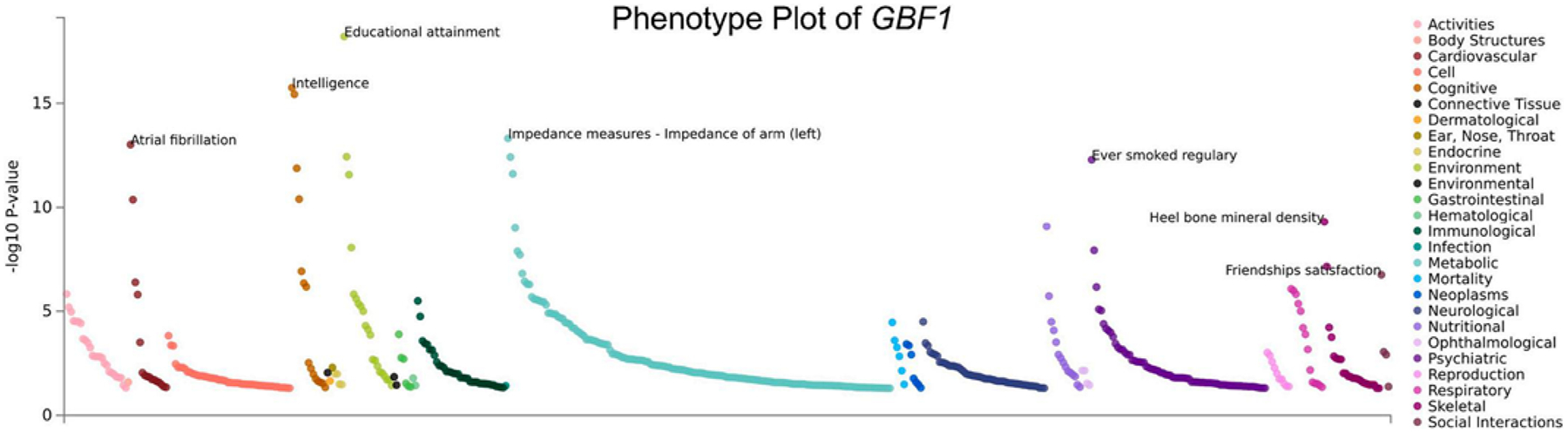
PhEWAS plot of gene GBF1. This plot shows phenotypic traits which are significantly correlated (p-values) with gene expression of the gene of interest. cg25435255, a significant site from the Female analytic group, is located within the gene region of GBF1. Phenotypic groups are listed on the right of the plot with corresponding color code.
Fig. 6.
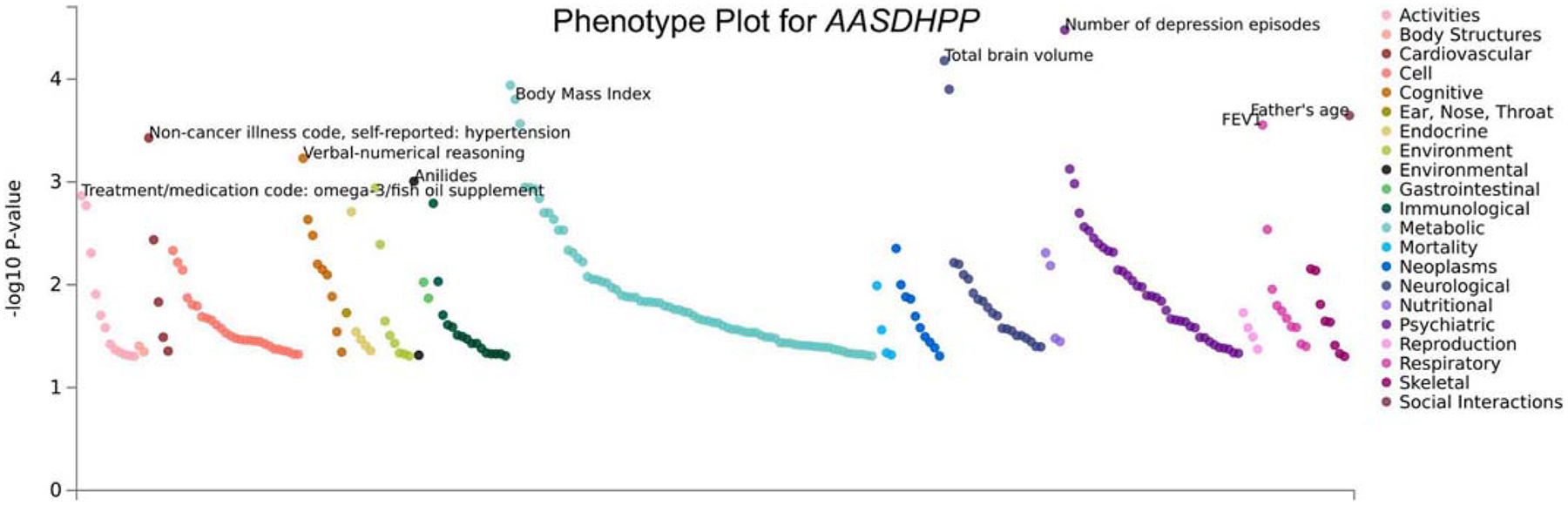
PhEWAS plot of gene AASDHPP. This plot shows phenotypic traits which are significantly correlated (p-values) with gene expression of the gene of interest, cg19292625, a significant site from the Male analytic group, is located within the gene region of AASDHPP. Phenotypic groups are listed on the right of the plot with corresponding color code.
4. Discussion
This study examined the relationship between fluoride exposure, estimated through the fluoride concentration of a spot urine sample, and DNA methylation in whole blood among adolescents at age 12 years. Among all participants, higher CUFsg concentrations were associated with differential methylation at one CpG loci (cg25435255) which annotates to GBF1. Sex-specific effects were evident with females exhibiting a larger number of differentially methylated loci (n = 7) compared to their male counterparts (n = 3) with cg25435255 being the most significantly differentially methylated CpG among females. The significant CpG sites associated with CUFsg concentrations map to genes that have been implicated in neurodevelopment/psychiatric outcomes (Watanabe et al., 2019; Lee et al., 2018; Okbay et al., 2016; Savage et al., 2018) as well as immunologic (Watanabe et al., 2019; Astle et al., 2016; Kanai et al., 2018), metabolic (Watanabe et al., 2019; Yengo et al., 2018) and reproductive (Watanabe et al., 2019; Day et al., 2017) endpoints. While further studies are needed, these data suggest fluoride may have some influence on neurodevelopmental processes and other developmental functions among adolescents in a sex-specific manner.
Animal studies have reported that fluoride exposure can impact the methylome. In rat models of skeletal fluorosis, DNA methylation has been shown to modulate the expression of the Cthrc1 gene, which plays a role in bone formation (Ding et al., 2023). Another study showed evidence of regulation of expression of bone growth genes, BMP-2 and BMP-7, by hypomethylation as a result of fluoride exposure (Ma et al., 2020). Our study did not observe CpGs in Cthrc1, BMP-2, or BMP-7 to be differentially methylated. However, it should be noted that most mouse studies have centered on embryo development and how fluoride-induced DNA methylation affects expression of imprinted genes, as well as the development of oocytes and embryos (Zhao et al., 2015; Zhu et al., 2014; Fu et al., 2014; Yin et al., 2015). Furthermore, fluoride exposures in animal studies far exceed typical human exposure.
Of the very few published human studies, all were conducted in China, cross-sectional or case-control designs, and only one utilized an epigenome wide approach screening differential methylation among children of similar age found in CCAAPS. The study examined the methylation status of 16 Chinese children (aged 8–12 years) using the Infinium-Methylation EPIC BeadChip Array and verified differentially methylated genes in 815 participants. Wang et al., found that fluoride exposure significantly altered the methylation status of genes CALCA and NNAT (Wang et al., 2021). NNAT, shown to regulate ACTH hormone levels, has reported functions in neurodevelopment. Specifically, fluoride exposure was negatively correlated with DNA methylation in NNAT which may be a molecular mechanism of neurotoxicity (Wang et al., 2021; Dec et al., 2017). Wang et al. identified a relevant DMR which contained the gene AASDHPPT. In our study among males, we identified a significantly differentially methylated CpG mapping to the same gene, AASDHPPT suggesting there may be some overlap in impacted biological processes. This was the only overlap we observed between the two studies.
Our EWAS of 272 U.S. adolescents found similar results regarding potential implications on neurodevelopment through the mapping of CpGs to genes associated with neurological processes and/or correlations between blood and brain tissue methylation. The most significantly differentially methylated CpG (cg25435255) identified in the total population and among females annotates to GBF1. Previously published genome-wide association studies have identified GBF1 and others to be associated with educational attainment, intelligence, and cognitive performance (Watanabe et al., 2019; Lee et al., 2018; Okbay et al., 2016; Savage et al., 2018). Further, we observed the methylation status of four CpG sites to be negatively correlated with methylation levels in three main regions of the brain: parietal cortex, temporal cortex, and prefrontal cortex.
Of the three CpG sites significantly associated with fluoride exposure among males, cgl9292625 maps to a gene (AASDHPPT) that has been implicated in psychiatric phenotypes (Watanabe et al., 2019). Interestingly, in previously published observational studies, early childhood exposure to fluoride was associated with lower IQ and internalizing symptoms in male but not female adolescents (Green et al., 2019; Adkins et al., 2022; Saeed et al., 2020). These findings suggest that fluoride exposure may be impacting males and females differently. Other genes mapping to CpG sites significantly associated with fluoride exposure have been implicated in metabolic (Watanabe et al., 2019; Yengo et al., 2018) and reproductive functions (Watanabe et al., 2019; Day et al., 2017). Sex differences are further demonstrated by the eFORGE analysis showing a significant enrichment of CpG site overlap in functional regions across several different tissues in the female group only, including fetal brain. Contrastingly, in the male group there were no tissue types with any overlap to the CpG sites of interest identified from blood. Potential mechansims related to sex-specific findings among children may also be related to fluoride impacting sex-steroid hormone levels and/or altering the course of pubertal development (e.g., age of menarche) (Malin et al., 2022; Bai et al., 2020). This is particularly relevant given our identification of differentially methylated CpGs located in genes relevant for reproductive outcomes.
This study has several strengths. To our knowledge it is the first and largest human study to be conducted in the US and among typically developing healthy adolescents to examine the impact of fluoride on the adolescent epigenome. Leveraging an extant cohort such as the CCAAPS study allows us the opportunity to interrogate several potential confounders including child sex, race, age, socioeconomic factors such as household income and parental psychiatric behaviors, and other neurotoxicant exposures such as tobacco smoke exposure limiting bias due to confounding. In 2015, the recommended fluoride level changed from 0.7 to 1.2 mg/L to 0.7 mg/L and the recommended fluoride concentration for community drinking water in Cincinnati is 0.7 mg/L. A study by Green et al. (Green et al., 2020) reported median CUFsg levels for children aged 4–6 years in Mexico and Canadian children aged 2–6 living in fluoridated communities to be 0.67 μg/mL and 0.64 μg/mL, respectively; given that the median fluoride exposure level in the CCAAPS study is 0.81 μg/mL, children in the CCAAPS study may be exposed to higher levels of fluoride. Although our sample size is limited for an EWAS, to our knowledge, there has been no other study reporting on urinary fluoride levels during early adolescence (~ age 12 years) and its impact on the methylome. Despite these strengths, there are some limitations. The study design is cross-sectional, at a single time point during early adolescence, making it difficult to draw conclusions regarding temporal effects of fluoride exposure on methylation. Further, it is likely that puberty may impact these findings given the impact hormones play in shaping the methylome during puberty (Thompson et al., 2018). We also only analyzed one spot urine fluoride sample for which the half-life is 4–12 h. Thus, we are unable to answer questions related to early life or chronic exposure and its impact on the methylome. To tease out the importance of timing of exposure and chronic exposure, future studies should consider other fluoride biomarkers such as teeth (Martinez et al., 2023). Lastly, this study does not explore how the methylation changes impact neurodevelopmental outcomes-although this will be a future direction of this work.
Taken together, these findings suggest that fluoride exposure is associated with differential methylation of the epigenome during early adolescence. The functional consequences of these changes are unclear; however, given prior research suggesting associations between fluoride and cognitive/neurodevelopmental outcomes in children, further studies are warranted to investigate the role of fluoride-related methylation on cognition and neurodevelopment.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2024.174916.
Supplementary Material
HIGH LIGHTS.
Fluoride has been linked to several neurodevelopmental conditions among children, yet the mechanisms involved are unclear.
Fluoride may elicit its neurotoxic effects by altering the epigenome.
Recent childhood fluoride exposure is associated with differential blood DNA methylation in a sex-specific manner.
DNA methylation changes may impact the expression of genes involved in neurodevelopment and other biological functions.
Acknowledgments
We thank the CCAAPS participants for their time and contribution to this research. Thank you to the Wadsworth Center, and specifically Drs. Patrick Parsons, Ken Aldous, and Mr. James Daly for analyzing serum cotinine measurements in the CCAAPS cohort.
Funding source
Funding for this project was provided by the National Institutes of Environmental Health Sciences (NIEHS) T32ES010957, P30 ES006096, R01 ES031054, R01 ES019890, R01 ESI 1170, R01 ES027224, ES031621, and the National Center for Advancing Translational Sciences (NCATS) UL1 TR001425. The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
CRediT authorship contribution statement
Anna K. Ruehlmann: Writing – original draft, Investigation, Formal analysis. Kim M. Cecil: Writing – review & editing, Project administration, Funding acquisition. Frank Lippert: Writing – review & editing, Resources, Methodology, Data curation. Kimberly Yolton: Writing – review & editing, Resources, Investigation, Funding acquisition, Data curation. Patrick H. Ryan: Writing – review & editing, Resources, Project administration, Investigation, Funding acquisition, Data curation. Kelly J. Brunst: Writing – review & editing, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
The data that has been used is confidential.
References
- Adkins EA, et al. , 2022. Fluoride exposure during early adolescence and its association with internalizing symptoms. Environ. Res 204 (Pt C), 112296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astle WJ, et al. , 2016. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell 167 (5) (p. 1415–1429 el9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylward LL, et al. , 2015. Biomonitoring equivalents for interpretation of urinary fluoride. Regul. Toxicol. Pharmacol 72 (1), 158–167. [DOI] [PubMed] [Google Scholar]
- Bai R, et al. , 2020. Associations of fluoride exposure with sex steroid hormones among U.S. children and adolescents, NHANES 2013–2016. Environ. Pollut 260, 114003. [DOI] [PubMed] [Google Scholar]
- Bashash M, et al. , 2017. Prenatal fluoride exposure and cognitive outcomes in children at 4 and 6–12 years of age in Mexico. Environ. Health Perspect 125 (9), 097017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck AT, Steer RA, Brown GK, 1996. Manual for the Beck Depression Inventory-II. Psychological Corporation, San Antonio, TX. [Google Scholar]
- Breeze CE, 2022. Cell type-specific signal analysis in epigenome-wide association studies. Methods Mol. Biol 2432, 57–71. [DOI] [PubMed] [Google Scholar]
- Chen YA, et al. , 2013. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 8 (2), 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K, Mondal NK, 2016. Dental fluorosis and urinary fluoride concentration as a reflection of fluoride exposure and its impact on IQ level and BMI of children of Laxmisagar, Simlapal Block of Bankura District, W.B., India. Environ. Monit. Assess 188 (4), 218. [DOI] [PubMed] [Google Scholar]
- Day FR, et al. , 2017. Genomic analyses identify hundreds of variants associated with age at menarche and support a role for puberty timing in cancer risk. Nat. Genet 49 (6), 834–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dec K, et al. , 2017. The influence of fluorine on the disturbances of homeostasis in the central nervous system Biol. Trace Elem Res 177 (2), 224–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, et al. , 2023. Screening of differentially methylated genes in skeletal fluorosis of rats with different types and involvement of aberrant methylation of Cthrc1. Environ. Pollut 332, 121931. [DOI] [PubMed] [Google Scholar]
- Edgar RD, et al. , 2017. BECon: a tool for interpreting DNA methylation findings from blood in the context of brain. Transl. Psychiatry 7 (8), el187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, et al. , 2014. Natrium fluoride influences methylation modifications and induces apoptosis in mouse early embryos. Environ. Sd. Technol 48 (17), 10398–10405. [DOI] [PubMed] [Google Scholar]
- Gao M, et al. , 2020. Association between low-to-moderate fluoride exposure and bone mineral density in Chinese adults: non-negligible role of RUNX2 promoter methylation. Ecotoxicol. Environ. Saf 203, 111031. [DOI] [PubMed] [Google Scholar]
- Green R, et al. , 2019. Association between maternal fluoride exposure during pregnancy and IQ scores in offspring in Canada. JAMA Pediatr 173 (10), 940–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R, et al. , 2020. Associations between urinary, dietary, and water fluoride concentrations among children in Mexico and Canada. Toxics 8 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon E, et al. , 2018. Characterizing genetic and environmental influences on variable DNA methylation using monozygotic and dizygotic twins. PLoS Genet 14 (8), el007544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, et al. , 2022. DNA methylation profiles of ovarian granular cells from fluorosis female patients suffering reproductive dysfunctions. Biol. Trace Elem. Res 200 (8), 3529–3536. [DOI] [PubMed] [Google Scholar]
- Houseman EA, et al. , 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert BR, et al. , 2016. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am. J. Hum. Genet 98 (4), 680–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphaus R, Reynolds C, 2008. Parenting Relationship Questionnaire. Pearson Clin Assess, San Antonio, TX. [Google Scholar]
- Kanai M, et al. , 2018. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet 50 (3), 390–400. [DOI] [PubMed] [Google Scholar]
- Lee JJ, et al. , 2018. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet 50 (8), 1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, et al. , 2012. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28 (6), 882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMasters GK, et al. , 2006. High prevalence of aeroallergen sensitization among infants of atopic parents. J. Pediatr 149 (4), 505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester BM, Conradt E, Marsit CJ, 2013. Epigenetic basis for the development of depression in children, Clin. Obstet. Gynecol 56 (3), 556–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, et al. , 2019. Effect of fluoride exposure on anxiety- and depression-like behavior in mouse. Chemosphere 215, 454–460. [DOI] [PubMed] [Google Scholar]
- Liu F, et al. , 2014. Fluoride exposure during development affects both cognition and emotion in mice. Physiol. Behav 124, 1–7. [DOI] [PubMed] [Google Scholar]
- Lu F, et al. , 2019. Fluoride related changes in behavioral outcomes may relate to increased serotonin. Physiol. Behav 206, 76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, et al. , 2020. The dose-time effects of fluoride on the expression and DNA methylation level of the promoter region of BMP-2 and BMP-7 in rats. Environ. Toxicol. Pharmacol 75, 103331. [DOI] [PubMed] [Google Scholar]
- Maitre L, et al. , 2022. Multi-omics signatures of the human early life exposome. Nat. Commun 13 (1), 7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malin AJ, Till C, 2015. Exposure to fluoridated water and attention deficit hyperactivity disorder prevalence among children and adolescents in the United States: an ecological association. Environ. Health 14, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malin AJ, et al. , 2022. Fluoride exposure and AGe of menarche: potential differences among adolescent girls and women in the United States. Expo. Health 14, 733–742. [Google Scholar]
- Malin AJ, et al. , 2024. Maternal urinary fluoride and child neurobehavior at age 36 months. JAMA Netw. Open 7 (5), e2411987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin EM, Fry RC, 2018. Environmental influences on the epigenome: exposure-associated DNA methylation in human populations. Annu. Rev. Public Health 39, 309–333. [DOI] [PubMed] [Google Scholar]
- Martinez M, et al. , 2023. Quantitative fluoride imaging of teeth using CaF emission by laser induced breakdown spectroscopy. J. Anal. At. Spectrom 38 (2), 303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Mier EA , et al. , 2003. Fluoride intake from foods, beverages and dentifrice by children in Mexico. Community Dent. Oral Epidemiol 31 (3), 221–230. [DOI] [PubMed] [Google Scholar]
- Martinez-Mier EA, et al. , 2011. Development of gold standard ion-selective electrode-based methods for fluoride analysis. Caries Res 45 (1), 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrotheodoros S, et al. , 2023. Negative parenting, epigenetic age, and psychological problems: prospective associations from adolescence to young adulthood. J. Child Psychol. Psychiatry 64 (10), 1446–1461. [DOI] [PubMed] [Google Scholar]
- McCartney DL, et al. , 2016. Identification of polymorphic and off-target probe binding sites on the Illumina Infinium MethylationEPIC BeadChip. Genom Data 9, 22–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, et al. , 2021. Effect of fluoride in drinking water on the level of 5-methylcytosine in human and rat blood. Environ. Toxicol. Pharmacol 81, 103511. [DOI] [PubMed] [Google Scholar]
- Min JL, et al. , 2018. Meffil: efficient normalization and analysis of very large DNA methylation datasets. Bioinformatics 34 (23), 3983–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min JL, et al. , 2021. Genomic and phenotypic insights from an atlas of genetic effects on DNA methylation. Nat. Genet 53 (9), 1311–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu L, Xu Z, Taylor JA, 2016. RCP: a novel probe design bias correction method for Illumina Methylation BeadChip. Bioinformatics 32 (17), 2659–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okbay A, et al. , 2016. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 533 (7604), 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, et al. , 2020. Aberrant DNA methylation of Cyclind-CDK4-p21 is associated with chronic fluoride poisoning. Chem. Biol. Interact 315, 108875. [DOI] [PubMed] [Google Scholar]
- Perlman SE, et al. , 2016. Exposure to secondhand smoke among nonsmokers in New York City in the context of recent tobacco control policies: current status, changes over the past decade, and national comparisons. Nicotine Tob. Res 18 (11), 2065–2074. [DOI] [PubMed] [Google Scholar]
- Peters TJ, et al. , 2015. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin 8, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenzi L, et al. , 2020. Maternal caregiving and DNA methylation in human infants and children: systematic review. Genes Brain Behav 19 (3), el2616. [DOI] [PubMed] [Google Scholar]
- Public Health Capacity and knowledge Management Unit: The State of community Water Fluoridation (CWF) across Canada, 2022. Quebec Region for the Office of the Chief Dental Officer of Canada.
- Rakyan VK, et al. , 2011. Epigenome-wide association studies for common human diseases. Nat. Rev. Genet 12 (8), 529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Arenas C, et al. , 2022. Identification of autosomal cis expression quantitative trait methylation (cis eQTMs) in children’s blood. Elife 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan PH, et al. , 2005. Is it traffic type, volume, or distance? Wheezing in infants living near truck and bus traffic. J. Allergy Clin. Immunol 116 (2), 279–284. [DOI] [PubMed] [Google Scholar]
- Saeed M, Malik RN, Kamal A, 2020. Fluorosis and cognitive development among children (6–14 years of age) in the endemic areas of the world: a review and critical analysis. Environ. Sd. Pollut. Res. Int 27 (3), 2566–2579. [DOI] [PubMed] [Google Scholar]
- Sanders AE, et al. , 2019. Association between water fluoridation and income-related dental caries of US children and adolescents. JAMA Pediatr 173 (3), 288–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage JE, et al. , 2018. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat. Genet 50 (7), 912–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, et al. , 2020. Fluoride exposure and CALCA methylation is associated with the bone mineral density of Chinese women. Chemosphere 253, 126616. [DOI] [PubMed] [Google Scholar]
- Thomas DB, et al. , 2016. Urinary and plasma fluoride levels in pregnant women from Mexico City. Environ. Res 150, 489–495. [DOI] [PubMed] [Google Scholar]
- Thompson EE, et al. , 2018. Global DNA methylation changes spanning puberty are near predicted estrogen-responsive genes and enriched for genes involved in endocrine and immune processes. Clin. Epigenetics 10, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till C, et al. , 2018. Community water fluoridation and urinary fluoride concentrations in a national sample of pregnant women in Canada. Environ. Health Perspect 126 (10), 107001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till C, et al. , 2020. Fluoride exposure from infant formula and child IQ in a Canadian birth cohort. Environ. Int 134, 105315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Usano M, et al. , 2020. In utero and childhood exposure to tobacco smoke and multi-layer molecular signatures in children. BMC Med 18 (1), 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, et al. , 2021. DNA methylation and fluoride exposure in school-age children: epigenome-wide screening and population-based validation. Ecotoxicol. Environ. Saf 223, 112612. [DOI] [PubMed] [Google Scholar]
- Wang F, et al. , 2023. Epidemiological analysis of drinking water-type fluorosis areas and the impact of fluorosis on children’s health in the past 40 years in China. Environ. Geochem. Health 45 (12), 9925–9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, et al. , 2019. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet 51 (9), 1339–1348. [DOI] [PubMed] [Google Scholar]
- Water Fluoridation Data & Statistics, 2019. Available from: https://www.cdc.gov/fluoridation/statistics/index.htm.
- Wu CX, et al. , 2018. Changes of DNA repair gene methylation in blood of chronic fluorosis patients and rats. J. Trace Elem. Med. Biol 50, 223–228. [DOI] [PubMed] [Google Scholar]
- Wu S, et al. , 2019. Aberrant methylation-induced dysfunction of pl6 is associated with osteoblast activation caused by fluoride. Environ. Toxicol 34 (1), 37–47. [DOI] [PubMed] [Google Scholar]
- Wu H, Eckhardt CM, Baccarelli AA, 2023. Molecular mechanisms of environmental exposures and human disease. Nat. Rev. Genet 24 (5), 332–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, et al. , 2016. ENmix: a novel background correction method for Illumina HumanMethylation450 BeadChip. Nucleic Acids Res 44 (3), e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, et al. , 2017. RELIC: a novel dye-bias correction method for Illumina Methylation BeadChip. BMC Genomics 18 (1), 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yengo L, et al. , 2018. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum. Mol. Genet 27 (20), 3641–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S, et al. , 2015. Adverse effects of high concentrations of fluoride on characteristics of the ovary and mature oocyte of mouse. PloS One 10 (6), e0l29594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, et al. , 2018. Threshold effects of moderately excessive fluoride exposure on children’s health: a potential association between dental fluorosis and loss of excellent intelligence. Environ. Int 118, 116–124. [DOI] [PubMed] [Google Scholar]
- Zhang Y, et al. , 2017. Do environmental fluoride exposure and ESRalpha genetic variation modulate methylation modification on bone changes in Chinese farmers? Chem. Res. Toxicol 30 (6), 1302–1308. [DOI] [PubMed] [Google Scholar]
- Zhao L, et al. , 2015. Sodium fluoride affects DNA methylation of imprinted genes in mouse early embryos. Cytogenet. Genome Res 147 (1), 41–47. [DOI] [PubMed] [Google Scholar]
- Zhu JQ, et al. , 2014. Sodium fluoride disrupts DNA methylation of H19 and Peg3 imprinted genes during the early development of mouse embryo. Arch. Toxicol 88 (2), 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that has been used is confidential.


